Abstract
The gene fgd, which codes for F420-dependent glucose-6-phosphate dehydrogenase (FGD), was cloned from Mycobacterium smegmatis, and its sequence was determined and analyzed. A homolog of FGD which has a very high similarity to the M. smegmatis FGD-derived amino acid sequence was identified in Mycobacterium tuberculosis. FGD showed significant homology with F420-dependent N5,N10-methylene-tetrahydromethanopterin reductase (MER) from methanogenic archaea and with several hypothetical proteins from M. tuberculosis and Archaeoglobus fulgidus, but FGD showed no significant homology with NADP-dependent glucose-6-phosphate dehydrogenases. Multiple alignment of FGD and MER proteins revealed four conserved consensus sequences. Multiple alignment of FGD with the hypothetical proteins also revealed portions of the same conserved sequences. Moderately high levels of FGD were expressed in Escherichia coli BL21(DE3) carrying fgd in pBluescript.
Each year, there are ∼10 million new cases of tuberculosis in the world (mostly in developing countries), one-third of which cause death (49). In the developed world, ∼50% of AIDS patients ultimately suffer from debilitating Mycobacterium avium infections (4, 26). However, there are major problems in the treatment of these diseases due to inherent or acquired drug resistance and to serious side effects of the drugs used (4, 5, 8, 22, 26). We have begun to examine coenzyme F420-dependent reactions in mycobacteria to determine if such reactions are important for this group of pathogens. If so, specific enzymatic steps can be identified as targets for chemotherapy. It is likely that drugs aimed at F420-related targets would act by mechanisms completely different from those of the more widely used drugs.
Coenzyme F420 is a two-electron transfer coenzyme. It was first discovered in methanogenic archaea (14, 15), where it is involved in several reactions in methane biosynthesis. At least five methanogenic enzymes are F420 dependent: F420-dependent methylene-tetrahydromethanopterin dehydrogenase (24, 39, 40), methylene-tetrahydromethanopterin reductase (MER) (34, 60), formate dehydrogenase (29, 53), F420-reducing hydrogenase (19, 28), and alcohol dehydrogenase (6, 61). In nonmethanogenic archaea, F420 is found in Halobacterium (13, 33), Thermoplasma (33), Sulfolobus (33), and Archaeoglobus (38) species. In the bacterial domain, F420 has been found in Streptomyces species (10, 12, 37), Anacystis nidulans (18), Nocardia aurantia (12), and several Mycobacterium species (12, 41, 47). Scenedesmus acutus, a green alga and a member of the domain Eucarya, also contains F420 (17). Some steps in tetracycline (37) and lincomycin (10) biosynthesis by Streptomyces species require F420. Coenzyme F420 is a component of the DNA repair photolyase in several microorganisms (16, 17, 30, 36).
Although the presence in Mycobacterium species of an unknown compound with spectral properties very similar to those of F420 was reported in 1960 (11), and F420 was clearly identified in these organisms in the 1980s (12, 41), any role of this coenzyme in these organisms remained unknown until recently. A study to determine the function of F420 in Mycobacterium smegmatis led us to the discovery of a novel glucose-6-phosphate dehydrogenase which specifically uses F420 as its electron acceptor (47). This enzyme was named FGD, for F420-dependent glucose-6-phosphate dehydrogenase. Not all F420-containing organisms possess FGD. So far, FGD has been found only in Mycobacterium and Nocardia species and in Gordona amarae (48).
We also observed that in addition to FGD, Mycobacterium species possess an NADP-dependent glucose-6-phosphate dehydrogenase (47, 48), which is consistent with previous reports (2, 3). NAD- or NADP-dependent glucose-6-phosphate dehydrogenases are commonly found in many organisms and are named ZWFs (for zwischenferment) (20). Many zwf genes from bacteria, yeasts, animals, and humans have been cloned and sequenced (21, 32, 42, 44, 51). The deduced amino acid sequences of these enzymes have been compared, and conserved regions have been identified. There is high homology between ZWF amino acid sequences from diverse sources. Conserved regions have been hypothesized to be sites for coenzyme (NAD or NADP) and substrate (glucose-6-phosphate) binding and for catalysis. We were interested to know the structural relationship between ZWFs and FGD since these two types of enzymes catalyze similar reactions, differing principally in the electron acceptor used. By using the NH2-terminal amino acid sequence of the purified FGD, a mixture of oligonucleotides was designed and used as a probe to isolate the gene for FGD. Here we describe the molecular characterization of fgd from M. smegmatis mc2155 and its functional expression in Escherichia coli.
(A preliminary version of some of this work has been presented in a poster format [46].)
MATERIALS AND METHODS
Chemicals and enzymes.
Restriction enzymes, T4 DNA ligase, and calf intestine alkaline phosphatase were purchased from New England Biolabs (Beverly, Mass.). The oligonucleotide labeling kit Genius no. 3, alkaline phosphatase-conjugated antidigoxigenin antibody, nitroblue tetrazolium (NBT), 5-bromo-4-chloro-3-indolylphosphate (BCIP), and DNase-free RNase were from Boehringer Mannheim Biochemicals (Indianapolis, Ind.). DNA molecular weight markers (1-kb ladder) and TRIzol were from Gibco-BRL Life Technologies (Grand Island, N.Y.). Qiagen tips (for plasmid preparation) and Qiaquick columns and accessories (for purification of DNA fragments from agarose gels) were from Qiagen Inc. (Chatsworth, Calif.). Coenzyme F420 was purified from Methanobacterium thermoautotrophicum Marburg, using 70% ethanol extraction at 4°C and DEAE and C18 column chromatography (45).
Bacterial strains, plasmids, media, and growth conditions.
M. smegmatis mc2155 (a gift from W. Jacobs, Jr., Albert Einstein College of Medicine, New York, N.Y.) was grown in Middlebrook 7H9 medium supplemented with 0.2% glycerol and 0.05% Tween 80. For cloning and sequencing, E. coli XL1-Blue and pBluescript II SK+ (Stratagene, La Jolla, Calif.) were used as recombinant host and vector, respectively. E. coli BL21(DE3) was used as a host for protein expression. E. coli strains were grown at 37°C on solid medium or in liquid Luria-Bertani (LB) medium. Ampicillin (100 μg/ml) was incorporated into LB medium to select for recombinants. For protein expression studies, E. coli strains were grown as described before (57), with 0.4 mM isopropyl-β-d-thiogalactopyranoside (IPTG) as an inducer.
