Abstract
Background:
Atopic dermatitis (AD) is an inflammatory disorder characterized by dominant type 2 inflammation leading to chronic pruritic skin lesions, allergic comorbidities and Staphylococcus aureus skin colonization and infections. S. aureus is thought to play a role in AD severity.
Objective:
We characterized the changes in the host-microbial interface in AD subjects following type 2 blockade with dupilumab.
Methods:
Participants (n=71) with moderate-severe AD were enrolled in a randomized (dupilumab vs placebo; 2:1), double-blind study at Atopic Dermatitis Research Network centers. Bioassays were performed at multiple timepoints: S. aureus and virulence factor quantification, 16s rRNA microbiome, serum biomarkers, skin transcriptomic analyses and peripheral blood T-cell phenotyping.
Results:
At baseline, 100% of participants were S. aureus colonized on the skin surface. Dupilumab treatment resulted in significant reductions in S. aureus after only 3 days (compared to placebo); 11 days before clinical improvement. Participants with the greatest S. aureus reductions had the best clinical outcomes, and these reductions correlated with reductions in serum CCL17 and disease severity. Reductions (10-fold) in S. aureus cytotoxins (day 7), perturbations in Th17 subsets (day 14), and increased expression of genes relevant for IL-17, neutrophil and complement pathways (day 7) were also observed.
Conclusion:
Blockade of IL-4 and IL-13 signaling, very rapidly (day 3) reduces S. aureus abundance in AD subjects, and this reduction correlates with reductions in the type 2 biomarker, CCL17 and measures of AD severity (excluding itch). Immunoprofiling and/or transcriptomics suggest a role for Th17, neutrophils and complement activation as potential mechanisms to explain these findings.
Keywords: Atopic dermatitis, dupilumab, interleukin-4, interleukin-13, interleukin-17, Staphylococcus aureus, microbiome, cytotoxins, barrier, type 2 immunity
Graphical Abstract
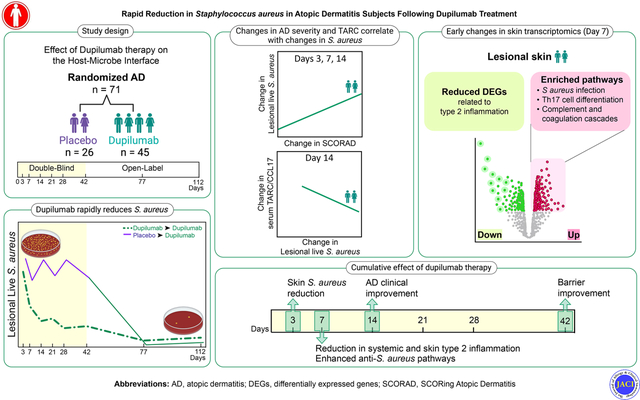
Capsule Summary:
Dupilumab reduces lesional S. aureus abundance and virulence factor production within three days. These changes likely occur through enhancement of innate immune responses linked to IL-17, as well as neutrophil and complement activation.
INTRODUCTION
Atopic dermatitis (AD), which affects 30 million people in the United States, is a chronic skin condition characterized by type 2 inflammation (i.e. elevated interleukin [IL]-4 and -13), skin barrier defects and colonization with Staphylococcus aureus.1 Skin infections represent common comorbidities with increased rates of both viral (herpes simplex virus [HSV], molluscum contagiosum and human papilloma virus) and bacterial infections, primarily caused by S. aureus.2 The Atopic Dermatitis Research Network (ADRN) is an NIH-funded, multi-institutional consortium with the overarching objective of improving our understanding of the observed susceptibility to cutaneous infections in patients with AD. To date, the ADRN uncovered genetic risk factors and novel immunologic abnormalities that contribute to the elevated rates of HSV infections.3–8 Most recently, the ADRN reported that 43% of AD patients (all severities) are colonized with S. aureus, based on skin swabs analyzed by a clinical microbiology laboratory.9 This S. aureus-colonized AD subset has greater skin barrier dysfunction, disease severity and allergen sensitization, increased serum IgE and Type 2 biomarkers than the non-colonized AD participants.
S. aureus secretes many virulence factors that may exacerbate atopic inflammation which may in part explain these findings. AD-associated S. aureus produces numerous cytotoxins.10 High cytotoxin levels appear to be required for S. aureus skin infections, and because many are pore-forming they enhance susceptibility to viral infections.11, 12 This is consistent with an earlier ADRN observation, that AD patients who have history of the HSV complication, eczema herpeticum, more commonly report a history of S. aureus skin infections than AD patients without a history of EH.9, 13, 14 Besides cytotoxins, all pathogenic strains of S. aureus produce superantigens; a large family of proteins that stimulate both T cells and epithelial cells.10 While cytotoxins act locally to kill immune cells and keratinocytes, superantigens act locally and systemically to alter structural cells and adaptive immunity. Superantigens can persist for months in tissues.15 The superantigens primarily associated with AD include the six-member enterotoxin (SE) gene cluster and SE-like Q.13, 16 Other potential S. aureus virulence factors include lipase and various proteases. S. aureus strains isolated from the skin produce more lipase and cytotoxins than strains infecting mucous membranes.17, 18 These virulence factors likely contribute to clinical infection, including impetigo, cellulitis and rarely sepsis as well as skin barrier defects in AD patients.19
Previous studies have shown topical and systemic anti-inflammatory treatments reduce S. aureus colonization in AD patients. However, the kinetics of the response and the molecular mechanisms involved in this reduction remain poorly understood.20 Pre-registration and post-marketing studies have demonstrated that the blockade of IL-4 and -13, achieved by treatment with dupilumab, a fully-humanized monoclonal antibody directed against IL-4Rα, leads to clinically meaningful improvements in disease severity by as early as two weeks and reduced non-viral skin infections.21, 22 To better understand the role that these type 2 cytokines play in the microbial environment on the skin surface, we investigated the clinical, host immune responses, microbiome, and S. aureus virulence factors and colonization kinetics in patients randomized to either dupilumab or placebo monotherapy.
METHODS
Study design
This was a National Institute of Allergy and Infectious Disea27ses (NIAID)-funded multi-center, randomized, double-blind, placebo-controlled (RDBPC) trial investigating the effect of 6 weeks of dupilumab treatment on measures of cutaneous microbial community structure, skin barrier biology, and circulating T cell profiles, followed by a 10-week open-label extension (OLE; ClinicalTrials.gov Identifier: NCT03389893 [ADRN09]). Adults (18–75 years) with moderate to severe AD were recruited from 8 US academic centers with AD expertise (Fig 1A) after protocol approval by the centralized Western Institutional Review Board Copernicus Group. Informed consent was obtained from the participants, and written assent was provided by the participants, as applicable, before participation. Dupilumab dosing followed package insert (600 mg loading dose, then 300 mg every 2 weeks). Fig 1B & E1 show the schedule of events during the study and the flow of enrollment and allocation into the study. The primary, secondary and exploratory endpoints, as well as the inclusion/exclusion criteria, are available on clinicaltrials.gov (NCT03389893).
Figure 1. US academic centers involved in AD subject recruitment and Study Schematic for the Atopic Dermatitis Research Network (ADRN) 09 study (NCT03389893).
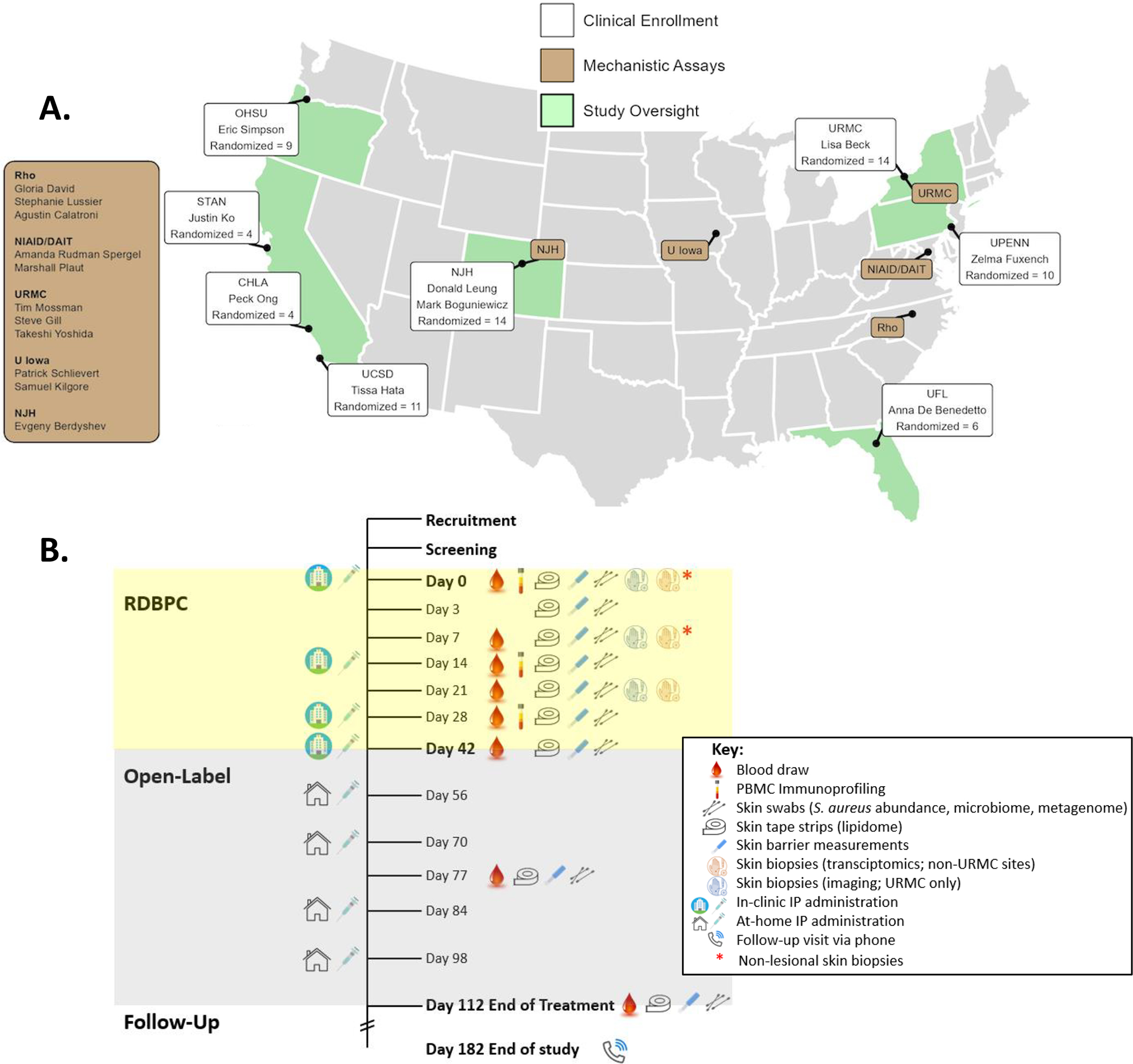
(A) A map of the US academic centers involved in clinical enrollment (white boxes) or mechanistic assays or study oversight (brown boxes). Each white box represents a single center with investigator(s) and enrollment totals. OHSU=Oregon Health & Science University, STAN=Stanford Medical Center, CHLA=Children’s Hospital of Los Angeles, UCSD=University of California San Diego, NJH=National Jewish Health, URMC=University of Rochester Medical Center, UPENN=University of Pennsylvania, UF=University of Florida. (B) High density sampling of the microbial, epithelial, and immune compartments was performed during the 6-week RDBPC phase (yellow) of the trial to characterize and quantify changes and their relationship to disease improvements, followed by a 10-week open-label extension (OLE; grey) phase with a safety assessment (by phone) 10 weeks after the OLE phase ended. ∗Timepoints when non-lesional skin biopsies were collected in addition to lesional biopsies for transcriptomics analysis.
Clinical assessments
Clinical evaluations included assessment of AD severity by Eczema Area and Severity Index (EASI), SCORing AD (SCORAD), validated Investigator Global Assessment (vIGA), pruritus Numerical Rating Scale (NRS) and Nottingham Eczema Severity Score (NESS).23 A detailed medical history was obtained, including concomitant medication use; allergy history; infection history; history of AD and other skin conditions; and observations of eyes/nose/throat, respiratory/lungs, heart, gastrointestinal, psychiatric, endocrine, and other conditions. A physical examination was performed which included vital signs (temperature and blood pressure), growth parameters (height and weight) and examination of the skin, hair, eyes, and lungs.
Additional methodological details including diagnostic criteria, case report forms, collection methodology (i.e. blood, skin swab and skin biopsy), skin barrier measurements, characterization of cutaneous microbial flora, RNA-Seq, peripheral blood immunoprofiling, and statistical analysis are listed in the Online Repository.
Statistical analysis
In the original power calculations, a sample size of approximately 84 participants (56 in the dupilumab arm and 28 in the placebo arm) was established to detect a 0.36 geometric mean ratio of S. aureus abundance between arms by Day 28 with 90% power and 5% type I error for a two-sample pooled t-test with log normal data. The final observed geometric mean ratio was below the one assumed, providing great power to primary and secondary hypotheses.
