Abstract
The market for illicit drugs has been reshaped by the emergence of more than 1100 new psychoactive substances (NPS) over the past decade, posing a major challenge to the forensic and toxicological laboratories tasked with detecting and identifying them. Tandem mass spectrometry (MS/MS) is the primary method used to screen for NPS within seized materials or biological samples. The most contemporary workflows necessitate labor-intensive and expensive MS/MS reference standards, which may not be available for recently emerged NPS on the illicit market. Here, we present NPS-MS, a deep learning method capable of accurately predicting the MS/MS spectra of known and hypothesized NPS from their chemical structures alone. NPS-MS is trained by transfer learning from a generic MS/MS prediction model on a large data set of MS/MS spectra. We show that this approach enables a more accurate identification of NPS from experimentally acquired MS/MS spectra than any existing method. We demonstrate the application of NPS-MS to identify a novel derivative of phencyclidine (PCP) within an unknown powder seized in Denmark without the use of any reference standards. We anticipate that NPS-MS will allow forensic laboratories to identify more rapidly both known and newly emerging NPS. NPS-MS is available as a web server at https://nps-ms.ca/, which provides MS/MS spectra prediction capabilities for given NPS compounds. Additionally, it offers MS/MS spectra identification against a vast database comprising approximately 8.7 million predicted NPS compounds from DarkNPS and 24.5 million predicted ESI-QToF-MS/MS spectra for these compounds.
Introduction
Over the past few decades, the illicit drug market has been reshaped by the proliferation of novel psychoactive substances (NPS). These compounds, which are often referred to as designer drugs, synthetic drugs, or bath salts, are designed to exert the same psychoactive effects as conventional drugs of abuse (e.g., methamphetamine, cocaine, and heroin). However, enterprising clandestine chemists introduce sufficient structural differences into NPS to ensure that they circumvent the legislative measures imposed around conventional drugs.1 The vast majority of the resulting compounds have never been tested in humans, and as a result, many NPS(s) have been associated with life-threatening toxidromes and fatalities.2 The resulting public health burden has led many countries to amend their drug laws to include known NPS. Paradoxically, however, these laws have only resulted in further proliferation of NPS analogues.3−5 As of the end of 2021, the European Monitoring Centre for Drugs and Drug Addiction (EMCDDA) is currently monitoring 830 NPS, with two new NPS emerging per week on average.6 Globally, as many as 1100 NPS are currently monitored by the United Nations Office of Drugs and Crime (UNODC) early warning advisory (EWA).7
The proliferation of NPS presents a major challenge for forensic chemistry and toxicology laboratories that are tasked with identifying these compounds. Mass spectrometry (MS) is the primary method used for NPS detection and identification, with most contemporary screening workflows relying on the diagnostic product ions produced by tandem mass spectrometry (MS/MS) for high-confidence NPS identification. The gold standard for identification of a putative NPS by MS/MS is through comparison to reference MS/MS data collected from a certified reference material (CRM), most commonly a synthetic reference standard. However, the expense of maintaining (and constantly extending) large collections of CRMs can be formidable.8 This financial burden is compounded by the rapid rate at which novel compounds emerge from, and disappear from, the illicit market. By the time a newly purchased CRM has been integrated into an NPS screening workflow, the NPS itself may already have disappeared from circulation. Moreover, for NPS that have just emerged on the illicit market, the CRMs necessary to detect and identify these compounds may not yet even be commercially available.
The limitations of workflows based on CRMs have led many forensic laboratories to complement these approaches by performing MS/MS database matching. This entails the use of in-house MS/MS databases that contain product and precursor ion data of known or theoretical substances for which the laboratory does not possess a CRM. This MS/MS information can be acquired from the scientific literature, monographs by SWGDRUG9 or NPS Discovery,10 and commercial MS/MS libraries (e.g., mzCloud11). MS/MS database matching allows forensic laboratories to monitor a much larger number of NPS samples in a more cost-effective way.
Of particular interest for forensic laboratories is the crowd-sourced MS/MS database, HighResNPS.12,13 HighResNPS represents, to our knowledge, the largest publicly available database of MS/MS data for NPS, which has been made available for direct implementation in NPS screening.13 However, there are often very limited MS/MS data on newly emerging NPS compounds. Approximately one-third of the compounds in the HighResNPS database have missing product ion information. Many of these compounds have recently been reported by different drug monitoring agencies but do not yet have published analytical data.
The difficulty of acquiring analytical data for emerging NPS has led to efforts to predict MS/MS spectra from the chemical structures of known or hypothesized NPS. Several studies have investigated the collision-induced dissociation (CID) pathways for established classes of NPS in order to enable manual prediction of MS/MS spectra for structurally related compounds.14−18 Unfortunately, this manual prediction of MS/MS spectra is limited in both throughput and accuracy. Recently, many are using a more viable solution to overcome the current dearth of experimental MS/MS spectra, which would be to use in silico MS spectral prediction models capable of accurately predicting MS/MS spectra directly from a given (known) structure. Programs such as MetFrag19 and MAGMa20 use heuristic or combinatorial fragment modeling techniques to predict the position and likelihood of bond breakages in molecules and the corresponding MS/MS spectra. Among the most accurate approaches to MS/MS prediction is an in-house previously described method, i.e., CFM-ID,21−25 which uses machine learning to predict CID pathways for small molecules. This approach renders the MS/MS spectra predicted by CFM-ID highly interpretable and explainable as each product ion in a predicted MS/MS spectrum is assigned a hypothetical fragment structure by the model. Beyond its interpretability, CFM-ID 4.0 is also a highly accurate tool, outperforming other popular in silico mass spectra tools when evaluated on the CASMI 2016 Cat3 data set.26
The NPS community has already started to explore the use of CFM-ID and in silico-predicted MS/MS spectra for MS/MS database matching. For instance, Polettini et al. applied CFM-ID 2.0 to predict and compare spectra acquired from 99 synthetic cannabinoids (SCs).27 More recently, we used CFM-ID 4.0 to predict MS/MS spectra for 8.9 million hypothetical NPS structures anticipated by a chemical language model, DarkNPS.28 Our findings indicated that the predicted MS/MS spectra could be used to help identify a substantial number of recently discovered NPSs. However, we also noted that the agreement between the observed MS/MS spectra and the predicted CFM-ID MS/MS spectra was highly variable, especially at higher collision energies.
