Rhodobacter sphaeroides has the capacity to grow by aerobic and anaerobic respiration and photosynthetically in the light under anaerobic conditions, as well as fermentatively. It can fix atmospheric nitrogen and carbon dioxide. It resembles other gram-negative members of the class Proteobacteria when growing aerobically, but a reduction in oxygen tension induces an intracellular differentiation of the inner membrane, leading to the formation of the intracytoplasmic membrane system (ICM). The ICM houses the integral membrane pigment-protein complexes constituting the photosystem (PS), comprised of the reaction center (RC) and two light-harvesting (LH) complexes. For an early review of ICM biosynthesis, see reference 44. The LH complexes are designated B800-850 (LHII) and B875 (LHI), based on their respective absorption maxima. The ratio of LHI to RC is fixed at approximately 15:1, whereas the ratio of LHII to the LHI-RC unit is variable, changing in a manner inverse to the incident light intensity. These three pigment-protein complexes are the spectral complexes (SC) of the R. sphaeroides PS. Detailed structural information about these complexes in several species of Rhodobacter is emerging (72).
The LH complexes capture light energy and direct that energy to the RC, where conversion of the excitation energy takes place and is intrinsically coupled with a cyclic flow of electrons, ultimately to the periplasmically localized cytochrome c2, which serves to rereduce the RC to allow a new cycle of electron flow (for further details, see references 42, 44, and 72).
Bacteriochlorophyll (Bchl) absorbs most of the light energy within the SCs and is critical to the assembly and final structure of the SCs (44, 84, 87). The carotenoids (Crt) have a minor role in absorbing light energy (15), but they function to protect the complexes against photo-oxidative damage, dissipate excess radiant energy, and help to maintain the structure and relative abundance of each SC (35, 50, 53).
A reduction in oxygen tension is both necessary and sufficient to induce synthesis of the ICM (reviewed in references 42 and 44), which is gratuitously produced under anaerobic dark growth conditions, in the presence of an alternate electron acceptor such as dimethyl sulfoxide (DMSO). Oxygen tension is the major environmental stimulus controlling PS induction, with variations in light intensity determining the cellular level of the ICM and the abundance of the different SCs. PS formation is tightly regulated, with checkpoints at all levels of information flow, from transcriptional through posttranslational. In the following sections, we will describe what we currently know about the regulatory processes controlling the formation and abundance of SCs in R. sphaeroides. We will present a working model for the regulation of PS formation in R. sphaeroides 2.4.1 which is based on the critical role of cellular redox carriers. For clarity, we will, throughout this review, define aerobic growth as that which occurs under highly oxygenic conditions, under which there are no detectable SCs present in wild-type membranes.
TRANSCRIPTIONAL REGULATION OF PS GENE EXPRESSION
Transcriptional regulation of PS genes, from an initial low background level, involves the coordinate action of several signal transduction pathways which result in the induction of PS gene expression following oxygen removal. Some of these pathways are specific to PS genes and others are more global, while still others are commonly recognized to be involved in cellular adaptation to anaerobiosis. Different PS genes are not all controlled by the same regulatory pathways, although these may overlap. We first describe those factors which appear to play a major role in sensing, signal generation, and transduction, as well as DNA binding. Thereafter, we describe other factors which have less pronounced effects and/or less obvious roles in PS gene transcription; these are believed to participate in fine tuning of transcriptional control. Finally, we described posttranscriptional processes essential to SC formation and abundance.
Sensors, signal generators, and transducers. (i) The Cco/Rdx redox signal-generating system.
The identification of CcoNOQP cytochrome c oxidase and RdxBH (redox) as components (Fig. 1) of a signal-generating pathway stemmed from the observation that inactivation of the ccoNOQP operon resulted in PS gene expression (93, 94) under aerobic conditions. The ccoNOQP operon encodes the cbb3-type cytochrome c oxidase of R. sphaeroides 2.4.1. Of the three respiratory oxidases present in R. sphaeroides, the cbb3 type is believed to have the highest affinity for oxygen and is the primary oxidase present under microaerobic conditions (26, 27, 37). Immediately downstream of the ccoNOQP operon is the rdxBHIS operon (70, 94). These operons are located at 443 kbp on chromosome I (94). (The locations of genes that map outside the PS gene cluster and are pertinent to this discussion are in boldface throughout the text.) The rdxB gene is a homolog of the rdxA gene, which maps to chromosome II (69). Both RdxB and RdxA are predicted to coordinate two [4Fe-4S] clusters (69), and these, together with other clusters of cysteine residues, may be involved in redox processes. They are both likely to be membrane bound, as has been shown for RdxA (69).
FIG. 1.
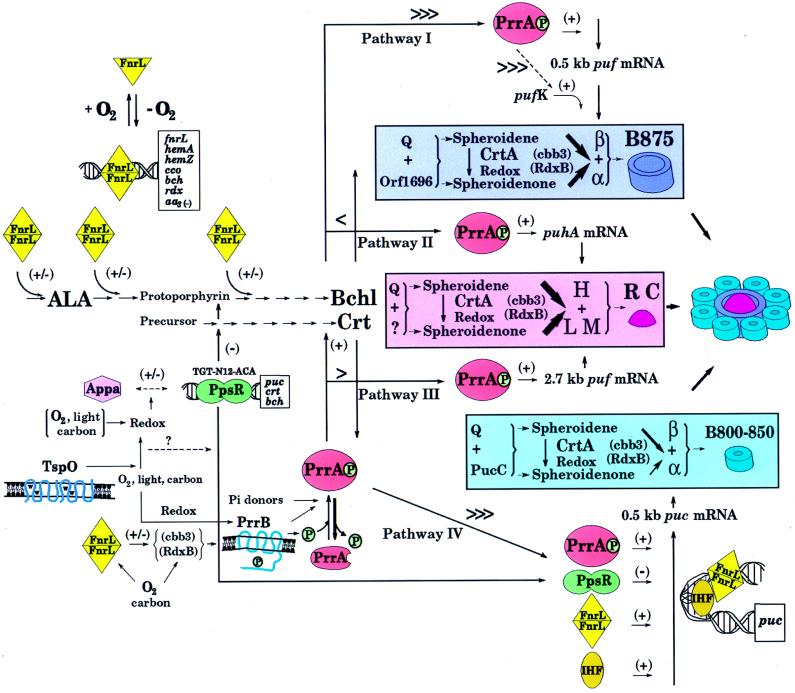
Most of the PS genes are arranged in a cluster on chromosome I of R. sphaeroides 2.4.1 (83). This region of approximately 60 kb begins with the pufKBALMX operon and extends, in a clockwise direction, to the pucBAC operon, which is 18 kb downstream of puhA. The pufB and -A genes encode structural polypeptides of the LHI SC, the pufL and -M genes encode two of the three structural proteins of the RC, and the two structural polypeptides of the LHII SC are encoded by pucB and -A, respectively (reviewed in reference 44). The Q gene, located upstream of the puf operon, together with the pufK and pucC genes, are not part of the PS, nor is orf1696. The pufX gene product is required for PS competence (4); it facilitates light-driven cyclic electron transfer (23). Between pufKBALMX and puhA, the latter of which codes for the largest structural polypeptide of the RC, are genes encoding enzymes catalyzing the biosynthesis of Crt and Bchl (1–3, 8, 9, 17, 52) and some genes that have a regulatory role in PS gene expression. Between puhA and the pucBAC operon is the cycA gene (19, 97), encoding the obligatory cytochrome c2 protein. In the related bacterium R. capsulatus, a similar arrangement of a portion of these genes extending from puf to puh (25), called the photosynthesis gene cluster, is present, and the DNA sequence of the entire R. capsulatus photosynthesis gene cluster has been determined (14). The PS gene cluster is defined as the region from 0 to 3,000 kbp on chromosome I of R. sphaeroides 2.4.1. In the text are presented the locations, in boldface, of those genes that map outside this cluster and are pertinent to the discussion presented here. For a detailed description of the above model of PS gene regulation, see the text. ALA, 5-Aminolevulinic acid.
