Highlights
-
•
We used spatial transcriptomics to determine the location and gene expression profile of cells within the adult rat patellar tendon.
-
•
We found two populations of tendon fibroblasts, one located in the tendon midsubstance, while the other localized with red blood cells, pericytes, and immune cells on the tendon periphery.
-
•
The top spatially expressed genes included multiple genes with known function in tendon: Col1a1, Dcn, Fmod, Sparc, and Comp.
-
•
Col1a1 and Col3a1 expression defines two different populations of fibroblasts, in the core and periphery of the tissue, respectively.
-
•
We also discovered a novel spatially regulated gene (AABR07000398.1) with no known function.
Keywords: Tendinopathy, Exercise, RNAseq, Transcriptomics
Abstract
Tendons are dense connective tissues with relatively few cells which makes studying the molecular profile of the tissue challenging. There is not a consensus on the spatial location of various cell types within a tendon, nor the accompanying transcriptional profile. In the present study, we used two male rat patellar tendon samples for sequencing-based spatial transcriptomics to determine the gene expression profile. We integrated our data with a mouse Achilles single cell dataset to predict the cell type composition of the patellar tendon as a function of location within the tissue. The spatial location of the predicated cell types suggested that there were two populations of tendon fibroblasts, one located in the tendon midsubstance, while the other localized with red blood cells, pericytes, and immune cells to the tendon peripheral connective tissue. Of the highest expressed spatially variable genes, there were multiple genes with known function in tendon: Col1a1, Col1a2, Dcn, Fmod, Sparc, and Comp. Further, a novel spatially regulated gene (AABR07000398.1) with no known function was identified. The spatial gene expression of tendon associated genes (Scx, Thbs4, Tnmd, Can, Bgn, Lum, Adamts2, Lox, Ppib, Col2a1, Col3a1, Col6a2) was also visualized. Both patellar tendon samples had similar expression patterns for all these genes. This dataset provides new spatial insights into gene expression in a healthy tendon.
Introduction
Tendons are dense connective tissues that contain a small population of cells that reside within a highly organized extracellular matrix primarily composed of collagen. Collagen molecules are secreted by these cells along the line of force[8], [22]. These molecules aggregate into fibrils, and fibrils aggregate into fibers. In the rat patellar tendon, there is no intrafascicular matrix and fibers are the largest unit of collagen; tendon fibroblasts are located between fibers [27]. Less than 5 % of the tendon volume is composed of cells [30] which makes characterizing the molecular biology of this tissue challenging.
The most common cell type in tendon is thought to be tenocytes, a type of specialized fibroblast responsible for maintaining the structure of the extracellular matrix. Tenocytes are found in-between collagen fibers and have elongated projections that spread through the matrix. Tendon can also contain a small immune cell population that increases upon injury[9], [18], [31]. Endothelial cells and pericytes, along with red blood cells (RBCs), have been reported within tendon [10], [18] and are best characterized around blood vessels in the paratenon and outside the tendon mid-substance[13].
Few studies have attempted to characterize the transcriptome of the multiple cell populations found in a tendon. The best unbiased report on gene expression in tendon cell types have been generated using single cell-RNA sequencing (scRNA-seq) [10], [18], [24]. These studies are limited by the lack of spatial resolution. Spatial transcriptomics has the capacity to address this gap by pairing histological and gene expression information. Here, we used spatial transcriptomics to provide the first thorough report of spatial gene expression in healthy, uninjured tendon.
Materials and methods
Experimental animals
All animal experiments were approved under UC Davis Institutional Animal Care and Use Committee (IACUC) protocol #22957. Male Sprague Dawley rats (3 months of age) were purchased from Charles River Laboratories (Wilmington, MA). Rats were housed under 12-hour light/dark cycle in pathogen-free conditions.
Sample preparation
Patellar tendons (PT) were dissected with patella bone and quadriceps muscle attached. Excess muscle was trimmed, the entire tissue rinsed in PBS, blotted dry, coated with OCT, then placed into a cryomold filled with optimal cutting temperature compound (OCT) (Tissue-Tek®, Finetek, USA). To ensure good longitudinal sections could be produced, the OCT-coated tendon was pressed flat into the cryomold which already contained a little OCT and carefully filled with OCT prior to freezing. OCT embedded tendon was immediately frozen in isopentane-cooled liquid nitrogen and stored on dry ice and −80 °C until processed.
Tissues were transferred to a Leica CM 3050S cryostat on dry ice and incubated to −20 °C at least 45 min prior to sectioning. Longitudinal sections (14 µM) were cut distal to proximal, starting at the anterior aspect of the patellar tendon. Section thickness from 8 to 30 µM were considered; it was decided that 14 µM had sufficient cellularity to perform spatial transcriptomics without being too difficult to section. Excess OCT and tissue surrounding the PT was brushed away with cryostat brushes. PT sections were placed on top of spatial tissue optimization or gene expression slides (10× Genomics, Pleasanton, CA, USA; Chemistry Spatial 3′ v1) and adhered by touching the tissue to the back of the slide. The tissue optimization and gene expression slides contain oligonucleotides with poly(dT) designed for mRNA capture with additional oligonucleotide sequencing primer, spatial barcode, and UMI on the gene expression slides. Each 6.5 mm × 6.5 mm capture area has 5000 spots (55 µM diameter) with 100 µM center to center distance. Slides were stored in 50 mL tubes at −80 °C until analysis.
To determine the optimal duration of permeabilization for mRNA capture, tissue sections were methanol fixed, hematoxylin and eosin (H&E) stained, and then permeabilized with 10X tissue permeabilization enzyme for 10, 13, 16, 19, 21, or 24 min. cDNA was synthesized with fluorescent nucleotides, tendon tissue enzymatically removed, and the fluorescent cDNA footprint imaged using a Leica DMi8. The strongest fluorescent signal was used to determine the optimal permeabilization time (21 min).
Library preparation and sequencing
Patellar tendon tissues on spatial gene expression slides were methanol fixed, H&E stained, then imaged under bright-field at 10X using a Lecia DMi8. Slides were placed in 50 mL tubes on ice and transported by bicycle within 10 min after imaging to the UC Davis Genome Center for permeabilization, library preparation, and sequencing. Library preparation was performed with 10X Visium Library Construction Kit (10x Visium Spatial Gene Expression Reagent Kit CG000239 Rev E). Libraries were paired end sequenced on a NovaSeq S4 200 and 87,840,443 reads (PF clusters) were generated.
Data analysis
Space Ranger (version 1.3.1) was used to process raw FASTQ files and H&E stained brightfield images. Genome alignment was performed with STAR within the Space Ranger pipeline against a custom reference genome Rnor_6.0 (GCA_000001895.4). Space Ranger generated unfiltered feature-barcode matrix and spatial image files that were read into R (R version 4.3.1) for downstream analysis using Seurat (version 4.3.0)[6], [17], [34], [39]. Individual spatial Seurat objects were created from each sample and with no filters applied for unique molecular identifier (UMI) counts, feature counts, or mitochondrial reads per spot. Samples were merged and spots with zero UMI counts were removed. SCT transformation was applied to data and principal component analysis (PCA) and Uniform Manifold Approximation and Projection for Dimension Reduction (UMAP) performed with 15 reduction dimensions. Samples were integrated with Harmony[25]. Differential expression analysis was performed using “FindAllMarkers” in Seurat on genes that were detected in at least 25 % of the population and with a log-scale fold change threshold of 0.2. Cluster determination was performed using default parameters that use a shared nearest neighbor modularity. Clustering resolutions of 0.2, 0.4, 0.6, 0.8, and 1 were considered and it was decided to use 0.4 resolution for subsequent analysis. The 0.4 resolution was selected since the clustering at 0.4 and 0.6 were nearly identical, indicating that this region gave the most reliable representation of the data. Cluster markers were determined considering upregulated genes only.
