Abstract
Introduction:
The global Mpox (MPX) disease outbreak caused by the Mpox virus (MPXV) in 2022 alarmed the World Health Organization (WHO) and health regulation agencies of individual countries leading to the declaration of MPX as a Public Health Emergency. Owing to the genetic similarities between smallpox-causing poxvirus and MPXV, vaccine JYNNEOS, and anti-smallpox drugs brincidofovir and tecovirimat were granted emergency use authorization by the United States Food and Drug Administration. The WHO also included cidofovir, NIOCH-14, and other vaccines as treatment options.
Areas covered:
This article covers the historical development of EUA-granted antivirals, resistance to these antivirals, and the projected impact of signature mutations on the potency of antivirals against currently circulating MPXV. Since a high prevalence of MPXV infections in individuals coinfected with HIV and MPXV, the treatment results among these individuals have been included.
Expert opinion:
All EUA-granted drugs have been approved for smallpox treatment. These antivirals show good potency against Mpox. However, conserved resistance mutation positions in MPXV and related poxviruses, and the signature mutations in the 2022 MPXV can potentially compromise the efficacy of the EUA-granted treatments. Therefore, MPXV-specific medications are required not only for the current but also for possible future outbreaks.
Keywords: Mpox, Cidofovir, Brincidofovir, Tecovirimat, OPG71, OPG57, F8L, C19L
1. Introduction
The 2022 Mpox (MPX) outbreak has spread to more than 110 countries with approximately 90,000 documented cases. Due to the volume of the outbreak, and lessons learned from the Coronavirus Disease 19 COVID-19 pandemic, the WHO quickly acted by declaring MPX a Public Health Emergency (PHE) of international concern on July 23, 2022. Subsequently, the United States Human Health Services Administration (US-HHS) declared MPX as a PHE on August 5, 2022. Similar actions were taken by health regulatory agencies of individual countries worldwide. Although the PHE status of MPX was lifted on Jan. 31, 2023, by the US-HHS, new cases of MPXV will likely be identified. Additionally, the emergence of a virus closely related to MPXV in future outbreaks cannot be ruled out.
MPX is a zoonotic disease. It is caused by infection with Mpox (formerly known as monkeypox) virus (MPXV). MPXV is transmitted via infected skin, body fluids, and respiratory droplets. Symptoms include Flu-like conditions and rashes. MPXV is a linear double-stranded DNA virus with a genome length of ~200 kb, which encodes ~200 proteins. It belongs to the order Chitovirales, the family Poxviridae, and the genus Orthopoxvirus. The other examples of the Orthopoxvirus genus are the cowpox virus (CPXV), vaccinia viruses (VACV), and the variola virus (VARV). VACV has been extensively studied since all smallpox vaccines have been derived from VACV.
MPXV was first discovered in 1958 when two outbreaks of pox-like non-fatal disease were identified in cynomolgus monkeys during the summer of 19581. These outbreaks occurred 51 and 62 days after the transport of monkeys from Singapore to Denmark, with 6% and 10% of the animals developing a pox-like disease, respectively. Additional MPX outbreaks were reported in 1968 from various countries (Panama, India, France, the USA, the Netherlands, Trinidad, Brazil, and Indonesia) in non-human primates2, 3. Concurrently, a pox-like disease was reported in humans, although the possibility of MPXV infections was ruled out at the time.4 The first documented human MPXV infection was reported in 1970 in a 9-month old boy from the Democratic Republic of Congo5.
Many sporadic outbreaks of MPXV have since been reported in different countries. Most of these were travel-related cases and restricted to travelers without any secondary transmission6–11. The first exported cases of MPXV infections were identified in the Midwestern USA in 2003, with 37 confirmed and 10 suspected infections8, 12. This outbreak was associated with exotic pocket pets (Prairie dogs) that were imported from Ghana. It is believed that the MPXV reservoir was the Gambian pouched rat, which transmitted the virus to the Prairie dogs. Therefore, while initially seemingly appropriate, the long-used name monkeypox is a misnomer as MPXV infects many animals13, 14, and a definite MPXV reservoir has yet to be conclusively identified7, 15.
2. MPXV genome replication cycle and therapeutic targets
In principle, all viral and host cell proteins that participate in viral replication can be antiviral therapeutic targets. However, as discussed in the following sections, only a few viral and/or host proteins routinely turn out to be viable therapeutic targets.
2.1. MPXV genome replication cycle
Details of the MPXV replication cycle steps have not been established. Therefore, other well-studied and closely related poxviruses, such as VACV, must be used as a surrogate for our understanding of the MPXV replication cycle. Two distinct forms of infectious poxvirus virions can infect a host cell: (i) a mature virion (MV), and (ii) an extracellular enveloped virion (EV) (Fig. 1). MV has a single membrane, whereas the EV has an additional outer membrane16. The additional EV outer membrane is disrupted prior to fusion, rendering EV similar to MV at the point of entry into the host cell16. A multitude (20–30) of VACV proteins constitute the MV membrane, while the EV has ~6 additional proteins within the outer membrane. Entry and fusion of the MVs and EVs involve multiple viral and cell-surface proteins17–19. In addition, the attachment of MV and EV differs significantly16. For example, proteinase treatment disrupts the binding of MV but not EV16, 20. Thus, poxvirus entry and fusion are multiplayer and complex processes, making it challenging to select feasible antiviral targets from many viral proteins.
Figure 1. Replication cycle of MPXV and steps targeted by CDV, BCV and tecovirimat.
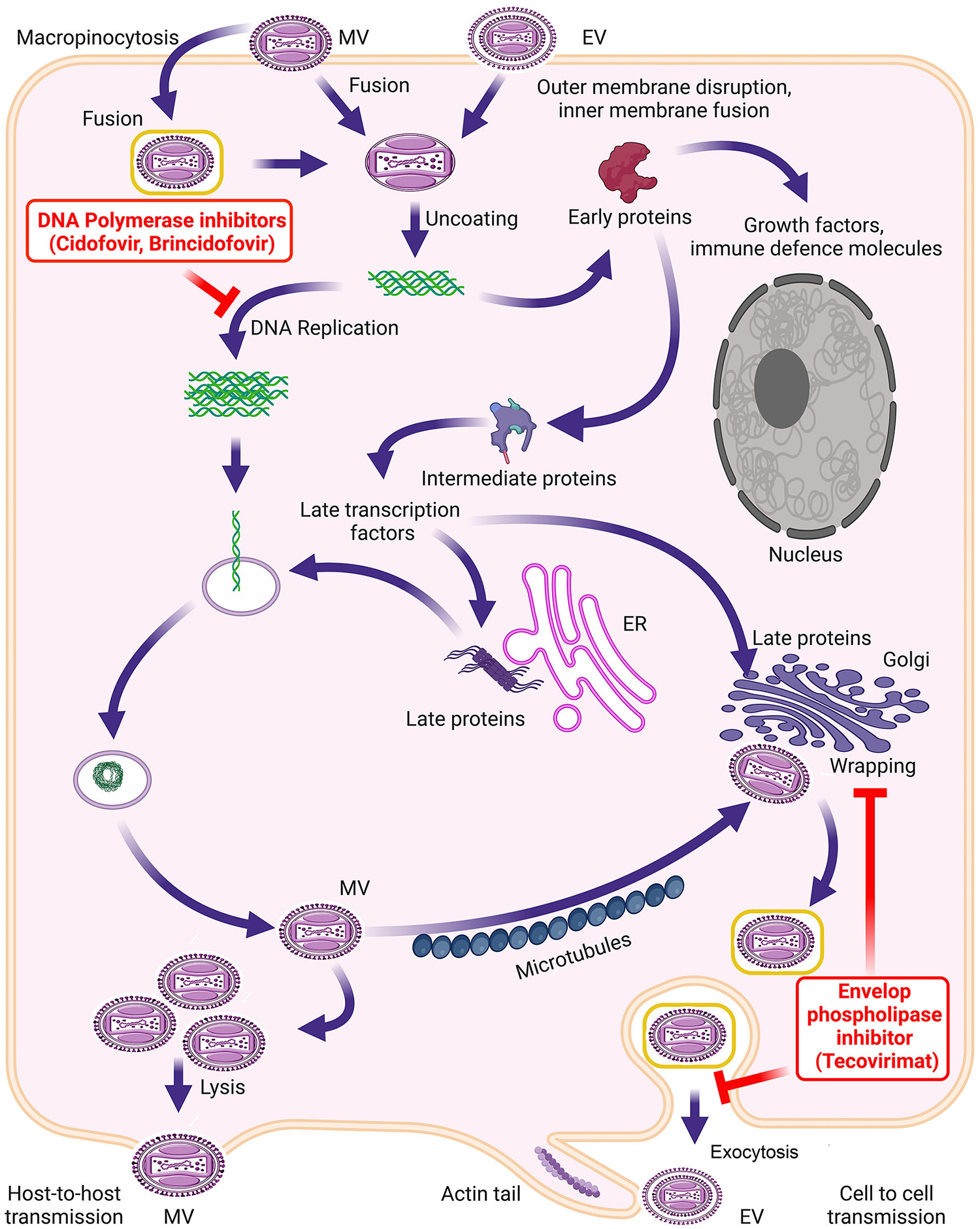
– The orthopoxviruses enter the cell either directly by binding to the cell surface receptors or through micropinocytosis. Following fusion, the core containing linear dsDNA genome and proteins required for early transcriptions are released into the cytosol. The replication of the dsDNA genome at this step serves as the template for intermediate and late gene transcription. CDV and BCV target the replication of the dsDNA genome. Late proteins and viral genomes assemble to form the MV. Some MVs acquire trans-Golgi and/or endosomal membranes to become triple-membrane-wrapped particles. The membrane biogenesis is mediated by the orthopoxvirus phospholipase protein (e.g., MPXV C19L), which is targeted by tecovirimat. The wrapped virions are transported to the cell surface, where the outer membrane fuses with the plasma membrane, and the virion is released as an EV.