The pre-specified primary endpoint, S. aureus abundance (log10 transformed) and secondary endpoints were analyzed using a mixed model for repeated measures using an unstructured covariance matrix with treatment, baseline endpoint value (randomization/Day 0), clinical site, baseline disease severity as measured by EASI (≥ 21.1 vs. < 21.1) and treatment by-day interactions included as covariates. The comparisons between placebo vs dupilumab were generated using least-square means at each discrete time. Similarly, the comparison for the open-label portion of the study between day 42 vs day 77 or day 112 within arms was provided using least square means.
All tests of treatment effects were performed at a 2-sided alpha level of 0.05, and no adjustments for multiplicity were made, except in the flow cytometry data in Supplemental Fig E5B. All statistical computations were performed using SAS software Version 14.1 (SAS Institute, Inc., Cary, NC, USA) and results displayed using R (Version 4.2.0) and the ggplot2 library.
RESULTS
Characteristics of study population
Of the 82 participants screened, 72 participants with moderate to severe AD met inclusion/exclusion criteria and were randomized, and 71 (ages 18–65 years) were analyzed, with 45 participants randomized to dupilumab and 26 to placebo arms (approximate 2:1 ratio; Fig E1). Demographics and baseline phenotypic and endotypic characteristics were balanced between the groups, with the exception of serum lipocalin-2 and lesional TEWL (Tables 1 and E1). Lipocalin-2 levels were higher, and basal TEWL values were lower in the dupilumab group (p=0.02 and 0.058; respectively). But differences in baseline values on study entry were accounted for in the repeated measures modeling. More males were enrolled than females (66% vs 34%), and the mean age was 36.9 years. The population was racially diverse with participants self-reporting as White (48%), Asian (20%), and Black (15%). Hispanics (11%) were slightly underrepresented compared to US census. Body Mass Index (BMI) was typical of the US average of 26.9.24 The majority (≥ 62%) of the participants had severe AD as measured by disease status on study entry-EASI (≥ 21.1), SCORAD (≥49) or IGA (=4), as well as by NESS (12–15) (92%).23, 25, 26 The two treatment arms were well matched for severity metrics.
Table 1.
Demographics and Baseline Characteristics
| Dupilumab, N = 45 | Placebo, N = 26 | Overall, N = 71 | |
|---|---|---|---|
| Study Site | |||
| NJH | 9 (20%) | 5 (19%) | 14 (20%) |
| URMC | 9 (20%) | 5 (19%) | 14 (20%) |
| UCSD | 7 (16%) | 4 (15%) | 11 (15%) |
| OHSU | 6 (13%) | 3 (12%) | 9 (13%) |
| UPENN | 6 (13%) | 3 (12%) | 9 (13%) |
| UFL | 4 (8.9%) | 2 (7.7%) | 6 (8.5%) |
| CHLA | 2 (4.4%) | 2 (7.7%) | 4 (5.6%) |
| STAN | 2 (4.4%) | 2 (7.7%) | 4 (5.6%) |
| Gender (Female) | 17 (38%) | 7 (27%) | 24 (34%) |
| Age at Screening | |||
| Mean (SD) | 36.6 (15.6) | 37.3 (15.6) | 36.9 (15.5) |
| Range | 18, 65 | 18, 65 | 18, 65 |
| Race | |||
| White | 21 (47%) | 13 (50%) | 34 (48%) |
| Asian | 7 (16%) | 7 (27%) | 14 (20%) |
| Black or African American | 8 (18%) | 3 (12%) | 11 (15%) |
| More than One Race | 5 (11%) | 1 (3.8%) | 6 (8.5%) |
| Unknown or not reported | 3 (6.7%) | 2 (7.7%) | 5 (7.0%) |
| American Indian or Alaska Native | 1 (2.2%) | 0 (0%) | 1 (1.4%) |
| Ethnicity | |||
| Not Hispanic or Latino | 39 (87%) | 22 (85%) | 61 (86%) |
| Hispanic or Latino | 4 (8.9%) | 4 (15%) | 8 (11%) |
| Unknown or Not Reported | 2 (4.4%) | 0 (0%) | 2 (2.8%) |
| BMI at Screening (kg/m2) | |||
| Mean (SD) | 27.5 (5.7) | 25.8 (4.9) | 26.9 (5.5) |
| Range | 18.6, 41.5 | 17.6, 36.0 | 17.6, 41.5 |
| IGA | |||
| Moderate | 16 (36%) | 11 (42%) | 27 (38%) |
| Severe | 29 (64%) | 15 (58%) | 44 (62%) |
| EASI at Baseline | |||
| EASI < 21.1 | 16 (36%) | 10 (38%) | 26 (37%) |
| EASI ≥ 21.1 | 29 (64%) | 16 (62%) | 45 (63%) |
| EASI (0–72) | |||
| Mean (SD) | 28.44 (12.16) | 28.55 (10.89) | 28.48 (11.63) |
| Range | 16.10, 58.80 | 16.00, 50.25 | 16.00, 58.80 |
| SCORAD (0–103) | |||
| Mean (SD) | 62.11 (11.70) | 59.89 (10.86) | 61.30 (11.38) |
| Range | 42.44, 91.41 | 43.97, 90.14 | 42.44, 91.41 |
| Pruritus NRS (0–10) | |||
| Mean (SD) | 6.69 (2.17) | 7.04 (1.48) | 6.82 (1.94) |
| Range | 1.00, 10.00 | 4.00, 10.00 | 1.00, 10.00 |
| NESS (3–15) | |||
| Mean (SD) | 13.51 (1.49) | 13.58 (1.21) | 13.54 (1.38) |
| Range | 10.00, 15.00 | 11.00, 15.00 | 10.00, 15.00 |
| Lesional S. aureus (rCFUs) OR (CFUs) | |||
| Negative | 1 (2.2%) | 0 (0%) | 1 (1.4%) |
| Positive | 44 (98%) | 26 (100%) | 70 (99%) |
| Non-lesional S. aureus (rCFUs) OR (CFUs) | |||
| Negative | 0 (0%) | 0 (0%) | 0 (0%) |
| Positive | 45 (100%) | 26 (100%) | 71 (100%) |
| S. aureus (rCFU/cm2 Log10) Lesional | |||
| Mean (SD) | 3.64 (1.52) | 3.14 (2.14) | 3.46 (1.77) |
| Range | 0.00, 6.04 | 0.00, 6.30 | 0.00, 6.30 |
| S. aureus (rCFU/cm2 Log10) Non-lesional | |||
| Mean (SD) | 2.12 (1.50) | 1.89 (1.18) | 2.03 (1.39) |
| Range | 0.00, 5.01 | 0.00, 4.04 | 0.00, 5.01 |
| Ever been diagnosed with EH? (Yes) | 1 (2.2%) | 1 (3.8%) | 2 (2.8%) |
| Ever had a Staph infection? (Yes) | 15 (33%) | 9 (35%) | 24 (34%) |
| Known allergy to animals? (Yes) | 30 (67%) | 19 (73%) | 49 (69%) |
| Pets living in home? (Yes) | 26 (58%) | 10 (38%) | 36 (51%) |
| Anyone smoke in home? (Yes) | 11 (24%) | 3 (12%) | 14 (20%) |
Data are presented as mean (standard deviation) for continuous variables or as number (%) for categorical variables.
Using quantitative measures to detect S. aureus on both lesional and non-lesional skin, all participants were colonized at study entry. Furthermore, 34% had a history of staphylococcal skin infections and almost 3% had a history of eczema herpeticum. There were no significant differences in most endotypic features, including serum biomarkers (except lipocalin-2), circulating cell numbers, skin barrier measures (except basal lesional TEWL) and measures of S. aureus abundance in these two populations on study entry (Table E1).
Clinical improvement
A greater absolute reduction in SCORAD (p=0.04) was observed in the dupilumab group compared to placebo by as early as day 14 and remained lower at all later timepoints (Fig 2). For all other severity measures (EASI, Pruritus NRS and IGA) a statistically significant separation of the dupilumab versus placebo groups was first achieved after 21 days of treatment. The dupilumab group continued to improve during OLE (i.e. from day 42 to days 77 and 112; p≤0.005) for each of the severity measures. The placebo group experienced significant (p<0.001) clinical improvement in all severity measures only during OLE when participants were receiving dupilumab treatment, comparing day 42 (day 0 of dupilumab treatment for this group) to days 77 (five weeks later) and 112 (10 weeks later). Safety was similar to what was observed in pivotal trials with no serious adverse events related to study drug or procedures (Table E2).
Figure 2. Dupilumab treatment improves all measures of AD severity.
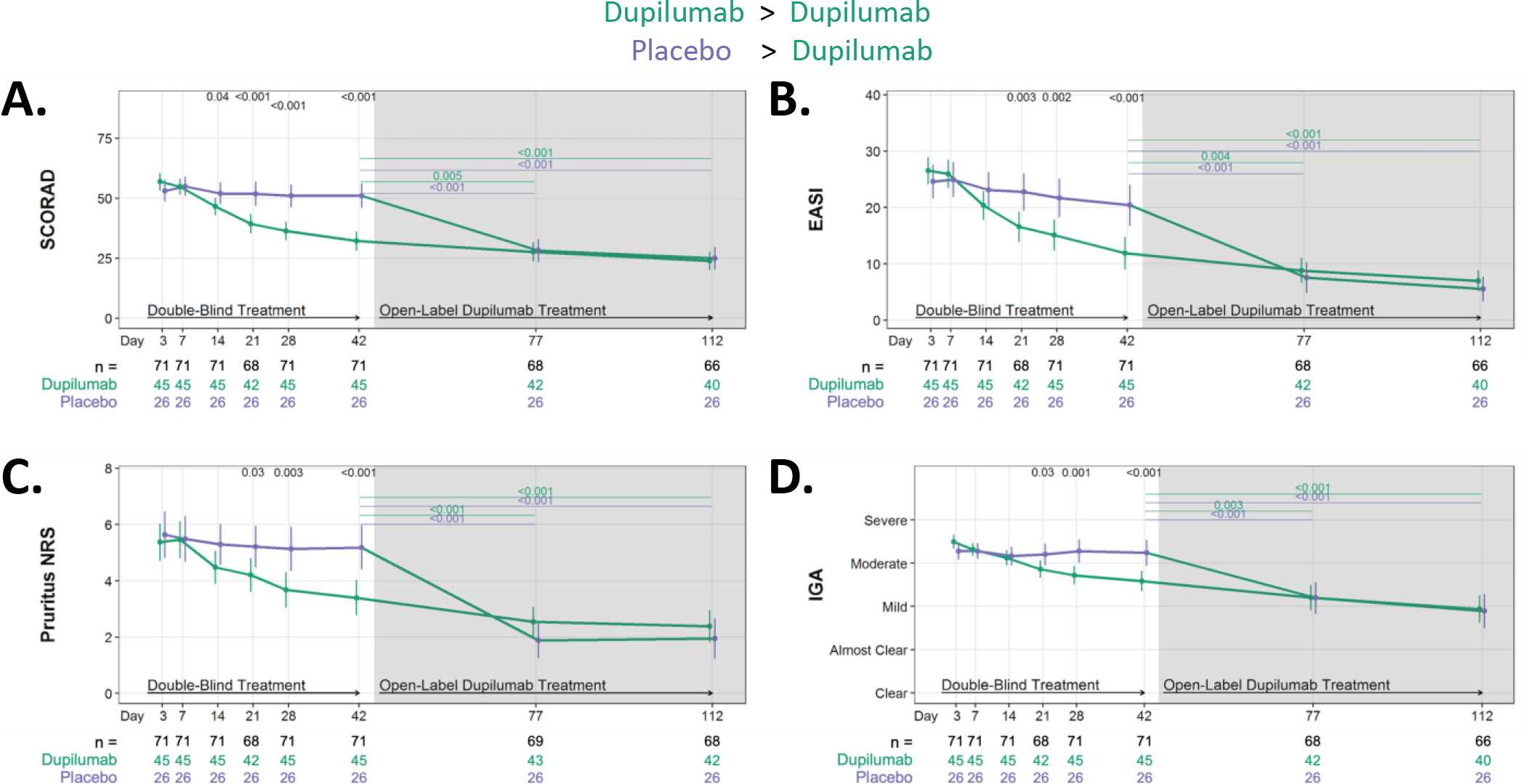
Absolute changes in AD severity measures during RDBPC phase (up to day 42 [6 weeks]) and during the OLE phase (day 42 – day 112 or [6 – 16 weeks]). (A) Absolute reductions in SCORing AD (SCORAD), (B) Eczema Area and Severity Index (EASI), (C) Pruritus Numerical Rating Scale (NRS) and (D) Investigator Global Assessment (IGA). Data for A, C, and D are shown as the means and 95% CIs adjusted for clinical site and EASI (≥ 21.1 or < 21.1), and the severity measure at day 0. Data for B is shown as the means and 95% CIs adjusted for clinical site and EASI at day 0. The number of participants with evaluable data at each timepoint are noted below the X axis (with the total population denoted in black, dupilumab-randomized participants in green and placebo-randomized in purple).
