SUMMARY
Atmospheric chemosynthesis is a recently proposed form of chemoautotrophic microbial primary production. The proposed process relies on the oxidation of trace concentrations of hydrogen (≤530 ppbv), carbon monoxide (≤90 ppbv), and methane (≤1,870 ppbv) gases using high-affinity enzymes. Atmospheric hydrogen and carbon monoxide oxidation have been primarily linked to microbial growth in desert surface soils scarce in liquid water and organic nutrients, and low in photosynthetic communities. It is well established that the oxidation of trace hydrogen and carbon monoxide gases widely supports the persistence of microbial communities in a diminished metabolic state, with the former potentially providing a reliable source of metabolic water. Microbial atmospheric methane oxidation also occurs in oligotrophic desert soils and is widespread throughout copiotrophic environments, with established links to microbial growth. Despite these findings, the direct link between trace gas oxidation and carbon fixation remains disputable. Here, we review the supporting evidence, outlining major gaps in our understanding of this phenomenon, and propose approaches to validate atmospheric chemosynthesis as a primary production process. We also explore the implications of this minimalistic survival strategy in terms of nutrient cycling, climate change, aerobiology, and astrobiology.
KEYWORDS: atmospheric chemosynthesis, microbial ecology, astrobiology, nutrient cycling, climate change, aerobiology, trace gases, hydrogenase, primary production, foundational science
THE PROPOSED NICHE FOR BACTERIA THAT LIVE ON TRACE GASES
In desert soils, water and inorganic electron donors are often scarce (1–5), limiting photoautotrophs (6–10) and the chemoautotrophs that use inorganic electron donors present within the soil from non-atmospheric sources (11–13) to low abundances and activities. This restricts carbon and energy inputs from these well-established primary production strategies. Despite the harsh conditions, microbial communities in these arid environments are frequently abundant and diverse. In these ecosystems, alternative energy acquisition strategies supplement carbon and energy inputs and aid microbial persistence. These strategies include trace gas oxidation (14–16) and heterotrophic activities fueled by nutrient inputs from aeolian deposition (17). Until recently, an understanding of what processes support primary production in such communities was largely unknown.
Hydrogen (H2), carbon monoxide (CO), and methane (CH4) concentrations fluctuate within the atmosphere due to extensive biological and chemical cycling, with average global mixing ratios of 531 (18, 19), 90 (20–23), and 1,857 (24) ppbv, respectively. These gases are ubiquitously present at the interface of air and soil, diffuse readily into the soil and microbial cells, and produce electron yields upon oxidation for input into diverse biochemical reactions, including the electron transport chain (25–29). As a result, the oxidation of these gases represents the use of a highly reliable energy source, particularly for microbial communities that are starved of alternative energy sources, such as those inhabiting dry and oligotrophic desert soils.
Within terrestrial microbiomes, the activity of high-affinity hydrogenases, carbon monoxide dehydrogenases, and methane monoxygenases, capable of oxidizing trace gases, has been predominantly linked to microbial survival strategies rather than cellular growth (15, 30–33). In cultivated taxa, the link between trace gas oxidation and microbial survival has been supported by the upregulation of these enzymes during nutrient starvation, particularly in late-stage exponential and stationary growth phases (26, 30, 34–36). However, given the low abundance of well-characterized primary producers in extreme environments, particularly oligotrophic soil niches, and the prevalence of diverse microbiomes that include trace gas oxidizing bacteria, it has been hypothesized that the energy liberated from trace gas oxidation also supports microbial primary production in these environments through a process coined “atmospheric chemosynthesis” (14, 15, 31, 37).
ATMOSPHERIC CHEMOSYNTHESIS: THE PROPOSED PATHWAYS
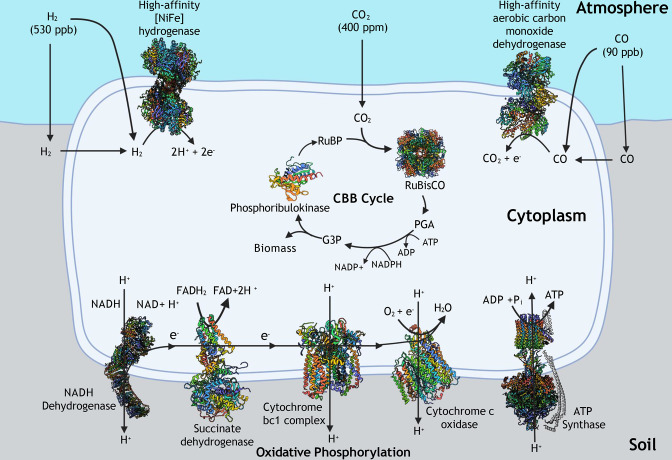
During atmospheric chemosynthesis, high-affinity [NiFe]-hydrogenases (forms 1h, 1l, 1m, and 2a) and carbon monoxide dehydrogenases are proposed to oxidize trace H2 and CO, respectively (Fig. 1). These [NiFe]-hyrogenases have a substantially higher affinity for H2 [Michaelis constant (Km) = 30–200 nM], compared to other forms (Km > 500 nM) which are unable to conduct hydrogen oxidation at trace atmospheric levels (38). Similarly, a large group of microbial carbon monoxide dehydrogenases have a low affinity for CO (Km > 400 nM), making them unable to oxidize CO at atmospheric levels (39). Comparatively, higher affinity carbon monoxide dehydrogenases (Km < 400 nM) are required for this to proceed (30). The high activity of these enzymes liberates electrons for input into the electron transport chain, resulting in the production of ATP (40–42). It is hypothesized that, due to the rapid activity of the hydrogenases and carbon monoxide dehydrogenases observed during activity studies, sufficient ATP is produced to surpass cellular maintenance energy requirements, and is directed into carbon fixation pathways, allowing inorganic carbon to be fixed into microbial biomass (Fig. 1) (14, 37, 43).
Fig 1.
The proposed pathway for trace gas-driven carbon fixation. Representative protein structures used were obtained from the Research Collaboratory for Structural Bioinformatics (RCSB) Protein Data Bank (PDB); 1h [NiFe]-hydrogenase from Cupriavidus necator H16 (PDB ID = 5AA5), carbon monoxide dehydrogenase from Oligotropha carboxidovorans (PDB ID = 1N5W), RuBisCO from C. necator (PDB ID = 1BXN), phosphoribulokinase from Cereibacter sphaeroides (PDB ID = 1A7J), NADH dehydrogenase from Thermus thermophilus (PDB ID = 6I1P), succinate dehydrogenase with ubiquinone bound from Escherichia coli (PDB ID = 1NEK), cytochrome bc1 complex from Paracoccus denitrificans PD1222 (PDB ID = 2YIU), cytochrome c oxidase from Rhodobacter sphaeroides (PDB ID = 1M56), and ATP synthase from Paracoccus denitrificans (PDB ID = 5DN6). During atmospheric chemosynthesis, trace gases are oxidized by high-affinity hydrogenases and carbon monoxide dehydrogenases. Electrons liberated drive oxidative phosphorylation, which produces ATP, which is subsequently input into the RuBisCO form IE-driven Calvin-Benson-Bassham (CBB) cycle. The orientation and size of proteins are not to scale, and the CBB cycle displayed is simplified. C. necator was selected as the model organism for 1h [NiFe]-hydrogenase as although lower affinity, higher affinity variations of this enzyme have yet to be structurally modeled through pure enzyme studies. Inspired by references (14, 40).
The Calvin-Benson-Bassham (CBB) cycle is the microbial carbon fixation pathway most associated with atmospheric chemosynthesis. This is due to the widespread and abundant detection of CBB cycle enzyme markers alongside trace gas oxidation genes within soils (44), and derived metagenome-assembled genomes (MAGs) (31, 37, 45, 46). In particular, these genetic markers co-occur in numerous soils that rapidly oxidize trace gases and have shown significant increases in carbon fixation upon trace gas stimulation (14, 43, 47). For example, in Robinson Ridge, East Antarctica, soils oxidized H2 at 3.49 nmol/h/g and CO at 0.42 nmol/h/g, with 1h [NiFe]-hydrogenase (hhyL), carbon monoxide dehydrogenase (coxL), and RuBisCO form IE (rbcL1E) gene expression confirmed. Furthermore, in this study, these genetic markers were detected in 43%, 13%, and 30% of the MAGs examined, respectively (14). In contrast, genetic indicators of alternative microbial carbon fixation pathways are frequently far less abundant.
Despite the dominance of CBB cycle genes widely associated with atmospheric chemosynthesis in oligotrophic desert soils, trace hydrogen oxidation has been more recently linked to microbial carbon fixation through the reverse tricarboxylic acid (rTCA) cycle (48). In this case, the metabolically flexible genus Nitrospira supplements nitrite-dependent growth using electrons derived from atmospheric H2 oxidation by high-affinity group 2a [NiFe]-hydrogenase (48). In contrast to oligotrophic cold desert soils, where 2a [NiFe]-hydrogenase has a low relative prevalence (43, 47), this enzyme is widely detected in marine surface waters, with its expression linked to mixotrophic growth in the marine bacteria Sphingopyxis alaskensis RB2256 (49). Mixotrophy is the simultaneous usage of heterotrophic and autotrophic processes, thereby involving multiple different inorganic and organic carbon and energy sources (50).
Links between the microbial oxidation of atmospheric methane and carbon fixation offer another alternative atmospheric chemosynthetic pathway. In this case, oxygen-dependent particulate methane monooxygenase oxidizes methane to methanol, which is then oxidized to formaldehyde by periplasmic methanol dehydrogenase (51, 52). After being transported into the cell, the formaldehyde is oxidized to formate, which is then oxidized to CO2 or incorporated into microbial biomass through the reductive glycine pathway and the serine cycle (51, 52).
Atmospheric chemosynthesis is predicted to complement heterotrophic energy and carbon acquisition pathways. For example, bacteria within the phylum Eremiobacterota encode glycoside hydrolase, complete glycolysis and pentose phosphate pathways, and a complete tricarboxylic acid (TCA) cycle, while MAGs from six candidate Eremiobacterota genera are putatively atmospheric chemosynthetic, co-encoding a high-affinity [NiFe]-hydrogenase, RuBisCO form IE and a complete CBB cycle (53). This indicates a mixotrophic lifestyle, with the genetic potential for atmospheric chemosynthesis alongside heterotrophy (53). Indeed, the first Eremiobacterota isolate, Vulcanimicrobium alpinus, recently obtained from a dark, oligotrophic, volcanic cave ecosystem rich in CO2, is a highly metabolically flexible aerobic anoxygenic photoheterotrophic bacterium. This strain appears to require elevated CO2 concentrations (>5%) for growth, even when cultivated under heterotrophic conditions (54). Despite this finding, V. alpinus was able to grow when incubated under the light in organic carbon-rich 6 cm stab cultures, despite being sealed to surrounding air and receiving no headspace amendments throughout the 30-day incubation (54). Further characterization studies are essential to clarify the underlying growth strategies of this novel taxa, including its capacity for atmospheric chemosynthesis alongside heterotrophic and photosynthetic mechanisms, and under what environmental conditions each is activated. Moreover, the isolation and characterization of additional Eremiobacterota cultures are vital for fully understanding this metabolically flexible phylum, including the potential for atmospheric chemosynthesis.
