Summary
The dynamics of immunity to infection in infants remain obscure. Here, we used a multi-omics approach to perform a longitudinal analysis of immunity to SARS-CoV-2 in infants and young children by analyzing blood samples and weekly nasal swabs collected before, during, and after infection with Omicron and Non-Omicron variants. Infection stimulated robust antibody titers that, unlike in adults, showed no sign of decay for up to 300 days. Infants mounted a robust mucosal immune response characterized by inflammatory cytokines, IFNα, and Th17 and neutrophil markers (IL17, IL8, CXCL1). The immune response in blood was characterized by upregulation of activation markers on innate cells, no inflammatory cytokines, but several chemokines and IFNα. The latter correlated with viral load and expression of interferon-stimulated genes (ISGs) in myeloid cells measured by single-cell multi-omics. Together, these data provide a snapshot of immunity to infection during the initial weeks and months of life.
Introduction
Infants and young children are born with an immune system that differs in composition and functionality from adults1-3 and undergoes profound maturation during the initial weeks and months of life1,3. While previous studies have described this maturation process in healthy infants1, a detailed system-wide, longitudinal analysis of the immune response to an infection in infants has yet to be undertaken. Here, we address this knowledge gap by assessing immunity to SARS-CoV-2 early after birth. In contrast to adults, infants and children develop mild symptoms after infection4, although severe cases and deaths have been observed.5 While previous publications primarily described immune responses to COVID-19 in older children (median age five years) with a relatively mature immune system6-9, little is known about how the immature immune system responds to SARS-CoV-2 infection during the first weeks and months of life. Several key questions arise in this context: 1) Given the nascency of the adaptive immune system in this age group2,3, to what extent do infants and young children develop durable antibody responses and T and B-cell memory to the SARS-CoV-2 virus? 2) In light of the mild course of pediatric COVID-19, what are the hallmarks of innate immune activation compared to that observed in adults? 3) Studies in older children and adults reported autoantibodies and lasting epigenomic changes after COVID-1910-12. How does SARS-CoV-2 infection impact the maturing infant immune system in the long term? To answer these questions, we used a multi-omics approach, including a comprehensive single-cell RNA-seq and ATAC-seq analysis, and profiled immunity to SARS-CoV-2 infection in a longitudinal cohort of infants and young children during the first weeks and months of life.
Results
Study cohort
We obtained pediatric COVID-19-infected and healthy control blood samples from infants and young children enrolled in the IMPRINT cohort at the Cincinnati Children’s Hospital Medical Center. All infants and young children were tested weekly for SARS-CoV-2, and healthy controls tested negative from birth to sampling. Overall, we analyzed 125 samples from 54 infected infants and young children (including 27 infants with paired pre-infection samples) and 27 additional matched control infants and young children (Figure 1a). Our cohort contains samples from infants and young children infected with different SARS-CoV-2 variants: 32 infants and young children were infected with pre-Omicron variants, and 22 were infected with Omicron variants (Figure 1a, DataS1). Samples in the pre-Omicron cohort were collected longitudinally, with paired samples from before, during, and after infection (Figure 1a). Samples in the Omicron cohort were collected in a cross-sectional manner. The age at infection for the combined cohort was 1 to 47 months (median age 9 months), and 56% of pediatric patients were male (DataS1). In addition, we obtained 62 blood samples from 48 adult COVID-19 patients and ten healthy controls from the Hope Clinic at Emory University in Atlanta and the Stanford University Medical Center (DataS1), and 47 blood samples from 41 mothers with mild COVID-19 (including three mothers with paired pre-infection samples) and three matched controls (DataS1). The median age in the adult cohort was 59 years; 48% of adult patients were male; the median age in the mother cohort was 33 years. For profiling of mucosal immunity, we collected 159 samples from weekly nasal swabs, including from 38 infants with COVID-19 (acute or convalescent; including two paired pre-infection samples) and 20 matched pediatric controls as well as 19 mothers with COVID-19 and 19 matched controls. Details on patient demographics, disease severity, and assay distribution can be found in the supplementary materials (DataS1; Figure S1a).
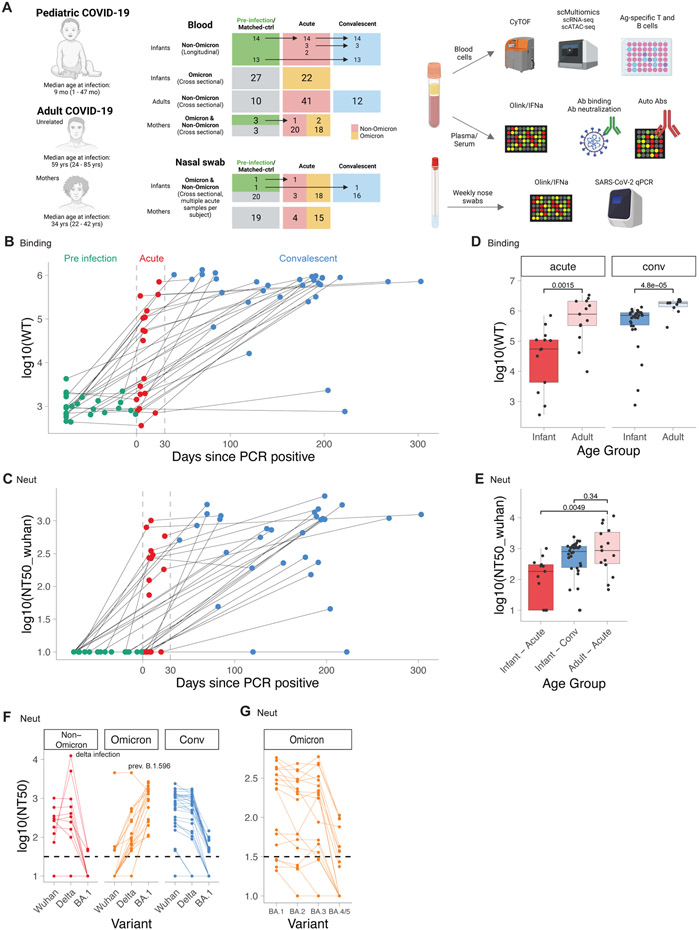
Figure 1. Durable antibody response in infants and young children with COVID-19.
A) Study layout. B-G) Antibody binding and neutralization titers of infants and adults with COVID-19. B) Antibody binding titers to WT strain in longitudinal samples from infants taken before (Pre, n=27), during (Acute, n=19), and after (Conv, n=30) infection. Dotted lines indicate days 0 and 30 post PCR+. C) Neutralizing antibody titers to Wuhan strain. D) Binding titers to WT strain in adults (Acute, n=13; Conv n=10) and infants (Acute n=13, Conv n=30). E) Neutralizing titers to Wuhan strain in adults (Acute n=15) and infants (Acute n=13, Conv n=30). F, G) Neutralizing titers against different variants in infants (Non-Omicron n=13, Omicron n=18, Conv n=30). D-G) Shown are only samples > 5 days post-infection. Statistical comparisons with Wilcoxon rank sum test. See also Figure S1
Robust and persistent antibody response
Whilst recent studies have documented the kinetics of the antibody response to SARS-CoV-2 infection in adults, there is currently a scarcity of information in the infant population13. The availability of samples obtained before, during, and following SARS-CoV-2 infection permitted us to perform a longitudinal analysis of the magnitude and kinetics of the antibody response during the first weeks and months of life. We thus measured binding and neutralizing antibody titers against SARS-CoV-2 variants, including the Omicron variants BA.1, BA.2, BA.3, and BA.4/5. For the antibody analysis, we analyzed a subset of 128 samples from 55 infants and young children and 29 adults with polymerase chain reaction (PCR)-confirmed SARS-CoV-2 infection (CoV-2+). While pre-infection binding and neutralization titers were low or nonexistent in infants and young children, respectively, anti-Spike antibodies emerged about 4-5 days after first testing CoV-2+ (Figure 1b,c, Figure S1b,c), in line with previous studies in adults.14 Strikingly, during acute infection, there was a wide variation in the magnitude of the neutralizing and binding antibody titers (Figure 1b,c, Figure S1b,c). During the convalescent phase, 28 out of 30 infants and young children developed robust antibody responses (Figure 1b,c), which were durably maintained for the entire observation period of up to 300 days (Figure 1b,c). This contrasts with previous studies in adults that demonstrated a marked decay in the antibody response during the initial months following infection.15-17 However, the magnitude of the antibody binding titers in infants and young children was significantly lower than in adults during both acute infection and convalescence (Figure 1d,e).
We next assessed the breadth of the antibody response against SARS-CoV-2 variants. Infection with non-Omicron variants induced binding and neutralization antibody responses against all measured pre-Omicron variants but bound and neutralized Omicron variants poorly (Figure 1f, Figure S1d,e). In contrast, infants and young children infected with the Omicron variant developed robust antibody binding and neutralizing titers against Omicron variants (Figure S1d,e, Figure 1f). Within the group of Omicron-infected infants and young children, subjects infected with BA.1 showed substantially reduced neutralizing titers for BA.4/5 (Figure 1g).
Autoantibody response
Previous studies reported autoantibodies against type I IFNs as a driver of severe COVID-19 infection in adults11 and autoantibodies against multiple self-antigens as a critical feature of Multisystem Inflammatory Syndrome in Children (MIS-C).6,10,18,19 Whether autoantibodies against IFNs are also a key feature of COVID-19 infection during the first weeks and months of life is unknown. To test this, we analyzed plasma samples from 77 infants and young children, and 25 adults, including maternal serum taken close to the time of birth, using a custom ELISA against IFNα2. One of 37 infants, during acute infection with the omicron variant, demonstrated high IFNα2-binding antibody levels (Figure S1f), in line with previous reports showing high auto-antibody levels in a small fraction of individuals with asymptomatic and mild disease.20 Longitudinal analysis of paired infant samples from before, during, and after infection showed a small increase in absorbance after infection compared to pre-infection samples (Figure S1g). Similarly, we observed an increase in absorbance in acute infection samples compared to serum taken from matched mothers close to the time of birth (Figure S1h).
Kinetics of T and B-cell responses
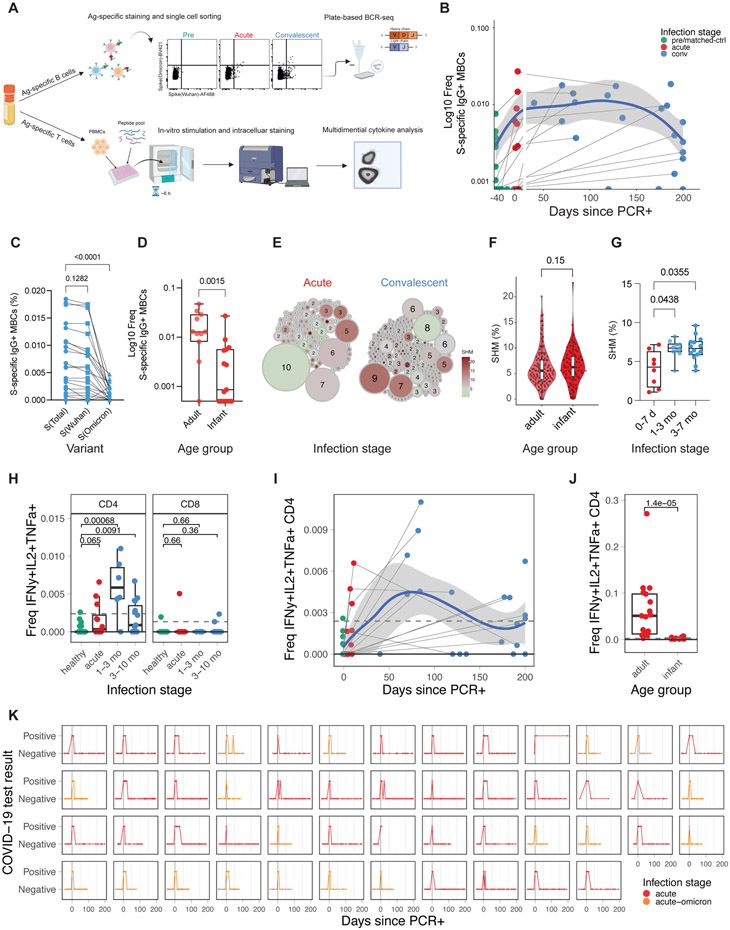
To further define the adaptive immune response to SARS-CoV-2 infection in infants and young children, we determined the frequency of SARS-CoV-2-specific memory B and T-cells in 77 PBMC samples from 34 infants and young children, and 19 adults (Figure 2a, Figure S2a). As expected, during the first week of infection, S-specific MBCs were few but emerged rapidly ten days after infection (Figure 2b). S-specific IgG+ MBCs in infants and young children were maintained for approximately six months after infection but appeared to decline afterward (Figure 2b, Figure S2b). This observation differs from studies in adults where MBCs were maintained for more than eight months without signs of decline15-17. Consistent with the serological data (Figure 1d,e), in convalescent infants and young children infected with non-Omicron variants, we observed a significantly lower frequency of Omicron S-binding MBCs than that of Wuhan S-binding MBCs (Figure 2c). Furthermore, the magnitude of the MBC response in infants during acute infection was lower than in adults (Figure 2d; Figure S2b,c). Interestingly, we observed low but detectable frequencies of S-specific class-switched MBCs in several pre-infection samples from infants and young children (Figure S2b), suggesting the existence of cross-coronavirus-reactive B-cells21. To further dissect memory B-cell maturation, we sorted S-specific IgG+ MBCs and analyzed their immunoglobulin B-cell receptor sequences. Expanded clones with relatively low somatic hypermutation rates (SHM) emerged during acute infection, characteristic of a primary B-cell response (Figure 2e, left). During convalescence, S-specific MBC clones harbored higher SHM rates (Figure 2e, right). Interestingly, the SHM rates in infants and young children were at least comparable, if not higher, than in adults (Figure 2f). To evaluate the maturation of the B-cell response in each individual infant, we calculated the mean SHM rate of all IGHV genes isolated from each infant. In agreement with the observation at the single-cell level, the SHM rate increased in individuals over time (Figure 2g), suggesting continuous B-cell evolution over the course of months, as has been shown in adults22.
Figure 2. Transient memory B and T-cell response to COVID-19 infection in infants and young children.
A) Experiment overview. B) Frequency of SARS-CoV-2 spike-specific IgG+ memory B-cells as a proportion of CD20+ B-cells (infants and young children: pre n=12, acute n=12, conv n=21). Blue line indicates average; shaded areas indicate 5th to 95th percentiles. C,D) Frequency of spike-specific IgG+ memory B-cells in C) infants and young children at convalescent phase and D) at acute phase in adults (mild/moderate infection n=11) and infants and young children (non-omicron n=12, omicron n=3). E) Clonality analysis of sorted spike-specific IgG+ memory B-cells in infants and young children (n=220). Circle size indicates the number of IGHV sequences; color represents the mean IGHV somatic hypermutation rate. F) Somatic hypermutation rates of IGHV genes in single sorted spike-specific IgG+ memory B-cells at acute phase. G) Mean somatic hypermutation rate of all cloned IGHV genes in indicated infant samples. H) Fraction of spike-specific multifunctional T-cells (IFNg+, IL-2+, TNFa+) at different infection stages. I) Kinetics of multifunctional CD4+ T-cell response. J) Comparison of multifunctional CD4+ T-cell response during the acute phase of infection in infants and young children and adults. K) Longitudinal overview of weekly COVID-19 test results from infant nasal swabs (Non-Omicron n=32, Omicron n=18). Statistical comparisons were conducted with Wilcoxon rank sum test. Solid line indicates median healthy response; dashed line indicates 3x median healthy response. See also Figure S2
With respect to T-cells, we observed an increase of multifunctional Th1-type CD4+ T-cells (IL-2, IFN-γ, TNFα triple-positive) in infants and young children post-infection (Figure 2h), while single-positive CD4+ T-cell responses did not exceed the LOD threshold (Figure S2d). Multifunctional CD4+ T-cells were induced during acute infection, peaked during the first three months of convalescence, and decayed during late convalescence with relatively low frequencies around 200 days post-PCR+ (Figure 2h,i, Figure S2e). Notably, the magnitude of WT and Omicron responses was similar in multifunctional CD4+ T-cells (Figure S2f). Compared to adults, infant multifunctional CD4 T-cell responses were reduced by roughly two orders of magnitude (Figure 2j).
One reason for the observed durable antibody and MBC response could be prolonged viral persistence in infants and young children, as proposed by several studies23,24. To examine if viruses persisted locally in mucosal airway, we analyzed weekly data from longitudinal SARS-CoV-2 nasal swab testing from every infant in our cohort. Tests returned positive results only during the initial 25 days post-infection (Figure 2k), providing no evidence for viral persistence beyond late acute infection. Likewise, we did not detect elevated levels of Spike protein in convalescent plasma using a sensitive electrochemiluminescent ELISA (data not shown).
Kinetics of the plasma cytokine responses
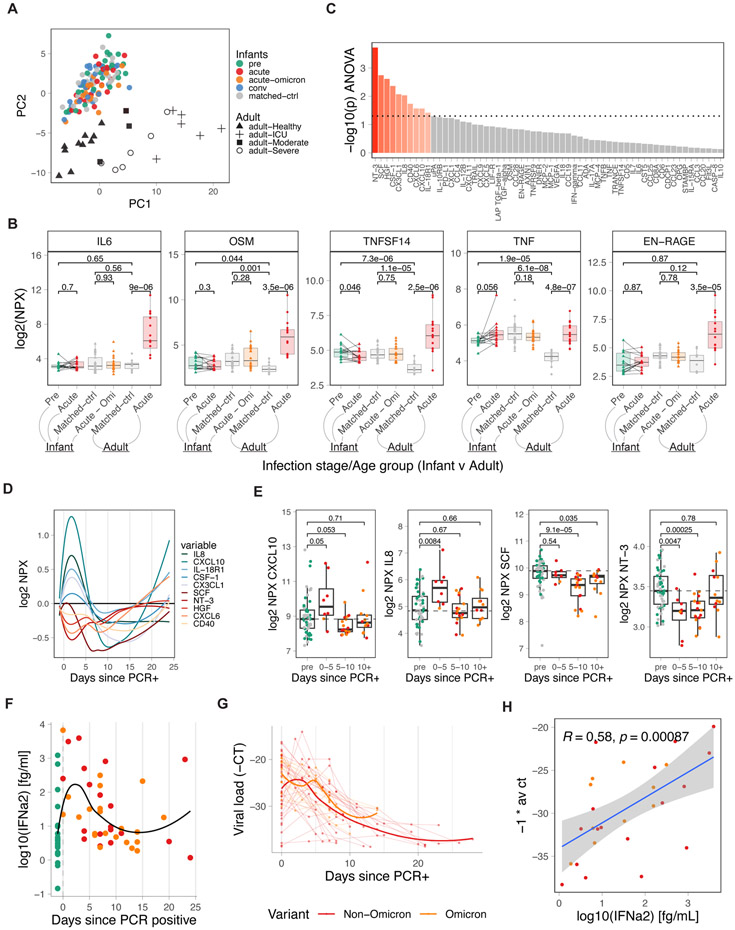
Previous studies have highlighted a dysregulated innate immune system, including the induction of inflammatory plasma cytokine responses, during COVID-19, which was associated with a severe course of disease25-27. We measured the concentration of 92 analytes in 146 plasma samples from 77 infants and young children and 25 adults25. Principal Component Analysis (PCA) revealed a separation of infant and adult samples in PC2, with PC1 separating adults by disease severity (Figure 3a). Infected infants and young children, who all had mild symptoms and in whom there was no significant increase in pro-inflammatory molecules such as IL-6, OSM, TNF, and EN-RAGE (Figure 3b), previously shown to be elevated in adult COVID-19 patients25-27, were occupying the same space as healthy infant samples (Figure 3a). Of note, in a previous study, older children (median age 5 years) with predominantly mild disease were shown to upregulate inflammatory markers, such as IL-6, similar to adults6. In contrast, in our cohort, IL-6 expression in infants and young children was still reduced when compared to mild and moderate adult samples only (Figure S3a). Importantly, PC2 was able to resolve differences between healthy adults and infants, and this was largely driven by cytokines and chemokines such as IL-17A, IL-17C, IL-12B, TNFRSF9 (4-1BB), CD8A, CD5, CD6, CCL20 (CCR6 ligand), CCL19 (CCR7 ligand), and TNFSF14 (Figure S3b,c), which were all elevated in healthy infants compared to healthy adults. Many of these markers are associated with T-cells and Th17 polarization. Longitudinal analysis revealed a transient upregulation for a subset of chemokines (CXCL10, IL8, IL-18R1, CSF-1, CX3CL1) during days zero to five, followed by a transient reduction starting around day five after turning PCR+ (Figure 3c-e). Interestingly, in a couple of subjects, we observed an increase in several cytokines late during infection (Figure 3d,e). To further validate the differences in the cytokine profile of infants from that of adults with mild COVID-19 symptoms, we compared the plasma levels of inflammatory cytokines in the infant cohort to a cohort of 41 mothers with mild or asymptomatic disease. Whilst there were elevated levels of IL-6 and CXCL10 in adults, infants had notably higher levels of IL-8, CXCL1, and CX3CL1 (Figure S3d). In contrast to the adults with mild/moderate disease in Figure S3a, mothers in Figure S3d were not seeking medical attention and were not hospitalized.