Reduction in S. aureus colonization and cytotoxin levels
All participants were colonized with S. aureus on the skin as determined by both fem A qPCR (Fig 3A) or viable counts (Fig 3C). Dupilumab rapidly reduced S. aureus abundance in lesional skin as measured by both rCFUs (Fig 3A) and viable CFUs (Fig 3C) with a separation (approximately 7.5-fold) from placebo as early as day 3 (p=0.02) which continued at later timepoints. The reduction (approximately 100-fold) in viable S. aureus (plate counts) plateaued after 28 days of dupilumab treatment, with no further reductions observed in OLE (day 42 to day 112). The placebo group had a similar trajectory in the OLE phase (when receiving dupilumab), with a remarkable drop of almost two logs in viable S. aureus observed as early as 35 days into treatment (at day 77) with no further reductions observed by day 112 (Fig 3C). qPCR quantification of S. aureus, which measures both viable and nonviable bacteria, was still dropping even in OLE for the dupilumab randomized group (Fig 3A).
Figure 3. Dupilumab treatment rapidly reduces S. aureus abundance and cytotoxin production on the skin surface of AD participants.
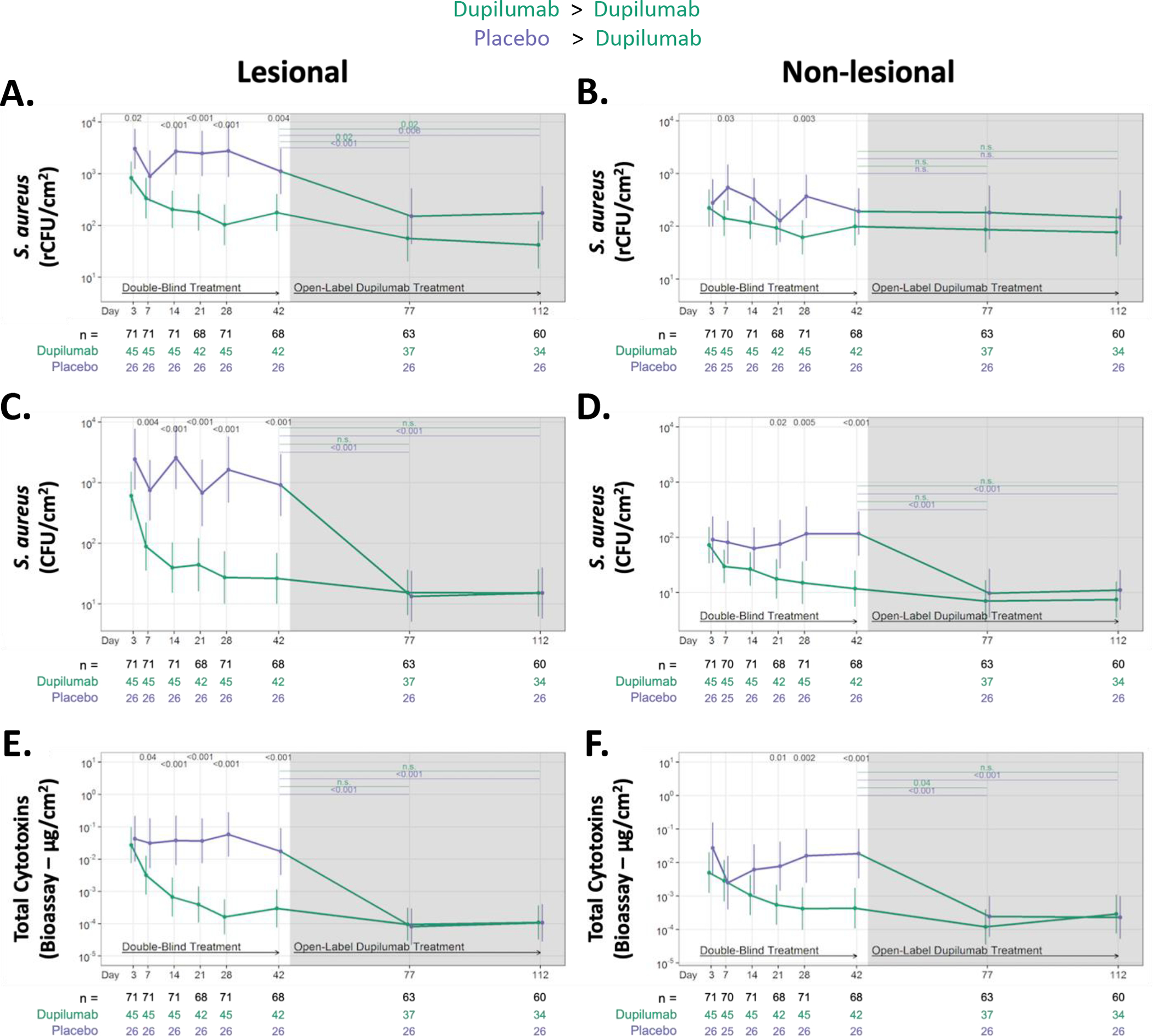
Absolute changes in S. aureus abundance and cytotoxin production during RDBPC (up to day 42 [6 weeks]) and OLE phases (6 – 16 weeks) on lesional (left) and non-lesional (right) skin. S. aureus abundance was measured using qPCR for the femA gene (rCFU/cm2) in both lesional skin (A) and non-lesional skin (B) and by quantitative culture techniques (CFU/cm2) in both lesional skin (C) and non-lesional skin (D). Total S. aureus cytotoxin levels (μg/cm2), measured by bioassay from skin swabs of both lesional (E) and non-lesional skin (F), are shown. Data are shown as geometric means and 95% CIs and are adjusted for clinical site, disease severity at day 0, as measured by EASI ≥ 21.1 or < 21.1, and S. aureus abundance on lesional skin at day 0. The number of participants with evaluable data at each timepoint are noted below the X axis (with the total population denoted in black, dupilumab-randomized participants in green and placebo-randomized in purple). rCFU, relative colony-forming units
Nonlesional skin had lower S. aureus CFU abundance (>1.7 logs lower; Fig 3D and Table E1) than lesional skin on study entry. Dupilumab treatment reduced S. aureus CFUs on non-lesional skin after 21 days, which was similar to what was observed in placebo-randomized participants during OLE. Treatment with dupilumab had a more modest impact on viable S. aureus than on rCFUs (Fig 3B and D).
S. aureus cytotoxins, measured directly from swabs, were significantly reduced (10-fold) in lesional skin of the dupilumab group by day 7, reaching maximal reduction (100-fold) by day 28 (Fig 3E). Consistent with the lower S. aureus counts on non-lesional skin, the non-lesional cytotoxin reductions were more modest, with dupilumab differentiated from the placebo group after 21 days of treatment (Fig 3F). Notably, reductions continued until day 77 (11 weeks). Similar changes were observed in the placebo-randomized group when they entered OLE. No differences in S. aureus superantigens, the four major S. aureus proteases or lipase (data not shown) from lesional and non-lesional skin swabs were observed with dupilumab versus placebo treatment (Fig E2).
Relationship of S. aureus reduction to AD severity
To evaluate the association between the change in S. aureus abundance with the change in AD severity measured by SCORAD on days 3, 7, and 14, a post hoc analysis was undertaken. A repeated measures correlation showed a statistically significant correlation (0.39, 95% CI [0.20, 0.56] p<0.001) in lesional skin of the dupilumab group while there was no association (0.05 95% CI [−0.23, 0.32] p=0.72) in the placebo treatment arm (Fig E3). A similar result was observed with other AD severity measurements, such as EASI, vIGA but not for pruritus NRS (Fig E3). These results suggest that dupilumab-treated participants with the most significant early reduction in lesional S. aureus are likely to have the greatest improvement in AD severity at early timepoints.
Changes in microbial diversity and relative abundance of different genera and species
Dupilumab treatment increased the Shannon microbial α-diversity in lesional skin (but not non-lesional) as early as 3 days, with near maximal effect by day 28 (Fig 4A and B). Similar changes were observed for the placebo group during OLE, when they were receiving dupilumab treatment and the maximal effect was observed at day 77 (five weeks) into dupilumab treatment. Fig 4C shows relative abundance of the most common bacterial genera in dupilumab (upper graphs) and placebo (lower graphs) in both lesional and non-lesional skin over the course of the study. Staphylococcus (genus level) was more abundant in lesional skin than non-lesional skin on study entry (day 0) as expected. The reduction in relative abundance of lesional and non-lesional Staphylococcus was most prominent in the first 28 days of dupilumab treatment. The placebo group showed no change in relative abundance of Staphylococcus until they entered dupilumab OLE (days 77, 112). The changes in the most common bacterial species are shown in Fig 4D. Greater relative abundance of S. aureus was observed in lesional compared with non-lesional skin on study entry, followed by S. epidermidis. Dupilumab treatment resulted in a progressive reduction in the relative abundance of S. aureus (both lesional and non-lesional skin) through day 28 and remained relatively stable through day 112, which was similar to changes observed from S. aureus rCFUs (Fig 3A and B). During dupilumab treatment, several species increased, including S. epidermidis, Cutibacterium acne, S. hominis and Micrococcus luteus. Placebo-randomized participants had stable bacterial species composition until OLE when the changes seen at day 77 and 112 mirrored those observed in the dupilumab group during RDBPC phase.
Figure 4. Measures of the cutaneous microbial community changed rapidly in response to dupilumab treatment.
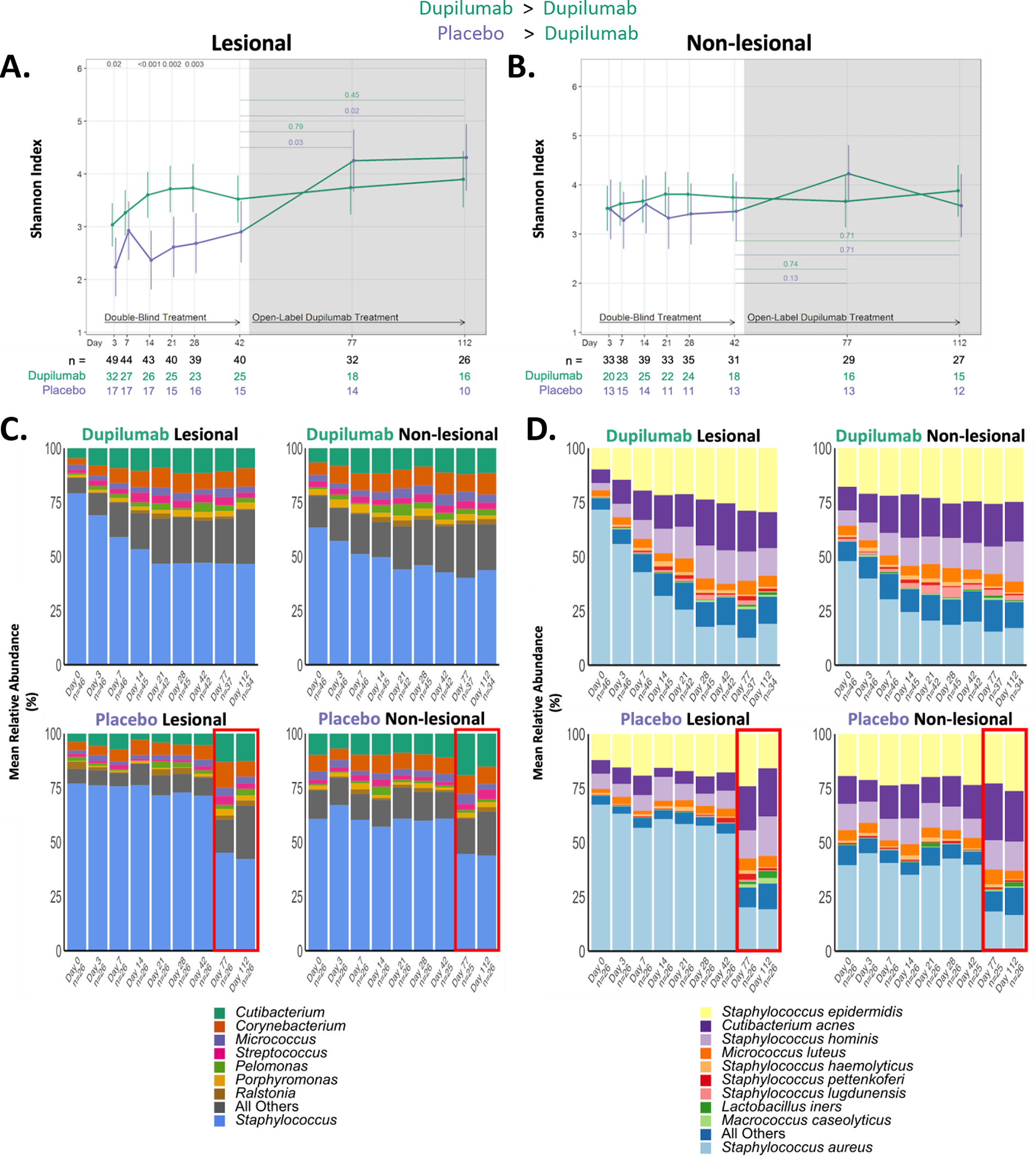
(A) Longitudinal changes in Shannon diversity observed from 16S rRNA analysis of lesional and (B) non-lesional skin in the placebo and dupilumab-treated groups. The number of participants with evaluable data at each timepoint are noted below the X axis (with the total population denoted in black, dupilumab-randomized participants in green and placebo-randomized in purple). (C & D) Relative abundance of the most dominant genera (C) and species (D) in dupilumab and placebo randomized groups. Only the most abundant OTUs are presented. A decrease in the relative abundance of S. aureus is observed in the dupilumab treatment groups. The red box in the placebo-randomized group (C & D) illustrates that for those two timepoints (Day 77 and 112) participants originally randomized to placebo were now receiving dupilumab (as part of the OLE phase of the study).