GAPS IN OUR FOUNDATIONAL UNDERSTANDING OF ATMOSPHERIC CHEMOSYNTHESIS
Trace gas oxidation can now be viewed as an almost ubiquitous process in terrestrial microbiomes, with rates quantified in numerous and diverse soils (55–58), including deserts (14, 15, 30, 31, 43, 46, 47), forests (59, 60), and volcanic soils (61–63). However, trace gas oxidation is generally slower in soils from temperate desert regions compared to their colder counterparts, often requiring wetting that mimics rare rainfall events for rapid oxidation to be observed (31, 46, 47). For example, in unwetted Negev Desert soils, H2 oxidation rates have been reported as 0.009–0.035 nmol/h/g, and in Australian dryland soil, H2 and CO oxidation rates have been reported as 0.00064 nmol/h/g and 0.0061 nmol/h/g, respectively (31). Comparatively, 6.3–623.9 H2 nmol/mol/h/g and 388 0–2.6 CO nmol/mol/h/g have reported in unwetted soils from the Antarctic, high Arctic, and Tibetan Plateau (43). Furthermore, few studies directly link rapid trace gas oxidation rates with significant increases in microbial carbon fixation (p < 0.05); such observations are restricted to cold deserts throughout the Antarctic, including the Vestfold Hills (43), the Windmill Islands (14), the McMurdo Dry Valleys (43), and high Arctic soils (43), with temperate deserts limited to Israel (47). Therefore, while trace gas oxidation has been established as a widespread mechanism for sustaining microbial survival, it remains unclear how pervasive atmospheric chemosynthesis is as a growth strategy in terrestrial ecosystems.
It is also unclear whether trace gas oxidation supports atmospheric chemosynthetic growth in other, more diverse environments. The recent identification of trace gas oxidizing microorganisms in marine ecosystems (63) demonstrates the potentially under-investigated ecological significance of these microorganisms in aquatic environments. There is a definite need to extend studies of the diversity and function of trace gas chemosynthetic microorganisms into diverse environmental reservoirs, including aquatic habitats (oceans, rivers, lakes, and wetlands), endolithic environments (inside rocks), lithic niches (the deep subterranean biosphere), and the aerosphere.
Rapid improvements in sequencing technologies over the past 5 years have expanded our understanding of atmospheric chemosynthesis. Genetic markers for trace gas oxidation, including those encoding RuBisCOs, high-affinity hydrogenases, and carbon monoxide dehydrogenases, are ubiquitous in soils globally, including throughout the Antarctic (14, 37, 44), Arctic (44), Tibetan Plateau (44), Israel (47), Namibia (46), the Americas (55, 64–66), and Australasia (31, 32, 46). In soils, the expression of RuBisCO form IE has been detected alongside key markers for trace gas oxidation; group 1h [NiFe]-hydrogenase and aerobic carbon monoxide dehydrogenase (14). Through extensive MAG analysis, numerous bacterial phyla have been shown to have the genetic capacity for atmospheric chemosynthesis. Relevant marker genes, in particular rbcL1E (encoding RuBisCO Form IE), coxL (encoding the large subunit of carbon monoxide dehydrogenase), and hhySL (encoding small and large subunits of high-affinity hydrogenase) co-occur in Actinobacteriota, Ca. Dormibacterota, Eremiobacterota, Chloroflexota, Firmicutes, Deinococcota, and Verrucomicrobiota (14, 43).
The taxonomic diversity of microorganisms performing atmospheric chemosynthesis may extend beyond seven phyla, particularly if additional unidentified enzymes and pathways are involved. For example, RuBisCO form IE and the CBB cycle are typically linked with atmospheric chemosynthesis, as they frequently co-occur and are co-expressed alongside trace gas oxidation markers in microbiomes that demonstrate significant increases in microbial carbon fixation activity with H2 supplementation at atmospherically relevant levels (14, 37, 43, 47). However, other yet to be identified forms of RuBisCO may also be involved in the process. RuBisCO forms IC and ID are of particular interest due to their close phylogenetic relationship with form IE, their association with light-independent primary production and their co-detection with 1h and/or 1l [NiFe]-hydrogenases within trace gas oxidizing microbiomes (67) and associated MAGs, including the uncultivated Chloroflexota class Ellin6529 (37). Furthermore, novel forms of high-affinity hydrogenases continue to be uncovered in diverse taxa and environments (37, 38, 43, 49, 68), suggesting that their distribution and ecological significance require further elucidation. To fully understand the atmospheric chemosynthetic pathway, RuBisCO form IE and associated trace gas oxidative proteins must be purified and structurally and enzymatically characterized so that any distinguishing features and activities can be determined. Only then can genetic marker detection be implemented to accurately assess the taxonomic distribution of microorganisms capable of atmospheric chemosynthesis.
Critically, while whole microbiome activity assays indicate the occurrence of atmospheric chemosynthesis, there is currently no unequivocal evidence linking the processes of trace gas oxidation and carbon fixation in individual microorganisms. Although MAG analysis has revealed microbial taxa that exhibit the genetic capacity for atmospheric chemosynthesis within these whole microbiomes, further studies are required to confirm that these microorganisms perform atmospheric chemosynthesis in pure culture. Furthermore, a selection of MAGs with atmospheric chemosynthetic potential have cultured representatives, which are heterotrophic or chemoautotrophic, having been isolated under conditions rich in organic carbon or inorganic electron donors (43). It is vital that these isolated microorganisms are studied under conditions conducive to atmospheric chemosynthesis, namely trace H2 and CO exposure under dark incubation, to confirm that they conduct this growth mechanism upon nutrient starvation.
Numerous approaches to validate this process rely upon the isolation of putatively atmospheric chemosynthesis microorganisms. Alternatively, atmospheric chemosynthesis could be validated and characterized in environmental microbiomes or enrichment cultures through the application of more sophisticated techniques, including DNA stable isotope probing (SIP), metatranscriptomic or metaproteomic analysis. In the case of DNA SIP, soils or enrichments would be incubated under conditions that inhibit photoautotrophs and stimulate atmospheric chemosynthetic activity, through the inclusion of trace gases alongside 13CO2. Following incubation, labeled and unlabeled DNA fractions would each be characterized using community profiling and metagenomic approaches. This approach would require substantial optimization, particularly regarding incubation time and soil or enrichment selection. However, if effectively implemented with appropriate controls and replication, this technique could prove invaluable for validating atmospheric chemosynthesis, and identifying taxa that undertake this process even within difficult-to-culture “microbial dark matter”. Perhaps more simply, metatranscriptomics or proteomics approaches could be applied under similar incubation conditions without radio isotopically labeling the headspace CO2. Conducting this experiment under varying conditions (e.g., different combinations of atmospheric gases at different concentrations) and multiple timepoints could effectively identify the atmospheric chemosynthesis pathway and characterize the conditions in which this survival strategy is activated. Furthermore, with a binning approach, omics techniques could continue to identify atmospheric chemosynthetic taxa within complex microbiomes.
Applying expression-based omics strategies to soil microbiomes has traditionally been technically challenging (69) due to RNA instability (70, 71), the co-extraction of complex organics (72) which impair molecular analysis (69), and considerable community complexity (73, 74). In the case of oligotrophic desert soils, the application of RNA and protein extraction methods is additionally challenging, due to low biomass and activities yielding lower concentrations of mRNA. However, improvements in sequencing technologies and methodological approaches have improved outlooks in this area. These improvements include the snap-freezing of samples or the use of RNA preservation solutions (75–77), incorporation of humic and fulvic acid separation and mRNA enrichment steps (69), and analysis against custom reference databases obtained using metagenomic analysis of identical or highly similar samples alongside public databases (78, 79). Recent transcriptomic studies have been successfully conducted on hyper-arid soils from the Namib Desert (80, 81). Furthermore, a recent preliminary metaproteomics study conducted on low-biomass and low-activity soil from Casey station in East Antarctica identified significant expression of 295 proteins, including RuBisCO form IE and group 1h [NiFe]-hydrogenase (82). These successes encourage further application of omics techniques to hyper-arid deserts. In addition, pure culture expression-based studies are vital for directly linking trace gas oxidation and microbial carbon fixation processes in individual taxa, potentially allowing atmospheric chemosynthesis to be validated and quantified under a range of environmental conditions.
THE MULTIDISCIPLINARY IMPLICATIONS OF ATMOSPHERIC CHEMOSYNTHESIS
The survival and growth strategies of microbiomes in extreme environments could have broad-reaching implications upon nutrient cycling, the minimum requirements for life, aerobiology and aeolian dispersal, and informing microbial cultivation for biodiscovery. Therefore, it is vital that our understanding of these mechanisms is further developed.
Uncovering roles in global nutrient cycling
Microbiomes have a fundamental role in nutrient cycling (83–85). On a global scale, microbial cycling of gaseous compounds, such as carbon dioxide, hydrogen, carbon monoxide, and nitrogen is a critical element of biogeochemical cycling (86–88). It is becoming clear that bacteria performing atmospheric chemosynthesis may function as a prominent carbon sink within desert ecosystems, and there is a need to assess and quantify this metabolic functionality in more diverse and under-investigated global ecosystems.
Within desert ecosystems, atmospheric chemosynthesis is thought to provide organic carbon to higher order, heterotrophic assemblages through the trophic web (15, 16, 31). Soil heterotrophs oxidize organic carbon as a source of energy through cellular respiration, with a proportion of the resulting simpler organic compounds incorporated into new microbial biomass through biosynthesis (89). Greater proportions of organic carbon assimilated into biomass equates to a higher carbon use efficiency (CUE) (90). A study of 23 taxonomically diverse heterotrophic bacterial isolates from temperate forest soils found that 26%–81% of consumed carbon was used for growth, averaging ~60% across the conditions studied (91). This suggests that in organic carbon-rich soils, heterotrophs assimilate and release CO2 at comparable rates (91). Comparatively, mixotrophs that supplement their energy input through atmospheric gas oxidation theoretically require less organic carbon for energy. This allows soil mixotrophs to more efficiently allocate carbon derived from consumed organic carbon matter into new microbial biomass, as opposed to being terminally respired and released into the atmosphere (92). A mixotrophic lifestyle potentially makes them a more effective carbon sink than their heterotrophic counterparts, particularly in organic carbon-limited environments. To verify this, it would be necessary to quantify and compare rates of microbial carbon fixation and respiration in identical soil microbiomes from various environments, with and without exposure to trace atmospheric gases. This objective poses significant technical challenges, as the metabolic products of respiration (CO2 and H2O) are commonly consumed by autotrophic carbon assimilation processes (89, 93). Instead, activity studies that trace and quantify the incorporation of 13C-labeled substrates into microbial biomass, or the incorporation of 18O from labeled water into newly formed microbial DNA, alongside the release of respiratory CO2 are most widely used to measure CUE (94–96). To determine the impact of trace gas oxidation upon CUE, this method should be applied to soil microbiomes from a range of environments, with and without trace gas exposure. We propose that metabolite-independent methods, including metatranscriptomic and metaproteomic analysis, should be implemented in conjunction with this research. These techniques could verify and quantify the expression of respiratory and carbon assimilatory pathways, alongside observed carbon and energy fluxes, for comparative analysis within whole microbiomes and isolated bacteria.
Guiding conservation frameworks to include microbes in changing environments
Terrestrial microbiomes play a substantial role in global carbon cycling, and in some cold desert environments such as the high Arctic, this role is rapidly changing due to climate change (97–99). It remains unclear how this warming will impact microbial ecology and the carbon cycle in Antarctica, particularly in under-studied regions, and in this case, in relation to photosynthetic and atmospheric chemosynthetic primary producers.
