Abstract
Bioengineering of photorespiratory bypasses is an effective strategy for improving plant productivity by modulating photosynthesis. In previous work, two photorespiratory bypasses, the GOC and GCGT bypasses, increased photosynthetic rates but decreased seed-setting rate in rice (Oryza sativa), probably owing to excess photosynthate accumulation in the stem. To solve this bottleneck, we successfully developed a new synthetic photorespiratory bypass (called the GMA bypass) in rice chloroplasts by introducing Oryza sativa glycolate oxidase 1 (OsGLO1), Cucurbita maxima malate synthase (CmMS), and Oryza sativa ascorbate peroxidase7 (OsAPX7) into the rice genome using a high-efficiency transgene stacking system. Unlike the GOC and GCGT bypass genes driven by constitutive promoters, OsGLO1 in GMA plants was driven by a light-inducible Rubisco small subunit promoter (pRbcS); its expression dynamically changed in response to light, producing a more moderate increase in photosynthate. Photosynthetic rates were significantly increased in GMA plants, and grain yields were significantly improved under greenhouse and field conditions. Transgenic GMA rice showed no reduction in seed-setting rate under either test condition, unlike previous photorespiratory-bypass rice, probably reflecting proper modulation of the photorespiratory bypass. Together, these results imply that appropriate engineering of the GMA bypass can enhance rice growth and grain yield without affecting seed-setting rate.
Key words: photorespiratory bypass, photosynthesis, rice, grain yield, seed-setting rates
Bioengineering a novel photorespiratory bypass is a viable and feasible approach for substantially improving plant productivity. The light-dependent GMA bypass can significantly improve photosynthetic rates and grain yields of rice under greenhouse and field conditions without affecting seed-setting rate.
Introduction
The global population is increasing dramatically and is expected to reach 10 billion by the end of 2050, leading to growing demands for crop production (von Caemmerer et al., 2012; Ort et al., 2015). Improving photosynthesis has been proposed as a promising approach to increase the productivity of important agronomic crops (Zhu et al., 2010; Evans, 2013; Nolke et al., 2014; Long et al., 2015). Reducing CO2 or energy loss associated with photorespiration could potentially increase gross photosynthesis by 12%–55% (Peterhansel et al., 2010; Walker et al., 2016), and engineering and repressing photorespiration could therefore be a practical strategy for enhancing photosynthesis (South et al., 2019; Roell et al., 2021).
Recently, synthetic biology–based approaches have been used to engineer photorespiratory bypasses and improve photosynthetic efficiency in C3 plants. Three well-known bypasses developed to reduce photorespiratory losses are the Escherichia coli glycolate catabolic bypass (Kebeish et al., 2007), synthetic glycolate oxidative cycle bypass (Maier et al., 2012), and AP3 bypass (South et al., 2019; Cavanagh et al., 2021). However, all these bioengineering approaches have been carried out in dicot plants. More efforts need to be made in major crops, particularly in monocot grain crops such as rice (Oryza sativa).
In our previous work, we established a GOC bypass in rice chloroplasts by introducing three rice genes encoding glycolate oxidase 3 (OsGLO3), oxalate oxidase3 (OsOXO3), and catalase (OsCATC) (Shen et al., 2019). The GOC bypass significantly increased photosynthetic rates and grain yield under field conditions. However, the grain yields of GOC rice were unstable in different cultivation seasons and decreased by up to 16% in the fall owing to a decrease in seed-setting rate. Subsequently, a GCGT bypass consisting of the rice protein OsGLO1 and three E. coli proteins, EcCAT, glyoxylate carboligase (EcGCL), and tartronic semialdehyde reductase (EcTSR) (Wang et al., 2020), was established in rice chloroplasts. It significantly increased photosynthesis but still produced a reduced seed-setting rate. More efforts are needed to engineer a new effective bypass using plant native genes to enhance grain yield without affecting seed-setting rate in rice.
Given that source–sink flow plays a central role in developing seeds, it is necessary to increase its balance to improve grain yield without reducing seed-setting rate. In GCGT plants, leaf productivity (source) increased, but the translocation of assimilated carbohydrates (flow) decreased dramatically compared with wild-type (WT) plants, leading to a “traffic jam” of photosynthates in the stem, which subsequently reduced seed-setting rate (Wang et al., 2020). Excess accumulation of photosynthates in the stem and decreased seed-setting rate in rice suggest that the use of strong constitutive promoters to drive target gene expression may not be suitable for photorespiratory bypass engineering (Shen et al., 2019; Wang et al., 2020), especially for expression of rate-limiting genes. Furthermore, the high overexpression levels of the protein driven by the constitutive promoter could result in a decrease in plant growth (Lopez-Calcagno et al., 2019). Hence, inducible promoters or tissue-specific promoters may be better for engineering photorespiration to improve plant growth and yield (Chen et al., 2020; Wang et al., 2020).
Here, we engineered a novel photorespiratory bypass in rice chloroplasts by introducing three plant native enzymes, OsGLO1, Cucurbita maxima malate synthase (CmMS), and rice ascorbate peroxidase 7 (OsAPX7), and named it the GMA bypass. OsGLO1 was driven by a green tissue–specific and light-inducible rice Rubisco small subunit promoter (pRbcS) (Nomura et al., 2000), which efficiently and dynamically induced the expression of OsGLO1 largely in response to light intensity, producing a mild increase in photosynthates in GMA rice. Seed-setting rate was unimpaired and grain yield was increased in GMA rice compared with WT plants under greenhouse and field conditions.
Results and discussion
Establishment of a functional GMA bypass in rice chloroplasts
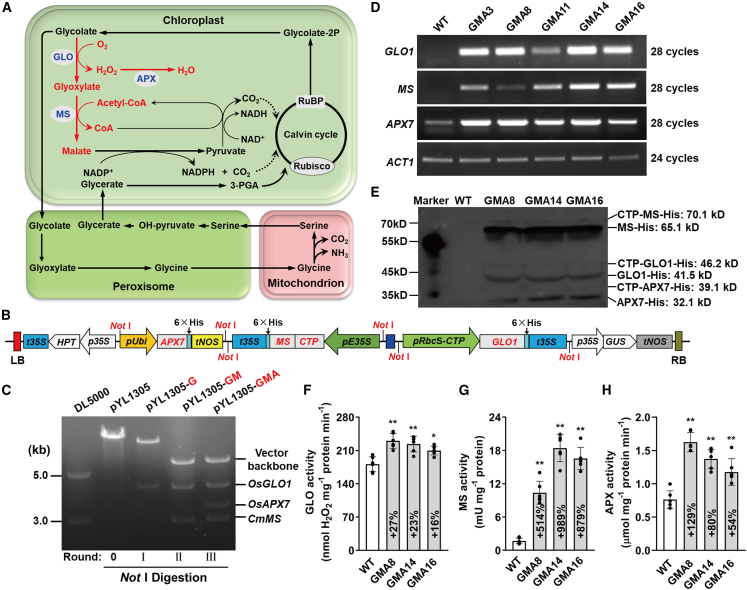
Three enzymes, OsGLO1, CmMS, and OsAPX7, were used to construct the GMA bypass in rice chloroplasts. Glycolate was metabolized in this bypass, leading to release of CO2 directly in the chloroplasts (Figure 1A). We previously demonstrated that inhibition of OsGLO1 expression led to dramatic accumulation of glycolate (Xu et al., 2009); OsGLO1 was the most abundant isoform at different developmental stages and the major contributor to GLO activity in rice leaves (Zhang et al., 2017). Therefore, OsGLO1 was recruited for the GMA bypass. Ascorbate peroxidase (APX) is more suitable than CAT for H2O2 quenching in the chloroplasts and prevents the release of O2 during H2O2 scavenging (Teixeira et al., 2004), which may contribute to an increased CO2/O2 ratio in chloroplasts. OsAPX7 displayed a higher transcript level than other chloroplastic APX isoforms, even with a high H2O2 level (Teixeira et al., 2006; Li et al., 2015). Thus, OsAPX7 was used instead of OsCAT in the GMA bypass to detoxify H2O2 in chloroplasts. OsGLO1 is the first enzyme that functions in the GMA bypass, and its expression is closely associated with diversion of glycolate into the bypass. To properly control metabolite distribution to the GMA bypass, a light-inducible pRbcS promoter was used to drive OsGLO1 expression. To avoid gene silencing probably caused by homologous promoters (Fagard and Vaucheret, 2000), we used another two constitutive promoters, the CaMV35S promoter (pE35S) and the maize UBIQUITIN promoter (pUbi), to drive CmMS and OsAPX7, respectively. An effective rice SSU chloroplast transit peptide (CTP; D00643) was used to direct OsGLO1 and CmMS into rice chloroplasts (Jang et al., 2002; Shen et al., 2017). OsAPX7 was directed by its CTP (Xu et al., 2013).