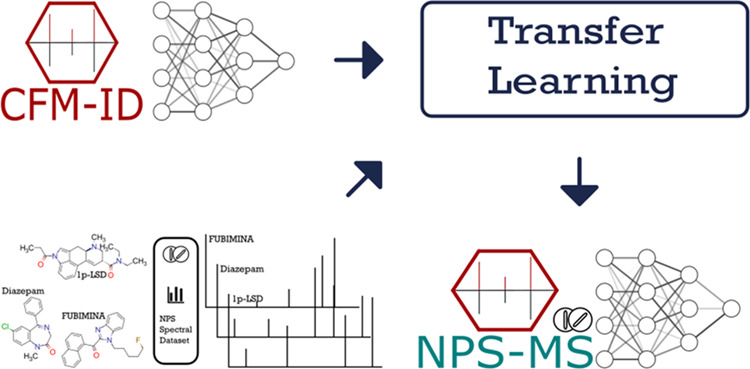
Because CFM-ID was developed to be a very general MS/MS spectral prediction tool, we investigated the possibility of developing a more specialized version of CFM-ID that could perform substantially better on the task of predicting MS/MS spectra specifically for NPS. Here, we describe this new version of CFM-ID for NPS, which we have dubbed NPS-MS. NPS-MS was trained exclusively on experimental QTOF MS/MS spectra acquired from NPS using a technique called “transfer learning.”29 We evaluate the accuracy of NPS-MS for both MS/MS prediction and compound identification and find that it is substantially more accurate than both earlier versions of CFM-ID and other widely used tools such as MetFrag19 and SIRIUS 4.30 In this manuscript, we refer to the MS/MS prediction task as “compound-to-mass spectrum” or “C2MS” and the compound identification task as “mass spectrum-to-compound” or “MS2C.” We anticipate that the performance of NPS-MS is sufficiently good that it could be routinely and reliably used for the putative identification of newly emerging NPS, as well as for the identification of never-before-seen NPS.
Methods
MS/MS Spectra Data Sets
To train and evaluate NPS-MS, we assembled a data set comprising 1872 MS/MS spectra for 624 NPS, obtained from the Institute for Legal Medicine (Johannes Gutenberg University, Mainz, Germany, n = 585 NPS) and ChemCentre (Perth, Australia, n = 39 NPS). The MS/MS data were acquired using two independent Agilent Technologies quadrupole time-of-flight mass spectrometer (QTOF) systems. Both systems were operated in positive electrospray ionization (ESI+) with collision energies of 10, 20, and 40 eV applied for CID experiments of the protonated precursor ion [M + H]+. For each MS/MS spectrum, we obtained product ion mass-to-charge ratios (m/z) and percent relative intensities (% rel. int.) after removing m/z values (peaks) with a relative intensity less than 3%. The instances were then randomly shuffled and split into a training set of 494 compounds (1482 spectra) and a held-out test set of 130 compounds (390 spectra) at approximately an 80:20 ratio.
NPS-MS Model Training
The architecture of the NPS-MS model is based on that previously described for CFM-ID 4.0.23 Conceptually, for the C2MS task, given an input molecule, CFM-ID first employs a combinatorial bond cleavage approach to enumerate all theoretically possible fragments. The output of this procedure is a molecular fragmentation graph, in which each node represents a theoretically possible fragment from the parent molecule with one bond cleavage, and each edge (also known as transition) between nodes encodes the chance that one fragment directly produces another fragment through a fragmentation event. The probability of each transition is estimated by parameters that CFM-ID learns from its training data set of known molecules and their associated MS/MS spectra. Finally, CFM-ID uses the fragmentation graph and associated transition probability estimates to reconstruct the corresponding MS/MS spectrum for the input molecule. The parameters of CFM-ID are learned from a training data set of known molecule-MS/MS spectrum pairs, using expectation maximization (EM) to attempt to optimize a negative-log-likelihood loss function.22,23
We trained NPS-MS using a machine learning technique called transfer learning.29 Transfer learning entails the reuse of a pretrained model to address a new task or problem. Here, we used the pretrained model from CFM-ID 4.0, which was trained on a large and diverse data set of generic MS/MS spectra. We then fine-tuned the learnable parameters of CFM-ID 4.0 by then training it on a smaller data set of MS/MS spectra obtained from authentic standards of NPS, as described above. We fine-tuned NPS-MS by freezing all but the last layer of the neural network in CFM-ID 4.0 during the retraining process. For comparison, we also trained a model with an identical architecture to CFM-ID 4.0 from scratch on the NPS-MS/MS spectra data set. We refer to this model as “NPS-MS De Novo.” For both models, training was limited to 30 iterations of the EM algorithm, and within each iteration, the neural network training was limited to 600 minibatches.
Model Evaluation
We evaluated the performance of NPS-MS on two tasks. In the first task, we applied NPS-MS to predict the MS/MS spectra of known NPS, and then we directly evaluated the accuracy of the MS/MS spectrum predictions (C2MS Tasks). In the second task, we used NPS-MS to identify known and unknown NPS from the experimentally acquired MS/MS spectra. That is, we first applied NPS-MS to predict MS/MS spectra for every molecule within a data set of known NPS chemical structures and then searched experimentally acquired MS/MS spectra against this data set of predicted spectra to match the experimentally acquired spectra to chemical structures (MS2C Tasks).
The primary evaluation metric in the C2MS task was the Dice coefficient, which quantifies the similarity between experimental and predicted MS/MS spectra by measuring the ratio between the number of matched peaks and the total number of peaks. We also used the dot product (also known as the cosine similarity) as a secondary measure of MS/MS spectral similarity. This method computes the cosine of the angle between the unit vectors obtained from two MS/MS spectra and, therefore, considers both the m/z values of each peak as well as their relative intensities. In addition, we also measured the precision and recall between the peaks in the experimental and predicted MS/MS spectra to further understand the performance of NPS-MS and the baseline models to which it was compared. Precision measures the number of peaks in the predicted spectra that are also present in the experimental spectra. Conversely, recall measures the percentage of peaks in the experimental spectra that are correctly predicted.