Molecular biology techniques.
M. smegmatis chromosomal DNA was purified as described by Husson et al. (27). Recombinant plasmids from E. coli XL1-Blue were purified by using Qiagen tips as instructed by the manufacturer. Restriction enzymes and DNA-modifying enzymes were used according to the manufacturer’s protocols. Standard molecular biology protocols for cloning, subcloning, and protein expression in E. coli (52) were used throughout.
Southern and colony hybridization was performed as specified by the manufacturer (Boehringer), using a completely degenerate oligonucleotide, 5′GCRAAYTGYTCNGCNGANGCYTTRTANCCNAGYTT3′ (R = A + G, Y = C + T, N = A + G + C + T), complementary to the NH2-terminal amino acid sequence from residues 3 to 15 of FGD (LKLGYKASAEQFA). This 35-mer was labeled at its 3′ end by digoxigenin-ddUTP, using a Genius labeling kit. Prehybridization and hybridization were at 50°C, and posthybridization washes were at room temperature. Hybridizing bands or colonies were detected by alkaline phosphatase-conjugated antidigoxigenin antibody and colorimetric substrates NBT and BCIP.
A combination of subcloning and primer walking methods was used for DNA sequencing. Universal primer (SK+) was used for the first round of sequencing. DNA sequencing was done at the University of Iowa DNA Facility by using an Applied Biosystems model 373A DNA sequencer.
Insertional mutagenesis of cloned fgd was conducted by using a kanamycin resistance (Kanr) cassette (Pharmacia Biotech, Piscataway, N.J.).
Computer analysis of sequences.
Sequence comparisons of the derived amino acid sequences to entries in the protein database were performed by using the BLASTP program of the National Center for Biotechnology Information (NCBI) (1). When appropriate, sequences were compared with each other by using the BestFit and PileUp programs of the Genetics Computer Group package and the ClustalW and Pima programs at the Human Genome Center at the Baylor College of Medicine (54, 58).
Detection of FGD activity in recombinant E. coli.
Cell extracts of E. coli strains were prepared by cell breakage in a French pressure cell (5,000 lb/in2). The resulting cell lysates were centrifuged at 10,000 × g for 15 min. The proteins from these extracts were precipitated with ammonium sulfate at 90% saturation (0°C) and resolubilized in 20 mM Tris HCl (pH 7.0). These solutions were assayed to determine FGD activities and protein content as described before (47).
Nucleotide sequence accession number.
The nucleotide sequence data reported here appear in the GenBank nucleotide sequence database under accession no. AF041061.
RESULTS AND DISCUSSION
Cloning, subcloning, and sequencing of M. smegmatis fgd.
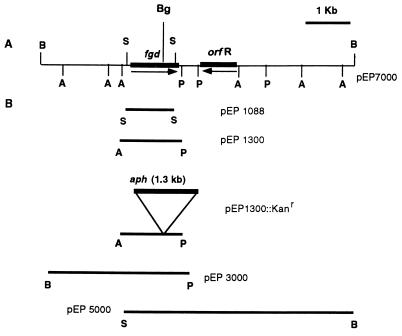
Southern analysis of M. smegmatis DNA that was completely digested with BamHI, using the degenerate oligonucleotide complementary to the NH2-terminal sequence of FGD, showed a positive hybridization signal of ca. 7 kb. Accordingly, M. smegmatis DNA was digested completely with BamHI and electrophoresed in a 0.8% agarose gel; DNA fragments of ca. 6 to 8 kb were recovered and purified by using Qiaquick columns. These DNA fragments were ligated into BamHI-digested, dephosphorylated pBluescript II SK+. The ligation mixture was used to transform competent cells of E. coli XL1-Blue, and recombinants were screened for an fgd clone by colony hybridization. Two colonies out of ca. 400 screened showed positive hybridization signals, both of which carried plasmids with inserts of the same size and restriction patterns. One of these two recombinant strains was preserved and used for further work, and the corresponding plasmid was designated pEP7000. The restriction map of the 7-kb fragment obtained by using various restriction enzymes is shown in Fig. 1A. Shown in Fig. 1B are the maps of various subclones described in more detail below.
FIG. 1.
Restriction maps of pEP7000 and various subclones. Symbols: B, BamHI; Bg, BglII; P, PstI; A, AccI; S, SmaI; aph, aminoglycoside phosphotransferase, which imparts Kanr; fgd, F420-dependent glucose-6-phosphate dehydrogenase gene; orfR, possible regulatory gene. Restriction sites at the ends of the subclone fragments indicate the boundary of M. smegmatis DNA in the plasmid inserts, not necessarily the restriction sites used for the final subcloning step (see text for descriptions of construction).
The 1.1-kb SmaI fragment was subcloned into the SmaI site of pBluescript, giving plasmid pEP1088. This plasmid was used for the first round of sequencing. Further sequencing was performed by primer walking using pEP7000. The gene for FGD was located ca. 2 kb away from one end of the 7-kb insert of the primary clone and in the same direction as and downstream of the T7 promoter of the vector.
In addition to pEP1088, three more subclones were developed by using pBluescript II SK+ (Fig. 1B): pEP1300, carrying the 1.3-kb AccI/PstI fragment (fgd with 244 bp upstream and 36 bp downstream); pEP3000, carrying the 3-kb PstI fragment of pEP7000 (fgd with ca. 2,000 bp upstream and 36 bp downstream; the downstream PstI site is shown in Fig. 1B, and the upstream PstI site is in the pBluescript multicloning site); and pEP5000, containing the equivalent of the 5-kb SmaI/BamHI fragment of pEP7000, which was constructed by the ligation of the 0.8-kb product of an EcoRI/BglII double digest of pEP1088 and the 7.3-kb fragment of an EcoRI/BglII double digest of pEP7000 (fgd with 250 bp upstream and approximately 3.9 kb downstream; the EcoRI site is upstream of fgd in the multicloning site, and the BglII site is in fgd). These plasmids were used for enzyme expression studies.
