Abstract
Despite significant advances in targeting mutant KRAS, tumor resistance to KRAS inhibitors (KRASi) remains a major barrier to progress. Here, we report proteostasis reprogramming as a key convergence point of multiple KRASi-resistance mechanisms. Inactivation of oncogenic KRAS downregulated both the heat shock response and the IRE1α branch of the unfolded protein response, causing severe proteostasis disturbances. However, IRE1α was selectively reactivated in an ER stress–independent manner in acquired KRASi-resistant tumors, restoring proteostasis. Oncogenic KRAS promoted IRE1α protein stability via ERK-dependent phosphorylation of IRE1α, leading to IRE1α disassociation from HRD1 E3-ligase. In KRASi-resistant tumors, both reactivated-ERK and hyperactivated-AKT restored IRE1α phosphorylation and stability. Suppression of IRE1α overcame resistance to KRASi. This study reveals a druggable mechanism that leads to proteostasis reprogramming and facilitates KRASi resistance.
One Sentence Summary:
ER stress-independent unconventional phosphorylation of IRE1α is a convergence point of multiple KRAS inhibitor resistance mechanisms.
KRAS is one of the most frequently mutated genes in human cancer, especially in pancreatic ductal adenocarcinoma (PDAC), non–small cell lung cancer (NSCLC), and colorectal carcinoma (CRC) (1–5). Oncogenic KRAS engages multiple effector pathways to drive tumorigenesis, notably the RAF/MEK/ERK (MAPK) and phosphatidylinositol 3-kinase (PI3K)/AKT pathways (6–10). Small molecules that directly target the KRAS G12C mutation, including sotorasib and adagrasib (11–13), have shown encouraging therapeutic efficacies in clinical trials (14–16). The US Food and Drug Administration (FDA) granted accelerated approval for sotorasib and adagrasib to treat patients with KRASG12C-mutant NSCLC. However, resistance to these KRAS inhibitors is almost inevitable resulting from the activation of compensatory pathways, e.g. EGF, FGF, AURKA, or SOS1, or the acquisition of new mutations, e.g. KRAS, NRAS, BRAF, EGFR, or FGFR2 (15–35). Although inhibitors targeting KRAS mutations other than G12C are in clinical trials (36), similar bypass of KRAS dependence has been demonstrated in preclinical studies using genetically engineered mouse models (GEMMs) of lung and pancreatic cancer (37–39). Due to these clinical challenges, understanding the mechanisms that mediate the resistance to KRAS inhibitors is imperative to develop more effective therapies to prevent the recurrence of KRAS-driven cancers.
The ability to overcome an imbalanced protein homeostasis or “proteostasis” network is instrumental to maintain cancer cell survival and circumvent various insults that impair the quality of protein synthesis and folding (40, 41). Cells use compartment-specific stress sensors to monitor and maintain a high-fidelity proteome. Cytosolic proteins are monitored by the heat shock response (HSR) (42), whereas transmembrane and secreted proteins are monitored in the endoplasmic reticulum (ER) by the unfolded protein response (UPR) (43–45). When the HSR is triggered by stress stimuli, heat shock factors (HSFs) become activated and transactivate genes encoding chaperones and other factors of the proteostasis network (46). HSF1 is a master regulator of the proteotoxic stress response and has been implicated in mediating proteome fidelity in cancer cells (47). The UPR is a three-branched stress response that is activated upon disruption of ER homeostasis. The UPR is mediated by the ER transmembrane sensors inositol-requiring enzyme 1α (IRE1α), activated transcription factor 6 (ATF6), and protein kinase RNA-like ER kinase (PERK) (48). IRE1α is the most ancient and conserved member of the mammalian UPR sensory triad (49, 50). Under ER stress conditions, IRE1α undergoes oligomerization and trans-autophosphorylation to activate its RNase domain. This results in excision of 26 nucleotides from unspliced XBP1 (XBP1u) mRNA, and a frame shift mutation to produce the mature, spliced XBP1 (XBP1s) encoding a potent transcriptional activator (fig. S1A) (51, 52). Mutant RAS was traditionally viewed as an inducer of general UPR through stressing ER during oncogenic transformation of non-malignant cells (53). Genetic screens revealed the synthetic lethal interaction between mutant RAS and IRE1α in yeast (54). Yet it remains unclear how the proteostasis network is orchestrated by oncogenic KRAS, or how the proteostasis reprogramming mechanisms occur that bypass KRAS addiction and allow for acquired resistance to KRAS inhibitors.
Here, we report that proteostasis is dynamically altered upon oncogenic KRAS inhibition. We identified the IRE1α-mediated reprogramming of proteostasis as an essential mechanism that facilitates resistance to KRAS–MAPK inhibition. Importantly, we elucidated the biochemical basis for the ER stress–independent post-translational modification and regulation of IRE1α by both oncogenic KRAS signaling and KRASi resistance signaling, which serves as a therapeutic vulnerability that can be targeted to overcome resistance to KRAS–MAPK inhibition.
Oncogenic KRAS Inactivation Reprograms Proteostasis
To understand the impacts of oncogenic KRAS on proteostasis, we used primary mouse PDAC cells derived from our previously generated, doxycycline (Dox)-inducible, KrasG12D-driven PDAC mouse model (55), which are hereafter designated as iKras cells. Dox withdrawal turned off KrasG12D expression in iKras cells (fig. S1B), resulting in the inactivation of downstream MAPK signaling (fig. S1C), decreased 5-bromodeoxyuridine (BrdU) incorporation (fig. S1D) and increased cell apoptosis (fig. S1E). We monitored protein aggregation upon KrasG12D inactivation using PROTEOSTAT, a molecular rotor dye that specifically intercalates into the cross-beta spines of the quaternary protein structures found in misfolded and aggregated proteins (Fig. 1A). As a positive control, the treatment of iKras cells with the proteasome inhibitor MG132 significantly induced PROTEOSTAT signal (fig. S1F). The inactivation of KrasG12D by Dox withdrawal induced protein aggregation (Fig. 1, B and C). However, 30 days after Dox withdrawal, iKras cells displayed restored proteostasis (Fig. 1, B and C) and resumed cell growth (fig. S1D, E and G), at levels comparable to those observed for parental cells (iKrasP). These cells resistant to KrasG12D inactivation are hereafter designated as iKrasR cells. Continuous KrasG12D inactivation was required to maintain the resistance phenotypes as reactivation of KrasG12D for extended periods (12 days) partially reversed the resistance of iKrasR cells to Dox withdrawal (fig. S1, H to K). Dox administration had no impact on cell growth or proteostasis in LSL-KrasG12D cells lacking Dox-inducible KrasG12D expression (fig. S1, L to P). In the iKras cells, staining with Congo red (CR) or thioflavin T (ThT), which recognize misfolded protein aggregates (56), or detection of lysine 48 (K48)-linked polyubiquitin levels, which tag misfolded or aggregated proteins for proteasome-mediated degradation (57), independently confirmed the restoration of proteostasis upon acquired resistance to KrasG12D inactivation (fig. S1, Q to S).
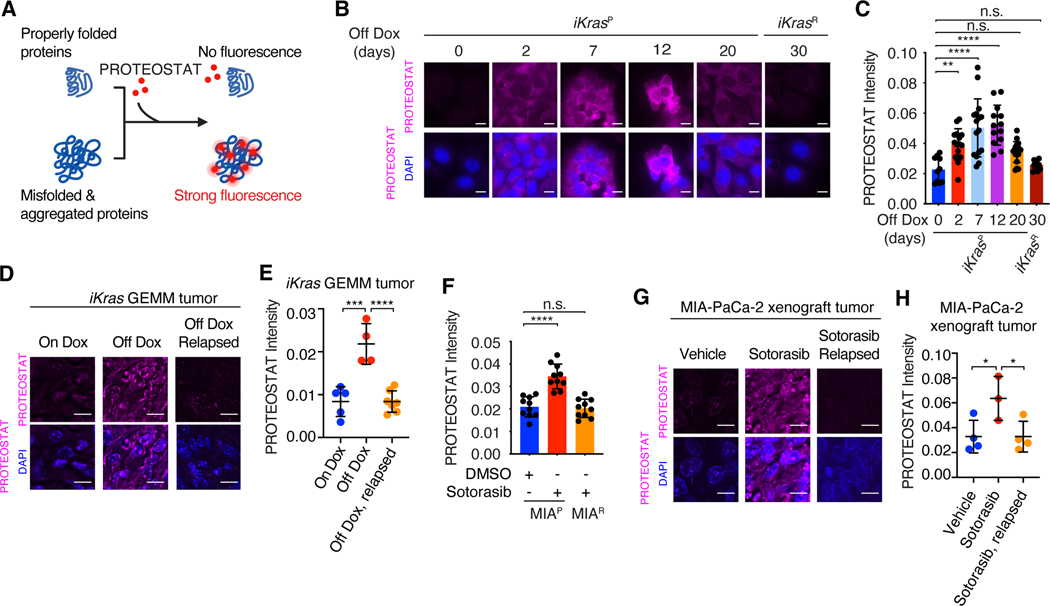
Fig. 1. Oncogenic KRAS inactivation reprograms proteostasis.
(A) Schematic illustration of labeling and detection of misfolded and aggregated proteins with PROTEOSTAT dye. Upon intercalation into the cross-beta spine typically found in misfolded and aggregated proteins, PROTEOSTAT dye emits strong fluorescence. (B and C) Representative images (B) and quantification (C) of PROTEOSTAT (magenta) and DAPI (blue) staining in iKrasP cells at different time points after KrasG12D inactivation by Dox-withdrawal (Off Dox) until the cells acquired resistance to KrasG12D inactivation (iKrasR cell). (D and E) Representative images (D) and quantification (E) of PROTEOSTAT (magenta) staining in spontaneous tumors from the Dox-inducible, KrasG12D-driven PDAC mouse model (iKras GEMM) treated with doxycycline (Dox, 2g/L, n=5), Dox withdrawal for 3 days (n=4) or relapsed after 30 weeks of Dox-withdrawal (n=7). (F) Quantification of PROTEOSTAT intensity in parental MIA-PaCa-2 (MIAP) cells treated with DMSO or 30nM sotorasib for 2 days or in sotorasib-resistant MIA-PaCa-2 (MIAR) cells treated with 30nM sotorasib. MIAR cells were generated in vitro by continued sotorasib treatment until the cells acquired resistance. (G and H) Representative images (G) and quantification (H) of PROTEOSTAT (magenta) and DAPI (blue) staining in MIA-PaCa-2 xenograft tumors treated with vehicle (n=4), sotorasib (30mg/kg for 1 day, n=3), or relapsed after 9 weeks of sotorasib treatment (30mg/kg, n=4). Data represent average fluorescence intensity of PROTEOSTAT/cell from each image (C and F) or tumor (E and H) and are presented as mean ± SD from n≥10 images. Scale bar: 20μm. Ordinary one-way ANOVA with Dunnett’s multiple comparisons test (C, E, F and H) was used to calculate P values. n.s., not significant, * P<0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001.
We also used the iKras GEMM to examine in vivo proteostasis reprogramming (55). KrasG12D inactivation by Dox withdrawal resulted in rapid tumor regression, but 70% of tumors relapsed after 9–47 weeks (37). Notably, PROTEOSTAT staining of iKras GEMM tumors at different stages of KrasG12D inactivation revealed increased protein aggregation in regressing parental tumors and restored proteostasis in relapsed tumors (Fig. 1, D and E, and fig. S2A), which grow independently of KrasG12D expression.
To further investigate the impacts of oncogenic KRAS inhibition on proteostasis, we used a KRASG12C inhibitor, sotorasib (12), to treat MIA-PaCa-2 human PDAC cells and H358 human NSCLC cells harboring the KRASG12C mutation. Sotorasib effectively suppressed the activation of MEK/ERK (fig. S2, B and C), leading to cell growth inhibition in both MIA-PaCa-2 and H358 cell lines (fig. S2, D and E). Both MIA-PaCa-2 and H358 cells also displayed increased PROTEOSTAT staining and K48-linked polyubiquitination levels in response to sotorasib treatment, suggesting enhanced protein aggregation (Fig. 1F and fig. S2, F to H). However, after MIA-PaCa-2 and H358 cells gained resistance to sotorasib (fig. S2, D and E), they also exhibited restored proteostasis (Fig. 1F and fig. S2, F to H). Similar as iKrasR cells, continuous sotorasib treatment was required to maintain the resistance of MIA-PaCa-2R cells to KRASi (fig. S2, I and J). MIA-PaCa-2 xenograft tumors were initially sensitive to sotorasib treatment but relapsed after 6 weeks (fig. S2K). PROTEOSTAT staining revealed increased protein aggregation after initial sotorasib treatment and restored proteostasis in relapsed tumors (Fig. 1, G and H). These data demonstrate that oncogenic KRAS inhibition induces protein aggregation and severely disrupts proteostasis. However, KRASi-resistant cancer cells able to grow in the absence of mutant KRAS gain the capacity to overcome associated proteotoxic stress and regain proteostasis.
Oncogenic KRAS Inactivation Differentially Impacts the Key Nodes of Proteostasis Regulatory Network
Proteostasis is maintained by an integrated network that includes translation, protein quality control mechanisms that regulate the content and quality of the proteome, and protein degradation pathways, such as the ubiquitin/proteasome system and the autophagy/lysosome system, which eliminate misfolded or aggregated proteins (42, 43, 58). KrasG12D inactivation by Dox withdrawal in iKras cells did not alter overall proteasomal activity (fig. S3, A and B). We observed increased LC3 cleavage and LAMP1 levels upon KrasG12D inactivation (fig. S3C), indicating elevated autophagy and lysosomal activities, which unlikely caused increased protein aggregation. Next, we examined the protein quality control and stress response pathways that monitor and regulate protein folding, including the UPR and HSR. KrasG12D inactivation in iKrasP cells markedly reduced IRE1α protein levels, Xbp1 splicing, and the expression levels of the XBP1s targets Edem1 and Sec61a1 (Fig. 2A and fig. S3, D to F). As a control, Dox treatment had no impacts on IRE1α levels in the constitutively activated LSL-KrasG12D cells (fig. S3G). By contrast, ATF6 was barely affected, and phospho-PERK was negatively correlated with IRE1α/XBP1 in response to KrasG12D inactivation (Fig. 2A). Resolving protein aggregation with tauroursodeoxycholic acid (TUDCA), a chemical chaperone that promotes protein folding and stimulates molecular chaperone function (59, 60), completely blunted KrasG12D inactivation-induced phospho-PERK (fig. S3, H to J), indicating that PERK was activated as a result of the disrupted proteostasis by KRASi. PERK is one of the four kinases (GCN2, PERK, HRI and PKR) that phosphorylate eIF2α and regulate the Integrated Stress Response (ISR) (61). Unexpectedly, eIF2α and its downstream ATF4 followed an opposite pattern as that of phosphor-PERK upon KrasG12D inactivation a(Fig. 2A). Perk deletion had no effects on their levels (fig. S3K). Genetic deletion of each ISR kinases demonstrated that phospho-eIF2α and ATF4 were dependent on GCN2 which was activated in iKrasP cells (Fig. 2A and fig. S3K). These data establish GCN2 as a regulator of ISR in this context. Notably, IRE1α/XBP1, but not GCN2 or eIF2α, was selectively restored in iKrasR cells (Fig. 2A and fig. S3, D to F). Consistently, sotorasib treatment reduced IRE1α protein levels in MIA-PaCa-2 and H358 cells, but IRE1α was restored following the acquisition of sotorasib resistance (Fig. 2, B and C, and fig. S2, D and E). IRE1α immunostaining in the parental, KrasG12D-extincted, relapsed iKras GEMM tumors (Fig. 2D), and sotorasib-treated MIA-PaCa-2 human xenograft tumors (Fig. 2E) all independently confirmed the in vivo pattern of acute IRE1α suppression followed by reactivation in relapsed tumors. We also confirmed the continuous in vivo suppression of phospho-GCN2, phosphor-eIF2α, and ATF4 by sotorasib treatment in MIA-PaCa-2 xenograft tumors (fig. S3L). Collectively, these data show that acute oncogenic KRAS inactivation inhibits the IRE1α branch of the UPR and GCN2-regulated ISR. However, only IRE1α is reactivated in the KRASi-resistant tumors.