Figure 3. IFN-driven plasma cytokine response to COVID-19 infection in infants and young children.
A) PCA analysis of Olink-based plasma cytokines from COVID-19-infected and healthy infants and young children and adults, including matched controls (adult n=10, infant n=27), pre-infection (infant n=27), acute (adult n=15, infant n=19), acute-omicron (infant n=18), and convalescent (n=30) time points, colored by infections stage. Shape indicates disease severity in adult samples. B) Comparison of key inflammatory mediators during COVID-19 infection. Only paired samples are shown for infant pre and acute (n=14). C) ANOVA analysis of time-dependent, infection-associated changes in plasma cytokines. Shown is the p-value for the top 60 analyzed cytokines. P-values < 0.05 are indicated in red. D) Kinetics of indicated plasma cytokines. E) Time-dependent changes in plasma levels of key cytokines. F) Plasma IFNα2 levels in infants and young children relative to the first positive COVID-19 test (healthy n=27, acute n=19, acute-omicron n=22). G) Viral load in nasal swabs of COVID-19-infected infants and young children. Shown are all −1 * Ct values since the first positive COVID-19 test for each infant (acute n=32, acute-omicron n=18). H) Correlation between plasma IFNα2 levels and viral load (−1 * Ct). Statistical comparisons were conducted with the Wilcoxon rank sum test (E) and the paired and unpaired t-test (B). Correlation analyses were conducted using Spearman correlation. Lines were fitted using the loess (D) and linear regression (H) approaches. See also Figure S3
Furthermore, we measured the concentration of IFNα2 in plasma. SARS-CoV-2 infection induced robust but transient levels of plasma IFNα2 during the first five to ten days after testing positive (Figure 3f), similar to what was previously observed in adults25. Notably, IFNα2 levels were comparable between infants and young children infected with Omicron and Non-Omicron variants (Figure S3e). Next, we measured viral loads using PCR in nasal swabs. Kinetic analysis revealed a peak viral load around day zero to five after initial positive testing, with most tests turning negative again after day 20 (Figure 3g). Interestingly, IFNα2 levels correlated with viral loads (Figure 3h). In contrast to previous studies showing elevated IFNα2 levels in infant plasma relative to adults, our analysis indicates that infants, young children, and adults exhibited similar IFNα2 responses in plasma with one cohort of adults (with mild, moderate, and severe disease) showing slightly elevated levels relative to infants (Figure S3f, g)6. Comparing nasal swab data during early infection, revealed similar viral load in adults and infants and young children (Figure S3h). Thus, we identified a distinct cytokine response in plasma of SARS-CoV-2-infected infants and young children that is characterized by negligible induction of pro-inflammatory mediators and a transient elevation in IFNα2 levels and chemokines (Figure 3c). In particular, the chemokines IL-8, CXCL1, and CX3CL1, key mediators in neutrophil and myeloid cell recruitment and activation28, were induced at even higher levels than that observed in adults with mild symptoms (Figure S3d).
Analysis of peripheral blood leukocytes by mass cytometry
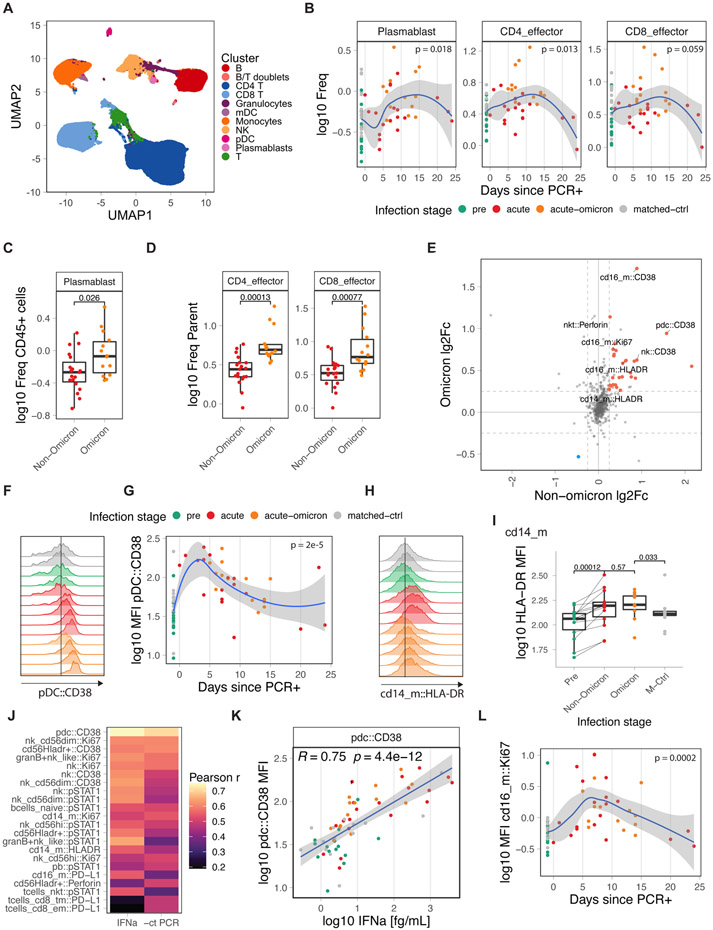
To obtain insights into the cellular dynamics of the immune response to SARS-CoV-2 in infants and young children, we profiled 75 PBMC samples from 47 infants and young children with cytometry by time of flight (CyTOF) using a panel of 39 antibodies against cell surface markers and intracellular signaling molecules (DataS1). Unsupervised clustering analysis revealed 11 cell types (Figure 4a), which we further subtyped by manual gating (DataS1, Figure S4a). In line with Figure 2, frequencies of plasmablasts and CD4 T-cells were increased after infection (Figure 4b) as previously observed in adults25. Interestingly, plasmablasts and effector T-cell frequencies were higher in infants and young children infected with the Omicron variant than WT or Delta variants (Figure 4c,d).
Figure 4. Immune cell activation during COVID-19 infection.
A) UMAP overview of cell clusters identified by CyTOF (n: pre=14, acute=19, acute-omicron=14, conv=14, matched-ctrl=14). B) Frequency of plasmablasts and effector T-cells as a proportion of total CD45+ and total T-cells, respectively. C, D) Comparison of plasmablast and effector T-cell frequencies in infants and young children. E) Average log fold-change of marker expression levels in healthy and infected samples. Markers that are significantly changed in Omicron and Non-Omicron cases are colored. F) Distribution of CD38 expression in pDCxs in representative samples. G) Kinetics of CD38 expression in pDCs. H) Distribution of HLA-DR expression in classical monocytes (CD14_m) in representative samples. I) HLA-DR expression in classical monocytes (CD14_m). J) Pearson r for correlations between CyTOF marker expression and plasma IFNα2 levels or viral load (−1 * Ct). K) Scatter graph plotting plasma IFNα2 levels against CD38 expression in pDCs. L) Kinetics of Ki67 expression in non-classical monocytes (cd16_m). Statistical comparisons were conducted with the Wilcoxon rank sum test. Correlation analyses were conducted using Pearson correlation. Lines were fitted using the loess (B,G,L) and linear regression (K) approaches. See also Figure S4
With respect to the innate immune system, cells from infection with both Omicron and pre-Omicron variants displayed elevated levels of activation, effector, and proliferation markers, including CD38, HLA-DR, perforin, and Ki67, on pDCs, NK cell subsets, and monocytes (Figure 4e, Figure S4b). In adults, we previously observed functional impairment of innate immune cells during COVID-19, which was characterized by reduced frequencies of pDCs in PBMCs, lower expression of pS6 in pDCs, a downstream mediator of mTOR signaling, and reduced levels of HLA-DR expression in monocytes and myeloid dendritic cells25,29. In infants and young children, we did not observe such impairment (Figure S4c-e). In fact, compared to two historical cohorts of adults with mild/moderate or severe COVID-1925, infants and young children demonstrated elevated levels of HLA-DR upregulation and no pDC-associated impairment (Figure S4f-h). During infection, CD38 and HLA-DR expression levels on pDCs and mDCs and classical monocytes (cd14_m), respectively, peaked early (Figure 4f-i, Figure S4d) and strongly correlated with IFNα2 plasma levels and viral load (Figure 4j,k). Of note, a recent study in individuals infected with influenza reported that CD38 upregulation is associated with IFNα production by pDCs in-vitro30. Phosphorylated STAT1 levels, a direct downstream mediator of IFNα2 signaling and an essential regulator of antiviral immunity31 correlated with IFNα2 levels in various cell types (Figure 4j), suggesting that IFNα could be a potential driver of the observed innate activation32,33. Finally, we observed the concerted upregulation of a set of activation markers (HLA-DR, CD38, Ki67) on non-classical monocytes (cd16_m) (Figure 4e,l).
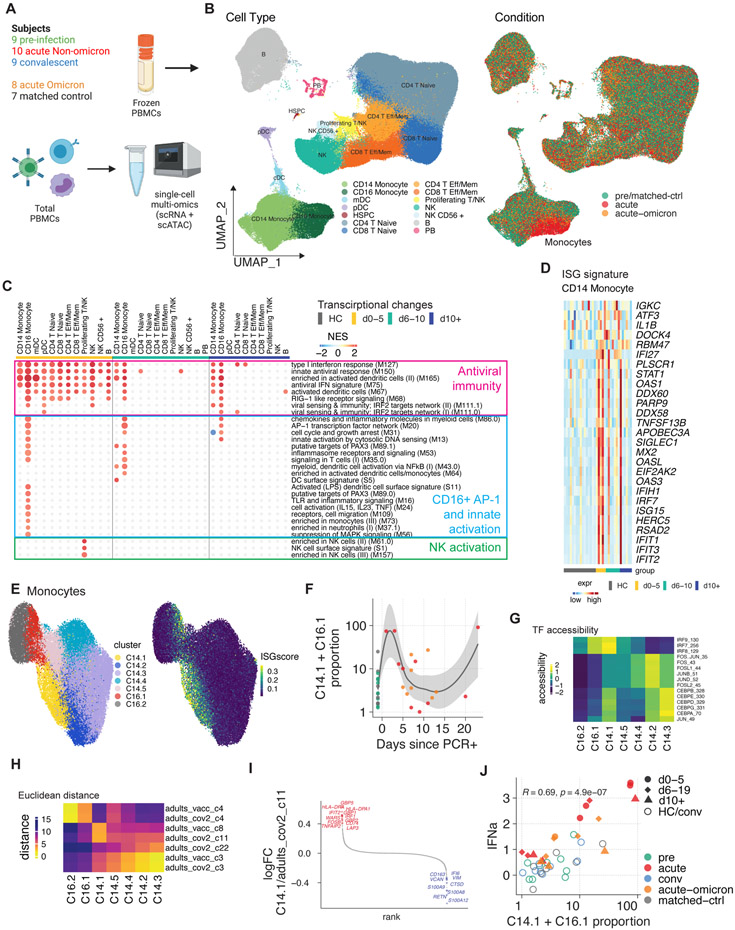
Single-cell transcriptional landscape
To gain further mechanistic insights into the activation state of individual immune cells, we used a single-cell multi-omics approach and determined paired gene expression and chromatin accessibility profiles in individual cells in 43 samples from before, during, and after infection with SARS-CoV-2 in infants and young children (Figure 5a). Using dimensionality reduction and clustering approaches, we constructed a map of the transcriptomic and epigenomic landscape (Figure 5b). Global analysis of differentially expressed genes demonstrated the induction of antiviral and interferon-related pathways in many cell types on days zero to five after PCR+ (Figure 5c, Figure S5a). While this signature waned rapidly in most cells, monocytes expressed elevated levels of antiviral and interferon-related genes for more than ten days post-PCR+ (Figure 5c). Analysis of the driver genes revealed enrichment of interferon-stimulated genes (ISG), including STAT1, MX2, IRF7, and multiple members of the IFIT and OAS families (Figure 5d).
Figure 5 – Single-cell multi-omics analysis of immunity to COVID-19 infection in infants and young children.
A) Cartoon of the conducted experiment. B) UMAP representation of PBMCs from all analyzed samples, colored by cell type (left) and infection stage (right; convalescent samples not shown). C) Pairwise comparison of genes from healthy (n=16) and COVID-19–infected infants and young children at different times during acute infection (D0-5: n=5, D5-10: n=7, D10+: n=6) was conducted for each cluster. DEGs were analyzed for the enrichment of BTMs. Ring plot shows an abridged representation of enriched pathways in each cluster. Size indicates the number of samples with enrichment; colors indicate the normalized enrichment score. Full ring plot in Figure S5a. D) Expression of ISGs enriched in CD14+ monocytes in C) (magenta box). E) UMAP representation of monocyte subclustering analysis. F) Kinetics of CD14.1 and C16.1 monocyte subsets. G) Chromatin accessibility for selected TFs in different monocyte subsets. H) Integrated analysis of monocyte clusters from this study and from adult COVID-19 patients25 and adult subjects immunized with the COVID-19 vaccine35. Shown is the Euclidean distance between infant and adult monocyte subsets. I) DEGs determined between infant C14.1 and adult COVID-19-infection C11 monocyte clusters are plotted and ranked by fold change. J) Spearman’s Rho correlation analysis between plasma IFNα2 levels and fraction of interferon-experienced monocytes (bottom). See also Figure S5
Next, we asked how the observed acute-phase ISG response in monocytes is regulated at the single-cell level. Subclustering analysis revealed two subsets of CD14+ and CD16+ monocytes (C14.1, C16.1) that emerge early during the acute phase and are characterized by elevated ISG expression (Figure 5e, f). Using the chromatin accessibility data from our multi-omics dataset, we identified regulatory features associated with the observed monocyte subsets: C14.1 monocytes were characterized by high chromatin accessibility at IRF loci and reduced accessibility at AP-1 loci (Figure 5g). These data suggest that a subset of ISG-expressing cells with elevated IRF accessibility drives the observed acute antiviral response in monocytes on a single-cell level. Previously, we identified a subset of monocytes with elevated IRF and reduced AP-1 accessibility after vaccination with an AS03-adjuvanted influenza vaccine34, and we showed that an ISG-expressing monocyte subset emerges rapidly after secondary immunization with the Pfizer/BioNTech COVID-19 vaccine, BNT162b235. In addition, our previous work with acute COVID-19 infection in adults has demonstrated a monocyte cluster expressing high levels of ISGs25. To determine whether C14.1 and C16.1 are similar to the aforementioned monocyte subsets elicited by vaccination or COVID-19 infection in adults, we integrated scRNA-seq data from the present study with data from our previous studies on COVID-19 vaccination35 and infection25 in adults and constructed a joined monocyte landscape (Figure S5b). In this joint space, infant C14.1 cells overlapped with both adult ISG-expressing cells (C8, adult Pfizer/BioNTech vaccine study35) and a cluster of IFN-activated monocytes (C11, adult COVID-19 study25), as demonstrated by UMAP and proximity analysis using Euclidean distance (Figure 5h, Figure S5b). Direct comparison of gene expression signatures revealed that infant C14.1 monocytes express elevated levels of ISGs (MX1,2, ISG15, IFI1, IFI44) and antigen presentation markers (HLA-DR) and reduced levels of inflammatory and monocyte activation markers (GBP5, S100A8, FCGR1A) (Figure 5i, Figure S5c). Correlation analysis demonstrated a strong association between ISG expression and plasma IFNα2 levels in all immune cell subsets (Figure S5d). Monocytes stood out as the cell type with the strongest correlation and the highest expression of ISGs, indicating that these cells are most sensitive to type I IFN signaling. In line with these findings, we observed a strong correlation between IFNα2 and ISG-expressing monocyte subsets (C14.1, C16.1) on a single-cell level (Figure 5j).
To perform a more detailed analysis of the transcriptional response in infants versus adults with mild COVID-19 symptoms, we integrated our single-cell multi-omics dataset with four historical datasets containing data on adult patients with mild/moderate and severe disease25,29,36. This single-cell analysis revealed that during the initial stage of infection, both infant and adult CD14+ monocytes exhibited comparable ISG upregulation (Figure S5e). However, adult CD14+ monocytes demonstrated a trend towards elevated expression levels of ISGs on days 5-10 and beyond day 10 (Figure S5e). On a single gene level, infection-induced ISG expression in CD14 monocytes correlated well between infants and young children, and adults with mild COVID-19 (Figure S5f). Interestingly, while IFI27, a biomarker for viral infection37-39, was induced more strongly in adults, infants and young children displayed stronger induction of nucleic acid detectors such as (DDX58/RIG-1, DDX60, IFIH1/MDA5, ZBP1, PARP9) as previously described7. To validate these findings, we conducted bulk RNA-seq on 90 Tempus tube samples from healthy and infected infants and young children and compared the CD14+ monocyte ISG signature to a historical bulk RNA-seq dataset (Inflammatrix) from adults with mild/moderate COVID-1940,41. In line with the single-cell data, infants and young children showed a shorter duration of ISG expression, returning to baseline levels by day six after PCR+ (Figure S5g). Finally, global, BTM-based analysis of infection-induced bulk gene expression demonstrated overall concordance between infants and young children and adults with mild/moderate disease during the first five days of infection (Inflammatix: R = 0.49, p = 2.5x10−11). However, several BTMs associated with antiviral immunity and chemokines (M75, M68, M27.0, M29), adaptive immune activation and proliferation (M46, M4.6, M4.10, M4.7), and mitochondrial energy metabolism (M187) were more strongly upregulated in adults (Figure S5h). This could be caused by elevated baseline ISG levels in immune cells from infants that we observed when comparing healthy infant samples from our dataset with public datasets with healthy adult samples (data not shown) and which was previously reported by others in human and non-human primates7,42.
Single-cell epigenetic landscape
Next, we determined global changes in chromatin accessibility during infection, using single-cell ATAC-seq. TF motif-based analysis revealed extensive chromatin rearrangements in monocytes and NK cells early during infection (Figure 6a). In line with Figure 5g, global IRF accessibility was increased in CD14+ monocytes after infection (Figure 6a). Especially, CD16+ monocytes displayed increased chromatin accessibility at gene loci associated with inflammation and immune activation, including AP-1, NF-B, and T-box family members (Figure 6a). Next, we calculated gene-level chromatin accessibility values (gene scores) in CD16+ monocytes, aggregating the chromatin accessibility at multiple loci associated with a specific gene43. Differential gene score analysis followed by overrepresentation analysis demonstrated enrichment of inflammatory and cytokine-related BTM gene modules in differentially accessible genes during late infection in these cells (Figure 6b). Similarly, gene expression data demonstrated a prolonged activation signature with AP-1, PAX-3, and other inflammation-related BTMs in these cells (Figure 5c, Figure 6c) that was driven by monocyte activation and inflammation-related genes, including AP-1 TF genes (CD83, NFKBIA, NLRP3, FOSB, JUN, FOSL2, TLR4) (Figure 6c). CD16+ monocytes also upregulated IL-8 (CXCL8) and CXCL10 expression, suggesting they might be contributing to the observed plasma chemokine response (Figure 3d,e). While many of these genes were already upregulated early after infection (Figure 6c), single-cell and bulk gene expression analyses indicated a subset of primarily inflammatory genes (CD83, NFKBIA, CXCL8, IL1B, NLRP3) that were elevated in some samples also during the later phase of infection (Figure 6c,d). In addition, NK cells showed increased accessibility at T-box, and AP-1 TF loci and differential gene expression analysis demonstrated the enrichment of BTMs and genes associated with NK cell function early after infection (Figure 5c, Figure S5i).
Figure 6. Single-cell multi-omics analysis of CD16+ monocyte activation.
