Summary
Studies of the relationships between environmental exposures and adverse health outcomes often rely on a two-stage statistical modeling approach, where exposure is modeled/predicted in the first stage and used as input to a separately fit health outcome analysis in the second stage. Uncertainty in these predictions is frequently ignored, or accounted for in an overly simplistic manner when estimating the associations of interest. Working in the Bayesian setting, we propose a flexible kernel density estimation (KDE) approach for fully utilizing posterior output from the first stage modeling/prediction to make accurate inference on the association between exposure and health in the second stage, derive the full conditional distributions needed for efficient model fitting, detail its connections with existing approaches, and compare its performance through simulation. Our KDE approach is shown to generally have improved performance across several settings and model comparison metrics. Using competing approaches, we investigate the association between lagged daily ambient fine particulate matter levels and stillbirth counts in New Jersey (2011–2015), observing an increase in risk with elevated exposure 3 days prior to delivery. The newly developed methods are available in the R package KDExp.
Keywords: Environmental health, Kernel density estimation, Two-stage modeling, Uncertainty propagation
1. Introduction
Environmental health studies require the linking of environmental exposure information for each observation in the analysis (e.g., individual or time point) in order to estimate the association with adverse health outcomes. Because exposure data are typically not available at every spatial location and time period covered by the study, researchers often rely on predictions from a first-stage statistical model to fill in the spatiotemporal gaps. For example, several advanced statistical methods have been developed to interpolate ambient air pollution concentrations using monitoring data combined with estimates from deterministic models and other data sources (e.g., Fuentes and Raftery, 2005; Berrocal and others, 2010a,b; McMillan and others, 2010; Berrocal and others, 2012; Reich and others, 2014; Guan and others, 2019; Warren and others, 2021).
These methods not only yield point predictions of exposure at the relevant locations and times of interest but also provide measures of uncertainty. Because many of the methods are fitted within a Bayesian framework using Markov chain Monte Carlo (MCMC) techniques, samples from posterior predictive distributions (ppd) are also available. Incorporating this exposure uncertainty into the subsequent health analysis is important for correctly characterizing uncertainty in the association between exposure and health. Specifically, health observations linked with exposure estimates with higher uncertainties should contribute less to the overall health effect estimate. However, previous studies often ignore this uncertainty entirely which may impact inference for these associations, though the full implications of this approach are not currently clear. Several methods for propagating exposure uncertainty have been developed and used previously (e.g., Gryparis and others, 2009; Lee and Shaddick, 2010; Peng and Bell, 2010; Chang and others, 2011; Szpiro and others, 2011; Warren and others, 2012; Szpiro and Paciorek, 2013; Blangiardo and others, 2016; Lee and others, 2017; Huang and others, 2018), and we provide full details on many of them in Sections 2 and 6. Although prior work in this area has recommended an investigation and comparison of their performances (Lee and others, 2017), to our knowledge such an analysis has yet to be conducted.
In this work, we present a flexible framework for exposure uncertainty propagation, carry out a simulation study to compare its performance to existing methods, and apply several of the methods to better understand the relationship between population-level fine particulate matter (PM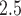 ) exposure and daily stillbirth counts using data from three counties in New Jersey (NJ), 2011–2015. The proposed framework uses kernel density estimation (KDE) with a Gaussian kernel function to specify prior distributions for the exposures within the health model. The resulting model fitting derivations suggest that this represents a hybrid between two existing approaches; allowing for more flexibility while also avoiding some of the limiting assumptions of those approaches. It is also shown to maintain computational efficiency for several common health outcome analysis types. Through simulation, we show that the new approach is flexible enough to accurately characterize uncertainty in the predictions, leading to improved estimation of the association of interest in the health analysis compared to existing approaches. Differences between the methods are also observed in the NJ stillbirth case study results, indicating the importance of selecting the optimal method in future applications.
) exposure and daily stillbirth counts using data from three counties in New Jersey (NJ), 2011–2015. The proposed framework uses kernel density estimation (KDE) with a Gaussian kernel function to specify prior distributions for the exposures within the health model. The resulting model fitting derivations suggest that this represents a hybrid between two existing approaches; allowing for more flexibility while also avoiding some of the limiting assumptions of those approaches. It is also shown to maintain computational efficiency for several common health outcome analysis types. Through simulation, we show that the new approach is flexible enough to accurately characterize uncertainty in the predictions, leading to improved estimation of the association of interest in the health analysis compared to existing approaches. Differences between the methods are also observed in the NJ stillbirth case study results, indicating the importance of selecting the optimal method in future applications.
2. Background
We specify that the primary epidemiological health outcome analysis of interest consists of 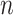 data points (e.g., individuals and time periods) where an exposure level (e.g., ambient air pollution) is assigned to each data point in order to examine its association with the outcome. For presentation purposes, we introduce the framework using a single exposure while noting that it is straightforward to extend the following results/derivations to accommodate multiple additive exposures. Statistical modeling of the exposures and health outcomes are assumed to take place in the Bayesian setting, as is common in the environmental health statistical methodology literature (e.g., Berrocal and others, 2010b; Chang and others, 2011).
data points (e.g., individuals and time periods) where an exposure level (e.g., ambient air pollution) is assigned to each data point in order to examine its association with the outcome. For presentation purposes, we introduce the framework using a single exposure while noting that it is straightforward to extend the following results/derivations to accommodate multiple additive exposures. Statistical modeling of the exposures and health outcomes are assumed to take place in the Bayesian setting, as is common in the environmental health statistical methodology literature (e.g., Berrocal and others, 2010b; Chang and others, 2011).
We assume that 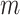 samples from the exposure ppd have been obtained based on the modeling and prediction of observed exposures in a first-stage Bayesian framework. While many modeling options are available, the end result is the same across all approaches; an
samples from the exposure ppd have been obtained based on the modeling and prediction of observed exposures in a first-stage Bayesian framework. While many modeling options are available, the end result is the same across all approaches; an 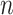 by
by 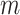 matrix of ppd samples (i.e.,
matrix of ppd samples (i.e., 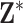 ) is obtained such that
) is obtained such that
 |
In this matrix, 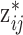 represents the
represents the 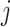 th ppd sample of exposure for health data point
th ppd sample of exposure for health data point  . In terms of notation, it is helpful to define the exposure matrix in terms of row and column vectors such that
. In terms of notation, it is helpful to define the exposure matrix in terms of row and column vectors such that
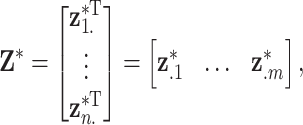 |
where 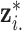 represents the complete set of exposure ppd samples for data point
represents the complete set of exposure ppd samples for data point 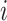 and
and 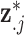 is the vector of exposures for all
is the vector of exposures for all 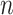 data points from the
data points from the 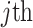 ppd sample.
ppd sample.
Within a column of 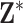 , the exposures for the different data points could be collected independently (i.e., from the marginal ppds) or jointly (i.e., from the joint ppd). Sampling from the marginal ppds, which may be necessary due to computational considerations when working with a large spatial/temporal domain, results in independence across the rows of the
, the exposures for the different data points could be collected independently (i.e., from the marginal ppds) or jointly (i.e., from the joint ppd). Sampling from the marginal ppds, which may be necessary due to computational considerations when working with a large spatial/temporal domain, results in independence across the rows of the 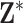 , whereas sampling from the joint ppd retains the correlation across the rows. In either case, trend in exposures across the rows may be present depending on the structure of the data points (e.g., air pollution concentrations across time). We note that independence across the columns of
, whereas sampling from the joint ppd retains the correlation across the rows. In either case, trend in exposures across the rows may be present depending on the structure of the data points (e.g., air pollution concentrations across time). We note that independence across the columns of 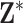 in practice is achieved through Monte Carlo sampling or MCMC sampling and thinning of the collected ppd samples.
in practice is achieved through Monte Carlo sampling or MCMC sampling and thinning of the collected ppd samples.
Once the exposure ppd samples are obtained from the first-stage modeling, they are used in a subsequent epidemiological health outcome analysis to determine their association with the outcome. This second-stage health model typically follows a regression framework of the form
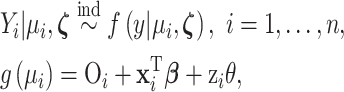 |
(2.1) |
where 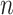 is the previously defined number of data points;
is the previously defined number of data points; 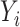 represents the health outcome for data point
represents the health outcome for data point 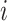 ;
; 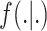 is the probability density function (pdf) of the outcome;
is the probability density function (pdf) of the outcome; 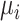 is the mean of this distribution with
is the mean of this distribution with 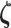 representing additional parameters that often define variance/dispersion or auxiliary variables used to improve the efficiency of posterior sampling;
representing additional parameters that often define variance/dispersion or auxiliary variables used to improve the efficiency of posterior sampling; 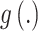 is a link function to connect the mean with a set of covariates;
is a link function to connect the mean with a set of covariates; 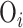 is an offset term sometimes used in the modeling of count data but will be zero otherwise; and
is an offset term sometimes used in the modeling of count data but will be zero otherwise; and 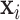 is a vector of covariates unrelated to the primary exposure of interest, including an intercept term, with
is a vector of covariates unrelated to the primary exposure of interest, including an intercept term, with 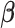 the vector of corresponding regression parameters.
the vector of corresponding regression parameters.
The true but unobserved exposure for data point 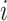 is denoted by
is denoted by 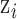 , where
, where 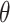 describes the association between exposure and outcome. In this work, we assume that the set of true exposures,
describes the association between exposure and outcome. In this work, we assume that the set of true exposures, 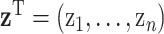 , follows the ppd derived in the first-stage analysis, though we relax this assumption in subsequent sensitivity analyses. However, we avoid the likely unrealistic assumptions made in some previous work that
, follows the ppd derived in the first-stage analysis, though we relax this assumption in subsequent sensitivity analyses. However, we avoid the likely unrealistic assumptions made in some previous work that 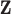 is included as one of the columns of
is included as one of the columns of  (i.e.,
(i.e., 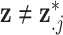 for any
for any 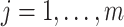 ) (e.g., Peng and Bell, 2010; Chang and others, 2011). Instead, we treat entries of
) (e.g., Peng and Bell, 2010; Chang and others, 2011). Instead, we treat entries of 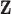 as unknown parameters in (2.1) with the first-stage ppd representing our current state of knowledge about their values. Therefore,
as unknown parameters in (2.1) with the first-stage ppd representing our current state of knowledge about their values. Therefore, 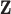 is thought to arise from the same process that produced the columns of
is thought to arise from the same process that produced the columns of 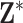 (possibly with additional noise in
(possibly with additional noise in 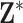 ) but is not actually observed in the finite set of
) but is not actually observed in the finite set of 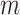 samples collected in the first stage. To complete the model specification, we assign weakly informative prior distributions for each of the introduced parameters in (2.1) with specific settings based on the likelihood choice.
samples collected in the first stage. To complete the model specification, we assign weakly informative prior distributions for each of the introduced parameters in (2.1) with specific settings based on the likelihood choice.
