Abstract
A novel predatory bacterium, strain LBG001T, has been isolated from Reynosa, Mexico. The 16S rRNA shares approximately 97 % sequence identity with many reported strains in the genus Bdellovibrio including the type strain Bdellovibrio bacteriovorus HD100T. Phylogenetic trees based on the 16S rRNA gene and on 30 concatenated housekeeping genes or core genes showed that LBG001T is on a separate branch from the B. bacteriovorus group. LBG0001T has a genome size of 3 582 323 bp with a G+C content of 43.1 mol %. The average nucleotide identity, average amino acid identity and digital DNA–DNA hybridization values with other members of the genus Bdellovibrio (<79, <72 and <17 %, respectively) qualifies the strain to represent a new species in the genus. Strain LBG001T formed visible plaques on all 10 tested Gram-negative bacterial species. The phenotypic characteristics, phylogenetic analysis and genomic taxonomic studies support the classification of the strain as representing a new species for which the name Bdellovibrio reynosensis sp. nov. is proposed. The type strain is LBG001T(=ATCC TSD-288T =CM-CNRG 0932T).
Keywords: Bdellovibrio reynosensis, genome comparison, genome sequence, new species, predatory bacteria
Introduction
Bdellovibrio species are obligate predators of Gram-negative bacteria. They are small, motile and comma-shaped with a single polar flagellum. They belong to the class Oligoflexia. This class contains other obligate predatory bacteria such as Bacteriovorax , Halobacteriovorax and Peredibacter , as well as the non-predatory bacteria Silvanigrella , Fluviispira and Oligoflexus [1–5]. While Halobacteriovorax is salt-tolerant and mostly isolated from marine environments, Bdellovibrio , Bacteriovorax and Peredibacter are isolated from soil and freshwater environments [4]. Of the predatory bacteria in the Oligoflexia group, Bdellovibrio is the most studied. The Bdellovibrio predation strategy can be periplasmic or epibiotic [6, 7]. Bdellovibrio bacteriovorus HD100T is the type strain for periplasmic predators and Bdellovibrio exovorus JSST is the type strain for epibiotic predators, which was recently separated from Bdellovibrio as a new genus Pseudobdellovibrio [8]. Isolation of Bdellovibrio species can be challenging due to low abundance and the need for a correct choice of prey and culture conditions. Most studies in the Bdellovibrio make use of the type strain B. bacteriovorus HD100T or 109J as they have a wide prey range and their genomic, phenotypic and life-cycle have been well studied.
Bdellovibrio species are increasingly being explored as alternatives to antibiotics and probiotics in agriculture, food and medicine. There is a need to isolate and characterize more Bdellovibrio species as their prey preference and predation efficiency differ. In our attempt to obtain Bdellovibrio for potential use in food preservation, we isolated a strain and herein confirm it to represent a new species in the genus Bdellovibrio . This study presents the phenotypic and genomic characteristics of the novel strain LBG001T.
Isolation and ecology
Strain LBG001T was isolated from soil using Klebsiella oxytocoa as the prey. The soil sample was collected at 26.0698623° N, −98.3135799° W, Reynosa, Tamaulipas, Mexico on 16 September, 2019. It was collected from the rhizosphere of an Agave plant in a dark agricultural soil. The samples were taken to the lab and 50 g soil was moved into 100 ml sterile HEPES buffer. The mixture was placed in a shaker at 150 r.p.m. for 2 h at 30 °C. Then it was filtered using a 0.45 µm sterile filter. The filtrate was used for co-culture with K. oxytoca to allow for the enrichment of predatory bacteria. The co-culture was set up in a shaker at 30 °C at 150 r.p.m. for 48 h. The culture was centrifuged at 1 308 g for 30 min at 4 °C and the supernatant was collected for the use of the double-layer agar plate technique [9]. A 100 µl serially diluted coculture supernatant was mixed with a 300 µl suspension of prepared K. oxytoca in 5 ml 55°C-molten diluted nutrient broth (DNB) top agar. The mixtures were immediately vortexed and plated on top of DNB bottom agar plates. Plates were incubated at 30 °C for 10 days. Plates were monitored daily for the presence of plaques. Plaques that appeared after 48 h were considered probable Bdellovibrio . Well-separated plaques were observed on plates at a dilution factor of 4. The plaques were then observed for differences in morphological characteristics. The plaques appeared small, circular and transparent. Therefore, a single representative plaque was chosen and purified for three more times to obtain a pure culture. The isolate was identified by PCR using the 16S rRNA gene-specific primers for Bdellovibrio [10]. The purified Bdellovibrio strain was cryopreserved at −80 °C in 20 % glycerol.