A) Pairwise comparison of TF motif accessibility (healthy: n=16; D0-5: n=5, D5-10: n=7, D10+: n=6). Color indicates differences in TF accessibility; non-significant changes (FDR>=0.001 or changed in less than three subjects) are grey. B) Enrichment of BTMs in differentially accessible gene scores in CD16+ monocytes at indicated time points. C) Expression of inflammation and AP-1-related genes enriched in CD16+ monocytes in Figure 5c (blue box). D) Kinetics of gene signature from (C) using bulk transcriptomics data (healthy: n=53, acute: n=19, acute-omicron: n=18). E) Integrated network analysis of plasma IFNα2 levels, BTM-based gene expression, TF motif accessibility, and CyTOF protein marker expression. Both line color and thickness indicate Spearman's rank correlation coefficient. See also Figure S6
Recent studies by us and others have shown that immunological events, including vaccines and infections, can have a lasting impact on the epigenomic landscape of the innate immune system34,44-46. To assess this during early life, we determined changes in chromatin accessibility in convalescent samples. To account for potential age-related changes in the epigenetic landscape, we compared the convalescent samples with samples from age- and sex-matched healthy controls. Using this approach, we observed reduced CEBP and AP-1 accessibility in CD14+ monocytes of convalescent infants and young children (Figure S6a). Importantly, sample-level analysis demonstrated profound interindividual heterogeneity in chromatin accessibility levels, with only a subset of infants and young children showing a reduction in CEBP and AP-1 accessibility (Figure S6b).
Finally, to better understand the relationship between the observed ISG response and the distinct innate activation and inflammation profiles in CD16+ monocytes, we integrated our sequencing-based single-cell multi-omics analysis with other measurements from this study and constructed a multi-omics network (Figure 6e). Our network integrated data on IFNα2 (Figure 3f), TF accessibility (Figure 6a), gene expression (Figure 5c), and CyTOF-based immune profiles (Figure 4). In line with our single-cell data, plasma IFNα2 levels were positively correlated with the expression of ISGs (BC4) and cell adhesion genes (BC7, BC12) (Figure 6e). However, no strong association with TF accessibility was observed (Figure 6e), indicating that gene loci associated with ISGs are already in an open chromatin confirmation at baseline. Indeed, analysis of gene tracks at key antiviral regions confirmed the presence of open chromatin regions at the promoter and distal loci before infection (Figure S6c). CD16+ monocytes upregulated an additional gene program during acute infection characterized by inflammation, innate activation, PAX3, and AP-1 BTMs (Figure 5c). Interestingly, IFNα2 was not associated with those genes (BC1) (Figure 6e). In fact, IFNα2 levels were negatively associated with AP-1 (MC2) TF accessibility (Figure 6e), suggesting the induction of a distinct and IFNα-independent inflammation program in CD16+ monocytes unique to infants and young children6,25.
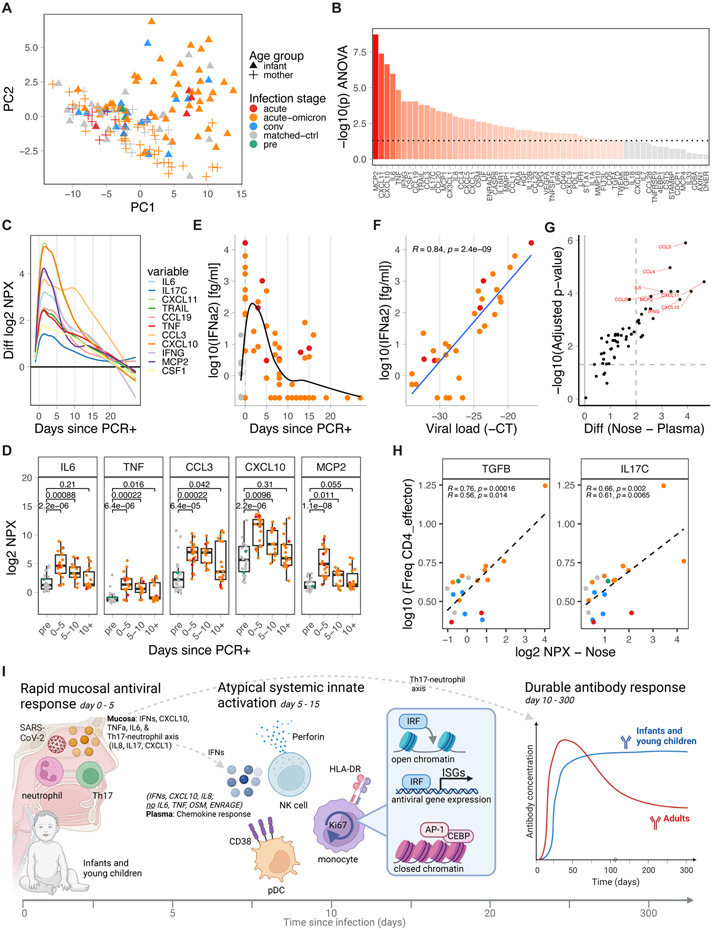
Mucosal immune response
Finally, to determine the mucosal immune response to SARS-CoV-2 in infants and adults, we studied 159 nasal swab samples from infants and young children and adults with mild symptoms of COVID-19 at different time points before, during, and after SARS-CoV-2 infection. Using PCA, we identified a group of infant nasal swab samples as mostly acute and distinct from healthy and convalescent samples (Figure 7a). Importantly, the samples from acutely infected mothers with mild symptoms were separated from those of acutely infected infants with mild symptoms (Figure 7a). In infants, ANOVA-based analysis found many inflammatory cytokines (IL6, TNF, IL17C, IL8 [CXCL8]) and chemokines (e.g., MCP1, MCP2, CCL3, CXCL1, CXCL5, CXCL10, CXCL11, CX3CL1) driving this effect (Figure 7b). These cytokines were immediately upregulated after infection, dropping to about 25% peak levels by day 10 post PCR+, and returning to baseline levels by day 20 (Figure 7c,d). Several chemokines and cytokines, including CX3CL1, CXCL10, IFNγ, and IL6, were correlated with viral load (Figure S7a). Especially MCP2, which is produced by macrophages and epithelial cells and attracts various immune cells to the nose, displayed an extremely strong correlation (r=0.87, Figure S7a). Similar to plasma, IFNα2 levels were raised during the first five days of infection (Figure 7e) and strongly correlated with viral load (Figure 7f).
Figure 7. Nasal immune response to SARS-CoV-2 infection in infants and young children.
A) PCA of nasal cytokines from SARS-CoV-2-infected and healthy infants and young children and mothers, including matched controls (infant n=20, mother n=19), pre-infection (infant n=2), acute (infant n=7, mother n=9), acute-omicron (infant n=45, mother n=40), and convalescent (infant n=17) time points, colored by infections stage. Shape indicates age group and vaccination status. B) Time-dependent, infection-associated changes analyzed by ANOVA. Shown is the p-value. P-values < 0.05 are indicated in red. C) Kinetics of top-upregulated nasal swab cytokines. D) Time-dependent changes in nasal swab levels of selected cytokines. E) Nasal swab IFNα2 levels in infants and young children relative to the first positive COVID-19 test (healthy n=22, non-omicron n=7, omicron n=45). F) Correlation between viral load (−1 * Ct) and nasal swab IFNα2 levels during acute infection (n=32). G) Difference in cytokine upregulation during early infection (d0-5) relative to healthy controls between plasma and nasal swab samples. H) Correlation between the frequency of CD4_effector cells and indicated mucosal cytokine levels using matched blood and nasal swab samples (+/− 5 days). I) Cartoon summary of the findings of this study. Statistical comparisons were conducted with the Wilcoxon rank sum test (D) and the unpaired t-test (G). Correlation analyses used Pearson (F, H) and Spearman correlation (H – lower stats). Lines were fitted using the loess (C, E) and linear model approach (F, H). See also Figure S7
The observed mucosal immune activation is in stark contrast to plasma cytokine levels which remained largely unchanged (Figure 3). Of the more than 40 cytokines significantly changed in nasal swabs (Figure 7b), only six were changed in plasma (Figure 3b, CSF1, CX3CL1, IL8, CD40, CXCL10, IL18R1). In line with these findings, correlation analyses revealed only modest relationships between nasal swabs and matched plasma samples (+/− 5 days) for a small number of cytokines (Figure S7b, IL6, OSM, TNFSF14). Similarly, we found only a modest relationship between nasal IFNα2 levels and matched plasma IFNα2 levels (+/− 5 days) (Pearson R = 0.44, p = 0.041). Directly compared to the plasma response, upregulation of nasal cytokines was highly increased for many markers, especially inflammatory chemokines, and cytokines (Figure 7g). Importantly, the abundance of CD4_effector T-cells (HLA-DR+CD38+) in blood correlated strongly with nasal markers associated with T-cell activation and Th17 polarization (Figure 7h, Figure S7c; IL17C, TGFB, IL7, TNFRSF9 [4-1BB]). In addition, Th17-associated cytokines (IL17C and IL1A) at the time of PCR+ (day 0) were associated with neutralizing antibody titers during late infection (>6 days; Figure S7d). Total protein levels in nasal swabs were not affected by infection (Figure S7e).
In contrast to infants and young children, nasal swabs from adult acute infection did not separate from healthy nasal swabs in PCA (Figure 7a). Maternal samples were mostly collected in 2022 when mothers had been vaccinated or previously exposed to COVID-19. Only a small group of five markers (CXCL11, CXCL10, MCP2, IFNγ, IFNα2) showed significant changes during acute infection (Figure S7f-j), and to a much lesser degree than in infants and young children (Figure S7k). This aligns with findings from a previous study that showed minimal changes in cytokine levels in nasal swabs from unvaccinated adults with COVID-1947. While adults show elevated levels of immunity and inflammation in the blood, infants and young children thus display profound immune activation in the nose.
Discussion
Here, we used a multi-omics approach to study immunity to SARS-CoV-2 infection in infants and young children. We observed robust and durable antibody responses against SARS-CoV-2. In contrast, prior studies in adults have shown a decay of antibody responses after COVID-19 infection with a half-life of approximately 120 days.15,17 However our data also suggest a potential vulnerability against emerging variants of concern (VOC): 1) serum neutralization titers against Omicron variants (BA.1, BA.2, BA.3, BA.4/5) were diminished in infants and young children infected with wildtype or Delta variants and vice versa, as also shown by others48; 2) memory B-cell responses, an essential source of affinity matured, cross-specific antibodies, were of limited duration and magnitude; 3) memory T-cell responses, which provide broad immunity that is less susceptible to mutational changes and VOC, were reduced compared to adults.
Three findings on the innate immune response are noteworthy. First, we observed stark differences between the mucosal and systemic immune responses in infants and young children. During the first five days of infection, in the infant's nasal mucosa there were high levels of type I and II IFNs, inflammatory cytokines (IL6, IL8, TNFα, IL17C), and various chemokines, in line with other reports49. While previous studies identified high levels of TNFa, IL-6, OSM, EN-RAGE, and other inflammatory mediators in plasma of older children6 and adults25-27 during acute COVID-19, we detected no increase in any of these cytokines in infants and young children. Secondly, the mucosal immune response in infants and young children was characterized by cytokines and chemokines associated with a Th17 response (IL17C, IL1A, IL6, TGFB), and mucosal levels of those cytokines correlated with activated CD4 T-cells and neutralizing antibody titers in blood. Previous studies demonstrated enhanced activity of Th17 and mucosa-associated T-cells in infants1,50 and a chemokine-dependent cross-talk between Th17 cells and neutrophils51, and a role for Th17 cells in supporting the development of antibody responses52. This suggests that the crosstalk between Th17 cells and neutrophils might play a role in orchestrating innate and adaptive immunity to SARS-CoV-2 in infants. Thirdly, our analysis revealed rapid activation of innate immunity. These results are in striking contrast to results in adults, especially in patients with severe COVID-19, showing major defects in myeloid cells and plasmacytoid dendritic cells during acute infection25,29,53. Taken together, these findings suggest that the rapid induction of mucosal immunity in the nasal tract might contribute to the mild course of disease in infants and young children by containing viral replication in the nose. These findings beg the important question of what causes the differences in mucosal immunity between adults versus infants and young children in the first place. Here, it is possible that the nascency of the infant microbiome54 or intrinsic ontogenic differences in the early immune system55 or the frequent presence of mucosal pathobionts in infants56 could contribute.
Finally, we observed enhanced accessibility of chromatin loci targeted by IRFs and reduced accessibility of AP-1 targeted loci, as well as traces of epigenetic imprinting, during convalescence (reduced levels of CEBP and AP-1 accessibility). These epigenetic changes were heterogeneous between individuals and contrast with previous adult studies demonstrating more consistent epigenomic changes34,44. One explanation for this difference could be that in infants frequent immune activation caused by vaccination could have blurred the epigenomic imprint of COVID-19.
In summary, our findings provide insights and a reference dataset into the dynamics of human immunity to an infection in early life and reveal a surprisingly robust and durable antibody response in the face of a potent mucosal immune activation, including Th17 and neutrophil-associated markers (Figure 7i). In contrast, there was a paucity of inflammatory markers in plasma. This apparent disconnect between the adaptive immune response and the pro-inflammatory cytokine response and the simultaneous occurrence of a Th17-neutrophil axis, suggests that a non-canonical pathway of innate activation might drive persistent humoral immunity in this special population in early life. This raises the prospect of devising vaccine adjuvants that target such non-canonical pathways of innate activation to stimulate persistent antibody responses, without the collateral immunopathology that often results from unwanted inflammation.
Limitations of the Study:
While we made all efforts to assemble a homogeneous and curated cohort, several factors, such as infections, vaccinations, microbiota, growing, and mother milk, could impact our results. With respect to the anti-IFN antibody analysis, a potential caveat is the possibility of a transfer of maternal immunoglobulins from mothers to infants before birth, as we lacked paired maternal pre-birth serum samples for the specific infants.
STAR Methods
RESOURCE AVAILABILITY
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Bali Pulendran (bpulend@stanford.edu).
Materials availability
This study did not generate new unique reagents.
Data and code availability
ScATAC-seq and scRNA-seq data and blood transcriptomics data have been deposited at GEO and are publicly available as of the date of publication. Accession numbers are listed in the key resources table. No original code was generated. All other code and scripts, and data reported in this paper are available from the lead contact upon request.
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Goat anti-Human IgG Fc Cross-Adsorbed Secondary Antibody | Invitrogen | Cat# PI31413 |
| anti-IgD (Clone IA6-2) PE | BD Biosciences | Cat# 555779 |
| anti-IgM (MHM-88) PerCP-Cy5.5 | BioLegend | Cat# 314512 |
| anti-CD20 (Clone 2H7) APC-H7 | BD Biosciences | Cat# 560734 |
| anti-CD27 (Clone O323) PE-Cy7 | BioLegend | Cat# 302838 |
| anti-CD14 (Clone M5E2) PE/Dazzle™ 594 | BioLegend | Cat# 301852 |
| anti-CD16 (Clone 3G8) BV605 | BioLegend | Cat# 302040 |
| anti-IgG (Clone G18-145) BV650 | BD Biosciences | Cat# 740596 |
| anti-CD3 (Clone UCHT1) BUV737 | BD Biosciences | Cat# 612750 |
| anti-IL-2 (clone MQ1-17H12) FITC | Biolegend | Cat# 500304 |
| anti-CXCR5 (clone MU5UBEE) PerCP-eF710 | Invitrogen | Cat# 46-9185-42 |
| anti-IL-4 (clone MP4-25D2) PE | BioLegend | Cat# 500810 |
| anti-CD45RA (clone 5H9) PE-CF594 | BD Biosciences | Cat# 565419 |
| anti-TNF- a (clone Mab11) PE-Cy7 | E-Bioscience | Cat# 25-7349-82 |
| anti-CD40L (clone 24-31) BV421 | Biolegend | Cat# 310824 |
| anti-TCR-gd (clone B1.1) BV506 | Biolegend | Cat# 331220 |
| anti-CD4 (clone OKT4) BV605 | Biolegend | Cat# 317438 |
| anti-CD3 (clone SP34-2) BV650 | BD Biosciences | Cat# 563916 |
| anti-CCR7 (clone G043H7) BV711 | Biolegend | Cat# 353228 |
| anti-CD127 (clone A019D5) BV785 | Biolegend | Cat# 351330 |
| anti-IL-21 (clone 3A3-N2) APC | BioLegend | Cat# 513008 |
| anti-IFN-g (clone 4S.B3) A700 | Biolegend | Cat# 502520 |
| anti-CD25 (clone BC96) APC-Cy7 | Biolegend | Cat# 302614 |
| anti-CXCR3 (clone 1C6/CXCR3) BUV395 | BD Biosciences | Cat# 565223 |
| anti-CD8 (clone RPA-T8) BUV563 | BD Biosciences | Cat# 612914 |
| anti-CCR6 (clone 11A9) BUV737 | BD Biosciences | Cat# 612780 |
| anti-CD69 (clone FN50) BUV805 | BD Biosciences | Cat# 748763 |
| Purified anti-human CD66b (G10F5) | BioLegend | Cat#305102; RRID: AB_314494 |
| Purified anti-human CD57 (HNK-1) | BioLegend | Cat#359602; RRID: AB_2562403 |
| Purified anti-human HLA-DR (L243) | BioLegend | Cat#307651; RRID: AB_2562826 |
| Anti-Human CD19 (HIB19) - 142Nd | Fluidigm | Cat#3142001B |
| Anti-Human CD127 (A019D5) - 143Nd | Fluidigm | Cat#3143012B |
| Anti-Human IL4 (MP4-25D2) - 144Nd | Fluidigm | Cat#3144010B |
| Anti-Human CD4(RPA-T4) - 145Nd | Fluidigm | Cat#3145001B |
| Anti-Human IgD (IA6-2) - 146Nd | Fluidigm | Cat#3146005B |
| Anti-Human CD20 (2H7) - 147Sm | Fluidigm | Cat#3147001B |
| Anti-Human CD34 (581) - 148Nd | Fluidigm | Cat#3148001B |
| Anti-pSTAT6 (18/P-Stat6) - 149Sm | Fluidigm | Cat#3149004A |
| Anti-pSTAT5 (47) - 150Nd | Fluidigm | Cat#3150005A |
| Anti-Human CD123 (6H6) - 151Eu | Fluidigm | Cat#3151001B |
| Purified anti-human CD370 (CLEC9A/DNGR1) (8F9) - 152Sm | BioLegend | Cat#353802; RRID: AB_10983070 |
| Anti-pSTAT1 (4a) - 153Eu | Fluidigm | Cat#3153005A |
| Histone H3K27ac antibody (mAb) (MABI 0309) | ActiveMotiv | Cat#39685; RRID: AB_2793305 |
| Anti-Human CD27 (L128) - 155Gd | Fluidigm | Cat#3155001B |
| Anti-Human CD45 (HI30) - 156Gd | Fluidigm | Cat#3156010B |
| Purified anti-human CD25 (M-A251) | BioLegend | Cat#356102; RRID: AB_2561752 |
| Anti-Human pSTAT3 (4/P-Stat3) - 158Gd | Fluidigm | Cat#3158005A |
| Anti-Human CD11c (Bu15) - 159Tb | Fluidigm | Cat#3159001B |
| Anti-Human CD14 (M5E2) - 160Gd | Fluidigm | Cat#3160001B |
| Anti-Ki-67 (B56) - 161Dy | Fluidigm | Cat#3161007B |
| Purified anti-human CD1c (L161) | BioLegend | Cat#331502; RRID: AB_1088995 |
| Purified anti-human TCRg/d (B1) | BioLegend | Cat#331202; RRID: AB_1089222 |
| Anti-Human Arginase-1 (658922) - 164Dy | Fluidigm | Cat#3164012B |
| Anti-pCREB (87G3) - 165Ho | Fluidigm | Cat#3165009A |
| Purified anti-human CD16 (B73.1) | BioLegend | Cat#360702; RRID: AB_2562693 |
| Anti-Human CD38 (HIT2) - 167Er | Fluidigm | Cat#3167001B |
| Anti-Human CD8 (SK1) - 168Er | Fluidigm | Cat#3168002B |
| Anti-Human CD45RA (HI100) - 169Tm | Fluidigm | Cat#3169008B |
| Anti-Human CD3 (UCHT1) - 170Er | Fluidigm | Cat#3170001B |
| Anti-Human/ Mouse Granzyme B (GB11) - 171Yb | Fluidigm | Cat#3171002B |
| Anti-Human CD15 (W6D3) - 172Yb | Fluidigm | Cat#3172021B |
| Anti-Perforin antibody (B-D48) | abcam | Cat#ab47225 |
| Purified anti-human IFNg (4S.B3) | BioLegend | Cat#502502; RRID: AB_315227 |
| Anti-Cross Phospho-S6 (N7-548) - 175Lu | Fluidigm | Cat#3175009A |
| Anti-Human CD56 (NCAM16.2) - 176Yb | Fluidigm | Cat#3176008B |
| Anti-Human CD274/PD-L1 (MIH1) - 209Bi | Fluidigm | Cat#3209014B |
| Bacterial and virus strains | ||
| rVSV-ΔG-GFP/nanoluciferase | PMID: 35025672 | N/A |
| Pseudovirus expressing SARS-CoV-2 variants | In-house | Han et al.13, Suthar et al.14, Cohen et al.15 |
| Biological samples | ||
| Chemicals, peptides, and recombinant proteins | ||
| rhIFN-α | Miltenyi | Cat# 130-093-874 |
| Recombinant SARS-CoV-2 Wuhan spike protein | SinoBiological™ | Cat# 40589-V27B-B |
| Recombinant SARS-CoV-2 Omicron spike protein | SinoBiological™ | Cat# 40589-V49H3-B |
| DMSO | Sigma | D2650-100ML |
| Peptide cocktail for T-cell stimulation | In-house | Tarke et al.79 |
| Critical commercial assays | ||
| V-plex COVID-19 panel 23 | Mesoscale Discovery | Cat# K15567U |
| S-PLEX Human IFN-α2a kit | Mesoscale Discovery | Cat # K151P3S |