A distinct structure of MV/EV particles is the core21, 22, which contains a linear dsDNA genome, virus-encoded enzymes, and factors required to transcribe early viral genes. Following entry and fusion, the uncoating of the viral core releases the genome into the cytosol, where the synthesis of early mRNA and genes begins, which is succeeded by DNA genome replication. The replicated DNA serves as the template for the synthesis of intermediate and late mRNAs. After the late genes’ translation, the assembly of new viral particles initiates, resulting in the formation of infectious MVs (Fig. 1). Some MVs are released from the cell by lysis, whereas a population of MVs acquires trans-Golgi and/or endosomal membranes to become triple membrane wrapped particles called wrapped virions (WVs)16. These virions are transported to the cell surface, where the outer membrane fuses with the plasma membrane, and the virion is released as an EV.
2.2. MPXV replication-associated proteins as therapeutic targets
The most sought-after antiviral targets for all viruses are the components of the nucleic acid replication machinery. To date, ~100 antivirals are available in the US. The majority of these drugs target Human Immunodeficiency Virus (HIV) (42), hepatitis C (18), hepatitis B (10), and herpesviruses (10). Fifteen out of 42 currently approved antiretrovirals target HIV-1 reverse transcriptase (HIV-1 RT), underlining that nucleic acid polymerase is the most important therapeutic target. In poxviruses, there are at least two nucleic acid replicating complexes: (i) a multi-subunit DNA-dependent RNA polymerase (vRNAP) complex and (ii) a multi-subunit DNA replication holoenzyme. The cryo-EM structure of the VACV vRNAP transcription complex provided the details of subunit arrangements and the mechanism of poxvirus transcription23. No antiviral drugs targeting vRNAP components have been approved for any poxvirus RNA transcription component.
VACV DNA genome replication is conducted by a holoenzyme consisting of multiple proteins22. An essential component of this holoenzyme is E9, the DNA-dependent DNA polymerase belonging to B family DNA polymerases. The MPXV genome encodes F8L (OPG71)24, also a B family DNA-dependent DNA polymerase25, which shares ~98% identity with VACV E9. The first B family DNA polymerase (RB69) structure showed an overall architecture of this class of enzymes26. This structure showed a canonical polymerase domain consisting of the Thumb, Palm, and Fingers subdomains, as seen in the structure of the Klenow Fragment (KF) of E. coli DNA polymerase I27. A notable difference between these polymerases is the relative position of the 3’ – 5’ exonuclease domain, which was ~180° opposite to that in KF relative to the polymerase active site. Subsequent crystal structures of the RB69 polymerase showed that residues of a β-hairpin positioned in the major groove of the template-primer played a role in the partitioning of primer to the 3’ – 5’ exonuclease site upon mismatch nucleotide incorporation26, 28–30. Indeed, a resistance mutation on topologically similar β-hairpin in poxviruses’ DNA polymerase showed the relevance of the resistance mechanism of nucleotide analogs mediated by 3’ – 5’ exonuclease function (discussed below).
Numerous structures of B family DNA polymerases have since been reported28, 31–39. The most notable viral DNA polymerase structures include the structure of unliganded herpes simplex virus 1 (HSV-1) DNA polymerase35, HSV1 DNA polymerase in complex with template-primer and 4-oxo-dihydroquinoline39, the crystal structure of VACV DNA polymerase E938, and a recently reported cryo-EM structure of the MPVX DNA polymerase holoenzyme40. Notably, the VACV E9 structure was the first to provide details of poxvirus-specific inserts38. Before the cryo-EM structure of MPXV DNA polymerase holoenzyme, a homology built MPXV F8L based on this structure, AlphaFold41, and ColabFold42, predicted structures of processivity factor A22R (OPG148), PCNA (Proliferation Cell Nuclear Antigen) ortholog G9R (OPG93), homology-derived MPXV uracil DNA glycosylase (OPG116) were used to assemble an MPXV replication complex43. This MPXV replication complex was guided by a low-resolution structure of the vaccinia virus DNA replication machinery44 and the structure of eukaryotic DNA polymerase δ bound to PCNA45. Mutations presented in the 2022 MPXV outbreak were mapped onto the assembled complex to predict the impact of these mutations on the replication of MPXV genome replication43.
At least 8 proteins participate in VACV DNA replication22 (Table 1) and are conserved between VACV and MPXV. The DNA polymerase, helicase/primase, UDG, processivity factor (A20R/A22R), and SSB are essential for viral replication16, 22. Host protein kinase VRK1 can complement the replication defect due to the B1R VACV kinase mutant46. Similarly, host DNA ligase I has been shown to substitute for VACV A50R47. The absence of the FEN1 family endonuclease results in the reduced mean size of the replicated DNA and packaged virion with little or no viral DNA48. VACV A20R and MPXV A22R are processivity factors, and SSB is an ssDNA coating protein; neither protein exhibits any enzymatic activity. In addition to these, H5R (a multifunctional phosphoprotein), A22R (A23R in MPXV, Holliday junction resolvase), and H6R (G7R in MPXV, a Topoisomerase) participate in the completion of poxvirus (VACV) genome replication21. Thus, DNA polymerase, helicase-primase, and topoisomerase remain attractive antiviral targets of MPXV infection.
Table 1.
Potential MPXV Therapeutic targets.
| Proteina | VACV | MPXV | Essential |
|---|---|---|---|
| Replication | |||
| DNA polymerase | E9L | F8L | Yes |
| Helicase–primase | D5R | E5R | Yes |
| UDG | D4R | E4R | Yes |
| Processivity factor | A20R | A22R | Yes |
| Protein kinase | B1R | B3R | Host-dependent |
| SSB | I3L | I3L | Yes |
| DNA ligase | A50R | A50R | Host-dependent |
| FEN1-like nuclease | G5R | G5R | Impaired |
SSB – single strand DNA binding protein
UDG – Uracil DNA glycosylase
2.2.1. DNA polymerase structure and the mechanism of CDV/BCV inhibition
As the poxvirus DNA polymerase (e.g., VACV E9 or MPXV F8L) is essential for viral replication21, 49, it is the most sought-after antiviral therapeutic target against MPX. The FDA-approved drug brincidofovir (BCV) has been granted EUA, while the WHO has included the parent compound cidofovir (CDV) (Fig. 2) for the treatment of MPX. CDV is an acyclic nucleoside phosphonate (ANP), which was developed by Antonin Holy as an antiviral compound50, 51. ANPs have been extremely successful antivirals as tenofovir disoproxil (TDF) and tenofovir alafenamide (TAF), the prodrugs of tenofovir are the integral part of antiretroviral therapy against HIV. BCV is a CDV prodrug converted into CDV-diphosphate (CDVpp) by cellular kinases, becoming a ready-to-use substrate for the viral DNA polymerase. CDV is a broad-spectrum inhibitor of dsDNA viruses’ DNA polymerases52. It inhibits viral DNA replication by multiple mechanisms: as a dCTP competitor, a nonobligate chain-terminator, resistant to 3’−5’ exonuclease (when present at the penultimate 3’OH position), an inhibitor of template-directed trans-lesion synthesis, and as a mutagen53–56 when present in the template strand. Since BCV is eventually metabolized to CDV-diphosphate (CDVpp), the inhibition mechanism of BCV is identical to CVD as far as the mutations in poxvirus DNA polymerases are concerned.
Figure 2.
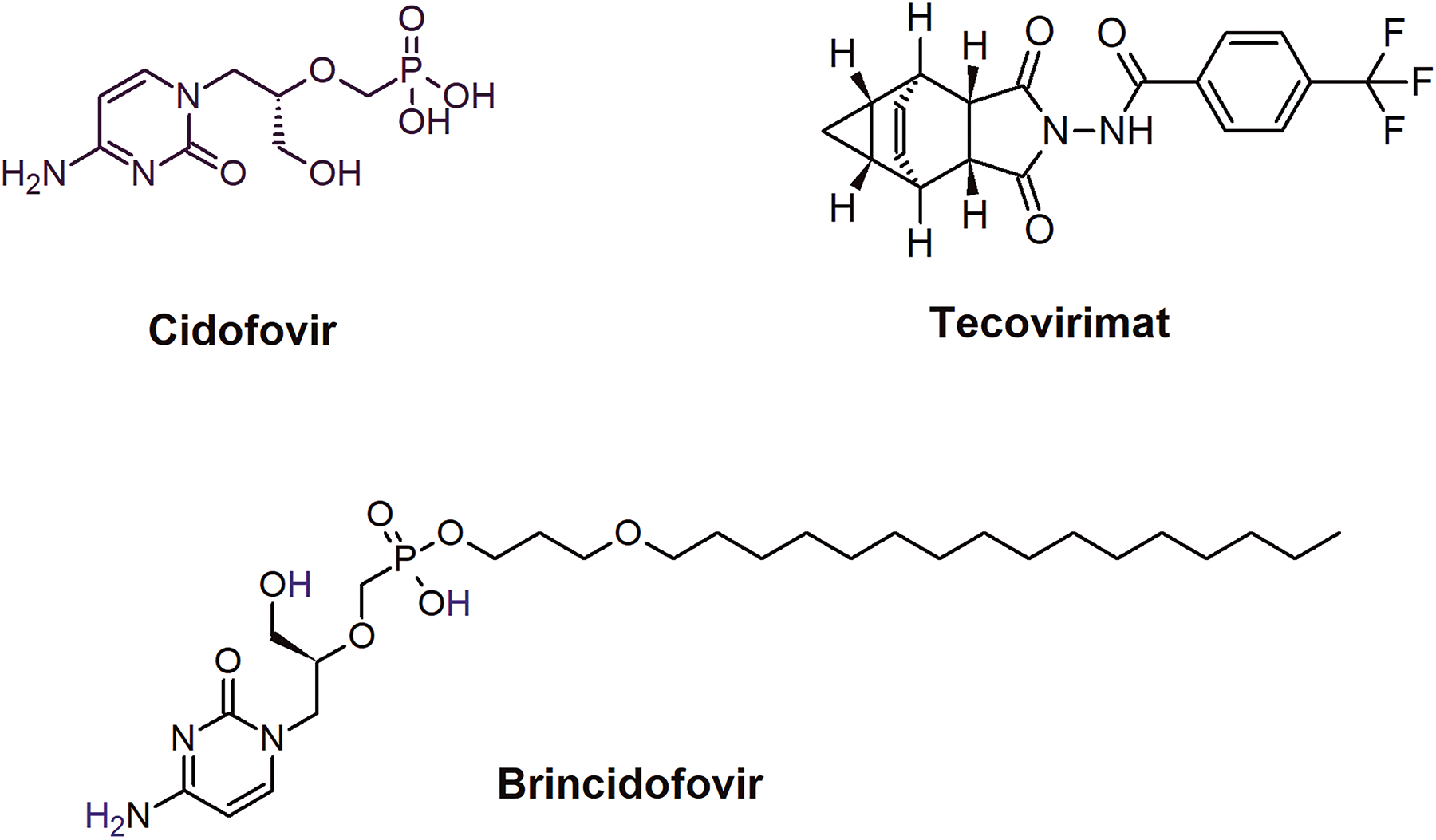
Chemical structures of CDV, BCV, and tecovirimat.
