Significance
Few recent advances in human medicine have been as influential as the finding that an imbalance (dysbiosis) of our resident microbial communities in the colon is linked to many chronic human illnesses. However, translating advances in microbiome research into clinical interventions requires a better understanding of the ecological causes of dysbiosis and the causative effects dysbiosis has on human disease. Recent progress in answering these questions suggests that the host determines which redox reactions are available for microbial growth by controlling the availability of respiratory electron acceptors. Dysbiosis is characterized by an increased availability of host-derived electron acceptors, which changes microbiota composition and function. These insights provide an alternative starting point for approaches to rebalance the microbiota.
Keywords: microbiota, dysbiosis, colitis, colorectal cancer
Abstract
The gut microbiota plays a role in many human diseases, but high-throughput sequence analysis does not provide a straightforward path for defining healthy microbial communities. Therefore, understanding mechanisms that drive compositional changes during disease (gut dysbiosis) continues to be a central goal in microbiome research. Insights from the microbial pathogenesis field show that an ecological cause for gut dysbiosis is an increased availability of host-derived respiratory electron acceptors, which are dominant drivers of microbial community composition. Similar changes in the host environment also drive gut dysbiosis in several chronic human illnesses, and a better understanding of the underlying mechanisms informs approaches to causatively link compositional changes in the gut microbiota to an exacerbation of symptoms. The emerging picture suggests that homeostasis is maintained by host functions that control the availability of resources governing microbial growth. Defining dysbiosis as a weakening of these host functions directs attention to the underlying cause and identifies potential targets for therapeutic intervention.
The human colon harbors a microbial community (microbiota) that is 100-fold larger than any other bacterial consortium inhabiting our body (1). Its magnitude makes the colonic microbiota a major source of microbial metabolites that influence human physiology (2–5). Fecal microbiota profiling reveals that a shift in the species composition, termed gut dysbiosis, is observed in diseases that include leading causes of human morbidity and mortality, such as cardiovascular disease (CVD) (6), diabetes (7, 8), colorectal cancer (CRC) (9), chronic kidney disease (CKD) (10), and inflammatory bowel disease (IBD) (11). These observations raise the intriguing prospect that the colonic microbiota plays a central role in human health (12).
Here, we examine the obstacles encountered in understanding microbiome homeostasis through analysis of the microbiota and their genes. We will then discuss how recent advances in the field of bacterial pathogenesis research delivered seminal insights into the ecological causes of dysbiosis, which are directly applicable to microbiome research. Finally, we will consider how understanding the ecological causes of dysbiosis opens avenues for establishing causative links to human disease and mitigating its adverse effects.
Omics, Omics, Everywhere, Nor Any Healthy Microbiome Link
Modern microbiome research is rooted in the innovation of culture-independent methods for profiling complex microbial communities, an approach powered by advances in high-throughput sequencing. Initially, this technology generated catalogs of microbial species names, which focused early work on the question of what a normal species composition of the human gut microbiota looks like. The first step in defining a “healthy” microbial community is the identification of core species that are common to the colonic microbiota of humans (13). However, determining abundant core species that define a healthy human fecal microbiota turned out to be challenging since this microbial community exhibits marked interpersonal differences in bacterial species content (14, 15). The finding that each person’s gut microbial community varies in the specific bacterial species present does not support the hypothesis that there is a healthy core gut microbiota defined by a set of microbial species that we all share (15).
The absence of core species in the human colonic microbiota makes it difficult to define gut homeostasis by profiling the microbiota composition. This impasse led some to suggest that a healthy human microbiome might not even exist (16). Dysbiosis is commonly defined as a decrease in microbial diversity, an absence of beneficial microbes or the presence of potentially harmful microorganisms (17). However, our inability to identify abundant core species that define a healthy human microbiome challenges the idea that gut dysbiosis can be defined based on changes in the species composition. Proponents of this standpoint go as far as to suggest that the term dysbiosis is a distraction from useful microbiome research (18). These limitations and controversies illustrate why microbiota profiling alone does not provide a straightforward path toward understanding the ecological causes of dysbiosis.
Although a healthy human gut microbiota cannot be defined by a set of core species, a core gut microbiome does exist at the level of physiological, often metabolic, functions. Metagenomic analysis of the human fecal microbiota reveals that despite interpersonal variation in species assemblages, there is an identifiable core of genes encoding various metabolic pathways, such as those involved in carbohydrate and amino acid metabolism (15). However, it is difficult to draw clear connections between metagenomic data and the health of a particular microbiome. These limitations persist although technology continues to advance as metatranscriptomics and metabolomics replace metagenomics. At the completion of the Human Microbiome Project in 2019 (19), a commentary summarized the state of the human microbiome field by remarking that despite generating 42 terabytes of multiomics data, “researchers don’t yet agree what constitutes a healthy microbiome or how to define an impaired one” (20). The inability to define a healthy human microbiota despite a wealth of high-throughput sequencing data evokes parallels to the notorious phrase from Samuel T. Coleridge’s Rime of the Ancient Mariner, “water, water, everywhere, nor any drop to drink”.
The question remains why cataloging bacterial species, their genes, and gene products does not provide a straightforward path for translating vast amounts of data into a better understanding of microbiome health. Since the microbiome is commonly defined as the collection of all microbes and their genes (21), it seems at first puzzling that a massive amount of data covering every aspect of the microbiome has not resulted in a better understanding of what it looks like during health. However, ecological theory suggests that microbes and their genes are only one part of the microbiome, which is defined ecologically as the microbiota and its environment, including the body part inhabited by the microbial community (22, 23). This alternative viewpoint suggests that microbiota profiling, metagenomics, and metatranscriptomics provide an incomplete picture of the microbiome since no direct measurements of the host environment are included in the analysis (24). As discussed below, incorporating the host environment into microbiome analysis does indeed provide the missing information to unravel the ecological causes of dysbiosis because this condition commonly features an increase in host-derived resources required for respiration, the most efficient driver of microbial growth. However, to review the origins of this conceptual advance, we must switch to the field of infectious disease research.
Pathogens Expose Causative Links between Host Physiology and Gut Microbiota Composition
Pathogen-Induced Dysbiosis.
During homeostasis, the colonic microbiota is dominated by bacteria belonging to the classes Clostridia (phylum Bacillota) and Bacteroidia (phylum Bacteroidota) (25). However, intestinal inflammation triggered by virulence factors of enteric pathogens, such as Salmonella enterica serovar (S.) Typhimurium (class Gammaproteobacteria; phylum Pseudomonadota), Citrobacter rodentium (class Gammaproteobacteria), or the parasite Toxoplasma gondii (phylum Apicomplexa), drives dysbiosis characterized by an increase in the abundance of bacteria belonging to the classes Gammaproteobacteria and Bacilli (phylum Bacillota) (26–28). The pioneering studies describing this phenomenon kicked off a new line of research seeking to understand how pathogen-indued inflammation changes the microbiota composition (29).
Tetrathionate.
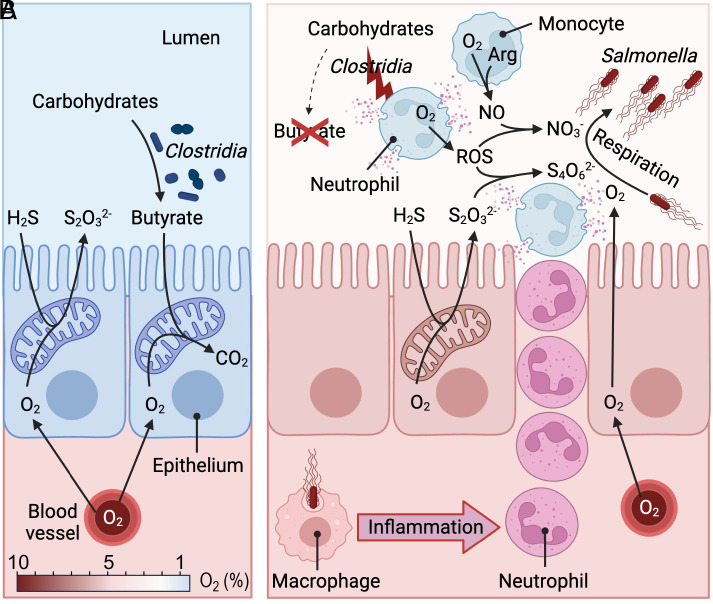
One beneficiary of gut inflammation is Salmonella Typhimurium because colitis triggered by the invasive pathogen promotes fecal–oral transmission by increasing its abundance in the feces (30). A breakthrough in understanding how colitis boosts growth of S. Typhimurium was the finding that phagocytes recruited into the intestinal lumen during gut inflammation provide a respiratory electron acceptor for the pathogen by oxidizing endogenous sulfur compounds to tetrathionate (S4O62−) (31) (Fig. 1). Tetrathionate has been used for the past century by clinical laboratories to enrich for S. Typhimurium in samples containing competing microbes (32). But in the colon, the use of tetrathionate as an electron acceptor for anaerobic respiration gives the pathogen an edge over bacteria that rely on fermentation for growth. Importantly, a combination of bacterial and host genetics made it possible to establish a causative link between the generation of host-derived tetrathionate during colitis and enhanced pathogen growth within the gut microbiota (31).
Fig. 1.
Pathogen-induced colitis increases the availability of host-derived respiratory electron acceptors to increase pathogen abundance in the colon. (A) During homeostasis, oxygen (O2)-consuming reactions maintain the colonic epithelium in a state of physiological hypoxia. (B) During Salmonella infection, invasion of the intestinal mucosa by the pathogen triggers transepithelial migration of neutrophils, resulting in a depletion of Clostridia and loss of epithelial hypoxia. NO, nitric oxide; ROS, reactive oxygen species; NO3−; nitrate; S2O32−, thiosulfate; H2S, hydrogen sulfide, S4O62−, tetrathionate; CO2, carbon dioxide; Arg, arginine. Created with BioRender.com.
Nitrate.
