ABSTRACT
Observational studies have shown that the gut microbiome is associated with frailty. However, whether these associations underlie causal effects remains unknown. Thus, this study aimed to assess the genetic correlation and causal relationships between the genetically predicted gut microbiome and frailty using linkage disequilibrium score regression (LDSC) and Mendelian Randomization (MR). Summary statistics for the gut microbiome were obtained from a genome-wide association study (GWAS) meta-analysis of the MiBioGen consortium (N = 18,340). Summary statistics for frailty were obtained from a GWAS meta-analysis, including the UK Biobank and TwinGene (N = 175,226). We used LDSC and MR analyses to estimate the genetic correlation and causality between the genetically predicted gut microbiome and frailty. Our findings indicate a suggestive genetic correlation between Christensenellaceae R-7 and frailty. Moreover, we found evidence for suggestive causal effects of twelve genus-level gut microbes on frailty using at least two MR methods. There was no evidence of horizontal pleiotropy or heterogeneity in the MR analysis. This study provides suggestive evidence for a potential genetic correlation and causal association between several genetically predicted gut microbes and frailty. More population-based observational studies and animal experiments are required to clarify this association and the underlying mechanisms.
KEYWORDS: Gut microbes, frailty, genetic correlation, mendelian randomization
Introduction
Frailty is a clinical syndrome related to aging that mainly manifests as a state of nonspecific vulnerability, reduced multisystem physiological reserves, and reduced resistance to stressors1. Frailty is a major public health challenge worldwide. A recent systematic review and meta-analysis involving 62 countries and regions showed that the combined prevalence of physical frailty in older adults was 12%2. The prevalence of frailty has created a serious burden on older adults, families, and society. Several meta-analyses have shown that frailty is associated with an increased risk of all-cause mortality, cause-specific mortality from cardiovascular disease (CVD), cancer, and respiratory illness3–5. In addition, frailty also increases the cost of medical care for older people6, resulting in catastrophic health expenditure7. A recent simulation prediction study in Japan showed that by 2043, it is estimated that 97 billion US dollars will be spent on frail care8. However, there are no specific drugs to prevent and treat frailty, and non-drug interventions such as nutritional interventions are still one of the main means of preventing and treating frailty9.
Although the pathophysiological mechanisms of frailty have not been fully elucidated, current studies identify inflammation as one of the core mechanisms10,11. In recent years, a new hypothesis regarding the origin of inflammation in the digestive tract, especially the imbalance of intestinal homeostasis, has attracted the attention of the academic community12. Previous studies have shown that intestinal ecological imbalance causes the transformation of gut microbiota to pathogenic bacteria and the reduction of microbial diversity13, thus increasing the permeability of the mucosal barrier and allowing bacteria and their products to enter the human body through the intestine, leading to systemic inflammation14–16. Thus, it can be inferred that the gut microbiota may be related to frailty. Some observational studies have preliminarily explored this relationship. A cross-sectional study of older adults in South Korea found that the frailty score was positively correlated with the Bacteroides and negatively correlated with Prevotella 17. A recent systematic review and meta-analysis based on observational studies showed that the species richness index and species diversity index of gut microbiota in frail older adults were significantly lower than those in non-frail18. A systematic review involving 10 case-control and one cohort study found that, compared with healthy older people, frail older adults exhibit decreased gut microbiota diversity and lower abundance of short chain fatty acids (SCFAs) producers19. Studies using mouse models also suggested that changes associated with aging-related disorders and frailty in gut microbiota involved a decrease in SCFAs producers such as Akkermansia 20–23. SCFAs, as epigenetic modifiers of DNA and histone proteins, can participate in regulating age-related aging-associated chronic low-grade inflammation, muscle loss, glucose and lipid metabolism, and other pathophysiological process of frailty24,25.
However, it should be noted that owing to the limitation that observational research mentioned above cannot infer the causal relationship, it is still unknown whether there is a causal relationship between gut microbiota and frailty. Therefore, the exploration of the causal relationship between the two needs to be deepened. In recent years, statistical methods based on genome-wide association study (GWAS) have been proposed to estimate the correlation and causality between traits. Linkage disequilibrium score regression (LDSC) can evaluate the genetic correlation from GWAS summary statistics and is not biased by sample overlap26. Furthermore, Mendelian randomization (MR) has attracted wide attention in the medical field by inferring the causal relationship between variables by means of the instrumental variable of genetic variation. Because genotype precedes phenotype and alleles are randomly assigned at conception, using genetic variation as an instrumental variable to estimate causality can avoid measurement bias, confounding bias, and reverse causality interference27. Therefore, this study assessed the genetic correlation and causal relationships between the genetically predicted gut microbiome and frailty using LDSC and MR.
Materials and methods
Study design
We conducted LDSC and two-sample MR to estimate the genetic correlation and causal relationships between the genetically predicted gut microbiome and frailty. An overview of the study design is presented in Figure 1 (by Figdraw).
Figure 1.
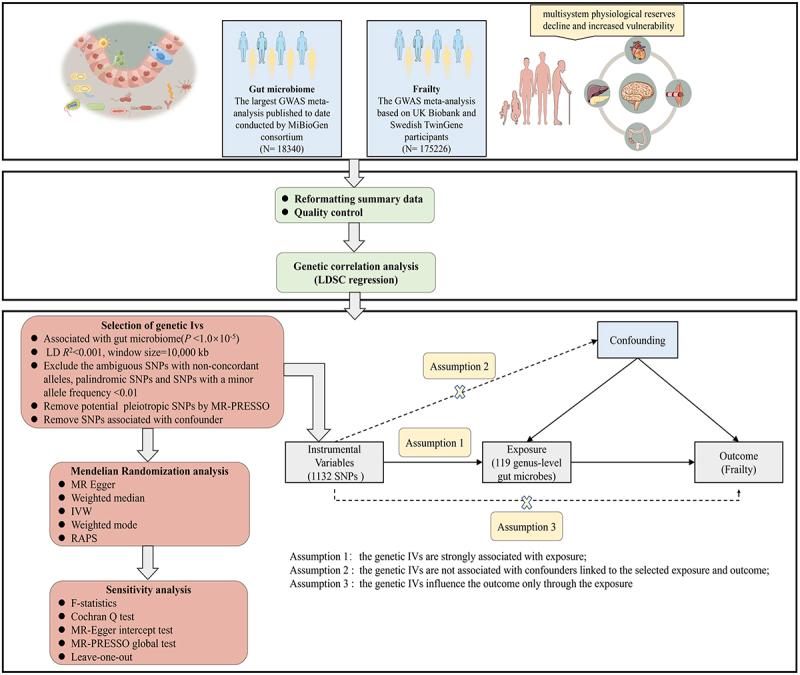
Overview of study design.
Data sources
Genetic variants associated with the gut microbiome were obtained from the largest GWAS meta-analysis published to date conducted by MiBioGen consortium28. This study coordinated 16S rRNA gene sequencing profiles and genotyping data from 18,340 participants from 24 cohorts from the USA, Canada, Israel, South Korea, Germany, Denmark, the Netherlands, Belgium, Sweden, Finland, and the UK. Most of participants had European ancestry (N = 13,266). Among them, genus was the lowest taxonomic level, and 131 genera (including 12 unknown genera) with a mean abundance higher than 1% were identified29.
We downloaded GWAS summary statistics for frailty via the GWAS catalog. This study included 175,226 individuals of European descent (164,610 UK Biobank individuals aged 60–70 years and 10,616 Swedish TwinGene individuals aged 41–87 years) and used the frailty index (FI) to measure frailty30. FI combines dozens of parameters, including symptoms, signs, disease status, and disability, and reflects the accumulation of potential health deficits during the life course31.
