Abstract
Imaging mass spectrometry (MS) allows a remarkable range of measurements including diagnosis of disease state of tissue based on detailed information on its chemical constituents, especially lipids and proteins. The recent emergence of ambient ionization allows imaging in the open environment without sample preparation. In this review, we briefly describe the history of imaging MS highlighting its main techniques and applications. We also demonstrate how the detailed molecular information obtained by imaging MS makes this technique suitable for a range of forensic and clinical applications with the potential to be successfully developed all the way to intra-surgical practice.
Molecular imaging is a topic of great current interest in mass spectrometry (MS). This is due to the enormous amount of detailed chemical information provided by imaging MS. What can we now achieve in imaging mass spectrometry? (1) We can spray a tissue section with charged aqueous droplets, record mass spectra as the spray is moved across the surface, and plot the data in the form of a 2D image for any compound of interest. This experiment (DESI imaging) is done without prior sample preparation in the open ambient environment. It gives information that allows diseased and healthy tissue to be discriminated; it even allows grades of some tumors to be assigned (Fig. 1). Chemical derivatization can be implemented by adding reagents to the spray solvent: they allow selective ionization and the special conditions in the microdroplets splashed from the surface accelerate reaction rates over bulk reaction rates by several orders of magnitude. It is reasonable to expect that these diagnostic experiments will soon be implemented in pathology laboratories within hospital surgical suites. (2) We can use fullerene ion beams under vacuum (SIMS) to image biological material on spatial scales below 100 nm, much below that available by any other general purpose molecular imaging method. Similar experiments can be done using the microscope instead of the microprobe mode, so that an entire object can be imaged rapidly, revealing the association between structural features and molecular components. (3) We can use laser beams, after suitable matrix application to the sample, to follow by MALDI the distribution of proteins in temporal sequence during embryonic development with the chemical information being precise down to protein variants and the spatial resolution being on the order of 100 microns. (4) We can record the pattern of chemicals left behind in a latent fingerprint, thereby identifying an individual both by the physical print and by its unique chemical characteristics. (5) We can identify specific diterpenoids in algae, record their distribution and provide information on their role as defense chemicals against pathogenic fungi.
Fig. 1.
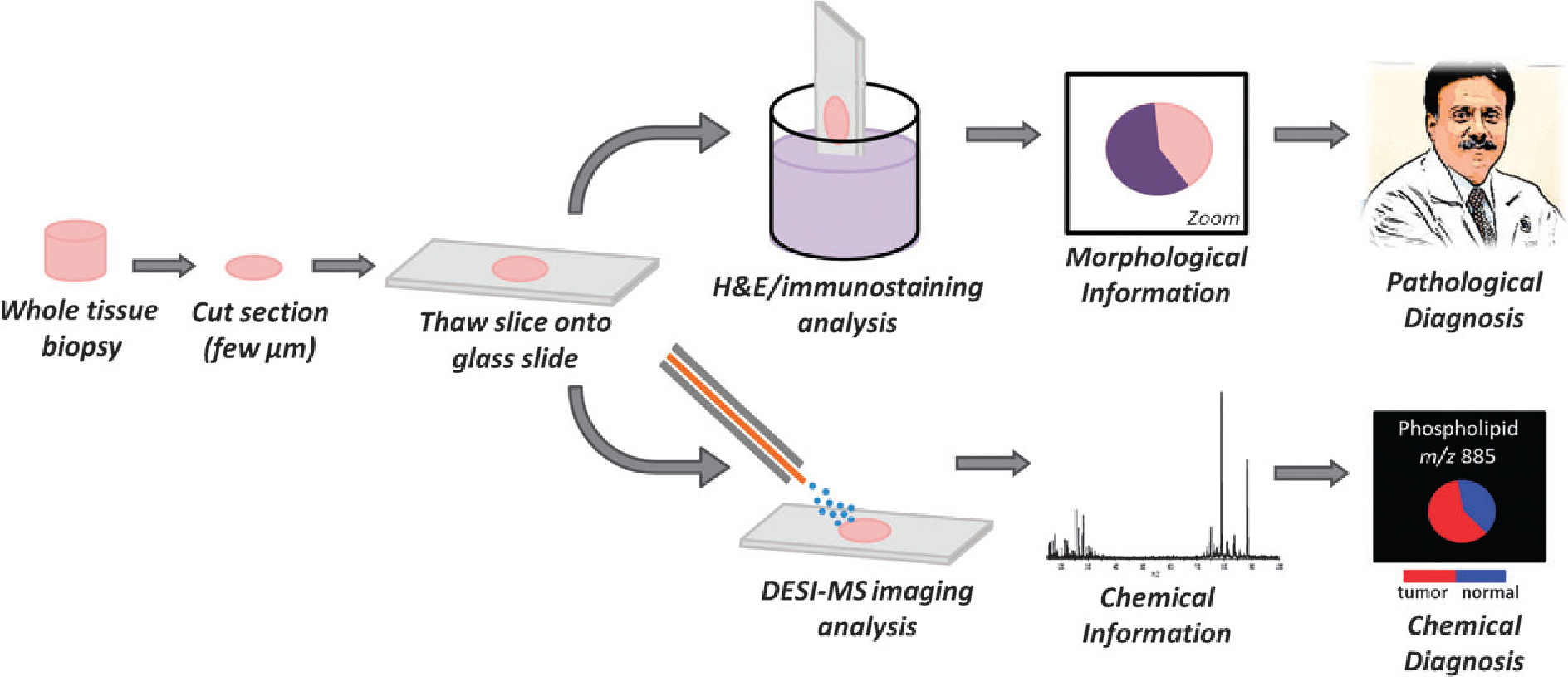
Overview of tissue processing to achieve a diagnosis by traditional pathological staining techniques and by DESI imaging mass spectrometry.