Under warming conditions, thawing events occur more frequently (100, 101), increasing microbial respiration rates, and resulting in the subsequent decomposition of ancient organic matter deposits (100–103). This process has the potential to release 92 (±17) Pg carbon into the atmosphere by 2100 (100). Rising temperatures and moisture availability are also expected to alter the carbon cycle in polar soils by increasing the biomass and activity of photosynthetic populations. Within terrestrial Antarctica, most studies on the effects of climate change have centered on the maritime regions of West Antarctica, where plant life is increasing (104–106). However, vascular plants capable of counteracting increased CO2 release from microbial decomposition are absent from most of the ice-free areas of the continent (107), and photosynthetic microorganisms are frequently limited to localized niches (6, 10, 14, 108, 109). In remote coastal regions in eastern Antarctica, where phototroph abundances are very low, atmospheric chemosynthesis may be the dominant mode of energy acquisition (14).
A warming Antarctic environment has the potential to alter the carbon balance and deselect microorganisms performing atmospheric chemosynthesis. For example, in cold oligotrophic soils at Mitchell Peninsula in the Windmill Islands of East Antarctica, phyla linked to this process, specifically Eremiobacterota and Ca. Dormibacterota, exist in far higher abundances (8.1% and 5.1%, respectively) than in temperate copiotrophic environments (110). In the dry Windmill Island soils, these taxa have been shown to decrease significantly alongside microbial community turnover when soil moisture levels exceeded 10%–12% (111). Given the projected increases in temperature, precipitation, and meltwater events across the Antarctic continent (97, 112), particularly the peninsula (113), it is critical that long-term monitoring of these soil communities is established. Furthermore, trace gas chemosynthetic taxa should be investigated across a broader array of threatened cold desert environments before the opportunity is lost.
Investigations into the impacts of increasing temperatures on photosynthetic and atmospheric chemosynthetic primary production are severely limited, both geographically and temporally (114). Studies that expand our understanding of the distribution and functional capacity of microorganisms performing atmospheric chemosynthesis across a broader Antarctic continental scale would allow us to further understand the impacts of climate change and microbial primary production dynamics. Such knowledge will be vital in the development of long-term observation systems and conservation strategies (115).
Redefining the minimum nutritional requirements for life
In the ongoing search for extra-terrestrial life, researchers have focused on planetary bodies that demonstrate potentially habitable conditions (116). While the requirements of habitability have remained controversial (117, 118), one condition that is widely regarded as vital is the presence of bioavailable water (119, 120). Although other requirements may limit extra-terrestrial life, including the presence of inorganic sources of energy, carbon, and trace elements, these are common throughout the universe (121). Comparatively, liquid water is far rarer (116, 121), making it the first variable considered when assessing a planet’s habitability. There is increasing evidence suggesting the presence of liquid water on extra-terrestrial bodies, including the interior of Enceladus (122–124), beneath the outer ice sheet of Europa (125, 126), and in the subsurface polar regions of Mars (127).
Despite a dependence upon bioavailable water, microbial life has been discovered in the most arid environments on Earth (128) with few reports of apparently abiotic habitats (13, 129). Within cold hyper-arid deserts, such as those of the McMurdo Dry Valleys and the Vestfold Hills in Antarctica, atmospheric chemosynthesis provides microbiomes with the energy required for bacterial growth and may also be a strategy for generating metabolic water (37, 130). This is because the microbial oxidation of trace hydrogen is hydrogenic (2H2 + O2 = 2H2O) (130).
In cold deserts, microbial trace H2 oxidation has been reported to be as rapid as 421.4 nmol/mol/h/g in unwetted soils (43), resulting in an estimated 0.2 mg of H2O produced per day, per gram of soil (130). Trace gas-dependent hydro-genesis could, therefore, be a significant source of water for microorganisms residing in hyper-arid ecosystems. In hot deserts, estimates of maximum H2 oxidation rates are theoretically sufficient to fulfil ecosystem water requirements (37); however, these rates have been primarily derived from wetted soils, which oxidize trace gases more rapidly than when dry. For example, wetting induces a 60-fold H2-uptake rate increase in Australian soils (31) and a 26-fold increase in Judea Hills and Negev Desert soils (47) with these increases not reflected in heat-killed controls. However, temporal moisture content fluctuations greatly influence the microbial oxidation of H2 in a non-linear manner (131), and the high levels of biochemical activity observed during these assays may only occur in situ during or after infrequent precipitation or snowmelt events. Therefore, the use of wetted soils to calculate H2 oxidation rates and theoretical water yields does not directly substantiate the hypothesis that H2 oxidation significantly contributes to cellular water budgets within arid and hyper-arid environments where water is naturally scarce. It is critical that the role and ecological significance of trace H2 oxidation in metabolic water generation are verified under in situ conditions, particularly extending to hot desert environments.
In addition to water, sources of energy and carbon are also vital for life. Subsurface environments within “habitable zones” are widely regarded as plausible extra-terrestrial habitats, as liquid water and redox disequilibria are more likely to exist, and the impacts of strong, mutagenic solar radiation are minimized. Within these environments, solar radiation could potentially be utilized by photosynthetic organisms, as this has been observed at intensities as low as 0.01 µmol m−2 s−1, which is ∼5 × 10−6 of the direct solar flux at Earth (121). However, trace gas oxidation and atmospheric chemosynthesis offer alternative mechanisms for energy liberation and light-independent growth, potentially allowing for the adaptation of microorganisms to more diverse extra-terrestrial niches.
Available data indicate that trace gas chemosynthetic microorganisms can survive by oxidizing atmospheric gases to support their carbon, energy, and possibly water requirements (31, 47). This means that planetary bodies with atmospheric H2 and CO or CO2 could be capable of supporting atmospheric chemosynthetic lifeforms (132), given that other abiotic factors fell within the biological requirements (116, 121, 133). The Martian polar subsurface is a candidate extra-terrestrial environment where trace gas oxidation and atmospheric chemosynthesis may occur. Dry permafrost ground in the north polar plains of Mars exhibits temperatures warmer than −18°C, water activity values >0.6, and a lower atmosphere rich in CO2 (95.32%), CO (0.6%), and H2 (15 ± 5 ppmv) with low levels of O2 (0.16%) available (134–137). Furthermore, in liquid environments across Mars, oxygen capable of driving hydrogen oxidation is dissolved at concentrations of ~2.5 × 10−6 mol m−3 to 2 mol m−3, and is particularly concentrated within polar regions due to lower temperatures (138).
Biosignatures of trace gas oxidation and carbon fixation processes offer new targets in the search for extra-terrestrial life. Further research is necessary first to confirm whether atmospheric chemosynthesis fulfils the energy, water, and carbon requirements of microbiomes within terrestrial analogs on Earth (139), such as the Atacama Desert (140, 141) and the McMurdo Dry Valleys (142–144), where the indicator genes have already been detected (43, 145). If trace gas oxidation and atmospheric chemosynthesis are viable sources of cellular water, energy, and carbon, it will become necessary to update the known detection limits of life (146) and incorporate a broader range of environments into our search for extra-terrestrial life.
Aerobiology and aeolian dispersal
Earth’s atmosphere is estimated to contain approximately 5 × 1022 microbial cells (147), and while bacteria have been detected in the mesosphere (148) and thermosphere (149), at altitudes as high as 400 km, the majority of this atmospheric biomass is within altitudes below 11 km (150, 151). Aerobiology is a relatively new discipline developed throughout the last century, historically focused on human health and food safety, particularly the airborne transmission of pollen, fungal spores, and pathogenic taxa (152). However, airborne bacteria are ecologically significant as they influence atmospheric composition (153) and climate events, including rainfall (154, 155), and are readily dispersed by wind to establish or alter microbiomes within new environments (151, 156, 157). As a result, there is a growing focus on the role of airborne microorganisms on microbial ecology and climate, with a dire need for studies generating more empirical data to support foundational science in this area.
Aeolian dispersal of microorganisms is of particular interest in the cold desert environment of Antarctica, where selection pressures faced in the atmosphere often resemble those in the terrestrial environment, and where geographical isolation and circumpolar currents limit microbial inputs from other sources (151). An important limitation on aeolian dispersal is the timeframe that microorganisms can remain dormant or metabolically active within the troposphere, although this duration is predicted to be longer over Antarctica compared to temperate environments (157). It was once believed that airborne microorganisms were inherently inert and were thus termed “spora” (148). However, studies have shown bacterial survival and growth within supercooled cloud droplets (158) and on airborne particles (159). The underlying metabolic processes that support airborne bacteria, and the proportion of airborne bacteria that remain active, are poorly understood. Organic material at oligotrophic concentrations may support heterotrophic communities (151, 160–162), and light exposure may support their photosynthetic counterparts (163–165). Actinobacteriota, a phylum widely linked to trace gas oxidation and atmospheric chemosynthesis, and Firmicutes and Verrucomicrobiota, which have recently been associated with these processes through metagenomic analysis (43), have been identified within outdoor aerobiomes, including above Antarctica (151, 156, 166, 167) and the high Arctic (168). However, aerobiological studies are highly limited in quantity and scope, restricting our ability to form concrete conclusions about the extent of this activity, the source of these microorganisms, and their taxonomic distribution throughout the broader aerosphere.
It is hypothesized here that a subset of airborne microorganisms may supplement energy needs through trace gas oxidation or use the energy to drive carbon fixation in the process of atmospheric chemosynthesis, thereby fulfilling their nutritional requirements through the uptake of almost exclusively atmospheric inputs. If this is the case, these microorganisms could also make up a substantial, uncharacterized component of aerobiology. Furthermore, if trace gas chemosynthetic bacteria do make up a significant proportion of the atmospheric microbiome, then they could be substantially influencing climatic conditions, nutrient cycling, and aeolian dispersal that warrants further investigation.
To fill this gap in our understanding, it is recommended that metagenomic and transcriptomic analyses are conducted on airborne microbial communities to identify potentially atmospheric chemosynthetic taxa and characterize their underlying functionality. Low microbial biomass in airborne environments (~1 × 104/m3) (160, 169) has previously restricted the use of omics technologies in aerobiology. However, improved study protocols that utilize longer sampling times and carefully designed controls, thorough DNA extraction protocols and particulate removal, and repair of damaged DNA (170, 171), as well as the development of more sensitive omics technologies (172, 173), have led to more successful analysis of air samples (170, 173, 174).
We recommend that future investigations into atmospheric chemosynthesis within airborne microbiomes focus on samples obtained from the troposphere over deserts. This is because the underlying terrestrial environment is likely to contain high abundances of trace gas oxidizing microorganisms and predicted atmospheric chemosynthetic taxa (14, 43, 47). Investigation into atmospheric microbiomes over Antarctica should be prioritized, due to the well-established significance of aeolian dispersal on the ecology within this region (175–177), higher estimated airborne residence times (157), and the existence of a proposed standardized protocol for Antarctic aerobiological sampling and analysis (178).
CONCLUSION
Atmospheric chemosynthesis is a potentially important microbial primary production process that appears to be widespread in the Earth’s desert soil environments. A growing body of evidence suggests that this is an ecologically significant process, with broad-reaching implications for global nutrient cycling and aerobiology, life detection limits and environmental habitability, and for the isolation of novel microorganisms. However, many issues remain unresolved: the global extent of atmospheric chemosynthesis, the true diversity of functional taxa, the validation of underlying metabolic pathways, and the time-dependent kinetics of these processes all remain as future research objectives. To fill these substantial gaps in our understanding, sophisticated methodologies should be implemented, including global phylogeographical studies in a broader array of microbiomes, DNA stable isotope probing, carbon flux experiments, expression-based “omics” surveys, and a wide array of relevant kinetic analyses, particularly targeting purified proteins linked to atmospheric chemosynthesis. The integration of such data will ultimately yield a much more complete understanding of atmospheric chemosynthetic metabolism and provide an accurate quantitative estimate of the contribution of trace gas chemotrophy to global ecosystem processes.