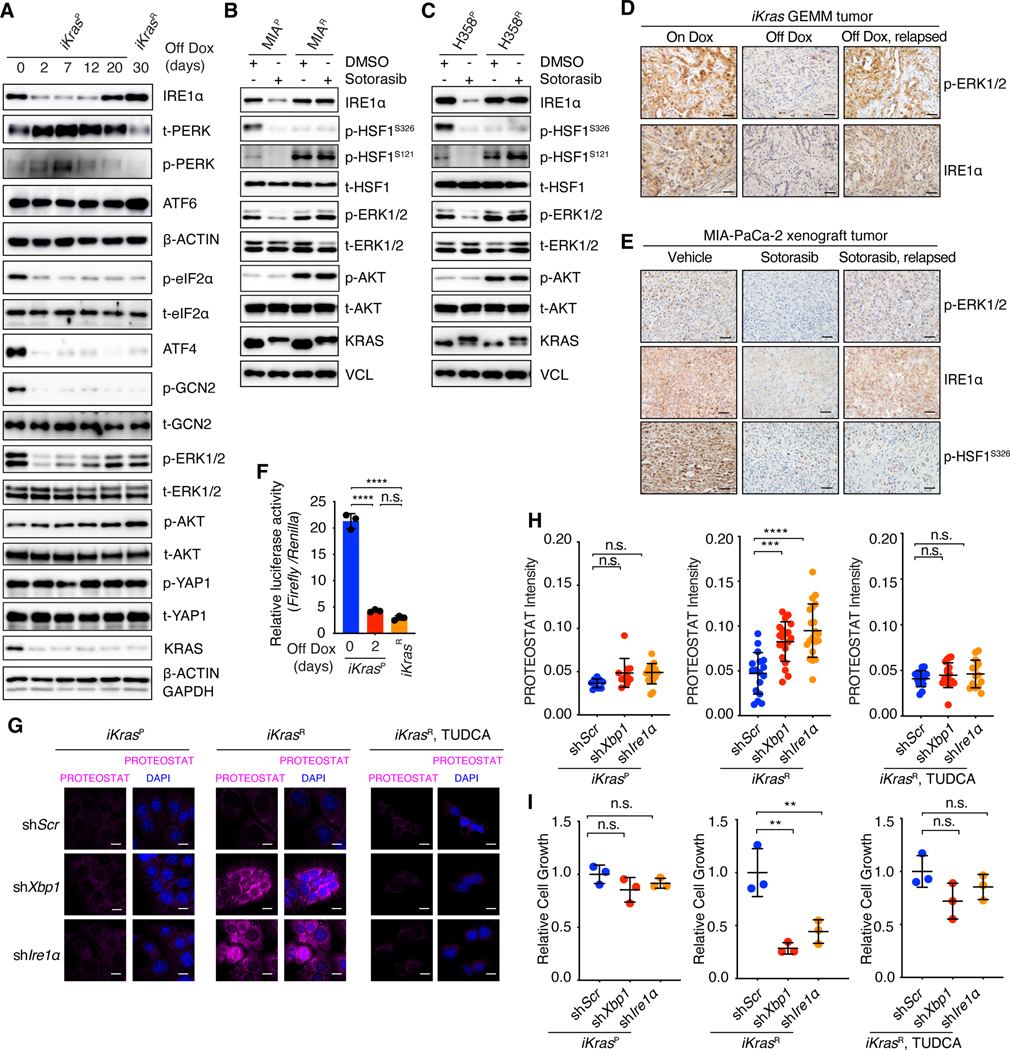
Fig. 2. Oncogenic KRAS inactivation differentially impacts the key nodes of the proteostasis regulatory network.
(A) Immunoblot with indicated antibodies in whole-cell lysates of iKrasP at different time points after KrasG12D inactivation by Dox-withdrawal (Off Dox) until the cells acquired resistance to KrasG12D inactivation (iKrasR cell). (B and C) Immunoblot with indicated antibodies in whole-cell lysates of parental or sotorasib-resistant MIA-PaCa-2 (B) or H358 (C) cells treated with DMSO or 30 nM sotorasib. (D) Immunohistochemical (IHC) staining with indicated antibodies in iKras GEMM tumors treated with doxycycline (On Dox), Dox withdrawal for 3 days (Off Dox), or relapsed after 30 weeks of Dox-withdrawal (Off Dox, relapsed). (E) IHC staining with indicated antibodies in MIA-PaCa-2 xenograft tumors treated with vehicle, sotorasib (30mg/kg for 1 day), or relapsed after 9 weeks of sotorasib treatment (30mg/kg). (F) Relative HSE luciferase activity in iKrasP or iKrasR cells cultured in the presence or absence of Dox for 2 days. Data are shown as mean ± SD, n=3. (G and H) Representative images (G) and quantification (H) of PROTEOSTAT (magenta) and DAPI (blue) staining in iKrasP or iKrasR cells infected with lentiviruses encoding scramble shRNA (shScr), Xbp1 shRNA (shXbp1) or Ire1 α shRNA (shIre1α). Cells were treated with 2.5mM TUDCA dissolved in water for 2 days as indicated. Data represent the average fluorescence intensity of PROTEOSTAT/cell from each image acquired and presented as mean ± SD from n=10 (On Dox), n=17 (Off Dox), or n=17 (Off Dox + TUDCA) images. (I) CCK-8 assay was used to quantify cell viability of iKras cells treated as in G and H. Data are presented as mean ± SD relative to shScr, n=3. Ordinary one-way ANOVA with Dunnett’s multiple comparisons test (H and I) or ordinary one-way ANOVA with Tukey’s multiple comparisons test (F) was used to calculate P values. n.s., not significant, **P < 0.01, ***P < 0.001, ****P < 0.0001. Scale bar: 40μm (D and E) or 20μm (G).
In contrast with our findings on IRE1α, sotorasib continuously suppressed HSF1 phosphorylation at serine residue 326 (S326), which is required for HSF1 activation (62), in both parental and sotorasib-resistant MIA-PaCa-2 and H358 cells (Fig. 2, B and C). The inhibitory phosphorylation of HSF1 at S121 (62) was increased in both sotorasib-resistant MIA-PaCa-2R and H358R cells (Fig. 2, B and C). As a result, sotorasib treatment markedly reduced the expression of HSF1 target genes, including HSPA6 and HSPA1B, in both parental and sotorasib-resistant MIA-PaCa-2 and H358 cells (fig. S3, M and N). We confirmed the in vivo suppression of HSF1-S326 phosphorylation by sotorasib treatment in MIA-PaCa-2 human xenograft tumors (Fig. 2E) and marked reduction of HSF1 luciferase reporter activities in sotorasib-resistant MIA-PaCa-2R cells (fig. S3, O and P). The genetic inactivation of KrasG12D also resulted in the considerable reduction of HSF1 luciferase reporter activities in both iKrasP and iKrasR cells (Fig. 2F). Collectively, these data demonstrate that oncogenic KRAS inactivation initially impairs both the ER and cytosolic protein quality control machinery, as well as the ISR. However, only IRE1α/XBP1, but not HSF1 or ISR, is restored in the KRASi-resistant tumors.
IRE1α/XBP1 is Required for Maintaining Proteostasis in KRASi-Resistant Cancer Cells
Next, we examined the necessity of IRE1α/XBP1 for proteostasis maintenance. In iKrasP cells, Ire1α and Xbp1 knockdown modestly induced protein aggregation (Fig. 2, G and H, and fig. S4A), with minimal effects on cell growth (Fig. 2I). By contrast, Ire1α and Xbp1 knockdown in the iKrasR cells led to profound protein aggregation and PERK phosphorylation (Fig. 2, G and H, and fig. S4, A to E), and significantly inhibited cell growth (Fig. 2I, and fig. S4F). These effects were rescued by TUDCA (Fig. 2, G to I, and fig. S4B), demonstrating the importance of IRE1α/XBP1 in the maintenance of proteostasis and cell survival. Depletion of Perk did not affect proteostasis and cell growth of iKrasR cells and had no impacts on Ire1α-depletion induced phenotypes (fig. S4, G to K), suggesting that PERK is dispensable for KRASi-resistant cancer cells. The kinase activity of IRE1α is required for its RNase activation (63). In contrast to WT IRE1α, neither kinase-dead IRE1αK599A nor RNase-dead IRE1αK907A mutant (63) was able to rescue Ire1α-depletion induced phenotypes in iKrasR cells (fig. S5, A to D), suggesting that IRE1α’s function depends on its catalytic RNase activity. In addition to XBP1s, IRE1α RNase also cleaves ER-localized RNAs through Regulated IRE1α-Dependent Decay (RIDD) pathway (64, 65). Although some RIDD targets were regulated by IRE1α (fig. S5, E and F), restoration of XBP1s completely rescued the Ire1α depletion-induced protein aggregation and cell growth defects in iKrasR cells (fig. S5, G to I). These data establish that IRE1α RNase activity and RNase-dependent XBP1 splicing drives proteostasis in KRASi-resistant cancer cells. Consistently, CRISPR-mediated Ire1α or Xbp1 knockout (fig. S5J) resulted in more severe protein aggregation in iKrasR cells than in iKrasP cells (fig. S5K). Taken together, we demonstrate that IRE1α/XBP1 is indispensable for the maintenance of balanced proteostasis in KRASi-resistant cancer cells.
The MAPK Pathway Regulates IRE1α/XBP1 in Parental KRAS-Driven Cancer Cells
We aimed to determine the mechanism through which oncogenic KRAS regulates IRE1α. MAPK and PI3K are two major effector pathways downstream of oncogenic KRAS (6). The MEK inhibitor trametinib and the ERK inhibitor SCH772984 both substantially reduced IRE1α protein levels in iKrasP cells (Fig. 3A). By contrast, the PI3K inhibitor pictilisib and the AKT inhibitor MK2206 had little impact on IRE1α levels in iKrasP cells (Fig. 3B). Examination of five additional KRAS-mutant cell lines confirmed that MEK/ERK inhibitors, but not PI3K/AKT inhibitors, reduced IRE1α protein levels (fig. S6, A to C). These effects were similar to what was observed in response to the genetic or pharmacological inhibition of oncogenic KRAS (Fig. 3F, and fig. S6D). Recent studies show that SHP2 is critical for KRASG12C cycling and ERK activation (30, 33, 66–69). Inhibition of SHP2 with SHP099 significantly suppressed ERK activity and reduced IRE1α levels in KRASG12C-mutant H358 cells (fig. S6E), whereas modest effect was observed in KRASG12D-mutant iKras cells due to the limited intrinsic GTPase activity of KRASG12D (fig. S6F). SHP099 also inhibited the growth of sotorasib-resistant H358R cells and MIA-PaCa-2R cells (fig. S6, G and H). Long-term treatment of H358 cells with SHP099 led to drug resistance and recovered proteostasis (fig. S6, I to K), accompanied with IRE1α restoration in the resistant cells (fig. S6L). Depletion of IRE1α in SHP2i-resistant cells resulted in marked protein aggregation and re-sensitized the cells to SHP099 (fig. S6, M to O). Another upstream activator of RAS signaling is EGFR, suppression of EGFR with gefitinib significantly reduced ERK and IRE1α levels in H358 cells (fig. S6P). In contrast, inhibition of MEK/ERK in non-malignant BEAS-2B lung epithelial cells with non-oncogenic RAS signaling barely affected IRE1α levels (fig. S6, Q and R). Using tissue microarrays, we found that IRE1α levels correlated with phospho-ERK levels in treatment-naïve PDAC patient samples (Fig. 3, C and D), and high expression of IRE1α was associated with higher histologic tumor grade (Fig. 3E). Collectively, these data demonstrate that oncogenic KRAS-mediated MAPK pathway activation leads to the activation of IRE1α in parental KRAS-mutant cancers.
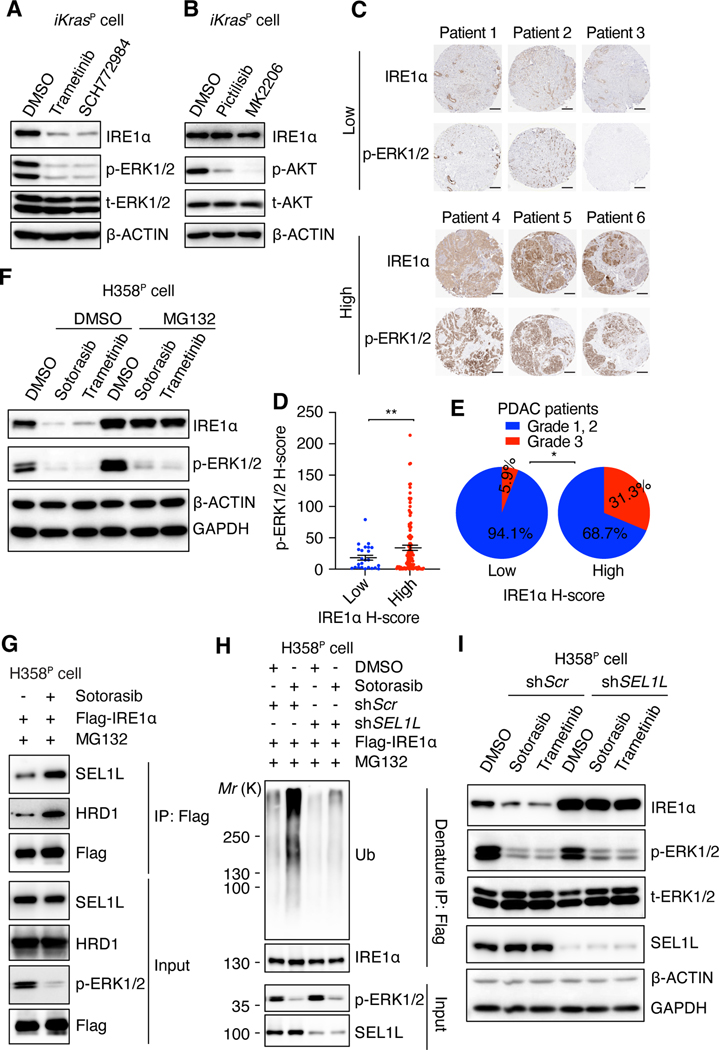
Fig. 3. KRAS-MAPK signaling stabilizes IRE1a through inhibiting SEL1L-HRD1 mediated IRE1a ubiquitination.
(A and B) Immunoblot with indicated antibodies in whole-cell lysates of iKrasP cells treated with DMSO, trametinib (MEK inhibitor, 20 nM), SCH772984 (ERK inhibitor, 1 μM), pictilisib (PI3K inhibitor, 1 μM), or MK2206 (AKT inhibitor, 2 μM) as indicated for 2 days. (C) Representative images of IHC staining of IRE1α and p-ERK1/2 in tissue microarray of treatment naïve tumors from PDAC patients. Scale bar: 200μm. (D) H-score of p-ERK1/2 in tissue microarray samples with distinct IRE1α intensities. Data are presented as mean ± SEM. (E) Proportion of patients with different tumor grades in PDAC patients with low or high IRE1α H-score. (F) Immunoblot with indicated antibodies in whole-cell lysates of H358P cells treated with DMSO, 30 nM sotorasib or 20 nM trametinib for 2 days. Cells were treated with DMSO or 1 μM MG132 for 12h before harvest. (G) Sotorasib promotes the interaction between IRE1α and SEL1L/HRD1. H358P cells expressing Flag-IRE1a were treated with DMSO or 30 nM sotorasib for 2 days and subjected to immunoprecipitation (IP) with anti-Flag M2 agarose beads. (H) Sotorasib promotes SEL1L-dependent IRE1α ubiquitination. H358P cells expressing Flag-IRE1α and shScr or shSEL1L were treated with DMSO or 30 nM sotorasib for 2 days and subjected to denature IP with anti-Flag M2 agarose beads. The immunoblot was probed with anti-ubiquitin (Ub) antibody to detect IRE1α ubiquitination. MG132 (1 μM) was added into the culture medium 12 h before harvest (G and H). (I) Immunoblot with indicated antibodies in whole-cell lysates of H358P cells infected with lentiviruses encoding shScr or shSEL1L and treated with DMSO, 30 nM sotorasib, or 20 nM trametinib for 2 days. 2-tailed, unpaired Student’s t test with Welch’s correction (D) or Fisher’s exact test (E) was used to calculate P values. * P<0.05, **P < 0.01.
MAPK Promotes IRE1α Protein Stability by Inhibiting SEL1L/HRD1-mediated IRE1α Ubiquitination
Sotorasib treatment did not downregulate IRE1α mRNA levels (fig. S7A), but it considerably reduced IRE1a protein abundance in H358 cells (Fig. 3F). Treatment with the proteasome inhibitor MG132 rescued both sotorasib- and trametinib-induced reductions in IRE1α protein levels in H358 cells (Fig. 3F). Similarly, the observed reduction in IRE1α protein levels in response to KrasG12D inactivation, trametinib, or SCH772984 treatment could be rescued by MG132 in iKrasP cells (fig. S7, B and C). These data demonstrate that the inhibition of oncogenic KRAS or MAPK promotes proteasome-mediated IRE1α degradation in parental KRAS-mutant cancers.
