The primary function of RNA polymerase (RNAP) is to produce RNA transcripts optimal for a given environmental or developmental state of a cell. RNAP accomplishes this selection by deciding whether or not to initiate transcription of a gene and then both how fast and how far to elongate the RNA chains. Because each decision depends on information input to RNAP and can produce results that influence subsequent decisions, RNAP’s function in the cell in many ways resembles an information processor. In this short review, we will (i) briefly summarize the transcription cycle from the perspective of RNAP as a cellular information processor, (ii) describe the structure of the transcription elongation complex (TEC) and the protein-nucleic acid contacts that function as intrinsic inputs to its regulatory decisions, and (iii) present a model of pausing and ρ-independent termination as an example of information processing by RNAP.
RNAP AS AN INFORMATION PROCESSOR
Because RNAP moves along a digital tape (the DNA), reads information from it (and from the nascent RNA chain), and then based on this information, either writes new information to an RNA chain or decides to halt transcription temporarily or permanently, it exhibits many similarities to the first theoretical computer (114), which was devised by the British mathematician Alan Turing and now is called a Turing machine. A Turing machine carries out computation by repeated logical operations in which, based on a series of rules and digital information it reads from a tape, it writes information back to the tape, potentially changes its logical rules for future operations, and moves to a new position on the tape (41). Since new RNAs can feed back on the activity of RNAP (directly or as translated proteins) and dictate both the selection of new start sites and its response to pause and termination signals, like a Turing machine and the CPU of a computer, RNAP is capable of logical operations. Unlike the digital logic of a computer, however, RNAP’s decisions are governed by competing rates and equilibria among alternative conformations and complexes and constitute the first step in the conversion of the fundamentally digital language of DNA into the analog circuitry of cellular metabolism.
To understand how RNAP makes regulatory decisions, we must describe the structures of the competing intermediates, the inputs that influence RNAP’s decision as to which among them is formed and the mechanisms and rates by which these intermediates interconvert in response to the various inputs. Regulatory decisions by RNAP rely on two types of information input to the TEC: intrinsic and extrinsic. Intrinsic inputs are discrete segments of RNA and DNA that interact with RNAP. These intrinsic interactions generate different TEC conformations, which vary in their capacity for RNA chain elongation from rapid nucleotide addition to halted chain elongation to termination. These different conformations are in turn the targets for a variety of extrinsic inputs that can enhance or inhibit chain elongation or termination (Table 1). These inputs are small molecules, proteins, and other RNAs that interact with the TEC and modulate its activity, either by directly contacting RNAP or via contacts to RNA or DNA. The combination of distinct TEC conformations and regulatory ligands whose affinities and actions can depend on the conformation and the position of the TEC confers enormous regulatory flexibility on the process of transcription. To illustrate how intrinsic and extrinsic inputs modify regulation by RNAP, we will first describe the transcriptional cycle. We will then direct our principal attention to the structure of the TEC and its response to intrinsic regulatory signals at related RNA hairpin-dependent pause and termination signals.
TABLE 1.
Regulatory inputs to the transcription complex
| Input (component of regulatory signal) | Target(s) | Effect(s) | References |
|---|---|---|---|
| Intrinsic | |||
| Nascent RNA structures | RNAP | Pausing, termination | 23, 85, 118, 121 |
| Upstream DNA (hybrid formation) | Replication primer formation, termination | 70 | |
| 3′-proximal RNA | RNAP | Pausing (10–11 nt); termination (U-rich 7–9 nt); arrest (U rich)a | 16, 37 |
| Template DNA strand | 3′-proximal RNA, RNAP | ? | 124 |
| Nontemplate DNA strand | Sigma factors, RNAP | Pausing | 95, 117 |
| Bases in active site | RNAP | Pausing | 1, 17 |
| Downstream DNA duplex | β′ N-terminal zinc finger, β C-terminal region (duplex clamp) | Pausing, termination | 62, 79, 111 |
| Extrinsic | |||
| ς | Nontemplate DNA | Pausing | 95 |
| ρ | Nascent RNA, RNAP? | Termination | 84, 93 |
| τ | Unknown | Termination | 10 |
| ppGpp | Unknown | Pausing | 11 |
| NusA | α C-terminal domain β or β′, RNA hairpins? | Pausing, termination antiterminationb | 35, 68, 93 |
| NusB | BoxA nascent RNA | Antiterminationb | 35, 93 |
| NusE | BoxA nascent RNA | Antiterminationb | 35, 93 |
| NusG | RNAP | Pausing, antiterminationb | 35, 93 |
| GreA | RNAP, nascent RNA? | Transcript cleavage, antiarrest, promoter escape | 8, 32, 42 |
| GreB | RNAP, nascent RNA? | Transcript cleavage, antiarrest, promoter escape | 8, 32, 42 |
| Gene-specific regulators | RNAP, DNA or nascent RNA | Various | 58 |
Arrest signals for RNAP II in eukaryotes are U-rich tracts (or corresponding dA tracts in template DNA). Arrest of bacterial RNAP has been detected in vitro (53) but remains of uncertain regulatory significance.
Nus factors assemble into an antitermination complex that modifies RNAP to resist termination signals (reviewed in reference 23), in combination with, for instance, λ N protein. Alone, Nus factors may exert different effects (e.g., NusA enhances termination). Cellular antiterminators functionally homologous to λ N or Q proteins are hypothesized but remain uncharacterized.
TRANSCRIPTION CYCLE AND INTERMEDIATES
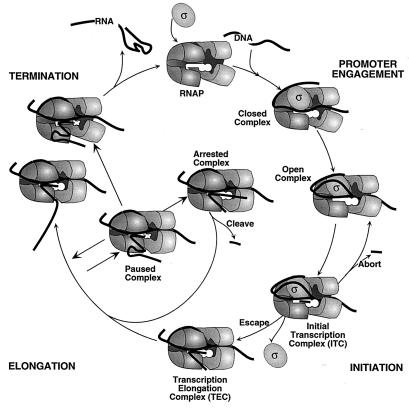
The regulatory intermediates of transcription are most easily depicted in a cycle (12). The cycle can be divided into major phases: promoter engagement, initiation, RNA chain elongation, and termination (Fig. 1). Each intermediate represents a decision point (or logical operation in our analogy) where inputs can alter the course of RNA synthesis.
FIG. 1.
The transcription cycle. Intermediates in the four phases of transcription (promoter engagement, promoter escape, RNA chain elongation, and transcript termination) are discussed in the text. Elongation is represented by growth of the RNA chain between two transcription elongation complexes. Termination and arrest are shown occurring from the paused transcription complex to reflect the likelihood that a transcriptional pause is the first step in each. However, not all paused complexes involve RNA secondary structures as depicted, especially those that lead to arrest, which likely would be blocked by an RNA hairpin that prevents reverse translocation of the RNA transcript (see the text).
Promoter engagement.
Promoter engagement encompasses several steps: promoter location and recognition by the RNAP holoenzyme (core enzyme complexed with one of several sigma factors), initial and reversible binding of RNAP to duplex promoter DNA (closed complex formation), and formation of an open complex in which ∼12 bp of DNA, including the transcription start site, are melted (Fig. 1). At least four important intrinsic inputs affect promoter engagement: the hexamer centered at position −10 upstream from the transcription start site (−10 region), the hexamer at position −35 (−35 region), the region of DNA between these two elements (spacer region), and a region located between −40 and −60 (the UP element). The mechanisms by which RNAP recognizes these promoter elements and positions itself at a promoter sequence were recently reviewed in detail (25). Briefly, RNAP holoenzyme initially binds to one face of double-stranded DNA at the promoter. Footprinting of these closed complexes indicate a protected region of 75 to 80 bp, extending from −55 to approximately +20 (87). The DNA strands are then melted, from −10 to +1 (26), forming open complexes. The process of open-complex formation may also be accompanied by major conformational changes in RNAP (25, 87, 97), which may include the closing of the jaw-like clamp that appears to be open in the holoenzyme but closed in the core polymerase (86).
Multiple closed- and open-complex intermediates form during promoter engagement (87). For some promoters, however, open-complex formation can be modeled by a simple two-step mechanism involving equilibrium between free RNAP and the closed complex followed by a rate-limiting isomerization to an open complex (71). This model must be applied with caution, though, since it can fail either because another step is rate limiting or because open complexes reverse to closed complexes rapidly.
The rates at which these steps occur are dictated by both extrinsic and intrinsic interactions. Extrinsic inputs include protein-protein contacts made by activators and repressors that bind in the vicinity of the promoter DNA and can modify the rates of either closed- or open-complex formation or promoter escape. In some cases, a single extrinsic factor can either inhibit or accelerate one or another of these steps with relatively subtle changes in contacts (63, 74, 107). Intrinsic contacts are made by the ς subunit of RNAP to the −10 and −35 regions of the DNA (reviewed in reference 36) and sometimes by the α-subunit C-terminal domain to the UP element (98). Open-complex stability can be further affected by the next step in the pathway, nucleoside triphosphate (NTP) binding, which in turn is affected by the spacer region between the −10 hexamer and the start site, as well as by the identity of nucleotide to be incorporated at the 5′ end of the RNA (33, 65).
Initiation.
After the open complex has bound the initiating NTPs, it becomes an initial transcription complex and can follow several alternative reaction pathways (Fig. 1): (i) the synthesis and release of short (2 to 8 nucleotides [nt]) RNA transcripts (abortive initiation); (ii) reiterative synthesis resulting in homopolymer extensions of the initial RNA transcripts (stuttering; not depicted in Fig. 1, see references 13 and 64); and (iii) release of ς, translocation away from the promoter, and formation of a TEC with loss of upstream DNA contacts, usually when the transcript is 8 to 9 nt in length (promoter escape). The relative rates of abortive initiation, stuttering, and promoter escape determine how fast productive RNA chain elongation begins and how soon the promoter is vacated to allow another RNAP molecule to bind. These rates appear to be determined principally by intrinsic interactions with promoter elements and with the initially transcribed 8 to 10 nt of RNA or corresponding DNA, but at least in vitro can also be modulated by extrinsic factors such as the transcript cleavage factors GreA and GreB, which stimulate escape from certain promoters by suppressing abortive initiation (32, 42). A better understanding of initiation will require learning the following: (i) to what extent do abortive initiation and stuttering occur in vivo and are they regulated at some promoters; (ii) what determines the relative rates of promoter escape, abortive initiation, and stuttering; and (iii) do the GreA and GreB transcript cleavage factors regulate promoter escape in vivo?
