Abstract
The molecular complex containing the phototaxis receptor sensory rhodopsin I (SRI) and transducer protein HtrI (halobacterial transducer for SRI) mediates color-sensitive phototaxis responses in the archaeon Halobacterium salinarum. One-photon excitation of the complex by orange light elicits attractant responses, while two-photon excitation (orange followed by near-UV light) elicits repellent responses in swimming cells. Several mutations in SRI and HtrI cause an unusual mutant phenotype, called orange-light-inverted signaling, in which the cell produces a repellent response to normally attractant light. We applied a selection procedure for intragenic and extragenic suppressors of orange-light-inverted mutants and identified 15 distinct second-site mutations that restore the attractant response. Two of the 3 suppressor mutations in SRI are positioned at the cytoplasmic ends of helices F and G, and 12 suppressor mutations in HtrI cluster at the cytoplasmic end of the second HtrI transmembrane helix (TM2). Nearly all suppressors invert the normally repellent response to two-photon stimulation to an attractant response when they are expressed with their suppressible mutant alleles or in an otherwise wild-type strain. The results lead to a model for control of flagellar reversal by the SRI-HtrI complex. The model invokes an equilibrium between the A (reversal-inhibiting) and R (reversal-stimulating) conformers of the signaling complex. Attractant light and repellent light shift the equilibrium toward the A and R conformers, respectively, and mutations are proposed to cause intrinsic shifts in the equilibrium in the dark form of the complex. Differences in the strength of the two-photon signal inversion and in the allele specificity of suppression are correlated, and this correlation can be explained in terms of different values of the equilibrium constant (Keq) for the conformational transition in different mutants and mutant-suppressor pairs.
The phototaxis receptor sensory rhodopsin I (SRI) is unusual among photoreceptors because of the color sensitivity of its signaling (32). Absorption of an orange photon triggers an attractant response (suppression of reversals in the direction of swimming) by the cell, whereas absorption of an orange photon followed by absorption of a near-UV photon triggers a repellent response (induction of reversals). Both the attractant and repellent signals are transmitted through the transducer protein HtrI, which is tightly bound to SRI (9, 15, 23, 27, 37).
Cyclic photochemical reactions produced by one-photon and two-photon excitation of SRI have been characterized by kinetic flash spectroscopy (1). Orange light converts the dark form of SRI (SR587 [subscript is λmax]) into a sequence of photointermediates, one of which is a long-lived (several seconds) near-UV-light-absorbing species, S373, identified as the attractant signaling state. S373 is photoreactive, and its excitation by a near-UV photon returns it more rapidly to SR587 via a pathway containing the long-lived species Sb510 (half-life, ∼80 ms; the superscript represents photoconversion “back” to SR587). Behavioral studies have shown that the one-photon reaction (orange light) generates attractant signals and that the S373 photoreaction to near-UV light generates repellent signals (32). White-light (i.e., orange light delivered together with near-UV light) stimulation of the dark-adapted SRI state SR587 produces a mixture of one-photon and two-photon cycle intermediates. The latter evidently dominate, since white light causes a repellent response.
Recently, a two-conformation equilibrium model has been proposed as a unified mechanism for ion pumping and sensory signaling by archaeal rhodopsins (31). In the model, two receptor conformers, assumed to be similar to the well-established closed and open cytoplasmic-channel conformers of the proton pump bacteriorhodopsin (BR), are responsible for both attractant and repellent signals from SRI: SR587 is proposed to exist in an equilibrium mixture of the two conformers, designated A (attractant) and R (repellent). The net effect of the one-photon cycle is to shift the equilibrium to the A conformer, which suppresses reversals, whereas the two-photon cycle shifts the equilibrium to the R conformer, which induces reversals. The repellent effect of white light is explained as an overall shift toward the R conformer in the mixture of photoproducts. One argument in favor of a metastable conformational equilibrium of SR587 is the existence of the “orange-light-inverted” phenotype resulting from some mutations affecting the SRI-HtrI complex. These mutants, D201N in SRI (22), several substitutions for H166 in SRI (38), and E56Q in HtrI (13), behave as if they are shifted extremely far into the A conformer in the dark, exhibiting a repellent response to both one-photon and two-photon activation.
Since conformational equilibria are generally sensitive to single-residue substitutions (8, 26, 36), one would expect to be able to isolate second-site suppressor mutations that restore attractant responses to orange light in the inverted mutants by restoring the dark equilibrium of the wild-type SRI-HtrI complex. Moreover, some suppressors should restore the one-photon attractant response by shifting the dark equilibrium strongly in favor of the R conformer. In such mutants an unusual phenotype is predicted, namely, that the two-photon repellent response would become inverted because further increase in the R conformer could not occur, due to the extreme initial bias toward the R conformer and because some A conformer is produced in white light by the excitation of SR587. Therefore, white light would elicit an attractant response rather than the wild-type repellent response.
To test these predictions, and to identify residues important in the SRI-HtrI interaction, we developed a selection procedure for one-photon attractant responses restored by suppressors of mutations causing the orange-light-inverted response. The behavioral phenotypes of mutants carrying such second-site suppressors confirm the predictions of the model and provide information regarding the conformationally sensitive regions of SRI and HtrI.
MATERIALS AND METHODS
Plasmids and recipient strain.
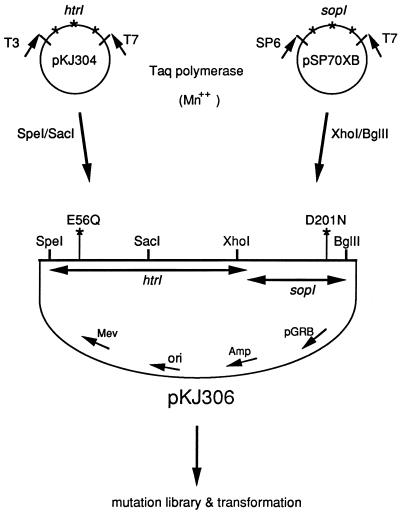
The plasmid pKJ306 is a shuttle vector that carries ampicillin and mevinolin resistance for selection in Escherichia coli and Halobacterium salinarum, respectively. The plasmid pKJ306 is a derivative of pKJ301 (28) in which XhoI and BglII restriction sites were introduced flanking the sopI gene. Pho81Wr− (BR− halorhodopsin− SRI− SRII− HtrI− HtrII−) is carotenoid deficient and restriction negative (25); it was used as the H. salinarum recipient in plasmid transformations. Wild-type or mutant SRI and HtrI were expressed from their native promoter in Pho81Wr− with plasmid pKJ306 (Fig. 1) and its mutated derivatives.
FIG. 1.
Random PCR mutagenesis scheme. Plasmids and steps in the preparation of randomly mutated libraries of sopI, which encodes the SRI apoprotein, and the portion of htrI encoding the 230-residue region of HtrI that is N terminal to the methylation and signaling domains (see Materials and Methods).
