Abstract
Many nitrogen fixation-associated genes in the soybean symbiont Bradyrhizobium japonicum are regulated by the transcriptional activator NifA, whose activity is inhibited by aerobiosis. NifA is encoded in the fixR-nifA operon, which is expressed at a low level under aerobic conditions and induced approximately fivefold under low-oxygen tension. This induction depends on a −24/−12-type promoter (fixRp1) that is recognized by the ς54 RNA polymerase and activated by NifA. Low-level aerobic expression and part of the anaerobic expression originates from a second promoter (fixRp2) that overlaps with fixRp1 and depends on an upstream DNA region (UAS) located around position −68 (H. Barrios, H. M. Fischer, H. Hennecke, and E. Morett, J. Bacteriol. 177:1760–1765, 1995). A protein binding to the UAS was previously postulated to act as an activator. This protein has now been purified, and the corresponding gene (regR) has been cloned. On the basis of the predicted amino acid sequence, RegR belongs to the family of response regulators of two-component regulatory systems. We identified upstream of the regR gene an additional gene (regS) encoding a putative sensor kinase. A regR mutant was constructed in which neither a specific UAS-binding activity nor fixRp2-dependent transcript formation and fixR′-′lacZ expression was detected in aerobically grown cells. Anaerobic fixR′-′lacZ expression was also decreased in regR mutants to about 10% of the level observed in the wild type. Similarly, regR mutants showed only about 2% residual nitrogen fixation activity, but unlike nodules induced by nifA mutants, the morphology of those nodules was normal, displaying no signs of necrosis. While regR mutants grew only slightly slower in free-living, aerobic conditions, they displayed a strong growth defect under anaerobic conditions. The phenotypic properties of regS mutants differed only marginally, if at all, from those of the wild type, suggesting the existence of a compensating sensor activity in these strains. The newly identified RegR protein may be regarded as a master regulator in the NifA-dependent network controlling nif and fix gene expression in B. japonicum.
Although the key enzyme of biological nitrogen fixation, the dinitrogenase complex, is highly conserved among diazotrophs, regulation of its synthesis greatly varies with respect to the effects of various environmental signals and the mode by which they are transduced to the level of gene expression (for reviews, see references 15 and 33). In the presence of combined nitrogen, free-living diazotrophs suppress expression of nif genes via the action of the ntr system. Moreover, nitrogen fixation genes are expressed only under microaerobic or anaerobic conditions in many diazotrophs, which prevents futile synthesis of the oxygen-labile dinitrogenase. For symbiotic nitrogen-fixing bacteria such as Rhizobium or Bradyrhizobium species, it turned out that the low-oxygen conditions prevailing in root nodules and in free-living microaerobic or anaerobic cultures are crucial for the synthesis of the nitrogen-fixing apparatus (for reviews, see references 7, 15, and 16).
The nitrogen-fixing root nodule symbiont of soybean, Bradyrhizobium japonicum, employs two oxygen-responsive cascades to control genes involved in symbiotic nitrogen fixation. The FixLJ-FixK2 cascade senses low-oxygen conditions by the FixLJ two-component regulatory pair. Active FixJ then activates fixK2, whose product, FixK2, is a positive regulator of genes required for microaerobic respiration (e.g., fixNOQP [2, 40, 45]), and one of two genes encoding an alternative RNA polymerase sigma factor, ς54 (29). This sigma factor forms a connection to the second cascade in which the NifA protein acts in concert with the ς54-RNA polymerase (36). Under low-oxygen tension, the NifA protein activates transcription from −24/−12 promoters that are associated with many nif and fix genes, usually by binding to upstream activation sites (UAS) and by catalyzing open promoter complex formation by the ς54-RNA polymerase bound at the core promoter (54). While all rhizobial NifA proteins are intrinsically oxygen or redox sensitive, the precise biochemical basis for this important property is not understood (15). Interestingly, NifA-mediated control in B. japonicum also includes genes not directly related to nitrogen fixation (e.g., the groESL genes encoding molecular chaperonins [18]). Furthermore, NifA seems to be required for an intact bacterium-plant interaction, as indicated by the necrotic phenotype of nodules induced by B. japonicum nifA mutants (17, 56).
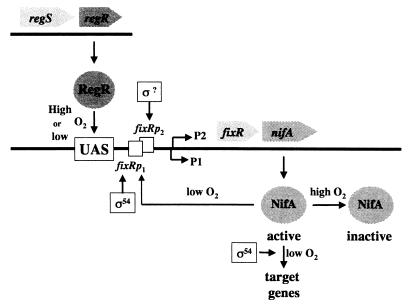
B. japonicum nifA is the promoter-distal gene of the fixR-nifA operon (58) (Fig. 1). Although FixR shows some sequence similarity to NAD-dependent dehydrogenases (4), this feature has not yet been of help in identifying its function. The Fix+ phenotype of nonpolar fixR mutants shows that fixR is not essential in conditions of symbiotic nitrogen fixation (58). The fixR-nifA operon is expressed not only under anaerobic but also under aerobic conditions despite the fact that NifA becomes rapidly inactivated under the latter conditions (30, 37). Anaerobic expression is approximately fivefold higher, and this induction depends on NifA itself and ς54 (5, 57) (Fig. 1). Low-level aerobic expression requires an upstream region located around position −68 (57). Detailed transcriptional analyses have shown that the fixR-nifA operon is preceded by two different overlapping promoters (5, 6) (Fig. 1). The first, designated fixRp1, is the ς54- and NifA-dependent −24/−12 promoter, which is responsible for the synthesis of the dominant transcript T1 under low-oxygen conditions. Transcription from the second promoter, fixRp2, depends on the upstream region located around position −68. The fixRp2 promoter is active in aerobically and anaerobically grown cells and leads to the synthesis of transcript T2 and the less-abundant transcript T1. The start site (P2) of transcript T2 is located just two nucleotides downstream of the start (P1) of T1 (Fig. 1). The nucleotide sequence in the −35/−10 region of fixRp2 looks rather dissimilar from that in B. japonicum housekeeping promoters (9), in contrast to a previous suggestion (5), so it remains unclear as to whether or not an alternative ς factor is required for its recognition.
FIG. 1.
Regulatory scheme and dual promoter of the B. japonicum fixR-nifA operon. P1 and P2 are the transcriptional start sites of transcripts T1 and T2 (see text) that originate from the overlapping promoters fixRp1 and fixRp2, respectively.
The findings that the −68 region is required for aerobic fixR-nifA expression and that a protein present in extracts of aerobically grown B. japonicum cells binds to a double-stranded oligonucleotide spanning this region led Thöny et al. (57) to raise the hypothesis that aerobic expression of this operon depends on an activator protein. However, several genetic approaches employed in our laboratory have so far failed to provide further support for this hypothesis. Therefore, we set out to purify the protein binding to the fixR-nifA UAS in order to eventually clone the respective gene and prove by mutational analysis that it is indeed the hitherto postulated activator. We report here that this approach has been successful and that it has resulted in the identification of a two-component regulatory system, termed RegSR, a new element in the NifA regulatory cascade of B. japonicum.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
The Escherichia coli and B. japonicum strains and plasmids used in this study are listed in Table 1. E. coli was grown in Luria-Bertani medium (35) at 37°C. B. japonicum strains were grown at 30°C, aerobically in PSY medium (48) supplemented with 0.1% (wt/vol) arabinose and anaerobically in YEM medium (11) supplemented with 10 mM KNO3. Appropriate concentrations of antibiotics were added as described previously (39). To measure aerobic and anaerobic growth of regS and regR mutants, antibiotic-free media were inoculated to an initial optical density at 600 nm (OD600) of 0.05 with stationary-phase precultures that had been washed with 0.9% NaCl to remove antibiotics present in the precultures. Growth was monitored by measuring the OD600 for 7 days (aerobic cultures) or 25 days (anaerobic cultures).