2.2.2. Resistance to CDV/BCV
CDV resistance mutations in multiple orthopoxviruses have been identified52, 57–62. These mutations are ΔK174 (deletion of K174), A314T, S338F, A613T61, A684V/T, T688A, T808M61, T831I, and S851Y (Fig. 3a). The wild-type residues are conserved in all poxvirus DNA polymerases studied to date. Therefore, the CDV resistance mechanism appears to be shared among orthopoxviruses61. The crystal structure of VACV E9 was the first solved structure that provided the topological positions of the CDV resistance mutations relative to the two active sites (polymerase and 3’−5’ exonuclease)38. However, a recently reported structure of MPXV DNA replication holoenzyme41 showed the proximity of CDV resistance mutations to the template-primer and dNTP substrate (Fig. 3a). While this cryo-EM structure is a breakthrough in poxvirus structural biology, it is not a complete DNA replication holoenzyme, as the helicase-primase, an integral part of the holoenzyme, is missing44.
Figure 3. The topological position of CDV/BCV resistance mutations in MPXV F8L.
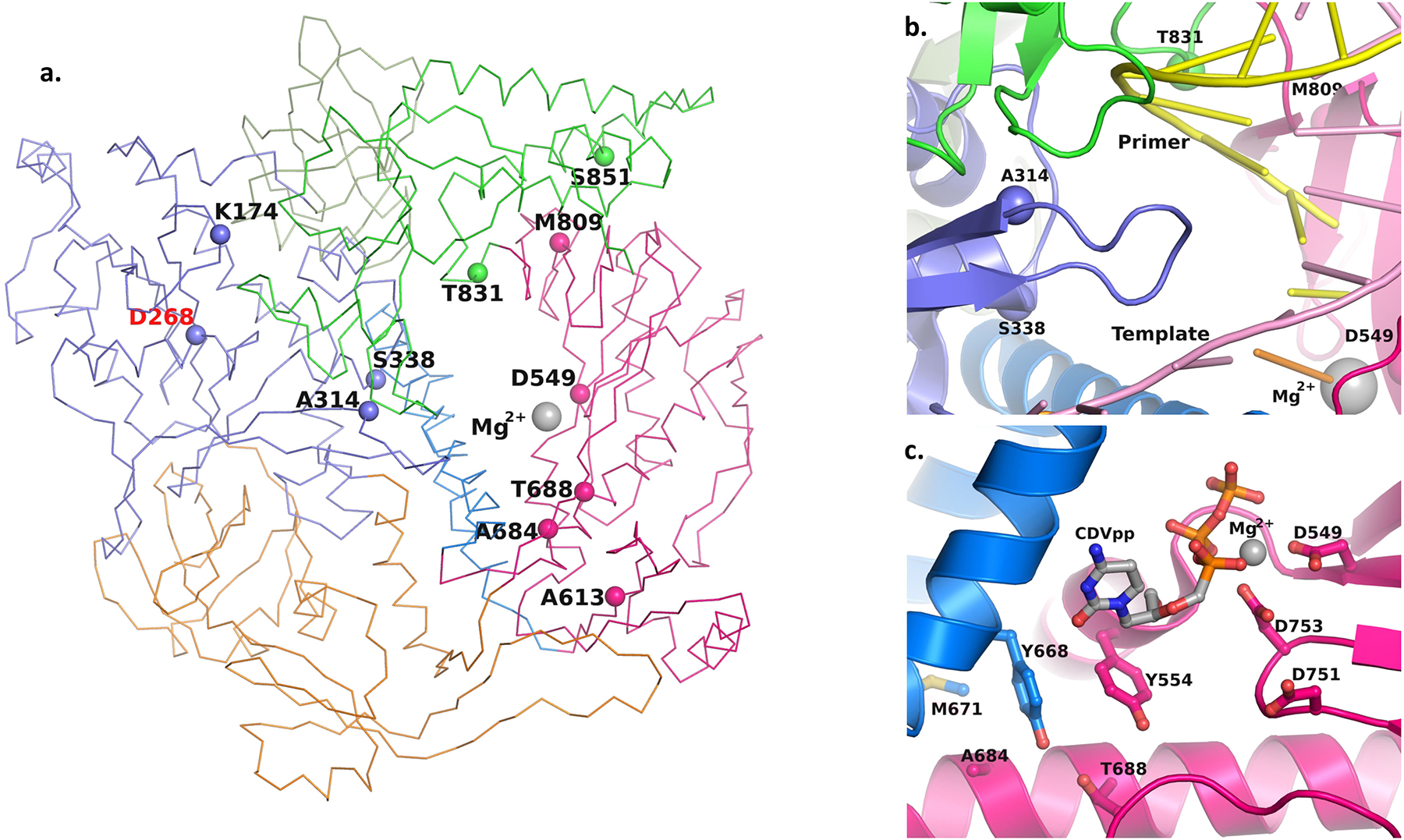
– Panel a shows Cα trace of F8L structure and topological position of the CDV/BCV resistance mutation positions in 3’ – 5’ exonuclease domain (purple), and subdomains fingers (blue), thumb (green) and palm (dark pink). The N-terminal domain of F8L is colored orange. The Cα atom of the resistance mutation residues is shown as a solid sphere. This structure was generated by homology modeling using a recently reported cryo-EM structure of MPXV holoenzyme40 (PDB entry 8HG1) as a template structure with Modeller117. Mg2+ and D549 represent the polymerase active site of F8L, whereas D268 represents 3’ – 5’ exonuclease active site. Panel b shows the resistance mutation position 314 on a β-hairpin located in the major groove of the template (light pink) and primer (yellow). Panel c shows the resistance mutation positions 684 and 688 and critical amino acids near these residues. The CDVpp was modeled using the dNTP substrate in PDB entry 8HG1. It is clear from this figure that the dNTP binding residues are in the proximity of resistance mutation positions 684 and 688. The α-helices and β-sheets are rendered as ribbons in panels b and c.
Three CDV resistance mutation positions (174, 314, and 338) are in the 3’−5’ exonuclease domain, whereas six (613, 684, 688, 808, 831, and 851), are in the polymerase domain (Fig. 3a). A CDV resistance mutation A314V/T has been identified in multiple poxviruses52. Mutation A314V in VACV DNA polymerase enhances the excision of CDV from the primer terminus52. A314 is part of a β-hairpin located within the major groove at the active site (Fig. 3b). Mutation A314V/T may change the interactions of this hairpin structure with the template-primer, resulting in the enhanced partitioning of the primer to the exonuclease site.
Two resistance mutation positions (684 and 688) are proximal to the CDVpp binding site (Fig. 3c). While these residues do not interact directly with the CDVpp (or dCTP), they do interact with critical dNTP binding pocket residues (Y554 and Y668) (Fig. 3c). Y554 is at the 5th position downstream of the first Motif A catalytic residue D549. A bulky residue is essential at this position for the exclusion of ribonucleotides (NTPs) to bind at the DNA polymerase site63–66. Y668 is at a topologically equivalent position to F762 of E. coli DNA polymerase I. Mutation F762Y enables E. coli DNA polymerase I to accept dideoxy nucleotide triphosphate substrates67. Therefore, mutations at residues 684 and 688 can alter the dNTP pocket by indirectly changing the interactions of Y554 and Y668 such that the polymerase discriminates against CDVpp. Another CDV resistance mutation at position 831 is close to the primer strand. All other CDV resistance mutation positions are not within the interacting distance to either template-primer or the dNTP substrate. However, these resistance mutations can indirectly interfere with the binding of CDVpp or the primer 3’OH position.
2.2.3. Mutations in F8L 2022 outbreak
A temporal analysis showed that two mutations in F8L (L108F and W411L) emerged during the 2022 outbreak43. Residue position 411 belongs to poxvirus-specific ‘insert 2’. Many phosophonoacetic acid (PAA) resistance mutations are in this insert68. Insert 2 is close to the Fingers subdomain which is involved in substrate binding. Therefore, W411 may affect the binding affinity of dNTP substrate resulting in the change in replication kinetics of the polymerase. Alternately, as seen in the reported MPXV replication holoenzyme40, L411 is exposed to the surface (Fig. 4a). For a hydrophobic residue to be exposed at the surface is highly unusual. It is possible that ‘insert 2’ also interacts with another factor, and this residue is buried at the interface of two proteins43. F8L amino acid position 108 is close to the ssDNA overhang of the template strand. As previously proposed, mutation L108F should increase the binding affinity of template-primer with F8L, which can enhance the processivity and/or polymerase strand-displacement DNA synthesis69. The phenylalanine residues at a topologically equivalent position in HIV-1 reverse transcriptase70, 71 and E. coli DNA polymerase I have been shown to interact with the template base in the ssDNA region and to contribute to strand-displacement DNA synthesis69, 72, 73. Therefore, it is possible that mutation L108F emerged to enhance the binding of template-primer with the polymerase and facilitate strand-displacement DNA synthesis.