ACKNOWLEDGMENTS
This work was supported by the Australian Government Research Training Program (RTP) Scholarship (awarded to D.T.), an Australian Research Council Future Fellowship (FT170100341), an ARC Discovery Project (DP220103430), and the Australian Antarctic Science project grant (4406) awarded to B.C.F. Biorender was used to generate the figure.
A.E.R. wrote the article and generated the figure. B.C.F., D.A.C., and D.Z.T. were involved in discussions of the content and the revision and editing of the manuscript.
Contributor Information
Belinda C. Ferrari, Email: b.ferrari@unsw.edu.au.
Corrella S. Detweiler, University of Colorado Boulder, Boulder, Colorado, USA
REFERENCES
- 1. Ortiz M, Bosch J, Coclet C, Johnson J, Lebre P, Salawu-Rotimi A, Vikram S, Makhalanyane T, Cowan D. 2020. Microbial nitrogen cycling in Antarctic soils. Microorganisms 8:1442. doi: 10.3390/microorganisms8091442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Zhang E, Czechowski P, Terauds A, Wong SY, Chelliah DS, Raven M, Tanaka MM, Ferrari BC. 2021. Tracing boundaries in Eastern Antarctica: multi-scale drivers of soil microbial communities across the hyperarid Vestfold hills. Microbiology. doi: 10.1101/2021.09.22.461446 [DOI]
- 3. Diaz MA, Li J, Michalski G, Darrah TH, Adams BJ, Wall DH, Hogg ID, Fierer N, Welch SA, Gardner CB, Lyons WB. 2020. Stable isotopes of nitrate, sulfate, and carbonate in soils from the transantarctic mountains, Antarctica: a record of atmospheric deposition and chemical weathering. Front Earth Sci 8. doi: 10.3389/feart.2020.00341 [DOI] [Google Scholar]
- 4. Lambrechts S, Willems A, Tahon G. 2019. Uncovering the uncultivated majority in Antarctic soils: toward a synergistic approach. Front Microbiol 10:242. doi: 10.3389/fmicb.2019.00242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Amils R. 2011. Chemoautotroph, p 288–289. In Gargaud M, Amils R, Quintanilla JC, Cleaves HJ, Irvine WM, Pinti DL, Viso M (ed), Encyclopedia of Astrobiology. Springer. doi: 10.1007/978-3-642-11274-4 [DOI] [Google Scholar]
- 6. Colesie C, Gommeaux M, Green TGA, Büdel B. 2014. Biological soil crusts in continental Antarctica: garwood valley, Southern Victoria land, and Diamond hill, Darwin mountains region. Antartic Sci 26:115–123. doi: 10.1017/S0954102013000291 [DOI] [Google Scholar]
- 7. Jung P, Schermer M, Briegel-Williams L, Baumann K, Leinweber P, Karsten U, Lehnert L, Achilles S, Bendix J, Büdel B.. 2019. Water availability shapes edaphic and lithic cyanobacterial communities in the Atacama Desert. J Phycol 55:1306-1318. [DOI] [PubMed] [Google Scholar]
- 8. Warren-Rhodes KA, Rhodes KL, Pointing SB, Ewing SA, Lacap DC, Gómez-Silva B, Amundson R, Friedmann EI, McKay CP. 2006. Hypolithic cyanobacteria dry limit of photosynthesis, and microbial ecology in the hyperarid Atacama desert. Microb Ecol 52:389–398. doi: 10.1007/s00248-006-9055-7 [DOI] [PubMed] [Google Scholar]
- 9. Rego A, Raio F, Martins TP, Ribeiro H, Sousa AGG, Séneca J, Baptista MS, Lee CK, Cary SC, Ramos V, Carvalho MF, Leão PN, Magalhães C. 2019. Actinobacteria and cyanobacteria diversity in terrestrial Antarctic microenvironments evaluated by culture-dependent and independent methods. Front Microbiol 10. doi: 10.3389/fmicb.2019.01018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Smith JJ, Tow LA, Stafford W, Cary C, Cowan DA. 2006. Bacterial diversity in three different Antarctic cold desert mineral soils. Microb Ecol 51:413–421. doi: 10.1007/s00248-006-9022-3 [DOI] [PubMed] [Google Scholar]
- 11. Valentín-Vargas A, Neilson JW, Root RA, Chorover J, Maier RM. 2018. Treatment impacts on temporal microbial community dynamics during phytostabilization of acid-generating mine tailings in semiarid regions. Sci Total Environ 618:357–368. doi: 10.1016/j.scitotenv.2017.11.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Huang M, Chai L, Jiang D, Zhang M, Zhao Y, Huang Y. 2019. Increasing aridity affects soil archaeal communities by mediating soil niches in semi-arid regions. Sci Total Environ 647:699–707. doi: 10.1016/j.scitotenv.2018.07.305 [DOI] [PubMed] [Google Scholar]
- 13. Dragone NB, Diaz MA, Hogg ID, Lyons WB, Jackson WA, Wall DH, Adams BJ, Fierer N. 2021. Exploring the boundaries of microbial habitability in soil. JGR Biogeosci 126. doi: 10.1029/2020JG006052 [DOI] [Google Scholar]
- 14. Ji M, Greening C, Vanwonterghem I, Carere CR, Bay SK, Steen JA, Montgomery K, Lines T, Beardall J, van Dorst J, Snape I, Stott MB, Hugenholtz P, Ferrari BC. 2017. Atmospheric trace gases support primary production in Antarctic desert surface soil. Nature 552:400–403. doi: 10.1038/nature25014 [DOI] [PubMed] [Google Scholar]
- 15. Leung PM, Bay SK, Meier DV, Chiri E, Cowan DA, Gillor O, Woebken D, Greening C, Stegen JC. 2020. Energetic basis of microbial growth and persistence in desert ecosystems. mSystems 5:e00495-19. doi: 10.1128/mSystems.00495-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Bay S, Ferrari B, Greening C. 2018. Life without water: how do bacteria generate biomass in desert ecosystems Microbiol Aust. 39:28. doi: 10.1071/MA18008 [DOI] [Google Scholar]
- 17. Fritsen CH, Grue AM, Priscu JC. 2000. Distribution of organic carbon and nitrogen in surface soils in the McMurdo Dry Valleys, Antarctica. Polar Biology 23:121–128. doi: 10.1007/s003000050017 [DOI] [Google Scholar]
- 18. Novelli PC, Lang PM, Masarie KA, Hurst DF, Myers R, Elkins JW. 1999. Molecular hydrogen in the troposphere: global distribution and budget. J Geophys Res 104:30427–30444. doi: 10.1029/1999JD900788 [DOI] [Google Scholar]
- 19. Schmidt U. 1974. Molecular hydrogen in the atmosphere. Tellus 26:78–90. doi: 10.1111/j.2153-3490.1974.tb01954.x [DOI] [Google Scholar]
- 20. Novelli PC, Masarie KA, Lang PM. 1998. Distributions and recent changes of carbon monoxide in the lower troposphere. J Geophys Res 103:19015–19033. doi: 10.1029/98JD01366 [DOI] [Google Scholar]
- 21. Petrenko VV, Martinerie P, Novelli P, Etheridge DM, Levin I, Wang Z, Blunier T, Chappellaz J, Kaiser J, Lang P, Steele LP, Hammer S, Mak J, Langenfelds RL, Schwander J, Severinghaus JP, Witrant E, Petron G, Battle MO, Forster G, Sturges WT, Lamarque J-F, Steffen K, White JWC. 2013. A 60 yr record of atmospheric carbon monoxide reconstructed from Greenland firn air. Atmos Chem Phys 13:7567–7585. doi: 10.5194/acp-13-7567-2013 [DOI] [Google Scholar]
- 22. Chi X, Winderlich J, Mayer JC, Panov AV, Heimann M, Birmili W, Heintzenberg J, Cheng Y, Andreae MO. 2013. Long-term measurements of aerosol and carbon monoxide at the ZOTTO tall tower to characterize polluted and pristine air in the Siberian taiga. Atmos. Chem. Phys 13:12271–12298. doi: 10.5194/acp-13-12271-2013 [DOI] [Google Scholar]
- 23. Khalil MAK, Rasmussen RA. 1990. The global cycle of carbon monoxide: trends and mass balance. Chemosphere 20:227–242. doi: 10.1016/0045-6535(90)90098-E [DOI] [Google Scholar]
- 24. Saunois M, Stavert AR, Poulter B, Bousquet P, Canadell JG, Jackson RB, Raymond PA, Dlugokencky EJ, Houweling S, Patra PK, Ciais P, Arora VK, Bastviken D, Bergamaschi P, Blake DR, Brailsford G, Bruhwiler L, Carlson KM, Carrol M, Castaldi S, Chandra N, Crevoisier C, Crill PM, Covey K, Curry CL, Etiope G, Frankenberg C, Gedney N, Hegglin MI, Höglund-Isaksson L, Hugelius G, Ishizawa M, Ito A, Janssens-Maenhout G, Jensen KM, Joos F, Kleinen T, Krummel PB, Langenfelds RL, Laruelle GG, Liu L, Machida T, Maksyutov S, McDonald KC, McNorton J, Miller PA, Melton JR, Morino I, Müller J, Murguia-Flores F, Naik V, Niwa Y, Noce S, O’Doherty S, Parker RJ, Peng C, Peng S, Peters GP, Prigent C, Prinn R, Ramonet M, Regnier P, Riley WJ, Rosentreter JA, Segers A, Simpson IJ, Shi H, Smith SJ, Steele LP, Thornton BF, Tian H, Tohjima Y, Tubiello FN, Tsuruta A, Viovy N, Voulgarakis A, Weber TS, van Weele M, van der Werf GR, Weiss RF, Worthy D, Wunch D, Yin Y, Yoshida Y, Zhang W, Zhang Z, Zhao Y, Zheng B, Zhu Q, Zhu Q, Zhuang Q. 2020. The global methane budget 2000–2017. Earth Syst Sci Data 12:1561–1623. doi: 10.5194/essd-12-1561-2020 [DOI] [Google Scholar]
- 25. Yonemura S, Yokozawa M, Kawashima S, Tsuruta H. 2000. Model analysis of the influence of gas diffusivity in soil on CO and H2 uptake. Tellus B 52:919–933. doi: 10.1034/j.1600-0889.2000.d01-2.x [DOI] [Google Scholar]
- 26. Berney M, Cook GM. 2010. Unique flexibility in energy metabolism allows mycobacteria to combat starvation and hypoxia. PLoS One 5:e8614. doi: 10.1371/journal.pone.0008614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Conrad R, Weber M, Seiler W. 1983. Kinetics and electron transport of soil hydrogenases catalyzing the oxidation of atmospheric hydrogen. Soil Biol Biochem 15:167–173. doi: 10.1016/0038-0717(83)90098-6 [DOI] [Google Scholar]
- 28. Greening C, Biswas A, Carere CR, Jackson CJ, Taylor MC, Stott MB, Cook GM, Morales SE. 2016. Genomic and metagenomic surveys of hydrogenase distribution indicate H2 is a widely utilised energy source for microbial growth and survival. ISME J 10:761–777. doi: 10.1038/ismej.2015.153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Morita RY. 1999. Is H2 the universal energy source for long-term survival Microb Ecol 38:307–320. doi: 10.1007/s002489901002 [DOI] [PubMed] [Google Scholar]
- 30. Cordero PRF, Bayly K, Man Leung P, Huang C, Islam ZF, Schittenhelm RB, King GM, Greening C. 2019. Atmospheric carbon monoxide oxidation is a widespread mechanism supporting microbial survival. ISME J 13:2868–2881. doi: 10.1038/s41396-019-0479-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Bay SK, Dong X, Bradley JA, Leung PM, Grinter R, Jirapanjawat T, Arndt SK, Cook PLM, LaRowe DE, Nauer PA, Chiri E, Greening C. 2021. Trace gas oxidizers are widespread and active members of soil microbial communities. Nat Microbiol 6:246–256. doi: 10.1038/s41564-020-00811-w [DOI] [PubMed] [Google Scholar]
