Abstract
Mutations that increase the low-level transcription of the Saccharomyces cerevisiae HIS4 gene, which results from deletion of the genes encoding transcription factors BAS1, BAS2, and GCN4, were isolated previously in SIT1 (also known as RPO21, RPB1, and SUA8), the gene encoding the largest subunit of RNA polymerase II (RNAPII). Here we show that sit1 substitutions cluster in two conserved regions of the enzyme which form part of the active site. Six sit1 mutations, affect region F, a region that is involved in transcriptional elongation and in resistance to α-aminatin. Four sit1 substitutions lie in another region involved in transcriptional elongation, region D, which binds Mg2+ ions essential for RNA catalysis. One region D substitution is lethal unless suppressed by a substitution in region G and interacts genetically with PPR2, the gene encoding transcription elongation factor IIS. Some sit1 substitutions affect the selection of transcriptional start sites at the CYC1 promoter in a manner reminiscent of that of sua8 (sua stands for suppression of upstream ATG) mutations. Together with previous findings which indicate that regions D and G are in close proximity to the 3′ end of the nascent transcript and that region F is involved in the translocation process, our results suggest that transcriptional activation by the sit1 mutations results from alteration of the RNAPII active center.
The structures and functions of eukaryotic RNA polymerases I, II, and III are well conserved during evolution (reviewed in references 6, 41, and 53). Structurally, these three enzymes contain two large subunits which are homologous to the two largest subunits, β and β′, of Escherichia coli RNA polymerase (RNAP) and which contain the active site of the enzyme (reviewed in references 6, 41, and 53). In addition to containing the two largest subunits, eukaryotic RNAPs also contain approximately 10 smaller subunits, some of which are present in both RNAPI and RNAPII or in all three enzymes (reviewed in references 41 and 53).
Mutations affecting the two largest subunits of Saccharomyces cerevisiae RNAPII were isolated by Arndt et al. (8) in a genetic selection for sit mutations (sit stands for suppressor of initiation of transcription defect) that can increase transcription of the HIS4 gene in a strain in which the BAS1, BAS2, and GCN4 genes were deleted. The last three of these genes encode transcription factors which activate transcription of HIS4 through specific DNA elements located in the promoter region of the gene (8). A yeast strain which lacks BAS1, BAS2, and GCN4 is auxotrophic for histidine because HIS4 is transcribed poorly (7, 8). Transcription of HIS4 can be increased by mutations in SIT1 (also known as RPO21, RPB1, and SUA8) and SIT2 (also known as RPO22 and RPB2), which encode the largest and second-largest subunits of RNAPII, respectively. Transcription of HIS4 in sit1 and sit2 mutant strains, as in a wild-type strain, requires the presence of a functional TATA element as well as a binding site for RAP1 in the promoter. The requirement for these two promoter elements most likely reflects a need for RAP1 to antagonize the repressing effect of chromatin and allow the general transcription machinery to gain access to the HIS4 promoter, at least in part through an interaction between TBP and the TATA box (22). RAP1 binds to the HIS4 promoter but does not efficiently stimulate its transcription in the absence of BAS1, BAS2, and GCN4 (8, 22).
One can envisage two general mechanisms by which the sit1 and sit2 mutations affect RNAPII in order to increase synthesis of HIS4 mRNAs. First, these mutations could increase the number of initiation complexes that assemble at or are recruited to the HIS4 TATA box. Results of experiments performed in vivo in which high levels of transcription were obtained by artificially tethering the RNAPII holoenzyme to promoter DNA suggested that recruitment of the holoenzyme to the promoter is a rate-limiting step in transcription (9).
Second, the sit1 and sit2 mutations could affect other steps that can be rate limiting following transcription initiation, such as an early block to elongation or premature termination (14, 28, 36, 39, 52). In this case, the sit1 mutations could increase the number of RNAPII molecules that reach the end of the transcription unit, without affecting the overall rate of initiation, by increasing the processivity of RNAPII during elongation. The increased processivity of the enzyme could affect either early elongation, by increasing the frequency at which the initiation complexes leave the HIS4 promoter and enter the elongation mode (promoter clearance), or later stages of elongation, by decreasing the rate of premature termination. As a first step toward understanding how the sit1 mutations affect transcriptional activation, we have identified regions of the largest subunit of RNAPII that are affected by these mutations.
MATERIALS AND METHODS
S. cerevisiae strains and yeast manipulations.
All sit1 strains used in this study were isolated as spontaneous His+ revertants of strain L3110 (MATa gcn4-2 bas1-2 bas2-2 ura3-52) as previously described (7, 8). The yeast strain YF2047 (MATα gcn4-2 bas1-2 bas2-2 rpo21::LEU2 leu2 trp1::hisG ura3-52 [pJS121; RPO21 on URA3 CEN/ARS plasmid]) was constructed in several steps. First, the TRP1 locus of L3110 was changed to trp1::hisG as previously described (1) to create yeast strain YF2037. YF2037 was then mated with yeast strain CY843 (MATα gcn4-2 bas1-2 bas2-2 ura3-52 leu2). The resulting diploid was sporulated, and tetrads were dissected. A Leu− Trp− haploid progeny, YF2044 (MATα gcn4-2 bas1-2 bas2-2 ura3-52 trp1::hisG leu2), was then transformed to uracil prototrophy with plasmid pJS121 (RPO21 URA3 CEN/ARS) (4). The RPO21 locus of this merodiploid strain was then disrupted (rpo21::LEU2) in one step to create strain YF2047. The yeast strain YDW383, which carries the sua8-1 mutation as well as an isogenic SUA8 wild-type strain, T16, have been described previously (13). YF2277 (MATα rpo21::ADE2 [pJS121]), a strain congenic with W303-1B (MATα ade2-1 trp1-1 leu2-3,112 his3-11,15 ura3-52 can1-100 ssd1-d) contains a disruption of the chromosomal RPO21 allele caused by a replacement by ADE2 of the RPO21 sequence from positions −718 to +660 (the A of the translation initiation codon is +1). YF2278 (MATα rpo21::ADE2 ppr2::hisG [pJS121]) is isogenic to YF2277 except for the disruption of PPR2 by hisG. This was introduced by using plasmid pJD3, which contains ppr2::hisG-URA3-hisG (a gift of C. M. Kane, University of California at Berkeley). YF2277 and YF2278 were generated by standard genetic techniques. Details of their construction are available upon request. Growth media used were as described previously (45). Yeast transformation was performed as described previously (26).
Plasmids.