Phylogeny
The Bdellovibrio strain was revived from the cryopreserved stock vial by predation on K. oxytoca as described previously [11]. The coculture was centrifuged at 1 308 g for 30 min at 4 °C to remove residual prey cells. The supernatant was filtered through a 0.45 µm filter. The filtrate was confirmed to be prey-free by streaking on a lysogeny broth agar plate, which was then incubated at 37 °C. The Bdellovibrio were collected by centrifugation at 36, 286 g at 4 °C for 30 min. DNA was extracted from the concentrated cells using the WizardGenomic DNA Purification kit (Promega) following the manufacturer’s instructions. Whole-genome sequencing was done by combining short-read and long-read data. The short-read data was generated using the Nextera XT DNA Library Prep kit (Illumina) and the NextSeq 550 sequencing instrument (Illumina). The long-read data was generated using the Native Barcoding Kit (Nanopore) and MinION instrument (Nanopore). A de novo assembly of reads was carried out on the patric platform and the online Medusa web server [12, 13]. The assembled genome was comprised of a single circular chromosome. This genome sequence is deposited on the GenBank with accession number CP093442.
We made use of the 16S rRNA gene, housekeeping genes and whole-genome sequences in our phylogenetic analysis. LBG001T encodes two identical 16 S rRNA genes separated by 804 827 bp. The 16S rRNA nucleotide sequences were first aligned with muscle and a neighbour-joining phylogenetic tree was reconstructed on mega X using default parameters at the bootstrap value of 1000 replicates. Bacteriovorax stolpii DSM 12778T was one of the outgroup members (See Table S1, available in the online version of this article for details). We further did a similar analysis based on concatenated sequences of 30 housekeeping genes on mega X (supplementary materials, Text 1) as well as that based on the concatenated sequences of orthologous core genes, which were obtained using the BPGA pipeline.
blast analysis of the 16S rRNA gene sequence on the EzBioCloud database (www.ezbiocloud.net/identify) identified the organism to be a member of the genus Bdellovibrio . The result reported its top match as B. bacteriovorus strain R0, the third being HD100T with a percentage identity of 97.63 and 97.14 respectively. The 16S rRNA gene sequence comparison was performed on the EMBL-EBI server. The percentage identity of ≤97 % across the tested members of this genus (Tables S1 and S2) suggests that the newly isolated strain LBG001T is a member of a new species in the genus Bdellovibrio [14, 15].
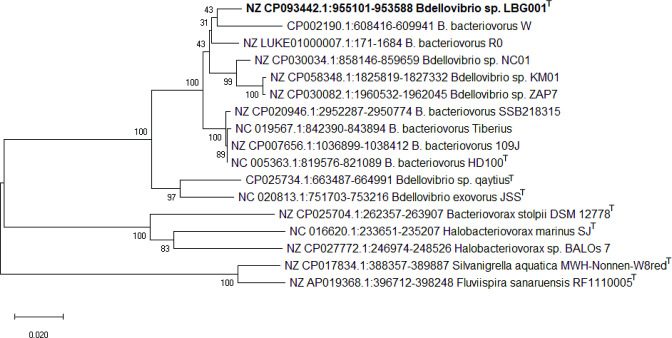
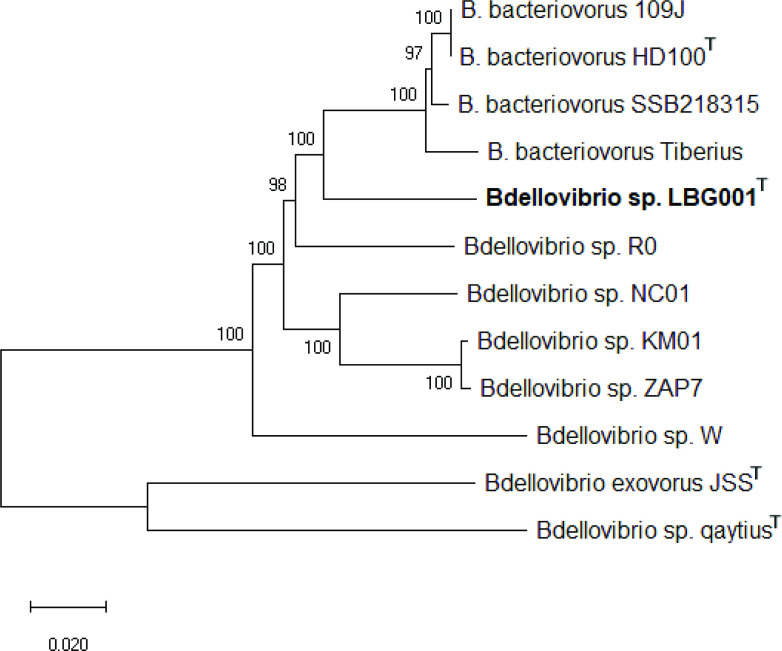
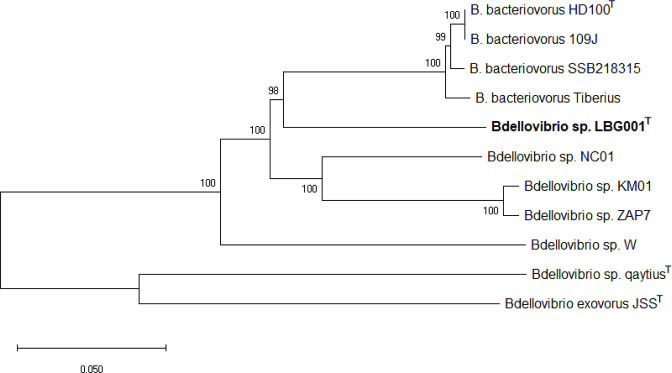
The 16S rRNA neighbour-joining phylogenetic tree showed LBG001T, B. bacteriovorus W and B. bacteriovorus R0 branching out far away from the B. bacteriovorus HD100T, supporting the proposal for the re-designation of B. bacteriovorus R0 and B. bacteriovorus W as Bdellovibrio sp. R0 and Bdellovibrio sp. W, respectively (Fig. 1). The redesignation of these strains is further confirmed by the concatenated 30 housekeeping genes (Fig. 2). The phylogenetic tree based on concatenated core genes showed that LBG001T does not belong to any known species of Bdellovibrio strains as it formed a distinct branch on the tree (Fig. 3). R0 did not occur in Fig. 3 due to its incomplete genome sequence.
