Abstract
Homologous recombination and post-replication repair facilitate restart of stalled or collapsed replication forks. The SRS2 gene of Saccharomyces cerevisiae encodes a 3′–5′ DNA helicase that functions both in homologous recombination repair and in post-replication repair. This study identifies and characterizes the SRS2 homolog in Neurospora crassa, which we call mus-50. A knockout mutant of N.crassa, mus-50, is sensitive to several DNA-damaging agents and genetic analyses indicate that it is epistatic with mei-3 (RAD51 homolog), mus-11 (RAD52 homolog), mus-48 (RAD55 homolog) and mus-49 (RAD57 homolog), suggesting a role for mus-50 in homologous recombination repair. However, epistasis evidence has presented that MUS50 does not participate in post-replication repair in N.crassa. Also, the N.crassa mus-25 (RAD54 homolog) mus-50 double mutant is viable, which is in contrast to the lethal phenotype of the equivalent rad54 srs2 mutant in S.cerevisiae. Tetrad analysis revealed that mus-50 in combination with mutations in two RecQ homologs, qde-3 and recQ2, is lethal, and this lethality is suppressed by mutation in mei-3, mus-11 or mus-25. Evidence is also presented for the two independent pathways for recovery from camptothecin-induced replication fork arrest: one pathway is dependent on QDE3 and MUS50 and the other pathway is dependent on MUS25 and RECQ2.
INTRODUCTION
Mechanisms of DNA repair are highly conserved from bacteria to higher eukaryotes. Studies in the model organism Saccharomyces cerevisiae have characterized several prototypical DNA repair pathways responsible for repairing specific kinds of damage (1–3). For example, proteins in the RAD3 epistasis group participate in the nucleotide excision repair pathway, which removes ultraviolet (UV)-radiation-induced pyrimidine dimers and other bulky DNA lesions. Proteins in the RAD52 epistasis group participate in homologous recombination (HR) repair of DNA double-strand breaks; and proteins in the RAD6 epistasis group are involved in post-replication repair (PRR), a mechanism for enhancing survival of cells with persistent unrepaired DNA damage.
The S.cerevisiae SRS2 gene encodes a structural homolog of bacterial 3′–5′ helicases UvrD and Rep. An srs2 mutation was first identified as a suppressor of the trimethoprim sensitivity of rad6 and rad18 mutants (4). UV-sensitivity of rad6 and rad18 mutants is also suppressed by srs2 in strains that are proficient in the RAD52 pathway (i.e. recombination repair) (4,5). In addition, srs2 mutations suppress methyl methanesulfonate (MMS)-sensitivity of rad5 and pol30-46 mutants that are deficient in the error-free RAD6- and RAD18-dependent repair pathway (6). A function of the wild-type SRS2 allele may be to suppress HR, because srs2 mutants have a hyper-recombination phenotype (7–9). It has been proposed on the basis of genetic studies that Srs2 facilitates switching from HR to PRR at stalled replication forks. This idea is supported by biochemical data indicating that Srs2 causes the dissociation of Rad51-nucleoprotein filaments from single-stranded DNA, thereby destroying strand-exchange intermediates during HR (10,11). The ability of the Srs2 protein to dislodge Rad51 from DNA appears to be especially important in Rad54- or Sgs1-deficient strains that may produce aberrant HR intermediates; thus, rad54Δ srs2Δ or sgs1Δ srs2Δ mutants are either not viable or grow extremely poorly (12,13). Rad54 is a protein of the SWI2/SNF2 family that promotes Rad51-mediated strand invasion and D-loop formation (14,15), and SGS1 encodes a DNA helicase with the same polarity as Srs2 (16–19). The double mutants rad54Δ srs2Δ or sgs1Δ srs2Δ are inviable. It is likely that this is due to aberrant or inappropriate HR, because they can be suppressed by mutations that block an early step in HR, such as rad51, rad52, rad55 and rad57 (20,21).
The Schizosaccharomyces pombe homolog of SRS2 shares a number of characteristics with S.cerevisiae SRS2, including similar mutant phenotype (i.e. mutagen sensitivity and hyper-recombination) and genetic interactions with rhp54 and rqh1 (homologs of S.cerevisiae RAD54 and SGS1, respectively) (22–24). However, in contrast to S.cerevisiae srs2, S.pombe srs2 does not suppress the UV-sensitivity of PRR mutants and is epistatic with mutants in PRR (24). Although SRS2 homologs have been identified in a number of organisms, they have not yet been reported in higher eukaryotes.
The RecQ helicase family is another conserved group of helicases (25). These helicases have 3′–5′ helicase activity, and RecQ homologs have been identified in numerous species. There is only one RecQ homolog in S.cerevisiae, but unlike Srs2 helicases, multiple RecQ homologs have been identified in higher eukaryotes. For example, four RecQ homologs were predicted from analysis of the complete Caenorhabditis elegans genomic sequence (26), and five human RecQ homologs have been identified, including BLM, WRN and RTS, which are associated with the genetic diseases Bloom syndrome, Werner syndrome and Rothmund–Thomson syndrome, respectively (27–29). These results suggest that RecQ genes have undergone several duplication events during the course of evolution.
The filamentous fungus Neurospora crassa is also a well-characterized model organism that has been used extensively for genetic studies of DNA repair. The DNA sequence of the entire ∼40 Mb N.crassa genome was completed recently (30). The N.crassa genome is approximately three times larger than that of the S.cerevisiae or S.pombe genomes (∼12 Mb each), and it may encode up to twice as many proteins as S.cerevisiae (∼6300) and S.pombe (∼4800). These characteristics suggest that N.crassa may be evolutionarily older than yeast.
Many repair-deficient mutants have been isolated and characterized in N.crassa. Genetic and molecular analyses of these mutants revealed that the uvs-6, mei-3, mus-11, mus-25, mus-48, mus-49, mus-51 and mus-52 genes are involved in recombinational repair and are homologs of S.cerevisiae RAD50, RAD51, RAD52, RAD54, RAD55, RAD57, YKU70 and YKU80, respectively [(31–34); Y. Murayama et al., unpublished data] (Table 1). The N.crassa mus-8, uvs-2 and mus-41 genes participate in PRR and are homologs of S.cerevisiae RAD6, RAD18 and RAD5, respectively [(35,36); A. Kato et al., unpublished data] (Table 1). Database searching identified two putative N.crassa RecQ homologs (qde-3 and recQ2), which were subsequently found to play roles in gene silencing and DNA repair (37–39). QDE3 is a large RecQ helicase with a molecular weight of 216.6 kDa, and RECQ2 is a small RecQ helicase of molecular weight 55.3 kDa. The qde-3 recQ2 double mutant is viable but hypersensitive to DNA-damaging agents. These results suggest that QDE3 and RECQ2 may play complementary roles in DNA repair (39).
Table 1.
Genes related to recombination repair and post-replication repair in N.crassa and S.cerevisiae
| S.cerevisiae | N.crassa |
|---|---|
| Recombination repair genes | |
| RAD50 | uvs-6 |
| RAD51 | mei-3 |
| RAD52 | mus-11 |
| RAD54 | mus-25 |
| RAD55 | mus-48 |
| RAD57 | mus-49 |
| YKU70 | mus-51 |
| YKU80 | mus-52 |
| SGS1 | qde-3 |
| recQ2 | |
| SRS2 | mus-50 |
| Post-replication repair genes | |
| RAD6 | mus-8 |
| RAD18 | uvs-2 |
| RAD5 | mus-41 |
This study characterizes a N.crassa homolog of yeast SRS2 called mus-50 and its interactions with N.crassa RecQ helicases. Mutants of mus-50 are mutagen-sensitive and epistatic to HR proteins including mei-3, mus-11, mus-48 and mus-49. The MMS-sensitivity of PRR-deficient mutants is not suppressed by mus-50. Genetic analysis showed that mus-50 is a synthetic lethal in combination with N.crassa qde-3 and recQ2 owing to aberrant HR. Evidence is also presented for the two independent subpathways for recovery from camptothecin (CPT)-induced replication fork arrest. One pathway is dependent on QDE3 and MUS50, and the other is dependent on MUS25 and RECQ2.