Figure 4. The topological position of signature mutations emerged in the 2022 outbreak in MPXV F8L.
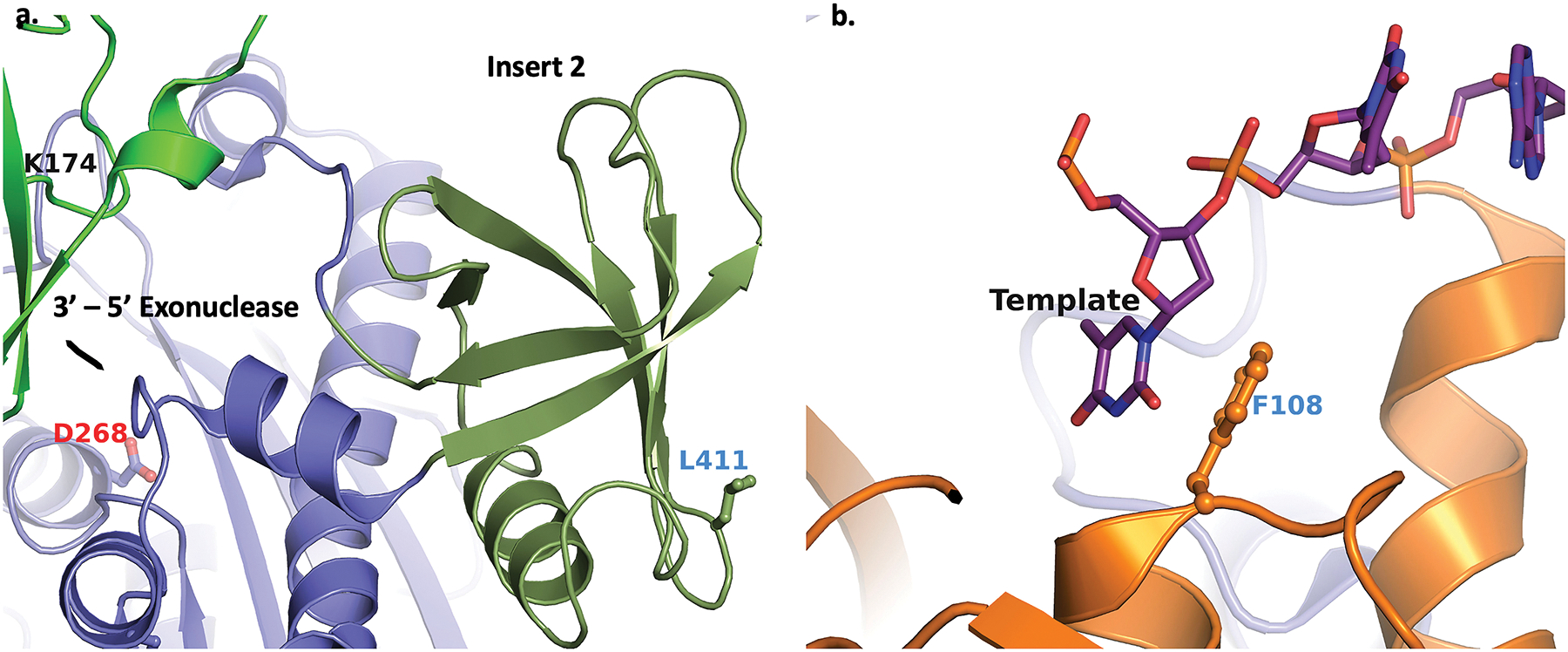
– Panel a depicts the signature mutation W411L in the poxvirus-specific ‘insert 2’ (rendered as forestgreen ribbons). The relative position of the 3’- 5’ exonuclease site (D268) is also a reference. Panel b shows that the 2022 MPXV signature mutation L108F (ball-and-stick) within the N-terminal domain (represented as orange ribbons) stacks against the template base as seen in the cryo-EM structure of MPXV DNA replication holoenzyme (PDB entry 8HG1).
3. ST-246 or tecovirimat
Tecovirimat (initially known as ST-246) (Fig. 2) was first reported in 2005 as an inhibitor of extracellular virus formation, and it protected mice from multiple orthopoxviruses including VACV, MPXV, camelpox, cowpox, mousepox, and VARV74. The G277C resistance mutation that emerged in the cowpox V061 gene suggested that ST-246 targeted the V061 gene74, an ortholog of F13L (VACV) and C19L (MPXV). F13L and C19L have phospholipase activity75, are palmitylated76, and participate in enveloping the intracellular MV to generate extracellular EV particle77, 78.
Tecovirimat is the most characterized among three poxvirus antivirals (CDV, BCV and ST-246). Its efficacy against MPXV infection has been tested in cell culture79–81, and animal models82–85 including non-human primates86–89. ST-246 showed strong inhibitory activity with good pharmacokinetics in all models (reviewed by Duraffour et al.90, and by Smee91). In vitro potency of tecovirimat (IC50 = 12.7 nM) was recently reported in cell-based assays using MPXV/France/IRBA2211i/2022 isolate92. Tecovirimat showed a synergistic effect when combined with BCV82. Co-administration of tecovirimat and vaccine ACAM2000 suggested that tecovirimat can be safely used after vaccination93.
Tecovirimat resistance mutations have been identified in F13L and its orthologs in different orthopoxviruses74, 94. Seven resistance mutations (F25V, H194N, G277C, D283Y, A290V, L315M , and I372N) and an insertion of an SVK triplet at positions 303–305 have been reported94. Most of these resistance mutations emerge in combination with other mutations94. Two resistance mutations, A290V, and L315M, were identified by next-generation sequencing of a clinical isolate obtained from an immunosuppressed patient with progressive vaccinia who received ST-246 and vaccinia immune globulin intravenous and BCV94, 95.
The structure of F13L or its orthologs has not been solved. However, a homology-derived model of MPXV C19L using the crystal structure of Phospholipase D from Streptomyces sp. as a template (PDB entry 1V0W)96 shows that there is only one pocket in Streptomyces sp. Phospholipase D or in the C19L homology model, where tecovirimat can be docked with a minimal conformational change in the protein. The best docked pose obtained based on the ‘Glide’ score (Schrödinger LLC, NY) is shown in Fig. 5. Most of the resistance mutations are around the docked tecovirimat. Three mutations emerged in the 2022 outbreak: V5A, S250N, and E353K (our unpublished results). The selection pressure of these mutations remains unknown. One of these mutations, S250N is in the vicinity of docked tecovirimat (Fig. 5). It is possible that S250N may impart some resistance to tecovirimat. A prodrug of tecovirimat, NIOCH-14 has been reported to have comparable efficacy in animal model97–99.
Figure 5. Docked tecovirimat in the homology-derived molecular model of C19L.
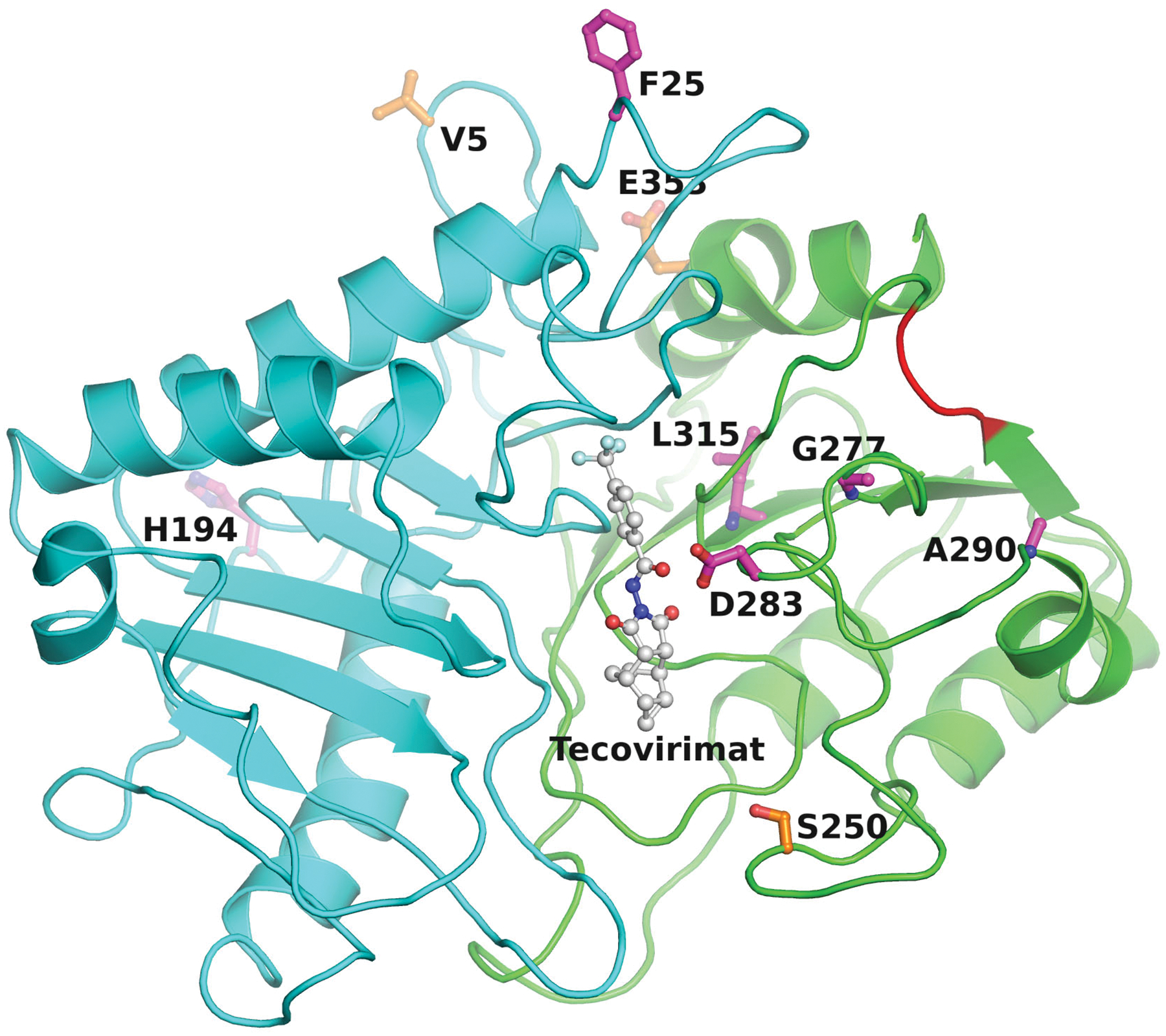
– This figure shows the most favorable docked pose of tecovirimat in the modeled structure of MPXV C19L phospholipase rendered as ribbons in cyan (N-terminal domain) and green (C-terminal domain). This figure also shows tecovirimat resistance mutation positions (in magenta balls-and-sticks) and signature mutations that emerged in the 2022 outbreak (in orange balls-and-sticks). Most of the tecovirimat resistance mutations are close to tecovirimat. The red loop represents the SVK triplet insertion at positions 303–305.