The finding that the host provides tetrathionate to boost growth of S. Typhimurium was soon followed by the finding that additional respiratory electron acceptors are generated during pathogen-induced colitis. Phagocytes recruited into the intestinal lumen during inflammation release superoxide (O2−) and nitric oxide radicals (NO) produced by phagocyte Nicotinamide Adenine Dinucleotide Phosphate (NADPH) oxidase (33) and inducible nitric oxide synthase (iNOS) (34), respectively. Superoxide and nitric oxide react to form peroxynitrite (ONOO−) (35, 36), an unstable bactericidal compound that decomposes to nitrate (NO3−) (37) (Fig. 1). Thus, a by-product of recruiting phagocytes into the intestinal lumen during S. Typhimurium infection is an elevated concentration of host-derived nitrate in the mucus layer (38, 39). Nitrate is used by S. Typhimurium as an electron acceptor for anaerobic respiration (40). By powering nitrate respiration, intestinal inflammation fuels growth of S. Typhimurium to increase the pathogen abundance in the feces (38, 39).
Host-derived nitrate is also generated when phagocytes migrate into the intestinal lumen in response to infection with the parasite Toxoplasma gondii (41). By increasing the nitrate concentration in the lumen of the small intestine, T. gondii–induced inflammation drives dysbiosis characterized by an elevated abundance of commensal Escherichia coli (class Gammaproteobacteria) because this commensal can accelerate its growth through anaerobic nitrate respiration (41). These data suggest that host-derived nitrate generated as a by-product of intestinal inflammation is an ecological driver of changes in the composition of gut-associated microbial communities during inflammation (42).
Oxygen.
S. Typhimurium–induced colitis results in a loss of anaerobiosis by increasing the diffusion of host-derived oxygen into the lumen. This process is initiated by luminal phagocytes that release antimicrobials during S. Typhimurium–induced colitis, thereby lowering the microbial density (43, 44) and altering the composition of the gut microbiota by reducing the abundance of Lachnospiraceae (class Clostridia) and Ruminococcaceae (class Clostridia) (45). Lachnospiraceae and Ruminococcaceae are the main producers of the short-chain fatty acid (SCFA) butyrate within the gut microbiota (46, 47). A reduced abundance of these butyrate-producing taxa during S. Typhimurium infection leads to a marked drop in the luminal butyrate concentration (48). Butyrate is an important signal regulating energy metabolism in colonic epithelial cells (49, 50). During gut homeostasis, butyrate-signaling preserves high mitochondrial oxygen (O2) consumption through oxidative phosphorylation, thereby maintaining the colonic surface in a state of physiological hypoxia (<1% O2) compared to normal levels of tissue oxygenation (3 to 10% O2) (51, 52) (Fig. 1A). Physiological hypoxia of the colonic epithelium limits diffusion of oxygen into the intestinal lumen to maintain anaerobiosis (53). Depletion of butyrate causes colonic epithelial cells to reduce mitochondrial oxygen consumption and shift their energy production toward a conversion of glucose into lactate (aerobic glycolysis), which increases epithelial oxygenation (51). This heightens the amount of oxygen emanating from the epithelial surface, thereby driving growth of S. Typhimurium in the gut lumen through aerobic respiration (48) (Fig. 1B).
Likewise, a disruption of anaerobiosis is observed during infection with the luminal pathogen C. rodentium, which increases epithelial oxygenation through a different mechanism than S. Typhimurium (54). C. rodentium uses its virulence factors to attach to and damage epithelial cells in the large intestine, which triggers an epithelial repair response (55). Epithelial cell numbers are increased by excessive cell division of undifferentiated epithelial cells, termed transit-amplifying (TA) cells, which expand from their usual location in the crypts to reach the epithelial surface (54). This regenerative response leads to a longitudinal increase in epithelial cell numbers in the crypts, which is visible as crypt elongation, a lesion known as colonic crypt hyperplasia (56, 57). Unlike differentiated epithelial cells that consume oxygen through oxidative phosphorylation in their mitochondria (49), TA cells obtain energy through aerobic glycolysis(58) and therefore do not exhibit hypoxia. By replacing differentiated epithelial cells with TA cells on the mucosal surface, C. rodentium–induced colonic crypt hyperplasia increases epithelial oxygenation, and the resulting surge of oxygen diffusing into the intestinal lumen fuels pathogen growth through aerobic respiration (54).
Fermentation vs. Respiration.
The discovery that infectious colitis increases the availability of exogenous respiratory electron acceptors explains an increase in the luminal abundance of enteric pathogens (31, 38, 48, 54), but the broader implications are that these changes in the host environment are an ecological driver of gut dysbiosis (41). To appreciate the role host-derived electron acceptors play in changing the composition of microbial communities, it is helpful to reflect on how their availability governs the abundance of bacteria that rely on fermentation for growth, i.e., obligately anaerobic bacteria, vs. bacteria that can support their grow by respiring oxygen or nitrate, i.e., facultatively anaerobic bacteria.
One basic tenet of microbial population biology is that taxa abundance is a function of the bacterial growth rate, as species with the shortest generation time come to dominate microbial communities. The bacterial growth rate is limited by energy metabolism, in particular the availability of adenosine triphosphate (ATP) (59). ATP is generated through redox reactions, in which the transfer of electrons from an electron donor, such as glucose, to an electron acceptor, such as oxygen, is coupled to oxidative phosphorylation or substrate-level phosphorylation. In general, the amount of ATP that can be generated from an electron donor, such as glucose, increases with the redox potential (E’0) of the electron acceptor. The redox potential is highest for oxygen (E’0 for O2/H2O = 820 mV), followed by nitrate (E’0 for NO3−/NO2− = 433 mV), and lower for endogenous electron acceptors used for fermentation, such as pyruvate (E’0 for pyruvate/lactate = −190 mV). As a result of this thermodynamic hierarchy of electron acceptors, the growth yield of Klebsiella aerogenes (class Gammaproteobacteria) on glucose under anaerobic conditions is almost doubled when nitrate is present and almost tripled when bacteria are cultured aerobically (60). As taxa abundance is a function of the amount of ATP generated through redox reactions, the influence host-derived electron acceptors exert over the microbiota composition is dominant over effects arbitrated by electron donors in the diet. By controlling the availability of host-derived electron acceptors, the host selects which metabolic traits can succeed in the gut environment by managing the top level in the hierarchy of factors determining the microbial growth rate. In practical terms, management of electron acceptor availability provides the host with a mechanism to control the abundance of obligately vs. facultatively anaerobic bacteria.
Since phylogeny is a good predictor of complex metabolic traits (61), a shift from obligately to facultatively anaerobic bacteria results in a class-level change in the gut microbiota composition. The ability to respire oxygen or nitrate is widely conserved among facultatively anaerobic bacteria, such as Gammaproteobacteria or Bacilli, but largely absent in obligately anaerobic bacteria, including Clostridia and Bacteroidia (62). Therefore, in the presence of host-derived oxygen or nitrate, Gammaproteobacteria and Bacilli are predicted to have the fastest growth rate, which explains their increased abundance during pathogen-induced colitis (26–28). Conversely, when oxygen and nitrate are not available, obligately anaerobic bacteria that specialize on fermentation exhibit the fastest growth rate, which drives the observed dominance of Clostridia and Bacteroidia during homeostasis (25).
Homeostasis vs. Dysbiosis.
Dysbiosis has traditionally been defined in terms of the changes in the bacterial species composition of the gut microbiota (17), but insights into the ecological causes of dysbiosis create a new paradigm, which shifts the focus away from bacterial species and toward understanding the underlying changes in host physiology (24). The host maintains homeostasis by restricting the availability of oxygen and nitrate, thereby regulating the composition of the colonic microbiota by controlling the luminal environment (55, 63, 64). Conversely, dysbiosis involves a change in the host environment characterized by an increased availability of host-derived electron acceptors (24, 63, 65). This environment fuels growth of facultatively anaerobic bacteria to generate a microbial signature of dysbiosis (66) that serves as a biomarker for the underlying alteration in host physiology (67). This paradigm shift suggests that dysbiosis represents a state of weakened host control over the microbial environment, whereas gut homeostasis defines a state where these host functions operate normally (24, 63, 65, 68).
Notably, an elevated availability of host-derived electron acceptors creates a microbial signature of dysbiosis that is not defined by the presence or prevalence of specific core species, but by the relative success of a metabolic trait. Facultatively anaerobic bacteria form a metabolic guild because they use the same resources, i.e., respiratory electron acceptors, in a similar way (69). This metabolic trait might be represented by different bacterial species in different individuals, but an increased abundance of facultatively anaerobic bacteria can be detected even in the face of marked interpersonal differences in bacterial species content.
Importantly, the microbial signature of dysbiosis generated during infection with enteric pathogens (26–28) is also observed in numerous noncommunicable human diseases (66, 67). For example, an increased abundance of facultatively anaerobic bacteria in the fecal microbiota is detected in patients with IBD (11), CRC (9), cancer cachexia (70), radiation enteritis during radiotherapy (71), CKD (10), type 1 diabetes (7), nonalcoholic steatohepatitis (72), graft vs. host disease (73), severe malnutrition (kwashiorkor) (74), chronic inflammation during aging (inflammageing) (75), and CVD (6). Furthermore, this microbial signature of dysbiosis is associated with chronic alcohol consumption (76), and with exposure to a diet that contains high levels of saturated fatty acids (77), two environmental risk factors for developing CVD (78, 79). By generating insights into the mechanistic underpinnings of this microbial signature of dysbiosis, infectious disease research offers a fresh starting point for understanding the ecological drivers of dysbiosis in many noninfectious human conditions.
Ecological Causes of Dysbiosis
Ulcerative Colitis.
Mechanistic insights into pathogen-induced dysbiosis have helped open new avenues for research into the causes of dysbiosis during IBD. IBD is an illness characterized by chronic mucosal inflammation of the digestive tract. Genetic and environmental risk factors are thought to trigger IBD by generating inappropriate mucosal immune activation that is driven by the gut microbiota. This consensus view predates modern microbiome research (80), but the underlying mechanisms are still obscure. One form of the disease, termed ulcerative colitis (UC), remains restricted to the colon, where it triggers dysbiosis characterized by an increased Gammaproteobacteria and decreased Clostridia abundance (81, 82). Insights into the mechanisms that drive dysbiosis during infection with enteric pathogens suggest that an elevated Gammaproteobacteria abundance is likely the result of a rise in host-derived respiratory electron acceptors (31, 38, 41, 48, 54, 83). This prediction raises two questions: i) is the electron acceptor hypothesis consistent with clinical observations and ii) does this hypothesis hold up to scrutiny when tested in animal models.
