Abstract
NF-κB inducing kinase (NIK) is vital for the induction of many immune responses, and as such, NIK dysregulation has been implicated in various inflammatory diseases and cancers. NIK has been pursued as a potential therapeutic target, and small-molecule inhibitors that bind the orthosteric site on NIK have been reported. However, despite the established chemical matter, NIK inhibitors have not yet reached the clinic. With the goal of developing allosteric NIK ligands using a fragment-based NMR screening approach, we report the identification and development of a series of allosteric, fragment-sized NIK ligands that bind with micromolar potency and good ligand efficiency.
Keywords: NF-κB inducing kinase, NIK, Allosteric, Fragments, Fragment-based drug discovery, FBDD
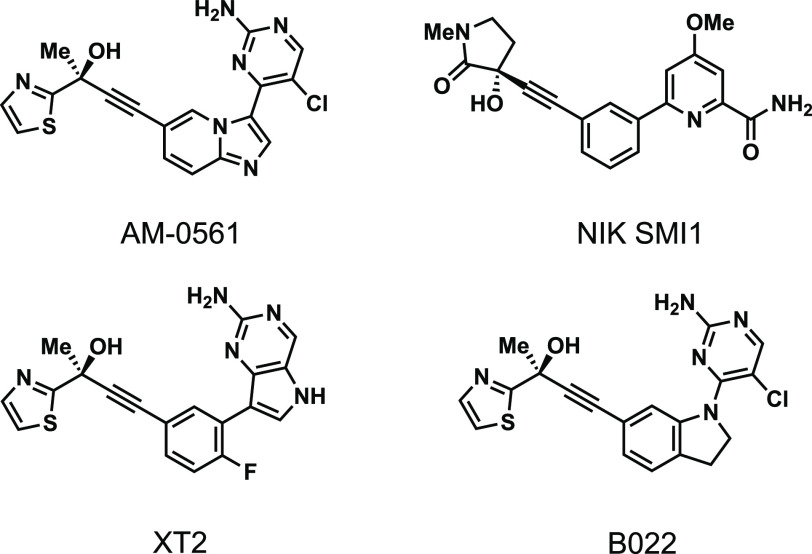
The non-canonical NF-κB signaling pathway is vital for the regulation of several immune responses such as apoptosis and inflammation;1 however, overactivation of the pathway has been implicated in the pathogenesis and disease progression of various immune disorders and blood cancers.2 As the central regulatory component of the pathway, NF-κB inducing kinase (NIK) has garnered significant attention as a drug target. Established NIK inhibitors target the ATP-binding site, and they share a propargyl alcohol motif that extends into a small hydrophobic pocket behind the ATP-binding site (Figure 1).3−6
Figure 1.
The therapeutic potential of inhibiting NIK has been established for a variety of human diseases. For example, the NIK inhibitor NIK SMI1 increased survival in a systemic lupus erythematosus murine model.4 Inhibition of NIK with B022 has been shown to rescue mice in a toxin-induced liver inflammation model,6 making NIK a promising target for diet-induced metabolic disorders. Additionally, mangiferin, a natural product NIK inhibitor, slows tumor growth and induces apoptosis in a melanoma mouse model.7 However, despite these promising experimental data, no NIK inhibitor has received FDA approval. Due to the inability of ATP-competitive NIK inhibitors to confer a clinically viable candidate, we elected to explore alternative strategies for the development of NIK-targeted ligands and inhibitors.
Targeting an allosteric site on NIK, as opposed to the highly conserved ATP-binding site, may provide a viable strategy for targeting this disease-relevant kinase. Fragment-based drug discovery (FBDD) approaches have been used to discover allosteric inhibitors of a variety of proteins,8−13 including kinases.14,15 Recently, a FBDD approach was used to discover novel allosteric binders of MEK1 that utilized screening conditions to bias for allosteric binding modes.16 We envisioned using a similar approach in our studies.
