Abstract
In recent years, various polymorphic loci and multicopy insertion elements have been discovered in the Mycobacterium tuberculosis genome, such as the direct repeat (DR) locus, the major polymorphic tandem repeats, the polymorphic GC-rich repetitive sequence, IS6110, and IS1081. These, especially IS6110 and the DR locus, have been widely used as genetic markers to differentiate M. tuberculosis isolates and will continue to be so used, due to the conserved nature of the genome of M. tuberculosis. However, little is known about the processes involved in generating these or of their relative rates of change. Without an understanding of the biological characteristics of these genetic markers, it is difficult to use them to their full extent for understanding the population genetics and epidemiology of M. tuberculosis. To address these points, we identified a cluster of 7 isolates in a collection of 101 clinical isolates and investigated them with various polymorphic genetic markers, which indicated that they were highly related to each other. This cluster provided a model system for the study of IS6110 transposition, evolution at the DR locus, and the effects of these on the determination of evolutionary relationships among M. tuberculosis strains. Our results suggest that IS6110 restriction fragment length polymorphism patterns are useful in grouping closely related isolates together; however, they can be misleading if used for making inferences about the evolutionary relationships between closely related isolates. DNA sequence analysis of the DR loci of these isolates revealed an evolutionary scenario, which, complemented with the information from IS6110, allowed a reconstruction of the evolutionary steps and relationships among these closely related isolates. Loss of the IS6110 copy in the DR locus was noted, and the mechanisms of this loss are discussed.
Tuberculosis is one of the most ancient infectious diseases of human beings, and it is still in a leading position among infectious diseases as a cause of morbidity and mortality (5). One of the factors hampering control of tuberculosis is the difficulty of differentiating among strains due to the conserved genome of Mycobacterium tuberculosis (9, 14), which has hindered understanding of the processes of the disease. In recent years, various polymorphic loci and multicopy insertion elements have been discovered in the genome, such as the direct repeat (DR) locus (19), the major polymorphic tandem repeats (21), the polymorphic GC-rich repetitive sequence (PGRS) (35), IS6110 (45), and IS1081 (10). These have been used as genetic markers to differentiate among M. tuberculosis strains in epidemiological studies. However, little is known about the processes involved in these genotypic changes or about the relative rates of these changes or of transposition events. Without an understanding of these aspects, it is difficult to use these markers to their full extent to understand the population genetics and epidemiology of M. tuberculosis. Indeed, little is known of the population genetics of insertion elements in any species.
Since bacteria reproduce by fission, all descendants should share acquired mutations with their recent common ancestors. It has been shown that the transposition rates of insertion elements are much higher than the rates of other heritable changes in the genome, such as nucleotide mutations (17, 36). Attempts to use insertion elements to establish phylogenetic relationships in Escherichia coli strains have shown that these markers were predictive only with closely related strains (25). Similarly, evolutionary relationships of Salmonella enteritidis isolates established with the Salmonella-specific IS200 are consistent with clonal lineages of recent origin and with phage-typing groups (38). IS6110 is a member of the IS3 family of insertion elements (27, 45) and is widely distributed throughout the M. tuberculosis complex (7, 20, 40, 44). It is currently the most widely used genetic marker for differentiating among M. tuberculosis strains (8, 20, 42, 44). As many as 25 copies are present in the genomes of clinical isolates of M. tuberculosis, although some strains without IS6110 copies have been identified (20, 27, 43). Typing schemes using IS6110 restriction fragment length polymorphisms (RFLPs) assume that the distribution of the IS6110 element in the genome is random; however, in the genome of M. tuberculosis H37Rv, IS6110 is restricted to about two-thirds of the genome around the DR locus (33), while there also seem to be particular IS6110 insertion hot spots, such as the DR locus (19) and the ipl locus (11). All these points lead to questioning of the appropriateness of the IS6110 element as a genetic marker for M. tuberculosis population genetic and phylogenetic analyses.
The DR region is a polymorphic locus in the genome of M. tuberculosis which comprises a cluster of directly repeating sequences of 36 bp, separated by unique spacer sequences of 36 to 41 bp (20). One repeat sequence and the following spacer sequence together have been termed a DVR (direct variable repeat) (18). The number of DVRs varies from strain to strain, allowing this locus to be used as a genetic marker to differentiate strains (18, 19). Two mechanisms have been proposed for the polymorphisms at this locus: homologous recombination between adjacent or distant DVRs and IS6110 transposition (18). The DR locus has become the second most important genetic marker for the differentiation of M. tuberculosis strains; however, little is known about its polymorphic changes over evolutionary time, and this limits its more informed use in population genetic and phylogenetic analyses.
To address these questions, we investigated a collection of 101 clinical isolates of M. tuberculosis and identified a cluster of 7 isolates which were highly related by a number of genotypic features. Intensive study of these seven isolates, in particular their IS6110 RFLP patterns, their specific IS6110 insertion sites, and polymorphisms at the DR locus, suggested that the overall IS6110 RFLP pattern is useful in grouping closely related isolates together; however, it can be misleading if used for making inferences about the evolutionary relationships between such closely related isolates. DNA sequence analysis of the DR loci of these isolates revealed an evolutionary scenario, which, when complemented with analysis of IS6110 copies, allowed the reconstruction of the evolutionary steps and phylogenetic relationships between these closely related isolates. Loss of the IS6110 copy in the DR locus was noted, and likely mechanisms for this loss are discussed.
MATERIALS AND METHODS
Strains.
A total of 104 isolates of M. tuberculosis complex were studied. These included 101 clinical isolates of M. tuberculosis from the Scottish Mycobacteria Reference Laboratory, Edinburgh, United Kingdom, collected in 1992 and 1995 (31); the M. tuberculosis type strain, H37Ra; the IS6110 fingerprinting reference strain, Mt14323 (42); and Mycobacterium bovis BCG (Pasteur). These strains and isolates were cultured in Middlebrook 7H9 broth in a 50-ml-volume centrifuge tube at 37°C for about 4 weeks. After it was ascertained that the cultures were free from other bacterial contamination, cells were harvested and heated to 80°C for 30 min to kill them and were stored at −20°C prior to DNA extraction.
IS6110 DNA Southern blot fingerprinting and analysis.