2.1. Existing approaches
Given 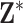 and the fully specified health model from (2.1), the question becomes how to efficiently utilize the information contained in the full set of ppd samples to accurately quantify uncertainty in the exposures when making inference on
and the fully specified health model from (2.1), the question becomes how to efficiently utilize the information contained in the full set of ppd samples to accurately quantify uncertainty in the exposures when making inference on 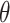 . A number of approaches, ranging in conceptual and computational complexity, have been proposed and we detail several of them below, with additional details given in Section 6. In Figure S1 of the Supplementary material available at Biostatistics online, we present an overview of the different approaches.
. A number of approaches, ranging in conceptual and computational complexity, have been proposed and we detail several of them below, with additional details given in Section 6. In Figure S1 of the Supplementary material available at Biostatistics online, we present an overview of the different approaches.
Plug-in exposures
The simplest approach used in previous work replaces 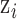 from (2.1) with
from (2.1) with 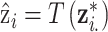 where
where 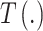 is a function of the input ppd samples (e.g., median, mean) (e.g., Warren and others, 2022). We refer to this as the Plug-in approach. Using only a summary measure of the ppd samples entirely ignores uncertainty in the exposures which may affect uncertainty estimation for
is a function of the input ppd samples (e.g., median, mean) (e.g., Warren and others, 2022). We refer to this as the Plug-in approach. Using only a summary measure of the ppd samples entirely ignores uncertainty in the exposures which may affect uncertainty estimation for 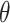 .
.
Multiple imputation
The multiple imputation (MI) approach attempts to incorporate uncertainty in the exposures by fitting the health model in (2.1) separately for each of the 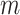 columns of
columns of 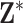 , replacing
, replacing 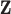 with
with 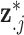 (Blangiardo and others, 2016). However, MI is known to produce a biased effect estimate given that it uses individual draws from a ppd which does not condition on the health outcome data (Little, 1992; Gryparis and others, 2009). During model fit
(Blangiardo and others, 2016). However, MI is known to produce a biased effect estimate given that it uses individual draws from a ppd which does not condition on the health outcome data (Little, 1992; Gryparis and others, 2009). During model fit 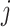 ,
, 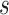 posterior samples (post-convergence and possibly thinned) from
posterior samples (post-convergence and possibly thinned) from 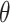 are collected and denoted as
are collected and denoted as 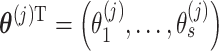 . After fitting the model to all columns of
. After fitting the model to all columns of  , posterior inference is conducted based on the combined samples across all
, posterior inference is conducted based on the combined samples across all 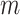 model fits;
model fits; 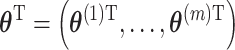 (Zhou and Reiter, 2010). Depending on how long it takes to fit the health model in (2.1), which is impacted by the likelihood choice and sample size, MI may be computationally demanding as
(Zhou and Reiter, 2010). Depending on how long it takes to fit the health model in (2.1), which is impacted by the likelihood choice and sample size, MI may be computationally demanding as 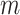 increases. This method also assumes that the columns of
increases. This method also assumes that the columns of 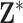 resemble
resemble 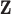 , which is not necessarily true and depends on the amount of variability in the first-stage ppd.
, which is not necessarily true and depends on the amount of variability in the first-stage ppd.
Multiple imputation approximation
The multiple imputation approximation (MIA) approach approximates the results from MI while only requiring a single fit of the health model in (2.1); representing a major computational improvement. Specifically, during each iteration of the MCMC algorithm developed for the health model in (2.1), MIA randomly selects a new column of exposures for the 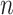 data points with replacement from
data points with replacement from 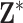 (i.e.,
(i.e., 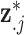 ) and completes a full sweep of the algorithm (i.e., collecting samples from all of the introduced model parameters) (Lee and others, 2017). Posterior inference for
) and completes a full sweep of the algorithm (i.e., collecting samples from all of the introduced model parameters) (Lee and others, 2017). Posterior inference for 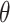 is made based on the
is made based on the 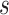 MCMC samples collected from this algorithm. However, similar to MI, MIA suffers from bias due to ignoring the health outcome data when selecting the column of exposures (Little, 1992), which can lead to attenuated effect estimates as observed by Lee and others (2017).
MCMC samples collected from this algorithm. However, similar to MI, MIA suffers from bias due to ignoring the health outcome data when selecting the column of exposures (Little, 1992), which can lead to attenuated effect estimates as observed by Lee and others (2017).
Discrete uniform prior distribution
Similar to MIA, the discrete uniform (DU) approach requires only a single fit of the health model in (2.1) and uses columns directly from 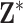 during model fitting. However, instead of randomly selecting a column during each MCMC iteration, DU incorporates the health data in the decision-making, resulting in the selection of more probable columns during posterior sampling. It does so by assigning a prior distribution to
during model fitting. However, instead of randomly selecting a column during each MCMC iteration, DU incorporates the health data in the decision-making, resulting in the selection of more probable columns during posterior sampling. It does so by assigning a prior distribution to 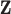 using the collected ppd samples in
using the collected ppd samples in 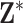 and carrying out full Bayesian inference for the health model in (2.1). Specifically, DU makes the likely unrealistic assumption that the true vector of exposures is contained in
and carrying out full Bayesian inference for the health model in (2.1). Specifically, DU makes the likely unrealistic assumption that the true vector of exposures is contained in 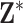 (i.e.,
(i.e., 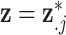 for some
for some 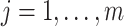 ). Based on this assumption, a prior distribution for
). Based on this assumption, a prior distribution for 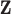 is specified such that
is specified such that
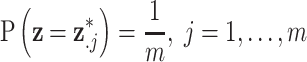 |
(Peng and Bell, 2010; Chang and others, 2011). Use of DU yields a semi-conjugate full conditional distribution for 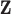 regardless of the choice for
regardless of the choice for 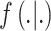 in (2.1), allowing for convenient updating during MCMC sampling. When
in (2.1), allowing for convenient updating during MCMC sampling. When 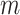 becomes large, a more computationally efficient Metropolis algorithm can be used to propose/evaluate columns from
becomes large, a more computationally efficient Metropolis algorithm can be used to propose/evaluate columns from 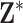 using a likelihood ratio calculation.
using a likelihood ratio calculation.
Multivariate normal prior distribution
Similar to DU, the multivariate normal (MVN) approach assigns a prior distribution to 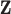 but avoids the assumption that
but avoids the assumption that 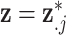 for some
for some 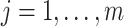 . Specifically, a MVN prior distribution for
. Specifically, a MVN prior distribution for 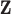 is specified such that
is specified such that
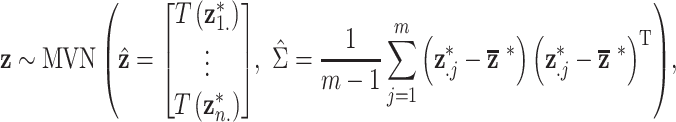 |
where 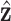 and
and 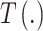 have been previously described, and
have been previously described, and 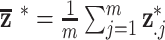 is the length
is the length 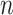 vector of average exposures across all
vector of average exposures across all 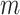 ppd samples (Warren and others, 2012; Lee and others, 2017).
ppd samples (Warren and others, 2012; Lee and others, 2017).
Updating 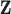 within an MCMC algorithm is straightforward (i.e., the vector has a standard, closed-form full conditional distribution) for multiple likelihood choices that cover a number of relevant health outcome data types, including Gaussian with identity link function (continuous outcome), Bernoulli with logit link function (binary outcome), and negative binomial with logit link function (count data). The latter two likelihood/link function results are made possible by the work of Polson and others (2013). Details for deriving this distribution are provided in Section S1 of the Supplementary material available at Biostatistics online. However, posterior sampling will be increasingly time consuming as
within an MCMC algorithm is straightforward (i.e., the vector has a standard, closed-form full conditional distribution) for multiple likelihood choices that cover a number of relevant health outcome data types, including Gaussian with identity link function (continuous outcome), Bernoulli with logit link function (binary outcome), and negative binomial with logit link function (count data). The latter two likelihood/link function results are made possible by the work of Polson and others (2013). Details for deriving this distribution are provided in Section S1 of the Supplementary material available at Biostatistics online. However, posterior sampling will be increasingly time consuming as  increases given the large dimension of the full conditional distribution covariance matrix. Additionally, MVN may struggle when the shape of the exposure ppd deviates substantially from normality (e.g., skewness, multiple modes).
increases given the large dimension of the full conditional distribution covariance matrix. Additionally, MVN may struggle when the shape of the exposure ppd deviates substantially from normality (e.g., skewness, multiple modes).
3. KDE prior distributions
We propose using univariate and multivariate KDE with a Gaussian kernel function to fully leverage the information contained in 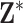 when estimating the health model in (2.1), detail its intuitive connections with existing approaches, and consider its computational requirements. We show that assigning prior distributions for
when estimating the health model in (2.1), detail its intuitive connections with existing approaches, and consider its computational requirements. We show that assigning prior distributions for 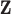 based on KDE results in a more flexible hybrid between DU and MVN, allowing us to avoid potentially problematic assumptions made by existing methods without significantly increasing the computational burden.
based on KDE results in a more flexible hybrid between DU and MVN, allowing us to avoid potentially problematic assumptions made by existing methods without significantly increasing the computational burden.
For the univariate version of KDE with a Gaussian kernel function (UKDE), the prior distributions for 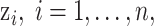 are specified independently as
are specified independently as
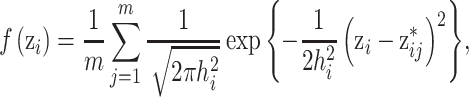 |
where 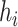 is the bandwidth variable that controls the level of smoothness of the density function. There are many methods available to select
is the bandwidth variable that controls the level of smoothness of the density function. There are many methods available to select 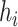 using the ppd samples in
using the ppd samples in 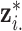 (e.g., Sheather and Jones, 1991) and density estimation results are often sensitive to its value. This prior distribution represents a mixture of
(e.g., Sheather and Jones, 1991) and density estimation results are often sensitive to its value. This prior distribution represents a mixture of 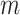 equally weighted normal distributions centered at the observed samples from the ppd (i.e.,
equally weighted normal distributions centered at the observed samples from the ppd (i.e., 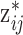 ) each with standard deviation
) each with standard deviation 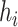 . This allows information from each individual ppd sample to be utilized when fitting the health model in (2.1) and thereby avoids relying on overly simplistic summaries of the samples used by existing methods (e.g., Plug-in, MVN).
. This allows information from each individual ppd sample to be utilized when fitting the health model in (2.1) and thereby avoids relying on overly simplistic summaries of the samples used by existing methods (e.g., Plug-in, MVN).