MATERIALS AND METHODS
Strains and plasmids
Neurospora crassa strains used in this study are listed in Table 2. C1-T10-37A and C1-T10-28a are wild-type strains closely related to the standard Oak Ridge wild type (40). S.cerevisiae W303-1A (MATa ade2-1 can1-100 his3-11, 15 leu2-3, 112 trp1-1 ura3-1) was used as a wild type and TH110-1A (MATa ade2-1 can1-100 his3-11, 15 leu2-3, 112 trp1-1 ura3-1 Δsrs2::HIS3) was used as an srs2 mutant. Escherichia coli strains DH-1 and XL-1 Blue were used for the amplification of plasmids. pBluescript SK+ (Stratagene) was used for general DNA manipulation. pCB1003 (41), carrying the E.coli hygromycin B resistance gene driven by the Aspergillus nidulans trpC promoter, was used as a vector in the transformation of N.crassa spheroplasts. pAS2-1 (Clontech) was a yeast expression vector.
Table 2.
Strains of N.crassa used in this study
| Strain/FGSC number | Genotype (allele number or isolation number) | Source/reference |
|---|---|---|
| C1-T10-37A | A | (40) |
| C1-T10-28a | a | (40) |
| FGSC4105A | A pan-2 (B36) | FGSC |
| C2-S2-8a | a pan-2 (OGW1) | Laboratory stock |
| K16-CR4-31A | A mus-50 (RIP1) | This study |
| K16-CR6-2a | a mus-50 (RIP1) | This study |
| K16-CR7-2A | A mus-50 (RIP1) pan-2 (B36) | This study |
| K16-CR12-2a | a mus-50 (RIP1) pan-2 (OGW1) | This study |
| FGSC2764A | A mei-3 | FGSC |
| FGSC6410a | a mus-11 | FGSC |
| YST-F1-5A | A mus-48::Hygr | Laboratory stock |
| YST-S5-9A | A mus-49::Hygr | Laboratory stock |
| FGSC6425A | A mus-25 | FGSC |
| 74OR-270-104a | a uvs-6 al-2 pan-2 cot-1 | (61) |
| FGSC5145a | a mus-8 | FGSC |
| K16-CR49-32a | a uvs-2 (RIP1) | Laboratory stock |
| KTO-R5-O4A | A mus-41 (RIP1) pan-2 (OGW1) | Laboratory stock |
| KTO-r-17A | A qde-3 (RIP1) | (38) |
| KTO-Q2-2A | A recQ2::Hygr | Laboratory stock |
| K16-CT6-2-6a | a mei-3 mus-50 (RIP1) | This study |
| K16-CT11-1-5 a | a mus-11 mus-50 (RIP1) | This study |
| K16-CT27-2-1A | A mus-48::Hygr mus-50 (RIP1) | This study |
| K16-CR33-8a | a mus-49::Hygr mus-50 (RIP1) | This study |
| K16-CR15-12a | a mus-25 mus-50 (RIP1) | This study |
| K16-CT20-2-4a | a uvs-6 mus-50 (RIP1) al-2 pan-2 cot-1 | This study |
| K16-CT3-1-1A | A mus-8 mus-50 (RIP1) | This study |
| K16-CR50-7a | a uvs-2 (RIP1) mus-50 (RIP1) pan-2 (B36) | This study |
| K16-CR9-46A | A mus-41 (RIP1) mus-50 (RIP1) pan-2 (OGW1) | This study |
| K16-CT7-1-1A | A qde-3 (RIP1) mus-50 (RIP1) | This study |
| K16-CB83-5a | a qde-3 (RIP1) mus-50 (RIP1) | This study |
| K16-CT13-1-1a | a recQ2::Hygr mus-50 (RIP1) | This study |
| K16-CR43-12A | A mei3 mus-25 | This study |
| K16-CR42-2A | A mei3 mus-25 mus-50 (RIP1) | This study |
| K16-CR35-11A | A mei-3 recQ2::Hygr mus-50 (RIP1) | This study |
| K16-CR34-17a | a mus-11 recQ2::Hygr mus-50 (RIP1) | This study |
| K16-CR36-10a | a mus-25 recQ2::Hygr mus-50 (RIP1) | This study |
FGSC, Fugal Genetics Stock Center.
DNA and RNA manipulation
Standard molecular techniques were carried out according to Sambrook et al. (42). DNA sequencing was achieved using the ALFexpress sequencer (Amersham Pharmacia Biotech). N.crassa genomic DNA was prepared from mycelia using a procedure described by Irelan et al. (43). N.crassa total RNA was extracted using RNAwiz (Ambion). PCR amplification was carried out with the Expand™ High-Fidelity PCR system (Roche Diagnostics Corp., Basel, Switzerland) according to the manufacturer's protocol. cDNA of mus-50 was cloned by PCR from a N.crassa cDNA library from the Fungal Genetics Stock Center (FGSC).
Gene disruption by Repeat Induced Point mutation
The Repeat Induced Point (RIP) mutation technique was used to disrupt the mus-50 gene (44). DNA fragments of mus-50 were generated by PCR using N.crassa genomic DNA as a template and the following two primers: mus50-5′ (5′-GGCTCAATATAGCTGCTCTTGC-3′) and mus50-3′ (5′-CTGCAGCCTCTTCCATCTTC-3′). PCR cycling conditions were as follows: initial denaturation at 94°C for 2 min, subsequent steps at 94°C for 15 s, annealing at 59°C for 30 s, extension at 72°C for 1.5 min, 10 cycles. Then, 94°C for 15 s, 59°C for 30 s, 72°C for 1.5 min (each cycle at 72°C for 5 s added), 25 cycles, final extension at 72°C for 5 min, hold at 4°C. The PCR product was integrated into pT7Blue (Novagen) to give pT7-mus50. pT7-mus50 was digested with BamHI and XbaI, and ligated to the BamHI- and XbaI-digested pCB1003 to yield the plasmid p1003-mus50. This plasmid was introduced into N.crassa wild-type strain and a hygromycin-resistant transformant was crossed to the wild-type strain to induce RIP mutation. Sequence analysis confirmed numerous GC-to-AT transitions at the mus50 gene locus.
Northern blots
RNA was isolated from germinating conidia, which were cultured at 30°C for 6 h with shaking. To induce gene expression, germinating conidia (1 × 107 ml−1) were irradiated with UV (100 J/m2) and cultured for the indicated time period. The BamHI–XbaI DNA fragment from pT7-mus50 was labeled with 32P using the Multiprime DNA Labeling system (Amersham Pharmacia Biotech) and used as a probe.
General genetic manipulation in N.crassa
Genetic procedures including cross and tetrad dissection were carried out according to the methods of Davis and de Serres (45). Transformation was performed as described by Vollmer and Yanofsky (46) and Tomita et al. (36), except that Trichoderma harzianum lysing enzyme (Sigma) was used as a substitute for Novozym.
Mutagen sensitivity
The survival of UV-irradiated N.crassa cells was measured as described previously (47). To measure the survival rate of hydroxyurea (HU)-, MMS-, CPT- or bleomycin (BLM)-treated N.crassa cells, 1 × 103 conidia were directly mixed with melted agar medium containing various concentrations of HU, MMS, CPT or BLM, and plated on Petri dishes. They were incubated at 30°C for 3 days, and the number of colonies were counted. The survival of mutagen-treated yeast cells was measured by a modification of the method of Prakash and Prakash (48). Stationary cells were washed and resuspended in sterile distilled water. To assay UV-sensitivity, yeast cell suspensions (1 × 106 cells/ml in 67 mM phosphate buffer, pH 7.0) were diluted and spread on the agar medium at a concentration of 300 cells per plate. These plates were UV-irradiated at various doses, and incubated at 30°C for 2 days. To assay MMS-sensitivity, 30 μl of MMS was added to 20 ml cell suspension (1 × 106 cells/ml in 67 mM phosphate buffer, pH 7.0) and incubated at 30°C with mild shaking. At 15 min intervals an aliquot was removed, diluted and plated at a concentration of 300 cells per plate. Plates were incubated at 30°C for 2 days and the number of colonies were counted. All the experiments were repeated at least three times, and standard deviation at each point was calculated.
