Abstract
The binding between receptor-activated nuclear factor-κB (RANK) and the RANK ligand (RANKL) during osteoclast development is an important target for drugs that treat osteoporosis. The leucine-rich repeat-containing G-protein-coupled receptor 4 (LGR4) acts as a negative regulator of RANK-RANKL that suppresses canonical RANK signaling during osteoclast differentiation. Therefore, LGR4 agonists may be useful in inhibiting osteoclastogenesis and effectively treating osteoporosis. In the present study, bone marrow-derived macrophages and a mouse model of RANKL-induced bone loss were used to investigate the effect of mutant RANKL (MT RANKL), which was previously developed based on the crystal structure of the RANKL complex. In the present study, the binding affinity of wild-type (WT) RANKL and MT RANKL to RANK and LGR4 was determined using microscale thermophoresis analysis, and the effect of the ligands on the AKT-glycogen synthase kinase-3β (GSK-3β)-nuclear factor of activated T cells, cytoplasmic, calcineurin-dependent 1 (NFATc1) signaling cascade was investigated using western blotting and confocal microscopy. In addition, the expression of LGR4 and the colocalization of LGR4 with MT RANKL were analyzed in a mouse model of RANKL-induced bone loss. The results showed that in osteoclast precursor cells, MT RANKL bound with high affinity to LGR4 and increased GSK-3β phosphorylation independently of AKT, resulting in the inhibition of NFATc1 nuclear translocation. In the mouse model, MT RANKL colocalized with LGR4 and inhibited bone resorption. These results indicated that MT RANKL may inhibit RANKL-induced osteoclastogenesis through an LGR4-dependent pathway and this could be exploited to develop new therapies for osteoporosis.
Keywords: leucine-rich repeat-containing G-protein-coupled receptor 4, receptor activator of nuclear factor-κB ligand, osteoclast, osteoporosis, receptor activator of nuclear factor-κB, glycogen synthase kinase-3β
Introduction
The receptor activator of nuclear factor-κB ligand (RANKL, also known as TNFSF11) is an essential cytokine that regulates osteoclast differentiation and function (1). RANKL-mediated regulation of osteoclast proliferation, differentiation and function dictate the degree of skeletal remodeling, a process that maintains calcium homeostasis and removes accumulated aged or weakened bone (2). Several pharmaceutical agents are used for treating osteoporosis, including bone antiresorptive agents (such as bisphosphonates, estrogen and denosumab) and drugs that stimulate bone formation (such as parathyroid hormone) (3-5). However, currently available drugs that promote bone formation either have side effects; for example, they can increase the risk of breast cancer and cardiovascular disease, or do not improve bone quality sufficiently to reduce fracture susceptibility (6). Therefore, the development of agents that minimize bone resorption may be beneficial for treating osteoporosis. Denosumab is a human monoclonal antibody against RANKL that blocks the binding of RANKL to its receptor RANK (also known as TNFRSF11A), thereby inhibiting osteoclast differentiation and activity, leading to suppression of bone resorption in osteoporosis and other bone-related disorders (7). The United States Food and Drug Administration-approved indications for denosumab include the prevention of skeletal-related events (e.g., bone pain and fractures) secondary to multiple myeloma or bone metastases from solid tumors, giant cell tumors of the bone, hypercalcemia related to malignancy, osteoporosis in postmenopausal women and men at high risk of fracture, glucocorticoid-induced osteoporosis and bone loss (8-11). However, its side effects include hypocalcemia, serious infections, skin reactions, inhibition of bone turnover and jaw necrosis (12,13). A recently reported side effect of denosumab is rebound resorption, which occurs when denosumab treatment is discontinued, especially in patients on long-term treatment (14).
The leucine-rich repeat-containing G protein-coupled receptor 4 (LGR4, also known as GPR48) is another potential target for inhibiting bone resorption, as activation of this receptor triggers a signaling pathway that inhibits RANK-RANKL signaling during osteoclastogenesis (15). Previous studies have indicated that LGR4 competes with RANK for binding to RANKL and suppresses canonical RANK signaling during osteoclast differentiation (15,16). LGR4 belongs to the LGR family of receptors; two other LGR family members, the thyroid-stimulating hormone receptor and follicle-stimulating hormone receptor, also regulate osteoclast differentiation and resorption (17,18). The binding of RANKL to LGR4 activates the Gαq-mediated glycogen synthase kinase-3β (GSK-3β) signaling pathway. When activated by RANKL, this pathway is crucial for osteoclast differentiation and the subsequent suppression of the expression and activity of nuclear factor of activated T cells, cytoplasmic, calcineurin-dependent 1 (NFATc1) during osteoclast development (15,16,19). In addition, RANKL-RANK-AKT-NFATc1 signaling may directly induce LGR4 expression. Notably, LGR4 is upregulated in severe pathological bone environments, such as osteoporosis, suggesting that therapies that target LGR4 would be ideal for rebalancing bone remodeling (20).
In our previous studies, mutant RANKL (MT RANKL), based on the wild-type RANKL (WT RANKL) sequence, was developed, and whether it bound to LGR4 and acted as an agonist was investigated. In in vitro and in vivo osteoporosis models, MT RANKL inhibited osteoclast differentiation and production, suggesting that crosstalk exists between RANKL, RANK, and LGR4 signaling (21,22). However, it is currently unknown whether MT RANKL triggers LGR4 downstream signaling pathways, such as GSK-3β and AKT.
Therefore, the present study aimed to demonstrate that MT RANKL binds to LGR4, instead of RANK, and that via this ligand, LGR4 negatively regulates the RANK signaling cascade during RANKL-induced osteoclast differentiation and bone remodeling. In addition, by clarifying the effect and mechanism of action of MT RANKL-LGR4 signaling on osteoclast differentiation and bone resorption, the study aimed to determine whether an agonist that binds LGR4 could be considered a pharmacological approach for treating osteoporosis.
Materials and methods
Chemicals and reagents
Unless otherwise indicated, all of the chemical reagents used in the present study were purchased from Millipore Sigma, and cell culture medium was purchased from Thermo Fisher Scientific, Inc.
Generation and purification of WT and MT RANKL, RANK and LGR4
WT RANKL and MT RANKL were generated as described previously (23). The genes encoding RANK and LGR4 were synthesized and codon-optimized for Escherichia coli expression by GeneArt Gene Synthesis (Thermo Fisher Scientific, Inc.), and then subcloned into the pETM-13 vector (EMBL) between the NcoI and XhoI restriction sites. This plasmid enables a histidine (His) tag to be placed at the C-terminus of the mini-protein. The recombinant protein was successfully overexpressed in E. coli BL21(DE3) cells (Thermo Fisher Scientific, Inc.), resulting in a yield of 70 mg/l. Briefly, an overnight starting culture of 10 ml was prepared for growth in 1 l of Luria Bertani medium containing kanamycin (50 μg/l), which was then induced with isopropyl β-D-1-thiogalactopyranoside (0.8 mM) at 16°C for 16 h. The protein was purified by ultrasonicating the bacterial cells at 20 KHz, at 5°C for 15 min, which were then resuspended in binding buffer [300 mM NaCl, 50 mM Tris-HCl, 10 mM imidazole, 2.5% (v/v) glycerol, pH 7.8] containing a protease inhibitor cocktail (Roche Diagnostics). The cell lysate was then centrifuged for 30 min at 14,000 × g and 5°C to remove the cell debris and inclusion bodies. The supernatant was centrifuged again for 30 min at 40,000 × g and 5°C after which, the resulting membrane pellet was resuspended in PBS lysis buffer [PBS, pH 8.0; 10% (w/v) glycerol, 1 mM DTT, 0.002% (w/v) phenylmethylsulfonyl fluoride (PMSF) and 10 mg/l DNase I (PanReac AppliChem); 5 ml PBS lysis buffer per 1 g cells] and the cell membranes were isolated by centrifugation at 34,000 × g and 5°C for 30 min. The membrane pellets were flash-frozen in liquid nitrogen and stored at −80°C. To solubilize the protein, the membranes were resuspended in buffer S [50 mM Tris/HCl, pH 7.8; 200 mM NaCl; 1.2% (w/v) FosCholine-16; 2.5 mM DTT; 0.002% (w/v) PMSF] and stirred at 700 rpm for 1 h. The cell lysate was ultracentrifuged at 230,000 × g and 5°C for 60 min. The resulting supernatant was loaded onto a 5 ml Ni-NTA HisTrap HP column (Cytiva) equilibrated in buffer C-P [buffer C (50 mM HEPES/NaOH, pH 7.6; 300 mM NaCl, 5% (w/v) glycerol; 5 ml lysis buffer C per 1 g cells) with 0.002% (w/v) PMSF]. The column was washed with 10 column volumes (CV) of buffer C-P, followed by 20 CV of buffer C-ATP [buffer C with an additional 50 mM KCl, 20 mM MgCl2, 10 mM ATP, and 0.002% (w/v) PMSF]. The column was further washed with 10 CV of buffer C-P containing 50 mM imidazole and 100 mM imidazole. The RANK and LGR4 proteins were eluted with 500 mM imidazole in the C-P buffer. The purified proteins were concentrated in a 50 kDa Amicon Ultra-15 concentrator (MilliporeSigma). The buffer was changed by desalting on a PD-10 column (Cytiva) for binding affinity measurement.