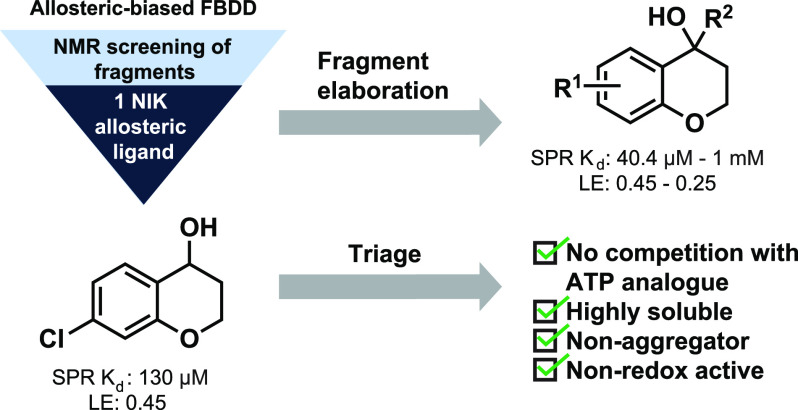
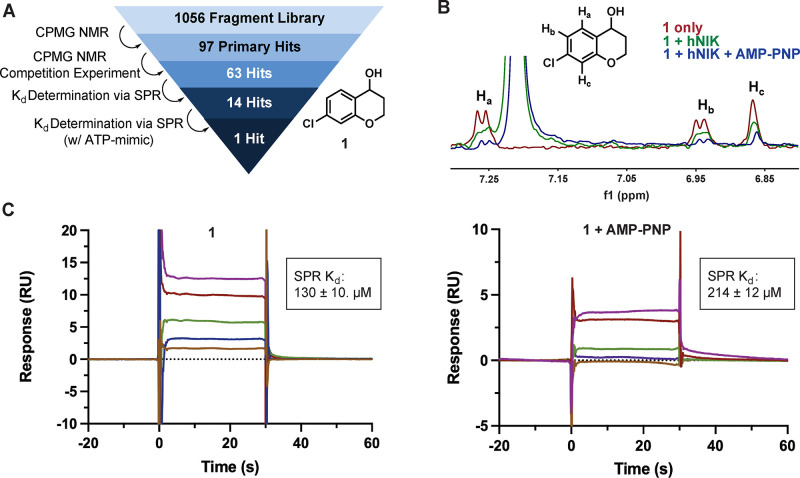
We report here the utilization of an in-house fragment library and NMR-based screening to develop allosteric NIK binders. The fragment library consisted of 1056 fragments purchased from the LifeChemicals high-solubility collection. Ligand-observed NMR, using a standard CPMG pulse sequence with T2 filtering, was employed as the primary screening method.17 The individual fragments (dissolved in DMSO-d6) were pooled into cocktails of five compounds and were screened for binding to NIK. To bias for allosteric-binding fragments, a non-hydrolyzable ATP analogue was included in each sample to saturate the NIK orthosteric ATP-binding site. Using this screening protocol, 97 primary hits were identified.
Utilizing the same pulse sequence as before, individual hit fragments were subjected to a competition experiment with AMP-PNP. Three separate NMR spectra were collected for each fragment (fragment alone, fragment with NIK, and fragment with NIK and AMP-PNP). Hits were carried on if the fragment had approximately 50% signal attenuation in the presence of NIK and the signal attenuation was either maintained or increased by adding AMP-PNP. If the signal attenuation was lost upon adding AMP-PNP, then the fragment was determined to be competing with the ATP analogue and rejected. The resulting 63 hits were identified as potential allosteric binders of NIK.
The ligand-observed NMR experiments gave qualitative binding data, but an orthogonal, quantitative experiment was needed to confirm the binding of the hit compounds. Surface plasmon resonance (SPR) with biotinylated NIK protein was chosen to further triage the hit fragments. The 63 hits were tested at five concentrations, and the responses were used to generate a binding curve. After SPR, 14 hits displayed a binding affinity of less than 500 μM. Those 14 hits were then evaluated by SPR in dose–response format in the presence of saturating AMP-PNP, and the response was used to graph a binding curve. After the full screening effort, one hit fragment displayed similar affinity with and without AMP-PNP. Taking all the results together, chromanol 1 was the sole hit from the allosteric bias fragment screening effort (Figure 2). Given the absence of known chemical matter for binding to an allosteric site on NIK, the low hit rate was not unexpected.
Figure 2.
(A) Overview of the allosteric-biased fragment-based screen. (B) CPMG NMR with T2 filtering competition experiment of 1. (C) SPR sensorgrams of 1 with and without AMP-PNP. Kd values shown represent the mean ± SEM of two determinations.
