Abstract
Erwinia amylovora was shown to secrete DspE, a pathogenicity factor of 198 kDa and a functional homolog of AvrE of Pseudomonas syringae pv. tomato. DspE was identified among the supernatant proteins isolated from cultures grown in an hrp gene-inducing minimal medium by immunodetection with a DspE-specific antiserum. Secretion required an intact Hrp pathway.
Erwinia amylovora causes the disease of apple, pear, and other members of Rosaceae known as fire blight. The dspE gene (4, 11) (“dspA” in reference 11) (“dsp” stands for “disease-specific”) of this gram-negative bacterium encodes an essential pathogenicity factor of 198 kDa and is homologous with avrE, one of at least two genes in the avirulence (avr) locus avrE of Pseudomonas syringae pv. tomato (4, 11, 21). Immediately downstream of dspE is dspF (4, 11) (“dspB” in reference 11), which encodes a protein physically similar to chaperones of virulence proteins of animal pathogenic bacteria such as Yersinia spp. and Shigella flexneri (32). dspF also has a homolog in the avrE locus, avrF (4). dspE and dspF together constitute the dspEF locus, which functions as the avrE locus does (19, 21), conferring avirulence (an inability to cause disease due to specific elicitation of plant defense responses) when heterologously expressed in the soybean pathogen P. syringae pv. glycinea (4).
Bacterial avr genes specifically limit host range in their native organism and when expressed in pathogenic bacteria different from the native organism (16). A corresponding resistance (R) gene in the plant is required for avr gene-dependent elicitation of plant defense responses. The avirulence phenomenon historically has been observed to affect interactions of pathovars of Pseudomonas and Xanthomonas spp. with cultivars of respective host plant species (10). avr genes have been observed also, however, to limit host range at the host species level (19, 36). In the absence of a corresponding plant R gene, some avr genes, including avrE (22), have been shown to play roles in virulence or pathogen fitness (10).
To function, avr genes typically require the Hrp pathway (12, 15, 18, 23), a specialized, type III secretion system conserved among phytopathogenic bacteria. The Hrp pathway components are encoded by hrp genes, required both for bacterial elicitation of the hypersensitive reaction (HR), a plant defense reaction, and for pathogenicity. Expression of hrp genes can be induced in vitro by growth in low-nutrient media (15, 35). Proteins known to traverse the Hrp pathway include harpins of P. syringae and Erwinia spp. and the harpin-like protein PopA of Ralstonia solanacearum (see reference 2 for a review of hrp genes and Hrp-secreted proteins). Harpins are glycine-rich proteins that, unlike Avr proteins, elicit the HR when introduced in solution into the apoplast (leaf intercellular space) (14, 34a). Purified harpins elicit the HR, and stimulate systemic resistance to a broad array of pathogens, in many plant species (27, 33). Genetic studies suggest an important role in virulence for harpins in several phytopathogenic bacteria (2, 14, 34a).
A number of virulence proteins of animal pathogenic bacteria such as Yersinia spp., Shigella flexneri, and Salmonella typhimurium are transported into host cells via type III pathways in a cell contact-dependent manner (8, 24, 26, 30). Recent evidence strongly suggests that Avr proteins similarly are translocated to the plant cell interior via the Hrp pathway (for further discussion, see reference 6). Four avr genes have been shown to elicit the hypersensitive response when expressed in plant cells (12, 20, 25, 28, 31). However, no natively expressed Avr protein has been reported outside of bacterial cells in culture or in planta; thus, direct evidence for Avr protein transit through the Hrp pathway has been lacking.
We report here that the product of the dspE gene, DspE, is secreted via the Hrp pathway by Erwinia amylovora cells grown in an hrp gene-inducing minimal medium (15).
