Abstract
Induction of the pluripotent cell mass termed callus from detached organs or tissues is an initial step in typical in vitro plant regeneration, during which auxin-induced ectopic activation of root stem cell factors is required for subsequent de novo shoot regeneration. While Arabidopsis (Arabidopsis thaliana) AUXIN RESPONSE FACTOR 7 (ARF7) and ARF19 and their downstream transcription factors LATERAL ORGAN BOUNDARIES DOMAIN (LBD) are known to play key roles in directing callus formation, the molecules responsible for activation of root stem cell factors and thus establishment of callus pluripotency are unclear. Here, we identified Arabidopsis WRKY23 and BASIC HELIX-LOOP-HELIX 041 (bHLH041) as a transcriptional activator and repressor, respectively, of root stem cell factors during establishment of auxin-induced callus pluripotency. We show that auxin-induced WRKY23 downstream of ARF7 and ARF19 directly activates the transcription of PLETHORA 3 (PLT3) and PLT7 and thus that of the downstream genes PLT1, PLT2, and WUSCHEL-RELATED HOMEOBOX 5 (WOX5), while LBD-induced removal of bHLH041 derepresses the transcription of PLT1, PLT2, and WOX5. We provide evidence that transcriptional activation by WRKY23 and loss of bHLH041-imposed repression act synergistically in conferring shoot-regenerating capability on callus cells. Our findings thus disclose a transcriptional mechanism underlying auxin-induced cellular reprogramming, which, together with previous studies, outlines the molecular framework of auxin-induced pluripotent callus formation for in vitro plant regeneration programs.
Auxin-induced WRKY23 accumulation and bHLH041 removal synergistically activate root stem cell genes and thus confer shoot-regenerating capacity on callus in Arabidopsis regeneration.
IN A NUTSHELL.
Background: Plant cells retain a remarkable capacity to regenerate new organs or entire individuals in the real world and under tissue culture conditions. A well-established in vitro plant regeneration procedure applicable for transgenic and biotechnological uses generally starts with the induction of pluripotent callus cells, which is required for subsequent de novo shoot or root regeneration. Recent studies in Arabidopsis (Arabidopsis thaliana) have revealed that auxin-induced ectopic activation of root stem cell factors within callus cells establishes the shoot-regenerating capability, and some auxin-signaling components involved in root development, such as AUXIN RESPONSE FACTOR (ARFs) and their downstream transcription factors LATERAL ORGAN BOUNDARIES DOMAIN (LBD), play critical roles in directing callus formation. However, the molecular link between these auxin-signaling components and activation of root stem cell factors during callus induction is missing.
Question: What are the factors responsible for activation of root stem cell factors to establish callus pluripotency in Arabidopsis for in vitro regeneration?
Findings: We identified the Arabidopsis transcription factors WRKY23 and bHLH041 as a transcriptional activator and repressor, respectively, of the expression of root stem cell genes during auxin-induced callus formation. Genetic and molecular evidence revealed that auxin-induced WRKY23 downstream of ARF7 and ARF19 directly activates the transcription of PLETHORA 3 (PLT3) and PLT7 and the downstream target genes of their encoded proteins PLT1, PLT2, and WUSCHEL-RELATED HOMEOBOX 5 (WOX5), while LBD induces the removal of bHLH041, alleviating the transcriptional repression of PLT1, PLT2, and WOX5. We also demonstrated that two transcriptional pathways synergize the shoot-regenerating capability of callus cells. These findings elucidate the transcriptional mechanism underlying callus pluripotency establishment, which links auxin signaling and cellular reprogramming during in vitro plant regeneration programs.
Next steps: It will be worth identifying the orthologs of WRKY23 and bHLH041 in crops and economically important plants and exploring whether such regulatory mechanisms are conserved, which would potentially boost regeneration-based transgene and gene editing in these species.
Introduction
A well-known feature of plant somatic cells is their remarkable capacity to regenerate a new organ or an entire plant under in vitro culture conditions (Birnbaum and Sanchez Alvarado 2008; Sugimoto et al. 2010; Sugimoto et al. 2011), during which the phytohormones auxin and cytokinin play a key role in determining cell fate transitions and regeneration programs (Skoog and Miller 1957; Duclercq et al. 2011; Ikeuchi et al. 2013). A typical in vitro plant regeneration generally starts with the induction of a pluripotent cell mass named callus from detached organs or tissues on auxin-rich callus-inducing medium (CIM); the subsequent incubation of newly formed calli on shoot-inducing medium (SIM) or root-inducing medium (RIM) with different ratios of auxin and cytokinin leads to the de novo production of shoots or roots, respectively (Skoog and Miller 1957; Valvekens et al. 1988; Che et al. 2002). Thus, auxin-induced callus formation has long been considered to reflect the mitotic activities of some differentiated cells that are reactivated, after which somatic cells are reprogrammed into pluripotent callus cells, which are required for subsequent de novo regeneration programs (Che et al. 2007; Gordon et al. 2007; Atta et al. 2009; Kareem et al. 2015).
Recent studies have begun to uncover the molecular characteristics of callus and regulation of auxin-induced callus formation during in vitro plant regeneration. In Arabidopsis (Arabidopsis thaliana), auxin-induced callus formation in multiple organs occurs from the pericycle or pericycle-like cells by a root developmental program; the root stem cell regulators, such as WUSCHEL-RELATED HOMEOBOX 5 (WOX5), PLETHORA 1 (PLT1), PLT2, SHORT-ROOT (SHR), and SCARECROW (SCR), are ectopically activated in the forming callus (Atta et al. 2009; Sugimoto et al. 2010; Kareem et al. 2015; Radhakrishnan et al. 2018). Consistent with this observation, several auxin-signaling components INDOLE-3-ACETIC ACID INDUCIBLE (IAA) and AUXIN RESPONSE FACTORs (ARFs) form modules involved in lateral root formation, such as IAA14/19–ARF7/19, playing a critical role in governing auxin-induced callus formation (Tian and Reed 1999; Fukaki et al. 2002; Tatematsu et al. 2004; Fukaki et al. 2005; Okushima et al. 2007; Uehara et al. 2008; Fan et al. 2012; Goh et al. 2012; Shang et al. 2016). Similarly, the auxin-inducible transcription factors LATERAL ORGAN BOUNDARIES DOMAIN (LBD), which act downstream of IAA14–ARF7/19 to control lateral root formation (Okushima et al. 2007; Lee et al. 2009; Lee et al. 2013; Lee et al. 2015; Lee, Kang, et al. 2017), are key factors in directing callus formation by forming a complex with BASIC LEUCINE ZIPPER 59 (bZIP59) (Fan et al. 2012; Xu, Cao, Xu, et al. 2018; Xu, Cao, Zhang, et al. 2018). Moreover, ARABIDOPSIS TRITHORAX-RELATED 2 (ATXR2) recruited by ARF7 and ARF19 can promote callus formation by activation of LBD gene expression (Lee, Park, and Seo 2017), while the calcium signaling module CALMODULIN–IQ-MOTIF CONTAINING PROTEIN (CaM–IQM) regulates callus formation by destabilizing the interaction of IAA and ARF7/19 (Zhang et al. 2022). Notably, the ectopic activation of root stem cell factors including PLTs and WOX5, which are considered to be root pluripotent transcription factors (Wang et al. 2022), represents the acquisition of cellular pluripotency or shoot-regenerating capability in the forming callus cells (Sugimoto et al. 2010; Kareem et al. 2015). Indeed, HISTONE ACETYLTRANSFERASE 1 (HAG1) promotes regeneration competence of callus cells by upregulation of several root-meristem regulator genes including WOX5, PLT1, and PLT2 (Kim et al. 2018), while the interaction of WOX5 with PLT1 and PLT2 to promote TRYPTOPHAN AMINOTRANSFERASE OF ARABIDOPSIS 1 (TAA1) expression is critical for maintenance of pluripotency in the middle layer of callus cells (Zhai and Xu 2021; Zhai et al. 2023). Importantly, PLT3, PLT5, and PLT7 function redundantly in conferring shoot-regenerating competence of callus cells by activating PLT1 and PLT2 expression, as calli derived from the explants of the plt3 plt5 plt7 triple mutant are largely defective in shoot-regenerating capability (Kareem et al. 2015). Intriguingly, the WOX11–LBD16 module promotes the acquisition of callus pluripotency by affecting transcription of PLT1 and PLT2 but not of PLT3 or PLT7 (Liu et al. 2018), suggesting that at least two molecular pathways are involved in activating root stem cell regulators and thus establishing cellular pluripotency (Sugiyama 2018). However, the molecules responsible for activation of these root pluripotent factors during auxin-induced callus formation are unclear.
Here, we reported that the Arabidopsis transcription factors WRKY23 and BASIC HELIX-LOOP-HELIX 041 (bHLH041) act as a transcriptional activator and repressor, respectively, downstream of IAA–ARFs to coordinate the activation of root stem cell genes during callus induction. We show that WRKY23, encoded by auxin-induced WRKY23 downstream of ARF7 and ARF19 directly activates the transcription of PLT3 and PLT7, whose encoding proteins in turn activate the transcription of the downstream genes PLT1, PLT2, and WOX5, and that LBD-induced removal of bHLH041 alleviates the transcriptional repression of PLT1, PLT2, and WOX5. We also provide evidence that transcriptional activation by WRKY23 and derepression by bHLH041 synergize the shoot-regenerating capability of callus cells. These findings uncover a transcriptional mechanism underlying cellular pluripotency establishment during in vitro plant regeneration programs.