Southern hybridization of M. smegmatis DNA, which had been cut with various restriction enzymes, with oligonucleotide probe that precisely complemented the 5′ end of fgd showed one hybridizing band from each restriction digest, which indicates that only one copy of fgd was present in the M. smegmatis genome. The sequence of fgd from M. smegmatis and its flanking regions are shown in Fig. 2. The gene fgd consisted of 1,008 bases, corresponding to 336 amino acid residues. The NH2-terminal amino acid sequence of purified FGD (47) precisely matched the deduced amino acid sequence of the NH2 terminus, with alanine as the first amino acid. This finding indicates that the initiation codon was GTG, which codes for valine. Thus, the ATG which immediately precedes the GTG is not translated. TGA was the stop codon. In E. coli, GTG rarely functions as an initiation codon, and when it does it is less efficient than ATG (23). However, Mycobacterium species use GTG as the initiation codon for many genes (9, 25, 35, 59). The G+C content of fgd was 65%, which is in the range of the G+C content of Mycobacterium chromosomes (62 to 70%). The FGD molecular mass predicted from the deduced amino acid sequence was 37,148 Da, slightly lower than the subunit molecular mass of ca. 40,000 Da determined for the purified M. smegmatis protein by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) (47). The theoretical pI value for FGD was 5.20.
FIG. 2.
Nucleotide sequence of fgd and some of its flanking regions, and deduced amino acid sequences of FGD and the possible regulatory protein ORFR. Underlined, NH2-terminal amino acid sequence determined from purified FGD; →, direction of fgd transcription; ←, direction of transcription of genes adjacent to fgd.
Comparative sequence analysis.
The complete amino acid sequence of FGD showed a very high homology (BLAST score, 1,592; probability, 4.4e−211) with a hypothetical protein (g1817673) from Mycobacterium tuberculosis (1). BestFit analysis of these two proteins showed 89% identity and 93% similarity, providing strong evidence that this is FGD in M. tuberculosis, given that FGD activity is present in M. tuberculosis (48). A BestFit analysis of the two fgd nucleotide sequences showed 80% identity.
While writing this report, we became aware of related work. A gene that was involved in the activation of an experimental antituberculosis drug called NAP (nitroimidazole pyran; also called PA-824) in Mycobacterium bovis BCG had been discovered and sequenced (55, 56). If this gene is deleted or mutated, M. bovis BCG becomes NAP resistant. The derived amino acid sequence of the product of the gene that complemented NAP resistance of M. bovis BCG was identical with that of FGD from M. tuberculosis (56). Thus, FGD is involved in some way in the activation of NAP.
No good sequence homology was observed when the complete amino acid sequences of ZWFs from E. coli, Leuconostoc mesenteroides, and human cells were individually compared with that of FGD by using the BestFit program. The percent identities of FGD with these ZWFs were 17 to 19%, much lower than the average percent identities seen between ZWFs themselves. For example, Rowland et al. showed that a multiple alignment of 13 known ZWF sequences from various organisms has a mean conservation of 39% (50). A multiple alignment of FGD from M. smegmatis with ZWF gene products from E. coli, L. mesenteroides, Saccharomyces cerevisiae, and human cells showed very little sequence conservation, and in particular key conserved regions found in all ZWFs were not present in the few areas of FGD that do align with ZWF (data not shown).
When the FGD sequence was compared by BLAST analysis with protein sequences in the NCBI databank, regions of this protein showed homology with the F420-dependent MER from several methanogens (7, 43, 60) and Archaeoglobus fulgidus (31). FGD also had regions that showed homology to at least eight hypothetical proteins from M. tuberculosis and one from A. fulgidus and to the product of the lmbY gene of unknown function from Streptomyces lincolnensis. Aside from the M. tuberculosis and M. bovis FGD homologs, an unknown hypothetical protein from M. tuberculosis (1877257) showed the highest similarity, with an identity of 37%. The percentage identities of FGD with MERs were 23 to 28%. The other group of M. tuberculosis hypothetical proteins had identities of 24 to 29%. These values were all higher than the percent identities seen between FGD and several ZWFs (17 to 19%). Similarities with other F420-utilizing enzymes were not seen in the BLAST analysis, except for the short NH2-terminal fragment of the F420-dependent alcohol dehydrogenase from Methanogenium liminatans (6); unfortunately, the full sequence of this protein is not yet known.
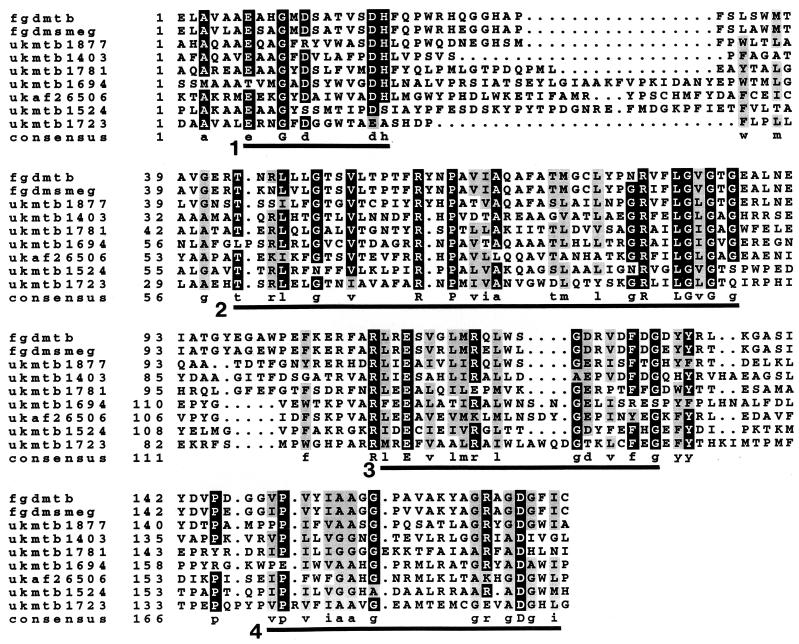
Both ClustalW and Pima multiple alignments of FGD were conducted with MER proteins, most of the homologous unknown proteins identified by the BLAST search, and four F420-using enzymes (methylene-tetrahydromethanopterin dehydrogenase, hydrogenase β subunit, formate dehydrogenase β subunit, and photolyase). FGD and MER showed significant homology in alignments using both programs, characterized by four conserved segments (identified by ClustalW) that are shown in Fig. 3. These segments were in the NH2-terminal two-thirds of the molecule. FGD-MER consensus sequence 2 identified MER and several of the mentioned unknown proteins in a BLAST search of the NCBI database. Sequence 4 identified MER and one unknown protein, while sequence 3 identified only MER. Sequence 1 did not identify homologous sequences, probably due to its short length. Consensus sequences 1 to 4 did not identify in a BLAST analysis any known enzymes that do not use F420 as a coenzyme. Areas with conserved amino acids in the carboxy one-third were less similar than the four identified in the NH2-terminal region and are not underlined in Fig. 3; they may be of significance, but their consensus sequences did not lead to the identification of any proteins in BLAST searches. Aside from MER and the alcohol dehydrogenase, no F420-utilizing enzymes that had similarities to FGD were detectable by ClustalW or Pima multiple alignments.