The structure of 1 was interesting due to the central chroman ring system. Although there are examples of chromanol structures in biologically active compounds, it is not overly common.18,19 Therefore, we thought it was necessary to triage hit compound 1 to rule out false positive results.
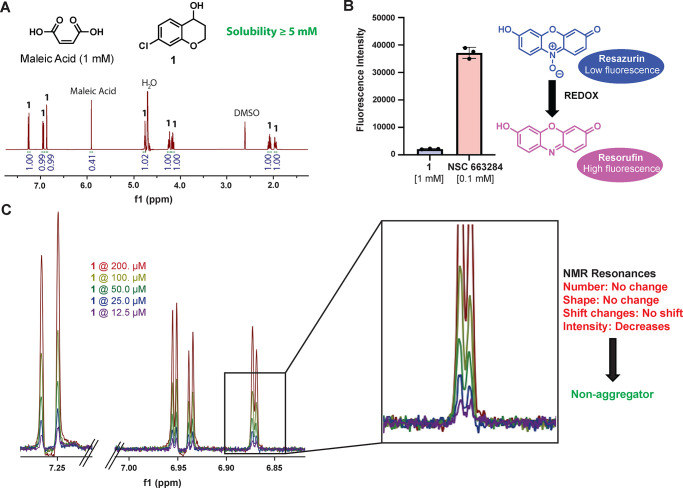
The solubility of 1 was determined by using a quantitative NMR solubility assay. The hit fragment was determined to have a solubility of at least 5 mM in an aqueous solution. To rule out redox activity contributing to false positive results, a plate-based redox assay was run with 1 and a known redox-active molecule (NSC 663284) as a positive control. Hit fragment 1 showed no redox activity in this assay. Finally, a NMR-based aggregation assay was conducted to ensure that 1 was not aggregating in solution to give false positives during screening. These triage experiments showed that 1 is a highly soluble, non-redox-active and non-aggregating fragment that is suitable to move forward and optimize (Figure 3).
Figure 3.
(A) qNMR solubility experiment with 1. (B) Resazurin redox assay with 1. (C) NMR aggregation experiment with 1.
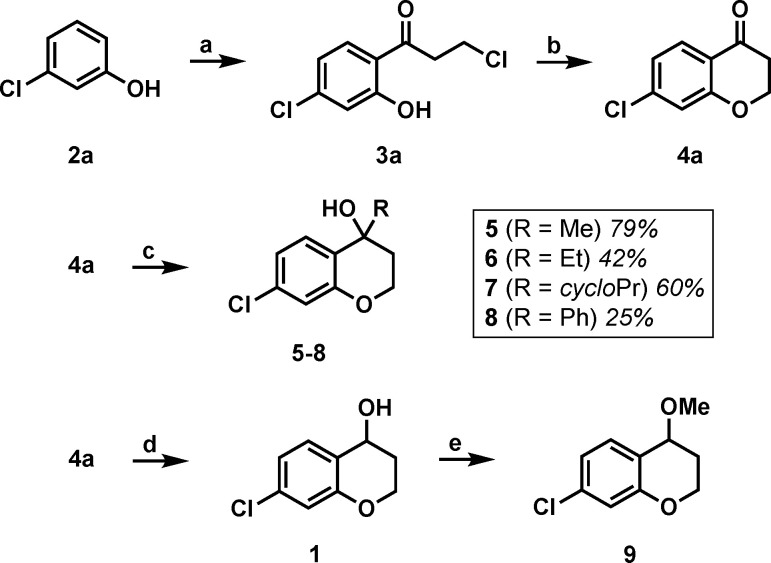
Hit fragment 1 was resynthesized along with a small library of analogues to test the tolerance of substituents on the 4-position of the chromanol structure. The synthesis began by acylating 3-chlorophenol to form 3a in moderate yield (48%): a phenolic ester was formed in situ, and upon adding AlCl3 a Fries phenolic ester rearrangement gave the desired product.20,21 Product 4a was subsequently afforded by an intramolecular cyclization with EtOH and K2CO3 in good yield (61%). Grignard addition to the ketone of 4a yielded tertiary alcohols 5–8 in varying yields (25–79%). Reduction of 4a with NaBH4 in MeOH yielded hit fragment 1 in high yield (75%). Product 1 was then converted to the methoxy derivative with iodomethane and NaH in THF in 68% yield (Scheme 1). Final compounds were tested by SPR for their binding affinity for NIK (Table 1).