All DNA extraction, digestion, and blotting techniques used were from the standardized protocol (42). The methods for making digoxigenin (DIG)-labelled probes (IS6110 probe, internal standard marker probe, DR probe, FL1 probe, IS1547 probe, and PGRS probe) and for detection of the probes are described below.
To make the DIG-labelled IS6110 DNA probe, 5 μl of M. bovis BCG (Pasteur) DNA solution (10 μg/ml) was added to a PCR tube which contained 40 μl of PCR mixture (50 mM KCl, 10 mM Tris-HCl [pH 8.0], 1.5 mM MgCl2, 5% glycerol, 225 μM [each] INS1 and INS2 primers [20], and 0.5 U of Taq polymerase). Five microliters of 10× DIG-dUTP/deoxynucleoside triphosphate mixture (Boehringer Mannheim GmbH, Mannheim, Germany) was added to the mixture, and the reaction was subjected to PCR at an annealing temperature of 65°C. A PvuII-digested supercoiled DNA ladder (Gibco-BRL, Life Technologies, Ltd., Paisley, United Kingdom) and HaeIII-digested φX174 DNA (Advanced Biotechnologies, London, United Kingdom) were DIG labelled by a randomly primed DNA labelling method (6). The working probe solutions were prepared by diluting the DIG-labelled PCR product in standard hybridization solution (5× SSC [1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate], 1.0% blocking reagent for nucleic acid hybridization, 0.1% N-lauroylsarcosine, 0.02% sodium dodecyl sulfate) to a concentration of 5 to 25 ng/ml.
Hybridization and detection were carried out according to the manufacturer’s instructions (Boehringer Mannheim). Briefly, the blotted membrane was hybridized with the DIG-labelled probe at 68°C overnight in a hybridization oven (Hybaid, Ltd., Middlesex, United Kingdom). The membrane was then washed, equilibrated, blocked, and incubated in anti-DIG antibody-alkaline phosphatase solution (1:10,000). CSPD solution (1%) (Boehringer Mannheim) was then pipetted over the membrane. The membrane was sealed in a plastic hybridization bag after removal of excess liquid, incubated, and then exposed to X-ray films. The X-ray films were developed in an RP X-Omat Processor, model M6B (Kodak Diagnostic Imaging, Rochester, N.Y.). To reprobe a membrane, the previous probe was stripped off by incubation in a 0.2 M NaOH solution.
Pairwise similarities of IS6110 fingerprint patterns were calculated by the Dice coefficient of similarity with GelCompar (version 3.0; Applied Maths, Kortrijk, Belgium). Clustering of the isolates by similarity to give a dendrogram was carried out with UPGMA (unweighted pair group method with arithmetic averages).
Primer oligonucleotide design and synthesis.
All primer oligonucleotides used in this study (Table 1) were designed with the software package OLIGO (version 5.0; National Bioscience, Inc., Plymouth, Minn.), synthesized on an Applied Biosystems (Warrington, United Kingdom) 291 DNA synthesizer, and purified by using oligonucleotide purification cartridges (Applied Biosystems).
TABLE 1.
PCR primers used in this study
| Primer | Sequence | Description |
|---|---|---|
| IS1 | 5′-CGGAGACGGTGCGTAAGTG-3′ | nt 192–210 in negative strand of X57835 |
| IS2 | 5′-GCTGCCTACTACGCTCAAC-3′ | nt 1272–1290 in X57835 |
| DR1 | 5′-AGGTTCGCGTCGATCAAGTCC-3′ | nt 2313–2333 in X57835 |
| DR2 | 5′-GGATGTGGTGCTGGATTTC-3′ | nt 2815–2833 in negative strand of X57835 |
| DR3 | 5′-CGAAATCCAGCACCACATC-3′ | nt 2815–2833 in X57835 |
| DR4 | 5′-GATCAAGTCCGGTCGTCAGA-3′ | nt 2324–2343 in negative strand of X57835 |
| DR5 | 5′-GCCCGCTAATCCCGCACAAGT-3′ | nt 25912–25932 in negative strand of Z81331 |
| DR6 | 5′-GGGACGGAAACTTTTCTGAAC-3′ | nt 25891–25911 in Z81331 |
| DR7 | 5′-GACGACCTCGGACAGCATCTC-3′ | nt 26439–26859 in negative strand of Z81331 |
| DR8 | 5′-CCCGCCCGGGGAGATGCTGTC-3′ | nt 27438–26449 in Z81331 |
| DR9 | 5′-CTGACGACTGGCGATTTACGA-3′ | nt 26723–26743 in negative strand of Z81331 |
| DR10 | 5′-GTTCCCGTCAGCGTCGTAAAT-3′ | nt 26710–26730 in Z81331 |
| FL1a | 5′-CGACACCGGCGCCACACTG-3′ | nt 267–285 in X95144 |
| FL1b | 5′-CATCCAATGCTCAACTATT-3′ | nt 1–19 in negative strand of X95144 |
| M1a | 5′-GCGCGTGGTAAGCATTCACAA-3′ | nt 14685–14705 in Z84725 |
| M1b | 5′-CGCGGTCCCGTGGTCATTTAC-3′ | nt 15011–15031 in negative strand of Z84725 |
| U16B7 | 5′-CATCATCAGCAGGCATTGTTA-3′ | nt 25453–25473 in Z81331 |
| L16B7 | 5′-CTCCCGGCGGGGTCATTTCAT-3′ | nt 29977–29997 in negative strand of Z81331 |
PCR and sequencing.
Semiarbitrarily primed PCR (11) was used to identify IS6110 flanking DNA sequences. Template DNA was sequenced by using an Applied Biosystems 377A automated DNA sequencer with a Prism Ready Mix Kit based on AmpliTaq DNA polymerase, FS (Applied Biosystems). The sequencing reaction mixtures, each containing 150 μg of template DNA, 3.2 pM one primer, and 9.5 μl of Prism Ready Mix, were subjected to 25 cycles of denaturation (at 96°C for 30 s), annealing (at 50°C for 15 s), and extension (at 60°C for 4 min).
Sequence analysis with computer.
DNA sequences identified in this study were compared to sequences in the GenBank and EMBL databases and in the M. tuberculosis DNA sequence database at the Sanger Centre, Cambridge, England, with the programs FASTA or PFASTA (32) and BLAST (2, 3, 23). For DNA sequence alignment, the programs GAP (30) and BESTFIT (37) in the GCG package (version 8, August 1994; Genetics Computer Group, Madison, Wis.) were used.