| Cell-ID 20-Plex Pd Barcoding Kit | Fluidigm | Cat#201060 |
| EQ™Four Element Calibration Beads | Fluidigm | Cat#201078 |
| Cell-ID™ Intercalator Ir | Fluidigm | Cat#201192A |
| One-Glo luciferase assay system | Promega | Cat# E6130 |
| Chromium NextGEM Chip J single Cell | 10x Genomics | 1000230 |
| Chromium NextGEM Single Cell Multiome ATAC + Gene Expression reagent bundle | 10x Genomics | 1000283 |
| Dual Index Kit TT Set A | 10x Genomics | 1000215 |
| Single Index Kit N Set A | 10x Genomics | 1000212 |
| Chromium X controller | 10x Genomics | 1000331 |
| NovaSeq 6000 S4 Reagent Kit V1.5 (200 cycles) | Illumina | 20028313 |
| Target 96 Inflammation Panel | Olink | |
| Bio-Rad protein assay kit II | Bio-Rad | 5000002 |
| Deposited data | ||
| scMultiome dataset - raw and processed data | This paper | GEO: GSE239799 |
| Blood transcriptomics dataset - raw and processed data | This paper | GEO: GSE239787 |
| Experimental models: Cell lines | ||
| Lenti-X 293T cells | Takara Bio | Cat. No. 632180 |
| 293T-ACE2-TMPRSS2 cells | In-house | Neerukonda et al.70 |
| Experimental models: Organisms/strains | ||
| Oligonucleotides | ||
| BCR primers | Liao HX et al. 75 | N/A |
| QIAseq SARS-CoV-2 Primer Panel | Qiagen Inc. | 333895 |
| QIAseq FX DNA Library Unique Dual Index Adapter Kit-A | Qiagen Inc. | 180479 |
| Recombinant DNA | ||
| pCAGGS Wuhan (SΔ19) | J. Craig Venter Institute – a gift from Dr. Gene S. Tan | N/A |
| pCAGGS Delta (B.1.617) | Sievers et al.69 | N/A |
| pCAGGS Omicron (B.1.529) | Sievers et al.69 | N/A |
| Software and algorithms | ||
| R 4.2.2; 4.1.1 | R Core Team102 | https://www.r-project.org/ |
| DESeq2 v1.38.3 | Love et al.101 | https://bioconductor.org/packages/release/bioc/html/DESeq2.html |
| Seurat (R package) v4.0.5 | Hao et al.,86 Hafemeister et al.87 | https://satijalab.org/seurat/ |
| harmony (R package) v0.1.1 | Korsunsky et al.88 | https://portals.broadinstitute.org/harmony/index.html |
| ggraph (R package) v2.1.0 | Pedersen et al.98 | https://ggraph.data-imaginist.com/index.html |
| igraph (R package) v1.3.1 | Csardi et al.97 | https://igraph.org/ |
| Complexheatmap (R package) v2.12.0; 2.8.0 | Gu104 | https://jokergoo.github.io/ComplexHeatmap-reference/book/ |
| ArchR (R package) v1.0.1 | Granja et al.43 | https://www.archrproject.com/ |
| chromVAR (R package) v1.20.2 | Schep et al.96 | https://bioconductor.org/packages/release/bioc/html/chromVAR.html |
| fgsea (R package) v1.24.0 | Korotkevich et al.89 | https://bioconductor.org/packages/release/bioc/html/fgsea.html |
| limma (R package) v3.54.2 | Ritchie et al.93 | https://bioconductor.org/packages/release/bioc/html/limma.html |
| pcaMethods (R package) v1.84.0 | Stacklies et al.81 | https://bioconductor.org/packages/release/bioc/html/pcaMethods.html |
| FlowSOM (R package) v2.0.0 | Van Gassen et al.83 | https://bioconductor.org/packages/release/bioc/html/FlowSOM.html |
| Other | ||
EXPERIMENTAL MODEL AND STUDY PARTICIPANT DETAILS
Cohort and study design
Specimens for this analysis were collected as part of the ongoing IMPRINT Influenza Cohort, an NIH-funded longitudinal birth cohort of healthy mothers and children in Cincinnati, Ohio. The Influenza IMPRINT Cohort received approval from Institutional Review Boards at Cincinnati Children’s Hospital Medical Center (2019-0629) and the University of Cincinnati Medical Center (SITE00000489). Pregnant women were screened for enrollment at three local delivery hospitals, and mothers were enrolled in the third trimester of pregnancy. A baseline serum specimen was collected from mothers at the time of enrollment. Starting at two weeks of life, research coordinators trained mothers to complete twice weekly SMS surveys to report the presence or absence of symptoms common to respiratory infections for their child and themselves. Mothers were also trained to collect midturbinate nasal swabs from their children and themselves, which were analyzed at an IMPRINT laboratory. Study participants were asked to collect at least one nasal swab each week. An additional swab was submitted if either the mother or the child was symptomatic of respiratory illness. Among children enrolled in this cohort, this project included 32 pre-omicron SARS-CoV2 cases and 22 omicron SARS-CoV2 cases. Cases were selected based on the timing of pre-infection, acute and convalescent blood collections, and availability of PBMCs at these time points. Pre-infection, acute, and convalescent nasal swabs from these cases were also selected. Pre-infection nasal swabs were the closest negative swabs collected before or on the date of the pre-blood collection. Acute and convalescent nasal swabs were most proximal to the acute and convalescent blood collections regardless of pathogen detections. Maternal enrollment sera from 30 cases were also selected for autoantibody transference analysis. A set of 30 children, age- and sex-matched to pre-Omicron child cases, were selected as controls; age-matching was based on the age of the case at the time of convalescence. Control nasal swabs were the closest negative swabs collected before or on the date of the control blood collection. Among COVID-19-positive mothers in the IMPRINT cohort, 4 pre-omicron cases and 15 omicron cases were included in this project. These positive mothers were not matched to child cases, and maternal cases were selected based on the timing of pre-infection and acute and blood collections, and availability of PBMCs at these time points. As was done for child cases, pre-infection, and acute nasal swabs were additionally selected. Pre-infection nasal swabs were the closest negative swabs collected before or on the date of the pre-blood collection. Acute nasal swabs were the swabs most proximal to the acute blood collection regardless of pathogen detections. Finally, COVID-19 negative swabs from 18 IMPRINT mothers were selected as additional controls. COVID-19 infections were considered symptomatic if the patient (pediatric and adult) experienced any symptom at least once, within −7/21+ days of their first COVID-19 positive swab, as reported on weekly surveys. Illness severity was evaluated by the presence of cough, fever or difficulty breathing, duration of cough and/or fever, and receipt of medical attention. The presence or absence of symptoms on the day of blood collection was determined by responses on the weekly survey completed closest to the blood collection date (−3/+3 days of blood collection). If a weekly survey response was unavailable, responses from the visit questionnaire were used (n=1). Demographic data is shown in DataS1.
Adult Cohort
The samples of the adult participants included in the study were collected in 2020 at the start of the pandemic, prior to the advent of vaccines. All samples were collected at the Hope Clinic of the Emory Vaccine Center or at Stanford University. Healthy controls were asymptomatic adults from whom samples were collected prior to the widespread circulation of SARS-CoV-2 in the community. Patients with COVID-19 were defined according to the original WHO guidance and positive SARS-CoV-2 RT-PCR testing by nasopharyngeal swabs as described in our original study 25. Patients with COVID-19 were classified as acute (less than 4 weeks from symptom onset or symptomatic at the time of sample collection) or convalescent (more than 4 weeks since symptom onset and asymptomatic or negative SARS-CoV-2 RT-PCR testing and asymptomatic). The severity of inpatient COVID-19 cases was classified based on the adaptation of the Sixth Revised Trial Version of the Novel Coronavirus Pneumonia Diagnosis and Treatment Guidance. Mild/moderate cases were defined as respiratory symptoms with radiological findings of pneumonia. Severe cases were defined as requiring supplemental oxygen. Critical cases were organ failure necessitating intensive care unit (ICU) care. The study received approval from the appropriate Institutional Review Board at Emory (#00022371) and Stanford University (#55689). All samples were collected after informed consent. Demographic data is shown in DataS1.
METHOD DETAILS
SARS-CoV-2 RT-PCR
Nasal swab samples underwent nucleic acid extraction using a custom protocol and the QIAamp 96 Virus Kit on the QIAGEN QIAcube HT instrument. Extracted RNA was run on Applied Biosystems 7500 or 7500 Fast instruments using CDC-developed real-time RT-PCR assays for Influenza 57,58, SARS-CoV-2 59,60, or Influenza and SARS CoV-2 in the Flu SC2 Multiplex assay 61,62. Nasal swabs were run on the influenza assay from November 2019-March 17, 2020. SARS-CoV-2 PCR was run for nasal swabs collected starting February 1, 2020, and was used in the lab through December 31, 2021. Starting on January 3, 2022, the Flu SC2 assay has been used for SARS-CoV-2 testing. In addition, all infant nasal swabs underwent multiplex detection of 21 respiratory pathogens using the Luminex NxTAG-RPP assay 63, including Influenza A and B. All samples positive for Influenza A and/or B underwent additional RT-PCR detection for Influenza A subtype and Influenza B lineage 57,58.
SARS-CoV-2 sequencing
Nasal swab samples underwent nucleic acid extraction using the QIAGEN Viral RNA Mini Kit (Qiagen, Inc) on the QIAGEN QIAcube Connect instrument following the manufacturer’s recommendations. RNA samples were subjected to direct sequencing using the modified ARTIC3 protocol ((https://artic.network/ncov-2019) with the addition of primer booster sets as implemented in the Qiagen QiaseqDirect protocol. RNA was subjected to random primed cDNA synthesis followed by amplification in two pools of multiplexed primer sets resulting in overlapping amplicons spanning the entire genome. Subsequently, 24 cycles of polymerase chain reaction were utilized to add dual index primers and amplify SARS-Cov-2 amplicons. DNA concentrations were normalized, samples were pooled and then subjected to sequencing to a depth of at least 100,000 reads per sample utilizing paired 150 nucleotide reads on an Illumina NextSeq 500 sequencing machine (Illumina, Inc).
Raw sequence data were demultiplexed and then aligned against the ancestral Wuhan-1 genome (Accession MN908947) 64 using bwa-mem 65. Samtools commands “sort”, “index”, “view”, and “mpileup” 66 were applied sequentially, and the ivar “consensus” command 67 was used to output a consensus sequence. Variant identification and lineage calling were performed with the software Phylogenetic Assignment of Named Global Outbreak Lineages (Pangolin) version 4.0.4 68.
Sample processing - blood
Peripheral blood was collected from study children at two and six weeks of life, every summer and acutely following “events.” An event included receipt of influenza or COVID-19 vaccines or having a nasal swab test positive for influenza or SARS-CoV2; participants completed an additional symptom survey at event visits. Up to 16mL of collected blood was deposited into sodium citrate Mononuclear Cell Preparation tubes (CPT) and promptly delivered to the laboratory for processing. After the initial spin of the CPT tubes, plasma was collected, and up to ten aliquots of plasma at 0.5mL were stored at −65°C or colder. Cells were collected, washed, and counted using the automated Vi-CELL XR Viability Analyzer. Aliquots were made at concentrations of either 2.0x106 or 5.0x106 cells in 1mL of Fetal Bovine Serum (FBS) +10% Dimethyl Sulfoxide (DMSO), depending on the number of cells obtained. Once aliquoted, the cryovials were placed into a Mr. Frosty cooler filled with isopropyl alcohol and placed into a −65°C or colder freezer for 24-72 hours. Samples were then placed into liquid nitrogen storage units for long-term storage.
Sample processing – nasal swabs
Mothers were given pre-labeled kits to collect nasal swab samples from their children. They were instructed to insert the swab into each nostril and rotate it a few times while ensuring it only touched the skin inside the nose. The swab was then placed into 3mL BD Universal Viral Transport Media (VTM) and refrigerated. VTM was designed for collecting and transporting clinical specimens that contained viruses from the collection site to the testing laboratory. After collection, the swab was kept refrigerated during transit by courier to the lab where it remained refrigerated until processing. Inside a biosafety cabinet, the sample was thoroughly vortexed before taking out 200μL for testing. The remaining volume was split into two more aliquots and frozen at −80°C.
Anti-Spike electrochemiluminescence (ECL) binding ELISA
Anti-Spike IgG titers were measured using V-plex COVID-19 panel 23 from Mesoscale Discovery (Cat #K15567U). The assay was performed as per the manufacturer’s instructions. Briefly, the multi-spot 96 well plates were blocked in 0.15 ml of blocking solution with shaking at 700 rpm at room temperature. After 30 min of incubation, 50 μl of plasma samples serially diluted in antibody diluent solution and serially diluted calibrator solution was added to each plate in the designated wells and incubated at room temperature for 2 h with shaking. Plasma samples were assayed at a 1:100 starting dilution and 6 additional 5-fold serial dilutions. After 2 h of incubation, the plates were washed, 50 μl of Sulfo-tag conjugated anti-IgG was added, and the plates were incubated at room temperature for 1 h. After incubation, the plates were washed, and 0.15 ml of MSD-Gold read buffer was added. The plates were immediately read using the MSD instrument. The Meso scale arbitrary light unit signal was used for calculating the area under curve (AUC) in Prism v.9.4.1.
Pseudovirus production and neutralization assay
VSV-based GFP/nanoluciferase-expressing SARS-CoV-2 pseudoviruses were produced as described previously 69. VSV-ΔG-GFP/nanoluciferase and plasmids encoding spike genes of SARS-CoV-2 Wuhan (SΔ19), Delta (B.1.617), and Omicron (B.1.529) were provided by Dr. Gene S. Tan (J. Craig Venter Institute, La Jolla, CA). To perform the neutralization assay, Vero E6-TMPRSS2-T2A-ACE2 cells (BEI Resources, NIAID; NR-54970) were seeded at a density of 1×104 per well in half area 96-well black opaque plates (Greiner Bio-One) and were grown overnight at 37°C in a 5% CO2 atmosphere. Serum samples were 5-fold serially diluted using the infection medium (DMEM supplemented with 2% FBS and 100 U/mL Penicillin-Streptomycin) in duplicates. Diluted serum samples were then mixed with an equal volume of Wuhan, Delta, or Omicron pseudoviruses, diluted in infection medium at an amount of 200-400 focus-forming units per well, followed by incubation at 37°C for 1 hour. Subsequently, immune complexes were added onto the monolayers of PBS-washed Vero E6-TMPRSS2-T2A-ACE2 cells and incubated at 37°C. At 18 hours post-incubation, supernatants were removed, cells were washed once with PBS, and nanoluciferase enzymatic activities were measured using the Nano-Glo Luciferase Assay System (Promega) and a SpectraMax iD3 multi-mode microplate reader. Percent inhibition values were calculated by subtracting the percent infection from 100. Non-linear curves and IC50 values were determined using GraphPad Prism.
Neutralization assay - Omicron subtyping
Human samples were evaluated in a qualified SARS-CoV-2 pseudovirion neutralization assay (PsVNA) using SARS-CoV-2 WA1/2020 strain and variants. SARS-CoV-2 neutralizing activity measured by PsVNA correlates with PRNT (plaque reduction neutralization test with authentic SARS-CoV-2 virus) in previous studies 70-72. Pseudovirions were produced as previously described 70. Briefly, human codon-optimized cDNA encoding SARS-CoV-2 spike glycoprotein of the WA1/2020 and variants was synthesized by GenScript and cloned into eukaryotic cell expression vector pcDNA 3.1 between the BamHI and XhoI sites. Pseudovirions were produced by co-transfection Lenti-X 293T cells with psPAX2(gag/pol), pTrip-luc lentiviral vector, and pcDNA 3.1 SARS-CoV-2-spike-deltaC19, using Lipofectamine 3000. The supernatants were harvested at 48h post-transfection and filtered through 0.45μm membranes, and titrated using 293T-ACE2-TMPRSS2 cells (HEK 293T cells that express ACE2 and TMPRSS2 proteins). Neutralization assays were performed as previously described 48,71-74. For the neutralization assay, 50 μL of SARS-CoV-2 S pseudovirions (counting ~200,000 relative light units) were pre-incubated with an equal volume of medium containing serial dilutions of samples at room temperature for 1h. Then 50 μL of virus-antibody mixtures were added to 293T-ACE2-TMPRSS2 cells (104 cells/50 μL) in a 96-well plate. The input virus with all SARS-CoV-2 strains used in the current study was the same (2 x 105 relative light units/50 μL/well). After a 3 h incubation, fresh medium was added to the wells. Cells were lysed 24 h later, and luciferase activity was measured using One-Glo luciferase assay system (Promega, Cat# E6130). The assay of each sample was performed in duplicate, and the 50% neutralization titer was calculated using Prism 9 (GraphPad Software). Controls included cells only, virus without any antibody and positive sera.
Auto-antibody analysis with ELISA
Elisas were conducted as previously described 11. Briefly, 96-well ELISA plates (MaxiSorp, Thermo Fisher Scientific) were coated by incubation overnight at 4°C with rhIFN-a2 (2 ug/ml; Miltenyi Biotec, reference number 130-108-984). Plates were then washed (PBS, 0.005% Tween 20), blocked by incubation with 5% nonfat milk powder in the same buffer, washed, and incubated with 1:50 dilutions of plasma from the patients or controls for 2 hours at room temperature (or with specific mAbs as positive controls). Each sample was tested once. Plates were thoroughly washed. Horseradish peroxidase–conjugated Fc-specific IgG fractions from polyclonal goat antiserum against human IgG were added to a final dilution of 1:4000. Plates were incubated for 1 hour at room temperature and washed. Substrate was added, and the optical density was measured. As positive control, serum from a patient with atypical mycobacterial infection was used who had high levels of IFNα2 auto-antibodies.
Spike-specific memory B-cell staining
Cryopreserved PBMCs were thawed and washed twice with 10 mL of FACS buffer (1 x PBS containing 2% FBS and 1 mM EDTA) and resuspended in 100 uL of 1x PBS containing Zombie UV live/dead dye at 1:200 dilution (BioLegend, 423108) and incubate at room temperature for 15 minutes. Following washing, cells were incubated with an antibody cocktail for 1 hour protected from light on ice. The following antibodies were used: IgD PE (BD Biosciences, 555779), IgM PerCP-Cy5.5 (BioLegend, 314512), CD20 APC-H7 (BD Biosciences, 560734), CD27 PE-Cy7 (BioLegend, 302838), CD14 PE/Dazzle™ 594 (BioLegend, 301852), CD16 BV605 (BioLegend, 302040), IgG BV650 (BD Biosciences, 740596), CD3 BUV737 (BD Biosciences, 612750) and Alexa Fluor 488-labeled Wuhan spike (SinoBiological, 40589-V27B-B), and BV421 labeled Omicron Spike (SinoBiological™, 40589-V49H3-B). All antibodies were used as the manufacturer's instruction and the final concentration of each probe was 0.1 ug/ml. Cells were washed twice in FACS buffer and immediately acquired on a BD FACS Aria III for acquisition and FlowJo for analysis.