PCR mutagenesis.
The plasmids pKJ304 and pSP70XB, carrying the SpeI/SacI (HtrI) and XhoI/BglII (SRI) fragments in pBluescript KS− (Stratagene, La Jolla, Calif.) and pSP70 (Promega, Madison, Wis.) were used as templates for random PCR mutagenesis (Fig. 1) with Taq polymerase (Promega). The htrI and sopI fragments were mutagenized and amplified under conditions of reduced fidelity by adding MnCl2 in the reaction mixture and increasing the polymerase extension time (3, 18). The reaction mixture contained 0.05 mM MnCl2, 1.5 mM MgCl2, 0.2 mM deoxynucleoside triphosphates, 50 mM KCl, 10 mM Tris (pH 9.0), and 0.1% Triton X-100, and the PCR was performed for 31 cycles at 95°C for 1 min, 50°C for 2 min, and 72°C for 3 min. The mutation frequency of the fragments used in this study was measured by sequencing and found to be 1 mutation per 300 bp. The frequency of transition and transversion mutations was about equal in the presence of Mn2+. The mutated sopI or htrI fragment was replaced into the htrI-sopI operon encoding the SRI D201N or HtrI E56Q mutation, and the mutant library was introduced into E. coli DH5α for amplification prior to transformation of H. salinarum. In general, two or three mutated sites were observed in the mutagenized gene in isolated suppressor mutants. Therefore, site-directed mutagenesis was carried out with the two-step megaprimer PCR method with Pfu polymerase (4, 13) in order to identify the site responsible for the suppression phenotype.
Isolation of suppressors.
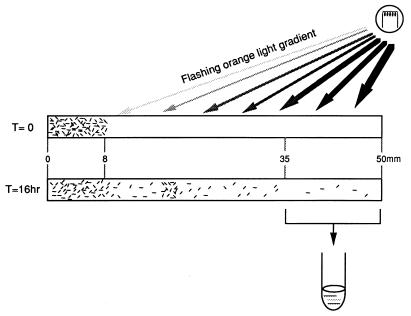
Halobacterial cells were transformed by the polyethylene glycol (PEG) method (5), except that PEG was first purified by using AG501-X8 resin (20/50 mesh; Bio-Rad, Hercules, Calif.). The cells with mutagenized plasmids were grown to early-stationary phase and diluted 1:10 with complex growth medium (CM; pH 6.0) (30), and 1.6-μl aliquots were loaded into flat capillaries (0.1 by 2 by 50 mm; Dynamics, Inc., Rockaway, N.J.) as described elsewhere (34). The capillary was positioned on a microscope slide so that an orange light (600 ± 20 nm; 1.4 × 105 ergs · cm−2 s−1, measured at the region of greatest intensity) applied near the distal end of the capillary produced a gradient along the capillary length (Fig. 2). Both ends of the capillary were sealed with paraffin. The light was delivered in 50- to 100-ms pulses every 10 s at 40°C. After 16 h, one-third of the distal end of the capillary was cut off and dropped into a culture tube containing 2 ml of CM with mevinolin (1 μg/ml). The cells were grown at 37°C for 4 to 5 days. The selection was repeated two or three times to enrich the suppressor population.
FIG. 2.
Selection scheme for suppressor mutants. Cells were loaded between 0 and 8 mm from the end of the capillary. A repetitively flashing orange-light gradient was delivered to the 8- to 50-mm region, and cells were harvested from the 35- to 50-mm region of the capillary after ∼16 h. The rationale is that cells carrying SRI D201N or HtrI E56Q will respond to the orange-light flash as a repellent stimulus and reverse their swimming direction at the frequency of the flashing light (0.1 Hz), which impedes their migration through the capillary. The time-averaged spatial gradient of orange light favors the migration of suppressed mutants that exhibit attractant phototaxis over that of nonresponding mutants. For details, see Materials and Methods.
Motion analysis.
The swimming behavior of cells was monitored by a computerized cell-tracking system (Motion Analysis, Santa Rosa, Calif.). Early-stationary-phase cultures were diluted 1:13 in fresh CM and incubated for 1 h at 37°C with agitation. Responses to orange, near-UV, and white-light photostimuli were monitored with infrared light (>700 nm). Stimuli were light at 600 nm in the infrared background, light at 400 nm in a >580-nm background, and light passed through a CS600 cyan-subtractive dichroic filter (“white-light” stimulus; 380 to 600 nm) (Corion, Franklin, Mass.) in an infrared background (>700 nm), respectively. Stimuli were delivered from a Nikon 100-W He/Xe short-arc lamp. All stimuli were saturating for wild-type cells. Data were analyzed on a Sun SPARC-IPC workstation (Sun Microsystems, Mountain View, Calif.).
Flash photolysis.
Flash-induced absorption changes were measured by a cross-beam spectrophotometer equipped with a 532-nm, 40-mJ/6-ns pulse Nd-YAG laser (Surelite I; Continuum, Santa Clara, Calif.). The pulse frequency was 0.08 Hz. Twenty transients were averaged for each trace at 18°C. The half-life of S373 reprotonation was calculated with SIGMAPLOT (Jandel, San Rafael, Calif.) by using a single exponential curve-fit.
RESULTS
Isolation of suppressor mutants.
Randomly mutagenized cells carrying either the E56Q substitution in HtrI or the D201N substitution in SRI, either of which produces an orange-light-inverted (repellent) response, were subjected to the capillary selection procedure (Fig. 2). Three hundred fifty single-colony isolates from the distal end of the capillary were screened for their orange-light-induced swimming behavior. Eighty-one (23%) of the cells contained suppressor mutations and exhibited orange-light-induced attractant responses like that of wild-type H. salinarum. One hundred nine (31%) were orange-light-blind cells, which upon further examination were found not to respond to any light stimuli used in this study. The blind mutants may have lost the plasmid or may carry mutations that disrupt SRI-HtrI expression or function, and they were not analyzed further. The remaining 160 (46%) isolates exhibited the unsuppressed, inverted phenotype of the parent D201N or E56Q strain.
The 81 mutants carrying suppressor mutations were analyzed by sequencing of their randomly mutagenized segments of htrI-sopI. Suppressor mutations were confirmed by making site-specific substitutions of each of the two to three mutations found per isolate and testing their phenotypes in the inverted-response mutant strain in which they were isolated. Thirteen distinct suppressor mutations were identified: 3 were in SRI, and 10 were in HtrI (Table 1). The three suppressors in SRI (A116T, N161D, and R215W) and five of the suppressors in HtrI (I61V, Q67L, V71F, N73Y, and R84N) were isolated with the HtrI E56Q mutant, and five of the HtrI suppressors (N53I, I64V, R70H, V71I, and E96A) were isolated with the SRI D201N mutant. Additionally, two residue substitutions made in a previous study (13) at positions in HtrI identified by the selection process, R84A and E96Q, were also found to be suppressor mutations (Table 1), bringing the total number of unique suppressor mutations to 15. In a separate study of the role of a cluster of Arg residues in SRI function, a triple mutant, R215A R216A R217A in SRI, had also been constructed. It was found to suppress HtrI E56Q, but photoactive SRI receptor was not expressed when the triple mutation was combined with the SRI-inverting mutations in SRI.