Elongation.
Once RNAP converts from initial transcription complex to TEC (i.e., escapes the promoter), it becomes stably associated with the RNA and DNA chains (most TECs resist dissociation in 1 M KCl or at 65°C) and can elongate the RNA chain 30 to 100 nt/s in vivo. These are average, not maximal, rates (see the next section). Two distinct types of translocations in the TEC must occur to allow this rapid movement: (i) translocation of the RNA 3′ end from position i+1 to i in the active site (modified from reference 30 to use the 3′-terminal nucleotide as the index position) as successive nucleotides are added (RNA 3′-end translocation; see Fig. 2) and (ii) translocation of DNA and RNA chains through RNAP (RNA and DNA translocation). The extent to which these two types of translocations are coupled has been a source of considerable controversy in the transcription field (reviewed in reference 54). RNAP could translocate the RNA 3′ end and RNA transcript and DNA chains in a monotonic manner, advancing 1 bp with each nucleotide addition (123). Alternatively, the addition of 2 or more nt could occur without the complete translocation of DNA and RNA through RNAP and be followed by chain translocation of 2 or more bp (discontinuous movement, sometimes called inchworming [14, 18]). Evidence for this model came from variably sized footprints of halted complexes (53, 80). New evidence allows the reinterpretation of these footprints within the context of a monotonic model by demonstrating that a potentially rigid RNAP can sometimes slide freely along the RNA and DNA chains and displace the 3′ end from the active site, resulting in larger or smaller footprints depending on the extent and frequency of sliding relative to cutting by footprinting reagents (49, 82, 88). Thus, a recent model for translocation postulates that the RNAP is distributed among all accessible positions by rapid sliding, with the occupancy of each template position dependent on its relative free energy (positional equilibrium [38, 50, 54]). In this view, the energy for directional translocation along DNA is derived when nucleotide addition shifts the positional equilibrium of the RNAP sliding back and forth along the RNA and DNA chains towards the forwardly translocated conformation (known as a thermal ratchet mechanism; reviewed in reference 34). The alternative possibility is that RNAP is a mechanoenzyme that usually maintains tight contacts to RNA and DNA and uses the energy of phosphodiester bond hydrolysis to generate internal movements (active locomotion or power-stroke mechanism; reviewed in reference 34).
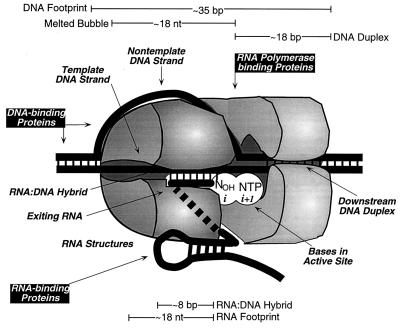
FIG. 2.
Structure and regulatory inputs of a transcription complex.
Although the rigid RNAP-sliding model is appealing, it cannot account for all the behaviors of RNAP; in particular, some internal flexibility of RNAP must exist to allow it to extend initial transcripts that are 5′ end cross-linked to the β subunit up to 8 nt (76). Three questions are particularly important to answer. (i) Are the two translocation cycles of RNAP tightly coupled, or can nucleotide addition occur without RNAP movement along DNA to produce significantly different conformations of the transcription complex at different template positions? (ii) If different conformations are possible, can they exist at a single template position; that is, can RNAP transcribe DNA in different phases such that only a fraction of elongating molecules reach a given template position in a particular conformation (see reference 18)? (iii) Does RNAP move by a thermal ratchet or power-stroke mechanism? Whether RNAP senses information as a rigid body that slides along the RNA and DNA chains nearly isoenergetically or undergoes significant internal movements between nucleic acid contacts or protein domains and requires energy input for translocation is a central issue for understanding how it processes the information in the nucleic acid chains.
Arrest, pausing, and termination.
RNA chain elongation is punctuated by certain sites where nucleotide addition is slowed by pause, arrest, and termination signals. Pause signals cause RNAP to isomerize from the rapidly elongating TEC to alternative conformations in which RNA chain extension is reversibly inhibited (by factors of 102 to 104). Arrest signals block elongation irreversibly in vitro; arrest has not yet been demonstrated to occur in vivo. Termination signals cause the release of RNA and DNA and can be positively or negatively regulated by a variety of extrinsic inputs (Table 1). At some terminators, these inputs can instruct the enzyme to read through the signal and continue transcription (antitermination).
Pausing and arrest both occur because the RNA 3′ OH and NTP substrate fail to maintain alignment in the active site, but may reflect different changes in TEC structure. In arrested transcription complexes, the RNA transcript appears to be threaded through the active site so that NTP binding is blocked, until 5 to 10 nt of 3′-proximal RNA is removed by hydrolytic cleavage. This cleavage reaction is catalyzed by RNAP’s active site and greatly stimulated by the GreA or GreB transcript cleavage factor (8, 83). Key questions about arrest are (i) does transcriptional arrest occur in vivo, and is it involved in the slow growth phenotype of ΔgreA ΔgreB strains; and (ii) how do the GreA and GreB cleavage factors, which contact both RNAP and the nascent RNA, stimulate transcript cleavage of arrested complexes?
There are several types of paused transcription complexes modulated by different intrinsic or extrinsic interactions. At some pause sites, elongation is slowed at unfavorable sequences, without mediating specific regulatory decisions. Other pauses are specific regulatory intermediates that both prevent transcription beyond a region where regulatory input is effective and put RNAP in the proper conformation to interact with regulatory molecules. An example of a regulatory pause is the class of hairpin-dependent pause sites that occur midway through the leader regions of all Escherichia coli or Salmonella amino acid biosynthetic operons that are regulated by attenuation (58). Pausing at these sites halts RNAP until a ribosome initiates synthesis of the leader peptide, which can cause attenuator readthrough during amino acid limitation, or until it eventually escapes spontaneously, which leads to superattenuation (58). The ribosome releases RNAP from the pause either by melting the RNA hairpin or through direct interaction with the enzyme. By slowing chain elongation to allow further rearrangements of the TEC, pausing likely is an initial step in arrest and termination (96). Finally, pausing allows surveillance for defective mRNAs by slowing RNAP at pause sites until it is released either by a translating ribosome or, if the RNA is not translatable, by the ρ termination factor (55). Thus, in our comparison of RNAP to an informational processor, pause signals instruct RNAP to halt and await regulatory input.
When RNAP encounters a termination signal, RNAP stops adding nucleotides to the RNA, separates the DNA-RNA hybrid, releases the newly synthesized transcript, and dissociates from the DNA template. Three types of termination signals for bacterial RNAP have been described to date: (i) intrinsic terminators (ρ-independent terminators) which require a stable RNA hairpin formed 7 to 9 nt from the terminated RNA 3′ end and immediately followed by at least 3 U residues, but no extrinsic factors (recently reviewed in references 85, 93, 96, and 116); (ii) ρ-dependent terminators, which depend on the presence of ρ factor, a hexameric RNA-binding protein with ATPase activity (recently reviewed in references 85 and 92); and (iii) persistent RNA-DNA hybrid terminators at which pairing of nascent RNA to the template just upstream from the TEC dissociates a complex containing 3′-proximal U-rich RNA (113).
We will return to a discussion of pausing and termination later by describing a model for the related mechanisms of hairpin-dependent pausing and ρ-independent termination.
STRUCTURE OF THE TEC
Both intrinsic and extrinsic inputs alter the behavior of RNAP (and thus its RNA transcript output) by modulating either the alignment of nucleotides and catalytic moieties in the active site or the contacts of RNAP to the DNA or RNA. We must first understand the fundamental mechanisms that govern nucleotide addition and TEC stability before we can meaningfully address how extrinsic inputs modify them. We will focus the remainder of this short review on the intrinsic inputs that govern RNA chain elongation, pausing, and termination by first describing relevant features of RNAP’s three-dimensional structure and RNAP’s two large subunits, β′ and β, where most intrinsic interactions during elongation occur. We will then detail the critical intrinsic inputs, how these interactions program RNAP for hairpin-dependent pausing and ρ-independent termination, and briefly describe how an extrinsic factor, NusA, can modify these intrinsic interactions.
Three-dimensional structure of RNAP.
Electron crystallographic methods have been used by Kornberg, Darst, and others to deduce low-resolution (16- to 25-Å resolution) structures of E. coli holo- and core RNAPs and Saccharomyces cerevisiae RNAPs I and II (21, 22, 86, 100). These structures reveal common features likely shared by all multisubunit RNAPs: (i) a large, ∼25-Å channel surrounded by “jaws” that have been suggested to close around the downstream duplex DNA in the transition from RNAP complexed with initiator proteins (ς in bacteria) to TEC (5, 86); (ii) other features that may accommodate single-stranded RNA (ssRNA) or DNA; and (iii) two tunnels that run through the enzyme from near the channel to the opposite side, one of which may function as the RNA exit tunnel (21, 48). This overall structure resembles the hand-like motif of DNA polymerases whose X-ray crystal structures have been determined to atomic resolution (for a recent review see reference 46). The correlation of RNAP’s structure with its function will require answers to the following questions: (i) what are the paths of the RNA and DNA chains within the RNAP structure; (ii) where are the β′, β, α, and ς subunits of RNAP located, particularly the conserved segments (see below); and (iii) which parts of the structure, if any, move during different stages of the transcription cycle?