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Relevant genotype or phenotype | Source or referencea |
|---|---|---|
| E. coli strains | ||
| DH5α | supE44 ΔlacU169 (φ80lacZΔM15) hsdR17 recA1 gyrA96 thi-1 relA1 | BRL |
| S17-1 | Smr SprhsdR (RP4-2 kan::Tn7 tet::Mu chromosomally integrated) | 55 |
| MC1061 | Δ(lacIPOZYA)X74 hsdR | 10 |
| UK198 | Nxrdam dcm | 23 |
| B. japonicum strainsb | ||
| 110spc4 | Spr wild type | 48 |
| A9 | Spr KmrnifA::aphII | 17 |
| 2408 | Spr SmrregS::Ω (same orientation) | This work |
| 2409 | Spr SmrregS::Ω (opposite orientation) | This work |
| 2408R | Spr Smr KmrfixR′-′lacZ regS::Ω (same orientation) | This work |
| 2409R | Spr Smr KmrfixR′-′lacZ regS::Ω (opposite orientation) | This work |
| 2410 | Spr KmrregS::aphII (same orientation) | This work |
| 2411 | Spr KmrregS::aphII (opposite orientation) | This work |
| 2426 | Spr SmrregR::Ω (opposite orientation) | This work |
| 2427 | Spr SmrregR::Ω (same orientation) | This work |
| 2426R | Spr Smr KmrfixR′-′lacZ regR::Ω (opposite orientation) | This work |
| 2427R | Spr Smr KmrfixR′-′lacZ regR::Ω (same orientation) | This work |
| 2428 | Spr KmrregR::aphII (same orientation) | This work |
| 2429 | Spr KmrregR::aphII (opposite orientation) | This work |
| 7276B | Spr KmrfixR′-′lacZ, chromosomally integrated | 57 |
| 7277C | Spr Kmr ΔUAS-fixR′-′lacZ, chromosomally integrated | 57 |
| Plasmids | ||
| pBluescript(SK+) | Apr, cloning vector | Stratagene |
| pUC18, pUC19 | Apr, cloning vectors | 41 |
| pUR2 | Apr, cloning vector | 49 |
| pSUP202 | Apr Cmr TcroriT from RP4 | 55 |
| pSUP202pol4 | Tcr (pSUP202) | 18 |
| pUC4-KIXX-PSP | Apr Kmr (pUC4-KIXX) aphII cassette with PmeI-SwaI-PacI linker in SmaI site | 31 |
| pHP45Ω | Apr Spr Smr (Ω cassette) | 46 |
| pSP72::regA | Apr (pSP72) R. capsulatus regA on an 857-bp BamHI-SalI fragment | 25 |
| pRJ2400 | Apr [pBluescript(SK+)] B. japonicum regSR on a 2.5-kb EcoRI-BamHI fragment | This work |
| pRJ2403 | Apr (pUC19) B. japonicum regSR on a 3.6-kb EcoRI fragment | This work |
| pRJ2408 | Spr Smr Tcr (pSUP202pol4) regS::Ω (same orientation) | This work |
| pRJ2409 | Spr Smr Tcr (pSUP202pol4) regS::Ω (opposite orientation) | This work |
| pRJ2410 | Kmr Apr Tcr (pSUP202pol4) regS::aphII (same orientation) | This work |
| pRJ2411 | Kmr Apr Tcr (pSUP202pol4) regS::aphII (opposite orientation) | This work |
| pRJ2426 | Spr Smr Apr Tcr (pSUP202) regR::Ω (opposite orientation) | This work |
| pRJ2427 | Spr Smr Apr Tcr (pSUP202) regR::Ω (same orientation) | This work |
| pRJ2428 | Kmr Apr Tcr (pSUP202) regR::aphII (same orientation) | This work |
| pRJ2429 | Kmr Apr Tcr (pSUP202) regR::aphII (opposite orientation) | This work |
| pRJ7211 | AprfixR′-′lacZ | 58 |
BRL, Bethesda Research Laboratories, Inc., Gaithersburg, Md.; Stratagene, La Jolla, Calif.
The genetic structure of the regS and regR mutant strains is depicted in Fig. 3C.
Purification of the fixR-nifA upstream binding protein (UBP).
About 20 to 25 g of aerobically grown B. japonicum wild-type cells (wet weight) was suspended in 100 ml of TEPDM buffer (50 mM Tris-HCl, pH 8; 1 mM EDTA; 1 mM phenylmethylsulfonyl fluoride; 2 mM dithiothreitol [DTT]; 25 mM MgCl2; 100 mM KCl) and disrupted by three passages through a French pressure cell at a pressure of 11,000 lb/in2. Cell debris and membranes were removed by two subsequent centrifugation steps at 4°C (30 min at 10,500 rpm [Sorvall SS-34 rotor] and 90 min at 35,000 rpm [Beckman SW55 Ti rotor]) yielding the crude protein extract.
All of the following purification steps were performed at 4°C except the high-pressure liquid chromatography (HPLC) procedures, for which the columns were cooled to 10°C. The crude extract was treated with RNase A (2.5 μg/ml) for 1 h and then loaded onto a 300-ml gravity-flow Sulfopropyl-Sepharose Fast Flow column (Pharmacia LKB Biotechnology, Uppsala, Sweden). After being washed with TEDM25 buffer (50 mM Tris-HCl, pH 8; 1 mM EDTA; 2 mM DTT; 25 mM MgCl2), the protein was eluted with a linear gradient of KCl (0.1 to 1.5 M) in TEDM25. Fractions exhibiting DNA binding activity in the gel retardation assay (eluting at 650 to 750 mM KCl) were pooled and diluted 1:4 with TEDM25 buffer. This protein solution was then applied to a second Sulfopropyl-Sepharose column (110-ml column volume) coupled to an HPLC system. Upon being washed with TEDM25 and eluted with a discontinuous KCl gradient in the same buffer, fractions from the 650 to 850 mM KCl eluate were pooled and concentrated by ultrafiltration to a volume of 1.5 ml (Amicon ultrafiltration cell equipped with a PM-10 membrane [10-kDa cutoff size]; Amicon, Beverly, Calif.). The concentrated solution was further fractionated by HPLC on a 30-ml Sephacryl S-300 gel filtration column (Pharmacia LKB Biotechnology) with TEDM25 as the buffer. Pooled fractions exhibiting DNA binding activity were then applied to a DNA affinity column prepared as follows. A 32-mer double-stranded oligonucleotide (see below) spanning the UBP binding site and containing single-stranded 5′-GATC-3′ overhanging ends was self-ligated to form 300- to 2,000-bp multimers which were then coupled to CNBr-activated Sepharose CL-2B (Pharmacia LKB Biotechnology). Protein solution (4.4 ml) from the previous step was diluted 1:1 with TEDM25 buffer, mixed with 1.7 μg of poly(dI-dC) per ml (Fluka, Buchs, Switzerland) and then loaded onto two consecutive 2-ml DNA-Sepharose columns equilibrated with TED (50 mM Tris-HCl, pH 8; 1 mM EDTA; 2 mM DTT) containing 50 mM KCl. Columns were washed with TED containing 200 mM KCl and then eluted with a KCl gradient of 250 to 750 mM in TED.
Protein analysis and immunoblotting.
Proteins were resolved by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) with 10 to 15% gels that were stained with silver or with Coomassie brilliant blue. For separation of low-molecular-weight proteins, 15% tricine–SDS–polyacrylamide gels were used as described by Schägger and von Jagow (52). If required, protein samples were concentrated by precipitation with trichloroacetic acid. Western blotting was done according to the method of Babst et al. (3). Proteins were detected by measuring the binding of a 500-fold dilution of rabbit anti-R. capsulatus RegA serum (kindly provided by G. Klug, Giessen, Germany) with a chemiluminescence detection kit (Boehringer GmbH, Mannheim, Germany). Proteins to be N-terminally sequenced were blotted on a polyvinylidene fluoride membrane (Millipore, Bedford, Mass.) and stained with Coomassie brilliant blue. Protein bands of interest were excised and subjected to automated Edman amino acid sequence analysis (P. James, Institute of Biochemistry, Eidgenössische Technische Hochschule, Zürich, Switzerland).
Gel retardation assay.
Protein fractions were tested for DNA binding activity in a gel retardation assay by using an HPLC-purified, double-stranded 32-bp oligonucleotide (5′-CATTCCGCGTGCGCGACATTAGGACGCAAAAC-3′) that spans the region from −83 to −52 upstream of the fixR-nifA transcription start site P2 (5). This oligonucleotide was end labeled with [γ-32P]ATP with T4 polynucleotide kinase and purified by gel filtration through Sepharose–NAP-10 columns (Pharmacia LKB Biotechnology). About 0.1 ng of labeled oligomer (ca. 60,000 cpm) was mixed with protein extracts preincubated with 1 μg of poly(dI-dC) per ml in binding buffer (12 mM HEPES, pH 7.9; 6 mM KCl; 3 mM MgCl2; 0.5 mM DTT; 4 mM Tris-HCl, pH 8; 6 mM EDTA; Stratagene, La Jolla, Calif.). Protein amounts ranged between 30 μg for crude extracts and 0.1 μg for highly enriched fractions. Protein-DNA mixtures were incubated for 5 min at room temperature, mixed with 0.2 volumes of loading dye (10% glycerol [vol/vol], 0.02% bromphenol blue [wt/vol] in water), and then loaded onto 6% nondenaturing polyacrylamide gels (cross-linker ratio of 29:1 in 1× TBE [89 mM Tris base, 89 mM boric acid, 2.5 mM EDTA]). Gels were run at 4°C, dried under vacuum, and exposed on a phosphorimager screen. To determine the specific binding activity of individual protein fractions, the ratio of the radioactivity detected in shifted bands originating from specific UBP-DNA complexes to the total radioactivity present in the lane was calculated and normalized to the amount of protein.
DNA work.