TABLE 1.
Suppression of orange-light-inverted response
| Inverted mutant tested | Suppressor mutationa
|
|
|---|---|---|
| SRI | HtrI | |
| H166A H166S (SRI) | R215W | R84N (R84A) |
| D201N (SRI) | R215W | N53I |
| N161D | I64V | |
| A116Tb | Q67L | |
| R70H | ||
| V71I | ||
| V71F | ||
| N73Y | ||
| R84N (R84A) | ||
| E96A (E96Q) | ||
| E56Q (HtrI) | R215W | N53I |
| N161D | I61V | |
| A116T | I64V | |
| (R215A R216A R217A)c | Q67L | |
| R70H | ||
| V71I | ||
| V71F | ||
| N73Y | ||
| R84N (R84A) | ||
| E96A | ||
Underlined mutations were originally isolated from the indicated orange-light-inverted mutant; those in parentheses were originally made by site-directed mutagenesis.
Delayed orange-light response.
This triple mutation did not produce SRI pigment when combined with SRI-inverting mutations.
Analysis of allele specificity.
A third residue at which substitutions cause an orange-light-inverted response, His166 in SRI, was identified (38) while this study was in progress. Substitutions of Ala or Ser for His166 produce a behavioral phenotype similar to that produced by the D201N substitution in SRI or the E56Q substitution in HtrI (38). Accordingly, the 15 suppressor mutations were tested for allele specificity by combining each of them with SRI D201N, SRI H166A, SRI H166S, and HtrI E56Q. The suppressor mutations fell into three classes based on this analysis. Class I (allele specific) comprised two mutations: I61V (HtrI) was specific for suppression of HtrI E56Q and did not suppress D201N, H166A, or H166S; E96Q (HtrI) suppressed only D201N. Class II (partially allele specific) comprised 11 mutations; each of these suppressed both D201N and E56Q, but not H166A or H166S (Table 1). Class III (non-allele specific, or “supersuppressors”) comprised three mutations; R215W in SRI and R84N or R84A in HtrI suppressed all four inverted-phenotype mutations.
Locations of suppressor mutations.
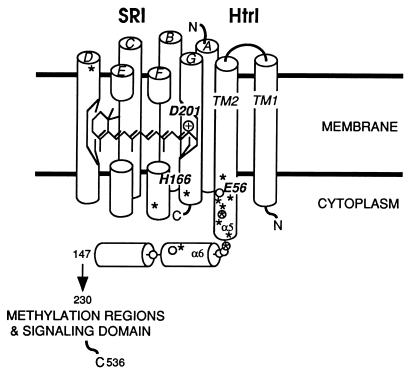
Two of the three suppressors in SRI are positioned near the cytoplasmic surface of the molecule, according to structural models of SRI based on the BR structure (11, 12, 19): SRI N161D, a class II mutation, is at the cytoplasmic end of helix F, and the supersuppressor SRI R215W (class III) is located at the cytoplasmic end of helix G (Fig. 3). The corresponding regions in BR undergo significant movement during its light-induced conformational change. The third mutation, SRI A116T (class II), is located at the periplasmic end of helix D (Fig. 3).
FIG. 3.
Positions of the residues in SRI and HtrI altered by suppressor mutations. Circled letters at the top represent specific helices. Asterisks indicate suppressor sites. Open circles indicate positions at which substitutions were observed to alter S373 lifetime in a previous study (13). α5 and α6 refer to homologous regions in eubacterial receptor/transducers (17). The relative arrangement of the transmembrane helices of SRI and HtrI was chosen arbitrarily.
All suppressor mutations in HtrI are located between residues 53 and 96 (Fig. 3). The location of this suppressor cluster adds to a body of observations suggesting that this region at the cytoplasmic end of the second HtrI transmembrane helix (TM2) is critical for SRI interaction.
One of the allele-specific (class I) mutations in HtrI, I61V, is only 5 residues C-terminal to the mutation that it suppresses, HtrI E56Q. This region is predicted to be α-helical, which would place these two mutations a little over one turn apart. The proximity and specificity of this pair of substitutions suggest that I61V may suppress by correction of a local structural change caused by E56Q. The other allele-specific mutation, HtrI E96Q, probably has a less direct effect. Another substitution at this position (E96A) is only partially allele specific (class II), a difference interpretable as a matter of degree rather than mechanism.
Swimming behavior of cells carrying suppressor mutations.
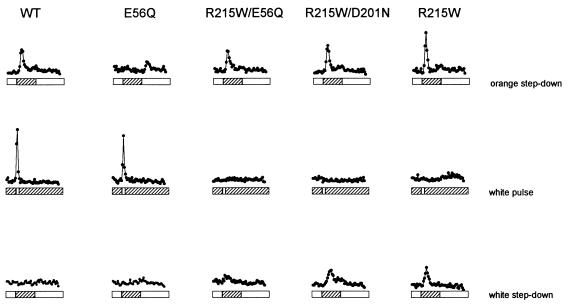
Cells carrying the suppressor mutations in an otherwise wild-type strain and in an orange-light-inverted mutant strain were analyzed for their responses to phototaxis stimuli. The reversal-frequency transients of the cell population are shown for the wild-type strain, the HtrI E56Q inverted mutant, and the SRI R215W suppressor in inverted mutants and in an otherwise wild-type strain (Fig. 4). A step-down in orange-light intensity was used to assess one-photon signaling, and a “white” pulse (380 to 600 nm) was used to assess two-photon signaling. A step-down in white-light intensity was also used to assess two-photon signaling.
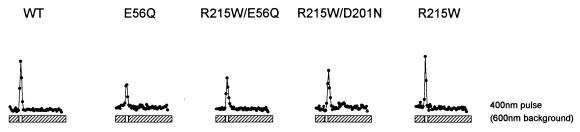
FIG. 4.
Phototaxis responses. Cells contained, from left to right, wild-type SRI and HtrI (WT); wild-type SRI plus HtrI mutant E56Q, SRI mutant R215W plus HtrI mutant E56Q, the double SRI mutant R215W D201N plus wild-type HtrI, and SRI mutant R215W plus wild-type HtrI. Two seconds after initiation of data acquisition, the cells were exposed to 4-s removal of orange light (600 nm; top row), a 100-ms pulse of white light (spanning the range 380 to 600 nm; middle row), or a 4-s removal of white light (bottom row). Traces represent population reversal-frequency transients collected by computerized motion analysis at pH 6.0 and 40°C.