Because we select a Gaussian kernel function, the full conditional distribution for each 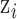 has a closed form for the likelihood/link function combinations mentioned previously in Section 2, allowing for convenient updates within an MCMC algorithm. Specifically, based on the health model in (2.1) the full conditional distribution for
has a closed form for the likelihood/link function combinations mentioned previously in Section 2, allowing for convenient updates within an MCMC algorithm. Specifically, based on the health model in (2.1) the full conditional distribution for  is a mixture of univariate normal distributions such that
is a mixture of univariate normal distributions such that
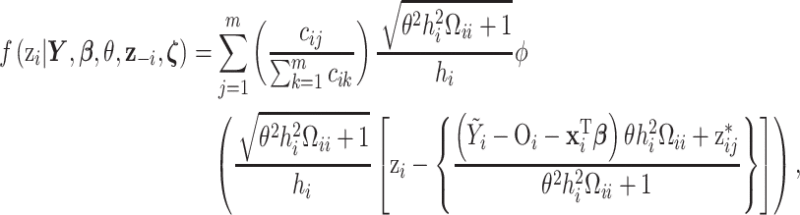 |
(3.2) |
where 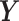 is the complete vector of
is the complete vector of 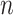 data points;
data points; 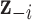 is the complete vector of true exposures with
is the complete vector of true exposures with 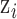 removed;
removed; 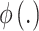 is the pdf of the standard univariate normal distribution;
is the pdf of the standard univariate normal distribution; 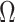 is an
is an 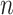 by
by 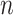 diagonal matrix with
diagonal matrix with
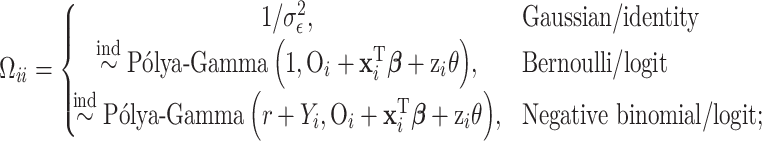 |
and
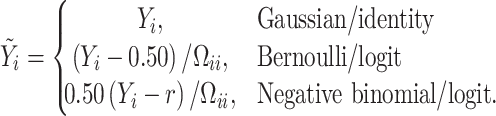 |
In the case of Gaussian distributed health outcome data, 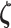 includes the error variance parameter
includes the error variance parameter 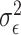 , and in the negative binomial case it includes the dispersion parameter
, and in the negative binomial case it includes the dispersion parameter 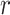 . The mixture weights in (3.2) are defined as
. The mixture weights in (3.2) are defined as
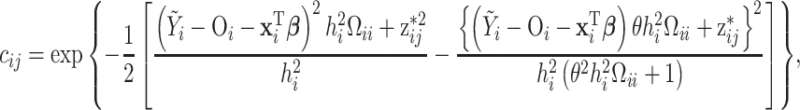 |
(3.3) |
where all remaining terms have been previously described. Further details for these derivations are provided in Section S1 of the Supplementary material available at Biostatistics online.
The form of the full conditional distribution in (3.2–3.3) suggests that the use of UKDE for specifying exposure prior distributions results in a hybrid approach, combining features of DU with those of MVN. Specifically, for updating 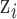 we sample from this mixture distribution in two steps.
we sample from this mixture distribution in two steps.
-
(1)
Choosing a probable observed exposure: We compute
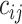 in (3.3), which depends on the health data and ppd exposure value
in (3.3), which depends on the health data and ppd exposure value 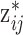 , for every
, for every 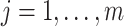 . Each
. Each 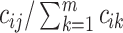 is computed and a random
is computed and a random 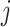 index corresponding to
index corresponding to 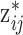 is selected based on these probabilities.
is selected based on these probabilities. -
(2)
Updating exposure based on selected value: Based on the selected
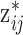 ppd sample, the true exposure
ppd sample, the true exposure 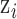 is drawn from the normal distribution in (3.2) whose mean depends on
is drawn from the normal distribution in (3.2) whose mean depends on 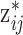 and none of the other collected ppd samples.
and none of the other collected ppd samples.
The first step resembles DU in that the health data are used to inform a probable ppd sample. However, once this choice is made, DU assigns it as the true exposure unlike UKDE. The second step resembles MVN since the true exposure is simulated from a distribution and is therefore, not required to be one of the observed columns of  . However, with MVN the prior mean of the exposures remains the same across all MCMC iterations for the corresponding full conditional distribution update (i.e.,
. However, with MVN the prior mean of the exposures remains the same across all MCMC iterations for the corresponding full conditional distribution update (i.e., 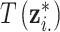 ). For UKDE, the mean in (3.2) changes each time a new
). For UKDE, the mean in (3.2) changes each time a new 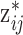 is selected.
is selected.
Therefore, UKDE represents a more flexible alternative to DU and MVN as it avoids the assumption that the true exposures are observed in 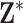 and allows the prior mean of the exposures to effectively vary in the full conditional distribution update across MCMC iterations. UKDE should also be better able to handle non-symmetric ppd behavior than MVN; although it does not directly account for correlation between the exposures corresponding to different data points. In Section S2 of the Supplementary material available at Biostatistics online, we extend the UKDE framework to the multivariate setting (i.e., MKDE) by specifying a prior distribution on the entire vector
and allows the prior mean of the exposures to effectively vary in the full conditional distribution update across MCMC iterations. UKDE should also be better able to handle non-symmetric ppd behavior than MVN; although it does not directly account for correlation between the exposures corresponding to different data points. In Section S2 of the Supplementary material available at Biostatistics online, we extend the UKDE framework to the multivariate setting (i.e., MKDE) by specifying a prior distribution on the entire vector 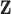 , detail similar connections with existing approaches, and note some limitations of MKDE due to the large dimension of the exposure data in most environmental health applications.
, detail similar connections with existing approaches, and note some limitations of MKDE due to the large dimension of the exposure data in most environmental health applications.
4. Simulation study
We design a simulation study to compare each of the methods detailed in Sections 2 and 3 with respect to estimating the association between exposure and health while accounting for exposure uncertainty.
4.1. Data generation
We begin by defining the health model based on (2.1) and using a Gaussian likelihood with identity link function, no offset term, and no intercept/covariates such that
 |
(4.4) |
for 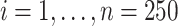 , where all terms have been previously described in Section 2. Two-dimensional spatial locations for the
, where all terms have been previously described in Section 2. Two-dimensional spatial locations for the 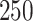 individuals in the study are simulated randomly within the unit square. We consider two different settings for the primary risk parameter,
individuals in the study are simulated randomly within the unit square. We consider two different settings for the primary risk parameter, 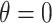 and
and 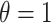 . These settings allow us to investigate the type I error rate and power of the competing approaches, respectively.
. These settings allow us to investigate the type I error rate and power of the competing approaches, respectively.
Next, we simulate exposure data for analysis. In each setting, we define a unique ppd and use it to create 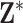 , which in this case represents a
, which in this case represents a 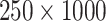 matrix of exposures (i.e.,
matrix of exposures (i.e., 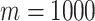 ). We define the true exposures by simulating an additional vector from the same ppd and assigning it to
). We define the true exposures by simulating an additional vector from the same ppd and assigning it to 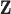 . The true exposures are used to simulate the health data from (4.4) and are not included in
. The true exposures are used to simulate the health data from (4.4) and are not included in 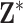 .
.
When defining the ppds, we vary several factors to test the performances of the competing methods across different settings. Specifically, we consider the correlation between exposures (uncorrelated, correlated), skewness of the exposure distribution (symmetric, skewed), and variability of the means of the marginal exposure distributions (low, high). Each column of 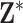 is simulated based on these settings such that
is simulated based on these settings such that
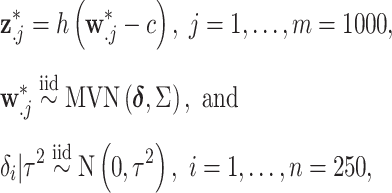 |
(4.5) |
where 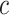 is the minimum simulated value across all
is the minimum simulated value across all 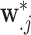 vectors and is used to ensure that the pretransformed exposures are
vectors and is used to ensure that the pretransformed exposures are 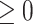 . We standardize
. We standardize 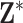 prior to analysis by subtracting the mean of the entire matrix from each element of
prior to analysis by subtracting the mean of the entire matrix from each element of 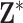 and dividing these centered values by the standard deviation of the entire matrix.
and dividing these centered values by the standard deviation of the entire matrix.
For uncorrelated exposures, we define 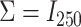 (i.e., the
(i.e., the 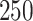 by
by 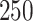 identity matrix) while for correlated data we define
identity matrix) while for correlated data we define 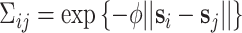 , where
, where  represents the Euclidean distance between individuals
represents the Euclidean distance between individuals 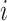 and
and 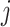 , and
, and 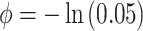 which allows for the correlation between two individuals to equal
which allows for the correlation between two individuals to equal 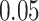 at a distance of
at a distance of 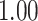 (recall that individuals are simulated within the unit square where the maximum possible distance between two individuals is
(recall that individuals are simulated within the unit square where the maximum possible distance between two individuals is 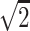 ). For symmetric data,
). For symmetric data,  is defined as the identity function while
is defined as the identity function while 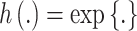 for skewed data. Finally,
for skewed data. Finally, 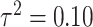 represents low variability in the means of the marginal ppds while
represents low variability in the means of the marginal ppds while 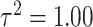 represents high variability. As
represents high variability. As 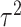 increases, these means become further separated and we expect improved performance across all methods since there will be less relative uncertainty in the exposure distributions. Full details on all considered settings are given in Table 1 while randomly selected simulated data sets from each setting are presented in Figures S2 and S3 of the Supplementary material available at Biostatistics online.
increases, these means become further separated and we expect improved performance across all methods since there will be less relative uncertainty in the exposure distributions. Full details on all considered settings are given in Table 1 while randomly selected simulated data sets from each setting are presented in Figures S2 and S3 of the Supplementary material available at Biostatistics online.
Table 1.
Simulation study settings for exposure posterior predictive distributions based on (4.5)
| Factor | Setting | Form |
|---|---|---|
| Associated | No |

|
| Yes |
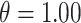
|
|
| Correlated | No |
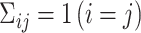
|
| Yes |
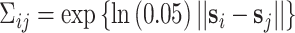
|
|
| Skewed | No |
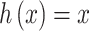
|
| Yes |
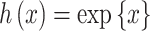
|
|
 Variance
Variance |
Low |
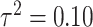
|
| High |
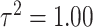
|
For each simulated data set, we generate a new set of spatial locations for the 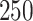 individuals and a unique set of exposures (i.e.,
individuals and a unique set of exposures (i.e., 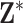 and
and 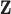 ). For every combination of factors in Table 1, we simulate 500 data sets and analyze each one using the approaches described in Sections 2 and 3.