Cell type predictions were performed on the integrated Seurat object with analysis anchored against a mouse Achilles single cell reference [10]. Data transfer was performed using rat homologs identified by Ensembl. The top upregulated genes in each cell type are presented.
Identification of spatially variable features was performed individually on each sample. The “FindSpatiallyVariableFeatures” function in Seuat was applied to transformed data (SCT assay) and spatially variability calculated using the markvariogram method. The top expressed spatially variable genes were determined by multiplying the spatial metric (Moran’s I) by the expression (Median Normalized Average Counts). Excel documents that contain the following information are available as sup. plemental files: Feature ID/Name, Moran’s I, P value, Adjusted p value, Feature Counts in Spots Under Tissue, Median Normalized Average Counts, Barcode, I Weighted Counts.
Results & discussion
This is the first report of a spatial transcriptomics profile for rat tendon. To date, there have been limited reports of spatial gene expression in healthy tendon. The first spatial transcriptomics data of tendon was presented in a short report that described the spatial expression pattern for four genes[2]. More recently, there was an excellent report using single-nuclear RNA seq that described four distinct types of nuclei present at the MTJ and the spatial transcriptomics data identified distinct muscle and tendon clusters[23]. Other reports on tendon have focused on the diseased tissue, which is more cellular and easier to permeabilize than healthy tendon[1], [16]. The technique of spatial transcriptomics was pioneered in brain and cancer tissue and is rarely applied to very fibrous tissues such as tendon given that the method requires a permeabilization step to release mRNA onto a barcoded poly (T) capture slide[37]. Furthermore, the tissue sections must adhere onto the capture slide completely flat, and it can be challenging to obtain high quality longitudinal sections of tendon because it is relatively thin and has dense parallel collagen fibers are notoriously difficult to section. Despite these challenges, we were able to obtain spatial transcriptomic information for two biological samples that were bioinformatically integrated to generate unbiased clusters and location of predicted cell types and identify the most spatially regulated genes.
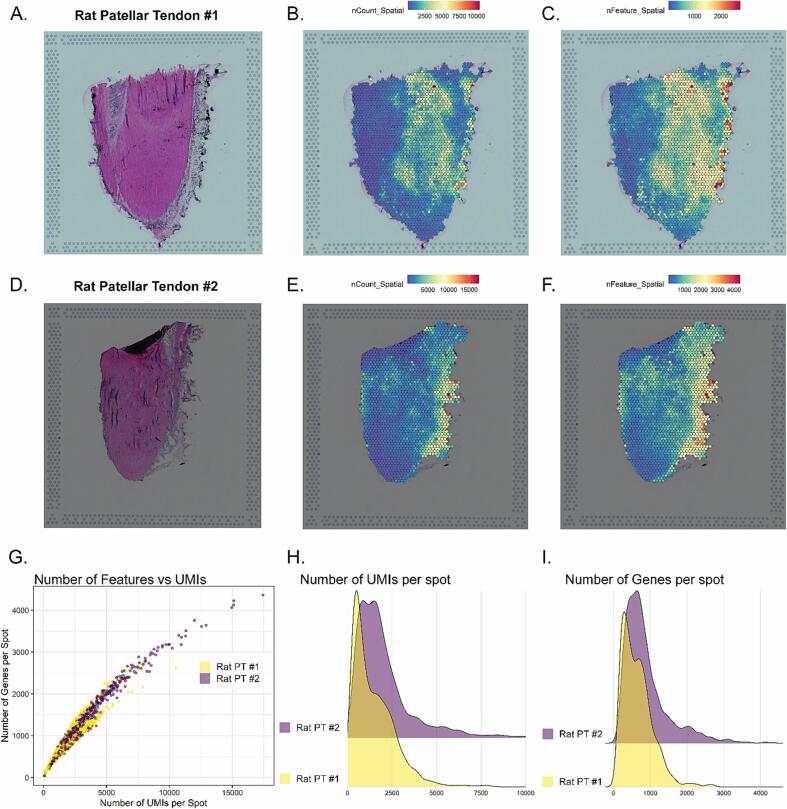
The tissue morphology and QC metrics were similar for both samples. Each spot on the Visium slide contains a spatial barcode and every piece of RNA captured has a unique molecular identifier (UMI). The spots with the most UMIs (labeled in figure as nCount) have the most RNA. For each sample, the strip of connective tissue surrounding the tendon had the most RNA which is not surprising given the relatively hypocellular nature of the tendon mid-substance (Fig. 1B, E and H). For PT #2, there was a small piece of patella bone as indicated by the dark semi-circle on top of the tendon which likely did not permeabilize as well as the tendon given the low UMI count. The average number of UMIs per spot was 40,312 for rat #1 and 64,438 for rat #2 (Table 1). The percentage of reads mapped to the genome was 93.8 % and 83.7 %, respectively, which is similar to mapping rates reported in other studies [11], [12]. There was a strong correlation (R2 = 0.97) between the number of genes per spot and number of UMIs per spot (Fig. 1G).
Fig. 1.
H&E Stains and QC Metrics. A&D) H&E stains of rat patellar tendon. B&E) Spatial projection of number of UMIs per spot. E&F) Spatial projection of number of genes per spot. G) Scatterplot of Number of genes per spot vs number of UMIs per spot. H) Ridge plot of Number of UMIs per spot for rat PT #1 and #2. I) Ridge plot of Number of Genes per spot for each sample.
Table 1.
QC Metrics.
| Sample | Number of Spots Under Tissue | UMIs per Spot, Mean | Genes per Spot, Mean |
|---|---|---|---|
| Sample 1 (DS128_10LA) | 2179 | 40,312 | 582 |
| Sample 2 (DS129_1LA) | 1624 | 64,438 | 716 |
Data taken from Space Ranger Web Browser (8/2/2023).