FIG. 3.
Multiple alignments of FGD with MER. Identical amino acids are highlighted in black, and similar amino acids are highlighted in grey. The solid bar below the consensus sequence indicates areas of significant similarity, corresponding to FGD-MER consensus sequences 1 through 4 as labeled adjacent to each bar. For presentation purposes, the eight NH2-terminal amino acids of Methanococcus jannaschii MER were not included in the alignment. ClustalW (58) was used to align the sequences, and Boxshade (0.9 setting) was used to determine the degree of residue shading. Sequences, from top to bottom: Methanococcus jannaschii MER (GenBank accession no. E64491), Methanobacterium thermoautotrophicum MER (S66529), A. fulgidus MER (2649522), Methanopyrus kandleri MER (1002714), M. tuberculosis FGD (e301455), and M. smegmatis FGD (AF041061).
When FGD was compared by multiple alignment to seven unknown proteins (six from M. tuberculosis and one from A. fulgidus) that had been identified in a BLAST search, consensus sequences similar to those of the FGD-MER alignments were found. Figure 4 presents an abbreviated alignment with the most relevant and most homologous sections shown. FGD-unknown consensus sequence 2 was the strongest in similarity to an FGD-MER consensus sequence and led to the identification of FGD, MER, and unknown proteins in a BLAST search. The other consensus sequences in this comparison were not as strong in their similarity to the FGD-MER comparison. We hypothesize that a conserved site important in F420-dependent catalysis is defined by one or more of the four FGD-MER consensus sequences in Fig. 3 and that other F420-dependent enzymes not similar to FGD (hydrogenase, formate dehydrogenase, methylene-tetrahydromethanopterin dehydrogenase, and photolyase) have a different type of F420-reactive site. We also hypothesize that the unknown hypothetical proteins in M. tuberculosis interact with F420; the number of these proteins (six) suggests that study of F420 metabolism in mycobacteria may be very fruitful in the discovery of novel enzymes. Proof of our hypotheses concerning conserved sites for interaction with F420 will require considerable further biochemical examination of MER, FGD, and the unknown proteins.
FIG. 4.
Multiple alignments of FGD and unknown hypothetical proteins initially identified by BLAST search with FGD. Identical amino acids are highlighted in black, and similar amino acids are highlighted in grey. The solid bar below the consensus sequence indicates areas of significant similarity which correspond to FGD-MER consensus sequences 1 through 4 shown in Fig. 3. For presentation purposes, short sections of NH2- or carboxy-terminal amino acid sequences of proteins are not included in the alignments. ClustalW (58) was used to align the sequences, and Boxshade (0.8 setting) was used to determine the degree of residue shading. Sequences, from top to bottom: M. tuberculosis FGD (e301455), M. smegmatis FGD (AF041061), M. tuberculosis unknown proteins (1877257, 1403414, 1781192, 1694883, 1524222, and 1723065), and A. fulgidus unknown protein (2650688).
Identity of sequences adjacent to fgd.
Analysis of the sequence downstream of fgd showed an open reading frame (called orfR) encoding a hypothetical protein (ORFR) of 254 amino acids which should be transcribed in the direction opposite that of fgd (Fig. 1). The DNA and corresponding protein sequences of ORFR are shown in Fig. 2. ORFR had good homology with several hypothetical regulatory proteins of the GntR family with helix-turn-helix motifs. The highest BLAST score (157, probability of 5.6e−18) was seen with a hypothetical regulatory protein in E. coli (accession no. P31453); there were 18 other hypothetical regulatory proteins with BLAST scores >100. Sequence comparison of ORFR to this protein with the BestFit program revealed an identity of 25% and similarity of 39%. The consensus sequence of the GntR family matched perfectly with the sequence of ORFR at positions 35 to 56 (EREMAETFAVSRSTLRSALLPL). The fact that BLAST analysis did not reveal a highly similar protein in M. tuberculosis (for which the complete genome is available) suggests this protein is not ubiquitous within the mycobacteria.
The sequence upstream of fgd showed an ORF encoding a protein of 248 amino acids which should be transcribed in the direction opposite of fgd. This protein showed high homology (BLAST score of 434, probability of 1.8e−101) to an unknown protein from M. tuberculosis (g1817672). The gene for this hypothetical protein, g1817672, was also located directly upstream of fgd (g1817673) in M. tuberculosis. The second-highest sequence similarity (BLAST score of 134, probability of 3.3e−10) was with a β-lactamase L1 precursor from Xanthomonas maltophilia. Most other proteins showing weak sequence similarities were hypothetical. Thus, it is not clear what this gene might code for in mycobacteria.
Expression of FGD in recombinant E. coli.
Initially, E. coli XL1-Blue strains containing recombinant plasmids pEP7000, pEP1300, and pEP1088 were assayed for FGD activity. The strains carrying pEP7000 and pEP1300 expressed very low FGD activities (0.001 μmol/min/mg of protein), and no detectable activity was observed with pEP1088. This low level of expression of pEP7000 and pEP1300 was thought to be a result of inefficient transcription of fgd in E. coli. To explore this possibility, pEP7000 and pEP1300 were transformed into E. coli BL21(DE3), a lysogen that contains a T7 RNA polymerase gene in the chromosome. As shown in Table 1, fgd was expressed at a ca. 15- to 523-fold-higher level in E. coli BL21(DE3) than in E. coli XL1-Blue, consistent with improved transcription in E. coli BL21(DE3).
