Abstract
Nucleotide excision repair (NER) is responsible for the removal of a variety of lesions from damaged DNA and proceeds through two subpathways, global repair and transcription-coupled repair. In Escherichia coli, both subpathways require UvrA and UvrB, which are induced following DNA damage as part of the SOS response. We found that elimination of the SOS response either genetically or by treatment with the transcription inhibitor rifampin reduced the efficiency of global repair of the major UV-induced lesion, the cyclobutane pyrimidine dimer (CPD), but had no effect on the global repair of 6-4 photoproducts. Mutants in which the SOS response was constitutively derepressed repaired CPDs more rapidly than did wild-type cells, and this rate was not affected by rifampin. Transcription-coupled repair of CPDs occurred in the absence of SOS induction but was undetectable when the response was expressed constitutively. These results suggest that damage-inducible synthesis of UvrA and UvrB is necessary for efficient repair of CPDs and that the levels of these proteins determine the rate of NER of UV photoproducts. We compare our findings with recent data from eukaryotic systems and suggest that damage-inducible stress responses are generally critical for efficient global repair of certain types of genomic damage.
Exposing DNA to short-wavelength UV light leads to the formation of two major lesions, the cyclobutane pyrimidine dimer (CPD) and the pyrimidine(6-4)pyrimidone photoproduct (6-4PP). These lesions, as well as those caused by other environmental agents, can have deleterious cellular effects if allowed to persist in the genome. Therefore, it is not surprising that cells possess a variety of mechanisms to recognize and remove damage from DNA efficiently. A common cellular process for repairing many types of bulky DNA lesions, including 6-4PPs and CPDs, is nucleotide excision repair (NER), which involves recognition, removal, and resynthesis of the damaged stretch of DNA. This process has been documented in several different organisms, but its molecular mechanism is best understood in the prokaryote Escherichia coli.
NER in E. coli requires six proteins: UvrA, UvrB, UvrC, UvrD, DNA polymerase I, and ligase (11, 37). In vivo, UvrA is present as both a monomer and a dimer, the latter complexing with UvrB to perform initial DNA damage recognition. This UvrA2B heterotrimer may carry out limited, ATP-dependent, processive scanning of the damaged region until the actual damage site is encountered (14). At this point, a conformational change occurs in the protein-DNA complex, leading to release of the UvrA dimer, stable UvrB-DNA binding, and a local bending and unwinding of the damaged region of DNA. UvrC then binds to the UvrB-DNA complex, unmasking the cryptic endonuclease activity of UvrB. In the case of UV photoproducts, this activity causes an incision to be made four bases 3′ to the lesion. A second incision is made by the UvrBC complex seven bases 5′ to the lesion. UvrD, also known as DNA helicase II, releases UvrC and the oligonucleotide between the dual incisions, leaving UvrB at a 12-base gap on one strand. DNA polymerase I fills the gap and dissociates the UvrB protein from the DNA. The repair process is completed by DNA ligase, which seals the nick.
NER can be separated into two subpathways, global repair and transcription-coupled repair. Global repair is the process by which most lesions are repaired regardless of their location in the genome. Transcription-coupled repair is characterized by the more rapid repair of lesions in the transcribed strand of an expressed gene than in the nontranscribed strand or in the rest of the genome (28, 29). In E. coli, both subpathways require the full set of NER proteins, but transcription-coupled repair additionally requires actively transcribing RNA polymerase (RNAP) and at least one additional factor, the transcription repair coupling factor, encoded by the mfd gene (27, 42). The latter factor is thought to recruit Uvr proteins to RNAP arrested at a lesion on the transcribed strand (41), resulting in rapid repair of the transcription-blocking lesion. In mfd mutants, this enhanced repair is abolished and repair of the transcribed strand occurs at the lower but efficient global rate (27).
In E. coli, DNA damage leads to the induction of a set of pleiotropic genes in a process known as the SOS response (46). This stress response is mediated by the RecA and LexA proteins, which operate as a coprotease and a transcriptional repressor, respectively. Upon damage to DNA, a signal (probably single-stranded DNA) that activates the RecA coprotease activity is generated. Activated RecA facilitates the cleavage of LexA repressor, which binds to the operators of several different genes, including uvrA, uvrB, and uvrD. Cleavage of LexA leads to their derepression and results in an increase in the amounts of these proteins in the cell, the extent of which is determined by the persistence of RecA activity and the strength of the inducible promoters (46). In the case of uvrA, uvrB, and uvrD, the derepression is rapid due to the relatively weak binding of LexA to their operator regions (1, 18, 38). The rapid induction of these three NER proteins suggests that they may be needed at higher levels to repair DNA damage efficiently and, ultimately, to allow the cell to recover from genomic injury.
Studies of the relevance of SOS-dependent protein induction to NER have produced conflicting results. Castellazzi et al. (5) showed that recA441 mutants, grown at 41°C to induce the SOS response, had an enhanced UV survival compared to an isogenic recA+ strain under the same conditions. The enhanced survival depended on de novo protein synthesis and the presence of wild-type uvrA. Similarly, Ganesan and Hanawalt (12) found enhanced repair of CPDs when cells possessing a thermolabile LexA repressor were grown at the restrictive temperature. The same study also showed that lexA+ cells with a deletion of recA had a significantly lower rate of CPD repair than an isogenic wild-type strain. These results suggest that the SOS response is required for efficient repair of CPDs and provide evidence for an inducible repair system dependent on wild-type excision repair activity. In contrast, Masek et al. (25) showed that cells carrying the SOS noninducible lexA3(Ind−) allele had only a small reduction in the rate of repair of CPDs induced by UV at 70 J/m2 compared to wild-type cells. Similarly, others have reported no reduction in the rate of CPD removal when cells were treated with the protein synthesis inhibitor chloramphenicol after UV (7, 22).
Our initial observation that treating wild-type cells with the RNAP inhibitor rifampin not only eliminated transcription-coupled repair but also reduced global repair of CPDs led us to undertake a detailed examination of the effects of SOS induction on NER of both CPDs and 6-4PPs. Our results revealed that efficient repair of CPDs in nontranscribed regions of the genome depended on the induction of the SOS response. However, neither transcription-coupled repair of CPDs nor global repair of 6-4PPs required SOS induction. We measured the basal and induced levels of UvrA and UvrB proteins and found that their SOS-dependent induction was coincident with the repair of CPDs. Correlating repair protein levels with the kinetics of both transcription-coupled and global repair of UV lesions illustrated that cells selectively repaired 6-4PPs and transcription-blocking lesions compared to global CPDs when NER capacity was low. We consider this damage recognition hierarchy and the implications of global stress responses for NER in both bacteria and eukaryotic systems.
MATERIALS AND METHODS
Bacterial strains.