The examples cited above help to make the point that imaging MS is at an exciting crossroads as it transitions from instrumentation research to applications, especially clinical applications (Fig. 1). In this brief review, we track the progress of chemical imaging mass spectrometry over the past decade. Some emphasis is given to the ambient ionization methods, invented over the past half dozen years.
At the start of the decade, imaging MS was dominated by two ionization techniques, secondary ion mass spectrometry (SIMS)1 and matrix assisted laser desorption ionization (MALDI).2 The advent of new ambient ionization techniques added to the imaging MS revolution by providing a means to analyze samples with little to no sample preparation, without the need of matrix application and in the open environment. Ambient techniques were initially developed to address challenges general to complex mixture analysis and specifically the enormous analytical effort required in traditional sample preparation.3 The earlier success of molecular imaging research using MALDI and SIMS facilitated the development of imaging applications of ambient ionization (Fig. 2). This expansion has generated excitement about possible future applications, especially those in medicine. In disease diagnosis, imaging MS is being explored as a complementary technique that provides sensitive and specific molecular information to augment the visual pathology observations of hematoxylin and eosin (H&E) and immunostained tissue sections (Fig. 1). The longer term hope for the ambient ionization techniques is that they will be able to provide real-time information in a surgical setting on specific diseases; a hope that is encouraged by mass spectrometric analysis of the molecules in surgical “smoke” released during electro-surgical operations.4
Fig. 2.
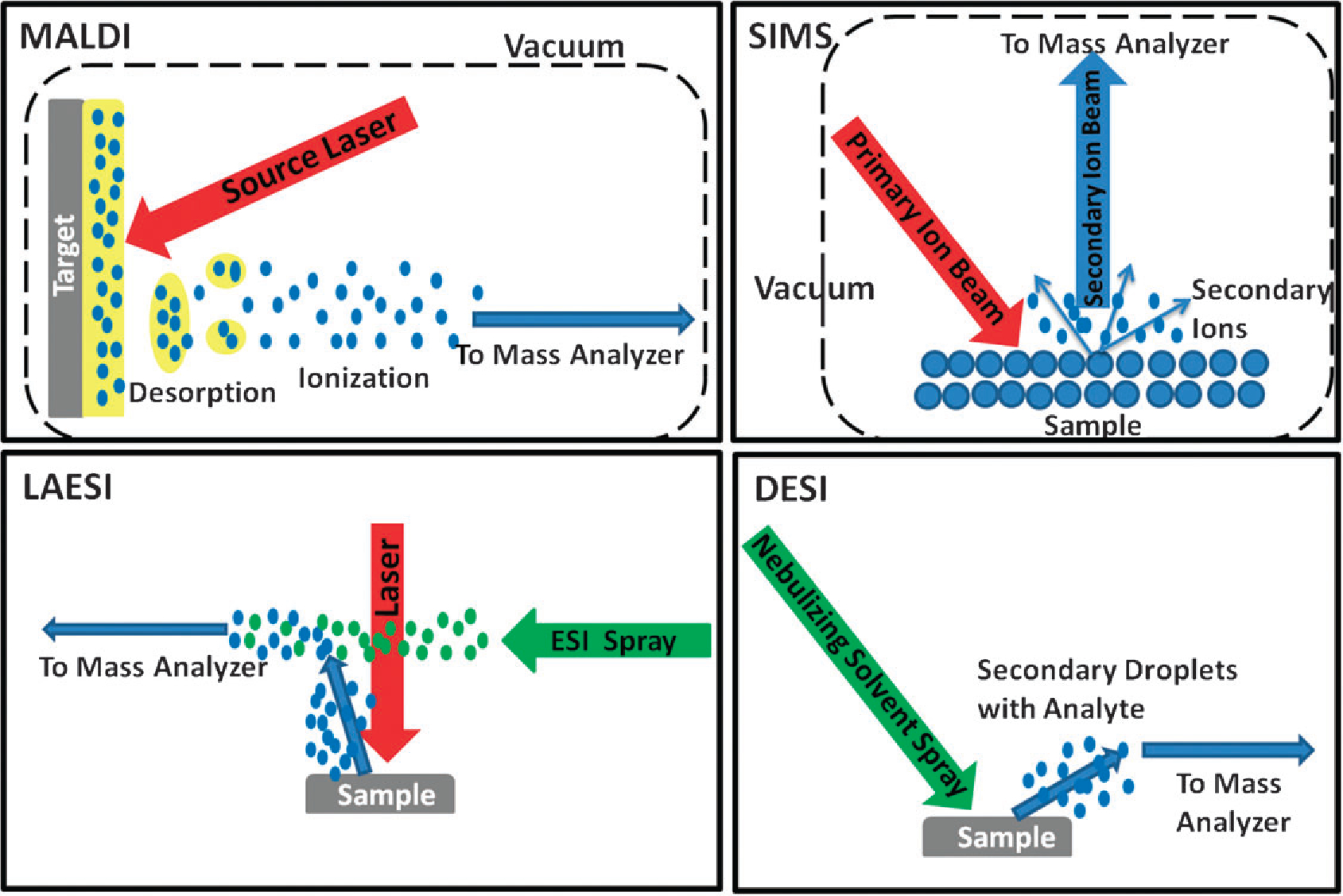
Simplified mechanistic diagrams for the four most commonly used ionization techniques in imaging mass spectrometry; matrix assisted laser desorption ionization (MALDI), secondary ion mass spectrometry (SIMS), laser ablation electrospray ionization (LAESI) and desorption electrospray ionization (DESI).