- 32. Peach HACG. 2019. Investigation into the microbial ecology and persistence of soil Taxa through trace-gas oxidation in Tongariro National Park. Master of Science (MSc). The University of Waikato. [Google Scholar]
- 33. Knief C, Dunfield PF. 2005. Response and adaptation of different methanotrophic bacteria to low methane mixing ratios. Environ Microbiol 7:1307–1317. doi: 10.1111/j.1462-2920.2005.00814.x [DOI] [PubMed] [Google Scholar]
- 34. Islam ZF, Cordero PRF, Feng J, Chen Y-J, Bay SK, Jirapanjawat T, Gleadow RM, Carere CR, Stott MB, Chiri E, Greening C. 2019. Two chloroflexi classes independently evolved the ability to persist on atmospheric hydrogen and carbon monoxide. ISME J 13:1801–1813. doi: 10.1038/s41396-019-0393-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Patrauchan MA, Miyazawa D, LeBlanc JC, Aiga C, Florizone C, Dosanjh M, Davies J, Eltis LD, Mohn WW. 2012. Proteomic analysis of survival of Rhodococcus jostii Rha1 during carbon starvation. Appl Environ Microbiol 78:6714–6725. doi: 10.1128/AEM.01293-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Meredith LK, Rao D, Bosak T, Klepac-Ceraj V, Tada KR, Hansel CM, Ono S, Prinn RG. 2014. Consumption of atmospheric hydrogen during the life cycle of soil-dwelling actinobacteria. Environ Microbiol Rep 6:226–238. doi: 10.1111/1758-2229.12116 [DOI] [PubMed] [Google Scholar]
- 37. Ortiz M, Leung PM, Shelley G, Jirapanjawat T, Nauer PA, Van Goethem MW, Bay SK, Islam ZF, Jordaan K, Vikram S, Chown SL, Hogg ID, Makhalanyane TP, Grinter R, Cowan DA, Greening C. 2021. Multiple energy sources and metabolic strategies sustain microbial diversity in Antarctic desert soils. Proc Natl Acad Sci U S A 118:e2025322118. doi: 10.1073/pnas.2025322118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Grinter R, Kropp A, Venugopal H, Senger M, Badley J, Cabotaje PR, Jia R, Duan Z, Huang P, Stripp ST, Barlow CK, Belousoff M, Shafaat HS, Cook GM, Schittenhelm RB, Vincent KA, Khalid S, Berggren G, Greening C. 2023. Structural basis for bacterial energy extraction from atmospheric hydrogen. Nature 615:541–547. doi: 10.1038/s41586-023-05781-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Conrad R, Meyer O, Seiler W. 1981. Role of carboxydobacteria in consumption of atmospheric carbon monoxide by soil. Appl Environ Microbiol 42:211–215. doi: 10.1128/aem.42.2.211-215.1981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Greening C, Berney M, Hards K, Cook GM, Conrad R. 2014. A soil actinobacterium scavenges atmospheric H-2 using two membrane-associated, oxygen-dependent [NiFe] hydrogenases. Proc Natl Acad Sci U S A 111:4257–4261. doi: 10.1073/pnas.1320586111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Cordero PRF, Grinter R, Hards K, Cryle MJ, Warr CG, Cook GM, Greening C. 2019. Two uptake hydrogenases differentially interact with the aerobic respiratory chain during mycobacterial growth and persistence. J Biol Chem 294:18980–18991. doi: 10.1074/jbc.RA119.011076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Schäfer C, Bommer M, Hennig SE, Jeoung J-H, Dobbek H, Lenz O. 2016. Structure of an actinobacterial-type [NiFe]-hydrogenase reveals insight into O2-tolerant H2 oxidation. Structure 24:285–292. doi: 10.1016/j.str.2015.11.010 [DOI] [PubMed] [Google Scholar]
- 43. Ray AE, Zaugg J, Benaud N, Chelliah DS, Bay S, Wong HL, Leung PM, Ji M, Terauds A, Montgomery K, Greening C, Cowan DA, Kong W, Williams TJ, Hugenholtz P, Ferrari BC. 2022. Atmospheric chemosynthesis is phylogenetically and geographically widespread and contributes significantly to carbon fixation throughout cold deserts. ISME J 16:2547–2560. doi: 10.1038/s41396-022-01298-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Ray AE, Zhang E, Terauds A, Ji M, Kong W, Ferrari BC. 2020. Soil microbiomes with the genetic capacity for atmospheric chemosynthesis are widespread across the poles and are associated with moisture. Front Microbiol 11:1936. doi: 10.3389/fmicb.2020.01936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Xu Y, Teng Y, Dong X, Wang X, Zhang C, Ren W, Zhao L, Luo Y, Greening C. 2021. Genome-resolved metagenomics reveals how soil bacterial communities respond to elevated H2 availability. Soil Biol Biochem 163:108464. doi: 10.1016/j.soilbio.2021.108464 [DOI] [Google Scholar]
- 46. Jordaan K, Lappan R, Dong X, Aitkenhead IJ, Bay SK, Chiri E, Wieler N, Meredith LK, Cowan DA, Chown SL, Greening C. 2020. Hydrogen-oxidizing bacteria are abundant in desert soils and strongly stimulated by hydration. mSystems 5:e01131-20. doi: 10.1128/mSystems.01131-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Bay SK, Waite DW, Dong X, Gillor O, Chown SL, Hugenholtz P, Greening C. 2021. Chemosynthetic and photosynthetic bacteria contribute differentially to primary production across a steep desert aridity gradient. ISME J 15:3339–3356. doi: 10.1038/s41396-021-01001-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Leung PM, Daebeler A, Chiri E, Hanchapola I, Gillett DL, Schittenhelm RB, Daims H, Greening C. 2022. A nitrite-oxidising bacterium constitutively consumes atmospheric hydrogen. ISME J 16:2213–2219. doi: 10.1038/s41396-022-01265-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Lappan R, Shelley G, Islam ZF, Leung PM, Lockwood S, Nauer PA, Jirapanjawat T, Chen Y-J, Kessler AJ, Williams TJ, Cavicchioli R, Baltar F, Cook PLM, Morales SE, Greening C. 2022. Molecular hydrogen is an overlooked energy source for marine bacteria. Microbiology. doi: 10.1101/2022.01.29.478295 [DOI] [PMC free article] [PubMed]
- 50. Ward BA. 2019. Mixotroph ecology: more than the sum of its parts. Proc Natl Acad Sci U S A 116:5846–5848. doi: 10.1073/pnas.1902106116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Tveit AT, Hestnes AG, Robinson SL, Schintlmeister A, Dedysh SN, Jehmlich N, von Bergen M, Herbold C, Wagner M, Richter A, Svenning MM. 2019. Widespread soil bacterium that oxidizes atmospheric methane. Proc Natl Acad Sci U S A 116:8515–8524. doi: 10.1073/pnas.1817812116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Greening C, Grinter R. 2022. Microbial oxidation of atmospheric trace gases. Nat Rev Microbiol 20:513–528. doi: 10.1038/s41579-022-00724-x [DOI] [PubMed] [Google Scholar]
- 53. Ji M, Williams TJ, Montgomery K, Wong HL, Zaugg J, Berengut JF, Bissett A, Chuvochina M, Hugenholtz P, Ferrari BC. 2021. Candidatus Eremiobacterota, a metabolically and phylogenetically diverse terrestrial phylum with acid-tolerant adaptations. ISME J 15:2692–2707. doi: 10.1038/s41396-021-00944-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Yabe S, Muto K, Abe K, Yokota A, Staudigel H, Tebo BM. 2022. Vulcanimicrobium alpinus gen. nov. sp. nov., the first cultivated representative of the candidate phylum “Eremiobacterota”, is a metabolically versatile aerobic anoxygenic phototroph. ISME COMMUN 2:120. doi: 10.1038/s43705-022-00201-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Quiza L, Lalonde I, Guertin C, Constant P. 2014. Land-use influences the distribution and activity of high affinity CO-oxidizing bacteria associated to type I-coxL genotype in soil. Front Microbiol 5:271. doi: 10.3389/fmicb.2014.00271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Kanno M, Constant P, Tamaki H, Kamagata Y. 2016. Detection and isolation of plant-associated bacteria scavenging atmospheric molecular hydrogen. Environ Microbiol 18:2495–2506. doi: 10.1111/1462-2920.13162 [DOI] [PubMed] [Google Scholar]
- 57. Chowdhury SP, Conrad R. 2010. Thermal deactivation of high-affinity H2 uptake activity in soils. Soil Biol Biochem 42:1574–1580. doi: 10.1016/j.soilbio.2010.05.027 [DOI] [Google Scholar]
- 58. Smith-Downey NV, Randerson JT, Eiler JM. 2008. Molecular hydrogen uptake by soils in forest, desert, and marsh ecosystems in California. J. Geophys. Res 113. doi: 10.1029/2008JG000701 [DOI] [Google Scholar]
- 59. Lalonde I, Constant P. 2016. Identification of unknown carboxydovore bacteria dominant in deciduous forest soil via succession of bacterial communities, coxL genotypes, and carbon monoxide oxidation activity in soil Microcosms. Appl Environ Microbiol 82:1324–1333. doi: 10.1128/AEM.03595-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Constant P, Chowdhury SP, Hesse L, Conrad R. 2011. Co-localization of atmospheric H2 oxidation activity and high affinity H2-oxidizing bacteria in non-axenic soil and sterile soil amended with Streptomyces SP. Soil Biol Biochem 43:1888–1893. doi: 10.1016/j.soilbio.2011.05.009 [DOI] [Google Scholar]
- 61. King GM, Weber CF. 2008. Interactions between bacterial carbon Monoxide and hydrogen consumption and plant development on recent volcanic deposits. ISME J 2:195–203. doi: 10.1038/ismej.2007.101 [DOI] [PubMed] [Google Scholar]
- 62. King GM. 2003. Contributions of atmospheric CO and hydrogen uptake to microbial dynamics on recent Hawaiian volcanic deposits. Appl Environ Microbiol 69:4067–4075. doi: 10.1128/AEM.69.7.4067-4075.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. King GM, Weber CF. 2007. Distribution, diversity and ecology of aerobic CO-oxidizing bacteria. Nat Rev Microbiol 5:107–118. doi: 10.1038/nrmicro1595 [DOI] [PubMed] [Google Scholar]
- 64. Dunfield KE, King GM. 2004. Molecular analysis of carbon Monoxide-Oxidizing bacteria associated with recent Hawaiian volcanic deposits. Appl Environ Microbiol 70:4242–4248. doi: 10.1128/AEM.70.7.4242-4248.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Lynch RC, Darcy JL, Kane NC, Nemergut DR, Schmidt SK. 2014. Metagenomic evidence for metabolism of trace atmospheric gases by high-elevation desert Actinobacteria. Front Microbiol 5:698. doi: 10.3389/fmicb.2014.00698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Weber CF, King GM. 2012. The phylogenetic distribution and ecological role of carbon monoxide oxidation in the genus Burkholderia. FEMS Microbiol Ecol 79:167–175. doi: 10.1111/j.1574-6941.2011.01206.x [DOI] [PubMed] [Google Scholar]