E. coli K-12 HL108 is a thyA deoC derivative of W3110 (2). Strain HL942 is a lexA3(Ind−) derivative of HL108. The relevant genotype of GW1010 is recA441 sfiA11 (17). We constructed strain HL940 by P1 transduction of the lexA71(Def) allele (20) from strain JL1470 (obtained from J. W. Little) into GW1010. We obtained strain DM1187 from David Mount via Graham Walker. The relevant genotype of DM1187 is recA441 sfiA11 lexA51(Def) (32).
Cell growth and DNA preparation.
HL108 and HL942 were grown at 37°C in Davis minimal salts supplemented with 0.4% glucose and 10 mg of thymine per ml. To label the DNA, cells were grown in medium containing 1 μCi of [3H]thymine per ml in addition to the nonradioactive thymine. All recA441 strains were grown at 32°C in the above medium additionally supplemented with 0.2% Casamino Acids and 1 mg of vitamin B1 per ml. Cultures were grown to saturation and subcultured in fresh medium supplemented with 1 mM isopropyl-β-d-thiogalactoside (IPTG). Cultures were grown to mid-log phase (approximately 3 × 108 cells/ml) as measured by optical density at 600 nm. Cells were collected by filtration on 0.45-μm-pore-size Millipore filters, washed with prewarmed Davis medium, and resuspended in TE (10 mM Tris, 1 mM EDTA [pH 7.5]) at one-fourth the initial volume of the culture. This suspension was incubated in a shaking water bath for 3 min, during which time the EDTA rendered cells permeable to rifampin (21, 35). Concentrated medium was added to the cells to produce 1× final concentration, and incubation was continued for 10 min. Rifampin (Calbiochem) was dissolved in dimethyl sulfoxide at a concentration of 50 mg/ml and added to the cultures to a final concentration of 50 μg/ml, a dose that inhibits all transcription as measured by [3H]uracil incorporation (data not shown). Control cultures were treated with dimethyl sulfoxide alone. After 10 min of incubation, cells were collected on Millipore filters, washed with warm Davis medium, and resuspended in Davis medium containing 1 mM IPTG. Cells were UV irradiated with a dose of 40 J/m2 and placed in a flask containing growth supplements. Samples of the culture were removed at various times and mixed with an equal volume of ice-cold NET (100 mM NaCl, 10 mM Tris [pH 8.0], 10 mM EDTA) buffer. Cells were pelleted by centrifugation at 4°C and resuspended in TE (pH 8.0). A sample of cells was taken for preparation of cell extracts for Western blotting. The remaining cells were lysed by addition of lysozyme (to 1 mg/ml) and RNase A (to 100 mg/ml) and incubation for 15 min at 37°C. Proteinase K (to 100 mg/ml) and Sarkosyl (to 0.5%) were then added, and the mixture was incubated at 50°C for 1 h. The DNA was extracted with phenol-chloroform and precipitated with 2.5 M ammonium acetate and 2 volumes of 95% ethanol. Purified DNA was resuspended in TE (pH 8.0). A portion of each DNA sample was incubated with SstII and ApaI restriction enzymes as instructed by the manufacturer (Gibco-BRL). The remaining DNA was lightly sonicated in a Branson sonifier, and the concentration was determined by fluorometry using Hoechst 33258 (4). The radioactivity in 3H-labeled DNA was quantified by scintillation spectrometry.
Repair of CPDs in lactose operon.
The frequency of CPDs in the individual strands of the lactose operon was determined by an established method (27, 28). DNA restricted with ApaI and SstII was quantified as described above, and 300 ng of each sample was treated or mock treated with T4 endonuclease V in NET buffer containing 1 mg of bovine serum albumin per ml. Samples were electrophoresed overnight in alkaline gels containing 1% agarose. DNA was transferred to Hybond N+ membranes by Southern blotting and hybridized with 32P-labeled probes. Strand-specific RNA probes were generated according to the protocol of Promega, using plasmid pZH10 (28) as a template. Detection was performed with a Bio-Rad phosphorimager and its associated Molecular Analyst software. The frequency of CPDs per 6.6-kb restriction fragment was calculated from the percentage of fragments with no CPDs (zero class), using the Poisson expression (−ln of the zero class = average number of dimers per fragment).
Immunoassay for global NER.
The repair of CPDs and 6-4PPs was measured by using an immunoassay (19). Following denaturation by boiling, 50 ng (CPD) or 500 ng (6-4PP) of each DNA sample was loaded in triplicate onto a Hybond N+ membrane, using a slot blot apparatus. The membrane was incubated for 2 h in the presence of a mouse antibody against either CPDs (TDM-2) or 6-4PPs (64M-2) diluted 1:2,000 in phosphate-buffered saline (PBS) (antibodies were a generous gift of Toshio Mori [31]). Horseradish peroxidase-conjugated secondary antibodies were used at a dilution of 1:5,000 and detected with an enhanced chemiluminescence detection system (Amersham) and subsequent phosphorimager (Bio-Rad) analysis. Following detection, the amount of 3H-labeled DNA loaded in each slot was confirmed by scintillation counting.
Western blots.
Extracts were made from equal amounts of cells following UV irradiation and incubation (see above) as described previously (36). The cells were washed and concentrated in a small volume of water. A sample of cells was lysed by freeze-thawing, and the amount of DNA was determined by fluorometry as described elsewhere (4). To the remaining cell suspension, an equal volume of 2× sodium dodecyl sulfate (SDS) loading buffer (100 mM Tris [pH 6.8], 200 mM dithiothreitol, 4% SDS, 0.2% bromophenol blue, 20% glycerol) was added, and the mixture was boiled for 5 min. The resulting extracts were vortexed and centrifuged to pellet DNA. Equal volumes of extract were loaded onto SDS–7.5% polyacrylamide gels and electrophoresed, and the proteins were transferred to nitrocellulose membranes. Known amounts of purified UvrA and UvrB (gift of B. Van Houten) were run on each gel and used as standards for protein quantification. Membranes were incubated for 1 h with polyclonal rabbit antibodies against UvrA (1:32,000 dilution in PBS) or UvrB (1:16,000 in PBS) followed by incubation in horseradish peroxidase-conjugated anti-rabbit antibodies (1:10,000 in PBS). Proteins were detected by enhanced chemiluminescence (Amersham), and their amounts were quantified by phosphorimager analysis and associated software.
RESULTS
Inhibition of transcription reduces global repair of CPDs but not 6-4PPs.