rRNA, ribosomal RNA; OTU, operational taxonomic unit
Reduction in CCL17 and other Type 2 biomarkers
Dupilumab led to a rapid drop in the type 2 chemokine, CCL17 (TARC) in the serum, which was significantly different from the placebo group by day 7 (p=0.04; Fig 5A). CCL17 dropped precipitously and significantly (p<0.001) through day 21 and then more slowly thereafter. Noting that the trajectory for CCL17 reduction mirrored that observed for S. aureus CFU abundance in lesional skin (Fig 3C), we examined the correlation between reduction in serum TARC in both dupilumab- and placebo-randomized participants and their log reductions in lesional S. aureus CFUs at early timepoints. Only the dupilumab-treated participants demonstrated greater reductions in this type 2 serum biomarker correlated with greater reductions in lesional S. aureus, which was most significant (p=0.01) at day 14 but was seen as early as day 7 (p=0.04; Fig 5B).
Figure 5. Reductions in type 2 serum and tissue biomarkers as a function of dupilumab treatment.
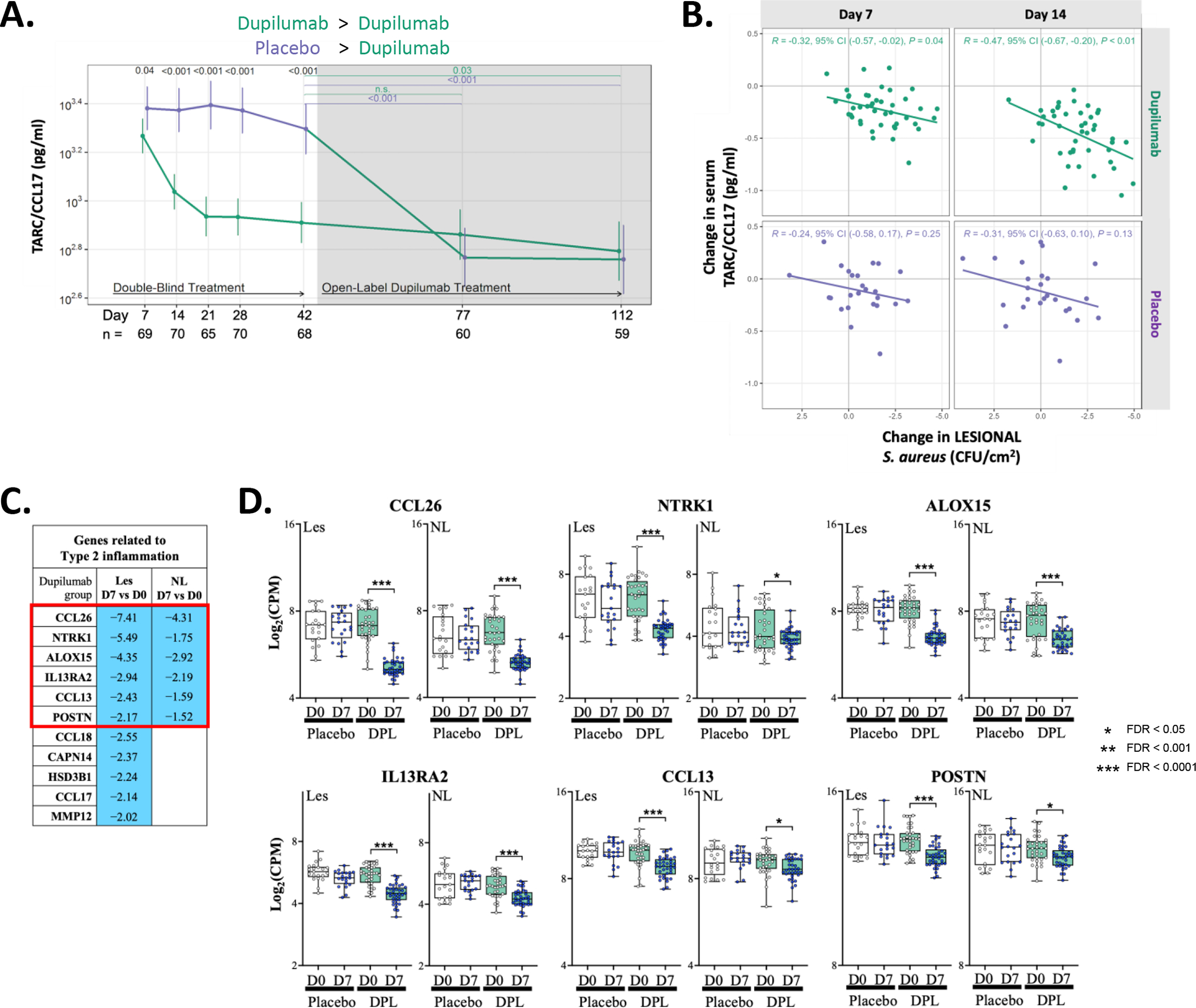
(A) Longitudinal changes in serum TARC/CCL17 during RDBPC and OLE phases of the study. The number of participants with evaluable data at each timepoint are noted below the X axis (with the total population denoted in black, dupilumab-randomized participants in green and placebo-randomized in purple). (B) Reductions in serum CCL17 significantly correlated with reductions in S. aureus lesional CFU abundance at both day 7 (p=0.04) and day 14 (p=0.01) in dupilumab-treated participants but there was no correlation observed at either timepoint in placebo-treated participants. (C) Differentially expressed genes (DEGs) related to type 2 inflammation (with a FDR of <0.05 and with a fold change > 2.0 in lesional skin) is shown comparing day 7 to day 0 in the dupilumab-treated group. Blue indicates reduced expression at day 7 in the dupilumab-treated group. (D) Graphical illustration of the genes reduced in both Les and NL skin (Red Box in [C]). Placebo data is shown in open box plots and dupilumab in shades of green.
CFU, colony forming units; FDR, false discovery rate; les, lesional skin; NL, nonlesional skin
Several additional serum biomarkers were measured; soluble CD25 (sIL-2R), LDH, NGAL (lipocalin-2) and calprotectin (heterodimer S100A8/S100A9) (Fig E4). Dupilumab treatment resulted in a drop in sCD25 and LDH (p=0.03) at day 42, but no changes were observed in acute inflammation and neutrophil activation markers, NGAL or calprotectin, which were only measured at early timepoints.27–31
To better understand dupilumabs impact on the skin transcriptome, we performed RNA-Seq collected from skin biopsies of lesional (dupilumab n=36/37 and placebo n=21/21, day 0/7; respectively) and non-lesional skin (dupilumab n=34/36 and placebo n=21/20, day 0/7; respectively) at study entry (day 0) and at day 7. Table E3 lists the top 50 differentially expressed genes (DEGs; up and down-regulated as a function of dupilumab treatment) identified by DESeq2 analysis (FC≥1.25; FDR <0.05). If we use a (FC)>2.0 we observe that of the 27 downregulated DEGs in dupilumab-treated lesional skin, eleven (or 40.7%) were genes related to type 2 inflammation, including CCL17 (Fig 5C), and six of these (CCL26/eotaxin-3, NTRK1, ALOX15, IL13RA2, CCL13/MCP-4 and POSTN) were also reduced in non-lesional skin, although not all at FC≥2. The expression of these six genes are shown in Fig 5D for days 0 and 7. The 16 additional downregulated DEGs from lesional skin biopsies (day 7) were AC243829.4, CERS1, COL6A5, COL6A6, DRAIC, FAM124B, FRMD5, HSD3BP4, LRAT, LURAP1L, MMP3, PPP1R3C, SLC16A14, SLC5A5, SLC9A3 and TREML2.
Transcriptomic DEG and pathway analysis relevant for S. aureus host responses
To further investigate mechanisms underlying the dramatic drop in S. aureus observed by day 3 in dupilumab-treated participants, we examined expression of different functional gene classes relevant for establishment and/or persistence of S. aureus skin colonization (Fig 6A) at the first post-baseline biopsy timepoint, day 7. These include genes 1) with antimicrobial activity, 2) that are innate immune receptors and signaling partners, 3) that are relevant for the Th17 pathway and/or neutrophil production/function, 4) host ligands for microbial surface components recognizing adhesive matrix molecules (MSCRAMMs), 5) found in the complement system and 6) epidermal barrier genes. Some of the antimicrobial peptides (AMPs) thought to be active against S. aureus were either not affected (DEFA1, DEFA1B, DEFA3, DEFA4, DEFA5, DEFA6, DEFB1, DEFB4A, DEFB103B) by dupilumab treatment or paradoxically had reduced expression (CAMP). Only CCL20/MIP-3α and LYZ were upregulated in both lesional and non-lesional skin after dupilumab treatment. Of the TLR and related genes, only TLR7 was upregulated in both lesional and non-lesional skin at day 7. Five Th17/neutrophil related genes (CLEC5A, CX3CR1/Fractalkine Receptor, VN1, ITGAL and LYZ) were upregulated in both lesional and non-lesional skin of dupilumab participants. Twenty-four additional Th17/neutrophil genes were also upregulated with dupilumab treatment, but only in non-lesional skin. LCN2 (lipocalin), also known as NGAL, was elevated in non-lesional skin (day 7 compared to day 0), but we did not observe increases in sera at this same timepoint (day 7; Fig E4C). Similarly, we observed increases in S100A8 and S100A9, which dimerize to form the alarmin calprotectin. We did not observe significant changes in serum calprotectin after dupilumab treatment at early timepoints (days 7 and 14; Fig E4D). There was little change in 21 host ligands for MSCRAMMs at Day 7; however, 7/30 complement genes were increased in lesional (n=5) or non-lesional (n=2) skin with dupilumab treatment. Lastly, very few barrier genes of relevance for AD were modulated by dupilumab treatment with only SERPINB9 (a serine proteinase inhibitor) increasing in both lesional and non-lesional skin samples.
Figure 6. Transcriptomics changes in lesional and non-lesional skin in response to dupilumab treatment.