The first question deserves particular attention. Although passage of the downstream duplex DNA through the jaws of the channel offers a compelling physical picture for how RNAP could become clamped onto the DNA during RNA chain elongation (79), it offers no ready explanation for establishment and maintainence of the separation of the DNA strands during open-complex formation and RNA chain elongation. Other arrangements of the nucleic acid chains in the existing structures are possible. For instance, the downstream DNA duplex could be positioned in one of the grooves that runs across the front face of the yeast RNAP structure (see Fig. 4 in reference 21) and the RNA-DNA hybrid held between the jaws in the prominent central channel. This would offer a ready explanation for DNA strand separation if the nontemplate strand passed outside of the jaws. One version of such a model is suggested by Kim et al. (48). Learning if any of these ideas is correct must await higher-resolution crystals and extensive structural work on ternary transcription complexes, which should be possible given their great stability. In the meantime, we caution against overinterpreting the features evident in structures of RNAPs and have deliberately depicted two major channels in models of transcription complexes to avoid implying a particular hypothesis (see Fig. 1, 2, and 4).
FIG. 4.
Mechanism of hairpin-dependent pausing and ρ-independent termination. The four components of the signals are color coded: magenta, pause or terminator hairpin; green, 3′-proximal RNA; purple, bases in the active site; and blue, downstream DNA duplex. The insert shows the hairpin, 3′-proximal RNA, and active-site base components of archetypical pause and termination signals (the his leader pause and the terminator found in the trp operon attenuation control region). See the text for a description of the mechanism.
Contacts in RNAP subunits.
The elongating form of RNAP consists of the β and β′ subunits and a dimer of α subunits. β and β′ appear to make most of the important DNA and RNA contacts during elongation and are responsible for RNAP’s catalytic activity (Fig. 3). β and β′ homologs are evident in all multisubunit RNAPs based on colinear sequence similarities, designated A-I for β and A-H for β′ (Fig. 3) (2, 45, 126). As a framework for describing RNAP’s intrinsic inputs, we will summarize the known contacts of RNA and DNA to RNAP’s β and β′ subunits (Fig. 3). Many of these contacts, which were deduced from cross-linking or genetic analysis, correspond to sequences within both the β and β′ subunits of RNAP that are known to influence pausing and termination (44, 57, 110, 120).
FIG. 3.
Sites of regulatory inputs to RNAP large subunits. The β and β′ subunits are shown in opposite orientation to simplify depiction of known contacts to RNA and DNA. Conserved regions (A to I for β and A to H for β′) are shaded in blue (2, 45, 126). Regions that tolerate deletion of disruption are indicated by wavy lines (104, 105). Rifampin (rifampicin) and streptolydigin resistance regions, represented by orange and yellow, respectively, are based on the locations of known amino acid changes that allow growth on the antibiotics (40, 43, 102, 106). Other features shown are (i) the location of the catalytic Mg2+ ions that bind to the Asp side chains in the highly conserved sequence DFDGD at β′460–464 (27, 128); (ii) a Cys4 Zn-finger-like-motif in β′ possibly involved in forming the downstream DNA clamp (79) and in antitermination (20); and (iii) sites of contact to 3′-proximal RNA or DNA, exiting RNA or RNA hairpins, active-site bases, and the downstream DNA duplex that are described in the text (69, 75–79, 82, 118). The pause hairpin and upstream RNA contacts are depicted in a single block because the same bases may contact the β′ or β subunit depending on whether or not a hairpin has formed (118). The active-site contact to β′-region G is indicated with a dotted line because the RNA 3′ end makes this contact only in an arrested transcription complex (69).
RNAP also makes contacts to several metal ions, including two Zn2+ ions, one bound to β and one bound to β′ (122), and two or more Mg2+ ions that are required for full activity (reference 77 and references therein; 127, 128). The Mg2+ ions catalyze nucleotide addition (see the next section) and may be involved in DNA melting (109). The β′ Zn2+ in bacteria and both the β and β′ Zn2+ ions in eukaryotes are complexed by Cys residues in an apparent Zn-finger-like arrangement (Fig. 3). The Zn2+-finger-like motifs, which are near the N terminus of β′ and the C terminus of β (in eukaryotes), were shown to interact genetically (125) and may be responsible for closing RNAP’s clamp around DNA (56, 79). The site of Zn2+ binding to β in bacteria is unknown.
Interestingly, downstream-to-upstream contacts in the TEC mostly move C to N terminal in β and N to C terminal in β′ (Fig. 3). Additionally, the similar locations of some mutations and split sites, the likely genesis of β and β′ by gene duplication (120), and the viability of a strain in which the C terminus of β is fused to the N terminus of β′ (103) all are consistent with a model in which key contacts in the TEC may occur at the interface of these two weakly homologous but oppositely oriented large subunits (56). Surprisingly, essentially nothing is known about the secondary or tertiary structures of any conserved segment of β and β′; so far they have defied the increasingly powerful structure prediction and folding algorithms based on sequence similarities. Thus, improving understanding of structure in the conserved regions is a key objective for current study of the β and β′ subunits.
INTRINSIC INPUTS
Multiple intrinsic inputs govern RNA chain elongation: (i) an active site that aligns the RNA 3′ end and incoming NTP, (ii) 10 to 18 bp of duplex DNA held by RNAP in a clamp-like structure at the front edge of the complex, (iii) an ∼18 bp melted region of DNA within which the nontemplate strand appears to be channeled to the outside of the TEC, (iv) an ∼8-bp RNA-DNA hybrid that positions the RNA 3′ end in the active site 1 to 3 nt from the point of DNA strand separation, (v) at least one region of contact to single-stranded RNA upstream from the hybrid (an exit tunnel), and (vi) an interaction site for nascent RNA secondary structures.
Active site.
RNAP’s active site catalyzes an SN2-type nucleophilic displacement of the NTP β-γ pyrophosphate moiety by the RNA 3′ oxygen (30). It closely resembles the active sites of DNA polymerase, reverse transcriptases, and T7 RNA polymerase, all of which catalyze the same reaction using two Mg2+ ions that are positioned by coordination bonds to the carboxylates of Asp or Glu residues and that direct the 3′ OH and α phosphate into a trigonal bipyramidyl transition state (46, 108). In RNAP, Asp side chains in the highly conserved sequence DFDGD at β′ positions 460 to 464 (β′460–464) are proposed to chelate the Mg2+ ions (27, 128), whereas the α phosphate on the RNA 3′ terminal nucleotide is near βK1065 and βH1237 (75). The 3′ base is near β515–660 (the “Rif” region), β1100, and β′460 in the reactive conformation, but relocates to near β′940 after arrest (7, 69, 75). Two observations may indicate the involvement of additional metal ions in positioning nucleotides in the active site: (i) the [Mg2+] dependence of DNA strand melting is consistent with binding of three rather than two Mg2+ ions (109), and (ii) nuclear magnetic resonance measurements suggest that the β-bound Zn2+ ion may contact the 3′ base in the active site (6, 29). The major mechanistic implications of these results make confirmation by alternative methods critical objectives for future study.
Ultimately, positioning of the RNA 3′ end and NTP in the active site determines most regulatory decisions made by the TEC. Several sets of interactions may connect this event to different parts of RNAP. The catalytic Mg2+ ions are connected through a coordination network with the 3′ oxygen and NTP phosphates in the transition state (46). Thus, NTP binding and proper location of the RNA 3′ OH may be cooperative because the coordination bond network stabilizes their reactive alignment; improper positioning may inhibit nucleotide addition. Further, contacts to the bases, the ribose ring, the reactive groups (3′ OH and α phosphate), and the complementary template nucleotides may be over 15 Å apart and involve several different portions of β and β′, based both on cross-linking (Fig. 3) and the recent mapping of multiple free-radical cleavages in both subunits resulting from Fe2+ being substituted for the active-site Mg2+ (77). Thus, regulatory interactions outside the active site may ultimately be transduced to altered nucleotide addition through this set of interconnections.
Downstream DNA duplex.
About 18 bp of duplex DNA downstream from the active site are protected by RNAP. The sequence of this downstream DNA influences recognition of at least some pause sites and terminators (62, 111) through interactions unrelated to its ease of melting (62). Recently, Nudler et al. found that the downstream DNA duplex cross-links to β1230–1272 and β′30–102, that this interaction confers salt resistance to the TEC, and that the substitution of Ala for two Cys residues in the β′ Zn-finger-like motif caused the TEC to terminate indiscriminately shortly after initiation (79). Clerget et al. have found that other substitutions in the β′ Zn-finger-like motif abrogate HK022 put-dependent antitermination (20). It seems likely that these segments of β and β′ comprise the downstream DNA clamp and that interactions between them hold RNAP on DNA similarly to the clamps of the DNA replication processivity factors (51, 52). Key questions now are (i) does the downstream DNA clamp actually contact the DNA template molecule nonionically (79), or does salt resistance reflect principally hydrophobic protein-protein interactions that hold the clamp on the DNA despite electrostatic interactions; (ii) how do certain DNA sequences in the clamp alter the properties of the TEC at pause sites and terminators; and (iii) what drives the opening and closing of the downstream DNA clamp?
Nontemplate DNA.
The DNA strands separate only 1 to 3 nt downstream of the site of polymerization in halted complexes. The upstream portion of the nontemplate strand appears to lie on the outside of RNAP with the bases exposed, based on its accessibility to nucleases and hydroxyl radical, and to rejoin the template strand just as the latter exits the TEC (60, 61, 117). No formal reannealing site in RNAP may be necessary. The transcription bubble may be propagated nearly isoenergetically by dissociation of 1 bp as the nontemplate strand enters its exit channel in concert with the formation of a new base pair at the upstream edge of the complex (117). This arrangement makes the exposed nontemplate strand bases attractive targets for regulatory factor interaction. Interestingly, Ring et al. recently reported the first example of such an interaction: stimulation of promoter-proximal pausing at +16 of the λ late operon by ς binding to this segment of nontemplate strand DNA (95). Sequence-specific contacts to core RNAP itself have not been reported and are less likely in the upstream portion if the bases are oriented outwards, but could occur in the exit channel. Key questions are (i) which parts of core RNAP contact the nontemplate strand; (ii) are the bases in the upstream portion of the melted nontemplate strand exposed, as suggested by sensitivity to micrococcal nuclease (117), and are they the target of regulatory factors; and (iii) since nuclease digestion of the nontemplate strand leads to backtracking of RNAP (117), how does this region of DNA contribute to the lateral stability of the TEC?
RNA-DNA hybrid.