TABLE 1.
Specific activities of FGD expressed from recombinant plasmids in E. coli BL21(DE3)
| E. coli BL21(DE3) carrying: | Mean FGD sp
act (μmol of F420 reduced/min/mg of protein) ± SD
|
|||
|---|---|---|---|---|
| 2 ha
|
15
hb
|
|||
| No IPTG | With IPTG | No IPTG | With IPTG | |
| pEP7000 | 0.014 ± 0.002 | 0.018 ± 0.002 | 0.030 ± 0.003 | 0.011 ± 0.001 |
| pEP5000 | 0.011 ± 0.002 | 0.010 ± 0.001 | 0.017 ± 0.002 | 0.005 ± 0.001 |
| pEP3000 | 0.173 ± 0.018 | 0.254 ± 0.012 | 0.410 ± 0.014 | 0.187 ± 0.015 |
| pEP1300 | 0.238 ± 0.014 | 0.277 ± 0.024 | 0.523 ± 0.033 | 0.224 ± 0.022 |
| pEP1088 | 0 | 0 | 0.015 ± 0.001 | 0 |
| pEP1300::Kanr | 0 | 0 | 0 | 0 |
Cells were harvested 2 h after addition of IPTG.
Cells were harvested 15 h after addition of IPTG.
Expression was higher with pEP1300 than with pEP7000. Induction with IPTG increased expression only slightly. The specific activity of FGD in E. coli BL21(DE3) carrying pEP1300 in the absence of IPTG increased as the culture aged and was highest at the stationary phase. We do not understand why IPTG did not result in a higher level expression, but this may have been due to inefficient translation of fgd in E. coli BL21(DE3). In these constructs, translation was expected to initiate from the mycobacterial sequence; however, the sequence upstream of fgd did not show strong homology with typical Shine-Dalgarno consensus sequences. It has been reported before that there is always a basal level of T7 RNA polymerase in E. coli BL21(DE3) without IPTG induction (57). This basal level of T7 RNA polymerase might be enough to express fgd at the levels which were observed in noninduced cells. Thus, the translation rate was possibly limiting for FGD expression in E. coli BL21(DE3). This argument might also explain why the increase in fgd message by IPTG induction did not result in increased FGD levels. It is also possible that FGD expression is tightly regulated in E. coli.
In pEP1088, a construct used for initial sequencing, fgd was truncated at its 3′ end (deletion of 82 bp, corresponding to deletion of 28 amino acids at the FGD carboxy terminus). FGD was expressed from this construct in BL21(DE3), although the level was only 3% of that from pEP1300. This result indicates that this carboxy-terminal portion of FGD is not essential for catalysis.
Despite high FGD activities in BL21(DE3) cells, when extracts of recombinant E. coli BL21(DE3) were examined by SDS-PAGE, no intense band for FGD at ∼40,000 Da was seen, indicating that FGD was not overexpressed. The formation of inclusion bodies by overexpression of FGD was not thought to be responsible for the lack of overexpression, since (i) E. coli BL21(DE3) carrying pEP1300 grown at 25°C (with and without IPTG) had FGD activities that were twofold lower than those in the corresponding 37°C cultures, (ii) microscopic (phase-contrast) examination did not show any sign of inclusion bodies in cells which were grown in the presence of IPTG, and (iii) SDS-PAGE of SDS-lysed whole cells (which would include proteins in inclusion bodies) did not reveal bands indicative of overexpression.
To clarify the differences in the levels of FGD expression from pEP7000 and pEP1300, we created two more plasmids (pEP3000 and pEP5000 [Fig. 1]). As shown in Table 1, the expression of fgd from pEP3000 was approximately the same as with pEP1300. Similarly, pEP5000 and pEP7000 gave comparable results: ca. 15- to 25-fold lower activities than pEP3000 and pEP1300. Since the level of expression from pEP5000 was comparable to that from pEP7000, and that from pEP3000 was comparable to the level of expression from pEP1300, it is possible that the sequences downstream of fgd, or the corresponding gene products, were responsible for the reduced expression of FGD in E. coli. This regulation might be due to a putative repressor protein encoded by orfR that is immediately downstream of fgd; ORFR has high sequence homology with regulatory proteins, as discussed above. We have no data on the ability of this gene to affect transcription in M. smegmatis. It is possible that in E. coli, ORFR binds to a putative mycobacterial promoter region upstream of fgd or to the T7 promoter region further upstream. Further experimental evidence is needed to determine the true reason for the large differences in expression seen with the different constructs.
The cloned FGD was partially purified from E. coli by using ammonium sulfate precipitation (the active fraction was recovered in the supernatant after treatment with ammonium sulfate at 60% of saturation and then precipitated by ammonium sulfate at 90% saturation) followed by desalting with a Centriplus-10 filter and finally F420-affinity column chromatography (47). The protein at this stage of purity had the same denatured molecular mass, and temperature and pH optima for FGD activities, as the native enzyme from M. smegmatis. Thus, the enzyme can be expressed in a fully functional form in E. coli and does not need Mycobacterium-specific cofactors or other mycobacterial proteins for activity.
Insertional mutagenesis of cloned fgd in E. coli was conducted by using the Kanr gene (aph, for aminoglycoside phosphotransferase). The 1.3-kb BamHI fragment of the Kanr cassette was inserted into the BglII site of fgd in pEP1300, resulting in pEP1300::Kanr. No FGD activity was detected in E. coli BL21(DE3) carrying pEP1300::Kanr.
ACKNOWLEDGMENTS
This work was supported by U.S. Department of Agriculture grant 4132008 to the Biotechnology Byproduct Consortium.
We are appreciative of M. smegmatis mc26, provided by William Jacobs, Jr. We thank Biswarup Mukhopadhyay for helpful discussions. We also thank Kendall Stover and Paul Warrener of PathoGenesis Corporation for sharing their unpublished data.