IRE1α is a bona-fide substrate of the SEL1L/HRD1 ER-associated degradation (ERAD) complex (70), which is composed of the E3 ubiquitin ligase HRD1 and its adapter protein SEL1L (71). The SEL1L/HRD1 complex ubiquitinates and targets IRE1α for proteasomal degradation in multiple cell types (70, 72, 73). Sotorasib treatment significantly enhanced the association between IRE1α and the SEL1L/HRD1 complex (Fig. 3G), resulting in increased IRE1α ubiquitination in H358 cells (Fig. 3H). Sotorasib also promoted the interaction of IRE1α with p97 and NPL4 (fig. S7, D and E), which deliver the ubiquitinated ERAD substrates to the proteasome for degradation (74). SEL1L knockdown reduced sotorasib- or trametinib-induced IRE1α ubiquitination and restored IRE1α protein levels (Fig. 3, H and I, fig. S7F), leading to the prevention of KRAS-MAPK inhibition induced protein aggregation (fig. S7G). Inhibition of p97 with CB5083 (75) also rescued sotorasib-induced IRE1α degradation (fig. S7I). Consistently, Sel1l or Hrd1 depletion in iKrasP cells blocked the induction of IRE1α degradation and prevented protein aggregation in response to KrasG12D extinction and trametinib or SCH772984 treatment (fig. S7, J to P). These data demonstrate that oncogenic KRAS-mediated MAPK activation stabilizes IRE1α protein by preventing SEL1/HRD1-mediated ubiquitination and subsequent proteasomal degradation.
ERK Directly Interacts with and Phosphorylates IRE1α
Oncogenic KRAS-MAPK did not directly regulate ERAD complex expression in iKrasP or H358 cells (Fig. 3I and fig. S7, J, M and Q). It is well established that phosphorylation often interferes with protein-protein interactions and thus regulates protein ubiquitination and stability. We tested whether MAPK might promote IRE1α phosphorylation, resulting in IRE1α disassociation from the SEL1L/HRD1 complex. Indeed, the expression of a constitutively activated MEK construct (MEKDD) dramatically enhanced IRE1α phosphorylation in 293T cells detected by an anti-phospho-MAPK substrate motif antibody (fig. S8A). In contrast, sotorasib treatment significantly reduced IRE1α phosphorylation levels compared with control H358 cells (Fig. 4A). Co-immunoprecipitation (co-IP) assay demonstrated that ERK interacted with IRE1α in 293T cells and H358 cells (Fig. 4B, and fig. S8B). Furthermore, GST pull-down and in vitro kinase assays confirmed that both ERK1 and ERK2 directly interacted with and phosphorylated IRE1α in vitro (Fig. 4, C and D, and fig. S8, C to H). Depletion of ERK1 or ERK2 in H358 cells revealed that both ERK1 and ERK2 regulated IRE1α phosphorylation (fig. S8I). IRE1α possesses three putative ERK binding D-motifs (Fig. 4E) (76). Deletion of the D-motif at amino acids 687–701 largely disrupted the binding between ERK and IRE1α (fig. S8J). Collectively, these data demonstrate that ERK directly interacts with and phosphorylates IRE1α.
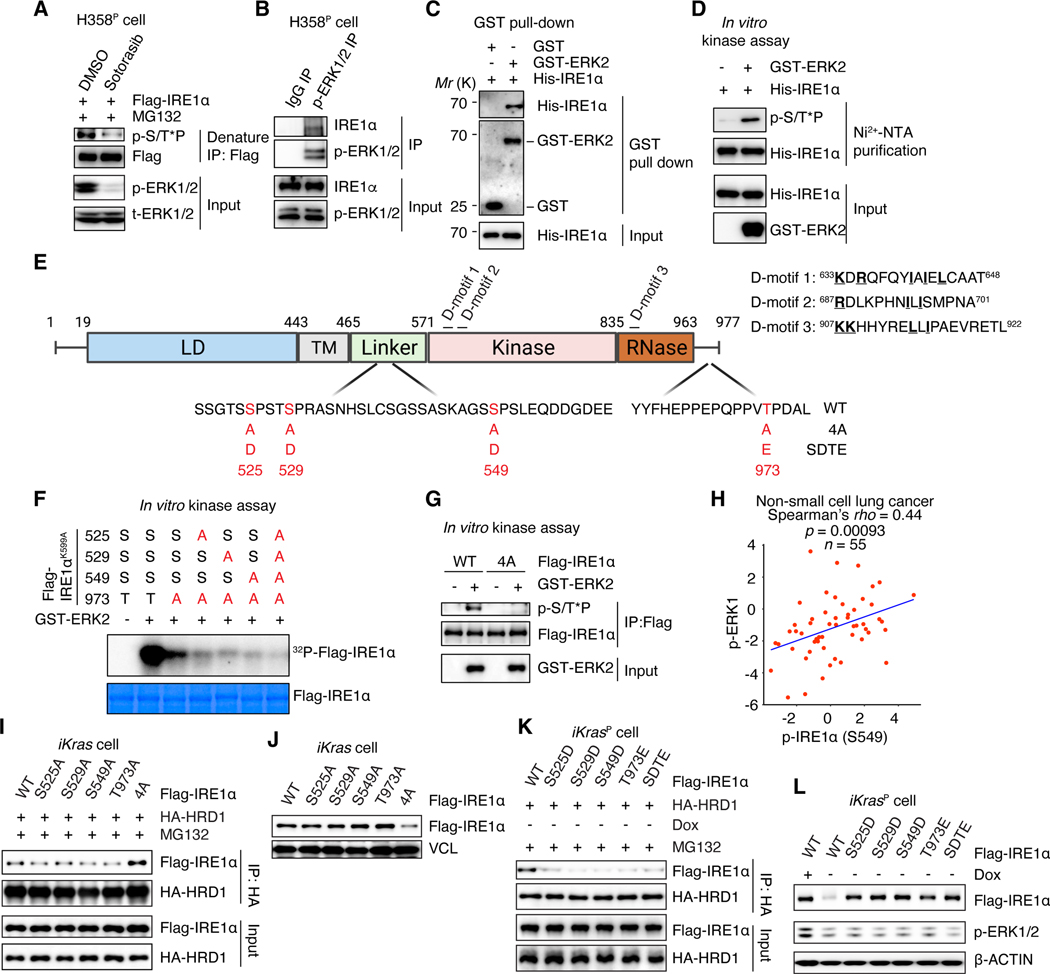
Fig. 4. ERK directly phosphorylates and stabilizes IRE1α.
(A) H358P cells expressing Flag-IRE1a were treated with DMSO or 30 nM sotorasib for 2 days and subjected to denature IP with anti-Flag M2 agarose beads. The immunoblot was probed with anti-phospho-MAPK substrates motif (S/T*P) antibody (p-S/T*P) to detect IRE1α phosphorylation. (B) Whole-cell lysate of H358P cells were subjected to co-IP with rabbit anti-p-ERK1/2 antibody or normal rabbit IgG. (C) GST pull-down assay was performed using recombinant His-tagged IRE1α and GST-ERK2 protein. (D) In vitro kinase assay was performed using recombinant GST-ERK2 and His-IRE1α. After phosphorylation reaction, the proteins were denatured by 8M urea buffer and subjected to purification of His-IRE1α with Ni2+-NTA agarose to detect IRE1α phosphorylation using anti-p-S/T*P antibody. (E) Schematic illustration of human IRE1α protein domains, three putative ERK binding D-motifs and ERK phosphorylation sites at Ser525, Ser529, Ser549, and Thr973 (red). Phospho-deficient (4A) and phospho-mimetic (SDTE) mutations of IRE1α are shown. LD: luminal domain. TM: transmembrane domain. (F) In vitro [g-32P] ATP kinase assay using different Flag-tagged IRE1α mutants and GST-ERK2. The IRE1α phosphorylation was detected by autoradiography. One-Step Blue Protein Stain was used to detect IRE1α protein loading. (G) In vitro kinase assay using equal amount of Flag-tagged WT or phospho-deficient IRE1α mutant proteins (4A) and GST-ERK2. (H) Spearman correlation between p-IRE1α (at S549) and p-ERK1 (at Y204) in 55 patients with non-small cell lung cancer. (I and K) Whole-cell lysates of iKras cells expressing HA-HRD1 together with WT or mutant IRE1α cultured in the absence of Dox were subjected to IP with anti-HA agarose beads to detect IRE1α interaction with HRD1. MG132 (1 μM) was added into the culture medium 12h before harvest (A, I and K). (J and L) Immunoblot of WT or mutant IRE1α in whole-cell lysates of iKras cells cultured in the presence or absence of Dox for 2 days.
Sequence analysis showed that human IRE1α contains four putative ERK phosphorylation sites at S525, S529, S549, and T973, consistent with the minimal ERK substrate motif pS/T*P (Fig. 4E). Mass spectrometry analysis of IRE1α protein purified from control or MEKDD-expressing 293T cells confirmed the ERK-dependent phosphorylation of IRE1α at S525, S529, S549, and T973 (fig. S9, A to C). We mutated the identified phospho-serine or - threonine amino acids (aa) to alanine (A) and performed an in vitro kinase assay using [γ−32P] ATP. The kinase-dead form of IRE1αK599A was used as a backbone to exclude the effects of IRE1α autophosphorylation. ERK was able to directly phosphorylate kinase-dead autophosphorylation-deficient IRE1αK599A (Fig. 4F). The T973A mutation significantly reduced but did not eliminate ERK-dependent IRE1αK599A phosphorylation (Fig. 4F). Additional mutation of S525, S529 or S549 to A together with T973A further decreased the IRE1α phosphorylation (Fig. 4F). The simultaneous mutation of all four sites largely abolished the ERK-dependent IRE1αK599A phosphorylation (Fig. 4F). In agreement, mutation of these four S/T to A (designated as 4A mutant) diminished ERK-dependent phosphorylation on WT IRE1α (Fig. 4G, and fig. S9D). The IRE1α mutations did not affect IRE1α binding with ERK (fig. S9, D and E). Collectively, these data identify S525, S529, S549 and T973 as ERK phosphorylation sites on IRE1α. Importantly, analysis of Clinical Proteomic Tumor Analysis Consortium (CPTAC) datasets in patients with NSCLC (77) showed a statistically significant correlation between IRE1α-S549 phosphorylation and ERK phosphorylation in treatment-naïve NSCLC patients (Fig. 4H, and fig. S9F). The peptides containing S525, S529, and T973 were not covered in the CPTAC datasets and could not be evaluated in patients.
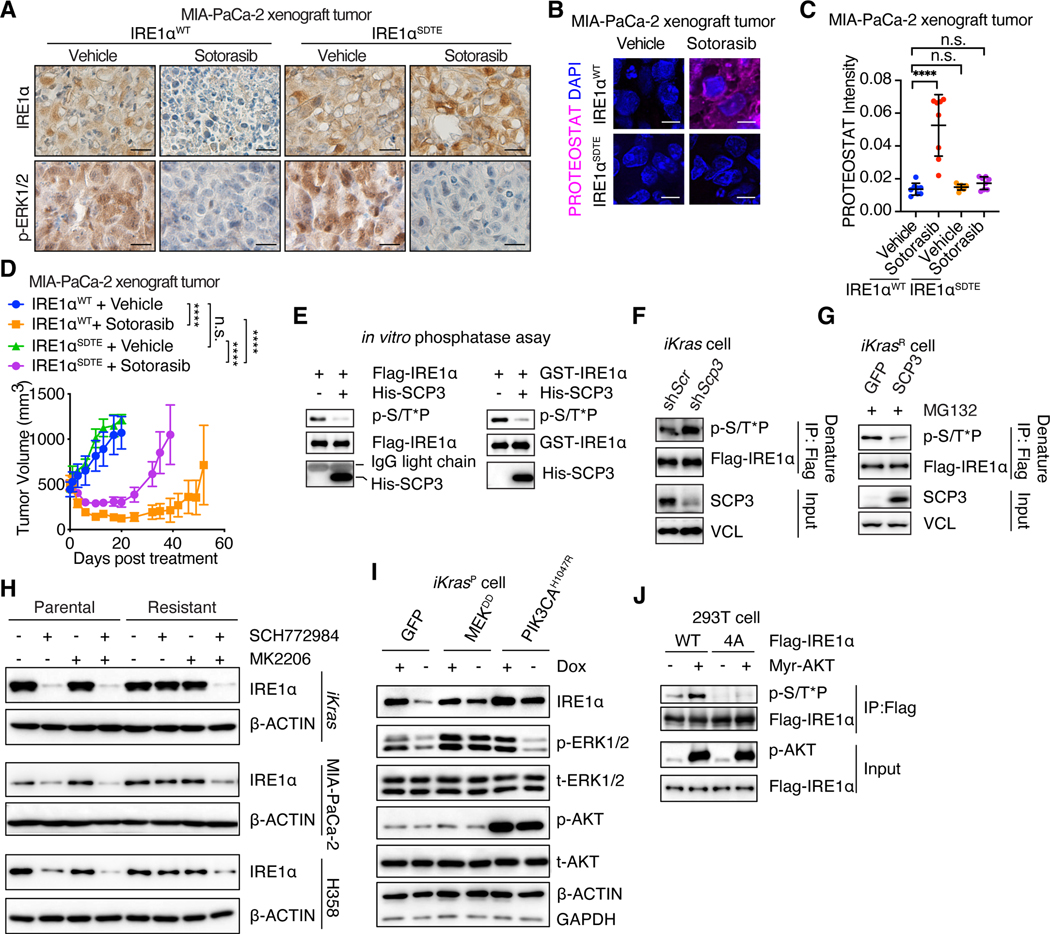
To determine the functional significance of IRE1α phosphorylation sites, we generated loss-of-function mutation for each site. Single site mutation was insufficient to promote IRE1α interaction with HRD1 and did not alter IRE1α levels in iKras cells (Fig. 4, I and J). Simultaneous mutation of all four sites promoted IRE1α interaction with HRD1 and its degradation (Fig. 4, I and J). In vitro pull-down assays confirmed that ERK-mediated IRE1α phosphorylation reduced IRE1α interaction with HRD1/SEL1L and that phospho-deficient IRE1α4A mutant bound to HRD1 regardless of ERK presence (fig. S10, A to E). By contrast, gain-of-function phospho-mimetic mutation for each individual site (S525D, S529D, S549D, or T973E) disrupted IRE1α interaction with HRD1 (Fig. 4K), leading to the stabilization of IRE1α protein in the absence of ERK (Fig. 4L). Similar effects were observed for IRE1αSDTE mutant with gain-of-function mutation for all four sites (Fig. 4, K and L, and fig. S10F). IRE1αSDTE was resistant to sotorasib- or SCH772984- promoted protein degradation in MIA-PaCa-2 cells (fig. S10, G and H). As a result, sotorasib failed to induce protein aggregation in IRE1αSDTE expressing MIA-PaCa-2 tumors (Fig. 5, A to C). These tumors became partially resistant to sotorasib-induced anti-tumor effects (Fig. 5D). In line with these data, the IRE1αSDTE mutant rescued IRE1α-depletion induced protein aggregation, phospho-PERK and cell growth defects in iKrasR cells (fig. S10, I to L). Single-site phospho-deficient IRE1α mutant also rescued these phenotypes due to the presence of the other three phosphorylated sites (fig. S10, L to O). The phospho-deficient IRE1α4A mutant failed to restore IRE1α protein levels and was unable to rescue these phenotypes (fig. S10, L to O). Collectively, these data demonstrate that IRE1α phosphorylation at S525, S529, S549, and T973 inhibits IRE1α association with the ERAD complex, leading to enhanced stability, maintaining proteostasis.
Fig. 5. Multiple pathways converge to restore IRE1α in KRASi-resistant cancer cells.