UC patients exhibit increased concentrations of nitric oxide in luminal gas and elevated nitrate levels in their feces (84, 85). This increase in the luminal concentration of nitrate in the large intestine is recapitulated in mouse models of genetically or chemically induced colitis (86). Nitrate generated during colitis is host-derived because its production can be abrogated either by chemical inhibitors of iNOS or by genetic deletion of the murine Nos2 gene, which encodes iNOS (86). The use of bacterial genetics reveals that increased levels of host-derived nitrate during colitis are causatively linked to an elevated abundance of commensal E. coli in the gut microbiota (86). Collectively, these data support the notion that a heightened availability of host-derived nitrate is one of the ecological causes of dysbiosis in UC (42).
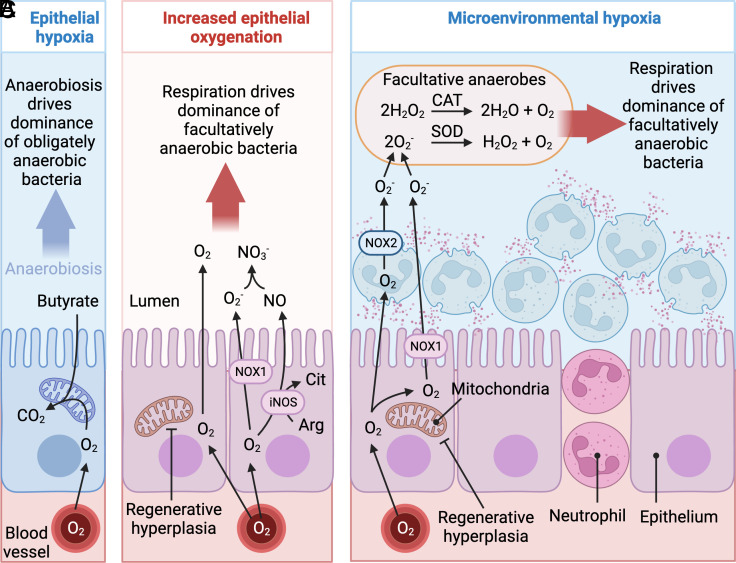
Direct measurements of oxygen levels in the colon of UC patients are not available. However, maintenance of epithelial hypoxia requires high mitochondrial oxygen consumption (51) (Fig. 2A). Notably, UC patients exhibit reduced mitochondrial function in colonic epithelial cells compared to healthy controls (87–89), which is consistent with the idea that epithelial oxygenation becomes elevated during UC. Experimental support for the oxygen hypothesis comes from a mouse model of dextran sulfate sodium (DSS)-induced colitis. Metagenomic sequencing reveals that metabolic pathways involved in aerobic respiration are overrepresented in the colonic microbiota of mice with DSS-induced colitis (90). Oxygen availability can become elevated during DSS-induced colitis through two mechanisms. The first is a consequence of DSS-induced endoplasmatic reticulum stress in epithelial cells, which results in epithelial injury (91, 92). DSS-induced epithelial injury stimulates regenerative hyperplasia, a process that eliminates physiological hypoxia of the colonic epithelium (93, 94) (Fig. 2B). In turn, elevated epithelial oxygenation increases diffusion of oxygen into the lumen to boost growth of commensal E. coli by aerobic respiration (90, 93, 94).
Fig. 2.
The ecological causes for an intestinal domination of facultatively anaerobic bacteria in the fecal microbiota during UC. (A) During homeostasis, mitochondrial oxygen (O2) consumption maintains the colonic epithelium in a state of physiological hypoxia. (B) Epithelial injury during UC triggers regenerative hyperplasia to increase diffusion of oxygen into the intestinal lumen. (C) Neutrophil transepithelial migration during UC leads to depletion of microenvironmental oxygen by the phagocyte NADPH oxidase (NOX2), which generates microenvironmental hypoxia at the mucosal surface. O2−, superoxide; CO2, carbon dioxide; H2O2, hydrogen peroxide; H2O, water; Arg, arginine; Cit, citrulline; NO, nitric oxide; ROS, reactive oxygen species; NO3−; nitrate; iNOS, inducible nitric oxide synthase, NOX1, epithelial NADPH oxidase, CAT, catalase, SOD, superoxide dismutase. Created with BioRender.com.
The second mechanism offering access to oxygen during DSS-induced colitis is a consequence of mucosal abnormalities that are linked to neutrophil transepithelial migration. During the respiratory burst of neutrophils, the phagocyte NADPH oxidase (NOX2) generates superoxide radicals by rapidly depleting microenvironmental oxygen (O2 + NADPH → O2− + NADP+). This process generates a microenvironmental hypoxia in areas of the mucosal surface that are affected by transmigration of neutrophils (95). Commensal E. coli can use superoxide dismutase to disproportionate superoxide radicals to hydrogen peroxide and oxygen (2O2− + 2H+ → H2O2 + O2), followed by catalase-mediated conversion of hydrogen peroxide to oxygen and water (2H2O2 → O2 + 2H2O). Oxygen liberated by these reactions can then be used for aerobic respiration, a process contributing to growth of commensal E. coli in the colon of mice with colitis (96) (Fig. 2C).
In summary, existing clinical data and the available experimental evidence from animal models point to an increased accessibility of host-derived oxygen and nitrate in the intestinal lumen as a cause for the elevated abundance of facultatively anaerobic bacteria in the feces of UC patients. This body of work suggests that the ecological causes of dysbiosis during UC represent those previously implicated in driving dysbiosis during infection with enteric pathogens.
Exposure to Antibiotics.
Since inflammation is a stereotypic host response, it is perhaps not surprising that the ecological causes of dysbiosis are similar for infectious colitis and UC. However, an elevated abundance of facultatively anaerobic bacteria in the fecal microbiota is a signature of dysbiosis that is also linked to antibiotic therapy (97, 98), which does not induce colitis.
Clinically, the use of broad-spectrum antibiotics perturbs the normal gut microbiota and sets the stage for intestinal domination by endogenous Gammaproteobacteria or Bacilli (99). Of special concern are carbapenem-resistant Enterobacteriaceae (class Gammaproteobacteria) and vancomycin-resistant Enterococcus faecalis (class Bacilli) because in hematopoietic cell transplantation recipients, an intestinal domination by these antibiotic-resistant opportunistic pathogens is a source of bloodstream infections with high mortality rates (98–100).
An increase in the abundance of Gammaproteobacteria and Bacilli is recapitulated in mice after oral administrations of antibiotics (101–103). The phenomenon has been studied since the 1950s using streptomycin treatment of mice followed by exogenous administration of commensal or pathogenic Enterobacteriaceae (104–110). Early work in this model shows that a perturbation of the microbiota with streptomycin depletes the SCFAs acetate, propionate, and butyrate, which is associated with an increase in the luminal redox potential (110). More recent data unveil that streptomycin treatment results in a loss of epithelial hypoxia (51) and an induction of mucosal Nos2 expression (111). The mechanism underlying these changes is an antibiotic-mediated depletion of SCFAs, which triggers a metabolic reprogramming of differentiated colonic epithelial cells to increase the availability of oxygen and nitrate (53).
Depletion of SCFAs generates two distinct changes in host physiology that participate in this metabolic reprogramming of epithelial cells. The first is a loss of epithelial signaling through PPAR-γ (peroxisome proliferator-activated receptor gamma), a nuclear receptor that is synthesized by differentiated epithelia cells of the colon (112, 113). During homeostasis, Clostridia-derived butyrate activates epithelial PPAR-γ-signaling (114), which results in suppression of iNOS synthesis (115) and activation of mitochondrial bioenergetics (49, 116). By depleting butyrate, streptomycin treatment reduces epithelial PPAR-γ-signaling, resulting in elevated epithelial Nos2 expression and an increase in the nitrate concentrations in the mucus layer (117).
The second signal required for a metabolic reprogramming of the epithelium during streptomycin treatment is linked to a reduction in the colonic pool of regulatory T cells (Tregs). Microbiota-derived SCFAs signal through G-protein coupled receptors in host cells to maintain the regulatory T cell pool in the colonic mucosa (118–121). By depleting SCFAs, streptomycin treatment reduces the numbers of Tregs in the colonic mucosa (118, 121). A reduction in the numbers of these immunosuppressive cells triggers low-grade mucosal inflammation (111), which generates a type I interferon-dependent signal needed by epithelial cells to undergo metabolic reprogramming (122). Neither a reduction in the numbers of colonic Tregs, nor a loss of epithelial PPAR-γ-signaling alone are sufficient for eliminating epithelial hypoxia in the colon. But by combining these two changes in host physiology, a streptomycin-mediated depletion of SCFAs reduces mitochondrial oxygen consumption in the epithelium to increase the diffusion of oxygen into the colonic lumen (117).
Collectively, data from the streptomycin-treated mouse model suggest that a depletion of microbiota-derived SCFAs during antibiotic therapy triggers a metabolic reprogramming of colonic epithelial cells that increases the availability of host-derived oxygen and nitrate in the intestinal lumen. Importantly, this increase in the availability of host-derived electron acceptors drives an intestinal domination by commensal E. coli, which is causatively linked to their ability to respire oxygen and nitrate (111, 117). Thus, the ecological causes of dysbiosis during antibiotic therapy are explained by some of the same environmental changes that increase the abundance of facultatively anaerobic bacteria during infection-induced colitis.
A Common Driver of Dysbiosis in Noncommunicable Diseases.