Nucleotide sequence accession numbers.
The sequences reported here have been assigned the following EMBL accession numbers: fl1::IS6110, X94955 and X94956; IS1547, Y13407; DR sequence from isolate 86, Y14045; DR sequence from isolate 149, Y14046; DR sequence from isolate 257, Y14047; DR sequence from isolate 191, Y14048; and DR sequence from isolate 93, Y14049.
RESULTS
Heterogeneity of IS6110 insertion sites in the M. tuberculosis isolates.
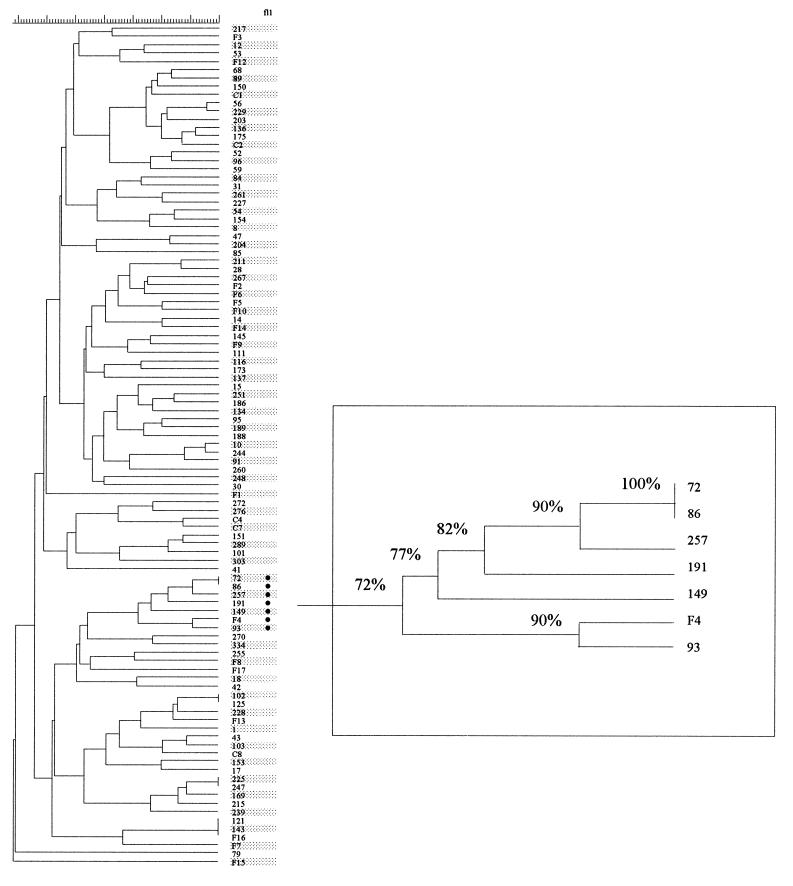
Following Southern blotting and hybridization with the IS6110 probe to PvuII-digested DNA of the 101 isolates, the resultant fingerprint patterns were imported into the computer package GelCompar, where they were normalized to a standard ladder coloaded with each sample to allow accurate comparative alignment. Dice coefficients of similarity were calculated for all pairwise comparisons, and a dendrogram was constructed (Fig. 1). A wide diversity of RFLP patterns were apparent in the IS6110 fingerprints of the 101 isolates; the number of copies ranged from 2 to 16, with a mean of 8.3. Dice coefficients of similarity for all pairwise comparisons of the isolates in the whole collection ranged from 0 to 100%, with a mean of 41%.
FIG. 1.
Dendrogram of IS6110 fingerprints of 101 isolates of M. tuberculosis, constructed with GelCompar and based on Dice coefficients and UPGMA clustering. In column fl1, the symbol (•) indicates isolates with fl1::IS6110. The graph in the box is an enlarged branch of the isolates carrying fl1::IS6110 and shows the Dice similarity coefficients between the isolates.
Isolates with the fl1::IS6110 allele.
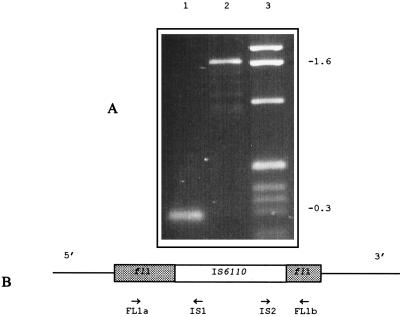
fl1::IS6110 is a genomic insertion site of IS6110, identified by the semiarbitrary PCR technique described in our previous publication (11), and was used as a genetic marker to identify the cluster of isolates studied here. Analysis of the genomic sequences flanking IS6110 in fl1::IS6110 indicated that this IS6110 had inserted into a long open reading frame, which has been postulated to be a dihydrofolate reductase gene (folA) in M. tuberculosis (31a). In fl1::IS6110, the insertion was located two-thirds of the way along the coding sequence. Isolates in the study collection carrying the fl1::IS6110 genotype were identified by PCR with three primer combinations. Primers FL1a and IS1 determined the presence of the left end (5′ end) of the IS6110 element in the fl1::IS6110 sequence, giving a product of 302 bp. Primers IS2 and FL1b determined the presence of the right end (3′ end) of the IS6110 element in the fl1::IS6110 sequence, giving a product of 250 bp. Primers FL1a and FL1b gave a product of 286 bp in isolates without the IS6110 element in fl1 and a product of 1,645 bp when IS6110 was present (Fig. 2). Among the 101 isolates tested, 7 isolates (7%) which carried fl1::IS6110 were identified, and these formed one cluster in the dendrogram derived from IS6110 fingerprints (Fig. 1).
FIG. 2.
PCR amplification of the fl1::IS6110 allele. (A) PCR products obtained with primers FL1a and FL1b from an fl1+ strain (lane 1) and an fl1::IS6110 strain (lane 2). Lane 3, 1-kb ladder. (B) IS6110 (open box) and fl1 (shaded boxes), with the locations of the primers used and directions of their extension.
The presence of fl1::IS6110 in these isolates could be due to the sharing of a recent common ancestor or to coincidence, so in order to clarify this specific IS6110 insertion site, the DR, IS1547, and PGRS RFLP patterns in these isolates were also examined.