Single cell BCR-seq
SARS-CoV-2 spike specific memory B-cells gated on singlet CD3− CD14− CD16− CD20+ IgM− IgD− CD27low/high IgG+ spikes+ were single-cell sorted into individual wells of 96-well plates containing 16 μl of lysis buffer per well using a FACS Aria III. The lysis buffer was composed of 20 U RNAse inhibitor (Invitrogen), 5 mM DTT (Invitrogen), 4 ul 5x RT buffer (Invitrogen), 0.0625 ul Igepal (Sigma), 10 ug/ml Carrier RNA (Applied Biosystems). The 96-well plates went through a quick freeze-thaw cycle, and 0.5 ug Oligo(dT)18 (Thermo Scientific), 0.5 mM dNTP mix (Invitrogen), and 200 U Superscript IV (Invitrogen) was added in a total volume of 4 ul followed by thorough mixing and spinning. The reverse transcription was performed as follows: 10 min at 42 °C, 10 min at 23 °C, 20 min at 50 °C, 5 min at 55 °C, 10 min at 80 °C and finally cooling to 4 °C. Ig heavy chain and light chain (kappa/lambda) variable gene fragments were amplified by nested PCR (HotStarTaq DNA Polymerase, QIAGEN) using primer cocktails 75,76 at a concentration of 250 nM per primer. The PCR mix consisted of 2.5 ul 10x PCR buffer, 0.5 ul 10 mM dNTP mix (Invitrogen), 0.5 ul 25 mM MgCl2, 5 ul Q-solution, 1 U HotStarTaq, 0.5 ul 5’ and 3’ primers and 2.5 ul of cDNA. Water was added up to a total volume of 25 ul. The 2nd round PCR products was evaluated on 2% agarose gels, purified using QIAquick spin columns (Qiagen) and sequenced using 2nd round PCR reverse primers. The sequences were analyzed using the online IMGT/HighV-QUEST tool. IGHV and IGLV nucleotide sequences were aligned against their closest germlines and the somatic hypermutation rate was calculated based on the IMGT v-identity output. The average mutation rate was calculated by dividing the sum of all somatic hypermutation rates by the number of sequences used for the analysis in each individual. The Change-O toolkit v.1.0.0. and SHazaM R package were used for B-cell clonality analysis 77.
T-cell stimulation and intracellular cytokine staining assay
Antigen-specific T-cell responses were measured using the intracellular cytokine staining assay as previously described 78. Live frozen PBMCs were revived, counted, and resuspended at a density of 2 million live cells per ml in complete RPMI (RPMI supplemented with 10% FBS and antibiotics). The cells were rested for 6 h at 37 °C in a CO2 incubator. At the end of 6 h, the cells were centrifuged, resuspended at a density of 10 million cells per ml in complete RPMI, and 100 μl of cell suspension containing 1 million cells was added to each well of a 96-well round-bottomed tissue culture plate. Each sample was treated with two or three conditions depending on cell numbers: no stimulation or a peptide pool spanning the Spike protein of the ancestral Wu strain or Omicron BA.1 variant (where cell numbers permitted) in the presence of anti-CD28 (1 μg ml−1; clone CD28.2, BD Biosciences) and anti-CD49d (clone 9F10, BD Biosciences), as well as anti-CXCR3 (DataS1). The peptide pools were 15-mer peptides with 10-mer overlaps spanning the entire Spike protein sequence of each variant 79. Each peptide pool contained 253 peptides and was resuspended in DMSO at a concentration of 1 mg/ml. PBMCs were stimulated at a final concentration of 1 μg/ml of each peptide in the final reaction with an equimolar amount of DMSO [0.5% (v/v) in 0.2-ml total reaction volume] as a negative control. The samples were incubated at 37°C in CO2 incubators for 2 hours before the addition of brefeldin A (10 μg ml−1). The cells were incubated for an additional 4 hours. The cells were washed with PBS and stained with Zombie ultraviolet (UV) fixable viability dye (BioLegend). The cells were washed with PBS containing 5% FBS before adding a surface antibody cocktail (Data S2). The cells were stained for 20 min at 4 °C in 100-μl volume. Subsequently, the cells were washed, fixed, and permeabilized with cytofix/cytoperm buffer (BD Biosciences) for 20 min. The permeabilized cells were stained with intracellular cytokine staining antibodies (Data S2) for 20 min at room temperature in 1× perm/wash buffer (BD Biosciences). Cells were then washed twice with perm/wash buffer and once with staining buffer before acquisition using the BD Symphony Flow Cytometer and the associated BD FACS Diva software. All flow cytometry data were analyzed using Flowjo software v.10 (BD Bioscience). DMSO background was subtracted from all samples and the positivity threshold was defined as 3x the median of peptide stimulated samples from healthy control infants.
Quantitation of human IFN-α2a
Human IFN-α2a was measured using an S-PLEX Human IFN-α2a kit from Mesoscale Discovery (Cat # K151P3S). The assay was performed as per the manufacturer’s instructions. Briefly, the uncoated 96 well plates were washed 3 times with 150 μl per well of 1xMSD wash buffer and coated with 50 μl of coating solution per well with shaking at 700 rpm at room temperature. After 1 hour of coating, the plates were washed, and 25 μl of blocking solution was added to each well, followed by adding 25 μl of neat plasma or nasal swab samples or serially diluted calibrator solution to each plate in the designated wells and incubated at room temperature for 1.5 h with shaking. After calibrator and sample incubation, the plates were washed, and 50 μl of TURBO-BOOST antibody solution was added, and the plates were incubated at room temperature for 1 h. After TURBO-BOOST antibody incubation, the plates were washed and 50 μl of enhance solution was added, and the plates were incubated at room temperature for 30 minutes. After enhance solution incubation, the plates were washed, 50 μl of TURBO-TAG detection solution was added, and the plates were incubated with shaking at 27 °C for 1 hour. After TURBO-TAG dection incubation, the plates were washed gently, 150 μl of MSD-Gold read buffer A was added, and the plates were immediately read using the MSD instrument. The IFN-α concentrations were determined by the calibration curves established by fitting the signals from the calibrators using a 4-parameter logistic model with a 1/Y2 weighting in Prism v.9.4.1.
Olink
Cytokines in plasma and nasal swab samples were measured using Olink multiplex proximity extension assay (PEA) inflammation panel (Olink proteomics: www.olink.com) according to the manufacturer’s instructions as described before 80. The PEA is a dual-recognition immunoassay, in which two matched antibodies labeled with unique DNA oligonucleotides simultaneously bind to a target protein in solution. This brings the two antibodies into proximity, allowing their DNA oligonucleotides to hybridize, serving as template for a DNA polymerase-dependent extension step. This creates a double-stranded DNA ‘barcode’ that is unique for the specific antigen and quantitatively proportional to the initial concentration of target protein. The hybridization and extension are immediately followed by PCR amplification and the amplicon is then finally quantified by microfluidic qPCR using Fluidigm BioMark HD system. Normalized Protein eXpression (NPX) values were used for downstream analysis after initial QC filtering. PCA analysis was conducted with the R package “pcaMethods” 81.
Quantitation of protein in nasal swabs by Bradford assay
Total protein concentration in nasal swabs was determined by the Bradford assay using Bio-Rad protein assay kit II (Bio-Rad, 5000002). The assay was performed as per the manufacturer’s instructions. Briefly, the dye reagent was 5-fold diluted with deionized water. 10 μl of nasal swabs at 1:200 dilution and serially diluted protein standards were added to each plate and incubated with 200 μl of diluted dye reagent for 5 minutes at room temperature. After incubation, the plates were immediately read using an absorbance reader set to 595 nm.
CyTOF analysis of PBMC samples
1 million peripheral blood mononuclear cells (PBMCs) were fixed using 2% PFA for 30 min at RT and washed. Fixed PBMCs were then resuspended in freezing media (10% DMSO + 90% FBS), transferred to cryovials, and stored at −80°C until read for downstream processing and staining. As previously described 25, fixed frozen PBMCs were thawed in a 37°C water bath and gently resuspended using 1 mL CSM (PBS supplemented with 2% BSA, 2mM EDTA and 0.1 % sodium azide) and transferred to 15 mL conical tubes containing 9 mL CSM. Samples were washed twice using CSM and counted. Cells were then permeabilized and barcoded using the Cell-ID™ 20-Plex Pd Barcoding Kit (Fluidigm, catalog # 201060). Post-barcoding, cells were washed, pooled into one barcode composite, and counted. The pooled composite was then stained for 30 min at RT with a pre-titrated surface antibody cocktail (DataS1). After surface staining, cells were washed twice with CSM and fixed in 4% PFA (freshly prepared paraformaldehyde in PBS) for 10 min at RT. Cells were then washed with CSM and permeabilized with 100% cold MeOH (Sigma), and kept overnight at −80°C. The next day, cells were washed with CSM and counted. Intracellular staining was then performed for 30 min at RT with a pre-titrated intracellular antibody cocktail (DataS1), followed by two CSM washes. Finally, cells were stained with iridium-containing DNA intercalator (Fluidigm) for 20 min at RT, washed first with CSM and then with MilliQ water. The washed cells were resuspended in MilliQ water supplemented with 1x EQ four element calibration beads (Fluidigm) and acquired on Helios mass cytometer (Fluidigm). The raw FCS files were normalized and concatenated using the Fluidigm software. The normalized .fcs files were then processed in FlowJo software v10 (BD Biosciences) for debarcoding. Briefly, the normalized .fcs file was used to gate single cells based on DNA content and event length in FlowJo. The single cells were then reimported and debarcoded using Helios software version 7.0.5189. The debarcoded samples were analyzed using FlowJo or R version 1.2.1335 for downstream analysis and visualization.
Single-cell multi-omics experiments
Single-cell ATAC and RNA-seq libraries were prepared using the Chromium Single Cell Multiome ATAC + Gene Expression platform (10X Genomics, Pleasanton, CA). Briefly, cryopreserved PBMCs were thawed and processed for single nuclei multi-omics analysis according to the manufacturer’s instructions (10x Genomics, CG000365 Rev B). Nuclei were obtained by incubating PBMCs for 3.10 minutes in freshly prepared Lysis buffer and washed and resuspended in chilled diluted nuclei buffer (10x Genomics, 2000153). About 9,000 cells were targeted for each experiment. Prepared nuclei were subjected to transposition of open chromatin regions. Next, transposed nuclei, reverse transcription Master Mix, barcoded Gel Beads, and Partitioning Oil were partitioned into single-cell GEMs (Gel Bead in EMulsions) using the 10X Chromium Controller and Next GEM Chip J. Within each GEM, Gel Beads are dissolved and poly-adenlyated (poly-A) mRNA transcripts are captured by uniquely barcoded poly(dt)VN oligos. Simultaneously, accessible chromatin fragments are captured by a separate oligo containing a spacer, unique barcode, and Illumina P5 adaptor sequence. The GEMs were then incubated in a C1000 Touch Thermal Cycler (BioRad) to produce barcodedDNA from the transposed DNA, and full-length cDNA. GEMs are then broken to release and pool single-cell fractions within each sample. Pooled fractions are purified using silane magnetic beads and subjected to PCR amplification to generate sufficient mass for library construction. Next, P7 and a sample index are added to transposed DNA via PCR to generate ATAC libraries. Finally, cDNA is enzymatically fragmented before the addition of P5, P7, i7 and i5 indexes, and TruSeq Read 2 via End Repair, A-tailing, Adaptor Ligation, and PCR resulting in gene expression libraries. Quantitation of gene expression and ATAC libraries was performed using Bioanalyzer High Sensitivity DNA Analysis (Agilent). Libraries were combined into gene expression and ATAC pools and sequenced on an Illumina NovaSeq 6000 system using the read lengths recommended by 10X: 28bp (read 1), 90bp (read 2), 10bp (i7 index), and 10bp (i5 index) for gene expression libraries and 50bp (read 1), 49bp (read 2), 8bp (i7 index) and 24bp (i5 index) for ATAC libraries. CellRanger v.3.1.0 (10xGenomics) was used to demultiplex raw sequencing data and quantify transcript levels against the 10x Genomics GRCh38 reference v.3.0.0.
Bulk RNA-seq
Blood was collected into Tempus Blood RNA tubes (Applied Biosystems) and the RNA was extracted using the MagMAX for Stabilized Blood Tubes RNA Isolation Kit, compatible with Tempus Blood RNA Tubes (ThermoFisher Scientific). RNA quality was assessed using a TapeStation 4200 (Agilent) and then 200 nanograms of total RNA was used as input for cDNA synthesis and library preparation using the KAPA RNA HyperPrep kit with RiboErase (HMR) Globin (Roche) according to the manufacturer’s instructions. Libraries were validated by capillary electrophoresis on a TapeStation 4200 (Agilent), pooled at equimolar concentrations, and sequenced with PE100 reads on an Illumina NovaSeq 6000, yielding ~60 million reads per sample on average.
QUANTIFICATION AND STATISTICAL ANALYSIS
CyTOF Analysis
High-dimensional analysis of phospho-CyTOF data was performed using a previously described R-based pipeline 82. In brief, the raw .fcs files were imported into R, and the data were transformed to normalize marker intensities using arcsinh with a cofactor of 5. For visualization, another transformation was applied that scales the expression of all values between 0 and 1 using percentiles as the boundary. Cell clustering was performed with 4,000 cells randomly selected from each sample using FlowSOM 83 and ConsensusClusterPlus 84. The transformed matrix was used as an input for FlowSOM, and cells were separated into 20 clusters. To obtain reproducible results (avoid random start), a seed was set for each clustering. The 20 clusters were manually annotated based on the lineage marker expression and were merged to produce the final clusters. The clusters were visualized in two-dimensional space using UMAP 85. In parallel, the data were manually gated to identify 34 immune cell subpopulations that were not well-distinguished in UMAP and used for all quantification purposes. For comparison with historical CyTOF data from adult COVID-19 patients, log2 fold changes were calculated by dividing values of individual patients during acute infection with the mean value of all healthy controls and subsequently log2 transforming the result.
Single-cell gene expression analysis
The single cell RNA-seq data was processed with Seurat (v4.0.5) 86. We removed cells with less than 800 or greater than 6,000 detected genes, less than 1,000 or great than 60,000 mRNA reads, or greater than 15% mitochondrial reads. Normalization and feature selection were performed using sctransform 87. Clusters were identified with Seurat SNN graph construction on Harmony 88-corrected PCA embeddings followed by Louvain community detection algorithm. After identification of major cell types, the clustering process was repeated on each cell type separately to get refined clusters, e.g., monocyte sub-clustering, and to remove doublets that are not identifiable in the first-round clustering of all the cells. Differentially expressed (DE) genes were identified using Wilcoxon Rank Sum test in Seurat. For comparison between two groups (e.g., group A vs. group B), two modes of DE analyses were performed: 1) DEall: all the samples in group A vs. all the samples in group B; 2) DEeach: each sample in group A vs. all the samples in group B. For each mode of DE analysis, genes with p-value ≤ 0.01 were ranked by log2 fold change and used as input in gene-set enrichment analysis (GSEA) analysis implemented in the fgsea R package 89. Enrichment was assessed with gene lists in Blood Transcriptomic Modules 90. For each comparison between two groups, enriched gene sets were filtered according to the following criteria: 1) the gene set was enriched in DEall comparison (adjust p value ≤ 0.05) 2) the gene set was enriched in at least two DEeach comparisons (adjust p value ≤ 0.05). Only gene sets satisfying both criteria were kept. Selected gene sets shown in the main figures were manually curated to select gene sets relevant to immunology and often enriched in several cell types across multiple DE comparisons.
The ISG score calculation
The ISG score was calculated as the geometric mean of 33 top differentially expressed genes between acute infection and healthy controls (adjusted p value < 0.05, average log2 fold change >= 0.5) across all cell types in 8 BTM terms that are involved in antiviral interferon response (M165, M75, M150, M127, M67, M68, M111.1, M111.0).
Monocyte single-cell gene expression integration
The single-cell gene expression data of monocytes from the previous adult COVID-19 infections study 91 and the Pfizer vaccine study 92 were integrated with monocytes in the current study using the Seurat integration workflow with reciprocal PCA algorithm. The normalized expression data in the integrated assay was then used to calculate the distance between clusters and the ISG score in the integrated data. The Euclidean distance between clusters was calculated using the average expression of the top 20 marker genes in each cluster. The folder change between C14.1, adults_cov2_c11, and adults_vacc_c8 clusters was calculated using limma 93.
Single-cell chromatin accessibility analysis
The single-cell ATAC data was processed with ArchR (v1.0.1) 94. The cell type annotations were transferred from the single-cell RNA-seq analysis. Transcription factor (TF) binding motifs were annotated using JASPAR 2016 transcription factor binding database 95. The per-cell motif deviations and scores were calculated using chromVAR 96. The difference in motif deviations between two groups was tested using the Wilcoxon Rank Sum test. Adjusted p values were computed using Bonferroni correction. Similar to DE gene analysis, differential accessibility motifs between two groups (e.g., group A and group B) are defined according to the following criteria: 1) the TF motif deviations are significantly different between all cells in group A and group B (adjust p value ≤ 0.001 for comparisons between acute infection and healthy controls, and 0.0001 for comparisons between convalescent and healthy controls). 2) when comparing cells in each sample in group A vs. all cells in group B, the TF motif deviations are significantly different in at least three comparisons between acute infection and healthy controls, and at least two comparisons between convalescent and healthy controls (adjust p value ≤ 0.05). The differential gene score analysis was performed using the “getMarkerFeatures” function in ArchR with Wilcoxon Rank Sum test. Genes with FDR ≤ 0.1 were used as input in gene set overrepresentation analysis for enrichment of Blood Transcriptomic Modules with hypergeometric test. The tracks of antiviral regions were generated using the “plotBrowserTrack” function in ArchR.
Correlation network
The correlation networks were computed using four data types: 1) transcriptomics, in which the average scores of BTM clusters in each cell type in each sample were used as features; 2) epigenomics, in which the average deviations of TF motif clusters in each cell type in each sample were used as features; 3) proteomics/CyTOF, in which the average level of proteins in each cell type in each sample were used as features; 4) Plasma cytokine levels in monocytes and dendric cells; 5) IFNa2 levels. The BTM score in each cell was calculated as the geometric mean of the expression of all the measured genes in the BTM gene sets. The BTM scores were then aggregated by each sample and each cell type, and hierarchical clustering (Euclidean distance, ward.D2 agglomeration method) was applied to identify BTM clusters. The TF motif clusters were identified in a similar way. Spearman’s rho correlation was used in all the correlation calculations. The correlation networks were identified and visualized using igraph 97 implemented in the ggraph package 98. Fruchterman-Reingold layout was used.
Adult and infant immune response comparison
Four scRNA-seq datasets from adults25,29,36 were collected and processed in a similar way as the infant dataset. For each dataset, we identified differentially expressed genes in monocytes in each patient compared with all the healthy samples in that dataset using Wilcoxon Rank Sum test in Seurat. Genes with p-value more than 0.05 were treated as unchanged. The fold change of ISG genes was averaged in each patient and then compared between age group and severity.
Bulk RNA-seq analysis
Alignment was performed using STAR version 2.7.3a 99 and transcripts were annotated using a composite genome reference which included GRCh38 Ensembl release 100 and SARS-CoV-2 (GCF_009858895.2,ASM985889v3). Transcript abundance estimates were calculated internal to the STAR aligner using the algorithm of htseq-count 100. ENSEMBL IDs were filtered to remove low/non-expressed transcripts (<5 reads in >50% of samples). Gene-level counts were created by averaging counts from all ENSEMBL IDs mapping to the same gene symbol (IDs mapping to multiple symbols were discarded), using the bioMart package. Gene counts were normalized using the estimateSizeFactors function of DESeq2101. To evaluate monocyte signatures expression during infection, scores for each signature were computed as the average of all genes within the signature. For comparison with adult cohorts, gene expression data from GEO dataset GSE152641 (Inflammatix cohort) were used. BTM fold changes were computed as the average fold change of all genes within each BTM between COVID-19 subjects and healthy controls.
Figures
All analysis was performed in R 4.1.1. if not stated differently102. Figures were generated using ggplot2 103, and Complexheatmap 104. Information on specific statistical tests used and sample sizes can be found in the figure legends.