More recent work reveals that a rise in the concentration of host-derived respiratory electron acceptors in the colon is not limited to antibiotic therapy or UC but also explains an elevated fecal abundance of facultatively anaerobic bacteria in several other human diseases. For instance, an altered gut epithelial metabolism and host-derived nitrate boost luminal growth of Klebsiella oxytoca (class Gammaproteobacteria) in a mouse model of cancer cachexia (70). Similarly, in a mouse model of CRC, an increase in the abundance of Gammaproteobacteria can be functionally linked to an impaired ability of the host to limit the availability of oxygen and nitrate during intestinal inflammation (93, 123). Furthermore, an elevated abundance of E. coli in the colonic microbiota during high fat intake (77), can be explained by a rise in host-derived oxygen and nitrate in the colon (124). Finally, in a mouse model of graft vs. host disease, loss of physiological hypoxia in the colonic epithelium is linked to dysbiosis characterized by an increased abundance of Gammaproteobacteria in the fecal microbiota (125).
The emerging concept that metabolic reprogramming of the colonic epithelium shapes the microbial environment to favor growth of facultatively anaerobic bacteria (53, 65, 67, 126) provides a promising starting point for studying human diseases in which the ecological causes for gut dysbiosis remain unknown. Low-hanging fruits for testing this concept are conditions associated with reduced mitochondrial function, which is indicative of increased epithelial oxygenation (51, 117). For example, mitochondrial dysfunction associated with senescence is a feature of inflammageing (127), but it remains to be tested whether it triggers a loss of epithelial hypoxia in the colon to explain the increased abundance of Gammaproteobacteria in the feces of patients (75). Similarly, mitochondrial dysfunction of colonic epithelial cells is observed in a mouse model of type 1 diabetes (128), but its potential impact on epithelial hypoxia and the overgrowth of Gammaproteobacteria in the fecal microbiota of patients remain unexplored (7). Finally, consumption of excessive quantities of alcohol triggers mitochondrial abnormalities in the colonic epithelium of patients (129), but it has never been examined whether a loss of epithelial hypoxia is an ecological driver of the increased Enterobacteriaceae abundance in the fecal microbiota of patients with alcohol dependence (76).
To summarize, by incorporating the host environment into microbiome analysis, infectious disease research identifies ecological causes of dysbiosis that are germane for understanding the mechanistic underpinnings of gut dysbiosis in a broad spectrum of noncommunicable human diseases. In the following paragraphs, we will consider how this information helps improve our understanding of potential consequences of dysbiosis.
Causative Effects of Dysbiosis on Disease
Metabolism-Based Editing of the Microbiota: IBD.
Insights into the ecological causes of dysbiosis suggest that this condition is secondary to an underlying defect in the host that weakens functions involved in controlling the microbial environment (53, 65, 67, 68). However, changes in microbiota composition and function during dysbiosis can contribute to disease. For example, antibiotic therapy can induce remission in IBD, suggesting that the microbiota contributes to intestinal inflammation (130). Similarly, transfer of dysbiotic microbial communities exacerbates intestinal inflammation in mouse models of IBD (131–133).
A prominent microbial signature of dysbiosis in IBD is a bloom of Enterobacteriaceae in the fecal microbiota (134). Enterobacteriaceae are proinflammatory because their lipopolysaccharide is a potent inducer of innate immune pathways. The proinflammatory nature of Enterobacteriaceae might be relevant because IBD is associated with decreased barrier function (135) and increased bacterial translocation to the mesenteric lymph nodes (136). However, to establish that an increased abundance of Enterobacteriaceae is causatively linked to a worsening of symptoms, it is necessary to demonstrate that a targeted approach for reducing their abundance (i.e., without changing the prevalence of other taxa) provides symptom relieve.
A possible strategy for normalizing the abundance of Enterobacteriaceae during colitis is to selectively block the metabolic pathways that enhance their growth during gut inflammation. Metagenomic sequencing shows that metabolic functions linked to aerobic and anaerobic respiration are overrepresented during DSS-induced colitis in mice (90). In Enterobacteriaceae, a subset of terminal reductases involved in these metabolic pathways require insertion of a molybdenum (Mo)-containing cofactor (molybdopterin) into their active sites (137). E. coli mutants deficient for molybdopterin biosynthesis can no longer take advantage of the increased availability of respiratory electron acceptors to boost their growth in the murine large intestine during colitis (86). Tungsten (W) can replace Mo in molybdopterin, rendering this cofactor inactive in Enterobacteriaceae (138). In mouse models of colitis, sodium tungstate (Na2WO4) administration selectively blunts the expansion of the Enterobacteriaceae population, whereas other major taxonomic families remain unchanged (139). These findings illustrate that elucidating the ecological causes of an elevated Enterobacteriaceae abundance during colitis (86, 90) directly informs the development of strategies for precision editing of the gut microbiota to normalize the abundance of this taxon during colitis (139).
Notably, when germ-free mice are engrafted with gut microbiota from IBD patients with active flares and inflammation is induced by treatment with DSS, tungstate administration selectively reduces the intestinal Enterobacteriaceae load and decreases markers of mucosal inflammation (139). Similarly, in mice with an intact microbiota, tungstate administration reduces the severity of DSS-induced colitis by normalizing the abundance of Enterobacteriaceae (139–141). In contrast, DSS-induced inflammation in germ-free mice does not respond to tungstate administration (139), demonstrating that tungstate reduces intestinal inflammation by acting on the microbiota. Thus, precision editing of the gut microbiota identifies a bloom of Enterobacteriaceae as a signature of dysbiosis that is causatively linked to an exacerbation of intestinal inflammation during IBD.
Metabolism-Based Editing of the Microbiota: CRC.
Gut dysbiosis is associated with CRC (142), the third most diagnosed cancer worldwide (143). Only about 20% of CRC cases can be attributed to familial history (144), pointing to environmental factors as important contributors to tumorigenesis. A prime candidate for these environmental factors is the presence of pathobionts in the gut microbiota. For example, colibactin-producing E. coli (9), toxin-producing Bacteroides fragilis (phylum Bacteroidota) (145, 146) and Fusobacterium nucleatum (phylum Fusobacteriota) (147) can potentiate intestinal tumorigenesis in animal models. These data establish causative links between the gut microbiota and tumorigenesis, but the evidence they provide to connect enhanced tumor formation with an elevated pathobiont abundance during dysbiosis remains correlative.
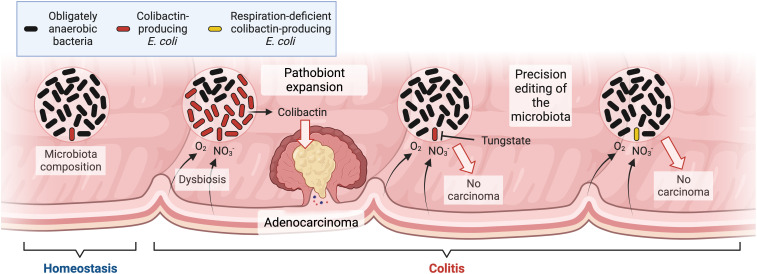
Colibactin-producing E. coli are a good example of a pathobiont with elevated abundance during dysbiosis. Intestinal inflammation is a risk factor for developing CRC (148, 149) and this condition correlates with an increase in the abundance of colibactin-producing E. coli in the gut microbiota (9) (Fig. 3). Colibactin is a genotoxin that alkylates DNA on adenine residues (150, 151) and induces double-strand breaks in cultured cells (152). Between 2% and 6.3% of human CRC cases have a mutational signature characteristic of colibactin (153), demonstrating that colibactin-producing E. coli have a direct role in the occurrence of oncogenic mutations in patients. Interleukin-10-deficient germ-free mice develop colitis and adenocarcinomas upon azoxymethane (AOM) treatment when they are monoassociated with colibactin-producing E. coli, but tumor formation is markedly reduced upon monoassociation with an E. coli mutant deficient for colibactin synthesis (9). Collectively, these data show that inflammation correlates with a dysbiotic expansion of a pathobiont with genotoxic potential.
Fig. 3.
Precision editing of the microbiota causatively links pathobiont expansion during dysbiosis to CRC formation. Colibactin-producing E. coli are minority species within the gut microbiota during homeostasis (Left). Colitis triggers dysbiosis by increasing the availability of host-derived oxygen (O2) and nitrate (NO3−). Oxygen and nitrate fuel growth of colibactin-producing E. coli to promote adenocarcinoma formation (Center), whereas blocking a bloom of the pathobiont through precision editing prevents carcinoma (Right). Created with BioRender.com.
The consequences of a dysbiotic expansion of colibactin-producing E. coli on tumor formation can be explored using precision editing of the gut microbiota. The ecological cause for an elevated abundance of E. coli in DSS/AOM-treated mice is an increased availability of host derived oxygen and nitrate (93, 123). One approach for precision editing of the microbiota is based on Enterobacteriaceae-free mice that are available from some commercial vendors (154). By engrafting littermates of Enterobacteriaceae-free mice with either colibactin-producing E. coli or E. coli mutants lacking pathways required for enhancing their abundance during colitis, it is possible to address the question whether the expansion of a pathobiont in the microbiota exacerbates disease (55). Work in this model shows that DSS/AOM-treated mice develop colitis and adenocarcinomas when they are engrafted with colibactin-producing E. coli, but tumor formation is markedly reduced upon engraftment with an aerobic respiration-deficient E. coli mutant, which can no longer enhance its abundance during inflammation (93). A second approach for selectively blocking an expansion of colibactin-producing E. coli during gut inflammation is tungstate administration, which reduces tumor formation in DSS/AOM-treated mice (123), thus causatively linking pathobiont expansion during dysbiosis to tumorigenesis (Fig. 3).
Collectively, these two approaches for precision editing of the gut microbiota provide complementary evidence that dysbiosis enhances tumorigenesis of the gut microbiota by specifically increasing the abundance of a pathobiont with genotoxic potential (93, 123). Each method for establishing a causative link between changes in the microbiota composition and CRC formation relies on information about the ecological causes of dysbiosis that were discovered through infectious disease research. In turn, the development of methods for precision editing of the microbiota provides a blueprint for investigating the role of dysbiosis in other diseases.
Changes in Microbial Metabolism during Dysbiosis.
The traditional definition of dysbiosis as a shift in the bacterial species configuration (17) focuses research on establishing links between compositional and functional changes in the gut microbiota. However, the concept that dysbiosis is characterized by an underlying change in host physiology (24, 63, 65, 68) raises the possibility that altering the growth conditions of the gut microbiota could shift microbial metabolism, an aspect of dysbiosis that might be missed by microbiota profiling.