Plasmid pJAY61, which was used for the isolation of sit1 alleles, was constructed in two steps. First, an integrating plasmid carrying URA3 as a selectable marker and the wild-type SIT1 gene on a HindIII-EcoRI genomic DNA fragment (the HindIII and EcoRI sites are located 1.6 kb upstream of the SIT1 translation initiation codon and 1.7 kb downstream of the translation termination codon, respectively) was constructed. Second, a deletion was introduced in the SIT1 open reading frame by removing the DNA sequences contained between an AvrII site (located 162 nucleotides downstream of the translation initiation codon) and a SnaB1 site (located 16 nucleotides downstream of the translation termination site) and replacing them with a BamHI linker (5′-GGGATCCC-3′) after filling in the protruding ends of the AvrII site with the Klenow fragment of DNA polymerase I. Plasmid pJA483 was created by subcloning a 5.7-kb EcoRI-HindIII fragment encompassing RPO21 into the EcoRI and HindIII sites of pFL39 (TRP1 CEN6 ARS) (15). Plasmids pM50 and pM107, which carry CYC1-lacZ and cyc1-5000-lacZ, respectively, cloned in a single-copy (URA3) plasmid, have been described previously (38).
DNA manipulation and sequencing.
All DNA manipulations were performed essentially as described previously (30). DNA sequencing of plasmid DNA was performed by the chain termination method (40) using primers synthesized on the basis of the SIT1 sequence.
PCR amplification of the DNA sequence encoding conserved region F.
The DNA sequence between nucleotides 2155 and 2907 (in the numbering system of Allison et al. [3]), which encodes the conserved region F of RPO21, was amplified by PCR from genomic DNA of several sit1 alleles by using the following two oligonucleotides: 5′-CGGGATCCGGTGTAGTAGAGAAAAAAAC-3′ and 5′-CGGGATCCTGAATAACGTTACCCAATG-3′. The resulting amplified fragments containing BamHI sites at both ends (generated by the two primers) were then cut with BamHI and cloned into the BamHI site of plasmid YEp24 (17). For each sit1 allele amplified and cloned, two independently isolated plasmids were sequenced. The amplified region F DNAs were also cut with PpuMI and XbaI and subcloned between the PpuMI and XbaI sites of pJA483 (RPO21 TRP1 CEN6 ARS).
PCR amplification of the DNA sequence encoding regions B, C, D, and E.
The DNA sequence between the SpeI and PpuMI sites of RPO21, which encodes conserved regions B, C, D, and E, was amplified by PCR using the following two oligonucleotides: 5′-AGAGCGAAAATTGGTGGTC-3′ and 5′-GCCTCTGCAATTGTCTCTG-3′. The amplified fragment was cut with SpeI and PpuMI and was subcloned into pJA483 (RPO21 TRP1 CEN6 ARS) cut with SpeI and PpuMI. For each sit1 allele, the DNA sequence between SpeI and PpuMI was determined in two independently isolated plasmids.
β-Galactosidase assays.
β-Galactosidase activity was measured as described by Miller (34). Cells were grown to an optical density (at 600 nm) of 0.6 in SC-Ura medium (45) with glucose as a carbon source. For each measurement, β-galactosidase activity was determined on three independent cultures.
RESULTS
Molecular cloning of sit1 alleles.
As a first step toward determining the mechanism by which mutations which affect RNAPII can increase transcription of HIS4 in the absence of the BAS1, BAS2, and GCN4 proteins, we set out to identify which regions of the largest subunits are altered by the sit1 mutations. A gap repair strategy (37) was used to clone five independent sit1 alleles (sit1-4, -5, -7, -8, and -9) as well as the wild-type SIT1 allele from the parental yeast strain, L3110 (7). In order to localize the sit1 mutations, two independently cloned copies of each sit1 allele were digested with several restriction endonucleases, each of which recognizes a different 4-bp site (AluI, HaeIII, TaqI, and Sau3A), in the hope that certain sit1 mutations would either create or destroy a site so as to give rise to a restriction fragment length polymorphism. A Sau3A restriction fragment was detected in the sit1-7 allele that was not present in any of the other mutant alleles or in the wild-type gene (data not shown). This Sau3A polymorphic fragment was cloned and sequenced. The mutation creating the new Sau3A site results in the replacement of glycine 730 with an aspartic acid residue in the conserved region F of the largest subunit of RNAPII (Table 1), one of eight regions of this polypeptide that have been conserved during evolution (27).
TABLE 1.
Nucleotide and amino acid sequence changes in sit1 alleles
| Region and allele | Nucleotide changea | Amino acid substitution |
|---|---|---|
| Region F | ||
| sit1-7 | G 2501 to A | Gly 730 to Asp |
| sit1-5 | G 2587 to C | Ala 759 to Pro |
| sit1-290 | G 2603 to T | Cys 764 to Phe |
| sit1-290 | G 2314 to A | Asp 668 to Asn |
| sit1-9 | G 2771 to A | Gly 820 to Asp |
| sit1-4 | C 2807 to T | Ala 832 to Val |
| sit1-246 | C 2807 to T | Ala 832 to Val |
| Region D | ||
| sit1-8D | A 1747 to T | Asn 479 to Tyr |
| sit1-252 | A 1771 to T | Met 487 to Leu |
| sit1-261 | A 1771 to T | Met 487 to Leu |
| sit1-278 | A 1646 to C | Asn 445 to Thr |
| Region G | ||
| sit1-8G | C 3539 to T | Ala 1076 to Val |
The positions of the nucleotide changes are given in the numbering system of Allison et al. (3).
Six sit1 alleles carry mutations affecting conserved region F of RPO21.
Additional sit1 mutants were then examined for the presence of mutations affecting region F (nucleotides 2200 to 2900 in the numbering system of Allison et al. [3]). When the other cloned sit1 alleles (sit1-4, -5, -8, and -9) were examined, region F mutations were found in sit1-4, sit1-5, and sit1-9 (Table 1 and Fig. 1). In the case of sit1 mutants for which the sit1 allele was not cloned, the DNA sequence encoding region F (between nucleotides 2200 and 2900) was amplified from genomic DNA by PCR and cloned into a plasmid and two independent plasmid isolates were sequenced. Of five sit1 mutants analyzed in this fashion (sit1-290, -246, -252, -261, and -278), two (sit1-290 and -246) were found to carry mutations that affect region F (Table 1 and Fig. 1). One of them (sit1-290) had two mutations, one in region F and one just upstream of region F (Table 1). Since four of the sit1 alleles tested (sit1-8, -252, -261, and -278) did not carry a mutation affecting region F, at least one other region of SIT1 can be altered so as to confer the Sit phenotype (see below).
FIG. 1.
Locations of sit1 amino acid substitutions. The amino acid sequences of regions F, D, and G of S. cerevisiae RNAPII (S. cerevisiae) are compared to those of other eukaryotes: Schizosaccharomyces pombe RNAPII (S. pombe), Arabidopsis thaliana RNAPII (A. thaliana), Caenorhabditis elegans RNAPII (C. elegans), D. melanogaster RNAPII (D. melano.), Mus musculus RNAPII (M. musculus), Sulfolobus acidocaldarius (S. acidocal.) subunit A or C as indicated, E. coli RNAP, and S. cerevisiae RNAPIII (S. c. RNAPIII). Residues identical to those of S. cerevisiae RNAPII are indicated by dots; gaps are indicated by hyphens. For each sit1 mutation, the substituted amino acid is indicated above the wild-type amino acid and the designation of the allele is given in parentheses. A second substitution in the sit1-290 polypeptide that is located upstream of region F is not shown (Table 1). Amino acid substitutions that confer resistance to the transcriptional inhibitor α-amanitin (Ama) in C. elegans (19), D. melanogaster (19), and M. musculus (10, 11) are also indicated. Single amino acid substitutions that affect elongation and termination by E. coli RNAP (49) are written below the sequence of the largest subunit of E. coli RNAP (E. coli mut.). Substitutions that confer streptolydigin resistance in E. coli (E. coli St1) (44) and Bacillus subtilis (B. sub. St1) (51) are indicated. The double substitutions in the S. cerevisiae RNAPIII C160-270 mutant (RNAPIII C160-270) (47) and C160-112 mutant (RNAPIII C160-112) (23) are indicated. The invariant Mg2+ binding motif NADFDGD in region D is underlined. Also indicated are the locations of the conditional lethal rpb1-17 substitution (43), the sua8-1 and sua8-2 substitutions (13), and the mutations (rpb1-501 and -502) that confer an Spt phenotype (25).