UV- and MMS-induced reversion assays
For reversion assay, two pantothenic acid-requiring strains, B36 and OGW1, were crossed with mus-50 to make pan-2 (B36) mus-50 and pan-2 (OGW1) mus-50 strains. The pan-2 (B36) and pan-2 (OGW1) alleles carry base substitution and frameshift mutations, respectively. Conidial suspensions (1 × 107 conidia/ml) were irradiated at various doses of UV and cell survival and reversion were measured as follows: (i) a portion of the conidial suspension was diluted to 104 conidia/ml and 0.1 ml samples of the diluted suspension were mixed with the medium supplemented with panthotenate (10 μg/ml) and plated on Petri dishes. Each plate was incubated at 30°C for 3 days and the number of colonies were counted. (ii) To determine the number of pan-2 revertants, 1 ml aliquot of conidial suspensions were mixed with the medium supplemented with panthotenate (0.04 μg/ml) and plated on Petri dishes. The number of revertants was counted and reversion frequency was calculated as the number of revertants per 1 × 107 survivors. MMS-induced reversion was carried out using a similar procedure as follows: conidial suspension (1 × 107 conidia/ml) was diluted to 104 conidia/ml, and 0.03 ml samples were mixed with medium containing various concentrations of MMS supplemented with panthotenate (10 μg/ml). Cells were plated, incubated at 30°C for 3 days and the number of colonies was counted. pan-2 revertants were identified by mixing 1 ml aliquot of the conidial suspensions in medium containing various concentrations of MMS supplemented with panthotenate (0.04 μg/ml). Cells were plated and revertants were counted after 3 days at 30°C. Reversion frequency was calculated as described above.
RESULTS
Identification and characterization of mus-50 mutant
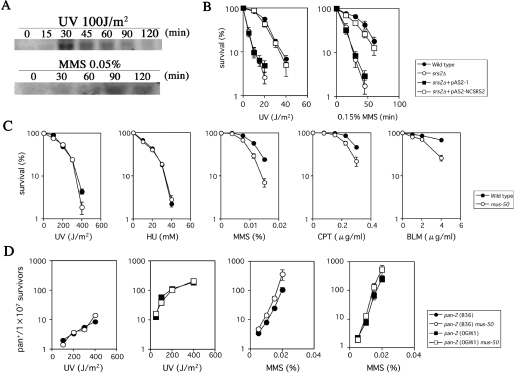
A putative homolog of S.cerevisiae SRS2 was identified by searching the Neurospora genome database (http://www.broad.mit.edu/annotation/fungi/neurospora/). One candidate Neurospora gene, NCU04733.1, was identified, which encodes an 1105 amino acid protein (DDBJ accession no. AB193440) with the seven consensus DNA helicase motifs that are characteristic of DNA helicases, including S.cerevisiae and S.pombe Srs2 (Figure 1A). The mRNA from this gene, which we tentatively call ncsrs2, was expressed at a low level in untreated cells, but induced significantly by UV or MMS (Figure 2A). The ncsrs2 gene was cloned into yeast expression vector pAS2-1 and the resulting plasmid, pAS2-NCSRS2, was introduced and expressed in S.cerevisiae srs2Δ mutant cells. The srs2Δ mutants with vector pAS2-1 or pAS2-NCSRS2 were exposed to UV or MMS and cell survival was evaluated (Figure 2B). The results show that the NCSRS2 expression vector fully complements the DNA damage sensitivity of S.cerevisiae srs2Δ, suggesting that NCSRS2 is functionally similar to yeast Srs2 in vivo.
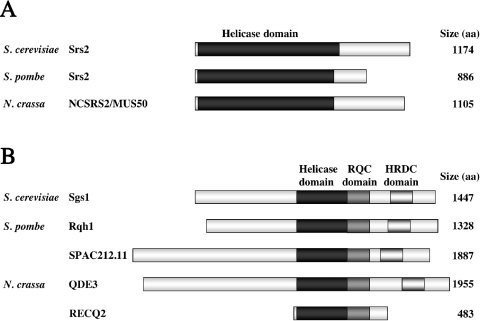
Figure 1.
Schematic alignment of Srs2 and RecQ helicases. The names of species and gene products are shown on the left-hand side, while protein size (number of amino acid residues) is shown on the right-hand side. (A) Schematic alignment of Srs2 helicases. Their conserved helicase domains are shown in black. (B) Schematic alignment of RecQ helicases. Their conserved helicase, RQC and HRDC domains are shown in black, dark gray and light gray, respectively.
Figure 2.
Characterization of mus-50. (A) Northern-blot analyses of mus-50 transcripts. Upper panel: germinating mycelia were irradiated with UV at a dose of 100 J/m2. At indicated time intervals after irradiation, a sample was collected and the level of mus-50 transcripts was analyzed by northern-blot hybridization. Lower panel: germinating mycelia were treated with 0.05% MMS. At indicated intervals after MMS treatment, a sample was collected and the level of mus-50 transcripts was analyzed by northern-blot hybridization. The 28S rRNA was estimated by ethidium bromide staining to confirm equal loading of all samples (data not shown). (B) Complementation of the UV- and MMS-sensitivities of the S.cerevisiae srs2Δ mutant by the N.crassa mus-50 gene. Stationary cells were UV-irradiated at the indicated dose. UV-irradiated cells were diluted and plated at 300 colonies per plate. Plates were incubated at 30°C for 2 days. To assay MMS-sensitivity, 30 μl of MMS was added to 20 ml of yeast suspension. Yeast suspensions were shaken continuously at 30°C for the indicated time period. Dilution, plating and incubation were as described above. Closed circles, wild type; open circles, srs2Δ; closed squares, srs2Δ+pAS2-1; and open squares, srs2Δ+pAS2-NCSRS2. The error bar of each point shows the standard deviation calculated from the data of those independent experiments. (C) Sensitivity to UV, HU, MMS, CPT and BLM of the wild-type and mus-50 strains. A conidial suspension was irradiated with UV at indicated dose or mixed with medium containing HU, MMS, CPT or BLM at indicated concentration. Colonies were counted after incubation at 30°C for 3 days. Closed circles, wild type and open circles, mus-50. The error bar of each point shows the standard deviation calculated from the data of those independent experiments. (D) Comparison of the UV- and MMS-induced pan2 reversion frequency. The B36 and OGW1 alleles of pan-2 have base substitution and frameshift mutations in the pan-2 locus, respectively. The y-axis indicates the number of revertants per 1 × 107 survivors. The x-axis indicates UV dose or MMS concentration. Closed circles, pan-2 (B36); closed squares, pan-2 (OGW1); open squares, pan-2 (B36) mus-50; and open squares, pan-2 (OGW1) mus-50. The error bar of each point shows the standard deviation calculated from the data of those independent experiments.
The function of the ncsrs2 gene in N.crassa was examined using targeted gene disruption by RIP mutation (44). The RIP mutation procedure effectively inactivated ncsrs2 by introducing numerous GC-to-AT transition mutations, as confirmed by DNA sequencing (data not shown). Furthermore, ncsrs2 mRNA was not detected by northern-blot analysis of mRNA from the RIPed strain even in cells exposed to UV irradiation (data not shown). These results indicate that the RIP mutation procedure effectively generated a null allele of ncsrs2. The mutagen sensitivity of this null ncsrs2 mutant was examined using quantitative dose–response survival curves (Figures 2C). The ncsrs2 mutant is sensitive to MMS, CPT and BLM, but it is not sensitive to UV or HU. In contrast, mutants in S.cerevisiae srs2 and S.pombe srs2 are sensitive to all these agents including UV and HU (24,49,50). Following established rules of genetic nomenclature in Neurospora (51), ncsrs2 was renamed mus-50, because it is differentially sensitive to some types of DNA damage. The mutation frequency of the mus-50 mutant is similar to wild-type N.crassa (Figure 2D). In contrast, srs2 mutants suppress UV-induced mutagenesis in S.cerevisiae (49).