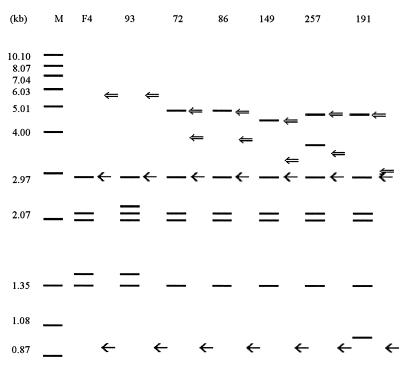
The DNAs of these isolates were digested with PvuII. Following Southern blotting, the membrane was probed sequentially with the IS6110 probe, the DR probe, and the FL1 probe (fl1 sequence amplified by primers FL1a and FL1b). The results are illustrated in Fig. 3. The numbers of IS6110-containing PvuII fragments were five or six, with four of them (3.00, 2.10, 2.06, and 1.35 kb) apparently common to all. The 3.00-kb fragment cohybridized to the FL1 probe, indicating that it carried the right-hand end of the IS6110 copy and the flanking 3′ end of the fl1 locus. The 2.10-kb fragment is probably analogous to an IS6110 insertion into an M. tuberculosis H37Rv DNA sequence (EMBL accession no. Z84725; sequence identification, MTCY4D9) between nucleotides (nt) 14840 and 14841, because an IS6110 insertion at this location of the IS6110-free DNA sequence Z84725 would give an IS6110-hybridizing PvuII fragment with a predicted length of 2.10 kb. In addition, PCR products of the expected lengths were obtained from all the isolates with primers (S1-M1, S2-M2) amplifying across the ends of an insertion at this genomic site (data not shown). Such an insertion has also been noted in another isolate (28) which carried the fl1::IS6110 allele. These observations indicate that the common bands in the IS6110 fingerprint patterns in different strains represent common IS6110 copies in terms of their locations in the genome, which presumably reflects the sharing of a recent common ancestor among these seven isolates.
FIG. 3.
IS6110, DR, and FL1 RFLP patterns of fl1::IS6110 isolates. Isolates F4, 93, 72, 86, 149, 257, and 191 were digested with PvuII and, following blotting, were probed sequentially with IS6110 (), DR (⇐), and FL1 (←) probes. Lane M, internal size standards, with DNA sizes marked in kilobase pairs.
Among the seven isolates, five had fragments of about 5 kb which cohybridized with both the IS6110 probe and the DR probe (Fig. 3), indicating that there was an IS6110 copy in the DR region in these isolates. There were no such cohybridizing fragments for isolates 93 and F4, indicating that the IS6110 copy in the DR region was missing in these isolates. Furthermore, all the 5-kb fragments showed greater length polymorphism than the other fragments, probably as a result of polymorphisms in the DR loci, as described below.
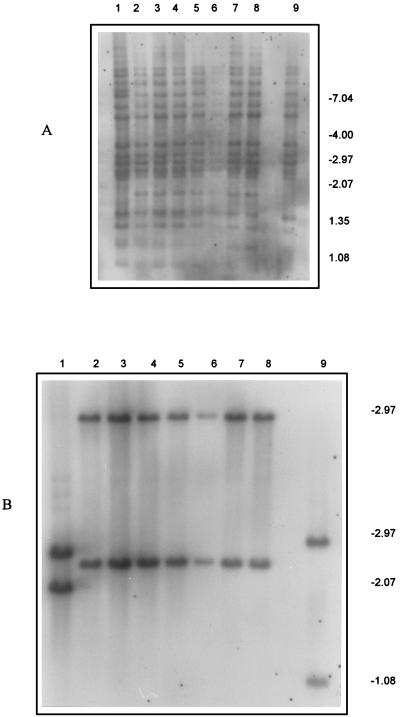
In addition to IS6110 insertion sites, IS1547 and PGRS were also used to assess the relatedness of these seven isolates. IS1547 is a new member of the IS900 family of insertion elements and is found in the M. tuberculosis complex (12, 22). A probe against the IS1547 DNA sequence (EMBL accession no. Y13470), which hybridizes to PvuII-digested genomic DNA, has detected one or two IS1547 copies in the M. tuberculosis isolates investigated so far, but these have highly variable RFLPs. However, all seven of the isolates carrying fl1::IS6110 had the same IS1547 RFLP pattern (Fig. 4). The PGRS is a tandem repeat of the consensus sequence CGGCGGCRR and is found in several mycobacterial species, including those of the M. tuberculosis complex (35). When used as a probe for restriction enzyme-digested DNA, it shows a high degree of RFLP and so can be used to distinguish different strains (43); however, the PGRS RFLP patterns of the isolates here were indistinguishable (Fig. 4).
FIG. 4.
Autoradiogram of PvuII-digested DNA from M. tuberculosis isolates hybridized with the PGRS probe (A) and the IS1547 probe (B). Lanes 1 through 9, isolates H37Ra, 72, 86, 93, 149, 191, 257, F4, and Mt14323, respectively. Sizes are indicated on the right, in kilobases.
Despite the extensive genotypic similarities among these isolates, there were some differences in terms of IS6110 copy number and their genomic locations. For instance, in addition to the four IS6110 copies common to all seven isolates, two isolates (F4 and 93) had a further common IS6110 copy (on a ca. 1.5-kb PvuII fragment), and one of these isolates (isolate 93) subsequently acquired another IS6110 copy. The other five isolates (isolates 72, 86, 149, 191 and 257) each had an additional IS6110 copy, and two of these (isolates 191 and 257) each carried yet a further element.
Polymorphism of the DR loci.