). For every combination of factors in Table 1, we simulate 500 data sets and analyze each one using the approaches described in Sections 2 and 3.
We also consider two additional sensitivity analyses to determine the impact of misspecifying the ppd (with respect to the true exposures) on the health effect estimation across the different methods. This misspecification may occur in practice due to the difficulty in choosing the most appropriate first-stage framework for modeling/predicting the exposures. To create misspecified ppd samples in our study, we first carry out the original algorithm detailed in (4.5) to define 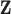 and
and 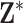 . However, we then add noise to each entry of
. However, we then add noise to each entry of 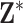 , but not to
, but not to 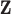 , such that
, such that  where
where 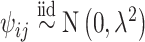 . This ensures that the ppd samples we analyze are from a different, and more variable, distribution than the distribution that generated
. This ensures that the ppd samples we analyze are from a different, and more variable, distribution than the distribution that generated 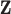 . As
. As 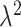 gets larger, the ppd samples provide less information about
gets larger, the ppd samples provide less information about 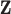 . As
. As 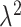 gets closer to zero, we reach our original limiting case where the ppd samples and true exposures arise from the same distribution. We select small (
gets closer to zero, we reach our original limiting case where the ppd samples and true exposures arise from the same distribution. We select small (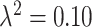 ) and large (
) and large (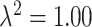 ) settings for the error variance and rerun the entire simulation study.
) settings for the error variance and rerun the entire simulation study.
4.2. Data analysis
The health model, which is shared across all methods, is given as
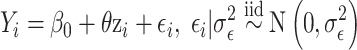 |
(4.6) |
with prior distributions 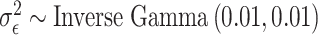 (all methods),
(all methods), 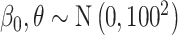 (MIA, MVN, DU, UKDE, MKDE), and
(MIA, MVN, DU, UKDE, MKDE), and 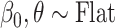 (Plug-in, MI). The decision to use different prior distributions for the regression parameters across the methods is made for computational reasons only, as the flat priors allow for more efficient Monte Carlo posterior sampling for Plug-in and MI, saving considerable computing time. We do not anticipate major changes in the results due to these differences given that neither distribution contains informative prior information about the parameters. For UKDE, we use the method from Sheather and Jones (1991) to select the bandwidth variables
(Plug-in, MI). The decision to use different prior distributions for the regression parameters across the methods is made for computational reasons only, as the flat priors allow for more efficient Monte Carlo posterior sampling for Plug-in and MI, saving considerable computing time. We do not anticipate major changes in the results due to these differences given that neither distribution contains informative prior information about the parameters. For UKDE, we use the method from Sheather and Jones (1991) to select the bandwidth variables 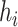 , while for MKDE we rely on Scott’s rule (Scott, 2015) and the sample covariance of
, while for MKDE we rely on Scott’s rule (Scott, 2015) and the sample covariance of 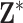 to select the full bandwidth matrix variable
to select the full bandwidth matrix variable 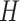 (i.e.,
(i.e., 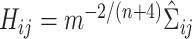 ). In early exploring of the data, we tested other standard methods for selecting the values of these variables and found similar estimates across the different approaches.
). In early exploring of the data, we tested other standard methods for selecting the values of these variables and found similar estimates across the different approaches.
From each method, we collect 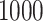 posterior samples with which to make inference. For the approaches where MCMC sampling is required, we run the algorithms for
posterior samples with which to make inference. For the approaches where MCMC sampling is required, we run the algorithms for 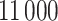 iterations, removing the first
iterations, removing the first 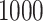 as a burn-in period, and thinning the remaining samples by a factor of
as a burn-in period, and thinning the remaining samples by a factor of 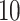 . For the Monte Carlo sampling approaches, we directly obtain
. For the Monte Carlo sampling approaches, we directly obtain 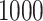 independent samples from the joint posterior distributions. We estimate
independent samples from the joint posterior distributions. We estimate 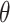 using the posterior mean and quantify uncertainty using the 95
using the posterior mean and quantify uncertainty using the 95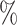 quantile-based, equal-tailed credible interval.
quantile-based, equal-tailed credible interval.
We compare the ability of each method to estimate 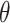 under the different simulation settings by estimating and comparing the bias and mean squared error (MSE) of the posterior mean, empirical coverage (EC) of the 95
under the different simulation settings by estimating and comparing the bias and mean squared error (MSE) of the posterior mean, empirical coverage (EC) of the 95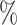 credible interval, and power/type I error rate. For reference, we also calculate these quantities for the model in (4.6) where the true exposures are used—an analysis not possible in practice.
credible interval, and power/type I error rate. For reference, we also calculate these quantities for the model in (4.6) where the true exposures are used—an analysis not possible in practice.
4.3. Results
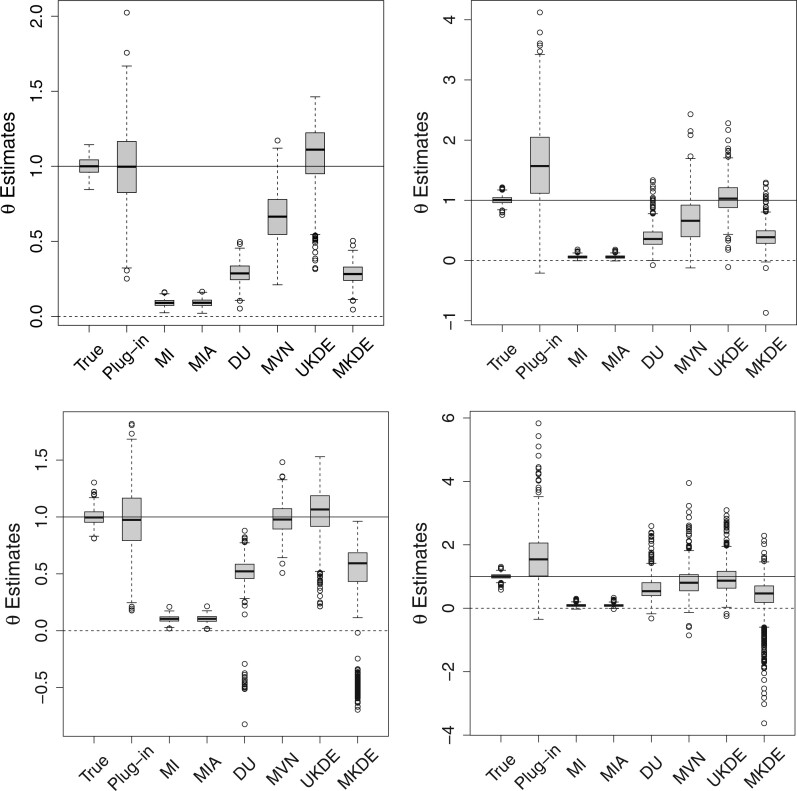
In Figure 1, we display boxplots of the 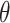 estimates across all 500 analyses for each method and correlation/skewness setting for the
estimates across all 500 analyses for each method and correlation/skewness setting for the 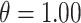 and
and 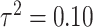 scenario. Similar boxplots for the other
scenario. Similar boxplots for the other 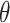 and
and 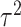 combinations are shown in Figures S4–S6 of the Supplementary material available at Biostatistics online. In Table 2, we present the simulation study results, also from the
combinations are shown in Figures S4–S6 of the Supplementary material available at Biostatistics online. In Table 2, we present the simulation study results, also from the 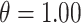 and
and 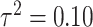 scenario. Similar results are displayed in Tables S1–S3 of the Supplementary material available at Biostatistics online for the other
scenario. Similar results are displayed in Tables S1–S3 of the Supplementary material available at Biostatistics online for the other 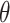 and
and 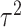 combinations. The
combinations. The 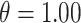 ,
, 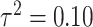 results suggest that MI and MIA perform very similarly overall as expected, and that both struggle to estimate
results suggest that MI and MIA perform very similarly overall as expected, and that both struggle to estimate 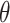 well across all settings. While DU outperforms MI and MIA, it tends to have a larger bias and MSE, and lower EC than some of the remaining approaches. These methods may only be appropriate when the centers of the ppds are well separated with low uncertainty such that each column of
well across all settings. While DU outperforms MI and MIA, it tends to have a larger bias and MSE, and lower EC than some of the remaining approaches. These methods may only be appropriate when the centers of the ppds are well separated with low uncertainty such that each column of 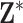 begins to resemble the true exposures
begins to resemble the true exposures 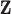 .
.
Fig. 1.
Posterior mean estimates of 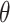 for each method and correlation/skewness setting across all 500 analyses for the
for each method and correlation/skewness setting across all 500 analyses for the 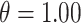 and
and 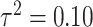 scenario (first panel: uncorrelated, not skewed; second panel: uncorrelated, skewed; third panel: correlated, not skewed; fourth panel: correlated, skewed). The solid horizontal line represents the true value of
scenario (first panel: uncorrelated, not skewed; second panel: uncorrelated, skewed; third panel: correlated, not skewed; fourth panel: correlated, skewed). The solid horizontal line represents the true value of 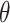 .
.
Table 2.