Anti-DspE antiserum was raised in rabbit to a polypeptide corresponding to the N-terminal half of DspE (DspE′) as follows. DspE′ was generated by expression of a truncated clone of the dspE gene in Escherichia coli DH5α (plasmid pCPP1244; Table 1) as previously described (4). It was isolated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) of cell lysate followed by electroelution in an Elutrap apparatus (Schleicher & Schuell, Nashua, N.H.) of the band corresponding to DspE′. Eluted protein was dialyzed against buffered saline (2.5 mM KPO4, 0.85% NaCl [pH 6.8]) and concentrated with Centricon ultrafiltration devices (Amicon Inc., Beverly, Mass.). Aliquots of 0.5 ml containing between 0.2 and 0.5 mg of protein were mixed with Freund’s incomplete adjuvant and injected into a female chinchilla rabbit, one aliquot each at 0, 4, and 6 weeks. Antiserum was harvested at 8 weeks and cross-adsorbed with boiled sonicates of E. coli DH5α cells carrying the expression vector (pCPP50; Table 1) and with trichloroacetic acid-precipitated supernatant proteins and boiled cell sonicates of hrp gene-induced dspE mutant cells derived from Erwinia amylovora Ea273 (4). The titer and specificity of the resulting antiserum preparation was ascertained by Western blots of cell lysates of E. coli DH5α(pCPP1244) and E. coli DH5α(pCPP50). The antiserum bound specifically to the band corresponding to DspE′ in lysates of E. coli DH5α(pCPP1244) and bound quantitatively at dilutions (of antiserum) up to 1:10,000 (data not shown).
TABLE 1.
Bacterial strains and plasmids used
| Strain or plasmid | Description | Source or reference |
|---|---|---|
| Strains | ||
| E. coli DH5α | F− φ80dlacZΔM15 Δ(lacZYA-argF)U169 endA1 recA1 hsdR17(rK− rM+) deoR thi-1 supE44 λ−gyrA96 relA1 | Life Technologies, Inc. (Gaithersburg, Md.) |
| Erwinia amylovora | ||
| Ea273 | Wild type | S. V. Beer |
| Ea273-K178 | Hrp secretion mutant; Tn10 mini-Km insertion in hrpJ operon | Z.-M. Wei (34) |
| Ea273dspEΔ1521 | Nonpolar dspE mutant; 1,521-bp in-frame deletion in 3′ half of dspE | 4 |
| Plasmids | ||
| pCPP50 | pINIII113-A2-based expression vector; Apr | 3 |
| pCPP1244 | 3.1-kb HpaI-BamHI clone containing 5′ half of dspE in pCPP50 | 4 |
| pCPP1259 | 7.7-kb HpaI-HindIII clone containing dspE and dspF in pCPP50 | 4 |
Erwinia amylovora Ea273, Ea 273-K178 (an hrp secretion mutant), and Ea273dspEΔ1521 (Table 1) were grown overnight at 28°C in Terrific Broth (29). For each strain, cells were harvested by centrifugation and resuspended to an optical density at 620 nm (OD620) of 0.3 in 15 ml of an hrp gene-inducing minimal medium (15) and grown at 20°C with shaking for 24 h, reaching an OD620 of ca. 1.1. Cells were harvested by centrifugation and resuspended in 0.125 volume of high-purity water. Seventy-five microliters of the cell suspension was combined with 25 μl of 4× NuPAGE SDS sample buffer (NOVEX, San Diego, Calif.) and heated to 100°C for 4 min. Culture supernatants were filter sterilized, followed by addition of phenylmethylsulfonyl fluoride (PMSF) (a serine protease inhibitor) and EDTA to 0.2 and 25 mM, respectively. Supernatants were concentrated 230-fold by using Centriprep and Centricon ultrafiltration devices. One-quarter volume of 4× NuPAGE SDS sample buffer was added to the concentrated supernatants, and the samples were heated to 100°C for 4 min. Proteins were separated by SDS-PAGE in a 4 to 12% gradient NuPAGE polyacrylamide gel by using 1× NuPAGE morpholinepropanesulfonic acid running buffer according to the manufacturer’s instructions. Transfer of the proteins to the Immobilon-P membrane (Millipore, Bedford, Mass.) was carried out by using 1× NuPAGE transfer buffer in a Bio-Rad Trans-Blot semidry transfer cell (Bio-Rad Laboratories, Hercules, Calif.), and immunodetection of blotted proteins was performed by using the Western-Star immunodetection system (Tropix, Bedford, Mass.) and anti-DspE antiserum.