The length and role of the RNA-DNA hybrid have been heatedly debated in the transcription field. In one model, the existence of a ∼12-bp RNA-DNA hybrid in the TEC is postulated based on RNA footprinting, the ability of the 5′ end of ≤12-nt nascent RNAs to cross-link to DNA during initiation, and the inhibition of self-cleavage of a nascent RNA hammerhead <12 nt from the 3′ end (summarized in reference 124). These results taken together with thermodynamical analysis led Yager and von Hippel to hypothesize that a 12-bp hybrid compensated energetically for the melted DNA bubble to produce a stable TEC and that weakening the hybrid by hairpin invasion and an rU · dA base pair at a ρ-independent terminator led to TEC instability and dissociation (124). In an alternative model, Chamberlin hypothesized that the hybrid was only 2 to 3 nt in length and was not the main source of TEC stability (14). This argument was based on the observation that all but 2 to 3 nt of RNA can be removed by RNase digestion without loss of elongation competence (91) and is also supported by the demonstration that TECs lacking the template DNA strand upstream from the active site are stable (79). Although it may not be the source of the resistance to dissociation, an ∼8-bp hybrid does appear to be present in TECs based on protection of template DNA bases from single-strand-specific reagents (47, 61) and RNA-DNA cross-linking (82). This hybrid may control the lateral stability of the TEC (resistance to backtracking along the RNA and DNA with RNA 3′-end displacement) (54 and see below) and thus play a central role in the response of the TEC to regulatory signals. Lateral stability is decreased by an unstable rU · dA base pair, destabilizing base analogs, or misincorporation and is increased by stabilizing base analogs (82).
Key questions about the RNA-DNA hybrid now are (i) is it a conventional A-form helix, or does its interaction with RNAP induce some other less stable structure (see Discussion in reference 61); (ii) what type of contacts occur between the hybrid and RNAP (Fig. 3); and (iii) what causes the RNA to separate from the DNA template strand after 8 bp?
RNA transcript exit.
In the TEC, 18 to 22 nt of RNA are partially protected from RNase digestion (61, 91, 119), suggesting interactions of ssRNA upstream of the 8-bp hybrid with RNAP. This segment of RNA, 9 to 20 nt upstream from the 3′ end, cross-links to both β and β′ (39), with at least portions of the β and β′ interactions mapping to β904–950 and β′1–81 (118). In the absence of NusA, RNA >14 nt from the 3′ end also can cross-link to the α-subunit C-terminal domain (apparently to the same surface that contacts upstream DNA at some promoters) (67). Thus, if the transcript exits RNAP through one of the tunnels visible in the three-dimensional structure, nt −9 to −13 may lie within it (upstream from the RNA-DNA hybrid and downstream from the α contact that occurs in the absence of NusA). Several lines of evidence are consistent with the idea that entry of nascent RNA into the exit channel confers stability to the TEC, perhaps, as suggested by Nudler et al. (79), by triggering the closing of the downstream DNA clamp: (i) abortive RNAs are typically 8 nt or less; (ii) ς release and the conversion to a stable TEC occurs when the RNA grows to >8 nt (13); (iii) occasionally stable complexes can be produced with short transcripts (∼6 nt) that are not completely complementary to the template, apparently because the upstream RNA shifts into the exit channel and allows re-pairing of the downstream RNA to the vacated template positions (primer shifting [9, 32, 101]). Rifampin, which binds 15 Å from the priming nucleotide (78) and blocks synthesis of RNAs >3 or 4 nt (72), may bind to amino acid side chains normally contacting the upstream portion of the RNA-DNA hybrid near the entrance to the RNA exit channel (Fig. 2 and 3) (78). Key questions about the interactions of the RNA transcript as it exits RNAP are (i) does RNA exit through an internal tunnel in the RNAP structure; (ii) if so, what type of RNA-protein interactions occur within the tunnel; and (iii) do these interactions stabilize the closed form of the downstream DNA clamp?
Nascent RNA hairpins.
RNA hairpins that form as the transcript emerges from the TEC are integral parts of some pause signals; are required at ρ-independent terminators (23, 94, 96), where they trigger dissociation of the transcription complex; and can function as antiterminators, which modify the transcription complex to block recognition of both pause sites and terminators, either alone (e.g., HK022 put [20]) or in association with proteins (e.g., λ N-nut contact) (23, 93). The key question about how hairpins modulate RNA chain growth is whether they act through direct contact with RNAP or, indirectly, by disrupting an interaction of ssRNA with RNAP or DNA.
Several lines of evidence favor direct interaction of both pause and terminator hairpins, but do not rule out indirect effects: (i) high salt concentration reduces pausing and dissociates halted TECs in which a hairpin exists (3, 15); (ii) salt effects at the his pause are ion specific, reversible by base substitution in the hairpin but not in the other three pause signal components, and appear to involve a region of RNAP that is easily disordered by chaotropes (15); (iii) the conservation of the size of most terminator hairpins (5- to 9-bp stem; 3- to 5-nt loop) and of sequences in the stem region may suggest direct hairpin-RNAP interaction in termination (19, 24, 121); and (iv) the his pause hairpin loop cross-links to β904–950 in a paused complex and loses the interaction with β′1–81 that is characteristic of TEC and involved in stabilizing the downstream DNA clamp (79, 118). However, recent experiments comparing the effects of antisense oligonucleotides suggest that hairpin-RNAP interactions play a direct role in pausing but that the indirect effect of disrupting ssRNA interaction with RNAP or DNA is sufficient to dissociate a preformed paused transcription complex when pairing extends to within 8 nt of the RNA 3′ end (4). A role for direct interaction at the pause is indicated because all oligonucleotides that pair to the pause RNA hairpin reduce pausing by factors of 10 or more, including DNA or RNA oligonucleotides that exactly recapitulate the 11-nt 3′-proximal spacing (4). We return to consideration of the possible direct and indirect effects of RNA hairpins in the following section. Other key questions about nascent RNA hairpins now are (i) what part of RNAP makes ionic contact to hairpins, (ii) what is the molecular basis of RNA hairpin-RNAP interactions, and (iii) what structural changes accompany hairpin formation at pause sites and ρ-independent terminators (see the next section)?
HAIRPIN-DEPENDENT PAUSING AND ρ-INDEPENDENT TERMINATION
To illustrate how subtle changes in the intrinsic interactions of RNA and DNA with RNAP alter its regulatory output, we will describe here a two-branchpoint, kinetic-competition mechanism by which RNAP may recognize a hairpin-dependent pause signal, the his leader pause, and a related regulatory signal, a ρ-independent terminator (Fig. 4) (16, 31, 119). Using enzyme kinetics and the known intrinsic interactions of the TEC, this mechanism provides a biochemical explanation for how RNAP accomplishes a computational event: deciding between two possible outcomes (pausing or termination) based on information it reads from the RNA and DNA chains.
Both hairpin-dependent pause signals and ρ-independent terminators depend on multiple and similar intrinsic inputs, cause a reduction in lateral stability of the TEC that correlates with pausing or termination efficiency, and require formation of a nascent RNA hairpin at a precise template position to trigger pausing or termination. The his leader pause signal consists of four different intrinsic inputs that cooperatively direct pausing: (i) the 5-bp-stem, 8-nt-loop pause RNA hairpin that forms 11 nt from the RNA 3′ end; (ii) the 3′-proximal region of RNA or DNA between the hairpin and 3′ end; (iii) alignment of the 3′-terminal nucleotide and the incoming NTP; and (iv) the downstream DNA duplex (Fig. 4) (16–18, 54). ρ-independent terminators also depend on (i) a stable RNA hairpin; (ii) a 7- to 9-nt 3′-proximal region that usually contains 6 to 8 U’s (rather than a 10- or 11-nt, less-U-rich 3′-proximal region found at a pause); (iii) the rate of nucleotide addition in the active site; and (iv) in cases where few U’s are present in the 3′-proximal region, a specific downstream DNA sequence (24, 73, 89, 90, 93, 96, 111, 121, 124).
When the TEC first encounters a pause or termination signal, intrinsic interactions appear to slow nucleotide addition to facilitate hairpin formation at the appropriate template position (decelerate, Fig. 4). This deceleration appears to be caused by increased blockage of the i+1 site (Fig. 2) by the nascent RNA because relative instability of the RNA-DNA hybrid and downstream DNA contacts lead to transient backtracking (49, 50, 54, 81, 82, 119). This deceleration puts the TEC at the first branchpoint in the kinetic mechanism, where isomerization to the paused TEC (pause, Fig. 4) competes with chain elongation (bypass, Fig. 4). The nature of the effects on the isomerization-versus-bypass competition of the downstream DNA, the RNA-DNA hybrid, and the bases in the active site remain important questions for future study.
At the second branchpoint in the mechanism, the paused TEC either escapes back to the elongation pathway by binding NTP substrate and reestablishing the active site geometry, or dissociates if the remaining interactions are too weak. All four components of the his pause signal additively slow escape from the pause, suggesting the presence of a single paused intermediate (16). The pause RNA 3′ end appears to be pulled upstream out of the active site because the his and trp leader paused complexes are resistant to transcript cleavage or pyrophosphorolysis (4). In contrast, backtracked TECs such as complexes approaching the his pause site or ρ-independent terminators and some other types of paused complexes are sensitive to GreA-stimulated transcript cleavage or pyrophosphorolysis (32), suggesting that the RNA is reverse translocated through the active site. Thus, the pause hairpin, once formed, appears to prevent backtracking by RNAP.
This kinetic mechanism can explain how small differences in the intrinsic interactions of RNAP with RNA and DNA can result in either pausing or termination. Once the paused intermediate forms, whether or not it persists long enough to escape or instead dissociates depends on the stability of the paused complex and the likelihood that the RNA 3′ end can reestablish a reactive alignment with the NTP in the active site. The short, U-rich 3′-proximal RNA at a terminator both destabilizes the complex and makes escape unlikely. The longer 3′-proximal RNA in the paused complex allows stable interaction of the pause hairpin with RNAP, retention of some ssRNA-RNAP interactions in the exit channel, and 3′ OH alignment in the active site.
