Abstract
We investigated the role in cell morphogenesis and pathogenicity of the Candida albicans GPR1 gene, encoding the G protein-coupled receptor Gpr1. Deletion of C. albicans GPR1 has only minor effects in liquid hypha-inducing media but results in strong defects in the yeast-to-hypha transition on solid hypha-inducing media. Addition of cAMP, expression of a constitutively active allele of the Gα protein Gpa2 or of the catalytic protein kinase A subunit TPK1 restores the wild-type phenotype of the CaGPR1-deleted strain. Overexpression of HST7, encoding a component of the mitogen-activated protein kinase pathway, does not suppress the defect in filamentation. These results indicate that CaGpr1 functions upstream in the cAMP–protein kinase A (PKA) pathway. We also show that, in the presence of glucose, CaGpr1 is important for amino acid-induced transition from yeast to hyphal cells. Finally, as opposed to previous reports, we show that CaGpa2 acts downstream of CaGpr1 as activator of the cAMP–PKA pathway but that deletion of neither CaGpr1 nor CaGpa2 affects glucose-induced cAMP signaling. In contrast, the latter is abolished in strains lacking CaCdc25 or CaRas1, suggesting that the CaCdc25-CaRas1 rather than the CaGpr1-CaGpa2 module mediates glucose-induced cAMP signaling in C. albicans.
INTRODUCTION
Candida albicans is the most prevalent opportunistic fungal pathogen in humans, causing various forms of candidiasis ranging from superficial mucosal infections to life-threatening systemic diseases, predominantly in patients with a compromised immune system (Odds, 1996). It is a pleiomorphic organism, undergoing reversible morphogenetic transitions between budding yeast, pseudohyphal, and hyphal growth forms. The ability to make the transition from the yeast form to the hyphal form has been considered a major requirement for virulence (Leberer et al., 1997; Lo et al., 1997; Saville et al., 2003). The yeast-to-hypha transition of C. albicans can be triggered in vitro by a variety of factors, including carbohydrates, amino acids, salts, pH changes, temperature increases, starvation, serum, and growth within a matrix (Ernst, 2000). These various hyphal inducers trigger a wide range of signal transduction pathways involved in morphogenesis. The two best studied pathways are the mitogen-activated protein kinase (MAPK) pathway and the cAMP-protein kinase A (PKA) pathway. The MAPK pathway, consisting of the kinases Cst20, Ste11, Hst7, and Cek1, regulates the activity of the transcription factor Cph1 (Liu et al., 1994; Köhler and Fink, 1996; Leberer et al., 1996; Sanchez-Martinez and Pérez-Martin, 2002), whereas the cAMP–PKA pathway, comprising CaCdc35, CaPde2, CaCap1, CaTpk1, CaTpk2, and CaBcy1, regulates the activity of the transcription factor Efg1 (Stoldt et al., 1997; Brown and Gow, 1999; Borges-Walmsley and Walmsley, 2000; Sonneborn et al., 2000; Bahn and Sundstrom, 2001; Bockmühl et al., 2001; Leberer et al., 2001; Rocha et al., 2001; Cassola et al., 2004; Doedt et al., 2004). Both the MAPK and cAMP–PKA pathways are activated by a common upstream factor, CaRas1 (Feng et al., 1999; Leberer et al., 2001), which is possibly activated by CaCdc25 as in Saccharomyces cerevisiae (Goldberg et al., 1993). Recently, Gpa2 has been described as part of the MAPK pathway in C. albicans (Sanchez-Martinez and Pérez-Martin, 2002), whereas in S. cerevisiae Gpa2 has been shown to act through the cAMP–PKA pathway (Kubler et al., 1997; Lorenz and Heitman, 1997).
Besides the MAPK and cAMP–PKA pathways, additional pathways influence the morphogenetic transition. The matrix embedding pathway regulates filamentous growth through the activity of the Czf1 transcription factor (Brown et al., 1999), the extracellular pH-induced pathway regulates morphology through the Rim101 transcription factor (Davis et al., 2000) and the Tup1-Nrg1–mediated pathway represses filamentation independent of Efg1 and Cph1 (Braun and Johnson, 1997; Braun et al., 2001; Murad et al., 2001).
Despite extensive studies and a growing number of identified genes involved in the process of morphogenesis, the early stages of inducer sensing are not well understood. An open fundamental question remaining to be answered is how the external signals are sensed and how the sensors activate the various signal transduction pathways. The molecular dissection of morphogenesis in C. albicans has been based on the strong evolutionary conservation of fungal signal transduction pathways. Many C. albicans signal transduction genes important for filamentous growth have been isolated using genetic screens in S. cerevisiae mutants affected in morphogenesis. In S. cerevisiae, different signal transduction pathways have been shown to be important for the morphogenetic transitions to pseudohyphal growth in diploid cells and invasive growth in haploid cells (Gagiano et al., 2002). These are the MAPK (pheromone) pathway, the cAMP–PKA pathway, the ammonium sensing pathway (Lorenz and Heitman, 1998), and the amino acid sensing pathway (Klasson et al., 1999; Pan et al., 2000). Deletion of GPR1, encoding a G protein-coupled receptor (GPCR), or GPA2, encoding its cognate Gα protein, which function upstream in the cAMP–PKA pathway, results in a defect in haploid invasive growth. On the other hand, deletion of SSY1 or PTR3, two components of the SPS amino acid sensing complex, results in enhanced invasive growth (Lorenz and Heitman, 1997; Klasson et al., 1999; Lorenz et al., 2000).
The C. albicans homologue of the S. cerevisiae Ssy1 amino acid sensor has recently been characterized. Like its Ssy1 homologue in S. cerevisiae, the CaCsy1 amino acid sensor is important for the induction of genes encoding amino acid permeases. Moreover, deletion of this gene results in a defect in filamentation in serum- and amino acid-based media (Brega et al., 2004). In an analogous way, we have investigated the function of CaGpr1, the putative sensor protein activating the cAMP–PKA pathway, in analogy with its function in S. cerevisiae. In the latter yeast, Gpr1 was shown to function upstream in the cAMP pathway (Xue et al., 1998) and to be required for glucose and sucrose activation of cAMP synthesis (Yun et al., 1997; Kraakman et al., 1999). Recent work using substituted cysteine accessibility method (SCAM) analysis has provided strong evidence for direct binding of glucose and sucrose as agonist ligands to Gpr1. Mannose acts as antagonist, whereas closely related sugars such as fructose and galactose do not interact with Gpr1 (Lemaire et al., 2004).
We show that the C. albicans Gpr1 homologue is important for cell and colony morphology on solid media and provide evidence that it functions upstream in the cAMP–PKA pathway. In contrast to a previous report (Sanchez-Martinez and Pérez-Martin, 2002), our results also strongly suggest that CaGpa2 acts in the cAMP–PKA pathway, downstream of CaGpr1, as is the case in S. cerevisiae. While this work was being prepared for publication, part of the results we present were published in a recent article (Miwa et al., 2004). They are consistent with our results.
MATERIALS AND METHODS
Candida Strains and Growth Conditions
All strains used in this work are described in Table 1. S. cerevisiae and C. albicans strains were grown in YPD (1% yeast extract, 2% Bacto-peptone, and 2% glucose) medium with the appropriate auxotrophic requirements.
Table 1.
Plasmids and C. albicans strains used in this study
| Strain | Genotype | Source |
|---|---|---|
| SC5314 | Wild type | Fonzi and Irwin (1993) |
| CAI4 | ura3:: imm434/ura3:: imm434 | Fonzi and Irwin (1993) |
| LDR1 | GPR1/gpr1Δ::hisG-URA3-hisG | This study |
| LDR2 | GPR1/gpr1Δ::hisG | This study |
| LDR8 | gpr1Δ::hisG/gpr1Δ::hisG-URA3-hisG | This study |
| LDR8–5 | gpr1Δ::hisG/gpr1Δ::hisG | This study |
| LR2 | gpr1Δ::hisG/gpr1Δ::hisG GPR1-URA3 | This study |
| NM2 | GPA2/gpa2Δ::hisG-URA3-hisG | This study |
| NM4 | GPA2/gpa2Δ::hisG | This study |
| NM6 | gpa2Δ::hisG/gpa2Δ::hisG-URA3-hisG | This study |
| NM8 | gpa2Δ::hisG/gpa2Δ::hisG | This study |
| NM29 | gpa2Δ::hisG/gpa2Δ::hisG GPA2-URA3 | This study |
| CCM12 | gpa2Δ::hisG/gpa2Δ::hisG-URA3-hisG | Sanchez-Martinez and Pérez-Martin (2002) |
| CCS14 | gpa2Δ::hisG/gpa2Δ::hisG | Sanchez-Martinez and Pérez-Martin (2002) |
| JKC131 | hst7Δ::hisG/hst7Δ::hisG-URA3-hisG | Köhler and Fink (1996) |
| MM1 | hst7Δ::hisG/hst7Δ::hisG GPR1/gpr1Δ::hisG-URA3-hisG | This study |
| MM2 | hst7Δ::hisG/hst7Δ::hisG | This study |
| MM5 | hst7Δ::hisG/hst7Δ::hisG gpr1Δ::hisG/gpr1Δ::hisG-URA3-hisG | This study |
| M231 | URA3-PCK1p::TPK1/tpk1Δ::hisG tpk2Δ::hisG/tpk2Δ::hisG | Bockmühl et al. (2001) |
| C52 | CAI4 + pCaEX (integrated) | This study |
| C53 | CAI4 + pCaEX/CaGPA2Q355L (integrated) | This study |
| C54 | LDR8–5 + pCaEX (integrated) | This study |
| C55 | LDR8–5 + pCaEX/CaGPA2Q355L (integrated) | This study |
| CR216 | cdc35::hisG/cdc35::hisG-URA3-hisG | Rocha et al. (2001) |
| CDH107 | ras1Δ::hisG/ras1Δ::hisG-URA3-hisG | Leberer et al. (2001) |
| BMY16 | cdc25::UAU1/CDC25 | Enloe et al. (2000) |
| BMY15 | cdc25::UAU1/cdc25::URA3 | Enloe et al. (2000) |
| C. albicans transformation vectors | ||
| pRC2312 | URA3-, LEU2-marked CaARS vector | Cannon et al. (1992) |
| pBI-HAHYD | PCK1p-HA-EFG1 fusion in pRC2312 | Stoldt et al. (1997) |
| pBI/His-C.TPK1 | PCK1p-His6-TPK1 fusion in pRC2312 | Bockmühl et al. (2001) |
| pBI-TPK | PCK1p-HA-TPK2 fusion in pRC2312 | Sonneborn et al. (2000) |
| PYPB1-ADHpr-HST7 | HST7 overexpression construct | Leberer et al. (1996) |
| pNM3 | GPR1-URA3 reintegration | This study |
| pNM11 | GPA2-URA3 reintegration | This study |
| pNM5 | GPR1::GFP fusion construct | This study |
| pCaEX | Care et al. (1999) | |
| pCaEX/CaGPA2Q355L | This study |
All strains have CAI4 as background except the cdc25 mutants that have BWP17 as background, which is a derivative of CAI4 (Wilson et al., 1999).
To determine the ability to undergo the yeast-to-hypha transition, budding C. albicans cells were grown overnight at 28°C with vigorous shaking in YPD medium. Ten to 50 cells were then incubated per plate for the indicated times at 30 or 37°C on different media. Solid Spider medium contains 1% (wt/vol) nutrient broth, 0.2% (wt/vol) K2HPO4, 1.35% (wt/vol) agar, and 1% (wt/vol) mannitol as a carbon source (Liu et al., 1994). Lee's medium is as described previously (Lee et al., 1975), and for serum plates, 10% (vol/vol) fetal calf serum (FCS) was added to YPD at 50°C after autoclaving. Cultivation under embedded growth conditions was performed as described in Brown et al. (1999). Synthetic low dextrose (SLD) medium is 0.17% YNB medium containing 0.5% (NH4)2 SO4 and 0.1% glucose.