Genetic IVs
We performed a series of selection criteria to filter eligible genetic IVs: (1) Since the number of eligible IVs less than the genome-wide significance threshold (p < 5 × 10−8) was extremely small, based on previous studies28,29,32,33, a relatively less stringent threshold (p < 1 × 10−5) was selected to capture potential sets of variants likely to be enriched for association and obtain more comprehensive results34. (2) We conducted a clumping procedure (R2 <0.001, window size = 10,000 kb) to exclude variants in strong linkage disequilibrium (LD) and ensure the independence of each SNP. (3) SNPs with a minor allele frequency of < 0.01, ambiguous SNPs with non-concordant alleles, and palindromic SNPs were excluded. (4) We applied a PhenoScanner35,36 search to identify all known phenotypes associated with genetic IVs (p < 5 × 10−8). If the genetic IV is associated with any other known phenotype, it would be excluded from subsequent MR analysis. We also refer to the largest GWAS published to date, which included 20 dietary habits such as raw vegetable intake, fresh fruit intake, and oily fish intake et al37. We removed genetic IVs associated with the aforementioned 20 dietary habits.
Statistical analysis
Genetic correlation analysis
We estimated the genetic correlation(rg) between gut microbiota and frailty using LDSC. GWAS summary statistics were filtered according to HapMap3 ref. Variants that were not SNPs (e.g., indels) and SNPs that were strand-ambiguous, repeated, and had a minor allele frequency (MAF) <0.01 were excluded. The LDSC examines the association between test statistics and linkage disequilibrium to quantify the contribution of inflation from a true polygenic signal or bias38. This method can evaluate genetic correlation from GWAS summary statistics and is not biased by sample overlap26. The z-scores of each variant from Trait 1 are multiplied by the z-scores of each variant from Trait 2. The genetic covariance was estimated by regressing this product against the LD score39. The genetic covariance normalized by SNP-heritability represents the genetic correlation. p < 0.0004 (0.05/119, after strict Bonferroni correction) was considered statistically significant. 0.0004 < p < 0.05 was considered to be suggestive evidence for potential genetic correlation.
MR analysis
Before the MR analysis, we calculated the F-statistic of the microbiome IVs to determine whether there was a weak IV bias40. An F-statistic <10 indicates a weak IV bias41. The formula for F-statistics is shown in Figure 2.
Figure 2.
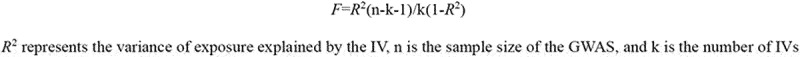
The formula for F statistics.
Note: *p < 0.05
In this study, we explored the causal relationship between the gut microbiome and frailty using five methods: inverse variance weighted (IVW), MR-Egger, weighted median, weighted mode, and Robust Adjusted Profile Score (RAPS). The IVW method is considered the most accurate and powerful method for estimating causal effects when all selected SNPs are valid IVs42.
A consistent casual effect of the gut microbiome on frailty across several methods could be more reliable33,43. In our study, the causal effect with an adjusted p < 0.0004 (0.05/119, after strict Bonferroni correction) in at least two analysis methods was considered significant. In at least two analysis methods, 0.0004 < p < 0.05, were considered to be suggestive evidence for potential causality. We applied the MR-Egger method to detect horizontal pleiotropy44. If pleiotropy was present, the analysis yielded an intercept of p < 0.05. The Cochran Q test was used to assess heterogeneity. Outlier variants and potential horizontal pleiotropy were assessed using the MR-PRESSO method45. We eliminated outliers based on this.
All statistical analyses were performed using the LDSC Version 1.0.1, “TwoSampleMR” package27 and the “MR-PRESSO” package in R version 4.2.0.
Results
LDSC regression analysis
We performed LDSC regression analysis to evaluate the genetic correlation between 119 genus-level gut microbes and frailty. Owing to limitations such as low heritability and sample size, some genera cannot be used for the above analysis. Finally, we obtained the estimations of genetic correlation between the 61 genera and frailty. As shown in Table 1 and Figure 3, LDSC showed a suggestive correlation between Christensenellaceae R-7 and frailty (rg= −0.212, p = 0.047). Detailed information regarding all genetic correlation results is listed in Table S1.
Table 1.
The genetic correlations between gut microbes and frailty.
| Trait1 | Trait2 | rg | SE | P-Value |
|---|---|---|---|---|
| Christensenellaceae R-7 | Frailty | −0.212 | 0.107 | 0.047 |
Figure 3.
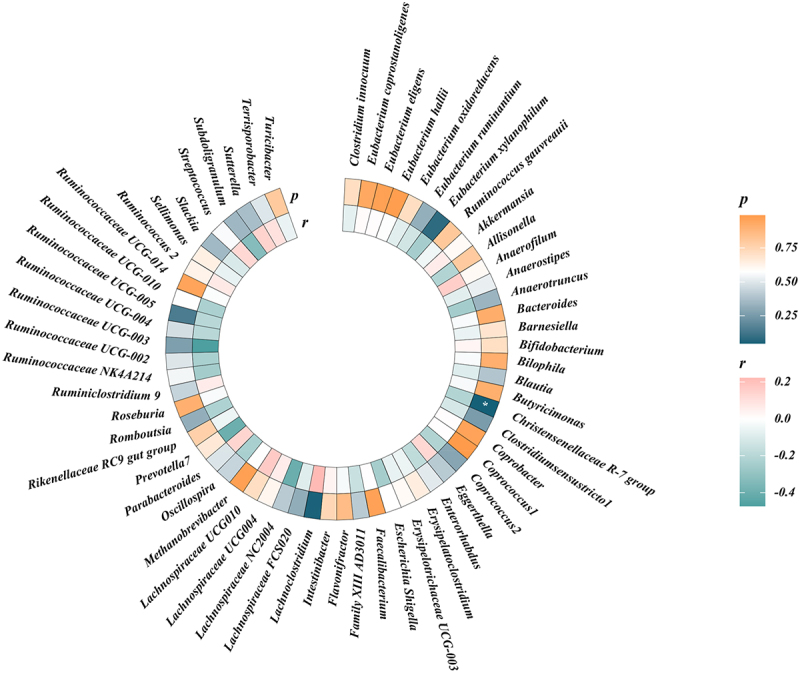
Circular heat map of suggestive genetic correlation between gut microbes and frailty.
MR analysis
According to the criteria of screening, 1132 SNPs were selected as IVs for 119 genus-level gut microbes. The F statistics of all selected IVs were > 10, indicating a small possibility of weak instrument bias. Details of all the selected IVs are shown in Table S2. 12 genus-level gut microbes were examined to have suggestive associations with frailty in at least two MR methods (Table 2 and Figures 4, 5, 6). Scatter plots displayed the associations of the SNP effects on 12 genus-level gut microbes against the SNP effects on frailty. The results of the estimates of causal associations between 119 genera and frailty are presented in Table S3.
Table 2.
Significant MR results of causal association between gut microbes and frailty.