The key role of spacing between the hairpin and RNA 3′ end in the pause-versus-termination decision is supported by several observations. First, the his pause signal can be converted to an inefficient ρ-independent terminator merely by extending the hairpin stem to within the 7- to 9-nt 3′-proximal spacing observed at terminators (16). Second, separation of the hairpin from a run of 5 U’s by 10 to 12 nt at a pause site in the pyrBI leader region leads to pausing, not termination (28). Finally, nascent RNA hairpins that extend to within 9 nt of the RNA 3′ end also destabilize halted transcription complexes at high salt concentrations or 20 mM EDTA (3).
Precisely how the paused intermediate is destabilized at a terminator remains uncertain but appears to involve both a weak RNA-DNA hybrid and a loss of ssRNA contact to the downstream DNA clamp. The requirement for a weak hybrid is evident because extension of base pairing to within 8 nt of the RNA 3′ end dissociates the TEC only at high salt concentrations unless three or more U’s immediately follow the hairpin (3, 4, 16). Thus, the U run may play two roles in termination: (i) decelerating the TEC to facilitate hairpin formation in the first step of the mechanism and (ii) creating a particularly weak hybrid once hairpin formation is complete. The requirement for loss of ssRNA-RNAP contact was suggested by Nudler et al. to explain how the downstream DNA clamp that appears to close during initiation when ssRNA-RNAP contact is established is reopened at a terminator (79). This suggestion is consistent with the loss of RNA-β′ interaction observed in the his paused transcription complex, where the loop region of the pause hairpin instead makes strong contacts to β-subunit region 904–950 (118), raising the question of whether the clamp may be at least partially destabilized in the paused transcription complex as well (Fig. 4). The recent report that ρ-independent terminators work while being transcribed as single-stranded DNA (115) is consistent with these roles of RNA-DNA and RNA-protein interactions, but this result probably cannot be compared directly to the thermodynamics of transcription of duplex DNA.
This mechanism also can explain how RNA hairpins can play either direct or indirect roles in pausing and termination, respectively (see the section on RNA hairpins above), and why reaction conditions can have different effects at different terminators (89). Stabilization of the paused intermediate against escape appears to involve direct RNA hairpin interaction with RNAP, which cannot be accomplished by pairing to oligonucleotides (4). However, dissociation of the paused complex at a terminator may require only the indirect effect of disrupting ssRNA-RNAP or RNA-DNA interaction by base pairing to within 8 nt of the RNA 3′ end (4). Reynolds et al. (89) interpreted the varying effects of solutes and NTP concentration on termination efficiency at different terminators to mean that termination must be a multistep mechanism in which different steps could be rate limiting. The mechanism described here with two branchpoints preceding termination is consistent with this idea. In principle, termination efficiency can be determined either by competition between pausing and escape at the first branchpoint or between dissociation and escape at the second branchpoint (Fig. 4).
Key questions about the intrinsic contributions to pausing and termination now under investigation are (i) does hairpin-RNAP interaction modulate formation of nascent RNA secondary structures; (ii) does hairpin-RNAP interaction play a direct allosteric role in active-site collapse, pull the RNA 3′ end out of the active site, or simply obscure NTP binding; (iii) does a terminator hairpin form within the RNA exit channel, pull the RNA through the exit channel making the RNA-DNA hybrid even smaller than 8 bp, or pull the RNA completely out of the exit channel; (iv) what are the exact sequence requirements for the contributions of 3′-proximal and downstream DNA sequences to pausing and termination, and do they differ between the two steps in the mechanism or between pausing and termination; (v) is release of RNA and DNA from the TEC ordered (RNA first or DNA first) or random; and (vi) do the contributions of the pause-versus-bypass and terminate-versus-escape pathways to termination efficiency differ at different terminators, as suggested by varying responses to ionic conditions (89).
EXTRINSIC INPUTS
We have described how interactions of RNAP with RNA and DNA can generate different conformations of the transcription complex. These conformations can serve as targets for further regulation by extrinsic transcription factors (Table 1). These factors can modify the rules by which RNAP responds to future intrinsic inputs, resembling the change in logical rules in a Turing computational device upon input of information. The key to a mechanistic understanding of these extrinsic inputs is to decipher how they alter the intrinsic interactions of RNA, DNA, and NTPs with RNAP to change its enzymatic properties.
To illustrate this point briefly, we consider the effect of the extrinsic factor NusA on pausing and termination (Table 1). E. coli NusA is a 55-kDa acidic protein that interacts with Rho, λ N, and RNA through one or more interaction regions and RNAP through contacts to the α-subunit C-terminal domain and either β′ or β (68, 93). NusA was discovered as a cellular factor necessary for λ N-dependent antitermination but enhances both pausing and ρ-independent termination in the absence of other cellular or phage proteins, is found in all prokaryotes and archaebacteria sequenced to date, and is essential in E. coli unless Rho activity is reduced by a mutation (130). With low affinity it competes for NTP binding and with higher affinity enhances pausing in a site-specific fashion (99, 112) that may involve interaction with RNA secondary structures (59, 66). NusA can also alter the 3′-end location of an arrested RNA (129). Despite its central role in the regulation of RNA chain elongation, however, precisely how NusA either enhances or inhibits pausing and termination depending on the presence of other factors remains unclear. Does it stabilize the paused intermediate simply by further stabilizing the RNA hairpin-RNAP interaction that already inhibits escape, or does it directly affect RNA 3′-end positioning? How are the effects of NusA reversed when λ N protein is present? Answering these questions will require both a fuller understanding of how intrinsic interactions of the transcription complex direct the pausing/termination mechanism (Fig. 4) and careful dissection of NusA’s effect on these interactions.
The same is true of the many other extrinsic factors that couple transcription elongation to cellular metabolism. By first understanding how intrinsic interactions in the transcription complex mediate its logical operations and then elucidating how extrinsic transcription factors modify these operations, we can build a comprehensive biochemical model of the information processing activities of RNAP.
REFERENCES
- 1.Aivazashvili V A, Bibilashvili R S, Vartikyan R M, Kutateladze T A. Relationship between the primary RNA structure and pulse elongation in vitro by Escherichia coli RNA polymerase. A model. Mol Biol. 1982;15:711–722. [PubMed] [Google Scholar]
- 2.Archambault J, Friesen J. Genetics of eukaryotic RNA polymerases I, II, and III. Microbiol Rev. 1993;57:703–724. doi: 10.1128/mr.57.3.703-724.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Arndt K M, Chamberlin M J. RNA chain elongation by Escherichia coli RNA polymerase. Factors affecting the stability of elongating ternary complexes. J Mol Biol. 1990;213:79–108. doi: 10.1016/S0022-2836(05)80123-8. [DOI] [PubMed] [Google Scholar]
- 4.Artsimovitch, I., and R. Landick. Submitted for publication.
- 5.Asturias F, Meredith G, Poglitsch C, Kornberg R. Two conformations of RNA polymerase II revealed by electron crystallography. J Mol Biol. 1997;272:536–540. doi: 10.1006/jmbi.1997.1273. [DOI] [PubMed] [Google Scholar]
- 6.Beal R, Pillai R, Chuknyisky P, Levy A, Tarien E, Eichhorn G. Structural studies on the active site of Escherichia coli RNA polymerase. 2. Geometrical relationship of the interacting substrates. Biochemistry. 1990;29:5994–6002. doi: 10.1021/bi00477a017. [DOI] [PubMed] [Google Scholar]
- 7.Borukhov S, Lee J, Goldfarb A. Mapping of a contact for the RNA 3′ terminus in the largest subunit of RNA polymerase. J Biol Chem. 1991;266:23932–23935. [PubMed] [Google Scholar]
- 8.Borukhov S, Sagitov V, Goldfarb A. Transcript cleavage factors from E. coli. Cell. 1993;72:459–466. doi: 10.1016/0092-8674(93)90121-6. [DOI] [PubMed] [Google Scholar]
- 9.Borukhov S, Sagitov V, Josaitis C A, Gourse R L, Goldfarb A. Two modes of transcription initiation in vitro at the rrnB P1 promoter of Escherichia coli. J Biol Chem. 1993;268:23477–23482. [PubMed] [Google Scholar]
- 10.Briat J F, Bollag G, Kearney C A, Molineux I, Chamberlin M J. Tau factor from Escherichia coli mediates accurate and efficient termination of transcription at the bacteriophage T3 early termination site in vitro. J Mol Biol. 1987;198:43–49. doi: 10.1016/0022-2836(87)90456-6. [DOI] [PubMed] [Google Scholar]
- 11.Cashel M, DR G, VJ H, D V. The stringent response. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella cellular and molecular biology. 2nd ed. Washington, D.C: ASM Press; 1996. pp. 1458–1496. [Google Scholar]
