Abstract
Background
Treatment resistance and tumor relapse are the primary causes of mortality in glioblastoma (GBM), with intratumoral heterogeneity playing a significant role. Patient-derived cancer organoids have emerged as a promising model capable of recapitulating tumor heterogeneity. Our objective was to develop patient-derived GBM organoids (PGO) to investigate treatment response and resistance.
Methods
GBM samples were used to generate PGOs and analyzed using whole-exome sequencing (WES) and single-cell karyotype sequencing. PGOs were subjected to temozolomide (TMZ) to assess viability. Bulk RNA sequencing was performed before and after TMZ.
Results
WES analysis on individual PGOs cultured for 3 time points (1–3 months) showed a high inter-organoid correlation and retention of genetic variants (range 92.3%–97.7%). Most variants were retained in the PGO compared to the tumor (range 58%–90%) and exhibited similar copy number variations. Single-cell karyotype sequencing demonstrated preservation of genetic heterogeneity. Single-cell multiplex immunofluorescence showed maintenance of cellular states. TMZ treatment of PGOs showed a differential response, which largely corresponded with MGMT promoter methylation. Differentially expressed genes before and after TMZ revealed an upregulation of the JNK kinase pathway. Notably, the combination treatment of a JNK kinase inhibitor and TMZ demonstrated a synergistic effect.
Conclusions
Overall, these findings demonstrate the robustness of PGOs in retaining the genetic and phenotypic heterogeneity in culture and the application of measuring clinically relevant drug responses. These data show that PGOs have the potential to be further developed into avatars for personalized adaptive treatment selection and actionable drug target discovery and as a platform to study GBM biology.
Keywords: glioblastoma, organoids, preclinical models, temozolomide resistance, tumor heterogeneity
Key Points.
Patient-derived glioblastoma organoids (PGOs) are stable representatives of the patient’s tumor.
PGOs display maintenance of genetic and phenotypic tumor heterogeneity.
PGOs can be used to identify novel treatment options and study treatment-resistance mechanisms.
Importance of the Study.
Tumor heterogeneity is recognized as one of the major determinants of treatment failure and tumor relapse in patients with glioblastoma (GBM). Many preclinical GBM models have limitations in adequately representing tumor heterogeneity and key GBM features with prognostic significance such as hypoxia. Novel in vitro models are needed which recapitulate tumor heterogeneity. This study addresses this gap by introducing a patient-derived GBM organoid (PGO) model that successfully maintains both the genetic and phenotypic characteristics of the original tumor. PGOs have significant translational potential to identify novel actionable targets, study mechanisms of primary and secondary resistance, and improve understanding of the complex biology of GBM including the influence of the tumor microenvironment.
Glioblastoma (GBM), grade 4 IDH1/2 wild-type astrocytoma, is the most common malignant primary brain tumor, with a global incidence of around 3 cases per 100 000 each year.1 The current standard-of-care treatment was established more than 15 years ago and consists of tumor resection, followed by chemoradiation and adjuvant chemotherapy with temozolomide (TMZ).2 However, despite this intensive multimodal treatment, the median survival of GBM patients remains only 15 months, which stresses the need for novel treatment options.1
According to the 2021 World Health Organization classification, GBM diagnosis is based on a combination of histological and molecular features.3 In daily clinical practice, methylguanine methyltransferase (MGMT) gene methylation is the only available predictive marker. MGMT promotor methylation abrogates expression in GBM is primarily due to promoter methylation and prevents DNA damage repair from alkylating agents, such as TMZ, and is therefore associated with a more favorable treatment response and greater median overall survival.4
More recently, extensive whole-genome and single-cell sequencing of GBM patient samples have identified driver mutations in GBM, resulting in novel classifications according to the transcriptomic5 and epigenetic characteristics.6 Moreover, intratumoral heterogeneity is frequently observed with multiple tumor clones residing in a single GBM,7 leading to heterogeneity in clinical subtypes.5,8 Furthermore, it has been established that GBM cells exist in cellular states that recapitulate neural progenitor-like (NPC-like), oligodendrocyte-progenitor-like (OPC-like), astrocyte-like (AC-like), and mesenchymal-like (MES-like) states. These cellular states have also been shown to coexist within 1 tumor.9 This extensive molecular and spatial heterogeneity creates multiple subclones that either respond or escape therapy to enable the development of treatment-refractory recurrent tumors.10 This inter- and intratumoral heterogeneity likely underlies the limited clinical effects of current homogenous treatment approaches.
Recently, organoid technology has enabled the development of in vitro 3-dimensional tissue models derived from adult human stem cells. These tissue organoids have been shown to retain the key architectural, phenotypic, and genetic characteristics of the primary tissue they are derived from11 organoid and have shown to maintain a cellular hierarchy and recapitulate intratumoral heterogeneity.12,13
So far, patient-derived glioblastoma cells have been cultured as spheroids,14 organoids,15–18 and explants13 each with their advantages and limitations.14
Here, we provide a detailed molecular and phenotypic analysis of 10 patient-derived primary GBM organoids (PGOs). We show that PGOs are genetically stable models over months of culture and largely retain key GBM driver mutations and copy number variations from matched patient tissue. Multiplex immunohistochemistry and single-cell karyotype sequencing show that PGOs maintain genetic and phenotypic intratumoral heterogeneity. Finally, we demonstrate the potential of PGOs to test the sensitivity to the standard-of-care treatment as well as targeted therapies and use PGOs to identify inhibition of the JNK pathway as a potential sensitizer to TMZ.
Together, GBM organoids recapitulate key features of human GBM composition and treatment response and are promising models for discovering novel disease mechanisms and actionable targets.
Materials and Methods
Patient Recruitment
The study protocol for collecting and processing tumor and blood samples from GBM patients, IDH1/2 wild-type astrocytoma grade 4, was approved by the Medical Ethics Committee Zuyderland (METC-Z) under study number 17-T-101. Experiments were registered under clinical trial numbers NCT04865315 and NCT04868396. The included patients were 18 years or older and were eligible for resection of a lesion suspect for GBM based on diagnostic magnetic resonance imaging. All patients provided informed consent prior to surgery. GBM samples were collected at Maastricht University Medical Center + (MUMC+) and Zuyderland (Heerlen). Following the neurosurgical procedure, tumor tissue was immediately processed. In addition, a peripheral arterial blood sample was taken during surgery. Other samples (1919, 1914, 2012.2, 3565, and 3128) were kindly provided by our collaborators.
Tumor Dissociation and Establishment of Primary (Organoid) Lines
Blood samples were centrifuged (3000G, 15 min at 4°C), after which serum was aspirated. The remaining blood was incubated with red blood cell (RBC) lysis buffer (155 mM NH4Cl, 10 mM KHCO3, 0.1 mM EDTA in H2O) for 10 min. Afterward, the samples were washed with cold phosphate-buffered saline (PBS) and centrifuged. The remaining white blood cells were suspended in a freezing medium (86% DMEM, 4% glucose, and 10% DMSO) and stored at −80 °C.
Tumor tissue was macroscopically dissected into approximately 1-mm-diameter pieces and nonviable tissue was removed. Part of the tumor was flash frozen in liquid nitrogen and stored. Subsequently, tumor pieces were incubated in RBC lysis buffer for 10 min while gently shaking at room temperature. After RBC lysis, tumor pieces were suspended in RPMI with the Tumor Dissociation Kit (Human, Miltenyi Biotec) and incubated at 37°C. During incubation, tumor pieces were manually sheared by pipetting. The total incubation time was dependent on the tumor tissue.