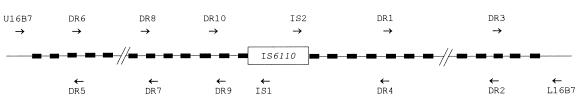
To investigate polymorphism at the DR locus, the primer pair U16B7–L16B7 was designed to amplify across the DR region (Fig. 5) and was used in combination with IS6110-specific primers (IS1 and IS2) to study all of the seven isolates. PCR with the primer pair U16B7–L16B7 amplified a 1.6-kb fragment from both isolate 93 and isolate F4, whereas PCR with primers U16B7–IS1 and IS2–L16B7 amplified fragments from the rest of the isolates, ranging from 1.4 to 2 kb with primers U16B7-IS1 and from 1.3 to 1.5 kb with primers IS2-L16B7. All these fragments were sequenced. To identify particular DVRs, the DNA sequences of the DR region of M. tuberculosis H37Rv (EMBL accession no. Z81331) and of M. bovis BCG (EMBL accession no. X57835) were used as references. The DVRs in strain H37Rv were numbered in order from the 5′ to the 3′ end, giving a total of 42 in the DR region, with the insertion of IS6110 in DVR24 (Fig. 6), while the numbering of M. bovis BCG was taken from reference 19. The DVRs in the sequenced PCR fragments here were enumerated by aligning them against the reference sequences Z81331 and X57835 with the BESTFIT program in the GCG package (16).
FIG. 5.
Schematic illustration of the PCR primers used in this study. The filled boxes represent the DR sequences, between which are spacer DNA sequences. The locations and directions of extension of the primers are indicated.
FIG. 6.
Schematic representation of the structures of the DR regions of M. tuberculosis isolates. A number and the hyphen following it represent one DVR. The DVRs are numbered from the 5′ end to the 3′ end of the DNA sequence. The DR structure of M. tuberculosis H37Rv (H37) (EMBL accession no. Z81331) is shown for reference. The numbers in parentheses, i.e., (25) and (26), represent DVR spacers identical to the spacers of DVR25 and DVR26 in M. bovis BCG (EMBL accession no. X57835) (19). IS6110 is inserted into DVR24, which is therefore shown on each side of the insertion.
The PCR fragments from isolates 86 and 72, obtained with the primer pair U16B7–IS1, were 2,077 bp and comprised a 5′-end DNA sequence flanking the DR region, 25 intact DVRs, the partial DR DNA sequence adjacent to the left-hand side of the IS6110, and part of the IS6110 up to the location of primer IS1. The PCR fragments from isolates 86 and 72, obtained with the primer pair IS2–L16B7, were 1,528 bp and comprised DNA sequence of the IS6110 from the location of primer IS2, the partial DVR, 18 intact DVRs, and DNA sequence flanking the right-hand side of the DR region (3′ end). Alignment of these DNA sequences against the DR regions of strain H37Rv and M. bovis BCG showed that the DNA sequence of the DR locus in isolates 86 and 72 had the same DR structure as that of strain H37Rv, except for two extra DVRs located between DVR20 and DVR21. These two extra DVRs matched DVR25 and DVR26 in the DR region in M. bovis BCG (19) (Fig. 6).
The same process of sequencing and identification was applied to the DR regions of the other isolates. In comparison with the DR structure of isolate 86, two DVRs (DVR9 and DVR10) were deleted in isolate 257, and eight DVRs (DVR5 to DVR13) were deleted in isolate 191. Isolates 93 and F4 not only had the same deletion of DVR5 to DVR13 as in isolate 191 but also had a 2,356-bp deletion which involved the IS6110 element in the DR region and DVR22 to -35. Isolate 149 has deletions in both the left-hand side (DVR7 to DVR 11) and the right-hand side (DVR35 to DVR37) of the DR region. In all cases, there was loss of intact DVR units, and there were no instances of other nucleotide changes. All these results are schematically illustrated in Fig. 6.
DISCUSSION
Seven of 101 isolates were found to have a particular IS6110 insertion (fl1::IS6110). These seven isolates were also found to share a number of other features, including four common IS6110 RFLP fragments which included the specific insertions fl1::IS6110 and MTCY4D9::IS6110, two copies of IS1547 with identical RFLP patterns, and indistinguishable PGRS RFLP patterns. All these common features suggest that these isolates are highly related to each other and are therefore derived from a common ancestor. Thus, they provide a well-described model system for the study of IS6110 transposition and the evolution of the DR locus in the genome of M. tuberculosis. The M. tuberculosis fl1::IS6110 and MTCY4D9::IS6110 mutations were found in isolates from patients from around the world; five strains were from British patients and two were from Indian patients, and these mutations have also been reported by Mendiola and colleagues (28) in an M. tuberculosis strain isolated from a patient in Guadeloupe, West Indies, in 1986. This dispersal suggests that these are not recent mutations and that this is not a recent clone.
IS6110 transposition.
Extensive use has been made of IS6110 as a probe to generate RFLP patterns for the assessment of isolate relatedness. In the IS6110 RFLP patterns of the seven isolates here which carried fl1::IS6110, five or six IS6110-containing bands were apparent; four of them were common to all of the isolates. Of these common bands, two have been confirmed to carry particular IS6110 insertions (fl1::IS6110 and MTCY4D9::IS6110). These common bands are presumably inherited from a common ancestor. Where there are a sufficient number of bands in common among isolates, IS6110 RFLP patterns are satisfactory for the tracing of infection sources and in epidemiological studies. Insertion elements have also been used to identify closely related isolates of E. coli (25) and S. enteritidis (38). However, IS6110 RFLP patterns are of limited use in making inferences about the evolutionary relationships among such closely related isolates. Here, for instance, it can be inferred that the common ancestor of the isolates in this cluster had the four IS6110 copies noted above plus an IS6110 copy in DR. These IS6110 copies form a framework or baseline for subsequent IS6110 changes in the isolates of this cluster. Based on this framework, isolates 93 and F4 have had a deletion of one of these five IS6110 copies, that in the DR region, and have acquired one additional IS6110 copy, with isolate F4 also gaining a further one. However, based solely on IS6110 RFLP patterns, it is impossible to infer whether the IS6110 deletion or addition occurred first. Similarly, isolates 191 and 257 have each acquired an additional IS6110 copy onto the framework, but it is not possible to infer which event occurred first. In this respect, IS6110 RFLP is akin to multilocus enzyme electrophoresis; it is a powerful tool for the determination of the genetic distances between strains and for the differentiation of closely related isolates, but it is poor for the determination of genetic descent (1).
IS6110 losses in the DR locus.