Supplementary Material
Figure S1. Humoral response to COVID-19 infection in infants, related to Figure 1
A) Summary of the conducted assays. B,C) Kinetics of developing binding (B) and neutralizing (C) antibody response in infants with COVID-19. D,E) Comparison of binding (D, Non-Omi n=13, Omi n=18, Conv n=30) and neutralizing (E, Non-O n=13, Omi n=18) titers between different variants in infants. F-H) ELISA for auto-Abs against IFNα2. G) Longitudinal development of auto-Abs in matched infant samples. H) auto-Ab levels in acute infant samples and matched maternal serum. Adult controls (n=10), adults with acute COVID-19 infection (n=15), infant controls who have not had COVID-19 (n=27), infants with acute omicron infection (n=18), and longitudinal samples from infants who had COVID-19 (pre-infection (n=27), acute infection (n=19), and convalescent samples (n=30)), and matched maternal samples taken at the time of birth (n=30) are shown. Statistical comparisons were conducted with Wilcoxon rank sum and Wilcoxon signed-rank test.
Figure S3. Cytokine response to COVID-19 infection in infants, related to Figure 3
A) Comparison of key inflammatory mediators during COVID-19 infection in infants and adults, stratified by severity. Infant infection was overall mild or asymptomatic. B) Comparison of differences in plasma cytokine levels between infants and adults at baseline. C) Plasma levels for top-upregulated cytokines from (B). D) Comparison of key inflammatory mediators during COVID-19 infection in infants and mothers, stratified by age and vaccine status (infants n=33, mothers n=18, vaccinated mothers n=23). E-G) Comparison of plasma IFNα2 levels between Non-Omicron (Non-O, n=19) and Omicron (Omi, n=22) infected infants (E), infants (n=41) and adults (n=15) (F), and infants (n=41), mothers (n=18), and vaccinated mothers, (n= 23) (G). H) Viral load in nasal swabs of SARS-CoV-2-infected infants and young children and mothers was estimated using RT-qPCR. Boxplot showing −1 * Ct values during the first 5 days of infection (infant n=50, mothers n=18, vaccinated mothers n=23). Statistical comparisons were conducted with the unpaired t-test (C) and Wilcoxon rank sum test (D-H).
Figure S2. Memory B and T-cell response, related to Figure 2
A) Gating strategy for SARS-CoV-2 spike-specific IgG+ memory B-cell staining and sorting. Gating was on singlets that were CD20+ and CD3− CD14− IgM− IgD− CD27low/+ IgG+. Sorted cells were Wuhan spike-AlexaFluor 488+ and/or Omicron spike-BV421+. B) Percentage of spike-specific IgG+ memory B-cells in healthy, acute, and convalescent infant individuals. The sample number for each group is indicated in brackets. C) Percentage of spike-specific IgG+ memory B-cells in adult individuals with mild, severe, and ICU symptoms and in adult convalescent individuals. The sample number for each group is indicated in brackets. D-F) T-cell cytokine production after stimulation with overlapping peptides. D) Fraction of responding T-cells at different infection stages. E) Fraction of multifunctional T-cells at different infection stages. F) Comparison of the multifunctional T-cell response after stimulation with WT and Omicron (Om) peptides. Statistical comparisons were conducted with Wilcoxon rank sum test. Solid line indicates median healthy response; dashed line indicates 3x median healthy response (Limit of detection [LOD]).
Figure S4. Cellular immune response to COVID-19 infection, related to Figure 4
A) Expression of CyTOF markers in all manually gated subsets. B) Expression levels of significantly changed markers in healthy and infected samples. Colors indicate the infection stage. C) Frequency of pDCs as a proportion of total CD45+ cells (left) and the expression of pS6 in pDCs for different infection stages. D) Kinetics of HLA-DR expression in cd14_m. E) Boxplots showing HLA-DR expression on mDCs for different infection stages. F-H) Log2 fold-changes of acute infection samples compared to healthy samples were calculated for CyTOF markers from this cohort and two historical cohorts of adult patients with COVID-19 (Atlanta: healthy n=17, mild n=4, severe n=13; Hong Kong: healthy n=45, mild n=39, severe n=15) 25. Boxplots show the differences in log2 fold-change for indicated markers. Correlation analyses were conducted using Spearman correlation. Statistical comparisons were conducted using Wilcoxon rank sum test.
Figure S7. Nasal immune response to SARS-CoV-2 infection in infants and young children and mothers, related to Figure 7
A) Correlation between viral load (−1 * Ct) and nasal swab cytokine levels during acute infection in infants (n=32). B) Pearson r for the correlation analysis between cytokine levels in nasal swabs and matched plasma samples (+/− 5 days; n=37). Red bars with p <= 0.05. C) Correlation between the frequency of CD4_effector cells and indicated mucosal cytokine levels using matched blood and nasal swab samples (+/− 5 days). D) Correlation between indicated nasal cytokine levels at time of first PCR+ (day 0) and neutralizing antibody levels at late infection (day 7 or later). E) Total protein concentration in nasal swab samples from healthy (mothers n=19, infants n=22), SARS-CoV-2 infected (mothers n=49, infants n=52), and convalescent individuals (infants n=17). F) Time-dependent, infection-associated changes in nasal swab cytokine levels of mothers were analyzed using ANOVA. Shown is the p-value. Cytokines with an ANOVA p-value < 0.05 are indicated in red. G) Kinetics of indicated maternal nasal swab cytokines. H,I) Time-dependent changes in nasal swab levels of key cytokines in maternal nasal swabs. J) IFNα2 levels in maternal nasal swabs. K) Comparison of IFNα2 levels in infants and mothers. Statistical comparisons were conducted with the Wilcoxon rank sum test (H, I, K), one-way ANOVA (E,F), and Sidak’s multiple comparison test (E). Correlation analyses were conducted using Pearson (A-D) and Spearman correlation (C – lower stats). Lines were fitted using the linear model (A,C,D) and loess (G,J) approaches.
Figure S6. Single-cell epigenomic analysis of immunity to COVID-19 infection in infants, related to Figure 6
A) Pairwise comparison of TF motif accessibility between convalescent and pre or matched-ctrl samples was conducted for each cluster. Color indicates difference in TF accessibility; non-significant changes (FDR>=0.0001 or changed in less than two subjects) are grey. Size indicates the number of samples with significant change. (pre: n=9, matched-ctrl: n=7, Conv: n=9) B) Sample-level accessibility of selected TFs from (A) in CD14+ monocytes. C) Chromatin accessibility at indicated time points in CD16+ monocytes.
Figure S5. Single-cell multi-omics analysis of immunity to COVID-19 infection in infants, related to Figure 5
A) Full ring plot from Figure 5c. Pairwise comparison of genes from healthy (n=16) and COVID-19–infected infants at different times during acute infection (D0-5: n=5, D5-10: n=7, D10+: n=6) was conducted for each cluster. DEGs were analyzed for the enrichment of BTMs. Ring plot shows an abridged representation of enriched pathways in each cluster. Size indicates the number of samples with enrichment; colors indicate the normalized enrichment score. B) Integrated analysis of monocyte clusters from this study and from adult COVID-19 patients 25 and adult subjects immunized with the COVID-19 vaccine 35. Colors indicate the study origin of cells (top), the cell cluster (middle), and the expression of ISGs (bottom). C) DEGs determined between infant C14.1 and adult COVID-19-infection C11 monocyte clusters are plotted and ranked by fold change. C) DEGs determined between infant C14.1 and adult vaccination C8 monocyte clusters are plotted and ranked by fold change. D) Correlation analysis between plasma IFNα2 levels and average ISG levels in each cell type. E) Infection-induced expression of ISG score in CD14 monocytes from infants and historical datasets with adult COVID-19 patients 25,29,36. Each dot represents an individual patient (infants: n=18; mild/moderate adults: N=178; severe adults: N=164). F) Correlation of infection-induced expression of ISGs in CD14 monocytes from infants (x-axis) and adults (y-axis). G) Infection-induced expression of ISG score in bulk transcriptomics data from infants and a historical dataset with adult COVID-19 patients (Inflammatix cohort) 40,41. Each dot represents an individual patient (Infant n=37, Adult Inflammatix n=36). H) Difference in COVID-19 vs healthy gene expression fold change between infants and adult COVID-19 patients (Inflammatix cohort). I) Expression of NK cell activation genes enriched in NK cells in Figure 5c (green box). Statistical comparisons were conducted using Wilcoxon rank sum test.
DataS1 – Demographic and disease information on human participants and details on cytometry experiments, related to STAR Methods.
Acknowledgments
We thank all study participants and the Cincinnati Children's Hospital Medical Center staff and faculty who conducted the clinical work with pediatric samples and isolated and banked specimens, especially Monica M McNeal and Brendon White. We thank the Hope Clinic and Emory Children Center staff and faculty, especially Nadine Rouphael and the Stanford Medical Center, for collecting adult samples; the Human Immune Monitoring Center (HIMC), the Parker Institute for Cancer Immunotherapy (PICI), and the Stanford FACS facility for maintenance and access to flow cytometers and FACS sorting; Stanford Functional Genomics Facility for technical assistance; Dhananjay Wagh for library preparation; John Coller for data analysis. Cartoons were created with BioRender.com.
Funding
This work was supported by NIH grants U01 AI144673-01 (principal investigator M.A.S.). Work in the laboratory of B.P. is supported in part by the NIH (R01 AI048638, U19 AI057266 and U19 AI167903), Bill and Melinda Gates Foundation, Open Philanthropy and the Violetta L. Horton and Soffer Endowments to B.P. F.W. is supported by the Deutsche Forschungsgemeinschaft (DFG, German Research Foundation) grants EXC2180-390900677 (Germany’s excellence strategy) and 503745673 (Emmy Noether Program). The sequencing data at Stanford were generated with instrumentation purchased with NIH funds (S10OD025212 and 1S10OD021763). Next-generation sequencing services were provided by the Emory NPRC Genomics Core, which is supported in part by NIH P51 OD011132. Sequencing data were acquired on an Illumina NovaSeq6000 funded by NIH S10 OD026799. The antibody neutralization work described in this manuscript was supported by FDA’s MCMi grant #OCET 2021-1565 and FDA’s Perinatal Health Center of Excellence (PHCE) project grants #GCBER005 and GCBER008 to S.K. The autoantibody work was supported by NIHs funds (R01 AI125197, RECOVER OTA-21-15B, and R38 HL143615) and philanthropic support from the Sean N Parker Center COVID-19 Research Fund and the Henry Gustav Floren Trust. This project has been funded in whole or in part with Federal funds from the National Institute of Allergy and Infectious Diseases, National Institutes of Health, Department of Health and Human Services, under Contract No. 75N93021C00016 to A.G. and 75N9301900065 to A.S. The funders had no role in study design, data collection, analysis, interpretation, writing, the decision to publish, or preparation of the manuscript. The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the U.S. Government.
Footnotes
Declaration of Interest
B.P. serves on the External Immunology Board of GSK and on the Scientific Advisory Board of Sanofi, Medicago, Boehringer-Ingelheim, Icosavax, and EdJen. F.W. is a consultant for Gilead. A.S. is a consultant for Gritstone Bio, Flow Pharma, Moderna, AstraZeneca, Qiagen, Fortress, Gilead, Sanofi, Merck, RiverVest, MedaCorp, Turnstone, NA Vaccine Institute, Emervax, Gerson Lehrman Group and Guggenheim. LJI has filed for patent protection for various aspects of T-cell epitope and vaccine design work.
Declaration of Generative AI and AI-assisted technologies in the writing process
During the preparation of this work, the authors used GrammarlyGO (Grammarly Inc.) and GPT-4 (openAI) to improve clarity and reduce redundancy in selected sentences. After using these tools/services, the authors reviewed and edited the content as needed and take full responsibility for the content of the publication.
ADDITIONAL RESOURCES
Clinical trials in this study were pre-registered at clinicaltrials.gov: NCT05436184.
References
- 1.Olin A, Henckel E, Chen Y, Lakshmikanth T, Pou C, Mikes J, Gustafsson A, Bernhardsson AK, Zhang C, Bohlin K, et al. (2018). Stereotypic Immune System Development in Newborn Children. Cell 174, 1277–1292.e14. 10.1016/j.cell.2018.06.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Simon AK, Hollander GA, and McMichael A (2015). Evolution of the immune system in humans from infancy to old age. Proc. R. Soc. B Biol. Sci 282, 20143085. 10.1098/rspb.2014.3085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sanchez-Schmitz G, and Levy O (2011). Development of Newborn and Infant Vaccines. Sci. Transl. Med 3, 90ps27–90ps27. 10.1126/scitranslmed.3001880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Castagnoli R, Votto M, Licari A, Brambilla I, Bruno R, Perlini S, Rovida F, Baldanti F, and Marseglia GL (2020). Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) Infection in Children and Adolescents: A Systematic Review. JAMA Pediatr. 174, 882–889. 10.1001/jamapediatrics.2020.1467. [DOI] [PubMed] [Google Scholar]
- 5.Provisional COVID-19 Deaths: Focus on Ages 0–18 Years ∣ Data ∣ Centers for Disease Control and Prevention. https://data.cdc.gov/NCHS/Provisional-COVID-19-Deaths-Focus-on-Ages-0-18-Yea/nr4s-juj3.
- 6.Sacco K, Castagnoli R, Vakkilainen S, Liu C, Delmonte OM, Oguz C, Kaplan IM, Alehashemi S, Burbelo PD, Bhuyan F, et al. (2022). Immunopathological signatures in multisystem inflammatory syndrome in children and pediatric COVID-19. Nat. Med 28, 1050–1062. 10.1038/s41591-022-01724-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Loske J, Röhmel J, Lukassen S, Stricker S, Magalhães VG, Liebig J, Chua RL, Thürmann L, Messingschlager M, Seegebarth A, et al. (2021). Pre-activated antiviral innate immunity in the upper airways controls early SARS-CoV-2 infection in children. Nat. Biotechnol, 1–6. 10.1038/s41587-021-01037-9. [DOI] [PubMed] [Google Scholar]
- 8.Yoshida M, Worlock KB, Huang N, Lindeboom RGH, Butler CR, Kumasaka N, Conde CD, Mamanova L, Bolt L, Richardson L, et al. (2021). Local and systemic responses to SARS-CoV-2 infection in children and adults. Nature, 1–10. 10.1038/s41586-021-04345-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dowell AC, Butler MS, Jinks E, Tut G, Lancaster T, Sylla P, Begum J, Bruton R, Pearce H, Verma K, et al. (2022). Children develop robust and sustained cross-reactive spike-specific immune responses to SARS-CoV-2 infection. Nat. Immunol 23, 40–49. 10.1038/s41590-021-01089-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Consiglio CR, Cotugno N, Sardh F, Pou C, Amodio D, Rodriguez L, Tan Z, Zicari S, Ruggiero A, Pascucci GR, et al. (2020). The Immunology of Multisystem Inflammatory Syndrome in Children with COVID-19. Cell 183, 968–981.e7. 10.1016/j.cell.2020.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bastard P, Rosen LB, Zhang Q, Michailidis E, Hoffmann H-H, Zhang Y, Dorgham K, Philippot Q, Rosain J, Béziat V, et al. (2020). Autoantibodies against type I IFNs in patients with life-threatening COVID-19. Science 370, eabd4585. 10.1126/science.abd4585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cheong J-G, Ravishankar A, Sharma S, Parkhurst CN, Nehar-Belaid D, Ma S, Paddock L, Fatou B, Karakaslar O, Thibodeau A, et al. (2022). Epigenetic Memory of COVID-19 in Innate Immune Cells and Their Progenitors. 2022.02.09.479588. 10.1101/2022.02.09.479588. [DOI] [Google Scholar]
- 13.Han MS, Um J, Lee EJ, Kim KM, Chang SH, Lee H, Kim YK, Choi YY, Cho EY, Kim DH, et al. (2022). Antibody Responses to SARS-CoV-2 in Children With COVID-19. J. Pediatr. Infect. Dis. Soc 11, 267–273. 10.1093/jpids/piac012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Suthar MS, Zimmerman MG, Kauffman RC, Mantus G, Linderman SL, Hudson WH, Vanderheiden A, Nyhoff L, Davis CW, Adekunle O, et al. (2020). Rapid Generation of Neutralizing Antibody Responses in COVID-19 Patients. Cell Rep. Med 1, 100040. 10.1016/j.xcrm.2020.100040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cohen KW, Linderman SL, Moodie Z, Czartoski J, Lai L, Mantus G, Norwood C, Nyhoff LE, Edara VV, Floyd K, et al. (2021). Longitudinal analysis shows durable and broad immune memory after SARS-CoV-2 infection with persisting antibody responses and memory B and T cells. Cell Rep. Med 2, 100354. 10.1016/j.xcrm.2021.100354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wheatley AK, Juno JA, Wang JJ, Selva KJ, Reynaldi A, Tan H-X, Lee WS, Wragg KM, Kelly HG, Esterbauer R, et al. (2021). Evolution of immune responses to SARS-CoV-2 in mild-moderate COVID-19. Nat. Commun 12, 1162. 10.1038/s41467-021-21444-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dan JM, Mateus J, Kato Y, Hastie KM, Yu ED, Faliti CE, Grifoni A, Ramirez SI, Haupt S, Frazier A, et al. (2021). Immunological memory to SARS-CoV-2 assessed for up to 8 months after infection. Science 371, eabf4063. 10.1126/science.abf4063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gruber CN, Patel RS, Trachtman R, Lepow L, Amanat F, Krammer F, Wilson KM, Onel K, Geanon D, Tuballes K, et al. (2020). Mapping Systemic Inflammation and Antibody Responses in Multisystem Inflammatory Syndrome in Children (MIS-C). Cell 183, 982–995.e14. 10.1016/j.cell.2020.09.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Porritt RA, Binek A, Paschold L, Rivas MN, McArdle A, Yonker LM, Alter G, Chandnani HK, Lopez M, Fasano A, et al. (2021). The autoimmune signature of hyperinflammatory multisystem inflammatory syndrome in children. J. Clin. Invest 131. 10.1172/JCI151520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bastard P, Gervais A, Le Voyer T, Rosain J, Philippot Q, Manry J, Michailidis E, Hoffmann H-H, Eto S, Garcia-Prat M, et al. (2021). Autoantibodies neutralizing type I IFNs are present in ~4% of uninfected individuals over 70 years old and account for ~20% of COVID-19 deaths. Sci. Immunol 6, eabl4340. 10.1126/sciimmunol.abl4340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yang F, Nielsen SCA, Hoh RA, Röltgen K, Wirz OF, Haraguchi E, Jean GH, Lee J-Y, Pham TD, Jackson KJL, et al. (2021). Shared B cell memory to coronaviruses and other pathogens varies in human age groups and tissues. Science 372, 738–741. 10.1126/science.abf6648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang Z, Muecksch F, Schaefer-Babajew D, Finkin S, Viant C, Gaebler C, Hoffmann H-H, Barnes CO, Cipolla M, Ramos V, et al. (2021). Naturally enhanced neutralizing breadth against SARS-CoV-2 one year after infection. Nature 595, 426–431. 10.1038/s41586-021-03696-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yonker LM, Gilboa T, Ogata AF, Senussi Y, Lazarovits R, Boribong BP, Bartsch YC, Loiselle M, Rivas MN, Porritt RA, et al. (2021). Multisystem inflammatory syndrome in children is driven by zonulin-dependent loss of gut mucosal barrier. J. Clin. Invest 131. 10.1172/JCI149633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Brodin P (2022). SARS-CoV-2 infections in children: Understanding diverse outcomes. Immunity 55, 201–209. 10.1016/j.immuni.2022.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Arunachalam PS, Wimmers F, Mok CKP, Perera RAPM, Scott M, Hagan T, Sigal N, Feng Y, Bristow L, Tak-Yin Tsang O, et al. (2020). Systems biological assessment of immunity to mild versus severe COVID-19 infection in humans. Science 369, 1210–1220. 10.1126/science.abc6261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Del Valle DM, Kim-Schulze S, Huang H-H, Beckmann ND, Nirenberg S, Wang B, Lavin Y, Swartz TH, Madduri D, Stock A, et al. (2020). An inflammatory cytokine signature predicts COVID-19 severity and survival. Nat. Med 26, 1636–1643. 10.1038/s41591-020-1051-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hadjadj J, Yatim N, Barnabei L, Corneau A, Boussier J, Smith N, Péré H, Charbit B, Bondet V, Chenevier-Gobeaux C, et al. (2020). Impaired type I interferon activity and inflammatory responses in severe COVID-19 patients. Science 369, 718–724. 10.1126/science.abc6027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Capucetti A, Albano F, and Bonecchi R (2020). Multiple Roles for Chemokines in Neutrophil Biology. Front. Immunol 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schulte-Schrepping J, Reusch N, Paclik D, Baßler K, Schlickeiser S, Zhang B, Krämer B, Krammer T, Brumhard S, Bonaguro L, et al. (2020). Severe COVID-19 Is Marked by a Dysregulated Myeloid Cell Compartment. Cell, S0092867420309922. 10.1016/j.cell.2020.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rahil Z, Leylek R, Schürch CM, Chen H, Bjornson-Hooper Z, Christensen SR, Gherardini PF, Bhate SS, Spitzer MH, Fragiadakis GK, et al. (2020). Landscape of coordinated immune responses to H1N1 challenge in humans. J. Clin. Invest 130, 5800–5816. 10.1172/JCI137265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ivashkiv LB, and Donlin LT (2014). Regulation of type I interferon responses. Nat. Rev. Immunol 14, 36–49. 10.1038/nri3581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Keskinen P, Ronni T, Matikainen S, Lehtonen A, and Julkunen I (1997). Regulation of HLA class I and II expression by interferons and influenza A virus in human peripheral blood mononuclear cells. Immunology 91, 421–429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bauvois B, Durant L, Laboureau J, Barthelemy E, Rouillard D, Boulla G, and Deterre P (1999). Upregulation of CD38 Gene Expression in Leukemic B Cells by Interferon Types I and II. J. Interferon Cytokine Res 19, 1059–1066. 10.1089/107999099313299. [DOI] [PubMed] [Google Scholar]