| Exposure | Method | N.snp | beta | 95%CI | p |
|---|---|---|---|---|---|
| Clostridium innocuum | Inverse variance weighted | 9 | 0.023 | (0.001,0.044) | 0.036 |
| Clostridium innocuum | RAPS | 9 | 0.024 | (0.001,0.046) | 0.043 |
| Eubacterium coprostanoligenes | Weighted median | 13 | 0.068 | (0.020,0.116) | 0.006 |
| Eubacterium coprostanoligenes | Inverse variance weighted | 13 | 0.054 | (0.019,0.090) | 0.003 |
| Eubacterium coprostanoligenes | RAPS | 13 | 0.055 | (0.016,0.094) | 0.005 |
| Eubacterium ruminantium | Inverse variance weighted | 18 | −0.027 | (−0.051,-0.003) | 0.028 |
| Eubacterium ruminantium | RAPS | 18 | −0.029 | (−0.050,-0.008) | 0.007 |
| Akkermansia | Inverse variance weighted | 10 | −0.042 | (−0.074,-0.011) | 0.009 |
| Akkermansia | RAPS | 10 | −0.044 | (−0.078,-0.010) | 0.011 |
| Bifidobacterium | Inverse variance weighted | 11 | 0.044 | (0.010,0.079) | 0.012 |
| Bifidobacterium | RAPS | 11 | 0.045 | (0.009,0.082) | 0.015 |
| Butyrivibrio | Inverse variance weighted | 13 | −0.019 | (−0.035,-0.003) | 0.020 |
| Butyrivibrio | RAPS | 13 | −0.020 | (−0.037,-0.003) | 0.021 |
| Catenibacterium | Inverse variance weighted | 4 | −0.031 | (−0.058,-0.003) | 0.030 |
| Catenibacterium | RAPS | 4 | −0.031 | (−0.061,-0.001) | 0.046 |
| Christensenellaceae R-7 group | Inverse variance weighted | 7 | −0.059 | (−0.107,-0.011) | 0.017 |
| Christensenellaceae R-7 group | RAPS | 7 | −0.061 | (−0.113,-0.008) | 0.023 |
| Defluviitaleaceae UCG-011 | Weighted median | 8 | −0.042 | (−0.082,-0.002) | 0.039 |
| Defluviitaleaceae UCG-011 | Inverse variance weighted | 8 | −0.034 | (−0.063,-0.005) | 0.024 |
| Defluviitaleaceae UCG-011 | RAPS | 8 | −0.035 | (−0.067,-0.004) | 0.027 |
| Howardella | Weighted median | 9 | 0.027 | (0.001,0.053) | 0.047 |
| Howardella | Inverse variance weighted | 9 | 0.024 | (0.004,0.045) | 0.019 |
| Howardella | RAPS | 9 | 0.025 | (0.003,0.046) | 0.028 |
| Ruminococcus 1 | Weighted median | 9 | 0.069 | (0.016,0.122) | 0.011 |
| Ruminococcus 1 | Inverse variance weighted | 9 | 0.054 | (0.013,0.096) | 0.010 |
| Ruminococcus 1 | RAPS | 9 | 0.057 | (0.016,0.098) | 0.007 |
| Allisonella | Inverse variance weighted | 8 | 0.032 | (0.007,0.057) | 0.012 |
| Allisonella | RAPS | 8 | 0.034 | (0.013,0.055) | 0.001 |
Figure 4.
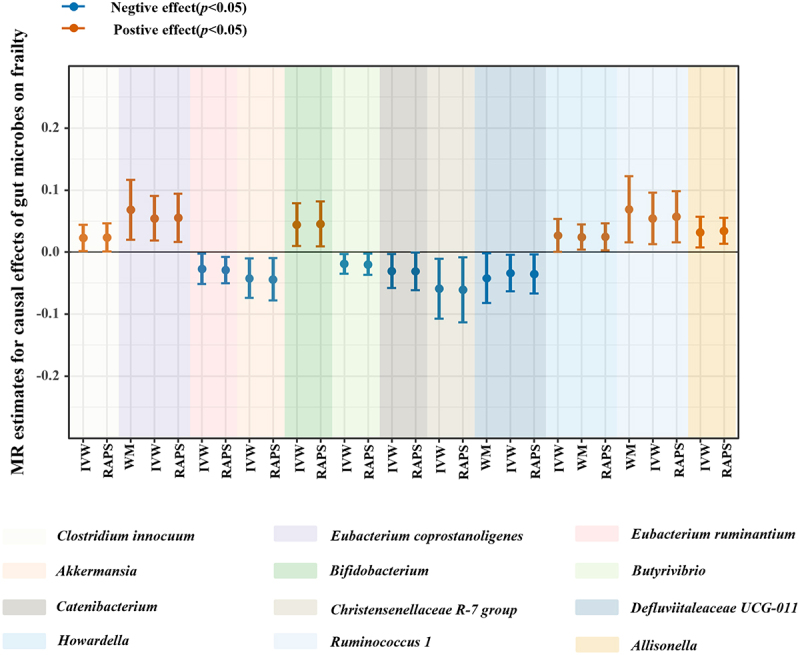
Suggestive causal effects of 12 genus-level gut microbes on frailty.
Figure 5.
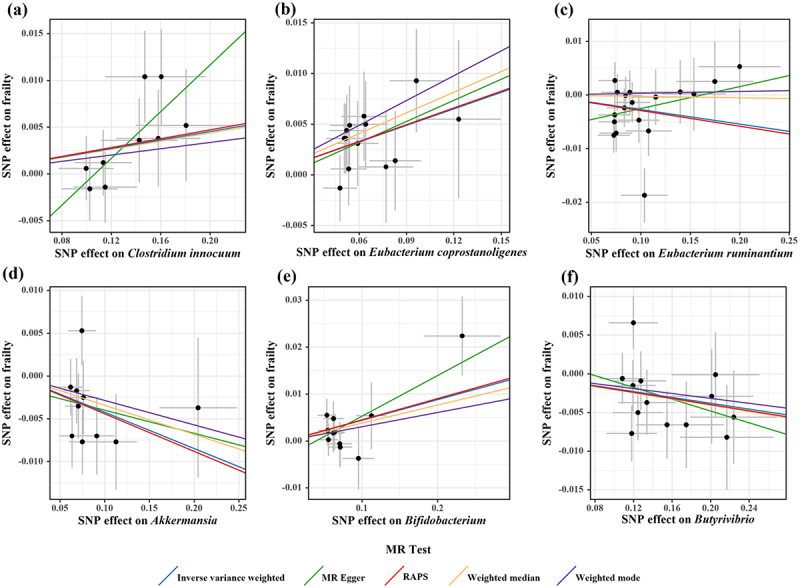
Scatter plots for causal effects of gut microbes on frailty.
Note: (a) Clostridium innocuum-frailty (b) Eubacterium coprostanoligenes-frailty (c) Eubacterium ruminantium-frailty (d) Akkermansia-frailty (e) Bifidobacterium-frailty(f) Butyrivibrio-frailty
Figure 6.
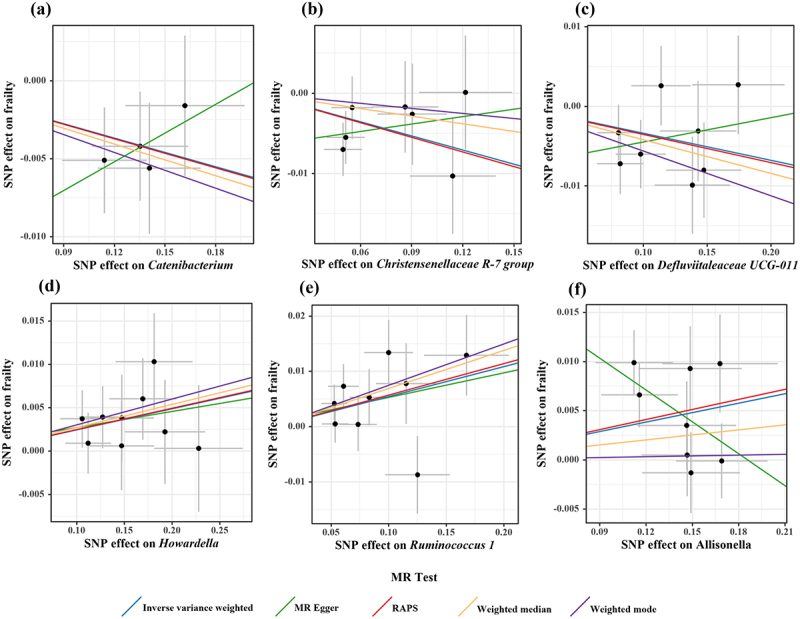
Scatter plots for causal effects of gut microbes on frailty.
Note: (a) Catenibacterium-frailty (b) Christensenellaceae R-7 group-frailty (c) Defluviitaleaceae UCG-011-frailty (d) Howardella-frailty (e) Ruminococcus 1-frailty(f) Allisonella-frailty
Sensitivity analysis
Through visual inspection of the scatter plot, there were potential outliers of the IVs of Eubacterium ruminantium, Akkermansia, Butyrivibrio, Defluviitaleaceae UCG-011, Ruminococcus 1, and Allisonella. However, according to the results of the radial MR-Egger intercept and MR-PRESSO global tests, there was no evidence of horizontal pleiotropy (Table S5). In addition, no significant heterogeneity was found in the Cochran’s Q test (Table S4).