Binding affinity measurement
The protein-binding affinity was measured using microscale thermophoresis (MST) (24). The MST experiments were performed using Monolith NT.115 systems (NanoTemper Technologies GmbH) and a red filter. Briefly, all dilutions were prepared to ensure that no other gradient (salt, glycerol, DMSO, etc.) was created during the buffer mixing. To minimize the adsorption of the sample to the material, 0.05% Tween 20 was added to PBS, which was used to dilute all of the receptors and ligands. To measure protein-protein binding, the receptor protein RANK or LGR4 and the ligand WT RANKL or MT RANKL were mixed with an equal volume of the fluorescent ligand spiperone-Cy5 (NanoTemper Technologies GmbH) to obtain final ligand concentrations of 0.125, 5, 7.5 and 12 nM. After incubation at 20°C for 1 h, the samples were loaded onto capillaries and the LED was set to 20% for 0.125 nM samples, and 1% for 5, 7.5 and 12 nM samples, using medium MST power. For the receptor titration assay, various concentrations of protein (10 mg/ml-5 μg/ml total protein) were mixed with a specific concentration of the fluorescent ligand, namely, spiperone-Cy5 was added at a final concentration of 5, 7.5 or 12 nM to each protein dilution point. The sample was incubated at 20°C for 1 h before capillary loading. The LED power was set to 1% and the MST power was set to medium. The intersection points in the binding curves were determined according to the manufacturer's protocol and equilibrium dissociation constant (Kd) values were obtained using Frobenius normalization (%) from the mean of three replicates.
Primary cell culture and in vitro osteoclast differentiation
A total of 20 female C57BL/6 mice (age, 5 weeks; weight, ~25 g; Orient Bio, Inc.) were housed under controlled 12-h light/dark cycle conditions, at a constant room temperature of 20±1°C and humidity of 40-60%. The mice were fed ad libitum for 1 week prior to euthanasia with CO2 at a displacement rate of 50%/min for use in the in vitro studies. Bone marrow cells were obtained from the tibiae and femurs of mice by flushing the bones with α-MEM (Gibco; Thermo Fisher Scientific, Inc.) using previously described methods (25). After the removal of red blood cells, bone marrow cells were resuspended in complete α-MEM containing 10% (v/v) fetal bovine serum (Gibco; Thermo Fisher Scientific, Inc.), 100 U/ml penicillin and 100 μg/ml streptomycin, and incubated at 37°C for 24 h in the presence of 10 ng/ml macrophage colony-stimulating factor (M-CSF; Cell Guidance Systems Ltd.). Non-adherent cells were collected and cultured with 30 ng/ml M-CSF for 3 days to generate bone marrow-derived macrophages (BMDMs). To generate pre-osteoclast lineage cells and osteoclast lineage cells, BMDMs (50,000/cm2) were cultured in complete α-MEM containing 30 ng/ml M-CSF and 75 ng/ml WT RANKL at 37°C for 72 h.
RNA interference
SMART pool mixtures of small interfering RNA (siRNA) targeting mouse LGR4 (cat. no. E-047291-00-0050, lot no. 210719) and Accell non-targeting mouse RNA (cat. no. D-001910-10-50, lot no. 3026176) were designed and synthesized by GE Healthcare Dharmacon, Inc. For siRNA transfection, BMDMs were seeded on a 24-well plate at a density of 100,000 cells/well, and after 24 h, the cells were transfected with each siRNA (20 pmol) at 37°C for 1 day using Lipofectamine® 2000 (Invitrogen; Thermo Fisher Scientific, Inc.) according to the manufacturer's protocol. After transfection, the BMDMs were cultured with M-CSF (30 ng/ml) and RANKL (75 ng/ml) for 3 days to induce differentiation into osteoclasts.
Tartrate-resistant acid phosphatase (TRAP) assay
After the BMDMs were cultured with WT RANKL or MT RANKL, the cells were washed once with PBS and fixed in 10% formalin (in PBS) at 5°C for 5 min. After three washes with distilled water, TRAP staining was performed for 30-40 min according to the manufacturer's instructions (Kamiya Biomedical Co.). The stained cells were examined under an ECLIPSE Ts2R inverted light microscope (Nikon Corporation) and images were captured using a digital camera (Nikon Corporation) with NIS-Elements imaging software (Nikon Corporation). The number of multinucleated cells was counted manually.
Reverse transcription-quantitative PCR (RT-PCR) analysis
The BMDMs were incubated with 30 ng/ml M-CSF and 75 ng/ml WT RANKL, or 75 ng/ml WT RANKL plus 75 ng/ml MT RANKL at 37.5°C for the indicated times (0, 1 and 2 days) in 6-well plates. Total RNA was extracted from the cells using TRIzol® reagent (Invitrogen; Thermo Fisher Scientific, Inc.). The cDNA was then obtained from 2 μg total RNA using ReverTra Ace qPCR RT master mix (Toyobo Life Science) according to the manufacturer's protocol. The mRNA expression levels were measured using qPCR and GAPDH was used as a control. qPCR was performed with a CFX Connect real-time PCR detection system (Bio-Rad Laboratories, Inc.) using a 20 μl reaction mixture containing 10 μl IQ SYBR Green supermix (Bio-Rad Laboratories, Inc.), 10 pmol forward primer, 10 pmol reverse primer and 1 μg cDNA. The sequences of the primers used to target TRAP, OSCAR, NFATc1 and GAPDH are presented as follows: TRAP, 5′-TAC CTG TGT GGA CAT GAC C-3′ (forward) and 5′-CAG ATC CAT AGT GAA ACC GC-3′ (reverse); OSCAR, 5′-CTG CTG GTA ACG GAT CAG CTC CCC AGA-3′ (forward) and 5′-CCA AGG AGC CAG AAC CTT CGA AAC T-3′ (reverse); NFATc1, 5′-GAG TAC ACC TTC CAG CAC CTT-3′ (forward) and 5′-TAT GAT GTC GGG GAA AGA GA-3′ (reverse) and GAPDH, 5′-TCA AGA AGG TGG TGA AGC AG-3′ (forward) and 5′-AGT GGG AGT TGC TGT TGA AGT-3′ (reverse). The amplification parameters consisted of an initial denaturation step at 95.8°C for 5 min followed by 40 cycles of three-step PCR (denaturation at 95.8°C for 1 min, annealing at 60.8°C for 30 sec and extension at 72.8°C for 1 min). The fluorescence resulting from the incorporation of SYBR Green dye into the double-stranded DNA produced during PCR was quantified using the 2−ΔΔCq method (26).
Western blot analysis
The BMDMs were serum-starved for 8 h and treated with 2 μg/ml WT RANKL or MT RANKL at 0, 5, 10 and 15 min intervals at 37°C. The cells were lysed in lysis buffer (50 mM Tris-HCl, pH 7.5; 1% NP-40; 150 mM NaCl; 0.02% sodium azide; 1 mg/ml pepstatin A; 2 mg/ml aprotinin; 20 mg/ml leupeptin; 150 mg/ml PMSF). After protein quantification using the BCA protein assay kit (Pierce; Thermo Fisher Scientific, Inc.), ~30 μg cell lysate was separated by SDS-PAGE 10% on gels and transferred onto a polyvinylidene difluoride membrane (Amersham; Cytiva). Each membrane was blocked for 30 min with a blocking solution containing 5% skim milk in Tris-buffered saline containing Tween-20 (TBST; 2.42 g/l Tris-HCl, 8 g/l NaCl, 0.1% Tween-20, pH 7.6) and rinsed with TBST. The membrane was then incubated overnight at 4°C with the following primary antibodies: Phosphorylated (p)-AKT (1:1,000; cat. no. 9271S; Cell Signaling Technology, Inc.), AKT (1:1,000; cat. no. 9272S; Cell Signaling Technology, Inc.), p-GSK-3β (Ser9, 1:1,000; cat. no. 9336S; Cell Signaling Technology, Inc.), GSK-3β (1:1,000; cat. no. 9315S; Cell Signaling Technology, Inc.), p-Src (1:1,000; cat. no. 2105; Cell Signaling Technology, Inc.), Src (1:1,000; cat. no. 2108; Cell Signaling Technology, Inc.), His Tag (1:1,000; cat. no. SAB1306082; MilliporeSigma), NFATc1 (1:1,000; cat. no. 8032; Cell Signaling Technology, Inc.), Histone H1 (1:1,000; cat. no. sc-393358; Santa Cruz Biotechnology, Inc.), β-Actin (1:1,000; cat. no. sc-47778; Santa Cruz Biotechnology, Inc.), RANK (1:1,000; cat. no. 4845S; Cell Signaling Technology, Inc.) and LGR4 (1:500; cat. no. MBS468030; MyBioSource, Inc.). A mouse monoclonal immunoglobulin G antibody specific for GAPDH (1:1,000; cat. no. 97166; Cell Signaling Technology, Inc.) was used as a control. After rinsing with TBST, the membrane was incubated at 4°C for 1 h with anti-rabbit (1:2,000; cat no. sc-2357; Santa Cruz Biotechnology, Inc.) or anti-mouse (1:2,000; cat. no. sc-525409; Santa Cruz Biotechnology, Inc.) horseradish peroxidase-conjugated secondary antibodies. The membrane was then rinsed with TBST, and the protein immunoreactivity was detected using an enhanced chemiluminescence detection kit (Amersham; Cytiva). Finally, the blot images were acquired using a chemiluminescence imaging system (Vilber Lourmat) and the densitometric semi-quantification of the detected bands was performed using Image J 1.52a (National Institutes of Health) after normalization to GAPDH.