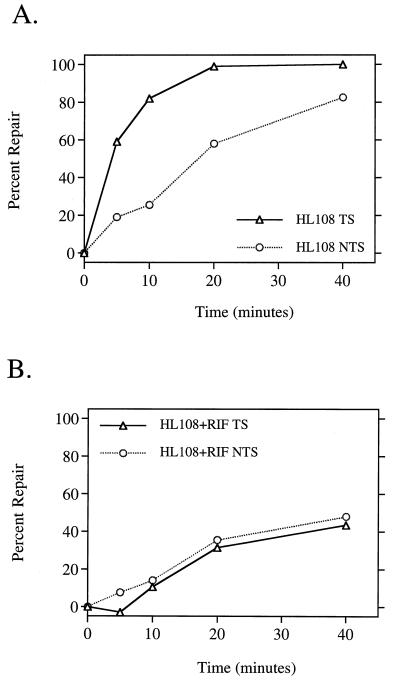
Treatment of EDTA-permeabilized E. coli with the antibiotic rifampin leads to a rapid cessation of both RNA and protein syntheses (34). This drug binds irreversibly near the active center of RNAP, specifically interacting with amino acid residues in the β subunit. This binding locks the RNAP in an abortive initiation cycle, preventing transcription elongation (15). Since transcription-coupled repair requires actively transcribing RNAP, we expected that rifampin treatment would eliminate all repair occurring by this subpathway. Wild-type HL108 cells rapidly removed CPDs from the transcribed strand of the IPTG-induced lactose operon (Fig. 1A), reaching 50% removal in the first 5 min after UV treatment. The nontranscribed strand was repaired more slowly, not reaching 50% repair until 20 min after UV. Treatment of the same cells with rifampin not only eliminated the strand bias as we had expected (Fig. 1B) but also dramatically decreased the rate of repair in both strands to a level significantly below the normal rate of the nontranscribed strand (compare Fig. 1A with Fig. 1B). Since the rate of repair of the nontranscribed strand is an indicator of the rate of global repair, this result suggested that rifampin treatment inhibited not only transcription-coupled repair but also global repair of CPDs. This surprising result led us to perform additional experiments to investigate whether rifampin treatment also affected the efficiency of repair of both CPDs and 6-4PPs throughout the genome.
FIG. 1.
Rifampin inhibits transcription-coupled repair and reduces the efficiency of repair of CPDs in the nontranscribed strand of the lac operon. Quantitative Southern hybridization of strand-specific RNA probes to the IPTG-induced lac operon was performed on DNA isolated from wild-type strain HL108 not treated (A) or treated with 50 μg of rifampin (RIF) per ml (B). The average number of CPDs was measured in each strand of the 6.6-kb restriction fragment in DNA isolated at the indicated times after UV irradiation with 40 J/m2. Each point represents the average repair calculated from three independent experiments. Δ, transcribed strand; ○, nontranscribed strand.
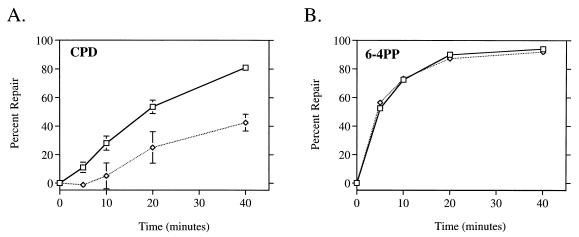
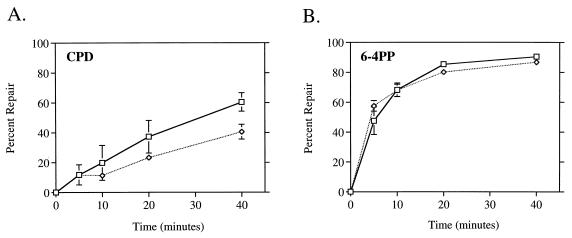
We measured the global repair of both CPDs and 6-4PPs in wild-type cells by using monoclonal antibodies specific for each lesion. Untreated cells removed CPDs at an efficient rate, achieving 50% removal in 20 min and more than 80% after 40 min (Fig. 2A). This global repair rate of CPDs was very similar to the kinetics of nontranscribed strand repair in the lactose operon (Fig. 1A). The same cells treated with rifampin exhibited a significant reduction in the global repair rate of CPDs (Fig. 2A). Only 20% of CPDs were repaired in 20 min, and less than 50% were repaired in 40 min. This result paralleled that obtained from the lactose operon of the rifampin-treated cells (Fig. 1B). Thus, rifampin not only directly eliminated transcription-coupled repair but, as shown by two independent assays, also significantly reduced the rate of global repair of CPDs. In contrast, removal of 6-4PPs was not affected by rifampin treatment (Fig. 2B). Both the rifampin-treated and untreated cells rapidly removed 6-4PPs from their genome, reaching 50% repair within 5 min after UV. No repair of CPDs or 6-4PPs was detected in an isogenic uvrA mutant strain (data not shown).
FIG. 2.
Rifampin inhibits global repair of CPDs but not 6-4PPs. Monoclonal antibodies specific for CPDs (A) and 6-4PPs (B) were used in an immunoassay with DNA isolated at the indicated times after UV irradiation with 40 J/m2. □, HL108 cells; ◊, HL108 cells treated with 50 μg of rifampin per ml. Points represent the average repair calculated from at least two immunoslot blots of samples from each of three independent biological experiments. Each error bar represents 1 standard deviation calculated from the averages of three independent experiments. Error bars not shown are obscured by the datum points.
Constitutive expression of the SOS response eliminates rifampin inhibition of CPD repair.
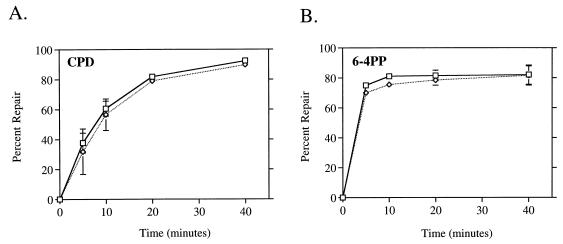
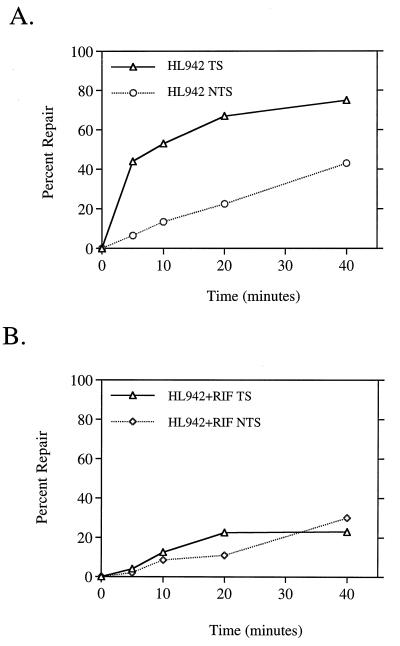
We hypothesized that the inhibitory effect of rifampin on global repair of CPDs was due to the drug’s indirect inhibition of protein synthesis. Since the NER recognition proteins UvrA and UvrB are induced as part of the SOS response following DNA damage, we reasoned that the inhibition of their induction by rifampin might account for the less efficient global repair of CPDs that we observed in treated wild-type cells. To test whether the presence of induced protein levels prior to UV circumvented the effect of rifampin observed in wild-type cells, we used cells that constitutively express high levels of SOS-regulated proteins, including UvrA and UvrB. We analyzed two strains, HL940 and DM1187, in which the SOS response is constitutively derepressed due to the lack of functional LexA repressor (20, 32). We found that global repair of CPDs in DM1187 (and HL940 [data not shown]) occurred more rapidly than in wild-type cells and was not affected by rifampin (Fig. 3A). Rifampin also had no effect on the rate of global repair of 6-4PPs in either lexA(Def) strain (Fig. 3B; HL940 data not shown). These results illustrate that the factor or factors necessary for efficient global repair of CPDs in wild-type cells are induced as part of the SOS response following DNA damage. It also demonstrates that highly efficient repair of both UV photoproducts can be attained even in the absence of actively transcribing RNAP.