After tumor dissociation, tumor cells were filtered (100 μM) and cultured as organoids (see below) or in regular culture flasks with and without poly-L-lysine (Sigma-Aldrich) coating (1:10 dilution in PBS).
Derivation and Culture of Primary (Organoid) Lines
The organoid derivation protocol and culture medium were derived from Hubert et al.17 Tumor cells were suspended in Cultrex RGF Basement Membrane Extract (R&D systems), and spheres of Cultrex (20 μL of Cultrex/cell suspension) were placed in a sterile plastic dish. Organoids were incubated at 37°C for 30 min to set and subsequently transferred into the medium and cultured on an orbital shaker. PGOs were cultured in Neurobasal Medium Complete (NBMc), consisting of NBM supplemented with 10 ng/mL EGF (R&D systems), 10 ng/mL FGF-2 (R&D systems), B27 minus vitamin A (Thermo Fisher), L-glutamine, sodium pyruvate (Thermo Fisher), antibiotic/antimycotic (Thermo Fisher), and amphotericin B (Thermo Fisher).
The frequency of passaging PGOs was highly patient specific. For dissociation, PGOs were manually sheared using a 1000 μL pipette tip, and placed on ice for 30–60 min to dissolve the Cultrex. Subsequently, organoids were incubated with TripLE (Thermo Fisher) for 10 min at 37°C after which they were manually sheared using a narrowed glass Pasteur pipette. Cells were resuspended in Cultrex and utilized for subsequent organoid culture.
Organoid Embedding and Multiplex Immunohistochemistry
Organoids were incubated with pimonidazole (40 mcg/mL) for 2 h and with EdU (ThermoFisher EdU Click-iT kit) for 45 min. Organoids were fixated with 4% paraformaldehyde for 2 h at room temperature and cryopreserved in OCT. Frozen sections were blocked using normal goat serum (5%) for 60 min. Sections were stained with primary antibodies overnight (Supplementary Methods) and subsequently with a suitable fluorescent secondary antibody and DAPI. Imaging was performed on a Leica DMI 4000 microscope.
For multiplex immunohistochemistry, organoids were fixated with 10% formalin for 2 h at room temperature. After that, organoids were embedded in a 3% agarose gel mold and processed for paraffin embedding to be used for Multiple Iterative Labeling by Antibody Neodeposition (Supplementary Methods).19 In short, tissue slides are used for repetitive cycling of immunofluorescent staining. Hereafter, single cells were identified to generate digital reconstructions. Combinations of markers were used to classify cells as either mesenchymal (MES): CD44+/Vimentin+, astrocyte-like (AC): GFAP+, oligodendrocyte-progenitor-like (OPC): OLIG2+/PDGFRA + or neural-progenitor-like (NPC): Nestin+/CD24+. Tumor cells that did not express any additional markers are reported as SOX2+. Blank cells are cells that do not express any of the tumor-related markers in our panel.
Protein Extraction and Western Blot Analysis
Organoids were manually sheared, washed, and incubated with RIPA buffer plus protease inhibitor for 15 min on ice. Afterward, the suspension was flash frozen in liquid nitrogen and thawed 3 times before centrifuging at 13 000G for 15 min. Protein samples were used for Western blot analysis as described in Supplementary Methods.
DNA Extraction and Whole-Exome Sequencing Analysis
DNA extraction was performed on the original tumor tissue (flash frozen after surgery), white blood cells, and cultured organoids. Organoids were cultured for at least 2 months and passaged multiple (minimum 3) times. DNA extraction was performed using the Sigma G1N350 GeneElute Mammalian Genomic DNA miniprep according to the manufacturer’s protocol.
DNA concentration was measured using Qubit DNA Assay Kit in Qubit 2.0 Fluorometer (Life Technologies, CA). DNA degradation and contamination were monitored on 1% agarose gels. The DNA was processed according to manufacturer protocols (Novogene). Bioinformatics analysis was performed as described in Supplementary Methods.
Single-Cell Karyotype Sequencing
Organoids were dissociated using Accumax (StemCell Technologies). Cells were sorted into single cells and plated into a 384-well plate. Karyotype single-cell libraries were prepared sequenced with a target depth of 16 M reads per plate according to local protocol (Single Cell Core, Hubrecht Institute Utrecht). The output was used for bioinformatics analysis (Supplementary Methods).
Cell Viability Assays, Statistical Analysis, and Organoid Treatment Schedules
PGOs were dissociated using TripLE (ThermoFisher) to determine cell viability, and 20–50 K cells were plated in a 96-well plate. Cells were let to rest for 72 h before the initiation of treatment.
Cells were treated with different concentrations of temozolomide (TMZ) (range 0–300 μM) for 5 days. Thirty μM TMZ was used as the human equivalent dose as measured in the cerebrospinal fluids of patients. The culture medium, including TMZ, was refreshed every 24 h. After 5 days of treatment, cells were rested for 48 h before performing CellTiterGlo 3D assay (Promega). CellTiterGlo 3D was performed as per the manufacturer’s instructions. The luminescent signal was measured using BMG Labtech FLUOstar Omega.
Radiotherapy treatment was performed using an X-ray cabinet (Philips 225 kV and 10 mA), and cells were irradiated with 2 Gy for either 1 day or 3 consecutive days. In addition, concurrent TMZ treatment was performed using 15 μM TMZ (human equivalent dose in concurrent chemoradiation).
Osimertinib was kindly provided by AstraZeneca. Cells were treated for 72 h with different concentrations (0–10 μM), after which a CellTiterGlo 3D assay was performed. Protein samples were prepared after culturing cells on poly-L-lysine-coated dishes and treated with 5 μM Osimertinib or DMSO for 24 h. Western blot analysis was performed as described before.
JNK and TMZ Combination Treatments
JNK inhibitor SP600125 was used for combination treatment with TMZ at a dose of 5 μM. Cells were treated with either 5 μM SP600125, 30 μM TMZ (human equivalent dose), or both. Additionally, to evaluate synergy scores, PGOs were treated with a ranging concentration of TMZ (0, 15.6, 31.25, 62.5, or 125 µM) with or without a fixed dose of SP600125 (5, 10, or 15 µM). Cell viability was measured after 7 days with Alamar Blue. DMSO was used as a control.
TMZ Resistance and RNA Sequencing Analysis
Organoids were treated with 30 μM TMZ for 5 consecutive days, after which organoids were dissociated, and regrown as organoids. After regrowth, TMZ treatment and passaging of the organoids were repeated. After the regrowth following the second TMZ cycle, RNA was extracted using Nucleospin RNA (Machery Nagel 740955.250).