Cell viability assay
Cell viability was evaluated using the MTT assay (MilliporeSigma). The BMDMs were seeded into 96-well plates at 5×103 cells/well in 200 μl medium and cultured for 1 day with MK2206 (0, 0.1, 0.2, 0.5, 1, 2, 5, and 10 μM; MilliporeSigma) or LiCl (0, 2, 5, 10, 15, 20, 25 and 30 mM; MilliporeSigma) at 37°C. For each experiment, the medium was removed and 20 μl MTT (50 μg/ml) was added to the wells. The cells were then incubated at 37°C for 4.5 h to allow the color to develop, and the formazan was solubilized with the addition of 50 μl dimethyl sulfoxide (Calbiochem; Merck KGaA). The optical density was measured at 570 nm using a microtiter plate reader (Bio-Rad Laboratories, Inc.).
Separation of nuclear and cytoplasmic fractions
After siRNA transfection for 24 h or treatment with LiCl for 8 h, the BMDMs were incubated with 75 ng/ml WT RANKL or 75 ng/ml MT RANKL for 24 h in 6-well plates. The cells were rinsed in PBS and collected in microtubes (Eppendorf). A 0.5-ml volume of Solution A (10 mM HEPES, 1.5 mM MgCl2, 10 mM KCl, 0.5 mM DTT, 0.05% NP40, pH 7.9) was then added, after which the cells were centrifuged at 805 × g for 10 min at 4°C. The supernatant, containing mostly cytoplasmic constituents, was then removed and transferred to another tube. To yield a nuclear pellet, 0.4 ml solution B [5 mM HEPES, 1.5 mM MgCl2, 0.2 mM EDTA, 0.5 mM DTT, 26% glycerol (v/v), 300 mM NaCl, pH 7.9] was added and the contents of the tube were mixed thoroughly and placed on a small rotator shaker for 15 min. Finally, the mixture was centrifuged at 24,000 × g for 20 min at 4°C. The supernatant containing proteins from the nuclear extract was removed and transferred carefully into a fresh tube. The nuclear and cytosolic extracts were frozen at −80°C in aliquots prior to western blot analysis. The protein concentration of each sample was determined using the BCA protein assay kit (Thermo Fisher Scientific, Inc.), and the cytosolic and nuclear fractions were subjected to western blot analysis. Histone-H1 and β-actin were used as loading controls for the nuclear and cytoplasmic fractions, respectively.
Animal study
A total of 20 female C57BL/6 mice (age, 5 weeks; weight, ~25 g; Orient Bio, Inc.) were housed under a controlled 12-h light/dark cycle, at a constant room temperature of 20±1°C) and humidity of 40-60%. The mice were fed ad libitum for 1 week prior to being randomly divided into four groups (n=5/group). The control group was intraperitoneally injected with PBS; the WT group was injected with WT RANKL (2 mg/kg) in PBS; and the WT + MT group was injected with WT RANKL (2 mg/kg) and MT RANKL (2 mg/kg) in PBS at 24-h intervals for 2 days. The MT group was also injected with MT RANKL (2 mg/kg) in PBS. The total injections were performed twice in each group. On day 3 after injection, all animals were euthanized with 50% CO2 and femur bone samples were collected for further study.
Micro-computed tomography (CT) imaging and data acquisition
Micro-CT scanning of the distal femurs of mice was initiated at the level of the growth plate using a Quantum GX micro-CT imaging system (PerkinElmer, Inc.) located at the Korea Basic Science Institute (Gwangju, Republic of Korea), according to the methods described in a previous study (23). The bone volume/tissue volume (BV/TV), trabecular separation (Tb. Sp.) and bone mineral density (BMD) of the femurs were calculated using the region of interest tool. Parameter values are shown as mean ± standard deviation (SD).
Histological and immunohistochemical analysis of bone specimens
Mouse femur tissues were fixed for 1 week in cold 4% formalin at 5°C. The bone tissue was then decalcified using 0.5 M EDTA, cut into 3-mm sections at the midpoint and embedded in paraffin. The paraffin-embedded tissue sections were stained with hematoxylin and eosin (H&E) using a staining kit (cat. no. ab245880; Abcam) according to the manufacturer's protocol, and images were acquired using an ECLIPSE Ts2R inverted light microscope (Nikon Corporation).
For the immunohistochemical study, bone sections were deparaffinized using three changes of xylene and then rehydrated using graded concentrations of ethanol solutions, ending with distilled water. For antigen retrieval, the slides were placed in 0.01 M citrate buffer (pH 6.0) and heated in a steamer for 30 min. The endogenous peroxidases were quenched by incubating the sections with 3% hydrogen peroxide for 20 min at room temperature and blockading was performed with 3% BSA (in PBS; cat. no. 9048-46-8; VWR; Avantor) at 4°C for 1 h. The sections were then incubated overnight at 4°C with a 1:50 dilution of the following appropriate primary antibodies: Anti-LGR4 (1:100; cat. no. PA5-67868; Invitrogen; Thermo Fisher Scientific, Inc.), ani-RANK (1:100; cat. no. PA5-88904; Invitrogen; Thermo Fisher Scientific, Inc.) or NFATc1 (1:100; cat. no. MA5-32686; Invitrogen; Thermo Fisher Scientific, Inc.). Subsequently, the sections were incubated at 20°C for 30 min with a biotin-labeled mouse anti-rabbit secondary antibody (1:200; cat. no. 31824; Invitrogen; Thermo Fisher Scientific, Inc.), washed in PBS, and incubated at 20°C for 30 min with a streptavidin-peroxidase conjugate (Dako; Agilent Technologies, Inc.). The reaction was developed for 5 min using 3,30-diaminobenzidine tetrahydrochloride (MilliporeSigma). The slides were counterstained with hematoxylin, dehydrated and a coverslip was added. Finally, immune-stained slides were imaged using an ECLIPSE Ts2R inverted light microscope (Nikon Corporation) and the immune-positive area was analyzed using ImageJ software.
Immunofluorescence analysis of bone specimens
Paraffin-embedded bone sections were prepared according to the aforementioned protocol and incubated overnight at 4°C with the following primary antibodies; Anti-LGR4 (1:100; cat. no. PA5-67868; Invitrogen; Thermo Fisher Scientific, Inc.), ani-RANK (1:100; cat. no. PA5-88904; Invitrogen; Thermo Fisher Scientific, Inc.) and anti-glutathione S-transferase (GST; 1:100; cat. no. 13-6700; Invitrogen; Thermo Fisher Scientific, Inc.). Subsequently, sections were stained at 4°C for 1 h using Alexa Fluor 594 goat anti-rabbit (1:500; cat. no. A11037; Invitrogen; Thermo Fisher Scientific, Inc.) and Alexa Fluor 594 goat anti–mouse secondary antibodies (1:500; cat. no. A11032; Invitrogen; Thermo Fisher Scientific, Inc.). After washing with PBST, immunolabeled cells were counter-stained with DAPI in Pro-Long Gold mounting solution (Invitrogen; Thermo Fisher Scientific, Inc.). Digital images were acquired using a TCS SP5 AOBS laser-scanning confocal microscope (Leica Microsystems GmbH) and co-localization of RANKL and LGR4, or RANKL and RANK was analyzed comparing the Pearson correlation coefficient between WT RANKL- and MT RANKL-treated groups.
Statistical analysis
All in vitro and in vivo studies were conducted at least in triplicate. All quantitative results are presented as the mean ± SD. The primary comparisons of cell-based data and data from all the animal studies were analyzed using a one-way or two-way analysis of variance with a Bonferroni multiple-comparisons test, or unpaired Student's t-test. All of the reported P-values are two-sided, and P<0.05 was considered to indicate a statistically significant difference. All statistical analyses were performed using GraphPad Prism version 7 (Dotmatics).
Results
Binding affinity of WT RANKL and MT RANKL for RANK and LGR4
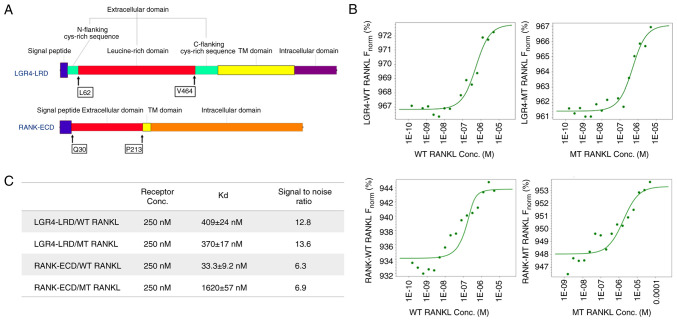
The extracellular domains of RANK and LGR4 used in the ligand-binding assays are shown in Fig. 1A. RANK and LGR4 were detected as His-tagged proteins of 24 kDa for RANK and 47 kDa for LGR4 (the impure form of LGR4 was detected above 47 kDa), respectively, by western blot analysis (Fig. S1).
Figure 1.