FIG. 3.
Constitutive expression of the SOS response results in rapid repair of CPDs and 6-4PPs and eliminates rifampin inhibition of genomic CPD repair. Monoclonal antibodies specific for CPDs (A) and 6-4PPs (B) were used in an immunoassay with DNA isolated from DM1187 lexA51(Def) cells at the indicated times after UV irradiation with 40 J/m2. □, untreated cells; ◊, cells treated with 50 μg of rifampin per ml. Points represent the average repair calculated from at least two immunoslot blots of samples from each of two independent biological experiments. Each error bar represents 1 standard deviation calculated from the averages of two independent experiments. Error bars not shown are obscured by the datum points.
The increase in the rate of global repair of CPDs in cells constitutively expressing the SOS response is also reflected in the rate of repair of CPDs in the nontranscribed strand of the induced lactose operon (Fig. 4A). This rate is not only higher than that for the normal nontranscribed strand (compare to Fig. 1A), but it is also indistinguishable from the rate of repair in the transcribed strand. This result shows that an increase in the efficiency of global repair can mask transcription-coupled repair, eliminating the ability to detect the normal bias for the transcribed strand. In the presence of rifampin, where transcription-coupled repair was directly inhibited, repair of both strands was rapid, with kinetics similar to those in the untreated cells (Fig. 4B). Therefore, even in the absence of any transcription, CPDs in both strands of the lactose operon were efficiently repaired.
FIG. 4.
Constitutive expression of the SOS response leads to rapid repair of CPDs in both strands of the lactose operon regardless of rifampin treatment. Transcription-coupled repair assays were performed on DNA isolated from DM1187 lexA51(Def) cells not treated (A) or treated with 50 μg of rifampin (RIF) per ml (B). For experimental details, see Materials and Methods and the legend to Fig. 1. Points represent the average repair calculated from two independent experiments. Δ, transcribed strand; ○, nontranscribed strand.
Cells unable to induce the SOS response do not repair CPDs efficiently.
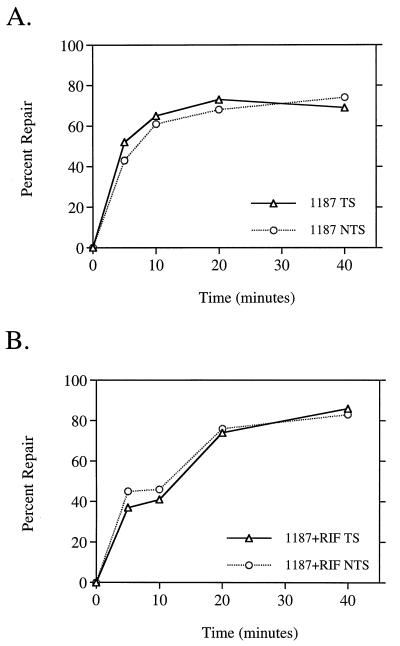
Because the data shown above suggested that the SOS response was necessary for efficient global repair of CPDs, we predicted that mutant cells unable to induce this response would show a lower rate of repair than wild-type cells. E. coli HL942 carries a lexA3(Ind−) mutation and is unable to induce the SOS response after DNA damage due to the presence of a noncleavable LexA repressor (32, 45). We found that these cells did repair CPDs less efficiently than the isogenic lexA+ cells; also, treatment of lexA3(Ind−) cells with rifampin yielded an additional impairment of global repair of CPDs, the resulting rate being almost exactly the same as that found for rifampin-treated wild-type cells (compare Fig. 5A to Fig. 2A). Based on the lack of an effect of rifampin on 6-4PP repair in wild-type cells, we predicted that the repair of these lesions would also be unaffected by the lexA3(Ind−) mutation. Indeed, our data showed the kinetics of global 6-4PP repair in these cells was the same with or without rifampin treatment (Fig. 5B) and exhibited no difference from the wild-type rate (Fig. 2B).
FIG. 5.
Global repair of CPDs, but not 6-4PPs, is attenuated in cells unable to induce the SOS response. Monoclonal antibodies specific for CPDs (A) and 6-4PPs (B) were used in an immunoassay with DNA isolated from HL942 lexA3(Ind−) cells at the indicated times after UV irradiation with 40 J/m2. □, untreated cells; ◊, cells treated with 50 μg of rifampin per ml. Points represent the average repair calculated from at least two immunoslot blots of samples from three independent biological experiments. Each error bar represents 1 standard deviation calculated from the averages of three independent experiments. Error bars not shown are obscured by the datum points.
We wanted to test whether elimination of the SOS response also inhibited transcription-coupled repair of CPDs in the induced lactose operon. We found that lexA3(Ind−) cells did perform transcription-coupled repair, removing 50% of the CPDs from the transcribed strand 5 min after UV (Fig. 6A). However, the rate of repair of the nontranscribed strand was as low as the global repair rate that we measured by the immunoassay (compare to Fig. 5A). Rifampin treatment eliminated the strand bias, and the repair of CPDs occurred at the low rate characteristic of rifampin-treated wild-type cells (Fig. 2A). We conclude from these results that induced levels of one or more of the proteins of the SOS response is required for efficient removal of CPDs from nontranscribed regions of the genome but not for repair of 6-4PPs or for transcription-coupled repair of CPDs.
FIG. 6.
Cells unable to induce the SOS response perform transcription-coupled repair of CPDs but exhibit a reduced rate of CPD repair in the nontranscribed strand in the presence or absence of rifampin. Transcription-coupled repair assays were performed on DNA isolated from HL942 lexA3(Ind−) cells not treated (A) or treated with 50 μg of rifampin (RIF) per ml (B). For experimental details, see Materials and Methods and the legend to Fig. 1. Points represent the average repair calculated from two independent experiments. Δ, transcribed strand; ○ and ⋄, nontranscribed strand.