REFERENCES
- 1.Altschul S F, Gish W, Miller W, Myers E W, Lipman D J. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 2.Bai N J, Pai M R, Murthy P S, Venkitasubramanian T A. Pathways of carbohydrate metabolism in Mycobacterium tuberculosisH37Rv. Can J Microbiol. 1975;21:1688–1691. doi: 10.1139/m75-247. [DOI] [PubMed] [Google Scholar]
- 3.Bai N J, Pai M R, Murthy P S, Venkitasubramanian T A. Pathways of glucose catabolism in Mycobacterium smegmatis. Can J Microbiol. 1976;22:1374–1380. doi: 10.1139/m76-201. [DOI] [PubMed] [Google Scholar]
- 4.Benson C A, Ellner J J. Mycobacterium aviumcomplex infections and AIDS: advances in theory and practice. Clin Infect Dis. 1993;17:7–20. doi: 10.1093/clinids/17.1.7. [DOI] [PubMed] [Google Scholar]
- 5.Bifani P J, Plikaytis B B, Kapur V, Stockbauer K, Pan X, Lutfey M L, Moghazeh S L, Eisner W, Daniel T M, Kaplan M H, Crawford J T, Musser J M, Kreiswirth B N. Origin and interstate spread of a New York City multidrug-resistant Mycobacterium tuberculosisclone family. JAMA. 1996;275:452–457. [PubMed] [Google Scholar]
- 6.Bleicher K, Winter J. Purification and properties of F420 and NADP-dependent alcohol dehydrogenases of Methanogenium liminatans and Methanobacterium palustre, specific for secondary alcohols. Eur J Biochem. 1991;200:43–51. doi: 10.1111/j.1432-1033.1991.tb21046.x. [DOI] [PubMed] [Google Scholar]
- 7.Bult C J, White O, Olsen G J, Zhou L, Fleischmann R D, Sutton G G, Blake J A, FitzGerald L M, Clayton R A, Gocayne J D, Kerlavage A R, Dougherty B A, Tomb J F, Adams M D, Reich C I, Overbeek R, Kirkness E F, Weinstock K G, Merrick J M, Glodek A, Scott J L, Geoghagen N S M, Weidman J F, Fuhrmann J L, Nguyen D, Utterback T R, Kelley J M, Peterson J D, Sadow P W, Hanna M C, Cotton M D, Roberts K M, Hurst M A, Kaine B P, Borodovsky M, Klenk H P, Fraser C M, Smith H O, Woese C R, Venter J C. Complete genome sequence of the methanogenic archaeon, Methanococcus jannaschii. Science. 1996;273:1058–1073. doi: 10.1126/science.273.5278.1058. [DOI] [PubMed] [Google Scholar]
- 8.Chin D P, Reingold A L, Stone E N, Vittinghoff E, Horsburgh C R, Simon E M, Yajko D M, Hadley W K, Ostroff S M, Hopewell P C. The impact of Mycobacterium aviumcomplex bacteremia and its treatment on survival of AIDS patients—a prospective study. J Infect Dis. 1994;170:578–584. doi: 10.1093/infdis/170.3.578. [DOI] [PubMed] [Google Scholar]
- 9.Cirillo J D, Weisbrod T R, Pascopella L, Bloom B R, Jacobs W R. Isolation and characterization of the aspartokinase and aspartate semialdehyde dehydrogenase operon from mycobacteria. Mol Microbiol. 1994;11:629–639. doi: 10.1111/j.1365-2958.1994.tb00342.x. [DOI] [PubMed] [Google Scholar]
- 10.Coats J H, Li G P, Kuo M-S, Yurek D A. Discovery, production, and biological assay of an unusual flavenoid cofactor involved in lincomycin biosynthesis. J Antibiot. 1989;42:472–474. doi: 10.7164/antibiotics.42.472. [DOI] [PubMed] [Google Scholar]
- 11.Cousins F B. The prosthetic group of a chromoprotein from mycobacteria. Biochim Biophys Acta. 1960;40:532–534. doi: 10.1016/0006-3002(60)91396-2. [DOI] [PubMed] [Google Scholar]
- 12.Daniels L, Bakhiet N, Harmon K. Widespread distribution of a 5-deazaflavin cofactor in Actinomyces and related bacteria. Syst Appl Microbiol. 1985;6:12–17. [Google Scholar]
- 13.deWit L E A, Eker A P M. 8-Hydroxy-5-deazaflavin-dependent electron transfer in the extreme halophile Halobacterium cutirubrum. FEMS Microbiol Lett. 1987;48:121–125. [Google Scholar]
- 14.Eirich L D, Vogels G D, Wolfe R S. Distribution of coenzyme F420and properties of its hydrolytic fragments. J Bacteriol. 1979;140:20–27. doi: 10.1128/jb.140.1.20-27.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Eirich L D, Vogels G D, Wolfe R S. Proposed structure for coenzyme F420 from Methanobacterium. Biochemistry. 1978;17:4583–4593. doi: 10.1021/bi00615a002. [DOI] [PubMed] [Google Scholar]
- 16.Eker A P M, Dekker R H, Berends W. Photo-reactivating enzyme from Streptomyces griseus: nature of the chromophoric cofactor. Photochem Photobiol. 1981;33:65–72. doi: 10.1111/j.1751-1097.1981.tb04298.x. [DOI] [PubMed] [Google Scholar]
- 17.Eker A P M, Hessels J K C, van de Velde J. Photo-reactivating enzyme from the green alga Scenedesmus acutus. Evidence for two different chromophores. Biochemistry. 1988;17:1758–1765. [Google Scholar]
- 18.Eker A P M, Kooiman P, Hessels J K C, Yasui A. DNA photoreactivating enzyme from the cyanobacterium Anacystis nidulans. J Biol Chem. 1990;265:8009–8015. [PubMed] [Google Scholar]
- 19.Fox J, Livingston D J, Orme-Johnson W H, Walsh C T. 8-Hydroxy-5-deazaflavin-reducing hydrogenase from Methanobacterium thermoautotrophicum. 1. Purification and characterization. Biochemistry. 1987;26:4219–4227. doi: 10.1021/bi00388a007. [DOI] [PubMed] [Google Scholar]
- 20.Fraenkel D G. Selection of E. colimutants lacking glucose-6-phosphate dehydrogenase or gluconate-6-phosphate dehydrogenase. J Bacteriol. 1968;95:1267–1271. doi: 10.1128/jb.95.4.1267-1271.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ganguli R, Ganguli N, Manning J. Isolation and characterization of the glucose-6-phosphate dehydrogenase gene of Drosophila melanogaster. Gene. 1985;35:91–101. doi: 10.1016/0378-1119(85)90161-1. [DOI] [PubMed] [Google Scholar]