For the MS2C task, we evaluated the accuracy of compound identification when searching in three different data sets of NPS chemical structures. Data Set #1 comprised the dedicated NPS compound candidate database, HighResNPS,13 consisting of 1922 compounds. Data Set #2 constructed a more diverse library of candidate structures by supplementing HighResNPS with the PubChem chemical structure database,31 comprising an additional 94.7 million compounds. Data Set #3 consisted of both the known NPS structures from HighResNPS, as well as a library of 8.9 million theoretical NPS compounds generated by DarkNPS,28 a chemical language model32−35 trained on known NPS, which we previously demonstrated to be capable of anticipating the structures of novel NPS that subsequently emerge on the illicit market.
For each NPS data set, the experimentally measured precursor ion m/z was used to filter the data set to generate a subset of potential candidates, using a window of ±10 ppm. The predicted MS/MS spectra of each candidate were then compared with the experimental MS/MS spectra to identify the top-scoring spectral matches. To avoid overestimating performance, we required that there were at least three candidates from each precursor ion. Each of the three NPS structural databases was preprocessed by removing all charged and zwitterionic chemicals. Stereochemistry was removed, and the resulting compounds merged into their base form. Spectral matches were scored and ranked based on the average of the Dice coefficient and dot product. The performance of each C2MS model was evaluated by using a cost score. In this setting, a cost score was assigned to each compound identification based on the rank relative to the correct answer, considering the possibility of equally ranked candidates. This cost score reflects the amount of expected MS/MS experiments required to reach the ground truth compound, given a list of identification results for a single task. Details of this score can be found in the Supporting Information (SI): Details for Cost Score.
In cases in which the top-ranked spectral match does not correspond to the ground truth NPS, we also assessed the structural similarity between the ground truth NPS and the top-ranked candidates. Structural similarity was quantified using the Tanimoto coefficient between ECFP4 fingerprints36,37 and the Euclidean distance between Continuous and Data-Driven Descriptors38 (CDDD) representations. The CDDD is a 512-dimensional continuous representation of a given compound generated by a deep learning model. If the top-ranked compound is the correct compound, the CDDD distance will be zero, whereas if multiple candidates are ranked equally, the Tanimoto coefficient and CDDD distances are averaged over all equally ranked candidates.
Baseline Models
To place the performance of NPS-MS on the compound-to-mass spectrum and mass spectrum-to-compound tasks in context, we compared NPS-MS to a series of literature baselines.
For the C2MS task, we compared NPS-MS and the version of NPS-MS trained without transfer learning (i.e., NPS-MS De Novo) to CFM-ID 2.022 and CFM-ID 4.0.23 (We did not consider CFM-ID 3.0,21 which was released in 2019, as it differed from CFM-ID 2.0 primarily in the incorporation of hand-crafted fragmentation rules to improve the handling of lipids.) For the MS2C task, we evaluated the performance of all four methods used in the C2MS task, as well as MetFrag19 and SIRIUS 4.30 Interestingly, MetFrag could only be evaluated against Data sets #1 and #2 since several structures in Data Set #3 cause it to crash. Only results using the 20 eV spectra are reported here, as MetFrag performed much better when using this collision energy compared to others. SIRIUS 4 was only evaluated against a subset of Data Set #2 since its underlying model (CSI:FingerID39) was trained on a number of NPS, some of which were present in our test set.
Acquisition of MS/MS Spectra for 3-Chloro-PCP
MS/MS spectra for 3-chloro-phencyclidine (3-Cl-PCP) at collision energies of 10, 20, and 40 eV were collected from an unknown powder seized in Denmark in January 2022, using a Waters Corporation LC-QTOF-MS in positive ionization mode, as previously described.40
Results and Discussion
A total of 1872 MS/MS spectra for 624 compounds, including both NPS themselves and their metabolites, were acquired using liquid chromatography-high resolution mass spectrometry (LC-HRMS) at collision energies of 10, 20, and 40 eV. Training and holdout test data sets were randomly selected, with the training and testing sets having 494 (1,482 spectra) and 130 (390 spectra) compounds, respectively. Details of the EMCDDA classification information for each set can be found in Figure S1.
NPS-MS Enables Accurate MS/MS Spectrum Prediction
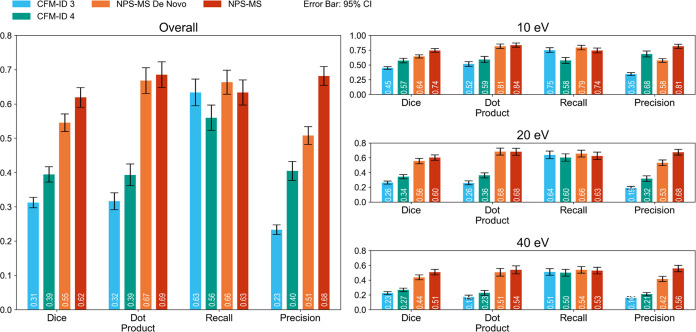
We initially assessed its ability to predict MS/MS spectra for molecules in the test set (i.e., the C2MS task). We compared NPS-MS to an identical model trained without transfer learning (NPS-MS De Novo), as well as two generic MS/MS prediction models (CFM-ID, versions 2.0 and 4.0). The performance of each model was quantified by using the average Dice coefficient and dot product of predicted MS/MS spectra against the held-out test set, as summarized in Figure 1. In general, models designed specifically to predict MS/MS spectra for NPS (NPS-MS and NP-MS De Novo) outperformed generic MS/MS prediction models (CFM-ID versions 2.0 and 4.0). Specifically, NPS-MS (Dice coefficient = 0.55; dot product = 0.67) and NPS-MS De Novo (Dice coefficient = 0.62; dot product = 0.69) outperformed CFM-ID 4.0 by margins of 44 and 59%, respectively, as quantified by the Dice coefficient and by margins of 72 and 77%, as quantified by the dot product. Notably, for 36 of the 390 MS/MS spectra in the test data set (9.2%), NPS-MS predictions achieved a perfect Dice coefficient of 1.0, as compared to just 10 (2.6%) for CFM-ID 4.0. Detailed discussion of individual predicted MS/MS is provided in the SI under Inspection of representative MS/MS spectra predicted by NPS-MS with Figure S2.