Uvr protein levels correlate with efficient global repair of CPDs.
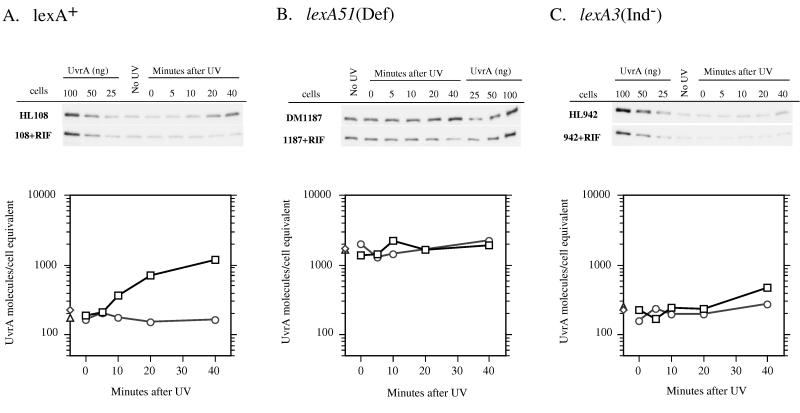
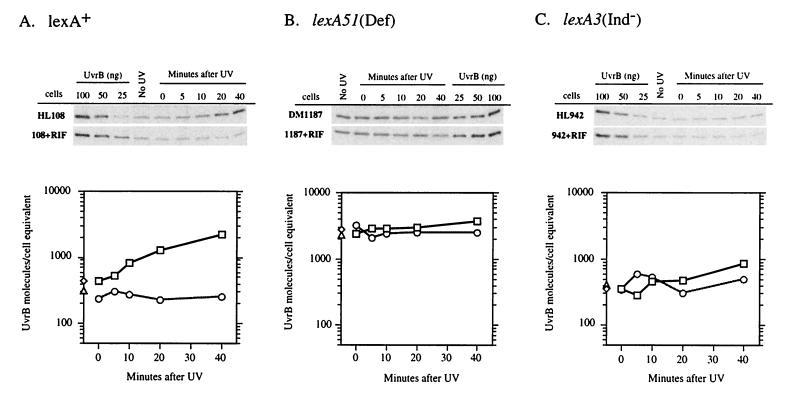
To account for the SOS-dependent variations in global repair of CPDs, our model predicts that one or both of the NER recognition proteins should be significantly induced in wild-type cells within the time period that repair was measured. Previous studies of mRNA levels (24) or β-galactosidase activity of tagged uvrA or uvrB (18, 38) suggested that these are among the most rapidly derepressed of all genes in the SOS response. We performed Western blotting on cell extracts from the lexA+, lexA3(Ind−), and lexA(Def) strains, using polyclonal antibodies raised against UvrA and UvrB (Fig. 7 and 8). We measured the rates of induction and relative amounts of these proteins in each genetic background. We found that lexA+ cells possessed approximately 200 UvrA and 400 UvrB molecules per cell equivalent of DNA (2.1 chromosomes [33]) prior to irradiation. Our Western blot analyses revealed an induction of both proteins after irradiation with a 40 J/m2 dose of UV (Fig. 7A and 8A). The time course of induction was relevant to the repair data, as both UvrA and UvrB were induced twofold in 10 min and over four- and threefold, respectively, in 20 min. Within 40 min, over 1,200 UvrA molecules (sixfold induction) and approximately 2,000 UvrB molecules (fivefold induction) were present per cell equivalent. By comparison, extracts from rifampin-treated lexA+ cells showed no evidence of induction of either protein, and the total number remained at or slightly below the basal level measured in the untreated cells. The lexA(Def) strains exhibited a constitutively high level of both proteins in the presence or absence of rifampin treatment (Fig. 7B and 8B). The agreement between the 1,000 to 2,000 UvrA molecules and 2,000 to 3,000 UvrB molecules per cell equivalent in the lexA(Def) strains and the levels of these proteins after 40 min of induction in wild-type cells suggests that these NER proteins reach maximal induction within 40 min after UV irradiation, coincident with completion of the majority of global repair of UV photoproducts. As expected, the lexA3(Ind−) cell extracts showed very limited induction of both proteins both in the presence and absence of rifampin treatment (Fig. 7C and 8C). We did find, however, that untreated lexA3(Ind−) cells showed a modest accumulation of both UvrA and UvrB and maintained a higher level of these proteins than the rifampin-treated cells.
FIG. 7.
Western blot analyses of UvrA protein levels in HL108 lexA+ (A), DM1187 lexA51(Def) (B), and HL942 lexA3(Ind−) (C) in the presence or absence of rifampin (RIF). Cultures were sampled prior to UV irradiation (No UV) and at the indicated times after UV irradiation with 40 J/m2. Cell extracts were prepared as described in Materials and Methods. Known amounts of purified UvrA protein were loaded onto each gel to generate a standard curve for quantitation. The Western blots shown were generated from phosphorimager scans and associated Molecular Analyst software. Associated graphs depict the average levels of UvrA/cell equivalent prior to irradiation and at each time point as determined from at least three independent experiments. These levels were calculated by using the UvrA standard curve and by measuring the amount of DNA in each cell extract to determine approximate cell number (assuming 2.1 chromosomes/cell). □, UV-irradiated cells (no rifampin); ○, UV-irradiated cells treated with rifampin; ◊, unirradiated cells (no rifampin); Δ, unirradiated cells treated with rifampin.
FIG. 8.
Western blot analyses of UvrB protein levels in HL108 lexA+ (A), DM1187 lexA51(Def) (B), and HL942 lexA3(Ind−) (C) in the presence or absence of rifampin (RIF). Experiments were performed as described in the legend to Fig. 7. □, UV-irradiated cells (no rifampin); ○, UV-irradiated cells treated with rifampin; ◊, unirradiated cells (no rifampin); Δ, unirradiated cells treated with rifampin.
DISCUSSION
Our results demonstrate that the DNA damage-inducible SOS response plays a role in upregulating NER capacity in E. coli. The existence of such an upregulation after DNA damage has been previously postulated for several reasons, including (i) the demonstration that uvrA, uvrB, and uvrD were under Rec/Lex control and were induced rapidly after DNA damage (1, 18, 24, 38); (ii) the dramatic increase in phage (5, 48) and bacterial (5, 12) survival if cells were preinduced by low fluences of UV or by genetic means; (iii) the reduced rate of CPD repair in recA mutants and the subsequent rapid rate of CPD repair when the SOS response was derepressed in the same cells (12); and (iv) the decrease in cell survival when protein synthesis treatments were given immediately following UV (13, 39, 40).