(i) A 4-s step-down of orange (600-nm) light in an infrared background.
The infrared (>700-nm) background light used for imaging the cells is beyond the absorption range of SRI and is accordingly nonactinic for phototaxis. The wild-type strain exhibited a peak in reversals induced by the disappearance of the orange light. The reversals are attributable to the decay of the intermediates of the one-photon cycle, the most prominent of which is S373 (Fig. 4). The re-formation of S373 when the light returned after 4 s suppressed reversals (evident in the higher-resolution data of reference 13). However, this effect was negligible within the signal/noise ratio of the low-spontaneous-reversal strains used here. The orange-light-inverted mutants exhibited the opposite behavior; slight suppression of reversals after the light was turned off, followed by an evident induction of reversals after the light returned (for example, HtrI E56Q in Fig. 4). The basis of the selection pressure used in this study, and the definition of the suppressor phenotype, is exemplified by the wild-type-like response to this stimulus shown by SRI mutant R215W with HtrI mutant E56Q and by SRI mutant R215W D201N with wild-type HtrI (Fig. 4).
(ii) A 100-ms pulse of white light (spanning 380 to 600 nm) in an infrared background.
This stimulus elicited the two-photon reaction of SRI and thus a reversal response from the wild-type strain. The E56Q mutant also exhibited a repellent response, as reported previously (13), demonstrating that the mutation that inverts the effect of S373 formation from attractant to repellent does not change the repellent effect of S373 photoreaction products. A significant finding from this study is that cells carrying any one of several suppressor mutations in the inverted-mutant strain or in the wild-type strain do not exhibit the two-photon repellent response (shown for SRI R215W in Fig. 4). The opposite response, a slight suppression of reversals, is evident in cells carrying the R215W suppressor in the SRI D201N mutant or wild-type strains. Therefore, R215W and many of the other suppressor mutations exhibit a new phenotype: inverted (attractant) responses to two-photon activation of SRI. This result is predicted by the metastable conformational-equilibrium model.
(iii) A 4-s step-down of white light in an infrared background.
The attractant effect of a pulse of white light in the R215W mutant was confirmed, and was more evident, when a long exposure to white light was interrupted, a procedure that produced reversal peaks in R215W mutants (Fig. 4). The response was strongest when R215W was present in an otherwise wild-type strain. This response was attenuated slightly by the SRI D201N mutation and more by the HtrI E56Q mutation. Wild-type and E56Q strains exhibited a slight suppression of reversals in response to this stimulus.
The behavioral analysis outlined above was conducted for all the suppressors present in SRI D201N, HtrI E56Q, and wild-type strains. The results are presented in Fig. 6 and 7 in terms of the phototaxis index, calculated as described previously (13) from reversal-frequency transients. A positive-response index results from an attractant response, and a negative index results from a repellent response. The values shown were reproducible to within ±0.1 index unit.
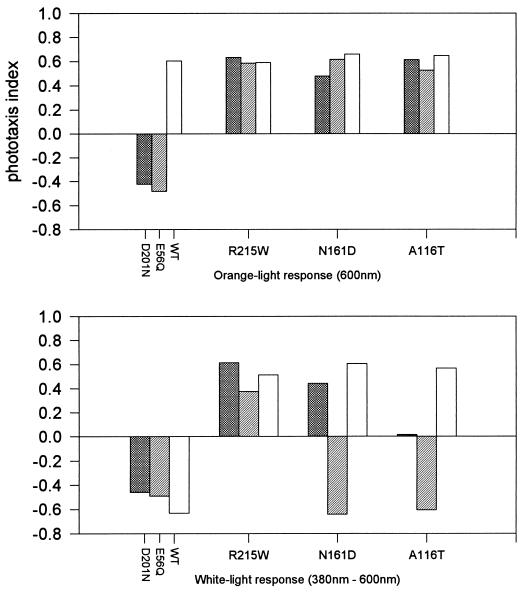
FIG. 6.
Phototaxis response indices from strains with suppressor mutations in SRI. Positive values indicate attractant responses, and negative values indicate repellent responses. D201N, SRI mutant D201N; E56Q, HtrI mutant E56Q; WT, wild type. For each of the three SRI suppressor mutations, the first bar shows the suppressor combined with the D201N mutation, the second bar shows the suppressor combined with the E56Q mutation, and the third bar shows the suppressor mutation alone in otherwise wild-type SRI. Photostimuli were delivered as described in Materials and Methods.
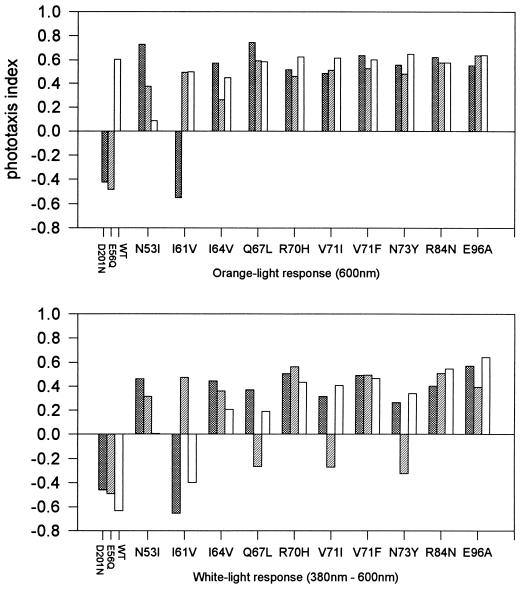
FIG. 7.
Phototaxis response indices from strains with suppressor mutations in HtrI. Notation is as explained for Fig. 6.
(iv) A 100-ms pulse of near-UV (400-nm) light in a constant orange-light background.
In previous work on SRI, the S373-mediated repellent response was typically assessed not by white light, as used above, but by a pulse of near-UV (400-nm) light in a constant orange-light background. The background light generates a steady-state presence of S373, which is excited by the 400-nm light to produce back photoreaction products (predominantly Sb510). This stimulus has two signal components, each of which induces reversals in a wild-type strain: the transient disappearance of S373 and the transient formation of Sb510. Therefore, the white-light stimulus is a more-specific test for two-photon signaling. However, because the UV pulse–orange-background protocol has been used so extensively, we included it in our analysis. The wild-type strain exhibited a strong reversal response (Fig. 5). The E56Q mutant exhibited a weaker reversal response, as was observed previously (13), presumably because the disappearance of S373 has a reversal-suppressing (inverted) effect in this mutant, which subtracts from reversal induction by S373 photoproducts. Consistent with this interpretation, the R215W-suppressed mutants had stronger responses than E56Q, but weaker responses than an otherwise wild-type strain containing R215W (Fig. 5). The near-UV stimulus in an orange background produced a repellent response in every mutant or double mutant assessed in this study. Since this stimulus elicits both removal of S373 and formation of Sb510, it is evident that if either of the two signals has a repellent effect, it dominates the response, and the net index is negative. Therefore, it is essential to use a white-light stimulus to detect the inversion of the two-photon stimulus to a positive index (attractant response).