Fig. 1.
Neighbour-joining tree reconstructed using 16S rRNA gene sequences.
Fig. 2.
Concatenated phylogenetic tree of 30 housekeeping genes showing the relatedness of Bdellovibrio reynosensis sp. nov. to the type strain B. bacteriovorus HD100T.
Fig. 3.
The concatenated phylogenetic tree of core genes shared by 11 complete whole-genome sequenced Bdellovibrio . Concatenated core genes were recovered from the BPGA analysis. The tree was reconstructed on mega X using default parameters at 1000 bootstraps.
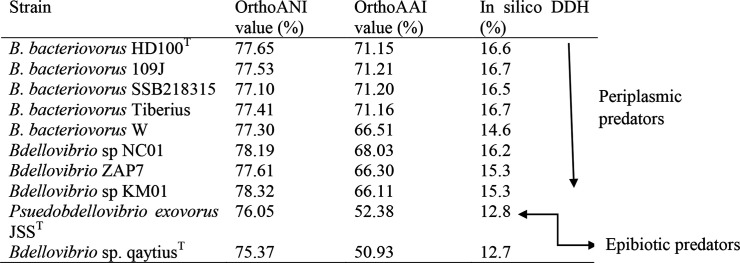
The digital DNA–DNA hybridization (DDH) values of LBG001T against the 10 most closely related species were calculated using the Genome-to-Genome Distance Calculator [16]. The genomic average nucleotide identity (ANI) and FastANI and genomic average amino acid identity (AAI) were also calculated for further investigation of species delineation [17, 18].
Based on microbial genome taxonomy, strains of the same species are expected to share a >95 % ANI, >95 % AAI and >70 % DDH [19]. The results of LBG001T with the other Bdellovibrio genomes are presented in Table 1 and Table 2. Our result (ANI, ≤79 %; FastANI, ≤80 %; AAI, ≤72 %; DDH, 17 %) confirms that LBG001T does not belong to the B. bacteriovorus and any other known species. Thus, strain LBG001T represents a novel species within the genus for which the name Bdellovibrio reynosensis sp. nov. is proposed.
Table 1.
In silico DNA–DNA hybridization (% DDH), average nucleotide identity (% ANI) and average amino acid identity (AAI) values between Bdellovibrio sp. LBG 001T and other Bdellovibrio strains
Table 2.
FastANI genome comparison results of LBG001Twith other Bdellovibrio strains
|
Genome (A) |
Genome (B) |
% Identity |
Genome (A) |
Genome (B) |
% Identity |
|---|---|---|---|---|---|
|
LBG001T |
109J |
78.802 |
109J |
LBG001T |
78.8218 |
|
LBG001T |
Tiberius |
78.8507 |
Tiberius |
LBG001T |
78.9863 |
|
LBG001T |
HD100T |
79.002 |
HD100T |
LBG001T |
78.8588 |
|
LBG001T |
SSB |
78.781 |
SSB |
LBG001T |
78.8058 |
|
LBG001T |
NC01 |
79.5106 |
NC01 |
LBG001T |
79.1514 |
|
LBG001T |
KM01 |
78.8541 |
KM01 |
LBG001T |
79.0788 |
|
LBG001T |
ZAP7 |
78.8428 |
ZAP7 |
LBG001T |
79.1001 |
|
LBG001T |
W |
78.3054 |
W |
LBG001T |
78.239 |
|
LBG001T |
ExovorusT |
77.5308 |
ExovorusT |
LBG001T |
77.563 |
|
LBG001T |
QaytiusT |
77.1479 |
QaytiusT |
LBG001T |
77.0045 |
Genome features
Genome features of LBG001T were analysed on patric [13], prophage analysis on phaster [20], genome island on islandviewer4 [21] and metabolic reconstruction on BlastKOALA KEGG [22]. LBG001T has a single circular chromosome with a size of 3 582 323 bp and a G+C content of 43.12 mol%. Comparative analysis of the genome features of LBG001T with other representative obligate periplasmic and epibiotic predatory bacteria B. bacteriovorus HD100T, Bdellovibrio sp. W, Bdellovibrio sp. NC01 and Pseudobdellovibrio exovorus are presented in Table 3. The LBG001T genome size is bigger than that of the epibiotic predator P. exovorus . But smaller than that of all periplasmic Bdellovibrio strains with a complete genome sequence except for W, which has a genome size of 3.01 Mb, while its G+C (mol%) content is the lowest among the periplasmic Bdellovibrio strains, similar to W genome (43.3 mol%). The LBG001T genome was predicted to contain one incomplete prophage (Fig. S1), with a region length of 29.4 kb. Five smaller genomic islands were identified (Fig. S2), of which the largest with a size of 25 936 bp contains ribosomal proteins (50S ribosomal protein L24, 50S ribosomal protein L28 and 30S ribosomal protein S18), hypothetical proteins and other enzymes. But this incomplete prophage is not located at the islands, indicating that this prophage could be ancient event by