Simulation study results for 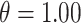 and
and 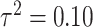 . Estimates are presented with the range of standard errors for a group of estimates provided in parentheses. Bold entries represent the optimal estimate across an entire row (i.e., closest to zero for bias, smallest for mean squared error (MSE), closest to 95 for empirical coverage (EC), and largest for power). All results are multiplied by 100 for presentation purposes
. Estimates are presented with the range of standard errors for a group of estimates provided in parentheses. Bold entries represent the optimal estimate across an entire row (i.e., closest to zero for bias, smallest for mean squared error (MSE), closest to 95 for empirical coverage (EC), and largest for power). All results are multiplied by 100 for presentation purposes
| Settings | Methods | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Metric | Corr | Skew | True | Plug-in | MI | MIA | DU | MVN | UKDE | MKDE |
| Bias | No | No | 0.15 |
 1.03
1.03
|
 90.95
90.95 |
 90.96
90.96 |
 71.26
71.26 |
 33.68
33.68 |
5.71 |
 71.65
71.65 |
| No | Yes | 0.30 | 60.77 |
 94.07
94.07 |
 94.07
94.07 |
 61.23
61.23 |
 30.73
30.73 |
4.59 |
 59.06
59.06 |
|
| Yes | No |
 0.05
0.05 |
 2.58
2.58 |
 89.66
89.66 |
 89.64
89.64 |
 50.49
50.49 |
 2.25
2.25
|
2.69 | -61.78 | |
| Yes | Yes | 0.00 | 62.14 |
 90.83
90.83 |
 90.83
90.83 |
-35.10 |
 15.02
15.02 |
 4.46
4.46
|
 73.23
73.23 |
|
| (0.27–0.40) | (1.21–3.24) | (0.11–0.21) | (0.12–0.21) | (0.31–1.80) | (0.61–2.33) | (0.99–2.25) | (0.31–3.57) | |||
| MSE | No | No | 0.36 | 7.29 | 82.79 | 82.80 | 51.25 | 14.17 | 5.22 | 51.80 |
| No | Yes | 0.46 | 89.38 | 88.56 | 88.57 | 40.90 | 24.01 | 7.89 | 38.99 | |
| Yes | No | 0.47 | 7.78 | 80.48 | 80.45 | 30.14 | 1.89 | 5.29 | 60.53 | |
| Yes | Yes | 0.82 | 116.24 | 82.72 | 82.72 | 28.44 | 29.36 | 25.41 | 117.26 | |
| (0.02–0.07) | (0.47–10.62) | (0.21–0.37) | (0.21–0.37) | (0.44–1.29) | (0.55–2.83) | (0.29–2.28) | (0.44–10.38) | |||
| EC | No | No | 96.00 | 97.00 | 0.00 | 0.00 | 0.00 | 58.80 | 93.00 | 0.00 |
| No | Yes | 94.80 | 80.40 | 0.00 | 0.00 | 2.40 | 49.40 | 86.00 | 4.00 | |
| Yes | No | 94.40 | 95.00 | 0.00 | 0.00 | 1.00 | 92.80 | 93.80 | 3.80 | |
| Yes | Yes | 95.00 | 81.60 | 8.00 | 8.20 | 30.60 | 46.00 | 64.80 | 19.80 | |
| (0.88–1.03) | (0.76–1.78) | (0.00–1.21) | (0.00–1.23) | (0.00–2.06) | (1.16–2.24) | (1.08–2.14) | (0.00–1.76) | |||
| Power | No | No | 100.00 | 92.60 | 0.00 | 0.00 | 54.80 | 88.40 | 94.80 | 52.40 |
| No | Yes | 100.00 | 70.80 | 0.00 | 0.00 | 67.20 | 54.60 | 98.20 | 70.00 | |
| Yes | No | 100.00 | 93.60 | 0.00 | 0.00 | 93.00 | 99.80 | 94.60 | 75.20 | |
| Yes | Yes | 100.00 | 70.20 | 0.00 | 0.00 | 85.60 | 78.80 | 89.00 | 63.60 | |
| (0.00–0.00) | (1.09–2.05) | (0.00–0.00) | (0.00–0.00) | (1.14–2.23) | (0.20–2.23) | (0.59–1.40) | (1.93–2.23) | |||
When the ppds are not skewed, Plug-in generally performs well overall which is an encouraging sign for past studies that have implemented this approach. However, Plug-in struggles greatly when the ppds become skewed, leading to elevated bias and MSE. MVN generally handles skewness better than Plug-in and has improved performance when the ppds are correlated. However, it tends to have lower EC, particularly when the ppds are independent and/or skewed.
MKDE performs similarly to DU across most metrics. It likely struggles due to the difficulty in selecting the bandwidth matrix variable 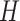 when the dimension of the data is large. Overall, UKDE has the best balance of performance compared to the other methods, especially when the ppds are skewed. For
when the dimension of the data is large. Overall, UKDE has the best balance of performance compared to the other methods, especially when the ppds are skewed. For 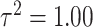 , several of the methods perform more similarly to each other and we anticipate that this trend will continue as the ppds become further separated and the relative error is effectively reduced.
, several of the methods perform more similarly to each other and we anticipate that this trend will continue as the ppds become further separated and the relative error is effectively reduced.
When 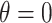 , we see that all methods perform as expected while MI, MIA, DU, and MKDE appear to be preferred in terms of bias and MSE primarily because they produce estimates of
, we see that all methods perform as expected while MI, MIA, DU, and MKDE appear to be preferred in terms of bias and MSE primarily because they produce estimates of 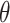 near zero regardless of its true value. Therefore, none of the considered methods are falsely identifying associations at a higher rate than expected; an important conclusion for previous studies that found evidence of significant associations between health and exposure using these techniques.
near zero regardless of its true value. Therefore, none of the considered methods are falsely identifying associations at a higher rate than expected; an important conclusion for previous studies that found evidence of significant associations between health and exposure using these techniques.
In Figures S7 (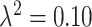 ) and S8 (
) and S8 (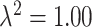 ) of the Supplementary material available at Biostatistics online, we display the boxplots for the misspecified ppd sensitivity analyses for each of the correlation/skewness settings and methods. In the
) of the Supplementary material available at Biostatistics online, we display the boxplots for the misspecified ppd sensitivity analyses for each of the correlation/skewness settings and methods. In the 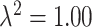 analysis, the posterior mean estimates of
analysis, the posterior mean estimates of 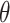 are generally more variable and pulled towards the null for most methods, with smaller deviations seen in the
are generally more variable and pulled towards the null for most methods, with smaller deviations seen in the 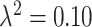 plots. However, the general pattern across the different methods remains consistent and suggests that UKDE performs relatively well even when the ppd is misspecified.
plots. However, the general pattern across the different methods remains consistent and suggests that UKDE performs relatively well even when the ppd is misspecified.
5. PM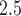 and stillbirth in New Jersey
and stillbirth in New Jersey
Stillbirth is generally defined as the loss of a fetus or baby before or during a delivery that occurs on or after 20 completed weeks of gestation (Centers for Disease Control and Prevention, 2022). Recent literature reviews suggest that exposure to ambient air pollution during pregnancy may be associated with an increased risk of stillbirth (Bekkar and others, 2020; Zhang and others, 2021), though further studies are needed to better understand this relationship. In this application, we examine the relationship between exposure to PM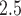 in the days prior to delivery and risk of stillbirth using a population-level time series approach in NJ, 2011–2015. We also investigate the impact of the different methods for accounting for exposure uncertainty in the health analysis, detailed in Sections 2 and 3, on the findings.
in the days prior to delivery and risk of stillbirth using a population-level time series approach in NJ, 2011–2015. We also investigate the impact of the different methods for accounting for exposure uncertainty in the health analysis, detailed in Sections 2 and 3, on the findings.
5.1. Data description
We obtain daily counts of fetal deaths and live birth across the three NJ counties located in the New York (NY)–White Plains–Wayne, NY–NJ Metropolitan Division (i.e., Bergen, Hudson, and Passaic) between 2011 and 2015 from the Division of Family Health Services in the NJ Department of Health. Similar to Warren and others (2022), we only include singleton deaths/births and those with a clinically estimated gestational age of 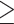 20 weeks. A fetal death occurring on or after 20 weeks of gestation is defined as a stillbirth. In Figure 2a, we display the study area as well as the proportion of stillbirths across time for this area.
20 weeks. A fetal death occurring on or after 20 weeks of gestation is defined as a stillbirth. In Figure 2a, we display the study area as well as the proportion of stillbirths across time for this area.
Fig. 2.
Description of the study area, health data, and exposure modeling/prediction for the New Jersey three county stillbirth and maximum daily 24-h PM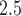 exposure analysis.
exposure analysis.
Observed air pollution data and model-derived estimates are both obtained from the United States Environmental Protection Agency (US EPA). Specifically, for each day in 2002–2015, we access 24-h average PM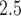 concentrations (micrograms per cubic meter (
concentrations (micrograms per cubic meter (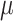 g/m
g/m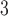 )) measured from all active monitors located in NJ, NY, Delaware, and Pennsylvania from the US EPA’s Air Quality System (AQS) (US EPA, 2022a). Model-derived daily estimates of 24-h average PM
)) measured from all active monitors located in NJ, NY, Delaware, and Pennsylvania from the US EPA’s Air Quality System (AQS) (US EPA, 2022a). Model-derived daily estimates of 24-h average PM concentrations (
concentrations (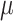 g/m
g/m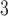 ) from the Community Multiscale Air Quality (CMAQ) model, a deterministic numerical air quality model, are obtained on a 12 km by 12 km grid across the same study area and time period (US EPA, 2022b). All data and estimates were downloaded from the US EPA’s Remote Sensing Information Gateway website (US EPA, 2022c). Figure 2b displays the locations of the AQS and CMAQ data from 2002 to 2015.
) from the Community Multiscale Air Quality (CMAQ) model, a deterministic numerical air quality model, are obtained on a 12 km by 12 km grid across the same study area and time period (US EPA, 2022b). All data and estimates were downloaded from the US EPA’s Remote Sensing Information Gateway website (US EPA, 2022c). Figure 2b displays the locations of the AQS and CMAQ data from 2002 to 2015.
We also obtain daily estimates of the minimum and maximum temperatures at a 1 km resolution across the three NJ counties between 2011 and 2015 from Daymet (Oak Ridge National Laboratory, 2022). The Daymet framework employs statistical modeling techniques to produce spatially temporally interpolated temperature estimates using observed ground-based data as input. On each study day, we average the estimates within the three counties to obtain a single daily average minimum/maximum temperature estimate for the region.
5.2. Stage 1: PM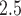 modeling and prediction
modeling and prediction
In the first stage of analysis, we use a hierarchical Bayesian framework for modeling and predicting the daily PM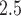 concentrations collected from the AQS using the closest CMAQ estimate as a predictor. Many of the air pollution monitors in the study region are not active on a given day and the network of monitors is only sparsely located across the study area (see Figure 2b). As a result, we use this model and the spatiotemporal completeness of the CMAQ estimates to predict the AQS data at unobserved locations and days. We then take the maximum of these predictions within the three NJ counties on each day to estimate the daily maximum of the 24-h PM
concentrations collected from the AQS using the closest CMAQ estimate as a predictor. Many of the air pollution monitors in the study region are not active on a given day and the network of monitors is only sparsely located across the study area (see Figure 2b). As a result, we use this model and the spatiotemporal completeness of the CMAQ estimates to predict the AQS data at unobserved locations and days. We then take the maximum of these predictions within the three NJ counties on each day to estimate the daily maximum of the 24-h PM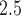 concentrations for the study area. This is used as the primary exposure of interest in the subsequent stillbirth epidemiological analysis.
concentrations for the study area. This is used as the primary exposure of interest in the subsequent stillbirth epidemiological analysis.
The model for the AQS PM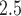 data uses the closest CMAQ estimate as a predictor within a flexible spatially and temporally varying regression coefficient framework, similar to the original downscaling work of Berrocal and others (2010b), such that
data uses the closest CMAQ estimate as a predictor within a flexible spatially and temporally varying regression coefficient framework, similar to the original downscaling work of Berrocal and others (2010b), such that
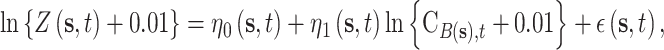 |
(5.7) |
where 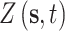 is the AQS PM
is the AQS PM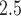 concentration measured at the monitor located at
concentration measured at the monitor located at 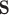 on day
on day 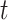 (i.e.,
(i.e., 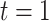 is January 1, 2002 and
is January 1, 2002 and 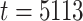 is December 31, 2015);
is December 31, 2015); 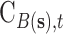 is the corresponding CMAQ estimate at the grid cell centroid located closest to
is the corresponding CMAQ estimate at the grid cell centroid located closest to 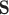 (i.e.,
(i.e., 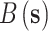 ) on day
) on day 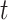 ; and
; and 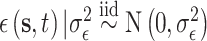 . We work on the log scale during modeling given that the PM
. We work on the log scale during modeling given that the PM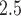 concentrations are
concentrations are 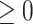 .