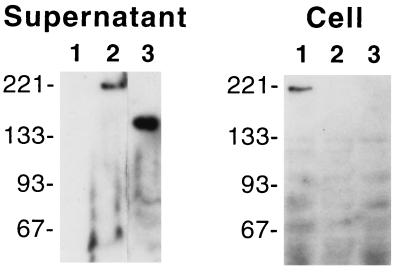
DspE was evident in culture supernatants of Ea273 and was not detected in culture supernatants of the Hrp pathway mutant strain Ea273-K178 (Fig. 1). Supernatant fractions were approximately 30-fold more concentrated than cell fractions based on culture volume. Nevertheless, DspE was evident in cells of Ea273-K178 but was not detected in cells of Ea273. Thus, the absence of DspE from the supernatant of Ea273-K178 was not due to a failure of the cells to produce the protein, and possible cell lysis during culture growth could not be the source of DspE detected in the culture supernatant of Ea273. Ea273dspEΔ1521 cells encode a DspE derivative of 145 kDa with an internal deletion corresponding to residues T1064 to V1570. This protein was present in the culture supernatant of this strain, suggesting that the missing residues are not required for secretion of DspE. Several faint bands of lower molecular weight were present in cell fractions. It is not clear whether these represent degradation products of DspE (and DspE′) or are due to residual cross-reactivity of the anti-DspE antiserum.
FIG. 1.
Immunodetection of DspE in a Western blot of culture supernatant and cell fractions. Cultures of the Erwinia amylovora Hrp pathway mutant strain Ea273-K178 (lanes 1), the wild-type strain Ea273 (lanes 2), and a partial dspE deletion mutant strain, Ea273dspEΔ1521 (lanes 3), were grown under hrp gene-inducing conditions. Molecular weight markers (BenchMark prestained protein ladder; Life Technologies, Gaithersburg, Md.) are indicated. Migration of proteins from the culture supernatant was slightly retarded due to the presence of the extracellular polysaccharide of Erwinia amylovora.
Our results contrast with results for other Avr proteins, which show detectable amounts only within the bacterial cell (7, 18, 37). Linked expression of DspE with the putative chaperone DspF (11) also sets apart DspE from characterized Avr proteins (other than AvrE). The question arises whether DspE and AvrE (and perhaps similar proteins in Pseudomonas and Xanthomonas spp.) are specifically released into the external medium, as are harpins, or Hrp pathway transit is less tightly regulated in Erwinia amylovora, in general, than in Pseudomonas and Xanthomonas spp. such that it can occur without the presumed requirement for contact with plant cells. Subtle differences between Erwinia amylovora and P. syringae in regulation of the Hrp secretion process are suggested by the observations that overexpression of harpin (HrpZ) in P. syringae shuts down Hrp secretion (1) yet overexpression of harpin (HrpN) in Erwinia amylovora does not (3a). Preliminary evidence suggests that, in addition to two harpins and DspE, Erwinia amylovora secretes several other proteins into the extracellular space via the Hrp pathway (17, 17a). If Erwinia amylovora indeed is unique among phytopathogenic bacteria in secreting through the Hrp pathway proteins such as DspE in vitro, these proteins could be characterized by N-terminal sequencing. A reverse-genetic approach then should yield novel genes whose products are involved in interactions with plants.
Secretion of DspE in culture suggests that, in planta, the protein may be released into the apoplast, possibly indicating a plant extracellular or cell-surface-associated target in apple, pear, and other host plants. However, secretion of DspE in culture does not necessarily preclude Hrp-mediated translocation of the protein to the plant cell interior during an attempted colonization by Erwinia amylovora. Virulence proteins of animal pathogenic bacteria that are transported into host cells also are secreted into the extracellular space under certain culture conditions (8, 9); one of these, in fact, was discovered to be structurally related to AvrRxv of Xanthomonas campestris pv. vesicatoria (13). We determined whether a cell-free preparation of DspE and DspF would elicit the HR when introduced into soybean leaves. E. coli DH5α containing plasmid pCPP1259 for DspE and DspF expression (Table 1) and, as a control, E. coli DH5α carrying the expression vector pCPP50 (Table 1) were grown to an OD620 of 0.5 in Luria-Bertani medium. Isopropylthio-β-d-galactoside was added to 0.1 mM to induce expression, and cells were further incubated until reaching an OD620 of 1.2. Cells were then harvested by centrifugation and lysed by sonication in 10 mM 2-(N-morpholino)ethanesulfonic acid (MES) buffer (pH 6.8)–0.1 mM PMSF. Following the removal of cellular debris by centrifugation, sonicates were infiltrated into the primary leaves of soybean plants (cultivar Harosoy) at full strength and dilutions ranging from 1:2 to 1:5. Despite the presence of large amounts of DspE and DspF in the sonicates (as ascertained by SDS-PAGE and staining with Coomassie blue), the HR did not occur (data not shown). Although the possibility exists that HR elicitation was blocked by interaction of DspE with the putative chaperone DspF, the data suggest that the target, or receptor, of DspE in soybean may be inside the plant cell. Determining the location of DspE (and DspF) in planta will be an important step toward understanding both the virulence and avirulence functions of the dspEF locus.