Plasmids for Disruption and Reintegration of CaGPR1 and CaGPA2
The plasmids for disruption of CaGPR1 and CaGPA2 were based on vector pMB7A (Fonzi and Irwin, 1993). The CaGPR1 gene, including promoter and terminator sequences, was amplified by polymerase chain reaction (PCR) from SC5314 genomic DNA with primers GPR1for1 (5′-GCgaattcACACCGTGACAGAAATTGATG-3′) and GPR1rev1 (5′-GCaagcttTGAATACCAATCAGTACTTGC-3′) and cloned into an EcoRI/HindIII-restricted pUC19 to obtain pUC19/CaGPR1. This plasmid was digested with PstI and NcoI to replace the GPR1 coding sequence with the PstI/AflII, hisG-URA3-hisG–containing fragment from pMB7A. The final plasmid, pUC19/Cagpr1::hisG-URA3-hisG, was digested with DraIII to generate the 6.3-kb fragment containing 1134 base pairs 5′ and 880 base pairs 3′ CaGPR1 homologous sequences.
For site-directed reintegration of CaGPR1, the CaGPR1 gene and its promoter were amplified with primers CaGPR1FOR (5′-ATGGCGGAAGTGGTACCAGTGG-3′) and CaGPR1REV (5′-TATGGGATCCTCATTTGAAGGTG-3′) and cloned into the blunted XbaI-digested pMB7-A plasmid, resulting in a new plasmid called pNM3.
The CaGPA2 gene, including promoter and terminator sequences, was amplified by PCR from SC5314 genomic DNA with primers CaGPA2R (5′-GCggatccTGCTAAACCTAATTGAGCATC-3′) and CaGPA2F (5′-GCgtcgacCCATCTAATAGCAATTGTCATTG-3′) and cloned into BamHI/SalI-restricted pUC19 to obtain pUC19/CaGPA2. This plasmid was digested with StyI and then treated with Klenow polymerase to replace the GPA2 coding sequence with the PvuII/StuI, hisG-URA3-hisG containing fragment from pMB7A. The final plasmid, pUC19/Cagpa2::hisG-URA3-hisG, was digested with AflII/HindIII to generate the 5.3-kb fragment containing 581 base pairs 5′ and 681 base pairs 3′ CaGPA2 homologous sequences. The plasmid for directed reintegration of CaGPA2 was constructed from pMB7-A in two steps by replacement of hisG sequences with CaGPA2 and GPA2 3′-flanking region. In the first step, CaGPA2 with adjusted 5′-flanking region was amplified by PCR by using genomic DNA of SC5314 as template with primers CaGPA2F and RIGPA2R (5′-ATAAgctagcTATGGAGTTTTAATATGGTGC-3′) and cloned into pMB7A cut with SalI/NheI to create pNM10. For the second step, a 625-base pair PvuII/SacI fragment of pUC19/CaGPA2 composed of sequences from the 3′ terminus of GPA2 (53 nucleotides) and adjusted 3′-flanking region was inserted into AfeI/SacI-digested pNM10 to obtain pNM11. The latter plasmid was digested with AflII and HindIII, releasing a 4.4-kb fragment comprised of CaGPA2 and CaURA3 flanked by GPA2 5′ (577 base pairs) and 3′ (572 base pairs) sequences and transformed into the gpa2Δ/gpa2Δ strain.
Deletion of CaGPR1 and CaGPA2
Disruption of the two allelic copies of CaGPR1 and of CaGPA2 was obtained using the two-step procedure described by Fonzi and Irwin (1993). The disruption cassettes obtained by DraIII digestion of pUC19/Cagpr1::hisG-URA3-hisG and by AflII/HindIII digestion of pUC19/Cagpa2::hisG-URA3-hisG were transformed into the ura- CAI4 strain. Transformants were selected on SD medium without uridine, and integration of the cassette into the respective loci was confirmed by PCR by using primers GPR1diagORF (5′-GGCGGTCATCATGGAGGT-3′), and GPR1diagFLS (5′-CAACCCACAACCCACCAC-3′), DIAGHISG (5′-GCGTAAGCGGGTGTTGTC-3′) for CaGPR1, and primers DIAGPA2FOR (5′-TGGATAATCGGTAATATCTT-3′) and DIAGPA2REV (5′-CTAATCAACCTTCAAGCTATA-3′) for GPA2. Correct disruption also was confirmed by Southern blot analysis by using 898-base pair XbaI/PstI fragment of pUC19/CaGPR1 and 576-base pair HindIII/StyI fragment of pUC19/CaGPA2 as probes for, respectively, GPR1 and GPA2. Spontaneous ura- derivatives were then selected on medium containing 5-fluoroorotic acid. Clones that had lost the URA3 gene by intrachromosomal recombination, mediated by the hisG repeats, were screened by Southern analysis. The second allele was then deleted by repeating the same procedure.
To reintroduce a wild-type copy of the GPR1 gene in the gpr1Δ/gpr1Δ mutant, this strain was grown on 5-fluoroorotic acid to obtain a ura- derivative and then transformed with a KpnI restriction fragment of pNM3 containing the CaGPR1-URA3-hisG sequence. Site-directed reintegration was confirmed by PCR and Southern analysis. To reintroduce a wild-type copy of the GPA2 gene in the gpa2Δ/gpa2Δ mutant, a ura- derivative was transformed with a 4.4-kb AflII/HindIII restriction fragment of pNM11.
Other Plasmids
Candida expression plasmids that we have used are summarized in Table 1. For two-hybrid analysis, pGBT9 and pGAD424 were used. The C-terminal part of CaGPR1 was isolated with primers CaGPR1CTAIL (5′-TCGAATTCCCTTGGAGAAACACTGCC-3′) and CaGPR1REV (5′-TATGGGATCCTCATTTGAAGGTG-3′). The EcoRI and BamHI sites are underlined. The complete CaGPA2 gene was isolated from genomic DNA with the use of two oligonucleotides: 5′ ATggatccCCATGGGTTCTTGTGCTTCG 3′ and 5′ TAActgcagTGCATAAAACTATAAAATACCAC 3′. The BamHI and PstI are in small letters and the start and stop codons are underlined.
The constitutively active allele of GPA2, GPA2Q355L, was constructed by amplification of the GPA2 gene with primers 5′-ATggatccCCATGGGTTCTTGTGCTTCG-3′ and 5′-TAActgcagTGCATAAAACTATAAAATACCAC-3′ to obtain a 1520-base pairs product. The BamHI/PstI-digested PCR product was ligated into a BamHI/PstI-digested pUC18 vector. The GPA2 mutated allele was obtained by site-directed mutagenesis with oligonucleotides GPA2MUT1 (5′-GATGTTGGTGGTCTAAGGTCAGAAAGAAAAAAATGGATCAATTG-3′) and GPA2MUT2 (5′-CAATTGATCCATTTTTTTCTTTCTGACCTTAGACCACCAACATC-3′). The mutation was confirmed by sequencing. The mutated GPA2 allele was then cut out of pUC18 by using BamHI and PstI and ligated into the pCaEX vector (Care et al., 1999) digested with the same restriction enzymes. The resulting plasmid, pCaEX/GPA2Q355L, was linearized with BglII for transformation into C. albicans strains.
Construction of the GPR1-Green Fluorescent Protein (GFP) Fusion
We constructed a shuttle vector expressing the GPR1-GFP fusion construct on a replicative plasmid. By using pMG1602 (Gerami-Nejad et al., 2001) as a template, the GFP-URA3 fragment was PCR-amplified using primers GPR1FPFO and GPR1FPRE (5′-GATAAAGCAGATGATAATGATGATGGAGGATTGGATTTGATGGATTTCTCAAAAAAGGACCCCCAATGGGTGGTGGTTCTAAAGGTGAAGAATTATT-3′ and 5′-TTGAAAAAATTCTTTGATTTTATGGACTCCTCATTTGAAGGTGTTTTATATACTGTGAAAAATTGTGATTCTAGAAGGACCACCTTTGATTG-3′, respectively), digested with PpuMI and PvuII, and ligated into PpuMI/BsaBI digested pNM3 to produce plasmid pNM4y. The 4.2-kb KpnI/KpnI fragment of plasmid pNM4 containing in frame fused GPR1 and GFP was inserted into the unique KpnI restriction site of pRC2312 (Cannon et al., 1992), resulting in autonomously replicating pNM5y. The wild-type strain (CAI4) and an ura- derivative of gpr1Δ/gpr1Δ were transformed with these plasmids by using a protoplast transformation protocol (Cregg et al., 1985). Transformants were isolated on SC-ura.
Fluorescent and Laser Scanning Microscopy
Strains were grown overnight in minimal medium with 2% glucose, inoculated into fresh minimal or YPD media to a starting OD600 of 1.0, and grown at 30°C with shaking for 2–4 h. Before image acquisition, cells were washed and resuspended in 0.5× minimal medium. Slides for microscopy were prepared by adding 5 μl of washed culture to a slide and affixing the coverslip with rubber cement. Confocal microscopy was performed using a LSM 510 U (Carl Zeiss, Zaventem, Belgium). Endocytosis of fluorescent dye FM4-64 was performed as described by Wiederkehr et al., (2000). Cells were pregrown in minimal medium containing 0.67% YNB with ammonium sulfate (MM) and 0.05% glucose + 1% glycerol as a carbon source. After FM4-64 internalization step, cells were resuspended in prewarmed YPglycerol, MM/2% glucose, MM/1% glycerol, MM/1% glycerol/10 mM alanine, or MM/1% glycerol/10 mM methionine in the presence or absence of cycloheximide (10 μg/ml). The protein internalization was followed over a 90-min time course with a Zeiss Axioplan 2 fluorescence microscope, and images were scanned with a Axio-Cam HRm camera by using Axiovision 3.0 software (Carl Zeiss, Thornwood, NY). Enhanced green fluorescent protein was excited at 488 nm or at 514 nm (for yellow isoform of GFP) and detected using the appropriate filters: BP 505–530 or LP 530.
Determination of cAMP Level
For determination of intracellular cAMP levels, cells were grown according to the procedure of Alspaugh et al. (2002). Cells were grown in YPD at 30°C for 20 h. Cells were resuspended in YPglycerol at OD600 = 0.05 and further incubated at 30°C for 20 h. Cells were collected and washed with 10 mM MES, 0.1 mM EDTA, pH 6. Cells (900 mg) were resuspended in 24 ml of buffer and incubated at 37°C for 2 h. After this incubation, either 100 mM glucose or 10% serum (final concentrations) was added to the cells. Sampling, extraction, and cAMP determination were performed as described previously for S. cerevisiae (Colombo et al., 1998).
Invasion Assay
To test the ability of C. albicans wild-type and GPR1 mutant strains to adhere to and invade human tissue, the fungal cells were embedded in DMEM and positioned on top of the Caco2 cells or the three-dimensional epithelium model as described by Dieterich et al. (2002). The experiment was conducted with at least three independent skin equivalents for each strain. The strains were incubated for 40 h on the reconstituted tissue before processing for histology ensued. From each tissue sample, one representative slide was taken for quantification of hyphal and yeast or pseudohyphal cells.
Virulence assays in mice were performed essentially as described previously (Van Dijck et al., 2002). Female BALB/c mice weighing 20 g were inoculated in the lateral caudal vein with 106 C. albicans cells suspended in 200 μl of saline. Survival was scored over a period of 40 d. A group of 10 mice per condition were tested. The morphology of the Candida cells in the kidneys, liver, spleen, and lungs was determined by periodic acid shiff (PAS) staining of tissue sections. Briefly, after perfusion with saline of the mice, the different organs were collected, washed in phosphate-buffered saline, and stored in 70% ethanol. After embedding in Paraffin, 7-μm-thick coupes were placed overnight at 37°C. These coupes were then stained with PAS reagent according to the manufacturer (Sigma-Aldrich, St. Louis, MO). Alternatively, C. albicans cellular morphology in the kidneys was performed as described by Csank et al. (1997) with minor modifications. Two days after infection, mice were killed and the kidneys were solubilized in 10% potassium hydroxide solution at 50°C for 1.5 h. After centrifugation at 1500 × g for 10 min, alkaline-resistant sediment was washed once with water and resuspended in water solution of 5 μg/ml Calcofluor white. Cells were viewed with a Zeiss Axioplan 2 fluorescence microscope, and images were scanned with an Axio-Cam HRm camera by using Axiovision 3.0 software (Carl Zeiss).