.
The spatially and temporally varying intercept and slope parameters are represented by 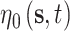 and
and 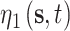 , respectively, and allow the association between the CMAQ estimates and AQS data to flexibly change across space and time if appropriate. They are modeled as a function of spatial and temporal covariates such that
, respectively, and allow the association between the CMAQ estimates and AQS data to flexibly change across space and time if appropriate. They are modeled as a function of spatial and temporal covariates such that
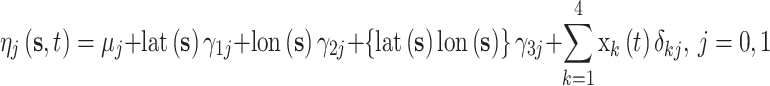 |
(5.8) |
where 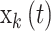 corresponds to the
corresponds to the 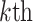 column of the B-spline basis matrix for a polynomial spline with four degrees of freedom (df) on day
column of the B-spline basis matrix for a polynomial spline with four degrees of freedom (df) on day 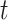 , and
, and 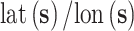 are the latitude/longitude at spatial location
are the latitude/longitude at spatial location 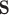 , respectively.
, respectively.
We complete the model by specifying non/weakly informative prior distributions for the introduced model parameters. Specifically, we choose flat prior distributions for all of the regression parameters (i.e., 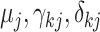 ), and
), and 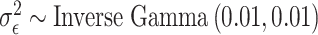 ; resulting in an efficient closed-form Monte Carlo sampling algorithm for obtaining samples from the joint posterior distribution of the model parameters. We use it to collect
; resulting in an efficient closed-form Monte Carlo sampling algorithm for obtaining samples from the joint posterior distribution of the model parameters. We use it to collect 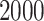 independent posterior samples in total.
independent posterior samples in total.
Next, we use composition sampling to generate independent posterior predictive samples of 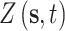 at each of the nine CMAQ grid cell locations within the three NJ counties on each day of the study. On each day and for every joint ppd sample, we calculate the maximum of the predictions across the three counties. In total, we obtain a
at each of the nine CMAQ grid cell locations within the three NJ counties on each day of the study. On each day and for every joint ppd sample, we calculate the maximum of the predictions across the three counties. In total, we obtain a 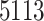 (i.e., number of days) by
(i.e., number of days) by  matrix of maximum 24-h average PM
matrix of maximum 24-h average PM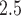 ppd samples, denoted by
ppd samples, denoted by 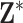 , and use it as the exposure for the stillbirth epidemiological analysis.
, and use it as the exposure for the stillbirth epidemiological analysis.
5.3. Stage 2: Modeling stillbirth and PM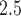
Given 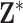 from Stage 1, we next turn to the stillbirth epidemiological analysis. We model the total number of stillbirths occurring across the three NJ counties on a specific day as a function of time-varying predictors (i.e., day of the week, long-term trend), meteorological variables (i.e., maximum/minimum temperature), and lagged PM
from Stage 1, we next turn to the stillbirth epidemiological analysis. We model the total number of stillbirths occurring across the three NJ counties on a specific day as a function of time-varying predictors (i.e., day of the week, long-term trend), meteorological variables (i.e., maximum/minimum temperature), and lagged PM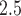 exposure. The model is given as
exposure. The model is given as
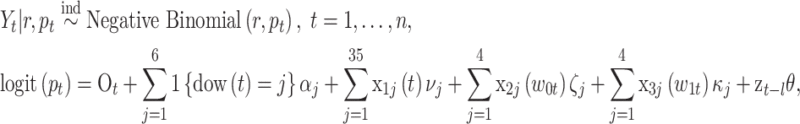 |
(5.9) |
where 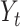 is the number of stillbirths occurring on day
is the number of stillbirths occurring on day 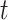 with
with 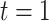 corresponding to January 1, 2011 and
corresponding to January 1, 2011 and 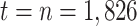 to December 31, 2015;
to December 31, 2015; 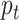 is the probability parameter that controls the magnitude of counts on day
is the probability parameter that controls the magnitude of counts on day 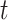 ;
; 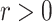 represents the dispersion parameter with small values indicating overdispersion in the data;
represents the dispersion parameter with small values indicating overdispersion in the data; 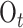 is the offset variable representing the log of the total number of births occurring on day
is the offset variable representing the log of the total number of births occurring on day 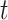 ;
; 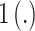 is an indicator function;
is an indicator function; 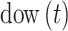 is the day of week that day
is the day of week that day 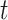 occurred on with Saturday (i.e.,
occurred on with Saturday (i.e., 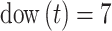 ) serving as the reference category;
) serving as the reference category; 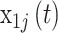 ,
, 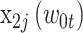 , and
, and 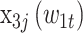 are the
are the 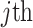 columns of the B-spline basis matrices for a natural cubic spline with 35, 4, and 4 df for study day, minimum temperature (i.e.,
columns of the B-spline basis matrices for a natural cubic spline with 35, 4, and 4 df for study day, minimum temperature (i.e., 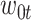 ), and maximum temperature (i.e.,
), and maximum temperature (i.e., 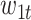 ), respectively; and
), respectively; and 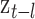 is the true but unobserved maximum 24-h average PM
is the true but unobserved maximum 24-h average PM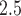 exposure
exposure  days prior to day
days prior to day 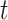 .
.
We choose 35 df for the long-term time trend based on selecting 7 df for each of the 5 study years as in Samet and others (2000) while noting that Peng and others (2006) found reduced bias in effect estimation with more aggressive smoothing in similar time series modeling. We consider daily lags from 2 to 6 days (i.e., 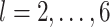 ), similar to previous stillbirth and air pollution modeling work (Faiz and others, 2013; Sarovar and others, 2020; Enebish and others, 2022) and the estimated timing of 48 h between fetal death and delivery (Gardosi and others, 1998).
), similar to previous stillbirth and air pollution modeling work (Faiz and others, 2013; Sarovar and others, 2020; Enebish and others, 2022) and the estimated timing of 48 h between fetal death and delivery (Gardosi and others, 1998).
Using the model in (5.9) and  , we test several of the existing methods from Section 2 for propagating exposure uncertainty in the health analysis along with the newly developed UKDE. MKDE is not considered given its poor performance in simulation and long computing time for the large analysis data set. MI is not applied due to its lengthy run time (i.e., requires fitting (5.9) in an MCMC framework
, we test several of the existing methods from Section 2 for propagating exposure uncertainty in the health analysis along with the newly developed UKDE. MKDE is not considered given its poor performance in simulation and long computing time for the large analysis data set. MI is not applied due to its lengthy run time (i.e., requires fitting (5.9) in an MCMC framework 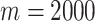 times) and its overall similarity with MIA in the simulation study results. Because the model that uses the true exposures is not possible in practice, we additionally fit a full joint version of the model (Joint) where Stages 1 and 2 are fit simultaneously within a single hierarchical Bayesian framework. We separately fit each method and exposure lag (
times) and its overall similarity with MIA in the simulation study results. Because the model that uses the true exposures is not possible in practice, we additionally fit a full joint version of the model (Joint) where Stages 1 and 2 are fit simultaneously within a single hierarchical Bayesian framework. We separately fit each method and exposure lag (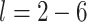 ) and make inference on
) and make inference on 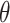 , the parameter that describes the association between maximum PM
, the parameter that describes the association between maximum PM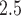 exposure and stillbirth risk.
exposure and stillbirth risk.
The prior distributions for the parameters in (5.9) are chosen as 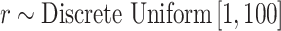 and
and 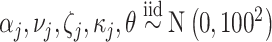 . From all methods, we collect
. From all methods, we collect 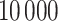 samples from the joint posterior distributions after discarding the first
samples from the joint posterior distributions after discarding the first 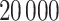 prior to convergence and thinning the remaining
prior to convergence and thinning the remaining 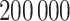 by a factor of
by a factor of 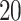 to reduce posterior autocorrelation. We assessed convergence by visually inspecting traceplots of individual parameters and monitoring Geweke’s diagnostic; neither tool suggested any obvious signs of nonconvergence across all model fits.
to reduce posterior autocorrelation. We assessed convergence by visually inspecting traceplots of individual parameters and monitoring Geweke’s diagnostic; neither tool suggested any obvious signs of nonconvergence across all model fits.
5.4. Results
In Figure 2b, we display daily predictions of the maximum PM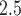 exposures from the three NJ counties in 2011–2015 along with a histogram of the ppd samples on a single day (August 1, 2011; other days were similar). The level of skewness in the ppd samples resembles that from the simulation study due to the log transformation used in (5.7). In Figure S9 of the Supplementary material available at Biostatistics online, we show a scatterplot of these predictions and the observed AQS data (daily maximum of the PM
exposures from the three NJ counties in 2011–2015 along with a histogram of the ppd samples on a single day (August 1, 2011; other days were similar). The level of skewness in the ppd samples resembles that from the simulation study due to the log transformation used in (5.7). In Figure S9 of the Supplementary material available at Biostatistics online, we show a scatterplot of these predictions and the observed AQS data (daily maximum of the PM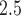 AQS concentrations across all of NJ). The plot shows that the model is generally predicting well with respect to the observed data.
AQS concentrations across all of NJ). The plot shows that the model is generally predicting well with respect to the observed data.
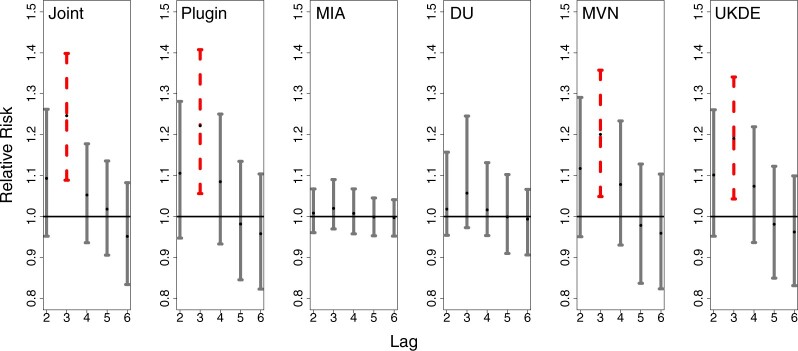
In Figure 3, we show results from the stillbirth analyses across all considered methods and lags. Specifically, we present posterior inference (i.e., posterior means and quantile-based equal-tailed credible intervals) for 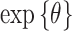 across all analyses, resulting in a relative risk interpretation. Because we standardize
across all analyses, resulting in a relative risk interpretation. Because we standardize 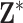 by subtracting off the median and dividing by the interquartile range (IQR) prior to each analysis, the estimates represent the relative risk of stillbirth for an IQR increase in exposure during a given lag (IQR was 10.22
by subtracting off the median and dividing by the interquartile range (IQR) prior to each analysis, the estimates represent the relative risk of stillbirth for an IQR increase in exposure during a given lag (IQR was 10.22 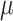 g/m
g/m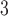 across all lags).
across all lags).