Scheme 1. Synthesis of 1, 5–9.
Reagents and conditions: (a) 3-chloropropionyl chloride, AlCl3, 90 °C, 2 h, 48%; (b) K2CO3, EtOH, rt, 24 h, 61%; (c) RMgBr, THF, 0 → 70 °C, 24 h, 25–79%; (d) NaBH4, MeOH, 0 °C, 2 h, 75%; (e) MeI, NaH, THF, rt, 3 h, 68%.
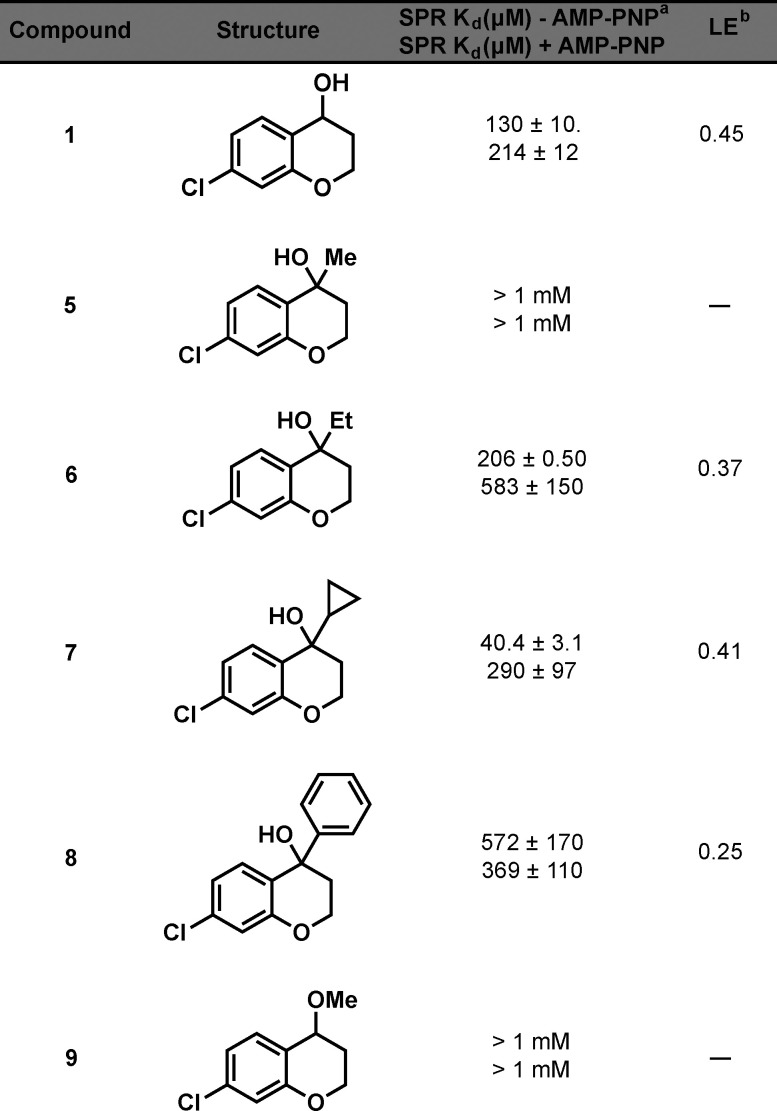
Table 1. NIK Affinity and LE of 1, 5–9.
Kd values shown represent the mean ± SEM of two determinations.
LE = 1.37pKd/HA (heavy atom count).
Interestingly, all of the tertiary alcohols except for methyl analogue 5 bind NIK with a Kd of less than 1 mM. The cyclopropyl analogue 7 improved upon the binding affinity of the parent hit by approximately 3-fold. The necessity for the hydroxyl was shown by inactive methoxy analogue 9. These data suggest that fragment elaboration on the 4-position of the chromanol structure is permitted.