- 12.Chamberlin M. RNA polymerase—an overview. In: Losick R, Chamberlin M, editors. RNA polymerase. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1976. pp. 17–68. [Google Scholar]
- 13.Chamberlin M, Hsu L. RNA chain initiation and promoter clearance by E. coli RNA polymerase. In: Lin E C C, Lynch A S, editors. Regulation of gene expression in E. coli. R. G. Austin, Tex: Landes Co.; 1996. pp. 7–25. [Google Scholar]
- 14.Chamberlin M J. New models for the mechanism of transcription elongation and its regulation. 1995. pp. 1–21. . The Harvey lectures. Wiley-Liss, New York, N.Y. [PubMed] [Google Scholar]
- 15.Chan C, Landick R. Effects of neutral salts of transcript elongation and pausing suggest the his leader pause RNA hairpin interacts with an easily disordered region of RNA polymerase. J Mol Biol. 1997;268:37–53. doi: 10.1006/jmbi.1997.0934. [DOI] [PubMed] [Google Scholar]
- 16.Chan C, Wang D, Landick R. Spacing from the transcript 3′ end determines whether a nascent RNA hairpin interacts with RNA polymerase to prolong pausing or triggers termination. J Mol Biol. 1997;268:54–68. [Google Scholar]
- 17.Chan C L, Landick R. Dissection of the his leader pause site by base substitution reveals a multipartite signal that includes a pause RNA hairpin. J Mol Biol. 1993;233:25–42. doi: 10.1006/jmbi.1993.1482. [DOI] [PubMed] [Google Scholar]
- 18.Chan C L, Landick R. New perspectives on RNA chain elongation and termination by E. coli RNA polymerase. In: Conaway R C, Conaway J W, editors. Transcription: mechanisms and regulation. New York, N.Y: Raven Press; 1994. pp. 297–321. [Google Scholar]
- 19.Cheng S-W, Lynch E C, Leason K R, Court D L, Shapiro B A, Friedman D I. Functional importance of sequence in the stem-loop of a transcription terminator. Science. 1992;254:1205–1207. doi: 10.1126/science.1835546. [DOI] [PubMed] [Google Scholar]
- 20.Clerget M, Jin D J, Weisberg R A. A zinc-binding region in the beta′ subunit of RNA polymerase is involved in antitermination of early transcription of phage HK022. J Mol Biol. 1995;248:768–780. doi: 10.1006/jmbi.1995.0259. [DOI] [PubMed] [Google Scholar]
- 21.Darst S A, Edwards A M, Kubalek E W, Kornberg R D. Three-dimensional structure of yeast RNA polymerase II at 16 Å resolution. Cell. 1991;66:121–128. doi: 10.1016/0092-8674(91)90144-n. [DOI] [PubMed] [Google Scholar]
- 22.Darst S A, Kubalek E W, Kornberg R D. Three-dimensional structure of Escherichia coli RNA polymerase holoenzyme determined by electron crystallography. Nature. 1989;340:730–732. doi: 10.1038/340730a0. [DOI] [PubMed] [Google Scholar]
- 23.Das A. Control of transcription termination by RNA-binding proteins. Annu Rev Biochem. 1993;62:893–930. doi: 10.1146/annurev.bi.62.070193.004333. [DOI] [PubMed] [Google Scholar]
- 24.d’Aubenton Carafa Y, Brody E, Thermes C. Prediction of Rho-independent Escherichia coli transcription terminators. A statistical analysis of their RNA stem-loop structures. J Mol Biol. 1990;216:835–858. doi: 10.1016/s0022-2836(99)80005-9. [DOI] [PubMed] [Google Scholar]
- 25.deHaseth P, Zupanic M, Record M., Jr RNA polymerase promoter interactions: the comings and goings of RNA polymerase. J Bacteriol. 1998;180:3019–3025. doi: 10.1128/jb.180.12.3019-3025.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.deHaseth P L, Helmann J D. Open complex formation by Escherichia coli RNA polymerase: the mechanism of polymerase-induced strand separation of double helical DNA. Mol Microbiol. 1995;16:817–824. doi: 10.1111/j.1365-2958.1995.tb02309.x. [DOI] [PubMed] [Google Scholar]
- 27.Dieci G, Hermann-Le Denmat S, Lukhtanov E, Thuriaux P, Werner M, Sentenac A. A universally conserved region of the largest subunit participates in the active site of RNA polymerase III. EMBO J. 1995;14:3766–3776. doi: 10.1002/j.1460-2075.1995.tb00046.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Donahue J P, Turnbough C L., Jr Nucleotide-specific transcriptional pausing in the pyrBI leader region of Escherichia coli K-12. J Biol Chem. 1994;269:18185–18191. [PubMed] [Google Scholar]
- 29.Eichhorn G, Chuknyisky P, Butzow J, Beal R, Garland C, Janzen C, Clark P, Tarien E. A structural model for fidelity in transcription. Proc Natl Acad Sci USA. 1994;91:7613–7617. doi: 10.1073/pnas.91.16.7613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Erie D A, Yager T D, von Hippel P H. The single-nucleotide addition cycle in transcription: a biophysical and biochemical perspective. Annu Rev Biophys Biomol Struct. 1992;21:379–415. doi: 10.1146/annurev.bb.21.060192.002115. [DOI] [PubMed] [Google Scholar]
- 31.Farnham P J, Platt T. A model for transcription termination suggested by studies on the trp attenuator in vitro using base analogs. Cell. 1980;20:739–748. doi: 10.1016/0092-8674(80)90320-7. [DOI] [PubMed] [Google Scholar]
- 32.Feng G, Lee D N, Wang D, Chan C L, Landick R. GreA-induced transcript cleavage in transcription complexes containing Escherichia coli RNA polymerase is controlled by multiple factors, including nascent transcript location and structure. J Biol Chem. 1994;269:22282–22294. [PubMed] [Google Scholar]
- 33.Gaal T, Bartlett M, Ross W, Turnbough C, Jr, Gourse R. Transcription regulation by initiating NTP concentration: rRNA synthesis in bacteria. Science. 1997;278:2092–2097. doi: 10.1126/science.278.5346.2092. [DOI] [PubMed] [Google Scholar]
- 34.Gelles J, Landick R. RNA polymerase as a molecular motor. Cell. 1998;93:13–16. doi: 10.1016/s0092-8674(00)81140-x. [DOI] [PubMed] [Google Scholar]
- 35.Greenblatt J, Nodwell J R, Mason S W. Transcriptional antitermination. Nature. 1993;364:401–406. doi: 10.1038/364401a0. [DOI] [PubMed] [Google Scholar]
- 36.Gross C, Lonetto M, Losick R. Bacterial sigma factors. In: McKnight S, Yamamoto K, editors. Transcriptional regulation. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1996. pp. 129–176. [Google Scholar]
- 37.Gu W, Wind M, Reines D. Increased accommodation of nascent RNA in a product site on RNA polymerase II during arrest. Proc Natl Acad Sci USA. 1996;93:6935–6940. doi: 10.1073/pnas.93.14.6935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Guajardo R, Sousa R. A model for the mechanism of polymerase translocation. J Mol Biol. 1997;265:8–19. doi: 10.1006/jmbi.1996.0707. [DOI] [PubMed] [Google Scholar]
- 39.Hanna M M, Meares C F. Topography of transcription: path of the leading end of nascent RNA through the Escherichia coli transcription complex. Proc Natl Acad Sci USA. 1983;80:4238–4242. doi: 10.1073/pnas.80.14.4238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Heisler L M, Suzuki H, Landick R, Gross C A. Four contiguous amino acids in the β subunit of Escherichia coli RNA polymerase define the target for streptolydigin resistance. J Biol Chem. 1993;268:25369–25375. [PubMed] [Google Scholar]
- 41.Hopcroft J. Turing machines. Sci Am. 1985;52:86–98. [Google Scholar]
- 42.Hsu L, Vo N, Chamberlin M. Escherichia coli transcript cleavage factors GreA and GreB stimulate promoter escape and gene expression in vivo and in vitro. Proc Natl Acad Sci USA. 1995;92:11588–11592. doi: 10.1073/pnas.92.25.11588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jin D J, Gross C. Mapping and sequencing of mutations in the Escherichia coli rpoB gene that lead to rifampicin resistance. J Mol Biol. 1988;202:45–58. doi: 10.1016/0022-2836(88)90517-7. [DOI] [PubMed] [Google Scholar]
- 44.Jin D J, Walter W A, Gross C A. Characterization of the termination phenotypes of rifampicin-resistant mutants. J Mol Biol. 1988;202:245–253. doi: 10.1016/0022-2836(88)90455-x. [DOI] [PubMed] [Google Scholar]
- 45.Jokerst R S, Weeks J R, Zehring W A, Greenleaf A L. Analysis of the gene encoding the largest subunit of RNA polymerase II in Drosophila. Mol Gen Genet. 1989;215:266–275. doi: 10.1007/BF00339727. [DOI] [PubMed] [Google Scholar]
- 46.Joyce C, Steitz T. Polymerase structures and function: variations on a theme? J Bacteriol. 1996;177:6321–6329. doi: 10.1128/jb.177.22.6321-6329.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kainz M, Roberts J. Structure of transcription elongation complexes in vivo. Science. 1992;255:838–841. doi: 10.1126/science.1536008. [DOI] [PubMed] [Google Scholar]
- 48.Kim T-K, Lagrange T, Wang Y-H, Griffith J, Reinberg D, Ebright R. Trajectory of DNA in the RNA polymerase II transcription preinitiation complex. Proc Natl Acad Sci USA. 1997;94:12268–12273. doi: 10.1073/pnas.94.23.12268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Komissarova N, Kashlev M. Arrest of transcription: E. coli RNA polymerase translocates backward leaving the 3′ end of the RNA intact and extruded. Proc Natl Acad Sci USA. 1997;94:1755–1760. doi: 10.1073/pnas.94.5.1755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Komissarova N, Kashlev M. RNA polymerase switches between inactivated and activated states by translocating back and forth along the DNA and the RNA. J Biol Chem. 1997;272:15329–15338. doi: 10.1074/jbc.272.24.15329. [DOI] [PubMed] [Google Scholar]
- 51.Kong X-P, Onrust R, O’Donnell M, Kuriyan J. Three-dimensional structure of the beta subunit of E. coli DNA polymerase III holoenzyme: a sliding DNA clamp. Cell. 1992;69:425–437. doi: 10.1016/0092-8674(92)90445-i. [DOI] [PubMed] [Google Scholar]
- 52.Krishna T, Kong X, Gary S, Burgers P, Kuriyan J. Crystal structure of the eukaryotic DNA polymerase processivity factor PCNA. Cell. 1994;79:1233–1243. doi: 10.1016/0092-8674(94)90014-0. [DOI] [PubMed] [Google Scholar]