4. Efficacy of CDV, BCV, and tecovirimat against currently circulating MPXV
The FDA has not approved CDV for treating MPXV infections. Instead, it has been approved for cytomegalovirus retinitis in HIV-infected patients. However, CDV has shown antiviral activity against molluscum contagiosum in HIV-infected patient100. BCV has been approved for smallpox treatment as an oral drug. The efficacy studies of these drugs against currently circulating MPXV viruses are limited. In a recent report, the efficacy of CDV, BCV and tecovirimat was evaluated using 12 patient isolates in relevant cell models (human foreskin fibroblasts and human foreskin keratinocytes)101. The IC50 of tecovirimat, cidofovir, and brincidofovir was 4000 nmol, 80 μmol, and 600 nmol, respectively101, and were reportedly within the range of therapeutic concentrations in plasma101. In another study, the plaque formation assay was used to evaluate the potency of tecovirimat, and CDV using a patient isolate (MPXV/France/IRBA2211i/2022). The results showed that tecovirimat IC50 was 12.7 nM whereas CDV IC50 was 30.4 μM, suggesting that tecovirimat was ~2400-fold more potent than CDV. This difference in Mpox inhibition92 does not corroborate with the results of Bojkova et al.101, where CDV is only 80-fold less potent than tecovirimat.
5. Efficacy of CDV, BCV, and tecovirimat MPXV in the context of HIV co-infection
Clinical studies have shown a significant prevalence (20% - 74%) of MPXV infection among HIV-infected patients102–109. Additionally, most (95% – 98%) MPXV infections among these clinical studies involved men with sex men (MSM) and bisexuals, suggesting that MSM and bisexual individuals may be highly susceptible to MPXV infection. Therefore, an MPXV treatment strategy for this group of individuals must be in place. CDV has broad-spectrum antiviral activities110, 111. However, only a few studies have been reported where orthopoxvirus infections have been treated with CDV, BCV and tecovirimat as described below. Early studies demonstrated that lipid esters of CDV (including BCV) were effective as prophylaxis in mice infected with cowpox or vaccinia virus112. CDV treatment cleared recalcitrant molluscum contagiosum, a poxvirus, in an AIDS patient100. Recently, a patient coinfected with HIV, MPXV, and primary syphilis was successfully treated with CDV113. There are a few examples of successful MPXV treatment by tecovirimat among patients coinfected with MPXV and HIV114, 115. One report showed that one of the two patients (Patient A) coinfected with HIV and ocular MPXV infection suffered profound visual impairment despite treatment with tecovirimat115. In contrast, the other patient (Patient B) recovered from the ocular MPXV infection after treatment with tecovirimat115. It is unclear why CDV or BCV was not prescribed to Patient A. In a 28-year-old patient infected with HIV and MPXV, the treatment with tecovirimat for 14 days resulted in decreased skin lesions and decreased MPXV viral load without any adverse events114.
6. Summary
Here we presented our opinion on the feasibility of EUA-granted antivirals for the treatment of MPX. We discussed the impact of resistance mutations to these antivirals, the probable effect of signature mutations in the 2022 outbreak on the antiviral targets at the atomic level, the potency of these antivirals on currently circulating MPXV, and the latest results on the outcomes of EUA-granted MPXV treatments in the cases of MPXV and HIV co-infections. The limited number of clinical trials show that all three drugs: CDV, BCV, and tecovirimat appear to have good efficacy against currently circulating MPXV viruses. Future clinical results will provide a clearer picture of the antiviral activities of these compounds. It is almost certain that resistance mutations will emerge as the treatment becomes widely available or the population that has received treatment becomes significant. Since the resistance mutations of the three drugs have been evaluated in MPXV-related poxviruses, analogous mutations in MPXV can also be reasonably deduced. However, new resistance mutations may emerge as the current MPXV circulates among various conditions, including HIV infected/immunocompromised patients. An additional challenge is that only three currently available anti-MPXV drugs (as access to NIOCH-14 is limited). Therefore, more drugs targeting F8L and C19L are needed for the current outbreak or for possible future outbreaks with variants of MPXV. Thus, broad-spectrum inhibitors such as cytosine arabinoside (ara-C, also known as cytarabine) and 9-β-D-arabinofuranosyladenine (ara-A) that have shown antiviral activities should also be considered116. Due to the reported high prevalence of MPX among MSM and bisexual individuals, individuals from this community must be advised to adopt safe practices and treatment strategies to control the spread of MPXV118.
Article highlights.
The 2022 Mpox outbreak and the volume of infections showed that poxviruses remain a constant threat to global health. Signature mutations in currently circulating Mpox viruses (MPXV) may be contributing in unknown ways to the outbreak. Therefore, the role of these mutations in viral replication needs to be established.
There are no Mpox-specific treatments available. Limited studies indicate that the treatments that have been granted emergency use authorization (EUA) have good efficacy against MPXV. However, additional, and more robust studies are needed to establish the potency of these drugs against MPXV.
Nucleic acid polymerases are the most sought-after antiviral targets. Considering their essential role in virus replication, research to discover poxvirus DNA polymerase inhibitors is needed. Additionally, other components of viral DNA replication holoenzyme, such as DNA helicase, should be extensively characterized so that new antivirals can be developed against such targets.
The current MPXV was most prevalent among a specific group of individuals (men who have sex with men, and bisexuals). Many of these individuals are coinfected with HIV and MPXV. Therefore, drugs targeting MPXV and HIV coinfection need to be developed.
Acknowledgments
K. Singh acknowledges the computation facilities of the Molecular Interactions Core at the University of Missouri, Columbia, MO 65212. We thank the laboratories that have generously deposited sequences into the NCBI database. We also thank numerous laboratories that have enormously contributed to poxvirus research, but we could not cite their work.
Funding
K. Singh was partially funded by the Bond Life Sciences Center (Early Concept grant), a subcontract from Emory University (5R37AI076119), and the University of Missouri startup support.
Footnotes
Declaration of competing interest
CLL is co-founder and Chief Scientific Officer of Shift Pharmaceuticals, Overland Park, KS, USA. KS is a consultant for Sanctum Therapeutics Corporation, Sunnyvale, CA, USA.
Data Sharing
Genomic sequences used in this study were obtained from NCBI GenBank.
References
- 1.•.Magnus v P, Andersen EK, Petersen KB, Birch-Andersen A. A Pox-Like disease in cynomolgus monkeys. Acta Pathologica Microbiologica Scandinavica 1959;146:156–76. [Google Scholar]; Landmark paper that reported the first Mpox-like disease among animals.
- 2.Arita I, Henderson DA. Smallpox and monkeypox in non-human primates. Bull World Health Organ 1968;39(2):277–83. [PMC free article] [PubMed] [Google Scholar]
- 3.Parker S, Buller RM. A review of experimental and natural infections of animals with monkeypox virus between 1958 and 2012. Future Virol 2013. Feb 1;8(2):129–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.McNulty WP Jr., Lobitz WC Jr., Hu F, Maruffo CA, Hall AS. A pox disease in monkeys transmitted to man. Clinical and Histological Features. Arch Dermatol 1968. Mar;97(3):286–93. [PubMed] [Google Scholar]
- 5.•.Ladnyj ID, Ziegler P, Kima E. A human infection caused by monkeypox virus in Basankusu Territory, Democratic Republic of the Congo. Bull World Health Organ 1972;46(5):593–7. [PMC free article] [PubMed] [Google Scholar]; First report of Mpox in humans.