- 67. Nanba K, King GM, Dunfield K. 2004. Analysis of Facultative Lithotroph distribution and diversity on volcanic deposits by use of the large subunit of Ribulose 1,5-Bisphosphate Carboxylase/Oxygenase. Appl Environ Microbiol 70:2245–2253. doi: 10.1128/AEM.70.4.2245-2253.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Islam ZF, Welsh C, Bayly K, Grinter R, Southam G, Gagen EJ, Greening C. 2020. A widely distributed Hydrogenase Oxidises atmospheric H2 during bacterial growth. ISME J 14:2649–2658. doi: 10.1038/s41396-020-0713-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Wang Y, Hayatsu M, Fujii T. 2012. Extraction of bacterial RNA from soil: challenges and solutions. Microb. Environ 27:111–121. doi: 10.1264/jsme2.ME11304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Gallego Romero I, Pai AA, Tung J, Gilad Y. 2014. RNA-Seq: impact of RNA degradation on transcript quantification. BMC Biol 12:42. doi: 10.1186/1741-7007-12-42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Lu W, Zhou Q, Chen Y. 2022. Impact of RNA degradation on next-generation sequencing Transcriptome data. Genomics 114:110429. doi: 10.1016/j.ygeno.2022.110429 [DOI] [PubMed] [Google Scholar]
- 72. Stevenson FJ. 1994. Humus chemistry: genesis, composition, reactions. John Wiley & Sons. [Google Scholar]
- 73. Conesa A, Madrigal P, Tarazona S, Gomez-Cabrero D, Cervera A, McPherson A, Szcześniak MW, Gaffney DJ, Elo LL, Zhang X, Mortazavi A. 2016. A survey of best practices for RNA-Seq data analysis. Genome Biol. 17:13. doi: 10.1186/s13059-016-0881-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Fierer N. 2017. Embracing the unknown: disentangling the complexities of the soil microbiome. Nat Rev Microbiol 15:579–590. doi: 10.1038/nrmicro.2017.87 [DOI] [PubMed] [Google Scholar]
- 75. Passow CN, Kono TJY, Stahl BA, Jaggard JB, Keene AC, McGaugh SE. 2019. Nonrandom RNAseq gene expression associated with RNAlater and flash freezing storage methods. Mol Ecol Resour 19:456–464. doi: 10.1111/1755-0998.12965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Hurt RA, Qiu X, Wu L, Roh Y, Palumbo AV, Tiedje JM, Zhou J. 2001. Simultaneous recovery of RNA and DNA from soils and sediments. Appl Environ Microbiol 67:4495–4503. doi: 10.1128/AEM.67.10.4495-4503.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Mettel C, Kim Y, Shrestha PM, Liesack W. 2010. Extraction of mRNA from soil. Appl Environ Microbiol 76:5995–6000. doi: 10.1128/AEM.03047-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Jouffret V, Miotello G, Culotta K, Ayrault S, Pible O, Armengaud J. 2021. Increasing the power of interpretation for soil metaproteomics data. Microbiome 9:195. doi: 10.1186/s40168-021-01139-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Carvalhais LC, Dennis PG, Tyson GW, Schenk PM. 2012. Application of metatranscriptomics to soil environments. J Microbiol Methods 91:246–251. doi: 10.1016/j.mimet.2012.08.011 [DOI] [PubMed] [Google Scholar]
- 80. Gunnigle E, Frossard A, Ramond J-B, Guerrero L, Seely M, Cowan DA. 2017. Diel-scale temporal dynamics recorded for bacterial groups in Namib desert soil. Sci Rep 7:40189. doi: 10.1038/srep40189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. León-Sobrino C, Ramond J-B, Maggs-Kölling G, Cowan DA. 2019. Nutrient acquisition, rather than stress response over diel cycles, drives microbial transcription in a hyper-arid namib desert soil. Front Microbiol 10:1054. doi: 10.3389/fmicb.2019.01054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Wencaslao A. 2021. Digging deep: a metaproteomics approach to elucidate the survival mechanisms expressed in polar soil. Bachelor of Biotechnology (Honours). University of New South Wales, Sydney, NSW. [Google Scholar]
- 83. Marschner P, Rengel Z. 2007. Nutrient cycling in terrestrial ecosystems, . . Springer-Verlag, Berlin, Heidelberg. doi: 10.1007/978-3-540-68027-7 [DOI] [Google Scholar]
- 84. Arrigo KR. 2005. Marine microorganisms and global nutrient cycles. Nature 437:349–355. doi: 10.1038/nature04159 [DOI] [PubMed] [Google Scholar]
- 85. Saleem M. 2015. Microbiome community Ecology, . In Microbiome community Ecology: Fundamentals and applications. Springer International Publishing, Cham. doi: 10.1007/978-3-319-11665-5 [DOI] [Google Scholar]
- 86. Stewart KJ, Brummell ME, Coxson DS, Siciliano SD. 2013. How is nitrogen fixation in the high Arctic linked to greenhouse gas emissions Plant Soil 362:215–229. doi: 10.1007/s11104-012-1282-8 [DOI] [Google Scholar]
- 87. Ota M, Mamet SD, Muller AL, Lamb EG, Dhillon G, Peak D, Siciliano SD. 2020. Could cryoturbic diapirs be key for understanding ecological feedbacks to climate change in high Arctic polar deserts. JGR Biogeosciences 125. doi: 10.1029/2019JG005263 [DOI] [Google Scholar]
- 88. Brummell ME, Farrell RE, Hardy SP, Siciliano SD. 2014. Greenhouse gas production and consumption in high Arctic deserts. Soil Biology and Biochemistry 68:158–165. doi: 10.1016/j.soilbio.2013.09.034 [DOI] [Google Scholar]
- 89. Jurtshuk P. 1996. Bacterial metabolism. In Baron S (ed), Medical Microbiology. University of Texas Medical Branch at Galveston. [PubMed] [Google Scholar]
- 90. Tao F, Huang Y, Hungate BA, Manzoni S, Frey SD, Schmidt MWI, Reichstein M, Carvalhais N, Ciais P, Jiang L, Lehmann J, Wang Y-P, Houlton BZ, Ahrens B, Mishra U, Hugelius G, Hocking TD, Lu X, Shi Z, Viatkin K, Vargas R, Yigini Y, Omuto C, Malik AA, Peralta G, Cuevas-Corona R, Di Paolo LE, Luotto I, Liao C, Liang Y-S, Saynes VS, Huang X, Luo Y. 2023. Microbial carbon use efficiency promotes global soil carbon storage. Nature 618:981–985. doi: 10.1038/s41586-023-06042-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Pold G, Domeignoz-Horta LA, Morrison EW, Frey SD, Sistla SA, DeAngelis KM. 2020. Carbon use efficiency and its temperature sensitivity covary in soil bacteria. mBio 11:e02293-19. doi: 10.1128/mBio.02293-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Carini P. 2021. Hazardous gases sustain microbes underfoot. Nat Microbiol 6:145–146. doi: 10.1038/s41564-020-00855-y [DOI] [PubMed] [Google Scholar]
- 93. Alberts B. 2002. Chloroplasts and Photosynthesis, molecular biology of the cell. 4th ed. Garland Science, New York. [Google Scholar]
- 94. Zheng Q, Hu Y, Richter A, Wanek W.. 2017. Carbon use efficiency (CUE) and biomass turnover of soil microbial communities as affected by bedrock, land management and soil temperature and moisture, abstr April 01
- 95. Hu J, Huang C, Zhou S, Kuzyakov Y.. 2022. Nitrogen addition to soil affects microbial carbon use efficiency: meta-analysis of similarities and differences in 13C and 18O approaches. Global Change Biol 28:4977-4988. [DOI] [PubMed] [Google Scholar]
- 96. Qu L, Wang C, Bai E. 2020. Evaluation of the 18O-H2O incubation method for measurement of soil microbial carbon use efficiency. Soil Biol Biochem 145:107802. doi: 10.1016/j.soilbio.2020.107802 [DOI] [Google Scholar]
- 97. Naylor D, Sadler N, Bhattacharjee A, Graham EB, Anderton CR, McClure R, Lipton M, Hofmockel KS, Jansson JK. 2020. Soil microbiomes under climate change and implications for carbon cycling. Annu. Rev. Environ. Resour 45:29–59. doi: 10.1146/annurev-environ-012320-082720 [DOI] [Google Scholar]
- 98. Jansson JK, Hofmockel KS. 2020. Soil microbiomes and climate change. Nat Rev Microbiol 18:35–46. doi: 10.1038/s41579-019-0265-7 [DOI] [PubMed] [Google Scholar]
- 99. Singh BK, Bardgett RD, Smith P, Reay DSJNRM. 2010. Microorganisms and climate change: terrestrial feedbacks and mitigation options. Nat Rev Microbiol 8:779–790. doi: 10.1038/nrmicro2439 [DOI] [PubMed] [Google Scholar]
- 100. Schuur EAG, McGuire AD, Schädel C, Grosse G, Harden JW, Hayes DJ, Hugelius G, Koven CD, Kuhry P, Lawrence DM, Natali SM, Olefeldt D, Romanovsky VE, Schaefer K, Turetsky MR, Treat CC, Vonk JE. 2015. Climate change and the permafrost carbon feedback. Nature 520:171–179. doi: 10.1038/nature14338 [DOI] [PubMed] [Google Scholar]
- 101. Natali SM, Holdren JP, Rogers BM, Treharne R, Duffy PB, Pomerance R, MacDonald E. 2021. Permafrost carbon feedbacks threaten global climate goals. Proc Natl Acad Sci U S A 118:e2100163118. doi: 10.1073/pnas.2100163118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Crowther TW, Todd-Brown KEO, Rowe CW, Wieder WR, Carey JC, Machmuller MB, Snoek BL, Fang S, Zhou G, Allison SD, Blair JM, Bridgham SD, Burton AJ, Carrillo Y, Reich PB, Clark JS, Classen AT, Dijkstra FA, Elberling B, Emmett BA, Estiarte M, Frey SD, Guo J, Harte J, Jiang L, Johnson BR, Kröel-Dulay G, Larsen KS, Laudon H, Lavallee JM, Luo Y, Lupascu M, Ma LN, Marhan S, Michelsen A, Mohan J, Niu S, Pendall E, Peñuelas J, Pfeifer-Meister L, Poll C, Reinsch S, Reynolds LL, Schmidt IK, Sistla S, Sokol NW, Templer PH, Treseder KK, Welker JM, Bradford MA. 2016. Quantifying global soil carbon losses in response to warming. Nature 540:104–108. doi: 10.1038/nature20150 [DOI] [PubMed] [Google Scholar]
- 103. Zimov SA, Schuur EAG, Chapin FS III. 2006. Permafrost and the global carbon budget. Science 312:1612–1613. doi: 10.1126/science.1128908 [DOI] [PubMed] [Google Scholar]
- 104. Smith RIL. 1994. Vascular plants as bioindicators of regional warming in Antarctica. Oecologia 99:322–328. doi: 10.1007/BF00627745 [DOI] [PubMed] [Google Scholar]
- 105. Day TA, Ruhland CT, Grobe CW, Xiong F. 1999. Growth and reproduction of Antarctic vascular plants in response to warming and UV radiation reductions in the field. Oecologia 119:24–35. doi: 10.1007/s004420050757 [DOI] [PubMed] [Google Scholar]
- 106. Hughes I. 2000. Biological consequences of global warming: Is the signal already apparent. Trends Ecol Evol 15:56–61. doi: 10.1016/s0169-5347(99)01764-4 [DOI] [PubMed] [Google Scholar]
- 107. Smith RL. 2003. The enigma of Colobanthus quitensis and Deschampsia antarctica in Antarctica.