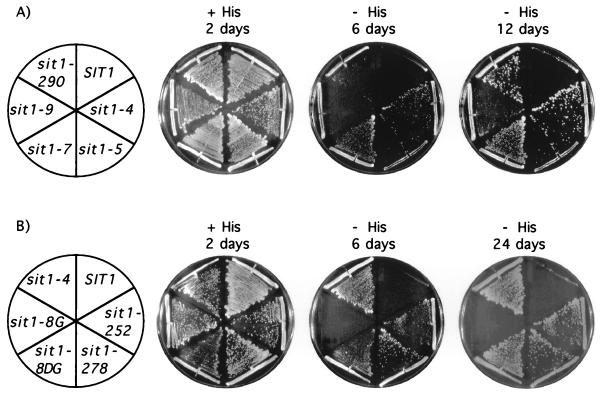
In order to determine whether the region F mutations that were identified are both necessary and sufficient to cause the Sit phenotype, the DNA fragments encoding each mutant region F (nucleotides 2224 to 2844 [PpuMI-XbaI fragment]) were subcloned individually into an otherwise wild-type SIT1 gene (sit1-246 was not used for this analysis since it carries the same mutation as sit1-4). The reconstructed sit1 mutant alleles were tested for the ability to support growth on medium lacking histidine (the Sit phenotype) by two different assays. In the first assay, which relies on the ability of the sit1 alleles to complement a disrupted allele of SIT1 (sit1::LEU2), reconstructed sit1 alleles carried on a single-copy plasmid were introduced into the diploid yeast strain YF2201 (MATa/MATα sit1::LEU2/SIT1 gcn4-2/gcn4-2 bas1-2/bas1-2 bas2-2/bas2-2 ura3-52/ura3-52 leu2/leu2 trp1::hisG/trp1::hisG). Following sporulation and tetrad dissection, haploid progeny that inherited the plasmid-borne reconstructed sit1 allele and the chromosomal disrupted SIT1 allele were tested for growth on medium lacking histidine. Reconstructed alleles sit1-4, -5, -7, -9, and -290 were capable both of supporting yeast cell growth and of bringing about the Sit phenotype (Fig. 2A).
FIG. 2.
Region F and D substitutions are necessary to confer the Sit phenotype. (A) Region F substitutions. Reconstructed sit1 alleles carrying a region F mutation or the wild-type SIT1 alleles were was assayed for the ability to confer the Sit phenotype by introduction into the diploid strain YF2201 (MATa/MATα sit1::LEU2/SIT1 gcn4-2/gcn4-2 bas1-2/bas1-2 bas2-2/bas2-2 ura3-52/ura3-52 leu2/leu2 trp1::hisG/trp1::hisG). The resulting transformants were then sporulated, 10 or more tetrads were dissected, and spores were allowed to germinate on YPD medium (45). Viable Trp+ (sit1 allele on plasmid) Leu+ (sit1::LEU2) haploid progeny were then tested by streaking onto solid SD medium (45) containing (+ His) or lacking (− His) histidine. Cells were allowed to grow for the indicated number of days at 30°C. sit1-9 required a longer incubation to show visible single colonies. (B) Region D substitutions. Reconstructed sit1 alleles carrying region D mutations or the wild-type SIT1 alleles were assayed for the ability to confer the Sit phenotype as described above. Cells were allowed to grow on solid SD medium (45) containing (+ His) or lacking (− His) histidine for the indicated number of days at 30°C. sit1-8G did not grow on medium lacking histidine, even when incubation was extended to 24 days. A region F substitution (sit1-4) is shown for comparison.
The second assay that was used to test for the Sit phenotype is based on our observation that mutant sit1 alleles (either reconstructed alleles or alleles derived entirely from a mutant strain), but not the wild-type SIT1 allele, can confer a semidominant Sit phenotype in yeast strain YF2047 (MATα gcn4-2 bas1-2 bas2-2 sit1::LEU2 ura3-52 leu2 trp1::hisG [pJS121; SIT1 on URA3 CEN/ARS plasmid]). This observation was surprising, because the Sit phenotype was shown previously to be recessive (8). It may be that the semidominant character of the Sit phenotype is influenced by the background of the strain. Alternatively, the degree of dominance of the Sit phenotype may depend on the relative level of expression of the mutant and wild-type alleles, which may be influenced by whether these alleles are expressed from their endogenous chromosomal location (8) or from episomal plasmids (this study). Regardless of the explanation, this assay was useful as an independent method of confirming that all five sit1 mutations affecting region F (sit1-4, -5, -7, -9, and -290) are sufficient to confer a Sit phenotype (data not shown).
Mapping the sit1 mutation in the sit1-8 allele.
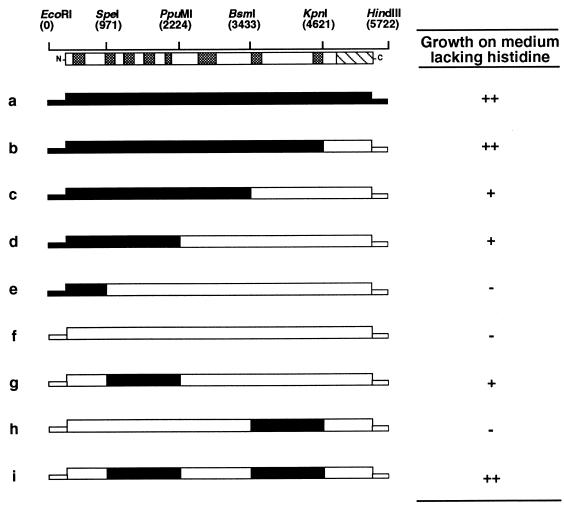
The above analysis indicated that in some sit1 alleles a region of the largest subunit other than region F is altered so as to bring about the Sit phenotype. Hybrid genes were constructed that contained various portions of sit1-8 and wild-type SIT1 (Fig. 3) in order to localize the mutation in the sit1-8 allele, which does not contain a region F mutation (see above). These chimeric genes were tested by the two assays described above for the ability to support growth on medium lacking histidine. As shown in Fig. 3, the smallest region of sit1-8 that was capable of conferring a His+ phenotype in an assay that relied on the semidominance of the Sit phenotype was located between nucleotides 971 and 2224 (hybrid g). The nucleotide sequence of this region was determined, and a mutation was found that replaces Asn 479 by Tyr in conserved region D of the largest subunit (Table 1 and Fig. 1). This mutation is referred to as sit1-8D. By the same assay, it was also noticed that yeast cells carrying a chimeric gene containing only the sit1-8D mutation (hybrid g) did not grow as well on medium lacking histidine as did cells carrying the entire sit1-8 allele (hybrid a). This enhancement of growth was associated with a second region of sit1-8, which was mapped to a region between nucleotides 3433 and 4621 (compare hybrid b with hybrid c and hybrid g with hybrid i). This region, however, was not sufficient by itself to bring about the Sit phenotype (hybrid h). The region located between nucleotides 3433 and 4621 was sequenced, and a mutation was found that results in the replacement of alanine 1076 by valine in conserved region G of the largest subunit (Table 1 and Fig. 1). The presence of both the sit1-8D and sit1-8G mutations was confirmed in other, independently isolated sit1-8 alleles (data not shown).