Figure 1.
Performance on the compound-to-mass spectrum (C2MS) task. The NPS-MS De Novo model is trained from scratch on the NPS training data set, whereas the NPS-MS model is trained via transfer learning from a CFM-ID 4.0 base model that is subsequently fine-tuned on the NPS training data set. Bars display mean scores for each metric with error bars indicating the 95% confidence interval. Left, the overall performance of each model averaged over three different collision energies (10, 20, and 40 eV). Right, the performance of each model on MS/MS spectra collected at each individual collision energy.
In addition, we observed that the mean recall values of CFM-ID and NPS-MS were essentially identical. Instead, the improved performance of NPS-MS over CFM-ID can be attributed to a nearly 3-fold increase in the precision of NPS-MS. In other words, although CFM-ID can correctly predict true-positive peaks that appear in the experimentally acquired MS/MS spectra of known NPS, it also predicts many false-positive peaks.
NPS-MS Enables Accurate Compound Identification from Experimental MS/MS Spectra
Having shown that NPS-MS affords a substantial increase in the accuracy of the MS/MS spectrum prediction, we next asked whether these more accurate MS/MS spectra would, in turn, enable more accurate compound identification. In this task, we first used NPS-MS to predict MS/MS spectra for a data set of NPS chemical structures and then searched the experimentally measured MS/MS spectra against this database of predicted spectra.
We evaluated the performance of NPS-MS when using three different chemical structure databases of varying sizes and diversity as input. Data Set #1 comprised the 1922 compounds in the HighResNPS database. Data Set #2 supplemented HighResNPS with 94.7 million compounds from PubChem. Data Set #3 supplemented HighResNPS with 8.9 million hypothetical NPS structures anticipated by DarkNPS.28 We compared the performance of six different MS2C models, including MetFrag19 and SIRIUS 4,30 in addition to the four models evaluated on the C2MS task. The performance of each method was quantified using the cost score, an extension of the top-k accuracy that is robust to the presence of ties (Methods). In addition, we quantified the structural similarity between the top-ranked candidate nominated by each method and the ground truth NPS using the Tanimoto coefficient and the CDDD distance.38
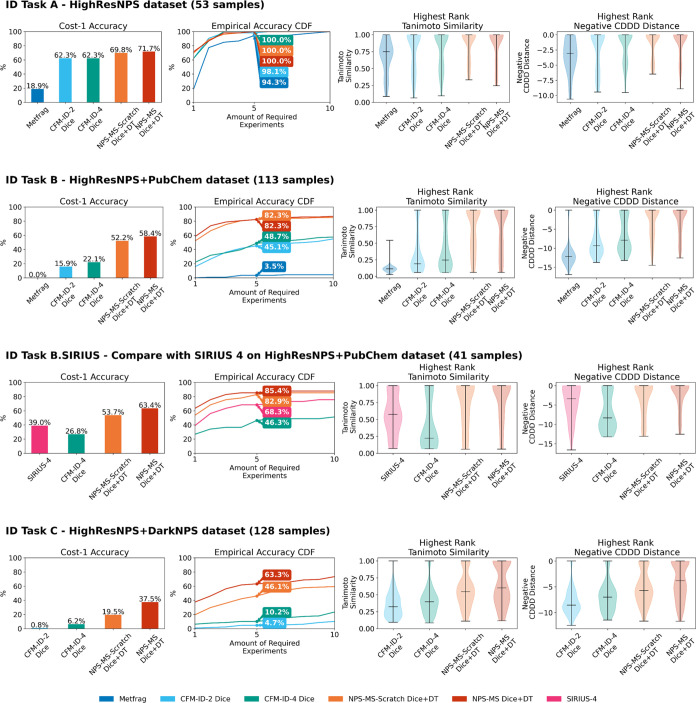
The number of test compounds used in evaluating the Data sets #1, #2, and #3 were 53, 113, and 128, respectively. The median number of candidates for each parent or precursor ion was 4, 8,061, and 2,414, respectively. A set of histograms displaying the number of candidates in each MS2C task is provided in Figure S3. Figure 2 summarizes the performance metrics for the MS2C experiments, including the cost-1 accuracy, the cost-1 to cost-10 cumulative distribution function (CDF), the Tanimoto coefficient, and the CDDD distance.
Figure 2.
Performance on the mass spectrum-to-compound (MS2C) tasks. ID Task A: MS2C identification task on HighResNPS data set (Data Set #1). ID Task B: MS2C identification task on HighResNPS + PubChem data set (Data Set #2). ID Task B. SIRIUS: MS2C identification task was performed on a subset of Data Set #2 to enable comparison with SIRIUS 4. ID Task C: MS2C identification task on HighResNPS + DarkNPS data set (Data Set #3). For each task, shown from left to right are far left, cost-1 score; middle left, CDF of cost-1 to cost-10 score; middle right, Tanimoto coefficient between the highest-ranked candidate and the ground truth structure; far right, negative CDDD distance of the highest-ranked candidate and the ground truth structure.
In Data Set #1, NPS-MS correctly identified the ground truth NPS as the highest-ranked candidate in 69.8% (n = 37) of cases. This represented an improvement of 12.0 and 15.4% over CFM-ID versions 2.0 and 4.0, respectively, as well as an improvement of 270% over MetFrag. The MS2C performance difference between each method diminished rapidly with increasing cost scores. This can likely be attributed to the small number of candidates per experimental MS/MS spectrum in this data set, which renders compound identification far less challenging than in more diverse databases such as Data sets #2 and #3.