Extracted RNA from pre-and post-TMZ treatment PGOs was further processed for RNA sequencing by Novogene Sequencing. For all samples, the total number of reads ranged between 40 and 43 million reads and the percentage of mapping was >95% for all samples. In addition, genes that were upregulated in the TMZ-treated PGOs were entered into the DAVID database for gene enrichment analysis.20,21
Statistical Analysis
Graphs were generated using GraphPad Prism (v9) by normalizing the luminescence counts of the treatment conditions to the vehicle control. For dose–response curves, an area under the curve (AUC) was calculated and AUC values were compared using an unpaired t-test. For comparison of different treatments, the response to treatment, as measured by cell viability was compared between patients using an unpaired t-test. To determine the synergy of drug interactions, Bliss scores were computed according to the formula Eab = Ea + Eb × (100 − Ea). A Bliss score of 10 or higher is considered to represent a synergistic interaction.22
Results
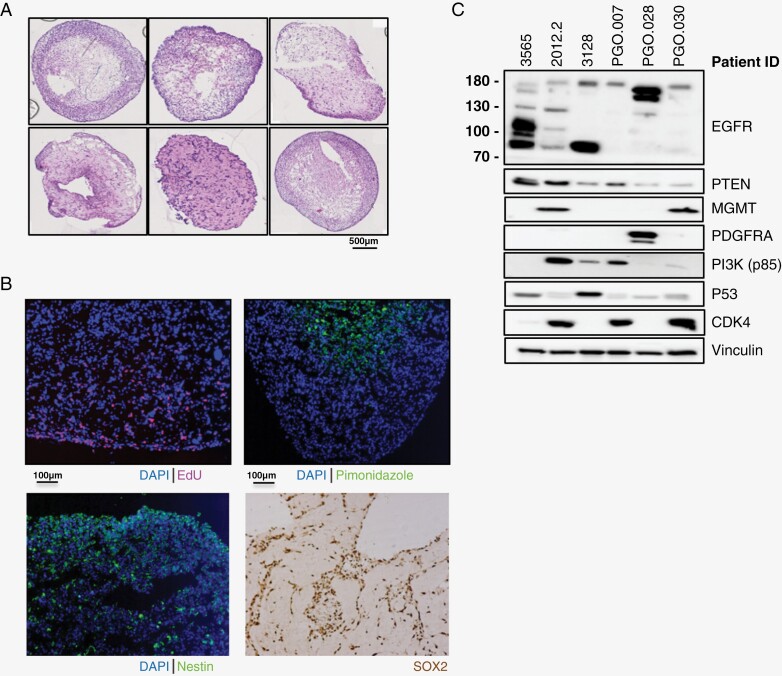
In total, 32 patients were included in this study, from which long-term organoid cultures were established from 10 patients (Supplementary Materials 1). The success rate of established organoids increased toward the end of the study as 5 out of 6 final collected tumors were successfully established as PGOs (83% success rate). H&E staining of PGOs shows tumor cells with a glial appearance and cell proliferation, as well as areas of necrosis and different morphological growth patterns. (Figure 1A).
Figure 1.
Development of a patient-derived glioblastoma (GBM) organoid (PGO) platform and characterization of key GBM features. (A) Representative H&E stainings of PGOs. Scale bar is 500 uM. (B) Representative immunofluoresence imaging of PGOs for proliferation (EdU, top-left), hypoxia (pimonidazole, top-right), Nestin (bottom-left) and SOX2 (bottom-right). Nuclei are stained with DAPI. Scale bar is 100 uM. (C) Western blot characterization of key proteins involved in GBM tumorigenesis in PGOs (N = 6). Data is shown for epidermal growth factor receptor (EGFR), phosphatase and tensin homolog (PTEN), methylguanine methyltransferase (MGMT), platelet-derived growth factor receptor alpha (PDGFRA), phosphoinositide 3-kinase (PI3K), tumor protein 53 (p53), and cyclin-dependent kinase 4 (CDK4).
PGOs display a proliferative rim (EdU+) and hypoxic core (pimonidazole), a common feature of GBM (Figure 1B). PGOs also express GBM markers Nestin and SOX2 (Figure 1B). Western blot analysis showed typical perturbations of GBM driver genes such as PTEN loss, TP53 overexpression, and epidermal growth factor receptor (EGFR) truncation and amplification (Figure 1C). The set of PGOs was derived from both MGMT-methylated (N = 6) and MGMT-unmethylated (N = 4) patients. These data show that our PGOs retain relevant genetic and phenotypic features of GBM and represent a heterogeneous group of GBM.
Whole-Exome Sequencing Analysis of Patient-Derived GBM Organoids
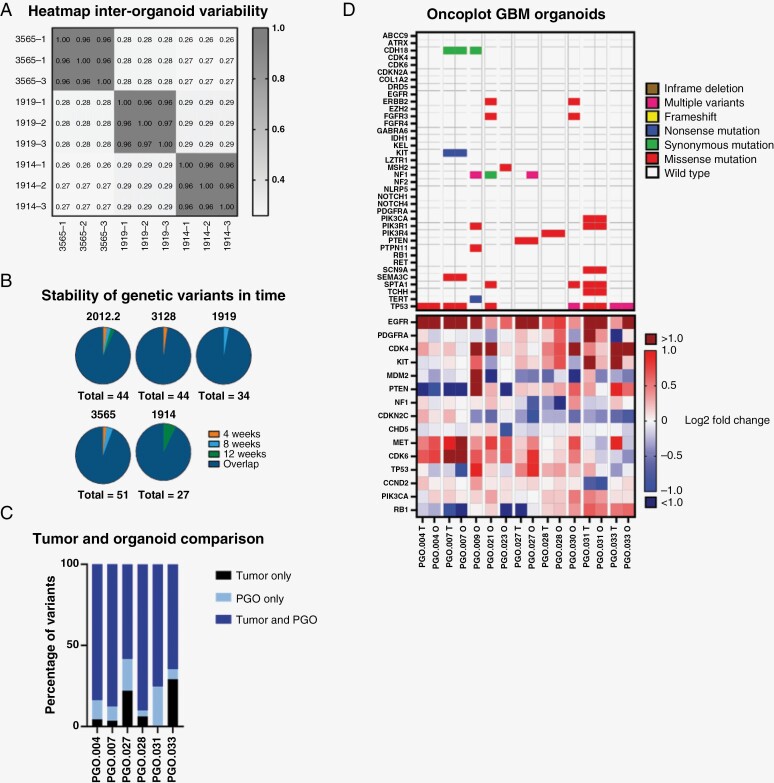
Next, we investigated whether genetic variants were stable during in vitro expansion of the PGOs. The genetic stability was determined based on variants found in 465 genes (Supplementary Materials 2), selected based on the COSMIC database.23 First, we performed whole-exome sequencing (WES) on 3 parallel cultured individual organoids derived from the same patient (N = 3), which showed high inter-organoid correlation (Spearman’s correlation coefficient >0.96 for all 3 samples) (Figure 2A; Supplementary Materials 3).PGOs from the same patient analyzed at different time points (4, 8, or 12 weeks) in culture showed most identified variants (range from 92.3% to 97.7%) were stable and identified at all time points (Figure 2B; Supplementary Materials 4).
Figure 2.
Genetic characterization of patient-derived GBM organoids (PGOs). (A) Heatmap of variants identified by whole-exome sequencing (WES) of 3 individual PGOs derived from the same patient (N = 3). Pearson correlation coefficients are reported between each sample. (B) Venn diagram of variants identified by WES of PGOs derived from the same patient (N = 5) at different time points in culture (4 weeks, 8 weeks, and 12 weeks). 2012.2: 98% overlap, 2% 4 weeks only; 3128: 98% overlap, 2% 4 weeks only; 1919: 97% overlap, 3% 8 weeks only; 3565: 94% overlap, 2% 4 weeks only, 4% 8 weeks only; 1914: 93% overlap, 7% 12 weeks only. (C) Comparison between coding variants found by WES in PGOs and their corresponding parental tumors. (D) Oncoplot of top relevant mutations (top panel) and copy number variations (CNVs, bottom panel) in GBM based on WES in the tumor (T) and organoid (O).