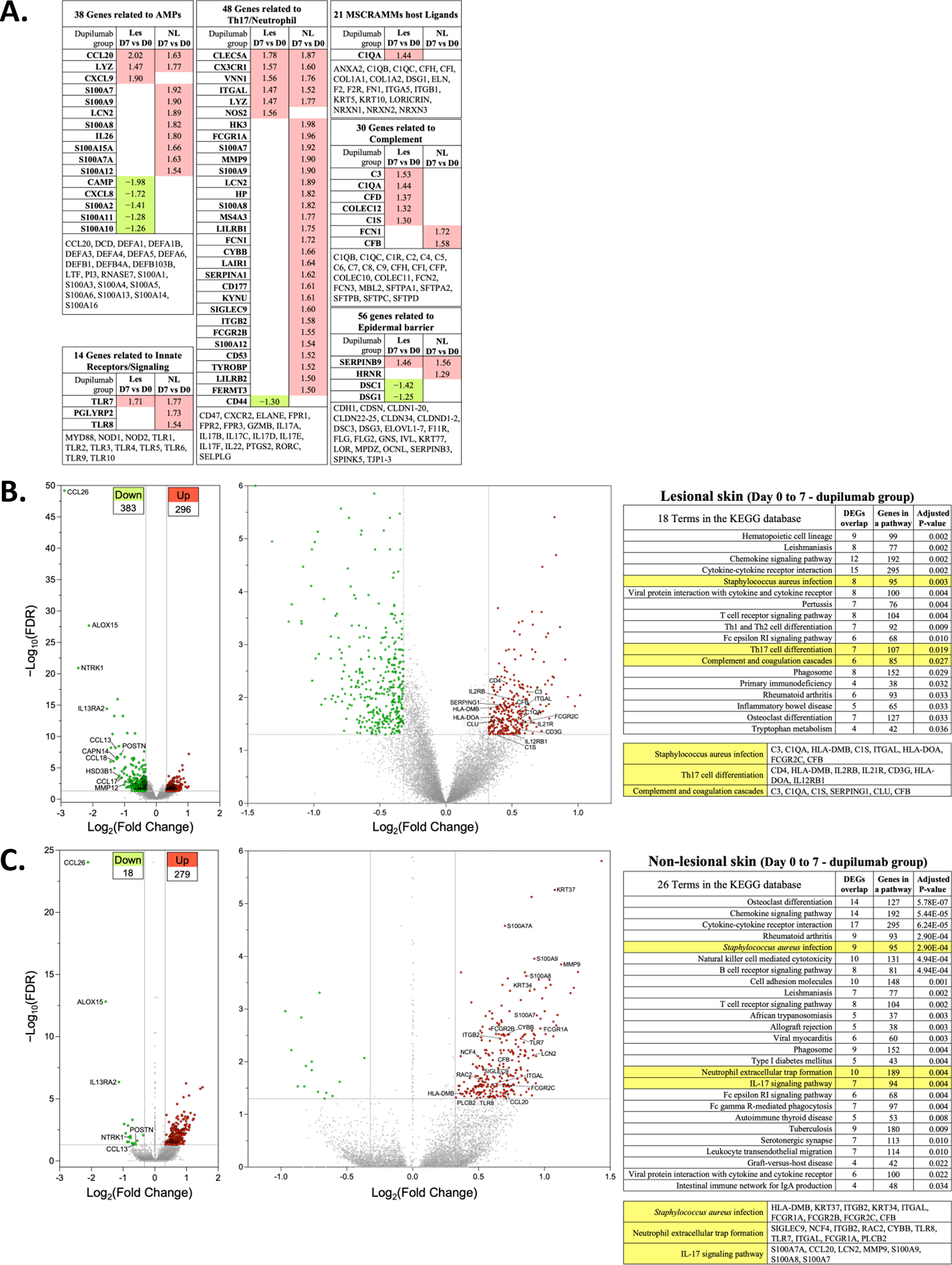
(A) DEGs with FDR of <0.05 and a log FC >1.25 in les and NL skin are shown. They are separated into functional groups: antimicrobial proteins (AMPs), Innate Receptors/Signaling, Th17/Neutrophil, microbial surface components recognizing adhesive matrix molecules (MSCRAMMs), complement and epidermal barrier. The genes listed at the bottom of each of these are those that are not significantly altered in their expression. (B) Volcano plot showing increased (n=296) or decreased (n=383) DEG from lesional skin of dupilumab treated subjects (Day 7 vs Day 0). The graphs show log2 FC in gene expression of lesional skin (Day 7) over lesional skin (Day 0) plotted against negative log10 FDR. (C) Volcano plot showing upregulated (n=279) or downregulated (n=18) DEGs from non-lesional skin of dupilumab treated subjects (Day 7 vs Day 0). Genes represented in red are upregulated by >1.25-fold in Day 7 lesional or non-lesional skin (FDR < 0.05). Genes represented in green are downregulated by >1.25-fold in Day 7 lesional or non-lesional skin (FDR < 0.05). The left most volcano plot is all of the DEG and the right most volcano plot reduces the y axis to enable labelling of some of the upregulated DEGs. The table lists the KEGG pathways with a few highlighted in yellow because of their potential relevance to dupilumab-induced S. aureus reductions. We list the DEGs in these pathways that are dysregulated in our samples. DEG, differentially expressed genes; FDR, false discovery rate; les, lesional skin; NL, nonlesional skin; KEGG, Kyoto Encyclopedia of Genes and Genomes
The Day 7 DEGs identified in lesional and non-lesional skin, with significant differences between dupilumab (or placebo) groups on day 7 compared to day 0 (FDR<0.05 and FC>1.25) were functionally annotated using the KEGG database.32–34 Volcano plots of lesional skin, highlight 296 upregulated and 383 downregulated DEGs in the dupilumab group (Fig 6B) and 279 upregulated and only 18 downregulated DEGs in non-lesional skin (Fig 6C). (The top 50 up- and down-regulated DEGs based on an FDR value were listed in Table E3). The upregulated DEGs were enriched in 18 pathways in lesional and 26 pathways in non-lesional skin (Fig 6B and C). Pathways that might suggest a mechanism by which dupilumab reduces S. aureus are shaded in yellow. Both lesional and non-lesional were enriched for genes in the S. aureus infection pathway. Lesional was also enriched for Th17 cell differentiation and complement and coagulation cascade pathways and non-lesional for neutrophil extracellular trap formation and IL-17 signaling pathway.
Changes in skin barrier function
There was a modest reduction in basal lesional TEWL in the dupilumab group at day 42 (6 weeks), with no further improvement when this group received 10 additional weeks of OLE treatment (Fig 7A). Although the dupilumab group appeared to have a significant reduction in lesional TEWL at day 3, this was not seen at subsequent timepoints (day 7, 14, 21 or 28). The group, initially randomized to placebo, was found to have reductions in TEWL after 10 weeks of dupilumab OLE treatment (day 112). As noted earlier, the placebo group had higher lesional TEWL on study entry than the dupilumab group (p=0.058; Table E1) but this was accounted for statistically.
Figure 7. Dupilumab treatment modestly affects lesional and non-lesional barrier function at later timepoints.
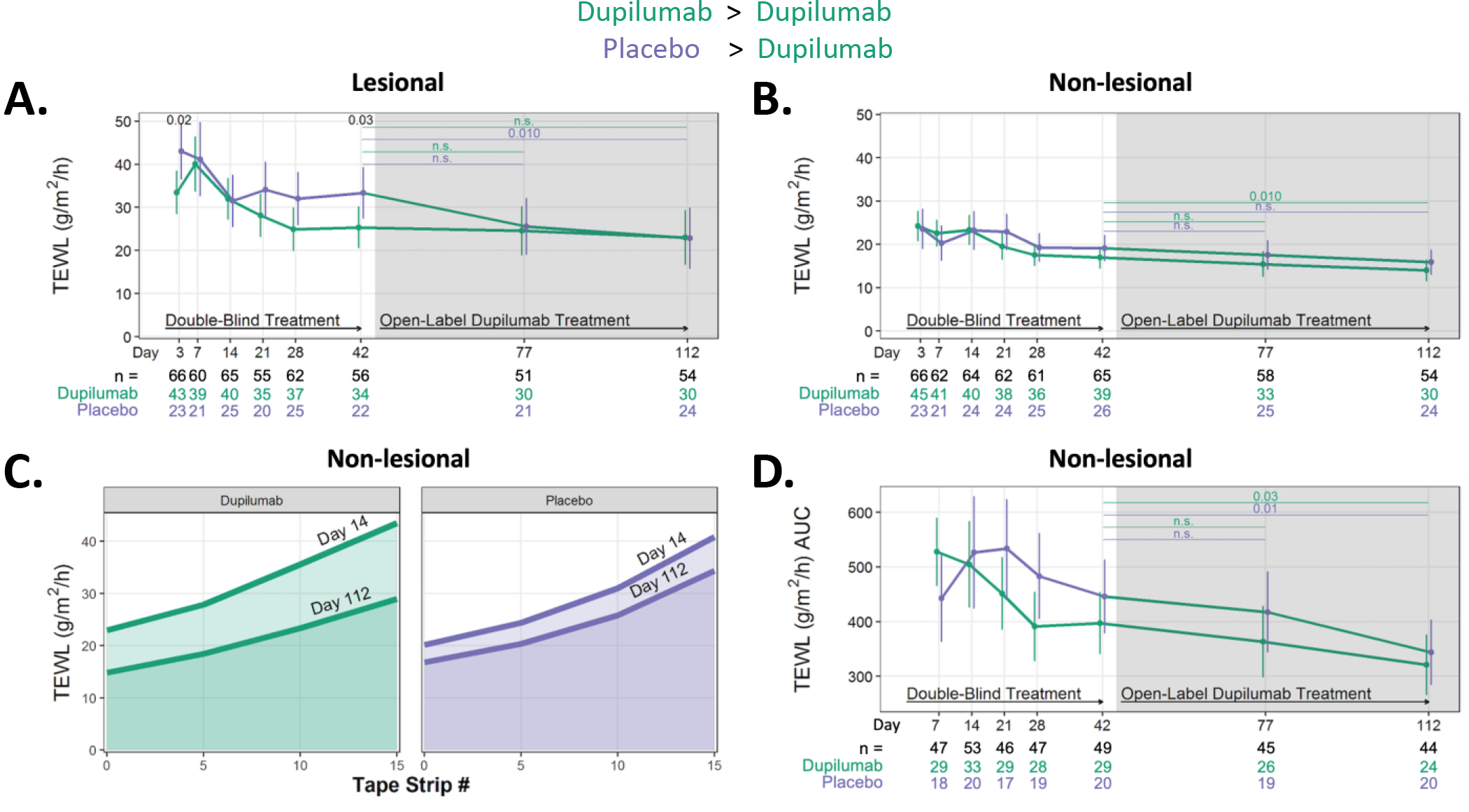
(A) Changes in basal TEWL measurements in les and (B) NL skin sites. (C) The mean TEWL values obtained at NL skin site at Days 14 and 112 following five, ten and fifteen tape strips are shown for the dupilumab (green) and placebo (purple)-randomized populations to visualize how the AUC is calculated. (D) The longitudinal changes in TEWL AUC at NL skin sites.
AUC, area under the curve; les, lesional skin; NL, nonlesional skin; TEWL, transepidermal water loss.
Not surprisingly, TEWL was higher in lesional than non-lesional skin on study entry, indicating greater barrier dysfunction in AD lesions (Table E1). Non-lesional skin barrier integrity was measured using two approaches, basal TEWL (as was done for lesional skin; Fig 7B) as well as by the stratum corneum (SC) integrity assay (Fig 7C and D). There were no differences in basal TEWL between the two treatment groups during the double-blind phase of the study; moreover, the placebo group did not experience a change in TEWL even during OLE (day 42 to 112) (Fig 7B). The dupilumab-randomized group experienced a reduction in TEWL from day 42 to day 112, suggesting that even the more modest barrier defect in non-lesional (compared to lesional) skin can improve with longer dupilumab treatment (≥ 6 weeks). AUC is graphically depicted in Fig 7C. The only significant reduction in AUC was observed in the dupilumab and placebo groups when comparing their day 42 (6 week) value to day 112 (16 weeks).
Changes in IL-17 CD4+ cell clusters in PBMC
In addition to Th17 cell-derived cytokines, which induce expression of neutrophil chemoattractants and AMPs,35–37 IL-26 released by Th17 cells has a direct antimicrobial activity against S. aureus.38 Therefore, we investigated the possible role of peripheral blood Th17 cells in S. aureus reductions with dupilumab therapy.
The clinical response to dupilumab varied among participants. Thus, to facilitate comparisons between the number of cells in the IL-17A high (IL-17+) sub-population and the cutaneous bacterial load, the clinical responses were assessed by calculating the slope (by linear regression) of disease severity scores (SCORAD) in dupilumab-treated participants from day 0 to day 42. The flow gating strategy used unsupervised SWIFT clustering to isolate cell populations more precisely than can be achieved by cell gating (Fig. E5A).39, 40 Participants with SCORAD slopes below −0.7 were classified as high responders (21 participants; Fig E5B [red]), and above −0.7 were classified as low responders (24 participants; Fig, E5B [black]). The number of cells in five IL-2lo, IL-17+ T cell clusters generally increased from day 0 to 14, in the high responder, but not in the low responder or placebo groups (Fig E5B). However, these differences were not significant after Benjamini-Hochberg 41 correction.
These clusters were then compared to the S. aureus CFUs on skin swabs taken from lesional or non-lesional skin at different time points in the dupilumab-treated group. The changes in frequency of these five IL-2lo, IL-17+ clusters were compared to the levels of S. aureus CFUs in skin swabs at all time points from 0 to 112 days. Fig E5C shows the Spearman correlation coefficients for each comparison of changes in a Th17 cluster and S. aureus burden at all timepoints. Star symbols represent significant values (p<0.05). Bacterial counts in non-lesional skin showed similar trends (Fig E5C [bottom row]), but were not as significant, possibly due to wide variability among non-lesional skin sites and their lower S. aureus burden on study entry. The change in the proportion of cluster 1584 (day 0 to 14) correlated significantly with CFU values (lesional skin) at days 3, 7, 14 and 21, in high responder dupilumab participants; this cluster increased more in participants with higher bacterial loads at these timepoints. Compared to other IL-17+ clusters, 1584 expressed higher levels of CLA and Ki-67, and lower levels of IL-2, GM-CSF, and IFN-γ (Fig E5D). This may represent a skin-homing, recently-proliferated Th17 population that is elevated because of the higher levels of S. aureus. The normalized cell counts for the twelve Th17 clusters in all samples can be found in Table E4. As the T cells were evaluated from day 0 to 14, it is not surprising that the correlations with bacterial load are most obvious at earlier timepoints.