Suitability of microscale thermophoresis as a sensitive method was presented to determine WT RANKL and MT RANKL binding affinities. (A) Protein sequences of the LGR4 and RANK receptor proteins used in the present study. The LGR4 receptor sequence from L62 to V464 is the extracellular domain that represents the ligand-binding domain. The ligand-binding domain of RANK is from Q30 to P213. (B) Concentration of WT RANKL/MT RANKL used in titration experiments ranged from 11.5 nM to 50 μM, and the concentration of the labeled LGR4 and RANK receptors was constant at 250 nM. The y-axis shows affinity analysis data using Frobenius normalization [F norm (%)]. (C) Binding affinities (Kd values) of LGR4 and RANK to WT RANKL and MT RANKL. Data are presented as the mean ± SD of each data point calculated from three independent thermophoresis measurements. Kd, dissociation constant; LGR4, leucine-rich repeat-containing G-protein-coupled receptor 4; MT, mutant; RANK, receptor-activated nuclear factor-κB; RANKL, RANK ligand; WT, wild-type; ECD, extracellular domain; LRD, leucine rich domain.
MST assays were carried out to determine the binding affinities of WT RANKL and MT RANKL for RANK and LGR4 (Fig. 1B and C). The MST measurements showed that the Kd for the binding of WT RANKL to RANK was 33.3±9.2 nM and that for binding of MT RANKL to RANK was 1.62±0.057 μM, indicating that the affinity of MT RANKL for RANK was ~48.7-fold lower than the affinity of WT RANKL for RANK. The Kd values for binding of WT RANKL to LGR4 (409±24 nM) and MT RANKL for LGR4 (370±17 nM) did not show much difference.
These results showed that MT RANKL bound strongly to LGR4 but not to RANK, whereas WT RANKL bound with equal affinity to LGR4 and RANK. This finding suggested that MT RANKL may stimulate the LGR4 signaling cascade without stimulating the canonical RANK signaling cascade.
Effect of MT RANKL on the RANK-RANKL and LGR4-RANKL signaling cascades
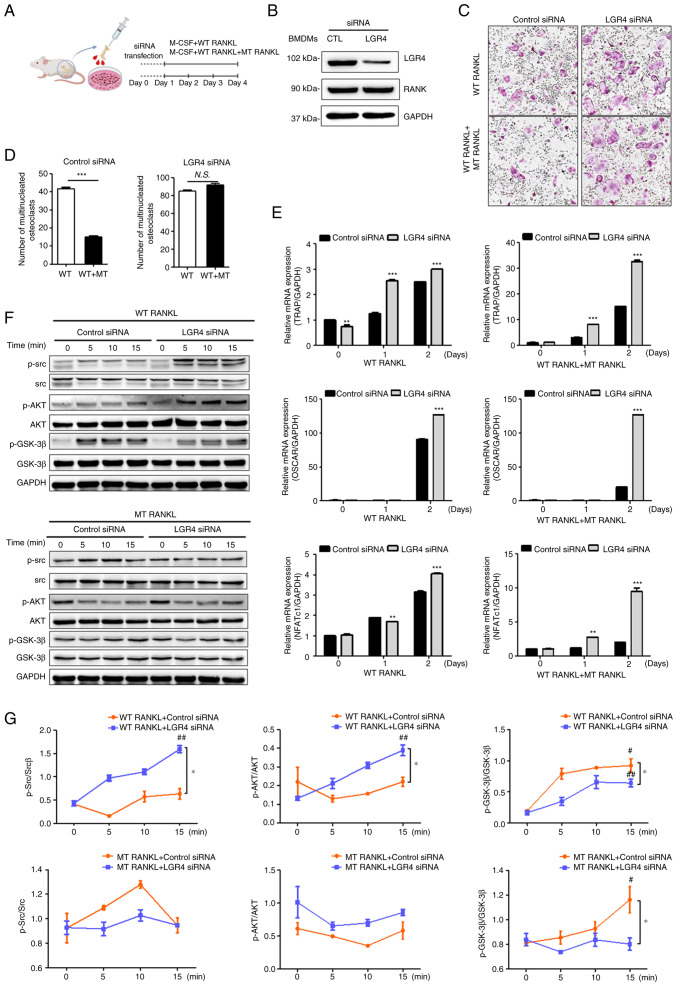
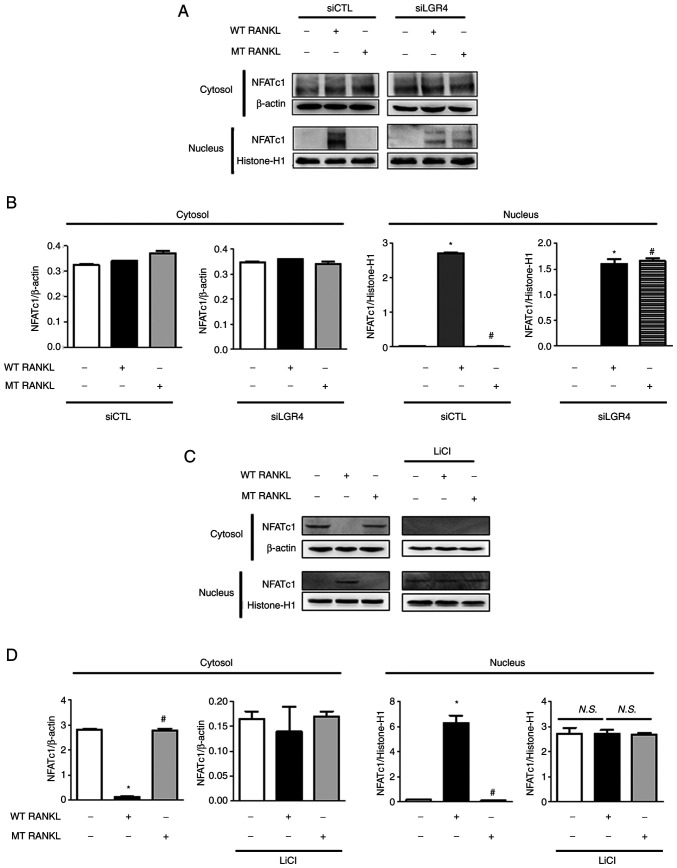
To investigate the effect of MT RANKL on the RANKL-induced LGR4 signaling cascade on osteoclastogenesis in vitro, BMDMs were transfected with LGR4 siRNA (Fig. 2A). The knockdown of LGR4 expression in LGR4 siRNA-transfected BMDMs was confirmed using western blotting, which also showed that RANK expression was unaffected (Fig. 2B).
Figure 2.
Effect of MT RANKL on osteoclast differentiation in vitro. (A) Schedule for treating control or LGR4 siRNA-transfected BMDMs with WT RANKL or WT RANKL + MT RANKL. (B) Western blotting of LGR4 and RANK expression in LGR4 siRNA-transfected BMDMs. LGR4 expression was markedly lower in LGR4 siRNA-treated BMDMs. (C) A representative image of BMDMs stained for TRAP (red) following treatment of control siRNA- or LGR4 siRNA-transfected BMDMs with WT RANKL (75 ng/ml) or WT RANKL (75 ng/ml) + MT RANKL (75 ng/ml). Magnification, ×100; scale bar, 20 μm. (D) Number of multinucleated TRAP-positive cells (≥3 nuclei). ***P<0.001. (E) Osteoclast-related gene expression in control siRNA- or LGR4 siRNA-transfected BMDMs. BMDMs were exposed to WT RANKL (75 ng/ml) or WT RANKL (75 ng/ml) + MT RANKL (75 ng/ml) for 2 days. Gene expression was determined by reverse transcription-quantitative PCR and normalized to the expression of GAPDH. **P<0.01 and ***P<0.001 vs. Control siRNA. (F) Western blot analysis of RANK and LGR4 signaling cascades in control siRNA- or LGR4 siRNA-transfected BMDMs in the presence of WT RANKL (2 μg/ml) or MT RANKL (2 μg/ml). GAPDH was used as a loading control. BMDM, bone marrow-derived macrophage. (G) Densitometric value of p-Src/Src, p-AKT/AKT and p-GSK-3β/GSK-3β, as determined by western blot analysis. Results are representative of three separate experiments that had comparable results. *P<0.05 Control siRNA vs.LGR4 siRNA at 15 min; #P<0.05 vs. WT RANKL + Control siRNA at 0 min, ##P<0.05 vs. MT RANKL+ Control siRNA at 0 min. GSK-3β, glycogen synthase kinase-3β; LGR4, leucine-rich repeat-containing G-protein-coupled receptor 4; M-CSF, macrophage colony-stimulating factor; MT, mutant; NFATc1, nuclear factor of activated T cells, cytoplasmic, calcineurin-dependent 1; N.S., not significant; p-, phosphorylated; RANK, receptor-activated nuclear factor-κB; RANKL, RANK ligand; siRNA, small interfering RNA; TRAP, tartrate-resistant acid phosphatase; WT, wild-type.
The numbers of BMDMs that differentiated into mature, TRAP-positive, multinucleated osteoclasts were subsequently counted (Fig. 2C). In the presence of WT RANKL, MT RANKL decreased the number of TRAP-positive cells in control siRNA-transfected BMDMs, but not in LGR4 siRNA-transfected BMDMs. In addition, the number of osteoclast cells was significantly decreased in the WT RANKL- and MT RANKL-treated BMDMs in the presence of control siRNA (Fig. 2D) but not in the presence of LGR4 siRNA.