- 108. Ferrari BC, Bissett A, Snape I, van Dorst J, Palmer AS, Ji M, Siciliano SD, Stark JS, Winsley T, Brown MV. 2016. Geological connectivity drives microbial community structure and connectivity in polar, terrestrial ecosystems. Environ Microbiol 18:1834–1849. doi: 10.1111/1462-2920.13034 [DOI] [PubMed] [Google Scholar]
- 109. Schostag M, Stibal M, Jacobsen CS, Bælum J, Taş N, Elberling B, Jansson JK, Semenchuk P, Priemé A. 2015. Distinct summer and winter bacterial communities in the active layer of svalbard permafrost revealed by DNA- and RNA-based analyses. Front Microbiol 6:399. doi: 10.3389/fmicb.2015.00399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Ji M, van Dorst J, Bissett A, Brown MV, Palmer AS, Snape I, Siciliano SD, Ferrari BC. 2016. Microbial diversity at Mitchell Peninsula, Eastern Antarctica: a potential biodiversity “Hotspot". Polar Biol 39:237–249. doi: 10.1007/s00300-015-1776-y [DOI] [Google Scholar]
- 111. Zhang E. 2021. Diversity, drivers and dispersal of East Antarctic soil Microbiota. , UNSW, Sydney. [Google Scholar]
- 112. Robinson SA, Klekociuk AR, King DH, Pizarro Rojas M, Zúñiga GE, Bergstrom DM. 2020. The 2019/2020 summer of Antarctic heatwaves. Glob Chang Biol 26:3178–3180. doi: 10.1111/gcb.15083 [DOI] [PubMed] [Google Scholar]
- 113. Siegert M, Atkinson A, Banwell A, Brandon M, Convey P, Davies B, Downie R, Edwards T, Hubbard B, Marshall G, Rogelj J, Rumble J, Stroeve J, Vaughan D. 2019. The Antarctic peninsula under a 1.5C global warming scenario. Front. Environ. Sci 7:102. doi: 10.3389/fenvs.2019.00102 [DOI] [Google Scholar]
- 114. Bradford MA, Wieder WR, Bonan GB, Fierer N, Raymond PA, Crowther TWJNCC. 2016. Managing uncertainty in soil carbon feedbacks to climate change. Nature Clim Change 6:751–758. doi: 10.1038/nclimate3071 [DOI] [Google Scholar]
- 115. Cavicchioli R, Ripple WJ, Timmis KN, Azam F, Bakken LR, Baylis M, Behrenfeld MJ, Boetius A, Boyd PW, Classen AT, Crowther TW, Danovaro R, Foreman CM, Huisman J, Hutchins DA, Jansson JK, Karl DM, Koskella B, Mark Welch DB, Martiny JBH, Moran MA, Orphan VJ, Reay DS, Remais JV, Rich VI, Singh BK, Stein LY, Stewart FJ, Sullivan MB, van Oppen MJH, Weaver SC, Webb EA, Webster NS. 2019. Scientists’ warning to humanity: microorganisms and climate change. Nat Rev Microbiol 17:569–586. doi: 10.1038/s41579-019-0222-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116. Merino N, Aronson HS, Bojanova DP, Feyhl-Buska J, Wong ML, Zhang S, Giovannelli D. 2019. Living at the extremes: extremophiles and the limits of life in a planetary context. Front Microbiol 10:1785. doi: 10.3389/fmicb.2019.01785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117. Hallsworth JE, Yakimov MM, Golyshin PN, Gillion JLM, D’Auria G, de Lima Alves F, La Cono V, Genovese M, McKew BA, Hayes SL, Harris G, Giuliano L, Timmis KN, McGenity TJ. 2007. Limits of life in Mgcl2-containing environments: chaotropicity defines the window. Environ Microbiol 9:801–813. doi: 10.1111/j.1462-2920.2006.01212.x [DOI] [PubMed] [Google Scholar]
- 118. Payler SJ, Biddle JF, Sherwood Lollar B, Fox-Powell MG, Edwards T, Ngwenya BT, Paling SM, Cockell CS. 2019. An ionic limit to life in the deep subsurface. Front Microbiol 10:426. doi: 10.3389/fmicb.2019.00426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119. Steinle L, Knittel K, Felber N, Casalino C, de Lange G, Tessarolo C, Stadnitskaia A, Sinninghe Damsté JS, Zopfi J, Lehmann MF, Treude T, Niemann H. 2018. Life on the edge: active microbial communities in the Kryos MgCl2-brine Basin at very low water activity. ISME J 12:1414–1426. doi: 10.1038/s41396-018-0107-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120. Kopparapu RK. 2018. The habitable zone: The Climatic limits of Habitability, p 2981–2993. In Deeg HJ, Belmonte JA (ed), Handbook of Exoplanets. Springer International Publishing. doi: 10.1007/978-3-319-55333-7 [DOI] [Google Scholar]
- 121. McKay CP. 2014. Requirements and limits for life in the context of Exoplanets. Proc Natl Acad Sci USA 111:12628–12633. doi: 10.1073/pnas.1304212111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122. Waite Jr JH, Lewis WS, Magee BA, Lunine JI, McKinnon WB, Glein CR, Mousis O, Young DT, Brockwell T, Westlake J, Nguyen M-J, Teolis BD, Niemann HB, McNutt Jr RL, Perry M, Ip W-H. 2009. Liquid water on enceladus from observations of ammonia and 40Ar in the plume. Nature 460:487–490. doi: 10.1038/nature08153 [DOI] [Google Scholar]
- 123. Postberg F, Kempf S, Schmidt J, Brilliantov N, Beinsen A, Abel B, Buck U, Srama R. 2009. Sodium salts in E-ring ice grains from an ocean below the surface of Enceladus. Nature 459:1098–1101. doi: 10.1038/nature08046 [DOI] [PubMed] [Google Scholar]
- 124. Schmidt J, Brilliantov N, Spahn F, Kempf S. 2008. Slow dust in Enceladus' plume from condensation and wall collisions in Tiger stripe fractures. Nature 451:685–688. doi: 10.1038/nature06491 [DOI] [PubMed] [Google Scholar]
- 125. Paganini L, Villanueva GL, Roth L, Mandell AM, Hurford TA, Retherford KD, Mumma MJ. 2020. A measurement of water vapour amid a largely quiescent environment on Europa. Nat Astron 4:266–272. doi: 10.1038/s41550-019-0933-6 [DOI] [Google Scholar]
- 126. Rathbun JA, Musser GS, Squyres SW. 1998. Ice diapirs on Europa: implications for liquid water. Geophys Res Lett 25:4157–4160. doi: 10.1029/1998GL900135 [DOI] [Google Scholar]
- 127. Lauro SE, Pettinelli E, Caprarelli G, Guallini L, Rossi AP, Mattei E, Cosciotti B, Cicchetti A, Soldovieri F, Cartacci M, Di Paolo F, Noschese R, Orosei R. 2021. Multiple Subglacial water bodies below the South pole of Mars unveiled by new MARSIS data. Nat Astron 5:63–70. doi: 10.1038/s41550-020-1200-6 [DOI] [Google Scholar]
- 128. Goordial J, Davila A, Greer CW, Cannam R, DiRuggiero J, McKay CP, Whyte LG. 2017. Comparative activity and functional ecology of permafrost soils and lithic niches in a hyper‐arid polar desert. Environ Microbiol 19:443–458. doi: 10.1111/1462-2920.13353 [DOI] [PubMed] [Google Scholar]
- 129. Cary SC, McDonald IR, Barrett JE, Cowan DA. 2010. On the rocks: the microbiology of Antarctic dry valley soils. Nat Rev Microbiol 8:129–138. doi: 10.1038/nrmicro2281 [DOI] [PubMed] [Google Scholar]
- 130. Cowan DA, Cary SC, DiRuggiero J, Eckardt F, Ferrari B, Hopkins DW, Lebre PH, Maggs-Kölling G, Pointing SB, Ramond J-B, Tribbia D, Warren-Rhodes K. 2023. Follow the water: microbial water acquisition in desert soils. Microorganisms 11:1670. doi: 10.3390/microorganisms11071670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131. Bertagni MB, Paulot F, Porporato A. 2021. Moisture fluctuations modulate Abiotic and biotic limitations of H2 soil uptake. GBC 35. doi: 10.1029/2021GB006987 [DOI] [Google Scholar]
- 132. Cowan DA, Ferrari BC, McKay CP. 2022. Out of thin air? Astrobiology and atmospheric chemotrophy. Astrobiology 22:225–232. doi: 10.1089/ast.2021.0066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133. Camprubí E, de Leeuw JW, House CH, Raulin F, Russell MJ, Spang A, Tirumalai MR, Westall F. 2019. The emergence of life. Space Sci Rev 215:56. doi: 10.1007/s11214-019-0624-8 [DOI] [Google Scholar]
- 134. Weiss BP, Yung YL, Nealson KH. 2000. Atmospheric energy for subsurface life on Mars? Proc Natl Acad Sci USA 97:1395–1399. doi: 10.1073/pnas.030538097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135. Williams DR. 2004. Mars fact sheet. Available from: http://nssdcgsfc nasa gov/planetary/factsheet/marsfact.html
- 136. Franz HB, Trainer MG, Malespin CA, Mahaffy PR, Atreya SK, Becker RH, Benna M, Conrad PG, Eigenbrode JL, Freissinet C, Manning HLK, Prats BD, Raaen E, Wong MH. 2017. Initial SAM calibration gas experiments on Mars: quadrupole mass spectrometer results and implications. Planet Space Sci 138:44–54. doi: 10.1016/j.pss.2017.01.014 [DOI] [Google Scholar]
- 137. Krasnopolsky VA, Feldman PD. 2001. Detection of molecular hydrogen in the atmosphere of Mars. Science 294:1914–1917. doi: 10.1126/science.1065569 [DOI] [PubMed] [Google Scholar]
- 138. Stamenković V, Ward LM, Mischna M, Fischer WW. 2018. O2 solubility in Martian near-surface environments and implications for aerobic life. Nature Geosci 11:905–909. doi: 10.1038/s41561-018-0243-0 [DOI] [Google Scholar]
- 139. Marlow JJ, Martins Z, Sephton MA. 2011. Organic host analogues and the search for life on Mars. Int J Astrobiol 10:31–44. doi: 10.1017/S1473550410000303 [DOI] [Google Scholar]
- 140. Azua-Bustos A, González-Silva C, Fairén AG. 2022. The Atacama desert in northern Chile as an analog model of Mars. Front Astron Space Sci 8:8. doi: 10.3389/fspas.2021.810426 [DOI] [Google Scholar]
- 141. Warren-Rhodes KA, Lee KC, Archer SDJ, Cabrol N, Ng-Boyle L, Wettergreen D, Zacny K, Pointing SB, NASA Life in the Atacama Project Team . 2019. Corrigendum: subsurface microbial Habitats in an extreme desert Mars-analog environment. Front Microbiol 10:2129. doi: 10.3389/fmicb.2019.02129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142. Marinova MM, McKay CP, Heldmann JL, Goordial J, Lacelle D, Pollard WH, Davila AF. 2022. Climate and energy balance of the ground in university valley, Antarctica. Antarctic Science 34:144–171. doi: 10.1017/S0954102022000025 [DOI] [Google Scholar]
- 143. Heldmann JL, Pollard W, McKay CP, Marinova MM, Davila A, Williams KE, Lacelle D, Andersen DT. 2013. The high elevation dry valleys in Antarctica as analog sites for subsurface ice on Mars. Planet Space Sci 85:53–58. doi: 10.1016/j.pss.2013.05.019 [DOI] [Google Scholar]