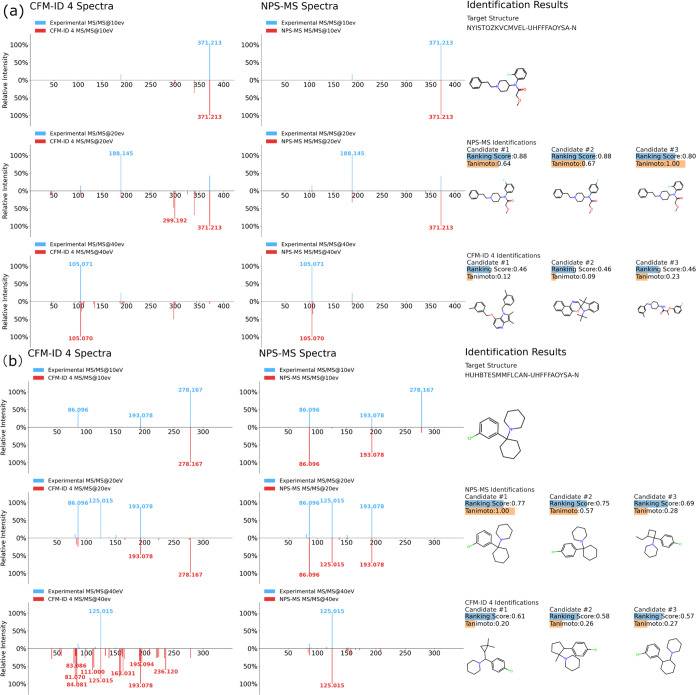
In Data Set #2, NPS-MS achieved a 58.4% cost-1 score, an improvement of 165 and 136% over CFM-ID 3.0 and 4.0, respectively, and 250% better than MetFrag. This performance improvement was observed consistently for varying cost scores: for instance, NPS-MS achieved an 80% improvement in the cost-5 score compared to CFM-ID 4.0. Moreover, in cases in which NPS-MS failed to correctly identify the ground truth NPS, the top-ranked candidate was generally structurally similar to the correct compound. For example, the top-3 candidates predicted by NPS-MS from Data Set #2 based on the MS/MS spectra of ocfentanil (i.e., 2-fluoromethoxyacetyl fentanyl; 2-methoxy-N-(1-phenethylpiperidin-4-yl)-N-2-fluorophenylacetamide) were all clearly structurally related to ocfentanil (Figure 3a). In this example, the correct structure was the third-highest scoring candidate, while the top-2 candidates were its 3-fluoro (Tc = 0.64) and 4-fluoro (Tc = 0.67) isomers. In contrast, the top-3 candidates given by CFM-ID 4.0 hardly resembled the correct structure, with Tanimoto coefficients of just 0.12, 0.09, and 0.23.
Figure 3.
(a). Identification of the designer fentanyl derivative ocfentanil in an MS2C task. The top-3 candidates predicted by NPS-MS from Data Set #2 based on the spectra of ocfentanil were all structurally related to ocfentanil. The correct structure was ranked as the third-best candidate, and the top-2 candidates were its 3-fluoro (Tc = 0.64) and 4-fluoro (Tc = 0.67) isomers. In contrast, the top-3 candidates given by CFM-ID 4.0 display little resemblance to the correct structure, with Tanimoto coefficients of 0.12, 0.09, and 0.23. (b) Retrospective application of NPS-MS to identify an unknown NPS detected in a seized powder. Left, MS/MS spectra of 3-Cl-PCP predicted by CFM-ID 4.0 at 10, 20, and 40 eV. Middle, MS/MS spectra of 3-Cl-PCP predicted by NPS-MS at 10, 20, and 40 eV. Right, the top-3 compounds identified by NPS-MS and CFM-ID 4.0 in a MS2C identification task when searching the experimentally acquired 3-Cl-PCP spectra against a database of novel chemical structures anticipated by DarkNPS.
We also evaluated the performance of SIRIUS 4 within a subset of Data Set #2 after removing compounds that were used to train SIRIUS 4. As seen in Figure 2, SIRIUS 4 outperformed CFM-ID 4.0, but NPS-MS predictors performed substantially better than SIRIUS 4. We also observed that despite its generally good accuracy, SIRIUS 4 nominated structurally very different compounds in several cases. This resulted in both the Tanimoto coefficient and the CDDD distance of SIRIUS 4 compounds having a much greater spread compared to NPS-MS.
Finally, we evaluated the performance of NPS-MS when searching against a database of 8.9 million predicted NPS structures anticipated by our previously described chemical language model, DarkNPS (Data Set #3). This data set presents a unique challenge for compound identification, since the DarkNPS compounds are far less chemically diverse than the candidates in PubChem (Data Set #2), and thus more difficult to distinguish. Consequently, the performance of both CFM-ID and NPS-MS was generally lower than in Data sets #1 and #2, with NPS-MS achieving a cost-1 score of 25.8%. However, this nonetheless represented an improvement of 416% compared with CFM-ID 4.0. NPS-MS was also 474% better than CFM-ID 4.0 in terms of the cost-5 score. Notably, NPS-MS also achieved a substantial improvement over NPS-MS De Novo in this data set (cost-1 score, 20.3%). The improved identification performance of NPS-MS over NPS-MS De Novo can be attributed to the use of transfer learning, which allowed NPS-MS to inherit more general knowledge about bond fragmentation captured by CFM-ID 4.0.
NPS-MS Enables Identification of NPS without Reference Materials in Forensic Samples
In January 2022, the Danish Customs Agency (Toldstyrelsen) seized an unknown powder that was then submitted to the Section of Forensic Chemistry (Retskemisk Afdeling) at the University of Copenhagen for analysis. Routine analysis using a Waters Corporation LC-QTOF-MS revealed the main constituent of the powder to be the novel arylcyclohexylamine 3-chloro-phencyclidine (3-Cl-PCP), which had first emerged on the illicit market in December 2020. Although the identity of the powder was determined without the use of NPS-MS, we believed this would be a useful example to simulate the applicability of NPS-MS in a forensic chemistry context, given that this NPS was unknown until very recently and consequently could not have been identified using a workflow that relied exclusively on CRMs.