Mutations in either ccoNOQP or the rdxB gene generate mutants which, under aerobic growth conditions, produce SCs, as well as high levels of expression of pucBAC and pufKBALMX. Uncoupling of PS gene expression from the presence of oxygen in Rdx and Cco mutant strains involves the activation of the two-component PrrBA regulatory system (71). Thus, a pathway involving cbb3 through its interaction with oxygen serves as an oxygen “sensor” to generate an “inhibitory signal” through RdxB to, presumably, PrrB (see below), which results in a lack of PrrA activation. This inhibitory signal may be through the membrane portion of PrrB. However, no physical interaction between cbb3 and PrrB has been demonstrated. This is analogous to the Arc system of Escherichia coli (40). The fact that the levels of SCs and PS gene expression in cbb3 and RdxB mutant strains are also substantially increased relative to those of the wild type, even under anaerobic conditions, strongly affirms the likely functioning of these redox carriers under such conditions. Under these same conditions, the activation of PS gene expression in these mutant strains is dependent upon the Prr system. FnrL (see below) is indirectly involved in modulating the activity of PrrA by regulating the level of ccoN gene expression in response to oxygen levels (∼10-fold increase following a high-to-low O2 transition; 66) and, presumably, rdxB as well.
(ii) The redox-responsive two-component Prr system.
Prr (photosynthetic response regulator) is a signal transduction system (Fig. 1) involved in the activation of PS gene expression (20–22). The prrA gene product functions as a response regulator, and the prrB gene product functions as a sensor histidine kinase/phosphatase. A third gene, prrC, located upstream of prrA, encodes a membrane-associated protein. This genetic region is located outside of the region of PS genes, 1149 kbp, on chromosome I of R. sphaeroides 2.4.1. The prrCA genes form a transcriptional unit, although prrA has its own promoter and prrB is divergently transcribed from prrCA. Prr is similar to the R. capsulatus regBA-and-senC system, which is also involved in regulation of PS gene expression (64, 79), as well as the Rhizobium meliloti actRS system, which is involved in acid tolerance (86). PrrBA regulates transcription of puhA, cycA, and the puc and puf operons, as well as some of the genes involved in Bchl and Crt synthesis and/or accumulation. The expression of genes involved in CO2 and N2 fixation is also affected by the Prr system (41). Inactivation of prrA causes a photosynthesis-negative phenotype, as the result of a nearly total shutdown of PS gene expression. Mutants defective in prrB are photosynthesis negative when grown at low-to-medium light intensities but grow photosynthetically at high light intensities, albeit with severely reduced levels of SCs. PrrC inactivation also leads to a small reduction in SC formation, but cells remain photosynthetically competent.
PrrA is thought to be cytoplasmically located. The carboxy-terminal end of PrrA lacks similarities with other DNA-binding proteins. Nonetheless, PrrA acts either by binding DNA directly (45) or by interacting with another protein(s) that binds DNA. When prrA is present in multicopy in aerobically growing wild-type or PrrB− cells, such cells aberrantly produce spectral complexes and have increased puc expression despite the inhibitory signal emanating from cbb3 (22). This further suggests that other phosphodonors are active in activating PrrA (20, 30, 71) and that PrrB may possess an intrinsic phosphatase activity (20). Thus, the cellular levels of PrrA must be carefully regulated (22). Additionally, there is some evidence to suggest that different PS genes require different levels of active PrrA for their activation (20).
In R. capsulatus, a truncated form of the PrrB homolog RegB lacking the membrane-associated domain phosphorylates the PrrA homolog RegA in vitro (39). A point mutation involving a Leu-to-Pro change at position 78 of the PrrB protein (PRRB78) confers an oxygen-insensitive phenotype on the cell (21); i.e., PS gene expression is “full on” under both aerobic and anaerobic conditions. This region of the protein is believed to be located in a nonmembranous cytoplasmic loop far removed from the active histidine.
The role of PrrC, SenC in R. capsulatus (11), in PS gene expression is much more subtle than that of PrrA or PrrB. PrrC shares amino acid sequence similarity with the yeast ScoI and ScoII proteins (10) believed to be involved in the assembly of mitochondrial cytochrome c oxidase subunits. PrrC appears to be membrane localized, based on prrC′-′phoA fusion analyses (21), with the carboxy-terminal region of the protein located in the periplasmic space.
(iii) AppA.
The appA (activation of photopigment and puc expression) gene resides at approximately 1285 kbp clockwise on chromosome I; i.e., approximately 1.2 Mb from the PS gene cluster (28). Deletion of appA results in greatly diminished SC formation, which correlates with decreased expression of the corresponding PS genes (28) (Fig. 1). Low levels of SC result in a prolonged lag in the growth of an AppA null mutant when it is shifted from aerobic to anaerobic light conditions, and this is followed by reduced photosynthetic growth rates (31). However, anaerobic dark growth is normal (31).
AppA consists of 450 amino acids and shows no homology with proteins of known function. Following overexpression of several truncated forms of the protein, specific ligand binding has been observed. The approximately 120-residue amino-terminal domain has been shown to bind flavin. The approximately 70-residue carboxy-terminal domain contains an unusual cysteine-rich motif which is capable of binding iron or, more likely, an Fe:S center. The central region of AppA appears to bind a heme. Removal of either the amino- or carboxy-terminal portion of AppA result in only slight impairment of AppA function, as judged by complementation of an AppA null mutant (32).
The activity associated with AppA is currently unknown. However, AppA affects PS gene expression independently of both the Prr and FnrL systems (31). Existing data strongly suggest that AppA antagonizes PpsR repressor activity (31) (see below). Whether the role of AppA is limited to PpsR alone has yet to be elucidated.
DNA-binding factors. (i) Anaerobiosis activator/repressor FnrL.
The fnrL gene encodes the R. sphaeroides 2.4.1 homolog of Fnr (Fig. 1), the E. coli anaerobic regulatory protein. It is located at approximately 443 kbp on chromosome I, in close proximity to the ccoNOQP and rdxBHIS operons of R. sphaeroides 2.4.1 (94). FnrL− mutant strains are unable to grow anaerobically, either photosynthetically or in the dark using DMSO as an electron acceptor, and such mutants have no detectable SCs under conditions of ICM induction (94).
FnrL has 11 of 22 amino acid identities with E. coli Fnr within the region corresponding to the helix-turn-helix DNA-binding domain. Residues that are thought to form direct amino acid-base contacts (54) are identical between the two. Therefore, we suggest that the recognition sequence for binding by FnrL resembles the FNR (fumarate nitrate regulator) consensus sequence, TTGAT-N4-ATCAA. Sequences similar or identical to this consensus sequence have been identified upstream of several genes involved in tetrapyrrole and Bch1 biosynthesis, including hemA (68), hemZ (94), hemF (recently renamed hemN) (18) and bchE (28, 95), as well as the puc operon (56). Under conditions of reduced oxygen tension, FnrL activates expression of hemA (94) and the puc operon (95). These results provide good evidence that the target sequence for regulation by FnrL is the FNR consensus sequence.
In contrast to R. sphaeroides, an FnrL mutant strain of R. capsulatus which has been shown to contain an fnrL homolog is unaffected in its ability to grow photosynthetically with many of the genes described above, lacking upstream FNR-binding sequences (92). However, like R. sphaeroides FnrL mutant strains, an FnrL mutant of R. capsulatus is incapable of anaerobic growth in the dark with DMSO (92). For both R. sphaeroides and R. capsulatus, the DMSO-inducible cytochrome c (encoded by dorC) and DMSO reductase, encoded by dorA (65), are not induced in the mutant strains (93). It appears that the role of FnrL in dor operon (65) expression is mediated through the FnrL-dependent regulation of dorS. DorS, which encodes a redox-responsive sensor histidine kinase, regulates its cognate transducer, the product of the dorR gene (65).
Based on the presence of upstream FNR consensus sequences (94), the ccoNOQP operon, the rdxBHIS operon, and the ctaD and ctaABC genes (24, 36) encoding subunits of the aa3 cytochrome oxidase are likely to be regulated, in whole or part, by FnrL in R. sphaeroides (94). Thus, FnrL could, by affecting the abundance of these redox carriers, control the flow of electrons through the respective terminal oxidases and the Rdx redox center. This has been shown to be true for the ccoN gene. In addition to directly effecting puc operon transcription due to the presence of an upstream FNR-binding sequence (95), FnrL could also indirectly affect puc operon transcription (95, 96) through its regulation of ccoNOQP and, thus, the signal reaching Prr (71).