RESULTS
Gpr1 Is Important for Hyphal Formation in C. albicans
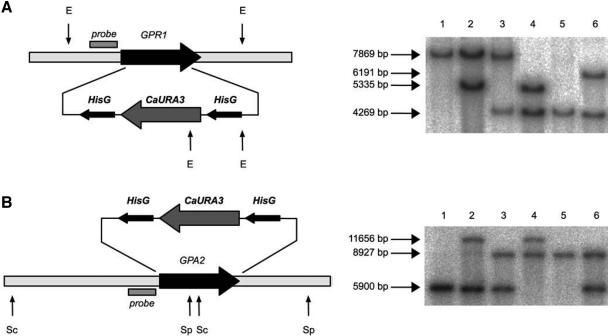
We have previously reported that the C. albicans genome contains one homologue of the S. cerevisiae G protein-coupled receptor Gpr1 (Versele et al., 2001). The C. albicans Gpr1 protein also contains seven membrane-spanning domains but has a longer extracellular N-terminal domain and a shorter third intracellular loop compared with its S. cerevisiae homolog. The open reading frame of CaGpr1 encodes a protein of 823 amino acids. To construct a CaGPR1 deletion strain, the C. albicans CAI4 strain was transformed with the hisG-URA3-hisG cassette flanked by 5′ and 3′ CaGPR1 gene sequences by using the URA-blaster method (Figure 1A). C. albicans cells deleted for both alleles of GPR1 were viable, and their growth rate at 30 and 37°C did not significantly differ compared with wild-type cells (our unpublished data). A CaGPR1 reintegration construct was transformed into the gpr1Δ/gpr1Δ ura- C. albicans strain. Homologous integration also was confirmed by PCR and Southern blot analysis (Figure 1A). The presence of CaGPR1 mRNA in the reintegrant strain was checked by Northern blot analysis (our unpublished data). CaGPR1 is expressed constitutively but at a very low level. No differential expression was observed in all the growth conditions tested (our unpublished data).
Figure 1.
Deletion strategy for C. albicans GPR1 and GPA2. (A) Genetic organization of the CaGPR1 locus. The CaGPR1 open reading frame (black arrow) was replaced with the URA-blaster cassette as described in Materials and Methods. Southern blot analysis of EcoRI-digested C. albicans genomic DNA probed with part of the CaGPR1 promoter as indicated on the figure. Lanes 1, CAI4; 2, LDR1 (GPR1/gpr1Δ::hisG-URA3-hisG); 3, LDR1–6 (GPR1/gpr1Δ::hisG); 4, LDR8 (gpr1Δ::hisG/gpr1Δ::hisG-URA3-hisG); 5, LDR8–5 (gpr1Δ::hisG/gpr1Δ::hisG); and 6, LR2 (gpr1Δ::hisG/gpr1Δ::hisG-riGPR1-URA3). (B) Genetic organization of the CaGPA2 locus. The CaGPA2 open reading frame (black arrow) was replaced with the URA-blaster cassette as described in Materials and Methods. Southern blot analysis of ScaI + SpeI-digested C. albicans genomic DNA probed with part of the CaGPA2 promoter as indicated on the figure. Lanes 1, CAI4; 2, NM2 (GPA2/gpa2Δ::hisG-URA3-hisG); 3, NM4 (GPA2/gpa2Δ::hisG); 4, NM6 (gpa2Δ::hisG/gpa2Δ::hisG-URA3-hisG); 5, NM8 (gpa2Δ::hisG/gpa2Δ::hisG); and 6, NM29 (gpa2Δ::hisG/gpa2Δ::hisG-riGPA2-URA3). Restriction endonucleases E, EcoRI; Sc, ScaI; and Sp, SpeI.
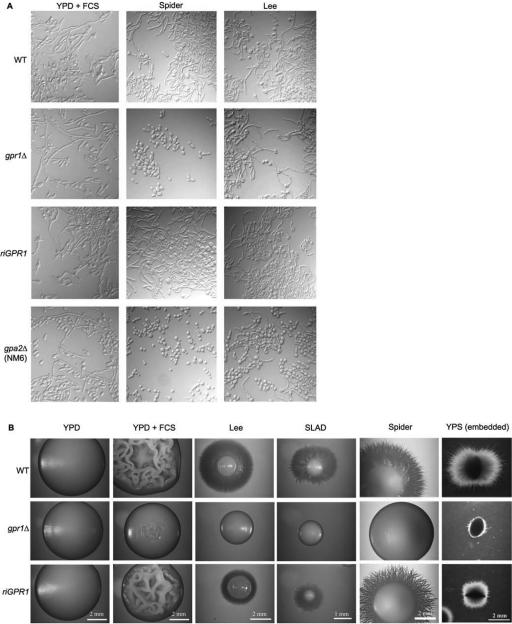
To study the role of CaGPR1 in the morphological transition from yeast to hyphae, the appropriate strains were grown in several media (solid and liquid) known to induce the morphological switch (including Spider, SLAD, serum, and Lee medium), and morphology was monitored over time. SLAD, serum, and LEE media all contain glucose as the carbon source. In Spider medium, the carbon source is mannitol. SLAD medium is a low nitrogen-containing medium that induces mainly pseudohyphae, and LEE is a medium rich in amino acids that induces true hyphae (Sudbery et al., 2004). Deletion of both alleles of CaGPR1 has no effect on germ tube formation in nearly all liquid media tested (our unpublished data). There was also no difference in morphology after prolonged (for 24- to 48-h) incubations. The only clear difference between wild-type and gpr1 mutant was observed when cells were incubated in liquid Spider medium at 37°C. Five hours after the shift from minimal medium, there was a clear defect in hyphal growth in the gpr1Δ/gpr1Δ mutant (Figure 2A). In contrast to the other media, Spider medium does not contain glucose, but it is unclear whether this is the reason for the absence of hyphae after prolonged incubation. CaGPR1 deletion caused a severe defect in hypha formation during surface growth on different solid media (Figure 2B). Whereas the wild-type strain formed wrinkled colonies (pseudohyphal and mycelial cells) on YPD containing 10% FCS and developed peripheral long hyphae invading the agar on Spider, SLAD and Lee media, the gpr1Δ/gpr1Δ strain formed smooth colonies consisting only of budding yeast cells. Reintegration of a wild-type copy of CaGPR1 resulted in an intermediate phenotype, similar to the morphology of the heterozygous strain (our unpublished data).
Figure 2.
Defects in hyphae formation caused by deletion of CaGPR1. Results for the wild-type strain SC5314, the gpr1Δ/gpr1Δ, the reintegrated strain, and the gpa2Δ/gpa2Δ strain (only in A) are shown. (A) Log phase cells were incubated in the different liquid media at 37°C. Cells were photographed after 5 h of incubation in the test medium. (B) Approximately 10 cells were plated on different solid media. Time of incubation and temperature were as follows: YPD + FCS, 3 d at 37°C; SLAD, 7 d at 30°C; YPD, 4 d at 30°C; LEE, 5 d at 30°C; and embedded growth, 3 d at 25°C.
In addition, deletion of CaGPR1 has a profound effect under conditions of embedded growth within the agar matrix. These experiments are generally performed at lower temperatures (Brown et al., 1999). When incubated at 25°C, the heterozygous mutant has a minor impairment in hyphal development compared with the wild-type strain, whereas the homozygous deletion strain is strongly reduced in hyphal formation (Figure 2B). At higher temperatures, the effect caused by deletion of CaGPR1 became less pronounced. At 30°C, formation of hyphae was only delayed compared with the wild-type (our unpublished data). After 24 h at 30°C, colonies of the gpr1Δ/gpr1Δ strain consisted of only yeast-like cells, whereas the yeast:hypha ratio for wild-type was ∼90:10. After 72 h, this ratio (yeast:hypha) was estimated as 10:90 and 45:55 for wild-type and gpr1Δ/gpr1Δ strain, respectively. The hyphae of the homozygous mutant were much shorter and colonies consisted of much more yeast-like cells in comparison with the wild-type strain (our unpublished data). Reintegration of one GPR1 allele restored hypha development in the tested conditions.
Deletion of CaGPR1 Slightly Affects Virulence and Invasion in Human Tissue
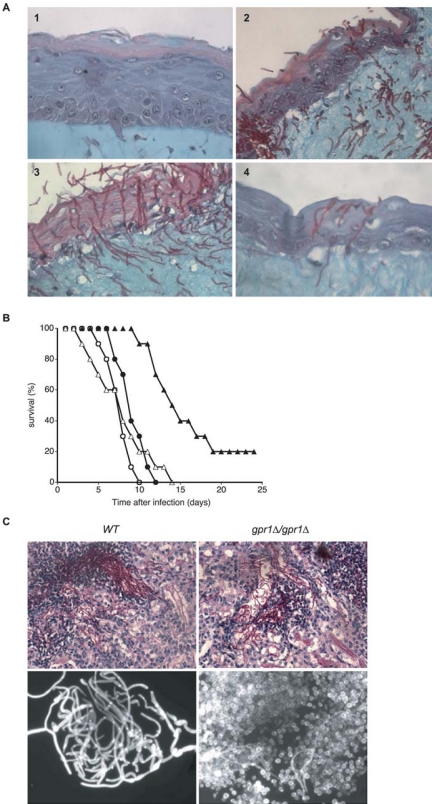
The results obtained in vitro in embedded conditions and on solid media (defect in yeast-to-hypha transition) led us to perform in vivo adhesion and invasion assays by using human cell and tissue models. First, wild-type and gpr1 null cells adhere similarly to Caco2 epithelial cells. Two hours after addition of the C. albicans cells, ∼90% of these adhere to the epithelial cells (our unpublished data). We also tested the potential of the different strains to invade human tissue. For this, we used a reconstructed skin model system (Dieterich et al., 2002). We confirmed that the wild-type SC5314 strain penetrates the protective layer of keratinocytes and invades the epithelial cell layers into the matrix, leading to severe damage of the model system (Figure 3A). The gpr1Δ/gpr1Δ strain is also able to penetrate tissue; however, the amount of pseudohyphal cells or yeast-type cells in the keratinocytic cell layer is significantly higher. The reintegrant strain shows a similar phenotype as the wild type. It forms hyphae that penetrate the epidermal layer and invade into the collagen/fibroblast matrix (Figure 3A). We have determined the number of yeast/pseudohyphal-like cells and true hyphae in the tissues, 40 h after infection in three independent experiments. For the gpr1Δ/gpr1Δ strain, between 34 and 45% (41% on average) of the cells were counted as yeast/pseudohyphal cells and between 55 and 66% (59% on average) as hyphae. For the gpr1Δ/gpr1Δ + GPR1 strain, between 10 and 13% (11% on average) of the cells were counted as yeast/pseudohyphal cells and between 87 and 90% (89% on average) as hyphae. The Sc5314 wild-type strain showed for all three sections <5% yeast/pseudohyphal cells.
Figure 3.
Virulence assays. (A) Infection of the reconstructed skin equivalent. Infection of a reconstructed skin equivalent with SC5314 (3), gpr1Δ/gpr1Δ (2), and gpr1Δ/RiGPR1 (4) for 40 h at 37°C. The undisturbed reconstructed skin is shown in 1. Magnification, 400×. (B) Survival curves of mice (female BALB/c, 20g, 10 mice/group) systemically infected with 106 cells of the C. albicans wild-type (•), CaGPR1/Cagpr1Δ (○), Cagpr1Δ/Cagpr1Δ strain (▴), and Cagpr1Δ/Cagpr1Δ +RiCaGPR1 (▵). (C) Top, PAS staining of kidney sections from mice infected with wild-type or gpr1Δ/gpr1Δ strains. Objective, 20×. Bottom, Calcofluor white staining of C. albicans cells isolated from solubilized kidneys of mice infected with wild-type or gpr1Δ/gpr1Δ strains. Objective, 40×.