Standard protocols were used for recombinant DNA techniques and Southern blotting (50). B. japonicum chromosomal DNA was isolated as described by Hahn and Hennecke (22). Heterologous hybridizations were carried out in 6× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate) at 56°C with probes that were 32P labeled by nick translation. Washing steps were performed at 58°C in 6× SSC. We used digoxigenin-labeled probes generated by PCR for homologous hybridizations. Hybridizations at 68°C in 2× SSC, washings, and chemiluminescence detection were done according to the manufacturer’s manual (Boehringer GmbH). Double-stranded plasmid DNA was sequenced by the chain termination method of Sanger et al. (51) with DNA sequencers (models 373A and 377; Applied Biosystems, Foster City, Calif.). DNA and deduced protein sequences were analyzed with the GCG software package (version 8; Genetics Computer Group, Madison, Wis.) or with the National Center for Biotechnology Information BLAST network server (http://www.ncbi.nlm.nih.gov/BLAST/). In the search for putative transmembrane domains in RegS, we used the services of the ISREC TMpred server (http://ulrec3.unil.ch/software/TMPRED_form.html).
Construction of B. japonicum regR and regS mutant strains.
For construction of regR mutations, the 3.6-kb EcoRI insert of pRJ2403 was cloned into vector pUR2, and a BamHI linker was inserted into the blunt-ended NdeI site located immediately upstream of regR. Subsequently, the 0.5-kb BamHI fragment spanning almost the entire regR gene was replaced by the 2-kb Ω cassette of pHP45Ω (Smr Spr) or the 1.7-kb aphII (Kmr) cassette of pUC4-KIXX-PSP. The resulting 5.1- and 4.8-kb EcoRI inserts containing the mutated regR gene were cloned into the vector pSUP202, yielding plasmids pRJ2426/pRJ2427 and pRJ2428/pRJ2429, respectively, which differ from each other with respect to the type and orientation of the inserted cassette (Table 1). To mutate regS, the 2.5-kb EcoRI-BamHI insert of pRJ2400 was cloned into pUC18. regS was then disrupted by insertion of the 2-kb SmaI Ω fragment (Smr Spr) or the 1.7-kb SmaI aphII fragment (Kmr) derived from pHP45Ω and pUC4-KIXX-PSP, respectively, into the blunt-ended regS internal NotI site. The mutated regS gene constructs were cloned as XbaI-EcoRI fragments into pSUP202pol4, yielding plasmids pRJ2408/pRJ2409 (both orientations of the Ω cassette) and pRJ2410/pRJ2411 (both orientations of the aphII cassette). All pSUP202 derivatives were introduced by conjugation (22) into B. japonicum strain 110spc4 (wild type). Furthermore, plasmids pRJ2408, pRJ2409, pRJ2426, and pRJ2427 were mobilized into B. japonicum 7276B, which carries a chromosomally integrated fixR′-′lacZ fusion. Marker exchange mutants resulting from double crossovers were selected by their resistance to streptomycin or kanamycin and screened for their sensitivity to tetracycline. The genetic structure of the mutants was verified by appropriate Southern blot hybridization of chromosomal DNA. The numbers and relevant characteristics of all constructed strains are listed in Table 1.
fixR-nifA transcriptional mapping.
Transcription from the two fixR-nifA promoters was studied with primer extension experiments. RNA was isolated from 10 ml of aerobically grown culture (OD600 = 0.8), from 20 ml of anaerobically grown culture of strains 7276B and 7277C (OD600 = 0.4), and from 100 ml of anaerobically grown culture of strain 2426R (OD600 = 0.1) by the hot phenol procedure described previously (3). RNA samples were further purified by using RNeasy columns (Qiagen AG, Basel, Switzerland). To detect the fixR′-′lacZ transcripts, the lac4B primer (5′-ATTAAGTTGGGTAACGCCAGGGTTTTCC-3′) was elongated with Superscript reverse transcriptase (Gibco-BRL Life Technologies, Gaithersburg, Md.). As an internal control, the 16S rRNA primary transcript was reverse transcribed by using the PBj16S oligonucleotide as a primer (5). At least 100,000 cpm of radiolabeled lac4B primer and 2,000 cpm of PBj16S primer were used per reaction. Primer extensions were performed according to a protocol slightly modified from that described by Babst et al. (3). Primer extension products were purified by a phenol extraction and 10-min treatment with RNase A (100 μg/ml) followed by ethanol precipitation.
regR transcriptional mapping.
Transcription of the regR gene was investigated with primer extension experiments with strains 110spc4 (wild type) and 2409 (regS mutant) and with the primers PEregR2 (5′-AATCAACCACGGCGAATGCCGGTGCCGCCTT-3′) and PEregR3 (5′-AGAAACGGCTTGTCGTCCTCCACGATGAGAAG-3′). Experiments were conducted as described for the fixR-nifA transcriptional mapping. RNA from strain 2426 (the regR deletion mutant), to which both primers cannot anneal, served as a control to test the specificity of the transcription signal.
β-Galactosidase assays and plant infection tests.
β-Galactosidase activity of B. japonicum strains harboring the fixR′-′lacZ fusion was measured in cells grown aerobically for 2 days and anaerobically for 6 days as described previously (35, 58). The symbiotic phenotypes of regR and regS mutants were determined in soybean infection tests (20, 22). Nodule ultrastructure was analyzed by transmission electron microscopy (R. Hermann, Institute for Cell Biology, Eidgenössische Technische Hochschule) (56).
Nucleotide sequence accession number.
The nucleotide sequence of the B. japonicum regSR genes has been deposited in the EMBL Nucleotide Sequence Database under accession number AJ006100.
RESULTS
Purification of the fixR-nifA UBP and cloning of the corresponding gene.
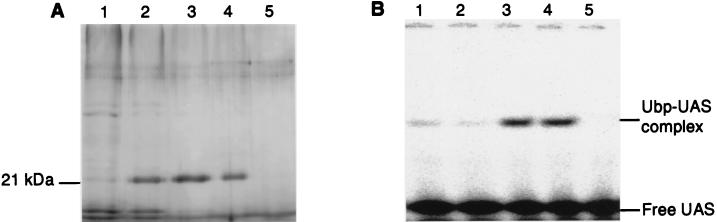
Following the purification protocol specified in Materials and Methods, we could enrich the fixR-nifA UBP from B. japonicum crude extracts by a factor of ca. 3,000, as determined from the specific UAS binding activity detected in the gel retardation assay (Fig. 2). When subjected to SDS-PAGE, the purified protein sample showed one dominant protein band with a relative molecular weight of ca. 21,000 (Fig. 2A) (28). The pooled final fractions from five parallel purification series (ca. 1 μg of protein) were precipitated with trichloroacetic acid, separated on a 15% tricine–SDS–polyacrylamide gel, and blotted on a polyvinylidene fluoride membrane. The prominent 21-kDa protein was then subjected to N-terminal amino acid sequence analysis. The resulting sequence of 20 amino acids, NAIAELNEQTDRSLLIVEDD, displayed striking similarities to the N termini of RegA (11 identical amino acids [44, 47, 53]), PrrA (10 identical amino acids [13]), and ActR (12 identical amino acids [59]), the response regulators of Rhodobacter capsulatus, Rhodobacter sphaeroides, and Sinorhizobium meliloti, respectively. A Western blot showed that the purified UBP cross-reacted with an anti-R. capsulatus RegA antibody (data not shown).
FIG. 2.
Purified UBP. (A) Silver-stained SDS–13% PAGE gel of protein fractions taken after elution from the DNA-Sepharose affinity column, the last purification step (see Materials and Methods). One microgram of protein from the fractions that were eluted with 250 mM (lane 1), 300 mM (lane 2), 350 mM (lane 3), and 400 mM KCl (lane 4) and 0.5 μg of protein from the fraction that was eluted with 450 mM KCl (lane 5) were loaded. The prominent 21-kDa bands in fractions 2 to 4 were excised and subjected to N-terminal amino acid sequencing as described in Materials and Methods. (B) Gel retardation assay of the samples shown in panel A. Protein (0.1 μg) was mixed with 0.1 ng of 32P-labeled double-stranded 32-bp oligonucleotide spanning the fixR-nifA −68 region (UAS) and incubated in the presence of poly(dI-dC) as a nonspecific competitor. The binding products were separated on a 6% nondenaturing polyacrylamide gel and visualized by phosphorimager analysis.