FIG. 5.
Phototaxis responses to near-UV light. Wild-type and mutant strains are as described for Fig. 4. Cells were exposed to a 100-ms pulse of near-UV light (400 nm) in a constant orange-light background.
(v) SRI suppressor mutations.
Strains carrying each of the three suppressor mutations in SRI in each of three genetic backgrounds (SRI D201N, HtrI E56Q, and wild-type) showed a suppressor phenotype after stimulation by orange light (Fig. 6, top panel). Cells carrying the H166A or H166S substitutions in SRI produced repellent responses to orange light that were not suppressed by N161D or A116T but were suppressed by R215W (data not shown).
Stimulation with white light revealed the inverted (attractant) response caused by R215W in each strain background (Fig. 6, bottom panel; also shown in Fig. 4). N161D and A116T did not cause inverted responses when combined with HtrI E56Q but did when combined with SRI D201N. These data allow a ranking to be made of the three suppressor mutants based on their ability to invert the two-photon response in different strains; from highest to lowest, the order of efficiency is R215W, N161D, A116T. Similarly, the two-photon data rank E56Q above D201N in its ability to resist two-photon response inversion by the SRI suppressors. An important observation is that R215W, the mutation that produced the most dominant two-photon response inversion, is also the most effective suppressor; i.e., R215W is a supersuppressor (class III), whereas the other two suppressors are in class II and failed to restore normal responses to H166A or H166S mutants. The correlation of rank order from two-photon inversion effects with that of suppression efficiency provides support for a metastable-equilibrium model, and it can be interpreted in terms of the relative values of the conformational-equilibrium constant (Keq) of the mutants (see Discussion).
(vi) HtrI suppressor mutations.
The class II, partially allele-specific suppressor HtrI N53I inverted the two-photon response of the D201N or E56Q mutant, and it decreased responses to all stimuli in an otherwise wild-type strain (Fig. 7). Four of the remaining class II HtrI suppressors (I64V, R70H, V71F, and E96A) and the class III suppressor R84N each inverted the two-photon response in all strains tested. The class II suppressors Q67L, V71I, and N73Y inverted the two-photon response of a wild-type or D201N mutant strain but not that of an E56Q mutant. These apparent differences in the strength of the suppressor mutations are explained in the conformational-equilibrium model as different values of the Keq for the transition between the two signaling conformations (i.e., E56Q is shifted more toward one conformational extreme than D201N, and V71F is shifted more than V71I toward the other conformer). The class I I61V suppressor only affects E56Q, and it also only inverts the two-photon response of E56Q. The N53I suppressor is unusual in that this mutant protein supported little or no response to any stimuli when it was expressed in a wild-type strain, but allowed good responses when expressed in inverted-mutant strains.
Effects of suppressor mutations on S373 decay in the SRI photocycle.
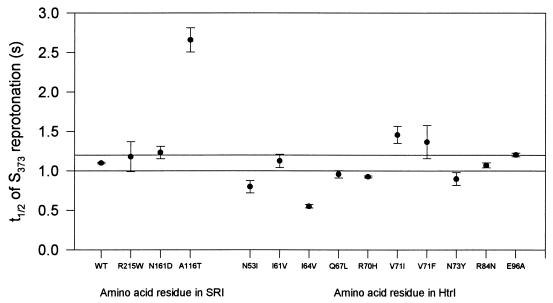
In a previous study, seven mutations between positions 70 and 108 of HtrI were shown to alter the half-life of the S373 intermediate (13). This effect was interpreted to mean that this region of HtrI is important in coupling HtrI to SRI. Of the 10 suppressor mutations in HtrI, most perturbed the S373 half-life, most notably I64V, which accelerated S373 decay, and V71I, which retarded it (Fig. 8). These two suppressors thus extend the list of residues in this cytoplasmic region at the end of TM2 that are involved in determining the stability of photocycle intermediates of SRI.
FIG. 8.
Half-lives (t1/2) of S373 in membranes from cells containing suppressor mutations in an otherwise wild-type strain. The half-life of the decay of S373 to SR587 was measured with excitation at 590 nm, at pH 6.8 and 18°C. Twenty flash photolysis transients were averaged for each determination. Error bars are ±1 standard error of the mean for three determinations, and horizontal lines span 2 standard deviations of the mean for membranes from wild-type cells.
Cells carrying the SRI A116T mutation combined with D201N exhibited a delayed orange-light step-down reversal response (data not shown). This delay is probably attributable to the very short half-life of 5.4 s (at 18°C) of this double-mutant protein. The half-life of S373 decay in the D201N protein alone and in the A116T protein alone is 2.7 s in each case, and therefore the retardation effects appear to be additive.
We detected no difference in the S373 decay rate of the HtrI N53I mutant protein expressed in an otherwise wild-type strain, which mediates little or no photoresponse, in contrast to the behavior of the N53I protein expressed in inverted-mutant strains, which showed photoresponses and slow S373 decay.
DISCUSSION
The results presented above identify 15 distinct mutations responsible for suppression of defects in one-photon signaling by the SRI-HtrI complex. Three results argue that the majority of these mutations modify the conformation of the SRI-HtrI interface. (i) Most of the mutations (3 in SRI and 10 in HtrI) are not allele specific, suggesting that they alter overall protein conformation. (ii) All of the non-allele-specific mutations are capable of both intragenic and extragenic complementation, demonstrating that they can correct the effects of mutation in the companion protein, and the algebraic additivity of phenotypes in double mutants argues that the mutations affect the same functional domain. (iii) The complex phenotypes of cells carrying the SRI suppressor mutations, which exhibit alterations of both one-photon and two-photon signaling, are the same for SRI and HtrI suppressors, suggesting that the general conformational changes generated by these mutations are the same.