We also performed virulence assays using a mouse model of systemic infection. Deletion of both alleles of CaGPR1 resulted in diminished virulence (Figure 3B). In three independent experiments, 20% of the mice infected with the gpr1Δ/gpr1Δ strain survived to the 40-d endpoint, whereas all mice infected with the wild-type strain were dead after ∼10 d. Mice injected with the heterozygous or reintegrant strain also died after ∼10 d. Attenuation of pathogenicity could be caused by a defect in morphological and metabolic adaptation to the new environment because the growth rate of the gpr1Δ/gpr1Δ strain in YPD and SD media at 30 and 37°C is not diminished in comparison with the growth rate of strains with reintegrated CaGPR1. We have investigated this by analyzing the morphology of the Candida cells in the infected tissues. Two days after infection, tissues were dissected and histology was performed either on tissue sections or in solubilized tissue (Figure 3C). Two days after infection, the number of hyphae that can be found in a tissue section after staining with PAS is much lower in the gpr1Δ/gpr1Δ strain compared with the wild type. However, true hyphae can still be found in the gpr1Δ/gpr1Δ mutant (Figure 3C). We also have performed calcofluor white staining of cells (which stains the bud scars and the septa fluorescent) after solubilization of infected tissue. Figure 3C (bottom) clearly shows that the number of yeast-like cells is much higher in the gpr1Δ/gpr1Δ mutant compared with the wild type, where there are nearly exclusively hyphae. These data, together with the data obtained with the reconstructed skin system clearly show that a gpr1Δ/gpr1Δ mutant is still able to invade tissue but that this strain is also in vivo clearly affected in hyphae formation, which might be the cause of its lower virulence. The observed change in morphology (more yeast-like cells) in the tissue sections for the gpr1Δ/gpr1Δ mutant also is observed under agar invasion conditions shown in Figure 4A.
Figure 4.
CaGPR1 is situated upstream of the cAMP–PKA pathway. (A) The LDR8–5 strain (gpr1Δ/gpr1Δ) was transformed with overexpression constructs of EFG1, CaTPK1, and CaTPK2. The gpr1 mutant was transformed with the empty plasmid as a negative control, and the same plasmid was transformed into the CAI4 strain as a positive control. The transformants were grown on solid SCAA medium for 5 d at 37°C. Top, top view of the colony morphology. Middle, hyphal growth within the agar. Bottom, individual cells taken from within the agar. (B) LDR8–5 (gpr1Δ/gpr1Δ) and MM2 (hst7Δ/hst7Δ) strains were transformed with the empty vector (pYPB1-ADHpt) or with a HST7 overexpression construct. JKC131 and LDR8 also were plated as controls. Transformants were grown at 30°C for 6 d on Spider medium and LEE medium. (C) CAI4 and LDR8–5 (gpr1Δ/gpr1Δ) were transformed with the overexpression construct for GPA2Q355L or with the empty plasmid. Transformants were grown on Spider and LEE medium at 25°C for 14 d. (D) SC5314, LDR8 (gpr1Δ/gpr1Δ), JKC131 (hst7Δ/hst7Δ), and MM5 (gpr1Δ/gpr1Δ hst7Δ/hst7Δ) cells were grown under embedded conditions for 5 d at 25°C or on Spider medium for 3 d at 37°C.
C. albicans Gpr1 Signals through the cAMP–PKA Pathway
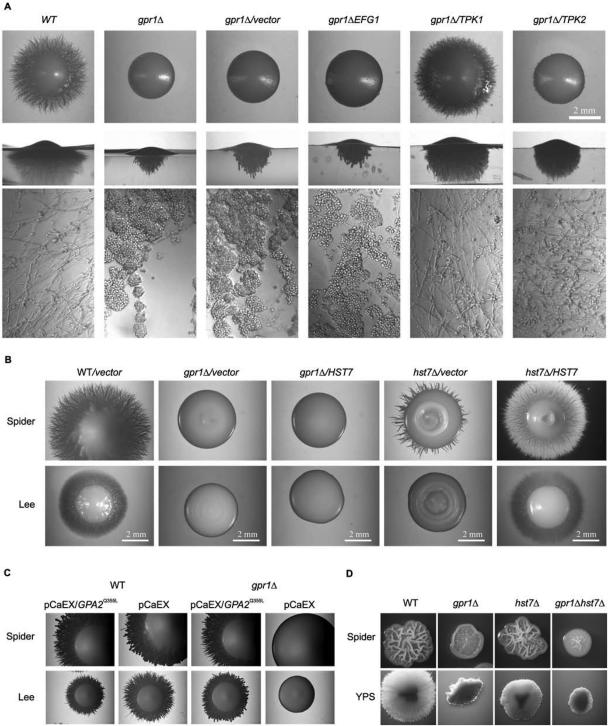
Two main signaling pathways are important for the yeast-to-hypha transition in C. albicans: the MAPK and cAMP–PKA pathways. To identify possible downstream components of Gpr1, the gpr1Δ/gpr1Δ strain was transformed with replicating plasmids containing the TPK1, TPK2, EFG1, or HST7 gene under control of a strong promoter. The first three were under the control of the PCK1 promoter, and HST7 was under control of the ADH1 promoter. The transformants with overexpression of TPK1, TPK2, and EFG1 were grown on SCAA medium, which strongly induces the PCK1 promoter (Leuker et al., 1997). The CAI4 wild-type strain transformed with the empty plasmid forms colonies with radial hyphae that grow into the agar on this medium. A gpr1Δ/gpr1Δ mutant forms smooth colonies but also grows into the agar (Figure 4A). However, 90% of the cells that are growing within the agar are yeast-like cells and only 10% form pseudohyphae. Overexpression of CaTPK1 in a gpr1Δ/gpr1Δ strain restored the wild-type phenotype. Such transformants formed true hyphae (Figure 4A). CaTPK2 overexpression also restored morphogenesis but to a lesser extent. EFG1 overexpression, using a plasmid that was previously shown to be functional (Stoldt et al., 1997) did not suppress the morphogenetic defect of the gpr1Δ/gpr1Δ mutant in this medium (see discussion for possible explanation). Overexpression of HST7 was unable to suppress the filamentation defect of the gpr1Δ/gpr1Δ strain on Spider and LEE medium, whereas it was clearly complementing the morphogenetic defect on these media of the hst7Δ/hst7Δ strain (Figure 4B). Based on previous observations that overexpression of HST7 induces yeast-to-hypha transition through the MAPK cascade (Csank et al., 1998), these data suggest that Gpr1 acts upstream of the cAMP–PKA cascade and most likely not upstream of the MAPK cascade.
Further experiments supported this conclusion. First, the CaGPR1 deletion phenotype also was suppressed by expression of a constitutively active CaGPA2 allele (GPA2Q355L) (Figure 4C). Second, deletion of HST7 had no effect for cells grown in Spider medium at 37°C or under embedded conditions. However, double deletion of HST7 and CaGPR1 resulted in a phenotype that is clearly different from that of the single hst7Δ/hst7Δ strain, indicating that CaGpr1 and Hst7 are functioning through different pathways (Figure 4D). However, it cannot be completely excluded that CaGpr1 also functions upstream of Hst7 because additional deletion of HST7 in a gpr1 mutant has only little effect on colony morphology. Third, addition of 5 mM cAMP under embedded growth conditions restored the wild-type phenotype of gpr1Δ/gpr1Δ colonies (Figure 5A). This suppression only occurred at 37°C not at 26°C (our unpublished data). Although hyphal formation in the gpr1Δ/gpr1Δ mutant is only delayed at 37°C (similar to 30°C), the cell morphology is clearly different in the absence (more pseudohyphae, more condensed) than in the presence (true hyphae) of cAMP (Figure 5B). As a control, we also have included strains, deleted in other components of the cAMP–PKA pathway. Cdc35Δ/cdc35Δ and ras1Δ/ras1Δ mutants have a stronger phenotype under embedded conditions, compared with the gpr1Δ/gpr1Δ mutant but in these strains as well, addition of cAMP restored wild-type hyphal formation. The morphogenetic defect of a tpk1 tpk2 double mutant (TPK1 is under the control of the PCK1 promoter, which is repressed in sucrose-containing medium) is not suppressed by addition of cAMP (Figure 5A).
Figure 5.
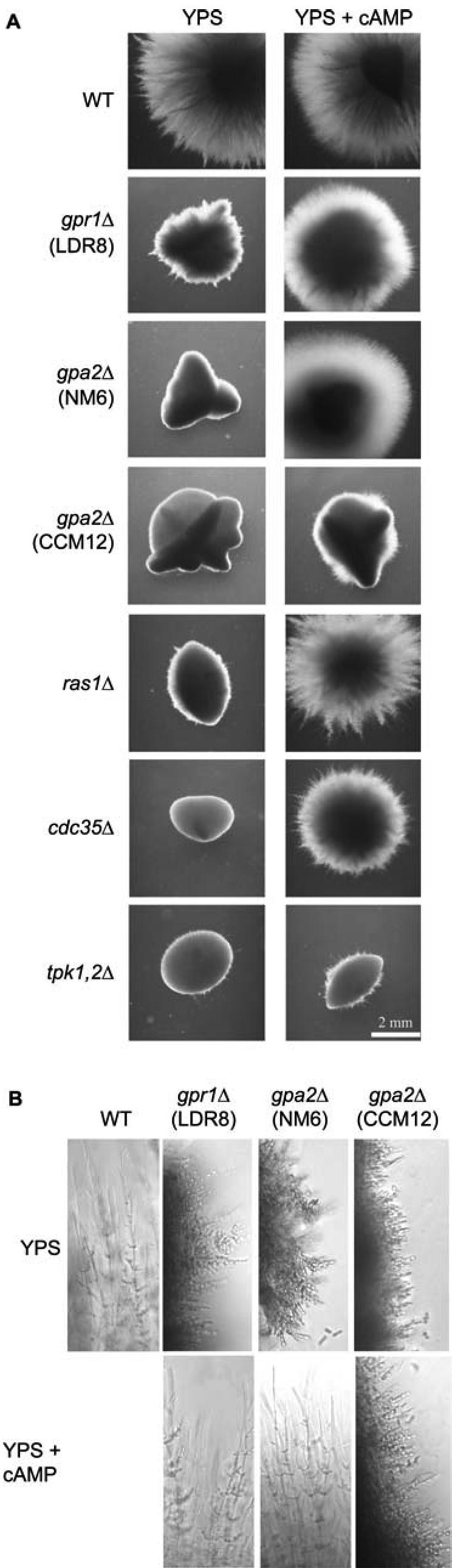
Addition of exogenous cAMP suppresses the phenotype of a gpr1 mutant. (A) Cells from SC5314 (WT), LDR8 (gpr1Δ/gpr1Δ), CCM12 (gpa2Δ/gpa2Δ), NM6 (gpa2Δ/gpa2Δ), CR216 (cdc35Δ/cdc35Δ), CDH107 (ras1Δ/ras1Δ), and M231 (tpk1Δ/PCK1p-TPK1/tpk2Δ/tpk2Δ) were incubated under embedded conditions in media without or with cAMP (final concentration 5 mM). The plates were incubated at 37°C for 3 d. (B) Colonies were isolated from the agar plates incubated at 37°C, and cell morphology was analyzed using the fluorescence microscope with phase contrast settings (magnification, 200×).