Epistasis analyses of mus-50 with recombination repair genes
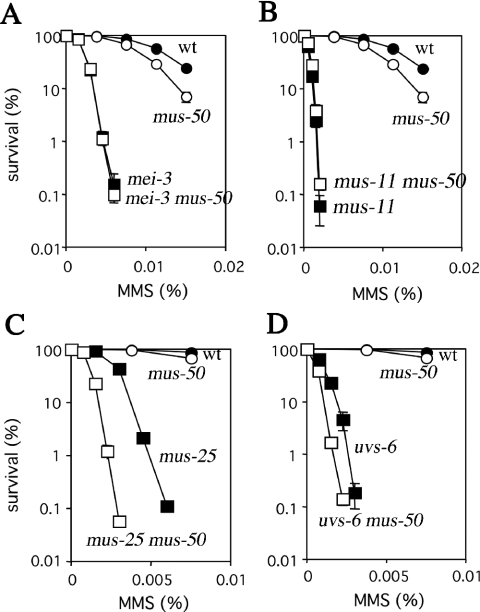
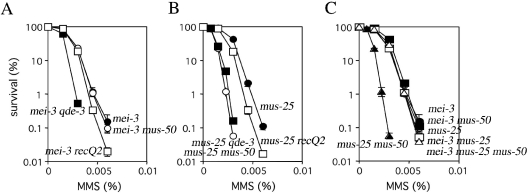
Genetic interactions between mus-50 and recombination repair genes were examined by measuring MMS-sensitivity of single or double mutants carrying mus-50 and mei-3, mus-11, mus-48, mus-49, mus-25 or uvs-6 (Table 1). The MMS-sensitivity of mei-3 and mus-11 was the same in mus-50 wild-type and mus-50 mutant backgrounds (Figure 3A and B). The same genetic effects were seen between mus-50 and mus-48 or mus-49 (data not shown). These results indicate that mus-50 is epistatic with these genes and may participate in the same biological functions as these genes.
Figure 3.
Epistasis analysis of mus-50 and recombination repair defective. Cells were scored for MMS-sensitivity as described in the legend to Figure 2C. The error bar of each point shows the standard deviation calculated from the data of those independent experiments. (A) Closed circles, wild type; open circles, mus-50; closed squares, mei-3; and open squares, mei-3 mus-50. (B) Closed circles, wild type; open circles, mus-50; closed squares, mus-11; and open squares, mus-11 mus-50. (C) Closed circles, wild type; open circles, mus-50; closed squares, mus-25; and open squares, mus-25 mus-50. (D) Closed circles, wild type; open circles, mus-50; closed squares, uvs-6; and open squares, uvs-6 mus-50.
S.cerevisiae Rad54 belongs to the SWI2/SNF2 family of chromatin remodeling proteins (14,15). One possible function of Rad54 might be to alter accessibility to the Rad51-DNA nucleofilament. rad54 is a synthetic lethal with S.cerevisiae and S.pombe srs2, such that S.cerevisiae rad54 srs2 and S.pombe rhp54 srs2 are not viable. However, the N.crassa homolog of rad54, mus-25, is viable as a double mutant with mus-50, and N.crassa mus-25 mus-50 is more sensitive to MMS than either parental single mutant (Figure 3C). This suggests that mus-50 is not epistatic with mus-25 in N.crassa, and that N.crassa may express a novel recombination repair pathway that is independent of MUS25 and MUS50.
Saccharomyces cerevisiae Rad50, Xrs2 and Mre11 proteins function in a complex that protects and processes the termini of DNA breaks and which is required for recombination between sister chromatids (52,53). Mutants in rad50, xrs2 and mre11 grow extremely poorly when combined with srs2Δ (9,21). In contrast, the N.crassa uvs-6 mus-50 double mutant is not defective for growth and is more sensitive to MMS than either parental single mutant (Figure 3D), suggesting that mus-50 is not epistatic with uvs-6.
N.crassa mus-50 does not suppress the mutagen sensitivity of PRR-deficient mutants
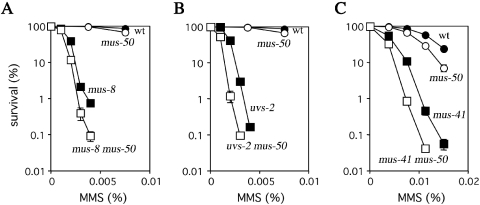
Mutations in srs2 suppress the UV- and MMS-sensitivity of PRR-deficient mutants such as S.cerevisiae rad18 or rad6. In contrast, S.pombe srs2 does not suppress or exacerbate the UV-sensitivity of PRR-deficient mutants (i.e. double mutants have same sensitivity as parental single mutants) (24). N.crassa mus-8 is the homolog of S.cerevisiae RAD6, so the role of mus-50 in PRR was determined by comparing the MMS-sensitivity of N.crassa mus-8, mus-50 and mus-8 mus-50 (Figure 4A). The results showed that double mutant is more sensitive than the mus-8 single mutant, indicating that mus-8 is not epistatic to mus-50. Similarly, two other N.crassa PRR genes, uvs-2 (RAD18 homolog) and mus-41 (RAD5 homolog) are not epistatic to mus-50 (Figure 4B and C). These results suggest that MUS50 is not directly involved in PRR in N.crassa.
Figure 4.
Epistasis analysis of mus-50 and PRR defective. Cells were scored for MMS-sensitivity as described in the legend to Figure 2C. The error bar of each point shows the standard deviation calculated from the data of those independent experiments. (A) Closed circles, wild type; open circles, mus-50; closed squares, mus-8; and open squares, mus-8 mus-50. (B) Closed circles, wild type; open circles, mus-50; closed squares, uvs-2; and open squares, uvs-2 mus-50. (C) Closed circles, wild type; open circles, mus-50; closed squares, mus-41; and open squares, mus-41 mus-50.
mus-50 is a synthetic lethal as a triple mutant with two N.crassa RecQ homologs
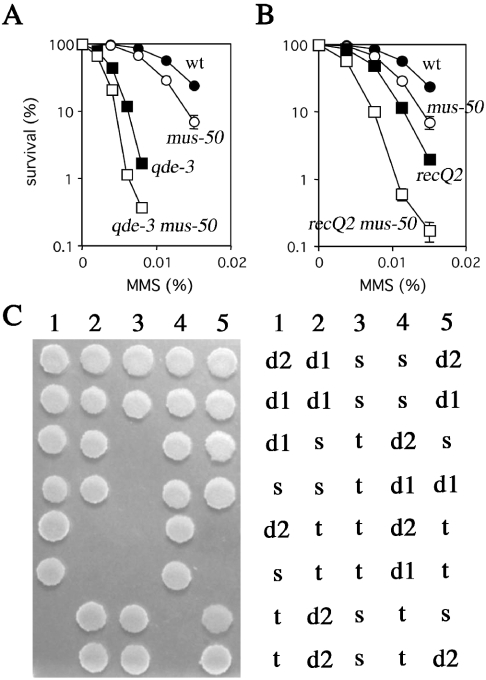
Double mutants sgs1 srs2 in S.cerevisiae and rqh1 srs2 in S.pombe are lethal or poorly viable (13,22). Analogous interaction between the corresponding genes in N.crassa was investigated in this study. In N.crassa, there are two RecQ helicase homologs, qde-3 and recQ2 (37–39) (Figure 1B), and double mutants qde-3 mus-50 and recQ2 mus-50 were constructed and characterized. These double mutants grow normally (data not shown) and are more sensitive to MMS than either parental single mutant (Figure 5A and B). However, qde-3 recQ2 mus-50 triple mutants are lethal, as shown by tetrad dissection after crossing the double mutant strains with each other (Figure 5C). This result suggests that MUS50, QDE3 and RECQ2 play redundant roles in repairing spontaneous DNA damage during vegetative growth.
Figure 5.
Genetic interactions between mus-50 and RecQ helicase mutants. (A) Epistasis analysis of mus-50 and qde-3 mutant. Cells were scored for MMS-sensitivity as described in the legend to Figure 2C. Closed circles, wild type; open circles, mus-50; closed squares, qde-3; and open squares, qde-3 mus-50. The error bar of each point shows the standard deviation calculated from the data of those independent experiments. (B) Epistasis analysis of mus-50 mutant and recQ2 mutant. Cells were scored for MMS-sensitivity as described in the legend to Figure 2C. Closed circles, wild type; open circles, mus-50; closed squares, recQ2; and open squares, recQ2 mus-50. The error bar of each point shows the standard deviation calculated from the data of those independent experiments. (C) Tetrad analysis of spores from a cross between qde-3 mus-50 and recQ2 mus-50 double mutant strains. In the N.crassa eight ascospores are produced in an ascus. After dissected spores were incubated for 1 week at 25°C, spot test was carried out to identify the genotypes of the progeny. mus-50 single mutant (s), qde-3 mus-50 double mutant (d1), recQ2 mus-50 double mutant (d2) and qde-3 recQ2 mus-50 triple mutant (t).