- 34.Wimmers F, Donato M, Kuo A, Ashuach T, Gupta S, Li C, Dvorak M, Foecke MH, Chang SE, Hagan T, et al. (2021). The single-cell epigenomic and transcriptional landscape of immunity to influenza vaccination. Cell. 10.1016/j.cell.2021.05.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Arunachalam PS, Scott MKD, Hagan T, Li C, Feng Y, Wimmers F, Grigoryan L, Trisal M, Edara VV, Lai L, et al. (2021). Systems vaccinology of the BNT162b2 mRNA vaccine in humans. Nature 596, 410–416. 10.1038/s41586-021-03791-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Su Y, Chen D, Yuan D, Lausted C, Choi J, Dai CL, Voillet V, Duvvuri VR, Scherler K, Troisch P, et al. (2020). Multi-Omics Resolves a Sharp Disease-State Shift between Mild and Moderate COVID-19. Cell 183, 1479–1495.e20. 10.1016/j.cell.2020.10.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tang BM, Shojaei M, Parnell GP, Huang S, Nalos M, Teoh S, O’Connor K, Schibeci S, Phu AL, Kumar A, et al. (2017). A novel immune biomarker IFI27 discriminates between influenza and bacteria in patients with suspected respiratory infection. Eur. Respir. J 49. 10.1183/13993003.02098-2016. [DOI] [PubMed] [Google Scholar]
- 38.Shojaei M, Shamshirian A, Monkman J, Grice L, Tran M, Tan CW, Teo SM, Rodrigues Rossi G, McCulloch TR, Nalos M, et al. (2023). IFI27 transcription is an early predictor for COVID-19 outcomes, a multi-cohort observational study. Front. Immunol 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gao J, Zhu X, Wu M, Jiang L, Wang F, and He S (2021). IFI27 may predict and evaluate the severity of respiratory syncytial virus infection in preterm infants. Hereditas 158, 3. 10.1186/s41065-020-00167-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zheng H, Rao AM, Dermadi D, Toh J, Murphy Jones L, Donato M, Liu Y, Su Y, Dai CL, Kornilov SA, et al. (2021). Multi-cohort analysis of host immune response identifies conserved protective and detrimental modules associated with severity across viruses. Immunity 54, 753–768.e5. 10.1016/j.immuni.2021.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Thair SA, He YD, Hasin-Brumshtein Y, Sakaram S, Pandya R, Toh J, Rawling D, Remmel M, Coyle S, Dalekos GN, et al. (2021). Transcriptomic similarities and differences in host response between SARS-CoV-2 and other viral infections. iScience 24, 101947. 10.1016/j.isci.2020.101947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Langel SN, Garrido C, Phan C, Travieso T, Kirshner H, DeMarco T, Ma Z-M, Reader JR, Olstad KJ, Sammak RL, et al. (2022). Dam–Infant Rhesus Macaque Pairs to Dissect Age-Dependent Responses to SARS-CoV-2 Infection. ImmunoHorizons 6, 851–863. 10.4049/immunohorizons.2200075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Granja JM, Corces MR, Pierce SE, Bagdatli ST, Choudhry H, Chang HY, and Greenleaf WJ (2021). ArchR is a scalable software package for integrative single-cell chromatin accessibility analysis. Nat. Genet 53, 403–411. 10.1038/s41588-021-00790-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Arts RJW, Moorlag SJCFM, Novakovic B, Li Y, Wang S-Y, Oosting M, Kumar V, Xavier RJ, Wijmenga C, Joosten LAB, et al. (2018). BCG Vaccination Protects against Experimental Viral Infection in Humans through the Induction of Cytokines Associated with Trained Immunity. Cell Host Microbe 23, 89–100.e5. 10.1016/j.chom.2017.12.010. [DOI] [PubMed] [Google Scholar]
- 45.Brauns E, Azouz A, Grimaldi D, Xiao H, Thomas S, Nguyen M, Olislagers V, Duc IV, Cano CO, Marmol VD, et al. (2022). Functional reprogramming of monocytes in patients with acute and convalescent severe COVID-19. JCI Insight 7. 10.1172/jci.insight.154183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lee A, Scott MKD, Wimmers F, Arunachalam PS, Luo W, Fox CB, Tomai M, Khatri P, and Pulendran B (2022). A molecular atlas of innate immunity to adjuvanted and live attenuated vaccines, in mice. Nat. Commun 13, 549. 10.1038/s41467-022-28197-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Smith N, Goncalves P, Charbit B, Grzelak L, Beretta M, Planchais C, Bruel T, Rouilly V, Bondet V, Hadjadj J, et al. (2021). Distinct systemic and mucosal immune responses during acute SARS-CoV-2 infection. Nat. Immunol 22, 1428–1439. 10.1038/s41590-021-01028-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tang J, Novak T, Hecker J, Grubbs G, Zahra FT, Bellusci L, Pourhashemi S, Chou J, Moffitt K, Halasa NB, et al. (2022). Cross-reactive immunity against the SARS-CoV-2 Omicron variant is low in pediatric patients with prior COVID-19 or MIS-C. Nat. Commun 13, 2979. 10.1038/s41467-022-30649-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pierce CA, Sy S, Galen B, Goldstein DY, Orner E, Keller MJ, Herold KC, and Herold BC (2021). Natural mucosal barriers and COVID-19 in children. JCI Insight 6. 10.1172/jci.insight.148694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Black A, Bhaumik S, Kirkman RL, Weaver CT, and Randolph DA (2012). Developmental regulation of Th17-cell capacity in human neonates. Eur. J. Immunol 42, 311–319. 10.1002/eji.201141847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pelletier M, Maggi L, Micheletti A, Lazzeri E, Tamassia N, Costantini C, Cosmi L, Lunardi C, Annunziato F, Romagnani S, et al. (2010). Evidence for a cross-talk between human neutrophils and Th17 cells. Blood 115, 335–343. 10.1182/blood-2009-04-216085. [DOI] [PubMed] [Google Scholar]
- 52.Mitsdoerffer M, Lee Y, Jäger A, Kim H-J, Korn T, Kolls JK, Cantor H, Bettelli E, and Kuchroo VK (2010). Proinflammatory T helper type 17 cells are effective B-cell helpers. Proc. Natl. Acad. Sci 107, 14292–14297. 10.1073/pnas.1009234107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Silvin A, Chapuis N, Dunsmore G, Goubet A-G, Dubuisson A, Derosa L, Almire C, Hénon C, Kosmider O, Droin N, et al. (2020). Elevated Calprotectin and Abnormal Myeloid Cell Subsets Discriminate Severe from Mild COVID-19. Cell 182, 1401–1418.e18. 10.1016/j.cell.2020.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.de Jong SE, Olin A, and Pulendran B (2020). The Impact of the Microbiome on Immunity to Vaccination in Humans. Cell Host Microbe 28, 169–179. 10.1016/j.chom.2020.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Brodin P (2022). Immune-microbe interactions early in life: A determinant of health and disease long term. Science 376, 945–950. 10.1126/science.abk2189. [DOI] [PubMed] [Google Scholar]
- 56.Watkins TA, Cheemarla NR, Hänsel K, Green AB, Amat JAR, Lozano R, Dudgeon SN, Landry ML, Schulz WL, and Foxman EF (2023). High burden of viruses and bacterial pathobionts drives heightened nasal innate immunity in children with and without SARS-CoV-2. 2023.06.17.23291498. 10.1101/2023.06.17.23291498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.CDC Human Influenza Virus Real-Time RT-PCR Diagnostic Panel Influenza A Subtyp-ing Kit: Instructions for Use.
- 58.CDC Human Influenza Virus Real-Time RT-PCR Diagnostic Panel Influenza B Lineage Genotyping Kit: Instructions for Use. [Google Scholar]
- 59.2019-novel Coronavirus (2019-nCoV) Real-Time rRT-PCR Panel Primers and Probes.
- 60.Real-Time RT-PCR Panel for Detection of 2019-novel Coronavirus: Instructions for Use.
- 61.Research Use Only CDC Influenza SARS-CoV-2 (Flu SC2) Multiplex Assay Primers and Probes.
- 62.CDC Influenza SARS-CoV-2 (Flu SC2) Multiplex Assay: Instructions for Use.
- 63.Gonsalves S, Mahony J, Rao A, Dunbar S, and Juretschko S (2019). Multiplexed detection and identification of respiratory pathogens using the NxTAG® respiratory pathogen panel. Methods 158, 61–68. 10.1016/j.ymeth.2019.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wu F, Zhao S, Yu B, Chen Y-M, Wang W, Song Z-G, Hu Y, Tao Z-W, Tian J-H, Pei Y-Y, et al. (2020). A new coronavirus associated with human respiratory disease in China. Nature 579, 265–269. 10.1038/s41586-020-2008-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Li H (2012). Exploring single-sample SNP and INDEL calling with whole-genome de novo assembly. Bioinformatics 28, 1838–1844. 10.1093/bioinformatics/bts280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R, and 1000 Genome Project Data Processing Subgroup (2009). The Sequence Alignment/Map format and SAMtools. Bioinformatics 25, 2078–2079. 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Grubaugh ND, Gangavarapu K, Quick J, Matteson NL, De Jesus JG, Main BJ, Tan AL, Paul LM, Brackney DE, Grewal S, et al. (2019). An amplicon-based sequencing framework for accurately measuring intrahost virus diversity using PrimalSeq and iVar. Genome Biol. 20, 1–19. 10.1186/s13059-018-1618-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.O’Toole Á, Scher E, Underwood A, Jackson B, Hill V, McCrone JT, Colquhoun R, Ruis C, Abu-Dahab K, Taylor B, et al. (2021). Assignment of epidemiological lineages in an emerging pandemic using the pangolin tool. Virus Evol. 7, veab064. 10.1093/ve/veab064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Sievers BL, Chakraborty S, Xue Y, Gelbart T, Gonzalez JC, Cassidy AG, Golan Y, Prahl M, Gaw SL, Arunachalam PS, et al. (2022). Antibodies elicited by SARS-CoV-2 infection or mRNA vaccines have reduced neutralizing activity against Beta and Omicron pseudoviruses. Sci. Transl. Med 14, eabn7842. 10.1126/scitranslmed.abn7842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Neerukonda SN, Vassell R, Herrup R, Liu S, Wang T, Takeda K, Yang Y, Lin T-L, Wang W, and Weiss CD (2021). Establishment of a well-characterized SARS-CoV-2 lentiviral pseudovirus neutralization assay using 293T cells with stable expression of ACE2 and TMPRSS2. PLOS ONE 16, e0248348. 10.1371/journal.pone.0248348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Ravichandran S, Coyle EM, Klenow L, Tang J, Grubbs G, Liu S, Wang T, Golding H, and Khurana S (2020). Antibody signature induced by SARS-CoV-2 spike protein immunogens in rabbits. Sci. Transl. Med 12, eabc3539. 10.1126/scitranslmed.abc3539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Tang J, Ravichandran S, Lee Y, Grubbs G, Coyle EM, Klenow L, Genser H, Golding H, and Khurana S (2021). Antibody affinity maturation and plasma IgA associate with clinical outcome in hospitalized COVID-19 patients. Nat. Commun 12, 1221. 10.1038/s41467-021-21463-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Bellusci L, Grubbs G, Srivastava P, Nemeth MJ, Griffiths EA, Golding H, and Khurana S (2022). Neutralization of SARS-CoV-2 Omicron after vaccination of patients with myelodysplastic syndromes or acute myeloid leukemia. Blood 139, 2842–2846. 10.1182/blood.2022016087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Ravichandran S, Tang J, Grubbs G, Lee Y, Pourhashemi S, Hussaini L, Lapp SA, Jerris RC, Singh V, Chahroudi A, et al. (2021). SARS-CoV-2 immune repertoire in MIS-C and pediatric COVID-19. Nat. Immunol 22, 1452–1464. 10.1038/s41590-021-01051-8. [DOI] [PubMed] [Google Scholar]
- 75.Liao H-X, Levesque MC, Nagel A, Dixon A, Zhang R, Walter E, Parks R, Whitesides J, Marshall DJ, Hwang K-K, et al. (2009). High-throughput isolation of immunoglobulin genes from single human B cells and expression as monoclonal antibodies. J. Virol. Methods 158, 171–179. 10.1016/j.jviromet.2009.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Tiller T, Meffre E, Yurasov S, Tsuiji M, Nussenzweig MC, and Wardemann H (2008). Efficient generation of monoclonal antibodies from single human B cells by single cell RT-PCR and expression vector cloning. J. Immunol. Methods 329, 112–124. 10.1016/j.jim.2007.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Gupta NT, Vander Heiden JA, Uduman M, Gadala-Maria D, Yaari G, and Kleinstein SH (2015). Change-O: a toolkit for analyzing large-scale B cell immunoglobulin repertoire sequencing data: Table 1. Bioinformatics 31, 3356–3358. 10.1093/bioinformatics/btv359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Arunachalam PS, Feng Y, Ashraf U, Hu M, Walls AC, Edara VV, Zarnitsyna VI, Aye PP, Golden N, Miranda MC, et al. (2022). Durable protection against the SARS-CoV-2 Omicron variant is induced by an adjuvanted subunit vaccine. Sci. Transl. Med 14, eabq4130. 10.1126/scitranslmed.abq4130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Tarke A, Coelho CH, Zhang Z, Dan JM, Yu ED, Methot N, Bloom NI, Goodwin B, Phillips E, Mallal S, et al. (2022). SARS-CoV-2 vaccination induces immunological T cell memory able to cross-recognize variants from Alpha to Omicron. Cell 185, 847–859.e11. 10.1016/j.cell.2022.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Assarsson E, Lundberg M, Holmquist G, Björkesten J, Thorsen SB, Ekman D, Eriksson A, Dickens ER, Ohlsson S, Edfeldt G, et al. (2014). Homogenous 96-Plex PEA Immunoassay Exhibiting High Sensitivity, Specificity, and Excellent Scalability. PLOS ONE 9, e95192. 10.1371/journal.pone.0095192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Stacklies W, Redestig H, Scholz M, Walther D, and Selbig J (2007). pcaMethods—a bioconductor package providing PCA methods for incomplete data. Bioinformatics 23, 1164–1167. 10.1093/bioinformatics/btm069. [DOI] [PubMed] [Google Scholar]
- 82.Nowicka M, Krieg C, Crowell HL, Weber LM, Hartmann FJ, Guglietta S, Becher B, Levesque MP, and Robinson MD (2019). CyTOF workflow: differential discovery in high-throughput high-dimensional cytometry datasets. 10.12688/f1000research.11622.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Van Gassen S, Callebaut B, Van Helden MJ, Lambrecht BN, Demeester P, Dhaene T, and Saeys Y (2015). FlowSOM: Using self-organizing maps for visualization and interpretation of cytometry data. Cytometry A 87, 636–645. 10.1002/cyto.a.22625. [DOI] [PubMed] [Google Scholar]
- 84.Wilkerson MD, and Hayes DN (2010). ConsensusClusterPlus: a class discovery tool with confidence assessments and item tracking. Bioinformatics 26, 1572–1573. 10.1093/bioinformatics/btq170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.McInnes L, Healy J, and Melville J (2020). UMAP: Uniform Manifold Approximation and Projection for Dimension Reduction. ArXiv180203426 Cs Stat. [Google Scholar]
- 86.Hao Y, Hao S, Andersen-Nissen E, Mauck WM, Zheng S, Butler A, Lee MJ, Wilk AJ, Darby C, Zager M, et al. (2021). Integrated analysis of multimodal single-cell data. Cell 184, 3573–3587.e29. 10.1016/j.cell.2021.04.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Hafemeister C, and Satija R (2019). Normalization and variance stabilization of single-cell RNA-seq data using regularized negative binomial regression. Genome Biol. 20, 296. 10.1186/s13059-019-1874-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Korsunsky I, Millard N, Fan J, Slowikowski K, Zhang F, Wei K, Baglaenko Y, Brenner M, Loh P-R, and Raychaudhuri S (2019). Fast, sensitive and accurate integration of single-cell data with Harmony. Nat. Methods 16, 1289–1296. 10.1038/s41592-019-0619-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Korotkevich G, Sukhov V, Budin N, Shpak B, Artyomov MN, and Sergushichev A (2021). Fast gene set enrichment analysis. 060012. 10.1101/060012. [DOI] [Google Scholar]
- 90.Li S, Rouphael N, Duraisingham S, Romero-Steiner S, Presnell S, Davis C, Schmidt DS, Johnson SE, Milton A, Rajam G, et al. (2014). Molecular signatures of antibody responses derived from a systems biology study of five human vaccines. Nat. Immunol 15, 195–204. 10.1038/ni.2789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Arunachalam PS, Wimmers F, Mok CKP, Perera RAPM, Scott M, Hagan T, Sigal N, Feng Y, Bristow L, Tak-Yin Tsang O, et al. (2020). Systems biological assessment of immunity to mild versus severe COVID-19 infection in humans. Science 369, 1210–1220. 10.1126/science.abc6261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Arunachalam PS, Scott MKD, Hagan T, Li C, Feng Y, Wimmers F, Grigoryan L, Trisal M, Edara VV, Lai L, et al. (2021). Systems vaccinology of the BNT162b2 mRNA vaccine in humans. Nature 596, 410–416. 10.1038/s41586-021-03791-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, and Smyth GK (2015). limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 43, e47. 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Granja JM, Corces MR, Pierce SE, Bagdatli ST, Choudhry H, Chang HY, and Greenleaf WJ (2021). ArchR is a scalable software package for integrative single-cell chromatin accessibility analysis. Nat. Genet 53, 403–411. 10.1038/s41588-021-00790-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Mathelier A, Fornes O, Arenillas DJ, Chen C, Denay G, Lee J, Shi W, Shyr C, Tan G, Worsley-Hunt R, et al. (2016). JASPAR 2016: a major expansion and update of the open-access database of transcription factor binding profiles. Nucleic Acids Res. 44, D110–D115. 10.1093/nar/gkv1176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Schep AN, Wu B, Buenrostro JD, and Greenleaf WJ (2017). chromVAR: inferring transcription-factor-associated accessibility from single-cell epigenomic data. Nat. Methods 14, 975–978. 10.1038/nmeth.4401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Csardi G, and Nepusz T (2006). The igraph software package for complex network research. InterJournal Complex Syst. 1695, 1–9. [Google Scholar]
- 98.Pedersen TL, and RStudio (2022). ggraph: An Implementation of Grammar of Graphics for Graphs and Networks. [Google Scholar]
- 99.Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, and Gingeras TR (2013). STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21. 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Anders S, Pyl PT, and Huber W (2015). HTSeq—a Python framework to work with high-throughput sequencing data. Bioinformatics 31, 166–169. 10.1093/bioinformatics/btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Love MI, Huber W, and Anders S (2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 1–21. 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.R Core Team (2020). R: A Language and Environment for Statistical Computing (R Foundation for Statistical Computing).