C. albicans Gpa2 Interacts with CaGpr1 and Also Activates the cAMP–PKA Pathway
We have performed two-hybrid analysis by using the C-terminal domain of CaGpr1 and the complete CaGpa2 protein. As a control, we used the homologous S. cerevisiae proteins that have previously been shown to interact physically (Kraakman et al., 1999). When the two-hybrid analysis was performed in one direction (CaGpr1 fused to the Gal4 DNA binding domain), there was always activation of the reporter constructs, already in the absence of CaGpa2. In the other direction, however, activation of the reporter constructs required the presence of CaGpa2 (our unpublished data). The fact that CaGpr1 regulates the cAMP–PKA pathway, that CaGpr1 interacts with CaGpa2, and that a Cagpr1Δ/Cagpr1Δ phenotype can be rescued by expression of a GPA2Q355L allele contradicts recently published results indicating that CaGpa2 is not involved in the cAMP pathway but rather in a MAPK signaling pathway (Sanchez-Martinez and Pérez-Martin, 2002). We have received the CaGPA2 deletion strain (CCM12) from these authors and confirmed the phenotypes reported for this strain. Contrary to the CaGPR1 deletion strain, the CaGPA2 deletion strain CCM12 cannot be suppressed by exogenous addition of cAMP (Figure 5A). To obtain independent confirmation, we have made new CaGPA2 deletion strains (see Materials and Methods). Heterozygous, homozygous, and reintegrated strains have been constructed using the URA-blaster strategy in both a wild-type and a Cagpr1Δ/Cagpr1Δ background. Figure 1B shows a Southern blot confirming correct deletion and reintegration of CaGPA2. We have repeated the morphogenesis experiments with the original and the new CaGPA2 deletion strains in liquid and on solid hypha-inducing media. In liquid Spider medium, the morphology was identical for both strains, but in LEE medium the published CCM12 strain was defective in hyphae formation, whereas the NM6 Cagpa2Δ/Cagpa2Δ mutant was not (Figure 2A). On solid medium, there was also no difference between the two strains (our unpublished data). However, the most striking difference was observed under embedded growth conditions in the presence of cAMP. The CCM12 Cagpa2Δ/Cagpa2 mutant strain maintained its strong defect in hyphae formation in the presence of cAMP, whereas the NM6 Cagpa2Δ/Cagpa2Δ mutant strain was rescued by cAMP similarly to the Cagpr1Δ/Cagpr1 mutant strain (Figure 5, A and B). We also transformed the new CaGPA2 deletion strain with HST7 and TPK1 overexpression constructs. Consistent with the previous results, overexpression of TPK1 but not that of HST7 suppressed the morphogenesis defect of the NM6 strain (our unpublished data).
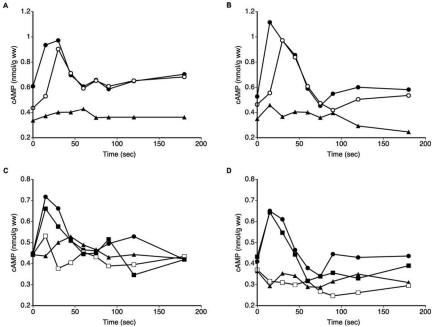
Induction of cAMP Accumulation by Glucose and Serum Is Independent of CaGpr1, but Dependent on CaRas1 and CaCdc25
It was shown in S. cerevisiae that Gpr1 is required for rapid glucose- and sucrose-induced cAMP signaling. The addition of glucose to yeast cells grown on a nonfermentable carbon source, such as glycerol, triggers a rapid transient increase in the intracellular cAMP level (Yun et al., 1997; Kraakman et al., 1999). The glucose-induced cAMP burst observed in C. albicans cells is not as pronounced as in S. cerevisiae cells (Rocha et al., 2001). We measured changes in cAMP levels in response to glucose (100 mM) but also after addition of known morphological transition inducers such as serum [10% (vol/vol)], N-acetylglucosamine (100 mM), and proline (100 mM), because hyphal morphogenesis has been reported to be accompanied by a raise in intracellular cAMP levels. For the wild-type strain and with glucose and serum as inducers, an early increase in cAMP levels was observed, followed by a gradual reduction (Figure 6, A and B). N-Acetylglucosamine and proline had no effect on the intracellular cAMP level (our unpublished data). Unexpectedly, no noticeable difference was found in the response of the wild type and the gpr1Δ/gpr1Δ strain to glucose or serum, except that the gpr1Δ/gpr1Δ strain had a lower basal cAMP level compared with the wild type (Figure 6, A and B). In S. cerevisiae, glucose-induced accumulation of cAMP is also dependent on the Ras1,2 (Mbonyi et al., 1988; Jiang et al., 1998) and Cdc25 (Munder and Küntzel, 1989; Van Aelst et al., 1990) proteins, and recent work has shown that it is associated with an increase in the GTP content on the Ras proteins (Colombo et al., 2004). Therefore, we also have determined the increase in cAMP after addition of glucose and serum in the Caras1Δ/Caras1Δ and Cacdc25Δ/Cacdc25Δ strains. Interestingly, the results show that both glucose and serum are unable to trigger an increase in cAMP in the Caras1Δ/Caras1Δ strain as well as the Cacdc25Δ/Cacdc25Δ strain (Figure 6, C and D). This suggests that glucose-induced activation of cAMP synthesis in C. albicans is mainly mediated by the CaCdc25-CaRas1 module and not by the CaGpr1-CaGpa2 module. The similarity between the results obtained with glucose and serum supports a previous conclusion (Hudson et al., 2004) that glucose is the main factor in serum affecting Candida morphogenesis.
Figure 6.
Glucose- and serum-induced cAMP signaling. Wild-type (•), gpr1Δ/gpr1Δ (○), ras1Δ/ras1Δ (▴), cdc25Δ/CDC25 (▪), and cdc25Δ/cdc25Δ (□) strains incubated at 37°C were supplemented with 100 mM glucose (A and C) or 10% serum (B and D). All strains have been tested in at least three independent experiments. A representative experiment is shown.
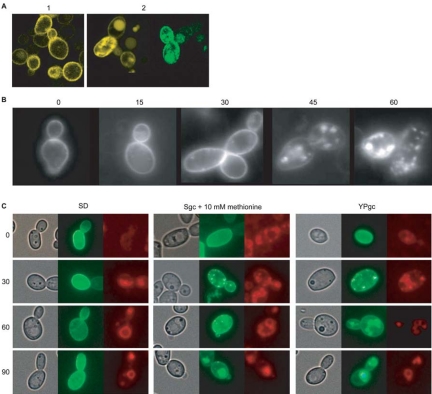
The CaGpr1 Receptor Is Rapidly Internalized in Rich Medium
To investigate the cellular localization of CaGpr1, we constructed an expression vector containing the CaGPR1 promoter and coding sequence fused with GFP optimized for C. albicans codon use (Gerami-Nejad et al., 2001). The plasmid carrying GPR1::GFP was introduced into the Cagpr1Δ/Cagpr1Δ and wild-type C. albicans strains. Isolated URA+ prototrophic transformants were grown on YPD and SD plates or in liquid media. Gpr1 was predicted to be an integral membrane protein consisting of seven putative transmembrane helices. Confocal laser scanning microscopy of C. albicans expressing a Gpr1::GFP fusion protein confirmed its localization in the plasma membrane (Figure 7A, 1). However, in cells grown in YPD medium fluorescence also was observed in intracellular vesicles and most probably in the vacuoles (Figure 7A, 2). We found that rapid internalization of Gpr1::GFP was initiated by transfer of cells from minimal glucose-containing medium to YPD. In such case, the fluorescence occurred already after 15 min in endocytotic vesicles and subsequently after 2 h in the vacuole. To exclude de novo synthesis of the receptor, these experiments also were performed in the presence of cycloheximide (Figure 7B). Ligand-induced internalization of the receptor is a well-known mechanism for rapid inactivation of G protein-coupled receptors. Hence, the rich medium might contain the ligand of the receptor as opposed to the minimal medium. This was confirmed by incubating the cells in synthetic glycerol (Sgc) and YPglycerol (YPgc). Whereas in Sgc medium there was no internalization, similar to SD medium, in YPgc medium there was a rapid internalization of the receptor (Figure 7C). We suggest that some component of peptone could activate Gpr1 that causes internalization of the receptor and such mechanism is characteristic for rapid inactivation of G protein-coupled receptors. In a first attempt to identify this component, we have repeated the internalization experiment by adding amino acids present in peptone to cells incubated in minimal glucose medium. Whereas most of the amino acids had no effect, there was a very rapid internalization upon addition of alanine (our unpublished data) or methionine (Figure 7C). These experiments also were performed in the presence of cycloheximide to differentiate between receptor internalization and de novo biosynthesis. Costaining of the cells using FM4-64 confirmed the vacuolar localization.
Figure 7.
Visualization by confocal laser scanning and normal fluorescence microscopy of a CaGpr1-GFP fusion construct. (A) CaGpr1 is membrane localized when cells are growing on minimal (without amino acids) glucose-containing medium (1). Incubation of cells in YPD medium results in intracellular localization of CaGpr1 (2). (B) Transition from minimal glucose containing medium to YPD results in a rapid internalization (15-min time intervals) of the CaGpr1 receptor. (C) Gpr1-GFP localization and visualization of endocytic membranes and vacuoles by staining with fluorescent dye FM4-64. For dye absorption, cells were incubated in the presence of 40 μM FM4-64 for 12 min at 24°C. The cells were washed three times with cold minimal medium to remove surface-bound dye and resuspended in prewarmed fresh media as indicated on the top of the panels, each containing cycloheximide (10 μg/ml) and incubated further for 90 min at 28°C. The cell samples were taken at the time points as indicated on the left of the figure and visualized by fluorescent microscopy. Objective, 40×.
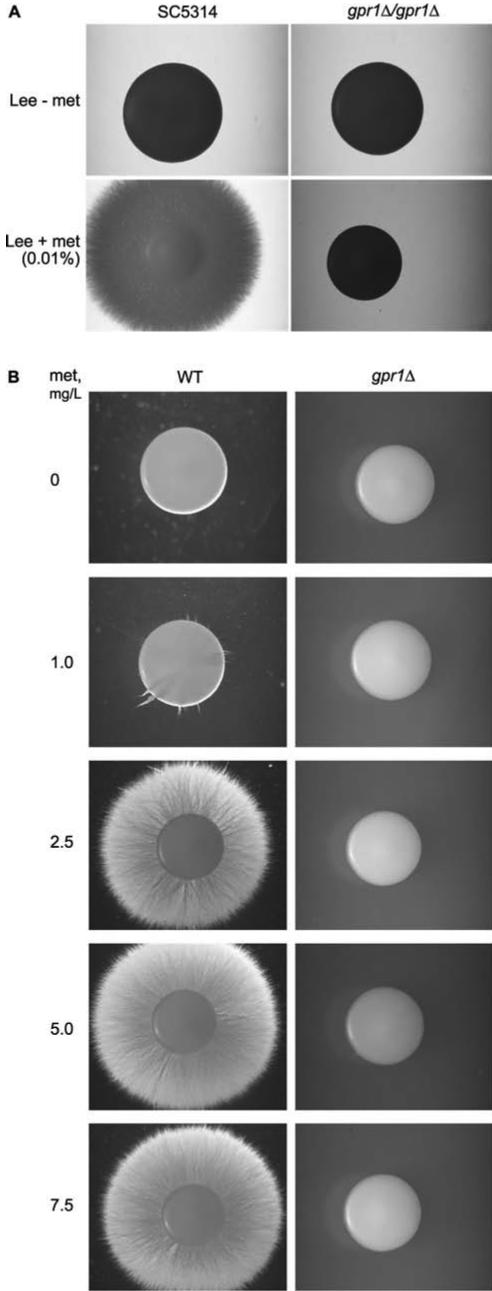
The Hyphal Morphogenesis Defect of the C. albicans Gpr1 Receptor Is Mediated via the Amino Acids Proline and Methionine
In a further attempt to identify the possible ligand for CaGpr1, we have used defined Lee medium. It contains seven amino acids, four salts, biotine, ornithine, and glucose as a carbon source. As shown in Figure 2B, there is a strong defect in hyphae formation of the CaGPR1 deletion strain in this medium as opposed to the wild-type strain. Subsequently, we have used a series of modified Lee medium in which the different constituents were either absent or present in various combinations. The main observation in these experiments was that proline or methionine is required for hypha induction in the wild-type strain. Figure 8A shows the results obtained for Lee medium in the presence and absence of methionine as the sole amino acid (of the 7, biotine and ornithine are still present, but they did not have any effect). In the absence of methionine both wild-type and gpr1Δ/gpr1Δ mutants form smooth colonies. Addition of low concentrations of methionine results in hypha formation in the wild-type but not in the gpr1Δ/gpr1Δ mutant. Similar results were obtained with proline but not with alanine (our unpublished data). Hence, it seems that receptor internalization and hyphal morphogenesis are not necessarily linked to each other. Similar discrepancies between sugar-specific rapid Gpr1-dependent cAMP signaling or pseudohyphal growth induction also has been observed in S. cerevisiae. Maltose, for instance, is a strong stimulator of pseudohyphal growth, but it does not stimulate cAMP signaling, despite the fact that both phenomena are Gpr1 dependent (Lorenz et al., 2000; Rolland et al., 2000). To determine whether the effect of methionine observed in the amino acid-based Lee medium is also present in other media, we have performed experiments in synthetic defined medium containing 0.1% glucose (SLD). In this case, there was also a clear effect of proline (our unpublished data) and methionine (Figure 8B) on colony morphology. In the absence of methionine, both wild-type and gpr1Δ/gpr1Δ strains form smooth colonies. Addition of methionine (at various concentrations) induced filamentation in the wild-type strain but not in the Cagpr1Δ/Cagpr1Δ mutant (Figure 8B). A concentration as low as 16.7 μM (2.5 mg/l) methionine fully induced hyphae formation in the wild-type strain, despite the presence of large concentrations of ammonium in the medium. Although these results do not allow yet to identify unequivocally the ligand(s) of CaGpr1, they do reveal an important role for micromolar concentrations of specific amino acids as triggers for hyphal morphogenesis either through Gpr1 or at least in concert with a pathway controlled by CaGpr1.