We first asked whether NPS-MS was capable of accurately predicting the MS/MS spectra of 3-Cl-PCP, which was not part of either the training or test data sets. We also applied NPS-MS to predict the MS/MS spectra of 2-Cl-PCP and 4-Cl-PCP, two other known chlorinated PCP derivatives (Figure S4a). Inspection of the resulting predicted spectra revealed excellent correspondence between the predicted and experimentally acquired 3-Cl-PCP spectra with an average Dice coefficient of 0.66 and an average dot product of 0.72. Moreover, for all three PCP derivatives, the major m/z peaks at each collision energy were correctly predicted, with the lone exception being m/z 125.0153 for 2-Cl-PCP. This corresponds to the 2-chlorophenyl cation, [C6H4Cl]+. This application also underscores the interpretability of NPS-MS, with each predicted m/z value assigned a corresponding fragment ion structure (Figure S4b). While we are aware that the proposed structures for some of the product ions are unlikely to exist, potentially undergoing rearrangement or cyclization (e.g., structures 5, 8, 10, and 13), the major product ions (i.e., 1, 2, and 3) that were proposed, are close to what is likely to exist in the gas-phase based on the proposed pathways for other phencyclidine-related compounds.41
As a further test of NPS-MS, we asked whether searching the experimentally acquired spectra for 3-Cl-PCP against Data Set #3 would have led to the identification of this unknown NPS, given that 3-Cl-PCP was among the novel NPS structures anticipated by DarkNPS. To this end, we applied NPS-MS to predict MS/MS spectra for a total of 1582 compounds in Data Set #3 with a precursor mass within 10 ppm of 3-Cl-PCP. Strikingly, among these 1582 candidates, 3-Cl-PCP correctly emerged as the top-ranked candidate (Figure 3b). Moreover, the second-ranked candidate was the structurally related derivative 4-Cl-PCP. Interestingly, the third-ranked compound was not 2-Cl-PCP but a structurally related isomer, wherein the cyclohexane ring is replaced with a 2-methylcyclopentane ring (i.e., 1-[1-(3-chlorophenyl)-2-methylcyclopentyl]piperidine). In contrast, the candidates suggested by CFM-ID 4.0, while structurally related to 3-Cl-PCP, did not include the correct compound. Instead, the top-ranked compound was the 2,2-dimethylcyclopropyl derivative with the chlorine atom at position 4 (i.e., 1-[1-(4-chlorophenyl)-2,2-dimethylcyclopropyl]piperidine). Collectively, these observations highlight the potential applications of NPS-MS to identify previously unknown NPS within real forensic samples.
Conclusions
Here, we present NPS-MS, and we demonstrate that it can accurately predict MS/MS spectra for both known and predicted NPS chemical structures. By leveraging transfer learning from a generic MS/MS prediction model, we show that NPS-MS can generate remarkably accurate MS/MS spectra in a variety of C2MS tests and substantially outperforms generic models on this task. Moreover, we demonstrate that this highly accurate MS/MS spectrum prediction, in turn, enables substantially more accurate identification of known NPS on a variety of MS2C tasks, even when searching in databases comprising millions of chemical structures. NPS-MS can be also used in conjunction with DarkNPS28 to search experimentally acquired MS/MS spectra against unknown NPS structures anticipated by a chemical language model, and in a retrospective evaluation, we show that NPS-MS would have enabled the identification of a previously unknown PCP derivative without the use of any reference materials. Collectively, these results open the possibility of using NPS-MS to identify emerging NPSs for which reference MS/MS spectra are not yet available or even never-before-seen NPS, within law enforcement seizures or biological samples. The improvement in prediction accuracy for NPS-MS can be largely attributed to the fact that it was optimized to work with NPS-like compounds. Future research could explore the potential of transfer learning to enhance CFM-ID for other specific subdomains, specifically, the relationship between prediction performance and structure diversity for targeted data sets.
Acknowledgments
This work was supported by funding from Genome Canada, Genome British Columbia, Genome Alberta (project 284MBO), and Estonian Research Council (PUTJD903). NSERC (Natural Sciences and Engineering Research Council of Canada), AMII (Alberta Machine Intelligence Institute), and computational resources provided by the Digital Research Alliance of Canada and Advanced Research Computing at the University of British Columbia.
Data Availability Statement
NPS-MS is available as a web server at https://nps-ms.ca/. This web server provides MS/MS spectra prediction capabilities for the given NPS compounds. Additionally, it offers MS/MS spectra identification against a vast database comprising approximately 8.6 million predicted NPS compounds from DarkNPS28 and 24.5 million predicted ESI-QToF-MS/MS spectra for these compounds. The Docker image for NPS-MS spectra prediction, named “wishartlab/cfmid:nps-ms_1.0.0,″ can be found on Docker Hub.
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acs.analchem.3c02413.
Additional materials showing the examples of NPS-MS predicted spectra and details of evaluation metrics, as well as additional figures showing the composition of the training data set, the number of candidate compounds in each MS2C task, and details for 3-Cl-PCP case study (PDF)
The authors declare no competing financial interest.
Supplementary Material
References
- Hill S. L.; Thomas S. H. L. Clinical Toxicology of Newer Recreational Drugs. Clin. Toxicol. 2011, 49 (8), 705–719. 10.3109/15563650.2011.615318. [DOI] [PubMed] [Google Scholar]
- Baumann M. H.; Volkow N. D. Abuse of New Psychoactive Substances: Threats and Solutions. Neuropsychopharmacology 2016, 41 (3), 663–665. 10.1038/npp.2015.260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varì M. R.; Mannocchi G.; Tittarelli R.; Campanozzi L. L.; Nittari G.; Feola A.; Umani Ronchi F.; Ricci G. New Psychoactive Substances: Evolution in the Exchange of Information and Innovative Legal Responses in the European Union. Int. J. Environ. Res. Public Health 2020, 17 (22), 8704. 10.3390/ijerph17228704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reuter P.; Pardo B. New Psychoactive Substances: Are There Any Good Options for Regulating New Psychoactive Substances?. Int. J. Drug Policy 2017, 40, 117–122. 10.1016/j.drugpo.2016.10.020. [DOI] [PubMed] [Google Scholar]
- Grafinger K. E.; Bernhard W.; Weinmann W. Scheduling of New Psychoactive Substance the Swiss Way: A Review and Critical Analysis. Sci. Justice 2019, 59 (4), 459–466. 10.1016/j.scijus.2019.03.005. [DOI] [PubMed] [Google Scholar]
- European Monitoring Centre for Drugs and Drug Addiction . European Drug Report 2021: Trends and Developments. 2021. https://www.emcdda.europa.eu/publications/edr/trends-developments/2021_en (accessed February 22, 2022).