Discussion
To the best of our knowledge, this is the first study to explore the genetic correlation and potential causality between the gut microbes and frailty by using GWAS summary statistics. Our findings indicate a suggestive genetic correlation between Christensenellaceae R-7 and frailty. Moreover, we found evidence for suggestive causal effects of twelve genus-level gut microbes on frailty in the MR analysis. These results will help us further explore the role of the gut microbes in aging and provide references for the development of future interventions and potential therapeutic targets.
Eubacterium coprostanoligenes are known to be associated with cholesterol metabolism46. Early animal experiments found that feeding Eubacterium coprostanoligenes significantly decreased blood cholesterol concentrations in mice47 and rabbits48. A recent intervention experiment in mice also found that supplementation with a mixture of Opuntia ficus-indica, Theobroma cacao, and Acheta domesticus increased the abundance of Eubacterium coprostanoligenes in obese mice, thereby reducing serum cholesterol levels49. Meanwhile, a large-scale study based on the UK Biobank found that lower levels of total, low-density lipoprotein (LDL), and high-density lipoprotein (HDL) cholesterol were associated with a higher prevalence of frailty50. It can be deduced from this that the higher the abundance of Eubacterium coprostanoligenes, the lower cholesterol level may be, which may increase the risk of frailty. However, it is worth noting that there is not yet sufficient evidence to support the association between Eubacterium coprostanoligenes and lower blood cholesterol concentrations in humans. Considering the racial differences between humans and animals such as rabbits and mice, future studies need to further explore whether the positive effect of Eubacterium coprostanoligenes on frailty is mediated by cholesterol concentration in human subjects.
Allisonella was associated with decreased bone mineral density and bone metabolic indicators in postmenopausal women51. Additionally, it was more abundant in individuals with a high inflammatory index52. Interleukin 6 (IL-6) and tumor necrosis factor-alpha (TNF-α) have been recognized as the biomarkers of frailty53,54. The proinflammatory properties of Allisonella seem to be a possible explanation for its suggestive association with frailty. In addition, this study found that a higher abundance of Bifidobacterium may increase the risk of frailty, similar to a previous observational study18. Bifidobacterium is considered to be a physiologically beneficial bacterium that can maintain intestinal homeostasis, regulate immune function, and reduce the growth of harmful bacteria55. Surprisingly, an increased abundance of Bifidobacterium was found in both Parkinson’s patients56 and frail older people18. A previous case-control study also found a significantly higher proportion of Bifidobacterium in patients with active inflammatory bowel than in healthy controls57. Given that direct evidence is currently lacking, further exploration of the relationship between Bifidobacterium and frailty is warranted in the future.
Moreover, researchers analyzed the gut microbiota characterization of 29 subjects using Illumina MiSeq sequencing and found that the abundance of Howardella in the prediabetic group was significantly higher than that in healthy subjects58. Recent studies have pointed out that diabetes and concomitant impaired glucose homeostasis and dysregulated nutrient-sensing participate in pathways linked to the metabolism of aging, which may weaken physiological reserves and lead to frailty59. This may be one of the potential mechanisms underlying the suggestive association between Howardella and frailty. A study of 85 community-dwelling adults suggested that the module of co-occurring microbial genera composed of Ruminococcus, Eggerthela, and Coprobacillus was positively correlated with the frailty index, and the association remained robust after correction for body mass index (BMI), subject age, antibiotic use, and other confounding factors60. Another study reported that Ruminococcus 1 was associated with lower adjusted body weight in older men61. This further implies that the cooperation between Ruminococcus 1 and other virulent symbionts may be involved in chronic inflammation62 and the subsequently acceleration of the aging process and frailty trajectory60.
In addition to the above five gut microbiota, we identified six genera that may be associated with a reduced risk of frailty. The first was Eubacterium ruminantium. A previous study involving 27 hospitalized elderly patients showed that the abundance of Eubacterium ruminantium in participants with frailty was lower than that in the non-frailty group63. The second was Akkermansia. The relative abundance of Akkermansia genus was higher in chronic kidney disease patients with sarcopenia64 and community-dwelling older adults with frailty65 but lower in cirrhotic patients with sarcopenia66. Although the results of few population-based studies are inconsistent, MR results suggested that Akkermansia may have a potential negative effect on frailty. These further supports previous animal experiments21,22. The third was Butyrivibrio. There is currently no observational study that has found an association between Butyrivibrio and frailty. However, a recent cohort study found that increased abundance of Butyrivibrio was associated with 1-year improvement in insulin status among elderly Mediterranean population at high cardiovascular risk.67 In general, Eubacterium ruminantium 68, 69Akkermansia 21 and Butyrivibrio 70 are believed to be new generation of “possibly helpful microbe” with the ability to produce SCFAs. SCFAs may prevent or alleviate frailty in the following ways: First, they regulate the differentiation, recruitment, and activation of immune cells, and reduce the secretion of inflammatory cytokines to play an anti-inflammatory role71,72. Second, they promote cognitive function by influencing the integrity of microglia and microglia-related activation involved in neuroinflammation, inducing the secretion of glucagon-like peptide 1 (GLP1) and peptide YY (PYY)73. Third, they regulate the synthesis and degradation of muscle proteins to maintain muscle quality and function74. In addition, they mediate the nuclear erythroid 2-related factor 2 (Nrf2)-related pathway, which can reduce oxidative and mitochondrial stress to delay aging75.
We found that a higher abundance of Catenibacterium may be associated with a lower risk of frailty. A recent study on patients with chronic liver diseases found that the abundance of Catenibacterium was lower in the sarcopenia group76. However, other studies have not obtained such significant results77. As a gram-positive anaerobic bacteria78, Catenibacterium typically has proinflammatory property and is highly abundant in obesity and infectious diseases79,80. However, our study implied that frailty seems to be associated with a lower risk of frailty. This finding may need further validation in experimental studies, as only four instrumental variables may have implicit associations.
We also found a suggestive genetic correlation and causal association between a higher abundance of Christensenellacea R-7 and a lower risk of frailty, which further corroborates the findings of previous observational studies. A cross-sectional study including 35 Italian community dwellers over 70 years old reported that the abundance of Christensenellaceae in physical frailty and sarcopenia (PF&S) group was significantly lower than non-PF&S group81. It seems to be gradually being identified as a potential biomarker of longevity in studies on the characteristics of the gut microbiome from centenarians82,83. Age-related adipose tissue dysfunction can lead to the infiltration of immune cells, secretion of proinflammatory cytokines and chemokines, and increased senescence-associated secretory phenotype84. Chronic low-grade inflammation, insulin resistance, metabolic disturbances, and redistribution of adipose tissue caused by the above pathophysiological changes are considered core processes of frailty85,86. As a butyrate-producing bacterium, Christensenellaceae R-7 is associated with lower insulin resistance87, reduced visceral adipose tissue accumulation, immune regulation, and improved metabolic health88. This appears to be the underlying mechanism of its suggestive causal effect on frailty.
Finally, a higher abundance of Defluviitaleaceae UCG-011 may be associated with a lower risk of frailty. A previous study found that, after dietary intervention in mice with cognitive impairment induced by a high-fat diet, the abundance of Defluviitaleaceae UCG-011 increased89. After herbal interventions in mice with depression and cognitive decline induced by chronic mild stress, the abundance of Defluviitaleaceae UCG-011 also increased90. This implies that Defluviitaleaceae UCG-011 may participate in the protection of cognitive function as a component of the gut-brain axis.