FIG. 3.
Localization of mutations in the sit1-8 allele. Schematic representation of the SIT1 locus (EcoRI-HindIII fragment) showing the positions (in the numbering system of Allison et al. [3]) of various endonuclease sites that were used in the construction of the chimeric genes. The encoded SIT1 protein is diagrammed below the restriction map. The gray boxes represent regions (A to H) of the polypeptide that are most conserved evolutionarily (27), and the diagonally striped box represents the carboxy-terminal domain. The structures of the chimeric genes are indicated by open and filled boxes representing wild-type (RPO21) and mutant (sit1-8) sequences, respectively. Each hybrid gene is designated by a letter (a to i). The ability of each chimeric gene to confer a semidominant Sit phenotype was assayed by introducing it into yeast strain YF2047 (MATα gcn4-2 bas1-2, bas2-2 ura3-52 leu2 trp1::hisG sit1::LEU2 [pJS121; SIT1 on URA3 CEN/ARS plasmid] [4]) and testing the resulting transformants for the ability to grow on solid SD medium (45) lacking histidine: ++, growth rate similar to that of a cell carrying the entire sit1-8 allele; +, growth rate slower than that of a cell carrying the entire sit1-8 allele; −, absence of growth.
Chimeric genes carrying both the sit1-8D and sit1-8G mutations or carrying either mutation alone were also tested for the ability to confer the Sit phenotype by the assay that relies on complementation of the sit1::LEU2 allele. In this assay, hybrid g, which carries only the sit1-8D mutation, was unable to support growth of haploid cells even when histidine was present in the growth medium, indicating that the sit1-8D mutation is lethal. In contrast, hybrid i (sit1-8DG), which carries both the sit1-8D and sit1-8G mutations, was able to complement the sit1::LEU2 allele and was also able to confer the Sit phenotype (Fig. 2B). Hybrid h, which carries only the sit1-8G mutation, was capable of complementing the sit1::LEU2 allele but was unable to confer the Sit phenotype (Fig. 2B). Taken together these results suggest that the sit1-8D allele encodes a mutant subunit that assembles into an RNAPII complex which is capable of transcribing HIS4 in the absence of BAS1, BAS2, and GCN4. However, this mutant RNAPII is not able to support cell growth unless it is modified further by the sit1-8G substitution, perhaps because genes that are either required or toxic for cell growth are aberrantly expressed (see Discussion).
In other experiments (see below), we found that the sit1-8D is not always lethal. This finding that the sit1-8D allele is capable of supporting cell growth in certain genetic backgrounds provides further evidence that this allele encodes a partially functional subunit. It also suggests that the pleiotropic phenotypes imposed by sit1 mutations can vary according to the genetic background in which they are expressed (see Discussion).
Identification of other region D mutations.
Other sit1 mutant strains which did not carry a region F mutation (sit1-252, -261, and -278) were then tested for the presence of a region D mutation. For each of these mutants, a portion of SIT1 encoding conserved regions B, C, D, and E (nucleotides 971 to 2224 [SpeI-PpuMI fragment]) was amplified and subcloned into an otherwise wild-type SIT1 gene. When tested for the Sit phenotype by the two assays described above, all three reconstructed sit1 alleles were capable of supporting growth on medium lacking histidine. DNA sequencing revealed that two alleles, sit1-252 and sit1-261, carry the same mutation which changes methionine 487 to leucine (Table 1 and Fig. 1). The mutation in the sit1-278 allele replaces asparagine 445 by threonine (Table 1 and Fig. 1). The Sit phenotype imposed by region D substitutions is shown in Fig. 2B.
Effect of sit1 mutations on start site selection.
Amino acid substitutions located in and near conserved region D of the largest subunit were identified previously that suppress the reduction in expression of CYC1 brought about by the insertion of a nonfunctional AUG codon into the 5′ untranslated sequence of the gene (Sua phenotype [Sua stands for suppression of upstream ATG]) (38). Mutations in SUA8 affect the position of the transcription start site at many yeast promoters in such a way that downstream start sites are favored relative to upstream ones (13). It was reported previously that some sit1 mutations have a similar effect in that they increase the number of transcripts that start downstream of the HIS4 translation initiation codon (8). This observation and the fact that some of the mutations characterized in this study change amino acids in region D which are also changed by sua8 mutations (sua8-1 and sit1-278 change asparagine 445 to Ser and Thr, respectively) prompted us to determine whether sit1 mutations can confer an alteration of start site selection at CYC1. Yeast strains carrying either the sit1-8 (regions D and G), the sit1-278 (region D), the sit1-4 (region F), the sit1-5 (region F), or the wild-type SIT1 gene were transformed separately with a plasmid carrying a CYC1-lacZ reporter gene that either contains (cyc1-5000 allele; plasmid pM107 [38]) or lacks (wild type CYC1; plasmid pM50 [38]) a nonfunctional ATG codon in the CYC1 5′ untranslated region. As controls, the same two plasmids were also introduced into a strain carrying the sua8-1 mutation (YDW383 [13]) and an isogenic SUA8 wild-type strain (T16 [13]). For each strain, the levels of β-galactosidase activity expressed from the CYC1 (wild-type) allele or from the cyc1-5000 allele were measured (Table 2). The ratio of the level of expression of the cyc1-5000 allele to that of the wild-type CYC1 gene increases in strains with sua8 mutations. A low ratio, such as that measured in strains carrying a wild-type SIT1 gene (SUA8 or SIT1 in Table 2), indicates that most of the transcripts that are initiated at the cyc1-5000 promoter start upstream of the nonfunctional ATG. In contrast, a high ratio, such as that measured in a sua8-1 mutant strain, indicates that a substantial proportion of cyc1-5000 transcripts start downstream of the nonfunctional ATG and hence that the mutation affects the position of the transcriptional start site.
TABLE 2.