(ii) PpsR.
The upstream regions of many bch and crt genes and operons, as well as the puc operon from both R. sphaeroides and R. capsulatus, contain the same dyad symmetry motif (Fig. 1), TGT-N12-ACA (52, 56, 61). The localization of this motif downstream of, or overlapping with, putative ς70-type promoters of PS genes and studies of the cis regulatory elements of the puc operon (56, 59) suggest that this sequence can function as a repressor-binding site. This repressor-encoding gene, designated ppsR (photopigment suppression) in R. sphaeroides (73; Fig. 1), was identified because of its ability to suppress photopigment production in wild-type cells when provided in extra copy. Inactivation of ppsR results in constitutive expression of both the photopigment genes and puc, which triggers production of SCs, even under aerobic conditions, thus making such mutants genetically unstable under these conditions (31, 74).
ppsR is localized within the region of PS genes, at approximately 26 kbp on chromosome I. PpsR contains a putative helix-turn-helix motif at the carboxy terminus which resembles those found in several known DNA-binding transcription factors (74). The ability of PpsR to act as a repressor has been demonstrated genetically in E. coli (74) and in Paracoccus denitrificans (29). Point mutations in the putative DNA-binding region result in decreased repression (31, 56, 59). It has been shown that the PpsR protein overexpressed in E. coli and P. denitrificans specifically retards DNA fragments containing the sequence TGT-N12-ACA (32). The PpsR homolog from R. capsulatus, designated CrtJ, has been purified recently and shown to bind to this motif as a dimer (75). Cooperative binding of CrtJ to two binding motifs positioned in the regulatory regions of several putative CrtJ/PpsR target genes, has also been indicated, which correlates well with earlier genetic studies (29).
R. sphaeroides ppsR gene expression is largely independent of growth conditions, and PpsR repressor activity does not respond directly to oxygen deprivation. Therefore, other cellular factors are required to communicate to PpsR the state of oxygen availability and changes in growth conditions (31, 32). Further, PpsR functions as a repressor not only under aerobic but also under anaerobic photosynthetic conditions, where it regulates LHII SC abundance, depending on light intensity (31). Because it is unlikely that PpsR senses both oxygen and light directly, we believe that PpsR must respond to an integral signal generated by changes in either of these environmental stimuli, e.g., either redox poise or a redox carrier. This view is complemented by the in vitro analysis of the R. capsulatus repressor protein, whose DNA-binding capacity has been shown to depend on redox-active agents (75).
The nature and regulation of the redox sensitivity of PpsR remain largely unknown (Fig. 1). Disruption of the ppsR gene in an AppA null mutant background completely overcomes the impairment(s) imposed by the appA null mutation. Furthermore, a large number of suppressors of the appA null mutation, showing improved photosynthetic growth, contain point mutations in ppsR, and partial suppression of the appA null mutation can be achieved by titration of PpsR in vivo (31, 32). AppA significantly decreases PpsR-mediated repression when these genes are simultaneously expressed in P. denitrificans (31). Therefore, we believe that AppA serves to regulate PpsR repressor activity by communicating a redox signal to PpsR which is ultimately derived from oxygen- and/or light-generated redox responses. The signal could affect either the redox state of PpsR or its dimerization or both. The overall role of PpsR in both oxygen and light control of SC formation, as well as its specificity toward PS genes, makes it a major player in PS gene regulation.
(iii) Spb.
The hvrA gene of R. capsulatus, located immediately downstream of the response regulator regA, is involved in regulation of RC and LHI SC gene expression (12). The hvrA gene encodes a small, 104-residue DNA-binding factor. Analogous to hvrA, a homolog from R. sphaeroides, termed spb, is positioned immediately downstream of prrA (77). Both Spb (Sphaeroides puf-binding protein) and HvrA bind upstream of the puhA gene and the puf operon under various conditions (49, 80). The amino-terminal regions of Spb and HvrA bear similarity to the HU-like proteins from E. coli and bacteriophages (49, 77). These proteins often have multiple targets and therefore are involved in transcriptional regulation of a variety of processes. Conforming with this idea are recent observations that HvrA is also involved in the ammonium regulation of nitrogen fixation in R. capsulatus (62). Thus, these proteins may be involved in PS gene expression by facilitating the binding of other transcription factors, e.g., PrrA, RNA polymerase, etc., and their role in light regulation may be a reflection of their abundance as a function of light intensity. It was also found that the amino terminus of Spb has some homology to the leucine zipper motif of the eukaryotic transcription factor c-JUN. This motif is proposed to play a role in protein dimerization (80). Because it is difficult to describe a more specific role for these factors, they have not been included in Fig. 1.
(iv) IHF.
The integration host factor (IHF) of E. coli is a global regulatory protein which acts by binding DNA targets and bending the DNA so that looping occurs. This brings together noncontiguous regions of the DNA, which interact through DNA-binding regulatory proteins (88). The genes for the two subunits of the R. sphaeroides IHF heterodimer have been cloned (78). Interestingly, the himA gene is located at approximately position 1760 kbp on chromosome I, while the himD (hip) gene is located at approximately 830 kbp on chromosome II. In vitro studies have shown that IHF of E. coli binds to the regulatory region of the puc operon (58). Additionally, mutations that alter the consensus IHF-binding site in this region abolish binding in vitro and negatively affect puc expression in vivo.
Modulators of PS gene transcription.
The tspO gene (formerly crtK, or orf160 in R. capsulatus), located within the crt gene cluster on chromosome I, encodes a 17-kDa protein (tryptophan-rich sensory protein) which is characterized by an unusually high content of aromatic amino acids, especially l-tryptophan. TspO (Fig. 1) has been localized to the outer membrane of R. sphaeroides 2.4.1 (90) and is present at severalfold higher levels in photosynthetic than in aerobically grown cells. Results of cross-linking studies suggest that TspO may form a homodimer within the bacterial membrane (91).
A TspO mutant has higher levels of Crt and Bchl than do wild-type cells when strains are grown aerobically or semiaerobically. The effect of TspO on pigment production occurs at the level of transcription of those same crt and bch biosynthetic genes which constitute the PpsR regulon (90). TspO also negatively affects the expression of the puc operon under both aerobic and photosynthetic conditions but has no effect on the expression of the puf operon. The available evidence implicates TspO as a both oxygen- and light-responsive modulator of PS gene expression with the same spectrum of regulatory targets as the PpsR repressor (29, 90). How a signal is transmitted from outer membrane-localized TspO to the cell interior is unknown. However, the quantitative effect of TspO upon PS gene expression is not as strong as that of PpsR, which suggests that there must be other elements or factors interposed between TspO and the PpsR regulon.
Useful information about the physiological activity of TspO may be drawn from studies on its mammalian homolog (91), the 18-kDa outer membrane component (pk18) of the mitochondrial peripheral benzodiazepine receptor (MBR). Although the precise biological role of MBR is not yet understood, this receptor was shown to be involved in the regulation of steroidogenesis, heme biosynthesis, mitochondrial respiratory control, and other metabolic activities (51). The rat pk18 gene, when expressed in R. sphaeroides in the TspO− background, functionally substituted for the bacterial homolog, negatively affecting the expression of crtI and puc. Additionally, when the pk18 gene was expressed in R. sphaeroides, it retained its quantitative binding properties towards a variety of MBR ligands. These results suggest that TspO is both evolutionarily and functionally related to the mammalian MBR and indicate the possibility that in both bacterial and mammalian cells, these proteins are involved in similar metabolic activities (91).
How TspO might “sense” and “transmit” an oxygen- or light-generated signal remains a mystery, although no ligand has been found to be associated with TspO. Very recent results suggest that TspO may be involved in regulating the entry of specific small molecules through the outer membrane in response to oxygen and light. Mutants with single changes of the five conserved tryptophans have either lost or retain partial TspO activity.