Figure 1.
Establishment of the GMA photorespiratory bypass in rice.
(A) The GMA bypass (in red) is integrated with the plant native metabolic pathway (black), which includes the chloroplast, peroxisome, and mitochondrion. RuBP, ribulose-1,5-bisphosphate; 3-PGA, 3-phosphoglycerate; GLO, OsGLO1, rice glycolate oxidase 1; MS, CmMS, Cucurbita maxima malate synthase; APX, OsAPX7, Oryza sativa ascorbate peroxidase 7. The three enzymes introduced into rice chloroplasts are highlighted in blue.
(B) Structure of the multi-gene expression cassette. LB, left border; t35S, CaMV 35S terminator; HPT, hygromycin phosphotransferase; p35S, 35S promoter; pUbi, ubi promoter; tNOS, nos terminator; CTP, chloroplast transit peptide; pE35S, CaMV 35S enhanced promoter; pRbcS-CTP, Rubisco small subunit promoter fused with CTP; GUS, β-glucuronidase; RB, right border.
(C) Digestion analysis of the different rounds of acceptor constructs with NotI.
(D) Expression analysis of photorespiratory bypass genes by semi-quantitative RT–PCR.(E) Immunoblot analysis was performed using total protein extracted from leaves.
(F–H) Activities of OsGLO1, CmMS, and OsAPX7 in rice leaves. Error bars indicate means ± SD (n = 6). Asterisks indicate a significant difference between WT and GMA rice (∗P < 0.05; ∗∗P < 0.01; the same convention is used below).
Next, we synthesized the multi-gene stacking construct pYL1305-GMA, which simultaneously expressed all three genes (Figure 1B). NotI digestion verified the successful insertion of these three expression cassettes into the acceptor vector pYL1305 (Figure 1C).
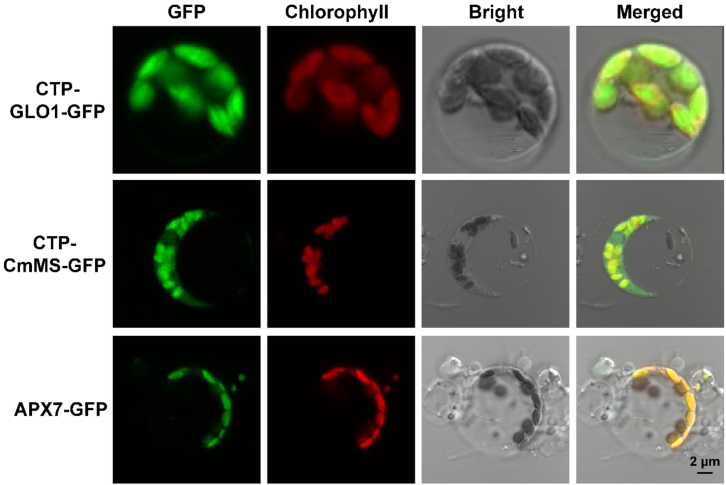
Several independent homozygous lines were identified, and three independent homozygous lines (GMA 8, 14, and 16) were used for subsequent analyses. OsGLO1, CmMS, and OsAPX7 showed higher expression at the transcriptional (Figure 1D) and protein (Figure 1E) levels and exhibited higher enzyme activity (Figures 1F–1H) in GMA lines than in the WT. Despite high GLO background activity in WT plants (Xu et al., 2006, 2009), GLO activity was 16%–27% higher in GMA plants than in WT plants (Figure 1F). Malate synthase (MS) and APX activities increased significantly by 5.1- to 9.9-fold and 0.54- to 1.29-fold, respectively, in GMA plants compared with WT plants (Figures 1G and 1H). Transient expression analysis in rice protoplasts also demonstrated that the three proteins were effectively targeted into chloroplasts (Figure 2). These results strongly indicated that the three enzymes were successfully targeted into the chloroplasts and were functional in transgenic rice.
Figure 2.
Subcellular targeting analysis of photorespiratory bypass genes by transient expression in rice protoplasts.
GFP, GFP fluorescence; Chlorophyll, chlorophyll autofluorescence; Bright, bright-field image; Merged, superposition of GFP, chlorophyll, and bright-field signals; CTP-GLO1-GFP, rice protoplasts were transformed with a plasmid harboring the CTP-GLO1-GFP construct; CTP-CmMS-GFP, rice protoplasts were transformed with a plasmid harboring the CTP-CmMS-GFP construct; APX7-GFP, rice protoplasts were transformed with a plasmid harboring the APX7-GFP construct.
GMA rice shows increased photosynthetic rate and capacity
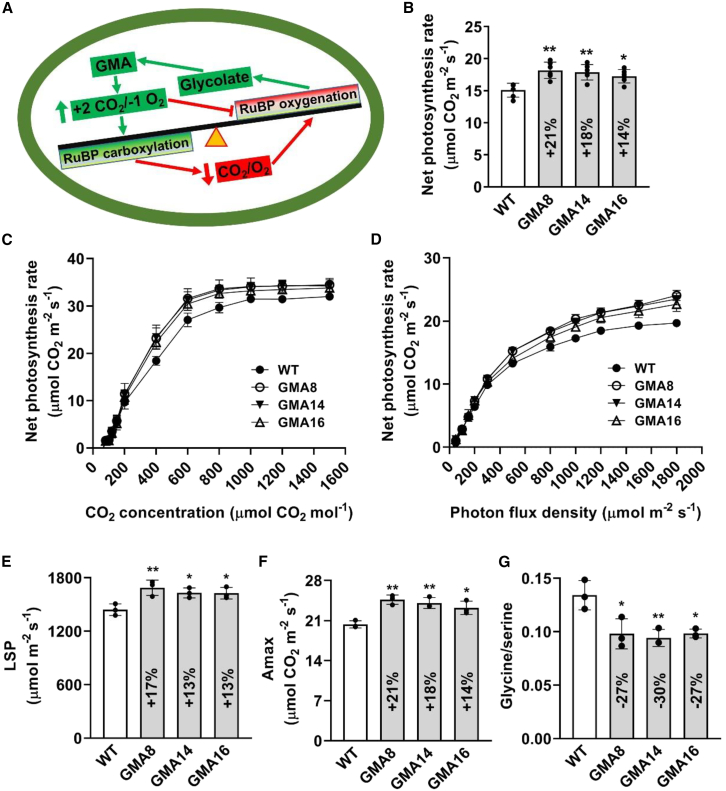
Theoretically, one molecule of glycolate would produce two molecules of CO2 and consume one molecule of O2 through the GMA bypass in the chloroplast. Therefore, the GMA bypass causes a direct increase in the chloroplast CO2/O2 ratio, which tends to promote the carboxylation reaction of Rubisco and enhances photosynthesis (Figure 3A). In brief, as long as glycolate flows into the GMA bypass, it will enhance the CO2/O2 ratio, thus increasing photosynthesis and suppressing photorespiration (Figure 3A). To verify that this occurred, we measured gas exchange in GMA and WT plants at the tillering stage. Compared with the WT plants, GMA rice showed a significant increase in net photosynthetic rate (PN), with an average increase of 14% to 21% in the three independent lines (Figure 3B). CO2-response and light-response curves were also analyzed to confirm the enhanced photosynthesis. In the CO2-response curves, GMA plants had higher PN values when the intracellular CO2 concentration was above 200 ppm, and the maximum difference occurred at about 400 ppm (Figure 3C). In the light-response curves, PN increased with increasing photon flux density (PFD) in both GMA and WT plants, and a difference in PN between WT and GMA plants emerged when the PFD was above 300 μmol m−2 s−1 (Figure 3D). In addition, the light saturation point (LSP) and light-saturated photosynthetic rate (Amax) increased by 13%–17% and 14%–21%, respectively, in GMA plants relative to the WT (Figures 3E and 3F). All these results demonstrated that photosynthetic performance was enhanced in GMA rice.