While an earlier version of this paper was under review, Gaudriault et al. (11) reported an independent characterization of the dsp locus of Erwinia amylovora. They reported also that when wild-type Erwinia amylovora cells were grown on a solid minimal medium and then washed from the plates and subsequently removed from the wash liquid by centrifugation, the liquid contained a protein corresponding in molecular weight to DspE (DspA). The protein was absent from wash liquids of both a dspA mutant and an Hrp pathway mutant. Although not definitive, their data provide the first published indication that DspE (DspA) is an Hrp-secreted protein. The authors also observed that the protein was absent from the wash supernatant of a dspF (dspB) mutant, bolstering the prediction that DspF serves as a molecular chaperone to DspE.
Acknowledgments
We thank Amy O. Charkowski, Jihyun F. Kim, and Alan Collmer for critical review of the manuscript, Kent Loeffler for photography, and the Laboratory Animal Care Facility of the Cornell School of Veterinary Medicine for services and advice regarding the DspE antiserum.
This work was supported by grants from Eden Bioscience Corporation (Bothell, Wash.) and the Cornell Center for Advanced Technology in Biotechnology, which is sponsored by the New York State Science and Technology Foundation and industrial partners.
REFERENCES
- 1.Alfano J R, Bauer D W, Milos T M, Collmer A. Analysis of the role of the Pseudomonas syringae pv. syringae HrpZ harpin in elicitation of the hypersensitive response in tobacco using functionally non-polar hrpZ deletion mutations, truncated HrpZ fragments, and hrmA mutations. Mol Microbiol. 1996;19:715–728. doi: 10.1046/j.1365-2958.1996.415946.x. [DOI] [PubMed] [Google Scholar]
- 2.Alfano J R, Collmer A. Bacterial pathogens in plants: life up against the wall. Plant Cell. 1996;8:1683–1698. doi: 10.1105/tpc.8.10.1683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bogdanove A J. Ph.D. dissertation. Ithaca, N.Y: Cornell University; 1997. [Google Scholar]
- 3a.Bogdanove, A. J., and S. V. Beer. Unpublished observations.
- 4.Bogdanove A J, Kim J F, Wei Z-M, Kolchinsky P, Charkowski A O, Conlin A K, Collmer A, Beer S V. Homology and functional similarity of an hrp-linked pathogenicity locus, dspEF, of Erwinia amylovora and the avirulence locus avrE of Pseudomonas syringae pathovar tomato. Proc Natl Acad Sci USA. 1998;95:1325–1330. doi: 10.1073/pnas.95.3.1325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bogdanove A J, Wei Z-M, Zhao L, Beer S V. Erwinia amylovora secretes harpin via a type III pathway and contains a homolog of yopN of Yersinia spp. J Bacteriol. 1996;178:1720–1730. doi: 10.1128/jb.178.6.1720-1730.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bonas U, Van den Ackerveken G. Recognition of bacterial avirulence proteins occurs inside the plant cell: a general phenomenon in resistance to bacterial diseases? Plant J. 1997;12:1–7. doi: 10.1046/j.1365-313x.1997.12010001.x. [DOI] [PubMed] [Google Scholar]
- 7.Brown I, Mansfield J, Irlam I, Conrads S J, Bonas U. Ultrastructure of interactions between Xanthomonas campestris pv. vesicatoria and pepper including immunocytochemical localization of extracellular polysaccharides and the AvrBS3 protein. Mol Plant-Microbe Interact. 1993;6:376–386. [Google Scholar]
- 8.Collazo C M, Galán J E. The invasion-associated type III system of Salmonella typhimurium directs the translocation of Sip proteins into the host cell. Mol Microbiol. 1997;24:747–756. doi: 10.1046/j.1365-2958.1997.3781740.x. [DOI] [PubMed] [Google Scholar]
- 9.Cornelis G R, Wolf-Watz H. The Yersinia Yop virulon: a bacterial system for subverting eukaryotic cells. Mol Microbiol. 1997;23:861–867. doi: 10.1046/j.1365-2958.1997.2731623.x. [DOI] [PubMed] [Google Scholar]