Other elements in PS gene expression.
Several additional factors have been observed to effect PS gene expression, but their precise role is presently unknown. The ppa (photopigment activation) gene (also known as ppsS and orf192) is positioned immediately upstream of ppsR (29). A role for Ppa in controlling PpsR repressor activity has been proposed but not yet tested (74). Thus, we have not included Ppa in Fig. 1. A locus at 2110 kbp on chromosome I, identified by virtue of its effect on expression of the puc operon (76), has been named mgpS (modulator of genes for photosynthesis) and contains an open reading frame of ∼2.8 kb. The N-terminal portion of mpgS shares similarity with a number of RNA helicase proteins which are involved in RNA modifications (76), suggesting that mpgS may have an additional role in posttranscriptional regulation of PS gene expression. The rpgS1 gene (regulator for photosystem genes; 77) a histonelike protein located approximately 500 bp clockwise from prrA, encodes a protein with similarity to the functionally unidentified orf5 gene from R. capsulatus (13). The rpgS1 gene is predicted to encode a protein of 187 amino acids which contains a putative helix-turn-helix motif located at its amino-terminal end (77).
Regulation of PS gene expression at the message level.
As is evident from the previous sections, transcriptional regulation of PS gene expression plays a crucial role in PS formation in R. sphaeroides 2.4.1. However, it has been observed that mRNAs corresponding to the structural polypeptides of the SCs are produced in excess, relative to the actual levels of the complexes present under a variety of growth conditions (34, 55, 67, 81). This provides evidence of posttranscriptional regulation. Such complexity of regulation is not surprising, given the fact that PS development requires coordination of a variety of diverse processes, involving protein and pigment biosynthetic pathways, SC assembly, and adaptation between alternative cellular steady states.
In the related organism R. capsulatus, PS genes are organized into superoperons (reviewed in references 5, 6, and 48). It is thought that there are multiple transcription initiations at several start sites within the superoperons which result in differences in message levels for genes and operons within the superoperons. However, degradation of larger transcripts is also considered to contribute to the disproportionate representation of gene messages in the cell (reviewed in reference 48). Some data also suggest the possibility that degradation of large transcripts is regulated by oxygen tension (46, 47).
While certain large transcripts indicative of a similar superoperon organization have been detected in R. sphaeroides (63), their physiological role, if any, is not known. In the case of the puc and puf operons of R. sphaeroides, multiple transcripts with differing half-lives are also detected. The longer puc and puf transcripts have been shown to be products of transcriptional readthrough of terminatorlike sequences within those operons (55, 57).
The first gene of the R. sphaeroides puf operon is the pufK gene, encoding a 20-amino-acid-residue polypeptide. It has been demonstrated that pufK is translated and that its expression is elevated severalfold under high-light PS conditions, compared to that in aerobically grown cells (33). An interesting feature of the pufK gene is the high occurrence of rare codons, with 9 of the 20 codons of pufK among those rarely used by R. sphaeroides 2.4.1. Studies suggest that translation through these rarely used codons serves to “gate” the entry of ribosomes to the immediate downstream pufBA cistrons (Fig. 1) and, as such, can affect the cellular abundance of the α and β polypeptides of the LHI complex (33).
THE ROLE OF BCHL AND CRT
Bchl and Crt are major nonprotein structural components of the SCs whose synthesis is regulated in response to oxygen availability and/or light intensity (Fig. 1). The availability of Bchl has been shown to be of critical importance and the limiting component for the formation and stability of the RC and the LH SCs (87). The puf-encoded RC-M and RC-L polypeptides, as well as the puhA-encoded RC-H protein, are detected at low levels under aerobic growth conditions but are rapidly degraded due to the absence of Bchl, both in R. sphaeroides and in R. capsulatus (16, 84, 85, 87).
Unlike Bchl, Crt are not essential components of the RC or the B875 complex, and mutants defective in Crt biosynthesis genes are photosynthetically competent. In contrast, the formation of the LHII complex requires colored Crt (intermediate products of the Crt biosynthetic pathway downstream of neurosporene), and it has been proposed that this requirement may be at the level of assembly (53). The type of Crt accumulated is also important in determining the relative stoichiometry between the two LH complexes. Spheroidenone (SO), the end product of the Crt biosynthetic pathway, is predominantly associated with the RC and LHI complexes, whereas spheroidene (SE), the penultimate product of the Crt pathway, is more abundant in the LHII complex (38, 89). The relative amounts of these two major Crt pigments are regulated in response to oxygen tension and/or light intensity, apparently through the localized cellular redox state (Fig. 1), which affects the CrtA-catalyzed conversion of SE to SO (89).
The Cco/RdxB putative oxygen-sensing, redox signal-generating pathway has been demonstrated to affect the regulation, through the Prr regulon (71), of the type of Crt accumulated under photosynthetic and diazotrophic growth conditions. Presumably, the localized redox poise generated through these pathways affects CrtA activity (70). Thus, the signal generated by the cbb3 oxidase-RdxB system is transferred not only to transcription factors through the Prr pathway (see the section on sensors, signal generators, and transducers) but also to another pathway(s) involved in the control of the conversion of SE to SO under photosynthetic conditions. The recently discovered gene likely to encode the first enzyme in the isoprenoid biosynthetic pathway (82) and the next downstream gene in the biosynthetic pathway, mapping to the PS gene cluster, are of major importance and are predicted to be highly regulated. These data provide further evidence that Cco and RdxB are “functional” as redox carriers under anaerobic, as well as aerobic, growth conditions, perhaps interacting with a light-generated redox intermediate.
Conditions which limit Bchl availability (such as high light illumination or high oxygen tension) are also characterized by low SE and high SO levels and correspond to limited LHII formation (89). Therefore, the net result is that available Bchl is incorporated into the RC and LHI complexes (Fig. 1), which appear to have no preference for which Crt is used in the assembly process (Fig. 1). In this way, changes in environmental conditions (to which Bchl levels and Crt accumulation are evidently sensitive) can be rapidly reflected by changes in LHII complex formation and this, in turn, may result in the de facto regulation of RC and LHI complex assembly and ultimate abundance (89).
ASSEMBLY FACTORS
Genes have been identified whose proposed role in the regulation of PS formation is at the level of SC assembly (Fig. 1). One of these genes is the monocistronic Q gene, located immediately upstream of the puf operon of R. sphaeroides, encoding a 187-amino-acid polypeptide (34). Single amino acid substitutions in the Q gene product have dramatic consequences for the formation of SCs, e.g., loss of LHI complex formation ability for one group of mutants, loss of LHII complex formation ability for another group, and altered levels of both SCs relative to those in wild-type cells for a third group, despite little or no changes in specific mRNA levels or Bchl and Crt availability. In the absence of the Q gene product, the cells are unable to grow under PS conditions. Based on these studies (34), the Q gene product is proposed to be involved in the assembly of the SCs. However, in R. capsulatus, the structure of the Q gene and the function of its product have been interpreted differently (7).
orf1696, located immediately upstream of puhA (14) of R. sphaeroides 2.4.1 (Fig. 1), has been shown to be required for LHI complex formation (81). The pucC gene product (Fig. 1) is required for LHII complex formation (43, 57, 85). These genes are proposed to encode complex-specific assembly factors (CSAFs). These two factors, together with the product of a third gene, which is called orf428 in R. capsulatus (14) but is not known to participate in SC formation (8), are similar to each other. This similarity may indicate common interactions with other factors, as part of an overall assembly hierarchy for the SCs. One of these interactions may be with the Q gene product (81). Thus, the specific role of the Q gene product is thought to be through the insertion of Bchl into the developing SCs (Fig. 1) in a process involving interaction(s) with the CSAFs (Fig. 1) (34). The difference in the affinities of the Q polypeptide for the CSAFs would then account for the preferential assembly of the RC complex, followed by the LHI complex, and finally the LHII complex when Bchl is plentiful.
Some of the more conventional chaperone genes have also been identified in R. sphaeroides; e.g., there are two groE operons (60). However, the role (if any) of these and/or other chaperones in SC formation has not yet been defined.