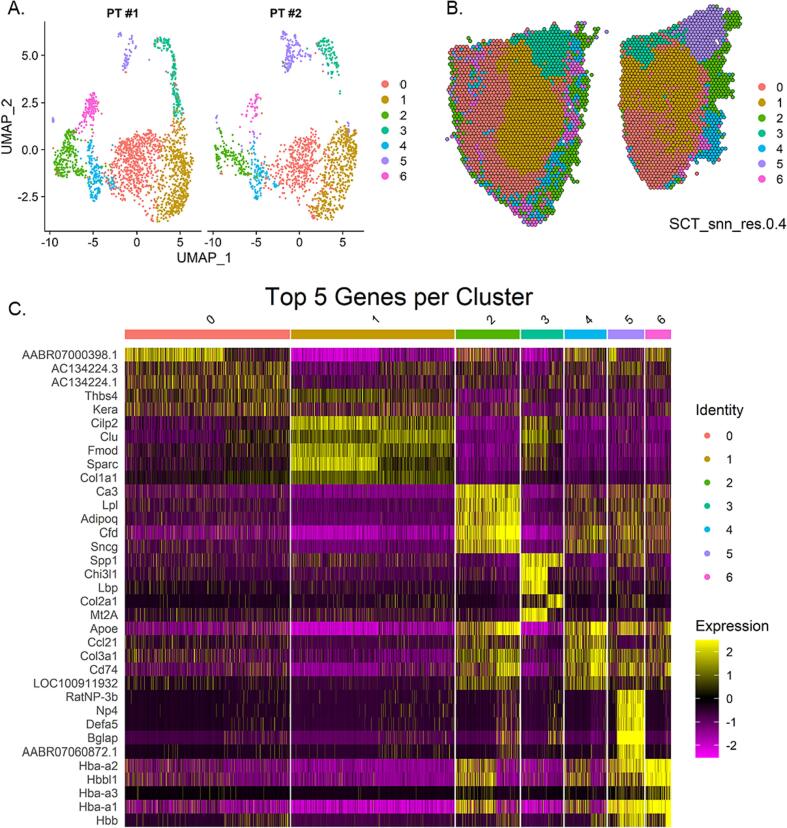
For cluster determination, the patellar tendon samples were merged and integrated together. We assumed that each patellar tendon should have similar cell populations because there was no difference in the sex, age, and morphology of the patellar tendons. Thus, integration should increase the sample size and power of clustering. Clusters identified with the same number represent the same cell population in each sample. There were seven clusters identified by unsupervised clustering at a resolution of 0.4 and 0.6 and these clusters were nearly identical (Fig. 2A). We chose the resolution to match the resolution used by a previously published tendon single cell RNA-seq dataset [10]. Higher resolutions produced an increased number of clusters with a minimal number of spots that did not correspond to histologically distinct tissue areas. The spatial projections of the clusters demonstrated that the two largest clusters (Cluster 0 and 1) were in the main tendon substance (Fig. 2) (Fig. 2B). Cluster 1 was characterized by expression of tendon-associated genes (Fmod, Sparc, Col1a1) and two genes (Cilp2 and Clu) that have been previously identified as markers of tendon fibroblasts [10] (Fig. 2C). Cluster 0 had an expression profile distinct from the other cluster and may represent spots of a mixed cell population that includes some fibroblasts. There was expression of fibroblast markers (Thbs4, Kera); however, expression of the other genes (AABR0700398.1, AC134224.3, AC134224.3) were not characteristic of a specific cell type [10](Fig. 2C). Hemoglobin gene expression was highest in cluster 6, suggesting that red blood cells were within this cluster (Fig. 2C). The spatial distribution of cluster 6 was not restricted to a specific area and was dispersed throughout the tendon periphery, to a larger extent in PT#1 than PT#2 (Fig. 2B). Cluster 5 contained unique genes encoding for defensins or defensin precursors (RatNP-3b, Np4, Defa5) (Fig. 2C). Defensins can be produced by neutrophils or endothelial cells which suggests that this cluster contained either or both of those cell types [42]. Clusters 2 and 4 had relatively similar expression patters and on the UMAP were in close proximity (Fig. 2A and C). Cluster 3 was spatially located near the enthesis and a subset of spots within this cluster was marked by Col2a1, which is typically expressed in fibrocartilage/cartilage [44] (Fig. 2B and C). The unsupervised clustering analysis displayed in Fig. 2 is limited by the resolution of the spots. Each spot can contain 1–10 cells and it is possible that many clusters contain multiple cell types. It is possible to deconvolute each spot; however, Seurat recommends single cell integration methods over deconvolution methods to determine underlying cell types.
Fig. 2.
Dimensionality Reduction and Clustering. A) UMAP of integrated samples displaying 7 clusters identified in the merged and integrated PT #1 & PT #2 samples. B) Visualization of integrated clusters projected onto each sample identity. PT#1 is on the left and PT#2 is on the right. C) Heatmap of the top three upregulated (sorted by adjusted p value) genes per cluster. Expression is the log2 fold change of the cluster relative to all other clusters and is derived from the SCT normalized, scaled data.
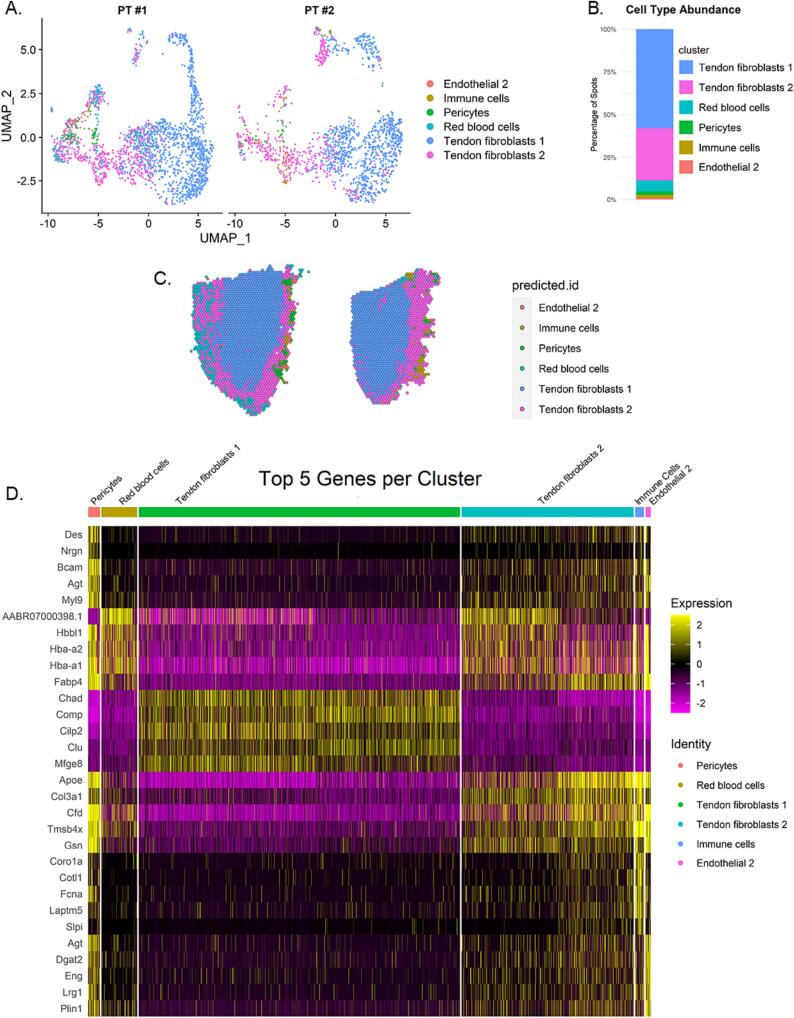
To understand the spatial distribution of previously characterized cell types within a tendon, we performed cell-type classifications by anchoring the data to a single-cell reference [10]. Interestingly, the single cell dataset identified 7 main cell types in the mouse Achilles tendon, yet we identified 5 distinct cell types in our samples (endothelial cells, immune cells, pericytes, red blood cells, and tendon fibroblasts) (Fig. 3). There were multiple populations of tendon fibroblasts that were distinguishable by spatial location and comprised 85 % of the overall population (Table 2, Fig. 3B). Tendon fibroblasts 1 comprised 57.4 % of the cells and were located directly in the tendon mid-substance, whereas tendon fibroblasts 2 were less prevalent (26.9 %) and located toward the periphery of the tendon (Fig. 3C and D). UMAP visualization of the clusters showed a major cluster that was formed primarily of tendon fibroblasts 1 and 2 (Fig. 3A). Tendon fibroblasts 1 were defined by expression of Chad, Comp, Cilp2, Clu, Ecrg4, Mfge8, Fmod, Cst3, Dcn, Col1a1, and Sparc (these genes all have equal p-values; however, only the first five are shown on the heatmap in Fig. 3D). Tendon fibroblasts 2 were characterized by expression of Apoe1, Col3a11, Cfd2, Tmsb4x2, Gsn1.