- 10.Dangl J L. The enigmatic avirulence genes of phytopathogenic bacteria. Curr Top Microbiol Immunol. 1994;192:99–118. doi: 10.1007/978-3-642-78624-2_5. [DOI] [PubMed] [Google Scholar]
- 11.Gaudriault S, Malandrin L, Paulin J-P, Barny M-A. DspA, an essential pathogenicity factor of Erwinia amylovora showing homology with AvrE of Pseudomonas syringae, is secreted via the Hrp secretion pathway in a DspB-dependent way. Mol Microbiol. 1997;26:1057–1069. doi: 10.1046/j.1365-2958.1997.6442015.x. [DOI] [PubMed] [Google Scholar]
- 12.Gopalan S, Bauer D W, Alfano J R, Loniello A O, He S Y, Collmer A. Expression of the Pseudomonas syringae avirulence protein AvrB in plant cells alleviates its dependence on the hypersensitive response and pathogenicity (Hrp) secretion system in eliciting genotype-specific hypersensitive cell death. Plant Cell. 1996;8:1095–1105. doi: 10.1105/tpc.8.7.1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hardt W-D, Galán J E. A secreted Salmonella protein with homology to an avirulence determinant of plant pathogenic bacteria. Proc Natl Acad Sci USA. 1997;94:9887–9892. doi: 10.1073/pnas.94.18.9887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.He S Y, Huang H C, Collmer A. Pseudomonas syringae pv. syringae harpinPss: a protein that is secreted via the Hrp pathway and elicits the hypersensitive response in plants. Cell. 1993;73:1255–1266. doi: 10.1016/0092-8674(93)90354-s. [DOI] [PubMed] [Google Scholar]
- 15.Huynh T V, Dahlbeck D, Staskawicz B J. Bacterial blight of soybean: regulation of a pathogen gene determining host cultivar specificity. Science. 1989;345:1374–1377. doi: 10.1126/science.2781284. [DOI] [PubMed] [Google Scholar]
- 16.Keen N T. Gene-for-gene complementarity in plant-pathogen interactions. Annu Rev Genet. 1990;24:447–463. doi: 10.1146/annurev.ge.24.120190.002311. [DOI] [PubMed] [Google Scholar]
- 17.Kim J F. Ph.D. dissertation. Ithaca, N.Y: Cornell University; 1997. [Google Scholar]
- 17a.Kim J F, Zumoff C H, Beer S V. HrpW, a new harpin of Erwinia amylovora, is a member of a family of pectate lyases. Phytopathology. 1997;87:S52. [Google Scholar]
- 18.Knoop V, Staskawicz B J, Bonas U. Expression of the avirulence gene avrBs3 from Xanthomonas campestris pv. vesicatoria is not under the control of hrp genes and is independent of plant factors. J Bacteriol. 1991;173:7142–7150. doi: 10.1128/jb.173.22.7142-7150.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kobayashi D Y, Tamaki S J, Keen N T. Cloned avirulence genes from the tomato pathogen Pseudomonas syringae pathovar tomato confer cultivar specificity on soybean. Proc Natl Acad Sci USA. 1989;86:157–161. doi: 10.1073/pnas.86.1.157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Leister R T, Ausubel F M, Katagiri F. Molecular recognition of pathogen attack occurs inside of plant cells in plant disease resistance specified by the Arabidopsis genes RPS2 and RPM1. Proc Natl Acad Sci USA. 1996;93:15497–15502. doi: 10.1073/pnas.93.26.15497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lorang J M, Keen N T. Characterization of avrE from Pseudomonas syringae pv. tomato: a hrp-linked avirulence locus consisting of at least two transcriptional units. Mol Plant-Microbe Interact. 1995;8:49–57. doi: 10.1094/mpmi-8-0049. [DOI] [PubMed] [Google Scholar]
- 22.Lorang J M, Shen H, Kobayashi D, Cooksey D, Keen N T. avrA and avrE in Pseudomonas syringae pv. tomato PT23 play a role in virulence on tomato plants. Mol Plant-Microbe Interact. 1994;7:508–515. [Google Scholar]