Mutation of mei-3, mus-11 or mus-25 suppresses the lethality of qde-3 recQ2 mus-50
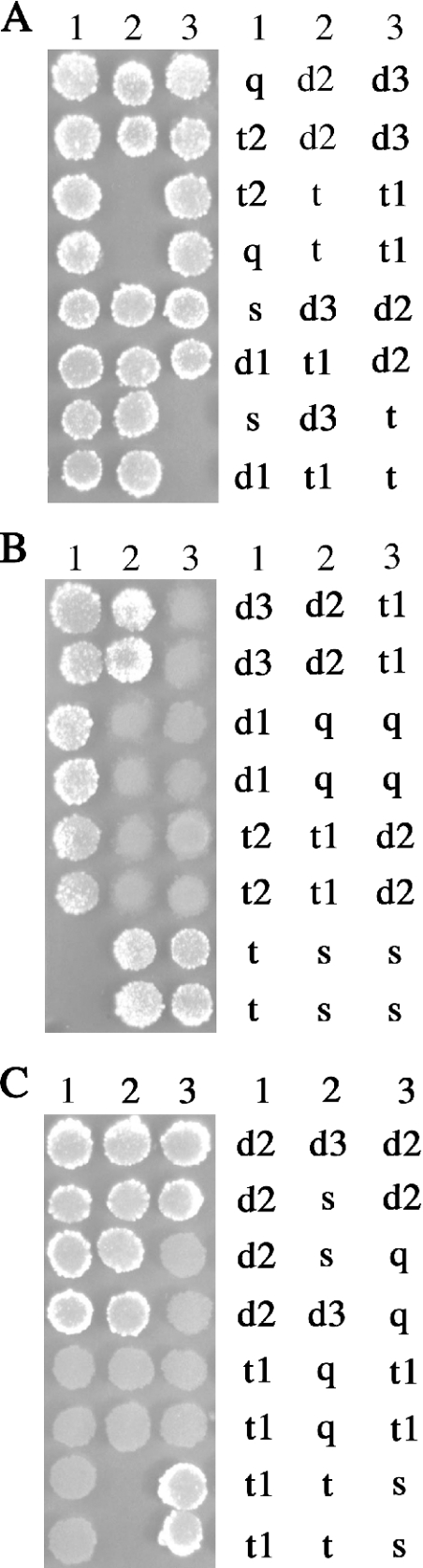
In S.cerevisiae, the lethality of the sgs1Δ srs2Δ double mutant is suppressed by the deletion of RAD51, RAD55 or RAD57, whose functions are in the early stage of HR (20). The possibility of a similar effect in N.crassa was examined by crossing a qde-3 mus-50 mutant to one of the following triple mutants: mei-3 recQ2 mus-50, mus-11 recQ2 mus-50 or mus-25 recQ2 mus-50. Tetrad analyses revealed that mei-3 qde-3 recQ2 mus-50 and mus-11 qde-3 recQ2 mus-50 quadruple mutants are viable, indicating that mei-3 and mus-11 (homologs of S.cerevisiae RAD51 and RAD52, respectively) suppress the synthetic lethality of qde-3 recQ2 mus-50 (Figure 6A and B). Surprisingly, mutations in mus-25 (homolog of S.cerevisiae RAD54), which acts in the later stage of HR, also suppress the synthetic lethality of the qde-3 recQ2 mus-50 triple mutant (Figure 6C). These results suggest that the lethality of qde-3 recQ2 mus-50 triple mutant is due to the formation of irreversible recombination intermediates.
Figure 6.
The synthetic lethality of the qde-3 recQ2 mus-50 triple mutant strain is rescued by the deletion of mei-3, mus-11 or mus-25. (A) Tetrad analysis of spores from a cross between qde-3 mus-50 and mei-3 recQ2 mus-50 mutant strains. mus-50 single mutant (s), qde-3 mus-50 double mutant (d1), recQ2 mus-50 double mutant (d2), mei-3 mus-50 double mutant (d3), qde-3 recQ2 mus-50 triple mutant (t), mei-3 qde-3 mus-50 triple mutant (t1), mei-3 recQ2 mus-50 triple mutant (t2) and mei-3 qde-3 recQ2 mus-50 quadruple mutant (q). (B) Tetrad analysis of spores from a cross between qde-3 mus-50 and mus-11 recQ2 mus-50 mutant strains. mus-50 single mutant (s), qde-3 mus-50 double mutant (d1), recQ2 mus-50 double mutant (d2), mus-11 mus-50 double mutant (d3), qde-3 recQ2 mus-50 triple mutant (t), mus-11 qde-3 mus-50 triple mutant (t1), mus-11 recQ2 mus-50 triple mutant (t2) and mus-11 qde-3 recQ2 mus-50 quadruple mutant (q). (C) Tetrad analysis of spores from a cross between qde-3 mus-50 and mus-25 recQ2 mus-50 mutant strains. mus-50 single mutant (s), recQ2 mus-50 double mutant (d2), mus-25 mus-50 double mutant (d3), qde-3 recQ2 mus-50 triple mutant (t), mus-25 qde-3 mus-50 triple mutant (t1) and mus-25 qde-3 recQ2 mus-50 quadruple mutant (q).
MEI3-dependent HR pathway requires MUS50, QDE3, RECQ2 and MUS25
The above results suggest that MUS50 is involved in MEI3-dependent homologous recombination and that MUS50 is functionally redundant to QDE3 and RECQ2. Previous studies also show that mei-3 is epistatic to mus-25 (33). To investigate whether these helicase proteins function in MEI3-dependent homologous recombination, the genetic relationship between these genes was analyzed in greater detail. Epistasis analysis based on MMS-sensitivity revealed that mei-3 is epistatic to mus-50 but not to qde-3 and recQ2 (Figure 7A). On the other hand, there is no epistatic relationship between mus-25 and mus-50, qde-3 or recQ2 (Figure 7B). In addition, the synergistic effect of mus-25 and mus-50 on MMS-sensitivity was suppressed by a mutation in mei-3, suggesting that MEI3 acts upstream of MUS25 and MUS50 in the repair of MMS-induced DNA damage (Figure 7C). The complexity of these results and epistatic relationships may reflect the fact that MMS induces a number of different biologically relevant types of cellular damage/stress.
Figure 7.
Epistasis analysis of mei-3, mus-25, mus-50, qde-3 and recQ2 for MMS-sensitivity. Cells were scored for MMS-sensitivity as described in the legend to Figure 2C. The error bar of each point shows the standard deviation calculated from the data of those independent experiments. (A) Epistasis analysis of mei-3 and mus-50, qde-3 and recQ2. Closed circles, mei-3; open circles, mei-3 mus-50; closed squares, mei-3 qde-3; and open squares, mei-3 recQ2. (B) Epistasis analysis of mus-25 and mus-50, qde-3 and recQ2. Closed circles, mus-25; open circles, mus-25 mus-50; closed squares, mus-25 qde-3; and open squares, mus-25 recQ2. (C) Epistasis analysis of mei-3, mus-50 and mus-25. Closed circles, mei-3; open circles, mei-3 mus-50; closed squares, mus-25; open squares, mei-3 mus-25; closed triangles, mus-25 mus-50; and open triangles, mei-3 mus-25 mus-50.
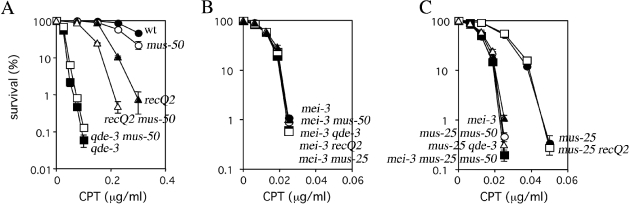
CPT causes type I topoisomerase-mediated single-strand nicks that lead to replication fork collapse (54,55). We investigated genetic interactions between recombination repair genes in the repair of CPT-induced DNA damage. Epistasis analysis revealed that qde-3 but not recQ2 is epistatic to mus-50, suggesting that MUS50 and QDE3 cooperate in the repair of CPT-induced replication fork collapse (Figure 8A). Epistasis analysis between mei-3 and mus-50, qde-3, recQ2 or mus-25 revealed that mei-3 is epistatic to mus-50, qde-3, recQ2 and mus-25, indicating that MUS50, QDE3, RECQ2 and MUS25 function together in a MEI3-dependent pathway for the repair of CPT-induced DNA damage (Figure 8B). The results also show that mus-25 is epistatic to recQ2 but not mus-50 and qde-3 (Figure 8C). Furthermore, the CPT sensitivities of mus-25 mus-50 and mus-25 qde-3 were similar to the mei-3 single mutant (Figure 8C). These observations suggest that MEI3-mediated repair of CPT-induced replication fork collapse has two downstream subpathways: one pathway utilizes MUS50 and QDE3 and the other pathway utilizes RECQ2 and MUS25. This hypothesis is also supported by the observation that the mei-3 mus-25 mus-50 triple mutant is as sensitive to CPT as the mei-3 single mutant (Figure 8C).