Results
WRKY23 and bHLH041 are respectively accumulated and abolished in forming callus
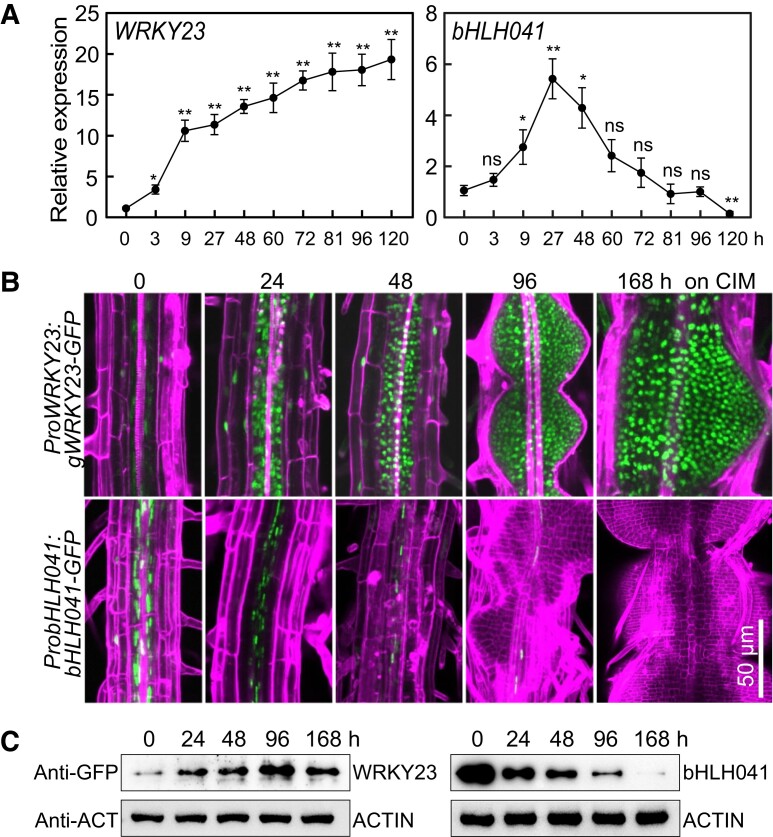
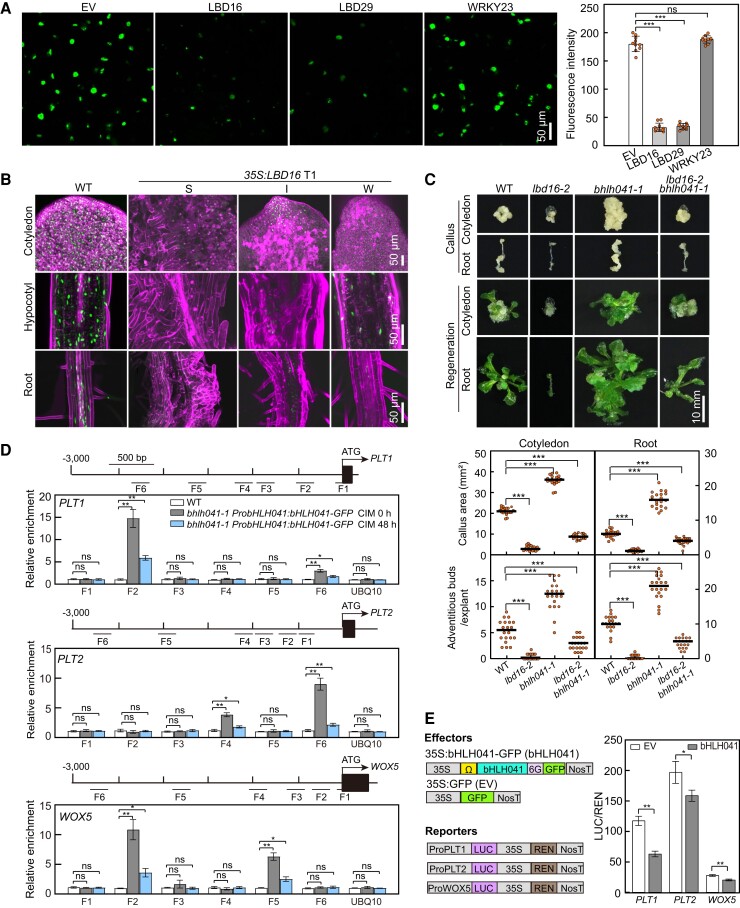
As previous studies have suggested that two molecular pathways are involved in activation of PLT3 and PLT7 and LBD-induced PLT1 and PLT2 transcription, respectively (Kareem et al. 2015; Liu et al. 2018; Sugiyama 2018), we wished to identify the transcription factors responsible for activation of these root stem cell genes during callus formation on CIM. To this end, we focused on genes whose expression patterns are correlated with those of PLT3 and PLT7 in a transcriptomic profiling of Arabidopsis explants on CIM and the potential LBD29-regulated genes whose transcript and/or protein accumulation are possibly associated with activation of PLT1 and PLT2. In an Arabidopsis ATH1 GeneChip database of transcript levels from root explants incubated on CIM (Che et al. 2006), we noticed that the dynamics of WRKY23, which encodes a member of the plant-specific WRKY family of transcription factors (Prát et al. 2018), was similar to those of PLT3 and PLT7 (Supplemental Fig. S1A). Reverse transcription quantitative PCR (RT-qPCR) analysis showed that WRKY23 transcript levels are elevated (close to 20-fold after 5 d) after incubation of seedlings on CIM (Fig. 1A). Next, we examined WRKY23 accumulation with transgenic ProWRKY23:gWRKY23-GFP seedlings harboring a transgene consisting of the WRKY23 promoter driving the WRKY23 genomic coding region cloned in-frame and upstream of the green fluorescent protein (GFP) sequence, when incubated on CIM. Although we detected WRKY23-GFP in the nuclei of pericycle or pericycle-like cells and other cells, its abundance markedly increased in the callus cells forming from roots, hypocotyls, and cotyledons after seedlings were incubated on CIM (Fig. 1B; Supplemental Fig. S1B). Among the transcription factor genes potentially regulated by LBD29 (Xu, Cao, Xu, et al. 2018), the transcript abundance of bHLH041, which encodes a member of the bHLH family (Toledo-Ortiz et al. 2003), was rapidly induced by chemically induced LBD29 but dramatically declined afterward (Xu, Cao, Xu, et al. 2018) (Supplemental Fig. S1C). We confirmed by RT-qPCR analysis that bHLH041 transcript levels are initially induced by incubation on CIM but decline later (Fig. 1A). Surprisingly, with the transgenic ProbHLH041:bHLH041-GFP seedlings, we observed that bHLH041 accumulates ubiquitously in the nuclei of all cells in roots, hypocotyls, and cotyledons; however, after seedlings were incubated on CIM, bHLH041 abundance gradually declined in these cells and bHLH041 completely disappeared in the forming callus (Fig. 1B; Supplemental Fig. S1D). Immunoblotting analysis confirmed that WRKY23 and bHLH041 abundance increases and declines by incubation on CIM, respectively (Fig. 1C). These observations suggest that WRKY23 and bHLH041 might participate in auxin-induced callus formation.
Figure 1.
Accumulation of WRKY23 and bHLH041 during callus induction. A) Relative WRKY23 and bHLH041 transcript levels in response to CIM. Seven-day-old wild type (WT) seedlings incubated in liquid CIM for the indicated times were used for RNA isolation and RT-qPCR analysis. Data are shown as means ± Sd (n = 3 biological replicates). *P < 0.05, **P < 0.01 by t-test. B, C) Accumulation of WRKY23 and bHLH041 during callus induction. Five-d-old transgenic seedlings carrying a ProWRKY23:gWRKY23-GFP or ProbHLH041:bHLH041-GFP transgene were incubated on CIM for the indicated times. GFP signals from the mature zone of primary roots were overlaid with signals stained with propidium iodide in B), and the total proteins from seedlings were immunoblotted by anti-GFP and anti-ACTIN antibodies for 2 biological replicates in C). Scale bar, 50 µm.
WRKY23 and bHLH041 respectively promote and inhibit callus formation and shoot regeneration
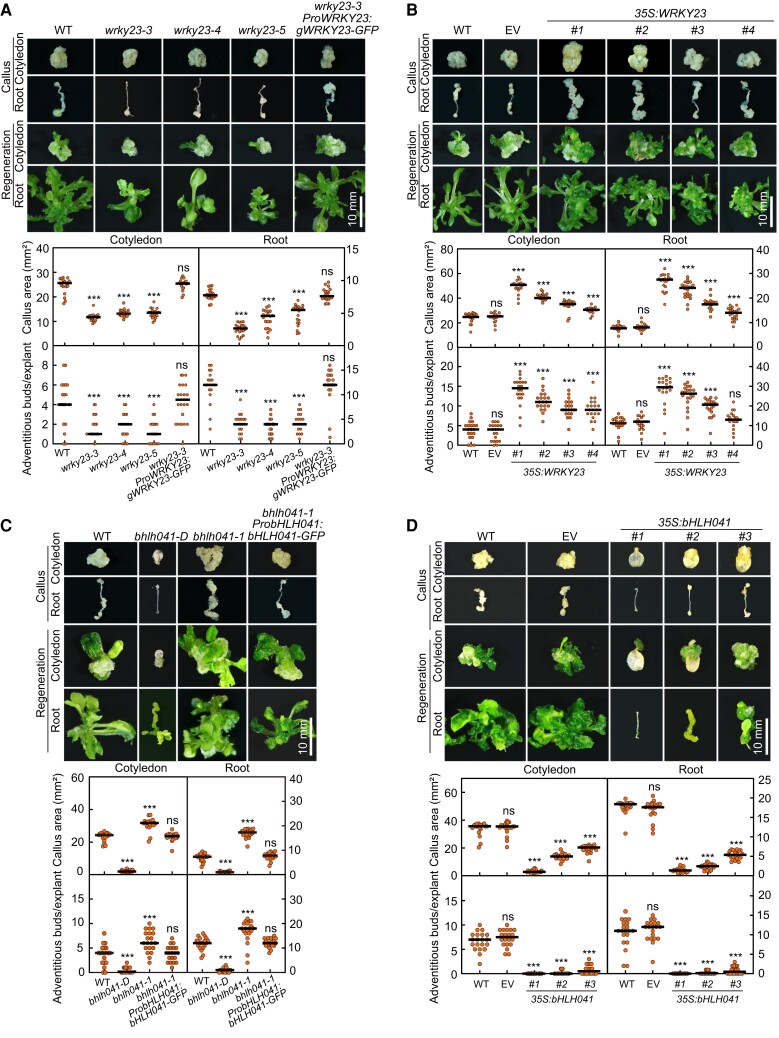
As WRKY23 has been reported to act downstream of ARF7 and ARF19 to regulate root development (Grunewald et al. 2012), we reasoned that WRKY23 might be involved in auxin-induced callus formation and possibly shoot regeneration. To test this idea, we generated 3 allelic mutants of WRKY23, namely wrky23-3, wrky23-4, and wrky23-5, by clustered regularly interspaced short palindromic repeat (CRISPR)/CRISPR-associated nuclease 9 (Cas9)-mediated gene editing (Supplemental Fig. S2A) and examined their callus-forming and shoot-regenerating phenotypes. When compared to wild type (WT), loss of WRKY23 function not only attenuated callus formation from cotyledon and root explants when incubated on CIM but also dampened shoot regeneration from the derived calli after incubation on SIM (Fig. 2A). Moreover, such callus-forming and shoot-regenerating defect in wrky23-3 explants and derived callus could be rescued by introduction of the ProWRKY23:gWRKY23-GFP construct, confirming that these phenotypes can be attributed to the loss of WRKY23 (Fig. 2A). We also generated transgenic 35S:WRKY23 plants overexpressing WRKY23 from the cauliflower mosaic (CaMV) 35S promoter and examined their callus-forming and shoot-regenerating phenotype. The overexpression of WRKY23 promoted callus formation and shoot regeneration from cotyledon and root explants and their derived callus in a WRKY23 transcript level-dependent manner (Fig. 2B; Supplemental Fig. S2B). These observations demonstrate that WRKY23 is a positive regulator of callus formation and shoot regeneration. In addition, wrky23 and 35S:WRKY23 seedlings developed slightly shorter or longer primary roots with fewer or more lateral root initiates when compared to WT, respectively (Supplemental Fig. S2C), while their cotyledons were of comparable size among the 3 genotypes (Supplemental Fig. S2D).
Figure 2.