(A) IHC staining of p-ERK1/2 and IRE1α in shRNA-resistant IRE1αWT- or IRE1αSDTE-transduced, endogenous IRE1α-depleted MIA-PaCa-2 tumors treated with vehicle or sotorasib (100mg/kg) for 4 days. Scale bar: 40μm. (B-C) Representative images (B) and quantification (C) of PROTEOSTAT (magenta) and DAPI (blue) staining in MIA-PaCa-2 tumors as in (A). Data represent average fluorescence intensity of PROTEOSTAT/cell from each image acquired and are presented as mean ± SD from n=8 independent images. Scale bar: 20μm. (D) Tumor volume quantification of MIA-PaCa-2 tumors as in (A). (E) In vitro phosphatase assay. Phosphorylated Flag-IRE1α protein purified from MEKDD-expressing 293T cells (Left panels) or recombinant IRE1α protein phosphorylated by recombinant ERK2 in vitro (Right panels) were subjected to in vitro phosphatase assay with recombinant SCP3. (F-G) iKras cells expressing Flag-IRE1α were infected with shRNA targeting Scr or Scp3 (F), GFP- or SCP3- expressing lentivirus (G). The whole cell lysates were subjected to denature IP with anti-Flag M2 agarose beads, followed by immunoblot with anti-pS/T*P antibody to detect IRE1α phosphorylation. (H) Immunoblot of IRE1α in whole-cell lysates of parental and KRAS inhibition-resistant cells treated with DMSO, 2μM MK2206 and/or 1 μM SCH772984 for 2 days (MIA-PaCa-2 and H358 cells) or 14 days (iKras cells). (I) Immunoblot of IRE1α in whole-cell lysates of iKrasP cells expressing GFP, MEKDD, or PIK3CAH1047R in the presence or absence of Dox for 2 days. (J) 293T cells expressing IRE1αWT or IRE1α4A in the presence or absence of myr-AKT were subjected to IP with anti-Flag M2 agarose beads followed by immunoblot to detect IRE1α phosphorylation. Ordinary one-way ANOVA with Dunnett’s multiple comparisons test (C), and Two-way ANOVA with Bonferroni’s multiple comparisons test (D) were used to calculate P values. n.s., not significant, **P < 0.01, ***P < 0.001, ****P < 0.0001.
A screen of 32 serine/threonine phosphatases in 293T cells revealed that expression of SCP3 significantly reduced MEKDD-induced IRE1α phosphorylation (fig. S11, A to C). In vitro phosphatase assays and co-IP experiments confirmed that SCP3 interacted with and directly dephosphorylated IRE1α (Fig. 5E, and fig. S11D). Scp3 silencing increased IRE1α phosphorylation and overexpression of SCP3 reduced IRE1α phosphorylation in iKras cells (Fig. 5, F and G). Similarly, SCP3 deletion significantly slowed down sotorasib-induced IRE1α dephosphorylation in H358 cells (fig. S11E). These data identified SCP3 as the phosphatase regulating IRE1α phosphorylation, although KRAS did not directly alter SCP3 levels or activities (fig. S11, F to I). Collectively, these analyses establish a mechanism of IRE1α regulation by oncogenic KRAS.
Multiple Pathways Converge to Reactivate IRE1α in KRASi-Resistant Cancer Cells
Next, we sought to determine how IRE1α evades oncogenic KRAS inhibition and determine the reactivation mechanism in KRASi-resistant cells. Oncogenic KRAS was efficiently suppressed by Dox withdrawal in iKrasR cells (Fig. 2A, and fig. S1B), and most KRAS proteins were bound by sotorasib in sotorasib-resistant H358 (H358R) and MIA-PaCa-2 (MIA-PaCa-2R) cells, similar to observations in parental cells (Fig. 2, B and C). Furthermore, silencing of KRAS in H358R cells did not hinder IRE1α reactivation (fig. S12A). These data exclude the possibility that the inefficient inhibition of oncogenic KRAS drives IRE1α reactivation in these resistant cells. eIF2α phosphorylation inhibits global protein synthesis (61, 78). Consistent with the inactivated phospho-eIF2α in KRASi-resistant cells (Fig. 2A), we observed increased global protein synthesis in iKrasR cells evidenced by enhanced puromycin incorporation compared with that in iKrasP cells (fig. S12B). However, inhibition of protein synthesis with cycloheximide did not affect IRE1α/XBP1s in iKrasR cells (fig. S12, C). Furthermore, ER stress sensing-deficient IRE1αD2M mutant (79) was similarly restored in KRASi-resistant cells as that of WT IRE1α (fig. S12, E and F), and successfully rescued IRE1α-depletion induced phenotypes (fig. S12, G to J). These data demonstrate that IRE1α is reactivated in KRASi-resistant cells in an ER stress independent manner.
Recent studies report reactivated ERK and AKT as sotorasib-resistant mechanisms in patients (18, 20–23). Indeed, we observed the reactivation of phospho-ERK and the hyperactivation of phospho-AKT in sotorasib-resistant MIA-PaCa-2R and H358R cells compared with their respective parental cells (Fig. 2, B and C). Unexpectedly, the inhibition of reactivated ERK by SCH772984 treatment was insufficient to suppress IRE1α levels in MIA-PaCa-2R, H358R, and iKrasR cells (Fig. 5H), as well as in MIA-PaCa-2R and iKrasR tumors in vivo (fig. S13, A and B). Similarly, the suppression of AKT by MK2206 treatment had no effect on IRE1α levels in KRASi-resistant cells (Fig. 5H). However, the simultaneous suppression of both ERK and AKT successfully blunted IRE1α reactivation in MIA-PaCa-2R, H358R, and iKrasR cells (Fig. 5H), and in MIA-PaCa-2R and iKrasR tumors in vivo (fig. S13, A and B). Consistently, the hyperactivation of either the MEK/ERK pathway, through expression of MEKDD, or the PI3K/AKT pathway, through the expression of constitutively active PIK3CAH1047R or myr-AKT, resulted in IRE1α restoration in the absence of oncogenic KRAS in iKras cells (Fig. 5I and fig. S13C). Interestingly, myr-AKT also promoted WT, but not phospho-deficient 4A mutant, IRE1α phosphorylation at serine and threonine residues (Fig. 5J), suggesting that these phosphorylation sites are regulated by both ERK and hyperactivated AKT in KRASi-resistant cells. In agreement, the activation of either MEK/ERK, through MEKDD expression, or the hyperactivation of AKT, through myr-AKT expression, was sufficient to disrupt the interaction of the SEL1L/HRD1 E3 ligase complex with IRE1α (fig. S13D). By contrast, the simultaneous suppression of both ERK and AKT, but not ERK or AKT alone, significantly promoted IRE1α interaction with the SEL1L/HRD1 E3 ligase complex in sotorasib-resistant MIA-PaCa-2R cells (fig. S13E). YAP1 also drives resistance of certain tumors to KRASi (37, 38). However, deletion of Yap1 in iKrasR_YAP1 cells derived from YAP1-amplified GEMM tumors escaping KRASG12D addiction did not impact IRE1α and proteostasis (fig. S14, A to F). Instead, IRE1α and proteostasis was dependent on ERK and AKT in these cells despite reduced ERK activity compared with that in parental iKras cells (fig. S14, G to J). Collectively, these data demonstrate that both reactivated ERK and hyperactivated AKT converge through IRE1α phosphorylation at S525, S529, S549, and T973 to prevent ubiquitination-mediated proteasomal degradation of IRE1α in KRASi-resistant cancer cells. Blocking either individual pathway is not sufficient to inhibit IRE1α due to functional redundancy and compensation by the other pathway.
Next, we sought to understand the mechanism that activates ERK and AKT in the KRASi-resistant cells. Receptor tyrosine kinases (RTKs) activation is one of the most common mechanisms driving sotorasib resistance in patients (19–21) and RTKs are known to activate ERK and AKT (17, 18, 30, 80). Array analysis of 49 RTKs in parental and sotorasib-resistant H358 and MIA-PaCa-2 models revealed the induction of multiple and distinct sets of RTKs in each model (fig. S15, A, B, and E). In H358 model, EGFR, ErbB2, ErbB3, FGFR3, and VEGFR2 were significantly induced in the resistant cells (fig. S15, A and B). Inhibiting these RTKs with combined sapitinib, AZD4547 and axitinib, but not individual inhibitor alone, completely suppressed ERK reactivation in H358R cells (fig. S15, C and D). Blocking FGFR3 with AZD4547 largely abolished AKT hyperactivation (fig. S15, C and D). These upregulated RTKs had to be simultaneously suppressed to completely blunt both ERK and AKT, resulting in the abrogation of IRE1α restoration in H358R cells (fig. S15D). Similar to the H358 model, blocking multiple, but not individual, upregulated RTKs (including EGFR, ErbB2, VEGFR, PDGFRb and DDR2) completely suppressed both ERK and AKT, leading to the abrogation of IRE1α restoration in MIA-PaCa-2R cell (fig. S15, F and G). Treatment of MIA-PaCa-2R tumors with combined RTK inhibitors, including sapitinib, axitinib and VU6015929, confirmed the inactivation of ERK and AKT in vivo, leading to the suppression of IRE1α, marked induction of protein aggregation, and reduced tumor growth (fig. S15, H to K). However, they were not well tolerated in the tumor-bearing mice, causing rapid drop of body weight and early lethality (fig. S15L). Collectively, these data demonstrate that multiple and diverse sets of RTKs drive ERK and AKT activation in different KRASi-resistant tumors, which subsequently converge on IRE1α to re-establish proteostasis.
IRE1α Inhibition Sensitizes Oncogenic KRAS-Driven Tumors to a MEK Inhibitor
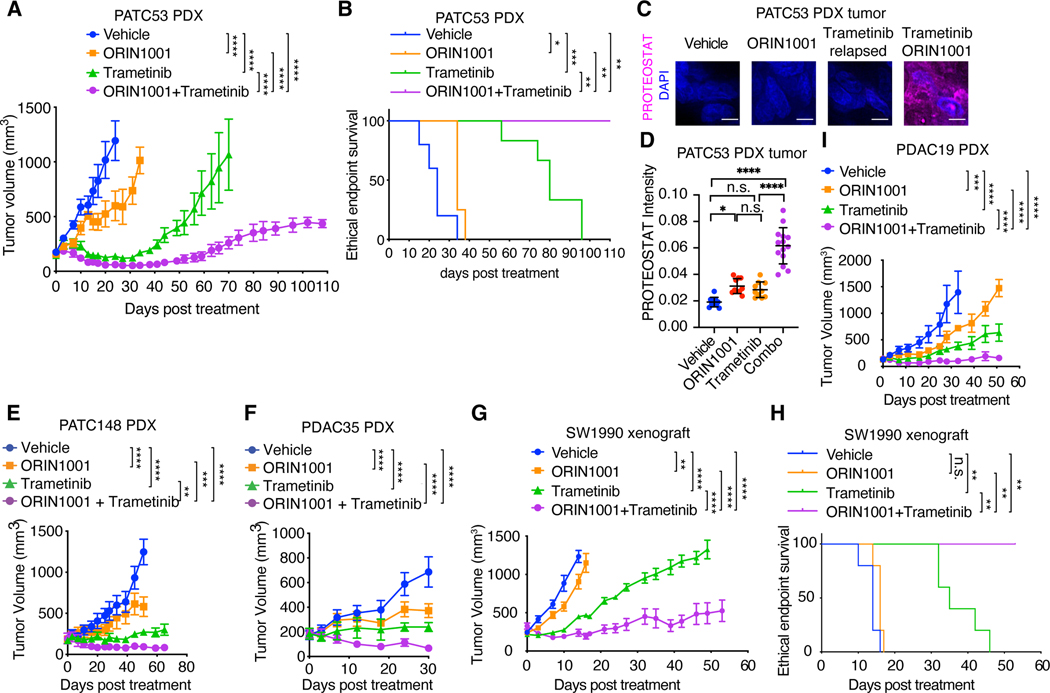
Although the simultaneous suppression of the MAPK and PI3K pathways, or diverse upstream RTKs, effectively inhibits IRE1α, the heterogeneous resistance mechanisms in different patients (18–23) and dose-limiting, on-target toxicity of these inhibitors (81, 82) limits their clinical applications for intervening in IRE1α-mediated proteostasis reprogramming in treatment-resistant tumors. Therefore, we directly targeted the IRE1α/XBP1 pathway in KRAS-driven cancers in combination with KRASG12C or MEK inhibitor. Although MEK inhibitor trametinib or Xbp1 knockout alone both modestly impeded iKras tumor growth in vivo, the loss of Xbp1 significantly enhanced the response of iKras xenograft tumors to trametinib and induced marked protein aggregation (fig. S16, A to D). Treatment of iKras tumors with a highly selective IRE1α RNase inhibitor, ORIN1001 (83–86), recapitulated the effects of the Xbp1 knockout and markedly enhanced the sensitivity of the iKras tumors to trametinib treatment with significant induction of protein aggregation (fig. S16, A, and E to G). In a cohort of PDAC patient-derived xenograft (PDX) models (fig. S16N), ORIN1001 also significantly enhanced the sensitivity of the KRASG12D-mutant PATC53 (Fig. 6, A to D), PATC148 (Fig. 6E, and fig. S16, H and I), PDAC35 (Fig. 6F, and fig. S16, J and K), SW1990 (Fig. 6, G and H, and fig. S16, L and M), and KRASG12V-mutant PDAC19 PDX (Fig. 6I) tumors to trametinib treatment and potently induced protein aggregation in the combination therapy-treated tumors. Collectively, these in vivo data demonstrate that IRE1α/XBP1 inhibition dramatically enhanced the response of KRAS-mutant PDAC tumors to trametinib treatment.
Fig. 6. IRE1α inhibition sensitizes oncogenic KRAS-driven tumors to MEK inhibition.
(A) Tumor volume quantification of established PATC53 PDX tumors in SCID/beige mice treated with vehicle (n=5), IRE1α RNase inhibitor ORIN1001 (n=4), MEK inhibitor trametinib (n=6), or ORIN1001 plus trametinib (n=4). (B) Kaplan-Meier survival curve of PATC53 PDX tumor-bearing mice under treatment as indicated in (A). (C-D) Representative images (C) and quantification (D) of PROTEOSTAT (magenta) and DAPI (blue) staining in endpoint PATC53 xenograft tumors treated as in (A). Data represent average fluorescence intensity of PROTEOSTAT/cell from each image acquired and are presented as mean ± SD from n=10 independent images. Scale bar: 20μm. (E) Tumor volume quantification of established PATC148 PDX tumors in SCID/beige mice treated with vehicle (n=6), ORIN1001 (n=6), trametinib (n=4), or ORIN1001 plus trametinib (n=4). (F) Tumor volume quantification of established PDAC35 PDX tumors in SCID/beige mice treated with vehicle (n=5), ORIN1001 (n=4), trametinib (n=4), or ORIN1001 plus trametinib (n=4). (G) Tumor volume quantification of established SW1990 PDAC xenograft tumors in SCID/beige mice treated with vehicle (n=6), ORIN1001 (n=6), trametinib (n=5), or ORIN1001 plus trametinib (n=4). (H) Kaplan-Meier survival curve of SW1990 PDAC xenograft tumor-bearing mice under treatment as indicated in (G). (I) Tumor volume quantification of established PDAC19 PDX tumors in SCID/beige mice treated with vehicle, ORIN1001, trametinib, or ORIN1001 plus trametinib (n=4). ORIN1001: 150mg/kg. Trametinib: 1mg/kg. Data are presented as mean ± SEM (A, E to G, I) or mean ± SD (D). Two-way ANOVA with Bonferroni’s multiple comparisons test (A, E to G, I), log-rank (Mantel-Cox) test (B and H), or ordinary one-way ANOVA with Dunnett’s multiple comparisons test (D) was used to calculate P values. n.s., not significant, * P<0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001.