However, this study has some limitations. First, the gut microbiome may be influenced by demographic factors, diet, or drugs et al. Most of them have heterogeneity, inter-individual variability, and low heritability (representing the variance explained by genetics), which decreases the statistical efficacy and robustness of the results. Second, although most individuals in the GWAS meta-analysis of the gut microbiome were of European descent, there was still the possibility of interference by a small number of participants from other races, which may cause minimal bias and affect the universality of the results. Moreover, to obtain more comprehensive results and conduct horizontal pleiotropy detection and sensitivity analysis, selected genetic IVs did not reach the traditional GWAS significance threshold (p < 5 × 10−8), which may increase the possibility of false positives.
Conclusion
In summary, this study provides evidence for a suggestive genetic correlation between the genetically predicted Christensenellaceae R-7 and frailty. Furthermore, MR analysis indicated suggestive causal effects of genetically predicted 12 genus-level gut microbes on frailty.
Supplementary Material
Acknowledgments
The authors thank the participants and investigators of all GWAS included in this work. The authors also thank the MiBioGen Consortium, UK Biobank, and Swedish TwinGene for making the summary data openly available. Thanks to Figdraw for providing the materials in Figure 1.
Funding Statement
This study was supported by the National Natural Science Foundation of China (Nos.81803910 and 81973615), Capital’s Funds for Health, and the Qi-Huang Scholar Chief Scientist Program of National Administration of Traditional Chinese Medicine Leading Talents Support Program (2021).
Disclosure statement
No potential conflict of interest was reported by the author(s).
Author contributions
Guanghui Cui, Shaojie Li, and Xuezhi Zhang contributed to study design. Guanghui Cui and Shaojie Li analyzed the data and drafted the manuscript. Yao Yang, Xiaofen Jia, Miaomiao Lin, Yue Feng, Zicheng Wang, Yingming Chu and Zongming Shi compiled the data and provided comments on the draft. Hui Ye and Xuezhi Zhang revised the manuscript. All authors have read and approved the final manuscript.
Data availability statement
The datasets analyzed in the current study can be downloaded from the website https://mibiogen.gcc.rug.nl/, https://www.ebi.ac.uk/gwas/downloads/summary-statistics.
Ethics approval
The datasets used in the current study were publicly available and ethical approval and informed consent were obtained prior to implementation. Therefore, our study did not require any additional informed consent or ethical approval.
Supplementary material
Supplemental data for this article can be accessed online at https://doi.org/10.1080/19490976.2023.2282795.
References
- 1.Rodríguez-Mañas L, Féart C, Mann G, Viña J, Chatterji S, Chodzko-Zajko W, Gonzalez-Colaço Harmand M, Bergman H, Carcaillon L, Nicholson C, et al. Searching for an operational definition of frailty: a Delphi method based consensus statement. The frailty operative definition-consensus conference project. J Gerontol A Biol Sci Med Sci. 2013;68(1):62–14. doi: 10.1093/gerona/gls119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.O’Caoimh R, Sezgin D, O’Donovan MR, Molloy DW, Clegg A, Rockwood K, Liew A.. Prevalence of frailty in 62 countries across the world: a systematic review and meta-analysis of population-level studies. Age Ageing. 2021;50(1):96–104. doi: 10.1093/ageing/afaa219. [DOI] [PubMed] [Google Scholar]
- 3.Kojima G, Iliffe S, Walters K. Frailty index as a predictor of mortality: a systematic review and meta-analysis. Age Ageing. 2018;47(2):193–200. doi: 10.1093/ageing/afx162. [DOI] [PubMed] [Google Scholar]
- 4.Peng Y, Zhong G-C, Zhou X, Guan L, Zhou L. Frailty and risks of all-cause and cause-specific death in community-dwelling adults: a systematic review and meta-analysis. BMC Geriatr. 2022;22(1):725. doi: 10.1186/s12877-022-03404-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yang X, Lupón J, Vidán MT, Ferguson C, Gastelurrutia P, Newton PJ, Macdonald PS, Bueno H, Bayés‐Genís A, Woo J. Impact of frailty on mortality and hospitalization in chronic heart failure: a systematic review and meta‐analysis. J Am Heart Assoc. 2018;7(23):e008251. doi: 10.1161/JAHA.117.008251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chi J, Chen F, Zhang J, Niu X, Tao H, Ruan H, Wang Y, Hu J. Impacts of frailty on health care costs among community-dwelling older adults: a meta-analysis of cohort studies. Arch Gerontol Geriatr. 2021;94:104344. doi: 10.1016/j.archger.2021.104344. [DOI] [PubMed] [Google Scholar]
- 7.Fan L, Hou X-Y, Liu Y, Chen S, Wang Q, Du W. Catastrophic health expenditure associated with frailty in community-dwelling Chinese older adults: a prospective cohort analysis. Front Public Health. 2021;9:718910. doi: 10.3389/fpubh.2021.718910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kasajima M, Eggleston K, Kusaka S, Matsui H, Tanaka T, Son B-K, Iijima K, Goda K, Kitsuregawa M, Bhattacharya J. Projecting prevalence of frailty and dementia and the economic cost of care in Japan from 2016 to 2043: a microsimulation modelling study. Lancet Public Health. 2022;7(5):e458–e468. doi: 10.1016/S2468-2667(22)00044-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sun X, Liu W, Gao Y, Qin L, Feng H, Tan H, Chen Q, Peng L, Wu IX. Comparative effectiveness of non-pharmacological interventions for frailty: a systematic review and network meta-analysis. Age Ageing. 2023;52(2):afad004. doi: 10.1093/ageing/afad004. [DOI] [PubMed] [Google Scholar]
- 10.Ferrucci L, Fabbri E. Inflammageing: chronic inflammation in ageing, cardiovascular disease, and frailty. Nat Rev Cardiol. 2018;15(9):505–522. doi: 10.1038/s41569-018-0064-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Soysal P, Stubbs B, Lucato P, Luchini C, Solmi M, Peluso R, Sergi G, Isik AT, Manzato E, Maggi S. Inflammation and frailty in the elderly: a systematic review and meta-analysis. Ageing Res Rev. 2016;31:1–8. doi: 10.1016/j.arr.2016.08.006. [DOI] [PubMed] [Google Scholar]
- 12.de Jong PR, González-Navajas JM, Jansen NJ, de Jong PR. The digestive tract as the origin of systemic inflammation. Crit Care. 2016;20(1):1–12. doi: 10.1186/s13054-016-1458-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gomaa EZ. Human gut microbiota/microbiome in health and diseases: a review. Antonie Van Leeuwenhoek. 2020;113(12):2019–2040. doi: 10.1007/s10482-020-01474-7. [DOI] [PubMed] [Google Scholar]
- 14.Thevaranjan N, Puchta A, Schulz C, Naidoo A, Szamosi J, Verschoor CP, Loukov D, Schenck LP, Jury J, Foley KP. Age-associated microbial dysbiosis promotes intestinal permeability, systemic inflammation, and macrophage dysfunction. Cell Host & Microbe. 2017;21(4):455–66. e4. doi: 10.1016/j.chom.2017.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zapata HJ, Quagliarello VJ. The microbiota and microbiome in aging: potential implications in health and age‐related diseases. J Am Geriatr Soc. 2015;63(4):776–781. doi: 10.1111/jgs.13310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mou Y, Du Y, Zhou L, Yue J, Hu X, Liu Y, Chen S, Lin X, Zhang G, Xiao H. Gut microbiota interact with the brain through systemic chronic inflammation: implications on neuroinflammation, neurodegeneration, and aging. Front Immunol. 2022;13:796288. doi: 10.3389/fimmu.2022.796288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lim MY, Hong S, Kim J-H, Nam Y-D, Le Couteur D. Association between gut microbiome and frailty in the older adult population in Korea. J Gerontol A Biol Sci Med Sci. 2021;76(8):1362–1368. doi: 10.1093/gerona/glaa319. [DOI] [PubMed] [Google Scholar]