OVERALL MODEL FOR THE REGULATION OF SC FORMATION
In the presence of oxygen, the R. sphaeroides cell membrane resembles that of other gram-negative bacteria. Energy for cellular metabolism is derived from aerobic respiratory chains, terminating at either one of two cytochrome c oxidases (the low oxygen affinity aa3 oxidase or the high oxygen affinity cbb3 oxidase or a bb3-type quinol oxidase (reviewed in reference 26). There is virtually no Bchl or Crt synthesized. There are low but detectable levels of transcription of the PS genes encoding the apoproteins of the SCs; in the absence of Bchl, these polypeptides turn over rapidly, although they are transported to the cell membrane and assume their proper topological orientation within the membrane.
When oxygen tension is reduced, transcription of PS genes is activated in response to three key regulatory systems, namely, the FnrL, Prr, and PpsR systems. We propose that these systems reflect change in the localized cellular redox poise or a critical redox intermediate(s) when oxygen tension is lowered. Yeliseev and Kaplan have demonstrated that TspO modulates this response (90). Similarly, the biosynthesis of Bchl and Crt ensues in response to these changes and SC formation can now occur.
The state of activation of the putative oxygen-labile [4Fe-4S] center of FnrL as a function of oxygen availability is thought to regulate the activity of FnrL. FnrL can act directly to activate specific PS gene transcriptions, which reflect critical points of control, in the tetrapyrrole and Bchl pathways, as well as puc operon expression. FnrL also affects PS gene expression through its regulation either positively or negatively of specific redox chains, e.g., cbb3 or aa3.
The Prr two-component regulatory system, in response to an altered signal emanating from the cbb3/RdxB redox pathway can begin to activate transcription of all or, certainly, most of the PS genes. Presumably, PrrB is no longer inhibited by this signal, to which it responds by activating PrrA in a mechanism believed to involve PrrB autophosphorylation and subsequent phosphorylation of PrrA. While the means by which PrrA regulates transcription of the PS genes is not precisely known, it may involve direct DNA binding or the formation of transcriptional complexes involving other protein factors, the net result being an increased expression of genes encoding all of the structural components of the SCs. The data also suggest that different levels of activated PrrA may be required to activate different subsets of PS genes.
In contrast to FnrL and Prr, which have a broad spectrum of gene targets, PpsR appears to repress specifically PS genes; i.e., most of the bch and crt genes, as well as the puc operon, which contain the repressor-binding sequence TGT-N12-ACA. Repression by PpsR, although relieved under anaerobic conditions, is still responsive to light under these conditions, and derepression is brought about, at least in part, by the action of the flavohemoprotein AppA, which acts either directly or indirectly on PpsR. The fact that PpsR is involved in PS gene control in response to both oxygen and light, perhaps through AppA, suggests that a different source of a redox-generated signal is involved in the transmission of this effect than in Prr-mediated regulation. However, this remains to be proven.
TspO modulates the rate of activation of PS gene derepression as oxygen tensions are decreased. Other modulators appear to include PrrC, Ppa, HvrA (Spb), and MgpS, as well as IHF. Overall, as the O2 tension decreases, there develops a cascade of both orderly and progressive events which serve to reinforce the induction of PS gene expression. This cascade must also involve subtle interactions between the system of activation and repression of PS gene expression, as oxygen tensions change and as light intensities vary under anaerobic conditions.
The net result of these transcriptional changes is that all of the components leading to SC formation are present at increased levels when oxygen tension is reduced, and the assembly process specific to each can take place. The total amounts of SC formed depend upon the cellular level of Bchl which, in turn, is determined not only by the activity of Bchl biosynthetic enzymes but also by the available pool of 5-aminolevulinic acid, and therefore, Bchl serves as the ultimate “governor” regulating cellular abundances of SCs. The Q protein, together with CSAFs, direct the flow of Bchl into the assembling SCs in a hierarchical fashion, with RC assembly first, followed by assembly of the B875 SC, and then assembly of the B800-850 SC. Usually, the mRNA levels, and the levels of apoproteins produced, are in excess of cellular needs. The abundance and composition (SE and SO) of the Crt present are also important for the ratio of LH complexes. Thus, posttranscriptional regulatory pathways serve to increase the ability of cells to respond rapidly to changes in oxygen availability and light intensity.
Although it has long been known that the amount of SC varies dramatically under anaerobic conditions, depending on the light intensity, the mechanism(s) of light control of PS formation is less well understood than that of oxygen control. We do know, however, that light regulation is achieved, at least in part, through the same transcriptional regulatory machinery which operates in response to changes in oxygen tension, e.g., PpsR/AppA (31) and HvrA (Spb) (49). It is also tempting to speculate that cbb3 and RdxB play some role(s), as well, in photosynthetically growing cells. This lends support to the idea that changes in light intensity are first transformed into changes in the cellular redox state. However, the presence of specific light-dependent regulators cannot be ruled out at present. We currently believe that light control of PS formation occurs mostly at posttranscriptional levels and involves rapid changes in Bchl levels and the cellular levels of SO and SE. Together with the available assembly factors, these elements will rapidly determine the abundance and kinds of SCs.
ACKNOWLEDGMENTS
All of the authors contributed equally to this work.
This work was supported by grant GM15590 from the U.S. Public Health Service.
ADDENDUM IN PROOF
We have recently observed that the ccoNOQP operon is negatively regulated by the two-component Prr system under aerobic growth conditions.
REFERENCES
- 1.Armstrong G A. Eubacteria show their true colors: genetics of carotenoid pigment biosynthesis from microbes to plants. J Bacteriol. 1994;176:4795–4802. doi: 10.1128/jb.176.16.4795-4802.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Armstrong G A, Alberti M, Leach F, Hearst J E. Nucleotide sequence, organization, and nature of the protein products of the carotenoid biosynthesis gene cluster of Rhodobacter capsulatus. Mol Gen Genet. 1989;216:254–268. doi: 10.1007/BF00334364. [DOI] [PubMed] [Google Scholar]
- 3.Bartley G E, Scolnik P A. Carotenoid biosynthesis in photosynthetic bacteria. J Biol Chem. 1989;264:13109–13113. [PubMed] [Google Scholar]
- 4.Barz W, Oesterhelt D. Photosynthetic deficiency of a pufX deletion mutant of Rhodobacter sphaeroides is suppressed by point mutations in the light-harvesting complex genes pufB or pufA. Biochemistry. 1994;33:9741–9752. doi: 10.1021/bi00198a045. [DOI] [PubMed] [Google Scholar]
- 5.Bauer C, Buggy J, Mosley C. Control of photosystem genes in Rhodobacter capsulatus. Trends Genet. 1993;9:56–60. doi: 10.1016/0168-9525(93)90188-N. [DOI] [PubMed] [Google Scholar]