One harmful metabolite exclusively derived from the gut microbiota is trimethylamine (TMA), which is produced during the catabolism of choline or carnitine, two nutrients abundant in red meat (155). Gene clusters encoding enzymes for choline or carnitine catabolism are present in phylogenetically diverse members of the gut microbiota, including representatives of the classes Gammaproteobacteria, Deltaproteobacteria (phylum Pseudomonadota), Coriobacteriia (phylum Actinomycetota), Bacilli, and Clostridia (156–158). Microbiota-derived TMA is absorbed by the host and converted by flavin monooxygenases in the liver to the uremic toxin trimethylamine-N-oxide (TMAO) (159). Plasma levels of TMAO are elevated in patients with CVD (155, 160, 161), CKD (162) and type 2 diabetes (163). Microbiota-derived TMAO accelerates disease in mouse models of atherosclerosis (155, 161) and CKD (164, 165). Conversely, targeted inhibition of microbiota-derived TMA production using a structural choline analog attenuates atherosclerosis (166) and slows progression of CKD in mouse models (167). Collectively, these data causatively link gut microbiota-derived TMA to leading causes of human morbidity and mortality.
Gut dysbiosis accompanies CVD, CKD, and type 2 diabetes, and in each disease, the fecal microbiota of patients commonly features an increase in the abundance of taxa within the Gammaproteobacteria, such as E. coli (6, 8, 10, 168). One unanswered question in the field is whether dysbiosis generates a “uremic microbiota” with increased production of metabolites, such as TMA, thereby heightening levels of uremic toxins in the plasma of patients (169). Compositional changes in the microbiota from patients with CKD do not result in increased uremic toxin production during in vitro anaerobic batch culture (170). But a limitation of this approach is that in vitro culture conditions do not resemble the gut environment of patients. Notably, nitrate is required for choline catabolism during in vitro anaerobic batch culture of E. coli (124). In a mouse model of high fat intake, dysbiosis features an elevated E. coli abundance (171) and a raised availability of host-derived oxygen and nitrate in the colonic lumen (124). Inoculation of Enterobacteriaceae-free mice with a TMA-producing E. coli strain increases TMAO levels in the serum when animals receive a high-fat diet supplemented with choline but not when animals are fed a low-fat chow supplemented with choline (124). Importantly, TMA production by Enterobacteriaceae is driven by host-derived nitrate because treatment with aminoguanidine, a chemical inhibitor of the host enzyme iNOS (172), blocks E. coli from increasing the TMAO serum levels in mice receiving high-fat diet supplemented with choline (124). Thus, host-derived nitrate powers choline catabolism of E. coli in vivo, a feature of dysbiosis that cannot be recapitulated during in vitro anaerobic batch culture in the absence of nitrate (170).
The emerging picture implies that an underlying change in host physiology (i.e., increased iNOS synthesis) can shift microbial metabolism (i.e., induce nitrate respiration-dependent choline catabolism in E. coli) to increase the production of uremic toxins during dysbiosis (124). This example illustrates how insights into the ecological causes of dysbiosis can directly inform approaches to tackle the question whether dysbiosis increases production of harmful metabolites by the gut microbiota.
Conclusions
At the dawn of modern microbiome research, massive amounts of data were generated when a theoretical framework seemed elusive, which made it challenging to approach questions using hypothesis-driven research (173). However, rather than rejecting hypothesis-driven research altogether (174), we suggest that understanding our microbiome requires supplementing large datasets and sophisticated algorithms to analyze them with basic microbiological concepts.
Studying how enteric pathogens manipulate host functions that control the growth conditions at mucosal surfaces has taught us that one of the ecological causes of dysbiosis in the large intestine is an increased availability of host-derived electron acceptors. This concept is relevant for understanding dysbiosis in many noninfectious human conditions because the underlying principles apply to microbial communities in general. Specifically, the environment selects for those microbes that use the redox reactions generating the greatest free energy, thus making electron acceptors the dominant drivers of microbial community composition in nature (175). Embracing this theoretical framework goes a long way in explaining the longitudinal and cross-sectional heterogeneity of the gut microbiota (176) and understanding the ecological causes of compositional changes during dysbiosis (53, 67).
The finding that changes in the microbiota composition can contribute to chronic human diseases has focused strategies to remediate dysbiosis on microbiota-based therapeutics, such as probiotics, prebiotics, precision editing of the microbiota, or fecal microbiota transplantation (177). However, the underlying cause for alterations in the microbiota composition is a change in the host environment, which represents an important component of the microbiome (22–24). Thus, in addition to targeting the microbes with microbiota-based therapeutics, dysbiosis can be remediated by targeting the organ in which the microbiota resides in with drugs to normalize host functions that control microbial growth on host surfaces (65). This concept provides a starting point for developing therapeutic strategies to remediate dysbiosis by targeting its underlying cause in the host. Exploring this strategy along with microbiota-based therapeutics might provide alternatives to traditional clinical interventions for a broad spectrum of chronic diseases, which makes this line of inquiry one of the most exciting areas in human microbiome research.
Acknowledgments
Author contributions
S.E.W. and A.J.B. wrote the paper.
Competing interests
US Patent for Tungstate treatment of the dysbiosis associated with gastrointestinal inflammation (Patent # 10,092,596).
Footnotes
Reviewers: E.P.S., Vanderbilt University Medical Center; and J.P.Z., University of Pennsylvania.
Data, Materials, and Software Availability
There are no data underlying this work.
References
- 1.Sender R., Fuchs S., Milo R., Revised estimates for the number of human and bacteria cells in the body. PLoS Biol. 14, e1002533 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Koh A., De Vadder F., Kovatcheva-Datchary P., Backhed F., From dietary fiber to host physiology: Short-chain fatty acids as key bacterial metabolites. Cell 165, 1332–1345 (2016). [DOI] [PubMed] [Google Scholar]
- 3.Levy M., Blacher E., Elinav E., Microbiome, metabolites and host immunity. Curr. Opin. Microbiol. 35, 8–15 (2017). [DOI] [PubMed] [Google Scholar]
- 4.Louis P., Hold G. L., Flint H. J., The gut microbiota, bacterial metabolites and colorectal cancer. Nat. Rev. Microbiol. 12, 661–672 (2014). [DOI] [PubMed] [Google Scholar]
- 5.McCarville J. L., Chen G. Y., Cuevas V. D., Troha K., Ayres J. S., Microbiota metabolites in health and disease. Annu. Rev. Immunol. 38, 147–170 (2020). [DOI] [PubMed] [Google Scholar]
- 6.Karlsson F. H., et al. , Symptomatic atherosclerosis is associated with an altered gut metagenome. Nat. Commun. 3, 1245 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ceccarani C., et al. , Proteobacteria overgrowth and butyrate-producing taxa depletion in the gut microbiota of glycogen storage disease type 1 patients. Metabolites 10, 133 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Qin J., et al. , A metagenome-wide association study of gut microbiota in type 2 diabetes. Nature 490, 55–60 (2012). [DOI] [PubMed] [Google Scholar]
- 9.Arthur J. C., et al. , Intestinal inflammation targets cancer-inducing activity of the microbiota. Science 338, 120–123 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hida M., et al. , Inhibition of the accumulation of uremic toxins in the blood and their precursors in the feces after oral administration of Lebenin, a lactic acid bacteria preparation, to uremic patients undergoing hemodialysis. Nephron 74, 349–355 (1996). [DOI] [PubMed] [Google Scholar]
- 11.Frank D. N., et al. , Molecular-phylogenetic characterization of microbial community imbalances in human inflammatory bowel diseases. Proc. Natl. Acad. Sci. U.S.A. 104, 13780–13785 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cani P. D., Gut microbiota–At the intersection of everything? Nat. Rev. Gastroenterol. Hepatol. 14, 321–322 (2017). [DOI] [PubMed] [Google Scholar]
- 13.Shade A., Handelsman J., Beyond the Venn diagram: The hunt for a core microbiome. Environ. Microbiol. 14, 4–12 (2012). [DOI] [PubMed] [Google Scholar]
- 14.Tap J., et al. , Towards the human intestinal microbiota phylogenetic core. Environ. Microbiol. 11, 2574–2584 (2009). [DOI] [PubMed] [Google Scholar]
- 15.Turnbaugh P. J., et al. , A core gut microbiome in obese and lean twins. Nature 457, 480–484 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yong E., There is no ‘healthy’ microbiome. The New York Times, 2 November 2014, Section SR, p. 4. [Google Scholar]
- 17.Petersen C., Round J. L., Defining dysbiosis and its influence on host immunity and disease. Cell Microbiol. 16, 1024–1033 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Olesen S. W., Alm E. J., Dysbiosis is not an answer. Nat. Microbiol. 1, 16228 (2016). [DOI] [PubMed] [Google Scholar]
- 19.Integrative HMPRNC, The integrative human microbiome project. Nature 569, 641–648 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Proctor L., Priorities for the next 10 years of human microbiome research. Nature 569, 623–625 (2019). [DOI] [PubMed] [Google Scholar]
- 21.Hooper L. V., Gordon J. I., Commensal host-bacterial relationships in the gut. Science 292, 1115–1118 (2001). [DOI] [PubMed] [Google Scholar]
- 22.Tipton L., Darcy J. L., Hynson N. A., A developing symbiosis: Enabling cross-talk between ecologists and microbiome scientists. Front. Microbiol. 10, 292 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Berg G., et al. , Microbiome definition re-visited: Old concepts and new challenges. Microbiome 8, 103 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tiffany C. R., Baumler A. J., Dysbiosis: From fiction to function. Am. J. Physiol. Gastrointest. Liver Physiol. 317, G602–G608 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Eckburg P. B., et al. , Diversity of the human intestinal microbial flora. Science 308, 1635–1638 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lupp C., et al. , Host-mediated inflammation disrupts the intestinal microbiota and promotes the overgrowth of Enterobacteriaceae. Cell Host. Microbe. 2, 204 (2007). [DOI] [PubMed] [Google Scholar]