- 6.Adler H, Gould S, Hine P, Snell LB, Wong W, Houlihan CF, et al. Clinical features and management of human monkeypox: a retrospective observational study in the UK. Lancet Infect Dis 2022. Aug;22(8):1153–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Simpson K, Heymann D, Brown CS, Edmunds WJ, Elsgaard J, Fine P, et al. Human monkeypox - After 40 years, an unintended consequence of smallpox eradication. Vaccine 2020. Jul 14;38(33):5077–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Reynolds MG, Yorita KL, Kuehnert MJ, Davidson WB, Huhn GD, Holman RC, et al. Clinical manifestations of human monkeypox influenced by route of infection. J Infect Dis 2006. Sep 15;194(6):773–80. [DOI] [PubMed] [Google Scholar]
- 9.Ng OT, Lee V, Marimuthu K, Vasoo S, Chan G, Lin RTP, et al. A case of imported Monkeypox in Singapore. Lancet Infect Dis 2019. Nov;19(11):1166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mauldin MR, McCollum AM, Nakazawa YJ, Mandra A, Whitehouse ER, Davidson W, et al. Exportation of monkeypox virus from the African continent. J Infect Dis 2022. Apr 19;225(8):1367–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Costello V, Sowash M, Gaur A, Cardis M, Pasieka H, Wortmann G, et al. Imported monkeypox from international traveler, Maryland, USA, 2021. Emerg Infect Dis 2022. May;28(5):1002–05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.•.Centers for Disease C, Prevention. Multistate outbreak of monkeypox--Illinois, Indiana, and Wisconsin, 2003. MMWR Morb Mortal Wkly Rep 2003. Jun 13;52(23):537–40. [PubMed] [Google Scholar]; Report of the first outbreak outside Africa.
- 13.Khodakevich L, Jezek Z, Kinzanzka K. Isolation of monkeypox virus from wild squirrel infected in nature. Lancet 1986. Jan 11;1(8472):98–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Khodakevich L, Jezek Z, Messinger D. Monkeypox virus: ecology and public health significance. Bull World Health Organ 1988;66(6):747–52. [PMC free article] [PubMed] [Google Scholar]
- 15.Sukhdeo S, Mishra S, Walmsley S. Human monkeypox: a comparison of the characteristics of the new epidemic to the endemic disease. BMC Infect Dis 2022. Dec 12;22(1):928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Moss B. Poxvirus cell entry: how many proteins does it take? Viruses 2012. May;4(5):688–707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chung CS, Hsiao JC, Chang YS, Chang W. A27L protein mediates vaccinia virus interaction with cell surface heparan sulfate. J Virol 1998. Feb;72(2):1577–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hsiao JC, Chung CS, Chang W. Cell surface proteoglycans are necessary for A27L protein-mediated cell fusion: identification of the N-terminal region of A27L protein as the glycosaminoglycan-binding domain. J Virol 1998. Oct;72(10):8374–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Vazquez MI, Esteban M. Identification of functional domains in the 14-kilodalton envelope protein (A27L) of vaccinia virus. J Virol 1999. Nov;73(11):9098–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Vanderplasschen A, Smith GL. A novel virus binding assay using confocal microscopy: demonstration that the intracellular and extracellular vaccinia virions bind to different cellular receptors. J Virol 1997. May;71(5):4032–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Greseth MD, Traktman P. The life cycle of the vaccinia virus genome. Annu Rev Virol 2022. Sep 29;9(1):239–59. [DOI] [PubMed] [Google Scholar]
- 22.Moss B. Poxvirus DNA replication. Cold Spring Harb Perspect Biol 2013. Sep 1;5(9). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.••.Grimm C, Hillen HS, Bedenk K, Bartuli J, Neyer S, Zhang Q, et al. Structural basis of poxvirus transcription: Vaccinia RNA polymerase complexes. Cell 2019. Dec 12;179(7):1537–50 e19. [DOI] [PubMed] [Google Scholar]; A landmark report presenting the VACV RNA polymerase complex.
- 24.Shchelkunov SN, Totmenin AV, Safronov PF, Mikheev MV, Gutorov VV, Ryazankina OI, et al. Analysis of the monkeypox virus genome. Virology 2002. Jun 5;297(2):172–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Braithwaite DK, Ito J. Compilation, alignment, and phylogenetic relationships of DNA polymerases. Nucleic Acids Res 1993. Feb 25;21(4):787–802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.••.Wang J, Sattar AK, Wang CC, Karam JD, Konigsberg WH, Steitz TA. Crystal structure of a pol alpha family replication DNA polymerase from bacteriophage RB69. Cell 1997. Jun 27;89(7):1087–99. [DOI] [PubMed] [Google Scholar]; The first report describing the structure of a DNA polymerase alpha.
- 27.••.Ollis DL, Brick P, Hamlin R, Xuong NG, Steitz TA. Structure of large fragment of Escherichia coli DNA polymerase I complexed with dTMP. Nature 1985 Feb 28-Mar 6;313(6005):762–6. [DOI] [PubMed] [Google Scholar]; A landmark paper reporting the very first structure of any DNA polymerase.
- 28.Berman AJ, Kamtekar S, Goodman JL, Lazaro JM, de Vega M, Blanco L, et al. Structures of Phi29 DNA polymerase complexed with substrate: the mechanism of translocation in B-family polymerases. EMBO J 2007. Jul 25;26(14):3494–505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Franklin MC, Wang J, Steitz TA. Structure of the replicating complex of a pol alpha family DNA polymerase. Cell 2001. Jun 1;105(5):657–67. [DOI] [PubMed] [Google Scholar]
- 30.Shamoo Y, Steitz TA. Building a replisome from interacting pieces: sliding clamp complexed to a peptide from DNA polymerase and a polymerase editing complex. Cell 1999. Oct 15;99(2):155–66. [DOI] [PubMed] [Google Scholar]
- 31.Hopfner KP, Eichinger A, Engh RA, Laue F, Ankenbauer W, Huber R, et al. Crystal structure of a thermostable type B DNA polymerase from Thermococcus gorgonarius. Proc Natl Acad Sci U S A 1999. Mar 30;96(7):3600–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hogg M, Osterman P, Bylund GO, Ganai RA, Lundstrom EB, Sauer-Eriksson AE, et al. Structural basis for processive DNA synthesis by yeast DNA polymerase ɛ. Nat Struct Mol Biol 2014. Jan;21(1):49–55. [DOI] [PubMed] [Google Scholar]
- 33.Swan MK, Johnson RE, Prakash L, Prakash S, Aggarwal AK. Structural basis of high-fidelity DNA synthesis by yeast DNA polymerase δ. Nat Struct Mol Biol 2009. Sep;16(9):979–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Perera RL, Torella R, Klinge S, Kilkenny ML, Maman JD, Pellegrini L. Mechanism for priming DNA synthesis by yeast DNA polymerase α. eLife 2013. Apr 2;2:e00482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.•.Liu S, Knafels JD, Chang JS, Waszak GA, Baldwin ET, Deibel MR Jr., et al. Crystal structure of the herpes simplex virus 1 DNA polymerase. J Biol Chem 2006. Jun 30;281(26):18193–200. [DOI] [PubMed] [Google Scholar]; A landmark paper reporting the first structure of a DNA virus DNA polymerase.
- 36.Savino C, Federici L, Johnson KA, Vallone B, Nastopoulos V, Rossi M, et al. Insights into DNA replication: the crystal structure of DNA polymerase B1 from the archaeon Sulfolobus solfataricus. Structure 2004. Nov;12(11):2001–8. [DOI] [PubMed] [Google Scholar]
- 37.Lancey C, Tehseen M, Raducanu VS, Rashid F, Merino N, Ragan TJ, et al. Structure of the processive human Pol δ holoenzyme. Nat Commun 2020. Feb 28;11(1):1109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.••.Tarbouriech N, Ducournau C, Hutin S, Mas PJ, Man P, Forest E, et al. The vaccinia virus DNA polymerase structure provides insights into the mode of processivity factor binding. Nat Commun 2017. Nov 13;8(1):1455. [DOI] [PMC free article] [PubMed] [Google Scholar]; A landmark paper reporting the very first structure of any poxvirus DNA polymerase.
- 39.Hayes RP, Heo MR, Mason M, Reid J, Burlein C, Armacost KA, et al. Structural understanding of non-nucleoside inhibition in an elongating herpesvirus polymerase. Nat Commun 2021. May 24;12(1):3040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.•.Peng Q, Xie Y, Kuai L, Wang H, Qi J, Gao GF, et al. Structure of monkeypox virus DNA polymerase holoenzyme. Science 2023. Jan 6;379(6627):100–05. [DOI] [PubMed] [Google Scholar]; Very first report on the structure of a poxvirus DNA holoenzyme.
- 41.•.Jumper J, Evans R, Pritzel A, Green T, Figurnov M, Ronneberger O, et al. Highly accurate protein structure prediction with AlphaFold. Nature 2021. Aug;596(7873):583–89. [DOI] [PMC free article] [PubMed] [Google Scholar]; A landmark report describing artificial intelligence to predict the 3D structure of proteins.
- 42.Mirdita M, Schutze K, Moriwaki Y, Heo L, Ovchinnikov S, Steinegger M. ColabFold: making protein folding accessible to all. Nat Methods 2022. Jun;19(6):679–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kannan SR, Sachdev S, Reddy AS, Kandasamy SL, Byrareddy SN, Lorson CL, et al. Mutations in the monkeypox virus replication complex: Potential contributing factors to the 2022 outbreak. J Autoimmun 2022. Dec;133:102928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sele C, Gabel F, Gutsche I, Ivanov I, Burmeister WP, Iseni F, et al. Low-resolution structure of vaccinia virus DNA replication machinery. J Virol 2013. Feb;87(3):1679–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zheng F, Georgescu RE, Li H, O’Donnell ME. Structure of eukaryotic DNA polymerase δ bound to the PCNA clamp while encircling DNA. Proc Natl Acad Sci U S A 2020. Dec 1;117(48):30344–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Boyle KA, Traktman P. Members of a novel family of mammalian protein kinases complement the DNA-negative phenotype of a vaccinia virus ts mutant defective in the B1 kinase. J Virol 2004. Feb;78(4):1992–2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Paran N, De Silva FS, Senkevich TG, Moss B. Cellular DNA ligase I is recruited to cytoplasmic vaccinia virus factories and masks the role of the vaccinia ligase in viral DNA replication. Cell Host Microbe 2009. Dec 17;6(6):563–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Senkevich TG, Koonin EV, Moss B. Predicted poxvirus FEN1-like nuclease required for homologous recombination, double-strand break repair and full-size genome formation. Proc Natl Acad Sci U S A 2009. Oct 20;106(42):17921–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Czarnecki MW, Traktman P. The vaccinia virus DNA polymerase and its processivity factor. Virus Res 2017. Apr 15;234:193–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.•.Holy A. Phosphonomethoxyalkyl analogs of nucleotides. Curr Pharm Des 2003;9(31):2567–92. [DOI] [PubMed] [Google Scholar]; First report of the ANP nucleotides that revolutionized antiviral therapy.