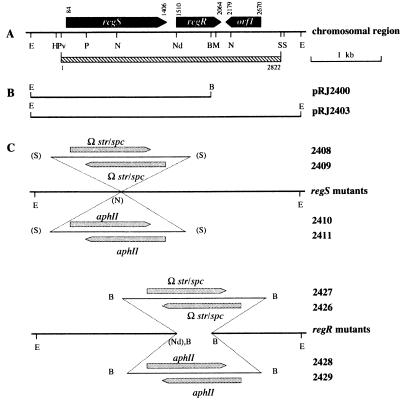
On the basis of these findings, we decided to use a 0.8-kb NdeI-HindIII fragment of the plasmid pSP72::regA (kindly provided by G. Klug, Giessen, Germany) containing the regA gene of R. capsulatus as a radioactive probe for hybridization of B. japonicum genomic DNA. A weak but reproducible hybridization to a 2.5-kb EcoRI-BamHI fragment was observed. EcoRI- plus BamHI-restricted chromosomal DNA of this size range was isolated from a preparative agarose gel and used for construction of a partial genomic library in the pBluescript(SK+) vector. Plasmids isolated from ca. 800 clones were analyzed by Southern blot hybridization with the regA probe. The dominantly hybridizing plasmid pRJ2400 contained a 2.5-kb EcoRI-BamHI insert (Fig. 3B). Sequence analysis revealed the presence of two open reading frames whose deduced products showed great similarity to RegB (38) and RegA, two-component regulatory proteins of R. capsulatus. Accordingly, the B. japonicum open reading frames were termed regS for the sensor gene and regR for the regulator gene (Fig. 3A). As it turned out that the 3′ end of regR was lacking on pRJ2400, we used the insert of this plasmid as a probe to subclone from a B. japonicum cosmid library a 3.6-kb EcoRI fragment that spans the complete regSR region (pRJ2403) (Fig. 3B). Its nucleotide sequence was determined, and the sequence of the regSR region, including an additional open reading frame (orf1) located on a 2,816-bp PvuII-SmaI fragment (Fig. 3A), was submitted to the EMBL Nucleotide Sequence Database.
FIG. 3.
Physical map of the B. japonicum regSR region and genetic structure of regS and regR mutations. (A) The physical map shows relevant restriction sites (B, BamHI; E, EcoRI; H, HindIII; M, MscI; N, NotI; Nd, NdeI; P, PstI; Pv, PvuII, S, SmaI), the location and orientation of regS and regR, and one additional open reading frame, orf1. Numbers at the top denote the nucleotide positions starting from the PvuII site upstream of regS. The hatched bar indicates the extent of the sequence submitted to the EMBL Nucleotide Sequence Database. (B) The inserts of plasmids pRJ2400 and pRJ2403 are depicted. (C) The structure of regS and regR mutations is shown along with the corresponding strain numbers. Horizontal arrows indicate the orientation of the inserted resistance cassettes, Ω (Smr Spr) or aphII (Kmr). Restriction sites in parentheses were lost during cloning.
We used the cloned regSR region (PvuII-MscI fragment) as a probe to look for possible homologs in the B. japonicum genome. No additional bands were detected in appropriate, low-stringency Southern blot hybridizations to B. japonicum total DNA.
Properties of the deduced regS, regR, and orf1 gene products.
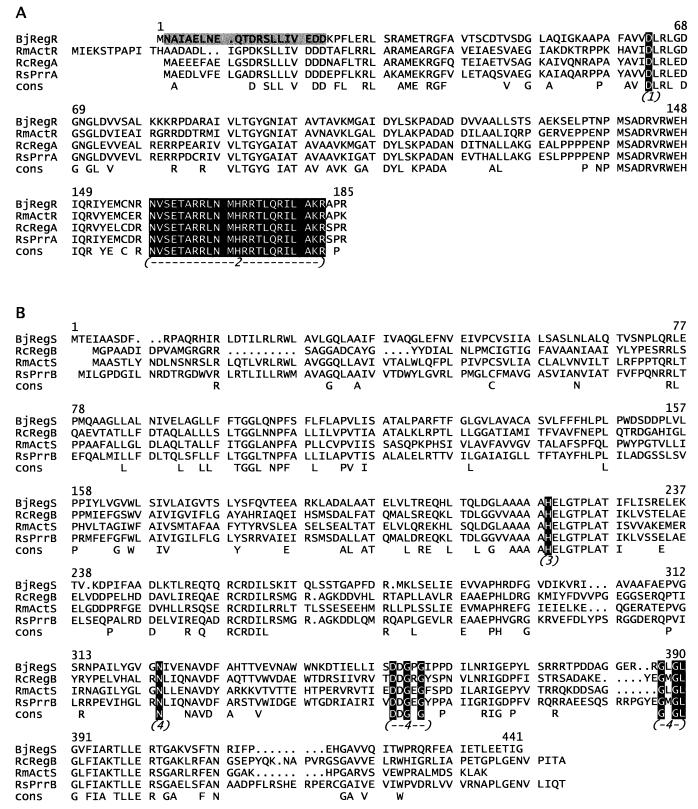
The predicted gene product of regR has 185 amino acids (Fig. 4A), resulting in a protein with a molecular mass of 20,160 Da and an isoelectric point of 9.29. The N terminus of the predicted RegR protein minus the N-formylmethionine is identical to the experimentally determined N-terminal amino acid sequence of purified UBP (Fig. 4A). The RegR sequence has all of the features of a response regulator belonging to the FixJ subfamily (60), including the putative phosphorylation site (Asp-63) and a proposed helix-turn-helix motif in a highly conserved region near the C terminus (sequence motif N159VSETARRLNMHRRTLQRILAK180; GCG Program HELIXTURNHELIX). RegR shows the highest degree of similarity to the response regulators ActR of S. meliloti (81% similar amino acids), RegA of R. capsulatus (80%), and RegA and PrrA of R. sphaeroides (76%). The regS gene codes for a putative histidine kinase consisting of 441 amino acids (Fig. 4B) with a predicted molecular mass of 48,077 Da and an isoelectric point of 5.14. RegS is most homologous to ActS of S. meliloti (65%) (59), RegB of R. capsulatus (59%) (38), and PrrB of R. sphaeroides (62%) (14). Sequence alignments to RegB, PrrB, and ActS (Fig. 4B) imply that the conserved autophosphorylation site in RegS is His-219, and the protein also contains the presumptive conserved kinase domains in the C terminus (60). The N terminus of RegS up to Val-183 is very hydrophobic, suggesting that RegS is membrane associated. However, no clearly defined transmembrane domains interrupted by hydrophilic loops are detectable. The open reading frame orf1 located downstream of, and divergently oriented to, regR encodes a protein consisting of 163 amino acids with no obvious sequence similarity to any database entry. We did not further analyze orf1.
FIG. 4.
Sequence alignments of the predicted B. japonicum RegR and RegS proteins to their homologs of R. capsulatus, R. sphaeroides, and S. meliloti. The consensus sequence (cons) in the bottom line was determined from amino acids identical in all four sequences. Putative essential domains are indicated by numbers in parentheses. Numbers above the sequence refer to positions in RegR and RegS. The sequences of RegA and RegB from R. sphaeroides, published in references 44 and 47, which are very similar to those of PrrA and PrrB, have been omitted for clarity. (A) Alignment of B. japonicum RegR (BjRegR) to S. meliloti ActR (RmActR [59]), R. capsulatus RegA (RcRegA [53]), and R. sphaeroides PrrA (RsPrrA [13]). The experimentally determined N terminus of Ubp (RegR) is shaded in gray. The presumptive phosphorylation site is indicated by “(1),” and the helix-turn-helix motif is indicated by “(2).” (B) Alignment of B. japonicum RegS (BjRegS) to S. meliloti ActS (RmActS [59]), R. capsulatus RegB (RcRegB [38]), and R. sphaeroides PrrB (RsPrrB [14]). The presumptive autophosphorylation site is indicated by “(3)”; the putative kinase domain, composed of three essential regions, is indicated by “(4).”
The regR and regS genes were mutated by deletion plus cassette insertion or by simple insertion of the Ω and aphII antibiotic resistance cassettes (Fig. 3C). The mutated genes were then integrated via homologous double crossover into the chromosomes of B. japonicum wild type and strain 7276B. The latter strain harbors a chromosomally integrated fixR′-′lacZ fusion and thus allows a test for potential consequences of regS and regR mutations on fixR-nifA expression.
Growth characteristics of regS and regR mutants.
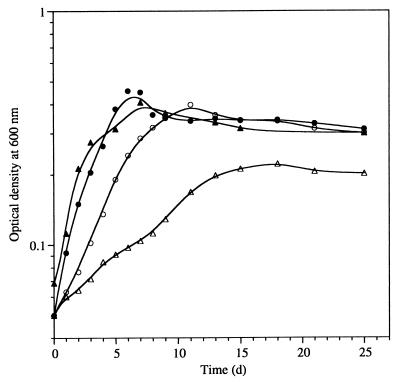
All regS and regR mutants were initially characterized by their growth behavior under aerobic conditions in PSY medium and under anaerobic conditions in YEM medium supplemented with KNO3. The aerobic growth rates of all regR mutants were slightly reduced compared with that of the wild type, and the mutants tended to synthesize higher levels of exopolysaccharides during the exponential growth phase. In contrast, all regS mutants grew like the wild type (data not shown). Under anaerobic conditions growth of all of the mutants was affected to different extents (Fig. 5). While regS mutants grew like the wild type, the generation time of the regR mutants 2426 and 2427 was threefold higher (6 days compared to 2 days for the wild type), and these mutants reached much lower final cell densities. Interestingly, regR mutants 2428 and 2429 did not grow at all under anaerobic conditions. Growth of the nifA mutant control strain A9 was only marginally slower than that of the wild type. Aerobic growth in YEM medium could not be assayed because of excessive exopolysaccharide production of all strains, including the wild type.