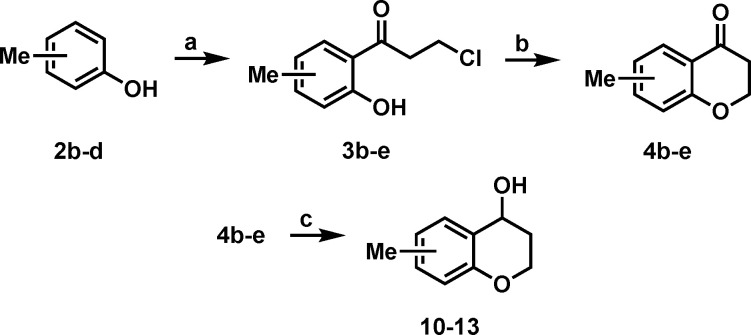
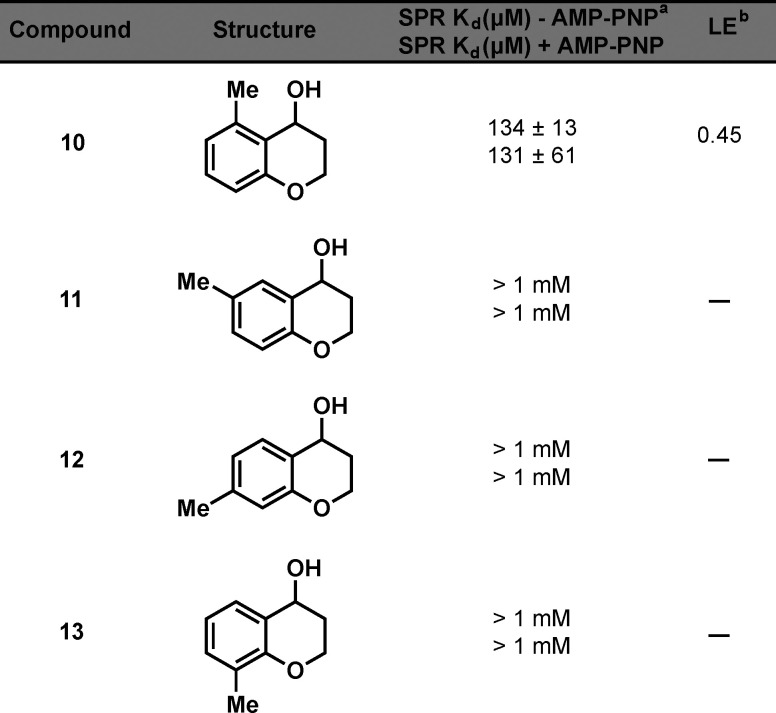
To investigate further growth vectors on hit fragment 1, compounds were synthesized with a methyl walk around the aromatic ring on the chromanol structure. The compounds were synthesized by utilizing chemistry analogous to that used for the synthesis of 1 (Scheme 2). Compounds 10–13 were then tested for their affinity for NIK by SPR (Table 2). We were surprised that the only fragment that showed binding was the 5-methyl derivative 10. Both series of compounds showed that substituents are tolerated if pointing up on the chromanol structure.
Scheme 2. Synthesis of 10–13.
Reagents and conditions: (a) 3-chloropropionyl chloride, AlCl3, 90–120 °C, 2–24 h, 19–72%; (b) K2CO3, EtOH, rt, 24 h, 28–73%; (c) NaBH4, MeOH, 0 °C, 2 h, 55−66%.
Table 2. NIK Affinity and LE of 10–13.
Kd values shown represent the mean ± SEM of two determinations.
LE = 1.37pKd/HA (heavy atom count).