Fig. 3.
Posterior mean and 95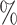 credible interval plots for
credible interval plots for 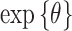 across the different models and daily lag periods for the New Jersey three county stillbirth and maximum daily 24-hour PM
across the different models and daily lag periods for the New Jersey three county stillbirth and maximum daily 24-hour PM exposure analysis. Dashed lines
indicate that the 95
exposure analysis. Dashed lines
indicate that the 95 credible intervals did not include one.
credible intervals did not include one.
Generally, the findings are in agreement with the simulation study results. MIA and, to a lesser extent, DU tend to pull the point estimates towards the null in comparison to the other approaches. Joint, Plug-in, MVN, and UKDE each suggest that elevated ambient levels of maximum 24-hour average PM exposure three days prior to delivery is associated with an increase in stillbirths. Specifically for UKDE, an IQR increase in exposure of 10.22
exposure three days prior to delivery is associated with an increase in stillbirths. Specifically for UKDE, an IQR increase in exposure of 10.22 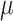 g/m
g/m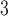 three days prior to delivery is associated with a
three days prior to delivery is associated with a 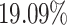 increase in stillbirths (
increase in stillbirths (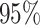 credible interval: 4.30–34.08). While the differences between Joint, Plug-in, MVN, and UKDE are subtle in this application, UKDE produces the shortest credible intervals among the two-stage approaches in this group followed by MVN and Plug-in, which may also be in agreement with the improved MSE performance of UKDE observed in the simulation study.
credible interval: 4.30–34.08). While the differences between Joint, Plug-in, MVN, and UKDE are subtle in this application, UKDE produces the shortest credible intervals among the two-stage approaches in this group followed by MVN and Plug-in, which may also be in agreement with the improved MSE performance of UKDE observed in the simulation study.
As a sensitivity analysis, we repeated several of the stillbirth analyses while randomly shuffling the order of the rows in 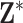 . This mixing breaks the temporal ordering of the exposures from the original analysis and we expect to estimate a null signal in
. This mixing breaks the temporal ordering of the exposures from the original analysis and we expect to estimate a null signal in 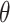 unless there are serious confounding issues that the model fails to capture. The results shown in Figure S10 of the Supplementary material available at Biostatistics online show no significant associations across all methods/lags, with point estimates near the null overall. This finding provides further evidence that population-level PM
unless there are serious confounding issues that the model fails to capture. The results shown in Figure S10 of the Supplementary material available at Biostatistics online show no significant associations across all methods/lags, with point estimates near the null overall. This finding provides further evidence that population-level PM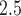 may play an important role in explaining stillbirth risk.
may play an important role in explaining stillbirth risk.
6. Discussion
In this work, we developed UKDE, a new framework for exposure uncertainty propagation in subsequent health outcome analyses, detailed its connection with existing approaches, derived its closed-form MCMC full conditional distributions, and created an R package for its implementation within several common epidemiological analyses
(KDExp; https://github.com/warrenjl/KDExp). Existing methods for quantifying this uncertainty were detailed within a unified framework, making it easier to compare the approaches. In a simulation study, we showed that UKDE had improved performance overall and particularly when the ppds were skewed. The multivariate extension of UKDE, MKDE, was consistently outperformed by the other methods, likely because of the difficulty associated with estimating high dimensional densities using multivariate KDE. The multiple imputation approaches (i.e., MI and MIA) produced estimates with substantial bias and are not recommended in this setting.
Several methods for measurement error correction that require modeling/prediction of the full set of exposure data were previously developed in the environment epidemiology setting. Fitting a joint model for the exposure and health data within a Bayesian framework naturally incorporates uncertainty from the predicted exposures into the health effect estimate but can be computationally intensive (Gryparis and others, 2009). The parametric bootstrap (Szpiro and others, 2011) first requires the fitting of the full two-stage analysis. Next, it simulates values from (i) the joint distribution of observed and unobserved exposures conditional on the estimated exposure modeling parameters and (ii) the distribution of the health data conditional on the simulated exposures and the estimate of the health effect parameter. The two-stage analysis is then repeated using the simulated data to obtain a bootstrap estimate of the health effect parameter. This process is repeated numerous times and the collected bootstrap estimates are used for bias correction and uncertainty quantification. Spatial simulation extrapolation (SIMEX) (Alexeeff and others, 2016) represents an extension of the original SIMEX framework for spatially correlated data, and serves to correct the asymptotic bias in the effect estimate due to a misspecified exposure model. However, estimating a standard error requires with-replacement resampling of the monitoring data and a repeating of the spatial SIMEX procedure, while the simulation step of spatial SIMEX requires knowledge about the spatial variance/covariance of the measurement error process which depends on external validation data in the form of held-out monitors.
In contrast, we focus on the setting where samples from the ppd obtained from a previously fit exposure model are available to define exposure prior distributions in the second stage health outcome analysis. A variant of this scenario is becoming increasingly common in air pollution epidemiology, as air pollution modeling becomes more specialized and separate from the health outcome modeling. For example, several exposure modeling groups publish air quality predictions that include or are capable of including measures of uncertainty (US EPA, 2022c; Di and others, 2019; Gong and others, 2021). Many epidemiologic studies use such output to investigate the health effects of air pollution in different geographic areas/times, populations, and health outcomes (Chang and others, 2012; Rushworth and others, 2014; Lim and others, 2018; Huang and others, 2021; Warren and others, 2022). Our findings suggest that providing actual ppd samples instead of summary statistics (i.e., ppd means and standard deviations) may be beneficial for better characterizing the exposure distribution using UKDE and could lead to improved effect estimation in future studies.
Thijssen and Wessels (2020) evaluated several density estimation techniques for performing sequential Bayesian inference, outside of the environmental health setting, using posterior samples collected from a first-stage analysis. However, the focus of these analyses was on making inference on a low-dimensional vector of parameters (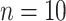 was the maximum considered in the study) that was shared across two or more data sets analyzed sequentially. The goals and assumptions of this type of analysis differ from those in the environmental health setting. In environmental health, the parameters shared across both modeling stages (i.e., exposures) are typically high dimensional (e.g., number of participants in a study), limiting the usefulness of some of the presented approaches (e.g., MKDE). Additionally, sequential analysis is primarily concerned with the estimation of the set of parameters included in both modeling stages/data sets, whereas in environmental health analyses the emphasis is on correctly characterizing uncertainty in the exposures to improve inference for parameters only included in the second-stage health outcome model (i.e., associations between exposure and health).
was the maximum considered in the study) that was shared across two or more data sets analyzed sequentially. The goals and assumptions of this type of analysis differ from those in the environmental health setting. In environmental health, the parameters shared across both modeling stages (i.e., exposures) are typically high dimensional (e.g., number of participants in a study), limiting the usefulness of some of the presented approaches (e.g., MKDE). Additionally, sequential analysis is primarily concerned with the estimation of the set of parameters included in both modeling stages/data sets, whereas in environmental health analyses the emphasis is on correctly characterizing uncertainty in the exposures to improve inference for parameters only included in the second-stage health outcome model (i.e., associations between exposure and health).
In our stillbirth data analysis in NJ, we found that elevated exposure to maximum 24-h average PM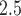 3 days prior to delivery was associated with an elevated risk of stillbirth (relative risk: 1.19 (1.04–1.34) for IQR increase). In a recent study based in Mongolia, Enebish and others (2022) identified multiple critical daily PM
3 days prior to delivery was associated with an elevated risk of stillbirth (relative risk: 1.19 (1.04–1.34) for IQR increase). In a recent study based in Mongolia, Enebish and others (2022) identified multiple critical daily PM exposure lags with respect to elevated stillbirth risk, including 3 days prior to delivery (odds ratio [OR]: 1.28 (1.00–1.62) for IQR increase). Another NJ-based analysis estimated an OR with a similar magnitude for an IQR increase in carbon monoxide exposure 2 days before delivery (OR: 1.20 (1.05–1.37)) (Faiz and others, 2013). Sarovar and others (2020) similarly identified a link between coarse particulate matter exposure two days before delivery and increased odds of stillbirth, while Mendola and others (2017) saw similar links between ozone and stillbirth (with similar magnitudes to our findings) on multiple days before delivery, including day 3. A recent meta-analysis found that stillbirth was positively associated with an increase in the ozone of 10
exposure lags with respect to elevated stillbirth risk, including 3 days prior to delivery (odds ratio [OR]: 1.28 (1.00–1.62) for IQR increase). Another NJ-based analysis estimated an OR with a similar magnitude for an IQR increase in carbon monoxide exposure 2 days before delivery (OR: 1.20 (1.05–1.37)) (Faiz and others, 2013). Sarovar and others (2020) similarly identified a link between coarse particulate matter exposure two days before delivery and increased odds of stillbirth, while Mendola and others (2017) saw similar links between ozone and stillbirth (with similar magnitudes to our findings) on multiple days before delivery, including day 3. A recent meta-analysis found that stillbirth was positively associated with an increase in the ozone of 10 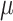 g/m
g/m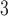 4 days before delivery but found no significant short-term effects for PM
4 days before delivery but found no significant short-term effects for PM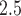 in pooled estimates (Zhang and others, 2021).
in pooled estimates (Zhang and others, 2021).
Future methods work in this area should focus on developing improved techniques for incorporating correlation between high-dimensional ppds. As methods for investigating the impact of pollution mixtures on health are becoming more common, extensions of the UKDE framework for more general application in these settings are needed. In our work, we showed that closed-form MCMC full conditional updates were still available with additive exposure models, but this does not necessarily hold for more complex interactions and hierarchical structures used for the exposures. Additionally, extensions of this work to accommodate critical exposure window identification/estimation (e.g., Warren and others, 2012) are also needed. Overall, UKDE is shown to be a promising framework for characterizing exposure uncertainty in environmental health studies, representing a more flexible hybrid approach between existing methods, and can be implemented in the R package KDExp for several common regression models.
Supplementary Material
Acknowledgments
Conflict of Interest: None declared.
Contributor Information
Saskia Comess, Emmett Interdisciplinary Program in Environment and Resources, Stanford University, 473 Via Ortega, Stanford, CA 94305, USA.