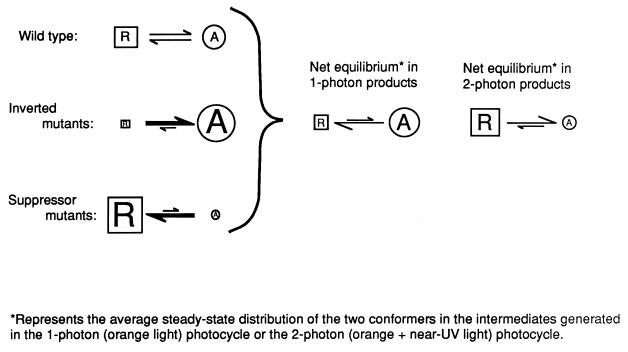
A model for the color-sensitive signaling by the SRI-HtrI complex that can explain color-sensitive signaling, the orange-light-inverted phenotype, and the phenotypes of its suppressors is based on a metastable equilibrium of two conformations of the SRI-HtrI interface (designated A and R in Fig. 9). The basic assumption of this model is that the wild-type SRI-HtrI complex is poised in an equilibrium mixture of the two conformers in the dark. This equilibrium is shifted toward the A conformer by formation of S373 and toward the R conformer by formation of Sb510. An increase in concentration of the A form produces an attractant signal, and an increase in the R form produces a repellent signal. In terms of this model, the orange-light-inverted responses caused by the D201N, E56Q, and His166 substitutions occur because these mutations destabilize the R state of HtrI associated with the dark species (SR587 of SRI), shifting the equilibrium far toward the A conformer. The one-photon products are assumed to contain some R form, and therefore repellent responses occur following orange-light stimulation. (An increased presence of the R conformer in the second half of the one-photon cycle is expected if the A and R conformers correspond to the open and closed cytoplasmic-channel conformers, respectively, of BR. In BR, the channel closes during the latter half of the photocycle, and in the dark state it is maintained closed by electrostatic and other constraints [16]. The inverting mutations are proposed to disrupt constraints that stabilize the R state in the dark, producing an extreme shift to A. The restoration of the R conformer in the photocycle would still occur, however, and would therefore produce a transient increase in R in the latter half of the photocycle.) Suppressor mutations in the SRI-HtrI complex restore the one-photon attractant response to orange-light-inverted mutants by shifting the conformational equilibrium back toward the R form. Suppressor mutations that overcompensate by shifting the equilibrium to generate the R form predominantly would invert white-light repellent responses to attractant responses because of the increase in A form in the white-light photoproducts (which include some S373). The model explains the concomitant inversion of the two-photon response upon restoration of the one-photon response by suppressor mutations, because these two responses result from opposing shifts in the same equilibrium.
FIG. 9.
Proposed conformational equilibria of a wild-type signaling complex, the signaling complex of an orange-light-inverted mutant, (such as SRI D201N or HtrI E56Q), and the signaling complex of supersuppressor strains, such as SRI R215W or HtrI R84N. A and R represent the two conformations of the SRI-HtrI interface in different spectral species of SRI. Light-induced transformations that increase the concentration of the A or R conformation suppress or induce reversals, producing attractant or repellent responses, respectively. The A form predominates in the one-photon (orange-light-induced) products, and the R form predominates in the two-photon (white-light-induced) products. In the wild-type SRI-HtrI complex, the one-photon reaction increases the amount of the A form, and therefore causes an attractant response, whereas the two-photon reaction increases the amount of the R form, causing a repellent response. The basis of the orange-light-inverted phenotype is proposed to be a mutation-induced shift of the equilibrium in the SR587 species toward the A form. Repellent responses are therefore elicited by either one-photon or two-photon activation, because of the greater amounts of the R form in the equilibrium mixtures. Suppressors restore the one-photon-induced attractant response by shifting the equilibrium back toward the R form in SR587. Supersuppressors shift excessively into the R form, thus restoring the one-photon attractant response but concomitantly inverting the two-photon repellent response to an attractant response.
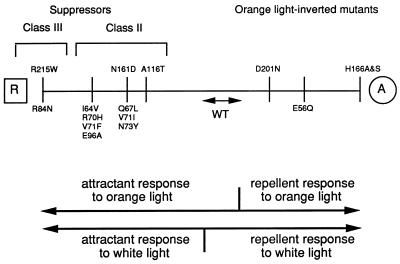
In terms of the model, the linear ranking of inverting and suppressing mutations (Fig. 10) represents different values of the Keq of the interconversion of A and R. The equilibrium is most strongly shifted to the R form in the class III suppressors SRI R215W and HtrI R84N and to the A form in the substitutions at SRI His166. Other mutations are ranked according to two criteria: their ability to suppress orange-light-inverted responses (i.e., whether they are in class II or class III), and their ability to invert the two-photon response in various backgrounds. These two properties are highly correlated. Class III supersuppressors are more effective at inverting the two-photon response than class II suppressors, which are partially allele specific. An important property of the conformational-equilibrium model is that it predicts this correlation, because the one-photon and two-photon responses are attributed to opposite shifts of the same equilibrium by a mechanism in which Keq is sensitive to the effects of mutations.
FIG. 10.
Relative bias of the dark equilibrium toward the R form (toward the left) or toward the A form (toward the right) of the wild-type (WT) signaling complex, the signaling complex of orange-light-inverted mutants, and the signaling complex of suppressed mutants. The range of values of Keq in which A and R are both present in sufficient amounts in the dark equilibrium mixture to yield wild-type behavior is placed at the center. Mutation-induced shifts of Keq for the dark equilibrium toward the R or A form lead to the indicated phenotypes, according to the model in Fig. 9 and as observed in this study. The relative ranking of Keq for different mutations is assigned on the basis of the abilities of mutations on the left to suppress the effects of those on the right and on the extent of inversion of the two-photon response produced by the mutation in wild-type or mutant strains.
The properties of the wild-type strain, the orange-light-inverted mutants, and strains containing class II and class III suppressors, all fit well into the framework of the model. The class I I61V suppressor in HtrI, which suppresses only E56Q, represents a special case. The proximity of residue 61 to Glu56 suggests that in this case the suppression is due to a direct effect on the local structure of HtrI. I61V inverts the white-light response of cells carrying E56Q, but it does not invert the response of cells carrying D201N or that of the wild-type strain, even though these are signaling complexes less shifted toward the A form than the E56Q complex. This result can be understood if I61V by itself does not shift the equilibrium much, but the structure of the E56Q I61V double-mutant protein produces a strong shift.
Other evidence supporting the model is that pH and temperature, which would be expected to affect the Keq of a protein conformational transition, have large effects on the behavioral responses. Low pH corrects the E56Q orange-light inversion (13), and over the range 25 to 45°C, lower temperatures also convert the behavior of the E56Q mutant to that of the wild-type strain (our unpublished data). Also supporting the existence of a metastable equilibrium is the observation that small changes in the side chains of the residues can have marked effects on the signaling phenotype. The suppressor mutations I61V, I64V, and V71I in HtrI show that, for example, addition or removal of only one methyl group of residues in the cytoplasmic portion of TM2 can greatly alter the function of the complex.
In terms of the metastable-equilibrium model proposed here, inverted responses result from subtle structural changes in the signaling complex that alter Keq. Inverted responses have also been reported in eubacterial cells containing modified taxis transducers (6, 14, 20, 21), and these may be explained by the same conformational-bias mechanism.