- 18.Almeida HM, Sardelia AV, Conway J, Duggal NA, Cavaglieri CR. Comparison between frail and non-frail older adults’ gut microbiota: a systematic review and meta-analysis. Ageing Res Rev. 2022;82:101773. doi: 10.1016/j.arr.2022.101773. [DOI] [PubMed] [Google Scholar]
- 19.Rashidah NH, Lim SM, Neoh CF, Majeed ABA, Tan MP, Khor HM, Tan AH, Rehiman SH, Ramasamy K. Differential gut microbiota and intestinal permeability between frail and healthy older adults: a systematic review. Ageing Res Rev. 2022;82:101744. doi: 10.1016/j.arr.2022.101744. [DOI] [PubMed] [Google Scholar]
- 20.Ma J, Liu Z, Gao X, Bao Y, Hong Y, He X, Zhu W, Li Y, Huang W, Zheng N, et al. Gut microbiota remodeling improves natural aging-related disorders through Akkermansia muciniphila and its derived acetic acid. Pharmacol Res. 2023;189:106687. doi: 10.1016/j.phrs.2023.106687. [DOI] [PubMed] [Google Scholar]
- 21.Shin J, Noh JR, Choe D, Lee N, Song Y, Cho S, Kang EJ, Go MJ, Ha SK, Chang DH, et al. Ageing and rejuvenation models reveal changes in key microbial communities associated with healthy ageing. Microbiome. 2021;9(1):240. doi: 10.1186/s40168-021-01189-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bárcena C, Valdés-Mas R, Mayoral P, Garabaya C, Durand S, Rodríguez F, Fernández-García MT, Salazar N, Nogacka AM, Garatachea N, et al. Healthspan and lifespan extension by fecal microbiota transplantation into progeroid mice. Nat Med. 2019;25(8):1234–1242. doi: 10.1038/s41591-019-0504-5. [DOI] [PubMed] [Google Scholar]
- 23.Lee J, Venna VR, Durgan DJ, Shi H, Hudobenko J, Putluri N, Petrosino J, McCullough LD, Bryan RM. Young versus aged microbiota transplants to germ-free mice: increased short-chain fatty acids and improved cognitive performance. Gut Microbes. 2020;12(1):1–14. doi: 10.1080/19490976.2020.1814107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Walsh ME, Bhattacharya A, Sataranatarajan K, Qaisar R, Sloane L, Rahman MM, Kinter M, Van Remmen H. The histone deacetylase inhibitor butyrate improves metabolism and reduces muscle atrophy during aging. Aging Cell. 2015;14(6):957–970. doi: 10.1111/acel.12387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Evans LW, Stratton MS, Ferguson BS. Dietary natural products as epigenetic modifiers in aging-associated inflammation and disease. Nat Prod Rep. 2020;37(5):653–676. doi: 10.1039/C9NP00057G. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bulik-Sullivan B, Finucane HK, Anttila V, Gusev A, Day FR, Loh P-R, Consortium R, Consortium PG, Patterson N, Robinson EB. 3 GCfANotWTCCC, Duncan L. An atlas of genetic correlations across human diseases and traits. Nat Genet. 2015;47(11):1236–1241. doi: 10.1038/ng.3406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hemani G, Zheng J, Elsworth B, Wade KH, Haberland V, Baird D, Laurin C, Burgess S, Bowden J, Langdon R. The MR-Base platform supports systematic causal inference across the human phenome. Elife. 2018;7:e34408. doi: 10.7554/eLife.34408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kurilshikov A, Medina-Gomez C, Bacigalupe R, Radjabzadeh D, Wang J, Demirkan A, Le Roy CI, Raygoza Garay JA, Finnicum CT, Liu X. Large-scale association analyses identify host factors influencing human gut microbiome composition. Nat Genet. 2021;53(2):156–165. doi: 10.1038/s41588-020-00763-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li P, Wang H, Guo L, Gou X, Chen G, Lin D, Fan D, Guo X, Liu Z. Association between gut microbiota and preeclampsia-eclampsia: a two-sample Mendelian randomization study. BMC Med. 2022;20(1):1–10. doi: 10.1186/s12916-022-02657-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Atkins JL, Jylhävä J, Pedersen NL, Magnusson PK, Lu Y, Wang Y, Hägg S, Melzer D, Williams DM, Pilling LC. A genome‐wide association study of the frailty index highlights brain pathways in ageing. Aging Cell. 2021;20(9):e13459. doi: 10.1111/acel.13459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rockwood K, Mitnitski A. Frailty in relation to the accumulation of deficits. J Gerontol A Biol Sci Med Sci. 2007;62(7):722–727. doi: 10.1093/gerona/62.7.722. [DOI] [PubMed] [Google Scholar]
- 32.Zhong H, Liu S, Zhu J, Wu L. Associations between genetically predicted levels of blood metabolites and pancreatic cancer risk. Int J Cancer. 2023;153(1):103–110. doi: 10.1002/ijc.34466. [DOI] [PubMed] [Google Scholar]
- 33.Liu X, Tong X, Zou Y, Lin X, Zhao H, Tian L, Jie Z, Wang Q, Zhang Z, Lu H. Mendelian randomization analyses support causal relationships between blood metabolites and the gut microbiome. Nat Genet. 2022;54(1):52–61. doi: 10.1038/s41588-021-00968-y. [DOI] [PubMed] [Google Scholar]
- 34.Sanna S, van Zuydam NR, Mahajan A, Kurilshikov A, Vich Vila A, Võsa U, Mujagic Z, Masclee AA, Jonkers DM, Oosting M, et al. Causal relationships among the gut microbiome, short-chain fatty acids and metabolic diseases. Nat Genet. 2019;51(4):600–605. doi: 10.1038/s41588-019-0350-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Staley JR, Blackshaw J, Kamat MA, Ellis S, Surendran P, Sun BB, Paul DS, Freitag D, Burgess S, Danesh J, et al. PhenoScanner: a database of human genotype–phenotype associations. Bioinformatics. 2016;32(20):3207–3209. doi: 10.1093/bioinformatics/btw373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kamat MA, Blackshaw JA, Young R, Surendran P, Burgess S, Danesh J, Butterworth AS, Staley JR, Kelso J. PhenoScanner V2: an expanded tool for searching human genotype–phenotype associations. Bioinformatics. 2019;35(22):4851–4853. doi: 10.1093/bioinformatics/btz469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cole JB, Florez JC, Hirschhorn JN. Comprehensive genomic analysis of dietary habits in UK Biobank identifies hundreds of genetic associations. Nat Commun. 2020;11(1):1467. doi: 10.1038/s41467-020-15193-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bulik-Sullivan BK, Loh P-R, Finucane HK, Ripke S, Yang J, Consortium SWGot PG, Patterson N, Daly MJ, Price AL, Neale BM. LD score regression distinguishes confounding from polygenicity in genome-wide association studies. Nat Genet. 2015;47(3):291–295. doi: 10.1038/ng.3211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wielscher M, Amaral AF, van der Plaat D, Wain LV, Sebert S, Mosen-Ansorena D, Auvinen J, Herzig K-H, Dehghan A, Jarvis DL. Genetic correlation and causal relationships between cardio-metabolic traits and lung function impairment. Genome Med. 2021;13(1):104. doi: 10.1186/s13073-021-00914-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Staiger DO, Stock JH. Instrumental variables regression with weak instruments. Mass., USA: National Bureau of Economic Research Cambridge; 1994. [Google Scholar]