Figure 8.
CaGpr1 is required for amino acid-induced yeast-to-hypha transition. (A) Wild-type and gpr1Δ/gpr1Δ cells were grown at 30°C for 8 d on Lee medium without and with methionine. (B) Yeast-to-hypha induction by micromolar concentrations of methionine (2.5 mg/l, 16.7 μM) in wild-type and gpr1Δ/gpr1Δ strains.
DISCUSSION
The cAMP–PKA pathway in S. cerevisiae has been used as a model to identify homologous components of this pathway in C. albicans to investigate in particular their possible involvement in hyphal morphogenesis and virulence. Similar to pseudohyphal growth in S. cerevisae, these studies have shown that the cAMP–PKA pathway in C. albicans plays a role in the control of morphogenesis (Brown and Gow, 1999; Borges-Walmsley and Walmsley, 2000). The Gpr1 G protein-coupled receptor has been identified in S. cerevisiae as an upstream component in the cAMP–PKA pathway (Yun et al., 1997; Xue et al., 1998) and was shown to mediate activation of cAMP synthesis by glucose and sucrose (Yun et al., 1998; Kraakman et al., 1999; Lemaire et al., 2004). It is the best characterized nutrient-sensing GPCR system (Holsbeeks et al., 2004). The existence of a C. albicans homologue of Gpr1 has been reported previously (Versele et al., 2001), but its function had not been studied. While this work was being prepared for publication, a report on the C. albicans Gpr1 homologue was published describing the importance of the protein for hyphal morphogenesis and claiming a similar role in activation of cAMP synthesis by glucose as in S. cerevisiae (Miwa et al., 2004).
Our results are largely consistent with those of Miwa et al. (2004), supporting the concept that CaGpr1 controls hyphal morphogenesis through stimulation of the cAMP–PKA pathway and not through the MAPK pathway. The main arguments are the following. The deletion of CaGpr1 blocks or severely delays the yeast-to-hypha transition on various solid hypha-inducing media. The morphogenesis defect of the Cagpr1 mutant can be rescued by addition of exogenous cAMP (Figure 5), by expression of the constitutively active Gpa2Q355L allele (Figure 4C), and by overexpression of CaTPK1 (Figure 4A) but not by overexpression of HST7 (Figure 4B) nor by overexpression of EFG1 (Figure 4A). The phenotypic defects of the Cagpr1 mutant strongly resemble those of the Catpk1 mutant, which is also defective in the yeast-to-hyphae transition on solid medium, whereas it shows only a very minor defect in liquid medium (Bockmühl et al., 2001).
The morphogenesis phenotypes of mutants in other components of the cAMP–PKA pathway are more severe. Deletion of CaRAS1, CaCDC35, CaCAP1, CaTPK2, and CaBCY1 causes a severe morphogenesis defect not only on solid medium but also in liquid medium, and they also show a partial growth defect (Feng et al., 1999; Bahn and Sundstrom, 2001; Bockmühl et al., 2001; Rocha et al., 2001; Cassola et al., 2004). The reason for this stronger defect is not clear. It can be due to a more severe down-regulation of the cAMP–PKA pathway and/or to inputs from other signaling pathways. Examples of both situations have been described in S. cerevisiae where, for instance, deletion of RAS2 and TPK1 or TPK2 produces stronger effects than deletion of RAS1 or TPK3, respectively. Also deletion of Gpr1 or Gpa2 in S. cerevisiae causes much less down-regulation of the cAMP–PKA pathway than deletion of the essential components Cdc25, Cdc35, Ras1 and 2, or Tpk1, 2, and 3. Activation of the PKA pathway by other nutrients, such as amino acids and phosphate, is not associated with an increase in the cAMP level and therefore apparently occurs at a point more downstream in the pathway (Thevelein and de Winde, 1999).
Our results are also consistent with a role for CaGpa2 as stimulator of the cAMP pathway similar to the role of Gpa2 in S. cerevisiae. This is different from the conclusion made by Sanchez-Martinez and Pérez-Martin (2002) that CaGpa2 acts upstream of the MAPK pathway instead of the cAMP pathway. Because our results obtained for Gpa2 fitted very well with the idea that Gpr1 acts through Gpa2 on the cAMP pathway, we have reinvestigated the connection between Gpa2 and the cAMP pathway. For this purpose, we constructed a new CaGPA2 deletion mutant and showed that its morphogenesis defect can be suppressed by exogenous cAMP and by overexpression of CaTPK1 but not HST7. This is different from the CaGPA2 deletion strain described by Sanchez-Martinez and Pérez-Martin (2002) of which we confirmed the phenotype reported. Our new results fit with a role for CaGpa2 as mediator of the stimulatory role of CaGpr1 on the cAMP pathway. Most probably, the CaGPA2 deletion strain constructed by Sanchez-Martinez and Pérez-Martin (2002) has an additional mutation that prevents rescue of the strain by stimulation of the cAMP pathway. This is supported by the additional deletion of CaGPR1 that we made in this CaGPA2 deletion strain. Whereas the phenotypes of the Cagpr1, Cagpa2 (our new strain), and our double Cagpr1 Cagpa2 mutant are very similar, there was a strong additional effect of deletion of CaGPR1 in the Cagpa2 strain made by Sanchez-Martinez and Pérez-Martin (2002). It did not adhere anymore to solid SLAD medium as opposed to the single deletion mutants (our unpublished data). When we compare the phenotypes of the two Cagpa2 deletion strains, we can conclude that the additional mutation must be responsible for the strong reduction in virulence and tissue invasiveness (our unpublished data) of the Cagpa2 strain made by Sanchez-Martinez and Pérez-Martin (2002).
What is the nature of the ligand that acts on CaGpr1? In S. cerevisiae, Gpr1 is known to be required for stimulation of cAMP synthesis by millimolar concentrations of glucose and sucrose. Other sugars also were claimed to stimulate cAMP synthesis in a Gpr1-dependent way (Yun et al., 1998; Lorenz et al., 2000), but this has been contradicted (Rolland et al., 2000; Lemaire et al., 2004). That sugars indeed directly interact with S. cerevisiae Gpr1 as ligands, similar to the wellknown interaction of other ligands with GPCRs, has only recently been substantiated. Studies with SCAM analysis have provided strong evidence for direct interaction of glucose and sucrose with Gpr1 in a cavity flanked by transmembrane domain VI (Lemaire et al., 2004). Other closely related sugars, such as fructose and galactose, neither act as agonist nor as antagonist. Mannose, however, was found to be a potent antagonist of both glucose- and sucrose-induced cAMP signaling. In our hands, glucose-induced cAMP signaling is much less pronounced in C. albicans compared with S. cerevisiae, and deletion of CaGPR1 or CaGPA2 seemed to affect the signaling. This contrasts with the data of Miwa et al. (2004), who report a much lower glucose-induced cAMP signal in the Cagpr1 and Cagpa2 deletion strains. Other evidence possibly arguing against glucose as the ligand is based on the rapid glucose-independent internalization of the CaGpr1 receptor when shifted from minimal to rich medium.
In S. cerevisiae, glucose-induced cAMP signaling also is affected by the Ras1 and Ras2 proteins (Mbonyi et al., 1988; Jiang et al., 1998) and by their guanine nucleotide exchange protein Cdc25 (Munder and Küntzel, 1989; Van Aelst et al., 1990), but an upstream glucose-sensing system for Cdc25 has not been identified. On the other hand, glucose phosphorylation also can stimulate cAMP synthesis to some extent in S. cerevisiae independent of Gpr1 and Gpa2 (Rolland et al., 2000). Recent work has shown that glucose causes an increase in the GTP content on the Ras proteins in a glucose phosphorylation-dependent way, creating a link between a potential glucose-sensing system and the Ras-Cdc25 module (Colombo et al., 2004). The importance of the glucose phosphorylation-dependent system is further underscored by the finding that glucose phosphorylation is essential for the stimulation of cAMP synthesis by the Gpr1-Gpa2 GPCR system (Beullens et al., 1988; Rolland et al., 2000). In C. albicans, the only Ras protein, CaRas1, also acts on adenylate cyclase (Feng et al., 1999; Leberer et al., 2001). Goldberg et al. (1993) have shown that CaCdc25 can functionally replace Cdc25 in S. cerevisiae, suggesting that CaCdc25 also might act as guanine nucleotide exchange protein for CaRas1 in C. albicans. Our present results (Figure 6) show that both CaRas1 and CaCdc25 are essential for glucose-induced stimulation of cAMP accumulation in C. albicans. This supports a function for Cdc25 in the cAMP pathway of C. albicans similar to its function in S. cerevisiae. Moreover, it indicates that in C. albicans the Cdc25-Ras module of the cAMP pathway is the mediator of glucose-induced cAMP signaling rather than the Gpr1-Gpa2 module. Hence, in contrast to the results of Miwa et al. (2004), we are bound to conclude that under the conditions used to measure glucose-induced cAMP signaling, the Gpr1-Gpa2 GPCR system does not function as the glucose sensor. It is very possible that the relationship between cAMP signaling and glucose phosphorylation is similar in C. albicans and S. cerevisiae. The effect of glucose phosphorylation on cAMP signaling, morphogenesis, or other targets of the cAMP–PKA pathway in C. albicans has not been studied yet.