The locations of the suppressor mutations suggest which regions of SRI and HtrI are important for coupling of the two proteins. Two of the three suppressors in SRI alter residues near the cytoplasmic surface of the protein, according to structural models of SRI based on the BR structure (11, 12, 19); N161D and R215W are at the cytoplasmic ends of helix F and helix G, respectively (Fig. 3). These sites are significant in view of BR structure and function. Recent studies indicate that SRI uses some of the same chemistry for signaling to HtrI that is used by BR for proton pumping (29, 31). This idea was strongly suggested by the observation that removal of HtrI from SRI by genetic deletion converted SRI into a BR-like electrogenic proton pump (2), whereas SRI complexed with HtrI does not carry out electrogenic ion transport (7, 29). Therefore, essential features of the BR mechanism must be conserved in SRI and influenced by interaction with HtrI. A crucial component of the pumping mechanism of BR is the light-triggered conformational change that opens a cytoplasmic channel for proton uptake during the second half of the pumping cycle (16). This conformational change has been visualized by crystallographic methods (33), and it is characterized by the movement first of helices G and B, followed by an outward tilt of the cytoplasmic end of helix F. The residues corresponding to SRI N161 (K172 in BR) and R215 (S226 in BR) are located in this conformationally active region of BR. An attractive hypothesis is that the contribution of the receptor to the conformational change from the R to the A form of the complex in the model is the same conformational change that opens the cytoplasmic channel of BR.
The third suppressor mutation in SRI, A116T, is located at the periplasmic end of helix D (Fig. 3). Expression of A116T in a wild-type background retards the reprotonation of the retinylidene Schiff base nearly threefold, as monitored by S373 decay in the SRI photocycle. This effect is much larger than that of HtrI mutations (Fig. 3), and it suggests that this site is more directly coupled to the photoactive site of SRI.
All suppressor mutations in HtrI are located between positions 53 and 96 (Fig. 3). This clustering must be significant, because our random mutagenesis of HtrI spanned residues 1 to 230. The suppressor sites are not restricted to a single face of the putative α-helix, but they are concentrated in a short region of the helix (Fig. 3). The location of this suppressor cluster adds to a body of observations that indicate that this region at the cytoplasmic end of TM2 is critical for interaction with SRI. Residues 1 to 147 in HtrI have been shown to contain the sites of interaction with SRI responsible for controlling the photocycle (25). Furthermore, in a previous study, seven mutations between positions 70 and 108 were shown to perturb the lifetime of the attractant signaling state S373, and the E56Q mutant protein was shown to exhibit altered signaling (13). Most of the HtrI suppressors also perturb the S373 lifetime, a finding that extends the number of sites in this region that have this modulating effect. The suppressor mutations cluster in HtrI within a region homologous to the linker region of the eubacterial chemotaxis receptor Tar, which cannot be removed without loss of chemotaxis (10, 35), and the linker region of EnvZ, which has been implicated in signaling (24).
ACKNOWLEDGMENTS
We thank Elena Spudich, Xue-Nong Zhang, Michael Manson, and Walther Stoeckenius for critical comments on the manuscript, and Claudia Ruiz for contributing to the construction of the pKJ306 plasmid.
This work was supported by National Institutes of Health grant R01-GM27750 (to J.L.S.).
ADDENDUM IN PROOF
A specific mechanism for inverted taxis responses has recently been suggested in which mutation or methylation repositions a critical transducer residue nearer to an active site on CheA so that stimuli that would normally shift the residue toward the site move it further away (B. L. Taylor and M. S. Johnson, FEBS Lett., in press). This mechanism is an example of a conformational-equilibrium bias as described here if one assumes that the positions of the critical residue are determined by conformations in equilibrium.
REFERENCES
- 1.Bogomolni R A, Spudich J L. The photochemical reactions of bacterial sensory rhodopsin-I: flash photolysis study in the one microsecond to eight second time window. Biophys J. 1987;52:1071–1075. doi: 10.1016/S0006-3495(87)83301-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bogomolni R A, Stoeckenius W, Szundi I, Perozo E, Olson K D, Spudich J L. Removal of transducer HtrI allows electrogenic proton translocation by sensory rhodopsin I. Proc Natl Acad Sci USA. 1994;91:10188–10192. doi: 10.1073/pnas.91.21.10188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cadwell R C, Joyce G F. Randomization of genes by PCR mutagenesis. PCR Methods Appl. 1992;2:28–33. doi: 10.1101/gr.2.1.28. [DOI] [PubMed] [Google Scholar]
- 4.Chen B, Przybyla A E. An efficient site-directed mutagenesis method based on PCR. BioTechniques. 1994;17:161–162. [PubMed] [Google Scholar]
- 5.Cline S W, Doolittle W F. Efficient transfection of the archaebacterium Halobacterium halobium. J Bacteriol. 1987;169:1341–1344. doi: 10.1128/jb.169.3.1341-1344.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dang C V, Niwano M, Ryu J-I, Taylor B L. Inversion of aerotactic response in Escherichia coli deficient in cheB protein methylesterase. J Bacteriol. 1986;166:275–280. doi: 10.1128/jb.166.1.275-280.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ehrlich B E, Schen C R, Spudich J L. Bacterial rhodopsins monitored with fluorescent dyes in vesicles and in vivo. J Membr Biol. 1984;82:89–94. doi: 10.1007/BF01870735. [DOI] [PubMed] [Google Scholar]
- 8.Faber H R, Matthews B W. A mutant T4 lysozyme displays five different crystal conformations. Nature. 1990;348:263–266. doi: 10.1038/348263a0. [DOI] [PubMed] [Google Scholar]
- 9.Ferrando-May E, Krah M, Marwan W, Oesterhelt D. The methyl-accepting transducer protein HtrI is functionally associated with the photoreceptor sensory rhodopsin I in the archaeon Halobacterium salinarium. EMBO J. 1993;12:2999–3005. doi: 10.1002/j.1460-2075.1993.tb05968.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gardina P J, Manson M D. Attractant signaling by an aspartate chemoreceptor dimer with one cytoplasmic domain. Science. 1996;274:425–426. doi: 10.1126/science.274.5286.425. [DOI] [PubMed] [Google Scholar]
- 11.Grigorieff N, Ceska T A, Downing K H, Baldwin J M, Henderson R. Electron-crystallographic refinement of the structure of bacteriorhodopsin. J Mol Biol. 1996;259:393–421. doi: 10.1006/jmbi.1996.0328. [DOI] [PubMed] [Google Scholar]