FIG. 5.
Anaerobic growth of B. japonicum wild type and nifA, regR, and regS mutants in YEM medium supplemented with KNO3. Symbols: •, wild type; ○, nifA mutant A9; ▴, regS mutant 2409; ▵, regR mutant 2426. Samples were taken from three parallel cultures of each strain, and growth was determined by measuring the OD600.
fixR-nifA UAS binding activity in extracts of regS and regR mutants.
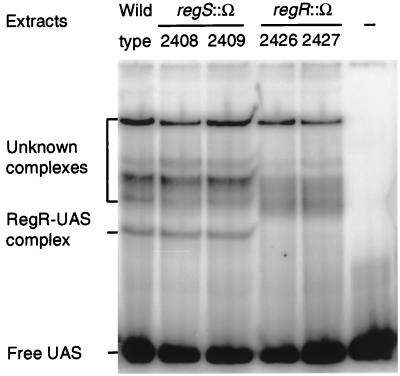
Crude protein extracts of aerobically grown regR and regS mutants were tested for binding activity to the fixR-nifA UAS in a gel retardation assay (Fig. 6). Extracts of regR mutants 2426 and 2427 reproducibly failed to form one of several protein-DNA complexes. The remaining, slower migrating complexes of unknown identity had been observed previously during RegR (UBP) purification, and their intensities varied. These additional complexes disappeared when enriched RegR preparations were used (Fig. 2B). Thus, it is likely that the complex formed by wild-type extracts but not by the extracts of regR mutants reflects the specific RegR-UAS complex. Interestingly, the regS mutations present in strains 2408 and 2409 had no effect on the formation of this complex.
FIG. 6.
UAS binding activity in extracts of regR and regS mutants analyzed by gel retardation. Crude extracts of aerobically grown cells of the strains indicated were prepared as described in Materials and Methods. Approximately 4 μg of protein was mixed with 0.1 ng of 32P-labeled double-stranded 32-bp oligonucleotide spanning the fixR-nifA −68 region (UAS) and incubated in the presence of poly(dI-dC) as a nonspecific competitor. The binding products were separated on a 6% nondenaturing polyacrylamide gel and visualized by phosphorimager analysis. The identity of the nonspecific, slow-migrating complexes is not known.
Effect of regS and regR mutations on fixR-nifA expression.
The presumed role of regR in the control of fixR-nifA expression was analyzed by monitoring the expression of a fixR′-′lacZ fusion present in regS and regR mutants and in the wild-type background at the levels of both β-galactosidase activity and mRNA. The results of the β-galactosidase activity tests are presented in Table 2. As known from previous studies (57), expression of fixR′-′lacZ in the wild-type background (strain 7276B) is about fivefold higher in anaerobically grown cells compared to aerobic cells, and the low expression level under aerobic conditions is dependent on the UAS located around position −68 (compare with strain 7277C). The regS mutations in strains 2408R and 2409R interfered only marginally, if at all, with the expression pattern of fixR′-′lacZ observed in the wild type. In contrast, the regR mutations present in strains 2426R and 2427R completely abolished aerobic fixR′-′lacZ expression, and anaerobic expression was reduced to ca. 10% of the level observed in the wild type.
TABLE 2.
Expression of chromosomally integrated fixR′-′lacZ fusions in B. japonicum regR and regS mutants
| Strain | Relevant genotype | β-Galactosidase activity (Miller U [mean ± SE])a
|
|
|---|---|---|---|
| Aerobic | Anaerobic | ||
| 110spc4 | Wild type | 1 ± 1 | 1 ± 1 |
| 7276B | fixR′-′lacZ | 246 ± 12 | 958 ± 63 |
| 7277C | fixR′-′lacZ ΔUAS | 11 ± 2 | 511 ± 43 |
| 2408R | fixR′-′lacZ regS::Ω | 270 ± 48 | 775 ± 100 |
| 2409R | fixR′-′lacZ regS::Ω | 170 ± 14 | 850 ± 144 |
| 2426R | fixR′-′lacZ regR::Ω | 3 ± 1 | 98 ± 36 |
| 2427R | fixR′-′lacZ regR::Ω | 1 ± 1 | 81 ± 14 |
Numbers are the mean values ± standard errors of at least three independent experiments. In each experiment at least two cultures of all strains were grown in parallel and assayed in duplicate. Bacteria were grown to mid exponential phase, i.e., for 2 days in PSY medium supplemented with 0.1% (wt/vol) arabinose in the case of aerobic cultures and for 6 days in YEM medium plus KNO3 in the case of anaerobic cultures.
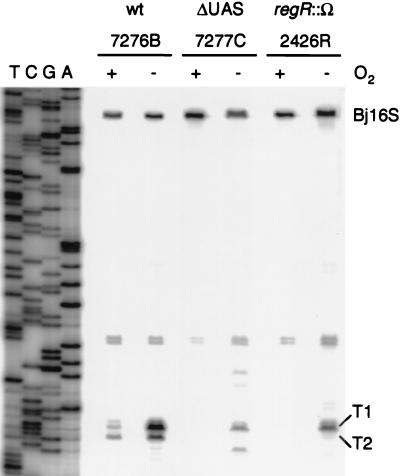
Next, we determined the effect of the regR mutation on the activity of the promoters fixRp1 and fixRp2 from which the fixR′-′lacZ reporter fusion is transcribed. The results of the corresponding primer extension experiments are shown in Fig. 7. The primer extension products derived from the fixR′-′lacZ fusion in the control strains 7276B and 7277C corresponded in length and abundance to those described previously by Barrios et al. (5). The dominant transcript under aerobic conditions (T2) originates from start point P2 (see also Fig. 1), and no transcript is detectable under these conditions in the UAS mutant strain 7277C. Under anaerobic conditions the major transcript (T1) is synthesized from start point P1 (see also Fig. 1), and this transcript is present also in strain 7277C. In agreement with the β-galactosidase activity tests, no transcript was found in aerobically grown cells of the regR mutant 2426R, and the intensity of the signal of transcript T1 as well as transcript T2 present in anaerobic cells was significantly reduced. Taken together, these results indicate that regR is absolutely required for aerobic fixR-nifA expression and that it also contributes to the expression of this operon under anaerobic conditions.
FIG. 7.
Primer extension analysis of the promoter fixRp1- and fixRp2-dependent fixR′-′lacZ transcripts in wild type and ΔUAS and regR mutants containing a chromosomally integrated fixR′-′lacZ fusion. Total RNA was purified from the indicated B. japonicum strains grown aerobically (+O2) in PSY medium or anaerobically (−O2) in YEM medium plus KNO3. Hybridization to RNA with the 32P-labeled oligonucleotides lac4B and PBj16S and reverse transcription of the fixR′-′lacZ mRNA and the 16S rRNA primary transcript, respectively, were performed as described in Materials and Methods. The products were separated on a 6% polyacrylamide gel next to a sequence ladder of plasmid pRJ7211 made with oligonucleotide lac4B. Transcripts T1, T2, and Bj16S (control) are marked. The origin of the unmarked reverse transcription products present in all lanes is not known; they had not been observed in similar, previous studies in which a shorter lacZ-specific oligonucleotide (lac4 [5]) was used.
Symbiotic phenotypes of regS and regR mutants.
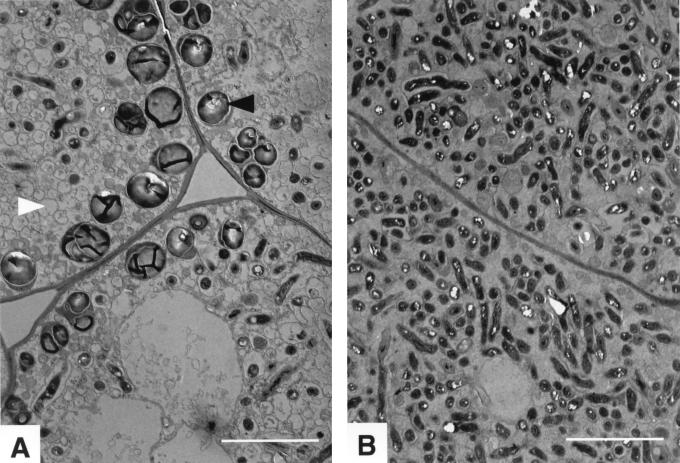
The ability of the regS::Ω and regR::Ω mutants to nodulate and to fix nitrogen symbiotically was examined in a plant infection test (Table 3). All mutants elicited nodules on soybean. The mutants produced about the same number of nodules as had the wild type, but mutant-elicited nodules were slightly smaller. However, nodules of regS mutants had an interior of slightly lighter pink color than wild-type nodules, and regR mutants produced nodules with a greenish interior. Necrotic nodule tissue as observed previously in nodules elicited by a nifA mutant (17, 56) was not found. The ultrastructure of nodules induced by regS mutants was indistinguishable from that of those induced by the wild type (data not shown). In contrast, only very few normally shaped bacteroids could be seen in nodules elicited by the regR mutants 2426 and 2427 (Fig. 8). Furthermore, more starch granules were present in these plant cells than in wild-type-infected cells. As with the nodulation phenotype, the regS and regR mutants also differed in their capacities to fix nitrogen. While regS mutants showed wild-type fixation activity, symbiotic nitrogen fixation of regR mutants was more than 97% reduced. Interestingly, however, it was not completely abolished as in nifA mutants.