SIMS was the first molecular ionization technique applied to surface imaging; it was followed by MALDI which allowed for the imaging of larger biomolecules. Both techniques traditionally require the sample to be examined under vacuum limiting manipulations and complementary experiments. Both are invasive but they are also generally applicable and label-free methods. SIMS analysis occurs on untreated sample surfaces, but SIMS is a “hard” ionization method, depositing relatively large amounts of internal energy and causing extensive fragmentation. This limits the mass range of the mass spectra and hence its applicability to more complex biomolecules.5 The advent of newer cluster ion sources increased secondary ion yields, reduced fragmentation and improved depth resolution, greatly facilitating biological applications.6 These applications have been highly varied and have provided information on a wide variety of biological and anthropomorphic questions: lipids in diseased human muscle tissue;7 exogenous compounds on latest fingerprints;8 surfactins excreted by bacterial colonies;9 and distributions of radiopharmaceutical compounds at the cellular level.10 Three-dimensional (3D) images with subcellular resolution have been constructed of freeze-dried glioblastomas, oocytes and thyroid tumor tissue.11,12 SIMS imaging data can be combined with that from other spectroscopic techniques to increase the amount of chemical information gained from a single tissue sample.13 The SIMS spatial resolution of approximately 100 nm provides extraordinary information at the subcellular level but, as for all techniques within imaging MS, this increased resolution comes at the cost of analysis time.14
MALDI has traditionally been used to image larger biomolecules such as proteins and peptides but is also effective for smaller molecules such as glycerophospholipids (GPs).15 In contrast to SIMS, sample preparation in MALDI imaging strongly affects the quality of the data obtained through such variables as the matrix chosen and the matrix application procedure. Consequentially, considerable effort has been and is still being made to develop ideal matrices for each chemical class including proteins, peptides, lipids, drugs and small metabolites and to develop matrix application procedures for obtaining consistently high quality images.16,17 One of the earliest protein imaging applications focused on probing the changes in human brain tumor xenograph sections, showing differences in protein expression in different parts of the tumor.18 Other types of cancer studied by MALDI imaging include human glioblastoma biopsies,19 human metastasis to the brain,20 human prostate cancer,21 and most recently, tumor margins in human renal carcinoma.22 3D molecular images of proteins can also be created using MALDI imaging of serial sections; MALDI 3D images of proteins associated with a mouse brain tumor in a mouse injected with glioma cells were created and coregistered with magnetic resonance imaging (MRI) 3D data.23 In addition to cancer research, MALDI imaging has also been used to study neurodegenerative diseases such as Alzheimer’s24 and Parkinson’s,25 to examine the proteome response in tissue to drug administration26 and to investigate membrane proteins in human ocular lens and retinal tissues.27 MALDI can also be used to image thin whole body tissue sections in order to map the distribution of a drug and its metabolites at differing dosages, providing detailed insights into therapeutic processes.28 The recent development of atmospheric pressure MALDI (AP-MALDI) allowed imaging of biological samples to take place at atmospheric pressure.29,30 MALDI-MS imaging provides spatial resolution as low as 8 μm,14,18 but most reported experiments employ a resolution of 250 mm to shorten the analysis time while still providing high quality data with extensive chemical information. The use of robotics has greatly increased the throughput of matrix application.31 Although MALDI imaging has been proposed for consideration in intraoperative diagnosis,18 the requirements of sample preparation, make direct laser ionization imaging using femtosecond lasers a simpler approach.