- United Nations Office on Drugs and Crime . UNODC Early Warning Advisory (EWA) on New Psychoactive Substances (NPS); UNODC: Geneva, 2023. https://www.drugsandalcohol.ie/37117 (accessed October 21, 2022).
- Pasin D.; Cawley A.; Bidny S.; Fu S. Current Applications of High-Resolution Mass Spectrometry for the Analysis of New Psychoactive Substances: A Critical Review. Anal. Bioanal. Chem. 2017, 409 (25), 5821–5836. 10.1007/s00216-017-0441-4. [DOI] [PubMed] [Google Scholar]
- Bowen B. P.; Northen T. R. Dealing with the Unknown: Metabolomics and Metabolite Atlases. J. Am. Soc. Mass Spectrom. 2010, 21 (9), 1471–1476. 10.1016/j.jasms.2010.04.003. [DOI] [PubMed] [Google Scholar]
- Krotulski A. J.; Papsun D.; Walton S.; Fogarty M.; Logan B.. NPS Discovery: Year in Review; Center for Forensic Science Research and Education, 2021. [Google Scholar]
- HighChem LLC. mzCloud—Advanced Mass Spectral Database. 2021, https://www.mzcloud.org.
- Davidsen A.; Mardal M.; Linnet K.; Dalsgaard P. W. How to Perform Spectrum-Based LC-HR-MS Screening for More than 1,000 NPS with HighResNPS Consensus Fragment Ions. PLoS One 2020, 15 (11), e0242224 10.1371/journal.pone.0242224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mardal M.; Andreasen M. F.; Mollerup C. B.; Stockham P.; Telving R.; Thomaidis N. S.; Diamanti K. S.; Linnet K.; Dalsgaard P. W. HighResNPS.Com: An Online Crowd-Sourced HR-MS Database for Suspect and Non-Targeted Screening of New Psychoactive Substances. J. Anal. Toxicol. 2019, 43 (7), 520–527. 10.1093/jat/bkz030. [DOI] [PubMed] [Google Scholar]
- Pasin D.; Cawley A.; Bidny S.; Fu S. Characterisation of Hallucinogenic Phenethylamines Using High-Resolution Mass Spectrometry for Non-Targeted Screening Purposes. Drug Test. Anal. 2017, 9 (10), 1620–1629. 10.1002/dta.2171. [DOI] [PubMed] [Google Scholar]
- Sekuła K.; Zuba D.; Lorek K. Analysis of Fragmentation Pathways of New-Type Synthetic Cannabinoids Using Electrospray Ionization. J. Am. Soc. Mass Spectrom. 2018, 29 (10), 1941–1950. 10.1007/s13361-018-2008-9. [DOI] [PubMed] [Google Scholar]
- Klingberg J.; Cawley A.; Shimmon R.; Fu S. Collision-Induced Dissociation Studies of Synthetic Opioids for Non-Targeted Analysis. Front. Chem. 2019, 7, 331. 10.3389/fchem.2019.00331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swanson K. D.; Shaner R. L.; Krajewski L. C.; Bragg W. A.; Johnson R. C.; Hamelin E. I. Use of Diagnostic Ions for the Detection of Fentanyl Analogs in Human Matrices by LC–QTOF. J. Am. Soc. Mass Spectrom. 2021, 32 (12), 2852–2859. 10.1021/jasms.1c00267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fornal E. Study of Collision-Induced Dissociation of Electrospray-Generated Protonated Cathinones: LC-Q/TOF Fragmentation of Cathinones. Drug Test. Anal. 2014, 6 (7–8), 705–715. 10.1002/dta.1573. [DOI] [PubMed] [Google Scholar]
- Ruttkies C.; Schymanski E. L.; Wolf S.; Hollender J.; Neumann S. MetFrag Relaunched: Incorporating Strategies beyond in Silico Fragmentation. J. Cheminf. 2016, 8 (1), 3 10.1186/s13321-016-0115-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ridder L.; van der Hooft J. J. J.; Verhoeven S. Automatic Compound Annotation from Mass Spectrometry Data Using MAGMa. Mass Spectrom. 2014, 3, S0033. 10.5702/massspectrometry.S0033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Djoumbou-Feunang Y.; Pon A.; Karu N.; Zheng J.; Li C.; Arndt D.; Gautam M.; Allen F.; Wishart D. S. CFM-ID 3.0: Significantly Improved ESI-MS/MS Prediction and Compound Identification. Metabolites 2019, 9 (4), 72. 10.3390/metabo9040072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen F.; Greiner R.; Wishart D. Competitive Fragmentation Modeling of ESI-MS/MS Spectra for Putative Metabolite Identification. Metabolomics 2015, 11 (1), 98–110. 10.1007/s11306-014-0676-4. [DOI] [Google Scholar]
- Wang F.; Liigand J.; Tian S.; Arndt D.; Greiner R.; Wishart D. S. CFM-ID 4.0: More Accurate ESI-MS/MS Spectral Prediction and Compound Identification. Anal. Chem. 2021, 93 (34), 11692–11700. 10.1021/acs.analchem.1c01465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen F.; Pon A.; Wilson M.; Greiner R.; Wishart D. CFM-ID: A Web Server for Annotation, Spectrum Prediction and Metabolite Identification from Tandem Mass Spectra. Nucleic Acids Res. 2014, 42 (W1), W94–W99. 10.1093/nar/gku436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang F.; Allen D.; Tian S.; Oler E.; Gautam V.; Greiner R.; Metz T. O.; Wishart D. S. CFM-ID 4.0—a Web Server for Accurate MS-Based Metabolite Identification. Nucleic Acids Res. 2022, 50, gkac383 10.1093/nar/gkac383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blaženović I.; Kind T.; Torbašinović H.; Obrenović S.; Mehta S. S.; Tsugawa H.; Wermuth T.; Schauer N.; Jahn M.; Biedendieck R.; Jahn D.; Fiehn O. Comprehensive Comparison of in Silico MS/MS Fragmentation Tools of the CASMI Contest: Database Boosting Is Needed to Achieve 93% Accuracy. J. Cheminf. 2017, 9 (1), 32 10.1186/s13321-017-0219-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polettini A. E.; Kutzler J.; Sauer C.; Guber S.; Schultis W. LC–QTOF-MS Presumptive Identification of Synthetic Cannabinoids without Reference Chromatographic Retention/Mass Spectral Information. II. Evaluation of a Computational Approach for Predicting and Identifying Unknown High-Resolution Product Ion Mass Spectra. J. Anal. Toxicol. 2020, 45 (5), 440–461. 10.1093/jat/bkaa127. [DOI] [PubMed] [Google Scholar]