In S. cerevisiae, Gpr1 is apparently also required to some extent for the basal activity of adenylate cyclase in the absence of stimulatory sugars. This is shown by the observation that double deletion of GPR1 and RAS2 or GPR1 and SCH9 is lethal in all nutrient media, also those without glucose or sucrose (Xue et al., 1998; Kraakman et al., 1999). None of the individual deletions, gpr1Δ, ras2Δ or sch9Δ is lethal. Our data show that the effects of CaGPR1 deletion can be observed on a variety of media, including media lacking glucose and sucrose (such as Spider medium). Hence, it seems likely that also in C. albicans CaGpr1 is to a significant extent required for basal adenylate cyclase activity. This raises the question whether a reduction observed in glucose-induced accumulation of cAMP in Cagpr1Δ and Cagpa2Δ strains might be due to a reduction in basal adenylate cyclase activity rather than to a function of CaGpr1 and CaGpa2 as receptor system for glucose. This also may explain the results obtained with the strains in which EFG1 was overexpressed. This transcription factor, a member of the conserved APSES class of proteins regulating morphogenetic processes in fungi, was unable to suppress the gpr1 mutant phenotype, although evidence with point mutants in which a putative PKA phosphorylation site in the Efg1 transcription factor was mutagenized demonstrated that Efg1 is downstream of PKA (Bockmühl and Ernst, 2001). A possible explanation may be that CaGpr1 activates Tpk1 through CaCdc35 and that Efg1 needs activation by Tpk1 to function properly. As a result, overexpression of Efg1 may not be able to suppress the phenotype caused by absence of CaGpr1. A similar suggestion was made to explain the inability of Efg1 in suppressing the Cacdc35 mutant (Rocha et al., 2001). It cannot be excluded that other signaling pathways regulate the activity of PKA, independent of the Gpr1-Gpa2 signaling pathway. In fact, in S. cerevisiae it has been shown that nutrient signals such as the presence of phosphate, ammonium, or amino acids in the medium may activate the PKA pathway without causing an increase in cAMP, although PKA activity and thus also a certain basal cAMP level is required (Donaton et al., 2003; Giots et al., 2003; Van Nuland, unpublished data). Our results obtained with the CaGpr1-dependent amino acid induced yeast-to-hypha transition may point to a similar mechanism. Deletion of CaGpr1 may reduce the basal cAMP level and therefore PKA activity to such an extent that amino acids can no longer trigger activation of the PKA pathway, resulting in defective morphogenesis. This may explain the difference observed between receptor internalization and hyphae formation. An alternative explanation is that the defect in amino acid-induced morphogenesis in the Cagpr1Δ/gpr1Δ mutant is due to the fact that these amino acids are ligands for CaGpr1. This would constitute an important difference in ligand specificity of ScGpr1 (millimolar levels of specific sugars) versus CaGpr1 (micromolar levels of specific amino acids) and therefore might seem less likely. Whatever the precise explanation, our results have shown a dramatic effect of micromolar concentrations of specific amino acids on hyphal morphogenesis and a clear interference of this effect with the cAMP–PKA pathway.
Acknowledgments
We thank J. F. Ernst, A. J. P Brown, C. Gale, G. Fink, J. Pérez Martin, M. Whiteway, and R. D. Cannon for plasmids and strains, and we also thank the organizers of the Woods Hole course on Molecular Mycology for stimulating discussions. We are very grateful for the excellent technical assistance of Cindy Colombo, Suzanne Marcelis, Ann Bouché, and Ann Manderveld. We are also grateful to Goedele Maertens for performing confocal laser scanning microscopy and to Nico Van Goethem for graphical support. Sequence data for C. albicans were obtained from the Stanford Genome Technology Center Web site at http://www-sequence.stanford.edu/group/candida. Sequencing of C. albicans was accomplished with the support of the National Institute of Dental and Craniofacial Research and the Burroughs Wellcome Fund. Annotations of C. albicans genes were obtained from the Candida database Web server (http://genolist.pasteur.fr/CandidaDB/). This work was supported by Interuniversity Attraction Poles Network P5/30 and the Research Fund of the Katholieke Universiteit Leuven (Concerted Research Actions) to J.M.T., the Fund for Scientific Research-Flanders (G.0242.04) to P.V.D., the Belgian Federal Science Policy Office for a research fellowship for Central and Eastern Europe to M.M.M., European Molecular Biology Organization for a short-term fellowship to L.D.R., and the European Community for a Marie-Curie intra-European fellowship to H.T. (MEIF-CT-2003-502296).
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E04-09-0780) on January 26, 2005.
References
- Alspaugh, J. A., Pukkila-Worley, R., Harashima, T., Cavallo, L. M., Funnell, D., Cox, G. M., Perfect, J. R., Kronstad, J. W., and Heitman, J. (2002). Adenylyl cyclase functions downstream of the Ga protein Gpa1 and controls mating and pathogenicity of Cryptococcus neoformans. Eukaryotic Cell 1, 75-84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bahn, Y.-S., and Sundstrom, P. (2001). CAP1, an adenylate cyclase-associated protein gene, regulates bud-hypha transitions, filamentous growth and cyclic AMP levels and is required for virulence of Candida albicans. J. Bacteriol. 183, 3211-3223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beullens, M., Mbonyi, K., Geerts, L., Gladines, D., Detremerie, K., Jans, A. W., and Thevelein, J. M. (1988). Studies on the mechanism of the glucose-induced cAMP signal in glycolysis and glucose repression mutants of the yeast Saccharomyces cerevisiae. Eur. J. Biochem. 172, 227-231. [DOI] [PubMed] [Google Scholar]
- Bockmühl, D. P., and Ernst, J. F. (2001). A potential phosphorylation site for an A-type kinase in the Efg1 regulator protein contributes to hyphal morphogenesis of Candida albicans. Genetics 157, 1523-1530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bockmühl, D. P., Krishnamurthy, S., Gerads, M., Sonneborn, A., and Ernst, J. F. (2001). Distinct and redundant roles of the two protein kinase A isoforms Tpk1 and Tpk2 in morphogenesis and growth of Candida albicans. Mol. Microbiol. 42, 1243-1257. [DOI] [PubMed] [Google Scholar]
- Borges-Walmsley, M. I., and Walmsley, A. R. (2000). cAMP signalling in pathogenic fungi: control of dimorphic switching and pathogenicity. Trends Microbiol. 8, 133-141. [DOI] [PubMed] [Google Scholar]
- Braun, B. R., and Johnson, A. D. (1997). Control of filament formation in Candida albicans by the transcriptional repressor TUP1. Science 277, 105-109. [DOI] [PubMed] [Google Scholar]
- Braun, B. R., Kadosh, D., and Johnson, A. D. (2001). NRGI, a repressor of filamentous growth in C. albicans, is down-regulated during filament induction. EMBO J. 20, 4753-4761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brega, E., Zufferey, R., and Mamoun, B. C. (2004). Candida albicans Csy1 is a nutrient sensor important for activation of amino acid uptake and hyphal morphogenesis. Eukaryot. Cell 3, 135-143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown, A.J.P., and Gow, N.A.R. (1999). Regulatory networks controlling Candida albicans morphogenesis. Trends Microbiol. 7, 333-338. [DOI] [PubMed] [Google Scholar]
- Brown, J.D.H., Giusani, A. D., Chen, X., and Kumamoto, C. A. (1999). Filamentous growth of Candida albicans in response to physical environmental cues, and its regulation by the unique CZF1 gene. Mol. Microbiol. 34, 651-662. [DOI] [PubMed] [Google Scholar]
- Cannon, R. D., Jenkinson, H. F., and Shepherd, M. G. (1992). Cloning and expression of Candida albicans ADE2 and proteinase genes on a replicative plasmid in C. albicans and in Saccharomyces cerevisiae. Mol. Gen. Genet. 235, 453-457. [DOI] [PubMed] [Google Scholar]
- Care, R. S., Trevethick, J., Binley, K. M., and Sudbery, P. E. (1999). The MET3 promoter: a new tool for Candida albicans molecular genetics. Mol. Microbiol. 34, 792-798. [DOI] [PubMed] [Google Scholar]
- Cassola, A., Parrot, M., Silberstein, S., Magee, B. B., Passeron, S., Giasson, L., and Cantore, M. L. (2004). Candida albicans lacking the gene encoding the regulatory subunit of protein kinase A displays a defect in hyphal formation and an altered localization of the catalytic subunit. Eukaryot. Cell 3, 190-199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colombo, S., et al. (1998). Involvement of distinct G-proteins, Gpa2 and Ras, in glucose- and intracellular acidification-induced cAMP signalling in the yeast Saccharomyces cerevisiae. EMBO J. 17, 3326-3341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colombo, S., Ronchetti, D., Thevelein, J. M., Winderickx, J., and Martegani, E. (2004). Activation state of the Ras2 protein and glucose-induced signalling in Saccharomyces cerevisiae. J. Biol. Chem. 279, 46715-46722. [DOI] [PubMed] [Google Scholar]
- Cregg, J. M., Barringer, K. J., Hessler, A. Y., and Madden, K. R. (1985). Pichia pastoris as a host system for transformations. Mol. Cell. Biol. 5, 3367-3385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Csank, C., Makris, C., Meloche, S., Schroppel, K., Rollinghoff, M., Dignard, D., Thomas, D. Y., and Whiteway, M. (1997). Derepressed hyphal growth and reduced virulence in a VH1 family-related protein phosphatase mutant of the human pathogen Candida albicans. Mol. Biol. Cell 8, 2539-2551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Csank, C., Schroppel, K., Leberer, E., Harcus, D., Mohamed, O., Meloche, S., Thomas, D. Y., and Whiteway, M. (1998). Roles of the Candida albicans mitogen-activated protein kinase homolog, Cek1p, in hyphal development and systemic candidiasis. Infect. Immun. 66, 2713-2721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis, D., Edwards, J.J.E., Mitchell, A. P., and Ibrahim, A. S. (2000). Candida albicans RIM101 pH response pathway is required for host-pathogen interactions. Infect. Immun. 68, 5953-5959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dieterich, C., Schandar, M., Noll, M., Johannes, F. J., Brunner, H., Graeve, T., and Rupp, S. (2002). In vitro reconstructed human epithelia reveal contributions of Candida albicans EFG1 and CPH1 to adhesion and invasion. Microbiology 148, 497-506. [DOI] [PubMed] [Google Scholar]
- Doedt, T., Krishnamurthy, S., Bockmühl, D. P., Tebarth, B., Stempel, C., Russell, C. L., Brown, A.J.P., and Ernst, J. F. (2004). APSES proteins regulate morphogenesis and metabolism in Candida albicans. Mol. Biol. Cell 15, 3167-3180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donaton, M. C., Holsbeeks, I., Lagatie, O., Van Zeebroeck, G., Crauwels, M., Winderickx, J., and Thevelein, J. M. (2003). The Gap1 general amino acid permease acts as an amino acid sensor for activation of protein kinase A targets in the yeast Saccharomyces cerevisiae. Mol. Microbiol. 50, 911-929. [DOI] [PubMed] [Google Scholar]
- Enloe, B., Diamond, A., and Mitchell, A. P. (2000). A single-transformation gene function test in diploid Candida albicans. J. Bacteriol. 182, 5730-5736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ernst, J. F. (2000). Regulation of dimorphism in Candida albicans. Contrib. Microbiol. 5, 98-111. [DOI] [PubMed] [Google Scholar]
- Feng, Q., Summers, E., Guo, B., and Fink, G. R. (1999). Ras signaling is required for serum-induced hyphal differentiation in Candida albicans. J. Bacteriol. 181, 6339-6346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fonzi, W. A., and Irwin, M. Y. (1993). Isogenic strain construction and gene mapping in Candida albicans. Genetics 134, 717-728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gagiano, M., Bauer, F. F., and Pretorius, I. S. (2002). The sensing of nutritional status and the relationship to filamentous growth in Saccharomyces cerevisiae. FEMS Yeast Res. 2, 433-470. [DOI] [PubMed] [Google Scholar]