To evaluate the effect of MT RANKL signaling via the LGR4-dependent pathway on the mRNA expression levels of osteoclastogenesis-related genes, RT-qPCR was used to investigate the expression of several osteoclast-specific genes in BMDMs treated with either WT RANKL, or both WT RANKL and MT RANKL (Fig. 2E). The results showed that on day 2 post-treatment, there was a significant increase in the mRNA expression levels of TRAP, OSCAR and NFATc1 in the LGR4 siRNA-transfected BMDMs treated with either WT RANKL, or both WT RANKL and MT RANKL, compared with those in the control siRNA-transfected BMDMs; these genes are markers of osteoclast differentiation and activity.
To evaluate the effect of MT RANKL on the LGR4-RANKL signaling cascade, the present study investigated whether treatment of BMDMs with WT RANKL or MT RANKL induced the phosphorylation of AKT, Src, and GSK-3β via RANK and LGR4 signaling cascades (Fig. 2F and G). In WT RANKL-treated BMDMs, transfection with LGR4 siRNA induced the obvious increase in Src and AKT phosphorylation, and a decrease in GSK-3β phosphorylation compared with the control siRNA. However, LGR4 siRNA in MT RANKL-treated BMDMs did not affect AKT phosphorylation compared with control siRNA. In addition, MT RANKL alone significantly increased the phosphorylation of GSK-3β compared with untreated BMDMs at 0 min, and LGR4 siRNA transfection decreased GSK-3β phosphorylation in MT RANKL-treated BMDMs compared with control siRNA.
Overall, these results demonstrated that in RANKL-treated BMDMs, MT RANKL may inhibit osteoclast differentiation and activity via the LGR4-dependent signaling pathway rather than via the RANK-mediated signaling.
Effect of MT RANKL on the AKT signaling cascade
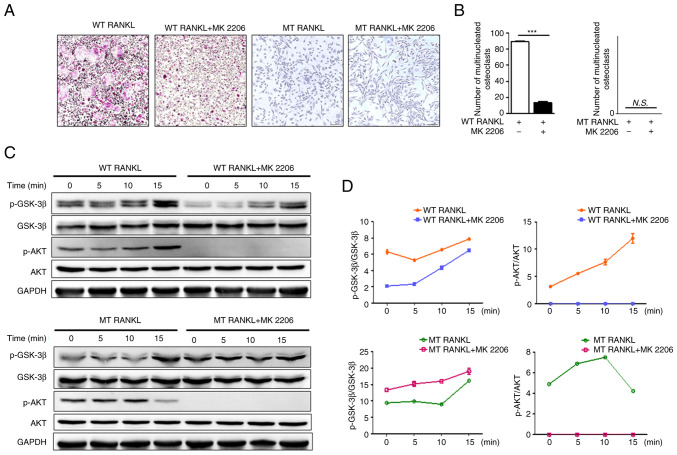
To evaluate the effects of WT RANKL and MT RANKL on the LGR4-mediated AKT signaling cascade, BMDMs were pretreated with the AKT inhibitor MK2206. Cell viability assays were performed to determine the optimal concentration of MK2206 (Fig. S2). Cell viability decreased with increases in MK2206 concentration; 0.2 μM MK2206 was the maximum concentration that did not affect BMDM cell viability; therefore, this concentration was used in subsequent experiments.
Treatment of BMDMs with WT RANKL in the presence of MK2206 significantly decreased the number of TRAP-positive multinuclear cells (Fig. 3A and B), but treatment with MT RANKL in the presence or absence of MK2206 did not affect the number of TRAP-positive multinuclear cells.
Figure 3.
Effect of MT RANKL on the AKT signaling cascade. (A) TRAP staining in the presence of WT RANKL (75 ng/ml) or MT RANKL (75 ng/ml) in BMDMs pretreated with MK2206 for 8 h before RANKL treatment. Magnification, ×100; scale bar, 20 μm. (B) Number of multinucleated TRAP-positive cells (≥3 nuclei). ***P<0.001. (C) Western blot analysis of RANK and LGR4 signaling cascades. BMDMs were exposed to WT RANKL (2 μg/ml) or MT RANKL (2 μg/ml) for 0, 5, 10 and 15 min after 8 h with or without MK2206 pretreatment. GAPDH was used as a loading control. (D) Densitometric value of p-GSK-3β/GSK-3β and p-AKT/AKT determined by western blot analysis. Results are representative of three separate experiments that had comparable results. BMDM, bone marrow-derived macrophage; GSK-3β, glycogen synthase kinase-3β; LGR4, leucine-rich repeat-containing G-protein-coupled receptor 4; MT, mutant; N.S., not significant; p-, phosphorylated; RANKL, receptor-activated nuclear factor-κB ligand; siRNA, small interfering RNA; TRAP, tartrate-resistant acid phosphatase; WT, wild-type.
Although WT RANKL slightly decreased GSK-3β phosphorylation in the presence of MK2206 (Fig. 3C and D), MT RANKL in MK2206-pretreated BMDMs exhibited increased levels of p-GSK-3β in compared with MT RANKL alone. In addition, MK2206 completely blocked phosphorylation of AKT in WT RANKL- and MT RANKL-treated BMDMs. These results suggested that the phosphorylation of GSK-3β by MT RANKL may be independent of AKT, since phosphorylation of GSK-3β still occurred in MT RANKL-treated BMDMS for which AKT phosphorylation was blocked, and MT RANKL alone did not have an effect on the osteoclastogenesis progress.
Effect of MT RANKL on the GSK-3β signaling cascade
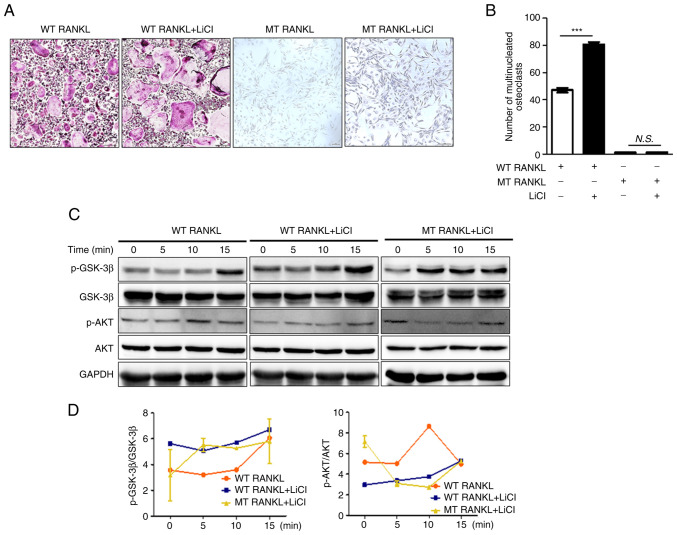
To investigate the LGR4-mediated GSK-3β signaling cascade, WT RANKL- or MT RANKL-treated BMDMs were pretreated with lithium chloride (LiCl), a powerful GSK-3β inhibitor (27,28). Cell viability assays were performed to determine the appropriate LiCl concentration (Fig. S3). LiCl concentration-dependently decreased cell viability; since 5 mM LiCl did not affect BMDM cell viability, subsequent experiments were performed using this concentration.
Treatment of LiCl-pretreated BMDMs with WT RANKL significantly increased the number of TRAP-positive multinuclear cell (Fig. 4A and B). However, MT RANKL did not affect the number of TRAP-positive multinuclear cells in the presence or absence of LiCl.
Figure 4.
Effect of MT RANKL on the GSK-3β signaling cascade. (A) TRAP staining results in the presence of WT RANKL (75 ng/ml) or MT RANKL (75 ng/ml) in BMDMs pretreated with LiCl for 8 h before RANKL treatment. Magnification, ×100; scale bar, 20 μm. (B) Number of multinucleated TRAP-positive cells (≥3 nuclei). ***P<0.001. (C) Western blot analysis of the RANK and LGR4 signaling cascades. BMDMs were exposed to WT RANKL (2 μg/ml) or MT RANKL (2 μg/ml) for 0, 5, 10 and 15 min after 8 h with or without LiCl pretreatment. GAPDH was used as a loading control. (D) Densitometric value of p-GSK-3β/GSK-3β and p-AKT/AKT was determined by western blot analysis. The results are representative of three separate experiments that had comparable results. BMDM, bone marrow-derived macrophage; GSK-3β, glycogen synthase kinase-3β; LGR4, leucine-rich repeat-containing G-protein-coupled receptor 4; LiCl, lithium chloride; MT, mutant; N.S., not significant; p-, phosphorylated; RANKL, receptor-activated nuclear factor-κB ligand; TRAP, tartrate-resistant acid phosphatase; WT, wild-type.
In addition, WT RANKL or MT RANKL slightly increased GSK-3β phosphorylation in a time-dependent manner in cells pretreated with LiCl, which indicated the successful inhibition of GSK-3β, although the level of AKT was not altered (Fig. 4C and D). These results suggested that the inhibition of GSK-3β may lead to an increase in osteoclastogenesis and MT RANKL could stimulate the GSK-3β phosphorylation.
Effect of MT RANKL on NFATc1 translocation to the nucleus
To evaluate the effect of MT RANKL on LGR4-dependent inhibition of NFATc1 nuclear translocation, the present study examined the nuclear and cytosolic localization of NFATc1 using western blotting and densitometric analysis of control siRNA- and LGR4 siRNA-transfected BMDMs (Fig. 5A and B). Nuclear NFATc1 was not detected in MT RANKL-treated BMDMs in the presence of control siRNA but was detected in BMDMs treated with WT RANKL or MT RANKL in the presence of LGR4 siRNA.