WRKY23 and bHLH041 regulate callus formation and shoot regeneration. A) Callus-forming and shoot-regenerating phenotypes of WT, wrky23-3, wrky23-4, wrky23-5, and wrky23-3 ProWRKY23:gWRKY23-GFP explants and derived calli. B) Callus-forming and shoot-regenerating phenotypes of WT, empty vector (EV), and 35S:WRKY23 explants and derived calli. Four independent 35S:WRKY23 transgenic lines named #1, #2, #3, and #4 were examined. C) Callus-forming and shoot-regenerating phenotypes of WT, bhlh041-D, bhlh041-1, and bhlh041-1 ProbHLH041:bHLH041-GFP explants and derived calli. D) Callus-forming and shoot-regenerating phenotypes of WT, EV, and 35S:bHLH041 explants and derived calli. Three independent 35S:bHLH041 transgenic lines named #1, #2, and #3 were examined. The cotyledon and root explants from 7-d-old seedlings were incubated on CIM for 21 d or for 7 d and subsequently on SIM for 14 d before being examined for callus formation and shoot regeneration, respectively. The callus area of each explant and the number of regenerating shoots from each callus were determined (n = 20 explants). Scale bars, 10 mm. Data are shown as means ± Sd. *P < 0.05, **P < 0.01, and ***P < 0.001 by t-test.
To test whether bHLH041 participates in the regeneration program, we obtained a loss-of-function and a gain-of-function mutant of bHLH041, bhlh041-1 (SAIL_330_F04), and bhlh041-D (SAIL_258_C53), in which a T-DNA was inserted in fourth exon or 5′ UTR of bHLH041 and leads to the disruption or overexpression of bHLH041, respectively (Supplemental Fig. S3A). After incubation on CIM, the cotyledon and root explants of bhlh041-1 displayed an enhanced callus-forming phenotype, whereas those of bhlh041-D had a callus-forming defect when compared to WT (Fig. 2C). We also observed a respective enhanced and dampened shoot regeneration phenotype in the calli derived from bhlh041-1 and bhlh041-D after being incubated on SIM (Fig. 2C). Moreover, the enhanced callus-forming and shoot-regenerating phenotype of bhlh041-1 was rescued by introduction of a transgene carrying the native bHLH041 promoter driving bHLH041 expression (Fig. 2C), and overexpression of bHLH041 strongly inhibited the callus formation and shoot regeneration from explants and derived calli (Fig. 2D; Supplemental Fig. S3B). Collectively, we conclude that bHLH041 negatively regulates callus formation and shoot regeneration. In addition, we observed that disruption or overexpression of bHLH041 resulted in an increase or decrease of lateral root numbers, respectively (Supplemental Fig. S3C). Notably, only dramatic overexpression of bHLH041 (several hundred-fold higher than WT) had an effect on cotyledon development (Supplemental Fig. S3D).
WRKY23 and bHLH041 are involved in the regulation of root stem cell genes
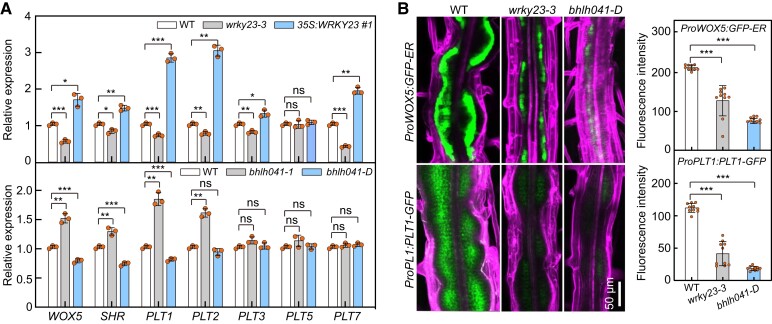
As WRKY23 and bHLH041 affect both callus formation and shoot regeneration, we investigated whether they participate in the activation of root stem cell genes during callus induction. First, we monitored the transcript abundance of WOX5, PLTs, and SHR and in the WT, wrky23-3, 35S:WRKY23, bhlh041-1, and bhlh041-D root explants incubated on CIM for 7 d. When compared to those in WT, the transcript levels of WOX5, SHR, PLT1, PLT2, PLT3, and PLT7 were lower in wrky23-3 but higher in 35S:WRKY23 explants (Fig. 3A). By contrast, we observed elevated and decreased transcript levels for WOX5, SHR, PLT1, and PLT2 in bhlh041-1 and bhlh041-D explants, respectively (Fig. 3A). Next, we visualized the cellular accumulation of PLT1 and WOX5, two root stem cell markers ectopically activated within callus cells (Sugimoto et al. 2010), in wrky23-3 and bhlh041-D plants harboring a ProPLT1:PLT1-YFP (yellow fluorescent protein) or ProWOX5:GFP-ER construct, respectively. We observed that disruption of WRKY23 or overexpression of bHLH041 leads to a drop in PLT1 accumulation and WOX5 expression levels in the forming callus (Fig. 3B). These observations indicate that WRKY23 and bHLH041 are involved in the regulation of root stem cell genes and thus establishment of callus pluripotency. We also noticed that PLT3 and PLT7 transcript levels are only regulated by WRKY23 but not by bHLH041, suggesting that they might execute their roles via different pathways.
Figure 3.
WRKY23 and bHLH041 are activator and repressor of root stem cell genes, respectively. A) Transcript abundance of root stem cell genes in root explants with forming callus from WT, wrky23-3, and 35S:WRKY23 (upper panel) and WT, bhlh041-1, and bhlh041-D (bottom panel). RT-qPCR analysis was performed with root explants incubated on CIM for 7 d (n = 3 biological replicates). B) Accumulation of WOX5 and PLT1 in the callus-forming roots of WT, wrky23-3, and bhlh041-D. Five-d-old transgenic seedlings carrying a ProWOX5:GFP-ER or ProPLT1:PLT1-YFP transgene were incubated on CIM for 4 d, and GFP fluorescent signals were visualized and quantified at the mature zone of primary roots (n = 10 seedlings). The GFP signals were overlaid with the cells stained with propidium iodide. Scale bar, 50 µm. Data are shown as means ± Sd. *P < 0.05, **P < 0.01, and ***P < 0.001 by t-test.
WRKY23 directly activates the transcription of PLT3 and PLT7
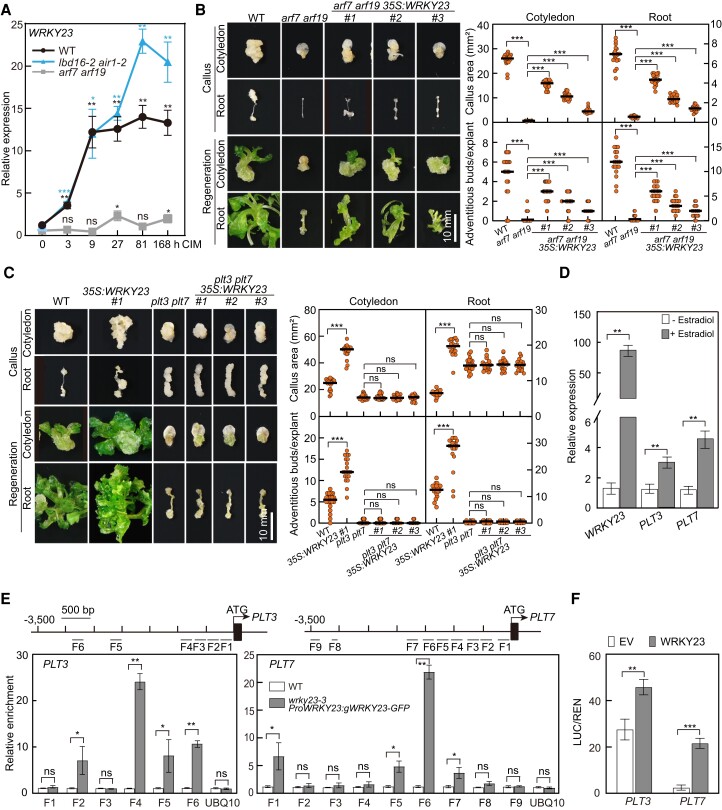
Both WRKY23 and LBDs have been shown to act downstream of ARF7 and ARF19 to regulate lateral root development and/or callus formation (Grunewald et al. 2008; Fan et al. 2012; Grunewald et al. 2012; Xu, Cao, Zhang, et al. 2018). Consistent with the notion that ARF-mediated auxin signaling is mainly through auxin-induced degradation of their repressor IAAs but not by alteration of their abundances (Wang et al. 2005; Lavy and Estelle 2016; Powers et al. 2019), we determined that only the transcript levels of ARF19 but not ARF7 show a slight response to CIM (Supplemental Fig. S4A). To clarify the possible inter-regulation between WRKY23 and LBDs, we first monitored the transcript levels of WRKY23 in WT, arf7 arf19, and lbd16-2 air1-2 (anthocyanin-impaired response 1-2, in which the bZIP59 gene is disrupted) seedlings on CIM. Consistent with previous observations (Grunewald et al. 2012), induction of WRKY23 by CIM was abolished in the arf7 arf19 but not in the lbd16-2 air1-2 mutant seedlings (Fig. 4A). Similarly, overexpression of LBD16 or LBD29 did not have a clear effect on WRKY23 transcript levels (Supplemental Fig. S4B), supporting the notion that WRKY23 acts downstream of ARF7 and ARF19 but not of LBDs. Interestingly, we observed that overexpression of WRKY23 results in higher transcript levels for LBD16 and LBD29 (Supplemental Fig. S4C), suggesting that WRKY23 has some degree of regulatory effect on LBDs. Furthermore, ectopic expression of WRKY23 indeed partially rescued the callus-forming and shoot-regenerating defect observed in arf7 arf19 explants and derived calli (Fig. 4B; Supplemental Fig. S5A). However, we failed to detect any binding activity of ARF7 toward the promoter region of WRKY23 by chromatin immunoprecipitation-qPCR (ChIP-qPCR) assay performed with transgenic ProARF7:ARF7-GFP seedlings treated with CIM; we also observed no transcriptional activity of ARF7 toward a ProWRKY23:LUC (firefly luciferase gene) reporter construct in a transcriptional activation assay in Arabidopsis protoplasts (Supplemental Fig. S5, B to D). These observations further support the idea that WRKY23 is an indirect target of ARF7 and ARF19 (Prát et al. 2018).
Figure 4.
WRKY23 acts downstream of ARF7 and ARF19 and directly activates transcription of PLT3 and PLT7. A) Relative WRKY23 transcript levels in response to CIM in arf7 arf19 and lbd16-2 air1-2 seedlings. Seven-day-old seedlings were incubated in liquid CIM for the indicated times, and the relative expression level at 0 h was set to 1; data are shown as means ± Sd (n = 3 biological replicates). B) Callus-forming and shoot-regenerating phenotypes of WT, arf7 arf19, and arf7 arf19 35S:WRKY23 explants and derived calli (n = 20 explants). Scale bar, 10 mm. C) Callus-forming and shoot-regenerating phenotypes of WT, 35S:WRKY23, plt3 plt7, and plt3 plt7 35S:WRKY23 explants (n = 20 explants) and derived calli. Scale bar, 10 mm. Three independent transgenic lines of arf7 arf19 35S:WRKY23 and plt3 plt7 35S:WRKY23 named #1, #2, and #3 were examined in B) and C), respectively. D) Induction of PLT3 and PLT7 transcript levels by WRKY23. Five-d-old transgenic ProXVE:WRKY23 seedlings were incubated in liquid B5 medium containing 10 μm 17-β-estradiol or DMSO (Mock) for 3 h, and relative expression levels are shown as means ± Sd (n = 3 biological replicates). E) ChIP-qPCR assay showing the WRKY23 binding activity to the PLT3 and PLT7 promoters. ChIP-qPCR was performed with 10-d-old WT and wrky23-3 ProWRKY23:gWRKY23-GFP seedlings incubated in liquid CIM for 48 h with an anti-GFP antibody, and the enrichment of PLT3 or PLT7 promoter fragments F-1 to F-9 are indicated. A fragment of the UBQ10 promoter was used as a negative control. Data are shown as means ± Sd (n = 3 biological replicates). F) Transcriptional activation of PLT3 and PLT7 by WRKY23. The activity of the ProPLT3:LUC or ProPLT7:LUC reporter was determined in Arabidopsis protoplasts cotransfected with an empty vector (EV), or 35S:Ω:WRKY23-GFP (WRKY23) construct, together with each LUC reporter. Data are shown as means of the original LUC/REN ratios × 100 ± Sd (n = 3 biological replicates). *P < 0.05, **P < 0.01, and ***P < 0.001 by t-test.