Isolates 93 and F4 did not have IS6110 copies in their DR loci; there are three possible reasons for this. Firstly, there may never have been an IS6110 in the DR region in these two isolates; however, this is unlikely, since the vast majority of the M. tuberculosis complex strains investigated so far, including all but these 2 of the 101 M. tuberculosis isolates examined in this study, carry one or two IS6110 copies there (13, 18, 19, 43). This absence of IS6110 in the DR locus is not the ancestral state in this cluster of isolates. Secondly, excision could also account for the loss, as has been observed with other insertion elements (15). For instance, IS1-mediated excision in E. coli removed more than 1 kb of flanking chromosome from either side of the element (34), while IS4 excision has been observed to delete chromosomal sequences from both sides of this element (24). Excision would typically involve the transposase of the insertion sequence and would not involve homologous recombination, so the losses here of intact DVR units would not be compatible with such a mechanism. Finally, homologous recombination has been proposed as a major mechanism for the generation of polymorphism in the DR region, since the deletion of DVRs often occurs in discrete units (18). Given that the loss of IS6110 from isolates 93 and F4 was associated with such a loss of intact, flanking DVR units and that there were no other nucleotide polymorphisms associated with this deletion, the most likely mechanism for the loss of the IS6110 copy in the DR locus is homologous recombination.
Evolutionary scenario in the DR locus.
As shown in Fig. 6, the changes in the DR loci of the investigated isolates are towards the loss of DVRs, and this trend has also been observed in other studies (18, 43). The intact nature of the DVRs in the DR sequences here, with the changes taking the form of deletion of whole DVRs, suggests that these losses are occurring by homologous recombination. The position around DVR9 and DVR10 is a hot spot for deletions in the isolates here and also in those analyzed by Groenen et al. (18), while the position around DVR35 also seems to be unstable. Whether these instabilities are an innate feature of these particular DVR sequences, of their locations relative to the internal IS6110, or of their locations relative to the ends of the DR sequence is unclear. Selection for the maintenance of an intact DR sequence does not seem to be strong, although no strains are reported to lack this locus entirely. If this locus has a role in replicon partitioning, as does a similar sequence in Haloferax spp. (29), then M. tuberculosis may be a useful study system for the clarification of the role of such sequences. Based on current knowledge of the DR locus, it can be hypothesized that the common ancestor of the M. tuberculosis complex of species had a DR structure similar to that in M. bovis BCG. Over evolutionary time, changes towards a decrease in the number of DVRs in the DR locus have taken place.
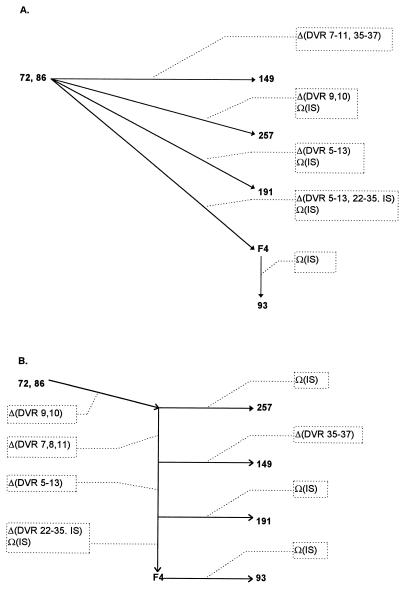
DNA sequence divergence has been widely used to trace evolutionary relationships among different species, or among different strains within a species (39, 41). Among the five different DR structures found in the isolates of this cluster (Fig. 6), those from isolates 72 and 86 showed the most intact structure, suggesting that the other isolates in this cluster originated from an ancestor which had the same DR structure as these. Whether the sequences of events leading to the alleles in the seven isolates here were dependent on or independent of each other cannot be inferred; however, the most parsimonious route would be one of dependent events, while the independent route could be accomplished in less evolutionary time, since events could happen simultaneously in independent lineages. These two extremes are illustrated in Fig. 7. In the scenario of dependent changes (Fig. 7B), deletion of DVR9 and DVR10 in the DR locus of isolate 86 would have led to the generation of the DR structure in an intermediate strain, such as isolate 257. After that, the further deletions of DVR7, -8, and -11 would have occurred to give an intermediate strain, which would have diverged to give isolate 149, with the loss of DVR35 through -37, and another lineage which had the further deletions of DVR5 to -6 and DVR11 to -13 at the left-hand side, as in isolate 191. The DR structure in isolates 93 and F4 would be due to the further deletion from DVR22 to DVR35, which included the IS6110. In the scenario of independent changes in the DR regions of isolates 149, 257, 191, F4, and 93, losses would have occurred independently from the intact DR regions of strains such as isolates 72 and 86 (Fig. 7A).
FIG. 7.
Deduced evolutionary scenarios for the seven closely related isolates, inferred from their DR locus polymorphisms and their IS6110 distributions. Solid lines with arrows represent routes; dotted lines and dotted boxes indicate changes of DRs and/or IS6110. Route A is based on the assumption that the structures of the DR loci of these isolates are the results of independent events, while route B assumes that they result from sequential events. Δ and Ω, deletion and insertion, respectively.
The correlation between DNA sequence divergence and divergence time has been used to calibrate molecular clocks and to estimate the time since particular taxa have diverged (4, 26). M. tuberculosis H37 was isolated in 1905, and in 1934 two variants, H37Rv and H37Ra, were isolated from it. PCR fragments of the same sizes are found across the DR region with the primer pairs U16B7–IS1 and IS2–L16B7 in both strains (data not shown), and DNA sequences from DVR24 to DVR38 (18) are found in both strains, suggesting a minimum time for change in the DR locus of 60 years in in vitro culture. On a molecular-clock model, the two extremes of the evolutionary model proposed above would give a minimum time for the evolution of these isolates of 120 or 300 years.
ACKNOWLEDGMENTS
We thank Allan Rayner and Gillian Harris at the Scottish Mycobacteria Reference Laboratory for bacteriological assistance; J. W. Dale, University of Surrey, for providing M. bovis BCG (Pasteur); and D. Gascoyne-Binzi, Department of Microbiology, University of Leeds, for providing the IS6110 fingerprinting strain Mt14323. DNA sequence databases and analysis benefited from SEQNET, the SERC facility (Daresbury, England).
This work was supported by an A.C.T.R. grant from Aberdeen University, an Endowment Research Grant from Aberdeen Royal Hospital NHS Trust, and a grant from Chest, Heart and Stroke Scotland.