Figure 5.
Effect of MT RANKL on NFATc1 nuclear translocation. (A) NFATc1 nuclear translocation in LGR4 siRNA-transfected BMDMs was analyzed in the cytoplasmic and nuclear fractions. Histone-H1 and β-actin were used as loading controls for the nuclear and cytoplasmic fractions, respectively. (B) Densitometric analysis of NFATc1 expression in the cytoplasmic and nuclear fractions of LGR4 siRNA-transfected BMDMs is presented as the mean ± standard deviation of three separate experiments. (C) NFATc1 nuclear translocation in BMDMs pretreated with LiCl for 8 h before RANKL treatment. (D) Densitometric analysis of NFATc1 expression in BMDMs pretreated with LiCl. The results are representative of three separate experiments that had comparable data. Data are presented as the mean ± standard deviation. *P<0.01 vs. control group; #P<0.01 vs. WT RANKL. BMDM, bone marrow-derived macrophage; CTL, control; LGR4, leucine-rich repeat-containing G-protein-coupled receptor 4; LiCl, lithium chloride; MT, mutant; NFATc1, nuclear factor of activated T cells, cytoplasmic, calcineurin-dependent 1; N.S., not significant; RANKL, receptor-activated nuclear factor-κB ligand; si, small interfering; WT, wild-type.
In addition, the expression levels of NFATc1 were detected in LiCl-treated BMDMs to investigate the effect of MT RANKL on GSK-3β-mediated inhibition of NFATc1 nuclear translocation (Fig. 5C and D). Nuclear NFATc1 was not detected in untreated BMDMs or MT RANKL-treated BMDMs, but was detected in BMDMs treated with WT RANKL or MT RANKL in the presence of LiCl, and in BMDMs treated with LiCl alone.
These results further support the hypothesis that LGR4 signaling serves a critical role in the GSK-3β-mediated inhibition of NFATc1 nuclear translocation in RANKL-treated BMDMs, suggesting that MT RANKL compensates for the inhibition of WT RANKL-RANK- NFATc1 signaling pathway via MT RANKL-LGR4-GSK-3β.
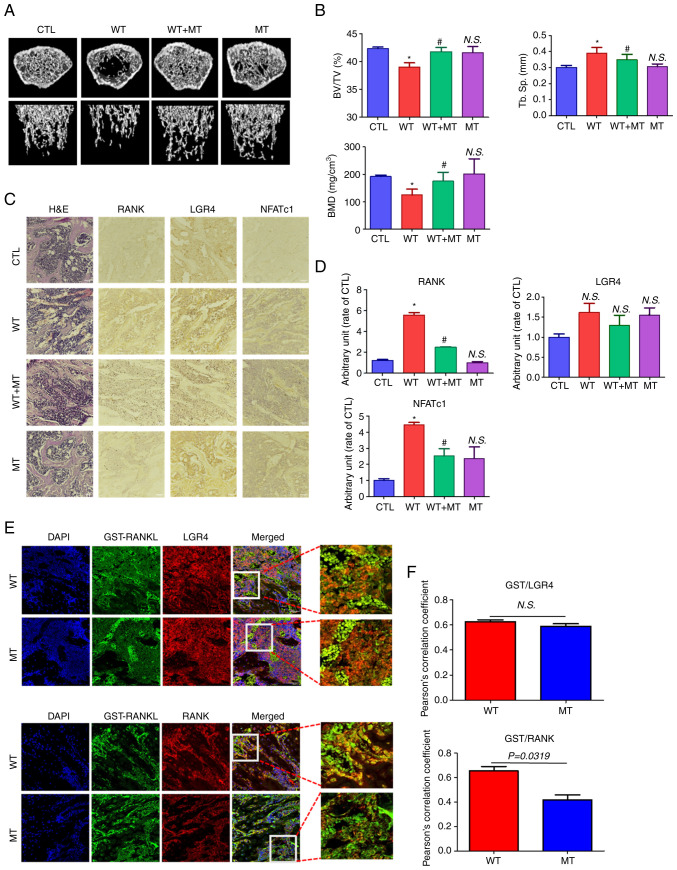
Effect of MT RANKL on bone loss in mice
To investigate the effect of MT RANKL on bone lysis, healthy mice were treated with MT RANKL in the presence or absence of WT RANKL and their femur bones were examined using micro-CT (Fig. 6A). Mice in the WT RANKL-treated group exhibited marked bone loss compared with the control (untreated) group, whereas those treated with WT RANKL + MT RANKL exhibited little bone loss. Bone loss in the MT RANKL-treated group was similar to that in the control group. The BV/TV, Tb. Sp and BMD scores were assessed using quantitative micro-CT (Fig. 6B). As expected, the BV/TV and BMD scores were lower in WT RANKL-treated mice than control mice, and significantly increased after MT RANKL treatment. The BV/TV, Tb.Sp and BMD scores in mice treated only with MT RANKL were similar to those in control mice. These results demonstrated the therapeutic effects of MT RANKL in a model of RANKL-induced bone loss.
Figure 6.
Effect of MT RANKL on RANKL-induced bone loss in a mouse model. (A) Representative micro-computed tomography images of the distal femurs of mice. (B) Measurements of BV/TV, Tb. Sp and BMD. Data are presented as the mean ± SD. *P<0.01 vs. control group; #P<0.01 vs. WT RANKL. (C) Immunohistochemistry staining of RANK, LGR4 and NFATc1 in femurs. Magnification, ×200; scale bar, 10 μm. (D) Densitometric analysis of immunohistochemistry. (E) Confocal microscopic images of the co-localization of GST-RANKL with LGR4 and RANK in WT RANKL- and MT RANKL-treated mice. Magnification, ×200; scale bar, 10 μm. (F) Pearson's correlation coefficient was calculated from the merged images of GST/LGR4 and GST/RANK. Data are presented as the mean ± SD of three independent measurements. BMD, bone mineral density; BV/TV, bone volume/tissue volume; CTL, control; GST, glutathione S-transferase; H&E, hematoxylin and eosin; LGR4, leucine-rich repeat-containing G-protein-coupled receptor 4; MT, mutant; NFATc1, nuclear factor of activated T cells, cytoplasmic, calcineurin-dependent 1; N.S., not significant; RANK, receptor-activated nuclear factor-κB; RANKL, RANK ligand; SD, standard deviation; Tb. Sp, trabecular spacing.
In addition, the immunopositive expression of RANK, LGR4 and NFATc1 was detected in H&E-stained histological sections of the femur (Fig. 6C and D). RANK and NFATc1 showed decreased immunopositive expression in mice treated with WT RANKL + MT RANKL compared with in WT RANKL-treated mice.
Finally, the co-localization of WT RANKL/MT RANKL as a GST-tagged RANKL with LGR4/RANK in WT RANKL- or MT RANKL-treated mice was assessed using confocal microscopy (Fig. 6E and F). GST-RANKL and LGR4 colocalized in mice treated with WT RANKL or MT RANKL, whereas GST-RANKL and RANK only colocalized in mice treated with WT RANKL. The correlation coefficient for GST-RANKL and LGR4 colocalization was not significantly different between WT RANK- or MT RANKL-treated mice, whereas the correlation coefficient of GST-RANKL and RANK colocalization was significantly higher in WT RANKL-treated mice than in WT RANKL-treated mice.
Taken together, these results demonstrated that MT RANKL could inhibit RANKL-induced bone lysis in a mouse model via the LGR4-dependent compensatory signaling pathway, and as such, MT RANKL may be a useful pharmaceutical agent in severe osteoporosis.
Discussion
Increasing evidence has indicated that the binding of RANKL to its receptor RANK, which drives the development of osteoclasts, is a key target for drugs that treat osteolytic bone diseases, including osteoporosis (29). Given the role of the RANKL-RANK signaling cascade in osteoclast development, the human monoclonal antibody denosumab, which targets RANKL, has been used as a clinical therapy for osteoporosis; furthermore, the use of denosumab validates RANKL as a therapeutic target (30-32). However, concerns have arisen regarding rebound bone resorption associated with hypercalcemia, parathyroid hyperplasia, severe BMD loss and multiple fractures after discontinuation of denosumab treatment (33-35). Scavenging of RANKL by denosumab in patients with osteoporosis leads to repeated regeneration of RANKL, resulting in rebound bone resorption that is more severe than that observed before treatment (14).
Because of the side effects associated with denosumab, the development of a competitive inhibitor of the RANKL-RANK signaling cascade may be an alternative approach for treating osteoporosis. LGR4 is another receptor for RANKL, and RANKL has a similar binding affinity for both RANK and LGR4, resulting in the negative regulation of RANKL-RANK signaling during osteoclastogenesis (36). Moreover, our previous studies showed that a novel MT RANKL acts as an agonist of LGR4, and inhibits the differentiation and activation of osteoclasts in RANKL-induced osteoclastogenesis in in vitro and in vivo models (15,22,23). Thus, agonist activation of LGR4-mediated signaling inhibits NFATc1 signaling through the intracellular LGR4-GSK-3β signaling pathway in osteoclast progenitor cells, and this pathway predominates over the canonical RANKL-RANK pathway, resulting in a block in osteoclast development (15,37).