IRE1α Inhibition Enhances the Responses of KRASG12C-Driven Tumors to Sotorasib
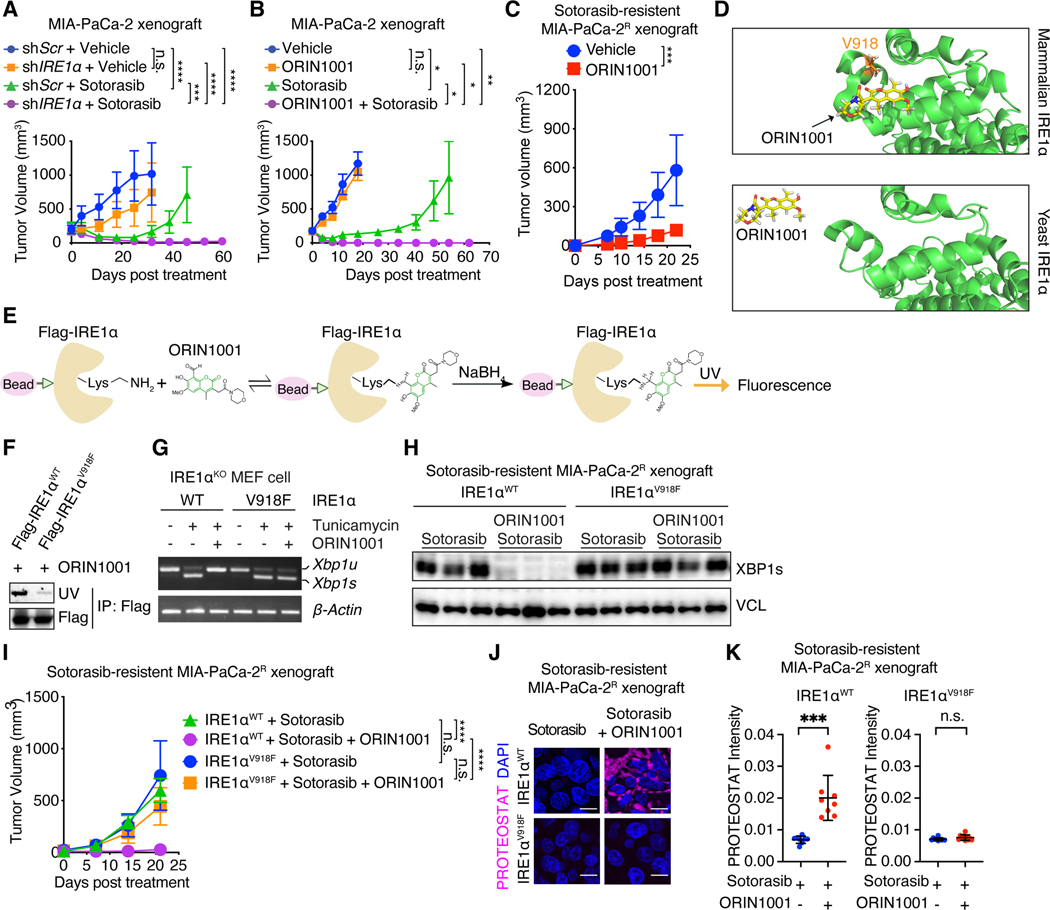
Next, we examined the effects of IRE1α inhibition on the response to sotorasib treatment in KRASG12C-driven tumors. IRE1α silencing modestly reduced MIA-PaCa-2 xenograft tumor growth but not to the extent observed with sotorasib treatment (Fig. 7A and fig. S17A). However, IRE1α deficiency significantly enhanced the response of these tumors to sotorasib treatment and considerably suppressed tumor relapse (Fig. 7A and fig. S17A). In contrast, PERK depletion had little impact on MIA-PaCa-2 tumor growth and response to sotorasib, as well as IRE1α-depletion induced tumor sensitivity to sotorasib (fig. S17, B to E), excluding the involvement of PERK in KRASi resistance. IRE1α inhibition with ORIN1001 treatment combined with sotorasib treatment also resulted in complete MIA-PaCa-2 tumor regression and long-term remission (Fig. 7B, and fig. S17, F and G). We did not observe significant bodyweight changes or signs of toxicity in the combination treatment group (fig. S17, H and I). Treatment of non-KRAS addicted MIA-PaCa-2R tumors with ORIN1001 alone also substantially impeded the tumor growth (Fig. 7C), but did not result in complete response. The reduced efficacy with ORIN1001 alone was likely due to the absence of sotorasib which was required for long-term inhibition of KRASG12C (87) and rewiring of the proteostasis network to be IRE1α-centered.
Fig. 7. IRE1α inhibition enhances tumor responses to sotorasib.
(A) Tumor volume of MIA-PaCa-2 tumors transduced with dox-inducible shScr or shIRE1α and treated with doxycycline water, vehicle (n=5) or sotorasib (n=6). (B) Tumor volume of MIA-PaCa-2 tumors treated with vehicle (n=5), ORIN1001 (n=4), sotorasib (n=4), or both (n=5). (C) Tumor volume of sotorasib-resistant MIA-PaCa-2R tumors treated with vehicle (n=4) or ORIN1001 (n=7). (D) Docking modeling of ORIN1001 with IRE1α. V918 is critical for the formation of the shallow pocket at mammalian IRE1α RNase-active site for ORIN1001 binding. (E) Biochemical fluorescence assay detecting the binding between ORIN1001 and IRE1α in vitro. (F) Equal amount of Flag-IRE1αWT or Flag-IRE1αV918F protein purified from 293T cells was used to pull down ORIN1001 in vitro. UV transmission was used to detect ORIN1001 that is covalently bound to IRE1α protein in the SDS-PAGE. (G) Xbp1 splicing in Ire1α-knock out MEF cells expressing IRE1αWT or IRE1αV918F and treated with tunicamycin (5 μg/mL) and/or ORIN1001 (5 μM) for 6 hours as indicated. (H) Immunoblot of XBP1s in shRNA-resistant IRE1αWT or IRE1αV918F-transduced, endogenous IRE1α-depleted MIA-PaCa-2R tumors treated as indicated. (I) Tumor volume of established shRNA-resistant IRE1αWT- or IRE1αV918F- expressing, endogenous IRE1α-depleted, sotorasib-resistant MIA-PaCa-2R tumors treated as indicated. n=10. (J-K) Representative images (J) and quantification (K) of PROTEOSTAT and DAPI staining in MIA-PaCa-2R tumors treated as in (I). Data represent average fluorescence intensity of PROTEOSTAT/cell from each image and are presented as mean ± SD from n>10 independent images. Scale bar: 20μm. Sotorasib: 100mg/kg. ORIN1001: 300mg/kg. Data are presented as mean ± SEM (A, B, C, and I). Two-way ANOVA test with Bonferroni’s multiple comparisons test (A, B, C, and I) or ordinary one-way ANOVA with Dunnett’s multiple comparisons test (K) was used to calculate P values. n.s., not significant, * P<0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001.
ORIN1001 has over 100-fold mammalian enzyme selectivity over its yeast ortholog (83). Structure analysis of the mammalian and yeast enzymes revealed a critical residue (V918) in mammalian IRE1α that differs from the yeast enzyme and could be critical for ORIN1001 binding (Fig. 7D). Binding of ORIN1001 to IRE1α results in the formation of an imine that could be reduced and detected by UV-excited fluorescence (Fig. 7E). Indeed, whereas purified WT IRE1α protein directly bound to ORIN1001, mutation of valine to phenylalanine at V918 (V918F) abolished the binding (Fig. 7F). The V918F mutation did not affect the ability of ER stressor tunicamycin to induce XBP1 splicing, but completely abolished the response of IRE1α to ORIN1001 (Fig. 7G). Treatment of IRE1αV918F-expressing MIA-PaCa-2R tumors with ORIN1001 failed to inhibit XBP1s in vivo (Fig. 7H). These data identify IRE1αV918F as a drug-resistant mutant that retains intact RNase activity but is immune to ORIN1001. Importantly, the IRE1αV918-expressing, but not WT IRE1α-expressing, MIA-PaCa-2R tumors were completely immune to ORIN1001-induced sensitivity to sotorasib (Fig. 7I) and protein aggregation (Fig. 7, J and K). Collectively, these data confirm the on-target effects of ORIN1001 in vivo.
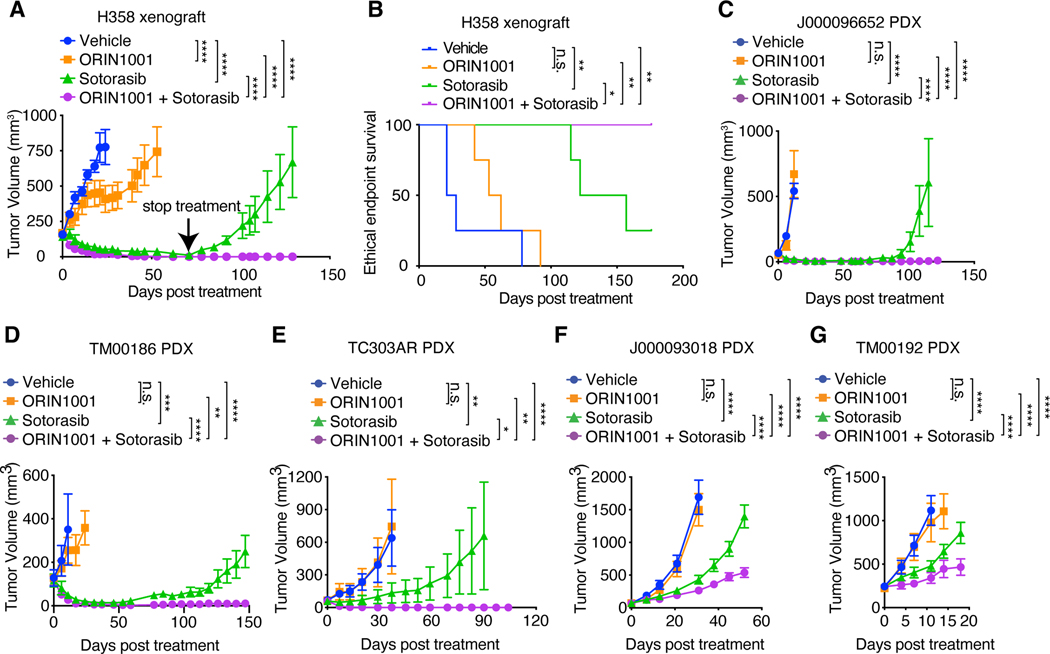
We also tested the therapeutic efficacy of combined ORIN1001 and sotorasib treatment in the H358 NSCLC model. The H358 tumors were highly sensitive to sotorasib treatment, and complete regression was observed within 70 days (Fig. 8A). However, the termination of sotorasib treatment resulted in rapid tumor relapse (Fig. 8A). Remarkably, tumors treated with combined sotorasib and ORIN1001 did not relapse after treatment termination (Fig. 8, A and B). KRAS inhibitor resistance mechanisms are heterogeneous in human patients (18–23). To assess the human relevance, we treated five KRASG12C-mutant NSCLC PDX models with sotorasib and ORIN1001. As shown in Fig. 8, C to G and fig. S18 A to E, ORIN1001 significantly sensitized all five PDX models to sotorasib. In three PDX models (J000096652, TM00186 and TC303AR), the tumors were initially sensitive to sotorasib treatment, but eventually all relapsed. Addition of ORIN1001 to sotorasib led to complete responses and prevented tumor relapse (Fig. 8, C to E). In the other two PDX models (J000093018 and TM00192), ORIN1001 also significantly enhanced the tumor responses to sotorasib, but the responses were not as striking as the other models (Fig. 8, F and G). Analysis of these five PDX models showed that sotorasib did not effectively inhibit MAPK in the J000093018 and TM00192 PDX models (fig. S18, F to J), leading to incomplete reprogramming of the proteostasis network and reduced efficacy (fig. S18, K and L). Collectively, these in vivo data demonstrate that IRE1α inhibition is effective in enhancing the response of KRASG12C-driven tumors to sotorasib. Combination therapy with ORIN1001 and sotorasib achieves complete responses and prevents tumor relapse in a significant portion of KRASG12C-driven tumors.
Fig. 8. IRE1α inhibition enhances the response of KRASG12C-driven tumors to sotorasib.
(A) Tumor volume quantification of established H358 tumors in SCID/beige mice treated with vehicle, ORIN1001 (150mg/kg), sotorasib (30mg/kg), or ORIN1001 plus sotorasib. (n=4). Treatment was stopped at day 71. (B) Kaplan-Meier survival curve of H358 tumor-bearing mice under different treatments as indicated in (A) from treatment start time. (C) Tumor volume quantification of established J000096652 PDX tumors in NSG mice treated with vehicle (n=6), ORIN1001 (300mg/kg, n=4), sotorasib (100mg/kg, n=7), or ORIN1001 plus sotorasib (n=8). Treatment was stopped at day 65. (D) Tumor volume quantification of established TM00186 PDX tumors in NSG mice treated with vehicle (n=6), ORIN1001 (300mg/kg, n=6), sotorasib (100mg/kg, n=7), or ORIN1001 plus sotorasib (n=9). (E) Tumor volume quantification of established TC303AR PDX tumors in NSG mice treated with vehicle (n=5), ORIN1001 (300mg/kg, n=5), sotorasib (100mg/kg, n=9), or ORIN1001 plus sotorasib (n=9). Treatment was stopped at day 53. (F) Tumor volume quantification of established J000093018 PDX tumors in NSG mice treated with vehicle (n=6), ORIN1001 (300mg/kg, n=6), sotorasib (100mg/kg, n=9), or ORIN1001 plus sotorasib (n=9). (G) Tumor volume quantification of established TM00192 PDX tumors in NSG mice treated with vehicle (n=6), ORIN1001 (300mg/kg, n=5), sotorasib (100mg/kg, n=9), or ORIN1001 plus sotorasib (n=9). Data are presented as mean ± SEM (A, C to G). Two-way ANOVA test with Bonferroni’s multiple comparisons test (A, C to G), log-rank (Mantel-Cox) test (B) was used to calculate P values. n.s., not significant, * P<0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001.
Concluding remarks
Most cancers require a balanced proteostasis network to maintain oncogenic growth. Therapeutic insults often disrupt proteostasis and induce proteotoxic stresses (88). How proteostasis network is orchestrated by driver oncogenes and the proteostasis reprogramming mechanisms that bypass oncogene addiction and allow for acquired resistance to targeted therapies remain largely unknown. We show that oncogenic KRAS is critical for protein quality control in tumor cells. Inhibition of oncogenic KRAS inactivates both cytosolic and ER protein quality control machinery by inhibiting the master regulators HSF1 and IRE1α. However, residual cancer cells that survive KRAS inhibition directly restore IRE1α through an ER stress–independent unconventional phosphorylation mechanism that re-establishes proteostasis and sustains acquired resistance to KRAS inhibition.
In contrast to what occurs in non-malignant cells (53), oncogenic KRAS activation resolves, rather than induces, ER stress in transformed cancer cells through oncogenic kinase-dependent phosphorylation of IRE1α. We identified four phosphorylation sites in IRE1α that are distinct from IRE1α autophosphorylation sites. The phosphorylation of IRE1α at these sites prevents IRE1α binding with the SEL1L/HRD1 E3 ligase complex, thus impairing the ubiquitination-dependent degradation of IRE1α and stabilizing the protein. Importantly, these sites are convergence points for multiple resistance pathways and function as central gatekeeper of the rewired proteostasis network in the KRASi-resistant tumors. Inactivation of these sites is sufficient to abolish the direct regulation of IRE1α by oncogenic signaling and collapse the re-established proteostasis to overcome resistance to KRAS inhibitor.
Despite the approval of sotorasib and adagrasib for the treatment of KRASG12C-mutant NSCLC patients, resistance to these inhibitors is rapid and almost inevitable (15, 18–29). The heterogeneous resistance mechanisms in patients and dose-limiting toxicity associated with targeting multiple resistance pathways, such as RTKs, MAPK and PI3K, remain a major barrier to progress. Our mechanistic study directly addressed these clinical challenges by revealing IRE1α-mediated proteostasis reprogramming as a convergence point for multiple heterogenous resistance mechanisms in response to KRAS–MAPK inhibition. ORIN1001 is a highly specific IRE1α RNase inhibitor (83) that demonstrates safety and tolerability in Phase I clinical trial (NCT03950570) despite the occurrence of some adverse effects (86, 89). ORIN1001 substantially enhanced the responses of KRAS-mutant lung or pancreatic cancer PDX models to sotorasib or trametinib. These data demonstrate that direct targeting IRE1α is a more effective and well-tolerated therapeutic strategy for reversing KRASi or MEKi resistance.
In summary, our study reveals the direct crosstalk between oncogenic signaling and the protein quality control machinery. This study elucidated a molecular mechanism that accounts for the proteostasis modulation observed in response to KRAS inhibition. It is noteworthy that the mechanisms of KRAS inhibitor resistance are heterogeneous in patients. Additional studies will be required to examine what proportion of KRAS-driven cancers that develop resistance to KRAS inhibitors use IRE1α-mediated mechanisms of resistance.
Materials and Methods
Cell culture and treatment
MIA-PaCa-2, SW1990, PaTu 8988T, 293T, H358, and BEAS-2B cells were obtained from the American Type Culture Collection (ATCC). Patient-derived PATC53 and PATC148 cells were a gift from Dr. Michael Kim at The University of Texas MD Anderson Cancer Center (90). iKras cells were derived from our previously generated, doxycycline (Dox)-inducible, KrasG12D-driven PDAC mouse model (tetO_LSL-KrasG12D/p53flox/+/p48-Cre/ROSA26-LSL-rtTA-IRES-GFP) (55). LSL-KrasG12D cells were derived from KrasG12D knock-in PDAC mouse model (LSL-KrasG12D/p53flox/+) as described previously (55). The cell lines used in this study are listed in Table S1. BEAS-2B cells were maintained in BEBM Bronchial Epithelial Cell Growth Basal Medium (Lonza, CC-3171) with growth factors and supplements from BEGM Bronchial Epithelial SingleQuots Kit (Lonza, CC-4175). Insulin and hEGF were withdrawn from the medium 48 hours before sample collection. H358, iKras, LSL-KrasG12D, and PATC148 cells were maintained in RPMI supplemented with 10% FBS serum (Gibco, 10437028) and 100μg/mL penicillin/streptomycin (Invitrogen, 15140163). H358R cells were generated by in vitro culture of H358 cells with increasing dose of sotorasib for 6 months until the cells acquired resistance to 30 nM sotorasib. Doxycycline (VWR, AAJ60579–22, 1μg/mL) was added to the iKras cell culture medium to maintain KRASG12D expression. 10% charcoal stripped FBS (VWR, 97065–304) was used to culture iKras cells for doxycycline-withdrawal experiments as previously described (55). iKrasR cells were generated by in vitro culture of parental iKras cells in the absence of Dox until the cells acquired resistance to KRASG12D inactivation. MIA-PaCa-2, SW1990, PaTu 8988T, 293T and PATC53 cells were maintained in DMEM supplemented with 10% FBS (Gibco, 10437028) and 100μg/mL penicillin and streptomycin (Invitrogen, 15140163). MIA-PaCa-2R cells were generated by in vitro culture of parental MIA-PaCa-2 cells with 30 nM sotorasib until the cells acquired resistance to KRAS inhibition. The chemicals used in this study are listed in Table S2.