TABLE 3.
Symbiotic phenotypes of regR and regS mutants compared to those of the wild type and the nifA mutant A9
| Strain | Relevant genotype | Characteristics (mean ± SE)a
|
||
|---|---|---|---|---|
| Nodule no. | Dry wt/nodule (mg) | Fix activity (% of wild type) | ||
| 110spc4 | Wild type | 31 ± 1 | 1.3 ± 0.06 | 100 ± 5 |
| A9 | nifAb | 59 | 0.3 | 0 |
| 2408 | regS | 36 ± 3 | 0.8 ± 0.05 | 69 ± 12 |
| 2409 | regS | 32 ± 3 | 1.0 ± 0.09 | 109 ± 8 |
| 2426 | regR | 28 ± 2 | 0.9 ± 0.05 | 2.3 ± 0.7 |
| 2427 | regR | 33 ± 2 | 0.8 ± 0.05 | 1.8 ± 0.4 |
Values are the means ± standard errors of at least 10 individual plants. Fixation (Fix) activity was measured as the amount of C2H2 reduced per minute per milligram of nodule weight (dry weight).
Values shown for the nifA mutant are taken from Fischer et al. (17) for comparison. The assay conditions were identical to those described here.
FIG. 8.
Electron micrographs showing the structure of soybean nodule cells infected by the regR mutant 2426 (A) and the wild-type 110spc4 (B). Black and white arrowheads in panel A mark starch granules and empty membrane vesicles, respectively. Bar = 5 μm.
Transcriptional mapping of regR.
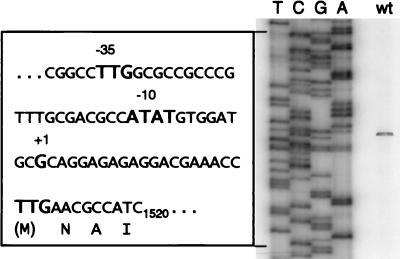
In all of the aforementioned tests it turned out that only regR but not regS mutations caused phenotypes distinguishable from those of the wild type. One possible interpretation of this phenomenon was that regR is expressed independently from regS. To test this inference, we performed extension experiments with primers that annealed to regR mRNA. In fact, the 5′ end of an mRNA species was detected that corresponded to a likely transcription start point in the regS-regR intergenic region (Fig. 9). This start point was located 20 nucleotides upstream of the regR open reading frame, and it was preceded by a −35/−10-type promoter region (Fig. 9). The same transcript was detectable with RNA from the regS mutant 2409 but not with RNA from the regR deletion strain 2426 (control), to which the primers cannot anneal (data not shown).
FIG. 9.
Transcriptional mapping of the regR promoter region. Total RNA was purified from B. japonicum wild type grown aerobically in PSY medium and used for primer extension. Hybridization to RNA with the 32P-labeled oligonucleotide PEregR2 and reverse transcription of the regR mRNA were performed as described in Materials and Methods. The products were separated on a 6% polyacrylamide gel next to a sequence ladder of plasmid pRJ2403 made with oligonucleotide PEregR2. The sequence of the indicated region (positions 1448 to 1520 of the database-submitted nucleotide sequence) is denoted in the box. It contains the transcription start point (+1), the putative −10 and −35 regions, and the probable translation start site (TTG).
DISCUSSION
We have identified a new two-component regulatory system, RegSR, of which at least the RegR protein is involved in the control of both aerobic and anaerobic expression of the fixR-nifA operon in B. japonicum. The existence of a positive regulator required for aerobic fixR-nifA expression had been implied by previous work of Thöny et al. (57, 58), in which a DNA region upstream of fixR-nifA (UAS) was shown to be necessary for expression of fixR′-′lacZ fusions. An as yet unknown protein from crude extracts could bind to this UAS in gel retardation experiments.
This predicted activator protein has now been discovered. Using different chromatography steps including a UAS affinity column, we could successfully enrich minute quantities of this cellular regulator in an active, DNA-binding form. The corresponding gene, regR, was cloned, and its predicted N-terminal amino acid sequence was shown to be identical to that of the purified DNA-binding protein. Functional characterization of the regR gene confirmed the regulatory model of Thöny et al. (57, 58). The regR mutants lacked specific fixR-nifA UAS binding activity, and aerobic fixR′-′lacZ expression was completely abolished in them. Moreover, anaerobic fixR′-′lacZ expression was also drastically reduced. The primer extension experiments documented that RegR is absolutely required for the synthesis of transcript T2 under all oxygen conditions and for the synthesis of transcript T1 under aerobic conditions and that RegR also contributes to the level of T1 in anaerobic cells. These results led us to propose an expanded model of the control of fixR-nifA expression in B. japonicum (Fig. 1). Regardless of the cellular oxygen status, RegR binds to the fixR-nifA UAS and activates transcription from the fixRp2 promoter, leading to a low level of NifA synthesis. The ς factor involved in this process remains to be identified. Under aerobic conditions the NifA protein is instantaneously degraded (37), but if the environment is anaerobic NifA activates its own expression via the ς54-dependent promoter fixRp1. Under these conditions, NifA also activates transcription of all of its other target genes, e.g., the nif and fix genes.
This model is in perfect agreement with the previous results of Thöny et al. (57) and Barrios et al. (5), who demonstrated the requirement of an intact UAS for fixRp2-dependent transcription. Under anaerobic conditions, residual NifA-dependent fixR-nifA expression was observed from the fixRp1 promoter. This means that some NifA protein must be synthesized in regR mutants enabling autoactivation of the operon upon switching to anaerobic conditions where NifA becomes active. The reduced level of fixR-nifA expression in regR mutants under anaerobic conditions may have two causes. First, the RegR-dependent transcripts are absent there. Second, the synthesis of the NifA-dependent transcript T1 originating from fixRp1 might be diminished because of a potential interference of the ς54-RNA polymerase with that RNA polymerase which, in the wild type, acts in concert with RegR but which may be unable to clear the promoter in the regR mutant. Indeed, Barrios et al. (5, 6) have found competition of the two RNA polymerase holoenzymes for the overlapping fixRp1 and fixRp2 promoter regions.
Another open reading frame, regS, was found upstream of the regR gene. The newly identified RegS and RegR two-component regulatory proteins exhibit the typical features of membrane-bound histidine kinases and soluble response regulators, respectively. The greatest degree of similarity was found with the ActSR system involved in acid tolerance in S. meliloti (59) and with RegBA and PrrBA of R. capsulatus and R. sphaeroides, respectively (13, 14, 38, 44, 47, 53). RegBA and PrrBA are integrated in a complex regulatory network which induces at the transcriptional level the synthesis of photosynthetic light-harvesting complexes, reaction centers, photosynthetic pigments, and cytochrome c2 in response to cellular oxygen deprivation (for a review, see reference 8). The signal that is transduced to RegR in B. japonicum remains to be identified. Given its involvement in both aerobic and anaerobic fixR-nifA expression it seems very unlikely to be oxygen per se. This notion is further supported by the absence in RegS of a heme-binding domain and cysteine motifs known to be involved in oxygen-sensing by the FixL protein of S. meliloti (1) and the Fnr protein of E. coli (26), respectively.
The recent findings that the PrrBA system in R. sphaeroides is required for transcriptional activation of the cbb operons I and II encoding two forms of ribulose 1,5-bisphosphate carboxylase-oxygenase (Rubisco) and, most strikingly, also for diazotrophic growth suggested a more global control function in fundamental processes such as photosynthesis and CO2 and N2 fixation (27, 47). Common to these processes is their requirement for reducing equivalents; thus, they are strictly dependent on the cellular redox state. In fact, mutations that affect electron transport in R. sphaeroides led to the induction of photosynthesis genes, possibly via activation of the PrrBA system by the accumulation of a critical redox intermediate (24, 42, 62). Similarly, activation of the ActSR system of S. meliloti by low pH may occur via sensing of the redox state of a pH-sensitive compound. If the RegSR system of B. japonicum were to be redox responsive, one must assume, however, that it is at least partially active under both aerobic and anaerobic conditions, since RegR-dependent expression of fixR-nifA was observed under both growth conditions.