- 6.Bauer C E, Bird T H. Regulatory circuits controlling photosynthesis gene expression. Cell. 1996;85:5–8. doi: 10.1016/s0092-8674(00)81074-0. [DOI] [PubMed] [Google Scholar]
- 7.Bauer C E, Marrs B L. Rhodobacter capsulatus puf operon encodes a regulatory protein (PufQ) for bacteriochlorophyll biosynthesis. Proc Natl Acad Sci USA. 1988;85:7074–7078. doi: 10.1073/pnas.85.19.7074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bollivar D W, Suzuki J Y, Beatty J T, Dobrowolski J M, Bauer C E. Directed mutational analysis of bacteriochlorophyll a biosynthesis in Rhodobacter capsulatus. J Mol Biol. 1994;237:622–640. doi: 10.1006/jmbi.1994.1260. [DOI] [PubMed] [Google Scholar]
- 9.Bollivar D W, Wang S, Allen J P, Bauer C E. Molecular genetic analysis of terminal steps in bacteriochlorophyll a biosynthesis: characterization of a Rhodobacter capsulatus strain that synthesizes geranylgeraniol-esterified bacteriochlorophyll a. Biochemistry. 1994;33:12763–12768. doi: 10.1021/bi00209a006. [DOI] [PubMed] [Google Scholar]
- 10.Buchwald P, Krummeck G, Rodel G. Immunological identification of yeast SCO1 protein as a component of the inner mitochondrial membrane. Mol Gen Genet. 1991;229:413–420. doi: 10.1007/BF00267464. [DOI] [PubMed] [Google Scholar]
- 11.Buggy J, Bauer C E. Cloning and characterization of senC, a gene involved in both aerobic respiration and photosynthesis gene expression in Rhodobacter capsulatus. J Bacteriol. 1995;177:6958–6965. doi: 10.1128/jb.177.23.6958-6965.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Buggy J J, Sganga M W, Bauer C E. Characterization of a light-responding trans-activator responsible for differentially controlling reaction center and light-harvesting-I gene expression in Rhodobacter capsulatus. J Bacteriol. 1994;176:6936–6943. doi: 10.1128/jb.176.22.6936-6943.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Buggy J J, Sganga M W, Bauer C E. Nucleotide sequence and characterization of the Rhodobacter capsulatus hvrB gene: HvrB is an activator of S-adenosyl-l-homocysteine hydrolase expression and is a member of the LysR family. J Bacteriol. 1994;176:61–69. doi: 10.1128/jb.176.1.61-69.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Burke D H, Alberti M, Armstrong G A, Hearst J E. The complete nucleotide sequence of the 46 kb photosynthesis gene cluster of Rhodobacter capsulatus. GenBank accession no. Z11165. 1992. [Google Scholar]
- 15.Cogdell R J. Carotenoids in photosynthesis. Phil Trans R Soc Lond B Biol Sci. 1978;284:569–579. [Google Scholar]
- 16.Cohen-Bazire G, Sistrom W R, Stanier R Y. Kinetic studies of pigment synthesis by non-sulfur purple bacteria. J Cell Comp Physiol. 1956;49:25–68. doi: 10.1002/jcp.1030490104. [DOI] [PubMed] [Google Scholar]
- 17.Coomber S A, Chaudhri M, Connor A, Britton G, Hunter C N. Localized transposon Tn5 mutagenesis of the photosynthetic gene cluster of Rhodobacter sphaeroides. Mol Microbiol. 1990;4:977–989. doi: 10.1111/j.1365-2958.1990.tb00670.x. [DOI] [PubMed] [Google Scholar]
- 18.Coomber S A, Jones R M, Jordan P M, Hunter C N. A putative anaerobic coproporphyrinogen III oxidase in Rhodobacter sphaeroides. I. Molecular cloning, transposon mutagenesis and sequence analysis of the gene. Mol Microbiol. 1992;6:3159–3169. doi: 10.1111/j.1365-2958.1992.tb01772.x. [DOI] [PubMed] [Google Scholar]
- 19.Donohue T J, McEwan A G, Kaplan S. Cloning, DNA sequence, and expression of the Rhodobacter sphaeroides cytochrome c2 gene. J Bacteriol. 1986;168:962–972. doi: 10.1128/jb.168.2.962-972.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Eraso J M, Kaplan S. Complex regulatory activities associated with the histidine kinase PrrB in expression of photosynthesis genes in Rhodobacter sphaeroides 2.4.1. J Bacteriol. 1996;178:7037–7046. doi: 10.1128/jb.178.24.7037-7046.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Eraso J M, Kaplan S. Oxygen-insensitive synthesis of the photosynthetic membranes of Rhodobacter sphaeroides: a mutant histidine kinase. J Bacteriol. 1995;177:2695–2706. doi: 10.1128/jb.177.10.2695-2706.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Eraso J M, Kaplan S. prrA, a putative response regulator involved in oxygen regulation of photosynthesis gene expression in Rhodobacter sphaeroides. J Bacteriol. 1994;176:32–43. doi: 10.1128/jb.176.1.32-43.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Farchaus J W, Barz W P, Grunberg H, Oesterhelt D. Studies on the expression of the pufX polypeptide and its requirement for photoheterotrophic growth in Rhodobacter sphaeroides. EMBO J. 1992;11:2779–2788. doi: 10.1002/j.1460-2075.1992.tb05345.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Flory J E, Donohue T J. Transcriptional control of several aerobically induced cytochrome structural genes in Rhodobacter sphaeroides. Microbiology. 1997;143:3101–3110. doi: 10.1099/00221287-143-10-3101. [DOI] [PubMed] [Google Scholar]
- 25.Fonstein M, Zheng S, Haselkorn R. Physical map of the genome of Rhodobacter capsulatus SB1003. J Bacteriol. 1992;174:4070–4077. doi: 10.1128/jb.174.12.4070-4077.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Garcia-Horsman J A, Barquera B, Rumbley J, Ma J, Gennis R B. The superfamily of heme-copper respiratory oxidases. J Bacteriol. 1994;176:5587–5600. doi: 10.1128/jb.176.18.5587-5600.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Garcia-Horsman J A, Berry E, Shapleigh J P, Alben J O, Gennis R B. A novel cytochrome c oxidase from Rhodobacter sphaeroides that lacks CuA. Biochemistry. 1994;33:3113–3119. doi: 10.1021/bi00176a046. [DOI] [PubMed] [Google Scholar]
- 28.Gomelsky M, Kaplan S. appA, a novel gene encoding a trans-acting factor involved in the regulation of photosynthesis gene expression in Rhodobacter sphaeroides 2.4.1. J Bacteriol. 1995;177:4609–4618. doi: 10.1128/jb.177.16.4609-4618.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gomelsky M, Kaplan S. Genetic evidence that PpsR from Rhodobacter sphaeroides 2.4.1 functions as a repressor of puc and bchF expression. J Bacteriol. 1995;177:1634–1637. doi: 10.1128/jb.177.6.1634-1637.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gomelsky M, Kaplan S. Isolation of regulatory mutants in photosynthesis gene expression in Rhodobacter sphaeroides 2.4.1 and partial complementation of a PrrB mutant by the HupT histidine-kinase. Microbiology. 1995;141:1805–1819. doi: 10.1099/13500872-141-8-1805. [DOI] [PubMed] [Google Scholar]
- 31.Gomelsky M, Kaplan S. Molecular genetic analysis suggesting interactions between AppA and PpsR in regulation of photosynthesis gene expression in Rhodobacter sphaeroides 2.4.1. J Bacteriol. 1997;179:128–134. doi: 10.1128/jb.179.1.128-134.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gomelsky, M., and S. Kaplan. Unpublished data.
- 33.Gong L, Kaplan S. Translational control of puf operon expression in Rhodobacter sphaeroides 2.4.1. Microbiology. 1996;142:2057–2069. doi: 10.1099/13500872-142-8-2057. [DOI] [PubMed] [Google Scholar]
- 34.Gong L, Lee J K, Kaplan S. The Q gene of Rhodobacter sphaeroides: its role in puf operon expression and spectral complex assembly. J Bacteriol. 1994;176:2946–2961. doi: 10.1128/jb.176.10.2946-2961.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Griffiths M, Stanier R Y. Some mutational changes in the photosynthetic pigment system of Rhodopseudomonas spheroides. J Gen Microbiol. 1956;14:2946–2961. doi: 10.1099/00221287-14-3-698. [DOI] [PubMed] [Google Scholar]
- 36.Hosler, J. Personal communication.