Figure 3.
GMA photorespiratory bypass increases photosynthetic rate and capacity in rice.
(A) Balance of Rubisco responses to CO2/O2 in GMA rice. RuBP, ribulose-1,5-bisphosphate.
(B) Net photosynthetic rate (PN) was measured at 800 μmol m−2 s−1.
(C) Response of PN to CO2 concentration at 1200 μmol m−2 s−1 photon flux density (PFD) and 30°C.
(D) Response of PN to light intensity at 30°C under normal air conditions.
(E and F) Light saturation point (LSP) and light-saturated photosynthetic rate (Amax) were fitted based on the light-response curves.
(G) Glycine/serine ratio in WT and GMA lines. The amino acid values were normalized per gram FW in WT and GMA lines.
A lower photorespiratory rate results in a lower glycine/serine ratio (Novitskaya et al., 2002; Kebeish et al., 2007; Dalal et al., 2015; Shen et al., 2019). Here, the glycine/serine ratio decreased by about 27%–30% in GMA rice relative to WT plants (Figure 3G), consistent with a reduced photorespiratory rate in GMA plants. The light and dark reactions cooperate to complete photosynthesis. The Fv/Fm ratio was measured to evaluate the light reactions and photoinhibition. Consistent with reports on the GOC and GCGT bypasses (Shen et al., 2019; Wang et al., 2020), the Fv/Fm ratio did not differ between WT and GMA rice (Supplemental Figure 1).
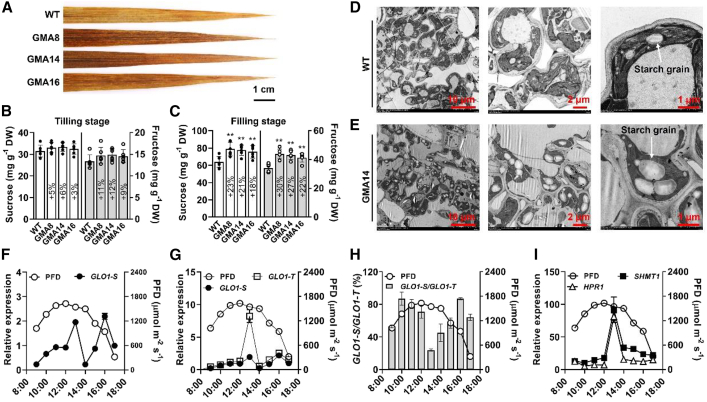
Metabolite content was analyzed at different developmental stages to evaluate the effects of the GMA bypass on primary metabolism. GMA rice accumulated more starch at the grain filling stage, as shown by darker staining (Figure 4A). These results were similar to those in other photorespiratory bypass plants (Maier et al., 2012; Nolke et al., 2014; Dalal et al., 2015; Wang et al., 2020). Sucrose and fructose contents in GMA plants were slightly higher than those in the WT at the early tillering stage (Figure 4B). By contrast, sucrose and fructose contents were 18%–23% and 22%–30% higher in leaves of the GMA lines than in those of WT plants at the filling stage (Figure 4C). We examined the ultrastructure of flag leaves by transmission electron microscopy (TEM) at the filling stage and found that GMA plants accumulated more starch grains than WT plants (Figures 4D and 4E). Collectively, these results strongly suggested that photosynthesis and photosynthate accumulation were improved in GMA rice.
Figure 4.
Photosynthetic carbohydrate content in leaves of GMA and WT plants and diurnal expression profiles of OsGLO1 and the key photorespiratory genes SHMT1 and HPR1 in GMA rice.
(A) Accumulation of starch assessed by iodine staining.
(B and C) Sucrose and fructose contents in leaves of WT and GMA lines.
(D and E) Transmission electron micrographs of WT and GMA leaves at the filling stage.
(F) Diurnal expression profile of transgene-specific OsGOL1 (GLO1-S) under ambient conditions.
(G) Comparative analysis of the diurnal expression profiles of GLO1-S and total OsGOL1 (GLO1-T) under ambient conditions.
(H) Diurnal changes in the GLO1-S/GLO1-T ratio.
(I) Diurnal expression profiles of two additional key photorespiratory genes, OsSHMT1 and OsHPR1.
OsGLO1 expression is generally controlled by light and probably contributes to the unimpaired setting rate
To address the setting rate defect in GOC and GCGT plants (Shen et al., 2019; Wang et al., 2020), we used the light-inducible promoter pRbcS to drive OsGLO1 expression in GMA plants. Theoretically, diversion of glycolate into the GMA bypass is closely associated with transgene-specific OsGLO1 (GLO1-S) expression in the chloroplast (Figure 1A). As shown in Figure 4F, the diurnal expression profile of GLO1-S in GMA plants displayed two peaks, one at 13:00 and one at 16:00. There was a valley at 14:00, similar to the midday depression in photosynthesis, suggesting that expression of GLO1-S was regulated not only by light but also by other factors such as stomatal closure (Barta and Loreto, 2006).
To further evaluate the role of pRbcS in the GMA bypass during the day, we analyzed the diurnal expression profiles of total OsGLO1 (GLO1-T, including the expression of GLO1-S and the native photorespiratory OsGLO1) and calculated the ratio of GLO1-S/GLO1-T. The GLO1-T diurnal expression profile was generally similar to that of GLO1-S (Figure 4G), and the GLO1-S/GLO1-T ratio was greater than 50%, except at 13:00 and 14:00 (Figure 4H). It is worth noting that the expression of GLO1-T drastically increased compared to GLO1-S at noon (Figure 4G), and the corresponding GLO1-S/GLO1-T ratio declined to its lowest level (less than 24%) at 13:00 (Figure 4H). Two other key photorespiratory genes, serine hydroxymethyl transferase (SHMT1, XM_015774325) and hydroxypyruvate reductase (HPR1, AK067642) (Figure 4I) (Peterhansel et al., 2010), showed expression profiles similar to that of GLO1-T, indicating that the photorespiratory rate was greatly enhanced at noon (Muraoka et al., 2000), and only a small fraction of glycolate could probably be diverted into the GMA bypass. Furthermore, the protein expression level of GLO1-S also indicated weak expression both at noon (12:00–14:00) and at nighttime (21:00) (Supplemental Figure 2), confirming the dynamic regulation of OsGLO1 protein levels. OsGLO1 contributes about 65% of GLO activity in rice leaves (Zhang et al., 2017), and GLO activity in chloroplasts is closely associated with the conversion of glycolate to glyoxylate (Fahnenstich et al., 2008). It is therefore reasonable to speculate that light-dependent OsGLO1 expression will play a critical role in determining the amount of glycolate directed into the GMA bypass and will probably limit excess photosynthate accumulation, especially at noon. Consistent with this prediction, sucrose and fructose accumulation (Figures 4D and 4E; sucrose, 18%–23% higher than WT; fructose, 22%–30% higher than WT) was lower in GMA plants than in GOC (sucrose, up to 30% higher than WT; fructose, up to 70% higher than WT) and GCGT plants (sucrose, up to 53% higher than WT; fructose, up to 80% higher than WT) at the filling stage (Shen et al., 2019; Wang et al., 2020).