phage attack. The ribosomal genes found in the genetic islands of LBG 001T were further analysed. Of the four ribosomal proteins associated with the genome islands, three were found to be part of the LBG001T core genome. The presence of the ribosomal proteins, sigma-54 dependent transcriptional regulator, DNA replication and repair protein RecF and DNA gyrase subunit A in the genome islands and being part of the LBG001T core genome suggest ancient gene transfer. Ancient lateral transfer of prey genes into Bdellovibrio genome have previously being established [23]. The core genome and pan-genome analysis of the most closely related strains with periplasmic predatory lifestyles are presented in Table 4. It is noticeable that LBG001T contains a high number of genome-specific genes (911).
Table 3.
A genome comparison of Bdellovibrio strains as indicaticated on the patric database
|
Genomic features |
LBG001T |
NC01 |
HD100T |
109J |
Tiberius |
SSB 218315 |
W |
QaytiusT |
ExovorusT |
|---|---|---|---|---|---|---|---|---|---|
|
CDS |
3395 |
3860 |
3592 |
3632 |
3737 |
3642 |
2872 |
3188 |
2649 |
|
tRNA |
33 |
34 |
36 |
35 |
35 |
34 |
34 |
31 |
34 |
|
rRNA |
4 |
4 |
4 |
3 |
6 |
3 |
6 |
2 |
3 |
|
Misc_RNA |
4 |
2 |
5 |
4 |
|||||
|
Repeat_region |
9 |
6 |
4 |
0 |
|||||
|
Protein features |
|||||||||
|
Hypothetical proteins |
1584 |
1875 |
1463 |
1532 |
1660 |
1554 |
1400 |
1780 |
1496 |
|
Proteins with functional assignments |
1811 |
1985 |
2129 |
2100 |
2077 |
2088 |
1472 |
1408 |
1153 |
|
Proteins with GO assignments |
494 |
536 |
617 |
613 |
620 |
510 |
541 |
473 |
501 |
|
Specialty genes |
|||||||||
|
Transporter |
11 |
10 |
64 |
63 |
54 |
5 |
18 |
||
|
Drug target |
1 |
1 |
1 |
1 |
1 |
1 |
1 |
||
|
Antibiotic resistance |
22 |
31 |
29 |
29 |
29 |
29 |
21 |
23 |
21 |
Table 4.
Pan-genome analysis using complete Whole-genome sequenced periplasmic Bdellovibrio
|
Organism name |
No. of core genes |
No. of accessory genes |
No. of unique genes |
No. of exclusively absent genes |
|---|---|---|---|---|
|
B. bacteriovorus 109J |
1481 |
1944 |
122 |
5 |
|
B. bacteriovorus HD100T |
1481 |
1954 |
61 |
3 |
|
B. bacteriovorus SSB218315 |
1481 |
1890 |
97 |
11 |
|
B. bacteriovorus Tiberius |
1481 |
1919 |
286 |
6 |
|
1481 |
414 |
790 |
271 |
|
|
Bdellovibrio LBG001T |
1481 |
952 |
911 |
39 |
|
Bdellovibrio sp KM01 |
1481 |
2036 |
172 |
6 |
|
Bdellovibrio sp. NC01 |
1481 |
1111 |
1126 |
19 |
|
Bdellovibrio sp. ZAP7 |
1481 |
2066 |
297 |
2 |
Metabolic reconstruction of the LBG001T genome supports a predatory lifestyle. The genome LBG001T has complete glycolysis, tricarboxylic acid and pentose phosphate pathway cycles as well as the complete pathways for histidine, cysteine and methionine metabolism, while its fatty acid elongation and fatty acid biosynthesis are incomplete. The LBG001T genome contains genes characterized as genetic signatures of predatory bacteria [24]. For example, it uses the mevalonate pathway for isoprenoid biosynthesis, unlike the non-predatory bacteria
Though Bdellovibrio is generally an obligate predatory bacterium, mutation at a region known as the host interaction locus (hit) has resulted in mutant strains that are facultative [25]. Such facultative mutants can be isolated and grow on complex media [9, 26]. The hit locus was first identified to be the switch from host-dependent to a host-independent Bdellovibrio [25] or to be the species B. bacteriovorus [27, 28]. The hit locus comprises bd0108 and the 3′ one-third of bd0109 in the Bdellovibrio genome HD100T [29]. Pairwise sequence alignment of bd0109 and phylogenetic analysis confirmed that this gene is well conserved in all Bdellovibrio , Bacteriovorax and Halobacteriovorax strains, suggesting bd0109 could be necessary for their predatory lifestyle.