In contrast, increases in cluster 1583 were correlated with increased S. aureus counts in dupilumab-treated participants who did NOT respond strongly to dupilumab (SCORAD), but did not change significantly in either placebo or high responder dupilumab participants. This correlation was seen with S. aureus counts at days 3, 7, 14, 21 and 28. Cluster 1583 expressed similar markers to 1584, except for lower CLA and Ki-67 (Fig E5D). Thus, dupilumab induced changes in PBMC sub-populations with the potential to produce IL-17, and these changes were correlated with the S. aureus counts on skin swabs at multiple time points.
DISCUSSION
This study found that all moderate to severe AD subjects were colonized by S. aureus, a bacterium known to worsen AD inflammation and increase the likelihood of infections.9 Inhibition of IL-4 and IL-13 signaling, achieved with dupilumab treatment resulted in a remarkable decrease in S. aureus colonization after only 3 days reaching a stable nadir after 28 days. Clinical and deep endotyping (skin microbial ecology, transcriptome, serum biomarkers, functional barrier assessments and PBMC immunoprofiling) of AD participants identified multiple molecular changes occurring at the host-microbe interface that may explain the early changes in S. aureus abundance and virulence factor production, including increased expression of neutrophil-, IL-17-, and complement-related genes, increased relative abundance of beneficial bacteria (observed in setting of greater microbial diversity)42 and increased clusters of skin-homing IL-17+ CD4+ cells in the circulation. These findings confirm the pleiotropic effects of type 2 cytokines in promoting AD comorbidities and highlight that inhibition of IL-4 and IL-13 enhances the host’s ability to fight S. aureus likely through a variety of mechanisms. Combined, dupilumab’s effects on the host-microbe interface substantially contribute to the clinical improvement and likely the reduction in bacterial skin infections observed in pivotal clinical trials. 22, 43
While S. aureus colonization rates are known to be elevated in patients with AD, our study found 100% S. aureus colonization in the patient population. Previous studies reported colonization rates between 28–99% on lesional skin.44 The higher rates in our study cannot be explained by geography and climate given that enrollment of patients was distributed across 8 different regions within the US (Fig 1A). The higher rates found in our study are likely due to the AD severity of our population and methodologic improvements in S. aureus detection. S. aureus was detected using two methods, qPCR and viable CFUs, performed in the laboratory of a S. aureus researcher (P.M.S.), which eliminated inter-laboratory variation. Mannitol salt plates were retained for longer than most laboratories because some S. aureus strains, grow slowly due to intrinsic metabolism differences or normal flora slowing their growth. Bright pink colonies on mannitol salt plates are often considered coagulase-negative staphylococci, but in our study some of these were catalase+, coagulase+, defining S. aureus. These data suggest that diagnostic microbiology laboratory assessments of S. aureus may underestimate the true abundance unless they include catalase and coagulase assays.
Dupilumab therapy not only reduced S. aureus colonization but also reduced virulence factor production. Cytotoxins quantified from skin swabs were significantly reduced while we observed no changes in the quantity of S. aureus superantigens, proteases or lipase. Cytotoxins, including α, β, γ, δ, ε, and phenol-soluble modulins are known initiators of inflammation.45–48 Furthermore, Type 2 cytokines enhance the necrotic responses to cytotoxins.49 Cytotoxins have been implicated in the maintenance of AD skin lesions, thus lending clinical credibility to our cytotoxin findings.11, 50 Superantigens have been widely implicated as potential mechanisms to explain AD flares, making it surprising to see no change in their levels with dupilumab therapy.51–53 We believe this is due to the fact that S. aureus superantigens can remain in tissue for up to 3 to 4 months. 15
The changes we observed in the microbial composition as a function of dupilumab treatment are consistent with previous studies, albeit our data identifies these changes much earlier (3 days) than previously realized (4 weeks).20, 54, 55 Dupilumab and tralokinumab, in contrast to less targeted anti-inflammatory therapies (i.e. azathioprine, mycophenolate, topical corticosteroids), have been shown to increase microbial diversity.55 We observed a decrease in the relative abundance of S. aureus and an increase in both coagulase-negative commensals (S. epidermidis, S. hominis and S. lugdunensis) and other commensals (Micrococcus luteus and Cutibacterium acnes). These bacteria can produce novel lantibiotics that selectively kill S. aureus.56, 57 Further, the ratio of S. epidermidis to S. aureus has been correlated to AD severity.58 Additional ADRN projects show that microbiome transplants utilizing these commensals have shown promise in reducing S. aureus in AD subjects.58
We found additional changes in innate host defenses with dupilumab therapy, starting with the broadest definition; namely skin barrier function. Biophysical measures of barrier function modestly improved with dupilumab, which was only observed in lesional skin after six weeks of treatment. This modest and delayed skin barrier improvement appears to contrast with some published literature which reported improvements in TEWL AUC by as early as two weeks.59 This may be explained by the fact that earlier reports were from non-randomized and often uncontrolled studies and compared barrier measures to baseline values. Notably, when we evaluated the change in lesional TEWL focusing only on the dupilumab group, we observed a significant reduction as early as day 14 (p=0.048) which became more significant with further treatment (Table E5; Fig 7A). This highlights the importance of a blinded, placebo-controlled study design as you can see that our placebo group also experienced improvements in barrier function during the RDBPC phase of the study. The somewhat delayed barrier improvement seen in our blinded and controlled study suggests that IL-4/-13 diminish barrier function indirectly; possibly by reducing S. aureus abundance and its cytotoxin production.60–62 Additionally, we and others have shown that S. aureus derived superantigens promote epidermal barrier dysfunction in vitro, but because superantigens can persist in tissues long after S. aureus bioburden is reduced – might provide another explanation for the delayed effect on barrier recovery in our participants (Fig E2).19, 63
While skin permeability is a gross measure of epidermal function, reductions in AMPs have been the prevailing hypothesis to explain AD susceptibility to S. aureus. Ong, et al. and others have reported decreased AMP (LL-37/CAMP and HBD2/DEFB4) expression in the lesional skin of AD, although some studies found elevated levels of AMPs in lesional skin64 and serum.65 Our transcriptomic analyses did not observe increased expression in the genes encoding for LL-37 or the defensin family (alpha or beta) members in the dupilumab-treated participants, but did demonstrate increases in other genes with anti-S. aureus actions (CCL20, LYZ, CXCL9, calprotectin, LCN2 and IL-26).66–70
With multi-parameter flow cytometry of SEB-stimulated PBMCs, we observed an increase in several IL-17+ IL-2lo CD4+T cell clusters at day 14 (compared to day 0) in the dupilumab-treated group. For some clusters, the magnitude of this increase correlated directly with the S. aureus counts in skin swabs, possibly because the persistence of S. aureus in some subjects may have led to continued stimulation and expansion of Th17 cells. Conversely, the rapid reduction of S. aureus counts as seen at day 3 may have led to a reduction in stimulation of Th17 cells. It is also possible that Th17 cells in the circulation are depleted by migration into the skin and deployment of their anti-S. aureus effector function, resulting in reduced S. aureus loads in subjects with lower Th17 cells in the circulation.
Recently, Leyva-Castillo, et al. found dupilumab’s ability to reduce S. aureus in a mouse model of AD was dependent on Th17 responses (Leyva-Castillo; manuscript submitted). We observed correlations between specific IL-17-producing CD4+T cell clusters and S. aureus load on the skin. Our transcriptomic data collected at much earlier timepoints (day 7) than earlier studies (day 28),71–73 suggest that dupilumab’s anti-S. aureus effects may be due to PMN-mediated killing. PMNs, critical for innate immunity, have downstream effects on S. aureus through the release of lysozyme (down-regulation of virulence factors) and complement activation (intracellular killing), both of which were up-regulated in our studies. Importantly, neutrophils have functional type I and II IL-4 receptors, and their signaling reduces a number of neutrophil effector functions such as chemotaxis and neutrophil extracellular trap (NET) formation.74 We hypothesize that this rapid dupilumab-mediated reduction in S. aureus is a consequence of the reducing the inhibitory actions of IL-4 and IL-13 on neutrophil functions; combined with enhanced Th17 mediated biology.75, 76 In fact, it has been hypothesized that dupilumab treatment may disturb the balance in the type 2 (IL-4 & IL-13)/type 3 (IL-17) axis77 and this may explain the reports of psoriasis and enthesitis, which are IL-17-mediated inflammatory diseases infrequently arising in dupilumab-treated AD patients.78–80
Lastly, we identified other changes in adaptive immunity with dupilumab therapy from our serum and peripheral blood immunophenotyping studies which help to explain our findings. The type 2 biomarker, CCL17/TARC was significantly decreased in skin (mRNA) and blood, and the reduction of serum CCL17/TARC levels correlated with the reduction in S. aureus. We previously showed a strong relationship between S. aureus colonization and serum CCL17/TARC and now confirm a direct linear relationship between the two endpoints in dupilumab-treated participants. Consistent with our observations, three children with the autosomal dominant hyper-IgE syndrome (caused by a mutation in STAT3), achieved good clinical improvement in their severe AD-like dermatitis, which was seen in the context of reduced in skin infections.81 Whether an expansion of skin-homing Th17 cells and activated neutrophils, critical for host defense against S. aureus, were responsible for this clinical improvement, was not addressed.
In summary, in our RDBPC trial, we report the novel discovery of dramatic changes in S. aureus colonization and virulence factor production after only 3 days of dupilumab therapy. The early reduction in S. aureus abundance (lesional skin) correlated with improvements in all measures of AD severity except itch. The magnitude of S. aureus reduction correlated with reductions in the type 2 biomarker, CCL17/TARC. A Cochrane review concluded that there is no clear benefit from commonly used antistaphylococcal interventions,82 but our study would suggest that therapies that can both reduce S. aureus abundance and reduce type 2 immunity may be highly beneficial. In other words, only reducing S. aureus may not provide sufficient clinical benefit. Our data suggest that this S. aureus reduction may be in part due to enhanced innate immune responses and IL-17 responses with downstream recruitment and activation of PMNs leading to S. aureus killing possibly through up-regulation of lysozyme and complement. Physiological measures of skin barrier improved more slowly and were not significantly different than placebo-treated subjects until 42 days, suggesting that this improvement might be in part due to changes in S. aureus. The extensive biosampling of multiple cutaneous and immune compartments allowed for a novel and comprehensive investigation into the impact dupilumab exerts on the host-microbe interface. Future studies are needed to determine whether S. aureus reduction persists upon treatment withdrawal. It will also be important to study whether S. aureus reductions in other type 2 driven diseases may also contribute to the clinical benefit seen with dupilumab treatment.
Supplementary Material
Clinical Implications:
Dupilumab leads to significant and rapid reductions in Staphylococcus aureus abundance (day 3) on atopic dermatitis skin. This reduction correlates with all metrics of AD improvement except itch.
ACKNOWLEDGEMENTS
The authors would like to acknowledge the helpful discussions with Wendy Davidson, MD, Joy Laurienzo-Panza, RN, BSN (NIAID) and Raif Geha, MD, PhD (Harvard) as well as assistance of Rebecca Field (OHSU), Miguel Villareal (Rho, Inc), Jonathan Rebhahn (URMC). A big thanks to our coordinators/Co-investigators who worked tirelessly to identify and enroll our participants (Alicia Papalia, Ashleigh Daniels, JoAnne Van Buskirk, Abigail Franco [URMC]; Kanwaljit Brar, Caroline Bronchick, Jessica Sussman, Joanne Streib, Marco Ramirez-Gama, Shannon Garcia, Shirley Palombi, Susan Leung, Trish Taylor [NJH], Christina Armstrong, Anna Ward [OHSU]; Mary Bohannon [UF]; Faiza Shafiq [UCSD]; Shelby Sparks, Josh Bryer, Anja Jones, Michelle Hampton [UPENN]; : Hanh Do, Katie Sum, Kristoffer Thordarson, Melissa Jenkins, Michelle Kim [Stanford]; and Elvira Lopez, Sydney Brown [CHLA]). Our biggest thanks to the participants who were so patient with the relatively long trial visits. The authors would also like to acknowledge Nicole Meiklejohn for her outstanding assistance preparing this manuscript.
Funding:
This work was supported by the National Institute of Allergy and Infectious Diseases (NIAID). Funding Mechanism: Grant 1 U19 AI117673-01; 1UM1AI151958 (ADRN Investigators), DAIT SACCC Grant Number: 1UM2AI117870-01 (Rho, Inc) and U19AI117673 (LAB). This work was also supported by the National Center for Advancing Translational Sciences (NCATS) Colorado CTSA Grant Number UL1 TR002535.