- 37.Hosler J P, Fetter J, Tecklenberg M J, Espe M, Lerma C, Ferguson-Miller S. Cytochrome aa3 of Rhodobacter sphaeroides as a model for mitochondrial cytochrome c oxidase: purification, kinetics, proton pumping and spectral analysis. J Biol Chem. 1992;267:24264–24272. [PubMed] [Google Scholar]
- 38.Hunter C N, Pennoyer J D, Sturgis J N, Farelly D, Niederman R A. Oligomerization states and associations of light-harvesting pigment-protein complexes of Rhodobacter sphaeroides as analyzed by lithium dodecyl sulfate-polyacrylamide gel electrophoresis. Biochemistry. 1988;27:3459–3467. [Google Scholar]
- 39.Inoue K, Kouadio J L, Mosley C S, Bauer C E. Isolation and in vitro phosphorylation of sensory transduction components controlling anaerobic induction of light harvesting and reaction center gene expression in Rhodobacter capsulatus. Biochemistry. 1995;34:391–396. doi: 10.1021/bi00002a002. [DOI] [PubMed] [Google Scholar]
- 40.Iuchi S, Cameron D C, Lin E C C. A second global regulator gene (arcB) mediating repression of enzymes in aerobic pathways of Escherichia coli. J Bacteriol. 1989;171:868–873. doi: 10.1128/jb.171.2.868-873.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Joshi H M, Tabita F R. A global two component signal transduction system that integrates the control of photosynthesis, carbon dioxide assimilation, and nitrogen fixation. Proc Natl Acad Sci USA. 1996;93:14515–14520. doi: 10.1073/pnas.93.25.14515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kaplan S, Arntzen C J. Govindjee (ed.), Photosynthesis: energy conversion by plants and bacteria. I. New York, N.Y: Academic Press, Inc.; 1982. Photosynthetic membrane structure and function; pp. 67–151. [Google Scholar]
- 43.Kiley P J, Kaplan S. Cloning, DNA sequence, and expression of the Rhodobacter sphaeroides light-harvesting B800-850-α and B800-850-β genes. J Bacteriol. 1987;169:3268–3275. doi: 10.1128/jb.169.7.3268-3275.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kiley P J, Kaplan S. Molecular genetics of photosynthetic membrane biosynthesis in Rhodobacter sphaeroides. Microbiol Rev. 1988;52:50–69. doi: 10.1128/mr.52.1.50-69.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kirndörfer M, Hebermehl M, Hemschemeir K, Jäger A, Ebel U, Klug G. Transcriptional regulation of puf and puc operon expression in Rhodobacter capsulatus by the DNA binding protein RegA and IHF, abstr. 69B. In: Peschek G A, Löffelhardt W, Schmetterer G, editors. IX International Symposium on Photosynthetic Prokaryotes. Book of Abstracts, Vienna, Austria. 1997. [Google Scholar]
- 46.Klug G. A DNA sequence upstream of the puf operon of R. capsulatus is involved in its oxygen-dependent regulation and functions as a protein binding site. Mol Gen Genet. 1991;226:167–176. doi: 10.1007/BF00273600. [DOI] [PubMed] [Google Scholar]
- 47.Klug G. Endonucleolytic degradation of puf mRNA in R. capsulatus is influenced by oxygen. Proc Natl Acad Sci USA. 1991;88:1765–1769. doi: 10.1073/pnas.88.5.1765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Klug G. Regulation of expression of photosynthesis genes in anoxygenic photosynthetic bacteria. Arch Microbiol. 1993;159:397–404. doi: 10.1007/BF00288584. [DOI] [PubMed] [Google Scholar]
- 49.Kouadio J-L, Bauer C. Diversity, Genetics and Physiology of Photosynthetic Prokaryotes. Bloomington: Indiana University; 1996. Structural and functional characterization of a dim light effector protein, HvrA, from R. capsulatus: a new member of the family of prokaryotic histone-like proteins, abstr. 41. [Google Scholar]
- 50.Krinsky N I. Non-photosynthetic functions of carotenoids. Phil Trans R Soc Lond B Biol Sci. 1978;284:581–590. [Google Scholar]
- 51.Krueger K E. Molecular and functional properties of mitochondrial benzodiazepine receptors. Biochim Biophys Acta. 1995;1241:453–470. doi: 10.1016/0304-4157(95)00016-x. [DOI] [PubMed] [Google Scholar]
- 52.Lang H P, Cogdell R J, Takaichi S, Hunter C N. Complete DNA sequence, specific Tn5 insertion map, and gene assignment of the carotenoid biosynthesis pathway of Rhodobacter sphaeroides. J Bacteriol. 1995;177:2064–2073. doi: 10.1128/jb.177.8.2064-2073.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lang H P, Hunter C N. The relationship between carotenoid biosynthesis and the assembly of the light-harvesting LH2 complex in Rhodobacter sphaeroides. Biochem J. 1994;298:197–205. doi: 10.1042/bj2980197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lazazzera B A, Bates D M, Kiley P J. The activity of the Escherichia coli transcription factor FNR is regulated by a change in oligomeric state. Genes Dev. 1993;7:1993–2005. doi: 10.1101/gad.7.10.1993. [DOI] [PubMed] [Google Scholar]
- 55.Lee J K, DeHoff B S, Donohue T J, Gumport R I, Kaplan S. Transcriptional analysis of puf operon expression in Rhodobacter sphaeroides 2.4.1 and an intercistronic transcription terminator mutant. J Biol Chem. 1989;264:19354–19365. [PubMed] [Google Scholar]
- 56.Lee J K, Kaplan S. cis-acting regulatory elements involved in oxygen and light control of puc operon transcription in Rhodobacter sphaeroides. J Bacteriol. 1992;174:1146–1157. doi: 10.1128/jb.174.4.1146-1157.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lee J K, Kiley P J, Kaplan S. Posttranscriptional control of puc operon expression of B800-850 light-harvesting complex formation in Rhodobacter sphaeroides. J Bacteriol. 1989;171:3391–3405. doi: 10.1128/jb.171.6.3391-3405.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lee J K, Wang S, Eraso J M, Gardner J, Kaplan S. Transcriptional regulation of puc operon expression in Rhodobacter sphaeroides. Involvement of an integration host factor-binding sequence. J Biol Chem. 1993;268:24491–24497. [PubMed] [Google Scholar]
- 59.Lee J K, Kaplan S. Transcriptional regulation of puc operon expression in Rhodobacter sphaeroides: analysis of cis-acting downstream regulatory sequence of the puf transcript. J Biol Chem. 1995;270:20453–20458. [PubMed] [Google Scholar]
- 60.Lee W T, Terlesky K C, Tabita F R. Cloning and characterization of two groESL operons of Rhodobacter sphaeroides: transcriptional regulation of the heat-induced groESL operon. J Bacteriol. 1997;179:487–495. doi: 10.1128/jb.179.2.487-495.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ma D, Cook D N, O’Brien D A, Hearst J E. Analysis of the promoter and regulatory sequences of an oxygen-regulated bch operon in Rhodobacter capsulatus by site-directed mutagenesis. J Bacteriol. 1993;175:2037–2045. doi: 10.1128/jb.175.7.2037-2045.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Masepohl B, Klipp W. Organization and regulation of genes encoding the molybdenum nitrogenase and alternative nitrogenase in Rhodobacter capsulatus. Arch Microbiol. 1996;165:80–90. [Google Scholar]
- 63.McGlynn P, Hunter C N. Genetic analysis of the bchC and bchA genes of Rhodobacter sphaeroides. Mol Gen Genet. 1993;236:227–234. doi: 10.1007/BF00277117. [DOI] [PubMed] [Google Scholar]
- 64.Mosley C S, Suzuki J Y, Bauer C E. Identification and molecular genetic characterization of a sensor kinase responsible for coordinately regulating light harvesting and reaction center gene expression in response to anaerobiosis. J Bacteriol. 1994;176:7566–7573. doi: 10.1128/jb.176.24.7566-7573.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Mouncey N J, Choudhary M, Kaplan S. Characterization of genes encoding dimethyl sulfoxide reductase of Rhodobacter sphaeroides 2.4.1T: an essential function encoded on chromosome II. J Bacteriol. 1997;179:7617–7624. doi: 10.1128/jb.179.24.7617-7624.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Mouncey N J, Kaplan S. Oxygen regulation of the ccoNOQP operon encoding the cbb3 oxidase in Rhodobacter sphaeroides 2.4.1T: involvement of the FnrL protein. J Bacteriol. 1998;180:2228–2231. doi: 10.1128/jb.180.8.2228-2231.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Neidle E L, Kaplan S. 5-Aminolevulinic acid availability and control of spectral complex formation in HemA and HemT mutants of Rhodobacter sphaeroides. J Bacteriol. 1993;175:2304–2313. doi: 10.1128/jb.175.8.2304-2313.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Neidle E L, Kaplan S. Expression of the Rhodobacter sphaeroides hemA and hemT genes, encoding two 5-aminolevulinic acid synthase isozymes. J Bacteriol. 1993;175:2292–2303. doi: 10.1128/jb.175.8.2292-2303.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Neidle E L, Kaplan S. Rhodobacter sphaeroides rdxA, a homolog of Rhizobium meliloti fixG, encodes a membrane protein which may bind cytoplasmic [4Fe-4S] clusters. J Bacteriol. 1992;174:6444–6454. doi: 10.1128/jb.174.20.6444-6454.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.O’Gara J P, Kaplan S. Evidence for the role of redox carriers in photosynthesis gene expression and carotenoid biosynthesis in Rhodobacter sphaeroides 2.4.1. J Bacteriol. 1997;179:1951–1961. doi: 10.1128/jb.179.6.1951-1961.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.O’Gara, J. P., J. M. Eraso, and S. Kaplan. Photosynthesis gene activation and the cbb3 oxidase of Rhodobacter sphaeroides 2.4.1. Submitted for publication.