Figure 8.
Epistasis analysis of mei-3, mus-25, mus-50, qde-3 and recQ2 for CPT-sensitivity. Cells were scored for CPT-sensitivity as described in the legend to Figure 2C. The error bar of each point shows the standard deviation calculated from the data of those independent experiments. (A) Epistasis analysis of mus-50, qde-3 and recQ2. Closed circles, wild type; open circles, mus-50; closed squares, qde-3; open squares, qde-3 mus-50; closed triangles, recQ2; and open triangles, recQ2 mus-50. (B) Epistasis analysis between mei-3 and mus-50, qde-3, recQ2, mus-25. Closed circles, mei-3; open circles, mei-3 mus-50; closed squares, mei-3 qde-3; open squares, mei-3 recQ2; and open triangles, mei-3 mus-25. (C) Epistasis analysis between mus-25 and mus-50, qde-3, recQ2 or between mei-3 and mus-25 mus-50. Closed circles, mus-25; open circles, mus-25 mus-50; closed squares, mus-25 qde-3; open squares, mus-25 recQ2; closed triangles, mei-3; and open triangles, mei-3 mus-25 mus-50.
DISCUSSION
This report demonstrates that the N.crassa SRS2 homolog mus-50 is functionally redundant to two RecQ family helicases, QDE3 and RECQ2, and that these two RecQ family helicases play different roles in HR in N.crassa. In contrast, S.cerevisiae has only one RecQ helicase, Sgs1. Although Rqh1 has been known as a RecQ homolog in S.pombe, a second RecQ homolog (SPAC212.11) was found and characterized recently (56). However, it has not been analyzed whether the SPAC212.11 is involved in the repair pathway of S.pombe. Therefore, this is the first report of multiple RecQ helicases with diverse recombination functions in a lower eukaryote.
Why is the phenotype of N.crassa mus-50 milder than the phenotype of yeast srs2?
In S.cerevisiae, srs2 mutations suppress the UV- and MMS-sensitivity of PRR-deficient mutants, such as rad6, rad18, rad5, ubc13 and mms2 (4,5,57), in strains that are proficient in recombination repair. The following explanation for these results was proposed: Srs2 interacts with collapsed replication forks and channels them from the HR repair pathway to the PRR pathway, thus preventing completion of aberrant HR events. In S.pombe, Srs2 may play a different role, because it is epistatic with PRR genes based on complementation of UV-sensitivity (24).
N.crassa mus-50 is functionally distinct from S.cerevisiae and S.pombe srs2. In particular, mus-50 is not sensitive to UV or HU and has different genetic interactions with PRR genes in N.crassa. N.crassa mus-50 is not epistatic with mus-8, uvs-2 or mus-41 based on MMS-sensitivity. These results suggest that N.crassa MUS50 is redundant to or has overlapping functions with another N.crassa protein. Since overexpression of Sgs1 partially suppresses some defects of srs2 mutants in S.cerevisiae (58), it seems possible that one of the N.crassa RecQ family helicases might be at least partially redundant in function to N.crassa MUS50. This hypothesis is supported by the following observations: (i) there are two RecQ homologs (qde-3 and recQ2) in N.crassa; (ii) qde-3 recQ2, qde-3 mus-50 and recQ2 mus-50 double mutants are viable; (iii) the qde-3 recQ2 mus-50 triple mutant is not viable; and (iv) the lethality of the triple mutant is suppressed by the mutation of mei-3. These data suggest that QDE3, RECQ2 and MUS50 share some level of redundancy and complement each other for cell viability. It appears likely that the redundant function of these helicases is to resolve collapsed recombination intermediates. If correct, this hypothesis explains the apparent dispensability of mus-50 for PRR in N.crassa, because qde-3 participates in both HR and PRR (38). If Srs2 helicase function is redundant to RecQ helicase activity, this also explains why Srs2 helicases are not conserved in higher eukaryotes that express multiple RecQ helicases.
Two subpathways for MEI3-mediated HR facilitate repair of CPT-induced replication fork collapse
The topoisomerase I inhibitor CPT inhibits topoisomerase I leading to single-strand nicks and eventual replication fork collapse (54,55). The sensitivity of mus-50 mutants to CPT indicates that MUS50 promotes the repair of CPT-induced collapsed replication forks. Epistasis analysis based on CPT-sensitivity demonstrated that there are two downstream subpathways involved in this process: one pathway requires MUS50 and QDE3 and the other pathway requires RECQ2 and MUS25. These results imply that (i) two RecQ helicases function in these two different recombination subpathways and (ii) multiple helicases participate in the MEI3-mediated recombination repair of collapsed replication forks.
The RecQ family is defined by the presence of seven highly conserved amino acid sequence motifs in the so-called helicase domain (25). Just C-terminal to this domain is the RQC (RecQ family C-terminal) domain, which is apparently unique to RecQ helicases and might have a role in directing specific protein–protein interactions. The C-terminal region of RecQ members also contains the HRDC (helicase RNase D C-terminal) domain, which has a probable role in DNA binding (59). QDE3 is a large RecQ helicase, which contains the highly conserved helicase domains, a RQC domain and an HRDC domain. In contrast, RECQ2 is smaller and does not contain an HRDC domain. This structural difference may provide variation in the binding affinities to DNA or protein of QDE3 and RECQ2. Thus, we can propose the hypothesis that QDE3 act in the early stage of MEI3-mediated HR, while RECQ2 may act in a later stage of HR (i.e. during MUS25-promoted D-loop formation and after MEI3 filament-mediated strand invasion). This hypothesis is supported by the fact that mus-25 is epistatic to recQ2 but not to mus-50 and qde-3 and that the mutation of mus-25 suppresses the synthetic lethality of the qde-3 recQ2 mus-50 triple mutant. Differences in the structure and binding activities of QDE3 and RECQ2 may determine their differential functions in MEI3-dependent HR.
Saccharomyces cerevisiae Sgs1 and Srs2 are thought to restrict or suppress HR, because sgs1 and srs2 mutants have a hyper-recombination phenotype and because Srs2 promotes dissociation of the Rad51-nucleoprotein filament (7–11,60). Thus, it is possible that MUS50 and QDE3 share this role in suppressing HR in its early stages and promoting alternate processing of recombination intermediates by other repair pathways. RECQ2 is not likely to function in this manner, since it is thought to act during the later stages of HR. Further analysis of these mutants and proteins will be required to confirm this hypothesis and to explore how the activities of these three helicases are regulated and coordinated.
Acknowledgments
We thank Hideo Shinagawa and Takashi Hishida for providing yeast strains. We thank George R. Hoffmann for his review of this paper. This works was partially supported by Rational Evolutionary Design of Advanced Biomolecules, Saitama Prefecture Collaboration of Regional Entities for the Advancement of Technological Excellence, Japan Science and Technology Agency. Funding to pay the Open Access publication charges for this article was provided by Japan Science and Technology Agency.
Conflict of interest statement. None declared.