Abbreviations:
- AD
atopic dermatitis
- ADRN
Atopic Dermatitis Research Network
- CBC
complete blood count
- AEC
absolute eosinophil count
- CHLA
Children’s Hospital of Los Angeles
- CRF
case report form
- CSC
clinical study consortium
- DEG
differentially expressed genes
- DAIT
Division of Allergy, Immunology and Transplantation
- EASI
Eczema Area and Severity Index
- EDC
electronic data capture
- FC
fold change
- FDR
false discovery rate
- IgE
immunoglobulin E
- IRB
institutional review board
- KEGG
Kyoto Encyclopedia of Genes and Genomes
- MSCRAMM
microbial surface components recognizing adhesive matrix molecules
- NESS
Nottingham Eczema and Severity Score
- NGAL
Neutrophil gelatinase-associated lipocalin
- NIAID
National Institute of Allergy and Infectious Diseases
- NIH
National Institutes of Health
- NJH
National Jewish Health
- NL
non-lesional skin
- NRS
numerical rating scale
- OHSU
Oregon Health & Science University
- OLE
Open Label Extension
- OTU
operational taxonomic unit
- PI
principal investigator
- rRNA
ribosomal ribonucleic acid
- SACCC
statistical and clinical coordinating center
- SCORAD
SCORing Atopic Dermatitis
- STAN
Stanford Medical Center
- UCSD
University of California San Diego
- UF
University of Florida
- UPENN
University of Pennsylvania
- URMC
University of Rochester Medical Center
- vIGA
validated Investigator Global Assessment
Footnotes
Conflicts of Interest: ADB – Consultant for dMed Biopharmaceutical Co, AbbVie, Sanofi-Aventis; Investigator for Kiniksa, Novartis, Incyte, and Pfizer. ELS – Consultant for AbbVie, Amgen, Arena Pharmaceuticals, Aslan Pharma, Boston Consulting Group, Collective Acumen, LLC (CA), Dermira, Eli Lilly, Evidera, Excerpta Medica, Forte Bio RX, Galderma, GlaxoSmithKline, Incyte, Janssen, Kyowa Kirin Pharmaceutical Development, Leo Pharm, Medscape LLC, Merck, Pfizer, Physicians World LLC, Regeneron, Roivant, Sanofi-Genzyme, Trevi therapeutics, Valeant, WebMD. Investigator for AbbVie, Amgen, Arcutis, Aslan, Castle Biosciences, Corevita, Dermavant, Dermira, Eli Lilly, Incyte, Kyowa Hakko Kirin, Leo Pharmaceuticals, Merck, Novartis, Pfizer, Regeneron, Sanofi, TARGET-DERM, Tioga, and Vanda. JK – Investigator and Consultant for Eli Lilly and Arena. Investigator for Abbvie and Regeneron. Consultant for Pfizer. MB - Investigator for Regeneron, Incyte. Consultant for Regeneron, Sanofi-Genzyme, Abbvie, Leo, Lilly, Pfizer, Janssen. PYO -Consultant for Incyte, Abbvie, Janssen; Investigator for Regeneron, Sanofi Genzyme, Leo, Incyte. DYML - Consultant for Genentech, LEO Pharma, and Incyte; Research grants from Sanofi Genzyme. LAB - Consultant for Allakos, Arena Pharmaceuticals, DermTech, Evelo Biosciences, Galderma, Incyte, Janssen, LEO Pharma, Merck, Numab Therapeutics, Pfizer, Rapt Therapeutics, Regeneron, Ribon Therapeutics, Sanofi/Genzyme, Sanofi-Aventis, Stealth BioTherapeutics, Trevi Therapeutics, Union Therapeutics and Xencor. DMC member – Novartis. Investigator for Abbvie, Astra-Zeneca, DermTech, Kiniksa, Pfizer, Regeneron, Ribon Therapeutics and Sanofi. TH - Investigator, Consultant for Eli Lilly and Arena. Investigator for Abbvie and Regeneron. Consultant for Pfizer. PMS, TY, SL, ZF AC, ALRS, MP, SAQ, SHK, LP, AG, GD, TM and SRG – No relevant conflicts of interest.
AKRS & MP-AKRS’ and MP’s co-authorship of this publication does not necessarily constitute endorsement by the National Institute of Allergy and Infectious Diseases, the National Institutes of Health or any other agency of the United States government.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Boguniewicz M, Leung DY. Recent insights into atopic dermatitis and implications for management of infectious complications. J Allergy Clin Immunol. 2010;125(1):4–13; quiz 4–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wang V, Boguniewicz J, Boguniewicz M, Ong PY. The infectious complications of atopic dermatitis. Ann Allergy Asthma Immunol. 2021;126(1):3–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Beck LA, Boguniewicz M, Hata T, Schneider LC, Hanifin J, Gallo R, et al. Phenotype of atopic dermatitis subjects with a history of eczema herpeticum. J Allergy Clin Immunol. 2009;124(2):260–9, 9.e1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Leung DY, Gao PS, Grigoryev DN, Rafaels NM, Streib JE, Howell MD, et al. Human atopic dermatitis complicated by eczema herpeticum is associated with abnormalities in IFN-γ response. J Allergy Clin Immunol. 2011;127(4):965–73.e1–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mathias RA, Weinberg A, Boguniewicz M, Zaccaro DJ, Armstrong B, Schneider LC, et al. Atopic dermatitis complicated by eczema herpeticum is associated with HLA B7 and reduced interferon-γ-producing CD8+ T cells. Br J Dermatol. 2013;169(3):700–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bin L, Edwards MG, Heiser R, Streib JE, Richers B, Hall CF, et al. Identification of novel gene signatures in patients with atopic dermatitis complicated by eczema herpeticum. J Allergy Clin Immunol. 2014;134(4):848–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Broccardo CJ, Mahaffey S, Schwarz J, Wruck L, David G, Schlievert PM, et al. Comparative proteomic profiling of patients with atopic dermatitis based on history of eczema herpeticum infection and Staphylococcus aureus colonization. J Allergy Clin Immunol. 2011;127(1):186–93, 93.e1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gao L, Bin L, Rafaels NM, Huang L, Potee J, Ruczinski I, et al. Targeted deep sequencing identifies rare loss-of-function variants in IFNGR1 for risk of atopic dermatitis complicated by eczema herpeticum. J Allergy Clin Immunol. 2015;136(6):1591–600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Simpson EL, Villarreal M, Jepson B, Rafaels N, David G, Hanifin J, et al. Patients with Atopic Dermatitis Colonized with Staphylococcus aureus Have a Distinct Phenotype and Endotype. J Invest Dermatol. 2018;138(10):2224–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Spaulding AR, Salgado-Pabón W, Kohler PL, Horswill AR, Leung DY, Schlievert PM. Staphylococcal and streptococcal superantigen exotoxins. Clin Microbiol Rev. 2013;26(3):422–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Moran MC, Cahill MP, Brewer MG, Yoshida T, Knowlden S, Perez-Nazario N, et al. Staphylococcal Virulence Factors on the Skin of Atopic Dermatitis Patients. mSphere. 2019;4(6). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bin L, Kim BE, Brauweiler A, Goleva E, Streib J, Ji Y, et al. Staphylococcus aureus α-toxin modulates skin host response to viral infection. J Allergy Clin Immunol. 2012;130(3):683–91.e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schlievert PM, Case LC, Strandberg KL, Abrams BB, Leung DY. Superantigen profile of Staphylococcus aureus isolates from patients with steroid-resistant atopic dermatitis. Clin Infect Dis. 2008;46(10):1562–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schlievert PM, Roller RJ, Kilgore SH, Villarreal M, Klingelhutz AJ, Leung DYM. Staphylococcal TSST-1 Association with Eczema Herpeticum in Humans. mSphere. 2021;6(4):e0060821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Davis CC, Kremer MJ, Schlievert PM, Squier CA. Penetration of toxic shock syndrome toxin-1 across porcine vaginal mucosa ex vivo: permeability characteristics, toxin distribution, and tissue damage. Am J Obstet Gynecol. 2003;189(6):1785–91. [DOI] [PubMed] [Google Scholar]
- 16.Merriman JA, Mueller EA, Cahill MP, Beck LA, Paller AS, Hanifin JM, et al. Temporal and Racial Differences Associated with Atopic Dermatitis Staphylococcus aureus and Encoded Virulence Factors. mSphere. 2016;1(6). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fischer AJ, Kilgore SH, Singh SB, Allen PD, Hansen AR, Limoli DH, et al. High Prevalence of Staphylococcus aureus Enterotoxin Gene Cluster Superantigens in Cystic Fibrosis Clinical Isolates. Genes (Basel). 2019;10(12). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schlievert PM, Osterholm MT, Kelly JA, Nishimura RD. Toxin and enzyme characterization of Staphylococcus aureus isolates from patients with and without toxic shock syndrome. Ann Intern Med. 1982;96(6 Pt 2):937–40. [DOI] [PubMed] [Google Scholar]
- 19.Moran MC, Pandya RP, Leffler KA, Yoshida T, Beck LA, Brewer MG. Characterization of Human Keratinocyte Cell Lines for Barrier Studies. JID Innov. 2021;1(2):100018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Callewaert C, Nakatsuji T, Knight R, Kosciolek T, Vrbanac A, Kotol P, et al. IL-4Ralpha Blockade by Dupilumab Decreases Staphylococcus aureus Colonization and Increases Microbial Diversity in Atopic Dermatitis. J Invest Dermatol. 2020;140(1):191–202 e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Canonica GW, Bourdin A, Peters AT, Desrosiers M, Bachert C, Weidinger S, et al. Dupilumab Demonstrates Rapid Onset of Response Across Three Type 2 Inflammatory Diseases. J Allergy Clin Immunol Pract. 2022;10(6):1515–26. [DOI] [PubMed] [Google Scholar]
- 22.Paller AS, Beck LA, Blauvelt A, Siegfried EC, Cork MJ, Wollenberg A, et al. Infections in children and adolescents treated with dupilumab in pediatric clinical trials for atopic dermatitis-A pooled analysis of trial data. Pediatr Dermatol. 2022;39(2):187–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Emerson RM, Charman CR, Williams HC. The Nottingham Eczema Severity Score: preliminary refinement of the Rajka and Langeland grading. Br J Dermatol. 2000;142(2):288–97. [DOI] [PubMed] [Google Scholar]
- 24.CDC/NCHS. Healthy weight, overweight, and obesity among U.S. adults. Atlanta, Georgia: Center for Disease Control. [Google Scholar]
- 25.Hon KL, Ma KC, Wong E, Leung TF, Wong Y, Fok TF. Validation of a self-administered questionnaire in Chinese in the assessment of eczema severity. Pediatr Dermatol. 2003;20(6):465–9. [DOI] [PubMed] [Google Scholar]
- 26.Gooderham MJ, Bissonnette R, Grewal P, Lansang P, Papp KA, Hong CH. Approach to the Assessment and Management of Adult Patients With Atopic Dermatitis: A Consensus Document. Section II: Tools for Assessing the Severity of Atopic Dermatitis. J Cutan Med Surg. 2018;22(1_suppl):10S–6S. [DOI] [PubMed] [Google Scholar]
- 27.Jarlborg M, Courvoisier DS, Lamacchia C, Martinez Prat L, Mahler M, Bentow C, et al. Serum calprotectin: a promising biomarker in rheumatoid arthritis and axial spondyloarthritis. Arthritis Res Ther. 2020;22(1):105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Buonafine M, Martinez-Martinez E, Jaisser F. More than a simple biomarker: the role of NGAL in cardiovascular and renal diseases. Clin Sci (Lond). 2018;132(9):909–23. [DOI] [PubMed] [Google Scholar]
- 29.Kamata M, Tada Y, Tatsuta A, Kawashima T, Shibata S, Mitsui H, et al. Serum lipocalin-2 levels are increased in patients with psoriasis. Clin Exp Dermatol. 2012;37(3):296–9. [DOI] [PubMed] [Google Scholar]
- 30.Shi H, Zuo Y, Yalavarthi S, Gockman K, Zuo M, Madison JA, et al. Neutrophil calprotectin identifies severe pulmonary disease in COVID-19. J Leukoc Biol. 2021;109(1):67–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ramos-Mozo P, Madrigal-Matute J, Vega de Ceniga M, Blanco-Colio LM, Meilhac O, Feldman L, et al. Increased plasma levels of NGAL, a marker of neutrophil activation, in patients with abdominal aortic aneurysm. Atherosclerosis. 2012;220(2):552–6. [DOI] [PubMed] [Google Scholar]
- 32.Kanehisa M, Goto S. KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000;28(1):27–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kanehisa M. Toward understanding the origin and evolution of cellular organisms. Protein Sci. 2019;28(11):1947–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kanehisa M, Furumichi M, Sato Y, Ishiguro-Watanabe M, Tanabe M. KEGG: integrating viruses and cellular organisms. Nucleic Acids Res. 2021;49(D1):D545–d51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wilson NJ, Boniface K, Chan JR, McKenzie BS, Blumenschein WM, Mattson JD, et al. Development, cytokine profile and function of human interleukin 17-producing helper T cells. Nat Immunol. 2007;8(9):950–7. [DOI] [PubMed] [Google Scholar]