To assess whether PGOs genetically resemble their parental tumor from, we performed WES on the tumor and corresponding PGOs. Figure 2D shows variants identified in PGOs and tumor tissue in GBM-relevant genes. We observed variants in well-known GBM driver genes. These include TP53 (6/10), NF1 (3/10), PTEN (1/10), and PIK3R1 (2/10). Copy number variants (CNVs) on commonly altered genes in GBM also largely corresponded between PGO and tumor (Figure 2D). This includes EGFR amplification (7/10) and loss of PTEN (3/10). The concordance in coding variants between PGOs and the parental tumor varied from 58% to 90% (N = 6; Figure 2C). From 4 patients, insufficient parental tumor material was available for WES. The complete list of somatic variants and CNVs is reported in Supplementary Materials 5.
Genetic and Phenotypic Intratumoral Heterogeneity
We used single-cell KaryoSeq to interrogate GBM heterogeneity in PGOs (N = 7) (Figure 3). All PGOs maintained significant genetic heterogeneity, shown by multiple subclones derived from a founder population. In addition, known key chromosomal aberrations, such as amplification of chromosome 7 (EGFR) and loss of chromosome 10 (PTEN), were observed.
Figure 3.
Phylogenetic trees derived from single-cell karyotype sequencing for different patient-derived glioblastoma (GBM) organoids (PGOs; N = 7). Losses and gains of (parts of) chromosomes are reported in the figure.
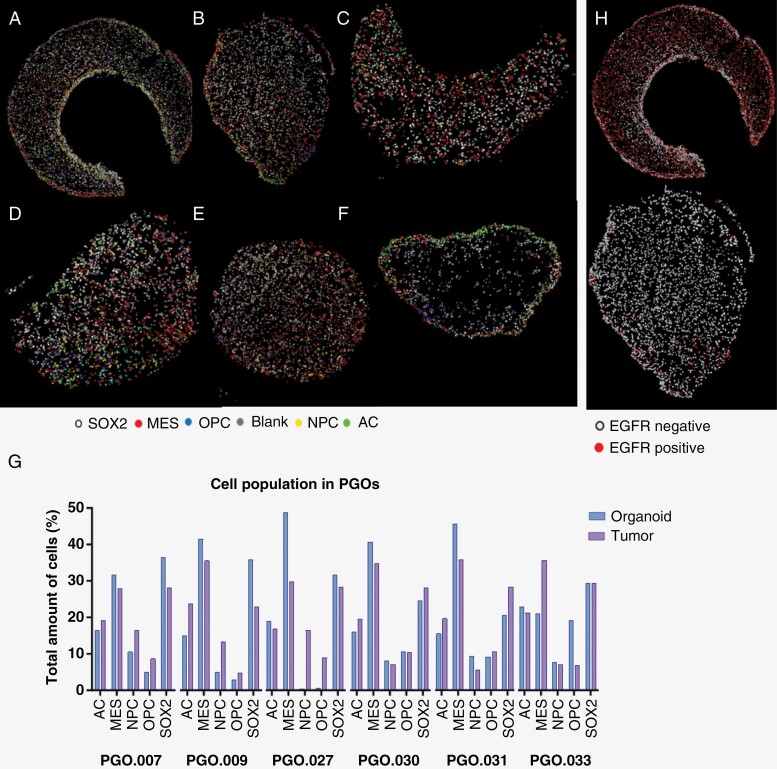
In addition to genetic heterogeneity, there is a high level of phenotypic plasticity where multiple subtypes can be present. To address this, we conducted multiplex immunohistochemistry on 6 PGOs. Digital reconstructions show the heterogeneity of different tumor cell subtypes within PGOs (Figure 4A–F). All PGOs showed MES-like cells as the most dominant cell population, with different distributions of the other cell subtypes. Overall, the proportions of the different cell types corresponded well between the PGO and parental tumor (Figure 4G). In PGO.027, we observed a complete loss of the NPC and OPC cells, which was retained in the other PGOs. (Figure 4G). Additionally, we observed marked differences in EGFR expression between PGOs showing molecular heterogeneity between PGOs (Figure 4H).
Figure 4.
Digital reconstructions of patient-derived glioblastoma organoid (PGO) slides after multiplex immunohistochemistry, cells were classified based on marker expression: MES (mesenchymal-like), OPC (oligodendrocyte-progenitor like), AC (astrocyte like), and NPC (neural progenitor cell like) or SOX2 (no specific classification). (A) PGO.007. (B) PGO.009. (C) PGO.027. (D) PGO.030. (E) PGO.031. (F) PGO.033. (G) Quantification of different cell populations in GBM organoids (blank bars) and their corresponding parental tumors (dashed bars). (H) Digital reconstructions of epidermal growth factor receptor (EGFR) expression heterogeneity within PGOs, representative examples are shown from a PGO with high EGFR and low EGFR expression.
Drug Sensitivity and Drug Resistance in GBM Organoids
The only marker to predict TMZ sensitivity in the clinic, MGMT promoter methylation status, was compared between PGO and parental tumor. When a discrepancy between MGMT expression on western blot and the clinical data occurred, we used multiplex ligation-dependent probe amplification (MLPA) to confirm MGMT-methylation status. Overall, MGMT-methylation status was retained in all but one patient (Supplementary Materials 6).
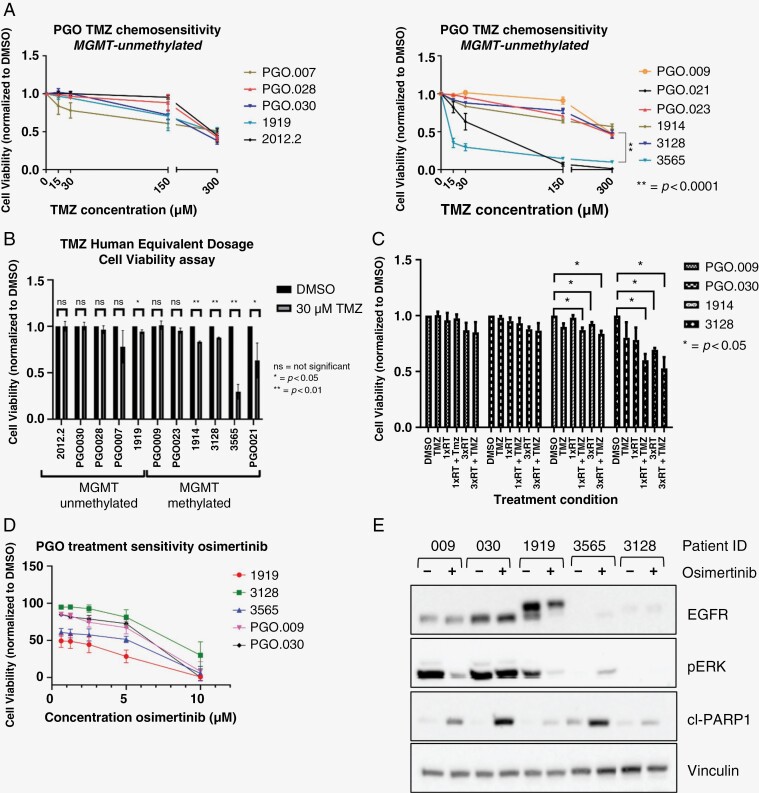
PGOs were treated with TMZ and mainly showed resistance at lower concentrations (15 and 30 μM) except for 2 MGMT-methylated PGOs (Figure 5A). AUC values of drug–response curves were analyzed and both 3565 and PGO.021 were statistically significantly more sensitive toward TMZ compared to other PGOs (P < .0001). PGO.009 was the most resistant PGO also compared to PGO.023 (P = .008), 3128 (P = .046), and 1914 (P = .002). None of the other PGOs were significantly different from the others. All MGMT-unmethylated PGOs were resistant to TMZ and only showed decreased cell viability at high concentrations (100 and 300 μM). The only statistically significant difference that was found was between PGO.007 and 2012.2 (P = .007). Next, PGOs were exposed to a clinically relevant 30-μM concentration. As suspected, MGMT-methylated PGOs responded more favorably when compared to MGMT-unmethylated PGOs. Only 1 MGMT-unmethylated PGO showed a significant decrease in cell viability. Two MGMT-methylated PGOs did not show a substantial reduction, whereas the other patients did (Figure 5B).