Since WRKY23 could promote the transcript levels of PLT3 and PLT7 as well as PLT1 and PLT2 (Fig. 3A) and as PLT3, PLT5, and PLT7 were shown to function redundantly in the transcriptional activation of PLT1 and PLT2 (Kareem et al. 2015), we tested whether PLT3 and/or PLT7 are targets of WRKY23. When WRKY23 was overexpressed in the plt3 plt7 double mutant, whose callus exhibits a defect in shoot regeneration (Kareem et al. 2015), the enhanced shoot-regeneration capacity observed in the 35S:WRKY23 calli was abolished (Fig. 4C; Supplemental Fig. S5E). Next, we generated transgenic plants carrying a chemically inducible OLexA-46 (also known as G1090:XVE) promoter driving WRKY23 expression (ProXVE:WRKY23) (Zuo et al. 2000) and observed that treatment with the inducer 17-β-estradiol leads to higher transcript levels for PLT3 and PLT7 (Fig. 4D). Furthermore, we performed a ChIP-qPCR assay with transgenic wrky23-3 ProWRKY23:gWRKY23-GFP seedlings incubated in liquid CIM, which revealed that WRKY23 can bind to the promoter regions of PLT3 and PLT7 but not to the PLT1, PLT2, and WOX5 promoters (Fig. 4E; Supplemental Fig. S5F). A transcriptional activation assay carried out in Arabidopsis protoplasts using ProPLT3:LUC and ProPLT7:LUC reporter constructs clearly showed that transiently expressing WRKY23 leads to a higher relative LUC activity derived from the 2 reporter constructs (Fig. 4F; Supplemental Fig. S5C). Collectively, we conclude that following the induction of its transcription by auxin, WRKY23 downstream of ARF7 and ARF19 directly activates PLT3 and PLT7 transcription.
LBD-induced bHLH041 disappearance alleviates the transcriptional repression of PLT1, PLT2, and WOX5
Since CIM-induced disappearance of bHLH041 results in the activation of PLT1, PLT2, and WOX5, which is suggested to contribute to LBD-triggered acquisition of callus pluripotency (Liu et al. 2018), we reasoned that bHLH041 might act downstream of the ARF7/19–LBD module to repress the transcription of PLT1, PLT2, and WOX5. Indeed, we determined that disruption of ARF7 and ARF19 completely blocks the CIM-induced disappearance of bHLH041 in the pericycle cells of root explants, strengthening the idea that bHLH041 is downstream of ARF7 and ARF19 (Supplemental Fig. S6A). Next, we investigated whether disappearance of bHLH041 is related to LBD accumulation. To this end, we coexpressed bHLH041-GFP with LBD16-FLAG, LBD29-FLAG, or WRKY23-FLAG in Nicotiana benthamiana leaves; we observed much lower GFP fluorescent signals for bHLH041-GFP in the presence of LBD16 or LBD29 but not WRKY23 (Fig. 5A). In agreement, the overexpression of LBD16 or LBD29 in ProbHLH041:bHLH041-GFP plants largely abolished bHLH041 accumulation in multiple organs (Fig. 5B; Supplemental Fig. S6B), demonstrating that LBD accumulation leads to the disappearance of bHLH041. Moreover, we generated the lbd16-2 bhlh041-1 double mutant: we determined that the callus-forming and shoot-regenerating defect of lbd16-2 is partially rescued by the introduction of the bhlh041-1 mutation (Fig. 5C), supporting the notion that bHLH041 is downstream of LBDs in controlling callus formation and regeneration capacity.
Figure 5.
LBD induces the removal of bHLH041 to alleviate bHLH041-mediated repression of PLT1, PLT2, and WOX5 transcription. A, B) Over-accumulation of LBD16 or LBD29 induces the disappearance of bHLH041. The fluorescent signals of bHLH041-GFP were visualized in N. benthamiana leaves transiently coexpressing bHLH041-GFP with an empty vector (EV), LBD16-FLAG (LBD16), LBD29-FLAG (LBD29), or WRKY23-FLAG (WRKY23) for 4 d (n = 10 leaves) A) and in transgenic Arabidopsis ProbHLH041:bHLH041-GFP seedlings harboring a 35S:LBD16 construct with a strong (S), intermediate (I), or weak (W) autonomous callus-forming phenotype B). Scale bars, 50 µm. C) Callus-forming and shoot-regenerating phenotypes of WT, lbd16-2, bhlh041-1, and lbd16-2 bhlh041-1, explants and derived calli (n = 20 explants). Scale bar, 10 mm. D) ChIP-qPCR assay of bHLH041 binding activity to the PLT1, PLT2, and WOX5 promoters. Ten-day-old WT and bhlh041-1 ProbHLH041:bHLH041-GFP seedlings incubated in liquid CIM for 0 and 48 h were assayed by ChIP-qPCR with anti-GFP antibody, and enrichments of the amplified F-1 to F-6 from PLT1, PLT2, or WOX5 promoters are indicated. A fragment of the UBQ10 promoter was used as a negative control. Data are shown as means ± Sd (n = 3 biological replicates). E) bHLH041 inhibits PLT1, PLT2, and WOX5 transcription. Left, diagrams of effector and reporter constructs used. Ω, translational enhancer. Arabidopsis protoplasts were cotransfected with the LUC reporter PLT1:LUC, PLT2:LUC, or WOX5:LUC with an empty vector (EV) or 35S:Ω:bHLH041-GFP construct (bHLH041) and incubated in CIM for 16 h. LUC and REN activities were determined and shown as means of original LUC/REN ratios × 100 ± Sd (n = 3 biological replicates). *P < 0.05, **P < 0.01, and ***P < 0.001 by t-test.
To further test whether the PLT1, PLT2, and WOX5 are targeted by bHLH041 during auxin-induced callus formation, we performed ChIP-qPCR analysis with bhlh041-1 ProbHLH041:bHLH041-GFP seedlings before and after incubation on CIM. We observed that bHLH041 can bind to specific regions of the PLT1, PLT2, and WOX5 promoters, with the binding enrichment decreasing after seedlings were incubated on CIM (Fig. 5D). We also conducted a transcriptional activity assay in Arabidopsis protoplasts incubated in CIM, using ProPLT1:LUC, ProPLT2:LUC, and ProWOX5:LUC reporter constructs and 35S:bHLH041 as effector construct. Cotransfecting each LUC reporter with the 35S:bHLH041 effector construct resulted in lower relative LUC activity from all reporters (Fig. 5E). Taken together, we conclude that LBD-induced disappearance of bHLH041 derepresses the transcription of PLT1, PLT2, and WOX5, thus coordinating the establishment of callus pluripotency.
WRKY23 and bHLH041 synergize shoot-regenerating capacity of callus
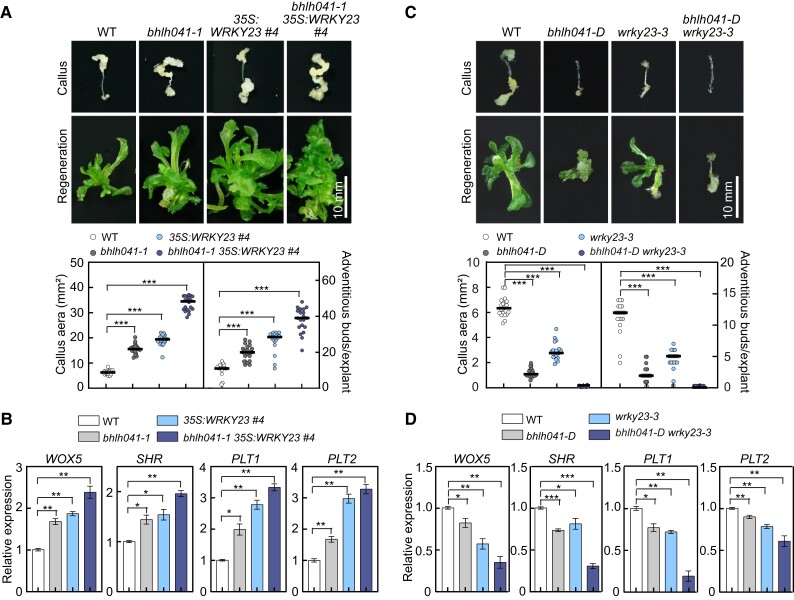
Since both the WRKY23–PLT3/7 and LBD–bHLH041 modules participate in the activation of PLT1, PLT2, and WOX5, we postulated that two modules synergize the shoot-regeneration capability of callus cells. To test this hypothesis, we crossed the 35S:WRKY23 line to the bhlh041-1 mutant and obtained bhlh041-1 35S:WRKY23 plants and examined the callus-forming phenotype of their root explants and the shoot-regenerating capability of their derived calli. When compared to those of 35S:WRKY23 and bhlh041-1, the callus-forming and shoot-forming phenotypes were additive in the bhlh041-1 35S:WRKY23 explants and derived calli (Fig. 6A). Consistent with this finding, the transcript levels of WOX5, SHR, PLT1, and PLT2 in the bhlh041-1 35S:WRKY23 root explants were higher than those of 35S:WRKY23 or bhlh041-1 root explants after being incubated on CIM (Fig. 6B). Next, we generated the bhlh041-D wrky23-3 double mutant and discovered that both the callus-forming and shoot-regenerating defects in the wrky23-3 root explants and derived calli are further enhanced by introduction of bhlh041-D (Fig. 6C) and that the transcript levels of WOX5, PLT1, PLT2, and SHR in bhlh041-D wrky23-3 are substantially lower than in the wrky23-3 or bhlh041-D root explants incubated on CIM (Fig. 6D). These observations demonstrate that transcriptional activation of WRKY23 and derepression by removal of bHLH041 synergistically establish callus pluripotency during in vitro regeneration programs.
Figure 6.