- 53.Krummel B, Chamberlin M J. Structural analysis of ternary complexes of Escherichia coli RNA polymerase. Individual complexes halted along different transcription units have distinct and unexpected biochemical properties. J Mol Biol. 1992;225:221–237. doi: 10.1016/0022-2836(92)90917-9. [DOI] [PubMed] [Google Scholar]
- 54.Landick R. RNA polymerase slides home: pause and termination site recognition. Cell. 1997;88:741–744. doi: 10.1016/s0092-8674(00)81919-4. [DOI] [PubMed] [Google Scholar]
- 55.Landick R, Carey J, Yanofsky C. Translation activates the paused transcription complex and restores transcription of the trp operon leader region. Proc Natl Acad Sci USA. 1985;82:4663–4667. doi: 10.1073/pnas.82.14.4663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Landick R, Roberts J. The shrewd grasp of RNA polymerase. Science. 1996;273:202–203. doi: 10.1126/science.273.5272.202. [DOI] [PubMed] [Google Scholar]
- 57.Landick R, Stewart J, Lee D. Amino acid changes in conserved regions of the β-subunit of Escherichia coli RNA polymerase alter transcription pausing and termination. Genes Dev. 1990;4:1623–1636. doi: 10.1101/gad.4.9.1623. [DOI] [PubMed] [Google Scholar]
- 58.Landick R, Turnbough C, Jr, Yanofsky C. Transcription attenuation. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Washington, D.C: ASM Press; 1996. pp. 1263–1286. [Google Scholar]
- 59.Landick R, Yanofsky C. Isolation and structural analysis of the Escherichia coli trp leader paused transcription complex. J Mol Biol. 1987;196:363–377. doi: 10.1016/0022-2836(87)90697-8. [DOI] [PubMed] [Google Scholar]
- 60.Lee D N, Feng G, Landick R. GreA-induced transcript cleavage is accompanied by reverse translocation to a different transcription complex conformation. J Biol Chem. 1994;269:22295–22303. [PubMed] [Google Scholar]
- 61.Lee D N, Landick R. Structure of RNA and DNA chains in paused transcription complexes containing Escherichia coli RNA polymerase. J Mol Biol. 1992;228:759–777. doi: 10.1016/0022-2836(92)90862-e. [DOI] [PubMed] [Google Scholar]
- 62.Lee D N, Phung L, Stewart J, Landick R. Transcription pausing by Escherichia coli RNA polymerase is modulated by downstream DNA sequences. J Biol Chem. 1990;265:15145–15153. [PubMed] [Google Scholar]
- 63.Li M, McClure W R, Susskind M M. Changing the mechanism of transcriptional activation by phage lambda repressor. Proc Natl Acad Sci USA. 1997;94:3691–3696. doi: 10.1073/pnas.94.8.3691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Liu C, Heath L, Turnbough C., Jr Regulation of pyrBI operon expression in Escherichia coli by UTP-sensitive reiterative RNA synthesis during transcriptional initiation. Genes Dev. 1994;8:2904–2912. doi: 10.1101/gad.8.23.2904. [DOI] [PubMed] [Google Scholar]
- 65.Liu J, Turnbough C L., Jr Effects of transcriptional start site sequence and position on nucleotide-sensitive selection of alternative start sites at the pyrC promoter in Escherichia coli. J Bacteriol. 1994;176:2938–2945. doi: 10.1128/jb.176.10.2938-2945.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Liu K, Hanna M M. NusA contacts nascent RNA in Escherichia coli transcription complexes. J Mol Biol. 1995;247:547–558. doi: 10.1006/jmbi.1994.0161. [DOI] [PubMed] [Google Scholar]
- 67.Liu K, Hanna M M. NusA interferes with interactions between the nascent RNA and the C-terminal domain of the alpha subunit of RNA polymerase in Escherichia coli transcription complexes. Proc Natl Acad Sci USA. 1995;92:5012–5016. doi: 10.1073/pnas.92.11.5012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Liu K, Zhang Y, Severinov K, Das A, Hanna M M. Role of Escherichia coli RNA polymerase alpha subunit in modulation of pausing, termination and anti-termination by the transcription elongation factor NusA. EMBO J. 1996;15:150–161. [PMC free article] [PubMed] [Google Scholar]
- 69.Markovtsov V, Mustaev A, Goldfarb A. Protein-RNA interactions in the active center of the transcription elongation complex. Proc Natl Acad Sci USA. 1996;93:3221–3226. doi: 10.1073/pnas.93.8.3221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Masukata H, Tomizawa J. A mechanism of formation of a persistent hybrid between elongation RNA and template DNA. Cell. 1990;62:331–338. doi: 10.1016/0092-8674(90)90370-t. [DOI] [PubMed] [Google Scholar]
- 71.McClure W R. Mechanism and control of transcription initiation in prokaryotes. Annu Rev Biochem. 1985;54:171–204. doi: 10.1146/annurev.bi.54.070185.001131. [DOI] [PubMed] [Google Scholar]
- 72.McClure W R, Cech C L. On the mechanism of rifampicin inhibition of RNA synthesis. J Biol Chem. 1978;253:8949–8956. [PubMed] [Google Scholar]
- 73.McDowell J C, Roberts J W, Jin D J, Gross C. Determination of intrinsic transcription termination efficiency by RNA polymerase elongation rate. Science. 1994;266:822–825. doi: 10.1126/science.7526463. [DOI] [PubMed] [Google Scholar]
- 74.Monsalve M, Mencia M, Rojo F, Salas M. Activation and repression of transcription at two different phage phi29 promoters are mediated by interaction of the same residues of regulatory protein p4 with RNA polymerase. EMBO J. 1996;15:383–391. [PMC free article] [PubMed] [Google Scholar]
- 75.Mustaev A, Kashlev M, Lee J Y, Polyakov A, Lebedev A, Zalenskaya K, Grachev M, Goldfarb A, Nikiforov V. Mapping of the priming substrate contacts in the active center of Escherichia coli RNA polymerase. J Biol Chem. 1991;266:23927–23931. [PubMed] [Google Scholar]
- 76.Mustaev A, Kashlev M, Zaychikov E, Grachev M, Goldfarb A. Active center rearrangement in RNA polymerase initiation complex. J Biol Chem. 1993;26:19185–19187. [PubMed] [Google Scholar]
- 77.Mustaev A, Kozlov M, Markovtsov V, Zaychikov E, Denissova L, Goldfarb A. Modular organization of the catalytic center of RNA polymerase. Proc Natl Acad Sci USA. 1997;94:6641–6645. doi: 10.1073/pnas.94.13.6641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Mustaev A, Zaychikov E, Severinov K, Kashlev M, Polyakov A, Nikiforov V, Goldfarb A. Topology of the RNA polymerase active center probed by chimeric rifampicin-nucleotide compounds. Proc Natl Acad Sci USA. 1994;91:12036–12040. doi: 10.1073/pnas.91.25.12036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Nudler E, Avetissova E, Markovstov V, Goldfarb A. Transcription processivity: RNA polymerase-DNA interactions holding together the elongation complex. Science. 1996;273:211–217. doi: 10.1126/science.273.5272.211. [DOI] [PubMed] [Google Scholar]
- 80.Nudler E, Goldfarb A, Kashlev M. Discontinuous mechanism of transcription elongation. Science. 1994;265:793–796. doi: 10.1126/science.8047884. [DOI] [PubMed] [Google Scholar]
- 81.Nudler E, Kashlev M, Nikiforov V, Goldfarb A. Coupling between transcription termination and RNA polymerase inchworming. Cell. 1995;81:351–357. doi: 10.1016/0092-8674(95)90388-7. [DOI] [PubMed] [Google Scholar]
- 82.Nudler E, Mustaev A, Lukhtanov E, Goldfarb A. The RNA:DNA hybrid maintains the register of transcription by preventing backtracking of RNA polymerase. Cell. 1997;89:33–41. doi: 10.1016/s0092-8674(00)80180-4. [DOI] [PubMed] [Google Scholar]
- 83.Orlova M, Newlands J, Das A, Goldfarb A, Borukhov S. Intrinsic transcript cleavage activity of RNA polymerase. Proc Natl Acad Sci USA. 1995;92:4596–4600. doi: 10.1073/pnas.92.10.4596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Platt T. Rho and RNA: models for recognition and response. Mol Microbiol. 1994;11:983–990. doi: 10.1111/j.1365-2958.1994.tb00376.x. [DOI] [PubMed] [Google Scholar]
- 85.Platt T. RNA structure in transcription elongation, termination, and antitermination. In: Simons R, Grunberg M, editors. RNA structure and function. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1997. pp. 541–574. [Google Scholar]
- 86.Polyakov A, Severinova E, Darst S. Three-dimensional structure of E. coli core RNA polymerase: promoter binding and elongation conformations of the enzyme. Cell. 1995;83:365–373. doi: 10.1016/0092-8674(95)90114-0. [DOI] [PubMed] [Google Scholar]
- 87.Record M, Jr, Reznikoff W, Craig M, McQuade K, Schlax P. Escherichia coli RNA polymerase (Eς70), promoters, and the kinetics of the steps of transcription initiation. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Washington, D.C: ASM Press; 1996. pp. 792–821. [Google Scholar]
- 88.Reeder T, Hawley D. Promoter proximal sequences modulate RNA polymerase II elongation by a novel mechanism. Cell. 1996;87:767–777. doi: 10.1016/s0092-8674(00)81395-1. [DOI] [PubMed] [Google Scholar]
- 89.Reynolds R, Bermúdez-Cruz R M, Chamberlin M J. Parameters affecting transcription termination by Escherichia coli RNA polymerase. Analysis of 13 rho-independent terminators. J Mol Biol. 1992;224:31–51. doi: 10.1016/0022-2836(92)90574-4. [DOI] [PubMed] [Google Scholar]
- 90.Reynolds R, Chamberlin M J. Parameters affecting transcription termination by Escherichia coli RNA polymerase. II. Construction and analysis of hybrid terminators. J Mol Biol. 1992;224:53–63. doi: 10.1016/0022-2836(92)90575-5. [DOI] [PubMed] [Google Scholar]
- 91.Rice G, Kane C, Chamberlin M. Footprinting analysis of mammalian RNA polymerase II along its transcript as an alternate view of transcription elongation. Proc Natl Acad Sci USA. 1991;88:1245–1249. doi: 10.1073/pnas.88.10.4245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Richardson J. Structural organization of transcription termination factor Rho. J Biol Chem. 1996;271:1251–1254. doi: 10.1074/jbc.271.3.1251. [DOI] [PubMed] [Google Scholar]