Howard H Chang, Department of Biostatistics and Bioinformatics, Rollins School of Public Health, Emory University, 1518 Clifton Rd., NE Atlanta, GA 30322, USA.
Joshua L Warren, Department of Biostatistics, Yale School of Public Health, Yale University, P.O. Box 208034, 60 College Street, New Haven, CT 06520, USA.
Supplementary material
Supplementary material is available at http://biostatistics.oxfordjournals.org. The R package KDExp and worked example are available at https://github.com/warrenjl/KDExp.
Funding
The National Institute of Environmental Health Sciences (NIEHS) of the National Institutes of Health (NIH) under R01 NIEHS ES028346.
References
- Alexeeff, S. E., Carroll, R. J. and Coull, B. (2016). Spatial measurement error and correction by spatial SIMEX in linear regression models when using predicted air pollution exposures. Biostatistics 17, 377–389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bekkar, B., Pacheco, S., Basu, R., Basu, R. and Denicola, N. (2020). Association of air pollution and heat exposure with preterm birth, low birth weight, and stillbirth in the US: a systematic review. JAMA Network Open 3, 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berrocal, V. J., Gelfand, A. E. and Holland, D. M. (2010a). A bivariate space-time downscaler under space and time misalignment. The Annals of Applied Statistics 4, 1942–1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berrocal, V. J., Gelfand, A. E. and Holland, D. M. (2010b). A spatio-temporal downscaler for output from numerical models. Journal of Agricultural, Biological, and Environmental Statistics 15, 176–197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berrocal, V. J., Gelfand, A. E. and Holland, D. M. (2012). Space-time data fusion under error in computer model output: an application to modeling air quality. Biometrics 68, 837–848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blangiardo, M., Finazzi, F. and Cameletti, M. (2016). Two-stage Bayesian model to evaluate the effect of air pollution on chronic respiratory diseases using drug prescriptions. Spatial and Spatio-temporal Epidemiology 18, 1–12. [DOI] [PubMed] [Google Scholar]
- Centers for Disease Control and Prevention. (2022). What is stillbirth? https://tools.cdc.gov/medialibrary/index.aspx#/media/id/218136. [Google Scholar]
- Chang, H. H., Peng, R. D. and Dominici, F. (2011). Estimating the acute health effects of coarse particulate matter accounting for exposure measurement error. Biostatistics 12, 637–652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang, H. H., Reich, B. J. and Miranda, M. L. (2012). Time-to-event analysis of fine particle air pollution and preterm birth: results from North Carolina, 2001-2005. American Journal of Epidemiology 175, 91–98. [DOI] [PubMed] [Google Scholar]
- Di, Q., Amini, H., Shi, L., Kloog, I., Silvern, R., Kelly, J., Sabath, M. B., Choirat, C., Koutrakis, P., Lyapustin, A.. and others. (2019). An ensemble-based model of PM2.5 concentration across the contiguous United States with high spatiotemporal resolution. Environment International 130, 104909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enebish, T., Warburton, D., Habre, R., Breton, C. and Tuvshindorj, N. (2022). The acute lag effects of elevated ambient air pollution on stillbirth risk in Ulaanbaatar, Mongolia. medRxiv. [Google Scholar]
- Faiz, A. S., Rhoads, G. G., Demissie, K., Lin, Y., Kruse, L. and Rich, D. Q. (2013). Does ambient air pollution trigger stillbirth? Epidemiology 24, 538–544. [DOI] [PubMed] [Google Scholar]
- Fuentes, M. and Raftery, A. E. (2005). Model evaluation and spatial interpolation by Bayesian combination of observations with outputs from numerical models. Biometrics 61, 36–45. [DOI] [PubMed] [Google Scholar]
- Gardosi, J., Mul, T.Mongelli, M. and Fagan, D. (1998). Analysis of birthweight and gestational age in anteparturn stillbirths. BJOG: An International Journal of Obstetrics & Gynaecology 105, 524–530. [DOI] [PubMed] [Google Scholar]
- Gong, W., Reich, B. J. and Chang, H. H. (2021). Multivariate spatial prediction of air pollutant concentrations with INLA. Environmental Research Communications 3, 101002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gryparis, A., Paciorek, C. J., Zeka, A., Schwartz, J. and Coull, B. A. (2009). Measurement error caused by spatial misalignment in environmental epidemiology. Biostatistics 10, 258–274. [DOI] [PMC free article] [PubMed] [Google Scholar]
-
Guan, Y., Reich, B. J., Mulholland, J. A. and Chang, H. H. (2019). Multivariate spectral downscaling for PM
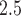 species. arXiv. [Google Scholar]
species. arXiv. [Google Scholar] - Huang, G., Lee, D. and Scott, E. M. (2018). Multivariate space-time modelling of multiple air pollutants and their health effects accounting for exposure uncertainty. Statistics in Medicine 37, 1134–1148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, W., Schinasi, L. H., Kenyon, C. C., Moore, K., Melly, S., Hubbard, R. A., Zhao, Y., Diez Roux, A. V., Forrest, C. B., Maltenfort, M.. and others. (2021). Effects of ambient air pollution on childhood asthma exacerbation in the Philadelphia Metropolitan Region, 2011–2014. Environmental Research 197. [DOI] [PubMed] [Google Scholar]
- Lee, D., Mukhopadhyay, S., Rushworth, A. and Sahu, S. K. (2017). A rigorous statistical framework for spatio-temporal pollution prediction and estimation of its long-term impact on health. Biostatistics 18, 370–385. [DOI] [PubMed] [Google Scholar]
- Lee, D. and Shaddick, G. (2010). Spatial modeling of air pollution in studies of its short-term health effects. Biometrics 66, 1238–1246. [DOI] [PubMed] [Google Scholar]
- Lim, C. C., Hayes, R. B., Ahn, J., Shao, Y., Silverman, D. T., Jones, R. R., Garcia, C. and Thurston, G. D. (2018). Association between long-term exposure to ambient air pollution and diabetes mortality in the US. Environmental Research 165, 330–336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Little, R. J. A. (1992). Regression with missing X’s: a review. Journal of the American Statistical Association 87, 1227–1237. [Google Scholar]
- McMillan, N. J., Holland, D. M., Morara, M. and Feng, J. (2010). Combining numerical model output and particulate data using Bayesian space–time modeling. Environmetrics 21, 48–65. [Google Scholar]
- Mendola, P., Ha, S., Pollack, A. Z., Zhu, Y., Seeni, I., Kim, S. S., Sherman, S. and Liu, D. (2017). Chronic and acute ozone exposure in the week prior to delivery is associated with the risk of stillbirth. International Journal of Environmental Research and Public Health 14, 731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oak Ridge National Laboratory. (2022). Daymet. https://daymet.ornl.gov/. [Google Scholar]
- Peng, R. D. and Bell, M. L. (2010). Spatial misalignment in time series studies of air pollution and health data. Biostatistics 11, 720–740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng, R. D., Dominici, F. and Louis, T. A. (2006). Model choice in time series studies of air pollution and mortality. Journal of the Royal Statistical Society: Series A (Statistics in Society) 169, 179–203. [Google Scholar]
- Polson, N. G., Scott, J. G. and Windle, J. (2013). Bayesian inference for logistic models using Pólya–Gamma latent variables. Journal of the American Statistical Association 108, 1339–1349. [Google Scholar]
- Reich, B. J., Chang, H. H. and Foley, K. M. (2014). A spectral method for spatial downscaling. Biometrics 70, 932–942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rushworth, A., Lee, D. and Mitchell, R. (2014). A spatio-temporal model for estimating the long-term effects of air pollution on respiratory hospital admissions in Greater London. Spatial and Spatio-temporal Epidemiology 10, 29–38. [DOI] [PubMed] [Google Scholar]
- Samet, J. M., Dominici, F., Curriero, F. C., Coursac, I. and Zeger, S. L. (2000). Fine particulate air pollution and mortality in 20 US cities, 1987–1994. New England Journal of Medicine 343, 1742–1749. [DOI] [PubMed] [Google Scholar]
- Sarovar, V., Malig, B. J. and Basu, R. (2020). A case-crossover study of short-term air pollution exposure and the risk of stillbirth in California, 1999–2009. Environmental Research 191, 110103. [DOI] [PubMed] [Google Scholar]
- Scott, D. W. (2015). Multivariate Density Estimation: Theory, Practice, and Visualization. New York: John Wiley & Sons. [Google Scholar]
- Sheather, S. J. and Jones, M. C. (1991). A reliable data-based bandwidth selection method for kernel density estimation. Journal of the Royal Statistical Society: Series B (Methodological) 53, 683–690. [Google Scholar]
- Szpiro, A. A. and Paciorek, C. J. (2013). Measurement error in two-stage analyses, with application to air pollution epidemiology. Environmetrics 24, 501–517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szpiro, A. A., Sheppard, L. and Lumley, T. (2011). Efficient measurement error correction with spatially misaligned data. Biostatistics 12, 610–623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thijssen, B. and Wessels, L. F. A. (2020). Approximating multivariate posterior distribution functions from Monte Carlo samples for sequential Bayesian inference. PLoS One 15, e0230101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- US EPA. (2022a). Air Quality System (AQS). https://www.epa.gov/aqs. [Google Scholar]
- US EPA. (2022b). CMAQ Models. https://www.epa.gov/cmaq/cmaq-models-0. [Google Scholar]
- US EPA. (2022c). RSIG-Related Downloadable Data Files. https://www.epa.gov/hesc/rsig-related-downloadable-data-files. [Google Scholar]
- Warren, J. L., Chang, H. H., Warren, L. K., Strickland, M. J., Darrow, L. A. and Mulholland, J. A. (2022). Critical window variable selection for mixtures: estimating the impact of multiple air pollutants on stillbirth. Annals of Applied Statistics 16:1633–1652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warren, J. L., Fuentes, M., Herring, A. H. and Langlois, P. H. (2012). Spatial-temporal modeling of the association between air pollution exposure and preterm birth: identifying critical windows of exposure. Biometrics 68, 1157–1167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warren, J. L., Miranda, M. L., Tootoo, J. L., Osgood, C. E. and Bell, M. L. (2021). Spatial distributed lag data fusion for estimating ambient air pollution. The Annals of Applied Statistics 15, 1–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, H., Zhang, X., Wang, Q., Xu, Y., Feng, Y., Yu, Z. and Huang, C. (2021). Ambient air pollution and stillbirth: an updated systematic review and meta-analysis of epidemiological studies. Environmental Pollution 278, 116752. [DOI] [PubMed] [Google Scholar]
- Zhou, X. and Reiter, J. P. (2010). A note on Bayesian inference after multiple imputation. The American Statistician 64, 159–163. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.