WRKY23 and bHLH041 synergize shoot-regenerating capability of callus. A) Callus-forming and shoot-regenerating phenotypes of WT, bhlh041-1, 35S:WRKY23, and bhlh041-1 35S:WRKY23 root explants and derived calli. B) Relative transcript levels of root stem cell genes in root explant-derived callus of the genotypes described in A). C) Callus-forming and shoot-regenerating phenotypes of WT, bhlh041-D, wrky23-3, and bhlh041-D wrky23-3 root explants and derived calli. D) Relative transcript levels of root stem cell genes in root explant-derived callus of the genotypes described in C). The callus area from each root explant on CIM and the number of regenerating shoots from each callus on SIM were determined (n = 20 explants) in A and C, Scale bars, 10 mm. RT-qPCR analysis in B) and D) was performed with the root explants incubated on CIM for 7 d, and data are shown as means ± Sd (n = 3 biological replicates). *P < 0.05, **P < 0.01, and ***P < 0.001 by t-test.
Discussion
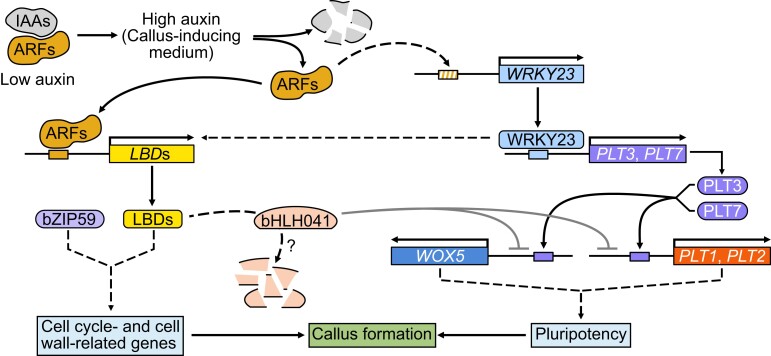
Although accumulating evidence demonstrates that several auxin-signaling components including IAA-ARFs and LBDs are critical regulators of auxin-induced callus formation (Fan et al. 2012; Shang et al. 2016; Xu, Cao, Zhang, et al. 2018) and that ectopic activation of root stem cell factors including PLTs and WOX5 are required for de novo shoot or root regeneration (Che et al. 2007; Atta et al. 2009; Sugimoto et al. 2010; Kareem et al. 2015), the molecular links between these auxin-signaling components and activation of root pluripotent factors during in vitro plant regeneration is unknown. Here, we defined Arabidopsis WRKY23 and bHLH041 as a transcriptional activator and repressor, respectively, as being responsible for activation of root stem cell factors to synergize the establishment of callus pluripotency and thus conferring shoot-regenerating capability on callus cells. These findings uncover the transcriptional mechanism underlying the acquisition of cellular pluripotency in auxin-induced callus formation, which links auxin signaling and cellular reprogramming during in vitro plant regeneration. These findings, together with previous works, outline a molecular framework for how callus formation and cellular reprogramming are governed in Arabidopsis in vitro regeneration. It is likely that, upon CIM treatment, ARF7- and ARF19-directed LBD accumulation in the pericycle or pericycle-like cells not only triggers the callus-forming program but also leads to the removal of bHLH041, which alleviates the transcriptional repression of PLT1, PLT2, and WOX5. At the same time, auxin-induced WRKY23 downstream of ARF7 and ARF19 directly activates the transcription of PLT3 and PLT7, whose encoded proteins target their downstream genes PLT1, PLT2, and WOX5. The two transcriptional regulatory modules coordinately establish the shoot-regenerating capability of callus cells (Fig. 7).
Figure 7.
A proposed molecular framework for auxin-induced callus formation. Upon CIM treatment, a high concentration of auxin activates LBDs and WRKY23 via the IAA–ARF7/19 signaling module in the pericycle or pericycle-like cells. LBDs together with their partner bZIP59 trigger callus formation and result in the removal of bHLH041, which alleviates the repression of transcription of PLT1, PLT2, and WOX5. At the same time, WRKY23 directly activates transcription of PLT3 and PLT7 and thus indirectly that of their downstream genes PLT1, PLT2, and WOX5. Two transcriptional regulations synergize the callus pluripotency establishment and thus shoot-regenerating capability.
Intriguingly, WRKY23 directly activates the transcription of PLT3 and PLT7 and thus indirectly that of PLT1, PLT2, and WOX5, while the LBD-induced removal of bHLH041 derepresses the transcription of PLT1, PLT2, and WOX5. Indeed, the WOX11–LBD16 module has been shown to promote the acquisition of pluripotency by callus cells via activation of PLT1 and PLT2 but not PLT3, PLT5, or PLT7 (Liu et al. 2018). The coexistence and coordination of transcriptional activation and derepression might be a mechanism to ensure that plant somatic cells acquire pluripotency during plant regeneration. WRKY23 is a unique auxin-responsive member of the Arabidopsis WRKY family to regulate root development (Grunewald et al. 2008, 2012), while bHLH041 belongs to a large family of bHLH transcription factors present in both plants and animals. It has been shown that different bHLH members function as transcriptional activators or repressors in directing cell fate changes (Toledo-Ortiz et al. 2003). For example, heterodimers of the bHLH factors GhTCE1 and GhTCEE1 promote transcriptional reprogramming during wound-induced callus formation in cotton (Gossypium hirsutum) (Deng et al. 2022). In animals, self-renewal, multipotency, and fate choice of neural progenitor cells (NPCs) are controlled by multiple bHLH factors with contradictory functions in promoting NPC proliferation and cell-cycle exit for differentiation (Imayoshi and Kageyama 2014). Importantly, callus formation in in vitro plant regeneration involves both reactivation of cellular mitotic activities and change of cell fates, which are concomitant (Zhao et al. 2001; Williams et al. 2003; Tessadori et al. 2007; Xu et al. 2012). It is likely that the regulatory mechanisms of callus formation and cell fate change are partly shared or overlap. In Arabidopsis, auxin-induced LBDs and their partner bZIP59 are sufficient to trigger the formation of pluripotent callus by mediating multiple cellular events, including cell wall metabolism and inhibition of the differentiation program (Fan et al. 2012; Xu, Cao, Xu, et al. 2018; Xu, Cao, Zhang, et al. 2018). Our finding that overexpression or disruption of bHLH041 also causes callus-forming phenotypes suggests that bHLH041 is involved in LBD-directed cellular mitotic activation. Similarly, although WRKY23 acts in parallel to bHLH041 by targeting PLT3 and PLT7, we also observed that WRKY23 has a regulatory effect on LBD transcript levels, suggesting that some extent of crosstalk exists between WRKY23 and LBD-regulated cellular mitotic activity (Fig. 7), which might explain why alteration of WRKY23 results in a callus-forming phenotype.
Although our work defines WRKY23 and bHLH041 as transcriptional activator and repressor to coordinate the establishment of callus pluripotency, the detailed molecular regulations of cellular pluripotency acquisition during auxin-induced callus formation need to be further clarified. First, as LBD transcription factors execute their roles at a transcriptional level, the molecular regulation behind LBD-induced removal of bHLH041 remains unclear. We speculate that other factors are involved in LBD-regulated bHLH041 stability in the forming callus. We also noticed that CIM treatment leads to a decrease in bHLH041 accumulation in other somatic cells besides the pericycle and pericycle-like cells; the biochemical or molecular regulation of bHLH041 stability in these cells needs to be further investigated. Second, we only characterized here how PLTs and WOX5 are regulated by WRKY23 and bHLH041, but we also observed that the transcript levels of SHR are regulated by WRKY23 and bHLH041 (Fig. 3). As the regulatory relationship of the PLT–WOX5 and SHR–SCR modules in root stem cell maintenance is not fully defined, how WRKY23 and bHLH041 regulate SHR and/or SCR remains to be elucidated. Furthermore, auxin-induced callus formation represents a type of cellular reprogramming in plant regeneration, during which genome-wide modifications, such as chromatin reorganization, heterochromatin redistribution, and epigenetic modification occur during the callus formation program (Ikeuchi et al. 2013). How WRKY23- and bHLH041-mediated transcriptional regulations are incorporated with these genome-wide modifications would be interesting. In addition, since WRKY23 and bHLH041 as well as the root stem cell factors are all transcription factors functioning in nuclei, whether they interact with any of these root stem cell factors remains unclear. It is also possible that some might function in a protein complex during the establishment and maintenance of callus pluripotency. Therefore, further work on these proteins will be helpful to understand the molecular basis of cellular reprogramming in in vitro plant regeneration.
Finally, as both WRKY23 and bHLH041 are involved in regulation of callus formation and shoot regeneration capability, it is likely that appropriate manipulation of their orthologs will be potential to improve the regeneration efficiency of crops and horticultural plants. Therefore, it is worth identifying the orthologs of WRKY23 and bHLH041 to clarify whether such molecular regulations by WRKY23 and bHLH041 are conserved in other plant species, which would benefit plant regeneration-based gene-editing and biotechnological practices.
Materials and methods
Plant materials and growth conditions
The Arabidopsis (A. thaliana) accession Col-0 was used as a WT and for all transgenic lines. The T-DNA insertion mutants bhlh041-1 (SAIL_330_F04), bhlh041-D (SAIL_258_C03), lbd16-2 (SALK_040739) (Fan et al. 2012), air1-2 (SALK_024459c) (Xu, Cao, Zhang, et al. 2018), plt3 (SALK_127417) (Kareem et al. 2015), and plt7 (SAIL_1167_C10) (Kareem et al. 2015) were obtained from the ABRC and verified by PCR analyses. The ProWOX5:GFP-ER and ProPLT1:PLT1-YFP marker lines and arf7 arf19 double mutant were described previously (Aida et al. 2004; Haecker et al. 2004; Fan et al. 2012). All seeds were surface-sterilized in 1% (w/v) sodium hypochlorite, rinsed 3 times with sterile water, and germinated on half-strength of MS medium (MS powder Coolaber, Beijing, China, 1% [w/v] sucrose, 0.55% [w/v] plant agar, and pH 5.7) at 22 ± 2 °C under long-day conditions (16-h light/8-h dark photoperiod) with incandescent lamps (spectrum: 400 to 700 nm; illumination intensity: 80 to 90 µmol m−2s−1); 7-d-old seedlings were transferred to soil and grown in a greenhouse under the same conditions.
Callus induction and shoot regeneration
For characterization of callus formation, cotyledon and root explants of 7-d-old seedlings were incubated on CIM (B5 medium [Coolaber, Beijing, China] supplemented with 2% [w/v] glucose, 0.5 g L–1 MES, 2.2 µm 2,4-dichlorophenoxyacetic acid [2,4-D], 0.2 µm kinetin, 0.25% [w/v] phytagel, and pH 5.7) as described (Che et al. 2006) for 21 d, and the formed callus was photographed, and callus area for each explant was quantified with ImageJ software (Shang et al. 2016). For examination of shoot regeneration from callus, the explants incubated on CIM for 7 d with forming callus were transferred onto SIM (MS medium [Coolaber, Beijing, China] supplemented with 1% [w/v] sucrose, 0.5 g L–1 MES, 0.25% [w/v] phytagel, 5.0 µm isopentenyladenine and 0.9 µm indole-3-acetic acid [IAA], and pH 5.7) (Che et al. 2006), and the leafy shoots produced from each callus were counted and photographed under a stereomicroscope at 14 d. To monitor transcript abundance of root stem cell marker genes, the root explants incubated CIM for 7 d were collected for RNA isolation, and 5-d-old seedlings harboring a ProPLT1:PLT1-YFP or ProWOX5:GFP-ER construct were incubated on CIM for 4 d for determination of GFP fluorescent signals. All experiments above were carried out by at least 3 independent biological replicates with more than 20 plants each time.