- 22.Goble M, Iseman M D, Madsen L A, Waite D, Ackerson L, Horsburgh C R. Treatment of 171 patients with pulmonary tuberculosis resistant to isoniazid and rifampin. N Engl J Med. 1993;328:527–532. doi: 10.1056/NEJM199302253280802. [DOI] [PubMed] [Google Scholar]
- 23.Gold L. Posttranscriptional regulatory mechanisms in Escherichia coli. Annu Rev Biochem. 1988;57:199–233. doi: 10.1146/annurev.bi.57.070188.001215. [DOI] [PubMed] [Google Scholar]
- 24.Hartzel P L, Zvilius G, Escalante-Semerena J G, Donnelly M I. Coenzyme F420 dependence of methylene-tetrahydromethanopterin dehydrogenase from Methanobacterium thermoautotrophicum. Biochim Biophys Acta. 1985;133:884–890. doi: 10.1016/0006-291x(85)91218-5. [DOI] [PubMed] [Google Scholar]
- 25.Honore N, Bergh S, Chanteau S, Docet-Populaire F, Eiglmeier K, Garnier T, Georges C, Launois P, Limpaiboon T, Newton S, Niang K, del Portillo P, Ramesh G R, Reddi P, Ridel P R, Sittisombut N, Wu-Hunter S, Cole S T. Nucleotide sequence of the first cosmid from the Mycobacterium lepraegenome project: structure and function of the Rif-Str regions. Mol Microbiol. 1993;7:207–214. doi: 10.1111/j.1365-2958.1993.tb01112.x. [DOI] [PubMed] [Google Scholar]
- 26.Horsburgh C R, Metchock B, Gordon S M, Havlik J A, McGowan J E, Thompson S E. Predictors of survival in patients with AIDS and disseminated Mycobacterium aviumcomplex disease. J Infect Dis. 1994;170:573–577. doi: 10.1093/infdis/170.3.573. [DOI] [PubMed] [Google Scholar]
- 27.Husson R N, James B E, Young R A. Gene replacement and expression of foreign DNA in mycobacteria. J Bacteriol. 1990;172:519–524. doi: 10.1128/jb.172.2.519-524.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jacobson F S, Daniels L, Fox J, Walsh C T, Orme-Johnson W H. Purification and properties of an 8-hydroxy-5-deazaflavin-reducing hydrogenase from Methanobacterium thermoautotrophicum. J Biol Chem. 1981;257:3385–3388. [PubMed] [Google Scholar]
- 29.Jones J, Stadtman T. Reconstitution of a formate-NADP oxidoreductase from formate dehydrogenase and a 5-deazaflavin linked NADP reductase from Methanococcus vannielii. J Biol Chem. 1980;256:656–663. [PubMed] [Google Scholar]
- 30.Kiener A, Husain I, Sancar A, Walsh C. Purification and properties of Methanobacterium thermoautotrophicumDNA photolyase. J Biol Chem. 1989;264:13880–13887. [PubMed] [Google Scholar]
- 31.Klenk H P, Clayton R A, Tomb J, White O, Nelson K E, Ketchum K A, Dodson R J, Gwill M, Hickey E K, Peterson J D, Richardson D L, Kerlavage A R, Graham D E, Kyrpides N C, Fleishmann R D, Quackenbush J, Lee N H, Sutton G G, Gill S, Kirkness E F, Dougherty B A, McKenney K, Adams M D, Loftus B, Peterson S, Reich C I, McNeil L K, Badger J H, Glodek A, Zhou L, Overbeek R, Gocayne J D, Weidman J F, McDonald L, Utterbach T, Cotton M D, Spriggs T, Artiach P, Kaine B P, Sykes S M, Sadow P W, D’Andrea K P, Bowman C, Fujii C, Garland S A, Mason T M, Olson G J, Fraser C M, Smith H O, Woese C R, Venter J C. The complete genome of the hyperthermophilic, sulfate-reducing archaeon Archaeoglobus fulgidus. Nature. 1997;390:364–370. doi: 10.1038/37052. [DOI] [PubMed] [Google Scholar]
- 32.Lee W T, Flynn T G, Lyons C, Levy H R. Cloning of the gene and amino acid sequence for glucose-6-phosphate dehydrogenase from Acetobacter hansenii (Acetobacter xylinum) J Biol Chem. 1991;266:13028–13034. [PubMed] [Google Scholar]
- 33.Lin X, White R. Occurrence of coenzyme F420and its γ-monoglutamyl derivative in nonmethanogenic archaebacteria. J Bacteriol. 1986;168:444–448. doi: 10.1128/jb.168.1.444-448.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ma K, Thauer R K. Purification and properties of N5,N10-methylenetetrahydromethanopterin reductase from Methanobacterium thermoautotrophicum(strain Marburg) Eur J Microbiol. 1990;191:187–193. doi: 10.1111/j.1432-1033.1990.tb19109.x. [DOI] [PubMed] [Google Scholar]
- 35.Madhusdan K, Nagaraja V. Mycobacterium smegmatis DNA gyrase: cloning and expression in Escherichia coli. Microbiology. 1995;141:3029–3037. doi: 10.1099/13500872-141-12-3029. [DOI] [PubMed] [Google Scholar]
- 36.Mayerl F, Piret J, Kiener A, Walsh C T, Yasui A. Functional expression of 5-deazaflavin-dependent DNA photolyase from Anacystis nidulans in Streptomyces coelicolor. J Bacteriol. 1990;172:6061–6065. doi: 10.1128/jb.172.10.6061-6065.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.McCormick J R D, Morton G O. Identity of cosynthetic factor 1 of Streptomyces aureofaciens and fragment FO from coenzyme F420 of Methanobacteriumsp. J Am Chem Soc. 1982;104:4014–4015. [Google Scholar]
- 38.Moller-Zinkan D, Borner G, Thauer R K. Function of methanofuran, tetrahydromethanopterin and coenzyme F420 in Archaeoglobus fulgidus. Arch Microbiol. 1989;152:362–368. [Google Scholar]
- 39.Mukhopadhyay B, Daniels L. Aerobic purification of N5,N10-methylenetetrahydromethanopterin dehydrogenase, separated from N5,N10-methenyltetrahydromethanopterin cyclohydrolase, from Methanobacterium thermoautotrophicumstrain Marburg. Can J Microbiol. 1989;35:499–507. doi: 10.1139/m89-077. [DOI] [PubMed] [Google Scholar]