However, the sequences of the bd0108 protein are not well conserved and have high diversity in different species, although the homologues of bd0109 are in the same operon. This small protein in LBG001T showed 43 % identity with bd0108 of HD100T, much lower than the comparison of bd0109 homologue between these two strains (87 % identity). There were no conserved regions of this small protein in Halobacteriovorax , Bacteriovorax and the epibiotic strain in Pseudobdellovibrio , while some regions are conserved in the periplasmic predation strains of Bdellovibrio . This small protein in LBG001T showed the highest identity (62 %) with a protein from a metagenomic sample strain. It also shares 52 % identity with that in strain NC01, but the conserved region only occurred in the first 42 amino acids. A sequence multi-alignment of the small protein in LBG001T with those in periplasmic predation members of B. bacteriovorus revealed a conserved region located at residues 34–42. The first 20 amino acids of all homologues are signal peptides although their signal sequences are completely different. Thus, all the members of this group contain these two genes but in a very different evolutionary manner: bd0109 homologues are conserved and can be considered an essential gene while bd0108 evolved much faster.
We could not conclude if LBG001T exists as host-independant mutants, as such strains have not been successfully isolated so far.
Genetic basis of predation
Orthologous genes related to predation in the LBG001T genome were identified by comparison with other previously identified Bdellovibrio essential predatory genes. Duncan and colleagues used transposon mutagenesis to identify 104 genes that are of importance in 109J predation of planktonic E. coli cells [30], of which 100 genes are present in LBG001 and 64 genes occur in the core genome of Bdellovibrio periplasmic predation strains. If all Bdellovibrio shares a similar mechanism of predation, the presence or absence of some predation genes might be the basis of different prey ranges or predation efficiency observed among Bdellovibrio strains. The genetic basis underlying the Bdellovibrio predation strategy remains incomplete, making the study of different species of interest.
Physiology
We tested the LBG001T prey range and the antibiotic susceptibility pattern. The double-layer plaque technique was used in investigating the prey range. The ability of the Bdellovibrio to form plaques on a prey lawn was considered positive for the prey bacteria under testing. The potential prey used in this study includes 10 Gram-negative bacteria Pseudomonas aeruginosa CDBB-B-1021 (ATCC 27853), Acinetobacter baumanii Jim01, Klebsiella pneumonia subspecies pneumonia B-969 (ATCC 13883), Proteus vulgaris ATCC 6380, Vibrio parahaemolyticus , Serratia marcens CDBB-B-1014 (ATCC 14756), Enterobacter aerogenes CDBB B-958 (ATCC 13048), Salmonella enterica subsp. enterica serovar Typhi CDBB-B-1101 (ATCC 7251), Klebsiella oxytoca B-968 (ATCC 13182), Escherichia coli DH5α and two Gram-positive bacteria: Staphylococcus aureus (ATCC25923), Enterococcus faecalis (ATCC 29212). Strain LBG001T formed plaques on lawn prey plates of all 10 Gram-negative bacteria but not the two Gram-positive bacteria, suggesting LBG001T has a wide Gram-negative prey range. LBG001T can prey on Proteus vulgaris ATCC 6380, different from the type strain B. bacteriovorus HD100T which has no ability to form plaques on Proteus vulgaris [5].
The antibiotic susceptibility testing for strain LBG001T on six antibiotics was carried out in liquid co-culture. Strain LBG001T was resistant to kanamycin, gentamycin and ampicillin but sensitive to streptomycin, tetracycline and chloramphenicol. Its susceptibility to streptomycin supports a previous assessment of all wild Bdellovibri o strains being susceptible to streptomycin [5].