Fig. 3.
Cell Type Classifications. A) UMAP of predicted cell types in each tendon based on scRNA seq dataset from mouse Achilles tendon [10]. B) Bar graph of cell type proportion from combined samples. C) Spatial projections of cell types. PT#1 is on the left and PT#2 is on the right D) Heatmap of top 5 genes (ordered by adjusted p-value) expressed by each cell type. Expression is the log2 fold change of the cluster relative to all other clusters and is derived from the SCT normalized, scaled data.
Table 2.
Cell type predictions based on scRNA-seq data from mouse Achilles tendon [10]. Total counts is the number of spots assigned to particular cell type. Percentage is relative to total number of spots assigned to cell types.
| Tendon Fibroblasts 1 | Tendon Fibroblasts 2 | Red Blood Cells | Pericytes | Immune Cells | Endothelial 2 | Total | |
|---|---|---|---|---|---|---|---|
| PT #1 (counts) | 1211 | 659 | 213 | 59 | 14 | 23 | 2179 |
| PT #1 (%) | 55.58 | 30.24 | 9.78 | 2.71 | 0.64 | 1.06 | 100.00 |
| PT #2 (counts) | 990 | 522 | 31 | 21 | 47 | 13 | 1624 |
| PT #2 (%) | 60.96 | 32.14 | 1.91 | 1.29 | 2.89 | 0.80 | 100.00 |
| PT #1&2 (counts) | 2201 | 1181 | 244 | 80 | 61 | 36 | |
| PT #1&2 (%) | 57.87 | 31.05 | 6.42 | 2.10 | 1.60 | 0.94 |
That the two populations of tendon fibroblasts identified were in different locations within the tendon is interesting (Fig. 3). We hypothesize two potential explanations for the distinct location of these fibroblasts. First, it is possible that as the cell types become more mixed towards the tendon periphery, these spots have a mixed cell population and are not as pure as the fibroblasts in the midsubstance. The other possibility is that tendon fibroblast cell population 1 forms the central core of the tissue and deposits Col1a1 along the line of force, while the second fibroblast population exists to produce circumferential collagen that wraps around the tendon. In support of the second hypothesis, in vivo there is a small population of paratenon cells surrounding the tendon which produce circumferential collagen that wraps around the tendon[13].
RBCs, immune cells, pericytes, and endothelial cells were identified in the loose connective tissue surrounding the tendon (Fig. 3C). Expression of hemoglobin genes Hba-a1 and Hba-a2 was higher in RBCs, but their expression was not restricted to RBCs (Fig. 3D). Immune cells made up less than 2 % of the total cell population. The markers originally used define the immune cell population in the scRNA-seq data set were CD45 (Ptprc), CD206 (Mrc1) for M2 macrophages, and Ncf1 for neutrophils. However, only Ptprc and Mrc1 were identified as significant immune cell markers in this dataset, but not within the top 5 (Fig. 3, Table 2).
To identify genes with spatially variable expression that are highly expressed in tendon, we ranked genes based on equally weighting the spatial variability score (Moran’s I) and expression levels (Table 3, Table 4). Of the top 20 ranked genes in each sample, 15 were shared between the two samples and we have visualized the top 12 in Fig. 4: Col1a1, Sparc, AABR07000398.1, Cst3, Dcn, Comp, Fmod, Col1a2, Cilp2, Gsn, Chad, and Clu (Table 3, Table 4, Fig. 4). For each gene, the spatial expression pattern was similar for both samples. As expected, Col1a1 had the highest expression levels and was robustly expressed throughout the tendon proper (Fig. 4A). The class I small leucine rich proteoglycans (SLRP) Dcn and Fmod that regulate collagen fibrillogenesis and organization had similar spatial expression patterns to Col1a1 (Fig. 4E and G) [14], [43]. Clu is a secreted chaperone with expression highest in the central region of the tendon (Fig. 4L). Sparc and Cst3 also had expression similar to Col1a1 (Fig. 4B and D). Sparc is a matricellular protein that has previously been shown to be made in response to mechanical load and is necessary to transduce the mechanical load signal for tendon growth. The increase in collagen I protein in response to load also requires Sparc [40]. The pattern of Sparc and Col1a1 expression in Fig. 4 suggest that in a healthy tendon load is passed predominantly through the middle of the tissue.
Table 3.
Top 20 spatially variable genes in rat PT #1.