The first of the ambient ionization techniques, desorption electrospray ionization (DESI)32,33 was introduced in 2004. Several other ambient imaging methods are now being applied to imaging MS including laser ablation electrospray ionization (LAESI),34–36 laser ablation flowing atmospheric pressure afterglow (LA-FAPA),37 low temperature plasma (LTP),38 infrared laser ablation metastable-induced chemical ionization (IR-LAMICI)39 and to some extent direct analysis in real time (DART).40 These methods allow imaging to take place in the open, ambient environment making the analysis simpler and offering the possibility of moving imaging MS outside the lab into clinical or forensic settings. Other closely related methods include surface sampling probe (SSP),41 probe electrospray ionization (PESI),42 and matrix assisted laser desorption electrospray ionization (MALDESI).43 All ambient ionization methods require two processes, molecular desorption from the surface and ionization. These steps can be accomplished separately, as in LAESI for example, or simultaneously, by means of the same agency, as in DESI. The methods are often characterized in terms of the desorption step—sprays are used in DESI, heat in DART and lasers in LAESI.
Each of the ambient ionization techniques has its own interesting physics and reaction chemistry but here we will illustrate these properties specifically for DESI. The vast majority of ions in DESI are produced through a ‘droplet pickup’ mechanism, where the initial micronsized droplets moving at more than 100 m s−1 impact a sample surface, wetting it and allowing analytes to dissolve. Subsequent droplet collisions at the surface produce analyte-containing secondary droplets from which ion formation occurs identically to ESI.44 The nature of the DESI mechanism allows for interesting chemistry to occur both at the wetted surface and in the progeny droplets.45 These secondary droplets form unique environments for chemical reactions allowing reactions to proceed at much increased rates.46
Imaging applications of DESI-MS32 have expanded rapidly (Fig. 3). Diagnosis of animal and human cancers has been achieved on the basis of the lipid profiles of thin tissue sections acquired by imaging DESI-MS.47–49 Also within the field of clinical diagnostics, different grades of human gliomas can be discriminated on the basis of the DESI-MS lipid profiles acquired during imaging experiments.50 Applications within the pharmaceutical industry include the imaging of drugs and their metabolites within tissues51 or whole animal sections.52 An advantage of spray based techniques such as DESI is the ability to add reagents to the solvent spray such that specific compounds present in the sample are targeted through chemical reactions. This reactive DESI-MS imaging experiment was used to derivatize and image cholesterol, which is not easily ionized under standard DESI-MS conditions.46
Fig. 3.
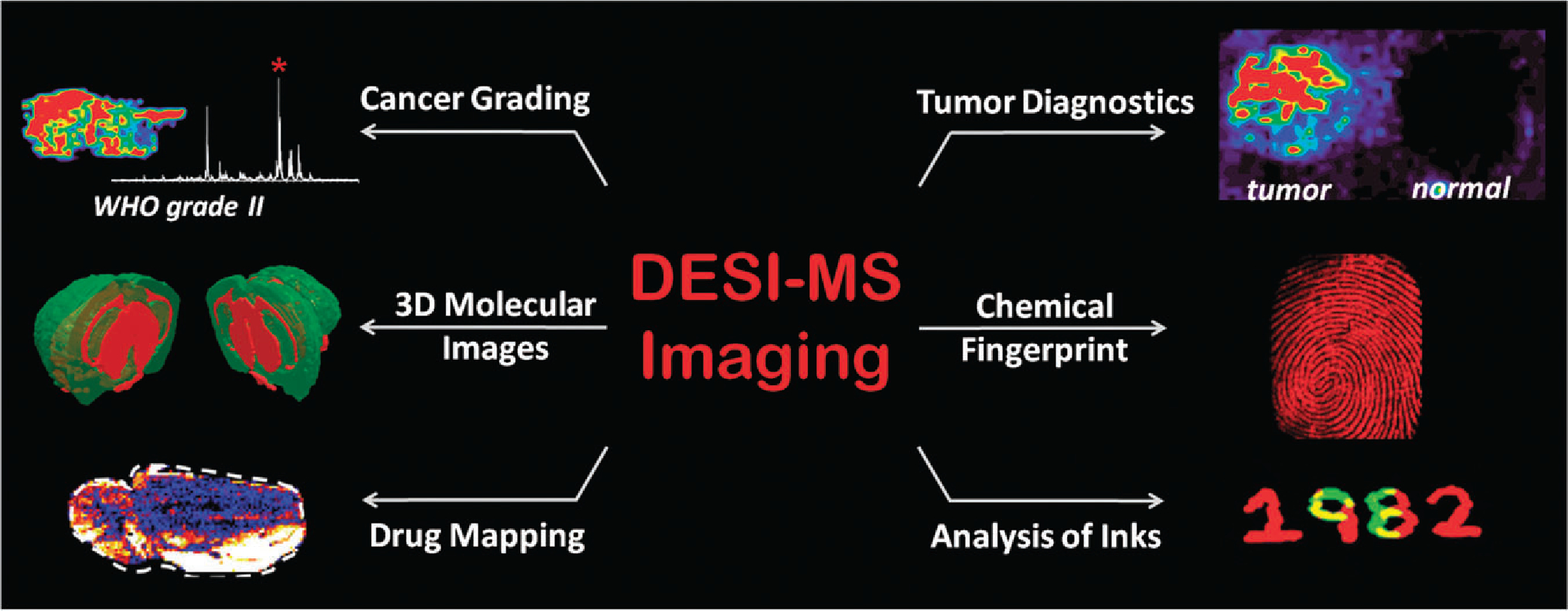
Overview of selected DESI-MS imaging applications, including disease diagnosis, 3D molecular image reconstruction, mapping of drugs in biological tissue and forensics applications.