Effects of sit1 mutations on expression of CYC1 and cyc1-5000
| Allele | Affected region(s) | Mean β-galactosidase activity (U)b
|
cyc1-5000/CYC1 ratio (%) | |
|---|---|---|---|---|
| CYC1 | cyc1-5000 | |||
| SUA8 | NAa | 53 (3) | 1.24 (0.01) | 2.3 |
| sua8-1 | D | 29 (5) | 6.3 (0.2) | 22 |
| SIT1 | NA | 15 (2) | 0.32 (0.05) | 2.1 |
| sit1-8 | D, G | 8 (1) | 0.38 (0.04) | 4.8 |
| sit1-278 | D | 4.04 (0.02) | 0.76 (0.04) | 19 |
| sit1-4 | F | 3.8 (0.7) | 0.32 (0.01) | 8.4 |
| sit1-5 | F | 5.1 (0.4) | 0.35 (0.07) | 6.9 |
NA, not applicable.
Standard deviations are indicated in parentheses.
Four observations were made. First, the level of expression of the wild-type CYC1 gene is reduced approximately two- to fourfold in sit1 mutant strains (sit1-8, -278, -4, and -5) compared to that of a wild-type SIT1 strain. This decrease in CYC1 expression is not surprising, since sit1 mutations were shown previously to affect the expression of many genes, including those, like CYC1, that are required for growth on nonfermentable carbon sources (8). Second, all four sit1 mutant strains confer an increase in the ratio of cyc1-5000 to CYC1 expression, albeit of variable magnitude: sit1-8 showed the smallest increase, whereas the increase conferred by sit1-278 was comparable to that imposed by the sua8-1 allele. Third, both region D and F mutations affect the expression ratio. Fourth, expression of the wild-type CYC1 gene in a wild-type SIT1 strain can be affected by the genetic background of the strain (compare SUA8 and SIT1 in Table 2). Whether this background difference is due to the absence of BAS1, BAS2, and GCN4 in one strain (SIT1) but not the other (SUA8) remains to be investigated. Although the sit1 alleles tested here have a feature of sua8 mutants (i.e., an alteration of start site selection that favors downstream start sites), we do not conclude that all of these alleles have a bona fide Sua phenotype. An Sua phenotype would require an absolute increase in the expression of cyc1-5000, such that this allele would be capable of supporting the growth of the strain when lactate is the carbon source (38). Our data speak only to the feature of altered start site selection that is shared by sua8 and these sit1 alleles.
Synthetic lethality of sit1-8D and ppr2.
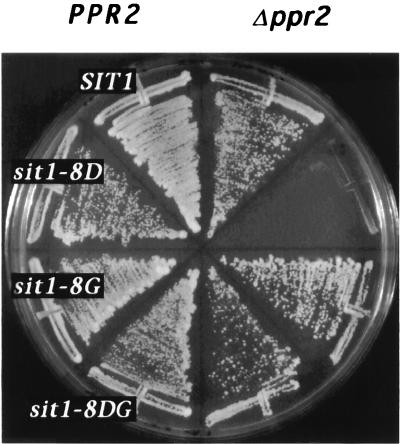
The clustering of sit1 mutations in regions of RNAPII which form part of its catalytic center suggest that these mutations may affect the elongation properties of the enzyme (see Discussion). We reasoned that if the sit1 mutations affect transcriptional elongation in vivo, this defect may be exacerbated by a deletion of the nonessential gene PPR2, which encodes transcription elongation factor IIS (TFIIS). This was tested by shuffling plasmids carrying the sit1 alleles (sit1-4, -5, -7, -9, -290, -252, -278, -8D, -8G, and -8DG) into a strain carrying a disruption of both SIT1 and PPR2 (YF2278) or, as a control, into a strain carrying only a disruption of SIT1 (YF2277). In these experiments, the ability of the various sit1 alleles to support growth in the absence or presence of TFIIS was tested by determining the ability of cells to grow on medium containing 5′-fluoroorotic acid, which selects for loss of the wild-type SIT1 maintenance plasmid. Two observations were made. First, in the genetic background used in these experiments, the sit1-8D mutation is not lethal (Fig. 4). Second, this mutation is, however, lethal when combined with a deletion of PPR2 (Fig. 4). To our knowledge, sit1-8D is the first mutation in a gene encoding a subunit of RNAPII, or for that matter in any gene, reported to be synthetically lethal with a PPR2 deletion. This synthetic lethality, between a sit1 mutation and a known elongation factor, lends further support to the notion that sit1 mutations affect transcriptional elongation in vivo. We also noted that this synthetic lethal phenotype could be suppressed by the sit1-8G substitution (Fig. 4), indicating that sit1-8G and PPR2 may perform a similar function. In this context, the sit1-8G substitution lies in a portion of the largest subunit which is close to, and may even be part of, a binding site for TFIIS (5, 50). None of the other sit1 mutations tested (sit1-4, -5, -7, -9, -290, -252, -278, -8G, and -8DG) were synthetically lethal in combination with a PPR2 deletion (Fig. 4, data not shown), perhaps because their effect on elongation is not as pronounced as that imposed by sit1-8D. The notion that the sit1-8D mutation imposes a more pronounced effect on transcription than the other sit1 alleles is supported by the observation that it is the only sit1 mutation which can be lethal.
FIG. 4.
Synthetic lethality of sit1-8D and ppr2. TRP1 CEN/ARS plasmids bearing SIT1 (RPO21) alleles (indicated by white letters on a black background) were introduced into YF2277 (MATα rpo21::ADE2 [pJS121, RPO21 on URA3, CEN/ARS]), shown on the left side of the plate or YF2278 (MATα rpo21::ADE2 ppr2::hisG [pJS121, RPO21 on URA3, CEN/ARS]), shown on the right side of the plate, and grown on SD medium (45) lacking tryptophan and containing 5′-fluoroorotic acid for 3 days at 30°C in order to select for loss of the URA3 maintenance plasmid (pJS121).
DISCUSSION
The results presented above, which indicate that both conserved regions D and F of the largest subunit are altered by the sit1 mutations, suggest that these mutations exert their effect by altering the overall structure of the RNAPII active center. Previous functional studies have suggested that regions D and F, together with other conserved regions of the largest and second-largest subunits of multimeric RNA polymerases, form the active site of the enzyme as broadly defined (23) to include the catalytic center together with the DNA and RNA binding sites (reviewed in references 6, 41, and 53). Region D contains a segment, NADFDGD, that is invariant among multimeric RNA polymerases (33) and is involved in binding Mg2+ ions, which are essential for RNA catalysis (the three aspartate residues that chelate Mg2+ are underlined) (54). Within the active site, region D is located very close to the transcription initiation site (54) and the 3′ end of the RNA transcript (16, 31). Cross-linking studies with E. coli RNAP indicated that region G is also in close proximity to the 3′ end of the transcript; while region G was the predominant cross-linking site within arrested complexes, region D was favored when cross-linking was performed on elongating complexes (31). Our finding that a region D mutation (sit1-8D) can be suppressed by a region G mutation (sit1-8G) is consistent with the notion that both regions participate in similar functions. Indeed, mutational studies have indicated that both regions are involved in the selection of the start site (13, 25; this study) and play a role in transcription elongation and termination (23, 49, 54). In S. cerevisiae, a mutant RNAPIII (C160-112) harboring a conditional lethal double substitution in region D (T to I at position 506 [T506I] and N509Y [Fig. 1]) was characterized biochemically (23). At a natural promoter, the mutant enzyme was as proficient as the wild-type enzyme in carrying the initiation reaction but was more prone to catalyze a slippage reaction during synthesis of the first few nucleotides. The mutant enzyme also had a markedly reduced elongation rate, which was attributable to increased pausing at intrinsic pausing sites. It was hypothesized that the double amino acid substitution could either affect the correct positioning of the 3′ end of the RNA transcript in the catalytic center of the enzyme or have a more direct effect on the formation of phosphodiester bonds (23). One of the two substitutions (N509Y) affects a residue of the invariant Mg2+-binding motif and was found to be lethal unless suppressed by the second region D substitution (T506I) (23). This is reminiscent of the sit1-8D substitution (N479Y), which changes the analogous residue in the invariant motif of RNAPII (Fig. 1) and can also be lethal unless suppressed by the sit1-8G substitution (A1076V).