| Feature ID | Feature Name | I | P value | Adjusted p value | Feature Counts in Spots Under Tissue | Median Normalized Average Counts | Barcodes Detected per Feature | I Weighted Counts |
|---|---|---|---|---|---|---|---|---|
| ENSRNOG00000003897 | Col1a1 | 0.7152737 | 0 | 0 | 341,200 | 119.34223 | 2179 | 85.362354 |
| ENSRNOG00000012840 | Sparc | 0.6195742 | 0 | 0 | 106,461 | 34.189836 | 2176 | 21.183139 |
| ENSRNOG00000047746 | AABR07000398.1 | 0.8869153 | 0 | 0 | 22,877 | 18.697837 | 2003 | 16.583397 |
| ENSRNOG00000005195 | Cst3 | 0.5887732 | 0 | 0 | 78,775 | 25.763724 | 2172 | 15.168989 |
| ENSRNOG00000004554 | Dcn | 0.558697 | 0 | 0 | 63,975 | 21.959311 | 2170 | 12.268602 |
| ENSRNOG00000048472 | Comp | 0.5783764 | 0 | 0 | 37,768 | 11.857946 | 2072 | 6.8583556 |
| ENSRNOG00000003183 | Fmod | 0.5267232 | 0 | 0 | 34,862 | 11.1208 | 2070 | 5.8575833 |
| ENSRNOG00000011292 | Col1a2 | 0.443539 | 0 | 0 | 29,211 | 10.248291 | 2115 | 4.5455167 |
| ENSRNOG00000020622 | Cilp2 | 0.5889624 | 0 | 0 | 26,027 | 7.5967707 | 1879 | 4.4742119 |
| ENSRNOG00000018991 | Gsn | 0.4518909 | 0 | 0 | 27,830 | 9.6611837 | 2056 | 4.3658006 |
| ENSRNOG00000003304 | Chad | 0.5342007 | 0 | 0 | 24,683 | 7.6421095 | 1938 | 4.0824204 |
| ENSRNOG00000016460 | Clu | 0.4956906 | 0 | 0 | 26,448 | 7.8603905 | 1973 | 3.8963214 |
| ENSRNOG00000023576 | Ecrg4 | 0.4569186 | 0 | 0 | 22,404 | 7.80542 | 2028 | 3.5664416 |
| ENSRNOG00000052564 | Gpx3 | 0.287671 | 0 | 0 | 32,922 | 11.287373 | 2140 | 3.2470504 |
| ENSRNOG00000029886 | Hba-a1 | 0.5023811 | 0 | 0 | 11,793 | 4.7980757 | 1487 | 2.4104627 |
| ENSRNOG00000017510 | Mfge8 | 0.4398014 | 0 | 0 | 15,103 | 4.6407002 | 1852 | 2.0409862 |
| ENSRNOG00000014288 | Fn1 | 0.366919 | 0 | 0 | 16,126 | 5.2466108 | 1920 | 1.9250811 |
| ENSRNOG00000034234 | Mt-co1 | 0.1580482 | 0 | 0 | 33,522 | 12.10943 | 2149 | 1.9138734 |
| ENSRNOG00000003172 | Serpinf1 | 0.3479317 | 0 | 0 | 13,385 | 4.2168183 | 1861 | 1.4671649 |
| ENSRNOG00000055962 | Bgn | 0.3878743 | 0 | 0 | 11,364 | 3.6116581 | 1779 | 1.4008695 |
Table 4.
Top 20 spatially variable genes in rat PT #2.
| Feature ID | Feature Name | I | P value | Adjusted p value | Feature Counts in Spots Under Tissue | Median Normalized Average Counts | Barcodes Detected per Feature | I Weighted Counts |
|---|---|---|---|---|---|---|---|---|
| ENSRNOG00000003897 | Col1a1 | 0.7787277 | 0 | 0 | 313,387 | 181.70393 | 1624 | 141.49789 |
| ENSRNOG00000012840 | Sparc | 0.6572676 | 0 | 0 | 73,798 | 41.072086 | 1624 | 26.995353 |
| ENSRNOG00000004554 | Dcn | 0.7274626 | 0 | 0 | 48,188 | 30.731519 | 1623 | 22.356031 |
| ENSRNOG00000005195 | Cst3 | 0.6904837 | 0 | 0 | 51,709 | 30.119332 | 1622 | 20.796909 |
| ENSRNOG00000011292 | Col1a2 | 0.6758884 | 0 | 0 | 47,203 | 27.704577 | 1622 | 18.725203 |
| ENSRNOG00000048472 | Comp | 0.7237457 | 0 | 0 | 35,179 | 21.558311 | 1612 | 15.602736 |
| ENSRNOG00000016460 | Clu | 0.6903348 | 0 | 0 | 25,057 | 15.52295 | 1612 | 10.716032 |
| ENSRNOG00000003183 | Fmod | 0.6017048 | 0 | 0 | 26,427 | 15.296939 | 1608 | 9.2042422 |
| ENSRNOG00000003304 | Chad | 0.7576799 | 0 | 0 | 16,459 | 11.060864 | 1522 | 8.3805938 |
| ENSRNOG00000047746 | AABR07000398.1 | 0.6344597 | 0 | 0 | 22,826 | 12.834411 | 1546 | 8.1429164 |
| ENSRNOG00000020622 | Cilp2 | 0.6022516 | 0 | 0 | 17,816 | 10.643428 | 1578 | 6.4100214 |
| ENSRNOG00000023576 | Ecrg4 | 0.5924794 | 0 | 0 | 14,253 | 8.8763458 | 1580 | 5.2590524 |
| ENSRNOG00000018991 | Gsn | 0.3219171 | 0 | 0 | 28,143 | 12.494644 | 1604 | 4.0222395 |
| ENSRNOG00000018454 | Apoe | 0.6575318 | 0 | 0 | 19,470 | 5.1316503 | 1170 | 3.374223 |
| ENSRNOG00000017510 | Mfge8 | 0.4717455 | 0 | 0 | 9028 | 5.6481093 | 1540 | 2.6644704 |
| ENSRNOG00000029886 | Hba-a1 | 0.6158559 | 0 | 0 | 12,373 | 4.2851155 | 942 | 2.6390138 |
| ENSRNOG00000012471 | Thbs4 | 0.4313857 | 0 | 0 | 10,628 | 5.8898541 | 1528 | 2.540799 |
| ENSRNOG00000003357 | Col3a1 | 0.4583044 | 0 | 0 | 15,037 | 5.2407713 | 1414 | 2.4018684 |
| ENSRNOG00000019607 | Bglap | 0.8533258 | 0 | 0 | 5229 | 2.7202679 | 505 | 2.3212747 |
| ENSRNOG00000033564 | Cfd | 0.6599834 | 0 | 0 | 12,894 | 3.2623052 | 930 | 2.1530672 |
Fig. 4.
Top Expressed Spatially Variable Genes. The top 12 shared expressed spatially variable genes listed in order of rank (Moran’s I * mean normalized average counts) as represented in Patellar Tendon #1. A-L) For each gene, PT#1 is on the left and PT#2 on the right. The visualization of expression is from SCT normalized data.
Interestingly, three genes named for their role in cartilage, cartilage oligomeric matrix protein (Comp), cartilage intermediate layer protein (Cilp2), and chondroadherin (Chad) were also identified as top spatially variable genes in PT #1 (Fig. 4F, I and K). They had expression patterns similar to Col1a1 (Fig. 4A, F, I and K). Comp has been shown to be expressed in tendon[36], while Cilp2 is expressed in joint (articular and meniscal), but not growth plate cartilage [5]. Chad is a SLRP primarily recognized for its ability to bind type II collagen and cell matrix adhesion in cartilage [7], [26]. However, it has been suggested to assist with type I collagen crosslinking because it can bind a conserved sequence in collagen I that is located near a crosslinking motif[32].
There were two genes in the weighted spatial expression rankings within the top 12 for both samples, AABR0700398.1 and Gsn, that had expression patterns opposite of Col1a1, meaning they were less expressed in the tendon midsubstance (Fig. 4C and I). AABR0700398.1 does not have a known function but was highly expressed in both PT samples (Fig. 4C, Table 2, Table 3). Gsn encodes an actin binding protein called gelsolin that, may play a role in cell shape and motility [15]. It does not have a known function in tendon.