The present study investigated the effects of the MT RANKL protein, in which the RANK binding site was modified using minimal amino acid changes, resulting in a protein that had a high binding affinity for LGR4 but a 500-fold lower binding affinity for RANK (22). The minimal change in the RANK-binding domain in MT RANKL means that if this protein is used in humans, RANKL homeostasis will be maintained without causing additional RANKL release, meaning that rebound resorption should not occur. Furthermore, the extracellular domain of LGR4 is known to have a higher binding affinity for MT RANKL than for RANK, implying that LGR4 -MT RANKL binding could have fewer physiological side effects than MT RANKL-RANK binding on osteoclast differentiation and activation. The potential reduction in side effects may be because MT RANKL competes with endogenous RANKL for binding to LGR4 and RANK.
In the mouse model, MT RANKL colocalized with only LGR4, whereas WT RANKL colocalized with RANK and LGR4. This finding suggested that in RANKL-induced osteoclastogenesis, WT RANKL interacts with RANK and LGR4, but MT RANKL interacts only with LGR4. Additional stimulation of LGR4 in RANKL-induced osteoclast precursor cells may trigger a negative regulatory signal that predominates over RANKL-RANK signaling and reduces NFATc1-related signaling. Luo et al (15) reported that the LGR4 extracellular domain acts as a molecular decoy receptor for RANKL binding both in vitro and in vivo, and inhibits RANKL-induced osteoclast activation. Another report showed that NFATc1 drives early osteoclast differentiation and causes osteoclast precursors to commit to the osteoclast lineage (38). The suppression of RANK signaling may lead to the upregulation of NFATc1 in differentiating osteoclast cells, as it has been suggested that targeting AKT signaling promotes IκBα degradation, resulting in the nuclear translocation of NFATc1. Moreover, LGR4 signaling prevents the inactivation of GSK-3β; active GSK-3β prevents the activation and nuclear translocation of NFATc1 (39,40). Thus, in the current study, signaling from LGR4-MT RANKL could inhibit AKT phosphorylation and stimulate GSK-3β phosphorylation, resulting in the inhibition of NFATc1 nuclear translocation that was independent of the RANK signaling cascade. In particular, MT RANKL signaling did not stimulate AKT phosphorylation and osteoclast development in BMDMs treated with an AKT inhibitor. The present study showed that MT RANKL, but not WT RANKL, acted as an LGR4 agonist and triggered LGR4-GSK-3β signaling (Fig. 7). A previous report showed that LGR4 expression is induced during osteoclast differentiation, resulting in RANKL-NFATc1 signaling (41). LGR4 expression is elevated in mature osteoclasts, and the LGR4-mediated signaling pathway is activated at a maximum level in mature osteoclasts, resulting in decreased RANKL-RANK signaling (15). This mechanism suggests that LGR4 could be used as a target for the development of novel osteoporosis therapies. Notably, mature osteoclasts ultimately undergo apoptosis in a RANKL-containing environment, suggesting that the existence of a RANKL-induced signaling pathway limits the maintenance of mature osteoclasts (42). The present study indicated that LGR4 may be a pivotal player in the negative-feedback mechanism that controls the activity of osteoclasts. The LGR4 signaling cascade activated by MT RANKL inactivated AKT and activated the GSK-3β signaling pathway, which resulted the inhibition of the activity of NFATc1 during osteoclast differentiation.
Figure 7.
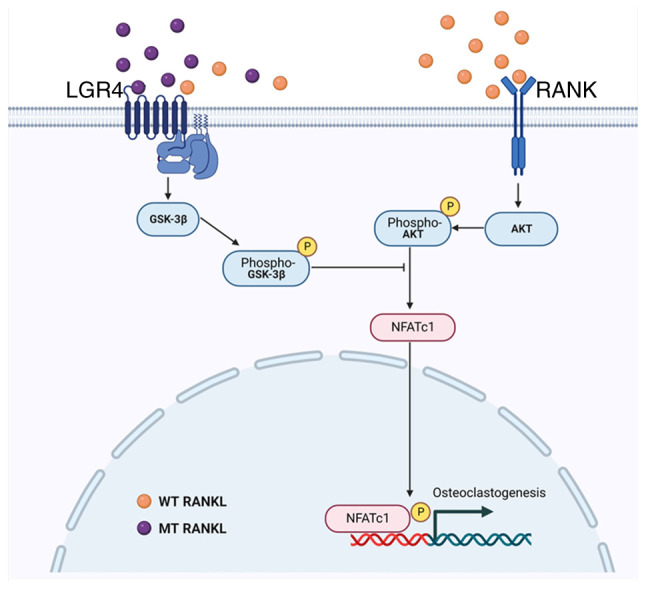
Schematic diagram of the inhibitory effect of MT RANKL on RANK-RANKL signaling during osteoclastogenesis. GSK-3β, glycogen synthase kinase-3β; LGR4, leucine-rich repeat-containing G-protein-coupled receptor 4; MT, mutant; NFATc1, nuclear factor of activated T cells, cytoplasmic, calcineurin-dependent 1; RANK, receptor-activated nuclear factor-κB; RANKL, RANK ligand; WT, wild-type.
In conclusion, MT RANKL, a novel agonist of LGR4, may activate an inhibitory signaling pathway during osteoclastogenesis that is different from the pathway activated by RANK-RANKL. MT RANKL could modulate the RANKL-AKT-NFATc1 signaling cascade through LGR4-induced GSK-3β phosphorylation and provides a negative-feedback mechanism to control osteoclast activity. The critical role of the RANK-RANKL protein interaction during osteoclast development means that it is an important target for drugs that treat osteoporosis. The results of the present study showed that in in vitro or in vivo experimental models of bone loss induced by RANKL, MT RANKL induced GSK-3β phosphorylation, and inhibited NFATc1 nuclear translocation and bone resorption. Furthermore, the results of this study suggested that MT RANKL, by activating a pathway that inhibits the effects of the RANKL-RANK signaling cascade, has the potential for treating osteoporosis.
Supplementary Data
Acknowledgments
Not applicable.
Funding Statement
The present study was supported by research funding from Chosun University (awarded in 2021).
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Authors' contributions
YJ and HL conducted the majority of the experiments, and YJ, HL and WL wrote the original manuscript. YC, SJ and EC performed the statistical analysis. YJ, HL and WL contributed to study conception and design. YJ and HL contributed to acquisition of data, and YC, SJ and HMS performed analysis and interpretation of data. HMS and WL reviewed and edited the original manuscript. BCK and YJK provided research materials and developed the methodology. YC and SJ confirm the authenticity of all the raw data. All authors read and approved the final manuscript.