Plasmid construction and Arabidopsis transformation
For generation of the ProWRKY23:gWRKY23-GFP, ProbHLH041:bHLH041-GFP, and ProARF7:ARF7-GFP constructs, a genomic WRKY23 fragment consisting of a 4,983-bp promoter fragment and a 1,399-bp coding region segment, a bHLH041 fragment with 2,025 bp of promoter and 1,578 bp of coding region, and ARF7 fragments with 2,489 bp of promoter and 3,498 bp of coding region fused in-frame and upstream of the GFP sequence were cloned into the pCAMBIA1300 vector by the multi-one-step seamless cloning approach, respectively. A 6-glycine linker sequence was inserted upstream of the GFP tag to minimize the influence of the tag on the fusion protein and optimize the stability of the target protein (Robinson and Sauer 1998; Funakoshi and Hochstrasser 2009). The full-length coding sequences of WRKY23 and bHLH041 were cloned into the pSuper1300 vector (Chinnusamy et al. 2003), to generate the 35S:WRKY23 and 35S:bHLH041 constructs, respectively. The full-length coding sequences of LBD16 and LBD29 were cloned into the pVIP96 vector to generate the 35S:LBD16 and 35S:LBD29 constructs, respectively (Hu et al. 2003). The coding sequence of WRKY23 was also cloned into the pER10 vector for generating the chemically inducible ProXVE:WRKY23 construct (Zuo et al. 2000). The primers used for the constructs are listed in Supplemental Data Set S1. All plasmids were verified by Sanger sequencing and introduced into Agrobacterium (Agrobacterium tumefaciens) strain EHA105 or ABI and transformed into Arabidopsis using the floral dip method (Clough and Bent 1998). At least 10 independent transgenic lines with a single T-DNA insertion were generated for each construct, and at least 3 lines of the T3 homozygotes were used for experimental examination.
CRISPR/Cas9 gene editing
To create the wrky23 allelic mutants, an Arabidopsis egg cell-specific promoter-controlled CRISPR/Cas9 gene-editing system was used as previously described (Wang et al. 2015). Briefly, the specific primers containing the 23-bp sequences with 2 PAM sites (5′-N20NGG-3′) corresponding to simple guide RNAs (sgRNAs) were manually designed against the first exon of WRKY23 and then checked by BLAST for evaluating their specificities on the TAIR website (https://www.arabidopsis.org/Blast/index.jsp). The pCBC-DT1T2 plasmid was used as a template to perform PCR amplification of the sgRNA sequences. The PCR products were cloned into the pHEE401 vector and introduced into Arabidopsis via the floral dipping method as above. The genomic fragments covering the mutation sites from the T1 and T2 transgenic plants were amplified by PCR and sequenced, and homozygous T3 plants without the editing vector were used for characterization. All primers used for generation of the constructs are listed in Supplemental Data Set S1.
Total RNA isolation and gene expression analysis
Total RNAs were isolated using an E.Z.N.A. Plant RNA Kit (OMEGA BioTek) according to the manufacturer's instructions. First-strand cDNA was synthesized with TransScript II All-in-One First-Strand cDNA Synthesis SuperMix (Invitrogen). qPCR was conducted with a SYBR Premix Ex Taq II kit (Takara, Dalian, China) as described previously (Fan et al. 2012), the amplified ACTIN2 transcript abundance was used as a normalization control, and the relative expression values were calculated using a modified 2−ΔΔCT method (Livak and Schmittgen 2001). For RT-PCR analysis, the transcript abundance of GLYCERALDEHYDE-3-PHOSPHATE DEHYDROGENASE C SUBUNIT 1 (GAPC) was used as an internal control. All RT-qPCR and RT-PCR analyses were performed with 3 independent biological replicates. The primers used are listed in Supplemental Data Set S1.
Protein extraction and immunoblotting
To monitor WRKY23 and bHLH041 accumulation during callus induction, total proteins from transgenic ProWRKY23:gWRKY23-GFP and ProbHLH041:bHLH041-GFP seedlings incubated on CIM for the indicated times were extracted with a Plant Total Protein Extraction Kit (Cwbio, Beijing, China), according to the manufacturer's instructions. The proteins were separated by a 12% SDS-PAGE and immobilized onto nitrocellulose membranes, and immunoblotting was performed with anti-GFP (MBL, lot: M048-3) (1:5000) and anti-ACTIN (EASYBIO, lot: 80790722) (1:5000) primary antibodies followed by horseradish peroxidase-labeled secondary antibody (Bioeasy) (1:10000) and subsequently detected with an ECL Super Sensitive Kit (DiNing). The immunoblotting signals were scanned with a Tanon5200 imaging system.
Confocal microscopy
To visualize accumulation of WRKY23-GFP, bHLH041-GFP, and root meristem markers, 5-d-old seedlings harboring a specific construct or marker incubated on CIM or infiltrated N. benthamiana leaves were mounted in 10 mg L–1 propidium iodide (Sigma) or distilled water, respectively, and GFP images were collected under an Olympus FV1000-MPE laser scanning microscope. The GFP signal was excited at 488 nm, and the emission was acquired between 500 and 550 nm. The propidium iodide signal was visualized by excitation with an argon laser at 488 nm and detected with a spectral detector set at >585 nm for emission. The GFP fluorescent signals of root stem cell marker lines were determined with the parameters at laser transmissivity: 80%, photomultiplier tube Voltage: 490 V, pinhole: 125 µm, objective lens magnification: 20×. The GFP-positive loci in each image were quantified with ImageJ software.
ChIP-qPCR assay
About 1 g of 10-d-old WT and transgenic seedlings harboring the respective GFP-fusion construct or seedlings incubated in liquid CIM for 12 or 48 h were cross-linked in 1% (w/v) formaldehyde, and the ChIP-qPCR assay was performed as described previously (Bowler et al. 2004), with minor modifications. Briefly, anti-GFP mAb-agarose (MBL) beads were used to immunoprecipitate the protein-DNA complex, and the precipitated DNA was purified for qPCR analysis. The isolated chromatin before precipitation was used as input control. Primers used for ChIP-qPCR are listed in Supplemental Data Set S1. The ChIP-qPCRs were performed with 3 biological and technical replicates.
Transient transcriptional activity assay
The promoter fragments of WRKY23 (4,983 bp), PLT3 (3,919 bp), PLT7 (5,154 bp), PLT1 (5,611 bp), PLT2 (5,315 bp), or WOX5 (4,534 bp) upstream of each translation start site were respectively amplified by PCR from Col-0 genomic DNA and cloned upstream of LUC into the TQ379 vector (Zhang et al. 2017), which harbors the Pro35S:REN (Renilla luciferase) cassette, to create the reporter constructs ProWRKY23:LUC, ProPLT3:LUC, ProPLT7:LUC, ProPLT1:LUC, ProPLT2:LUC, and ProWOX5:LUC, respectively. The full-length coding sequences of ARF7, WRKY23, and bHLH041 fused with a 6-glycine linker were respectively cloned upstream of the GFP sequence into p326-35S-cGFP vector with a Ω translational enhancer as effectors (Lin et al. 2016). The Ω translational enhancer (Gallie et al. 1989; Gallie and Kado 1989) was cloned upstream of ARF7, WRKY23, and bHLH041 to increase their final expression levels. All primers used for the generation of the constructs are listed in Supplemental Data Set S1.
Three-week-old Arabidopsis (Col-0) leaves were used for preparing protoplasts according to a published protocol (Yoo et al. 2007) for dual-luciferase reporter assays. The transfected protoplasts were cultured at 22 °C in the light for 4 h and then in the dark for 14 h. The protoplasts were then lysed with passive lysis buffer (Promega; E1910), and LUC and REN activities were quantified and measured with a luminometer (Promega GloMax Multi Jr). The relative LUC activity was calculated by normalizing to that of REN in 3 biological triplicates.
N. benthamiana infiltration
The full-length coding sequences of LBD16, LBD29, and WRKY23 were cloned into the binary vector pSuper1300-FLAG vector, and the full-length coding sequence of bHLH041 was cloned into the vector pSuper1300-GFP to generate LBD16-FLAG, LBD29-FLAG, WRKY23-FLAG, and bHLH041-GFP constructs, respectively. A 6-glycine linker sequence was also inserted upstream of FLAG or GFP tags, and all primers used for constructions are listed in Supplemental Data Set S1. Agrobacterium strain EHA105 harboring the different constructs cultured at 28 °C for 2 d was harvested by centrifugation at 3,500 rpm for 10 min at room temperature and resuspended in infiltration buffer (10 mm MES, 10 mm MgCl2, 150 μm acetosyringone, and pH 5.8) at a final OD600 = 1.0, and then mixed by the specific combination (v:v = 1:1) after 4 h. The mixed Agrobacterium cells were infiltrated into the leaves of 4-wk-old N. benthamiana plants using an injector as previously described (Sparkes et al. 2006). The fluorescent signals were examined under an Olympus FV1000-MPE laser scanning microscope after plants were kept in dark for 24 h and then in the light for 72 h. The signals were quantified by ImageJ software from 3 biological replicates.
Statistical analysis
All analyses were performed using GraphPad Prism v.8. Statistical analyses were performed using unpaired Student's t-test with Welch's correction (not assuming equal standard deviations) (*P < 0.05; **P < 0.01; ***P < 0.001). The values for N and the specific statistical test performed for each experiment are described in the figure legends and Supplemental Data Set S2.
Accession numbers
Sequence data in this article can be found in the Arabidopsis Genome initiative or GenBank/EMBL databases under the following accession numbers: WRKY23 (At2g47260), PLT1 (At3g20840), PLT2 (At1g51190), PLT3 (At5g10510), PLT5 (At5g57390), PLT7 (At5g65510), WOX5 (At3g11260), SHR (At4g37650), ARF7 (At5g20730), ARF19 (At1g19220), LBD16 (At2g42430), LBD29 (At3g58190), bZIP59 (At2g31370), bHLH041 (At5g56960), ACTIN2 (At3g18780), GPAC (At3g04120), and UBQ10 (At4g05320).
Supplementary Material
Acknowledgments
We are grateful to Drs Ben Scheres, Jim Haseloff, Nam-Hai Chua, Jia-Wei Wang, and Jingbo Jin for providing seeds or constructs used in this study. We acknowledge Huichao Liu and Kezhen Yang for help in preparing some plant samples and the solutions for the ChIP-qPCR experiment.
Contributor Information
Chongyi Xu, Key Laboratory of Plant Molecular Physiology, Institute of Botany, Chinese Academy of Sciences, China National Botanical Garden, Beijing 100093, China.
Pengjie Chang, Key Laboratory of Plant Molecular Physiology, Institute of Botany, Chinese Academy of Sciences, China National Botanical Garden, Beijing 100093, China; University of Chinese Academy of Sciences, Beijing 100049, China.
Shiqi Guo, Key Laboratory of Plant Molecular Physiology, Institute of Botany, Chinese Academy of Sciences, China National Botanical Garden, Beijing 100093, China; University of Chinese Academy of Sciences, Beijing 100049, China.