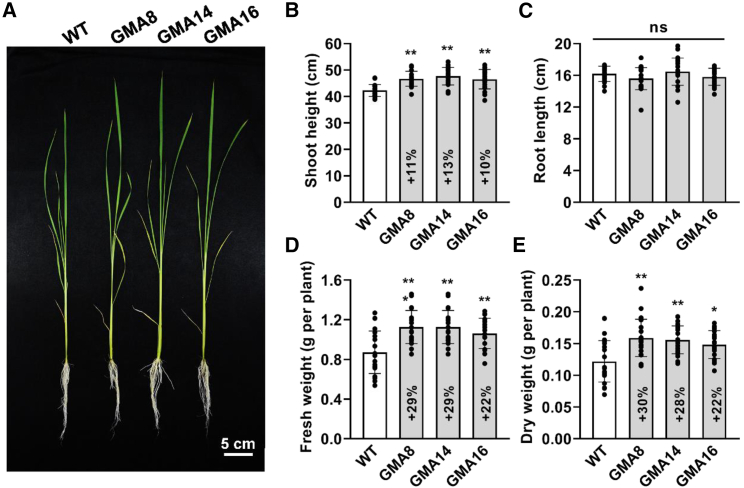
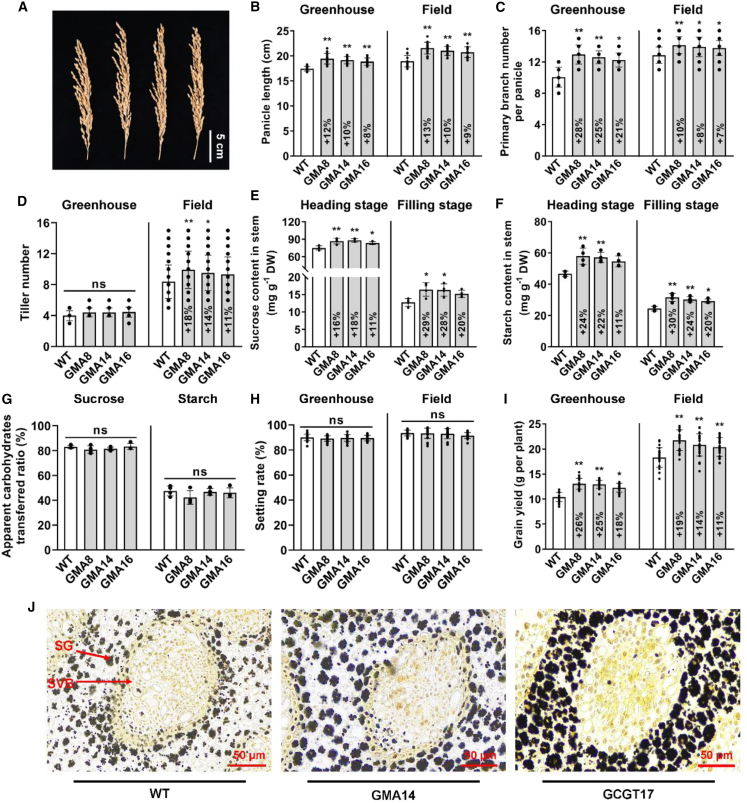
GMA rice shows marked increases in biomass and grain yield
GMA rice showed significantly more rapid growth than the WT at the seedling stage in a greenhouse experiment (Figure 5A). Although GMA and WT plants had similar root lengths, shoot height increased by 10%–13% in GMA rice (Figure 5B). Likewise, fresh weight (FW) and dry weight (DW) increased significantly by 22%–29% and 22%–30%, respectively, in GMA rice relative to the WT (Figures 5D and 5E), indicating that GMA transgenic rice grew faster than the WT at this stage. At the mature stage, the main panicle length of GMA rice was increased by 8%–12% and 9%–13% relative to the WT under greenhouse and field conditions, respectively (Figures 6A and 6B), and the primary branch number was enhanced by 21%–28% and 7%–10% (Figure 6C). The tiller number was 11%–18% higher in GMA plants than in the WT under field conditions in Shanghai (Figure 6D). We speculate that the increased photosynthates were quickly and efficiently transported to the grain in GMA rice. Consistent with this speculation, although the sucrose content was 11%–18% and 20%–29% higher in the GMA stem at the heading and filling stages (Figure 6E), respectively, and starch content in the GMA stem was 11%–24% and 20%–30% higher (Figure 6F), the apparent sucrose and starch transfer ratios were similar to those of the WT (Figure 6G). Not surprisingly, the seed-setting rate of GMA plants was comparable to that of WT plants (Figure 6H), and the grain yield per plant was 18%–26% and 11%–19% higher under greenhouse and field conditions, respectively, compared with the WT (Figure 6I). Moreover, the light-dependent GMA bypass increased tiller number and grain yield without affecting seed-setting rate in the field in Wuhan (Supplemental Figure 3). During the grain filling stage, under the same field conditions, GMA stems contained more starch than those of WT plants; however, the starch level was lower relative to GCGT plants (Figure 6J and Supplemental Figure 4). RNA sequencing (RNA-seq) data revealed that although sucrose transporter genes were unaffected, fewer genes involved in starch synthesis and degradation were upregulated in GMA plants than in GCGT plants (Supplemental Table 1), suggesting that the GMA bypass probably prevented a photosynthate traffic jam and thus enabled normal seed-setting rates in rice. In addition, we found that fewer differentially expressed genes (42) were present in GMA stems (vs. WT), and they were divided into fewer Kyoto Encyclopedia of Genes and Genomes (KEGG) categories (6) than those in GCGT stems (52/9; Supplemental Table 2), suggesting that the light-dependent GMA bypass has less influence on the balance of metabolites or energy in plants. Hence, plants containing the synthetic light-dependent GMA bypass accumulate more photosynthates but presumably transport them to grains in a timely manner, avoiding confounding effects on the balance of metabolites or energy, thereby improving growth and grain yield without affecting seed-setting rate in rice. However, it should be noted that many factors can affect seed-setting rate in rice (Zhou et al., 2011; Li et al., 2013; Liu et al., 2018; Xiang et al., 2019; Wang et al., 2021). For example, seed-setting rate is substantially influenced by aspects of the external environment such as temperature (Xu et al., 2014; Wang et al., 2016) and drought (Fu et al., 2014), and GOC rice showed various seed-setting rates in different planting seasons (Shen et al., 2019). How the photorespiratory bypass regulates seed-setting rate therefore remains an open question and requires further investigation.
Figure 5.
Phenotypic analysis of GMA and WT plants at the seedling stage in the greenhouse.
(A) Phenotypes of WT and GMA plants at the seedling stage.
(B) Shoot height.
(C) Root length.
(D) FW.
(E) DW.
Figure 6.
Yield traits of GMA lines at the mature stage and carbohydrate flow in the stem.