Microscopy
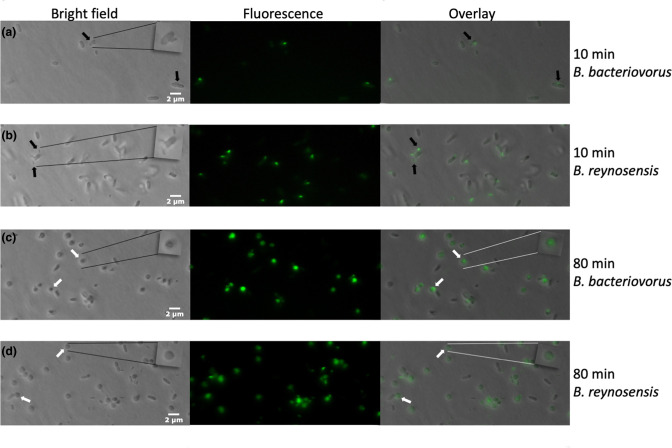
Fluorescently labelled LBG001T and B. bacteriovorus 109J were separately used to infect Klebsiella pneumoniae (ATCC 43816) for 10 and 80 min. The infected prey cells were examined on agarose pads on microscope slides by ×100 bright field and ×100 fluorescence microscopy using the FITC channel on a Revolve microscope (Echo Laboratories, RVL-100-B2). Details of the experimental protocol are in supplementary materials, Text 2. The results showed a prey-seeking LBG001T at 10 mins (Fig. 4), where LBG001T was attached to the prey, indicating the beginning of its attack. At 80 mins of infection, previously rod-shaped prey cells were seen as bdelloplasts, which are spherical prey cells that have been remodelled as the result of Bdellovibrio periplasmic invasion. The results for LBG001T were indistinguishable from predation by B. bacteriovorus 109J (Fig. 4), indicating that LBG001 is able to invade the prey cell in a periplasmic lifestyle.
Fig. 4.
Bdellovibrio predation of Klebsiella pneumoniae . B. bacteriovorus (a, c) and B. reynosensis sp. nov. (b, d) were fluorescently labelled, added to unlabeled K. pneumoniae resuspended in HEPES buffer, and incubated at 30 °C with aeration. After 10 and 80 min, cells were placed on agarose pads on slides and examined by bright field and fluorescence microscopy. Results were equivalent for both Bdellovibrio species. At 10 min, uninfected large, rod-shaped prey cells and small, extracellular but mostly attached fluorescent predator cells (filled arrows) are visible. At 80 min, prey cells have rounded into bdelloplasts with intracellular fluorescent predator (open arrows). Inset boxes in the upper right of some panels show enlarged images of example attached predator (a, b) and bdelloplasts (c, d).
Description of Bdellovibrio reynosensis sp. nov.
Bdellovibrio reynosensis (rey.no.sen’sis. N.L. masc. adj. reynosensis, pertaining to Reynosa, the location where the type strain was isolated).
The isolated bacterium is a Gram-negative rod-like bacterium with fast darting motility. It forms a circular plaque on a lawn plate of Klebsiella oxytoca (Fig. 5) and other Gram-negative bacteria by endobiotic predation. Plaques appear after 48 h of incubation at 30 °C. Plaque diameters increase with incubation time. Strain LBG001T grows in HEPES buffer in the presence of suitable prey, which can be set up at a temperature of 30 °C, pH 7, with or without agitation (with agitation is more suitable). LBG001T is resistant to kanamycin, gentamycin and ampicillin but sensitive to streptomycin, tetracycline and chloramphenicol.
Fig. 5.
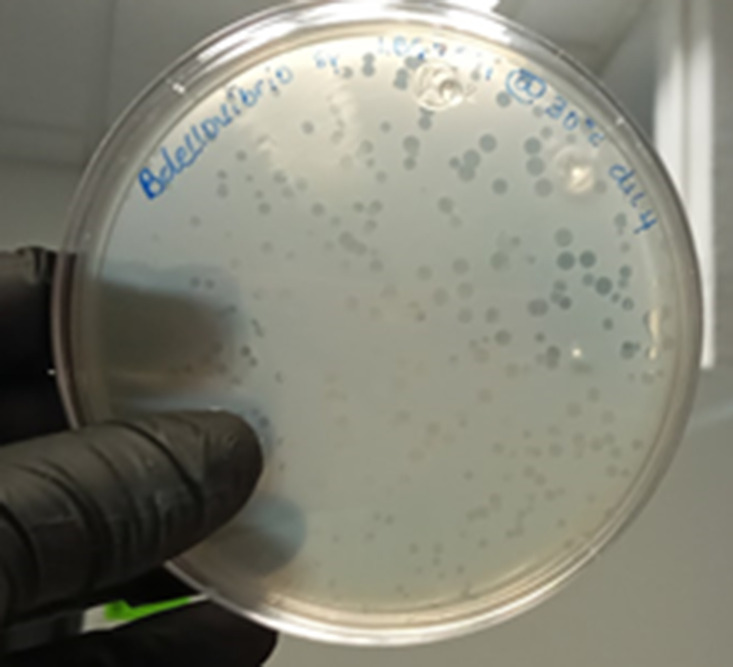
Bdellovibrio reynosensis LBG001T growing as plaques on a lawn plate of Klebsiella oxytocoa.
The type strain is LBG001T (ATCC TSD-288T=CM-CNRG 0932T). It was isolated from a soil sample collected at Reynosa, Mexico. The complete whole-genome sequence has a size of 3.58 Mb and a DNA G+C content of 43.1 mol%.
Supplementary Data
Funding information
This work was supported by Secretaría de Investigación y Posgrado del Instituto Politécnico Nacional, México (project numbers: 20202202, 20202200) and fund from Tufts University. Y.O.A. held scholarships from CONACyT.