Xiaona Yang, Key Laboratory of Plant Molecular Physiology, Institute of Botany, Chinese Academy of Sciences, China National Botanical Garden, Beijing 100093, China; University of Chinese Academy of Sciences, Beijing 100049, China.
Xinchun Liu, Key Laboratory of Plant Molecular Physiology, Institute of Botany, Chinese Academy of Sciences, China National Botanical Garden, Beijing 100093, China; University of Chinese Academy of Sciences, Beijing 100049, China.
Baofeng Sui, Key Laboratory of Plant Molecular Physiology, Institute of Botany, Chinese Academy of Sciences, China National Botanical Garden, Beijing 100093, China; University of Chinese Academy of Sciences, Beijing 100049, China.
Dongxue Yu, Key Laboratory of Plant Molecular Physiology, Institute of Botany, Chinese Academy of Sciences, China National Botanical Garden, Beijing 100093, China; University of Chinese Academy of Sciences, Beijing 100049, China.
Wei Xin, Key Laboratory of Plant Molecular Physiology, Institute of Botany, Chinese Academy of Sciences, China National Botanical Garden, Beijing 100093, China; University of Chinese Academy of Sciences, Beijing 100049, China.
Yuxin Hu, Key Laboratory of Plant Molecular Physiology, Institute of Botany, Chinese Academy of Sciences, China National Botanical Garden, Beijing 100093, China; National Center for Plant Gene Research, Beijing 100093, China.
Author contributions
Y.H. conceived the project; C.X. and Y.H. designed the experiments; C.X. performed most of the experiments; P.C., S.G., X.Y., X.L., B.S., D.Y., and W.X. contributed to the generation of constructs or transgenic Arabidopsis plants; and C.X. and Y.H. analyzed data and wrote the manuscript.
Supplemental data
The following materials are available in the online version of this article.
The following materials are available in the online version of this article.
Supplemental Figure S1 . Reponses of WRKY23 and bHLH041 to CIM.
Supplemental Figure S2 . Characterization of wrky23 and 35S:WRKY23 plants.
Supplemental Figure S3 . Characterization of bhlh041-1, bhlh041-D, and 35S:bHLH041 plants.
Supplemental Figure S4 . Relative ARF7 and ARF19 transcript levels in response to CIM and inter-regulation of WRKY23 and LBDs.
Supplemental Figure S5 . Genetic interactions of WRKY23, ARF7, ARF19, and PLTs.
Supplemental Figure S6 . Accumulation of bHLH041 in arf7 arf19 and 35S:LBD29 plants.
Supplemental Data Set S1 . Primers used in this study.
Supplemental Data Set S2. Summary of statistical analyses.
Funding
This work was supported by the Key Program of National Natural Science Foundation of China (grant no. 31830055) and the General Program of National natural Science Foundation of China (grant no. 32170317) and the Strategic Priority Research Program of Chinese Academy of Sciences (grant no. XDB27030102).
Data availability
The data underlying this article are available in the article and in its online supplementary material.
Dive Curated Terms
The following phenotypic, genotypic, and functional terms are of significance to the work described in this paper:
References
- Aida M, Beis D, Heidstra R, Willemsen V, Blilou I, Galinha C, Nussaume L, Noh YS, Amasino R, Scheres B. The PLETHORA genes mediate patterning of the Arabidopsis root stem cell niche. Cell 2004:119(1):109–120. 10.1016/j.cell.2004.09.018 [DOI] [PubMed] [Google Scholar]
- Atta R, Laurens L, Boucheron-Dubuisson E, Guivarc’h A, Carnero E, Giraudat-Pautot V, Rech P, Chriqui D. Pluripotency of Arabidopsis xylem pericycle underlies shoot regeneration from root and hypocotyl explants grown in vitro. Plant J. 2009:57(4):626–644. 10.1111/j.1365-313X.2008.03715.x [DOI] [PubMed] [Google Scholar]
- Birnbaum KD, Sanchez Alvarado A. Slicing across kingdoms: regeneration in plants and animals. Cell 2008:132(4):697–710. 10.1016/j.cell.2008.01.040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowler C, Benvenuto G, Laflamme P, Molino D, Probst AV, Tariq M, Paszkowski J. Chromatin techniques for plant cells. Plant J. 2004:39(5):776–789. 10.1111/j.1365-313X.2004.02169.x [DOI] [PubMed] [Google Scholar]
- Che P, Gingerich DJ, Lall S, Howell SH. Global and hormone-induced gene expression changes during shoot development in Arabidopsis. Plant Cell 2002:14(11):2771–2785. 10.1105/tpc.006668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Che P, Lall S, Howell SH. Developmental steps in acquiring competence for shoot development in Arabidopsis tissue culture. Planta 2007:226(5):1183–1194. 10.1007/s00425-007-0565-4 [DOI] [PubMed] [Google Scholar]
- Che P, Lall S, Nettleton D, Howell SH. Gene expression programs during shoot, root, and callus development in Arabidopsis tissue culture. Plant Physiol. 2006:141(2):620–637. 10.1104/pp.106.081240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chinnusamy V, Ohta M, Kanrar S, Lee BH, Hong X, Agarwal M, Zhu JK. ICE1: a regulator of cold-induced transcriptome and freezing tolerance in Arabidopsis. Genes Dev. 2003:17(8):1043–1054. 10.1101/gad.1077503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough SJ, Bent AF. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998:16(6):735–743. 10.1046/j.1365-313x.1998.00343.x [DOI] [PubMed] [Google Scholar]
- Deng J, Sun W, Zhang B, Sun S, Xia L, Miao Y, He L, Lindsey K, Yang X, Zhang X. GhTCE1-GhTCEE1 dimers regulate transcriptional reprogramming during wound-induced callus formation in cotton. Plant Cell 2022:34(11):4554–4568. 10.1093/plcell/koac252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duclercq J, Sangwan-Norreel B, Catterou M, Sangwan RS. De novo shoot organogenesis: from art to science. Trends Plant Sci. 2011:16(11):597–606. 10.1016/j.tplants.2011.08.004 [DOI] [PubMed] [Google Scholar]
- Fan M, Xu C, Xu K, Hu Y. LATERAL ORGAN BOUNDARIES DOMAIN transcription factors direct callus formation in Arabidopsis regeneration. Cell Res. 2012:22(7):1169–1180. 10.1038/cr.2012.63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukaki H, Nakao Y, Okushima Y, Theologis A, Tasaka M. Tissue-specific expression of stabilized SOLITARY-ROOT/IAA14 alters lateral root development in Arabidopsis. Plant J. 2005:44(3):382–395. 10.1111/j.1365-313X.2005.02537.x [DOI] [PubMed] [Google Scholar]
- Fukaki H, Tameda S, Masuda H, Tasaka M. Lateral root formation is blocked by a gain-of-function mutation in the SOLITARY-ROOT/IAA14 gene of Arabidopsis. Plant J. 2002:29(2):153–168. 10.1046/j.0960-7412.2001.01201.x [DOI] [PubMed] [Google Scholar]
- Funakoshi M, Hochstrasser M. Small epitope-linker modules for PCR-based C-terminal tagging in Saccharomyces cerevisiae. Yeast 2009:26(3):185–192. 10.1002/yea.1658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallie DR, Kado CI. A translational enhancer derived from tobacco mosaic virus is functionally equivalent to a Shine-Dalgarno sequence. Proc Natl Acad Sci U S A. 1989:86(1):129–132. 10.1073/pnas.86.1.129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallie DR, Lucas WJ, Walbot V. Visualizing mRNA expression in plant protoplasts: factors influencing efficient mRNA uptake and translation. Plant Cell 1989:1(3):301–311. 10.1105/tpc.1.3.301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goh T, Kasahara H, Mimura T, Kamiya Y, Fukaki H. Multiple AUX/IAA-ARF modules regulate lateral root formation: the role of Arabidopsis SHY2/IAA3-mediated auxin signalling. Philos Trans R Soc Lond B Biol Sci. 2012:367(1595):1461–1468. 10.1098/rstb.2011.0232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon SP, Heisler MG, Reddy GV, Ohno C, Das P, Meyerowitz EM. Pattern formation during de novo assembly of the Arabidopsis shoot meristem. Development 2007:134(19):3539–3548. 10.1242/dev.010298 [DOI] [PubMed] [Google Scholar]
- Grunewald W, De Smet I, Lewis DR, Löfke C, Jansen L, Goeminne G, Vanden Bossche R, Karimi M, De Rybel B, Vanholme B, et al. Transcription factor WRKY23 assists auxin distribution patterns during Arabidopsis root development through local control on flavonol biosynthesis. Proc Natl Acad Sci U S A. 2012:109(5):1554–1559. 10.1073/pnas.1121134109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grunewald W, Karimi M, Wieczorek K, Van de Cappelle E, Wischnitzki E, Grundler F, Inzé D, Beeckman T, Gheysen G. A role for AtWRKY23 in feeding site establishment of plant-parasitic nematodes. Plant Physiol. 2008:148(1):358–368. 10.1104/pp.108.119131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haecker A, Gross-Hardt R, Geiges B, Sarkar A, Breuninger H, Herrmann M, Laux T. Expression dynamics of WOX genes mark cell fate decisions during early embryonic patterning in Arabidopsis thaliana. Development 2004:131(3):657–668. 10.1242/dev.00963 [DOI] [PubMed] [Google Scholar]
- Hu Y, Xie Q, Chua NH. The Arabidopsis auxin-inducible gene ARGOS controls lateral organ size. Plant Cell 2003:15(9):1951–1961. 10.1105/tpc.013557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeuchi M, Sugimoto K, Iwase A. Plant callus: mechanisms of induction and repression. Plant Cell 2013:25(9):3159–3173. 10.1105/tpc.113.116053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imayoshi I, Kageyama R. bHLH factors in self-renewal, multipotency, and fate choice of neural progenitor cells. Neuron 2014:82(1):9–23. 10.1016/j.neuron.2014.03.018 [DOI] [PubMed] [Google Scholar]
- Kareem A, Durgaprasad K, Sugimoto K, Du Y, Pulianmackal AJ, Trivedi ZB, Abhayadev PV, Pinon V, Meyerowitz EM, Scheres B, et al. PLETHORA Genes control regeneration by a two-step mechanism. Curr Biol. 2015:25(8):1017–1030. 10.1016/j.cub.2015.02.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JY, Yang W, Forner J, Lohmann JU, Noh B, Noh YS. Epigenetic reprogramming by histone acetyltransferase HAG1/AtGCN5 is required for pluripotency acquisition in Arabidopsis. EMBO J. 2018:37(20):e98726. 10.15252/embj.201798726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavy M, Estelle M. Mechanisms of auxin signaling. Development 2016:143(18):3226–3229. 10.1242/dev.131870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee HW, Cho C, Kim J. Lateral organ boundaries domain16 and 18 act downstream of the AUXIN1 and LIKE-AUXIN3 auxin influx carriers to control lateral root development in Arabidopsis. Plant Physiol. 2015:168(4):1792–1806. 10.1104/pp.15.00578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee HW, Kang NY, Pandey SK, Cho C, Lee SH, Kim J. Dimerization in LBD16 and LBD18 transcription factors is critical for lateral root formation. Plant Physiol. 2017:174(1):301–311. 10.1104/pp.17.00013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee HW, Kim MJ, Kim NY, Lee SH, Kim J. LBD18 acts as a transcriptional activator that directly binds to the EXPANSIN14 promoter in promoting lateral root emergence of Arabidopsis. Plant J. 2013:73(2):212–224. 10.1111/tpj.12013 [DOI] [PubMed] [Google Scholar]