Our results are consistent with these observations and support the hypothesis that efficient NER requires upregulation of the constitutively low level of repair capacity present before UV in wild-type cells. We used a dose of 40 J/m2, which generates approximately 4,000 CPDs and 1,000 6-4PPs in the DNA of an E. coli cell containing two chromosomes (3, 30). We found that uninduced wild-type cells possessed approximately 200 UvrA and 400 UvrB molecules and upregulated these proteins five- to sixfold in the first 40 min following UV irradiation (Fig. 7 and 8). In the same period of time, we measured a high rate of global 6-4PP repair and a lower but efficient rate of global repair of CPDs in these cells (Fig. 2). The relative rates and extents of global repair that we measured agree with previous data obtained by using a similar procedure (19). Treatment of wild-type cells with rifampin prevented the upregulation of repair proteins (Fig. 7A and 8A) and led to a significant decrease in the rate of global CPD removal (Fig. 2A) but had no effect on the repair of 6-4PPs (Fig. 2B). These results are consistent with the idea that 6-4PPs in the DNA preferentially attract the limited resources available for repair under these conditions. Since the 6-4PP is a much better substrate than the CPD for Uvr proteins (44), the limited number of repair complexes are targeted to these lesions, which are then rapidly removed from the genome. Only upon completion of the majority of 6-4PP repair (between 10 and 20 min) is any substantial repair of CPDs observed in the presence of rifampin (compare Fig. 2A and B).
We used two strains carrying different lexA mutations to test our hypothesis that induced levels of repair enzymes are necessary for efficient CPD removal. Strain DM1187 possesses a lexA51(Def) allele that causes this strain to express high levels of SOS-regulated proteins constitutively. These cells possessed 1,000 to 2,000 UvrA and 2,000 to 3,000 UvrB molecules, levels which were equal to or above those that we measured in wild-type cells 40 min after UV (compare Fig. 7A and B with Fig. 8A and B). The fact that these cells not only repaired CPDs rapidly compared to wild-type cells but did so in both the presence and the absence of rifampin (Figure 3) confirms that the effect of rifampin on global repair of CPDs in wild-type cells was due to the inability to upregulate repair protein levels.
Cells with a lexA3(Ind−) mutation are unable to induce the SOS response and therefore should not be able to upregulate cellular NER capacity. We found that these cells had a greatly attenuated induction of both UvrA and UvrB after UV, although they possessed higher levels of both proteins than did cells treated with rifampin (Fig. 7C and 8C). In accordance with our hypothesis, the rate of in these cells was significantly lower than that in the isogenic wild-type cells but not as low as that in either cell type treated with rifampin (Fig. 2 and 5). The higher level of repair proteins in the untreated cells may allow for slightly more efficient global repair of CPDs compared to that in the rifampin-treated cells. The lexA3(Ind−) cells repaired 6-4PPs efficiently in both the presence and the absence of rifampin (Fig. 3B), confirming that these lesions are essentially unaffected by the induction level of the SOS response.
We have confirmed previous results (27, 28) showing rapid repair of CPDs in the transcribed strand of the induced lac operon of wild-type cells (Fig. 1A). By performing both strand-specific and global repair assays on DNA isolated from cells of differing repair capacities, we have also been able to verify that the rate of repair of CPDs in the nontranscribed strand is a consistent, valid estimate of the rate of global repair of CPDs (19, 27).
We found that the ability to detect transcription-coupled repair is affected by the extent to which a cell can perform global repair. The high level of repair proteins present in lexA(Def) cells eliminated the normal bias of repair to the transcribed strand by raising the rate of repair of the nontranscribed strand (Fig. 4A). The lack of an effect of rifampin on repair of either strand (Fig. 4B) suggests that the transcription-coupled repair of CPDs occurring in untreated lexA(Def) cells does not make a significant contribution to the overall removal of CPDs.
The lexA3(Ind−) cells performed transcription-coupled repair of CPDs (Fig. 6A) but had a reduced capacity for global repair. The transcription-coupled repair of CPDs that occurs in these cells may account for the more efficient removal of these lesions that we observed in the untreated cells relative to the rifampin-treated cells (Fig. 5A). This finding suggests that cells unable to induce NER proteins after UV depend on transcription-coupled repair for a larger fraction of total NER of CPDs. Transcription-coupled repair may be a critical process in cells with a limited capacity for global repair because it serves not only to clear transcription-blocking lesions but also to target NER proteins to lesions that would otherwise be poorly recognized. The effect of rifampin on NER of CPDs in wild-type cells may be due to a combination of the inhibition of SOS induction and the elimination of transcription-coupled repair.
Parallel evidence from eukaryotic systems suggests that a suboptimal cellular repair capacity may specifically compromise global repair of CPDs while having little effect on the rate of transcription-coupled repair of CPDs or the global repair of 6-4PPs. The xeroderma pigmentosum group A revertant XP129 exhibited normal repair of 6-4PPs and efficient repair of CPDs in the transcribed strand of the dihydrofolate reductase gene but was completely deficient in the global repair of CPDs (6, 23). Western blots of extracts from these cells revealed that the level of XP-A protein, the major NER recognition protein in mammalian cells, was only 30% of normal (16). In Saccharomyces cerevisiae, leaky or partial-function mutant alleles of the NER genes rad1, rad3, and rad14 showed only a small reduction in the normally high 6-4PP repair rate but exhibited a marked decrease in the rate of CPD repair (26).
Eukaryotic cells may also depend on damage-induced stress responses for efficient global repair of CPDs. Studies of UV-irradiated human fibroblasts homozygous for mutations in the p53 tumor suppressor gene indicated that a p53-dependent response was necessary for efficient global repair of CPDs (9, 10). In normal cells, p53 plays a central role in the cellular responses to DNA damage including cell cycle checkpoints and apoptosis. It is not clear how p53 is involved in NER, but part of its role could be to activate, stabilize, or induce NER proteins. Recent data from other investigators are consistent with these findings (8, 43). Others have shown that irradiation of Saccharomyces cerevisiae with a low dose of UV increases the rate of excision of CPDs from both inactive and active loci following a subsequent, higher dose (47). This enhancement of repair was eliminated by treatment with a protein synthesis inhibitor during incubation between the two doses, suggesting that yeast activate or upregulate the levels of repair proteins in response to DNA damage. The RAD2, RAD7, RAD16, and RAD23 genes all possess inducible promoters and may play a role in the proposed inducible response. Interestingly, the RAD7, RAD16, and RAD23 proteins are involved specifically in the repair of bulky lesions in nontranscribed regions of the genome (11), which our data suggest are the main targets for inducible NER in E. coli.