Aside from tissue imaging, DESI-MS has been used to image thin films of bacteria in order to characterize the natural products produced as well as to monitor metabolite exchange between adjacent bacteria strains.53 In another example, DESI-MS was used to image algal surfaces, uncovering potential natural products with anti-fungal properties produced by the algae.54 This study was important in that it elucidated the chemical structure of a previously unknown natural product, mapped its distribution and helped uncover its biological significance.54 In these and most other examples, the capabilities for secure identification of the imaged compounds provided by using tandem mass spectrometry often add value to the study. In cases where the mass spectrometer used for imaging allows exact mass measurements, this simplifies molecular identification.
In still other applications, DESI-MS was successfully applied to image the chemical profiles of many different types of inks for forensic applications55 and to image inks used in banknotes to detect counterfeit currency.56 Within the realm of forensic applications, latent fingerprints are readily imaged using DESI-MS,57 the required spatial resolution being achieved by DESI. Overlapping fingerprints could also be resolved using the distinct chemical signatures of each individual as well as being able to map chemically specific compounds of interest such as cocaine or explosives.57 3D models can also be constructed using DESI-MS imaging data, as shown for a mouse brain.58 The unique versatility of the chemically-based spray method of imaging of DESI-MS holds promise for the further expansion of applications. The modest spatial resolution is likely to remain the main limitation of the DESI method.
LAESI is another common ambient MS imaging method. LAESI uses laser desorption followed by the take-up of the ablated material into charged droplets produced by ESI.36 Production of dry ions in the course of droplet transfer into the mass spectrometer is therefore much like that which occurs in DESI and both methods are applicable to high mass compounds, including proteins. LAESI differs from the closely related electrospray-assisted laser desorption ionization (ELDI) method only in the choice of infrared laser wavelengths that allow biological materials to be imaged by absorption of laser power by water. Ablated tissue craters are typically between 200–400 μm in diameter and 40–50 μm in depth for focused mid-infrared laser beams with a wavelength of 2940 nm. LAESI has been used to image and identify metabolites in plant tissue.35 This ionization technique can also be used to construct three-dimensional distributions of metabolites in tissues.34 Recently, the imaging of lipids and small metabolites in rat brain was demonstrated at a resolution of 200 μm.59,60
Other ambient MS imaging techniques are beginning to see varied applications. For example, LA-FAPA uses a focused UV laser beam to ablate samples followed by helium atmospheric pressure afterglow ionization;37 it has been applied to the direct analysis of pharmaceuticals and food.61 The imaging capabilities of LA-FAPA have been demonstrated by mapping lidocaine and caffeine in animal and vegetable tissues respectively.37 In another ambient technique, LTP, a dielectric barrier discharge ionization of helium gas is employed to produce a microplasma, which is then directed at the sample promoting desorption and ionization.62,63 Art work requires non-destructive imaging for which the laser and plasma-based desorption methods are better suited than the spray methods; for example LTP, allows imaging inkpads on rice paper with a lateral resolution of 250 μm.38
The driving force behind the development of ambient MS imaging techniques, specifically DESI-MS, was to create fast, accurate and powerful bioanalytical methodologies that could expand disease diagnostic capabilities. These methods could then be easily translated to clinical and surgical settings, offering chemical information on disease state that could enhance patient survival and improve disease management. This goal was preceded by efforts at validating the applicability of the technique to diagnosis of cancer in human tissue, an ongoing task. The next objective is to translate the technology into a clinical setting. Currently, the most commonly used imaging techniques for in vivo diagnosis and staging in clinical oncology are ultrasound, computer tomography (CT), magnetic resonance imaging (MRI) and [18F]fluorodeoxyglucose-positron emission tomography (FDG-PET).64 These imaging technologies have been used for disease detection, diagnosis, treatment determination and surveillance of disease recurrence.65 Despite the extensive use of these technologies, the ability of these methods to facilitate early detection, precise characterization, and accurate localization of malignant disease could be improved (Table 1).65 Limitations of these techniques are related to spatial resolution and molecular specificity. For example, the spatial resolution routinely used in PET imaging (several millimeters) is not high enough to fully evaluate the pathological progression of cancer. From this perspective, ambient imaging MS techniques are poised to deliver technology that can be applied routinely in clinical and surgical settings, offering enhanced spatial resolution and chemical specificity not currently available. Note that the required invasive nature of these imaging methods is successfully juxtaposed against this necessary characteristic of surgery. Moreover, because ambient ionization techniques have characteristic ease and speed of execution, the approach might allow in situ analysis of tissue to assist in intraoperative surgical decision making. In vivo and in situ tissue analysis by mass spectrometry has been performed with a recently developed technique, rapid evaporative ionization mass spectrometry (REIMS).4 This technique interrogates the surgical smoke produced by electrothermal desorption for real-time identification of tissue features, which are compared to libraries of data to reach clinically relevant conclusions. New, even unexpected applications of MS imaging techniques in forensics, biomedicine and material sciences are likely to continue to emerge. To meet these unexpected needs strides must be taken to understand the underlying mechanism of these new ionization techniques and to understand the biological basis behind the images. As clinical applications advance, larger more diverse sample populations will be necessary for validation and with these come the demands of data reduction and data processing. With all opportunities come challenges but it seems likely that these will be overcome and imaging MS will become widely used in biomedicine.