REFERENCES
- 1.Achtman M. A surfeit of YATMs? J Clin Microbiol. 1996;34:1870. doi: 10.1128/jcm.34.7.1870-1870.1996. . (Letter.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Altschul S F. Amino acid substitution matrices from an information theoretic perspective. J Mol Biol. 1991;219:555–565. doi: 10.1016/0022-2836(91)90193-A. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Altschul S F, Gish W, Miller W, Myers E W, Lipman D J. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 4.Ayala F J, Barrio E, Kwiatowski J. Molecular clock or erratic evolution? A tale of two genes. Proc Natl Acad Sci USA. 1996;93:11729–11734. doi: 10.1073/pnas.93.21.11729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bloom B R. Tuberculosis. Back to a frightening future. Nature. 1992;358:538–539. doi: 10.1038/358538b0. [DOI] [PubMed] [Google Scholar]
- 6.Boehringer Mannheim GmBH. The DIG system user’s guide for filter hybridisation. Mannheim, Germany: Boehringer Mannheim GmBH; 1993. [Google Scholar]
- 7.Cave M D, Eisenach K D, McDermott P F, Bates J H, Crawford J T. IS6110: conservation of sequence in the Mycobacterium tuberculosis complex and its utilization in DNA fingerprinting. Mol Cell Probes. 1991;5:73–80. doi: 10.1016/0890-8508(91)90040-q. [DOI] [PubMed] [Google Scholar]
- 8.Cave M D, Eisenach K D, Templeton G, Salfinger M, Mazurek G, Bates J H, Crawford J T. Stability of DNA fingerprint pattern produced with IS6110 in strains of Mycobacterium tuberculosis. J Clin Microbiol. 1994;32:262–266. doi: 10.1128/jcm.32.1.262-266.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Collins D M, Erasmuson S K, Stephens D M, Yates G F, De Lisle G W. DNA fingerprinting of Mycobacterium bovis strains by restriction fragment analysis and hybridization with insertion elements IS1081 and IS6110. J Clin Microbiol. 1993;31:1143–1147. doi: 10.1128/jcm.31.5.1143-1147.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Collins D M, Stephens D M. Identification of an insertion sequence, IS1081, in Mycobacterium bovis. FEMS Microbiol Lett. 1991;67:11–15. doi: 10.1016/0378-1097(91)90435-d. [DOI] [PubMed] [Google Scholar]
- 11.Fang Z, Forbes K J. A Mycobacterium tuberculosis IS6110 preferential locus (ipl) for insertion into the genome. J Clin Microbiol. 1997;35:479–481. doi: 10.1128/jcm.35.2.479-481.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fang, Z., C. Doig, N. Morrison, B. Watt, and K. J. Forbes. Characterization of IS1547, a new member of the IS900 family in the Mycobacterium tuberculosis complex and its association with IS6110. Submitted for publication. [DOI] [PMC free article] [PubMed]
- 13.Fomukong N G, Dale J W, Osborn T W, Grange J M. Use of gene probes based on the insertion sequence IS986 to differentiate between BCG vaccine strains. J Appl Bacteriol. 1992;72:126–133. doi: 10.1111/j.1365-2672.1992.tb01813.x. [DOI] [PubMed] [Google Scholar]
- 14.Frothingham R, Hills H G, Wilson K H. Extensive DNA sequence conservation throughout the Mycobacterium tuberculosis complex. J Clin Microbiol. 1994;32:1639–1643. doi: 10.1128/jcm.32.7.1639-1643.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Galas D J, Chandler M. Bacterial insertion sequences. In: Berg D E, Howe M M, editors. Mobile DNA. Washington, D.C: American Society for Microbiology; 1989. pp. 109–162. [Google Scholar]
- 16.Genetics Computer Group. Program manual for the Wisconsin package, version 8, September 1994. Madison, Wis: Genetics Computer Group; 1994. [Google Scholar]
- 17.Green L, Miller R D, Dykhuizen D E, Hartl D L. Distribution of DNA insertion element IS5 in natural isolates of Escherichia coli. Proc Natl Acad Sci USA. 1984;81:4500–4504. doi: 10.1073/pnas.81.14.4500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Groenen P M, Bunschoten A E, van Soolingen D, van Embden J D. Nature of DNA polymorphism in the direct repeat cluster of Mycobacterium tuberculosis; application for strain differentiation by a novel typing method. Mol Microbiol. 1993;10:1057–1065. doi: 10.1111/j.1365-2958.1993.tb00976.x. [DOI] [PubMed] [Google Scholar]
- 19.Hermans P W, van Soolingen D, Bik E M, de Haas P E, Dale J W, van Embden J D. Insertion element IS987 from Mycobacterium bovis BCG is located in a hot-spot integration region for insertion elements in Mycobacterium tuberculosis complex strains. Infect Immun. 1991;59:2695–2705. doi: 10.1128/iai.59.8.2695-2705.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hermans P W, van Soolingen D, Dale J W, Schuitema A R, McAdam R A, Catty D, van Embden J D. Insertion element IS986 from Mycobacterium tuberculosis: a useful tool for diagnosis and epidemiology of tuberculosis. J Clin Microbiol. 1990;28:2051–2058. doi: 10.1128/jcm.28.9.2051-2058.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hermans P W, van Soolingen D, van Embden J D. Characterization of a major polymorphic tandem repeat in Mycobacterium tuberculosis and its potential use in the epidemiology of Mycobacterium kansasii and Mycobacterium gordonae. J Bacteriol. 1992;174:4157–4165. doi: 10.1128/jb.174.12.4157-4165.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hernandez Perez M, Fomukong N G, Hellyer T, Brown I N, Dale J W. Characterization of IS1110, a highly mobile genetic element from Mycobacterium avium. Mol Microbiol. 1994;12:717–724. doi: 10.1111/j.1365-2958.1994.tb01059.x. [DOI] [PubMed] [Google Scholar]
- 23.Karlin S, Altschul S F. Methods for assessing the statistical significance of molecular sequence features by using general scoring schemes. Proc Natl Acad Sci USA. 1990;87:2264–2268. doi: 10.1073/pnas.87.6.2264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Klaer R, Pfeifer D, Starlinger P. IS4 is still found at its chromosomal site after transposition to galT. Mol Gen Genet. 1980;178:281–284. doi: 10.1007/BF00270473. [DOI] [PubMed] [Google Scholar]