In conclusion, we have shown that E. coli depends on an inducible response to upregulate NER capacity and remove the major UV-induced lesion, the CPD, from the genome efficiently. Although other SOS-dependent proteins may be necessary for efficient global repair, induction of UvrA and UvrB in wild-type cells is coincident with efficient repair of CPDs. Basal levels of these enzymes are sufficient to repair 6-4PPs and to perform transcription-coupled repair of CPDs.
ACKNOWLEDGMENTS
We appreciate the helpful discussions and critical reading of the manuscript by Ann Ganesan, Justin Courcelle, and C. Allen Smith. We also thank Ben Van Houten for his generous gift of UvrA and UvrB proteins and antibodies and Toshio Mori for providing the CPD and 6-4PP antibodies.
This work was supported by a Cellular and Molecular Biology traineeship GM07276 and Outstanding Investigator grant CA44349 from the National Cancer Institute, NIH.
REFERENCES
- 1.Arthur H A, Eastlake P B. Transcriptional control of the uvrD gene of Escherichia coli. Gene. 1983;25:309–316. doi: 10.1016/0378-1119(83)90235-4. [DOI] [PubMed] [Google Scholar]
- 2.Bachmann B J. Derivations and genotypes of some mutant derivatives of Escherichia coli K-12. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Vol. 2. Washington, D.C: ASM Press; 1996. pp. 2460–2488. [Google Scholar]
- 3.Bohr V A, Smith C A, Okumoto D S, Hanawalt P C. DNA repair in an active gene: removal of pyrimidine dimers from the DHFR gene of CHO cells is much more efficient that in the genome overall. Cell. 1985;40:359–369. doi: 10.1016/0092-8674(85)90150-3. [DOI] [PubMed] [Google Scholar]
- 4.Brunk C F, Jones K C, James T W. Assay for nanogram quantities of DNA in cellular homogenates. Anal Biochem. 1979;92:497–500. doi: 10.1016/0003-2697(79)90690-0. [DOI] [PubMed] [Google Scholar]
- 5.Castellazzi M, Jacques M, George J. tif-stimulated deoxyribonucleic acid repair in Escherichia coli K-12. J Bacteriol. 1980;143:703–709. doi: 10.1128/jb.143.2.703-709.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cleaver J E, Cortes F, Karentz D, Lutze L H, Morgan W H, Player A N, Vuksanovic L, Mitchell D L. The relative biological importance of cyclobutane and (6-4) pyrimidine-pyrimidone dimer photoproducts in human cells: evidence from a xeroderma pigmentosum revertant. Photochem Photobiol. 1988;48:41–49. doi: 10.1111/j.1751-1097.1988.tb02784.x. [DOI] [PubMed] [Google Scholar]
- 7.Cooper P K. Characterization of long patch excision repair of DNA in ultraviolet-irradiated Escherichia coli: an inducible function under Rec-Lex control. Mol Gen Genet. 1982;185:189–197. doi: 10.1007/BF00330785. [DOI] [PubMed] [Google Scholar]
- 8.Eller M S, Maeda T, Magnoni C, Atwal D, Gilchrest B A. Enhancement of DNA repair in human skin cells by thymidine dinucleotides: evidence for a p53-mediated mammalian SOS response. Proc Natl Acad Sci USA. 1997;94:12627–12632. doi: 10.1073/pnas.94.23.12627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ford J M, Hanawalt P C. Expression of wild-type p53 is required for efficient global genomic nucleotide excision repair in UV-irradiated human fibroblasts. J Biol Chem. 1997;272:28073–28080. doi: 10.1074/jbc.272.44.28073. [DOI] [PubMed] [Google Scholar]
- 10.Ford J M, Hanawalt P C. Li-Fraumeni syndrome fibroblasts homozygous for p53 mutations are deficient in global DNA repair but exhibit normal transcription-coupled repair and enhanced UV resistance. Proc Natl Acad Sci USA. 1995;92:8876–8880. doi: 10.1073/pnas.92.19.8876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Friedberg E, Walker G C, Siede W. DNA repair and mutagenesis. Washington, D.C: American Society of Microbiology; 1995. [Google Scholar]
- 12.Ganesan A K, Hanawalt P C. Effect of a lexA41(Ts) mutation on DNA repair in recA(Def) derivatives of Escherichia coli K-12. Mol Gen Genet. 1985;201:387–392. doi: 10.1007/BF00331328. [DOI] [PubMed] [Google Scholar]
- 13.Ganesan A K, Smith K C. Requirement for protein synthesis in rec-dependent repair of deoxyribonucleic acid in Escherichia coli after ultraviolet or X irradiation. J Bacteriol. 1972;111:575–585. doi: 10.1128/jb.111.2.575-585.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Grossman L, Thiagalingam S. Nucleotide excision repair, a tracking mechanism in search of damage. J Biol Chem. 1993;268:16871–16874. [PubMed] [Google Scholar]
- 15.Jin D J, Walter W A, Gross C A. Characterization of the termination phenotypes of rifampicin-resistant mutants. J Mol Biol. 1988;202:245–253. doi: 10.1016/0022-2836(88)90455-x. [DOI] [PubMed] [Google Scholar]
- 16.Jones C J, Cleaver J E, Wood R D. Repair of damaged DNA by extracts from a xeroderma pigmentosum complementation group A revertant and expression of a protein absent in its parental cell line. Nucleic Acids Res. 1992;20:991–995. doi: 10.1093/nar/20.5.991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kenyon C J, Walker G C. DNA-damaging agents stimulate gene expression at specific loci in Escherichia coli. Proc Natl Acad Sci USA. 1980;77:2819–2823. doi: 10.1073/pnas.77.5.2819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kenyon C J, Walker G C. Expression of the E. coli uvrA gene is inducible. Nature. 1981;289:808–810. doi: 10.1038/289808a0. [DOI] [PubMed] [Google Scholar]
- 19.Koehler D, Courcelle J, Hanawalt P. Kinetics of pyrimidine(6-4)pyrimidone photoproduct repair in Escherichia coli. J Bacteriol. 1996;178:1347–1350. doi: 10.1128/jb.178.5.1347-1350.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Krueger J H, Elledge S J, Walker G C. Isolation and characterization of Tn5 insertion mutations in the lexA gene of Escherichia coli. J Bacteriol. 1983;153:1368–1378. doi: 10.1128/jb.153.3.1368-1378.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Leive L. Actinomycin sensitivity in Escherichia coli produced by EDTA. Biochem Biophys Res Commun. 1965;18:13–17. doi: 10.1016/0006-291x(65)90874-0. [DOI] [PubMed] [Google Scholar]