- 27.Stecher B., et al. , Salmonella enterica serovar typhimurium exploits inflammation to compete with the intestinal microbiota. PLoS Biol. 5, 2177–2189 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Heimesaat M. M., et al. , Gram-negative bacteria aggravate murine small intestinal Th1-type immunopathology following oral infection with Toxoplasma gondii. J. Immunol. 177, 8785–8795 (2006). [DOI] [PubMed] [Google Scholar]
- 29.Spiga L., Winter S. E., Using enteric pathogens to probe the gut microbiota. Trends Microbiol. 27, 243–253 (2019). [DOI] [PubMed] [Google Scholar]
- 30.Lawley T. D., et al. , Host transmission of Salmonella enterica serovar Typhimurium is controlled by virulence factors and indigenous intestinal microbiota. Infect. Immun. 76, 403–416 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Winter S. E., et al. , Gut inflammation provides a respiratory electron acceptor for Salmonella. Nature 467, 426–429 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Muller L., Un nouveau milieu d’enrichissement pour la recherche du Bacille Typhique at Paratyphique. C. R. Seances Soc. Biol. Ses. Fil. 89, 434–437 (1923). [Google Scholar]
- 33.Hohn D. C., Lehrer R. I., NADPH oxidase deficiency in X-linked chronic granulomatous disease. J. Clin. Invest. 55, 707–713 (1975). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Palmer R. M., Ashton D. S., Moncada S., Vascular endothelial cells synthesize nitric oxide from L-arginine. Nature 333, 664–666 (1988). [DOI] [PubMed] [Google Scholar]
- 35.Zhu L., Gunn C., Beckman J. S., Bactericidal activity of peroxynitrite. Arch. Biochem. Biophys. 298, 452–457 (1992). [DOI] [PubMed] [Google Scholar]
- 36.De Groote M. A., et al. , Genetic and redox determinants of nitric oxide cytotoxicity in a Salmonella typhimurium model. Proc. Natl. Acad. Sci. U.S.A. 92, 6399–6403 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Balagam B., Richardson D. E., The mechanism of carbon dioxide catalysis in the hydrogen peroxide N-oxidation of amines. Inorganic chem. 47, 1173–1178 (2008). [DOI] [PubMed] [Google Scholar]
- 38.Lopez C. A., et al. , Phage-mediated acquisition of a type III secreted effector protein boosts growth of salmonella by nitrate respiration. mBio 3, e00143-12 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lopez C. A., Rivera-Chavez F., Byndloss M. X., Baumler A. J., The periplasmic nitrate reductase NapABC supports luminal growth of Salmonella Enterica serovar typhimurium during colitis. Infect. Immun. 83, 3470–3478 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Stewart V., Nitrate respiration in relation to facultative metabolism in enterobacteria. Microbiol. Rev. 52, 190–232 (1988). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang S., et al. , Infection-induced intestinal dysbiosis is mediated by macrophage activation and nitrate production. mBio 10, e00935-19 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Winter S. E., Lopez C. A., Baumler A. J., The dynamics of gut-associated microbial communities during inflammation. Embo. Rep. 14, 319–327 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Barman M., et al. , Enteric salmonellosis disrupts the microbial ecology of the murine gastrointestinal tract. Infect. Immun. 76, 907–915 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sekirov I., et al. , Salmonella SPI-1-mediated neutrophil recruitment during enteric colitis is associated with reduction and alteration in intestinal microbiota. Gut. Microbes 1, 30–41 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gill N., et al. , Neutrophil elastase alters the murine gut microbiota resulting in enhanced Salmonella colonization. PLoS One 7, e49646 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Vital M., Howe A. C., Tiedje J. M., Revealing the bacterial butyrate synthesis pathways by analyzing (meta)genomic data. mBio 5, e00889 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Louis P., Flint H. J., Diversity, metabolism and microbial ecology of butyrate-producing bacteria from the human large intestine. FEMS Microbiol. Lett. 294, 1–8 (2009). [DOI] [PubMed] [Google Scholar]
- 48.Rivera-Chavez F., et al. , Depletion of butyrate-producing clostridia from the gut microbiota drives an aerobic luminal expansion of Salmonella. Cell Host. Microbe 19, 443–454 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Donohoe D. R., et al. , The microbiome and butyrate regulate energy metabolism and autophagy in the mammalian colon. Cell Metab. 13, 517–526 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Roediger W. E., Role of anaerobic bacteria in the metabolic welfare of the colonic mucosa in man. Gut 21, 793–798 (1980). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kelly C. J., et al. , Crosstalk between microbiota-derived short-chain fatty acids and intestinal epithelial HIF augments tissue barrier function. Cell Host. Microbe. 17, 662–671 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Furuta G. T., et al. , Hypoxia-inducible factor 1-dependent induction of intestinal trefoil factor protects barrier function during hypoxia. J. Exp. Med. 193, 1027–1034 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Litvak Y., Byndloss M. X., Baumler A. J., Colonocyte metabolism shapes the gut microbiota. Science 362, eaat9076 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lopez C. A., et al. , Virulence factors enhance Citrobacter rodentium expansion through aerobic respiration. Science 353, 1249–1253 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Rivera-Chavez F., Lopez C. A., Baumler A. J., Oxygen as a driver of gut dysbiosis. Free Radic. Biol. Med. 105, 93–101 (2017). [DOI] [PubMed] [Google Scholar]
- 56.Barthold S. W., Coleman G. L., Bhatt P. N., Osbaldiston G. W., Jonas A. M., The etiology of transmissible murine colonic hyperplasia. Lab. Anim. Sci. 26, 889–894 (1976). [PubMed] [Google Scholar]
- 57.Barthold S. W., Coleman G. L., Jacoby R. O., Livestone E. M., Jonas A. M., Transmissible murine colonic hyperplasia. Vet. Pathol. 15, 223–236 (1978). [DOI] [PubMed] [Google Scholar]
- 58.Fan Y. Y., et al. , A bioassay to measure energy metabolism in mouse colonic crypts, organoids, and sorted stem cells. Am. J. Physiol. Gastrointest. Liver Physiol. 309, G1–G9 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Stouthamer A. H., A theoretical study on the amount of ATP required for synthesis of microbial cell material. Antonie Van Leeuwenhoek 39, 545–565 (1973). [DOI] [PubMed] [Google Scholar]
- 60.Hadjipetrou L. P., Stouthamer A. H., ENergy production during nitrate respiration by Aerobacter Aerogenes. J. Gen. Microbiol. 38, 29–34 (1965). [DOI] [PubMed] [Google Scholar]
- 61.Martiny A. C., Treseder K., Pusch G., Phylogenetic conservatism of functional traits in microorganisms. ISME J. 7, 830–838 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Pascal Andreu V., et al. , gutSMASH predicts specialized primary metabolic pathways from the human gut microbiota. Nat. Biotechnol. 41, 1416–1423 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Rogers A. W. L., Tsolis R. M., Baumler A. J., Salmonella versus the Microbiome. Microbiol. Mol. Biol. Rev. 85, e00027-19 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Tsolis R. M., Baumler A. J., Gastrointestinal host-pathogen interaction in the age of microbiome research. Curr. Opin. Microbiol. 53, 78–89 (2020). [DOI] [PubMed] [Google Scholar]
- 65.Lee J. Y., Tsolis R. M., Baumler A. J., The microbiome and gut homeostasis. Science 377, eabp9960 (2022). [DOI] [PubMed] [Google Scholar]
- 66.Shin N. R., Whon T. W., Bae J. W., Proteobacteria: Microbial signature of dysbiosis in gut microbiota. Trends Biotechnol. 33, 496–503 (2015). [DOI] [PubMed] [Google Scholar]
- 67.Litvak Y., Byndloss M. X., Tsolis R. M., Baumler A. J., Dysbiotic Proteobacteria expansion: A microbial signature of epithelial dysfunction. Curr. Opin. Microbiol. 39, 1–6 (2017). [DOI] [PubMed] [Google Scholar]
- 68.Byndloss M. X., Pernitzsch S. R., Baumler A. J., Healthy hosts rule within: Ecological forces shaping the gut microbiota. Mucosal. Immunol. 11, 1299–1305 (2018). [DOI] [PubMed] [Google Scholar]
- 69.Wu G., Zhao N., Zhang C., Lam Y. Y., Zhao L., Guild-based analysis for understanding gut microbiome in human health and diseases. Genome Med. 13, 22 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Potgens S. A., et al. , Klebsiella oxytoca expands in cancer cachexia and acts as a gut pathobiont contributing to intestinal dysfunction. Sci. Rep. 8, 12321 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Wang Z., et al. , Gut microbial dysbiosis is associated with development and progression of radiation enteritis during pelvic radiotherapy. J. Cell Mol. Med. 23, 3747–3756 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Zhu L., et al. , Characterization of gut microbiomes in nonalcoholic steatohepatitis (NASH) patients: A connection between endogenous alcohol and NASH. Hepatology 57, 601–609 (2013). [DOI] [PubMed] [Google Scholar]
- 73.Montassier E., et al. , 16S rRNA gene pyrosequencing reveals shift in patient faecal microbiota during high-dose chemotherapy as conditioning regimen for bone marrow transplantation. Microb. Ecol. 67, 690–699 (2014). [DOI] [PubMed] [Google Scholar]
- 74.Pham T. P., et al. , Gut microbiota alteration is characterized by a proteobacteria and fusobacteria bloom in Kwashiorkor and a bacteroidetes paucity in Marasmus. Sci. Rep. 9, 9084 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 75.Shintouo C. M., et al. , Is inflammageing influenced by the microbiota in the aged gut? A systematic review. Exp. Gerontol. 141, 111079 (2020). [DOI] [PubMed] [Google Scholar]
- 76.Dubinkina V. B., et al. , Links of gut microbiota composition with alcohol dependence syndrome and alcoholic liver disease. Microbiome 5, 141 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Martinez-Medina M., et al. , Western diet induces dysbiosis with increased E coli in CEABAC10 mice, alters host barrier function favouring AIEC colonisation. Gut 63, 116–124 (2014). [DOI] [PubMed] [Google Scholar]