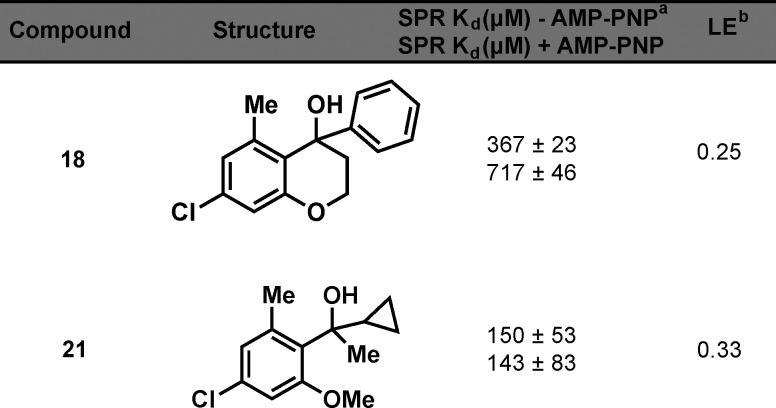
To test for a synergistic effect of the derivatives displaying micromolar binding affinity for NIK, a series of compounds were synthesized with the 7-chloro and 5-methyl groups, as well as varying substituents at the 4-position (Table S1). We were surprised that none of the analogues displayed enhanced affinity and only phenyl derivative 18 showed a Kd of less than 1 mM (Table 3). We hypothesize that steric bulk on that side of the chromanol structure, along with the rigidity of the ring system, was not optimal for binding.
Table 3. NIK Affinity and LE of 18 and 21.
Kd values shown represent the mean ± SEM of two determinations.
LE = 1.37pKd/HA (heavy atom count).
One possible solution is the addition of rotatable bonds on this fragment series, which may permit a more favorable conformation for binding. To test this hypothesis, we synthesized ring-opened analogues where the bond between C-2 and C-3 was disconnected and measured their binding affinity by SPR (Table S2). Only cyclopropyl 21 showed binding of less than 1 mM (Table 3); however, the affinity of 21 for NIK was similar to that of 1. Currently we are pursuing fragment–NIK co-structure studies to better understand the allosteric binding pocket on NIK and the binding mode of these chromanol-based fragments, which will better inform the rational design of new compounds.
In conclusion, we have reported chromanol analogues that bind to NIK in the micromolar range and do not show competition with the non-hydrolyzable ATP analogue (AMP-PNP). Growth of these chromanol compounds at the 4-position gave promising results, with fragment 7 showing a 3-fold improved binding affinity over parent hit fragment 1. This class of chromanol analogues provides evidence that NIK possesses allosteric binding sites that are amenable to small-molecule targeting. Ongoing efforts are focused on structurally enabling this project, as well as defining features on the chromanol chemotype that may be modified/elaborated to improve NIK-targeting potency. The development of high-affinity, NIK-targeting ligands even in the absence of enzymatic inhibition would enable therapeutic approaches to regulate this biomedically important kinase by targeted protein degradation, which is currently under investigation by our team.
Acknowledgments
We thank the Minnesota NMR Center and Todd Rappe for facilities and assistance with the CPMG NMR experiments.
Glossary
Abbreviations
- NIK
NF-κB inducing kinase
- SPR
surface plasmon resonance
- NMR
nuclear magnetic resonance
- ATP
adenosine triphosphate
- LE
ligand efficiency
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acsmedchemlett.3c00429.
Detailed description of synthesis and chemical characterization of compounds; materials and methods for CPMG NMR, SPR, qNMR, aggregation, and redox assays; supplementary SPR sensorgrams and data (PDF)
Author Contributions
The study was conceptualized and designed by J.J.A., M.J.G., and D.A.H. M.J.G. contributed to curation of the fragment collection and setup of the CPMG NMR assay. All syntheses and collection of experimental data were conducted by J.J.A. The manuscript was written by J.J.A. with editing from M.J.G. and D.A.H.
We gratefully acknowledge the NIH (P01-CA234228) and DOD (W81XWH-21-1-0674) for financial support. J.J.A. acknowledges support from the NIH Chemical Biology Interface Training Grant (T32-GM132029) and the American Chemical Society, Division of Medicinal Chemistry Predoctoral Fellowship program (2022–2023). The NIH (1S10OD021539-01) is acknowledged for support of the surface plasmon resonance instrument at the University of Minnesota. Mass spectrometry was performed at the Analytical Biochemistry Core Facility of the University of Minnesota Masonic Cancer Center, which is supported by the NIH (P30-CA77598).
The authors declare no competing financial interest.
Special Issue
Published as part of the ACS Medicinal Chemistry Lettersvirtual special issue “Celebrating the 60th Anniversary of the MIKIW Meeting-in-Miniature”.