Tumor inoculation and treatment
The inoculation and establishment of PDX or xenograft tumors was described previously (90). PDAC19 and PDAC35 PDX models were generated by Baylor College of Medicine PDAC PDX Core. TC303AR PDX was generated by MD Anderson Cancer Center PDX Core. J000096652, TM00186, J00093018, and TM00192 PDX models were purchased from Jackson Laboratory. For tumor fragments transplantation of PDX, 1mm3 fresh tumor fragments were transplanted into the lower flanks of 6-week-old immune-compromised SCID/Beige mice (Charles river, strain code 250, CB17.Cg-PrkdcscidLystbg-J/Crl, both female and male) or NSG mice (Jackson Laboratory, stain code 005557, NOD.Cg-Prkdcscid Il2rgtm1Wjl/SzJ, both female and male). For subcutaneous xenograft experiments, 1 × 106 PATC53, SW1990, MIA-PaCa-2, H358 or iKras cells suspended in 100 μL 50% Matrigel (Corning, #354230, in PBS) were injected subcutaneously into the lower flanks of 6-week-old SCID/Beige mice or Athymic Nude mice (Envigo, strain code 69, Hsd:Athymic Nude-Foxn1nu, female). Tumor growth was monitored using calipers and tumor volumes were calculated by the equation V (mm3) = L x W2/2, where L is the largest diameter and W is the perpendicular diameter. When tumors reached a volume of approximately 50–500 mm3, mice were randomized and treated with drugs as indicated. ORIN1001 was provided by Orinove Inc. and suspended in 1% microcrystalline cellulose in 50% sucrose and sonicated for 90 min in water bath sonicator (VWR Ultrasonic Cleaner, Model 97043–964) before dosing at 150 mg/kg or 300 mg/kg body weight via daily oral gavage (83).
Trametinib (1mg/kg) or sotorasib (30, 50 or 100 mg/kg) was formulated in the hydroxypropylmethylcellulose (HPMC)-Tween 80 buffer solution (0.5% HPMC and 0.2% Tween 80, pH 8.0) and administered via daily oral gavage as described previously (12). SCH772984 (50mg/kg) was formulated in 45% saline, 50% PEG 400 and 5% DMSO, and administered via daily intraperitoneal injection. MK2206 (120mg/kg) was formulated in 30% Captisol (Cydex) and administered by oral gavage every other day. All mice were maintained in accordance with Baylor College of Medicine Animal Care and Use Committee procedures and guidelines.
Protein aggregation detection assay
The PROTEOSTAT Aggresome detection kit (Enzo Life Sciences, ENZ-51035-K100), Congo red dye (CR, Sigma, 234610) or thioflavin T dye (ThT, Sigma, T3516) was used to detect misfolded or aggregated proteins in cells or tumor tissues (56, 72). The PROTEOSTAT aggresome detection assay was performed according to the manufacturer’s instructions. Briefly, the iKras cells seeded on glass slides were washed with PBS, fixed with 4% formaldehyde for 30 min at room temperature (RT), permeabilized with Permeabilizing Solution (0.5% Triton X-100, 3mM EDTA) for 30 min on ice with gentle shake, and stained with the PROTEOSTAT dye (1:20,000 dilution) for 30 min at RT. MIA-PaCa-2 or H358 cells were trypsinized from culture dishes followed by washing, fixation, permeabilization and staining as described above. Tumor sections were deparaffinized and rehydrated before staining. Samples were incubated with PROTEOSTAT dye (1:20,000 dilution) for 30 min at RT. Nuclei were counterstained with DAPI. Cells treated with 10 μM MG132 (provided in the PROTEOSTAT Aggresome detection kit) for 16 h was used as positive control. Samples stained with DAPI only were used as negative control. Congo red (CR) and thioflavin T (ThT) staining were performed as described previously (56). Briefly, the iKras cells seeded on glass slides were washed with PBS, fixed with 4% formaldehyde for 30 min at RT, permeabilized with Permeabilizing Solution (0.5% Triton X-100, 3mM EDTA) for 30 min on ice with gentle shake, and stained with 20 μM ThT or 50 nM CR dissolved in PBS for 30min at RT, followed by rinsing in PBS for 3 times. Nuclei were stained with DAPI.
Images (16-bit greyscale TIFFs) were analyzed using CellProfiler v2.2 (Broad Institute) as described previously (72). Briefly, the DAPI channel images were first smoothened with a median filter and nuclei were identified with automatic thresholding and a fixed diameter. The cell nuclei that touch the border of the image were eliminated for quantification. The cell nuclei that touch each other were separated with a watershed algorithm. Then, cell boundaries were identified by watershed gradient based on the dye signal of PROTEOSTAT, ThT or CR, using nuclei as a seed. Metrics for PROTEOSTAT, ThT or CR were extracted from the cells.
Cell viability assay
200 cells were seeded in 96-well plate and treated with different inhibitors as indicated in the figures. Cell viability was measured daily with CCK-8 kit (APExBIO, # K1018). Briefly, 10μL CCK-8 solution was added to each well and incubated for 1 hour at 37 °C. The absorbance at 450 nm was measured using BioTek Synergy HTX Multi-Mode microplate reader.
Colony formation assay
200 cells were seeded in 12-well plate and cultured for 5 days. Cells were washed with PBS, fixed with methanol and stained with 0.5% crystal violet. The colony number was counted and quantified.
BrdU incorporation Assay
For BrdU staining, cells in the logarithmic phase of proliferation were first labeled with BrdU at a final concentration of 10 μM for 30 min at 37 °C, followed by intracellular staining using the BrdU staining kit according to the manufacturer’s instructions (BD Biosciences, 559619). Flow cytometry data were collected using BD FACS Diva 8 on a BD LSR II or BD Fortessa analyzer. The acquired data were analyzed using the FlowJo 10 software.
Luciferase assay
For iKras cells, the HSF1 firefly luciferase reporter was constructed by cloning 4 copies of the heat shock element (HSE) followed by a minimal promoter sequence (5’-AGAGGGTATATAATGGAAGCTCGACTTCCAG-3’) (Promega, E375A) (all primer sequences are listed in Table S3) into the SacI and HindIII sites of the pGL3-basic luciferase reporter (Promega, E1751). The construct was verified by DNA sequencing. The iKrasP or iKrasR cells were seeded in 96-well plate at 800 cells/well and co-transfected with 100ng firefly luciferase reporter and 5ng Renilla luciferase plasmid (pRL-TK, Promega, E2241, used as internal control) using Lipofectamine 3000 (Thermo Fisher, L300008). The transfected iKrasPcells cultured in the presence or absence of Dox for 48 h or the transfected iKrasR cells cultured in the absence of Dox were heat shocked at 43 °C for 1 h and recovered overnight before measuring the luciferase activities using the Dual-luciferase Reporter Assay System (Promega, #E1910) according to the manufacturer’s instructions. For MIA-PaCa-2 cells, the same sequence (Promega, E375A) was cloned into the XhoI and BamHI sites of the pRRL-Luciferase plasmid (Addgene, 120798). The construct was verified by DNA sequencing. The firefly or pLenti.PGK.blast-Renilla_Luciferase (Addgene, 74444) plasmid was packaged into lenti-viruses and infected MIA-PaCa-2P or MIA-PaCa-2R cells. After selection with blasticidine (20 μg/mL), the cells were seeded in 96-well plate at 800 cells/well. The infected MIA-PaCa-2P cells cultured in the presence or absence of sotorasib for 48 h were heat shocked at 43 °C for 1 h and recovered overnight before measuring the luciferase activities using the Dual-luciferase Reporter Assay System (Promega, #E1910) according to the manufacturer’s instructions.
Proteasome activity assay
The Proteasome Activity Fluorometric Assay Kit (BioVision, K245) was used to detect proteasome activity according to the manufacturer’s instructions. Briefly, 1×106 iKras cells from different treatments were homogenized in a tight-fitting bounce homogenizer (Thomas Scientific, 1176F27) with 500μl 0.5% NP-40 in PBS. 10 μL Proteasome Substrate (AMC-peptide, provided in the kit) was added to 10 μL cell lysate from each treatment group or 10 μL positive control lysate (provided in the kit) and mixed. The reaction was performed at 37 °C and protected from light. The kinetics of fluorescence at excitation/emission = 350/440 nm were measured every 30 min using BioTek Synergy HTX Multi-Mode Plate Reader. Cells treated with 10μM MG132 (provided in the detection kit) for 16 h were used as negative control. The fluorescence signals were normalized against total protein abundance detected with BCA Protein Assay Kit (Thermo Fisher Scientific, 23225).
Plasmids, virus production, and infection
The plasmids used are listed in Table S4. The pRK5-Flag-IRE1α or pCDH-Flag-IRE1α was generated by cloning the full-length human IRE1α into pRK5 (Genentech) or pCDH (System Biosciences, CD511B-1) vector. The point mutations of IRE1α were introduced with Q5 Site-Directed Mutagenesis Kit (New England Biolabs, E0554). All the IRE1α plasmids are shRNA-resistant and listed in Table S4. Primers used for cloning are listed in Table S3. pHAGE-BRAFV600E, pHAGE-MEKDD, and pHAGE-PIK3CAH1047R plasmids were generated as described previously (91). The pCDH-HA-Myr-AKT plasmid was purchased from Addgene (# 46969) (92). pLVX-Flag-HA-HRD1 was generated as described previously (73). The shRNAs targeting mouse Xbp1, Ire1α, Perk, Gcn2, Hri, Pkr and Scp3 were cloned into pLKO.1-TRC (Addgene, 10878). The shRNAs targeting human XBP1, IRE1α, NcK, and PERK were cloned in pLKO.1-TRC (Addgene 10878) or pLKO-Tet-On (Addgene 21915) vector to generate constitutive or inducible constructs. The shRNA sequences are listed in Table S3. The pLKO.1 shScramble (Addgene 1864) or pLKO.1 Tet-On shScramble (same shRNA sequence as in pLKO.1 shScramble) was used as control. The p-GIPZ non-silencing shRNA control, p-GIPZ-MAPK1, p-GIPZ-MAPK3 were from Dharmacon Reagents. The Cas9-expressing plasmid lentiCas9-Blast was purchased from Addgene (#52962) (93). The gRNAs targeting Ire1α or Xbp1 were cloned into lentiGuide-Puro vector (Addgene, #52963). All gRNA sequences are listed in Table S3. Plasmids containing coding sequence of different phosphatases are listed in Table S4. To generate lentiviruses, 293T cells were co-transfected with psPAX2 and pMD2.G using Lipofectamine 3000 (Thermo Fisher Scientific, # L3000008). Lentiviruses were collected 48 and 72 hours after transfection and used for infecting cells in the presence of 8 μg/ml polybrene (Millipore Sigma, TR-1003-G) prior to puromycin selection (2 μg/ml, Millipore Sigma, P8833).
Generation of knock-out (KO) cells
To generate Ire1α or Xbp1 KO cells, iKras cells were first infected with lentiviruses encoding Cas9 (lentiCas9-Blast, Addgene 52962) (93) and selected with 10 μg/mL blasticidin (Santa Cruz, 3513–03-09). The Cas9-expressing cells were then infected with lentiviruses expressing two gRNAs targeting the same exon and selected with puromycin (2μg/mL, MilliporeSigma, P8833) to generate pooled KO cells. The gRNA sequences are listed in Table S3.
Immunohistochemical (IHC) Staining
Tumor specimens were fixed with freshly made 4% paraformaldehyde for 24 h, washed with PBS and stored in 70% ethanol until paraffin embedding. IHC staining was performed on 5 μm-thick paraffin sections. For p-ERK, p-HSF1, p-GCN2, p-eIF2α and YAP1 staining, 10mM sodium citrate buffer (pH 6.0) was used for antigen retrieval. For IRE1α, p-PERK, ATF4 and p-AKT IHC, 1mM EDTA buffer (pH 9.0) was used for antigen retrieval. Endogenous peroxidase was quenched with 3% H2O2 for 20 min followed by blocking with 3% normal goat serum. The following primary antibodies were used: IRE1α (1:20, Cell Signaling Technology, 3294); p-ERK (1:200, Cell Signaling Technology, 4376); and p-HSF1 (1:200, Life Technologies, BSM-52166R); p-PERK (1:25, Cell Signaling Technology, 3179); ATF4 (1:50, Santa Cruz, 390063); p-GCN2 (1:200, Thermo Fisher, PA5–105886); p-AKT (1:50, Cell Signaling Technology, 4060); p-eIF2α(1:50, Cell Signaling Technology, 9721); YAP1 (1:400, Cell Signaling Technology, 14074). Slides were incubated with ImmPRESS Excel HRP Goat Anti-Rabbit Polymer Reagent (Vector labs, MP-7451–15) for 30 minutes. Sections were developed with DAB+ solution (Dako, K3468) and counterstained with Harris Hematoxylin. The antibody used are listed in Table S5.
Tissue microarray (TMA) analysis
For quantifications of TMA staining, TMAs stained with anti-IRE1α or anti-p-ERK antibody were scanned using the Aperio scanner and analyzed with QuPath software (94). Detailed tutorials of the software can be found at https://qupath.readthedocs.io/en/stable/index.html. Briefly, images were preprocessed by automated ‘TMA dearraying’ and ‘stain’ vector estimation. Tissue sections were identified by running ‘simple tissue detection’. The ‘positive cell detection’ command was used to detect DAB staining intensity. The score compartment was set as ‘DAB OD mean’. Tumor cells and stromal cells were classified by ‘training object classifier’ based on annotations. The fraction score was calculated as the proportion of positively stained tumor cells (0%−100%). The intensity and fraction scores were then multiplied to obtain the H-score.
RNA extraction and real-time quantitative reverse-transcriptase PCR
Total RNA was extracted using TRIzol (Thermo Fisher Scientific, 15596026). Total RNA (1 μg) was converted to cDNA using the High-Capacity cDNA Reverse Transcription Kit (Applied Biosystems, 4368813), followed by qPCR on a QuantStudio 6 Flex Real-Time PCR System (Applied Biosystems). The sequences of all primers are listed in Table S3.
Detergent-insoluble aggregates detection
Detergent-insoluble aggregates detection was performed as described previously (47). Briefly, cells with different treatments were harvested by trypsinization. After washing with cold PBS, 1 × 106 cells were lysed with RIPA buffer (25 mM Tris-HCl pH7.6, 150 mM NaCl, 1% Triton X-100, 1% sodium deoxycholate, 0.1% SDS, 1.5 mM EDTA) supplemented with protease inhibitor cocktail (Roche, 14826500), phosphatase inhibitor cocktail (Sigma, 4906845001) and 10 mM N-Ethylmaleimide (Sigma, E3876). Protein lysates were cleared by centrifugation at 16,000 g for 20 min at 4 °C. The remaining insoluble pellets were then washed with RIPA buffer for three times to remove any remaining detergent-soluble proteins. The pellet containing detergent-insoluble aggregates was solubilized with urea buffer (8 M urea, 2% SDS, 50 mM DTT, 50 mM Tris-HCl pH7.4) for western blot analysis. 3 × 105 cells were directly lysed in urea buffer (8 M urea, 2% SDS, 50 mM DTT, 50 mM Tris-HCl pH7.4) serving as loading control.
Co-immunoprecipitation
The co-immunoprecipitation (co-IP) assay was performed as previously described (73). Briefly, 293T cells transfected with different plasmids or H358 cells infected with indicated viruses were lysed with lysis buffer (50mM Tris-HCl pH7.5, 150 mM NaCl, 1 mM EDTA, 1% Triton X-100) supplemented with protease inhibitor cocktail (Roche, 14826500) and phosphatase inhibitor cocktail (Sigma, 4906845001). Protein lysates were cleared by centrifugation at 12,000 g for 20 min at 4 °C. Supernatant was incubated with anti-Flag M2 beads (Sigma, F-2426) or anti-Myc beads (Sigma, E-6654) for 4h to overnight at 4 °C with gentle rotating. The beads were then washed once with lysis buffer, followed by additional three washes with wash buffer (50mM Tris-HCl pH7.5, 150 mM NaCl, 1 mM EDTA, 1% Triton X-100, 10% glycerol). The proteins were eluted by boiling in 2 × Laemmli sample buffer (65.8 mM Tris-HCl, pH 6.8, 2.1% SDS, 26.3% (w/v) glycerol, 355 mM b-mercaptoethanol, 0.01% bromophenol blue) and analyzed by SDS-PAGE and western blot.