- 41.Pierce BL, Ahsan H, VanderWeele TJ. Power and instrument strength requirements for Mendelian randomization studies using multiple genetic variants. Int J Epidemiol. 2011;40(3):740–752. doi: 10.1093/ije/dyq151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Burgess S, Butterworth A, Thompson SG. Mendelian randomization analysis with multiple genetic variants using summarized data. Genet Epidemiol. 2013;37(7):658–665. doi: 10.1002/gepi.21758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chen Y, Li C, Cheng S, Pan C, Zhang H, Zhang J, Zhang Z, Yao Y, Cheng B, Liu L, et al. The causal relationships between sleep-related phenotypes and body composition: a Mendelian randomized study. J Clin Endocrinol Metab. 2022;107(8):e3463–e3473. doi: 10.1210/clinem/dgac234. [DOI] [PubMed] [Google Scholar]
- 44.Bowden J, Davey Smith G, Burgess S. Mendelian randomization with invalid instruments: effect estimation and bias detection through Egger regression. Int J Epidemiol. 2015;44(2):512–525. doi: 10.1093/ije/dyv080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Verbanck M, Chen CY, Neale B, Do R. Detection of widespread horizontal pleiotropy in causal relationships inferred from Mendelian randomization between complex traits and diseases. Nat Genet. 2018;50(5):693–698. doi: 10.1038/s41588-018-0099-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kenny DJ, Plichta DR, Shungin D, Koppel N, Hall AB, Fu B, Vasan RS, Shaw SY, Vlamakis H, Balskus EP. Cholesterol metabolism by uncultured human gut bacteria influences host cholesterol level. Cell Host & Microbe. 2020;28(2):245–57. e6. doi: 10.1016/j.chom.2020.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Li L, Batt SM, Wannemuehler M, Dispirito A, Beitz DC. Effect of feeding of a cholesterol-reducing bacterium, Eubacterium coprostanoligenes, to germ-free mice. Lab Anim Sci. 1998;48:253–255. [PubMed] [Google Scholar]
- 48.Li L, Buhman K, Hartman P, Beitz D. Hypocholesterolemic effect of Eubacterium coprostanoligenes ATCC 51222 in rabbits. Lett Appl Microbiol. 1995;20(3):137–140. doi: 10.1111/j.1472-765X.1995.tb00410.x. [DOI] [PubMed] [Google Scholar]
- 49.Rosas-Campos R, Meza-Rios A, Rodriguez-Sanabria JS, De la Rosa-Bibiano R, Corona-Cervantes K, García-Mena J, Santos A, Sandoval-Rodriguez A, Armendariz-Borunda J. Dietary supplementation with Mexican foods, opuntia ficus indica, theobroma cacao, and acheta domesticus: improving obesogenic and microbiota features in obese mice. Front Nutr. 2022;9:987222. doi: 10.3389/fnut.2022.987222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chu W, Lynskey N, Iain-Ross J, Pell JP, Sattar N, Ho FK, Welsh P, Celis-Morales C, Petermann-Rocha F. Identifying the biomarker profile of pre-frail and frail people: a cross-sectional analysis from UK Biobank. Int J Environ Res Public Health. 2023;20(3):2421. doi: 10.3390/ijerph20032421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.He J, Xu S, Zhang B, Xiao C, Chen Z, Si F, Fu J, Lin X, Zheng G, Yu G. Gut microbiota and metabolite alterations associated with reduced bone mineral density or bone metabolic indexes in postmenopausal osteoporosis. Aging (Albany NY). 2020;12(9):8583. doi: 10.18632/aging.103168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Aranaz P, Ramos-Lopez O, Cuevas-Sierra A, Martinez JA, Milagro FI, Riezu-Boj JI. A predictive regression model of the obesity-related inflammatory status based on gut microbiota composition. Int J Obes (Lond). 2021;45(10):2261–2268. doi: 10.1038/s41366-021-00904-4. [DOI] [PubMed] [Google Scholar]
- 53.Cardoso AL, Fernandes A, Aguilar-Pimentel JA, de Angelis MH, Guedes JR, Brito MA, Ortolano S, Pani G, Athanasopoulou S, Gonos ES, et al. Towards frailty biomarkers: candidates from genes and pathways regulated in aging and age-related diseases. Ageing Res Rev. 2018;47:214–277. doi: 10.1016/j.arr.2018.07.004. [DOI] [PubMed] [Google Scholar]
- 54.Gonçalves RSd SA, Maciel ÁCC, Rolland Y, Vellas B, de Souto Barreto P. Frailty biomarkers under the perspective of geroscience: a narrative review. Ageing Res Rev. 2022;81:101737. doi: 10.1016/j.arr.2022.101737. [DOI] [PubMed] [Google Scholar]
- 55.Sharma M, Wasan A, Sharma RK. Recent developments in probiotics: an emphasis on Bifidobacterium. Food Biosci. 2021;41:100993. doi: 10.1016/j.fbio.2021.100993. [DOI] [Google Scholar]
- 56.Shen T, Yue Y, He T, Huang C, Qu B, Lv W, Lai H-Y. The association between the gut microbiota and Parkinson’s disease, a meta-analysis. Front Aging Neurosci. 2021;13:40. doi: 10.3389/fnagi.2021.636545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wang W, Chen L, Zhou R, Wang X, Song L, Huang S, Wang G, Xia B, Forbes BA. Increased proportions of Bifidobacterium and the Lactobacillus group and loss of butyrate-producing bacteria in inflammatory bowel disease. J Clin Microbiol. 2014;52(2):398–406. doi: 10.1128/JCM.01500-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yang J, Summanen PH, Henning SM, Hsu M, Lam H, Huang J, Tseng C-H, Dowd SE, Finegold SM, Heber D. Xylooligosaccharide supplementation alters gut bacteria in both healthy and prediabetic adults: a pilot study. Front Physiol. 2015;6:216. doi: 10.3389/fphys.2015.00216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Perazza LR, Brown‐Borg HM, Thompson LV. Physiological systems in promoting frailty. Compr Physiol. 2022;12:3575–3620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Maffei VJ, Kim S, Blanchard E IV, Luo M, Jazwinski SM, Taylor CM, Welsh DA. Biological aging and the human gut microbiota. J Gerontol A Biol Sci Med Sci. 2017;72:1474–1482. doi: 10.1093/gerona/glx042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Shardell M, Parimi N, Langsetmo L, Tanaka T, Jiang L, Orwoll E, Shikany JM, Kado DM, Cawthon PM, Masternak M. Comparing analytical methods for the gut microbiome and aging: gut microbial communities and body weight in the osteoporotic fractures in men (MrOS) study. J Gerontol A Biol Sci Med Sci. 2020;75(7):1267–1275. doi: 10.1093/gerona/glaa034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Yao Z-Y, Li X-H, Zuo L, Xiong Q, He W-T, Li D-X, Dong Z-F. Maternal sleep deprivation induces gut microbial dysbiosis and neuroinflammation in offspring rats. Zool Res. 2022;43(3):380. doi: 10.24272/j.issn.2095-8137.2022.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zhang L, Liao J, Chen Q, Chen M, Kuang Y, Chen L, He W. Characterization of the gut microbiota in frail elderly patients. Aging Clin Exp Res. 2020;32(10):2001–2011. doi: 10.1007/s40520-019-01385-2. [DOI] [PubMed] [Google Scholar]
- 64.Margiotta E, Caldiroli L, Callegari ML, Miragoli F, Zanoni F, Armelloni S, Rizzo V, Messa P, Vettoretti S. Association of sarcopenia and gut microbiota composition in older patients with advanced chronic kidney disease, investigation of the interactions with Uremic Toxins, inflammation and oxidative stress. Toxins (Basel). 2021;13(7):472. doi: 10.3390/toxins13070472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Xu Y, Wang Y, Li H, Dai Y, Chen D, Wang M, Jiang X, Huang Z, Yu H, Huang J, et al. Altered fecal microbiota composition in older adults with frailty. Front Cell Infect Microbiol. 2021;11:696186. doi: 10.3389/fcimb.2021.696186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ponziani FR, Picca A, Marzetti E, Calvani R, Conta G, Del Chierico F, Capuani G, Faccia M, Fianchi F, Funaro B, et al. Characterization of the gut-liver-muscle axis in cirrhotic patients with sarcopenia. Liver Int. 2021;41(6):1320–1334. doi: 10.1111/liv.14876. [DOI] [PubMed] [Google Scholar]