Figure 5.
Treatment sensitivity of patient-derived glioblastoma organoids (PGOs): standard-of-care treatment and proof-of-principle targeted therapy. (A) Dose–response curves temozolomide (TMZ) treatment in PGOs (N = 3, mean and SEM) in methylguanine methyltransferase (MGMT)-unmethylated (left) and MGMT-methylated (right) patients. Cell viability was measured using CellTiterGlo assay and compared to untreated control (DMSO). (B) Comparison of response to human equivalent dosage (30 uM) of TMZ inPGOs (N = 3, Mean and SD). Significance was calculated using unpaired t-test. (C) Bart chart of sensitivity of PGOs toward chemoradiation treatment (N = 3, Mean and SD). PGOs were treated with 15 uM TMZ, 1 time radiotherapy (RT) 1Gy or 3 times RT 1Gy radiation or a combination treatment (RT + TMZ). (D) Dose–response curve of osimertinib treatment (N = 3, Mean and SD) as measured by cell viability. (E) Western blot analysis of PGOs for EGFR (endothelial growth factor receptor) protein, pERK (phosphorylated extracellular regulated kinase), and cleaved PARP (Poly [ADP-ribose] polymerase 1) before and after 5 uM osimertinib treatment for 24 h. Vinculin was used as a loading control.
Moreover, PGOs were treated with concurrent chemoradiation, and overall patients showed low sensitivity toward this standard of care (Figure 5C). This was concordant with the clinical patient outcome as most patients showed progression of disease immediately after completing their first-line treatment (Supplementary Materials 1). Next, we investigated whether PGOs would respond to treatments that target commonly mutated genes. As a proof of principle, osimertinib, a third-generation tyrosine-kinase inhibitor targeting EGFR shown to cross the blood–brain barrier (BBB),24 was tested on PGOs. Previous preclinical research showed osimertinib to be effective in EGFRvIII positive GBM25 as well as EGFR-negative GBM.26 AUC values of the dose–response curves were analyzed (Figure 5D). 1919 was the most sensitive PGO, which was also statistically significant compared to 3128, PGO.009, and PGO.030 (P < .0001), as well as 3565 (P = .003). 3565 was the second most sensitive, which was also significant when compared to the other PGOs (vs. PGO.009: P = .018; vs. PGO.030: P = .0005, and vs 3128: P < .0001). 3128 was significantly more resistant when compared to PGO.009 (P = .03) and PGO.030 (P = .014). Osimertinib blocked the phosphorylation of EGFR (not shown) and downstream pERK/MEK signaling, with concomitant upregulation of apoptosis marker cleaved PARP (Figure 5E).
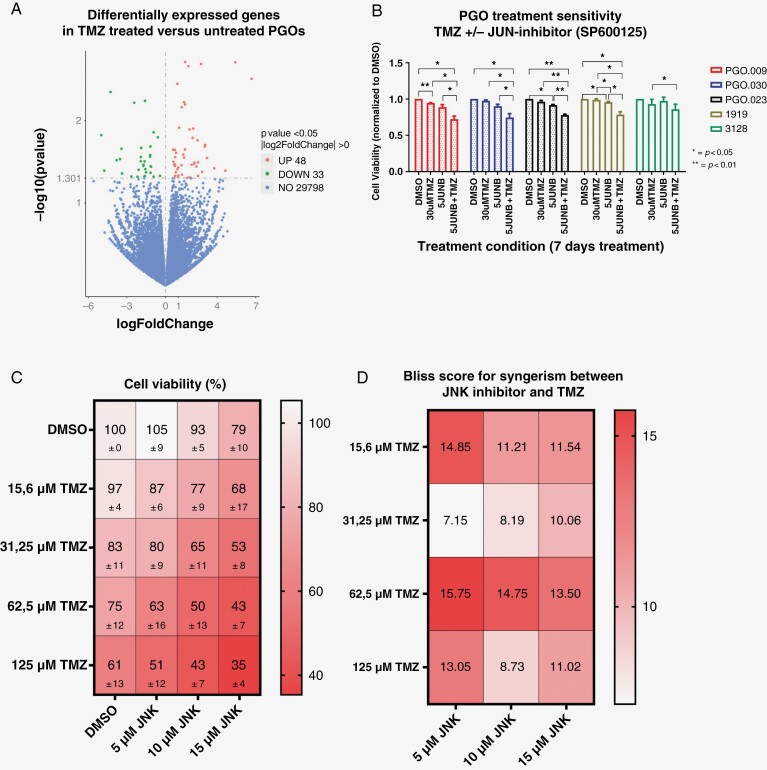
In addition, GBM patients invariably develop resistance to TMZ after which no relevant treatment options exist. We, therefore, asked whether we could use PGOs to identify novel treatment-resistance mechanisms to TMZ. PGOs were treated with 2 cycles of TMZ (30 μM), and bulk RNA sequencing was performed before and after treatment (Figure 6A). Gene-set enrichment analysis was conducted (Supplementary Materials 7) which reported a trend for negative enrichment for multiple pathways. The only trend for enrichment after TMZ treatment was found for mitotic spindle. Differential gene expression and pathway analysis showed the JNK-kinase pathway as the top hit upregulated after TMZ treatment (P-value <.05). To test whether interfering in the JNK pathway would increase TMZ sensitivity, PGOs were treated with the JNK inhibitor SP600125 and/or TMZ. In 4 out of 5 PGOs tested, the combined treatment was more effective (Figure 6B).
Figure 6.
Changes in patient-derived glioblastoma (GBM) organoid (PGO) gene expression upon temozolomide (TMZ) treatment. (A) Volcano plot of differentially expressed genes (identified by RNAseq) between TMZ-treated and TMZ-untreated PGOs (N = 3). (B) Dose–response curves of TMZ and JNK inhibitor combination treatment in PGOs (N = 5). PGOs were treated with either 30 uM TMZ, 5 uM SP600125, or a combination. (C) Cell viability results of PGOs treated with different dosages of TMZ and JNK inhibitor. Data are shown for 3 PGOs combined. (D) Bliss scores showing the interaction between different concentrations of TMZ and JNK inhibitor for 3 PGOs combined. A Bliss score >10 confirms a synergistic reacting when both treatment options are combined.