- 93.Richardson J, Greenblatt J. Control of RNA chain elongation and termination. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. Washington, D.C: ASM Press; 1996. pp. 822–848. [Google Scholar]
- 94.Richardson J P. Transcription termination. Crit Rev Biochem Mol Biol. 1993;28:1–30. doi: 10.3109/10409239309082571. [DOI] [PubMed] [Google Scholar]
- 95.Ring B, Yarnell W, Roberts J. Function of E. coli RNA polymerase ς factor ς70 in promoter-proximal pausing. Cell. 1996;86:485–493. doi: 10.1016/s0092-8674(00)80121-x. [DOI] [PubMed] [Google Scholar]
- 96.Roberts J W. Transcription termination and its control. In: Lin E, Lynch A, editors. Regulation of gene expression in E. coli. R. G. Austin, Tex: Landes Co.; 1996. pp. 27–44. [Google Scholar]
- 97.Roe J, Burgess R, Record M. Temperature dependence of the rate constants of the E. coli RNA polymerase lambda PR promoter interaction. Assignment of the kinetic steps corresponding to protein conformational change and DNA opening. J Mol Biol. 1985;184:441–453. doi: 10.1016/0022-2836(85)90293-1. [DOI] [PubMed] [Google Scholar]
- 98.Ross W, Gosink K, Salomon J, Igarashi K, Zou C, Ishihama A, Severinov K, Gourse R. A third recognition element in bacterial promoters: DNA binding by the alpha subunit of RNA polymerase. Science. 1993;262:1407–1413. doi: 10.1126/science.8248780. [DOI] [PubMed] [Google Scholar]
- 99.Schmidt M, Chamberlin M. Amplification and isolation of Escherichia coli nusA protein and studies of its effects on in vitro RNA chain elongation. Biochemistry. 1984;23:197–203. doi: 10.1021/bi00297a004. [DOI] [PubMed] [Google Scholar]
- 100.Schultz P, Célia H, Riva M, Sentenac A, Oudet P. Three-dimensional model of RNA polymerase I determined by electron microscopy of two-dimensional crystals. EMBO J. 1993;12:2601–2607. doi: 10.1002/j.1460-2075.1993.tb05920.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Severinov K, Goldfarb A. Topology of the product binding site in RNA polymerase revealed by transcript slippage at the phage lambda PL promoter. J Biol Chem. 1994;269:31701–31705. [PubMed] [Google Scholar]
- 102.Severinov K, Markov D, Sererinova E, Nikiforov V, Landick R, Darst S, Goldfarb A. Stretolydigin-resistant mutants in an evolutionarily conserved region of the β′ subunit of Escherichia coli RNA polymerase. J Biol Chem. 1995;270:23926–23929. doi: 10.1074/jbc.270.41.23926. [DOI] [PubMed] [Google Scholar]
- 103.Severinov K, Mooney R, Darst S, Landick R. Tethering of the large subunits of Escherichia coli RNA polymerase. J Biol Chem. 1997;272:24137–24140. doi: 10.1074/jbc.272.39.24137. [DOI] [PubMed] [Google Scholar]
- 104.Severinov K, Mustaev A, Kukarin A, Muzzin O, Bass I, Darst S, Goldfarb A. Structural modules of the large subunits of RNA polymerase. Introducing archaebacterial and chloroplast split sites in the beta and beta′ subunits of Escherichia coli RNA polymerase. J Biol Chem. 1997;271:27969–27974. doi: 10.1074/jbc.271.44.27969. [DOI] [PubMed] [Google Scholar]
- 105.Severinov K, Mustaev A, Severinova E, Bass I, Kashlev M, Landick R, Nikiforov V, Goldfarb A, Darst S. Assembly of functional Escherichia coli RNA polymerase containing β subunit fragments. Proc Natl Acad Sci USA. 1995;92:4591–4595. doi: 10.1073/pnas.92.10.4591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Severinov K, Soushko M, Goldfarb A, Nikiforov V. Rifampicin region revisited. New rifampicin-resistant and streptolydigin-resistant mutants in the beta subunit of Escherichia coli RNA polymerase. J Biol Chem. 1993;268:14820–14825. [PubMed] [Google Scholar]
- 107.Smith T, Sauer R. Dual regulation of open-complex formation and promoter clearance by Arc explains a novel repressor to activator switch. Proc Natl Acad Sci USA. 1996;93:8868–8872. doi: 10.1073/pnas.93.17.8868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Steitz T, Smerdon S, Jager J, Joyce C. A unified polymerase mechanism for nonhomologous DNA and RNA polymerase. Science. 1994;266:2022–2025. doi: 10.1126/science.7528445. [DOI] [PubMed] [Google Scholar]
- 109.Suh W, Ross W, Record M., Jr Two open complexes and a requirement for Mg2+ to open the lambda PR transcription start site. Science. 1993;259:358–361. doi: 10.1126/science.8420002. [DOI] [PubMed] [Google Scholar]
- 110.Tavromina P, Landick R, Gross C. Isolation and characterization of defective Escherichia coli RNA polymerase rpoB mutations. J Bacteriol. 1996;178:5263–5371. doi: 10.1128/jb.178.17.5263-5271.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Telesnitsky A, Chamberlin M. Terminator-distal sequences determine the in vitro efficiency of the early terminators of bacteriophages T3 and T7. Biochemistry. 1989;28:5210–5218. doi: 10.1021/bi00438a044. [DOI] [PubMed] [Google Scholar]
- 112.Theissen G, Pardon B, Wagner R. A quantitative assessment for transcriptional pausing of DNA-dependent RNA polymerases in vitro. Anal Biochem. 1990;189:254–261. doi: 10.1016/0003-2697(90)90117-r. [DOI] [PubMed] [Google Scholar]
- 113.Tomizawa J, Masukata H. Factor-independent termination of transcription in a stretch of deoxyadenosine residues in the template DNA. Genes Dev. 1987;1:217–226. doi: 10.1016/0092-8674(87)90131-0. [DOI] [PubMed] [Google Scholar]
- 114.Turing A. On computable numbers, with an application to the entscheidungs-problem. Proc Lond Math Soc. 1936;43:230–265. [Google Scholar]
- 115.Uptain S, Chamberlin M. Escherichia coli RNA polymerase terminates transcription efficiently at rho-independent terminators on single-stranded DNA templates. Proc Natl Acad Sci USA. 1997;94:13548–13553. doi: 10.1073/pnas.94.25.13548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Uptain S, Kane C, Chamberlin M. Basic mechanisms of transcript elongation and its regulation. Annu Rev Biochem. 1997;66:117–172. doi: 10.1146/annurev.biochem.66.1.117. [DOI] [PubMed] [Google Scholar]
- 117.Wang D, Landick R. Nuclease cleavage of the upstream half of the nontemplate strand DNA in an E. coli transcription elongation complex causes upstream translocation and transcriptional arrest. J Biol Chem. 1997;272:5989–5994. doi: 10.1074/jbc.272.9.5989. [DOI] [PubMed] [Google Scholar]
- 118.Wang D, Landick R. Preferential interaction of the his pause RNA hairpin with RNA polymerase β subunit residues 904-950 correlates with strong transcriptional pausing. Proc Natl Acad Sci USA. 1997;94:8433–8438. doi: 10.1073/pnas.94.16.8433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Wang D, Meier T, Chan C, Feng G, Lee D, Landick R. Discontinuous movements of DNA and RNA in E. coli RNA polymerase accompany formation of a paused transcription complex. Cell. 1995;81:341–350. doi: 10.1016/0092-8674(95)90387-9. [DOI] [PubMed] [Google Scholar]
- 120.Weilbaecher R, Hebron C, Feng G, Landick R. Termination-altering amino acid substitutions in the β′ subunit of Escherichia coli RNA polymerase identify regions involved in RNA chain elongation. Genes Dev. 1994;8:2913–2917. doi: 10.1101/gad.8.23.2913. [DOI] [PubMed] [Google Scholar]
- 121.Wilson K, von Hippel P. Transcription termination at intrinsic terminators: the role of the RNA hairpin. Proc Natl Acad Sci USA. 1995;92:8793–8797. doi: 10.1073/pnas.92.19.8793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Wu C, Wu F, Speckhard D. Subunit location of the intrinsic divalent metal ions in RNA polymerase from Escherichia coli. Biochemistry. 1977;16:5449–5454. doi: 10.1021/bi00644a008. [DOI] [PubMed] [Google Scholar]
- 123.Yager T D, von Hippel P H. Transcript elongation and termination in Escherichia coli. In: Neidhardt F C, Ingraham J L, Low K B, Magasanik B, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella typhimurium: cellular and molecular biology. Washington, D.C: American Society for Microbiology; 1987. pp. 1241–1275. [Google Scholar]
- 124.Yager T D, von Hippel P H. A thermodynamic analysis of RNA transcript elongation and termination in Escherichia coli. Biochemistry. 1991;30:1097–1118. doi: 10.1021/bi00218a032. [DOI] [PubMed] [Google Scholar]
- 125.Yano R, Nomura M. Suppressor analysis of temperature-sensitive mutations of the largest subunit of RNA polymerase I in Saccharomyces cerevisiae: a suppressor gene encodes the second-largest subunit of RNA polymerase I. Mol Cell Biol. 1991;11:754–764. doi: 10.1128/mcb.11.2.754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Young R A. RNA polymerase II. Annu Rev Biochem. 1991;60:689–715. doi: 10.1146/annurev.bi.60.070191.003353. [DOI] [PubMed] [Google Scholar]
- 127.Zaychikov E, Denissova L, Meier T, Gotte M, Heumann H. Influence of Mg2+ and temperature on formation of the transcription bubble. J Biol Chem. 1997;272:2259–2267. doi: 10.1074/jbc.272.4.2259. [DOI] [PubMed] [Google Scholar]
- 128.Zaychikov E, Martin E, Denissova L, Kozlov M, Markovstov V, Kashlev M, Heumann H, Nikiforov V, Goldfarb A, Mustaev A. Mapping of catalytic residues in the RNA polymerase active center. Science. 1996;271:107–109. doi: 10.1126/science.273.5271.107. [DOI] [PubMed] [Google Scholar]
- 129.Zhang Y, Hanna M. NusA changes the conformation of Escherichia coli RNA polymerase at the binding site for the 3′ end of the nascent RNA. J Bacteriol. 1994;176:1787–1789. doi: 10.1128/jb.176.6.1787-1789.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Zheng C, Friedman D. Reduced Rho-dependent transcription termination permits NusA-independent growth of Escherichia coli. Proc Natl Acad Sci USA. 1994;91:7543–7547. doi: 10.1073/pnas.91.16.7543. [DOI] [PMC free article] [PubMed] [Google Scholar]