REFERENCES
- 1.Friedberg E.C. Deoxyribonucleic acid repair in the yeast Saccharomyces cerevisiae. Microbiol. Rev. 1988;52:70–102. doi: 10.1128/mr.52.1.70-102.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Prakash S., Sung P., Prakash L. DNA repair genes and proteins of Saccharomyces cerevisiae. Annu. Rev. Genet. 1993;27:33–70. doi: 10.1146/annurev.ge.27.120193.000341. [DOI] [PubMed] [Google Scholar]
- 3.Friedberg E.C., Bardwell A.J., Bardwell L., Feaver W.J., Kornberg R.D., Svejstrup J.Q., Tomkinson A.E., Wang Z. Nucleotide excision repair in the yeast Saccharomyces cerevisiae: its relationship to specialized mitotic recombination and RNA polymerase II basal transcription. Philos. Trans. R. Soc. Lond. B Biol. Sci. 1995;347:63–68. doi: 10.1098/rstb.1995.0010. [DOI] [PubMed] [Google Scholar]
- 4.Lawrence C.W., Christensen R.B. Metabolic suppressors of trimethoprim and ultraviolet light sensitivities of Saccharomyces cerevisiae rad6 mutants. J. Bacteriol. 1979;139:866–876. doi: 10.1128/jb.139.3.866-876.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schiestl R.H., Prakash S., Prakash L. The SRS2 suppressor of rad6 mutations of Saccharomyces cerevisiae acts by channeling DNA lesions into the RAD52 DNA repair pathway. Genetics. 1990;124:817–831. doi: 10.1093/genetics/124.4.817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Broomfield S., Xiao W. Suppression of genetic defects within the RAD6 pathway by srs2 is specific for error-free post-replication repair but not for damage-induced mutagenesis. Nucleic Acids Res. 2002;30:732–739. doi: 10.1093/nar/30.3.732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Aguilera A., Klein H.L. Genetic control of intrachromosomal recombination in Saccharomyces cerevisiae. I. Isolation and genetic characterization of hyper-recombination mutations. Genetics. 1988;119:779–790. doi: 10.1093/genetics/119.4.779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Aguilera A., Klein H.L. Genetic and molecular analysis of recombination events in Saccharomyces cerevisiae occurring in the presence of the hyper-recombination mutation hpr1. Genetics. 1989;122:503–517. doi: 10.1093/genetics/122.3.503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rong L., Palladino F., Aguilera A., Klein H.L. The hyper-gene conversion hpr5-1 mutation of Saccharomyces cerevisiae is an allele of the SRS2/RADH gene. Genetics. 1991;127:75–85. doi: 10.1093/genetics/127.1.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Krejci L., Van Komen S., Li Y., Villemain J., Reddy M.S., Klein H., Ellenberger T., Sung P. DNA helicase Srs2 disrupts the Rad51 presynaptic filament. Nature. 2003;423:305–309. doi: 10.1038/nature01577. [DOI] [PubMed] [Google Scholar]
- 11.Veaute X., Jeusset J., Soustelle C., Kowalczykowski S.C., Le Cam E., Fabre F. The Srs2 helicase prevents recombination by disrupting Rad51 nucleoprotein filaments. Nature. 2003;423:309–312. doi: 10.1038/nature01585. [DOI] [PubMed] [Google Scholar]
- 12.Palladino F., Klein H.L. Analysis of mitotic and meiotic defects in Saccharomyces cerevisiae SRS2 DNA helicase mutants. Genetics. 1992;132:23–37. doi: 10.1093/genetics/132.1.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lee S.K., Johnson R.E., Yu S.L., Prakash L., Prakash S. Requirement of yeast SGS1 and SRS2 genes for replication and transcription. Science. 1999;286:2339–2342. doi: 10.1126/science.286.5448.2339. [DOI] [PubMed] [Google Scholar]
- 14.Eisen J.A., Sweder K.S., Hanawalt P.C. Evolution of the SNF2 family of proteins: subfamilies with distinct sequences and functions. Nucleic Acids Res. 1995;23:2715–2723. doi: 10.1093/nar/23.14.2715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tan T.L., Kanaar R., Wyman C. Rad54, a Jack of all trades in homologous recombination. DNA Repair (Amst.) 2003;2:787–794. doi: 10.1016/s1568-7864(03)00070-3. [DOI] [PubMed] [Google Scholar]
- 16.Gangloff S., McDonald J.P., Bendixen C., Arthur L., Rothstein R. The yeast type I topoisomerase Top3 interacts with Sgs1, a DNA helicase homolog: a potential eukaryotic reverse gyrase. Mol. Cell. Biol. 1994;14:8391–8398. doi: 10.1128/mcb.14.12.8391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Watt P.M., Louis E.J., Borts R.H., Hickson I.D. Sgs1: a eukaryotic homolog of E. coli RecQ that interacts with topoisomerase II in vivo and is required for faithful chromosome segregation. Cell. 1995;81:253–260. doi: 10.1016/0092-8674(95)90335-6. [DOI] [PubMed] [Google Scholar]
- 18.Lu J., Mullen J.R., Brill S.J., Kleff S., Romeo A.M., Sternglanz R. Human homologues of yeast helicase. Nature. 1996;383:678–679. doi: 10.1038/383678a0. [DOI] [PubMed] [Google Scholar]
- 19.Bennett R.J., Keck J.L., Wang J.C. Binding specificity determines polarity of DNA unwinding by the Sgs1 protein of S. cerevisiae. J. Mol. Biol. 1999;289:235–248. doi: 10.1006/jmbi.1999.2739. [DOI] [PubMed] [Google Scholar]
- 20.Gangloff S., Soustelle C., Fabre F. Homologous recombination is responsible for cell death in the absence of the Sgs1 and Srs2 helicases. Nature Genet. 2000;25:192–194. doi: 10.1038/76055. [DOI] [PubMed] [Google Scholar]
- 21.Klein H.L. Mutations in recombinational repair and in checkpoint control genes suppress the lethal combination of srs2Δ with other DNA repair genes in Saccharomyces cerevisiae. Genetics. 2001;157:557–565. doi: 10.1093/genetics/157.2.557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang S.W., Goodwin A., Hickson I.D., Norbury C.J. Involvement of Schizosaccharomyces pombe Srs2 in cellular responses to DNA damage. Nucleic Acids Res. 2001;29:2963–2972. doi: 10.1093/nar/29.14.2963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Maftahi M., Hope J.C., Delgado-Cruzata L., Han C.S., Freyer G.A. The severe slow growth of Δsrs2 Δrqh1 in Schizosaccharomyces pombe is suppressed by loss of recombination and checkpoint genes. Nucleic Acids Res. 2002;30:4781–4792. doi: 10.1093/nar/gkf581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Doe C.L., Whitby M.C. The involvement of Srs2 in post-replication repair and homologous recombination in fission yeast. Nucleic Acids Res. 2004;32:1480–1491. doi: 10.1093/nar/gkh317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bachrati C.Z., Hickson I.D. RecQ helicases: suppressors of tumorigenesis and premature aging. Biochem. J. 2003;374:577–606. doi: 10.1042/BJ20030491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jeong Y.S., Kang Y., Lim K.H., Lee M.H., Lee J., Koo H.S. Deficiency of Caenorhabditis elegans RecQ5 homologue reduces life span and increases sensitivity to ionizing radiation. DNA Repair (Amst.) 2003;2:1309–1319. doi: 10.1016/j.dnarep.2003.07.003. [DOI] [PubMed] [Google Scholar]
- 27.Ellis N.A., Groden J., Ye T.Z., Straughen J., Lennon D.J., Ciocci S., Proytcheva M., German J. The Bloom's syndrome gene product is homologous to RecQ helicases. Cell. 1995;83:655–666. doi: 10.1016/0092-8674(95)90105-1. [DOI] [PubMed] [Google Scholar]
- 28.Yu C.E., Oshima J., Fu Y.H., Wijsman E.M., Hisama F., Alisch R., Matthews S., Nakura J., Miki T., Ouais S., et al. Positional cloning of the Werner's syndrome gene. Science. 1996;272:258–262. doi: 10.1126/science.272.5259.258. [DOI] [PubMed] [Google Scholar]
- 29.Kitao S., Shimamoto A., Goto M., Miller R.W., Smithson W.A., Lindor N.M., Furuichi Y. Mutations in RECQL4 cause a subset of cases of Rothmund–Thomson syndrome. Nature Genet. 1999;22:82–84. doi: 10.1038/8788. [DOI] [PubMed] [Google Scholar]