- 72.Papiz M Z, Prince S M, Hawthornthwaite-Lawless A M, McDermott G, Freer A A, Isaacs N W, Cogdell R J. A model for the photosynthetic apparatus of purple bacteria. Trends Plant Sci. 1996;1:198–206. [Google Scholar]
- 73.Penfold R J, Pemberton J M. A gene from the photosynthetic gene cluster of Rhodobacter sphaeroides induces in trans suppression of bacteriochlorophyll and carotenoid levels in R. sphaeroides and in R. capsulatus. Curr Microbiol. 1991;23:259–263. [Google Scholar]
- 74.Penfold R J, Pemberton J M. Sequencing, chromosomal inactivation, and functional expression of ppsR, a gene which represses carotenoid and bacteriochlorophyll synthesis in Rhodobacter sphaeroides. J Bacteriol. 1994;176:2869–2876. doi: 10.1128/jb.176.10.2869-2876.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Ponnampalam S N, Bauer C E. DNA binding characteristics of CrtJ. A redox-responding repressor of bacteriochlorophyll, carotenoid, and light harvesting-II gene expression in Rhodobacter capsulatus. J Biol Chem. 1997;272:18391–18396. doi: 10.1074/jbc.272.29.18391. [DOI] [PubMed] [Google Scholar]
- 76.Sabaty M, Kaplan mgpS, a complex regulatory locus involved in the transcriptional control of the puc and puf operons in Rhodobacter sphaeroides 2.4.1. J Bacteriol. 1996;178:35–45. doi: 10.1128/jb.178.1.35-45.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Sen, P., J. M. Eraso, and S. Kaplan. Unpublished observations.
- 78.Sen, P., and S. Kaplan. Unpublished data.
- 79.Sganga M W, Bauer C E. Regulatory factors controlling photosynthetic reaction center and light-harvesting gene expression in Rhodobacter capsulatus. Cell. 1992;68:945–954. doi: 10.1016/0092-8674(92)90037-d. [DOI] [PubMed] [Google Scholar]
- 80.Shimada H, Wada T, Handa H, Ohta H, Mizoguchi H, Nishimura K, Masuda T, Shioi Y, Takamiya K. A transcription factor with a leucine-zipper motif involved in light-dependent inhibition of expression of the Puf operon in the photosynthetic bacterium Rhodobacter sphaeroides. Plant Cell Physiol. 1996;37:515–522. doi: 10.1093/oxfordjournals.pcp.a028974. [DOI] [PubMed] [Google Scholar]
- 81.Sockett R E, Donohue T J, Varga A R, Kaplan S. Control of photosynthetic membrane assembly in Rhodobacter sphaeroides mediated by puhA and flanking sequences. J Bacteriol. 1988;171:436–446. doi: 10.1128/jb.171.1.436-446.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Sprenger C A, Schörken U, Wiegert T, Grolle S, Graaf A A, Taylor V, Begley T P, Bringer-Meyer S, Sohm H. Identification of a thiamin-dependent synthase in Escherichia coli required for the formation of the 1-deoxy-d-δglucose-5-phosphate precursor to isoprenoids, thiamin and pyridoxol. Proc Natl Acad Sci USA. 1997;94:12857–12862. doi: 10.1073/pnas.94.24.12857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Suwanto A, Kaplan S. Physical and genetic mapping of the Rhodobacter sphaeroides 2.4.1 genome: genome size, fragment identification, and gene localization. J Bacteriol. 1989;171:5840–5849. doi: 10.1128/jb.171.11.5840-5849.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Takemoto J, Lascelles J. Coupling between bacteriochlorophyll and membrane protein synthesis in Rhodopseudomonas sphaeroides. Proc Natl Acad Sci USA. 1973;70:800–803. doi: 10.1073/pnas.70.3.799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Tichy H V, Oberle B, Stiehle H, Schiltz E, Drews G. Genes downstream from pucB and pucA are essential for formation of the B800-850 complex of Rhodobacter capsulatus. J Bacteriol. 1989;171:4914–4922. doi: 10.1128/jb.171.9.4914-4922.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Tiwari R P, Reeve W G, Dilworth M J, Glenn A R. Acid tolerance in Rhizobium meliloti strain WSM419 involves a two-component sensor-regulator system. Microbiology. 1996;142:1693–1704. doi: 10.1099/13500872-142-7-1693. [DOI] [PubMed] [Google Scholar]
- 87.Varga A R, Kaplan S. Synthesis and stability of reaction center polypeptides and implications for reaction center assembly in Rhodobacter sphaeroides. J Biol Chem. 1993;268:19842–19850. [PubMed] [Google Scholar]
- 88.Yang C-C, Nash H A. The interaction of E. coli IHF protein with its specific binding sites. Cell. 1989;57:869–880. doi: 10.1016/0092-8674(89)90801-5. [DOI] [PubMed] [Google Scholar]
- 89.Yeliseev A A, Eraso J M, Kaplan S. Differential carotenoid composition of the B875 and B800-850 photosynthetic antenna complexes in Rhodobacter sphaeroides 2.4.1: involvement of spheroidene and spheroidenone in adaptation to changes in light intensity and oxygen availability. J Bacteriol. 1996;178:5877–5883. doi: 10.1128/jb.178.20.5877-5883.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Yeliseev A A, Kaplan S. A sensory transducer homologous to the mammalian peripheral-type benzodiazepine receptor regulates photosynthetic membrane complex formation in Rhodobacter sphaeroides 2.4.1. J Biol Chem. 1995;270:21167–21175. doi: 10.1074/jbc.270.36.21167. [DOI] [PubMed] [Google Scholar]
- 91.Yeliseev A A, Krueger K E, Kaplan S. A mammalian mitochondrial drug receptor functions as a bacterial “oxygen” sensor. Proc Natl Acad Sci USA. 1997;94:5101–5106. doi: 10.1073/pnas.94.10.5101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Zeilstra-Ryalls J H, Gabbert K, Mouncey N J, Kaplan S, Kranz R G. Identification, cloning, sequencing, and analysis of the role of the fnrL gene of Rhodobacter capsulatus. J Bacteriol. 1997;179:7264–7273. doi: 10.1128/jb.179.23.7264-7273.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Zeilstra-Ryalls J H, Kaplan S. Aerobic and anaerobic regulation in Rhodobacter sphaeroides 2.4.1: the role of the fnrL gene. J Bacteriol. 1995;177:6422–6431. doi: 10.1128/jb.177.22.6422-6431.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Zeilstra-Ryalls J H, Kaplan S. Control of hemA expression in Rhodobacter sphaeroides 2.4.1: regulation through alterations in cellular redox state. J Bacteriol. 1996;178:985–993. doi: 10.1128/jb.178.4.985-993.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Zeilstra-Ryalls J H, Kaplan S. The role of the fnrL gene in photosystem gene expression and photosynthetic growth of Rhodobacter sphaeroides 2.4.1. J Bacteriol. 1998;180:1496–1503. doi: 10.1128/jb.180.6.1496-1503.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Zeilstra-Ryalls, J. H., and S. Kaplan. 1996. Unpublished observations.
- 97.Zsebo K M, Hearst J E. Genetic-physical mapping of a photosynthetic gene cluster from R. capsulatus. Cell. 1984;37:937–947. doi: 10.1016/0092-8674(84)90428-8. [DOI] [PubMed] [Google Scholar]