(A) Phenotype of the main panicle.
(B) Panicle length.
(C) Primary branch number per panicle.
(D) Tiller number.
(E and F) Sucrose and starch contents in stems (culms and sheaths).
(G) Apparent ratio of transferred stem carbohydrates.
(H) Setting rate.
(I) Grain yield per plant.
(J) Accumulation of starch in stems. SG, starch grain; SVB, small vascular bundle.
In conclusion, we established a new light-dependent photorespiratory GMA bypass in rice chloroplasts. Results showed that introduction of this light-dependent GMA bypass into rice chloroplasts is an efficient strategy for improving photosynthesis, thereby promoting growth and grain yield under field conditions without reducing seed-setting rate. This study provides an essential step toward increasing grain yield by establishing a photorespiratory bypass in rice chloroplasts.
Methods
Construction of multi-gene expression vector pYL1305-GMA and generation of GMA transformants
OsGLO1 (AK098878), OsAPX7 (XM_015780377), and pRbcS with CTP (pRbcS-CTP; AB067656) were cloned from rice leaves. CmMS (X56948) was amplified from pumpkin cotyledons. Unique restriction sites HindIII and SalI, ClaI and SacI, and KpnI and SpeI were introduced to the 5′ and 3′ ends of t35S, pRbcS, and OsGLO1, respectively. t35S, pRbcS, and OsGLO1 were then successively cloned into pYL322d1 by the conventional restriction-ligation method (Supplemental Figure 5A). The same strategy was used to construct two other donor vectors, pYL322d2-CmMS and pYL322d1-OsAPX7 (Supplemental Figures 5B and 5C). After three rounds of gene assembly, OsGLO1, CmMS, and OsAPX7 expression cassettes were inserted successively into the acceptor vector pYL1305 by the homologous recombination method mediated by the Cre/loxP system (Zhu et al., 2017), resulting in the final multi-gene expression vector pYL1305-GMA. The integration of each target gene in pYL1305-GMA was analyzed by NotI digestion. All primers are listed in Supplemental Table 3. The binary vector pYL1305-GMA was then transformed into the japonica rice variety ZH11 by Agrobacterium-mediated transformation. Homozygous lines were isolated in the T2 generation by examining segregation ratios of hygromycin B resistance in at least 50 seeds per line. PCR was then performed to confirm the presence of transgenes in individual plants. The azygous rice was used as the WT, which had been segregated through the T1 generation without the transformation construct and the selectable resistance marker.
Plant growth conditions
Pre-germinated rice seeds from the T1 generation were transferred to Kimura B nutrient solution (Ma and Takahashi, 1990) for 1 week and then transplanted into soil and grown in a greenhouse at 30°C/25°C under a light:dark cycle of 14 h:10 h and an irradiance of 800–1000 μmol photons m−2 s−1. The 21-day-oldseedlings were then transplanted to pots or experimental fields for further growth. The plants in pots were used for greenhouse experiments. For the field experiments, the GMA and WT plants were transplanted to paddy fields in Shanghai, China, and grown from May to October in 2017, 2018, and 2022. For field experiments, all plants were under routine management practices, well irrigated, and protected from pests to ensure meaningful field tests. Climate data for the growing seasons are provided in Supplemental Table 4 and were obtained from http://www.tianqi.com/. The three GMA lines and the WT were arranged in a completely randomized block design with a planting density of 25 × 25 cm per plant and approximately 16 m2 (4 × 4 m) per plot area. All experiments were carried out with three independent replications. The field data presented in the main text are average values based on the experiments in 2018 and 2022. The field experiment was also performed in Wuhan, China, in 2022.
Gene expression, protein detection, and enzyme activity assays
Total RNA was isolated from rice leaves using the TRIzol reagent (Takara Bio, Dalian, China) according to the manufacturer’s instructions. Gene expression analysis and western blot analysis were performed based on our previous study (Shen et al., 2019). Transgene-specific primers were designed for OsGLO1-S. The rice ACTIN1 (ACT1; KC140126) gene was used as an internal control.
GLO activity was assayed according to a previously reported method (Xu et al., 2009). Fresh leaf samples (0.1 g) were ground to powder with liquid nitrogen and homogenized in 1.0 ml of extraction buffer (100 mM phosphate, pH 8.0) at 4°C. The extracts were then centrifuged at 15 000 g for 15 min at 4°C. The supernatant was collected for the enzyme activity assay. The 1.5-ml reaction mixture was composed of 66 mM phosphate buffer (pH 8.0), 1 mM 4-amino-antipyrine, five units of horseradish peroxidase, 2 mM phenol, 0.1 mM FMN, 5 mM glycolate/glyoxylate, and 0.05 ml of enzyme extract. The reaction mixture was incubated at 30°C for 15 min in the dark. Distilled water substituted for the substrate in the blank. GLO activity was determined from H2O2 production by measuring the absorbance change at 520 nm for 1 min at 30°C.
MS activity was determined as described by Fahnenstich et al. (2008). Leaf samples were ground to powder in liquid nitrogen, homogenized in extraction buffer containing 1 mM MgCl2 and 50 mM Tris–HCl (pH 8.0), and clarified by centrifugation. Then 10 μl of crude extract was used directly for enzymatic measurements with a reaction buffer containing 50 mM Tris–HCl (pH 8.0), 5 mM MgCl2, 3 mM acetyl-CoA, and 1.6 mM 5,5-dithiobis 2-nitrobenzoic acid. After addition of 4 mM glyoxylate, the absorbance change was measured at 410 nm and 25°C. One unit of MS activity was defined as the amount of enzyme that catalyzed the production of 1 μmol of 4-nitrothiolate min−1 calculated by the Lambert–Beer law.
APX activity was measured as described by Singh and Sinha (2016). Fresh samples (0.15 g) were powdered in liquid nitrogen and homogenized in extraction buffer (100 mM phosphate buffer [pH 7.3], 1 mM EDTA, 1% polyvinylpyrrolidone, and 1 mM ascorbate). The extracts were then centrifuged at 12 000 rpm for 20 min at 4°C, and the supernatant was used for the APX activity assay. The 3-ml reaction medium contained 0.1 M phosphate buffer (pH 7.0), 0.5 mM ascorbic acid, 2.5 mM H2O2, and 100 μl of enzyme extract. The rate of H2O2-dependent ascorbic acid oxidation was determined by monitoring the decline in absorbance at 290 nm (extinction coefficient of 2.8 mM−1 cm−1). One unit of APX activity was expressed as the ascorbic acid oxidized by 0.001 g of extracted protein per minute.
Phenotypic characterization
Thirty-day-old seedlings were used to measure shoot FW and DW. DW was measured after 48 h at 80°C. Panicle length and primary branch number were analyzed at full maturity. Grain yield was measured by harvesting all full grains from each plant.
Gas exchange measurements and chlorophyll fluorescence analysis
PN, CO2-response curves, and light-response curves were measured with a portable photosynthesis system (LI-6400, LI-COR, NE, USA) based on Ely et al. (2021) at the tillering stage. PN was measured at a leaf temperature of 30°C in natural air using an irradiance of 800 μmol m−2 s−1. CO2-response curves were obtained at 1200 μmol m−2 s−1 PFD, a temperature of 30°C, and 60% relative humidity; CO2 concentration was gradually reduced from 1600 to 80 μmol mol−1. Light-response curves were obtained at a leaf temperature of 30°C, 60% relative humidity, and 400 μmol mol−1 CO2 concentration; PFD was gradually increased from 0 to 2200 μmol m−2 s−1. LSP and Amax were calculated with the software photo-assistant supplied by the manufacturer (LI-COR, NE, USA) based on the light-response and CO2-response curves. Chlorophyll fluorescence was measured with a PAM-2100 portable chlorophyll fluorometer (Walz, Effeltrich, Germany). The leaves were dark acclimated for 15 min before analysis of the maximum photochemical efficiency (Fv/Fm) of photosystem II (PSII).