Next, we titrated different concentrations of JNK inhibitor (5, 10, and 15 μM) with different concentrations of TMZ (15–125 μM) and measured viability 11 days after the start of treatment (Figure 6C). A Bliss score was calculated and a Bliss score of above 10 after treatment with different concentrations SP600125 and TMZ confirms a synergistic interaction. We observed strong synergistic interactions at different dose levels (Figure 6D) that differed between PGO (Supplemental Figure 8A). The most synergistic combinations were 15.6 or 125 µM TMZ with 5 µM SP600125, and 62.5 µM TMZ combined with 5, 10, or 15 µM SP600125. However, the combination treatment of 31.25 µM of TMZ with 5 or 10 µM SP600125 was not synergistic. Remarkably, each patient-derived GBM organoid line shows different optimal concentrations of TMZ and/or SP600125 (Supplementary Figure 8B). Interestingly, Kaplan–Meier analysis in the TCGA database shows a lower overall and progression-free survival for patients with high JNK expression, suggesting its role as an adverse prognostic factor (Supplementary Materials 9A). Consistently, JNK expression is also significantly higher in GBM than in normal brain (Supplemental Materials 9B).
Discussion
Here we describe the generation and validation of a GBM organoid model from patient-derived GBM cells. Overall, we showed that PGOs are genetically stable tumor cell models that maintain clinically relevant markers, such as MGMT-methylation status, somatic gene variants, CNVs, and exhibit phenotypic and genetic intratumoral heterogeneity. Furthermore, we demonstrate that PGOs can be used to identify actionable targets to enhance sensitivity to standard-of-care chemotherapy.
For most PGOs, there was substantial concordance between somatic variants and CNVs found between PGOs and the parental tumor, though not for all. One explanation is that tumor tissue and tissue for organoid generation are from distinct tumor regions. Hence not all subclones are likely to be represented in the material used for organoid derivation due to intratumoral heterogeneity. Moreover, the presence of stromal nontumor cells in the tumor biopsies, used for sequencing, may yield an underrepresentation of mutated and amplified GBM driver genes compared to the PGO which has also been observed by others.27,28 Whether the low concordance in some PGOs is due to sampling error, stromal contamination, or the inability of specific tumor cell clones to grow in vitro requires further investigation.
We observed that PGOs preserved genetic and phenotypic heterogeneity at the single-cell level. Using single-cell karyotype sequencing, we showed the existence of multiple subclones within PGOs while representing well-known GBM chromosomal alterations.1,3 To our knowledge, no previous study investigated chromosomal aneuploidy in PGOs. Furthermore, we show phenotypic heterogeneity within the PGOs using multiplex immunohistochemistry. Remarkably, PGOs maintained different cellular states after prolonged in vitro culture. Overall, the distribution of the different cell types corresponded well between PGO and parental tumor with the exception of 1 case with a complete loss of NPC and OPC cells. The reasons why some PGOs lose certain states while others maintain them are of interest and need to be understood at present. Culture medium and organoid growth methods may contribute to transcriptomic changes not reflective of the in vivo tumor microenvironment (TME) and promote the outgrowth or loss of specific subclones, which might also be patient specific as no variability was consistent among all PGOs.
To functionally test PGOs, we performed cell viability assays. Importantly, all but 1 patient retained the MGMT-methylation status. We showed that at a clinically relevant dosage, most PGOs respond as expected based on their MGMT-methylation status. Interestingly, PGO.023 did not show any response while being MGMT-methylated. This sample was taken from a patient with a TMZ pretreated recurrent GBM and harbored an MSH6 mutation, a well-known mechanism of TMZ resistance.29 Similarly, PGO.009 showed little response to TMZ despite the corresponding patient being classified as MGMT-methylated. Additional analysis of this PGO revealed the organoid to be MGMT-unmethylated. This discrepancy in MGMT-methylation status could be due to intratumoral heterogeneity of MGMT expression in the parental tumor.30 This indicates that regional sampling bias also influences PGO composition, which should be considered.
Additionally, we investigated whether PGOs could be used for drug screening. As a proof of principle, we set out to use a tyrosine-kinase inhibitor of EGFR (Osimertinib), which can cross the BBB. The PGO with the highest protein expression of EGFR (1919) was also the most sensitive. However, EGFR protein expression did not predict treatment sensitivity overall. The second most sensitive patient (3565) does not show any EGFR expression; however, it does show a clear increase in apoptosis protein cleaved PARP. All in all, these experiments show the potential of PGOs to be used for drug screening, contribute to identifying novel treatment options, and improve patient stratification.
We identified the JNK pathway as differentially expressed in TMZ-treated versus untreated PGOs and confirmed that JNK inhibition synergizes with TMZ treatment to reduce cell viability. The JNK pathway is highly active in the central nervous system (`) and is implicated in the response of the CNS to injury. JNK pathway activity correlates to GBM aggressiveness, infiltration, and progression.31 In GBM, it has been demonstrated that JNK kinase signaling is important for maintaining stemness32 and inhibition of either isoform JNK1 or JNK2 can diminish stemness of glioma stem cells.33 JNK expression is also notably increased in GBM compared to healthy brain tissue. Previous studies in GBM confirm that inhibition of the JNK pathway sensitizes cells to TMZ treatment.34,35JNK kinase has not been reported to be involved in TMZ resistance; however, a previous study comparing primary and recurrent GBM also reported enrichment of JNK family members36 This supports future research into the efficacy of JNK inhibition in recurrent GBM as a possible mechanism for regaining TMZ sensitivity or delaying progression. These findings show the potential of PGOs to study treatment-resistance mechanisms; however, whether the changes that occur after treatment in a PGO reflect the changes in vivo are subject to future research including PGOs derived from recurrent GBM.
Our GBM organoid model has particular strengths as a novel preclinical model to study GBM. First, PGOs can be propagated for a long time, bio-banked, and successfully regrown. Additionally, we have shown that GBM organoids can be used for multiple purposes, including studying genetic and phenotypic characteristics, performing drug screens, and identifying treatment-resistance mechanisms. The strengths of this study include the direct comparison between patient material, clinical features, and GBM organoids and the spatial and single-cell analysis to show the maintenance of tumor heterogeneity, one of the most important additive values of cancer organoids.
This study also has its limitations. First of all, the time it takes to establish GBM organoids is currently too long to be able to use them as patient avatars. Moreover, the absence of the TME, including stromal cells, immune cells, endothelial cells, and a brain extracellular matrix, is an obvious shortcoming of the current methods. The TME is also known to influence treatment effectiveness in GBM and therefore results from tumor cell-only models might not be representative of the actual in vivo response.37 The next step in optimizing the GBM organoid mode can be done by using co-culture systems38 to include specific parts of the TME within the organoids. The potential of this has been shown, both in GBM organoids as in different cancer types. Immune cells were either retained by preserving the original tissue architecture16 or co-culturing lymph node cells39 or peripheral blood mononuclear cells.38 Additionally, co-culture systems of patient-derived GBM cells together with human embryonic stem-cell-derived cerebral organoids have been developed to mimic invasion and GBM–brain cell interaction.40 Differences in treatment response between different patients could also be due to gender-specific differences, which have proven to be an important determinant in patient outcome.41
Importantly, single-cell genetic analysis of the parental tumor was not available to compare to the single-cell data of PGOs. Therefore, whether the intratumoral genetic heterogeneity observed in PGOs is similar to that of its parental tumor remains debatable. Additionally, the concordance of genetic makeup in PGOs compared to their parental tumor still has a large variance. These limitations also reflect that in their current state, PGOs are not yet suited to be used as direct patient avatars. However, given that GBM organoids maintain genetic and phenotypic heterogeneity, we still believe they form a superior model to traditional cell culture methods.