Table 1.
Characteristics of commonly used medical imaging techniques and imaging MS
| Method | Resolution | Chemical information | Molecular vs. anatomical | Exam time/min | Limitations | Label-free | Radiation |
|---|---|---|---|---|---|---|---|
|
| |||||||
| Imaging MS | nm-μm | Yes, unlimited | Molecular | 30–40 | Invasive | Yes | No |
| CT | mm | No | Anatomical | 5 | Low molecular specificity | Yes | Yes |
| MRI | mm-cm | No | Anatomical | 30 | Low molecular specificity | No | No |
| PET | mm | Yes, limited to specific functional groups | Molecular and Anatomical | 30–40 | Need for specific reagents to be administered intravenously | No | Yes |
| Ultrasound | mm-cm | No | Anatomical | 15 | Low reproducibility | Yes | No |
Acknowledgments
This work was supported by the National Institutes of Health (Grant 1 R21 EB00 9459-01).
Biographies
Allison Dill received BS degrees in biology and chemistry from Indiana University, Bloomington, Indiana. Following graduation she worked as an analytical chemist at Eli Lilly and Company. She is currently pursuing a PhD degree in analytical chemistry at Purdue University, West Lafayette, under the direction of Professor Graham Cooks. Her research interests involve tissue imaging using desorption electrospray ionization mass spectrometry for disease state characterization.
Livia Eberlin received her BS in chemistry in 2007 from the State University of Campinas, São Paulo, Brazil. She is currently pursuing her PhD degree at Purdue University, West Lafayette, under the direction of Prof. R. Graham Cooks. Her current research focuses on developments in ambient mass spectrometry, specifically desorption electrospray ionization imaging and its applications in biomedical research.
Demian Ifa received his BS in pharmacy from the State University of São Paulo (UNESP), Brazil. He received his MS in organic chemistry from the University of Rio de Janeiro (UFRJ) and his PhD in pharmacology from the University of São Paulo (USP), Brazil. He is an associate research scientist at the Aston Labs for Mass Spectrometry at Purdue University working on the development of desorption electrospray ionization and its applications to imaging and quantitation.
Graham Cooks received PhD degrees from the University of Natal (Frank Warren) and Cambridge University (Peter Sykes). His interests involve construction of mass spectrometers and their use in fundamental studies and applications. Early in his career contributed to the concept and implementation of tandem mass spectrometry and to desorption ionization, especially matrix-based methods. His interest in minimizing sample work-up and avoiding chromatography contributed to the development of the ambient ionization methods, including desorption electrospray ionization (DESI). These same interests also led to the construction of miniature ion trap mass spectrometers and their application to problems of trace chemical detection.
Notes and references
- 1.Winograd N, Appl. Surf. Sci, 2003, 203, 13–19. [Google Scholar]
- 2.Caprioli RM, Farmer TB and Gile J, Anal. Chem, 1997, 69, 4751–4760. [DOI] [PubMed] [Google Scholar]
- 3.Ifa DR, Wu C, Ouyang Z and Cooks RG, Analyst, 2010, 135, 669–681. [DOI] [PubMed] [Google Scholar]
- 4.Schafer KC, Denes J, Albrecht K, Szaniszlo T, Balog J, Skoumal R, Katona M, Toth M, Balogh L and Takats Z, Angew. Chem., Int. Ed, 2009, 48, 8240–8242. [DOI] [PubMed] [Google Scholar]
- 5.Walker AV, Anal. Chem, 2008, 80, 8865–8870. [DOI] [PubMed] [Google Scholar]
- 6.Vickerman JC, Surf. Sci, 2009, 603, 1926–1936. [Google Scholar]
- 7.Tahallah N, Brunelle A, De La Porte S and Laprevote O, J. Lipid Res, 2008, 49, 438–454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Szynkowska MI, Czerski K, Rogowski J, Paryjczak T and Parczewski A, Surf. Interface Anal, 2010, 42, 393–397. [Google Scholar]