- 67.Atzeni A, Nishi SK, Babio N, Belzer C, Konstanti P, Vioque J, Corella D, Castañer O, Vidal J, Moreno-Indias I, et al. Carbohydrate quality, fecal microbiota and cardiometabolic health in older adults: a cohort study. Gut Microbes. 2023;15(2):2246185. doi: 10.1080/19490976.2023.2246185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Hu S, Ma Y, Xiong K, Wang Y, Liu Y, Sun Y, Yang Y, Ma A. Ameliorating effects of vitamin K2 on dextran sulfate sodium-induced ulcerative colitis in mice. Int J Mol Sci. 2023;24(3):2986. doi: 10.3390/ijms24032986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Xu T, Huang W, Liang J, Zhong Y, Chen Q, Jie F, Lu B. Tuber flours improve intestinal health and modulate gut microbiota composition. Food Chem. 2021;12:100145. doi: 10.1016/j.fochx.2021.100145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Nielsen TS, Jensen BB, Purup S, Jackson S, Saarinen M, Lyra A, Sørensen JF, Theil PK, Knudsen KE. A search for synbiotics: effects of enzymatically modified arabinoxylan and Butyrivibrio fibrisolvens on short-chain fatty acids in the cecum content and plasma of rats. Food Funct. 2016;7(4):1839–1848. doi: 10.1039/C6FO00114A. [DOI] [PubMed] [Google Scholar]
- 71.Rodrigues HG, Sato FT, Curi R, Vinolo MA. Fatty acids as modulators of neutrophil recruitment, function and survival. Eur J Pharmacol. 2016;785:50–58. doi: 10.1016/j.ejphar.2015.03.098. [DOI] [PubMed] [Google Scholar]
- 72.Corrêa‐Oliveira R, Fachi JL, Vieira A, Sato FT, Vinolo MAR. Regulation of immune cell function by short‐chain fatty acids. Clin Transl Immunology. 2016;5(4):e73. doi: 10.1038/cti.2016.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Dalile B, Van Oudenhove L, Vervliet B, Verbeke K. The role of short-chain fatty acids in microbiota–gut–brain communication. Nat Rev Gastroenterol Hepatol. 2019;16(8):461–478. doi: 10.1038/s41575-019-0157-3. [DOI] [PubMed] [Google Scholar]
- 74.Liu C, Cheung WH, Li J, Chow SKH, Yu J, Wong SH, Ip M, Sung JJY, Wong RMY. Understanding the gut microbiota and sarcopenia: a systematic review. J Cachexia Sarcopenia Muscle. 2021;12(6):1393–1407. doi: 10.1002/jcsm.12784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.González-Bosch C, Boorman E, Zunszain P, Mann G. Short-chain fatty acids as modulators of redox signaling in health and disease. Redox Biol. 2021;47:102165. doi: 10.1016/j.redox.2021.102165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Yamamoto K, Ishizu Y, Honda T, Ito T, Imai N, Nakamura M, Kawashima H, Kitaura Y, Ishigami M, Fujishiro M. Patients with low muscle mass have characteristic microbiome with low potential for amino acid synthesis in chronic liver disease. Sci Rep. 2022;12(1):3674. doi: 10.1038/s41598-022-07810-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Ponziani FR, Picca A, Marzetti E, Calvani R, Conta G, Del Chierico F, Capuani G, Faccia M, Fianchi F, Funaro B. Characterization of the gut‐liver‐muscle axis in cirrhotic patients with sarcopenia. Liver Int. 2021;41(6):1320–1334. doi: 10.1111/liv.14876. [DOI] [PubMed] [Google Scholar]
- 78.Kageyama A, Benno Y. Catenibacterium mitsuokai gen. nov., sp. nov., a gram-positive anaerobic bacterium isolated from human faeces. Int J Syst Evol Microbiol. 2000;50(4):1595–1599. doi: 10.1099/00207713-50-4-1595. [DOI] [PubMed] [Google Scholar]
- 79.Lozupone CA, Li M, Campbell TB, Flores SC, Linderman D, Gebert MJ, Knight R, Fontenot AP, Palmer BE. Alterations in the gut microbiota associated with HIV-1 infection. Cell Host & Microbe. 2013;14(3):329–339. doi: 10.1016/j.chom.2013.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Insenser M, Murri M, Del Campo R, Martinez-Garcia MA, Fernandez-Duran E, Escobar-Morreale HF. Gut microbiota and the polycystic ovary syndrome: influence of sex, sex hormones, and obesity. J Clin Endocrinol Metab. 2018;103(7):2552–2562. doi: 10.1210/jc.2017-02799. [DOI] [PubMed] [Google Scholar]
- 81.Picca A, Ponziani FR, Calvani R, Marini F, Biancolillo A, Coelho-Júnior HJ, Gervasoni J, Primiano A, Putignani L, Del Chierico F. Gut microbial, inflammatory and metabolic signatures in older people with physical frailty and sarcopenia: results from the BIOSPHERE study. Nutrients. 2019;12(1):65. doi: 10.3390/nu12010065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Badal VD, Vaccariello ED, Murray ER, Yu KE, Knight R, Jeste DV, Nguyen TT. The gut microbiome, aging, and longevity: a systematic review. Nutrients. 2020;12(12):3759. doi: 10.3390/nu12123759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Santoro A, Ostan R, Candela M, Biagi E, Brigidi P, Capri M, Franceschi C. Gut microbiota changes in the extreme decades of human life: a focus on centenarians. Cell Mol Life Sci. 2018;75(1):129–148. doi: 10.1007/s00018-017-2674-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Stout MB, Justice JN, Nicklas BJ, Kirkland JL. Physiological aging: links among adipose tissue dysfunction, diabetes, and frailty. Physiol (Bethesda). 2017;32(1):9–19. doi: 10.1152/physiol.00012.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Clegg A, Hassan-Smith Z. Frailty and the endocrine system. Lancet Diabetes Endocrinol. 2018;6(9):743–752. doi: 10.1016/S2213-8587(18)30110-4. [DOI] [PubMed] [Google Scholar]
- 86.Kalinkovich A, Livshits G. Sarcopenic obesity or obese sarcopenia: a cross talk between age-associated adipose tissue and skeletal muscle inflammation as a main mechanism of the pathogenesis. Ageing Res Rev. 2017;35:200–221. doi: 10.1016/j.arr.2016.09.008. [DOI] [PubMed] [Google Scholar]
- 87.Chen Z, Radjabzadeh D, Chen L, Kurilshikov A, Kavousi M, Ahmadizar F, Ikram MA, Uitterlinden AG, Zhernakova A, Fu J, et al. Association of insulin resistance and type 2 diabetes with gut microbial diversity: a microbiome-wide analysis from population studies. JAMA Netw Open. 2021;4(7):e2118811–e. doi: 10.1001/jamanetworkopen.2021.18811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Tavella T, Rampelli S, Guidarelli G, Bazzocchi A, Gasperini C, Pujos-Guillot E, Comte B, Barone M, Biagi E, Candela M. Elevated gut microbiome abundance of Christensenellaceae, porphyromonadaceae and rikenellaceae is associated with reduced visceral adipose tissue and healthier metabolic profile in Italian elderly. Gut Microbes. 2021;13(1):1880221. doi: 10.1080/19490976.2021.1880221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Yang X, Zheng M, Hao S, Shi H, Lin D, Chen X, Becvarovski A, Pan W, Zhang P, Hu M. Curdlan prevents the cognitive deficits induced by a high-fat diet in mice via the gut-brain axis. Front Neurosci. 2020;14:384. doi: 10.3389/fnins.2020.00384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Huang H-S, Lin Y-E, Panyod S, Chen R-A, Lin Y-C, Chai LMX, Hsu C-C, Wu W-K, Lu K-H, Huang Y-J. Anti-depressive-like and cognitive impairment alleviation effects of gastrodia elata Blume water extract is related to gut microbiome remodeling in ApoE−/− mice exposed to unpredictable chronic mild stress. J Ethnopharmacol. 2023;302:115872. doi: 10.1016/j.jep.2022.115872. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets analyzed in the current study can be downloaded from the website https://mibiogen.gcc.rug.nl/, https://www.ebi.ac.uk/gwas/downloads/summary-statistics.