- Lee HW, Kim NY, Lee DJ, Kim J. LBD18/ASL20 regulates lateral root formation in combination with LBD16/ASL18 downstream of ARF7 and ARF19 in Arabidopsis. Plant Physiol. 2009:151(3):1377–1389. 10.1104/pp.109.143685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee K, Park OS, Seo PJ. Arabidopsis ATXR2 deposits H3K36me3 at the promoters of LBD genes to facilitate cellular dedifferentiation. Sci Signal. 2017:10(507):eaan0316. 10.1126/scisignal.aan0316 [DOI] [PubMed] [Google Scholar]
- Lin XL, Niu D, Hu ZL, Kim DH, Jin YH, Cai B, Liu P, Miura K, Yun DJ, Kim WY, et al. An Arabidopsis SUMO E3 ligase, SIZ1, negatively regulates photomorphogenesis by promoting COP1 activity. PLoS Genet. 2016:12(4):e1006016. 10.1371/journal.pgen.1006016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Hu X, Qin P, Prasad K, Hu Y, Xu L. The WOX11-LBD16 pathway promotes pluripotency acquisition in callus cells during de novo shoot regeneration in tissue culture. Plant Cell Physiol. 2018:59(4):734–743. 10.1093/pcp/pcy010 [DOI] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods 2001:25(4):402–408. 10.1006/meth.2001.1262 [DOI] [PubMed] [Google Scholar]
- Okushima Y, Fukaki H, Onoda M, Theologis A, Tasaka M. ARF7 and ARF19 regulate lateral root formation via direct activation of LBD/ASL genes in Arabidopsis. Plant Cell 2007:19(1):118–130. 10.1105/tpc.106.047761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powers SK, Holehouse AS, Korasick DA, Schreiber KH, Clark NM, Jing H, Emenecker R, Han S, Tycksen E, Hwang I, et al. Nucleo-cytoplasmic partitioning of ARF proteins controls auxin responses in Arabidopsis thaliana. Mol Cell. 2019:76(1):177–190.e175. 10.1016/j.molcel.2019.06.044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prát T, Hajný J, Grunewald W, Vasileva M, Molnár G, Tejos R, Schmid M, Sauer M, Friml J. WRKY23 Is a component of the transcriptional network mediating auxin feedback on PIN polarity. PLoS Genet. 2018:14(1):e1007177. 10.1371/journal.pgen.1007177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radhakrishnan D, Kareem A, Durgaprasad K, Sreeraj E, Sugimoto K, Prasad K. Shoot regeneration: a journey from acquisition of competence to completion. Curr Opin Plant Biol. 2018:41:23–31. 10.1016/j.pbi.2017.08.001 [DOI] [PubMed] [Google Scholar]
- Robinson CR, Sauer RT. Optimizing the stability of single-chain proteins by linker length and composition mutagenesis. Proc Natl Acad Sci U S A. 1998:95(11):5929–5934. 10.1073/pnas.95.11.5929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shang B, Xu C, Zhang X, Cao H, Xin W, Hu Y. Very-long-chain fatty acids restrict regeneration capacity by confining pericycle competence for callus formation in Arabidopsis. Proc Natl Acad Sci U S A. 2016:113(18):5101–5106. 10.1073/pnas.1522466113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skoog F, Miller CO. Chemical regulation of growth and organ formation in plant tissues cultured in vitro. Symp Soc Exp Biol. 1957:54(11):118–130. [PubMed] [Google Scholar]
- Sparkes IA, Runions J, Kearns A, Hawes C. Rapid, transient expression of fluorescent fusion proteins in tobacco plants and generation of stably transformed plants. Nat Protoc. 2006:1(4):2019–2025. 10.1038/nprot.2006.286 [DOI] [PubMed] [Google Scholar]
- Sugimoto K, Gordon SP, Meyerowitz EM. Regeneration in plants and animals: dedifferentiation, transdifferentiation, or just differentiation? Trends Cell Biol. 2011:21(4):212–218. 10.1016/j.tcb.2010.12.004 [DOI] [PubMed] [Google Scholar]
- Sugimoto K, Jiao Y, Meyerowitz EM. Arabidopsis regeneration from multiple tissues occurs via a root development pathway. Dev Cell. 2010:18(3):463–471. 10.1016/j.cell.2008.01.040 [DOI] [PubMed] [Google Scholar]
- Sugiyama M. Partnership for callusing. Nat Plants. 2018:4(2):69–70. 10.1038/s41477-018-0104-2 [DOI] [PubMed] [Google Scholar]
- Tatematsu K, Kumagai S, Muto H, Sato A, Watahiki MK, Harper RM, Liscum E, Yamamoto KT. MASSUGU2 encodes Aux/IAA19, an auxin-regulated protein that functions together with the transcriptional activator NPH4/ARF7 to regulate differential growth responses of hypocotyl and formation of lateral roots in Arabidopsis thaliana. Plant Cell 2004:16(2):379–393. 10.1105/tpc.018630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tessadori F, Chupeau MC, Chupeau Y, Knip M, Germann S, van Driel R, Fransz P, Gaudin V. Large-scale dissociation and sequential reassembly of pericentric heterochromatin in dedifferentiated Arabidopsis cells. J Cell Sci. 2007:120(Pt 7):1200–1208. 10.1242/jcs.000026 [DOI] [PubMed] [Google Scholar]
- Tian Q, Reed JW. Control of auxin-regulated root development by the Arabidopsis thaliana SHY2/IAA3 gene. Development 1999:126(4):711–721. 10.1242/dev.126.4.711 [DOI] [PubMed] [Google Scholar]
- Toledo-Ortiz G, Huq E, Quail PH. The Arabidopsis basic/helix-loop-helix transcription factor family. Plant Cell 2003:15(8):1749–1770. 10.1105/tpc.013839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uehara T, Okushima Y, Mimura T, Tasaka M, Fukaki H. Domain II mutations in CRANE/IAA18 suppress lateral root formation and affect shoot development in Arabidopsis thaliana. Plant Cell Physiol. 2008:49(7):1025–1038. 10.1093/pcp/pcn079 [DOI] [PubMed] [Google Scholar]
- Valvekens D, Van Montagu M, Van Lijsebettens M. Agrobacterium tumefaciens-mediated transformation of Arabidopsis thaliana root explants by using kanamycin selection. Proc Natl Acad Sci U S A. 1988:85(15):5536–5540. 10.1073/pnas.85.15.5536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang FX, Shang GD, Wang JW. Towards a hierarchical gene regulatory network underlying somatic embryogenesis. Trends Plant Sci. 2022:27(12):1209–1217. 10.1016/j.tplants.2022.06.002 [DOI] [PubMed] [Google Scholar]
- Wang S, Tiwari SB, Hagen G, Guilfoyle TJ. AUXIN RESPONSE FACTOR7 restores the expression of auxin-responsive genes in mutant Arabidopsis leaf mesophyll protoplasts. Plant Cell 2005:17(7):1979–1993. 10.1105/tpc.105.031096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang ZP, Xing HL, Dong L, Zhang HY, Han CY, Wang XC, Chen QJ. Egg cell-specific promoter-controlled CRISPR/Cas9 efficiently generates homozygous mutants for multiple target genes in Arabidopsis in a single generation. Genome Biol. 2015:16(1):144. 10.1186/s13059-015-0715-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams L, Zhao J, Morozova N, Li Y, Avivi Y, Grafi G. Chromatin reorganization accompanying cellular dedifferentiation is associated with modifications of histone H3, redistribution of HP1, and activation of E2F-target genes. Dev Dyn. 2003:228(1):113–120. 10.1002/dvdy.10348 [DOI] [PubMed] [Google Scholar]
- Xu C, Cao H, Xu E, Zhang S, Hu Y. Genome-wide identification of Arabidopsis LBD29 target genes reveals the molecular events behind auxin-induced cell reprogramming during callus formation. Plant Cell Physiol. 2018:59(4):744–755. 10.1093/pcp/pcx168 [DOI] [PubMed] [Google Scholar]
- Xu C, Cao H, Zhang Q, Wang H, Xin W, Xu E, Zhang S, Yu R, Yu D, Hu Y. Control of auxin-induced callus formation by bZIP59-LBD complex in Arabidopsis regeneration. Nat Plants. 2018:4(2):108–115. 10.1038/s41477-017-0095-4 [DOI] [PubMed] [Google Scholar]
- Xu K, Liu J, Fan M, Xin W, Hu Y, Xu C. A genome-wide transcriptome profiling reveals the early molecular events during callus initiation in Arabidopsis multiple organs. Genomics 2012:100(2):116–124. 10.1016/j.ygeno.2012.05.013 [DOI] [PubMed] [Google Scholar]
- Yoo SD, Cho YH, Sheen J. Arabidopsis mesophyll protoplasts: a versatile cell system for transient gene expression analysis. Nat Protoc. 2007:2(7):1565–1572. 10.1038/nprot.2007.199 [DOI] [PubMed] [Google Scholar]
- Zhai N, Pan X, Zeng M, Xu L. Developmental trajectory of pluripotent stem cell establishment in Arabidopsis callus guided by a quiescent center-related gene network. Development 2023:150(5):dev200879. 10.1242/dev.200879 [DOI] [PubMed] [Google Scholar]
- Zhai N, Xu L. Pluripotency acquisition in the middle cell layer of callus is required for organ regeneration. Nat Plants. 2021:7(11):1453–1460. 10.1038/s41477-021-01015-8 [DOI] [PubMed] [Google Scholar]
- Zhang TQ, Lian H, Zhou CM, Xu L, Jiao Y, Wang JW. A two-step model for de novo activation of WUSCHEL during plant shoot regeneration. Plant Cell 2017:29(5):1073–1087. 10.1105/tpc.16.00863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S, Yu R, Yu D, Chang P, Guo S, Yang X, Liu X, Xu C, Hu Y. The calcium signaling module CaM-IQM destabilizes IAA-ARF interaction to regulate callus and lateral root formation. Proc Natl Acad Sci U S A. 2022:119(27):e2202669119. 10.1073/pnas.2202669119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J, Morozova N, Williams L, Libs L, Avivi Y, Grafi G. Two phases of chromatin decondensation during dedifferentiation of plant cells: distinction between competence for cell fate switch and a commitment for S phase. J Biol Chem. 2001:276(25):22772–22778. 10.1074/jbc.M101756200 [DOI] [PubMed] [Google Scholar]
- Zuo J, Niu QW, Chua NH. An estrogen receptor-based transactivator XVE mediates highly inducible gene expression in transgenic plants. Plant J. 2000:24(2):265–273. 10.1046/j.1365-313x.2000.00868.x [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data underlying this article are available in the article and in its online supplementary material.