Author contribution
Y.O.A., strain isolation, experiments, writing the original draft; I.C.R.L., experiments; T.O.E., genomic data analysis; A.S.-V., experiments; D.V.C.E., project design and critical comments; A.C., genome sequencing and analysis, English editing; X.G., project design, data analysis and writing.
Conflicts of interest
The authors declare that there are no conflicts of interest.
Footnotes
Abbreviations: AAI, genomic average amino acid identity; ANI, average nucleotide identity; BPGA, bacterial pan genome analysis; dDDH, digital DNA–DNA hybridization; DNB, diluted nutrient broth; FastANI, fast alignment-free computation of whole-genome ANI.
Two supplementary figures and two supplementary tables are available with the online version of this article.
References
- 1.Hahn MW, Schmidt J, Koll U, Rohde M, Verbarg S, et al. Silvanigrella aquatica gen. nov., sp. nov., isolated from a freshwater lake, description of Silvanigrellaceae fam. nov. and Silvanigrellales ord. nov., reclassification of the order Bdellovibrionales in the class Oligoflexia, reclassification of the families Bacteriovoracaceae and Halobacteriovoraceae in the new order Bacteriovoracales ord. nov., and reclassification of the family Pseudobacteriovoracaceae in the order Oligoflexales . Int J Syst Evol Microbiol. 2017;67:2555–2568. doi: 10.1099/ijsem.0.001965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pitt A, Koll U, Schmidt J, Hahn MW. Fluviispira multicolorata gen. nov., sp. nov. and Silvanigrella paludirubra sp. nov., isolated from freshwater habitats. Int J Syst Evol Microbiol. 2020;70:1630–1638. doi: 10.1099/ijsem.0.003947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nakai R, Nishijima M, Tazato N, Handa Y, Karray F, et al. Oligoflexus tunisiensis gen. nov., sp. nov., a Gram-negative, aerobic, filamentous bacterium of a novel proteobacterial lineage, and description of Oligoflexaceae fam. nov., Oligoflexales ord. nov. and Oligoflexia classis nov. Int J Syst Evol Microbiol. 2014;64:3353–3359. doi: 10.1099/ijs.0.060798-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Williams HN, Chen H. Environmental regulation of the distribution and ecology of Bdellovibrio and like organisms. Front Microbiol. 2020;11:545070. doi: 10.3389/fmicb.2020.545070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Stolp H, Starr MP. Bdellovibrio bacteriovorus gen. et sp. n., a predatory, ectoparasitic, and bacteriolytic microorganism. Antonie van Leeuwenhoek. 1963;29:217–248. doi: 10.1007/BF02046064. [DOI] [PubMed] [Google Scholar]
- 6.Bratanis E, Andersson T, Lood R, Bukowska-Faniband E. Biotechnological potential of Bdellovibrio and like organisms and their secreted enzymes. Front Microbiol. 2020;11:662. doi: 10.3389/fmicb.2020.00662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pérez J, Moraleda-Muñoz A, Marcos-Torres FJ, Muñoz-Dorado J. Bacterial predation: 75 years and counting! Environ Microbiol. 2016;18:766–779. doi: 10.1111/1462-2920.13171. [DOI] [PubMed] [Google Scholar]
- 8.Waite DW, Chuvochina M, Pelikan C, Parks DH, Yilmaz P, et al. Proposal to reclassify the proteobacterial classes Deltaproteobacteria and Oligoflexia, and the phylum Thermodesulfobacteria into four phyla reflecting major functional capabilities. Int J Syst Evol Microbiol. 2020;70:5972–6016. doi: 10.1099/ijsem.0.004213. [DOI] [PubMed] [Google Scholar]
- 9.Jurkevitch E. Isolation and classification of Bdellovibrio and like organisms. Curr Protoc Microbiol. 2012;7:Unit7B.1. doi: 10.1002/9780471729259.mc07b01s26. [DOI] [PubMed] [Google Scholar]
- 10.Van Essche M, Sliepen I, Loozen G, Van Eldere J, Quirynen M, et al. Development and performance of a quantitative PCR for the enumeration of Bdellovibrionaceae . Environ Microbiol Rep. 2009;1:228–233. doi: 10.1111/j.1758-2229.2009.00034.x. [DOI] [PubMed] [Google Scholar]
- 11.Lambert C, Sockett RE. Laboratory maintenance of Bdellovibrio . Curr Protoc Microbiol. 2008;7:7B–2. doi: 10.1002/9780471729259.mc07b02s9. [DOI] [PubMed] [Google Scholar]
- 12.Bosi E, Donati B, Galardini M, Brunetti S, Sagot MF, et al. MeDuSa: a multi-draft based scaffolder. Bioinformatics. 2015;31:2443–2451. doi: 10.1093/bioinformatics/btv171. [DOI] [PubMed] [Google Scholar]