- Skinnider M. A.; Wang F.; Pasin D.; Greiner R.; Foster L. J.; Dalsgaard P. W.; Wishart D. S. A Deep Generative Model Enables Automated Structure Elucidation of Novel Psychoactive Substances. Nat. Mach. Intell. 2021, 3 (11), 973–984. 10.1038/s42256-021-00407-x. [DOI] [Google Scholar]
- Pan S. J.; Yang Q. A Survey on Transfer Learning. IEEE Trans. Knowl. Data Eng. 2010, 22 (10), 1345–1359. 10.1109/TKDE.2009.191. [DOI] [Google Scholar]
- Dührkop K.; Fleischauer M.; Ludwig M.; Aksenov A. A.; Melnik A. V.; Meusel M.; Dorrestein P. C.; Rousu J.; Böcker S. SIRIUS 4: A Rapid Tool for Turning Tandem Mass Spectra into Metabolite Structure Information. Nat. Methods 2019, 16 (4), 299–302. 10.1038/s41592-019-0344-8. [DOI] [PubMed] [Google Scholar]
- Bolton E. E.; Wang Y.; Thiessen P. A.; Bryant S. H. PubChem: Integrated Platform of Small Molecules and Biological Activities. Annu. Rep. Comput. Chem. 2008, 4, 217–241. 10.1016/S1574-1400(08)00012-1. [DOI] [Google Scholar]
- Flam-Shepherd D.; Zhu K.; Aspuru-Guzik A. Language Models Can Learn Complex Molecular Distributions. Nat. Commun. 2022, 13 (1), 3293 10.1038/s41467-022-30839-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skinnider M. A.; Stacey R. G.; Wishart D. S.; Foster L. J. Chemical Language Models Enable Navigation in Sparsely Populated Chemical Space. Nat. Mach. Intell. 2021, 3 (9), 759–770. 10.1038/s42256-021-00368-1. [DOI] [Google Scholar]
- Segler M. H. S.; Kogej T.; Tyrchan C.; Waller M. P. Generating Focused Molecule Libraries for Drug Discovery with Recurrent Neural Networks. ACS Cent. Sci. 2018, 4 (1), 120–131. 10.1021/acscentsci.7b00512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grisoni F.; Schneider G.. De Novo Molecular Design With. In Artificial Intelligence in Drug Design; Heifetz A., Ed.; Methods in Molecular Biology; Springer US: New York, NY, 2022; Vol. 2390, pp 207–232. 10.1007/978-1-0716-1787-8_9. [DOI] [PubMed] [Google Scholar]
- Rogers D.; Hahn M. Extended-Connectivity Fingerprints. J. Chem. Inf. Model. 2010, 50 (5), 742–754. 10.1021/ci100050t. [DOI] [PubMed] [Google Scholar]
- Bajusz D.; Rácz A.; Héberger K. Why Is Tanimoto Index an Appropriate Choice for Fingerprint-Based Similarity Calculations?. J. Cheminf. 2015, 7 (1), 20 10.1186/s13321-015-0069-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winter R.; Montanari F.; Noé F.; Clevert D.-A. Learning Continuous and Data-Driven Molecular Descriptors by Translating Equivalent Chemical Representations. Chem. Sci. 2019, 10 (6), 1692–1701. 10.1039/C8SC04175J. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dührkop K.; Shen H.; Meusel M.; Rousu J.; Böcker S. Searching Molecular Structure Databases with Tandem Mass Spectra Using CSI:FingerID. Proc. Natl. Acad. Sci. U.S.A. 2015, 112 (41), 12580–12585. 10.1073/pnas.1509788112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mollerup C. B.; Dalsgaard P. W.; Mardal M.; Linnet K. Targeted and Non-Targeted Drug Screening in Whole Blood by UHPLC-TOF-MS with Data-Independent Acquisition: Targeted and Non-Targeted Drug Screening in Whole Blood by UHPLC-TOF-MS. Drug Test. Anal. 2017, 9 (7), 1052–1061. 10.1002/dta.2120. [DOI] [PubMed] [Google Scholar]
- Michely J. A.; Manier S. K.; Caspar A. T.; Brandt S. D.; Wallach J.; Maurer H. H. New Psychoactive Substances 3-Methoxyphencyclidine (3-MeO-PCP) and 3-Methoxyrolicyclidine (3-MeO-PCPy): Metabolic Fate Elucidated with Rat Urine and Human Liver Preparations and Their Detectability in Urine by GC-MS, “LC-(High Resolution)-MSn” and “LC-(High Resolution)-MS/MS. Curr. Neuropharmacol. 2017, 15 (5), 692–712. 10.2174/1570159X14666161018151716. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
NPS-MS is available as a web server at https://nps-ms.ca/. This web server provides MS/MS spectra prediction capabilities for the given NPS compounds. Additionally, it offers MS/MS spectra identification against a vast database comprising approximately 8.6 million predicted NPS compounds from DarkNPS28 and 24.5 million predicted ESI-QToF-MS/MS spectra for these compounds. The Docker image for NPS-MS spectra prediction, named “wishartlab/cfmid:nps-ms_1.0.0,″ can be found on Docker Hub.