- 9.Brunelle A and Laprevote O, Anal. Bioanal. Chem, 2009, 393, 31–35. [DOI] [PubMed] [Google Scholar]
- 10.Chehade F, de Labriolle-Vaylet C, Moins N, Moreau MF, Papon J, Labarre P, Galle P, Veyre A and Hindie E, J. Nucl. Med, 2005, 46, 1701–1706. [PubMed] [Google Scholar]
- 11.Chandra S, Appl. Surf. Sci, 2004, 231, 467–469. [Google Scholar]
- 12.Mas S, Perez R, Martinez-Pinna R, Egido J and Vivanco F, Proteomics, 2008, 8, 3735–3745. [DOI] [PubMed] [Google Scholar]
- 13.Petit VW, Refregiers M, Guettier C, Jamme F, Sebanayakam K, Brunelle A, Laprevote O, Dumas P and Le Naour F, Anal. Chem, 2010, 82, 3963–3968. [DOI] [PubMed] [Google Scholar]
- 14.Koestler M, Kirsch D, Hester A, Leisner A, Guenther S and Spengler B, Rapid Commun. Mass Spectrom, 2008, 22, 3275–3285. [DOI] [PubMed] [Google Scholar]
- 15.Vidova V, Pol J, Volny M, Novak P, Havlicek V, Wiedmer SK and Holopainen JM, J. Lipid Res, 2010, 51, 2295–2302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Goodwin RJA, Scullion P, MacIntyre L, Watson DG and Pitt AR, Anal. Chem, 2010, 82, 3868–3873. [DOI] [PubMed] [Google Scholar]
- 17.Kaletas BK, van der Wiel IM, Stauber J, Dekker LJ, Guzel C, Kros JM, Luider TM and Heeren RMA, Proteomics, 2009, 9, 2622–2633. [DOI] [PubMed] [Google Scholar]
- 18.Stoeckli M, Chaurand P, Hallahan DE and Caprioli RM, Nat. Med, 2001, 7, 493–496. [DOI] [PubMed] [Google Scholar]
- 19.Chaurand P, Sanders ME, Jensen RA and Caprioli RM, Am. J. Pathol, 2004, 165, 1057–1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Johnson MD, Floyd JL and Caprioli RM, J. Neuropathol. Exp. Neurol, 2006, 65, 837–845. [DOI] [PubMed] [Google Scholar]
- 21.Schwamborn K, Krieg RC, Reska M, Jakse G, Knuechel R and Wellmann A, Int. J. Mol. Med, 2007, 20, 155–159. [PubMed] [Google Scholar]
- 22.Oppenheimer SR, Mi DM, Sanders ME and Caprioli RM, J. Proteome Res, 2010, 9, 2182–2190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Crecelius AC, Cornett DS, Caprioli RM, Williams B, Dawant BM and Bodenheimer B, J. Am. Soc. Mass Spectrom, 2005, 16, 1093–1099. [DOI] [PubMed] [Google Scholar]
- 24.Rohner TC, Staab D and Stoeckli M, Mech. Ageing Dev, 2005, 126, 177–185. [DOI] [PubMed] [Google Scholar]
- 25.Andersson M, Groseclose MR, Deutch AY and Caprioli RM, Nat. Methods, 2008, 5, 101–108. [DOI] [PubMed] [Google Scholar]
- 26.Reyzer ML, Caldwell RL, Dugger TC, Forbes JT, Ritter CA, Guix M, Arteaga CL and Caprioli RM, Cancer Res, 2004, 64, 9093–9100. [DOI] [PubMed] [Google Scholar]
- 27.Grey AC, Chaurand P, Caprioli RM and Schey KL, J. Proteome Res, 2009, 8, 3278–3283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Khatib-Shahidi S, Andersson M, Herman JL, Gillespie TA and Caprioli RM, Anal. Chem, 2006, 78, 6448–6456. [DOI] [PubMed] [Google Scholar]
- 29.Li Y, Shrestha B and Vertes A, Anal. Chem, 2008, 80, 407–420. [DOI] [PubMed] [Google Scholar]
- 30.Doroshenko VM, Laiko VV, Taranenko NI, Berkout VD and Lee HS, Int. J. Mass Spectrom, 2002, 221, 39–58. [Google Scholar]
- 31.Burnum KE, Frappier SL and Caprioli RM, Annu. Rev. Anal. Chem, 2008, 1, 689–705. [DOI] [PubMed] [Google Scholar]
- 32.Takats Z, Wiseman JM, Gologan B and Cooks RG, Science, 2004, 306, 471–473. [DOI] [PubMed] [Google Scholar]
- 33.Wiseman JM, Ifa DR, Song Q and Cooks RG, Angew. Chem., Int. Ed, 2006, 45, 7188–7192. [DOI] [PubMed] [Google Scholar]
- 34.Nemes P, Barton AA and Vertes A, Anal. Chem, 2009, 81, 6668–6675. [DOI] [PubMed] [Google Scholar]
- 35.Nemes P, Barton AA, Li Y and Vertes A, Anal. Chem, 2008, 80, 4575–4582. [DOI] [PubMed] [Google Scholar]
- 36.Nemes P and Vertes A, Anal. Chem, 2007, 79, 8098–8106. [DOI] [PubMed] [Google Scholar]
- 37.Shelley JT, Ray SJ and Hieftje GM, Anal. Chem, 2008, 80, 8308–8313. [DOI] [PubMed] [Google Scholar]
- 38.Liu YY, Ma XX, Lin ZQ, He MJ, Han GJ, Yang CD, Xing Z, Zhang SC and Zhang XR, Angew. Chem., Int. Ed, 2010, 49, 4435–4437. [DOI] [PubMed] [Google Scholar]