- 78.Shield K. D., Parry C., Rehm J., Chronic diseases and conditions related to alcohol use. Alcohol. Res. 35, 155–173 (2013). [PMC free article] [PubMed] [Google Scholar]
- 79.Schwab U., et al. , Effect of the amount and type of dietary fat on cardiometabolic risk factors and risk of developing type 2 diabetes, cardiovascular diseases, and cancer: A systematic review. Food Nutr. Res. 58, 10.3402/fnr.v58.25145 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Podolsky D. K., Inflammatory bowel disease. N. Engl. J. Med. 347, 417–429 (2002). [DOI] [PubMed] [Google Scholar]
- 81.Lepage P., et al. , Twin study indicates loss of interaction between microbiota and mucosa of patients with ulcerative colitis. Gastroenterology 141, 227–236 (2011). [DOI] [PubMed] [Google Scholar]
- 82.Machiels K., et al. , A decrease of the butyrate-producing species Roseburia hominis and Faecalibacterium prausnitzii defines dysbiosis in patients with ulcerative colitis. Gut 63, 1275–1283 (2014). [DOI] [PubMed] [Google Scholar]
- 83.Rigottier-Gois L., Dysbiosis in inflammatory bowel diseases: The oxygen hypothesis. ISME J. 7, 1256–1261 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Lundberg J. O., Hellstrom P. M., Lundberg J. M., Alving K., Greatly increased luminal nitric oxide in ulcerative colitis. Lancet 344, 1673–1674 (1994). [DOI] [PubMed] [Google Scholar]
- 85.Reinders C. A., et al. , Rectal nitric oxide and fecal calprotectin in inflammatory bowel disease. Scand. J. Gastroenterol. 42, 1151–1157 (2007). [DOI] [PubMed] [Google Scholar]
- 86.Winter S. E., et al. , Host-derived nitrate boosts growth of E. coli in the inflamed gut. Science 339, 708–711 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Roediger W. E., The colonic epithelium in ulcerative colitis: An energy-deficiency disease? Lancet 2, 712–715 (1980). [DOI] [PubMed] [Google Scholar]
- 88.Santhanam S., et al. , Mitochondrial electron transport chain complex dysfunction in the colonic mucosa in ulcerative colitis. Inflamm. Bowel. Dis. 18, 2158–2168 (2012). [DOI] [PubMed] [Google Scholar]
- 89.Sifroni K. G., et al. , Mitochondrial respiratory chain in the colonic mucosal of patients with ulcerative colitis. Mol. Cell Biochem. 342, 111–115 (2010). [DOI] [PubMed] [Google Scholar]
- 90.Hughes E. R., et al. , Microbial respiration and formate oxidation as metabolic signatures of inflammation-associated dysbiosis. Cell Host. Microbe 21, 208–219 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Laukens D., et al. , Tauroursodeoxycholic acid inhibits experimental colitis by preventing early intestinal epithelial cell death. Lab. Invest. 94, 1419–1430 (2014). [DOI] [PubMed] [Google Scholar]
- 92.Cao S. S., et al. , The unfolded protein response and chemical chaperones reduce protein misfolding and colitis in mice. Gastroenterology 144, 989–1000.e6 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Cevallos S. A., et al. , Increased epithelial oxygenation links colitis to an expansion of tumorigenic bacteria. mBio 10, e02244-19 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Cevallos S. A., et al. , 5-aminosalicylic acid ameliorates colitis and checks dysbiotic Escherichia coli expansion by activating PPAR-gamma signaling in the intestinal epithelium. mBio 12, e03227-20 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Campbell E. L., et al. , Transmigrating neutrophils shape the mucosal microenvironment through localized oxygen depletion to influence resolution of inflammation. Immunity 40, 66–77 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Chanin R. B., et al. , Epithelial-derived reactive oxygen species enable AppBCX-mediated aerobic respiration of Escherichia coli during intestinal inflammation. Cell Host. Microbe 28, 780–788.e5 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Vollaard E. J., Clasener H. A., Janssen A. J., Co-trimoxazole impairs colonization resistance in healthy volunteers. J. Antimicrob. Chemother. 30, 685–691 (1992). [DOI] [PubMed] [Google Scholar]
- 98.Ubeda C., et al. , Vancomycin-resistant Enterococcus domination of intestinal microbiota is enabled by antibiotic treatment in mice and precedes bloodstream invasion in humans. J. Clin. Invest. 120, 4332–4341 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Taur Y., et al. , Intestinal domination and the risk of bacteremia in patients undergoing allogeneic hematopoietic stem cell transplantation. Clin. Infect. Dis. 55, 905–914 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Tamburini F. B., et al. , Precision identification of diverse bloodstream pathogens in the gut microbiome. Nat. Med. 24, 1809–1814 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Sekirov I., et al. , Antibiotic-induced perturbations of the intestinal microbiota alter host susceptibility to enteric infection. Infect. Immun. 76, 4726–4736 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Hattori K., et al. , Gut microbiota prevents sugar alcohol-induced diarrhea. Nutrients 13, 2029 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Schubert A. M., Sinani H., Schloss P. D., Antibiotic-induced alterations of the murine gut microbiota and subsequent effects on colonization resistance against clostridium difficile. mBio 6, e00974 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Myhal M. L., Laux D. C., Cohen P. S., Relative colonizing abilities of human fecal and K 12 strains of Escherichia coli in the large intestines of streptomycin-treated mice. Eur. J. Clin. Microbiol. 1, 186–192 (1982). [DOI] [PubMed] [Google Scholar]
- 105.Bohnhoff M., Drake B. L., Miller C. P., Effect of streptomycin on susceptibility of intestinal tract to experimental Salmonella infection. Proc. Soc. Exp. Biol. Med. 86, 132–137 (1954). [DOI] [PubMed] [Google Scholar]
- 106.Saito K., Studies on the habitation of pathogenic Escherichia coli in the intestinal tract of mice. I. Comparative experiments on the habitation of each type of resistant pathogenic Escherichia coli under an administration of streptomycin. Paediatr. Jpn. 65, 385–393 (1961). [PubMed] [Google Scholar]
- 107.Bohnhoff M., Miller C. P., Enhanced susceptibility to Salmonella infection in streptomycin-treated mice. J. Infect. Dis. 111, 117–127 (1962). [DOI] [PubMed] [Google Scholar]
- 108.Miller C. P., Bohnhoff M., Changes in the mouse’s enteric microflora associated with enhanced susceptibility to salmonella infection following streptomycin treatment. J. Infect. Dis. 113, 59–66 (1963). [DOI] [PubMed] [Google Scholar]
- 109.Bohnhoff M., Miller C. P., Martin W. R., Resistance of the mouse’s intestinal tract to experimental salmonella infection. Ii. Factors responsible for its loss following streptomycin treatment. J. Exp. Med. 120, 817–828 (1964). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Meynell G. G., Antibacterial mechanisms of the mouse gut. II. The role of Eh and volatile fatty acids in the normal gut. Br. J. Exp. Pathol. 44, 209–219 (1963). [PMC free article] [PubMed] [Google Scholar]
- 111.Spees A. M., et al. , Streptomycin-induced inflammation enhances Escherichia coli gut colonization through nitrate respiration. mBio 4, e00935-13 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Lefebvre M., et al. , Peroxisome proliferator-activated receptor gamma is induced during differentiation of colon epithelium cells. J. Endocrinol. 162, 331–340 (1999). [DOI] [PubMed] [Google Scholar]
- 113.Tylichova Z., et al. , Activation of autophagy and PPARgamma protect colon cancer cells against apoptosis induced by interactive effects of butyrate and DHA in a cell type-dependent manner: The role of cell differentiation. J. Nutr. Biochem. 39, 145–155 (2017). [DOI] [PubMed] [Google Scholar]
- 114.Alex S., et al. , Short-chain fatty acids stimulate angiopoietin-like 4 synthesis in human colon adenocarcinoma cells by activating peroxisome proliferator-activated receptor gamma. Mol. Cell Biol. 33, 1303–1316 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Marion-Letellier R., Butler M., Dechelotte P., Playford R. J., Ghosh S., Comparison of cytokine modulation by natural peroxisome proliferator-activated receptor gamma ligands with synthetic ligands in intestinal-like Caco-2 cells and human dendritic cells–potential for dietary modulation of peroxisome proliferator-activated receptor gamma in intestinal inflammation. Am. J. Clin. Nutr. 87, 939–948 (2008). [DOI] [PubMed] [Google Scholar]
- 116.Duszka K., Oresic M., Le May C., Konig J., Wahli W., PPARgamma modulates long chain fatty acid processing in the intestinal epithelium. Int. J. Mol. Sci. 18, 2559 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Byndloss M. X., et al. , Microbiota-activated PPAR-g signaling inhibits dysbiotic Enterobacteriaceae expansion. Science 357, 570–575 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Atarashi K., et al. , Induction of colonic regulatory T cells by indigenous Clostridium species. Science 331, 337–341 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Arpaia N., et al. , Metabolites produced by commensal bacteria promote peripheral regulatory T-cell generation. Nature 504, 451–455 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Furusawa Y., et al. , Commensal microbe-derived butyrate induces the differentiation of colonic regulatory T cells. Nature 504, 446–450 (2013). [DOI] [PubMed] [Google Scholar]
- 121.Smith P. M., et al. , The microbial metabolites, short-chain fatty acids, regulate colonic Treg cell homeostasis. Science 341, 569–573 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Wilson R. P., et al. , STAT2 dependent Type I Interferon response promotes dysbiosis and luminal expansion of the enteric pathogen Salmonella Typhimurium. PLoS Pathog. 15, e1007745 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Zhu W., et al. , Editing of the gut microbiota reduces carcinogenesis in mouse models of colitis-associated colorectal cancer. J. Exp. Med. 216, 2378–2393 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Yoo W., et al. , High-fat diet–induced colonocyte dysfunction escalates microbiota-derived trimethylamine N-oxide. Science 373, 813–818 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Seike K., et al. , Ambient oxygen levels regulate intestinal dysbiosis and GVHD severity after allogeneic stem cell transplantation. Immunity 56, 353–368.e6 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Rath E., Haller D., Intestinal epithelial cell metabolism at the interface of microbial dysbiosis and tissue injury. Mucosal. Immunol. 15, 595–604 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Wiley C. D., et al. , Mitochondrial dysfunction induces senescence with a distinct secretory phenotype. Cell Metab. 23, 303–314 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Han C., Geng Q., Qin J., Li Y., Yu H., Activation of 5-hydroxytryptamine 4 receptor improves colonic barrier function by triggering mucin 2 production in a mouse model of type 1 diabetes. Am. J. Pathol. 192, 876–886 (2022). [DOI] [PubMed] [Google Scholar]