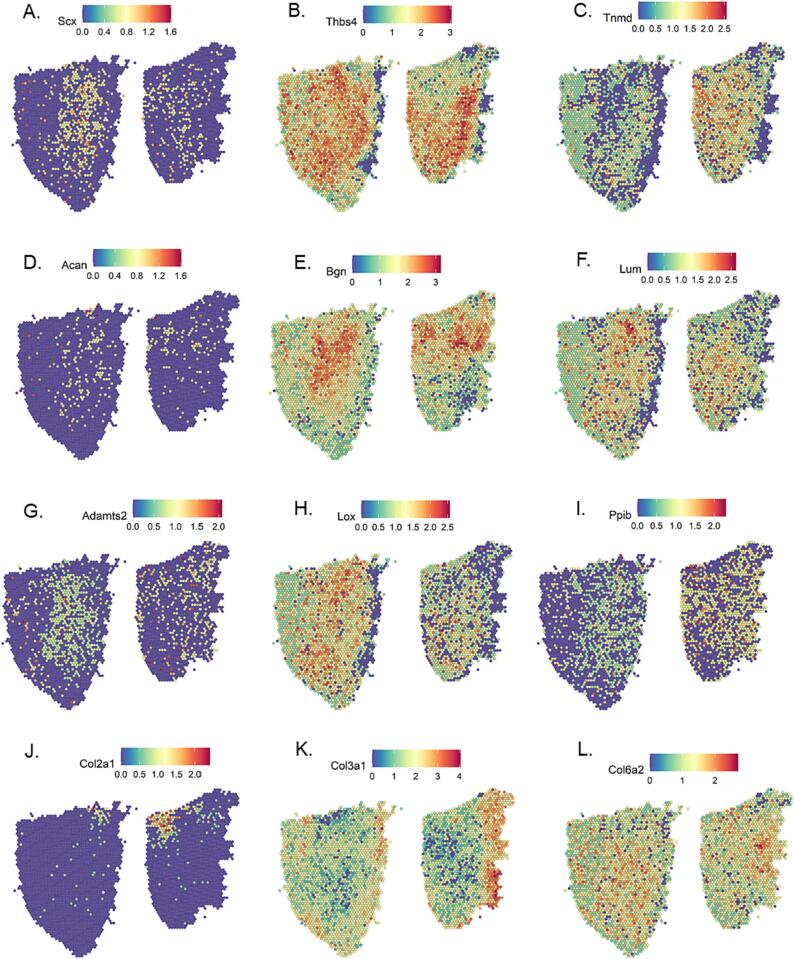
Lastly, we chose to visualize expression of genes that have a known function in tendon (Fig. 5). These genes were: Scx, Thbs4, Tnmd, Acan, Bgn, Lum, Adamts2, Lox, Ppib, Col2a1, Col3a1, and Col6a2. Scleraxis, thrombosponsin4, and tenomodulin are recognized to have higher expression in tendon/ligament than other tissues[21], [29], [35]. Scx expression was relatively low, which was not surprising given that these were adult tendons and scleraxis expression decreases in tendon development (Fig. 5A)[38]. The SLRPs biglycan and lumican had highest expression in the midsubstance (Fig. 5E and F). In contrast, the large proteoglycan aggrecan showed low expression and was not localized to a particular region (Fig. 5D).
Fig. 5.
Spatial Gene Expression of Selected Tendon-Associated Genes. A-L) For each gene, PT#1 is on the left and PT #2 is on the right. Note that the scale bars have different ranges for each gene. This is to allow for increased visualization of samples with low expression.
The spatial expression patterns of molecules (Ppib, Adamts2, Lox) used for type I collagen synthesis or crosslinking was similar to the pattern of Col1a1 itself (Fig. 5G – I, Fig. 4A). A single type I procollagen molecule must first undergo a series of post-translational modifications before the three chains can assemble into a procollagen molecule; this process requires Ppib [33]. The secreted procollagen is then cleaved both ends and at the N-terminal by ADAMTS2 [3]. These mature collagen fibrils are crosslinked by LOX and maintenance of normal fibril shape also requires LOX[19]. The overlapping expression patterns of Ppib, Adamts2, Lox and Col1a1 suggests tight spatial regulation of type I collagen homeostasis.
Besides, type I collagen there are other minor collagens in the tendon extracellular matrix. Type II collagen can occur as an adaptation to compression especially near the enthesis [4]. In our samples, Col2a1 expression was near but not in perfect alignment with the enthesis, possibly due to slight diffusion of RNA during permeabilization (Fig. 5K). Col3a1 expression was highest in the connective tissue in the tendon periphery (Fig. 5K). In healthy tendon, collagen III is present in minimal amounts[28]. However, the amount of type III collagen is increased in tendon scar tissue [41]and could possibly be due to migration of cells that express higher levels of type III collagen into the tendon proper. Lastly, collagen type VI is a non-fibrillar collagen that is required for collagen fibrillogenesis[20]and its expression was dispersed through the tendon (Fig. 5L).
In conclusion, we report the first complete spatial transcriptomics dataset for tendon from two healthy male rat patellar tendons. In our analysis, the cell clusters determined by unsupervised clustering match tendon morphology but are not representative of obvious cell types. When anchored to a single cell reference, the abundance and spatial location of the four major cell types identified in tendon match their expected location with tendon fibroblasts located throughout the tendon and immune cells, pericytes, and red blood cells located only in the tendon peripheral connective tissue. The gene expression visualization of the top spatially expressed genes included many genes that have a known function in tendon (Col1a1, Sparc, Dcn, Comp, Fmod, Col1a2) and their expression was throughout the midsubstance of the tendon proper, whereas other novel genes (AABR0700398.1 and Gsn) were found in the adjacent loose connective tissue. How these genes become spatially distinct and the role of AABR0700398.1 and Gsn remain to be determined.
Author Contributions.
DS, MM, and KB conceived the study. DS & KB collected data. DS analyzed data with help from the UC Davis Bioinformatics Core. DS drafted the manuscript. DS, MM, and KB revised the manuscript and approved the final version.
Declaration of competing interest
The authors declare the following financial interests/personal relationships which may be considered as potential competing interests: Keith Baar reports financial support was provided by Wu Tsai Foundation. Keith Baar reports financial support was provided by Human Performance Alliance.
Acknowledgements
UC Davis Genome Center – DNA Technologies and Expression Analysis Core performed the sequencing and was supported by NIH Shared Instrumentation Grant 1S10OD010786-01. Hannah Lyman and Blythe Durbin-Johnson at the UC Davis Bioinformatics Core for help with analysis and Suraj Pathak for technical assistance.
Funding
The current study was funded by the Joe and Clara Tsai Foundation (DS, MM, KB).
Data availability
Data will be made available on request.