- 23.Pirhonen M U, Lidell M C, Rowley D L, Lee S W, Jin S, Liang Y, Silverstone S, Keen N T, Hutcheson S W. Phenotypic expression of Pseudomonas syringae avr genes in E. coli is linked to the activities of the hrp-encoded secretion system. Mol Plant-Microbe Interact. 1996;9:252–260. doi: 10.1094/mpmi-9-0252. [DOI] [PubMed] [Google Scholar]
- 24.Rosqvist R, Magnusson K E, Wolf-Watz H. Target cell contact triggers expression and polarized transfer of Yersinia YopE cytotoxin into mammalian cells. EMBO J. 1994;13:964–972. doi: 10.1002/j.1460-2075.1994.tb06341.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Scofield S R, Tobias C M, Rathjen J P, Chang J H, Lavelle D T, Michelmore R W, Staskawicz B J. Molecular basis of gene-for-gene specificity in bacterial speck disease of tomato. Science. 1996;274:2063–2065. doi: 10.1126/science.274.5295.2063. [DOI] [PubMed] [Google Scholar]
- 26.Sory M P, Cornelis G R. Translocation of a hybrid YopE-adenylate cyclase from Yersinia enterocolitica into HeLa cells. Mol Microbiol. 1994;14:583–594. doi: 10.1111/j.1365-2958.1994.tb02191.x. [DOI] [PubMed] [Google Scholar]
- 27.Strobel N E, Ji C, Gopalan S, Kuc J A, He S Y. Induction of systemic acquired resistance in cucumber by Pseudomonas syringae pv. syringae 61 HrpZPss protein. Plant J. 1996;9:431–439. [Google Scholar]
- 28.Tang X, Frederick R D, Zhou J, Halterman D A, Jia Y, Martin G B. Initiation of plant disease resistance by physical interaction of AvrPto and Pto kinase. Science. 1996;274:2060–2063. doi: 10.1126/science.274.5295.2060. [DOI] [PubMed] [Google Scholar]
- 29.Tartof K D, Hobbs C A. Improved media for growing plasmid and cosmid clones. Bethesda Res Lab Focus. 1987;9:12. [Google Scholar]
- 30.Thirumalai K, Kim K-S, Zychlinsky A. IpaB, a Shigella flexneri invasin, colocalizes with interleukin-1β-converting enzyme in the cytoplasm of macrophages. Infect Immun. 1997;65:787–793. doi: 10.1128/iai.65.2.787-793.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Van Den Ackerveken G, Marois E, Bonas U. Recognition of the bacterial avirulence protein AvrBs3 occurs inside the host plant cell. Cell. 1996;87:1307–1316. doi: 10.1016/s0092-8674(00)81825-5. [DOI] [PubMed] [Google Scholar]
- 32.Wattiau P, Woestyn S, Cornelis G R. Customized secretion chaperones in pathogenic bacteria. Mol Microbiol. 1996;20:255–262. doi: 10.1111/j.1365-2958.1996.tb02614.x. [DOI] [PubMed] [Google Scholar]
- 33.Wei Z-M, Beer S V. Harpin from Erwinia amylovora induces plant resistance. Acta Hortic. 1996;411:223–225. [Google Scholar]
- 34.Wei Z-M, Beer S V. HrpI of Erwinia amylovora functions in secretion of harpin and is a member of a new protein family. J Bacteriol. 1993;175:7958–7967. doi: 10.1128/jb.175.24.7958-7967.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34a.Wei Z M, Laby R J, Zumoff C H, Bauer D W, He S Y, Collmer A, Beer S V. Harpin, elicitor of the hypersensitive response produced by the plant pathogen Erwinia amylovora. Science. 1992;257:85–88. doi: 10.1126/science.1621099. [DOI] [PubMed] [Google Scholar]
- 35.Wei Z-M, Sneath B J, Beer S V. Expression of E. amylovora hrp genes in response to environmental stimuli. J Bacteriol. 1992;174:1875–1882. doi: 10.1128/jb.174.6.1875-1882.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Whalen M C, Stall R E, Staskawicz B J. Characterization of a gene from a tomato pathogen determining hypersensitive resistance in non-host species and genetic analysis of this resistance in bean. Proc Natl Acad Sci USA. 1988;85:6743–6747. doi: 10.1073/pnas.85.18.6743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Young S A, White F F, Hopkins C M, Leach J E. AvrXa10 protein is in the cytoplasm of Xanthomonas oryzae pv. oryzae. Mol Plant-Microbe Interact. 1994;7:799–804. doi: 10.1094/mpmi-7-0799. [DOI] [PubMed] [Google Scholar]