Metabolite content determination
For carbohydrate content analysis, shoots, young fully expanded leaves, or stems (including the culm and sheath) were collected from plants of different developmental stages at 14:00–15:00. Starch accumulation in flag leaves at the filling stage was determined by iodine staining for 30 min in a buffer containing 5.7 mM iodine and 43.4 mM potassium iodide in 0.2 N HCl (Caspar et al., 1985). Sucrose and fructose contents were detected as described previously with slight modification (Tang, 1999). Plant samples were collected quickly and placed into an oven at 105°C for 30 min, then dried at 80°C for 48 h. Dry samples were ground and passed through a 1-mm mesh sieve. The 0.05-g leaf samples were digested with 4 ml of 80% ethyl alcohol in a water bath at 80°C for 30 min. The extract was centrifuged, and the supernatant was collected. This digestion was repeated three times, and the combined extracts were used for the soluble sugar assay. The residue after centrifugation was used for starch determination.
Starch accumulation in stems at the filling stage was determined by the iodine–potassium iodide (IKI) staining method (Demarco, 2017). The stems were cut crosswise with a single blade and fixed in a 70% formalin–acetic acid–alcohol (5% formalin, 5% glacial acetic acid, and 90% ethanol) fixative solution (FAA) (Servicebio, Wuhan, China) for 24 h at room temperature. They were then dehydrated with an ethanol series and encased in a wax block. The wax blocks were sliced to a thickness of 4 μm on an RM2016 ultramicrotome (Laika, Shanghai, China) with a glass knife, then stained with an IKI dye solution for 5–10 min until starch grains were a dark blue color. Stained sections were observed under a light microscope (Eclipse Ci; Nikon, Tokyo, Japan) and analyzed using CaseViewer software (https://www.3dhistech.com/caseviewer).
To determine sucrose content, the reaction mixture (0.4 ml of filter liquor, 200 μl of 2 N NaOH) was heated in a water bath at 100°C for 5 min and cooled quickly to room temperature. The mixture was combined with 2.8 ml of 30% HCl and 0.8 ml of 0.1% resorcinol and then incubated in a water bath at 80°C for 10 min. The absorbance was measured at 480 nm after cooling to room temperature. To determine fructose content, the reaction mixture (0.4 ml of filter liquor, 0.8 ml of 0.1% resorcinol, and 0.4 ml of H2O) was incubated in a water bath at 80°C for 10 min, and the absorbance was measured at 480 nm after cooling to room temperature. Sucrose and fructose contents were determined from a standard curve prepared using the corresponding standards.
The starch assay was performed according to Li et al. (2017) with minor modifications. In brief, the residue was dried at 80°C, 2 ml of distilled water was added, and the mixture was boiled for 60 min. Two milliliters of 9.36 M HClO4 was added to the tube and shaken for 15 min. The supernatant was collected after centrifugation for 20 min at 3000 rpm. The extraction was repeated three times by adding another 2 ml of 4.68 M HClO4 to the tube. The supernatants were pooled, and distilled water was added to 10 ml. An aliquot of the extract was used for the glucose assay with anthrone reagent, and absorbance was measured at 620 nm. Glucose content was multiplied by 0.9 to calculate the starch value (Pucher et al., 1932).
Free amino acid contents of flag leaves at the filling stage were assayed according to a previously described method (Xu et al., 2009) with a high-speed automatic amino acid analyzer (Hitachi 835-50; Tokyo, Japan). Fully expanded leaves (0.2 g) were harvested between 17:00 and 18:00, then immediately frozen in liquid nitrogen. The leaf samples were homogenized in 3 ml of 4% (w/v) sulfosalicylic acid and then kept at room temperature for 2 h. The homogenate was centrifuged at 12 000 g for 20 min. The supernatant was used for determination of free amino acids with the amino acid analyzer.
TEM assay
TEM was performed according to our previous report (Shen et al., 2019). Fresh leaves were cut into 2-mm2 pieces and fixed in 2.5% (v/v) glutaraldehyde and 0.2 M phosphoric acid buffer (PBS, pH 7.4) for 4 h at 4°C. The samples were then washed in wash buffer (0.1 M phosphate buffer, pH 7.4), and post-fixed for 7 h in 1% OsO4. After dehydration and embedding in Spurr’s resin, the ultrathin sections (70–80 nm) were cut using a Diatome diamond knife on an RMC Power Tome XL Ultramicrotome. Thin sections were collected onto copper grids, stained with 5% uranium acetate for 20 min, and counterstained with lead citrate for 10 min. The sections were examined and photographed on a Hitachi HT7700 transmission electron microscope (Hitachi, Tokyo, Japan) at 80 kV.
RNA-seq assay
At the filling stage, stems of WT, GMA, and GCGT plants were collected for RNA-seq using the analysis parameters described previously (Wang et al., 2020). Total RNA was extracted using the TRIzol reagent, and the quality of RNA was assessed using agarose gel electrophoresis and a NanoDrop 8000 spectrophotometer (NanoDrop, Wilmington, DE). Three micrograms of total RNA was used to construct sequencing libraries. Sequencing was performed on the Illumina Nova-Seq platform by Shanghai Hanyu Bio-Tech (China). Clean reads were obtained from the raw data by removing low-quality sequences, including low-quality reads, duplicated sequences, and poly-Ns. The clean reads were mapped against the O. sativa ssp. japonica cv. Nipponbare reference genome (http://rice.plantbiology.msu.edu). Differentially expressed genes (DEGs) in different comparisons were identified using the R package DEGSEQ V.1.20.0 with P value <0.05 and |fold change| > 2. KEGG enrichment analysis of the annotated DEGs was performed using Phyper (https://en.wikipedia.org/wiki/Hypergeometric_distribution) based on a hypergeometric test.
Funding
This work was supported by the National Natural Science Foundation of China (3110019, 32271757), the Natural Science Foundation of Henan Province (21010338), and the Gansu Provincial Science and Technology Major Projects (22ZD6NA007).
Author contributions
The experiments were conceived and designed by H.-W.X. and T.H. The experiments were performed by H.-W.X., H.-H.W., Y.-W.Z., X.-Y.Y., S.-F.L., C.-R.M., and B.Y. The data were analyzed by D.-Y.H., M.W., and T.H. The paper was written by H.-W.X. and T.H. All the authors have read the manuscript and approved it for submission.
Acknowledgments
We are very grateful to Prof. Xinxiang Peng (College of Life Sciences, South China Agricultural University) for providing GCGT rice seeds and for invaluable suggestions and comments on the manuscript. We thank Prof. Yaoguang Liu and Prof. Qinlong Zhu (College of Life Sciences, South China Agricultural University) for providing the multi-gene assembly and transformation vector system TGSII. No conflict of interest is declared.
Published: June 22, 2023
Footnotes
Published by the Plant Communications Shanghai Editorial Office in association with Cell Press, an imprint of Elsevier Inc., on behalf of CSPB and CEMPS, CAS.
Supplemental information is available at Plant Communications Online.
Contributor Information
Huawei Xu, Email: xhwcyn@163.com.
Tao Hu, Email: hut@lzu.edu.cn.
Supplemental information
Data availability
The RNA-seq data generated in this study can be found in ScienceDB (https://www.scidb.cn/en) as https://www.scidb.cn/anonymous/cllON3Zh.