Region F, the other region altered by sit1 mutations, forms a part of the enzyme active site which is targeted by certain inhibitors. Region F can be mutated to confer resistance to α-amanitin (reviewed in reference 6), which inhibits the translocation step of eukaryotic RNAPII (21), and to the antibiotic streptolydigin (44, 51), which inhibits the elongation phase of prokaryotic RNAP (18, 32, 48). A role for region F in transcriptional elongation and translocation is also supported by studies of mutant RNAPs. One Drosophila melanogaster RNAPII mutant (C4) bearing an α-amanitin resistance substitution in region F (R741H) (Fig. 1) was found to elongate more slowly in vitro and have an increased apparent Km for UTP (20). In yeast, a mutant RNAPIII (C160-270) that carried a double substitution in region F (D829A R830A) (Fig. 1) and was defective in the transition from abortive initiation to elongation and in its ability to exit from pause sites was isolated (47). The mutant enzyme also showed an increased RNA cleavage activity in halted ternary complexes. Finally, in E. coli, several region F mutations were isolated that increase termination in the trp operon leader region or decrease termination at a ρ-independent terminator in vivo (49). These mutations also affected termination efficiency at other ρ-independent terminators in vitro, although the magnitude and direction of the effect appeared to be dependent on the type of terminator used (49). The last of these observations raise the possibility that the effect of sit1 mutations on transcription varies from one gene to another. Indeed sit1 mutations can lead to both over- and underexpression of genes such as HIS4 and CYC1 (Table 2), respectively. These profound effects on gene expression most likely underlie the many pleiotropic phenotypes of sit1 mutants.
Further support for the notion that sit1 mutations alter the active site of RNAP comes from experiments in which the Mg2+ ion of the enzyme was replaced by Fe2+. Hydroxyl radicals generated by Fe2+ cleave protein and DNA in a 1-nm radius of the ion. In these experiments, DNA is cleaved immediately upstream of the transcriptional start site (54), indicating that the Mg2+ ion is near or at the catalytic center of the enzyme. In a similar fashion, cut sites in the RNAP peptide chain also identified residues that are in the active-site area (35). β′ was cleaved in three regions: conserved regions D, F, and G. These regions correspond to those in which the sit1 mutants and a suppressor to the growth defect of a sit1 mutation (sit1-8G) were identified.
Two lines of evidence suggest that the pleiotropic effects of sit1 alleles can vary according to the genetic background of the strain in which they are expressed. First, in at least one instance, we have shown that the Sit phenotype can be semidominant, whereas in other genetic backgrounds it is recessive. Second, we found that the sit1-8D substitution is lethal in one genetic background, whereas in another it is viable. Although we do not yet fully understand which set of genes is involved in these background differences, we nevertheless have gathered evidence to suggest that at least one gene, PPR2, which encodes the transcription elongation factor TFIIS, can affect the phenotype imposed by the sit1-8D substitution. We found that in a genetic background in which it is normally viable, the sit1-8D allele can be synthetically lethal in combination with a disruption of the nonessential gene PPR2 and that this synthetic phenotype can be suppressed by the sit1-8G substitution which lies in, or close to, a TFIIS-binding site in RNAPII (5, 50). These findings have two implications. First, they raise the possibility that some of the differences we observed between genetic backgrounds are due to differences in genes which encode proteins that can affect transcriptional elongation, either directly or indirectly. Further analysis of these genetic differences should reveal whether this hypothesis is correct. Second, and most importantly, the genetic connection between sit1-8D, sit1-8G, and a known transcription elongation factor lends further support to the hypothesis that sit1-8D affects the elongation properties of RNAPII in vivo.
Other than mutations that truncate the C-terminal domain of the largest subunit of RNAPII (2, 42), the sit1 mutations are the only mutations that implicate RNAPII directly in the process of transcriptional activation. Our finding that sit1 substitutions affect start site selection and occur in regions of RNAPII that form the enzyme active center implies that there is an intimate relationship between initiation and elongation and has implications for the mechanism of transcriptional activation. To our knowledge, our findings provide the first evidence in eukaryotes that alteration of the enzyme catalytic center can affect transcriptional activation. Previous studies of the mechanism of transcriptional activation have suggested that activation domains stimulate transcription by several mechanisms, including counteracting the repressing effect of chromatin, recruiting the RNAPII holoenzyme complex to the promoter through multiple contacts between activation domains and components of the RNAPII holoenzyme, and/or increasing the processivity of elongating RNAPII (reviewed in references 12 and 46). The results presented in this study provide genetic evidence that partial activation of transcription can also occur by alteration of the RNAPII active site and suggest that some transcription-regulatory proteins exert their effect by modifying, either directly or indirectly, the RNAPII active center. In E. coli, the conformation of the RNAP active center and in particular the way in which it contacts the nascent RNA and the DNA template are major determinants of the processivity of the enzyme (reviewed in reference 29), which can be regulated by antitermination factors such as the N and Q gene products of phage lambda (reviewed in reference 24). By analogy, eukaryotic regulatory proteins that are known to increase the processivity of RNAPII, such as human immunodeficiency virus type 1 Tat, yeast GAL4, human p53 and E2F1, and herpes simplex virus VP16 (14, 28, 52), could exert their effect by altering, either directly or indirectly, the structure or conformation of the enzyme active site during elongation. sit1 mutations, by directly affecting the RNAPII active center, could help stabilize a more processive form of the enzyme and in doing so partially bypass the need for promoter-bound transcriptional activators at HIS4.
ACKNOWLEDGMENTS
This work was supported by grants from the Medical Research Council (MRC) of Canada to J.G. and J.D.F. and by a grant from the National Cancer Institute of Canada to J.G. J.A. held a fellowship from the MRC.
We thank Ying Zou for technical assistance, Michael Hampsey and Caroline Kane for the gift of plasmids and yeast strains, and Shahrzad Nouraini for critical reading of the manuscript.