Supplementary Material
References
- Cheng J.; Feng X.; Li Z.; Zhou F.; Yang J.; Zhao Y. Pharmacological inhibition of NF-κB-inducing kinase (NIK) with small molecules for the treatment of human diseases. RSC Med. Chem. 2021, 12, 552. 10.1039/D0MD00361A. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zinatizadeh M. R.; Schock B.; Chalbatani G. M.; Zarandi P. K.; Jalali S. A.; Miri S. R. The Nuclear Factor Kappa B (NF-κB) signaling in cancer development and immune diseases. Genes Dis. 2021, 8, 287. 10.1016/j.gendis.2020.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demchenko Y. N.; Brents L. A.; Li Z.; Bergsagel L. P.; McGee L. R.; Kuehl M. W. Novel inhibitors are cytotoxic for myeloma cells with NFkb inducing kinase-dependent activation of NFkb. Oncotarget 2014, 5, 4554. 10.18632/oncotarget.2128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brightbill H. D.; Suto E.; Blaquiere N.; Ramamoorthi N.; Sujatha-Bhaskar S.; Gogol E. B.; Castanedo G. M.; Jackson B. T.; Kwon Y. C.; Haller S.; Lesch J.; Bents K.; Everett C.; Kohli P. B.; Linge S.; Christian L.; Barrett K.; Jaochico A.; Berezhkovskiy L. M.; Fan P. W.; Modrusan Z.; Veliz K.; Townsend M. J.; DeVoss J.; Johnson A. R.; Godemann R.; Lee W. P.; Austin C. D.; McKenzie B. S.; Hackney J. A.; Crawford J. J.; Staben S. T.; Ismaili M. H. A.; Wu L. C.; Ghilardi N. NF-κB inducing kinase is a therapeutic target for systemic lupus erythematosus. Nat. Commun. 2018, 9, 179. 10.1038/s41467-017-02672-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z.; Li X.; Su M.; Gao L.; Zhou Y.; Yuan B.; Lyu X.; Yan Z.; Hu C.; Zhang H.; Luo C.; Chen Z.; Li J.; Zhao Y. Discovery of a Potent and Selective NF-κB-Inducing Kinase (NIK) Inhibitor That Has Anti-inflammatory Effects in Vitro and in Vivo. J. Med. Chem. 2020, 63, 4388. 10.1021/acs.jmedchem.0c00396. [DOI] [PubMed] [Google Scholar]
- Ren X.; Li X.; Jia L.; Chen D.; Hou H.; Rui L.; Zhao Y.; Chen Z. A small-molecule inhibitor of NF-κB-inducing kinase (NIK) protects liver from toxin-induced inflammation, oxidative stress, and injury. FASEB J. 2017, 31, 711. 10.1096/fj.201600840R. [DOI] [PubMed] [Google Scholar]
- Takeda T.; Tsubaki M.; Sakamoto K.; Ichimura E.; Enomoto A.; Suzuki Y.; Itoh T.; Imano M.; Tanabe G.; Muraoka O.; Matsuda H.; Satou T.; Nishida S. Mangiferin, a novel nuclear factor kappa B-inducing kinase inhibitor, suppresses metastasis and tumor growth in a mouse metastatic melanoma model. Toxicol. Appl. Pharmacol. 2016, 306, 105. 10.1016/j.taap.2016.07.005. [DOI] [PubMed] [Google Scholar]
- Christopher J. A.; Aves S. J.; Bennett K. A.; Doré A. S.; Errey J. C.; Jazayeri A.; Marshall F. H.; Okrasa K.; Serrano-Vega M. J.; Tehan B. G.; Wiggin G. R.; Congreve M. Fragment and Structure-Based Drug Discovery for a Class C GPCR: Discovery of the mGlu5 Negative Allosteric Modulator HTL14242 (3-Chloro-5-[6-(5-fluoropyridin-2-yl)pyrimidin-4-yl]benzonitrile. J. Med. Chem. 2015, 58, 6653–6664. 10.1021/acs.jmedchem.5b00892. [DOI] [PubMed] [Google Scholar]
- Coutard B.; Decroly E.; Li C.; Sharff A.; Lescar J.; Bricogne G.; Barral K. Assessment of Dengue virus helicase and methyltransferase as targets for fragment-based drug discovery. Antiviral Res. 2014, 106, 61–70. 10.1016/j.antiviral.2014.03.013. [DOI] [PubMed] [Google Scholar]