- 51.De Clercq E, Holy A. Acyclic nucleoside phosphonates: a key class of antiviral drugs. Nat Rev Drug Discov 2005. Nov;4(11):928–40. [DOI] [PubMed] [Google Scholar]
- 52.Andrei G, Snoeck R. Cidofovir activity against poxvirus infections. Viruses 2010. Dec;2(12):2803–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Magee WC, Hostetler KY, Evans DH. Mechanism of inhibition of vaccinia virus DNA polymerase by cidofovir diphosphate. Antimicrob Agents Chemother 2005. Aug;49(8):3153–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Magee WC, Aldern KA, Hostetler KY, Evans DH. Cidofovir and (S)-9-[3-hydroxy-(2-phosphonomethoxy)propyl]adenine are highly effective inhibitors of vaccinia virus DNA polymerase when incorporated into the template strand. Antimicrob Agents Chemother 2008. Feb;52(2):586–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Magee WC, Evans DH. The antiviral activity and mechanism of action of (S)-[3-hydroxy-2-(phosphonomethoxy)propyl] (HPMP) nucleosides. Antiviral Res 2012. Nov;96(2):169–80. [DOI] [PubMed] [Google Scholar]
- 56.De Clercq E Vaccinia virus inhibitors as a paradigm for the chemotherapy of poxvirus infections. Clin Microbiol Rev 2001. Apr;14(2):382–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Becker MN, Obraztsova M, Kern ER, Quenelle DC, Keith KA, Prichard MN, et al. Isolation and characterization of cidofovir resistant vaccinia viruses. Virol J 2008. May 14;5:58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Smee DF, Sidwell RW, Kefauver D, Bray M, Huggins JW. Characterization of wild-type and cidofovir-resistant strains of camelpox, cowpox, monkeypox, and vaccinia viruses. Antimicrob Agents Chemother 2002. May;46(5):1329–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kornbluth RS, Smee DF, Sidwell RW, Snarsky V, Evans DH, Hostetler KY. Mutations in the E9L polymerase gene of cidofovir-resistant vaccinia virus strain WR are associated with the drug resistance phenotype. Antimicrob Agents Chemother 2006. Dec;50(12):4038–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Gammon DB, Snoeck R, Fiten P, Krecmerova M, Holy A, De Clercq E, et al. Mechanism of antiviral drug resistance of vaccinia virus: identification of residues in the viral DNA polymerase conferring differential resistance to antipoxvirus drugs. J Virol 2008. Dec;82(24):12520–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Farlow J, Ichou MA, Huggins J, Ibrahim S. Comparative whole genome sequence analysis of wild-type and cidofovir-resistant monkeypoxvirus. Virol J 2010. May 28;7:110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Duraffour S, Andrei G, Topalis D, Krecmerova M, Crance JM, Garin D, et al. Mutations conferring resistance to viral DNA polymerase inhibitors in camelpox virus give different drug-susceptibility profiles in vaccinia virus. J Virol 2012. Jul;86(13):7310–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Joyce CM. Choosing the right sugar: how polymerases select a nucleotide substrate. Proc Natl Acad Sci U S A 1997. Mar 4;94(5):1619–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Gao G, Orlova M, Georgiadis MM, Hendrickson WA, Goff SP. Conferring RNA polymerase activity to a DNA polymerase: a single residue in reverse transcriptase controls substrate selection. Proc Natl Acad Sci U S A 1997. Jan 21;94(2):407–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Cases-Gonzalez CE, Gutierrez-Rivas M, Menendez-Arias L. Coupling ribose selection to fidelity of DNA synthesis. The role of Tyr-115 of human immunodeficiency virus type 1 reverse transcriptase. J Biol Chem 2000. Jun 30;275(26):19759–67. [DOI] [PubMed] [Google Scholar]
- 66.Astatke M, Ng K, Grindley ND, Joyce CM. A single side chain prevents Escherichia coli DNA polymerase I (Klenow fragment) from incorporating ribonucleotides. Proc Natl Acad Sci U S A 1998. Mar 31;95(7):3402–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Tabor S, Richardson CC. A single residue in DNA polymerases of the Escherichia coli DNA polymerase I family is critical for distinguishing between deoxy- and dideoxyribonucleotides. Proc Natl Acad Sci U S A 1995. Jul 3;92(14):6339–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Taddie JA, Traktman P. Genetic characterization of the vaccinia virus DNA polymerase: cytosine arabinoside resistance requires a variable lesion conferring phosphonoacetate resistance in conjunction with an invariant mutation localized to the 3’−5’ exonuclease domain. J Virol 1993. Jul;67(7):4323–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.•.Singh K, Srivastava A, Patel SS, Modak MJ. Participation of the fingers subdomain of Escherichia coli DNA polymerase I in the strand displacement synthesis of DNA. J Biol Chem 2007. Apr 6;282(14):10594–604. [DOI] [PubMed] [Google Scholar]; Very first report on the polymerase kinetics of strand-displacement synthesis of DNA.
- 70.Fisher TS, Darden T, Prasad VR. Substitutions at Phe61 in the beta3-beta4 hairpin of HIV-1 reverse transcriptase reveal a role for the fingers subdomain in strand displacement DNA synthesis. J Mol Biol 2003. Jan 17;325(3):443–59. [DOI] [PubMed] [Google Scholar]
- 71.Rutvisuttinunt W, Meyer PR, Scott WA. Interactions between HIV-1 reverse transcriptase and the downstream template strand in stable complexes with primer-template. PLoS One 2008;3(10):e3561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Srivastava A, Singh K, Modak MJ. Phe 771 of Escherichia coli DNA polymerase I (Klenow fragment) is the major site for the interaction with the template overhang and the stabilization of the pre-polymerase ternary complex. Biochemistry 2003. Apr 8;42(13):3645–54. [DOI] [PubMed] [Google Scholar]
- 73.Datta K, Wowor AJ, Richard AJ, LiCata VJ. Temperature dependence and thermodynamics of Klenow polymerase binding to primed-template DNA. Biophys J 2006. Mar 1;90(5):1739–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.••.Yang G, Pevear DC, Davies MH, Collett MS, Bailey T, Rippen S, et al. An orally bioavailable antipoxvirus compound (ST-246) inhibits extracellular virus formation and protects mice from lethal orthopoxvirus Challenge. J Virol 2005. Oct;79(20):13139–49. [DOI] [PMC free article] [PubMed] [Google Scholar]; Very first report on the discovery of an anti-poxvirus compound that targeted p37.