References
- 1.Ackerman J.E., Best K.T., Muscat S.N., Pritchett E.M., Nichols A.E.C., Wu C.L., Loiselle A.E. Defining the spatial-molecular map of fibrotic tendon healing and the drivers of Scleraxis-lineage cell fate and function. Cell Reports. 2022;41(8) doi: 10.1016/j.celrep.2022.111706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Akbar, M., MacDonald, L., Crowe, L. A. N., Carlberg, K., Kurowska-Stolarska, M., Ståhl, P. L., Snelling, S. J. B., McInnes, I. B., & Millar, N. L. (2021). Single cell and spatial transcriptomics in human tendon disease indicate dysregulated immune homeostasis. In Annals of the Rheumatic Diseases (Vol. 80, Issue 11). https://doi.org/10.1136/annrheumdis-2021-220256. [DOI] [PMC free article] [PubMed]
- 3.Bekhouche, M., & Colige, A. (2015). The procollagen N-proteinases ADAMTS2, 3 and 14 in pathophysiology. In Matrix Biology (Vols. 44–46, pp. 46–53). https://doi.org/10.1016/j.matbio.2015.04.001. [DOI] [PubMed]
- 4.Benjamin M., Ralphs J.R. Fibrocartilage in tendons and ligaments–an adaptation to compressive load. J Anat. 1998;193(4):481–494. doi: 10.1046/j.1469-7580.1998.19340481.x. https://www.ncbi.nlm.nih.gov/pubmed/10029181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bernardo B.C., Belluoccio D., Rowley L., Little C.B., Hansen U., Bateman J.F. Cartilage intermediate layer protein 2 (CILP-2) is expressed in articular and meniscal cartilage and down-regulated in experimental osteoarthritis. Journal of Biological Chemistry. 2011;286(43) doi: 10.1074/jbc.M111.248039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Butler A., Hoffman P., Smibert P., Papalexi E., Satija R. Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nature Biotechnology. 2018;36(5) doi: 10.1038/nbt.4096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Camper L., Heinegård D., Lundgren-Åkerlund E. Integrin α2β1 is a receptor for the cartilage matrix protein chondroadherin. Journal of Cell Biology. 1997;138(5) doi: 10.1083/jcb.138.5.1159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Canty E.G., Starborg T., Lu Y., Humphries S.M., Holmes D.F., Meadows R.S., Huffman A., O’Toole E.T., Kadler K.E. Actin filaments are required for fibripositor-mediated collagen fibril alignment in tendon. J Biol Chem. 2006;281(50):38592–38598. doi: 10.1074/jbc.M607581200. [DOI] [PubMed] [Google Scholar]
- 9.De Micheli A.J., Laurilliard E.J., Heinke C.L., Ravichandran H., Fraczek P., Soueid-Baumgarten S., De Vlaminck I., Elemento O., Cosgrove B.D. Single-Cell Analysis of the Muscle Stem Cell Hierarchy Identifies Heterotypic Communication Signals Involved in Skeletal Muscle Regeneration. Cell Reports. 2020;30(10) doi: 10.1016/j.celrep.2020.02.067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.de Micheli A.J., Swanson J.B., Disser N.P., Martinez L.M., Walker N.R., Oliver D.J., Cosgrove B.D., Mendias C.L. Single-cell transcriptomic analysis identifies extensive heterogeneity in the cellular composition of mouse Achilles tendons. American Journal of Physiology - Cell Physiology. 2020;319(5) doi: 10.1152/ajpcell.00372.2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Disser N.P., Ghahramani G.C., Swanson J.B., Wada S., Chao M.L., Rodeo S.A., Oliver D.J., Mendias C.L. Widespread diversity in the transcriptomes of functionally divergent limb tendons. Journal of Physiology. 2020 doi: 10.1113/JP279646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Disser N.P., Piacentini A.N., De Micheli A.J., Schonk M.M., Yao V.J.H., Deng X.H., Oliver D.J., Rodeo S.A. Achilles Tendons Display Region-Specific Transcriptomic Signatures Associated With Distinct Mechanical Properties. American Journal of Sports Medicine. 2022;50(14) doi: 10.1177/03635465221128589. [DOI] [PubMed] [Google Scholar]
- 13.Dyment N.A., Liu C.F., Kazemi N., Aschbacher-Smith L.E., Kenter K., Breidenbach A.P., Shearn J.T., Wylie C., Rowe D.W., Butler D.L. The Paratenon Contributes to Scleraxis-Expressing Cells during Patellar Tendon Healing. PLoS ONE. 2013;8(3) doi: 10.1371/journal.pone.0059944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ezura Y., Chakravarti S., Oldberg Å., Chervoneva I., Birk D.E. Differential Expression of Lumican and Fibromodulin Regulate Collagen Fibrillogenesis in Developing Mouse Tendons. The Journal of Cell Biology. 2000;151(4):779–788. doi: 10.1083/jcb.151.4.779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Feldt J., Schicht M., Garreis F., Welss J., Schneider U.W., Paulsen F. Structure, regulation and related diseases of the actin-binding protein gelsolin. Expert Reviews in Molecular Medicine. 2019;20 doi: 10.1017/erm.2018.7. [DOI] [PubMed] [Google Scholar]
- 16.Fu W., Yang R., Li J. Single-cell and spatial transcriptomics reveal changes in cell heterogeneity during progression of human tendinopathy. BMC Biology. 2023;21(1) doi: 10.1186/s12915-023-01613-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hao Y., Hao S., Andersen-Nissen E., Mauck W.M., Zheng S., Butler A., Lee M.J., Wilk A.J., Darby C., Zager M., Hoffman P., Stoeckius M., Papalexi E., Mimitou E.P., Jain J., Srivastava A., Stuart T., Fleming L.M., Yeung B., Satija R. Integrated analysis of multimodal single-cell data. Cell. 2021;184(13) doi: 10.1016/j.cell.2021.04.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Harvey T., Flamenco S., Fan C.M. A Tppp3 + Pdgfra + tendon stem cell population contributes to regeneration and reveals a shared role for PDGF signalling in regeneration and fibrosis. Nature Cell Biology. 2019 doi: 10.1038/s41556-019-0417-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Herchenhan A., Uhlenbrock F., Eliasson P., Weis M., Eyre D., Kadler K.E., Magnusson S.P., Kjaer M. Lysyl Oxidase Activity Is Required for Ordered Collagen Fibrillogenesis by Tendon Cells. J Biol Chem. 2015;290(26):16440–16450. doi: 10.1074/jbc.M115.641670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Izu Y., Ansorge H.L., Zhang G., Soslowsky L.J., Bonaldo P., Chu M.-L., Birk D.E. Dysfunctional tendon collagen fibrillogenesis in collagen VI null mice. Matrix Biology. 2011;30(1):53–61. doi: 10.1016/j.matbio.2010.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jelinsky S.A., Archambault J., Li L., Seeherman H. Tendon-selective genes identified from rat and human musculoskeletal tissues. J Orthop Res. 2010;28(3):289–297. doi: 10.1002/jor.20999. [DOI] [PubMed] [Google Scholar]
- 22.Kapacee Z., Richardson S., Lu Y., Stargorg T., Holmes D., Baar K., Kadler K. Tension is required for fibripositor formation. Matrix Biology. 2008;27(4):371–375. doi: 10.1016/j.matbio.2007.11.006. [DOI] [PubMed] [Google Scholar]