- Orgován Z.; Ferenczy G. G.; Keserű G. M. Fragment-Based Approaches for Allosteric Metabotropic Glutamate Receptor (mGluR) Modulators. Curr. Top. Med. Chem. 2019, 19, 1768–1781. 10.2174/1568026619666190808150039. [DOI] [PubMed] [Google Scholar]
- Scott D. E.; Coyne A. G.; Hudson S. A.; Abell C. Fragmentbased approaches in drug discovery and chemical biology. Biochemistry 2012, 51, 4990–5003. 10.1021/bi3005126. [DOI] [PubMed] [Google Scholar]
- Tiefenbrunn T.; Forli S.; Happer M.; Gonzalez A.; Tsai Y.; Soltis M.; Elder J. H.; Olson A. J.; Stout C. D. Crystallographic fragment-based drug discovery: use of a brominated fragment library targeting HIV protease. Chem. Biol. Drug Des. 2014, 83, 141–148. 10.1111/cbdd.12227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vance N. R.; Gakhar L.; Spies M. A. Allosteric Tuning of Caspase-7: A Fragment-Based Drug Discovery Approach. Angew. Chem., Int. Ed. 2017, 56, 14443–14447. 10.1002/anie.201706959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C.; Zhang X.; Zhang N.; Zhou Y.; Sun G.; Zhao L.; Zhong R. Identification and Biological Evaluation of CK2 Allosteric Fragments through Structure-Based Virtual Screening. Molecules 2020, 25, 237. 10.3390/molecules25010237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoepfer J.; Jahnke W.; Berellini G.; Buonamici S.; Cotesta S.; Cowan-Jacob S. W.; Dodd S.; Drueckes P.; Fabbro D.; Gabriel T.; Groell J. M.; Grotzfeld R. M.; Hassan A. Q.; Henry C.; Iyer V.; Jones D.; Lombardo F.; Loo A.; Manley P. W.; Pellé X.; Rummel G.; Salem B.; Warmuth M.; Wylie A. A.; Zoller T.; Marzinzik A. L.; Furet P. Discovery of Asciminib (ABL001), an Allosteric Inhibitor of the Tyrosine Kinase Activity of BCR-ABL1. J. Med. Chem. 2018, 61, 8120–8135. 10.1021/acs.jmedchem.8b01040. [DOI] [PubMed] [Google Scholar]
- Di Fruscia P.; Edfeldt F.; Shamovsky I.; Collie G. W.; Aagaard A.; Barlind L.; Börjesson U.; Hansson E. L.; Lewis R. J.; Nilsson M. K.; Öster L.; Pemberton J.; Ripa L.; Storer R. I.; Käck H. Fragment-Based Discovery of Novel Allosteric MEK1 Binders. ACS Med. Chem. Lett. 2021, 12, 302–308. 10.1021/acsmedchemlett.0c00563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meiboom S.; Gill D. Modified Spin-Echo Method for Measuring Nuclear Relaxation Times. Rev. Sci. Instrum. 1958, 29, 688. 10.1063/1.1716296. [DOI] [Google Scholar]
- Vakalopoulos A.; Schmeck C.; Thutewohl M.; Li V.; Bischoff H.; Lustig K.; Weber O.; Paulsen H.; Elias H. Chromanol Derivatives – A Novel Class of CETP Inhibitors. Bioorg. Med. Chem. Lett. 2011, 21, 488. 10.1016/j.bmcl.2010.10.110. [DOI] [PubMed] [Google Scholar]
- Liu L.; Wang F.; Lu H.; Ren X.; Zou J. Chromanol 293B, an Inhibitor of KCNQ1 Channels, Enhances Glucose-Stimulated Insulin Secretion and Increases Glucagon-Like Peptide-1 Level in Mice. Islets 2014, 6, e962386 10.4161/19382014.2014.962386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fries K.; Finck G. Über Homologe des Cumaranons und ihre Abkömmlinge. Chem. Ber. 1908, 41, 4271. 10.1002/cber.190804103146. [DOI] [Google Scholar]
- Martin R. Uses of the Fries Rearrangement for the Preparation of Hydroxyarylketones. A Review. Org. Prep. Proced. Int. 1992, 24, 369. 10.1080/00304949209356226. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.