Ubiquitination and phosphorylation assay
The ubiquitination and phosphorylation assays were performed to detect IRE1α ubiquitination and phosphorylation. MIA-PaCa-2 or H358 cells infected with lentiviruses encoding control or Flag-IRE1α were treated with DMSO, sotorasib or trametinib as described in figure legend and lysed with RIPA buffer (25 mM Tris-HCl pH7.6, 150 mM NaCl, 1% Triton X-100, 1% sodium deoxycholate, 0.1% SDS, 1.5 mM EDTA) supplemented with protease inhibitor cocktail (Roche, 14826500), phosphatase inhibitor cocktail (Sigma, 4906845001) and 10 mM N-Ethylmaleimide (Sigma, E3876). Protein lysates were sonicated for 30s and cleared by centrifugation at 12,000 g for 20 min at 4 °C. Supernatant was incubated with anti-Flag M2 beads (Sigma, F-2426) for 4h at 4 °C with gentle rotating. The beads were then washed with RIPA buffer for three times. The immunoprecipitates were eluted and denatured by boiling for 10 min in denature buffer (50 mM Tris-HCl pH7.6, 1% SDS, 0.5 mM EDTA, 1 mM DTT) to disrupt the interactions between immunoprecipitated IRE1α and its interacting proteins. The denatured elutes were diluted 1:10 with lysis buffer (50mM Tris-HCl pH7.6, 150 mM NaCl, 1 mM EDTA, 1% Triton X-100) and subjected to a second-round immunoprecipitation with anti-Flag M2 beads (Sigma, F-2426) (4h at 4 °C) to remove all the interacting proteins and selectively pull down only IRE1α protein which allows for the specific analysis IRE1α ubiquitination and phosphorylation. The beads were then washed with RIPA buffer for three times. The proteins were eluted by boiling in 2 × Laemmli sample buffer (65.8 mM Tris-HCl pH 6.8, 2.1% SDS, 26.3% (w/v) glycerol, 355 mM β-mercaptoethanol, 0.01% bromophenol blue) and analyzed by SDS-PAGE and western blot.
GST pull-down assay
The GST pull-down assay was performed to detect interaction between ERK and IRE1α. Briefly, recombinant GST or GST-ERK2 protein purified from E.coli (SignalChem, M28–10G-20) was incubated with recombinant His-IRE1α protein purified from Sf9 cells (83) in RIPA buffer (25 mM Tris-HCl pH7.6, 150 mM NaCl, 1% Triton X-100, 1% sodium deoxycholate, 0.1% SDS, 1.5 mM EDTA) for 30 min before the GSH-Sepharose beads (GE Healthcare, 17075601) were added and rotation for 1 h at 4 °C . The beads were washed with RIPA buffer for three times. The proteins were eluted by boiling in 2 × Laemmli sample buffer and analyzed by SDS-PAGE and western blot.
Flag pull-down assay
293T cells transfected with plasmid expressing Flag-GFP or Flag-IRE1α were lysed in RIPA buffer (25 mM Tris-HCl pH7.6, 150 mM NaCl, 1% Triton X-100, 1% sodium deoxycholate, 0.1% SDS, 1.5 mM EDTA, supplemented with protease inhibitor cocktail and phosphatase inhibitor cocktail) 48 hours after transfection. Protein lysates were cleared by centrifugation at 12,000 g for 20 min at 4 °C. Supernatant was incubated with anti-Flag M2 beads (Sigma, F-2426) overnight at 4 °C with gentle rotating. The beads were then washed with RIPA buffer for three times. These preloaded Flag M2 beads with Flag-GFP or Flag-IRE1α were then incubated with purified GST-ERK2 (SignalChem, M28–10G-20) for 30 min before washing with wash buffer (50mM Tris-HCl pH7.5, 150 mM NaCl, 1 mM EDTA, 1% Triton X-100) for three times and elution by boiling in 2 × Laemmli sample buffer for 10 minutes. The eluents were analyzed by SDS-PAGE and western blot.
In vitro kinase assay
Flag-GFP or Flag-IRE1α proteins were purified from 293T cells as described above. The Flag M2 beads pre-loaded with Flag-GFP or Flag-IRE1α proteins were rinsed with kinase assay buffer I (SignalChem, K01–09, 25 mM MOPS pH7.2, 12.5 mM beta-glycerol-phosphate, 25 mM MgCl2, 5 mM EGTA, 2 mM EDTA, 0.25 mM DTT) and subjected to in vitro kinase assay in the kinase assay buffer I plus 0.4 mM cold ATP and 1.0 μg GST-ERK2 activated by MEK1 in vitro (SignalChem, M28–10G-20). The reaction was carried out at 30 °C for 30 min. The beads were then washed for three times with RIPA buffer (25 mM Tris-HCl pH7.6, 150 mM NaCl, 1% Triton X-100, 1% sodium deoxycholate, 0.1% SDS, 1.5 mM EDTA). The proteins were eluted by boiling in 2 × Laemmli sample buffer and analyzed by SDS-PAGE and western blot.
For in vitro kinase assay labeled with [g-32P] ATP, Flag-IRE1αK599A (kinase-dead form) or different mutated Flag-IRE1αK599A proteins expressed in 293T cells were bound to anti-Flag M2 beads as described above and incubated with 0.5 μg GST-ERK2 activated by MEK1 in vitro (SignalChem, M28–10G-20) in the presence of 1 μCi of [g-32P] ATP (PerkinElmer, NEG002A100UC) and 0.4 mM cold ATP in the kinase buffer I (SignalChem, K01–09) for 30 min at 30 °C. The reaction was stopped by boiling in 2 × Laemmli sample buffer for 10 minutes. The eluted proteins were resolved by SDS-PAGE and detected by autoradiography.
For in vitro kinase assay with recombinant His-IRE1α, 1.0 μg protein purified from Sf9 cells (83) and recombinant GST-ERK2 protein, the reaction was carried out in the kinase assay buffer I (SignalChem, K01–09) plus 0.4 mM cold ATP for 30 min at 30 °C. The reaction was stopped by adding 8 M urea buffer followed by purification of His-IRE1α with Ni-NTA agarose (QIAGEN, 30210). The proteins were eluted with 2 × Laemmli sample buffer by boiling for 10 minutes and analyzed by SDS-PAGE and western blot.
In vitro phosphatase assay
The in vitro phosphatase assay was performed as described previously (95). Phosphorylated Flag-IRE1α proteins were purified from 293T cells expressing MEKDD as described above. The Flag M2 beads pre-loaded with phosphorylated Flag-IRE1α proteins were rinsed with phosphatase assay buffer (40 mM Tris-HCl pH7.5, 20 mM KCl, 10 mM MnCl2, and 2 mM DTT) and subjected to in vitro phosphatase assay in the phosphatase assay buffer and 1.0 μg recombinant His-SCP3 protein (NOVUS, NBP1–99109). The reaction was carried out at 30 °C for 30 min. The beads were then washed for three times with RIPA buffer (25 mM Tris-HCl pH7.6, 150 mM NaCl, 1% Triton X-100, 1% sodium deoxycholate, 0.1% SDS, 1.5 mM EDTA). The proteins were eluted by boiling in 2 × Laemmli sample buffer and analyzed by SDS-PAGE and western blot. For in vitro kinase assay with recombinant His-IRE1α, 1.0 μg His-IRE1α protein was phosphorylated by recombinant GST-ERK2 protein in vitro as described above, and then subjected to in vitro phosphatase assay with SCP3 protein. 1.0 μg recombinant His-SCP3 protein (NOVUS, NBP1–99109) or Myc-SCP3 protein purified from H358 cells treated with DMSO or sotorasib (30 nM) were used for the in vitro phosphatase assay as indicated in the figures. The proteins were boiled in 2 × Laemmli sample buffer and analyzed by SDS-PAGE and western blot.
Phospho-RTK array
The Proteome Profiler Human Phospho-RTK Array Kit (R&D systems, ARY001B) was used to assess the phosphorylation status of 49 RTKs in H358 cells or MIA-PaCa-2 tumors treated with vehicle or sotorasib. The phospho-RTK array assay was performed according to the manufacturer’s instructions. Briefly, the H358 cells or MIA-PaCa-2 tumors were lysed in the Lysis Buffer 17 (provided in the kit) with protease inhibitors. The protein concentration was measured with Pierce™ BCA Protein Assay Kit (ThermoFisher Scientific, 23225). 1000 μg total protein lysate was incubated with the RTK array membrane overnight at 4°C. After washing, the anti-phospho-tyrosin-HRP detection antibody was incubated with the membrane, and chemiluminescence signal was detected with Amersham Imager 600 (GE Healthcare Lifer Sciences) and quantified with Image J.
Biochemical fluorescence assay to detect IRE1α interaction with ORIN1001
The in vitro binding assay to detect IRE1α with ORIN1001 was performed as described previously (96). Briefly, WT or V918F mutant Flag-IRE1α proteins were purified from 293T cells as described above. The Flag M2 beads pre-loaded with equal amount of Flag-IRE1αWT or Flag-IRE1αV918F proteins were incubated with 200 μM ORIN1001 in binding buffer (50 mM Tris-HCl pH7.5, 100 mM NaCl, 10% Glycerol, 1% Triton X-100, 50 mM EDTA) for 3 hours at 4°C. After that, 6 mM NaBH4 was added to reduce the imine (Schiff base) between IRE1α and ORIN1001 to stable amine. The beads were then extensively washed for three times with binding buffer and three times with RIPA buffer (25 mM Tris-HCl pH7.6, 150 mM NaCl, 1% Triton X-100, 1% sodium deoxycholate, 0.1% SDS, 1.5 mM EDTA). The proteins were eluted by boiling in 2 × Laemmli sample buffer and analyzed by SDS-PAGE. UV transmission (GE Healthcare Lifer Sciences) was used to detect ORIN1001 that is covalently bound to IRE1α protein in the SDS-PAGE.
Modelling of the IRE1α-ORIN1001 complex
Murine IRE1α and MKC9989 (PDB 4PL3) was used as a template to model ORIN1001 bound to murine IRE1α with Schrodinger software. Homology modelling of yeast IRE1α (PDB 3FBV) was also achieved and was superimposed manually onto the model of ORIN1001 with murine IRE1α.
Clinical Proteomic Tumor Analysis Consortium (CPTAC) data analysis
Correlation between IRE1α phosphorylation (S549) and ERK phosphorylation (Y204) in CPTAC NSCLC tumors were analyzed using the LinkedOmics platform (97) (http://www.linkedomics.org).
Western Blot
Whole cell lysates or immunoprecipitation samples were separated by SDS-PAGE and transferred to nitrocellulose membrane (Biorad, 1620112). The antibody used are listed in Table S5. The reagents and kits used in this study are listed in Table S2.
Statistics and reproducibility
Data are expressed as the mean ± SD or mean ± SEM as indicated in the figure legends; n is the number of independent biological replicates, unless specifically indicated otherwise in the figure legend. The respective n values are shown in the figure legends. No statistical method was used to pre-determine the sample sizes. For animal experiments, at least 4 biological replicates were included based on previously published work, preliminary studies as standard for this field of research. See figures legends for each experiment. Treatments were performed in a non-blinded manner by a research technician who was not aware of the objectives of the study because the colors of the drugs used are different from that of vehicle control. The results were quantified using GraphPad Prism 8. 2-tailed, unpaired Student’s t test with or without Welch’s correction was utilized to compare the differences between 2 groups as indicated in figure legends. One-way ANOVA with Dunnett’s or Tukey’s multiple comparison test was used to compare the differences among 3 or more groups as indicated in figure legends. Two-way ANOVA with Bonferroni’s multiple comparison test was used to calculate the significance difference for cell growth, tumor volume and body weight measurement over time. The log-rank (Mantel-Cox) test was used to test for the significant differences of survival between the groups. P < 0.05 was considered statistically significant. No samples or animals were excluded from the analysis.
Study approval
All protocols described in this study were approved by the Baylor College of Medicine Institutional Animal Care and Use Committee (AN6813).
Supplementary Material
Acknowledgements
We thank Dr. J. Rosen, Q. Zhang, and J. Xu for advice, discussion, and critical review of the manuscript.
Funding:
This work was supported by US Department of Defense Congressionally Directed Medical Research Programs (W81XWH1910524 to X.C., W81XWH1910035 to X.Lv.), the National Institutes of Health (R01CA270240, R37CA228304, P50CA186784, and R01HL146642 to X.C.; R01CA214793 and 5P01CA117969 to H.Y.; 1R01CA272744 to W.Y., R01DK115454 and R01GM142143 to A.C.; T32DK60445–17 to L.M., CA215591 and SPORE DRP CA126752 to Z.S., U24CA210954 to B.Z., U54CA224065 and U54CA224081 to J.A.R.), Cancer Prevention and Research Institute of Texas (RP230285 to X.C., RR140038 CPRIT Scholar in Cancer Research award to A.C., and RP160283 Baylor College of Medicine Comprehensive Cancer Training Program award to F.P., RR160027 CPRIT Scholar in Cancer Research award to B.Z.), and American Cancer Society (RSG-22–017-01-CCB to W.Y.). This work utilized the Aperio GT-450 digital pathology scanner in the Baylor College of Medicine Breast Center Pathology Core that was purchased with funding from the NIH S10 grant S10OD028671. This work was supported by the Cytometry and Cell Sorting Core at the Baylor College of Medicine with funding from the CPRIT Core Facility Support Award (CPRIT-RP180672), the NIH (CA125123, S10OD025251 and RR024574) and the assistance of J.M. Sederstrom. Imaging for this work was supported by the Integrated Microscopy Core at the Baylor College of Medicine with funding from NIH (DK56338 and CA125123) and CPRIT (RP150578). This work was supported by the DNA Sequencing Core at the Baylor College of Medicine with funding from the NIH support (R01HD100535) and the assistance of Yunping Lei and Paula Pimienta Ramirez.
Competing interests:
M.F.R. receives research funding from Pfizer and Genentech. M.F.R. is a consultant and receives consulting fees from: Novartis, Seagen, Macrogenics, and AstraZeneca. M.F.R. is the Principal Investigator of the clinical trial NCT03950570. X.C. reports previous research funding from Fosun Pharma. J.A.R. reports receiving a commercial research grant from The University of Texas MD Anderson Cancer Center, sponsored research agreement from Genprex, Inc, has ownership interest (including stock, patents, etc.) in Genprex, Inc., and is a consultant/advisory board member for Genprex, Inc. M.J.E. reports full time employment with AstraZeneca from March 7th 2022. M.J.E. holds patents and receives income from Veracyte on the PAM50-based products, Prosigna.
Data and materials availability:
All data are available in the manuscript or the supplementary material. Requests for materials should be addressed to X.C.