- 36.Furue M, Furue K, Tsuji G, Nakahara T. Interleukin-17A and Keratinocytes in Psoriasis. Int J Mol Sci. 2020;21(4). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cho JS, Pietras EM, Garcia NC, Ramos RI, Farzam DM, Monroe HR, et al. IL-17 is essential for host defense against cutaneous Staphylococcus aureus infection in mice. J Clin Invest. 2010;120(5):1762–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Meller S, Di Domizio J, Voo KS, Friedrich HC, Chamilos G, Ganguly D, et al. T(H)17 cells promote microbial killing and innate immune sensing of DNA via interleukin 26. Nat Immunol. 2015;16(9):970–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mosmann TR, Naim I, Rebhahn J, Datta S, Cavenaugh JS, Weaver JM, et al. SWIFT-scalable clustering for automated identification of rare cell populations in large, high-dimensional flow cytometry datasets, part 2: biological evaluation. Cytometry A. 2014;85(5):422–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Naim I, Datta S, Rebhahn J, Cavenaugh JS, Mosmann TR, Sharma G. SWIFT-scalable clustering for automated identification of rare cell populations in large, high-dimensional flow cytometry datasets, part 1: algorithm design. Cytometry A. 2014;85(5):408–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Benjamini Y, Hochberg Y. Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. Journal of the Royal Statistical Society: Series B (Methodological). 1995;57(1):289–300. [Google Scholar]
- 42.Nakatsuji T, Hata TR, Tong Y, Cheng JY, Shafiq F, Butcher AM, et al. Development of a human skin commensal microbe for bacteriotherapy of atopic dermatitis and use in a phase 1 randomized clinical trial. Nat Med. 2021;27(4):700–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Eichenfield LF, Bieber T, Beck LA, Simpson EL, Thaci D, de Bruin-Weller M, et al. Infections in Dupilumab Clinical Trials in Atopic Dermatitis: A Comprehensive Pooled Analysis. Am J Clin Dermatol. 2019;20(3):443–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Totté JE, van der Feltz WT, Hennekam M, van Belkum A, van Zuuren EJ, Pasmans SG. Prevalence and odds of Staphylococcus aureus carriage in atopic dermatitis: a systematic review and meta-analysis. Br J Dermatol. 2016;175(4):687–95. [DOI] [PubMed] [Google Scholar]
- 45.Niebuhr M, Mamerow D, Heratizadeh A, Satzger I, Werfel T. Staphylococcal α-toxin induces a higher T cell proliferation and interleukin-31 in atopic dermatitis. Int Arch Allergy Immunol. 2011;156(4):412–5. [DOI] [PubMed] [Google Scholar]
- 46.Niebuhr M, Scharonow H, Gathmann M, Mamerow D, Werfel T. Staphylococcal exotoxins are strong inducers of IL-22: A potential role in atopic dermatitis. J Allergy Clin Immunol. 2010;126(6):1176–83.e4. [DOI] [PubMed] [Google Scholar]
- 47.Breshears LM, Gillman AN, Stach CS, Schlievert PM, Peterson ML. Local Epidermal Growth Factor Receptor Signaling Mediates the Systemic Pathogenic Effects of Staphylococcus aureus Toxic Shock Syndrome. PLoS One. 2016;11(7):e0158969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gillman AN, Breshears LM, Kistler CK, Finnegan PM, Torres VJ, Schlievert PM, et al. Epidermal Growth Factor Receptor Signaling Enhances the Proinflammatory Effects of Staphylococcus aureus Gamma-Toxin on the Mucosa. Toxins (Basel). 2017;9(7). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kim J, Kim BE, Ahn K, Leung DYM. Interactions Between Atopic Dermatitis and Staphylococcus aureus Infection: Clinical Implications. Allergy Asthma Immunol Res. 2019;11(5):593–603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lomholt H, Andersen KE, Kilian M. Staphylococcus aureus clonal dynamics and virulence factors in children with atopic dermatitis. J Invest Dermatol. 2005;125(5):977–82. [DOI] [PubMed] [Google Scholar]
- 51.Strange P, Skov L, Lisby S, Nielsen PL, Baadsgaard O. Staphylococcal enterotoxin B applied on intact normal and intact atopic skin induces dermatitis. Arch Dermatol. 1996;132(1):27–33. [PubMed] [Google Scholar]
- 52.Bunikowski R, Mielke M, Skarabis H, Herz U, Bergmann RL, Wahn U, et al. Prevalence and role of serum IgE antibodies to the Staphylococcus aureus-derived superantigens SEA and SEB in children with atopic dermatitis. J Allergy Clin Immunol. 1999;103(1 Pt 1):119–24. [DOI] [PubMed] [Google Scholar]
- 53.Cardona ID, Cho SH, Leung DY. Role of bacterial superantigens in atopic dermatitis : implications for future therapeutic strategies. Am J Clin Dermatol. 2006;7(5):273–9. [DOI] [PubMed] [Google Scholar]
- 54.Lee SJ, Kim SE, Shin KO, Park K, Lee SE. Dupilumab Therapy Improves Stratum Corneum Hydration and Skin Dysbiosis in Patients With Atopic Dermatitis. Allergy Asthma Immunol Res. 2021;13(5):762–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Olesen CM, Ingham AC, Thomsen SF, Clausen ML, Andersen PS, Edslev SM, et al. Changes in Skin and Nasal Microbiome and Staphylococcal Species Following Treatment of Atopic Dermatitis with Dupilumab. Microorganisms. 2021;9(7). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Nakatsuji T, Chen TH, Narala S, Chun KA, Two AM, Yun T, et al. Antimicrobials from human skin commensal bacteria protect against Staphylococcus aureus and are deficient in atopic dermatitis. Sci Transl Med. 2017;9(378). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Severn MM, Williams MR, Shahbandi A, Bunch ZL, Lyon LM, Nguyen A, et al. The Ubiquitous Human Skin Commensal Staphylococcus hominis Protects against Opportunistic Pathogens. mBio. 2022;13(3):e0093022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Byrd AL, Deming C, Cassidy SKB, Harrison OJ, Ng WI, Conlan S, et al. Staphylococcus aureus and Staphylococcus epidermidis strain diversity underlying pediatric atopic dermatitis. Sci Transl Med. 2017;9(397). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Berdyshev E, Goleva E, Bissonnette R, Bronova I, Bronoff AS, Richers BN, et al. Dupilumab significantly improves skin barrier function in patients with moderate-to-severe atopic dermatitis. Allergy. 2022;77(11):3388–97. [DOI] [PubMed] [Google Scholar]
- 60.Hanel KH, Cornelissen C, Luscher B, Baron JM. Cytokines and the skin barrier. Int J Mol Sci. 2013;14(4):6720–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Basler K, Galliano MF, Bergmann S, Rohde H, Wladykowski E, Vidal YSS, et al. Biphasic influence of Staphylococcus aureus on human epidermal tight junctions. Ann N Y Acad Sci. 2017;1405(1):53–70. [DOI] [PubMed] [Google Scholar]
- 62.Beck LA, Cork MJ, Amagai M, De Benedetto A, Kabashima K, Hamilton JD, et al. Type 2 Inflammation Contributes to Skin Barrier Dysfunction in Atopic Dermatitis. JID Innov. 2022;2(5):100131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Brewer MG, Monticelli SR, Moran MC, Miller BL, Beck LA, Ward BM. Conditions That Simulate the Environment of Atopic Dermatitis Enhance Susceptibility of Human Keratinocytes to Vaccinia Virus. Cells. 2022;11(8). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Harder J, Dressel S, Wittersheim M, Cordes J, Meyer-Hoffert U, Mrowietz U, et al. Enhanced expression and secretion of antimicrobial peptides in atopic dermatitis and after superficial skin injury. J Invest Dermatol. 2010;130(5):1355–64. [DOI] [PubMed] [Google Scholar]
- 65.Kanda N, Watanabe S. Increased serum human β-defensin-2 levels in atopic dermatitis: relationship to IL-22 and oncostatin M. Immunobiology. 2012;217(4):436–45. [DOI] [PubMed] [Google Scholar]
- 66.Robinson KM, McHugh KJ, Mandalapu S, Clay ME, Lee B, Scheller EV, et al. Influenza A virus exacerbates Staphylococcus aureus pneumonia in mice by attenuating antimicrobial peptide production. J Infect Dis. 2014;209(6):865–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Yang D, Chen Q, Hoover DM, Staley P, Tucker KD, Lubkowski J, et al. Many chemokines including CCL20/MIP-3alpha display antimicrobial activity. J Leukoc Biol. 2003;74(3):448–55. [DOI] [PubMed] [Google Scholar]
- 68.Sharma-Kuinkel BK, Zhang Y, Yan Q, Ahn SH, Fowler VG Jr. Host gene expression profiling and in vivo cytokine studies to characterize the role of linezolid and vancomycin in methicillin-resistant Staphylococcus aureus (MRSA) murine sepsis model. PLoS One. 2013;8(4):e60463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kehl-Fie TE, Chitayat S, Hood MI, Damo S, Restrepo N, Garcia C, et al. Nutrient metal sequestration by calprotectin inhibits bacterial superoxide defense, enhancing neutrophil killing of Staphylococcus aureus. Cell Host Microbe. 2011;10(2):158–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Hansen BT, Maschkowitz G, Podschun R, Fickenscher H. The Kinocidin Interleukin-26 Shows Immediate Antimicrobial Effects Even to Multi-resistant Isolates. Front Microbiol. 2021;12:757215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Beck LA, Thaçi D, Hamilton JD, Graham NM, Bieber T, Rocklin R, et al. Dupilumab treatment in adults with moderate-to-severe atopic dermatitis. N Engl J Med. 2014;371(2):130–9. [DOI] [PubMed] [Google Scholar]
- 72.Mobus L, Rodriguez E, Harder I, Stolzl D, Boraczynski N, Gerdes S, et al. Atopic dermatitis displays stable and dynamic skin transcriptome signatures. J Allergy Clin Immunol. 2021;147(1):213–23. [DOI] [PubMed] [Google Scholar]
- 73.Guttman-Yassky E, Bissonnette R, Ungar B, Suarez-Farinas M, Ardeleanu M, Esaki H, et al. Dupilumab progressively improves systemic and cutaneous abnormalities in patients with atopic dermatitis. J Allergy Clin Immunol. 2019;143(1):155–72. [DOI] [PubMed] [Google Scholar]
- 74.Impellizzieri D, Ridder F, Raeber ME, Egholm C, Woytschak J, Kolios AGA, et al. IL-4 receptor engagement in human neutrophils impairs their migration and extracellular trap formation. J Allergy Clin Immunol. 2019;144(1):267–79.e4. [DOI] [PubMed] [Google Scholar]
- 75.Woytschak J, Keller N, Krieg C, Impellizzieri D, Thompson RW, Wynn TA, et al. Type 2 Interleukin-4 Receptor Signaling in Neutrophils Antagonizes Their Expansion and Migration during Infection and Inflammation. Immunity. 2016;45(1):172–84. [DOI] [PubMed] [Google Scholar]
- 76.Heeb LEM, Egholm C, Impellizzieri D, Ridder F, Boyman O. Regulation of neutrophils in type 2 immune responses. Curr Opin Immunol. 2018;54:115–22. [DOI] [PubMed] [Google Scholar]
- 77.Egholm C, Heeb LEM, Impellizzieri D, Boyman O. The Regulatory Effects of Interleukin-4 Receptor Signaling on Neutrophils in Type 2 Immune Responses. Front Immunol. 2019;10:2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Mirza FN, Wang A, Ramachandran SM, Damsky W, Cohen JM. Dupilumab-induced phenotype switch from atopic dermatitis to psoriasis is characterized by de novo interleukin-17A expression: a case report. Br J Dermatol. 2021;185(2):432–4. [DOI] [PubMed] [Google Scholar]
- 79.Parker JJ, Sugarman JL, Silverberg NB, Gonzalez ME, Ramien ML, Teng JMC, et al. Psoriasiform dermatitis during dupilumab treatment for moderate-to-severe atopic dermatitis in children. Pediatr Dermatol. 2021;38(6):1500–5. [DOI] [PubMed] [Google Scholar]
- 80.Bridgewood C, Wittmann M, Macleod T, Watad A, Newton D, Bhan K, et al. T Helper 2 IL-4/IL-13 Dual Blockade with Dupilumab Is Linked to Some Emergent T Helper 17Type Diseases, Including Seronegative Arthritis and Enthesitis/Enthesopathy, but Not to Humoral Autoimmune Diseases. J Invest Dermatol. 2022;142(10):2660–7. [DOI] [PubMed] [Google Scholar]
- 81.Staudacher O, Krüger R, Kölsch U, Thee S, Gratopp A, Wahn V, et al. Relieving job: Dupilumab in autosomal dominant STAT3 hyper-IgE syndrome. J Allergy Clin Immunol Pract. 2022;10(1):349–51.e1. [DOI] [PubMed] [Google Scholar]
- 82.Bath-Hextall FJ, Birnie AJ, Ravenscroft JC, Williams HC. Interventions to reduce Staphylococcus aureus in the management of atopic eczema: an updated Cochrane review. Br J Dermatol. 2010;163(1):12–26. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