REFERENCES
- 1.Alani E, Cao L, Kleckner N. A method for gene disruption that allows repeated use of URA3 selection in the construction of multiply disrupted yeast strains. Genetics. 1987;116:541–545. doi: 10.1534/genetics.112.541.test. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Allison L A, Ingles C J. Mutations in RNA polymerase II enhance or suppress mutations in GAL4. Proc Natl Acad Sci USA. 1989;86:2794–2798. doi: 10.1073/pnas.86.8.2794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Allison L A, Moyle M, Shales M, Ingles C J. Extensive homology among the largest subunits of eukaryotic and prokaryotic RNA polymerases. Cell. 1985;42:599–610. doi: 10.1016/0092-8674(85)90117-5. [DOI] [PubMed] [Google Scholar]
- 4.Archambault J, Drebot M A, Stone J C, Friesen J D. Isolation and phenotypic analysis of conditional-lethal, linker-insertion mutations in the gene encoding the largest subunit of RNA polymerase II in Saccharomyces cerevisiae. Mol Gen Genet. 1992;232:408–414. doi: 10.1007/BF00266244. [DOI] [PubMed] [Google Scholar]
- 5.Archambault J, Lacroute F, Ruet A, Friesen J D. Genetic interaction between transcription elongation factor TFIIS and RNA polymerase II. Mol Cell Biol. 1992;12:4142–4152. doi: 10.1128/mcb.12.9.4142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Archambault J, Friesen J D. Genetics of eukaryotic RNA polymerases I, II, and III. Microbiol Rev. 1993;57:703–724. doi: 10.1128/mr.57.3.703-724.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Arndt K T, Styles C, Fink G R. Multiple global regulators control HIS4 transcription in yeast. Science. 1987;237:874–880. doi: 10.1126/science.3303332. [DOI] [PubMed] [Google Scholar]
- 8.Arndt K T, Styles C A, Fink G R. A suppressor of a HIS4 transcriptional defect encodes a protein with homology to the catalytic subunit of protein phosphatases. Cell. 1989;56:527–537. doi: 10.1016/0092-8674(89)90576-x. [DOI] [PubMed] [Google Scholar]
- 9.Barberis A, Pearlberg J, Simkovich N, Farrell S, Reinagel P, Bamdad C, Sigal G, Ptashne M. Contact with a component of the polymerase II holoenzyme suffices for gene activation. Cell. 1995;81:359–368. doi: 10.1016/0092-8674(95)90389-5. [DOI] [PubMed] [Google Scholar]
- 10.Bartolomei M S, Corden J L. Clustered alpha-amanitin resistance mutations in mouse. Mol Gen Genet. 1995;246:778–782. doi: 10.1007/BF00290727. [DOI] [PubMed] [Google Scholar]
- 11.Bartolomei M S, Corden J L. Localization of an α-amanitin resistance mutation in the gene encoding the largest subunit of mouse RNA polymerase II. Mol Cell Biol. 1987;7:586–594. doi: 10.1128/mcb.7.2.586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bentley D L. Regulation of transcriptional elongation by RNA polymerase II. Curr Opin Genet Dev. 1995;5:210–216. doi: 10.1016/0959-437x(95)80010-7. [DOI] [PubMed] [Google Scholar]
- 13.Berroteran R W, Ware D E, Hampsey M. The sua8 suppressors of Saccharomyces cerevisiae encode replacements of conserved residues within the largest subunit of RNA polymerase II and affect transcription start site selection similarly to sua7 (TFIIB) mutations. Mol Cell Biol. 1994;14:226–237. doi: 10.1128/mcb.14.1.226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Blau J, Xiao H, McCracken S, O’Hare P, Greenblatt J, Bentley D. Three functional classes of transcriptional activation domains. Mol Cell Biol. 1996;16:2044–2055. doi: 10.1128/mcb.16.5.2044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bonneaud N, Ozier K O, Li G Y, Labouesse M, Minvielle S L, Lacroute F. A family of low and high copy replicative, integrative and single-stranded S. cerevisiae/E. coli shuttle vectors. Yeast. 1991;7:609–615. doi: 10.1002/yea.320070609. [DOI] [PubMed] [Google Scholar]
- 16.Borukhov S, Lee J, Goldfarb A. Mapping of a contact for the RNA 3′ terminus in the largest subunit of RNA polymerase. J Biol Chem. 1991;266:23932–23935. [PubMed] [Google Scholar]
- 17.Botstein D, Falco S C, Stewart S E, Brennan M, Scherer S, Stinchcomb D T, Struhl K, Davis R W. Sterile host yeasts (SHY): a eukaryotic system of biological containment for recombinant DNA experiments. Gene. 1979;8:17–24. doi: 10.1016/0378-1119(79)90004-0. [DOI] [PubMed] [Google Scholar]
- 18.Cassani G, Burgess R R, Goodman H M, Gold L. Inhibition of RNA polymerase by streptolydigin. Nat New Biol. 1971;230:197–200. doi: 10.1038/newbio230197a0. [DOI] [PubMed] [Google Scholar]
- 19.Chen Y, Weeks J, Mortin M A, Greenleaf A L. Mapping mutations in genes encoding the two large subunits of Drosophila RNA polymerase II defines domains essential for basic transcription functions and for proper expression of developmental genes. Mol Cell Biol. 1993;13:4214–4222. doi: 10.1128/mcb.13.7.4214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Coulter D E, Greenleaf A L. A mutation in the largest subunit of RNA polymerase II alters RNA chain elongation in vitro. J Biol Chem. 1985;260:13190–13198. [PubMed] [Google Scholar]
- 21.de Mercoyrol L, Job C, Job D. Studies on the inhibition by alpha-amanitin of single-step addition reactions and productive RNA synthesis catalyzed by wheat-germ RNA polymerase II. Biochem J. 1989;258:165–169. doi: 10.1042/bj2580165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Devlin C, Tice-Baldwin K, Shore D, Arndt K T. RAP1 is required for BAS1/BAS2- and GCN4-dependent transcription of the yeast HIS4 gene. Mol Cell Biol. 1991;11:3642–3651. doi: 10.1128/mcb.11.7.3642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dieci G, Hermann-Le Denmat S, Lukhtanov E, Thuriaux P, Werner M, Sentenac A. A universally conserved region of the largest subunit participates in the active site of RNA polymerase III. EMBO J. 1995;14:3766–3776. doi: 10.1002/j.1460-2075.1995.tb00046.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Greenblatt J, Nodwell J R, Mason S W. Transcriptional antitermination. Nature. 1993;364:401–406. doi: 10.1038/364401a0. [DOI] [PubMed] [Google Scholar]