References
- Barta C., Loreto F. The relationship between the methyl-erythritol phosphate pathway leading to emission of volatile isoprenoids and abscisic acid content in leaves. Plant Physiol. 2006;141:1676–1683. doi: 10.1104/pp.106.083063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caspar T., Huber S.C., Somerville C. Alterations in growth, photosynthesis, and respiration in a starchless mutant of Arabidopsis thaliana (L.) deficient in chloroplast phosphoglucomutase activity. Plant Physiol. 1985;79:11–17. doi: 10.1104/pp.79.1.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavanagh A.P., South P.F., Bernacchi C.J., Ort D.R. Alternative pathway to photorespiration protects growth and productivity at elevated temperatures in a model crop. Plant Biotechnol. J. 2022;20:711–721. doi: 10.1111/pbi.13750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J.H., Chen S.T., He N.Y., Wang Q.L., Zhao Y., Gao W., Guo F.Q. Nuclear-encoded synthesis of the D1 subunit of photosystem II increases photosynthetic efficiency and crop yield. Nat. Plants. 2020;6:570–580. doi: 10.1038/s41477-020-0629-z. [DOI] [PubMed] [Google Scholar]
- Dalal J., Lopez H., Vasani N.B., Hu Z., Swift J.E., Yalamanchili R., Dvora M., Lin X., Xie D., Qu R., Sederoff H.W. A photorespiratory bypass increases plant growth and seed yield in biofuel crop Camelina sativa. Biotechnol. Biofuels. 2015;8:175. doi: 10.1186/s13068-015-0357-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demarco D. 2017. Histochemical Analysis of Plant Secretory Structures; pp. 313–330. Histochemistry of single molecules: methods and protocols. [DOI] [PubMed] [Google Scholar]
- Ely K.S., Rogers A., Agarwal D.A., Ainsworth E.A., Albert L.P., Ali A., Anderson J., Aspinwall M.J., Bellasio C., Bernacchi C., et al. A reporting format for leaf-level gas exchange data and metadata. Ecol. Inform. 2021;61:101232. doi: 10.1016/j.ecoinf.2021.101232. [DOI] [Google Scholar]
- Evans J.R. Improving photosynthesis. Plant Physiol. 2013;162:1780–1793. doi: 10.1104/pp.113.219006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fagard M., Vaucheret H. (Trans) gene silencing in plants: How many mechanism? Annu. Rev. Plant Physiol. Plant Mol. Biol. 2000;51:167–194. doi: 10.1146/annurev.arplant.51.1.167. [DOI] [PubMed] [Google Scholar]
- Fahnenstich H., Scarpeci T.E., Valle E.M., Flügge U.I., Maurino V.G. Generation of hydrogen peroxide in chloroplasts of Arabidopsis overexpressing glycolate oxidase as an inducible system to study oxidative stress. Plant Physiol. 2008;148:719–729. doi: 10.1104/pp.108.126789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu J., Liu J., Cao Z.Q., Wang Z.Q., Zhang H., Yang J.C. Effects of alternate wetting and drying irrigation during grain filling on the seed-setting rate and grain weight of two super rice cultivars. Acta Agron. Sin. 2014;40:1056–1065. [Google Scholar]
- Jang I.C., Lee K.H., Nahm B.H., Kim J.K. Chloroplast targeting signal of a rice rbcS gene enhances transgene expression. Mol. Breeding. 2002;9:81–91. [Google Scholar]
- Kebeish R., Niessen M., Thiruveedhi K., Bari R., Hirsch H.J., Rosenkranz R., Stäbler N., Schönfeld B., Kreuzaler F., Peterhänsel C. Chloroplastic photorespiratory bypass increases photosynthesis and biomass production in Arabidopsis thaliana. Nat. Biotechnol. 2007;25:593–599. doi: 10.1038/nbt1299. [DOI] [PubMed] [Google Scholar]
- Li G., Pan J., Cui K., Yuan M., Hu Q., Wang W., Mohapatra P.K., Nie L., Huang J., Peng S. Limitation of unloading in the developing grains is a possible cause responsible for low stem non-structural carbohydrate translocation and poor grain yield formation in rice through verification of recombinant inbred lines. Front. Plant Sci. 2017;8:1369. doi: 10.3389/fpls.2017.01369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S., Li W., Huang B., Cao X., Zhou X., Ye S., Li C., Gao F., Zou T., Xie K., et al. Natural variation in PTB1 regulates rice seed setting rate by controlling pollen tube growth. Nat. Commun. 2013;4:2793. doi: 10.1038/ncomms3793. [DOI] [PubMed] [Google Scholar]
- Li Z., Su D., Lei B., Wang F., Geng W., Pan G., Cheng F. Transcriptional profile of genes involved in ascorbate glutathione cycle in senescing leaves for an early senescence leaf (esl) rice mutant. J. Plant Physiol. 2015;176:1–15. doi: 10.1016/j.jplph.2014.09.020. [DOI] [PubMed] [Google Scholar]
- Liu C., Xue Z., Tang D., Shen Y., Shi W., Ren L., Du G., Li Y., Cheng Z. Ornithine d-aminotransferase is critical for floret development and seed setting through mediating nitrogen reutilization in rice. Plant J. 2018;96:842–854. doi: 10.1111/tpj.14072. [DOI] [PubMed] [Google Scholar]
- Long S.P., Marshallcolon A., Zhu X.G. Meeting the global food demand of the future by engineering crop photosynthesis and yield potential. Cell. 2015;161:56–66. doi: 10.1016/j.cell.2015.03.019. [DOI] [PubMed] [Google Scholar]
- López-Calcagno P.E., Fisk S., Brown K.L., Bull S.E., South P.F., Raines C.A. Overexpressing the H-protein of the glycine cleavage system increases biomass yield in glasshouse and field-grown transgenic tobacco plants. Plant Biotechnol. J. 2019;17:141–151. doi: 10.1111/pbi.12953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma J., Takahashi E. Effect of silicon on the growth and phosphorus uptake of rice. Plant Soil. 1990;126:115–119. [Google Scholar]
- Maier A., Fahnenstich H., von Caemmerer S., Engqvist M.K.M., Weber A.P.M., Flügge U.I., Maurino V.G. Transgenic introduction of a glycolate oxidative cycle into A. thaliana chloroplasts leads to growth improvement. Front. Plant Sci. 2012;3:38. doi: 10.3389/fpls.2012.00038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muraoka H., Tang Y.H., Terashima I., Koizumi H., Washitani I. Contributions of diffusional limitation, photoinhibition and photorespiration to midday depression of photosynthesis in Arisaema heterophyllum in natural high light. Plant Cell Environ. 2000;23:235–250. [Google Scholar]
- Nolke G., Houdelet M., Kreuzaler F., Peterhansel C., Schillberg S. The expression of a recombinant glycolate dehydrogenase polyprotein in potato (Solanum tuberosum) plastids strongly enhances photosynthesis and tuber yield. Plant Biotechnol. J. 2014;12:734–742. doi: 10.1111/pbi.12178. [DOI] [PubMed] [Google Scholar]
- Nomura M., Katayama K., Nishimura A., Ishida Y., Ohta S., Komari T., Miyao-Tokutomi M., Tajima S., Matsuoka M. The promoter of rbcS in a C3 plant (rice) directs organ-specific, light-dependent expression in a C4 plant (maize), but does not confer bundle sheath cell-specific expression. Plant Mol. Biol. 2000;44:99–106. doi: 10.1023/a:1006461812053. [DOI] [PubMed] [Google Scholar]