- 30.Galagan J.E., Calvo S.E., Borkovich K.A., Selker E.U., Read N.D., Jaffe D., FitzHugh W., Ma L.J., Smirnov S., Purcell S., et al. The genome sequence of the filamentous fungus Neurospora crassa. Nature. 2003;422:859–868. doi: 10.1038/nature01554. [DOI] [PubMed] [Google Scholar]
- 31.Hatakeyama S., Ishii C., Inoue H. Identification and expression of the Neurospora crassa mei-3 gene which encodes a protein homologous to Rad51 of Saccharomyces cerevisiae. Mol. Gen. Genet. 1995;249:439–446. doi: 10.1007/BF00287106. [DOI] [PubMed] [Google Scholar]
- 32.Sakuraba Y., Schroeder A.L., Ishii C., Inoue H. A Neurospora double-strand-break repair gene, mus-11, encodes a RAD52 homologue and is inducible by mutagens. Mol. Gen. Genet. 2000;264:392–401. doi: 10.1007/s004380000342. [DOI] [PubMed] [Google Scholar]
- 33.Handa N., Noguchi Y., Sakuraba Y., Ballario P., Macino G., Fujimoto N., Ishii C., Inoue H. Characterization of the Neurospora crassa mus-25 mutant: the gene encodes a protein which is homologous to the Saccharomyces cerevisiae Rad54 protein. Mol. Gen. Genet. 2000;264:154–163. doi: 10.1007/s004380000303. [DOI] [PubMed] [Google Scholar]
- 34.Ninomiya Y., Suzuki K., Ishii C., Inoue H. Highly efficient gene replacements in Neurospora strains deficient for nonhomologous end-joining. Proc. Natl Acad. Sci. USA. 2004;101:12248–12253. doi: 10.1073/pnas.0402780101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Soshi T., Sakuraba Y., Kafer E., Inoue H. The mus-8 gene of Neurospora crassa encodes a structural and functional homolog of the Rad6 protein of Saccharomyces cerevisiae. Curr. Genet. 1996;30:224–231. doi: 10.1007/s002940050125. [DOI] [PubMed] [Google Scholar]
- 36.Tomita H., Soshi T., Inoue H. The Neurospora uvs-2 gene encodes a protein which has homology to yeast RAD18, with unique zinc finger motifs. Mol. Gen. Genet. 1993;238:225–233. doi: 10.1007/BF00279551. [DOI] [PubMed] [Google Scholar]
- 37.Cogoni C., Macino G. Posttranscriptional gene silencing in Neurospora by a RecQ DNA helicase. Science. 1999;286:2342–2344. doi: 10.1126/science.286.5448.2342. [DOI] [PubMed] [Google Scholar]
- 38.Kato A., Akamatsu Y., Sakuraba Y., Inoue H. The Neurospora crassa mus-19 gene is identical to the qde-3 gene, which encodes a RecQ homologue and is involved in recombination repair and postreplication repair. Curr. Genet. 2004;45:37–44. doi: 10.1007/s00294-003-0459-3. [DOI] [PubMed] [Google Scholar]
- 39.Pickford A., Braccini L., Macino G., Cogoni C. The QDE-3 homologue RecQ-2 co-operates with QDE-3 in DNA repair in Neurospora crassa. Curr. Genet. 2003;42:220–227. doi: 10.1007/s00294-002-0351-6. [DOI] [PubMed] [Google Scholar]
- 40.Tamaru H., Inoue H. Isolation and characterization of a laccase-derepressed mutant of Neurospora crassa. J. Bacteriol. 1989;171:6288–6293. doi: 10.1128/jb.171.11.6288-6293.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Carroll A.M., Sweigard J.A., Valent B. Improved vectors for selecting resistance to hygromycin. Fungal Genet. Newslett. 1994;41:22. [Google Scholar]
- 42.Sambrook J., Fritsch E.F., Maniatis T.M. Molecular Cloning: A Laboratory Manual, 2nd edn. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 43.Irelan J., Miao V., Selker E.U. Small scale DNA preps for Neurospora crassa. Fungal Genet. Newslett. 1993;40:24. [Google Scholar]
- 44.Selker E.U., Cambareri E.B., Jensen B.C., Haack K.R. Rearrangement of duplicated DNA in specialized cells of Neurospora. Cell. 1987;51:741–752. doi: 10.1016/0092-8674(87)90097-3. [DOI] [PubMed] [Google Scholar]
- 45.Davis R.H., de Serres F.J. Genetic and microbiological research techniques for Neurospora crassa. Methods Enzymol. 1970;17:79–143. [Google Scholar]
- 46.Vollmer S.J., Yanofsky C. Efficient cloning of Neurospora crassa. Proc. Natl Acad. Sci. USA. 1986;83:4869–4873. doi: 10.1073/pnas.83.13.4869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Inoue H., Ishii C. Isolation and characterization of MMS-sensitive mutants of Neurospora crassa. Mutat. Res. 1984;125:185–194. doi: 10.1016/0027-5107(84)90068-x. [DOI] [PubMed] [Google Scholar]
- 48.Prakash L., Prakash S. Isolation and characterization of MMS-sensitive mutants of Saccharomyces cerevisiae. Genetics. 1977;86:33–55. doi: 10.1093/genetics/86.1.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Aboussekhra A., Chanet R., Zgaga Z., Cassier-Chauvat C., Heude M., Fabre F. RADH, a gene of Saccharomyces cerevisiae encoding a putative DNA helicase involved in DNA repair. Characteristics of radH mutants and sequence of the gene. Nucleic Acids Res. 1989;17:7211–7219. doi: 10.1093/nar/17.18.7211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Huang M.E., de Calignon A., Nicolas A., Galibert F. POL32, a subunit of the Saccharomyces cerevisiae DNA polymerase delta, defines a link between DNA replication and the mutagenic bypass repair pathway. Curr. Genet. 2000;38:178–187. doi: 10.1007/s002940000149. [DOI] [PubMed] [Google Scholar]
- 51.Perkins D.D. Neurospora genetic nomenclature. Fungal Genet. Newslett. 1999;46:34–41. [Google Scholar]
- 52.Haber J.E. The many interfaces of Mre11. Cell. 1998;95:583–586. doi: 10.1016/s0092-8674(00)81626-8. [DOI] [PubMed] [Google Scholar]
- 53.Saeki T., Machida I., Nakai S. Genetic control of diploid recovery after gamma-irradiation in the yeast Saccharomyces cerevisiae. Mutat. Res. 1980;73:251–265. doi: 10.1016/0027-5107(80)90192-x. [DOI] [PubMed] [Google Scholar]
- 54.Liu L.F., Duann P., Lin C.T., D'Arpa P., Wu J. Mechanism of action of camptothecin. Ann. N Y Acad. Sci. 1996;803:44–49. doi: 10.1111/j.1749-6632.1996.tb26375.x. [DOI] [PubMed] [Google Scholar]
- 55.Nitiss J.L., Wang J.C. Mechanisms of cell killing by drugs that trap covalent complexes between DNA topoisomerases and DNA. Mol. Pharmacol. 1996;50:1095–1102. [PubMed] [Google Scholar]
- 56.Mandell J.G., Goodrich K.J., Bahler J., Cech T.R. Expression of a RecQ helicase homolog affects progression through crisis in fission yeast lacking telomerase. J. Biol. Chem. 2005;280:5249–5257. doi: 10.1074/jbc.M412756200. [DOI] [PubMed] [Google Scholar]
- 57.Ulrich H.D. The srs2 suppressor of UV sensitivity acts specifically on the RAD5- and MMS2-dependent branch of the RAD6 pathway. Nucleic Acids Res. 2001;29:3487–3494. doi: 10.1093/nar/29.17.3487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Mankouri H.W., Craig T.J., Morgan A. SGS1 is a multicopy suppressor of srs2: functional overlap between DNA helicases. Nucleic Acids Res. 2002;30:1103–1113. doi: 10.1093/nar/30.5.1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Janscak P., Garcia P.L., Hamburger F., Makuta Y., Shiraishi K., Imai Y., Ikeda H., Bickle T.A. Characterization and mutational analysis of the RecQ core of the bloom syndrome protein. J. Mol. Biol. 2003;330:29–42. doi: 10.1016/s0022-2836(03)00534-5. [DOI] [PubMed] [Google Scholar]
- 60.Watt P.M., Hickson I.D., Borts R.H., Louis E.J. SGS1, a homologue of the Bloom's and Werner's syndrome genes, is required for maintenance of genome stability in Saccharomyces cerevisiae. Genetics. 1996;144:935–945. doi: 10.1093/genetics/144.3.935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.de Serres F.J., Inoue H., Schupbach M.E. Mutagenesis at the ad-3A and ad-3B loci in haploid UV-sensitive strains of Neurospora crassa. I. Development of isogenic strains and spontaneous mutability. Mutat. Res. 1980;71:53–65. doi: 10.1016/0027-5107(80)90006-8. [DOI] [PubMed] [Google Scholar]