- 22.Lin C G, Kovalsky O, Grossman L. DNA damage-dependent recruitment of nucleotide excision repair and transcription proteins to the Escherichia coli inner membranes. Nucleic Acids Res. 1997;25:3151–3158. doi: 10.1093/nar/25.15.3151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lommel L, Hanawalt P C. Increased UV resistance of a xeroderma pigmentosum revertant cell line is correlated with selective repair of the transcribed strand of an expressed gene. Mol Cell Biol. 1993;13:970–976. doi: 10.1128/mcb.13.2.970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Markham B E, Harper J E, Mount D W, Sancar G B, Sancar A, Rupp W D, Kenyon C J, Walker G C. Analysis of mRNA synthesis following induction of the Escherichia coli SOS system. J Mol Biol. 1984;178:237–248. doi: 10.1016/0022-2836(84)90142-6. [DOI] [PubMed] [Google Scholar]
- 25.Masek F, Skorvaga M, Sedliakova M. Repression of damage-inducible (din) genes by the lexA3 mutation or by plasmid carrying the lexA gene; effect on pyrimidine dimer excision in UV-irradiated Escherichia coli. Gene. 1989;78:195–199. doi: 10.1016/0378-1119(89)90329-6. [DOI] [PubMed] [Google Scholar]
- 26.McCready S. Repair of 6-4 photoproducts and cyclobutane pyrimidine dimers in rad mutants of Saccharomyces cerevisiae. Mutat Res. 1994;315:261–273. doi: 10.1016/0921-8777(94)90037-x. [DOI] [PubMed] [Google Scholar]
- 27.Mellon I, Champe G. Products of DNA mismatch repair genes mutS and mutL are required for transcription-coupled nucleotide-excision repair of the lactose operon in Escherichia coli. Proc Natl Acad Sci USA. 1996;93:1292–1297. doi: 10.1073/pnas.93.3.1292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mellon I, Hanawalt P C. Induction of the Escherichia coli lactose operon selectively increases repair of its transcribed DNA strand. Nature. 1989;342:95–98. doi: 10.1038/342095a0. [DOI] [PubMed] [Google Scholar]
- 29.Mellon I, Spivak G, Hanawalt P C. Selective removal of transcription-blocking DNA damage from the transcribed strand of the mammalian DHFR gene. Cell. 1987;51:241–249. doi: 10.1016/0092-8674(87)90151-6. [DOI] [PubMed] [Google Scholar]
- 30.Mitchell D L, Nairn R S. The biology of the (6-4) photoproduct. Photochem Photobiol. 1989;49:805–819. doi: 10.1111/j.1751-1097.1989.tb05578.x. [DOI] [PubMed] [Google Scholar]
- 31.Mori T, Nakane M, Hattori T, Matsunaga T, Ihara M, Nikaido O. Simultaneous establishment of monoclonal antibodies specific for either cyclobutane dimers and (6-4) photoproducts from the same mouse immunized with ultraviolet-irradiated DNA. Photochem Photobiol. 1991;54:225–232. doi: 10.1111/j.1751-1097.1991.tb02010.x. [DOI] [PubMed] [Google Scholar]
- 32.Mount D W. A mutant of Escherichia coli showing constitutive expression of the lysogenic induction and error-prone DNA repair pathways. Proc Natl Acad Sci USA. 1977;74:300–304. doi: 10.1073/pnas.74.1.300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Neidhardt F C, Umbarger H E. Chemical composition of Escherichia coli. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Vol. 1. Washington, D.C: ASM Press; 1996. pp. 13–16. [Google Scholar]
- 34.Reid P, Speyer J. Rifampicin inhibition of ribonucleic acid and protein synthesis in normal and ethylenediaminetetraacetic acid-treated Escherichia coli. J Bacteriol. 1970;104:376–389. doi: 10.1128/jb.104.1.376-389.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rose J K, Mosteller R D, Yanofsky C. Tryptophan messenger ribonucleic acid elongation rates and steady-state levels of tryptophan operon enzymes under various growth conditions. J Mol Biol. 1970;51:541–550. doi: 10.1016/0022-2836(70)90007-0. [DOI] [PubMed] [Google Scholar]
- 36.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 37.Sancar A. DNA excision repair. Annu Rev Biochem. 1996;65:43–81. doi: 10.1146/annurev.bi.65.070196.000355. [DOI] [PubMed] [Google Scholar]
- 38.Schendel P F, Fogliano M, Strausbaugh L D. Regulation of the Escherichia coli K-12 uvrB operon. J Bacteriol. 1982;150:676–685. doi: 10.1128/jb.150.2.676-685.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sedliakova M, Prachar J, Masek F. Dependence of DNA dark repair on protein synthesis in Escherichia coli. Mol Gen Genet. 1977;153:23–27. doi: 10.1007/BF01035992. [DOI] [PubMed] [Google Scholar]
- 40.Sedliakova M, Slezarikova V, Pirsel M. UV-inducible repair. II. Its role in various defective mutants of Escherichia coli K-12. Mol Gen Genet. 1978;167:209–215. doi: 10.1007/BF00266914. [DOI] [PubMed] [Google Scholar]
- 41.Selby C P, Sancar A. Molecular mechanism of transcription-repair coupling. Science. 1993;260:53–58. doi: 10.1126/science.8465200. [DOI] [PubMed] [Google Scholar]
- 42.Selby C P, Witkin E M, Sancar A. Escherichia coli mfd mutant deficient in “mutation frequency decline” lacks strand-specific repair: in vitro complementation with purified coupling factor. Proc Natl Acad Sci USA. 1991;88:11574–11578. doi: 10.1073/pnas.88.24.11574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Smith M L, Fornace A J. p53-mediated protective responses to UV irradiation. Proc Natl Acad Sci USA. 1997;94:12255–12257. doi: 10.1073/pnas.94.23.12255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Svoboda D L, Smith C A, Taylor J S, Sancar A. Effect of sequence, adduct type, and opposing lesions on the binding and repair of ultraviolet photodamage by DNA photolyase and (A)BC excinuclease. J Biol Chem. 1993;268:10694–10700. [PubMed] [Google Scholar]
- 45.Walker G C. Mutagenesis and inducible responses to deoxyribonucleic acid damage in Escherichia coli. Microbiol Rev. 1984;48:60–93. doi: 10.1128/mr.48.1.60-93.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Walker G C. The SOS response of Escherichia coli. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Vol. 1. Washington, D.C: ASM Press; 1996. pp. 1400–1416. [Google Scholar]
- 47.Waters R, Zhang R, Jones N J. Inducible removal of UV-induced pyrimidine dimers from transcriptionally active and inactive genes of Saccharomyces cerevisiae. Mol Gen Genet. 1993;239:28–32. doi: 10.1007/BF00281597. [DOI] [PubMed] [Google Scholar]
- 48.Weigle J J. Induction of mutation in a bacterial virus. Proc Natl Acad Sci USA. 1953;39:628–636. doi: 10.1073/pnas.39.7.628. [DOI] [PMC free article] [PubMed] [Google Scholar]