- 13.Davis JJ, Wattam AR, Aziz RK, Brettin T, Butler R, et al. The PATRIC bioinformatics resource center: expanding data and analysis capabilities. Nucleic Acids Res. 2020;48:D606–D612. doi: 10.1093/nar/gkz943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kim M, Oh HS, Park SC, Chun J. Towards a taxonomic coherence between average nucleotide identity and 16S rRNA gene sequence similarity for species demarcation of prokaryotes. Int J Syst Evol Microbiol. 2014;64:346–351. doi: 10.1099/ijs.0.059774-0. [DOI] [PubMed] [Google Scholar]
- 15.Yarza P, Yilmaz P, Pruesse E, Glöckner FO, Ludwig W, et al. Uniting the classification of cultured and uncultured bacteria and archaea using 16S rRNA gene sequences. Nat Rev Microbiol. 2014;12:635–645. doi: 10.1038/nrmicro3330. [DOI] [PubMed] [Google Scholar]
- 16.Meier-Kolthoff JP, Klenk HP, Göker M. Taxonomic use of DNA G+C content and DNA-DNA hybridization in the genomic age. Int J Syst Evol Microbiol. 2014;64:352–356. doi: 10.1099/ijs.0.056994-0. [DOI] [PubMed] [Google Scholar]
- 17.Rodriguez-R LM, Konstantinidis KT. The enveomics collection: a toolbox for specialized analyses of microbial genomes and metagenomes. PeerJ Preprints. 2016 doi: 10.7287/peerj.preprints.1900v1. [DOI]
- 18.Jain C, Rodriguez-R LM, Phillippy AM, Konstantinidis KT, Aluru S. High throughput ANI analysis of 90K prokaryotic genomes reveals clear species boundaries. Nat Commun. 2018;9:1–8. doi: 10.1038/s41467-018-07641-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Thompson CC, Chimetto L, Edwards RA, Swings J, Stackebrandt E, et al. Microbial genomic taxonomy. BMC Genomics. 2013;14:1–8. doi: 10.1186/1471-2164-14-913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Arndt D, Grant JR, Marcu A, Sajed T, Pon A, et al. PHASTER: a better, faster version of the PHAST phage search tool. Nucleic Acids Res. 2016;44:W16–21. doi: 10.1093/nar/gkw387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bertelli C, Laird MR, Williams KP, Simon Fraser University Research Computing Group. Lau BY, et al. IslandViewer 4: expanded prediction of genomic islands for larger-scale datasets. Nucleic Acids Res. 2017;45:W30–W35. doi: 10.1093/nar/gkx343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kanehisa M, Sato Y, Morishima K. BlastKOALA and GhostKOALA: KEGG tools for functional characterization of genome and metagenome sequences. J Mol Biol. 2016;428:726–731. doi: 10.1016/j.jmb.2015.11.006. [DOI] [PubMed] [Google Scholar]
- 23.Gophna U, Charlebois RL, Doolittle WF. Ancient lateral gene transfer in the evolution of Bdellovibrio bacteriovorus. Trends Microbiol. 2006;14:64–69. doi: 10.1016/j.tim.2005.12.008. [DOI] [PubMed] [Google Scholar]
- 24.Pasternak Z, Pietrokovski S, Rotem O, Gophna U, Lurie-Weinberger MN, et al. By their genes ye shall know them: genomic signatures of predatory bacteria. ISME J. 2013;7:756–769. doi: 10.1038/ismej.2012.149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cotter TW, Thomashow MF. Identification of a Bdellovibrio bacteriovorus genetic locus, hit, associated with the host-independent phenotype. J Bacteriol. 1992;174:6018–6024. doi: 10.1128/jb.174.19.6018-6024.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Seidler RJ, Starr MP. Isolation and characterization of host-independent Bdellovibrios . J Bacteriol. 1969;100:769–785. doi: 10.1128/jb.100.2.769-785.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jurkevitch E, Minz D, Ramati B, Barel G. Prey range characterization, ribotyping, and diversity of soil and rhizosphere Bdellovibrio spp. isolated on phytopathogenic bacteria. Appl Environ Microbiol. 2000;66:2365–2371. doi: 10.1128/AEM.66.6.2365-2371.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schwudke D, Strauch E, Krueger M, Appel B. Taxonomic studies of predatory bdellovibrios based on 16S rRNA analysis, ribotyping and the hit locus and characterization of isolates from the gut of animals. Syst Appl Microbiol. 2001;24:385–394. doi: 10.1078/0723-2020-00042. [DOI] [PubMed] [Google Scholar]
- 29.Rendulic S, Jagtap P, Rosinus A, Eppinger M, Baar C, et al. A predator unmasked: life cycle of Bdellovibrio bacteriovorus from a genomic perspective. Science. 2004;303:689–692. doi: 10.1126/science.1093027. [DOI] [PubMed] [Google Scholar]
- 30.Duncan MC, Gillette RK, Maglasang MA, Corn EA, Tai AK, et al. High-throughput analysis of gene function in the bacterial predator Bdellovibrio bacteriovorus . mBio. 2019;10:1–12. doi: 10.1128/mBio.01040-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.