- 23.Karlsen A., Yeung C.Y.C., Schjerling P., Denz L., Hoegsbjerg C., Jakobsen J.R., Krogsgaard M.R., Koch M., Schiaffino S., Kjaer M., MacKey A.L. Distinct myofibre domains of the human myotendinous junction revealed by single-nucleus RNA sequencing. Journal of Cell Science. 2023;136(8) doi: 10.1242/jcs.260913. [DOI] [PubMed] [Google Scholar]
- 24.Kendal A.R., Layton T., Al-Mossawi H., Appleton L., Dakin S., Brown R., Loizou C., Rogers M., Sharp R., Carr A. Multi-omic single cell analysis resolves novel stromal cell populations in healthy and diseased human tendon. Scientific Reports. 2020;10(1):13939. doi: 10.1038/s41598-020-70786-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Korsunsky I., Millard N., Fan J., Slowikowski K., Zhang F., Wei K., Baglaenko Y., Brenner M., Loh P., ru, & Raychaudhuri, S. Fast, sensitive and accurate integration of single-cell data with Harmony. Nature Methods. 2019;16(12) doi: 10.1038/s41592-019-0619-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Larsson T., Sommarin Y., Paulsson M., Antonsson P., Hedbom E., Wendel M., Heinegard D. Cartilage matrix proteins. A basic 36-kDa protein with a restricted distribution to cartilage and bone. Journal of Biological Chemistry. 1991;266(30) doi: 10.1016/s0021-9258(18)54941-3. [DOI] [PubMed] [Google Scholar]
- 27.Lee A.H., Elliott D.M. Comparative multi-scale hierarchical structure of the tail, plantaris, and Achilles tendons in the rat. Journal of Anatomy. 2019;234(2):252–262. doi: 10.1111/joa.12913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Little D., Thompson J.W., Dubois L.G., Ruch D.S., Moseley M.A., Guilak F. Proteomic Differences between Male and Female Anterior Cruciate Ligament and Patellar Tendon. PLoS ONE. 2014;9(5):e96526. doi: 10.1371/journal.pone.0096526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mienaltowski M.J., Cánovas A., Fates V.A., Hampton A.R., Pechanec M.Y., Islas-Trejo A., Medrano J.F. Transcriptome profiles of isolated murine Achilles tendon proper- and peritenon-derived progenitor cells. Journal of Orthopaedic Research. 2019;37(6):1409–1418. doi: 10.1002/jor.24076. [DOI] [PubMed] [Google Scholar]
- 30.Moore M.J., De Beaux A. A quantitative ultrastructural study of rat tendon from birth to maturity. Journal of Anatomy. 1987;153 [PMC free article] [PubMed] [Google Scholar]
- 31.Noah A.C., Li T.M., Martinez L.M., Wada S., Swanson J.B., Disser N.P., Sugg K.B., Rodeo S.A., Lu T.T., Mendias C.L. Adaptive and innate immune cell responses in tendons and lymph nodes after tendon injury and repair. Journal of Applied Physiology. 2020;128(3) doi: 10.1152/japplphysiol.00682.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Paracuellos P., Kalamajski S., Bonna A., Bihan D., Farndale R.W., Hohenester E. Structural and functional analysis of two small leucine-rich repeat proteoglycans, fibromodulin and chondroadherin. Matrix Biology. 2017;63 doi: 10.1016/j.matbio.2017.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pyott S.M., Schwarze U., Christiansen H.E., Pepin M.G., Leistritz D.F., Dineen R., Harris C., Burton B.K., Angle B., Kim K., Sussman M.D., Weis M.A., Eyre D.R., Russell D.W., McCarthy K.J., Steiner R.D., Byers P.H. Mutations in PPIB (cyclophilin B) delay type I procollagen chain association and result in perinatal lethal to moderate osteogenesis imperfecta phenotypes. Human Molecular Genetics. 2011;20(8) doi: 10.1093/hmg/ddr037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Satija R., Farrell J.A., Gennert D., Schier A.F., Regev A. Spatial reconstruction of single-cell gene expression data. Nature Biotechnology. 2015;33(5) doi: 10.1038/nbt.3192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schweitzer R., Chyung J.H., Murtaugh L.C., Brent A.E., Rosen V., Olson E.N., Lassar A., Tabin C.J. Analysis of the tendon cell fate using Scleraxis, a specific marker for tendons and ligaments. Development. 2001;128(19):3855–3866. doi: 10.1242/dev.128.19.3855. [DOI] [PubMed] [Google Scholar]
- 36.Smith R.K.W., Zunino L., Webbon P.M., Heinegård D. The distribution of Cartilage Oligomeric Matrix Protein (COMP) in tendon and its variation with tendon site, age and load. Matrix Biology. 1997;16(5):255–271. doi: 10.1016/S0945-053X(97)90014-7. [DOI] [PubMed] [Google Scholar]
- 37.Ståhl, P. L., Salmén, F., Vickovic, S., Lundmark, A., Navarro, J. F., Magnusson, J., Giacomello, S., Asp, M., Westholm, J. O., Huss, M., Mollbrink, A., Linnarsson, S., Codeluppi, S., Borg, Å., Pontén, F., Costea, P. I., Sahlén, P., Mulder, J., Bergmann, O., … Frisén, J. (2016). Visualization and analysis of gene expression in tissue sections by spatial transcriptomics. In Science (Vol. 353, Issue 6294). https://doi.org/10.1126/science.aaf2403. [DOI] [PubMed]
- 38.Steffen D., Avey A., Mienaltowski M.J., Baar K. The rat Achilles and patellar tendons have similar increases in mechanical properties but become transcriptionally divergent during postnatal development. Journal of Physiology. 2023 doi: 10.1113/JP284393. [DOI] [PubMed] [Google Scholar]
- 39.Stuart T., Butler A., Hoffman P., Stoeckius M., Smibert P., Satija R., Hafemeister C., Papalexi E., Mauck Iii W.M., Hao Y. Comprehensive Integration of Single-Cell Data Resource Comprehensive Integration of Single-Cell Data. Cell. 2019;177 doi: 10.1016/j.cell.2019.05.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang T., Wagner A., Gehwolf R., Yan W., Passini F.S., Thien C., Weissenbacher N., Lin Z., Lehner C., Teng H., Wittner C., Zheng Q., Dai J., Ni M., Wang A., Papadimitriou J., Leys T., Tuan R.S., Senck S., Traweger A. Load-induced regulation of tendon homeostasis by SPARC, a genetic predisposition factor for tendon and ligament injuries. Science Translational Medicine. 2021;13(582) doi: 10.1126/scitranslmed.abe5738. [DOI] [PubMed] [Google Scholar]
- 41.Williams I.F., Heaton A., McCullagh K.G. Cell morphology and collagen types in equine tendon scar. Research in Veterinary Science. 1980;28(3):302–310. http://www.ncbi.nlm.nih.gov/pubmed/7414083 [PubMed] [Google Scholar]
- 42.Xu, D., & Lu, W. (2020). Defensins: A Double-Edged Sword in Host Immunity. In Frontiers in Immunology (Vol. 11). https://doi.org/10.3389/fimmu.2020.00764. [DOI] [PMC free article] [PubMed]
- 43.Zhang G., Ezura Y., Chervoneva I., Robinson P.S., Beason D.P., Carine E.T., Soslowsky L.J., Iozzo R.V., Birk D.E. Decorin regulates assembly of collagen fibrils and acquisition of biomechanical properties during tendon development. Journal of Cellular Biochemistry. 2006;98(6) doi: 10.1002/jcb.20776. [DOI] [PubMed] [Google Scholar]
- 44.Zhang T., Wan L., Xiao H., Wang L., Hu J., Lu H. Single-cell RNA sequencing reveals cellular and molecular heterogeneity in fibrocartilaginous enthesis formation. Elife. 2023;12 doi: 10.7554/elife.85873. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data will be made available on request.