- Novitskaya L., Trevanion S.J., Driscoll S., Foyer C.H., Noctor G. How does photorespiration modulate leaf amino acid contents? A dual approach through modelling and metabolite analysis. Plant Cell Environ. 2002;25:821–835. [Google Scholar]
- Ort D.R., Merchant S.S., Alric J., Barkan A., Blankenship R.E., Bock R., Croce R., Hanson M.R., Hibberd J.M., Long S.P., et al. Redesigning photosynthesis to sustainably meet global food and bioenergy demand. Proc. Natl. Acad. Sci. USA. 2015;112:8529–8536. doi: 10.1073/pnas.1424031112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterhansel C., Horst I., Niessen M., Blume C., kebeish R., Kürkcüoglu S., Kreuzaler F. Photorespiration. Arabidopsis Book. 2010;8:e0130. doi: 10.1199/tab.0130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pucher G.W., Leavenworth C.S., Vickery H.B. Determination of starch in plant tissues. Anal. Chem. 1948;20:850–853. [Google Scholar]
- Roell M.S., Schada von Borzykowski L., Westhoff P., Plett A., Paczia N., Claus P., Urte S., Erb T.J., Weber A.P.M. A synthetic C4 shuttle via the β-hydroxyaspartate cycle in C3 plants. Proc. Natl. Acad. Sci. USA. 2021;118 doi: 10.1073/pnas.2022307118. e2022307118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen B.R., Wang L.M., Lin X.L., Yao Z., Xu H.W., Zhu C.H., Teng H.Y., Cui L.L., Liu E.E., Zhang J.J., et al. Engineering a new chloroplastic photorespiratory bypass to increase photosynthetic efficiency and productivity in rice. Mol. Plant. 2019;12:199–214. doi: 10.1016/j.molp.2018.11.013. [DOI] [PubMed] [Google Scholar]
- Shen B.R., Zhu C.H., Yao Z., Cui L.L., Zhang J.J., Yang C.W., He Z.H., Peng X.X. An optimized transit peptide for effective targeting of diverse foreign proteins into chloroplasts in rice. Sci. Rep. 2017;7:46231. doi: 10.1038/srep46231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh P., Sinha A.K. A positive feedback loop governed by SUB1A1 interaction with MITOGEN-ACTIVATED PROTEIN KINASE3 imparts submergence tolerance in rice. Plant Cell. 2016;28:1127–1143. doi: 10.1105/tpc.15.01001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- South P.F., Cavanagh A.P., Liu H.W., Ort D.R. Synthetic glycolate metabolism pathways stimulate crop growth and productivity in the field. Science. 2019;363:eaat9077. doi: 10.1126/science.aat9077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang Z.C. Science Press; Beijing, China: 1999. Guide of Modern Plant Physiology Experiment. [Google Scholar]
- Teixeira F.K., Menezes-Benavente L., Galvão V.C., Margis R., Margis-Pinheiro M. Rice ascorbate peroxidase gene family encodes functionally diverse isoforms localized in different subcellular compartments. Planta. 2006;224:300–314. doi: 10.1007/s00425-005-0214-8. [DOI] [PubMed] [Google Scholar]
- Teixeira F.K., Menezes-Benavente L., Margis R., Margis-Pinheiro M. Analysis of the molecular evolutionary history of the ascorbate peroxidase gene family: inferences from the rice genome. J. Mol. Evol. 2004;59:761–770. doi: 10.1007/s00239-004-2666-z. [DOI] [PubMed] [Google Scholar]
- von Caemmerer S., Quick W.P., Furbank R.T. The development of C4 rice: current progress and future challenges. Science. 2012;336:1671–1672. doi: 10.1126/science.1220177. [DOI] [PubMed] [Google Scholar]
- Walker B.J., VanLoocke A., Bernacchi C.J., Ort D.R. The costs of photorespiration to food production now and in the future. Annu. Rev. Plant Biol. 2016;67:107–129. doi: 10.1146/annurev-arplant-043015-111709. [DOI] [PubMed] [Google Scholar]
- Wang L.M., Shen B.R., Li B.D., Zhang C.L., Lin M., Tong P.P., Cui L.L., Zhang Z.S., Peng X.X. A synthetic photorespiratory shortcut enhances photosynthesis to boost biomass and grain yield in rice. Mol. Plant. 2020;13:1802–1815. doi: 10.1016/j.molp.2020.10.007. [DOI] [PubMed] [Google Scholar]
- Wang L.Z., Wang L.M., Xiang H.T., Luo Y., Li R., Li Z.J., Wang C.Y., Meng Y. Relationship of photosynthetic efficiency and seed-setting rate in two contrasting rice cultivars under chilling stress. Photosynthetica. 2016;54:581–588. [Google Scholar]
- Wang T., Li Y., Song S., Qiu M., Zhang L., Li C., Dong H., Li L., Wang J., Li L. EMBRYO SAC DEVELOPMENT 1 affects seed setting rate in rice by controlling embryo sac development. Plant Physiol. 2021;186:1060–1073. doi: 10.1093/plphys/kiab106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang X., Zhang P., Yu P., Zhang Y., Yang Z., Sun L., Wu W., Khan R.M., Abbas A., Cheng S., Cao L. LSSR1 facilitates seed setting rate by promoting fertilization in rice. Rice. 2019;12:31. doi: 10.1186/s12284-019-0280-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu H., Zhang J., Zeng J., Jiang L., Liu E.E., Peng C., He Z., Peng X. Inducible antisense suppression of glycolate oxidase reveals its strong regulation over photosynthesis in rice. J. Exp. Bot. 2009;60:1799–1809. doi: 10.1093/jxb/erp056. [DOI] [PubMed] [Google Scholar]
- Xu H.W., Ji X.M., He Z.H., Shi W.P., Zhu G.H., Niu J.K., Li B.S., Peng X.X. Oxalate accumulation and regulation is independent of glycolate oxidase in rice leaves. J. Exp. Bot. 2006;57:1899–1908. doi: 10.1093/jxb/erj131. [DOI] [PubMed] [Google Scholar]
- Xu L., Carrie C., Law S.R., Murcha M.W., Whelan J. Acquisition, conservation, and loss of dual-targeted proteins in land plants. Plant Physiol. 2013;161:644–662. doi: 10.1104/pp.112.210997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu X.J., Li B., Liu S.Y., Zhang G.L. Effects of high temperature stress at heading stage on flowering habits and seed-setting rate in rice. Hybrid. Rice. 2014;29:57–62. [Google Scholar]
- Zhang Z.S., Li X.Y., Cui L.L., Meng S., Ye N.H., Peng X.X. Catalytic and functional aspects of different isozymes of glycolate oxidase in rice. BMC Plant Biol. 2017;17:135. doi: 10.1186/s12870-017-1084-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou S., Wang Y., Li W., Zhao Z., Ren Y., Wang Y., Gu S., Lin Q., Wang D., Jiang L., et al. Pollen semi-sterility1 encodes a kinesin-1-like protein important for male meiosis, anther dehiscence, and fertility in rice. Plant Cell. 2011;23:111–129. doi: 10.1105/tpc.109.073692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Q., Yu S., Zeng D., Liu H., Wang H., Yang Z., Xie X., Shen R., Tan J., Li H., et al. Development of "purple endosperm rice" by engineering anthocyanin biosynthesis in the endosperm with a high-efficiency transgene stacking system. Mol. Plant. 2017;10:918–929. doi: 10.1016/j.molp.2017.05.008. [DOI] [PubMed] [Google Scholar]
- Zhu X.G., Long S.P., Ort D.R. Improving photosynthetic efficiency for greater yield. Annu. Rev. Plant Biol. 2010;61:235–261. doi: 10.1146/annurev-arplant-042809-112206. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The RNA-seq data generated in this study can be found in ScienceDB (https://www.scidb.cn/en) as https://www.scidb.cn/anonymous/cllON3Zh.