In conclusion, PGOs provide a genetically stable model that represents the genetic makeup of the tumor it was derived from and maintains genetic and phenotypic heterogeneity. Additionally, GBM organoids can be used for drug screening, understanding treatment sensitivity and resistance mechanisms, and maybe developed further for patient stratification.
Supplementary Material
Contributor Information
Maikel Verduin, Department of Radiation Oncology (Maastro), GROW School for Oncology and Reproduction, Maastricht University Medical Centre, Maastricht, The Netherlands.
Linde Hoosemans, Department of Radiation Oncology (Maastro), GROW School for Oncology and Reproduction, Maastricht University Medical Centre, Maastricht, The Netherlands.
Maxime Vanmechelen, Laboratory for Precision Cancer Medicine, Translational Cell and Tissue Research Unit, Department of Imaging and Pathology, KU Leuven, Leuven, Belgium; LISCO—KU Leuven Institute for Single Cell Omics, KU Leuven, Leuven, Belgium.
Mike van Heumen, Department of Radiation Oncology (Maastro), GROW School for Oncology and Reproduction, Maastricht University Medical Centre, Maastricht, The Netherlands.
Jolanda A F Piepers, Department of Radiation Oncology (Maastro), GROW School for Oncology and Reproduction, Maastricht University Medical Centre, Maastricht, The Netherlands.
Galuh Astuti, Department of Pathology, Radboud University Medical Center, Nijmegen, The Netherlands.
Linda Ackermans, Department of Neurosurgery, School for Mental Health and Neuroscience (MHeNS), Maastricht University Medical Center, Maastricht, The Netherlands.
Olaf E M G Schijns, Department of Neurosurgery, School for Mental Health and Neuroscience (MHeNS), Maastricht University Medical Center, Maastricht, The Netherlands; Academic Center for Epileptology, Maastricht University Medical Center and Kempenhaeghe, Maastricht—Heeze, The Netherlands.
Kim R Kampen, Department of Radiation Oncology (Maastro), GROW School for Oncology and Reproduction, Maastricht University Medical Centre, Maastricht, The Netherlands; Laboratory for Disease Mechanisms in Cancer, Department of Oncology, KU Leuven, Leuven, Belgium.
Vivianne C G Tjan-Heijnen, Department of Medical Oncology, GROW School for Oncology and Reproduction, Maastricht University Medical Centre, Maastricht, The Netherlands.
Buys A de Barbanson, Barbanson Biotech.
Alida A Postma, Department of Radiology and Nuclear Medicine, School for Mental Health and Neuroscience, Maastricht University Medical Center, Maastricht, The Netherlands.
Danielle B P Eekers, Department of Radiation Oncology (Maastro), GROW School for Oncology and Reproduction, Maastricht University Medical Centre, Maastricht, The Netherlands.
Martijn P G Broen, Department of Neurology, Maastricht University Medical Center, Maastricht, The Netherlands.
Jan Beckervordersandforth, Department of Pathology, Maastricht University Medical Center, Maastricht, The Netherlands.
Katerina Staňková, Institute for Health Systems Science, Delft University of Technology, Delft, The Netherlands.
Frederik de Smet, Laboratory for Precision Cancer Medicine, Translational Cell and Tissue Research Unit, Department of Imaging and Pathology, KU Leuven, Leuven, Belgium; LISCO—KU Leuven Institute for Single Cell Omics, KU Leuven, Leuven, Belgium.
Jeremy Rich, University of Pittsburgh Medical Center Hillman Cancer Center, Pittsburgh, Pennsylvania, USA; Department of Neurology, University of Pittsburgh, Pittsburgh, Pennsylvania, USA.
Christopher G Hubert, Department of Biochemistry, School of Medicine, Case Western Reserve University, Cleveland, Ohio, USA.
Gregory Gimenez, Department of Pathology, Dunedin School of Medicine, University of Otago, Dunedin, New Zealand.
Aniruddha Chatterjee, Department of Pathology, Dunedin School of Medicine, University of Otago, Dunedin, New Zealand.
Ann Hoeben, Department of Medical Oncology, GROW School for Oncology and Reproduction, Maastricht University Medical Centre, Maastricht, The Netherlands.
Marc A Vooijs, Department of Radiation Oncology (Maastro), GROW School for Oncology and Reproduction, Maastricht University Medical Centre, Maastricht, The Netherlands.
Funding
PhD fellowship from Fonds Wetenschappelijk Onderzoek FWO (MvM# 11L0822N), KWF Kankerbestrijding (Dutch Cancer Society) Unique High Risk (16698/2018-1), and StopHersentumoren (SHT-RNA-PGO).
Conflict of interest statement
None declared.
Authorship statement
Conceptualization: M.Ve., M.Va, V.C.G.T-.H., F.d.S, A.H., M.A.V. Methodology: M.Ve., M.Va, L.H., M.v.H., J.A.F.P., G.A., B.A.B., F.d.S., J.R., C.G.H., K.R.K., G.G., A.H., A.C., M.A.V. Investigation: M.Ve., L.H., M.v.H., M.Va, J.A.F.P., G.A., B.A.B., K.R.K., J.B., F.S., G.G., M.A.V. Writing: M.Ve., M.Va, G.A., A.J., A.H., M.A.V. Funding acquisition: M.Ve., A.H., M.A.V. Resources: L.A., O.E.M.G.S., K.R.K., J.B., F.d.S., J.R., C.G.H., G.G., A.C., M.A.V. Supervision: V.C.G.T-.H., J.A.F.P, B.A.B, M.P.G.B, K.S., F.d.S., A.H., M.A.V.
Data availability
All data, so far as not already published in the manuscript, will be made publically available upon request to marc.vooijs@maastrichtuniversity.nl.