Aerobic growth of the regS and regR mutants was almost indistinguishable from that of the wild type, indicating that genes essential for heterotrophic aerobic growth do not belong to the RegSR regulon. In contrast, anaerobic growth of regR mutants was drastically retarded. This defect cannot solely be attributed to reduced fixR-nifA expression since growth was much less affected in the nifA mutant A9. Hence, one might speculate about the existence of other RegR-dependent targets whose products, unlike those of fixR-nifA, are required for anaerobic growth under nitrate-respiring conditions. By analogy with the critical role that PrrBA plays in CO2 fixation of R. sphaeroides (47), alternative targets for RegR control in B. japonicum might also include the CO2 fixation genes that enable B. japonicum to grow chemoautotrophically (reference 32 and references therein).
B. japonicum regR mutants were symbiotically defective, as indicated by the almost complete lack of nitrogen fixation activity and the altered nodule ultrastructure. Interestingly, however, the symbiotic defect was clearly less drastic than that described previously for nifA mutants (17, 56). This difference is possibly due to the residual level of nifA expression observed in regR mutants, which might lead to the synthesis of small amounts of NifA protein sufficient to suppress the plant defense reaction but insufficient for optimal expression of the nitrogen fixation genes. Electron micrographs of infected nodules imply that regR mutants fail to efficiently multiply and/or persist in plant cells. It is possible that this is a consequence of the impaired growth of regR mutants under oxygen-limiting conditions. Alternatively, a specific function required for the symbiotic lifestyle might be affected in these mutants. Although several lines of evidence clearly show that RegR activates the expression of nifA, we cannot rule out the formal possibility that the Fix− phenotype is caused by the hampered bacteroid development. It is interesting to note here that, in the wild type, the UAS- and RegR-dependent fixR-nifA transcript T2 is not detectable in 30-day-old nodules (5), indicating that the RegR-controlled functions are required during the earlier stages of the symbiotic interaction.
An intriguing observation was the striking phenotypic difference between regS and regR mutants. A similar phenomenon has been described for regB and regA mutants of R. capsulatus (38). The genetic linkage of the regSR genes along with the pronounced similarity of RegS and RegR to other bacterial histidine kinases and response regulators, respectively, would suggest that they are cognate two-component regulatory partners. In fact, recent in vitro phosphorylation experiments provided solid support for this assumption (12). Although we presently cannot exclude the possibility that RegR functions as a transcriptional activator in its nonphosphorylated form, we favor the idea that it is phosphorylated via cross talk by an alternative protein kinase in the regS mutant, i.e., this implies the existence of a second RegS-like protein in B. japonicum. Cross talk among two-component regulatory systems is well documented in vivo and in vitro (for a review, see reference 61). For example, a mutation in the prrB gene of R. sphaeroides could be partly complemented in vivo by overexpression of hupT, the sensor gene for regulation of the hydrogen uptake system (19). Similarly, cross talk was observed in B. japonicum between the nodulation regulatory proteins NodV and NwsB (21). Regardless of which mechanism is responsible for suppression of the effect of regS mutations, expression of regR seems to be little (if at all) affected by the mutations introduced into the regS gene. This was apparently not due to transcriptional outreading from the resistance gene cassette. Instead, we obtained evidence for the presence of a promoter located immediately upstream of regR, which was active not only in the wild type but also in a regS mutant background. This suggests that, even if regS and regR were to form an operon, a substantial amount of regR mRNA can be synthesized independently of the regS promoter. Further transcriptional analyses of the regSR region should clarify this point.
With the regR gene described in this study we have added a new element to the complex regulatory network controlling nif and fix gene expression in B. japonicum (Fig. 1). As with the FixLJ-FixK2 cascade, the NifA cascade now also includes a response regulator of a two-component regulatory system at the (currently known) top level. As long as the signal for the RegSR system is unidentified the physiological meaning of this additional control level remains speculative. Quite likely, it provides B. japonicum cells with the possibility to integrate into the regulatory circuits an additional external or internal signal. Moreover, given the global function of the RegSR homologs described in other bacteria, it seems possible that RegR forms a link between the nitrogen fixation system and other metabolic routes. It might be of interest to examine whether a RegR-like protein plays a role in nifA regulation in those diazotrophs in which this gene is expressed under aerobic conditions (e.g., Rhizobium etli [34] or Rhizobium leguminosarum bv. viciae [43]).
ACKNOWLEDGMENTS
We are indebted to G. Klug for the generous gift of the regA plasmid and anti-R. capsulatus RegA serum, R. Hermann for analyzing the nodule ultrastructure, P. James for N-terminal protein sequencing, and C. Kündig for preparation of the cosmid library available in our laboratory. We thank M. Göttfert and S. Röthlisberger for help with automated DNA sequencing.
This study was supported by a grant from the Swiss Federal Institute of Technology, Zürich, and the Swiss National Foundation for Scientific Research.
REFERENCES
- 1.Agron P G, Helinski D R. Symbiotic expression of Rhizobium meliloti nitrogen fixation genes is regulated by oxygen. In: Hoch J A, Silhavy T J, editors. Two-component signal transduction. Washington, D.C: American Society for Microbiology; 1995. pp. 275–287. [Google Scholar]
- 2.Anthamatten D, Scherb B, Hennecke H. Characterization of a fixLJ-regulated Bradyrhizobium japonicum gene sharing similarity with the Escherichia coli fnr and Rhizobium meliloti fixK genes. J Bacteriol. 1992;174:2111–2120. doi: 10.1128/jb.174.7.2111-2120.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Babst M, Hennecke H, Fischer H M. Two different mechanisms are involved in the heat shock regulation of chaperonin gene expression in Bradyrhizobium japonicum. Mol Microbiol. 1996;19:827–839. doi: 10.1046/j.1365-2958.1996.438968.x. [DOI] [PubMed] [Google Scholar]
- 4.Baker M E. Human placental 17β-hydroxysteroid dehydrogenase is homologous to NodG protein of Rhizobium meliloti. Mol Endocrinol. 1989;3:881–884. doi: 10.1210/mend-3-5-881. [DOI] [PubMed] [Google Scholar]
- 5.Barrios H, Fischer H M, Hennecke H, Morett E. Overlapping promoters for two different RNA polymerase holoenzymes control Bradyrhizobium japonicum nifA expression. J Bacteriol. 1995;177:1760–1765. doi: 10.1128/jb.177.7.1760-1765.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Barrios H, Grande R, Olvera L, Morett E. In vivo genomic footprinting, chemical probing and mutagenesis analyses reveal that the complex Bradyrhizobium japonicum fixR-nifA promoter region is differently occupied and melted by two distinct RNA polymerase holoenzymes. Proc Natl Acad Sci USA. 1998;95:1014–1019. doi: 10.1073/pnas.95.3.1014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Batut J, Boistard P. Oxygen control in Rhizobium. Antonie Leeuwenhoek. 1994;66:129–150. doi: 10.1007/BF00871636. [DOI] [PubMed] [Google Scholar]
- 8.Bauer C E, Bird T H. Regulatory circuits controlling photosynthesis gene expression. Cell. 1996;85:5–8. doi: 10.1016/s0092-8674(00)81074-0. [DOI] [PubMed] [Google Scholar]
- 9.Beck C, Marty R, Kläusli S, Hennecke H, Göttfert M. Dissection of the transcription machinery for housekeeping genes of Bradyrhizobium japonicum. J Bacteriol. 1997;179:364–369. doi: 10.1128/jb.179.2.364-369.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Casadaban M J, Martinez-Arias A, Shapira S K, Chou J. β-Galactosidase gene fusions for analyzing gene expression in Escherichia coli and yeast. Methods Enzymol. 1983;100:293–308. doi: 10.1016/0076-6879(83)00063-4. [DOI] [PubMed] [Google Scholar]
- 11.Daniel R M, Appleby C A. Anaerobic-nitrate, symbiotic and aerobic growth of Rhizobium japonicum: effects on cytochrome P450, other haemoproteins, nitrate and nitrite reductases. Biochim Biophys Acta. 1972;275:347–354. doi: 10.1016/0005-2728(72)90215-0. [DOI] [PubMed] [Google Scholar]
- 12.Emmerich, R., et al. Unpublished results.