- 39.Galhena AS, Harris GA, Nyadong L, Murray KK and Fernandez FM, Anal. Chem, 2010, 82, 2178–2181. [DOI] [PubMed] [Google Scholar]
- 40.Yew JY, Cody RB and Kravitz EA, Proc. Natl. Acad. Sci. U. S. A, 2008, 105, 7135–7140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kertesz V, Ford MJ and Van Berkel GJ, Anal. Chem, 2005, 77, 7183–7189. [DOI] [PubMed] [Google Scholar]
- 42.Chen LC, Yoshimura K, Yu Z, Iwata R, Ito H, Suzuki H, Mori K, Ariyada O, Takeda S, Kubota T and Hiraoka K, J. Mass Spectrom, 2009, 44, 1469–1477. [DOI] [PubMed] [Google Scholar]
- 43.Sampson JS, Hawkridge AM and Muddiman DC, J. Am. Soc. Mass Spectrom, 2008, 19, 1527–1534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Costa AB and Cooks RG, Chem. Phys. Lett, 2008, 464, 1–8. [Google Scholar]
- 45.Costa AB and Cooks RG, Chem. Commun, 2007, 3915–3917. [DOI] [PubMed] [Google Scholar]
- 46.Wu C, Ifa DR, Manicke NE and Cooks RG, Anal. Chem, 2009, 81, 7618–7624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Dill AL, Ifa DR, Manicke NE, Costa AB, Ramos-Vara JA, Knapp DW and Cooks RG, Anal. Chem, 2009, 81, 8758–8764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Eberlin LS, Dill AL, Costa AB, Ifa DR, Cheng L, Masterson T, Koch M, Ratliff TL and Cooks RG, Anal. Chem, 2010, 82, 3430–3434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Dill AL, Eberlin LS, Costa AB, Zheng C, Ifa DR, Cheng L, Masterson TA, Koch MO, Vitek O and Cooks RG, Chem.–Eur. J, 2010, in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Eberlin L, Dill A, Golby A, Ligon K, Wiseman J, Cooks R and Agar N, Angew. Chem., Int. Ed, 2010, 49, 5953–5956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wiseman JM, Ifa DR, Zhu YX, Kissinger CB, Manicke NE, Kissinger PT and Cooks RG, Proc. Natl. Acad. Sci. U. S. A, 2008, 105, 18120–18125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kertesz V, Van Berkel GJ, Vavrek M, Koeplinger KA, Schneider BB and Covey TR, Anal. Chem, 2008, 80, 5168–5177. [DOI] [PubMed] [Google Scholar]
- 53.Watrous J, Hendricks N, Meehan M and Dorrestein PC, Anal. Chem, 2010, 82, 1598–1600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lane AL, Nyadong L, Galhena AS, Shearer TL, Stout EP, Parry RM, Kwasnik M, Wang MD, Hay ME, Fernandez FM and Kubanek J, Proc. Natl. Acad. Sci. U. S. A, 2009, 106, 7314–7319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ifa DR, Gumaelius L, Eberlin LS, Manicke N and Cooks RG, Analyst, 2007, 132, 461–467. [DOI] [PubMed] [Google Scholar]
- 56.Eberlin LS, Haddad R, Neto RCS, Cosso RG, Maia DRJ, Maldaner AO, Zacca JJ, Sanvido GB, Romao W, Vaz BG, Ifa DR, Dill A, Cooks RG and Eberlin MN, Analyst, 2010, 135, 2533–2539. [DOI] [PubMed] [Google Scholar]
- 57.Ifa DR, Manicke NE, Dill AL and Cooks G, Science, 2008, 321, 805–805. [DOI] [PubMed] [Google Scholar]
- 58.Eberlin LS, Ifa DR, Wu C and Cooks RG, Angew. Chem., Int. Ed, 2010, 49, 873–876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Nemes P, Woods AS and Vertes A, Anal. Chem, 2010, 82, 982–988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Shrestha B, Nemes P, Nazarian J, Hathout Y, Hoffman EP and Vertes A, Analyst, 2010, 135, 751–758. [DOI] [PubMed] [Google Scholar]
- 61.Andrade FJ, Shelley JT, Wetzel WC, Webb MR, Gamez G, Ray SJ and Hieftje GM, Anal. Chem, 2008, 80, 2654–2663. [DOI] [PubMed] [Google Scholar]
- 62.Na N, Zhao MX, Zhang SC, Yang CD and Zhang XR, J. Am. Soc. Mass Spectrom, 2007, 18, 1859–1862. [DOI] [PubMed] [Google Scholar]
- 63.Harper JD, Charipar NA, Mulligan CC, Zhang X, Cooks RG and Ouyang Z, Anal. Chem, 2008, 80, 9097–9104. [DOI] [PubMed] [Google Scholar]
- 64.Barentsz J, Takahashi S, Oyen W, Mus R, De Mulder P, Reznek R, Oudkerk M and Mali W, J. Clin. Oncol, 2006, 24, 3234–3244. [DOI] [PubMed] [Google Scholar]
- 65.Kobayashi H, Longmire MR, Ogawa M, Choyke PL and Kawamoto S, Lancet Oncol, 2010, 11, 589–595. [DOI] [PMC free article] [PubMed] [Google Scholar]