- 12.Henderson R, Baldwin J M, Ceska T A, Zemlin F, Beckmann E, Downing K H. Model for the structure of bacteriorhodopsin based on high-resolution electron cryo-microscopy. J Mol Biol. 1990;213:899–929. doi: 10.1016/S0022-2836(05)80271-2. [DOI] [PubMed] [Google Scholar]
- 13.Jung K-H, Spudich J L. Protonatable residues at the cytoplasmic end of transmembrane helix-2 in the signal transducer HtrI control photochemistry and function of sensory rhodopsin I. Proc Natl Acad Sci USA. 1996;93:6557–6561. doi: 10.1073/pnas.93.13.6557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kihara M, Macnab R M. Cytoplasmic pH mediates pH taxis and weak-acid repellent taxis of bacteria. J Bacteriol. 1981;145:1209–1221. doi: 10.1128/jb.145.3.1209-1221.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Krah M, Marwan W, Vermeglio A, Oesterhelt D. Phototaxis of Halobacterium salinarium requires a signalling complex of sensory rhodopsin I and its methyl-accepting transducer HtrI. EMBO J. 1994;13:2150–2155. doi: 10.1002/j.1460-2075.1994.tb06491.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lanyi J K. Bacteriorhodopsin as a model for proton pumps. Nature. 1995;375:461–463. doi: 10.1038/375461a0. [DOI] [PubMed] [Google Scholar]
- 17.Le Moual H, Koshland D E., Jr Molecular evolution of the C-terminal cytoplasmic domain of a superfamily of bacterial receptors involved in taxis. J Mol Biol. 1996;261:568–585. doi: 10.1006/jmbi.1996.0483. [DOI] [PubMed] [Google Scholar]
- 18.Leung D W, Chen E, Goeddel D V. A method for random mutagenesis of a defined DNA segment using a modified polymerase chain reaction. Technique. 1989;1:11–15. [Google Scholar]
- 19.Lin S L, Yan B. Three-dimensional model of sensory rhodopsin I reveals important restraints between the protein and the chromophore. Protein Eng. 1997;10:197–206. doi: 10.1093/protein/10.3.197. [DOI] [PubMed] [Google Scholar]
- 20.Muskavitch M A, Kort E N, Springer M S, Goy M F, Adler J. Attraction by repellents: an error in sensory information processing by bacterial mutants. Science. 1978;201:63–65. doi: 10.1126/science.351803. [DOI] [PubMed] [Google Scholar]
- 21.Nishiyama S-I, Nara T, Homma M, Imae Y, Kawagishi I. Thermosensing properties of mutant aspartate chemoreceptors with methyl-accepting sites replaced singly or multiply by alanine. J Bacteriol. 1997;179:6573–6580. doi: 10.1128/jb.179.21.6573-6580.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Olson K D, Zhang X-N, Spudich J L. Residue replacements of buried aspartyl and related residues in sensory rhodopsin I: D201N produces inverted phototaxis signals. Proc Natl Acad Sci USA. 1995;92:3185–3189. doi: 10.1073/pnas.92.8.3185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Olson K D, Spudich J L. Removal of the transducer protein from sensory rhodopsin I exposes sites of proton release and uptake during the receptor photocycle. Biophys J. 1993;65:2578–2585. doi: 10.1016/S0006-3495(93)81295-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Park H, Inouye M. Mutational analysis of the linker region of EnvZ, an osmosensor in Escherichia coli. J Bacteriol. 1997;179:4382–4390. doi: 10.1128/jb.179.13.4382-4390.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Perazzona B, Spudich E N, Spudich J L. Deletion mapping of the sites on the HtrI transducer for sensory rhodopsin I interaction. J Bacteriol. 1996;178:6475–6478. doi: 10.1128/jb.178.22.6475-6478.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Royer W E, Jr, Pardanani A, Gibson Q H, Peterson E S, Friedman J M. Ordered water molecules as key allosteric mediators in a cooperative dimeric hemoglobin. Proc Natl Acad Sci USA. 1996;93:14526–14531. doi: 10.1073/pnas.93.25.14526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Spudich E N, Spudich J L. The photochemical reactions of sensory rhodopsin I are altered by its transducer. J Biol Chem. 1993;268:16095–16097. [PubMed] [Google Scholar]
- 28.Spudich E N, Zhang W, Alam M, Spudich J L. Constitutive signaling by the phototaxis receptor sensory rhodopsin II from disruption of its protonated Schiff base-Asp73 interhelical salt bridge. Proc Natl Acad Sci USA. 1997;94:4960–4965. doi: 10.1073/pnas.94.10.4960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Spudich J L. Protein-protein interaction converts a proton pump into a sensory receptor. Cell. 1994;79:747–750. doi: 10.1016/0092-8674(94)90064-7. [DOI] [PubMed] [Google Scholar]
- 30.Spudich J L, Spudich E N. Selection and screening methods for halophilic archaeal rhodopsin mutants. In: Robb F T, Place A R, Sowers K R, Schreier H J, DasSarma S, Fleischmann E M, editors. Archaea: a laboratory manual. Plainview, N.Y: Cold Spring Harbor Laboratory Press; 1995. pp. 23–28. [Google Scholar]
- 31.Spudich J L, Lanyi J K. Shuttling between two protein conformations: the common mechanism for sensory transduction and ion transport. Curr Opin Cell Biol. 1996;8:452–457. doi: 10.1016/s0955-0674(96)80020-2. [DOI] [PubMed] [Google Scholar]
- 32.Spudich J L, Bogomolni R A. Mechanism of colour discrimination by a bacterial sensory rhodopsin. Nature. 1984;312:509–513. doi: 10.1038/312509a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Subramaniam S, Gerstein M, Oesterhelt D, Henderson R. Electron diffraction analysis of structural changes in the photocycle of bacteriorhodopsin. EMBO J. 1993;12:1–8. doi: 10.1002/j.1460-2075.1993.tb05625.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sundberg S A, Bogomolni R A, Spudich J L. Selection and properties of phototaxis-deficient mutants of Halobacterium halobium. J Bacteriol. 1985;164:282–287. doi: 10.1128/jb.164.1.282-287.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tatsuno I, Homma M, Oosawa K, Kawagishi I. Signaling by the Escherichia coli aspartate chemoreceptor Tar with a single cytoplasmic domain per dimer. Science. 1996;274:423–424. doi: 10.1126/science.274.5286.423. [DOI] [PubMed] [Google Scholar]
- 36.Wolf A, Shaw E W, Oh B-H, Bondt H, Joshi A K, Ames G F-L. Structure/function analysis of the periplasmic histidine-binding protein. J Biol Chem. 1995;270:16097–16106. doi: 10.1074/jbc.270.27.16097. [DOI] [PubMed] [Google Scholar]
- 37.Yao V J, Spudich J L. Primary structure of an archaebacterial transducer, a methyl-accepting protein associated with sensory rhodopsin I. Proc Natl Acad Sci USA. 1992;89:11915–11919. doi: 10.1073/pnas.89.24.11915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhang X-N, Spudich J L. His-166 is critical for active site proton transfer and phototaxis signaling by sensory rhodopsin I. Biophys J. 1997;73:1516–1523. doi: 10.1016/S0006-3495(97)78183-9. [DOI] [PMC free article] [PubMed] [Google Scholar]