- 75.Baek SH, Kwak JY, Lee SH, Lee T, Ryu SH, Uhlinger DJ, et al. Lipase activities of p37, the major envelope protein of vaccinia virus. J Biol Chem 1997. Dec 19;272(51):32042–9. [DOI] [PubMed] [Google Scholar]
- 76.Grosenbach DW, Ulaeto DO, Hruby DE. Palmitylation of the vaccinia virus 37-kDa major envelope antigen. Identification of a conserved acceptor motif and biological relevance. J Biol Chem 1997. Jan 17;272(3):1956–64. [DOI] [PubMed] [Google Scholar]
- 77.Blasco R, Moss B. Extracellular vaccinia virus formation and cell-to-cell virus transmission are prevented by deletion of the gene encoding the 37,000-Dalton outer envelope protein. J Virol 1991. Nov;65(11):5910–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Husain M, Weisberg A, Moss B. Topology of epitope-tagged F13L protein, a major membrane component of extracellular vaccinia virions. Virology 2003. Apr 10;308(2):233–42. [DOI] [PubMed] [Google Scholar]
- 79.Duraffour S, Snoeck R, de Vos R, van Den Oord JJ, Crance JM, Garin D, et al. Activity of the anti-orthopoxvirus compound ST-246 against vaccinia, cowpox and camelpox viruses in cell monolayers and organotypic raft cultures. Antivir Ther 2007;12(8):1205–16. [PubMed] [Google Scholar]
- 80.Nalca A, Hatkin JM, Garza NL, Nichols DK, Norris SW, Hruby DE, et al. Evaluation of orally delivered ST-246 as postexposure prophylactic and antiviral therapeutic in an aerosolized rabbitpox rabbit model. Antiviral Res 2008. Aug;79(2):121–7. [DOI] [PubMed] [Google Scholar]
- 81.Smith SK, Olson VA, Karem KL, Jordan R, Hruby DE, Damon IK. In vitro efficacy of ST246 against smallpox and monkeypox. Antimicrob Agents Chemother 2009. Mar;53(3):1007–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Quenelle DC, Buller RM, Parker S, Keith KA, Hruby DE, Jordan R, et al. Efficacy of delayed treatment with ST-246 given orally against systemic orthopoxvirus infections in mice. Antimicrob Agents Chemother 2007. Feb;51(2):689–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Hutson CL, Kondas AV, Mauldin MR, Doty JB, Grossi IM, Morgan CN, et al. Pharmacokinetics and efficacy of a potential smallpox therapeutic, Brincidofovir, in a lethal monkeypox virus animal model. mSphere 2021. Feb 3;6(1). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Smith SK, Self J, Weiss S, Carroll D, Braden Z, Regnery RL, et al. Effective antiviral treatment of systemic orthopoxvirus disease: ST-246 treatment of prairie dogs infected with monkeypox virus. J Virol 2011. Sep;85(17):9176–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Grosenbach DW, Berhanu A, King DS, Mosier S, Jones KF, Jordan RA, et al. Efficacy of ST-246 versus lethal poxvirus challenge in immunodeficient mice. Proc Natl Acad Sci U S A 2010. Jan 12;107(2):838–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Jordan R, Goff A, Frimm A, Corrado ML, Hensley LE, Byrd CM, et al. ST-246 antiviral efficacy in a nonhuman primate monkeypox model: determination of the minimal effective dose and human dose justification. Antimicrob Agents Chemother 2009. May;53(5):1817–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Huggins J, Goff A, Hensley L, Mucker E, Shamblin J, Wlazlowski C, et al. Nonhuman primates are protected from smallpox virus or monkeypox virus challenges by the antiviral drug ST-246. Antimicrob Agents Chemother 2009. Jun;53(6):2620–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Grosenbach DW, Honeychurch K, Rose EA, Chinsangaram J, Frimm A, Maiti B, et al. Oral tecovirimat for the treatment of smallpox. N Engl J Med 2018. Jul 5;379(1):44–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Russo AT, Grosenbach DW, Brasel TL, Baker RO, Cawthon AG, Reynolds E, et al. Effects of treatment delay on efficacy of tecovirimat following lethal aerosol monkeypox virus challenge in cynomolgus Macaques. J Infect Dis 2018. Sep 22;218(9):1490–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Duraffour S, Andrei G, Snoeck R. Tecovirimat, a p37 envelope protein inhibitor for the treatment of smallpox infection. iDrugs 2010. Mar;13(3):181–91. [PubMed] [Google Scholar]
- 91.Smee DF. Progress in the discovery of compounds inhibiting orthopoxviruses in animal models. Antivir Chem Chemother 2008;19(3):115–24. [DOI] [PubMed] [Google Scholar]
- 92.Frenois-Veyrat G, Gallardo F, Gorge O, Marcheteau E, Ferraris O, Baidaliuk A, et al. Tecovirimat is effective against human monkeypox virus in vitro at nanomolar concentrations. Nat Microbiol 2022. Dec;7(12):1951–55. [DOI] [PubMed] [Google Scholar]
- 93.Russo AT, Berhanu A, Bigger CB, Prigge J, Silvera PM, Grosenbach DW, et al. Co-administration of tecovirimat and ACAM2000 in non-human primates: Effect of tecovirimat treatment on ACAM2000 immunogenicity and efficacy versus lethal monkeypox virus challenge. Vaccine 2020. Jan 16;38(3):644–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Duraffour S, Lorenzo MM, Zoller G, Topalis D, Grosenbach D, Hruby DE, et al. ST-246 is a key antiviral to inhibit the viral F13L phospholipase, one of the essential proteins for orthopoxvirus wrapping. J Antimicrob Chemother 2015. May;70(5):1367–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Lederman ER, Davidson W, Groff HL, Smith SK, Warkentien T, Li Y, et al. Progressive vaccinia: case description and laboratory-guided therapy with vaccinia immune globulin, ST-246, and CMX001. J Infect Dis 2012. Nov;206(9):1372–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Leiros I, McSweeney S, Hough E. The reaction mechanism of phospholipase D from Streptomyces sp. strain PMF. Snapshots along the reaction pathway reveal a pentacoordinate reaction intermediate and an unexpected final product. J Mol Biol 2004. Jun 11;339(4):805–20. [DOI] [PubMed] [Google Scholar]
- 97.Mazurkov OY, Kabanov AS, Shishkina LN, Sergeev AA, Skarnovich MO, Bormotov NI, et al. New effective chemically synthesized anti-smallpox compound NIOCH-14. J Gen Virol 2016. May;97(5):1229–39. [DOI] [PubMed] [Google Scholar]
- 98.Mazurkov OY, Shishkina LN, Bormotov NI, Skarnovich MO, Serova OA, Mazurkova NA, et al. Estimation of absolute bioavailability of the chemical substance of the anti-smallpox preparation NIOCH-14 in Mice. Bull Exp Biol Med 2020. Dec;170(2):207–10. [DOI] [PubMed] [Google Scholar]
- 99.Shishkina LN, Mazurkov OY, Bormotov NI, Skarnovich MO, Serova OA, Mazurkova NA, et al. Safety and pharmacokinetics of the substance of the anti-smallpox drug NIOCH-14 after oral administration to laboratory animals. Viruses 2023. Jan 11;15(1). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Ibarra V, Blanco JR, Oteo JA, Rosel L. Efficacy of cidofovir in the treatment of recalcitrant molluscum contagiosum in an AIDS patient. Acta Derm Venereol 2000. Jul-Aug;80(4):315–6. [DOI] [PubMed] [Google Scholar]
- 101.Bojkova D, Bechtel M, Rothenburger T, Steinhorst K, Zoller N, Kippenberger S, et al. Drug sensitivity of currently circulating Mpox viruses. N Engl J Med 2023. Jan 19;388(3):279–81. [DOI] [PubMed] [Google Scholar]
- 102.•.Thornhill JP, Palich R, Ghosn J, Walmsley S, Moschese D, Cortes CP, et al. Human monkeypox virus infection in women and non-binary individuals during the 2022 outbreaks: a global case series. Lancet 2022. Dec 3;400(10367):1953–65. [DOI] [PMC free article] [PubMed] [Google Scholar]; A landmark paper showing a high prevalence of the 2022 MPXV infections among men who have sex with men and bisexuals.
- 103.Sihuincha Maldonado M, Lucchetti AJ, Paredes Pacheco RA, Martinez Cevallos LC, Zumaeta Saavedra EU, Ponce Zapata LR, et al. Epidemiologic characteristics and clinical features of patients with monkeypox virus infection from a hospital in Peru between July and September 2022. Int J Infect Dis 2023. Apr;129:175–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Antoine M, Rubenstein E, Lascoux-Combe C, Chawki S, Zeggagh J, Pintado C, et al. Clinical Description of sixty-six cases of monkeypox virus (MKPV) infection among men who have sex with men (MSM) in an HIV/PrEP French clinic. J Acquir Immune Defic Syndr 2023. Mar 1;92(3):e11–e14. [DOI] [PubMed] [Google Scholar]
- 105.Palich R, Burrel S, Monsel G, Nouchi A, Bleibtreu A, Seang S, et al. Viral loads in clinical samples of men with monkeypox virus infection: a French case series. Lancet Infect Dis 2023. Jan;23(1):74–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Brites C, Deminco F, Sa MS, Brito JT, Luz E, Stocker A. The first two cases of monkeypox infection in MSM in Bahia, Brazil, and viral sequencing. Viruses 2022. Aug 23;14(9). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Vaughan AM, Cenciarelli O, Colombe S, Alves de Sousa L, Fischer N, Gossner CM, et al. A large multi-country outbreak of monkeypox across 41 countries in the WHO European Region, 7 March to 23 August 2022. Euro Surveill 2022. Sep;27(36). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Ferre VM, Bachelard A, Zaidi M, Armand-Lefevre L, Descamps D, Charpentier C, et al. Detection of monkeypox virus in anorectal swabs from asymptomatic men who have sex with men in a sexually transmitted infection screening program in Paris, France. Ann Intern Med 2022. Oct;175(10):1491–92. [DOI] [PubMed] [Google Scholar]
- 109.Vusirikala A, Charles H, Balasegaram S, Macdonald N, Kumar D, Barker-Burnside C, et al. Epidemiology of early monkeypox virus transmission in sexual networks of gay and bisexual men, England, 2022. Emerg Infect Dis 2022. Oct;28(10):2082–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.De Clercq E. Acyclic nucleoside phosphonates in the chemotherapy of DNA virus and retrovirus infections. Intervirology 1997;40(5–6):295–303. [DOI] [PubMed] [Google Scholar]
- 111.De Clercq E. Acyclic nucleoside phosphonates: past, present and future. Bridging chemistry to HIV, HBV, HCV, HPV, adeno-, herpes-, and poxvirus infections: the phosphonate bridge. Biochem Pharmacol 2007. Apr 1;73(7):911–22. [DOI] [PubMed] [Google Scholar]
- 112.Quenelle DC, Collins DJ, Wan WB, Beadle JR, Hostetler KY, Kern ER. Oral treatment of cowpox and vaccinia virus infections in mice with ether lipid esters of cidofovir. Antimicrob Agents Chemother 2004. Feb;48(2):404–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Labate L, Brucci G, Ciccarese G, Bruzzone B, Ricucci V, Stefanelli F, et al. Nasal monkeypox virus infection successfully treated with cidofovir in a patient newly diagnosed with HIV. Int J STD AIDS 2023. Mar;34(3):208–10. [DOI] [PubMed] [Google Scholar]
- 114.Viguier C, de Kermel T, Boumaza X, Benmedjahed NS, Izopet J, Pasquier C, et al. A severe monkeypox infection in a patient with an advanced HIV infection treated with tecovirimat: clinical and virological outcome. Int J Infect Dis 2022. Dec;125:135–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Cash-Goldwasser S, Labuda SM, McCormick DW, Rao AK, McCollum AM, Petersen BW, et al. Ocular Monkeypox - United States, July-September 2022. MMWR Morb Mortal Wkly Rep 2022. Oct 21;71(42):1343–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Schabel FM Jr. The antiviral activity of 9-beta-D-arabinofuranosyladenine (ara-A). Chemotherapia (Basel) 1968;13(6):321–38. [DOI] [PubMed] [Google Scholar]
- 117.Eswar N, Webb B, Marti-Renom MA, Madhusudhan MS, Eramian D, Shen MY, et al. Comparative protein structure modeling using Modeller. Curr Protoc Bioinformatics 2006. Oct;Chapter 5:Unit-5 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Kumar N, Acharya A, Gendelman HE, Byrareddy SN. The 2022 outbreak and the pathobiology of the monkeypox virus. J Autoimmun 2022. Jul;131:102855. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Genomic sequences used in this study were obtained from NCBI GenBank.