- Gerami-Nejad, M., Berman, J., and Gale, C. A. (2001). Cassettes for PCR-mediated construction of green, yellow, and cyan fluorescent protein fusions in Candida albicans. Yeast 18, 859-864. [DOI] [PubMed] [Google Scholar]
- Giots, F., Donaton, M. C., and Thevelein, J. M. (2003). Inorganic phosphate is sensed by specific phosphate carriers and acts in concert with glucose as a nutrient signal for activation of the protein kinase A pathway in the yeast Saccharomyces cerevisiae. Mol. Microbiol. 47, 1163-1181. [DOI] [PubMed] [Google Scholar]
- Goldberg, D., Marbach, I., Gross, E., Levitzki, A., and Simchem, G. (1993). A Candida albicans homolog of CDC25 is functional in Saccharomyces cerevisiae. Eur. J. Biochem. 213, 195-204. [DOI] [PubMed] [Google Scholar]
- Holsbeeks, I., Lagatie, O., Van Nuland, A., Van De Velde, S., and Thevelein, J. M. (2004). The eukaryotic plasma membrane as a nutrient-sensing device. Trends Biochem. Sci. 29, 556-564. [DOI] [PubMed] [Google Scholar]
- Hudson, D. A., Sciascia, Q. L., Sanders, R. J., Norris, G. E., Edwards, P.J.B., Sullivan, P. A. and Farley, P. C. (2004). Identification of the dialysable serum inducer of germ-tube formation in Candida albicans. Microbiology 150, 3041-3049. [DOI] [PubMed] [Google Scholar]
- Jiang, Y., Davis, C., and Broach, J. R. (1998). Efficient transition to growth on fermentable carbon sources in Saccharomyces cerevisiae requires signaling through the Ras pathway. EMBO J. 17, 6942-6951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klasson, H., Fink, G. R., and Ljungdahl, P. O. (1999). Ssy1p and Ptr3p are plasma membrane components of a yeast system that senses extracellular amino acids. Mol. Cell. Biol. 19, 5405-5416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Köhler, J. R., and Fink, G. R. (1996). Candida albicans strains heterozygous and homozygous for mutations in mitogen-activated protein kinase signaling components have defects in hyphal development. Proc. Natl. Acad. Sci. USA 93, 13223-13228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraakman, L., Lemaire, K., Ma, P., Teunissen, A.W.R.H., Donaton, M.C.V., Van Dijck, P., Winderickx, J., de Winde, J. H., and Thevelein, J. M. (1999). A Saccharomyces cerevisiae G-protein coupled receptor, Gpr1, is specifically required for glucose activation of the cAMP pathway during the transition to growth on glucose. Mol. Microbiol. 32, 1002-1012. [DOI] [PubMed] [Google Scholar]
- Kubler, E., Mosch, H. U., Rupp, S., and Lisanti, M. P. (1997). Gpa2p, a G-protein alpha-subunit, regulates growth and pseudohyphal development in Saccharomyces cerevisiae via a cAMP-dependent mechanism. J. Biol. Chem. 272, 20321-20323. [DOI] [PubMed] [Google Scholar]
- Leberer, E., Harcus, D., Broadbent, I. D., Clark, K. L., Dignard, D., Ziegelbauer, K., Schmidt, A., Gow, N. A., Brown, A. J., and Thomas, D. Y. (1996). Signal transduction through homologs of the Ste20p and Ste7p protein kinases can trigger hyphal formation in the pathogenic fungus Candida albicans. Proc. Natl. Acad. Sci. USA 93, 13217-13222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leberer, E., Harcus, D., Dignard, D., Johnson, L., Ushinsky, S., Thomas, D. Y., and Schröppel, K. (2001). Ras links cellular morphogenesis to virulence by regulation of the MAP kinase and cAMP signalling pathways in the pathogenic fungus Candida albicans. Mol. Microbiol. 42, 673-687. [DOI] [PubMed] [Google Scholar]
- Leberer, E., Ziegelbauer, K., Schmidt, A., Harcus, D., Dignard, D., Ash, J., Johnson, L., and Thomas, D. Y. (1997). Virulence and hyphal formation of Candida albicans require the Ste20p-like protein kinase CaCla4p. Curr. Biol. 7, 539-546. [DOI] [PubMed] [Google Scholar]
- Lee, K. L., Buckley, H. R., and Campbell, C. C. (1975). An amino acid liquid synthetic medium for the development of mycelial and yeast forms of Candida albicans. Sabouraudia 13, 148-153. [DOI] [PubMed] [Google Scholar]
- Lemaire, K., Van De Velde, S., Van Dijck, P., and Thevelein, J. M. (2004). Nutrients as ligand for the G protein coupled receptor Gpr1 in the yeast Saccharomyces cerevisiae. Mol. Cell 16, 293-299. [DOI] [PubMed] [Google Scholar]
- Leuker, C. E., Sonneborn, A., Delbrück, S., and Ernst, J. F. (1997). Sequence and promoter regulation of the PCK1 gene encoding phosphoenolpyruvate carboxykinase of the fungal pathogen Candida albicans. Gene 192, 235-240. [DOI] [PubMed] [Google Scholar]
- Liu, H., Kohler, J., and Fink, G. R. (1994). Suppression of hyphal formation in Candida albicans by mutation of a STE12 homolog [erratum appears in Science. 1995. 267, 17]. Science 266, 1723-1736 [DOI] [PubMed] [Google Scholar]
- Lo, H. J., Kohler, J. R., DiDomenico, B., Loebenberg, D., Cacciapuoti, A., and Fink, G. R. (1997). Nonfilamentous C. albicans mutants are avirulent. Cell 90, 939-949. [DOI] [PubMed] [Google Scholar]
- Lorenz, M. C., and Heitman, J. (1997). Yeast pseudohyphal growth is regulated by GPA2, a G protein α homolog. EMBO J. 16, 7008-7018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorenz, M. C., and Heitman, J. (1998). The MEP2 ammonium permease regulates pseudohyphal differentiation in Saccharomyces cerevisiae. EMBO J. 17, 1236-1247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorenz, M. C., Pan, X., Harashima, T., Cardenas, M. E., Xue, Y., Hirsch, J. P., and Heitman, J. (2000). The G protein-coupled receptor Gpr1 is a nutrient sensor that regulates pseudohyphal differentiation in Saccharomyces cerevisiae. Genetics 154, 609-622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mbonyi, K., Beullens, M., Detremerie, K., Geerts, L., and Thevelein, J. M. (1988). Requirement of one functional RAS gene and inability of an oncogenic ras variant to mediate the glucose-induced cyclic AMP signal in the yeast Saccharomyces cerevisiae. Mol. Cell. Biol. 8, 3051-3057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miwa, T., Takagi, Y., Shinozaki, M., Yun, C.-W., Schell, W. A., Perfect, J. R., Kumagai, H., and Tamaki, H. (2004). Gpr1, a putative G-protein-coupled receptor, regulates morphogenesis and hypha formation in the pathogenic fungus Candida albicans. Eukaryot. Cell 3, 919-931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munder, T., and Küntzel, H. (1989). Glucose-induced cAMP signaling in Saccharomyces cerevisiae is mediated by the CDC25 protein. FEBS Lett. 242, 341-345. [DOI] [PubMed] [Google Scholar]
- Murad, A. M., Leng., P., Straffon, M., Wishart, J., Macaskill, S., MacCallum, D., Schnell, N., Talibi, D., Marechal, D., Tekaia, F., d'Enfert, C., Gaillardin, C., Odds, F. C., and Brown, A. J. (2001). NRGI represses yeast-hypha morphogenesis and hypha-specific gene expression in Candida albicans. EMBO J. 20, 4742-4752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Odds, F. C. (1996). Epidemiological shifts in opportunistic and nosocomial Candida infections: mycological aspects. Int. J. Antimicrob. Agents 6, 141-144. [DOI] [PubMed] [Google Scholar]
- Pan, X., Harashima, T., and Heitman, J. (2000). Signal transduction cascades regulating pseudohyphal differentiation of Saccharomyces cerevisiae. Curr. Opin. Microbiol. 3, 567-572. [DOI] [PubMed] [Google Scholar]
- Rocha, C.R.C., Schröppel, K., Harcus, D., Marcil, A., Dignard, D., Taylor, B. N., Thomas, D. Y., Whiteway, M., and Leberer, E. (2001). Signalling through adenylyl cyclase is essential for hyphal growth and virulence in the pathogenic fungus Candida albicans. Mol. Biol. Cell 12, 3631-3643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rolland, F., de Winde, J. H., Lemaire, K., Boles, E., Thevelein, J. M., and Winderickx, J. (2000). Glucose-induced cAMP signalling in yeast requires both a G-protein coupled receptor system for extracellular glucose detection and a separable hexose kinase-dependent sensing process. Mol. Microbiol. 38, 348-358. [DOI] [PubMed] [Google Scholar]
- Sanchez-Martinez, C., and Pérez-Martin, J. (2002). Gpa2, a G-protein a subunit required for hyphal development in Candida albicans. Eukaryotic Cell 1, 865-874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saville, S. P., Lazzell, A. L., Monteagudo, C., and Lopez-Ribot, J. L. (2003). Engineered control of cell morphology in vivo reveals distinct roles for yeast and filamentous forms of Candida albicans during infection. Eukaryot. Cell 2, 1053-1060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonneborn, A., Bockmühl, D. P., Gerads, M., Kurpanek, K., Sanglard, D., and Ernst, J. F. (2000). Protein kinase A encoded by TPK2 regulates dimorphism of Candida albicans. Mol. Microbiol. 35, 386-396. [DOI] [PubMed] [Google Scholar]
- Stoldt, V. R., Sonneborn, A., Leuker, C. E., and Ernst, J. F. (1997). Efg1p, an essential regulator of morphogenesis of the human pathogen Candida albicans, is a member of a conserved class of bHLH proteins regulating morphogenetic processes in fungi. EMBO J. 16, 1982-1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sudbery, P., Gow, N., and Berman, J. (2004). The distinct morphogenic states of Candida albicans. Trends Microbiol. 12, 317-324. [DOI] [PubMed] [Google Scholar]
- Thevelein, J. M., and de Winde, J. H. (1999). Novel sensing mechanisms and targets for the cAMP-protein kinase A pathway in the yeast Saccharomyces cerevisiae. Mol. Microbiol. 32, 1002-1012. [DOI] [PubMed] [Google Scholar]
- Van Aelst, L., Boy-Marcotte, E., Camonis, J. H., Thevelein, J. M., and Jacket, M. (1990). The C-terminal part of the CDC25 gene product plays a key role in signal transduction in the glucose-induced modulation of cAMP level in Saccharomyces cerevisiae. Eur. J. Biochem. 193, 675-680. [DOI] [PubMed] [Google Scholar]
- Van Dijck, P., De Rop, L., Szlufcik, K., Van Ael, E., and Thevelein, J. M. (2002). Disruption of the Candida albicans TPS2 gene encoding trehalose-6-phosphate phosphatase decreases infectivity without affecting hypha formation. Infect. Immun. 70, 1772-1782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Versele, M., Lemaire, K., and Thevelein, J. M. (2001). Sex and sugar in yeast: two distinct GPCR systems. EMBO Rep. 2, 574-579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiederkehr, A., Avaro, S., Prescianotto-Baschong, C. Hagenauer-Tsapis, C., and Riezman, H. (2000). The F-box protein Rcy1p is involved in endocytotic membrane traffic and recycling out of an early endosome in Saccharomyces cerevisiae. J. Cell Biol. 149, 397-410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson, R. B., Davis, D., and Mitchell, A. P. (1999). Rapid hypothesis testing with Candida albicans through gene disruption with short homology regions. J. Bacteriol. 181, 1868-1874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue, Y., Batlle, M., and Hirsch, J. P. (1998). GPR1 encodes a putative G protein-coupled receptor that associates with the Gpa2p G subunit and functions in a Ras-independent pathway. EMBO J. 17, 1996-2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yun, C. W., Tamaki, H., Nakayama, R., Yamamoto, K., and Kumagai, H. (1997). G-protein coupled receptor from yeast Saccharomyces cerevisiae. Biochem. Biophys. Res. Commun. 240, 287-292. [DOI] [PubMed] [Google Scholar]
- Yun, C. W., Tamaki, H., Nakayama, R., Yamamoto, K., and Kumagai, H. (1998). Gpr1p, a putative G-protein coupled receptor, regulates glucose-dependent cellular cAMP level in yeast Saccharomyces cerevisiae. Biochem. Biophys. Res. Commun. 252, 29-33. [DOI] [PubMed] [Google Scholar]