- 25.Hekmatpanah D S, Young R A. Mutations in a conserved region of RNA polymerase II influence the accuracy of mRNA start site selection. Mol Cell Biol. 1991;11:5781–5791. doi: 10.1128/mcb.11.11.5781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ito H, Fukuda Y, Murata K, Kimura A. Transformation of intact yeast cells treated with alkali cations. J Bacteriol. 1983;153:163–168. doi: 10.1128/jb.153.1.163-168.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jokerst R S, Weeks J R, Zehring W A, Greenleaf A L. Analysis of the gene encoding the largest subunit of RNA polymerase II in Drosophila. Mol Gen Genet. 1989;215:266–275. doi: 10.1007/BF00339727. [DOI] [PubMed] [Google Scholar]
- 28.Krumm A, Hickey L B, Groudine M. Promoter-proximal pausing of RNA polymerase II defines a general rate-limiting step after transcription initiation. Genes Dev. 1995;9:559–572. doi: 10.1101/gad.9.5.559. [DOI] [PubMed] [Google Scholar]
- 29.Landick R, Roberts J W. The shrewd grasp of RNA polymerase. Science. 1996;273:202–203. doi: 10.1126/science.273.5272.202. [DOI] [PubMed] [Google Scholar]
- 30.Maniatis T, Fritsch E F, Sambrook J. Molecular cloning: a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1982. [Google Scholar]
- 31.Markovtsov V, Mustaev A, Goldfarb A. Protein-RNA interactions in the active center of transcription elongation complex. Proc Natl Acad Sci USA. 1996;93:3221–3226. doi: 10.1073/pnas.93.8.3221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.McClure W R. On the mechanism of streptolydigin inhibition of Escherichia coli RNA polymerase. J Biol Chem. 1980;255:1610–1616. [PubMed] [Google Scholar]
- 33.Memet S, Gouy M, Marck C, Sentenac A, Buhler J M. RPA190, the gene coding for the largest subunit of yeast RNA polymerase A. J Biol Chem. 1988;263:2830–2839. [PubMed] [Google Scholar]
- 34.Miller J. Experiments in molecular genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1972. [Google Scholar]
- 35.Mustaev A, Kozlov M, Markovtsov V, Zaychikov E, Denissova L, Goldfarb A. Modular organization of the catalytic center of RNA polymerase. Proc Natl Acad Sci USA. 1997;94:6641–6645. doi: 10.1073/pnas.94.13.6641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.O’Brien T, Hardin S, Greenleaf A, Lis J T. Phosphorylation of RNA polymerase II C-terminal domain and transcriptional elongation. Nature. 1994;370:75–77. doi: 10.1038/370075a0. [DOI] [PubMed] [Google Scholar]
- 37.Orr-Weaver T L, Szostak J W, Rothstein R J. Genetic applications of yeast transformation with linear and gapped plasmids. Methods Enzymol. 1983;101:228–245. doi: 10.1016/0076-6879(83)01017-4. [DOI] [PubMed] [Google Scholar]
- 38.Pinto I, Na J G, Sherman F, Hampsey M. cis- and trans-acting suppressors of a translation initiation defect at the cyc1 locus of Saccharomyces cerevisiae. Genetics. 1992;132:97–112. doi: 10.1093/genetics/132.1.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rougvie A E, Lis J T. The RNA polymerase II molecule at the 5′ end of the uninduced hsp70 gene of D. melanogaster is transcriptionally engaged. Cell. 1988;54:795–804. doi: 10.1016/s0092-8674(88)91087-2. [DOI] [PubMed] [Google Scholar]
- 40.Sanger F, Nicklen S, Coulson A R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sawadogo M, Sentenac A. RNA polymerase B (II) and general transcription factors. Annu Rev Biochem. 1990;59:711–754. doi: 10.1146/annurev.bi.59.070190.003431. [DOI] [PubMed] [Google Scholar]
- 42.Scafe C, Chao D, Lopes J, Hirsch J P, Henry S, Young R A. RNA polymerase II C-terminal repeat influences response to transcriptional enhancer signals. Nature. 1990;347:491–494. doi: 10.1038/347491a0. [DOI] [PubMed] [Google Scholar]
- 43.Scafe C, Martin C, Nonet M, Podos S, Okamura S, Young R A. Conditional mutations occur predominantly in highly conserved residues of RNA polymerase II subunits. Mol Cell Biol. 1990;10:1270–1275. doi: 10.1128/mcb.10.3.1270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Severinov K, Markov D, Severinova E, Nikiforov V, Landick R, Darst S A, Goldfarb A. Streptolydigin-resistant mutants in an evolutionarily conserved region of the beta′ subunit of Escherichia coli RNA polymerase. J Biol Chem. 1995;270:23926–23929. doi: 10.1074/jbc.270.41.23926. [DOI] [PubMed] [Google Scholar]
- 45.Sherman F, Fink G R, Hicks J B. Laboratory course manual for methods in yeast genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1986. [Google Scholar]
- 46.Stargell L A, Struhl K. Mechanisms of transcriptional activation in vivo—two steps forward. Trends Genet. 1996;12:311–315. doi: 10.1016/0168-9525(96)10028-7. [DOI] [PubMed] [Google Scholar]
- 47.Thuillier V, Brun I, Sentenac A, Werner M. Mutations in the α-amanitin conserved domain of the largest subunit of yeast RNA polymerase III affect pausing, RNA cleavage and transcriptional transitions. EMBO J. 1996;15:618–629. [PMC free article] [PubMed] [Google Scholar]
- 48.von der Helm K, Krakow J S. Inhibition of RNA polymerase by streptolydigin. Nat New Biol. 1972;235:82–83. doi: 10.1038/newbio235082a0. [DOI] [PubMed] [Google Scholar]
- 49.Weilbaecher R, Hebron C, Feng G, Landick R. Termination-altering amino acid substitutions in the beta′ subunit of Escherichia coli RNA polymerase identify regions involved in RNA chain elongation. Genes Dev. 1994;8:2913–2927. doi: 10.1101/gad.8.23.2913. [DOI] [PubMed] [Google Scholar]
- 50.Wu J, Awrey D E, Edwards A M, Archambault J, Friesen J D. In vitro characterization of mutant yeast RNA polymerase II with reduced binding for elongation factor TFIIS. Proc Natl Acad Sci USA. 1996;93:11552–11557. doi: 10.1073/pnas.93.21.11552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yang X, Price C W. Streptolydigin resistance can be conferred by alterations to either the beta or beta′ subunits of Bacillus subtilis RNA polymerase. J Biol Chem. 1995;270:23930–23933. doi: 10.1074/jbc.270.41.23930. [DOI] [PubMed] [Google Scholar]
- 52.Yankulov K, Blau J, Purton T, Roberts S, Bentley D L. Transcriptional elongation by RNA polymerase II is stimulated by transactivators. Cell. 1994;77:749–759. doi: 10.1016/0092-8674(94)90058-2. [DOI] [PubMed] [Google Scholar]
- 53.Young R A. RNA polymerase II. Annu Rev Biochem. 1991;60:689–715. doi: 10.1146/annurev.bi.60.070191.003353. [DOI] [PubMed] [Google Scholar]
- 54.Zaychikov E, Martin E, Denissova L, Kozlov M, Markovtsov V, Kashlev M, Heumann H, Nikiforov V, Goldfarb A, Mustaev A. Mapping of catalytic residues in the RNA polymerase active center. Science. 1996;273:107–109. doi: 10.1126/science.273.5271.107. [DOI] [PubMed] [Google Scholar]