Ethics approval and consent to participate
All of the experimental procedures involving animals were performed in compliance with institutional and governmental requirements, and were approved by the Institutional Animal Care and Use Committee (approval no. CIACUC2018-S0012-1) of Chosun University (Gwangju, Republic of Korea).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Hofbauer L, Kuhne C, Viereck V. The OPG/RANKL/RANK system in metabolic bone diseases. J Musculoskel Neuro Inter. 2004;4:268–275. [PubMed] [Google Scholar]
- 2.Wang L, You X, Zhang L, Zhang C, Zou W. Mechanical regulation of bone remodeling. Bone Res. 2022;10:16. doi: 10.1038/s41413-022-00190-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hanley DA, Adachi JD, Bell A, Brown V. Denosumab: Mechanism of action and clinical outcomes. Int J Clin Pract. 2012;66:1139–1146. doi: 10.1111/ijcp.12022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gowen M, Stroup GB, Dodds RA, James IE, Votta BJ, Smith BR, Bhatnagar PK, Lago AM, Callahan JF, DelMar EG, et al. Antagonizing the parathyroid calcium receptor stimulates parathyroid hormone secretion and bone formation in osteopenic rats. J Clin Invest. 2000;105:1595–1604. doi: 10.1172/JCI9038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Martin TJ. Bone biology and anabolic therapies for bone: Current status and future prospects. J Bone Metab. 2014;21:8–20. doi: 10.11005/jbm.2014.21.1.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sozen T, Ozisik L, Basaran NC. An overview and management of osteoporosis. Eur J Rheumatol. 2017;4:46–56. doi: 10.5152/eurjrheum.2016.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Miller PD. Denosumab: Anti-RANKL antibody. Curr Osteoporos Rep. 2009;7:18–22. doi: 10.1007/s11914-009-0004-5. [DOI] [PubMed] [Google Scholar]
- 8.Cadieux B, Coleman R, Jafarinasabian P, Lipton A, Orlowski RZ, Saad F, Scagliotti GV, Shimizu K, Stopeck A. Experience with denosumab (XGEVA(R)) for prevention of skeletal-related events in the 10 years after approval. J Bone Oncol. 2022;33:100416. doi: 10.1016/j.jbo.2022.100416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lei MM, Tavares E, Buzgo E, Lou U, Raje N, Yee AJ. Denosumab versus intravenous bisphosphonate use for hypercalcemia in multiple myeloma. Leuk Lymphoma. 2022;63:1–4. doi: 10.1080/10428194.2022.2115840. [DOI] [PubMed] [Google Scholar]
- 10.Terpos E, Jamotte A, Christodoulopoulou A, Campioni M, Bhowmik D, Kennedy L, Willenbacher W. A cost-effectiveness analysis of denosumab for the prevention of skeletal-related events in patients with multiple myeloma in four European countries: Austria, Belgium, Greece, and Italy. J Med Econ. 2019;22:766–776. doi: 10.1080/13696998.2019.1606002. [DOI] [PubMed] [Google Scholar]
- 11.Benlidayi IC. Denosumab in the treatment of glucocorticoid-induced osteoporosis. Rheumatol Int. 2018;38:1975–1984. doi: 10.1007/s00296-018-4106-1. [DOI] [PubMed] [Google Scholar]
- 12.Pittman K, Antill YC, Goldrick A, Goh J, de Boer RH. Denosumab: Prevention and management of hypocalcemia, osteonecrosis of the jaw and atypical fractures. Asia Pac J Clin Oncol. 2017;13:266–276. doi: 10.1111/ajco.12517. [DOI] [PubMed] [Google Scholar]
- 13.Gkoufa A, Angelousi A, Neonaki A, Athanasouli F, Cholongitas E. Severe symptomatic hypocalcemia associated with denosumab administration in a patient with decompensated cirrhosis and renal dysfunction. Ann Pharmacother. 2022;56:853–855. doi: 10.1177/10600280211050216. [DOI] [PubMed] [Google Scholar]
- 14.Anastasilakis AD, Makras P, Yavropoulou MP, Tabacco G, Naciu AM, Palermo A. Denosumab discontinuation and the rebound phenomenon: A narrative review. J Clin Med. 2021;10:152. doi: 10.3390/jcm10010152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Luo J, Yang Z, Ma Y, Yue Z, Lin H, Qu G, Huang J, Dai W, Li C, Zheng C, et al. LGR4 is a receptor for RANKL and negatively regulates osteoclast differentiation and bone resorption. Nat Med. 2016;22:539–546. doi: 10.1038/nm.4076. [DOI] [PubMed] [Google Scholar]
- 16.Takegahara N, Kim H, Choi Y. RANKL biology. Bone. 2022;159:116353. doi: 10.1016/j.bone.2022.116353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yue Z, Niu X, Yuan Z, Qin Q, Jiang W, He L, Gao J, Ding Y, Liu Y, Xu Z, et al. RSPO2 and RANKL signal through LGR4 to regulate osteoclastic premetastatic niche formation and bone metastasis. J Clin Invest. 2022;132:e144579. doi: 10.1172/JCI144579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jin Y, Yang Y. LGR4: A new receptor for a stronger bone. Sci China Life Sci. 2016;59:735–736. doi: 10.1007/s11427-016-5068-8. [DOI] [PubMed] [Google Scholar]
- 19.Luo W, Tan P, Rodriguez M, He L, Tan K, Zeng L, Siwko S, Liu M. Leucine-rich repeat-containing G protein-coupled receptor 4 (Lgr4) is necessary for prostate cancer metastasis via epithelial-mesenchymal transition. J Biol Chem. 2017;292:15525–15537. doi: 10.1074/jbc.M116.771931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Elango J, Bao B, Wu W. The hidden secrets of soluble RANKL in bone biology. Cytokine. 2021;144:155559. doi: 10.1016/j.cyto.2021.155559. [DOI] [PubMed] [Google Scholar]
- 21.Ko Y, Lee G, Kim B, Park M, Jang Y, Lim W. Modification of the RANKL-RANK-binding site for the immunotherapeutic treatment of osteoporosis. Osteoporos Int. 2020;31:983–993. doi: 10.1007/s00198-019-05200-6. [DOI] [PubMed] [Google Scholar]
- 22.Ko YJ, Sohn HM, Jang Y, Park M, Kim B, Kim B, Park JI, Hyun H, Jeong B, Hong C, Lim W. A novel modified RANKL variant can prevent osteoporosis by acting as a vaccine and an inhibitor. Clin Transl Med. 2021;11:e368. doi: 10.1002/ctm2.368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jang Y, Sohn HM, Ko YJ, Hyun H, Lim W. Inhibition of RANKL-induced osteoclastogenesis by novel mutant RANKL. Int J Mol Sci. 2021;22:434. doi: 10.3390/ijms22010434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Romain M, Thiroux B, Tardy M, Quesnel B, Thuru X. Measurement of protein-protein interactions through microscale thermophoresis (MST) Bio Protoc. 2020;10:e3574. doi: 10.21769/BioProtoc.3574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bartell SM, Kim HN, Ambrogini E, Han L, Iyer S, Ucer SS, Rabinovitch P, Jilka RL, Weinstein RS, Zhao H, et al. FoxO proteins restrain osteoclastogenesis and bone resorption by attenuating H2O2 accumulation. Nat Commun. 2014;5:3773. doi: 10.1038/ncomms4773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 27.Law SM, Zheng KK. Premise and peril of Wnt signaling activation through GSK-3β inhibition. iScience. 2022;25:104159. doi: 10.1016/j.isci.2022.104159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fan X, Xiong H, Wei J, Gao X, Feng Y, Liu X, Zhang G, He QY, Xu J, Liu L. Cytoplasmic hnRNPK interacts with GSK3β and is essential for the osteoclast differentiation. Sci Rep. 2015;5:17732. doi: 10.1038/srep17732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cao X. RANKL-RANK signaling regulates osteoblast differentiation and bone formation. Bone Res. 2018;6:35. doi: 10.1038/s41413-018-0040-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tokuyama N, Tanaka S. Updates of denosumab, anti-RANKL antibody for osteoporosis. Clin Calcium. 2014;24:85–91. In Japanese. [PubMed] [Google Scholar]
- 31.Cipriani C, Piemonte S, Colangelo L, De Martino V, Diacinti D, Ferrone F, Piazzolla V, Fassino V, Nieddu L, Minisola S, Pepe J. Inhibition of the RANKL with denosumab has no effect on circulating markers of atherosclerosis in women with postmenopausal osteoporosis: A pilot study. Endocrine. 2021;71:199–207. doi: 10.1007/s12020-020-02483-2. [DOI] [PubMed] [Google Scholar]
- 32.Lasco A, Morabito N, Basile G, Atteritano M, Gaudio A, Giorgianni GM, Morini E, Faraci B, Bellone F, Catalano A. Denosumab inhibition of RANKL and insulin resistance in postmenopausal women with osteoporosis. Calcif Tissue Int. 2016;98:123–128. doi: 10.1007/s00223-015-0075-5. [DOI] [PubMed] [Google Scholar]
- 33.Kim AS, Girgis CM, McDonald MM. Osteoclast recycling and the rebound phenomenon following denosumab discontinuation. Curr Osteoporos Rep. 2022;20:505–515. doi: 10.1007/s11914-022-00756-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Anastasilakis AD, Evangelatos G, Makras P, Iliopoulos A. Rebound-associated vertebral fractures may occur in sequential time points following denosumab discontinuation: Need for prompt treatment re-initiation. Bone Rep. 2020;12:100267. doi: 10.1016/j.bonr.2020.100267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Anastasilakis AD, Trovas G, Balanika A, Polyzos SA, Makras P, Tournis S. Progression of rebound-associated vertebral fractures following denosumab discontinuation despite reinstitution of treatment: Suppressing increased bone turnover may not be enough. J Clin Densitom. 2021;24:338–340. doi: 10.1016/j.jocd.2020.10.014. [DOI] [PubMed] [Google Scholar]
- 36.Luo J, Zhou W, Zhou X, Li D, Weng J, Yi Z, Cho SG, Li C, Yi T, Wu X, et al. Regulation of bone formation and remodeling by G-protein-coupled receptor 48. Development. 2009;136:2747–2756. doi: 10.1242/dev.033571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shi GX, Zheng XF, Zhu C, Li B, Wang YR, Jiang SD, Jiang LS. Evidence of the role of R-spondin 1 and its receptor lgr4 in the transmission of mechanical stimuli to biological signals for bone formation. Int J Mol Sci. 2017;18:564. doi: 10.3390/ijms18030564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lee Y, Kim HJ, Park CK, Kim YG, Lee HJ, Kim JY, Kim HH. MicroRNA-124 regulates osteoclast differentiation. Bone. 2013;56:383–389. doi: 10.1016/j.bone.2013.07.007. [DOI] [PubMed] [Google Scholar]
- 39.Cong F, Wu N, Tian X, Fan J, Liu J, Song T, Fu H. MicroRNA-34c promotes osteoclast differentiation through targeting LGR4. Gene. 2017;610:1–8. doi: 10.1016/j.gene.2017.01.028. [DOI] [PubMed] [Google Scholar]
- 40.Moon JB, Kim JH, Kim K, Youn BU, Ko A, Lee SY, Kim N. Akt induces osteoclast differentiation through regulating the GSK3beta/NFATc1 signaling cascade. J Immunol. 2012;188:163–169. doi: 10.4049/jimmunol.1101254. [DOI] [PubMed] [Google Scholar]
- 41.Wang M, Liu J, Zhu G, Chen X. Low levels of cadmium exposure affect bone by inhibiting Lgr4 expression in osteoblasts and osteoclasts. J Trace Elem Med Biol. 2022;73:127025. doi: 10.1016/j.jtemb.2022.127025. [DOI] [PubMed] [Google Scholar]
- 42.Manolagas SC, Parfitt AM. What old means to bone. Trends Endocrinol Metab. 2010;21:369–374. doi: 10.1016/j.tem.2010.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.