- 103.Villanueva RAM, and Chen ZJ (2019). ggplot2: Elegant Graphics for Data Analysis (2nd ed.). Meas. Interdiscip. Res. Perspect 17, 160–167. 10.1080/15366367.2019.1565254. [DOI] [Google Scholar]
- 104.Gu Z. (2022). Complex heatmap visualization. iMeta 1, e43. 10.1002/imt2.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Humoral response to COVID-19 infection in infants, related to Figure 1
A) Summary of the conducted assays. B,C) Kinetics of developing binding (B) and neutralizing (C) antibody response in infants with COVID-19. D,E) Comparison of binding (D, Non-Omi n=13, Omi n=18, Conv n=30) and neutralizing (E, Non-O n=13, Omi n=18) titers between different variants in infants. F-H) ELISA for auto-Abs against IFNα2. G) Longitudinal development of auto-Abs in matched infant samples. H) auto-Ab levels in acute infant samples and matched maternal serum. Adult controls (n=10), adults with acute COVID-19 infection (n=15), infant controls who have not had COVID-19 (n=27), infants with acute omicron infection (n=18), and longitudinal samples from infants who had COVID-19 (pre-infection (n=27), acute infection (n=19), and convalescent samples (n=30)), and matched maternal samples taken at the time of birth (n=30) are shown. Statistical comparisons were conducted with Wilcoxon rank sum and Wilcoxon signed-rank test.
Figure S3. Cytokine response to COVID-19 infection in infants, related to Figure 3
A) Comparison of key inflammatory mediators during COVID-19 infection in infants and adults, stratified by severity. Infant infection was overall mild or asymptomatic. B) Comparison of differences in plasma cytokine levels between infants and adults at baseline. C) Plasma levels for top-upregulated cytokines from (B). D) Comparison of key inflammatory mediators during COVID-19 infection in infants and mothers, stratified by age and vaccine status (infants n=33, mothers n=18, vaccinated mothers n=23). E-G) Comparison of plasma IFNα2 levels between Non-Omicron (Non-O, n=19) and Omicron (Omi, n=22) infected infants (E), infants (n=41) and adults (n=15) (F), and infants (n=41), mothers (n=18), and vaccinated mothers, (n= 23) (G). H) Viral load in nasal swabs of SARS-CoV-2-infected infants and young children and mothers was estimated using RT-qPCR. Boxplot showing −1 * Ct values during the first 5 days of infection (infant n=50, mothers n=18, vaccinated mothers n=23). Statistical comparisons were conducted with the unpaired t-test (C) and Wilcoxon rank sum test (D-H).
Figure S2. Memory B and T-cell response, related to Figure 2
A) Gating strategy for SARS-CoV-2 spike-specific IgG+ memory B-cell staining and sorting. Gating was on singlets that were CD20+ and CD3− CD14− IgM− IgD− CD27low/+ IgG+. Sorted cells were Wuhan spike-AlexaFluor 488+ and/or Omicron spike-BV421+. B) Percentage of spike-specific IgG+ memory B-cells in healthy, acute, and convalescent infant individuals. The sample number for each group is indicated in brackets. C) Percentage of spike-specific IgG+ memory B-cells in adult individuals with mild, severe, and ICU symptoms and in adult convalescent individuals. The sample number for each group is indicated in brackets. D-F) T-cell cytokine production after stimulation with overlapping peptides. D) Fraction of responding T-cells at different infection stages. E) Fraction of multifunctional T-cells at different infection stages. F) Comparison of the multifunctional T-cell response after stimulation with WT and Omicron (Om) peptides. Statistical comparisons were conducted with Wilcoxon rank sum test. Solid line indicates median healthy response; dashed line indicates 3x median healthy response (Limit of detection [LOD]).
Figure S4. Cellular immune response to COVID-19 infection, related to Figure 4
A) Expression of CyTOF markers in all manually gated subsets. B) Expression levels of significantly changed markers in healthy and infected samples. Colors indicate the infection stage. C) Frequency of pDCs as a proportion of total CD45+ cells (left) and the expression of pS6 in pDCs for different infection stages. D) Kinetics of HLA-DR expression in cd14_m. E) Boxplots showing HLA-DR expression on mDCs for different infection stages. F-H) Log2 fold-changes of acute infection samples compared to healthy samples were calculated for CyTOF markers from this cohort and two historical cohorts of adult patients with COVID-19 (Atlanta: healthy n=17, mild n=4, severe n=13; Hong Kong: healthy n=45, mild n=39, severe n=15) 25. Boxplots show the differences in log2 fold-change for indicated markers. Correlation analyses were conducted using Spearman correlation. Statistical comparisons were conducted using Wilcoxon rank sum test.
Figure S7. Nasal immune response to SARS-CoV-2 infection in infants and young children and mothers, related to Figure 7
A) Correlation between viral load (−1 * Ct) and nasal swab cytokine levels during acute infection in infants (n=32). B) Pearson r for the correlation analysis between cytokine levels in nasal swabs and matched plasma samples (+/− 5 days; n=37). Red bars with p <= 0.05. C) Correlation between the frequency of CD4_effector cells and indicated mucosal cytokine levels using matched blood and nasal swab samples (+/− 5 days). D) Correlation between indicated nasal cytokine levels at time of first PCR+ (day 0) and neutralizing antibody levels at late infection (day 7 or later). E) Total protein concentration in nasal swab samples from healthy (mothers n=19, infants n=22), SARS-CoV-2 infected (mothers n=49, infants n=52), and convalescent individuals (infants n=17). F) Time-dependent, infection-associated changes in nasal swab cytokine levels of mothers were analyzed using ANOVA. Shown is the p-value. Cytokines with an ANOVA p-value < 0.05 are indicated in red. G) Kinetics of indicated maternal nasal swab cytokines. H,I) Time-dependent changes in nasal swab levels of key cytokines in maternal nasal swabs. J) IFNα2 levels in maternal nasal swabs. K) Comparison of IFNα2 levels in infants and mothers. Statistical comparisons were conducted with the Wilcoxon rank sum test (H, I, K), one-way ANOVA (E,F), and Sidak’s multiple comparison test (E). Correlation analyses were conducted using Pearson (A-D) and Spearman correlation (C – lower stats). Lines were fitted using the linear model (A,C,D) and loess (G,J) approaches.
Figure S6. Single-cell epigenomic analysis of immunity to COVID-19 infection in infants, related to Figure 6
A) Pairwise comparison of TF motif accessibility between convalescent and pre or matched-ctrl samples was conducted for each cluster. Color indicates difference in TF accessibility; non-significant changes (FDR>=0.0001 or changed in less than two subjects) are grey. Size indicates the number of samples with significant change. (pre: n=9, matched-ctrl: n=7, Conv: n=9) B) Sample-level accessibility of selected TFs from (A) in CD14+ monocytes. C) Chromatin accessibility at indicated time points in CD16+ monocytes.
Figure S5. Single-cell multi-omics analysis of immunity to COVID-19 infection in infants, related to Figure 5
A) Full ring plot from Figure 5c. Pairwise comparison of genes from healthy (n=16) and COVID-19–infected infants at different times during acute infection (D0-5: n=5, D5-10: n=7, D10+: n=6) was conducted for each cluster. DEGs were analyzed for the enrichment of BTMs. Ring plot shows an abridged representation of enriched pathways in each cluster. Size indicates the number of samples with enrichment; colors indicate the normalized enrichment score. B) Integrated analysis of monocyte clusters from this study and from adult COVID-19 patients 25 and adult subjects immunized with the COVID-19 vaccine 35. Colors indicate the study origin of cells (top), the cell cluster (middle), and the expression of ISGs (bottom). C) DEGs determined between infant C14.1 and adult COVID-19-infection C11 monocyte clusters are plotted and ranked by fold change. C) DEGs determined between infant C14.1 and adult vaccination C8 monocyte clusters are plotted and ranked by fold change. D) Correlation analysis between plasma IFNα2 levels and average ISG levels in each cell type. E) Infection-induced expression of ISG score in CD14 monocytes from infants and historical datasets with adult COVID-19 patients 25,29,36. Each dot represents an individual patient (infants: n=18; mild/moderate adults: N=178; severe adults: N=164). F) Correlation of infection-induced expression of ISGs in CD14 monocytes from infants (x-axis) and adults (y-axis). G) Infection-induced expression of ISG score in bulk transcriptomics data from infants and a historical dataset with adult COVID-19 patients (Inflammatix cohort) 40,41. Each dot represents an individual patient (Infant n=37, Adult Inflammatix n=36). H) Difference in COVID-19 vs healthy gene expression fold change between infants and adult COVID-19 patients (Inflammatix cohort). I) Expression of NK cell activation genes enriched in NK cells in Figure 5c (green box). Statistical comparisons were conducted using Wilcoxon rank sum test.
DataS1 – Demographic and disease information on human participants and details on cytometry experiments, related to STAR Methods.
Data Availability Statement
ScATAC-seq and scRNA-seq data and blood transcriptomics data have been deposited at GEO and are publicly available as of the date of publication. Accession numbers are listed in the key resources table. No original code was generated. All other code and scripts, and data reported in this paper are available from the lead contact upon request.
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Goat anti-Human IgG Fc Cross-Adsorbed Secondary Antibody | Invitrogen | Cat# PI31413 |
| anti-IgD (Clone IA6-2) PE | BD Biosciences | Cat# 555779 |
| anti-IgM (MHM-88) PerCP-Cy5.5 | BioLegend | Cat# 314512 |
| anti-CD20 (Clone 2H7) APC-H7 | BD Biosciences | Cat# 560734 |
| anti-CD27 (Clone O323) PE-Cy7 | BioLegend | Cat# 302838 |
| anti-CD14 (Clone M5E2) PE/Dazzle™ 594 | BioLegend | Cat# 301852 |
| anti-CD16 (Clone 3G8) BV605 | BioLegend | Cat# 302040 |
| anti-IgG (Clone G18-145) BV650 | BD Biosciences | Cat# 740596 |
| anti-CD3 (Clone UCHT1) BUV737 | BD Biosciences | Cat# 612750 |
| anti-IL-2 (clone MQ1-17H12) FITC | Biolegend | Cat# 500304 |
| anti-CXCR5 (clone MU5UBEE) PerCP-eF710 | Invitrogen | Cat# 46-9185-42 |
| anti-IL-4 (clone MP4-25D2) PE | BioLegend | Cat# 500810 |
| anti-CD45RA (clone 5H9) PE-CF594 | BD Biosciences | Cat# 565419 |
| anti-TNF- a (clone Mab11) PE-Cy7 | E-Bioscience | Cat# 25-7349-82 |
| anti-CD40L (clone 24-31) BV421 | Biolegend | Cat# 310824 |
| anti-TCR-gd (clone B1.1) BV506 | Biolegend | Cat# 331220 |
| anti-CD4 (clone OKT4) BV605 | Biolegend | Cat# 317438 |
| anti-CD3 (clone SP34-2) BV650 | BD Biosciences | Cat# 563916 |
| anti-CCR7 (clone G043H7) BV711 | Biolegend | Cat# 353228 |
| anti-CD127 (clone A019D5) BV785 | Biolegend | Cat# 351330 |
| anti-IL-21 (clone 3A3-N2) APC | BioLegend | Cat# 513008 |
| anti-IFN-g (clone 4S.B3) A700 | Biolegend | Cat# 502520 |
| anti-CD25 (clone BC96) APC-Cy7 | Biolegend | Cat# 302614 |
| anti-CXCR3 (clone 1C6/CXCR3) BUV395 | BD Biosciences | Cat# 565223 |
| anti-CD8 (clone RPA-T8) BUV563 | BD Biosciences | Cat# 612914 |
| anti-CCR6 (clone 11A9) BUV737 | BD Biosciences | Cat# 612780 |
| anti-CD69 (clone FN50) BUV805 | BD Biosciences | Cat# 748763 |
| Purified anti-human CD66b (G10F5) | BioLegend | Cat#305102; RRID: AB_314494 |
| Purified anti-human CD57 (HNK-1) | BioLegend | Cat#359602; RRID: AB_2562403 |
| Purified anti-human HLA-DR (L243) | BioLegend | Cat#307651; RRID: AB_2562826 |
| Anti-Human CD19 (HIB19) - 142Nd | Fluidigm | Cat#3142001B |
| Anti-Human CD127 (A019D5) - 143Nd | Fluidigm | Cat#3143012B |
| Anti-Human IL4 (MP4-25D2) - 144Nd | Fluidigm | Cat#3144010B |
| Anti-Human CD4(RPA-T4) - 145Nd | Fluidigm | Cat#3145001B |
| Anti-Human IgD (IA6-2) - 146Nd | Fluidigm | Cat#3146005B |
| Anti-Human CD20 (2H7) - 147Sm | Fluidigm | Cat#3147001B |
| Anti-Human CD34 (581) - 148Nd | Fluidigm | Cat#3148001B |
| Anti-pSTAT6 (18/P-Stat6) - 149Sm | Fluidigm | Cat#3149004A |
| Anti-pSTAT5 (47) - 150Nd | Fluidigm | Cat#3150005A |
| Anti-Human CD123 (6H6) - 151Eu | Fluidigm | Cat#3151001B |
| Purified anti-human CD370 (CLEC9A/DNGR1) (8F9) - 152Sm | BioLegend | Cat#353802; RRID: AB_10983070 |
| Anti-pSTAT1 (4a) - 153Eu | Fluidigm | Cat#3153005A |
| Histone H3K27ac antibody (mAb) (MABI 0309) | ActiveMotiv | Cat#39685; RRID: AB_2793305 |
| Anti-Human CD27 (L128) - 155Gd | Fluidigm | Cat#3155001B |
| Anti-Human CD45 (HI30) - 156Gd | Fluidigm | Cat#3156010B |
| Purified anti-human CD25 (M-A251) | BioLegend | Cat#356102; RRID: AB_2561752 |
| Anti-Human pSTAT3 (4/P-Stat3) - 158Gd | Fluidigm | Cat#3158005A |
| Anti-Human CD11c (Bu15) - 159Tb | Fluidigm | Cat#3159001B |
| Anti-Human CD14 (M5E2) - 160Gd | Fluidigm | Cat#3160001B |
| Anti-Ki-67 (B56) - 161Dy | Fluidigm | Cat#3161007B |
| Purified anti-human CD1c (L161) | BioLegend | Cat#331502; RRID: AB_1088995 |
| Purified anti-human TCRg/d (B1) | BioLegend | Cat#331202; RRID: AB_1089222 |
| Anti-Human Arginase-1 (658922) - 164Dy | Fluidigm | Cat#3164012B |
| Anti-pCREB (87G3) - 165Ho | Fluidigm | Cat#3165009A |
| Purified anti-human CD16 (B73.1) | BioLegend | Cat#360702; RRID: AB_2562693 |
| Anti-Human CD38 (HIT2) - 167Er | Fluidigm | Cat#3167001B |
| Anti-Human CD8 (SK1) - 168Er | Fluidigm | Cat#3168002B |
| Anti-Human CD45RA (HI100) - 169Tm | Fluidigm | Cat#3169008B |
| Anti-Human CD3 (UCHT1) - 170Er | Fluidigm | Cat#3170001B |
| Anti-Human/ Mouse Granzyme B (GB11) - 171Yb | Fluidigm | Cat#3171002B |
| Anti-Human CD15 (W6D3) - 172Yb | Fluidigm | Cat#3172021B |
| Anti-Perforin antibody (B-D48) | abcam | Cat#ab47225 |
| Purified anti-human IFNg (4S.B3) | BioLegend | Cat#502502; RRID: AB_315227 |
| Anti-Cross Phospho-S6 (N7-548) - 175Lu | Fluidigm | Cat#3175009A |
| Anti-Human CD56 (NCAM16.2) - 176Yb | Fluidigm | Cat#3176008B |
| Anti-Human CD274/PD-L1 (MIH1) - 209Bi | Fluidigm | Cat#3209014B |
| Bacterial and virus strains | ||
| rVSV-ΔG-GFP/nanoluciferase | PMID: 35025672 | N/A |
| Pseudovirus expressing SARS-CoV-2 variants | In-house | Han et al.13, Suthar et al.14, Cohen et al.15 |
| Biological samples | ||
| Chemicals, peptides, and recombinant proteins | ||
| rhIFN-α | Miltenyi | Cat# 130-093-874 |
| Recombinant SARS-CoV-2 Wuhan spike protein | SinoBiological™ | Cat# 40589-V27B-B |
| Recombinant SARS-CoV-2 Omicron spike protein | SinoBiological™ | Cat# 40589-V49H3-B |
| DMSO | Sigma | D2650-100ML |
| Peptide cocktail for T-cell stimulation | In-house | Tarke et al.79 |
| Critical commercial assays | ||
| V-plex COVID-19 panel 23 | Mesoscale Discovery | Cat# K15567U |
| S-PLEX Human IFN-α2a kit | Mesoscale Discovery | Cat # K151P3S |
| Cell-ID 20-Plex Pd Barcoding Kit | Fluidigm | Cat#201060 |
| EQ™Four Element Calibration Beads | Fluidigm | Cat#201078 |
| Cell-ID™ Intercalator Ir | Fluidigm | Cat#201192A |
| One-Glo luciferase assay system | Promega | Cat# E6130 |
| Chromium NextGEM Chip J single Cell | 10x Genomics | 1000230 |
| Chromium NextGEM Single Cell Multiome ATAC + Gene Expression reagent bundle | 10x Genomics | 1000283 |
| Dual Index Kit TT Set A | 10x Genomics | 1000215 |
| Single Index Kit N Set A | 10x Genomics | 1000212 |
| Chromium X controller | 10x Genomics | 1000331 |
| NovaSeq 6000 S4 Reagent Kit V1.5 (200 cycles) | Illumina | 20028313 |
| Target 96 Inflammation Panel | Olink | |
| Bio-Rad protein assay kit II | Bio-Rad | 5000002 |
| Deposited data | ||
| scMultiome dataset - raw and processed data | This paper | GEO: GSE239799 |
| Blood transcriptomics dataset - raw and processed data | This paper | GEO: GSE239787 |
| Experimental models: Cell lines | ||
| Lenti-X 293T cells | Takara Bio | Cat. No. 632180 |
| 293T-ACE2-TMPRSS2 cells | In-house | Neerukonda et al.70 |
| Experimental models: Organisms/strains | ||
| Oligonucleotides | ||
| BCR primers | Liao HX et al. 75 | N/A |
| QIAseq SARS-CoV-2 Primer Panel | Qiagen Inc. | 333895 |
| QIAseq FX DNA Library Unique Dual Index Adapter Kit-A | Qiagen Inc. | 180479 |
| Recombinant DNA | ||
| pCAGGS Wuhan (SΔ19) | J. Craig Venter Institute – a gift from Dr. Gene S. Tan | N/A |
| pCAGGS Delta (B.1.617) | Sievers et al.69 | N/A |
| pCAGGS Omicron (B.1.529) | Sievers et al.69 | N/A |
| Software and algorithms | ||
| R 4.2.2; 4.1.1 | R Core Team102 | https://www.r-project.org/ |
| DESeq2 v1.38.3 | Love et al.101 | https://bioconductor.org/packages/release/bioc/html/DESeq2.html |
| Seurat (R package) v4.0.5 | Hao et al.,86 Hafemeister et al.87 | https://satijalab.org/seurat/ |
| harmony (R package) v0.1.1 | Korsunsky et al.88 | https://portals.broadinstitute.org/harmony/index.html |
| ggraph (R package) v2.1.0 | Pedersen et al.98 | https://ggraph.data-imaginist.com/index.html |
| igraph (R package) v1.3.1 | Csardi et al.97 | https://igraph.org/ |
| Complexheatmap (R package) v2.12.0; 2.8.0 | Gu104 | https://jokergoo.github.io/ComplexHeatmap-reference/book/ |
| ArchR (R package) v1.0.1 | Granja et al.43 | https://www.archrproject.com/ |
| chromVAR (R package) v1.20.2 | Schep et al.96 | https://bioconductor.org/packages/release/bioc/html/chromVAR.html |
| fgsea (R package) v1.24.0 | Korotkevich et al.89 | https://bioconductor.org/packages/release/bioc/html/fgsea.html |
| limma (R package) v3.54.2 | Ritchie et al.93 | https://bioconductor.org/packages/release/bioc/html/limma.html |
| pcaMethods (R package) v1.84.0 | Stacklies et al.81 | https://bioconductor.org/packages/release/bioc/html/pcaMethods.html |
| FlowSOM (R package) v2.0.0 | Van Gassen et al.83 | https://bioconductor.org/packages/release/bioc/html/FlowSOM.html |
| Other | ||