- 144. Salvatore MR, Levy JS. 2021. Chapter 11 - the Mcmurdo dry valleys of Antarctica: a geological, environmental, and ecological analog to the Martian surface and near surface, p 291–332. In Soare RJ, Conway SJ, Williams JP, Oehler DZ (ed), Mars geological enigmas: from the late Noachian epoch to the present day. doi: 10.1016/b978-0-12-820245-6.00011-2 [DOI] [Google Scholar]
- 145. Hwang Y, Schulze-Makuch D, Arens FL, Saenz JS, Adam PS, Sager C, Bornemann TLV, Zhao W, Zhang Y, Airo A, Schloter M, Probst AJ. 2021. Leave no stone unturned: individually adapted xerotolerant Thaumarchaeota sheltered below the boulders of the Atacama Desert hyperarid core. Microbiome 9:234. doi: 10.1186/s40168-021-01177-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146. Kasting JF, Kopparapu R, Ramirez RM, Harman CE. 2014. Remote life-detection criteria, habitable zone boundaries, and the frequency of earth-like planets around M and late K stars. Proc Nat Acad Sci USA 111:12641–12646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147. Flemming H-C, Wuertz SJNRM. 2019. Bacteria and archaea on earth and their abundance in biofilms. Nat Rev Microbiol 17:247–260. doi: 10.1038/s41579-019-0158-9 [DOI] [PubMed] [Google Scholar]
- 148. Imshenetsky AA, Lysenko SV, Kazakov GA. 1978. Upper boundary of the biosphere. Appl Environ Microbiol 35:1–5. doi: 10.1128/aem.35.1.1-5.1978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149. Grebennikova TV, Syroeshkin AV, Shubralova EV, Eliseeva OV, Kostina LV, Kulikova NY, Latyshev OE, Morozova MA, Yuzhakov AG, Zlatskiy IA, Chichaeva MA, Tsygankov OS. 2018. The DNA of bacteria of the world ocean and the earth in cosmic dust at the international space station. Sci World J 2018:7360147. doi: 10.1155/2018/7360147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150. DeLeon-Rodriguez N, Lathem TL, Rodriguez-R LM, Barazesh JM, Anderson BE, Beyersdorf AJ, Ziemba LD, Bergin M, Nenes A, Konstantinidis KT. 2013. Microbiome of the upper Troposphere: Species composition and prevalence, effects of tropical storms, and atmospheric implications. Proc Natl Acad Sci USA 110:2575–2580. doi: 10.1073/pnas.1212089110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151. Pearce DA, Bridge PD, Hughes KA, Sattler B, Psenner R, Russell NJ. 2009. Microorganisms in the atmosphere over Antarctica. FEMS Microbiol Ecol 69:143–157. doi: 10.1111/j.1574-6941.2009.00706.x [DOI] [PubMed] [Google Scholar]
- 152. Lancia A, Capone P, Vonesch N, Pelliccioni A, Grandi C, Magri D, D’Ovidio MC. 2021. Research progress on Aerobiology in the last 30 years: a focus on methodology and occupational health. Sustainability 13:4337. doi: 10.3390/su13084337 [DOI] [Google Scholar]
- 153. Deguillaume L, Leriche M, Amato P, Ariya PA, Delort A-M, Pöschl U, Chaumerliac N, Bauer H, Flossmann AI, Morris CE. 2008. Microbiology and atmospheric processes: chemical interactions of primary biological aerosols. Biogeosciences 5:1073–1084. doi: 10.5194/bg-5-1073-2008 [DOI] [Google Scholar]
- 154. Bauer H, Giebl H, Hitzenberger R, Kasper-Giebl A, Reischl G, Zibuschka F, Puxbaum H. 2003. Airborne bacteria as cloud condensation nuclei. J Geophys Res 108. doi: 10.1029/2003JD003545 [DOI] [Google Scholar]
- 155. Morris CE, Sands DC, Bardin M, Jaenicke R, Vogel B, Leyronas C, Ariya PA, Psenner R. 2008. Microbiology and atmospheric processes: an upcoming era of research on bio-meteorology. doi: 10.5194/bgd-5-191-2008 [DOI]
- 156. Bottos EM, Woo AC, Zawar-Reza P, Pointing SB, Cary SC. 2014. Airborne bacterial populations above desert soils of the McMurdo dry valleys, Antarctica. Microb Ecol 67:120–128. doi: 10.1007/s00248-013-0296-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157. Burrows SM, Butler T, Jöckel P, Tost H, Kerkweg A, Pöschl U, Lawrence MG. 2009. Bacteria in the global atmosphere – part 2: modeling of emissions and transport between different ecosystems. Atmos Chem Phys 9:9281–9297. doi: 10.5194/acp-9-9281-2009 [DOI] [Google Scholar]
- 158. Sattler B, Puxbaum H, Psenner R. 2001. Bacterial growth in supercooled cloud droplets. Geophys Res Lett 28:239–242. doi: 10.1029/2000GL011684 [DOI] [Google Scholar]
- 159. Dimmick RL, Wolochow H, Chatigny M, Microbiology E. 1979. Evidence that bacteria can form new cells in airborne particles. Appl Environ Microbiol 37:924–927. doi: 10.1128/aem.37.5.924-927.1979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160. Bauer H, Kasper-Giebl A, Löflund M, Giebl H, Hitzenberger R, Zibuschka F, Puxbaum H. 2002. The contribution of bacteria and fungal spores to the organic carbon content of cloud water, precipitation and aerosols. Atmos Res 64:109–119. doi: 10.1016/S0169-8095(02)00084-4 [DOI] [Google Scholar]
- 161. Herlihy LJ, Galloway JN, Mills A. 1987. Bacterial utilization of formic and acetic acid in Rainwater. Atmos Environ 21:2397–2402. doi: 10.1016/0004-6981(87)90374-X [DOI] [Google Scholar]
- 162. Limbeck A, Puxbaum H. 2000. Dependence of in‐cloud scavenging of polar organic aerosol compounds on the water solubility. J Geophys Res 105:19857–19867. doi: 10.1029/2000JD900123 [DOI] [Google Scholar]
- 163. Finlay BJ. 2002. Global dispersal of free-living microbial eukaryote species. Science 296:1061–1063. doi: 10.1126/science.1070710 [DOI] [PubMed] [Google Scholar]
- 164. Wiśniewska K, Śliwińska-Wilczewska S, Lewandowska A, Konik M. 2021. The effect of abiotic factors on abundance and photosynthetic performance of airborne cyanobacteria and microalgae isolated from the Southern Baltic sea region. Cells 10:103. doi: 10.3390/cells10010103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165. Marshall WA, Chalmers MO. 1997. Airborne dispersal of Antarctic terrestrial algae and Cyanobacteria. Ecography 20:585–594. doi: 10.1111/j.1600-0587.1997.tb00427.x [DOI] [Google Scholar]
- 166. Cameron RE, Morelli FA, Johnson RM. 1972. Bacterial species in soil and air of the Antarctic continent. Antarct J US 7(5):187–189. [Google Scholar]
- 167. Malard LA, Avila-Jimenez ML, Schmale J, Cuthbertson L, Cockerton L, Pearce DA. 2022. Aerobiology over the Southern ocean - implications for bacterial colonization of Antarctica. Environ Int 169:107492. doi: 10.1016/j.envint.2022.107492 [DOI] [PubMed] [Google Scholar]
- 168. Cuthbertson L, Amores-Arrocha H, Malard LA, Els N, Sattler B, Pearce DA. 2017. Characterisation of Arctic bacterial communities in the air above Svalbard. Biology 6:29. doi: 10.3390/biology6020029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169. Burrows SM, Elbert W, Lawrence MG, Pöschl U. 2009. Bacteria in the global atmosphere – part 1: Review and synthesis of literature data for different ecosystems. Atmos Chem Phys 9:9263–9280. doi: 10.5194/acp-9-9263-2009 [DOI] [Google Scholar]
- 170. Yooseph S, Andrews-Pfannkoch C, Tenney A, McQuaid J, Williamson S, Thiagarajan M, Brami D, Zeigler-Allen L, Hoffman J, Goll JB, Fadrosh D, Glass J, Adams MD, Friedman R, Venter JC, Moreno-Hagelsieb G. 2013. A Metagenomic framework for the study of airborne microbial communities. PLoS ONE 8:e81862. doi: 10.1371/journal.pone.0081862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171. Dommergue A, Amato P, Tignat-Perrier R, Magand O, Thollot A, Joly M, Bouvier L, Sellegri K, Vogel T, Sonke JE, Jaffrezo J-L, Andrade M, Moreno I, Labuschagne C, Martin L, Zhang Q, Larose C. 2019. Methods to investigate the global atmospheric microbiome. Front Microbiol 10. doi: 10.3389/fmicb.2019.00243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172. Cain AK, Barquist L, Goodman AL, Paulsen IT, Parkhill J, van Opijnen T. 2020. A decade of advances in transposon-insertion sequencing. Nat Rev Genet 21:526–540. doi: 10.1038/s41576-020-0244-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173. Calderón-Ezquerro MDC, Serrano-Silva N, Brunner-Mendoza C. 2021. Aerobiological study of bacterial and fungal community composition in the atmosphere of Mexico city throughout an annual cycle. Environ Pollut 278:116858. doi: 10.1016/j.envpol.2021.116858 [DOI] [PubMed] [Google Scholar]
- 174. Jaing C, Thissen J, Morrison M, Dillon MB, Waters SM, Graham GT, Be NA, Nicoll P, Verma S, Caro T, Smith DJ. 2020. Sierra Nevada sweep: metagenomic measurements of bioaerosols vertically distributed across the troposphere. Sci Rep 10:12399. doi: 10.1038/s41598-020-69188-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175. Nkem JN, Wall DH, Virginia RA, Barrett JE, Broos EJ, Porazinska DL, Adams BJ. 2006. Wind dispersal of soil invertebrates in the McMurdo dry valleys, Antarctica. Polar Biol 29:346–352. doi: 10.1007/s00300-005-0061-x [DOI] [Google Scholar]
- 176. Schulte NO, Khan AL, Smith EW, Zoumplis A, Kaul D, Allen AE, Adams BJ, McKnight DM. 2022. Blowin’ in the wind: dispersal, structure, and metacommunity dynamics of aeolian diatoms in the McMurdo sound region, Antarctica. J Phycol 58:36–54. doi: 10.1111/jpy.13223 [DOI] [PubMed] [Google Scholar]
- 177. Rosa LH, Pinto OHB, Convey P, Carvalho-Silva M, Rosa CA, Câmara PEAS. 2021. DNA metabarcoding to assess the diversity of airborne fungi present over Keller peninsula. Microb Ecol 82:165–172. doi: 10.1007/s00248-020-01627-1 [DOI] [PubMed] [Google Scholar]
- 178. Pearce DA, Alekhina IA, Terauds A, Wilmotte A, Quesada A, Edwards A, Dommergue A, Sattler B, Adams BJ, Magalhães C, Chu W-L, Lau MCY, Cary C, Smith DJ, Wall DH, Eguren G, Matcher G, Bradley JA, de Vera J-P, Elster J, Hughes KA, Cuthbertson L, Benning LG, Gunde-Cimerman N, Convey P, Hong SG, Pointing SB, Pellizari VH, Vincent WF. 2016. Aerobiology over Antarctica – a new initiative for atmospheric ecology. Front Microbiol 7:16. doi: 10.3389/fmicb.2016.00016 [DOI] [PMC free article] [PubMed] [Google Scholar]