- 129.Brozinsky S., Fani K., Grosberg S. J., Wapnick S., Alcohol ingestion-induced changes in the human rectal mucosa: Light and electron microscopic studies. Dis. Colon. Rectum. 21, 329–335 (1978). [DOI] [PubMed] [Google Scholar]
- 130.Khan K. J., et al. , Antibiotic therapy in inflammatory bowel disease: A systematic review and meta-analysis. Am. J. Gastroenterol. 106, 661–673 (2011). [DOI] [PubMed] [Google Scholar]
- 131.Schaubeck M., et al. , Dysbiotic gut microbiota causes transmissible Crohn’s disease-like ileitis independent of failure in antimicrobial defence. Gut 65, 225–237 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Couturier-Maillard A., et al. , NOD2-mediated dysbiosis predisposes mice to transmissible colitis and colorectal cancer. J. Clin. Invest. 123, 700–711 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Garrett W. S., et al. , Enterobacteriaceae act in concert with the gut microbiota to induce spontaneous and maternally transmitted colitis. Cell Host. Microbe. 8, 292–300 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Baldelli V., Scaldaferri F., Putignani L., Del Chierico F., The role of enterobacteriaceae in gut microbiota dysbiosis in inflammatory bowel diseases. Microorganisms 9, 697 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Sartor R. B., Microbial influences in inflammatory bowel diseases. Gastroenterology 134, 577–594 (2008). [DOI] [PubMed] [Google Scholar]
- 136.Ambrose N. S., Johnson M., Burdon D. W., Keighley M. R., Incidence of pathogenic bacteria from mesenteric lymph nodes and ileal serosa during Crohn’s disease surgery. Br. J. Surg. 71, 623–625 (1984). [DOI] [PubMed] [Google Scholar]
- 137.Gennis R. B., Stewart V., “Respiration” in Escherichia coli and Salmonella: Cellular and Molecular Biology, F. C. Neidhardt et al. , Eds. (ASM Press, Washington, D.C., ed. 2, 1996), vol. 1, pp. 217–261. [Google Scholar]
- 138.Gates A. J., et al. , Properties of the periplasmic nitrate reductases from Paracoccus pantotrophus and Escherichia coli after growth in tungsten-supplemented media. FEMS Microbiol. Lett. 220, 261–269 (2003). [DOI] [PubMed] [Google Scholar]
- 139.Zhu W., et al. , Precision editing of the gut microbiota ameliorates colitis. Nature 553, 208–211 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Qin Y. T., et al. , Colonic mucus-accumulating tungsten oxide nanoparticles improve the colitis therapy by targeting Enterobacteriaceae. Nano Today 39, 101234 (2021). [Google Scholar]
- 141.Yang J., et al. , Bionic regulators break the ecological niche of pathogenic bacteria for modulating dysregulated microbiome in colitis. Adv. Mater. 34, e2204650 (2022). [DOI] [PubMed] [Google Scholar]
- 142.Sobhani I., et al. , Microbial dysbiosis in colorectal cancer (CRC) patients. PLoS One 6, e16393 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Ferlay J., et al. , Cancer incidence and mortality worldwide: Sources, methods and major patterns in GLOBOCAN 2012. Int. J. Cancer 136, E359–E386 (2015). [DOI] [PubMed] [Google Scholar]
- 144.Rustgi A. K., The genetics of hereditary colon cancer. Genes. Dev. 21, 2525–2538 (2007). [DOI] [PubMed] [Google Scholar]
- 145.Wu S., et al. , A human colonic commensal promotes colon tumorigenesis via activation of T helper type 17 T cell responses. Nat. Med. 15, 1016–1022 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Toprak N. U., et al. , A possible role of Bacteroides fragilis enterotoxin in the aetiology of colorectal cancer. Clin. Microbiol. Infect. 12, 782–786 (2006). [DOI] [PubMed] [Google Scholar]
- 147.Kostic A. D., et al. , Fusobacterium nucleatum potentiates intestinal tumorigenesis and modulates the tumor-immune microenvironment. Cell Host. Microbe. 14, 207–215 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Ullman T. A., Itzkowitz S. H., Intestinal inflammation and cancer. Gastroenterology 140, 1807–1816 (2011). [DOI] [PubMed] [Google Scholar]
- 149.Grivennikov S. I., Inflammation and colorectal cancer: Colitis-associated neoplasia. Semin. Immunopathol. 35, 229–244 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Wilson M. R., et al. , The human gut bacterial genotoxin colibactin alkylates DNA. Science 363, eaar7785 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Xue M., et al. , Structure elucidation of colibactin and its DNA cross-links. Science 365, eaax2685 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Nougayrede J. P., et al. , Escherichia coli induces DNA double-strand breaks in eukaryotic cells. Science 313, 848–851 (2006). [DOI] [PubMed] [Google Scholar]
- 153.Pleguezuelos-Manzano C., et al. , Mutational signature in colorectal cancer caused by genotoxic pks(+) E. coli. Nature 580, 269–273 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Velazquez E. M., et al. , Endogenous Enterobacteriaceae underlie variation in susceptibility to Salmonella infection. Nat. Microbiol. 4, 1057–1064 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Wang Z., et al. , Gut flora metabolism of phosphatidylcholine promotes cardiovascular disease. Nature 472, 57–63 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Martinez-del Campo A., et al. , Characterization and detection of a widely distributed gene cluster that predicts anaerobic choline utilization by human gut bacteria. mBio 6, e00042-15 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Zhu Y., et al. , Carnitine metabolism to trimethylamine by an unusual Rieske-type oxygenase from human microbiota. Proc. Natl. Acad. Sci. U.S.A. 111, 4268–4273 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Rath S., Rud T., Pieper D. H., Vital M., Potential TMA-producing bacteria are ubiquitously found in mammalia. Front. Microbiol. 10, 2966 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Lang D. H., et al. , Isoform specificity of trimethylamine N-oxygenation by human flavin-containing monooxygenase (FMO) and P450 enzymes: Selective catalysis by FMO3. Biochem. Pharmacol. 56, 1005–1012 (1998). [DOI] [PubMed] [Google Scholar]
- 160.Tang W. H., et al. , Intestinal microbial metabolism of phosphatidylcholine and cardiovascular risk. N. Engl. J. Med. 368, 1575–1584 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Koeth R. A., et al. , Intestinal microbiota metabolism of L-carnitine, a nutrient in red meat, promotes atherosclerosis. Nat. Med. 19, 576–585 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Rhee E. P., et al. , A combined epidemiologic and metabolomic approach improves CKD prediction. J. Am. Soc. Nephrol. 24, 1330–1338 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Liu W., et al. , Elevated plasma trimethylamine-N-oxide levels are associated with diabetic retinopathy. Acta Diabetol. 58, 221–229 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Xie F., et al. , Dietary choline, via gut microbe- generated trimethylamine-N- oxide, aggravates chronic kidney disease-induced cardiac dysfunction by inhibiting hypoxia-induced factor 1alpha. Front. Physiol. 13, 996166 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Dong F., et al. , Trimethylamine N-oxide promotes hyperoxaluria-induced calcium oxalate deposition and kidney injury by activating autophagy. Free Radic. Biol. Med. 179, 288–300 (2022). [DOI] [PubMed] [Google Scholar]
- 166.Wang Z., et al. , Non-lethal inhibition of gut microbial trimethylamine production for the treatment of atherosclerosis. Cell 163, 1585–1595 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Zhang W., et al. , Inhibition of microbiota-dependent TMAO production attenuates chronic kidney disease in mice. Sci. Rep. 11, 518 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Xu K. Y., et al. , Impaired renal function and dysbiosis of gut microbiota contribute to increased trimethylamine-N-oxide in chronic kidney disease patients. Sci. Rep. 7, 1445 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Koppe L., Soulag C. O., The impact of dietary nutrient intake on gut microbiota in the progression and complications of chronic kidney disease. Kidney Int. 102, 728–739 (2022). [DOI] [PubMed] [Google Scholar]
- 170.Gryp T., et al. , Gut microbiota generation of protein-bound uremic toxins and related metabolites is not altered at different stages of chronic kidney disease. Kidney Int. 97, 1230–1242 (2020). [DOI] [PubMed] [Google Scholar]
- 171.Anitha M., et al. , Intestinal dysbiosis contributes to the delayed gastrointestinal transit in high-fat diet fed mice. Cell Mol. Gastroenterol. Hepatol. 2, 328–339 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Glyan’ko A. K., Initiation of nitric oxide (NO) synthesis in roots of etiolated seedlings of pea (Pisum sativum L.) under the influence of nitrogen-containing compounds. Biochemistry (Mosc) 78, 471–476 (2013). [DOI] [PubMed] [Google Scholar]
- 173.Tripathi A., et al. , Are microbiome studies ready for hypothesis-driven research? Curr. Opin. Microbiol. 44, 61–69 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Mazzocchi F., Could big data be the end of theory in science?: A few remarks on the epistemology of data-driven science. Embo. Rep. 16, 1250–1255 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Stahl D. A., Hullar M., Davidson S., “The structure and function of microbial communities” in Prokaryotes: A Handbook on the Biology of Bacteria, (Springer, ed. 3, 2006), vol. 1, pp. 299–327. [Google Scholar]
- 176.Miller B. M., Liou M. J., Lee J. Y., Baumler A. J., The longitudinal and cross-sectional heterogeneity of the intestinal microbiota. Curr. Opin. Microbiol. 63, 221–230 (2021). [DOI] [PubMed] [Google Scholar]
- 177.Peterson C. T., Sharma V., Elmen L., Peterson S. N., Immune homeostasis, dysbiosis and therapeutic modulation of the gut microbiota. Clin. Exp. Immunol. 179, 363–377 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
There are no data underlying this work.