References
- 1.Scheidig AJ, Burmester C, Goody RS, The pre-hydrolysis state of p21(ras) in complex with GTP: new insights into the role of water molecules in the GTP hydrolysis reaction of ras-like proteins. Structure 7, 1311–1324 (1999). [DOI] [PubMed] [Google Scholar]
- 2.Scheffzek K. et al. , The Ras-RasGAP complex: structural basis for GTPase activation and its loss in oncogenic Ras mutants. Science 277, 333–338 (1997). [DOI] [PubMed] [Google Scholar]
- 3.Bollag G, McCormick F, Differential regulation of rasGAP and neurofibromatosis gene product activities. Nature 351, 576–579 (1991). [DOI] [PubMed] [Google Scholar]
- 4.Li C. et al. , The G protein signaling regulator RGS3 enhances the GTPase activity of KRAS. Science 374, 197–201 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.McGrath JP, Capon DJ, Goeddel DV, Levinson AD, Comparative biochemical properties of normal and activated human ras p21 protein. Nature 310, 644–649 (1984). [DOI] [PubMed] [Google Scholar]
- 6.Downward J, Targeting RAS signalling pathways in cancer therapy. Nat. Rev. Cancer 3, 11–22 (2003). [DOI] [PubMed] [Google Scholar]
- 7.Simanshu DK, Nissley DV, McCormick F, RAS proteins and their regulators in human disease. Cell 170, 17–33 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cox AD, Fesik SW, Kimmelman AC, Luo J, Der CJ, Drugging the undruggable RAS: Mission possible? Nature reviews. Drug discovery 13, 828–851 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Encarnación-Rosado J, Kimmelman AC, Harnessing metabolic dependencies in pancreatic cancers. Nat. Rev. Gastroenterol. Hepatol. 18, 482–492 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pylayeva-Gupta Y, Grabocka E, Bar-Sagi D, RAS oncogenes: weaving a tumorigenic web. Nat. Rev. Cancer 11, 761–774 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ostrem JM, Peters U, Sos ML, Wells JA, Shokat KM, K-Ras(G12C) inhibitors allosterically control GTP affinity and effector interactions. Nature 503, 548–551 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Canon J. et al. , The clinical KRAS(G12C) inhibitor AMG 510 drives anti-tumour immunity. Nature 575, 217–223 (2019). [DOI] [PubMed] [Google Scholar]
- 13.Hallin J. et al. , The KRAS(G12C) Inhibitor MRTX849 Provides Insight toward Therapeutic Susceptibility of KRAS-Mutant Cancers in Mouse Models and Patients. Cancer Discov. 10, 54–71 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Skoulidis F. et al. , Sotorasib for Lung Cancers with KRAS p.G12C Mutation. N. Engl. J. Med. 384, 2371–2381 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hong DS et al. , KRAS(G12C) Inhibition with Sotorasib in Advanced Solid Tumors. N. Engl. J. Med. 383, 1207–1217 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Janne PA et al. , Adagrasib in Non-Small-Cell Lung Cancer Harboring a KRAS(G12C) Mutation. N. Engl. J. Med. 387, 120–131 (2022). [DOI] [PubMed] [Google Scholar]
- 17.Akhave NS, Biter AB, Hong DS, Mechanisms of Resistance to KRAS(G12C)-Targeted Therapy. Cancer Discov. 11, 1345–1352 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Awad MM et al. , Acquired Resistance to KRASG12C Inhibition in Cancer. N. Engl. J. Med. 384, 2382–2393 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li BT et al. , Largest evaluation of acquired resistance to sotorasib in KRAS p.G12C-mutated non–small cell lung cancer (NSCLC) and colorectal cancer (CRC): Plasma biomarker analysis of CodeBreaK100. J. Clin. Oncol. 40, 102–102 (2022). [Google Scholar]
- 20.Zhao Y. et al. , Diverse alterations associated with resistance to KRAS(G12C) inhibition. Nature 599, 679–683 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yaeger R. et al. , Molecular Characterization of Acquired Resistance to KRASG12C-EGFR Inhibition in Colorectal Cancer. Cancer Discov. 13, 41–55 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tanaka N. et al. , Clinical Acquired Resistance to KRAS(G12C) Inhibition through a Novel KRAS Switch-II Pocket Mutation and Polyclonal Alterations Converging on RAS-MAPK Reactivation. Cancer Discov. 11, 1913–1922 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tsai YS et al. , Rapid idiosyncratic mechanisms of clinical resistance to KRAS G12C inhibition. J. Clin. Invest. 132, (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Amodio V. et al. , EGFR blockade reverts resistance to KRAS G12C inhibition in colorectal cancer. Cancer Discov, (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Xue JY et al. , Rapid non-uniform adaptation to conformation-specific KRAS(G12C) inhibition. Nature 577, 421–425 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ryan MB et al. , Vertical Pathway Inhibition Overcomes Adaptive Feedback Resistance to KRAS(G12C) Inhibition. Clin. Cancer Res. 26, 1633–1643 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Brown WS et al. , Overcoming Adaptive Resistance to KRAS and MEK Inhibitors by Co-targeting mTORC1/2 Complexes in Pancreatic Cancer. Cell Rep Med 1, 100131 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Molina-Arcas M. et al. , Development of combination therapies to maximize the impact of KRAS-G12C inhibitors in lung cancer. Sci. Transl. Med. 11, eaaw7999 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Suzuki S. et al. , KRAS Inhibitor Resistance in MET-Amplified KRAS (G12C) Non-Small Cell Lung Cancer Induced By RAS- and Non-RAS-Mediated Cell Signaling Mechanisms. Clin. Cancer Res. 27, 5697–5707 (2021). [DOI] [PubMed] [Google Scholar]
- 30.Punekar SR, Velcheti V, Neel BG, Wong KK, The current state of the art and future trends in RAS-targeted cancer therapies. Nat. Rev. Clin. Oncol. 19, 637–655 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Longo DL, Rosen N, Targeting Oncogenic RAS Protein. N. Engl. J. Med. 387, 184–186 (2022). [DOI] [PubMed] [Google Scholar]
- 32.Thatikonda V. et al. (Cold Spring Harbor Laboratory, 2023). [Google Scholar]
- 33.Fedele C. et al. , SHP2 inhibition diminishes KRASG12C cycling and promotes tumor microenvironment remodeling. J. Exp. Med. 218, (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ho CSL et al. , HER2 mediates clinical resistance to the KRAS(G12C) inhibitor sotorasib, which is overcome by co-targeting SHP2. Eur. J. Cancer 159, 16–23 (2021). [DOI] [PubMed] [Google Scholar]
- 35.Ryan MB et al. , KRAS(G12C)-independent feedback activation of wild-type RAS constrains KRAS(G12C) inhibitor efficacy. Cell reports 39, 110993 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sattler M, Mohanty A, Kulkarni P, Salgia R, Precision oncology provides opportunities for targeting KRAS-inhibitor resistance. Trends in Cancer 9, 42–54 (2023). [DOI] [PubMed] [Google Scholar]
- 37.Kapoor A. et al. , Yap1 activation enables bypass of oncogenic Kras addiction in pancreatic cancer. Cell 158, 185–197 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shao DD et al. , KRAS and YAP1 converge to regulate EMT and tumor survival. Cell 158, 171–184 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fisher GH et al. , Induction and apoptotic regression of lung adenocarcinomas by regulation of a K-Ras transgene in the presence and absence of tumor suppressor genes. Genes Dev. 15, 3249–3262 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Joshi S. et al. , Adapting to stress - chaperome networks in cancer. Nat. Rev. Cancer 18, 562–575 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chen X, Cubillos-Ruiz JR, Endoplasmic reticulum stress signals in the tumour and its microenvironment. Nat. Rev. Cancer 21, 71–88 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Labbadia J, Morimoto RI, The biology of proteostasis in aging and disease. Annu. Rev. Biochem. 84, 435–464 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Walter P, Ron D, The unfolded protein response: from stress pathway to homeostatic regulation. Science 334, 1081–1086 (2011). [DOI] [PubMed] [Google Scholar]
- 44.Wang M, Kaufman RJ, Protein misfolding in the endoplasmic reticulum as a conduit to human disease. Nature 529, 326–335 (2016). [DOI] [PubMed] [Google Scholar]
- 45.Almanza A. et al. , Endoplasmic reticulum stress signalling - from basic mechanisms to clinical applications. FEBS J 286, 241–278 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gomez-Pastor R, Burchfiel ET, Thiele DJ, Regulation of heat shock transcription factors and their roles in physiology and disease. Nature reviews Molecular cell biology 19, 4–19 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tang Z. et al. , MEK guards proteome stability and inhibits tumor-suppressive amyloidogenesis via HSF1. Cell 160, 729–744 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Clarke HJ, Chambers JE, Liniker E, Marciniak SJ, Endoplasmic reticulum stress in malignancy. Cancer Cell 25, 563–573 (2014). [DOI] [PubMed] [Google Scholar]
- 49.Cox JS, Shamu CE, Walter P, Transcriptional induction of genes encoding endoplasmic reticulum resident proteins requires a transmembrane protein kinase. Cell 73, 1197–1206 (1993). [DOI] [PubMed] [Google Scholar]
- 50.Morl K, Ma W, Gething M-J, Sambrook J, A transmembrane protein with a cdc2+ CDC28-related kinase activity is required for signaling from the ER to the nucleus. Cell 74, 743–756 (1993). [DOI] [PubMed] [Google Scholar]
- 51.Yoshida H, Matsui T, Yamamoto A, Okada T, Mori K, XBP1 mRNA is induced by ATF6 and spliced by IRE1 in response to ER stress to produce a highly active transcription factor. Cell 107, 881–891 (2001). [DOI] [PubMed] [Google Scholar]
- 52.Calfon M. et al. , IRE1 couples endoplasmic reticulum load to secretory capacity by processing the XBP-1 mRNA. Nature 415, 92–96 (2002). [DOI] [PubMed] [Google Scholar]
- 53.Denoyelle C. et al. , Anti-oncogenic role of the endoplasmic reticulum differentially activated by mutations in the MAPK pathway. Nat. Cell Biol. 8, 1053–1063 (2006). [DOI] [PubMed] [Google Scholar]
- 54.Sustic T. et al. , A role for the unfolded protein response stress sensor ERN1 in regulating the response to MEK inhibitors in KRAS mutant colon cancers. Genome Med. 10, 90 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ying H. et al. , Oncogenic Kras maintains pancreatic tumors through regulation of anabolic glucose metabolism. Cell 149, 656–670 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Nilsson MR, Techniques to study amyloid fibril formation in vitro. Methods 34, 151–160 (2004). [DOI] [PubMed] [Google Scholar]
- 57.Galves M, Rathi R, Prag G, Ashkenazi A, Ubiquitin signaling and degradation of aggregate-prone proteins. Trends Biochem. Sci. 44, 872–884 (2019). [DOI] [PubMed] [Google Scholar]
- 58.Klaips CL, Jayaraj GG, Hartl FU, Pathways of cellular proteostasis in aging and disease. J. Cell Biol. 217, 51–63 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Cortez L, Sim V, The therapeutic potential of chemical chaperones in protein folding diseases. Prion 8, 197–202 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Engin F. et al. , Restoration of the unfolded protein response in pancreatic beta cells protects mice against type 1 diabetes. Sci. Transl. Med. 5, 211ra156 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Costa-Mattioli M, Walter P, The integrated stress response: From mechanism to disease. Science 368, eaat5314 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Guettouche T, Boellmann F, Lane WS, Voellmy R, Analysis of phosphorylation of human heat shock factor 1 in cells experiencing a stress. BMC Biochem. 6, 4 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Tirasophon W, Welihinda AA, Kaufman RJ, A stress response pathway from the endoplasmic reticulum to the nucleus requires a novel bifunctional protein kinase/endoribonuclease (Ire1p) in mammalian cells. Genes Dev. 12, 1812–1824 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Hollien J, Weissman JS, Decay of endoplasmic reticulum-localized mRNAs during the unfolded protein response. Science 313, 104–107 (2006). [DOI] [PubMed] [Google Scholar]
- 65.Lhomond S. et al. , Dual IRE1 RNase functions dictate glioblastoma development. EMBO Mol. Med. 10, e7929 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Fedele C. et al. , SHP2 Inhibition Prevents Adaptive Resistance to MEK Inhibitors in Multiple Cancer Models. Cancer Discov. 8, 1237–1249 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Mainardi S. et al. , SHP2 is required for growth of KRAS-mutant non-small-cell lung cancer in vivo. Nat. Med. 24, 961–967 (2018). [DOI] [PubMed] [Google Scholar]
- 68.Nichols RJ et al. , RAS nucleotide cycling underlies the SHP2 phosphatase dependence of mutant BRAF-, NF1- and RAS-driven cancers. Nat. Cell Biol. 20, 1064–1073 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ruess DA et al. , Mutant KRAS-driven cancers depend on PTPN11/SHP2 phosphatase. Nat. Med. 24, 954–960 (2018). [DOI] [PubMed] [Google Scholar]
- 70.Sun S. et al. , IRE1α is an endogenous substrate of endoplasmic-reticulum-associated degradation. Nat. Cell Biol. 17, 1546–1555 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Brodsky JL, Cleaning up: ER-associated degradation to the rescue. Cell 151, 1163–1167 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Liu X. et al. , Notch-induced endoplasmic reticulum-associated degradation governs mouse thymocyte beta-selection. Elife 10, e69975 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Xu L. et al. , Protein quality control through endoplasmic reticulum-associated degradation maintains haematopoietic stem cell identity and niche interactions. Nat. Cell Biol. 22, 1162–1169 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Meyer H, Bug M, Bremer S, Emerging functions of the VCP/p97 AAA-ATPase in the ubiquitin system. Nat. Cell Biol. 14, 117–123 (2012). [DOI] [PubMed] [Google Scholar]
- 75.Anderson J, Daniel et al. , Targeting the AAA ATPase p97 as an Approach to Treat Cancer through Disruption of Protein Homeostasis. Cancer Cell 28, 653–665 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Roskoski R Jr., ERK1/2 MAP kinases: structure, function, and regulation. Pharmacol. Res. 66, 105–143 (2012). [DOI] [PubMed] [Google Scholar]
- 77.Satpathy S. et al. , A proteogenomic portrait of lung squamous cell carcinoma. Cell 184, 4348–4371 e4340 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.You K. et al. , QRICH1 dictates the outcome of ER stress through transcriptional control of proteostasis. Science 371, eabb6896 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Liu CY, Xu Z, Kaufman RJ, Structure and intermolecular interactions of the luminal dimerization domain of human IRE1alpha. J. Biol. Chem. 278, 17680–17687 (2003). [DOI] [PubMed] [Google Scholar]
- 80.Buscail L, Bournet B, Cordelier P, Role of oncogenic KRAS in the diagnosis, prognosis and treatment of pancreatic cancer. Nat. Rev. Gastroenterol. Hepatol. 17, 153–168 (2020). [DOI] [PubMed] [Google Scholar]
- 81.Tolcher AW et al. , Antitumor activity in RAS-driven tumors by blocking AKT and MEK. Clin. Cancer Res. 21, 739–748 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Chung V. et al. , Effect of Selumetinib and MK-2206 vs Oxaliplatin and Fluorouracil in Patients With Metastatic Pancreatic Cancer After Prior Therapy: SWOG S1115 Study Randomized Clinical Trial. JAMA Oncol 3, 516–522 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Zhao N. et al. , Pharmacological targeting of MYC-regulated IRE1/XBP1 pathway suppresses MYC-driven breast cancer. J. Clin. Invest. 128, 1283–1299 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Le Reste PJ et al. , Local intracerebral inhibition of IRE1 by MKC8866 sensitizes glioblastoma to irradiation/chemotherapy in vivo. Cancer Lett. 494, 73–83 (2020). [DOI] [PubMed] [Google Scholar]
- 85.Logue SE et al. , Inhibition of IRE1 RNase activity modulates the tumor cell secretome and enhances response to chemotherapy. Nature communications 9, 3267 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Gabrail NY et al. , A phase 1/2 trial of ORIN1001, a first-in-class IRE1 inhibitor, in patients with advanced solid tumors. J. Clin. Oncol. 39, 3080–3080 (2021). [Google Scholar]
- 87.Salmon M. et al. , Kras oncogene ablation prevents resistance in advanced lung adenocarcinomas. J. Clin. Invest. 133, (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Mahler HC, Friess W, Grauschopf U, Kiese S, Protein aggregation: pathways, induction factors and analysis. J. Pharm. Sci. 98, 2909–2934 (2009). [DOI] [PubMed] [Google Scholar]
- 89.Rimawi MF et al. , Early efficacy evaluation of ORIN1001, a first in class IRE1 alpha inhibitor, in advanced solid tumors. J. Clin. Oncol. 41, 1092–1092 (2023).36493335 [Google Scholar]
- 90.Kim MP et al. , Generation of orthotopic and heterotopic human pancreatic cancer xenografts in immunodeficient mice. Nat. Protoc. 4, 1670–1680 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Yan L. et al. , Targeting Glucose Metabolism Sensitizes Pancreatic Cancer to MEK Inhibition. Cancer Res. 81, 4054–4065 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Cheng Z. et al. , Inhibition of BET bromodomain targets genetically diverse glioblastoma. Clin. Cancer Res. 19, 1748–1759 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Sanjana NE, Shalem O, Zhang F, Improved vectors and genome-wide libraries for CRISPR screening. Nat Methods 11, 783–784 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Bankhead P. et al. , QuPath: Open source software for digital pathology image analysis. Sci. Rep. 7, 1–7 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Wrighton KH et al. , Small C-terminal domain phosphatases dephosphorylate the regulatory linker regions of Smad2 and Smad3 to enhance transforming growth factor-beta signaling. J. Biol. Chem. 281, 38365–38375 (2006). [DOI] [PubMed] [Google Scholar]
- 96.Cross BC et al. , The molecular basis for selective inhibition of unconventional mRNA splicing by an IRE1-binding small molecule. Proc. Natl. Acad. Sci. U. S. A. 109, E869–878 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Vasaikar SV, Straub P, Wang J, Zhang B, LinkedOmics: analyzing multi-omics data within and across 32 cancer types. Nucleic Acids Res. 46, D956–D963 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data are available in the manuscript or the supplementary material. Requests for materials should be addressed to X.C.