References
- 1. Wen PY, Weller M, Lee EQ, et al. Glioblastoma in adults: a Society for Neuro-Oncology (SNO) and European Society of Neuro-Oncology (EANO) consensus review on current management and future directions. Neuro-Oncol. 2020;22(8):1073–1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Stupp R, Mason WP, van den Bent MJ, et al. ; European Organisation for Research and Treatment of Cancer Brain Tumor and Radiotherapy Groups. Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma. N Engl J Med. 2005;352(10):987–996. [DOI] [PubMed] [Google Scholar]
- 3. Louis DN, Perry A, Wesseling P, et al. The 2021 WHO Classification of Tumors of the Central Nervous System: a summary. Neuro-Oncol. 2021;23(8):1231–1251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Hegi ME, Diserens AC, Gorlia T, et al. MGMT gene silencing and benefit from temozolomide in glioblastoma. N Engl J Med. 2005;352(10):997–1003. [DOI] [PubMed] [Google Scholar]
- 5. Lee JK, Wang J, Sa JK, et al. Spatiotemporal genomic architecture informs precision oncology in glioblastoma. Nat Genet. 2017;49(4):594–599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Ceccarelli M, Barthel FP, Malta TM, et al. ; TCGA Research Network. Molecular profiling reveals biologically discrete subsets and pathways of progression in diffuse glioma. Cell. 2016;164(3):550–563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Verhaak RG, Hoadley KA, Purdom E, et al. ; Cancer Genome Atlas Research Network. Integrated genomic analysis identifies clinically relevant subtypes of glioblastoma characterized by abnormalities in PDGFRA, IDH1, EGFR, and NF1. Cancer Cell. 2010;17(1):98–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Patel AP, Tirosh I, Trombetta JJ, et al. Single-cell RNA-seq highlights intratumoral heterogeneity in primary glioblastoma. Science. 2014;344(6190):1396–1401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Neftel C, Laffy J, Filbin MG, et al. An integrative model of cellular states, plasticity, and genetics for glioblastoma. Cell. 2019;178(4):835–849.e21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Qazi MA, Vora P, Venugopal C, et al. Intratumoral heterogeneity: pathways to treatment resistance and relapse in human glioblastoma. Ann Oncol. 2017;28(7):1448–1456. [DOI] [PubMed] [Google Scholar]
- 11. Clevers H. Modeling development and disease with organoids. Cell. 2016;165(7):1586–1597. [DOI] [PubMed] [Google Scholar]
- 12. Rossi G, Manfrin A, Lutolf MP.. Progress and potential in organoid research. Nat Rev Genet. 2018;19(11):671–687. [DOI] [PubMed] [Google Scholar]
- 13. LeBlanc VG, Trinh DL, Aslanpour S, et al. Single-cell landscapes of primary glioblastomas and matched explants and cell lines show variable retention of inter- and intratumor heterogeneity. Cancer Cell. 2022;40(4):379–392.e9. [DOI] [PubMed] [Google Scholar]
- 14. Souberan A, Tchoghandjian A.. Practical review on preclinical human 3D glioblastoma models: advances and challenges for clinical translation. Cancers (Basel). 2020;12(9):2347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Sundar SJ, Shakya S, Barnett A, et al. Three-dimensional organoid culture unveils resistance to clinical therapies in adult and pediatric glioblastoma. Transl Oncol. 2022;15(1):101251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Jacob F, Salinas RD, Zhang DY, et al. A patient-derived glioblastoma organoid model and biobank recapitulates inter- and intra-tumoral heterogeneity. Cell. 2020;180(1):188–204.e22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Hubert CG, Rivera M, Spangler LC, et al. A three-dimensional organoid culture system derived from human glioblastomas recapitulates the hypoxic gradients and cancer stem cell heterogeneity of tumors found in vivo. Cancer Res. 2016;76(8):2465–2477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Sundar SJ, Shakya S, Recinos V, Hubert CG.. Maintaining human glioblastoma cellular diversity ex vivo using three-dimensional organoid culture. J Vis Exp. 2022;(186):e63745. [DOI] [PubMed] [Google Scholar]
- 19. Bosisio FM, Van Herck Y, Messiaen J, et al. Next-generation pathology using multiplexed immunohistochemistry: mapping tissue architecture at single-cell level. Front Oncol. 2022;12:918900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Huang da W, Sherman BT, Lempicki RA.. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4(1):44–57. [DOI] [PubMed] [Google Scholar]
- 21. Huang da W, Sherman BT, Lempicki RA.. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009;37(1):1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Zheng S, Wang W, Aldahdooh J, et al. SynergyFinder plus: toward better interpretation and annotation of drug combination screening datasets. Genom Proteom Bioinformatics. 2022;20(3):587–596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Tate JG, Bamford S, Jubb HC, et al. COSMIC: the catalogue of somatic mutations in cancer. Nucleic Acids Res. 2019;47(D1):D941–D947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Colclough N, Chen K, Johnstrom P, et al. Preclinical comparison of the blood-brain barrier permeability of osimertinib with other EGFR TKIs. Clin Cancer Res. 2021;27(1):189–201. [DOI] [PubMed] [Google Scholar]
- 25. Chagoya G, Kwatra SG, Nanni CW, et al. Efficacy of osimertinib against EGFRvIII+ glioblastoma. Oncotarget. 2020;11(22):2074–2082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Chen C, Cheng CD, Wu H, et al. Osimertinib successfully combats EGFR-negative glioblastoma cells by inhibiting the MAPK pathway. Acta Pharmacol Sin. 2021;42(1):108–114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Tiriac H, Belleau P, Engle DD, et al. Organoid profiling identifies common responders to chemotherapy in pancreatic cancer. Cancer Discov. 2018;8(9):1112–1129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. de Witte CJ, Espejo Valle-Inclan J, Hami N, et al. Patient-derived ovarian cancer organoids mimic clinical response and exhibit heterogeneous inter- and intrapatient drug responses. Cell Rep. 2020;31(11):107762. [DOI] [PubMed] [Google Scholar]
- 29. Singh N, Miner A, Hennis L, Mittal S.. Mechanisms of temozolomide resistance in glioblastoma - a comprehensive review. Cancer Drug Resist. 2021;4(1):17–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Parker NR, Hudson AL, Khong P, et al. Intratumoral heterogeneity identified at the epigenetic, genetic and transcriptional level in glioblastoma. Sci Rep. 2016;6:22477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. de Los Reyes Corrales T, Losada-Perez M, Casas-Tinto S.. JNK pathway in CNS pathologies. Int J Mol Sci. 2021;22(8):3883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Yoon CH, Kim MJ, Kim RK, et al. c-Jun N-terminal kinase has a pivotal role in the maintenance of self-renewal and tumorigenicity in glioma stem-like cells. Oncogene. 2012;31(44):4655–4666. [DOI] [PubMed] [Google Scholar]
- 33. Kitanaka C, Sato A, Okada M.. JNK signaling in the control of the tumor-initiating capacity associated with cancer stem cells. Genes Cancer. 2013;4(9-10):388–396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Vo VA, Lee JW, Lee HJ, et al. Inhibition of JNK potentiates temozolomide-induced cytotoxicity in U87MG glioblastoma cells via suppression of Akt phosphorylation. Anticancer Res. 2014;34(10):5509–5515. [PubMed] [Google Scholar]
- 35. Ohba S, Hirose Y, Kawase T, Sano H.. Inhibition of c-Jun N-terminal kinase enhances temozolomide-induced cytotoxicity in human glioma cells. J Neurooncol. 2009;95(3):307–316. [DOI] [PubMed] [Google Scholar]
- 36. Wang L, Jung J, Babikir H, et al. A single-cell atlas of glioblastoma evolution under therapy reveals cell-intrinsic and cell-extrinsic therapeutic targets. Nat Cancer. 2022;3(12):1534–1552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Zhang X, Ding K, Wang J, Li X, Zhao P.. Chemoresistance caused by the microenvironment of glioblastoma and the corresponding solutions. Biomed Pharmacother. 2019;109:39–46. [DOI] [PubMed] [Google Scholar]
- 38. Dijkstra KK, Cattaneo CM, Weeber F, et al. Generation of tumor-reactive T cells by co-culture of peripheral blood lymphocytes and tumor organoids. Cell. 2018;174(6):1586–1598.e12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Votanopoulos KI, Mazzocchi A, Sivakumar H, et al. Appendiceal cancer patient-specific tumor organoid model for predicting chemotherapy efficacy prior to initiation of treatment: a feasibility study. Ann Surg Oncol. 2019;26(1):139–147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Linkous A, Balamatsias D, Snuderl M, et al. Modeling patient-derived glioblastoma with cerebral organoids. Cell Rep. 2019;26(12):3203–3211.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Carrano A, Juarez JJ, Incontri D, Ibarra A, Guerrero Cazares H.. Sex-specific differences in glioblastoma. Cells. 2021;10(7):1783. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data, so far as not already published in the manuscript, will be made publically available upon request to marc.vooijs@maastrichtuniversity.nl.