- 40.Mukhopadhyay B, Purwantini E, Pihl T, Reeve J N, Daniels L. Cloning, sequencing and transcriptional analysis of the coenzyme-F420-dependent methylene H4MPT dehydrogenase gene from Methanobacterium thermoautotrophicum and functional expression in E. coli. J Biol Chem. 1995;270:2827–2832. doi: 10.1074/jbc.270.6.2827. [DOI] [PubMed] [Google Scholar]
- 41.Naraoka T, Mamoi K, Fukasawa K, Goto M. Isolation and identification of a naturally occurring 7,8-didemethyl-8-hydroxy-5-deazariboflavin derivative from Mycobacterium avium. Biochim Biophys Acta. 1984;797:377–380. [Google Scholar]
- 42.Nogae I, Johnston M. Isolation and characterization of the ZWF1 gene of Saccharomyces cerevisiae, encoding glucose-6-phosphate dehydrogenase. Gene. 1990;96:161–169. doi: 10.1016/0378-1119(90)90248-p. [DOI] [PubMed] [Google Scholar]
- 43.Nolling J, Pihl T, Reeve J. Cloning, sequencing, and growth phase-dependent transcription of the coenzyme F420-dependent N5-N10-methylene-tetrahydromethanopterin reductase-encoding genes from Methanobacterium thermoautotrophicum delta H and Methanopyurus kandleri. J Bacteriol. 1995;177:7238–7244. doi: 10.1128/jb.177.24.7238-7244.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Persico M G, Viglietto G, Martini G, Toniolo G, Paonessa G, Moscatelli C, Dono R, Vulliamy T, Luzzato L, D’Urso M. Isolation of human glucose-6-phosphate dehydrogenase (G6PD) cDNA clones: primary structure of the protein and unusual 5′ non-coding region. Nucleic Acids Res. 1986;6:2511–2522. doi: 10.1093/nar/14.6.2511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Purwantini E. Coenzyme F420: factors affecting its purification from Methanobacterium thermoautotrophicum and its conversion to F390, and effect of temperature on the spectral properties of coenzyme F420 and related compounds. Master’s thesis. Iowa City, Iowa: University of Iowa; 1992. [Google Scholar]
- 46.Purwantini E, Bair T, Daniels L. Presented at the 97th Annual Meeting of the American Society for Microbiology. 1997. [Google Scholar]
- 47.Purwantini E, Daniels L. Purification of a novel coenzyme F420-dependent glucose-6-phosphate dehydrogenase from Mycobacterium smegmatis. J Bacteriol. 1996;178:2861–2866. doi: 10.1128/jb.178.10.2861-2866.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Purwantini E, Gillis T, Daniels L. Presence of F420-dependent glucose-6-phosphate dehydrogenase in Mycobacterium and Nocardia species, but absence in Streptomyces and Corynebacteriumspecies and methanogenic Archaea. FEMS Microbiol Lett. 1997;146:129–134. doi: 10.1111/j.1574-6968.1997.tb10182.x. [DOI] [PubMed] [Google Scholar]
- 49.Raviglione M C, Snider J D E, Kochi A. Global epidemiology of tuberculosis. JAMA. 1995;273:220–226. [PubMed] [Google Scholar]
- 50.Rowland P, Basak A K, Grover S, Levy H R, Adams M J. The three-dimensional structure of glucose-6-phosphate dehydrogenase from Leuconostoc mesenteroidesrefined at 2.0 A resolution. Structure. 1994;2:1073–1087. doi: 10.1016/s0969-2126(94)00110-3. [DOI] [PubMed] [Google Scholar]
- 51.Rowley D L, Wolf R E., Jr Molecular characterization of the Escherichia coli K-12 zwfgene encoding glucose-6-phosphate dehydrogenase. J Bacteriol. 1991;173:968–977. doi: 10.1128/jb.173.3.968-977.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Press; 1989. [Google Scholar]
- 53.Schauer N L, Ferry J G. FAD requirement for the reduction of F420 by formate dehydrogenase from Methanobacterium formicicum. J Bacteriol. 1983;155:467–472. doi: 10.1128/jb.155.2.467-472.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Smith R F, Smith T F. Pattern-induced multi-sequence alignment (PIMA) algorithm employing secondary structure-dependent gap penalties for comparative protein modelling. Protein Eng. 1992;5:35–41. doi: 10.1093/protein/5.1.35. [DOI] [PubMed] [Google Scholar]
- 55.Stover C K. Discovery of a new drug series for the treatment of tuberculosis. Presented at the 97th Annual Meeting of the American Society for Microbiology. 1997. [Google Scholar]
- 56.Stover, C. K., and P. Warrener. 1997. Personal communication.
- 57.Studier F W, Rosenberg A H, Dunn J J, Dubendorff J W. Use of T7 RNA polymerase to direct expression of cloned genes. Methods Enzymol. 1990;185:60–89. doi: 10.1016/0076-6879(90)85008-c. [DOI] [PubMed] [Google Scholar]
- 58.Thompson J D, Higgens D G, Gibson T J. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, positions-dependent gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tuckman D, Donnely R J, Zhao F X, Jacobs W R, Jr, Connell N D. Interruption of the phosphoglucose isomerase gene results in glucose auxotrophy in Mycobacterium smegmatis. J Bacteriol. 1997;179:2724–2730. doi: 10.1128/jb.179.8.2724-2730.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Vaupel M, Thauer R K. Coenzyme F420-dependent N5,N10-methylenetetrahydromethanopterin reductase (Mer) from Methanobacterium thermoautotrophicum. Eur J Biochem. 1995;231:773–778. doi: 10.1111/j.1432-1033.1995.0773d.x. [DOI] [PubMed] [Google Scholar]
- 61.Widdel F, Wolfe R S. Expression of secondary alcohol dehydrogenase in methanogenic bacteria and purification of the F420-specific enzyme from Methanogenium thermophilumstrain TCI. Arch Microbiol. 1989;152:322–328. [Google Scholar]