- 25.Lawrence J G, Dykhuizen D E, DuBose R F, Hartl D L. Phylogenetic analysis using insertion sequence fingerprinting in Escherichia coli. Mol Biol Evol. 1989;6:1–14. doi: 10.1093/oxfordjournals.molbev.a040531. [DOI] [PubMed] [Google Scholar]
- 26.Leitner T, Escanilla D, Franzen C, Uhlen M, Albert J. Accurate reconstruction of a known HIV-1 transmission history by phylogenetic tree analysis. Proc Natl Acad Sci USA. 1996;93:10864–10869. doi: 10.1073/pnas.93.20.10864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.McAdam R A, Hermans P W, van Soolingen D, Zainuddin Z F, Catty D, van Embden J D, Dale J W. Characterization of a Mycobacterium tuberculosis insertion sequence belonging to the IS3 family. Mol Microbiol. 1990;4:1607–1613. doi: 10.1111/j.1365-2958.1990.tb02073.x. [DOI] [PubMed] [Google Scholar]
- 28.Mendiola M V, Martin C, Otal I, Gicquel B. Analysis of the regions responsible for IS6110 RFLP in a single Mycobacterium tuberculosis strain. Res Microbiol. 1992;143:767–772. doi: 10.1016/0923-2508(92)90104-v. [DOI] [PubMed] [Google Scholar]
- 29.Mojica F J, Ferrer C, Juez G, Rodriguez-Valera F. Long stretches of short tandem repeats are present in the largest replicons of the Archaea Haloferax mediterranei and Haloferax volcanii and could be involved in replicon partitioning. Mol Microbiol. 1996;17:85–93. doi: 10.1111/j.1365-2958.1995.mmi_17010085.x. [DOI] [PubMed] [Google Scholar]
- 30.Needleman S B, Wunsch C D. A general method applicable to the search for similarities in the amino acid sequence of two proteins. J Mol Biol. 1970;48:443–453. doi: 10.1016/0022-2836(70)90057-4. [DOI] [PubMed] [Google Scholar]
- 31.Olson E S, Forbes K J, Watt B, Pennington T H. Population genetics of Mycobacterium tuberculosis complex in Scotland analysed by pulsed-field gel electrophoresis. Epidemiol Infect. 1995;114:153–160. doi: 10.1017/s0950268800052006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31a.Patki, A. H., and J. W. Dale. EMBL accession no. X59271.
- 32.Pearson W R, Lipman D J. Improved tools for biological sequence comparison. Proc Natl Acad Sci USA. 1988;85:2444–2448. doi: 10.1073/pnas.85.8.2444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Philipp W J, Nair S, Guglielmi G, Lagranderie M, Gicquel B, Cole S T. Physical mapping of Mycobacterium bovis BCG Pasteur reveals differences from the genome map of Mycobacterium tuberculosis H37Rv and from M. bovis. Microbiology. 1996;142:3135–3145. doi: 10.1099/13500872-142-11-3135. [DOI] [PubMed] [Google Scholar]
- 34.Reif H J, Saedler H. IS1 is involved in deletion formation in the gal region of E. coli K12. Mol Gen Genet. 1975;137:17–28. doi: 10.1007/BF00332538. [DOI] [PubMed] [Google Scholar]
- 35.Ross B C, Raios K, Jackson K, Dwyer B. Molecular cloning of a highly repeated DNA element from Mycobacterium tuberculosis and its use as an epidemiological tool. J Clin Microbiol. 1992;30:942–946. doi: 10.1128/jcm.30.4.942-946.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sawyer S A, Hartl D L. Population genetics of polymorphism and divergence. Genetics. 1992;132:1161–1176. doi: 10.1093/genetics/132.4.1161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Smith D, Waterman R. Adv. Appl Math. 1980;2:482–489. [Google Scholar]
- 38.Stanley J, Jones C S, Threlfall E J. Evolutionary lines among Salmonella enteritidis phage types are identified by insertion sequence IS200 distribution. FEMS Microbiol Lett. 1991;66:83–89. doi: 10.1016/0378-1097(91)90425-a. [DOI] [PubMed] [Google Scholar]
- 39.Tamames J, Casari G, Ouzounis C, Valencia A. Conserved clusters of functionally related genes in two bacterial genomes. J Mol Evol. 1997;44:66–73. doi: 10.1007/pl00006122. [DOI] [PubMed] [Google Scholar]
- 40.Thierry D, Cave M D, Eisenach K D, Crawford J T, Bates J H, Gicquel B, Guesdon J L. IS6110, an IS-like element of Mycobacterium tuberculosis complex. Nucleic Acids Res. 1990;18:188. doi: 10.1093/nar/18.1.188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Thummler F, Algarra P, Fobo G M. Sequence similarities of phytochrome to protein kinases: implication for the structure, function and evolution of the phytochrome gene family. FEBS Lett. 1995;357:149–155. doi: 10.1016/0014-5793(94)01327-w. [DOI] [PubMed] [Google Scholar]
- 42.van Embden J D A, Cave M D, Crawford J T, Dale J W, Eisenach K D, Gicquel B, Hermans P, Martin C, McAdam R, Shinnick T M, Small P M. Strain identification of Mycobacterium tuberculosis by DNA fingerprinting: recommendations for a standardized methodology. J Clin Microbiol. 1993;31:406–409. doi: 10.1128/jcm.31.2.406-409.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.van Soolingen D, de Haas P E, Hermans P W, Groenen P M, van Embden J D. Comparison of various repetitive DNA elements as genetic markers for strain differentiation and epidemiology of Mycobacterium tuberculosis. J Clin Microbiol. 1993;31:1987–1995. doi: 10.1128/jcm.31.8.1987-1995.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.van Soolingen D, Hermans P W, de Haas P E, Soll D R, van Embden J D. Occurrence and stability of insertion sequences in Mycobacterium tuberculosis complex strains: evaluation of an insertion sequence-dependent DNA polymorphism as a tool in the epidemiology of tuberculosis. J Clin Microbiol. 1991;29:2578–2586. doi: 10.1128/jcm.29.11.2578-2586.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zainuddin Z F, Dale J W. Polymorphic repetitive DNA sequences in Mycobacterium tuberculosis detected with a gene probe from a Mycobacterium fortuitum plasmid. J Gen Microbiol. 1989;135:2347–2355. doi: 10.1099/00221287-135-9-2347. [DOI] [PubMed] [Google Scholar]