- 13.Eraso J M, Kaplan S. prrA, a putative response regulator involved in oxygen regulation of photosynthesis gene expression in Rhodobacter sphaeroides. J Bacteriol. 1994;176:32–43. doi: 10.1128/jb.176.1.32-43.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Eraso J M, Kaplan S. Oxygen-insensitive synthesis of the photosynthetic membranes of Rhodobacter sphaeroides: a mutant histidine kinase. J Bacteriol. 1995;177:2695–2706. doi: 10.1128/jb.177.10.2695-2706.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fischer H M. Genetic regulation of nitrogen fixation in rhizobia. Microbiol Rev. 1994;58:352–386. doi: 10.1128/mr.58.3.352-386.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fischer H M. Environmental regulation of rhizobial symbiotic nitrogen fixation genes. Trends Microbiol. 1996;4:317–320. doi: 10.1016/0966-842x(96)10049-4. [DOI] [PubMed] [Google Scholar]
- 17.Fischer H M, Alvarez-Morales A, Hennecke H. The pleiotropic nature of symbiotic regulatory mutants: Bradyrhizobium japonicum nifA gene is involved in control of nif gene expression and formation of determinate symbiosis. EMBO J. 1986;5:1165–1173. doi: 10.1002/j.1460-2075.1986.tb04342.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fischer H M, Babst M, Kaspar T, Acuña G, Arigoni F, Hennecke H. One member of a groESL-like chaperonin multigene family in Bradyrhizobium japonicum is co-regulated with symbiotic nitrogen fixation genes. EMBO J. 1993;12:2901–2912. doi: 10.1002/j.1460-2075.1993.tb05952.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gomelsky M, Kaplan S. Isolation of regulatory mutants in photosynthesis gene expression in Rhodobacter sphaeroides 2.4.1 and partial complementation of a PrrB mutant by the HupT histidine-kinase. Microbiology. 1995;141:1805–1819. doi: 10.1099/13500872-141-8-1805. [DOI] [PubMed] [Google Scholar]
- 20.Göttfert M, Hitz S, Hennecke H. Identification of nodS and nodU, two inducible genes inserted between the Bradyrhizobium japonicum nodYABC and nodIJ genes. Mol Plant-Microbe Interact. 1990;3:308–316. doi: 10.1094/mpmi-3-308. [DOI] [PubMed] [Google Scholar]
- 21.Grob P, Hennecke H, Göttfert M. Cross-talk between the two-component regulatory systems NodVW and NwsAB of Bradyrhizobium japonicum. FEMS Microbiol Lett. 1994;120:349–354. [Google Scholar]
- 22.Hahn M, Hennecke H. Localized mutagenesis in Rhizobium japonicum. Mol Gen Genet. 1984;193:46–52. [Google Scholar]
- 23.Henrich B, Schmidtberger B. Positive-selection vector with enhanced lytic potential based on a variant of ΦX174 phage gene E. Gene. 1995;154:51–54. doi: 10.1016/0378-1119(94)00839-k. [DOI] [PubMed] [Google Scholar]
- 24.Horne I M, Pemberton J M, McEwan A G. Photosynthesis gene expression in Rhodobacter sphaeroides is regulated by redox changes which are linked to electron transport. Microbiology. 1996;142:2831–2838. [Google Scholar]
- 25.Inoue K, Kouadio J L K, Mosley C S, Bauer C E. Isolation and in vitro phosphorylation of sensory transduction components controlling anaerobic induction of light harvesting and reaction center gene expression in Rhodobacter capsulatus. Biochemistry. 1995;34:391–396. doi: 10.1021/bi00002a002. [DOI] [PubMed] [Google Scholar]
- 26.Jordan P A, Thomson A J, Ralph E T, Guest J R, Green J. FNR is a direct oxygen sensor having a biphasic response curve. FEBS Lett. 1997;416:349–352. doi: 10.1016/s0014-5793(97)01219-2. [DOI] [PubMed] [Google Scholar]
- 27.Joshi H M, Tabita F R. A global two component signal transduction system that integrates the control of photosynthesis, carbon dioxide assimilation, and nitrogen fixation. Proc Natl Acad Sci USA. 1996;93:14515–14520. doi: 10.1073/pnas.93.25.14515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kaspar T. Reinigung und Charakterisierung des postulierten Regulatorproteins für die aerobe fixR-nifA-Expression in Bradyrhizobium japonicum. Ph.D. thesis. Zürich, Switzerland: Eidgenössische Technische Hochschule; 1997. [Google Scholar]
- 29.Kullik I, Fritsche S, Knobel H, Sanjuan J, Hennecke H, Fischer H M. Bradyrhizobium japonicum has two differentially regulated, functional homologs of the ς54 gene (rpoN) J Bacteriol. 1991;173:1125–1138. doi: 10.1128/jb.173.3.1125-1138.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kullik I, Hennecke H, Fischer H M. Inhibition of Bradyrhizobium japonicum nifA-dependent nif gene activation by oxygen occurs at the NifA protein level and is irreversible. Arch Microbiol. 1989;151:191–197. [Google Scholar]
- 31.Kündig C, Hennecke H, Göttfert M. Correlated physical and genetic map of the Bradyrhizobium japonicum 110 genome. J Bacteriol. 1993;175:613–622. doi: 10.1128/jb.175.3.613-622.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.McClung C R, Chelm B K. A genetic locus essential for formate-dependent growth of Bradyrhizobium japonicum. J Bacteriol. 1987;169:3260–3267. doi: 10.1128/jb.169.7.3260-3267.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Merrick M J. Regulation of nitrogen fixation genes in free-living and symbiotic bacteria. In: Stacey G, Burris R H, Evans H J, editors. Biological nitrogen fixation. New York, N.Y: Chapman & Hall; 1992. pp. 835–876. [Google Scholar]
- 34.Michiels J, D’hooghe I, Verreth C, Pelemans H, Vanderleyden J. Characterization of the Rhizobium leguminosarum biovar phaseoli nifA gene, a positive regulator of nif gene expression. Arch Microbiol. 1994;161:404–408. doi: 10.1007/BF00288950. [DOI] [PubMed] [Google Scholar]
- 35.Miller J H. Experiments in molecular genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1972. [Google Scholar]
- 36.Morett E, Buck M. In vivo studies on the interaction of RNA polymerase-ς54 with the Klebsiella pneumoniae and Rhizobium meliloti nifH promoters: the role of NifA in the formation of an open promoter complex. J Mol Biol. 1989;210:65–77. doi: 10.1016/0022-2836(89)90291-x. [DOI] [PubMed] [Google Scholar]
- 37.Morett E, Fischer H M, Hennecke H. Influence of oxygen on DNA binding, positive control, and stability of the Bradyrhizobium japonicum NifA regulatory protein. J Bacteriol. 1991;173:3478–3487. doi: 10.1128/jb.173.11.3478-3487.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mosley C S, Suzuki J Y, Bauer C E. Identification and molecular genetic characterization of a sensor kinase responsible for coordinately regulating light harvesting and reaction center gene expression in response to anaerobiosis. J Bacteriol. 1994;176:7566–7573. doi: 10.1128/jb.176.24.7566-7573.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Narberhaus F, Krummenacher P, Fischer H M, Hennecke H. Three disparately regulated genes for ς32-like transcription factors in Bradyrhizobium japonicum. Mol Microbiol. 1997;24:93–104. doi: 10.1046/j.1365-2958.1997.3141685.x. [DOI] [PubMed] [Google Scholar]
- 40.Nellen-Anthamatten, D., P. Rossi, O. Preisig, I. Kullik, M. Babst, H. M. Fischer, and H. Hennecke. Bradyrhizobium japonicum FixK2, a crucial distributor in the FixLJ-dependent regulatory cascade for the control of low-oxygen-inducible genes. Submitted for publication. [DOI] [PMC free article] [PubMed]
- 41.Norrander J, Kempe T, Messing J. Construction of improved M13 vectors using oligonucleotide-directed mutagenesis. Gene. 1983;26:101–106. doi: 10.1016/0378-1119(83)90040-9. [DOI] [PubMed] [Google Scholar]
- 42.O’Gara J P, Kaplan S. Evidence for the role of redox carriers in photosynthesis gene expression and carotenoid biosynthesis in Rhodobacter sphaeroides 2.4.1. J Bacteriol. 1997;179:1951–1961. doi: 10.1128/jb.179.6.1951-1961.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Patschkowski T, Schlüter A, Priefer U B. Rhizobium leguminosarum bv. viciae contains a second fnr/fixK-like gene and an unusual fixL homologue. Mol Microbiol. 1996;21:267–280. doi: 10.1046/j.1365-2958.1996.6321348.x. [DOI] [PubMed] [Google Scholar]
- 44.Phillips-Jones M K, Hunter C N. Cloning and nucleotide sequence of regA, a putative response regulator gene of Rhodobacter sphaeroides. FEMS Microbiol Lett. 1994;116:269–276. doi: 10.1111/j.1574-6968.1994.tb06714.x. [DOI] [PubMed] [Google Scholar]
- 45.Preisig O, Zufferey R, Thöny-Meyer L, Appleby C A, Hennecke H. A high-affinity cbb3-type cytochrome oxidase terminates the symbiosis-specific respiratory chain of Bradyrhizobium japonicum. J Bacteriol. 1996;178:1532–1538. doi: 10.1128/jb.178.6.1532-1538.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Prentki P, Krisch H M. In vitro insertional mutagenesis with a selectable DNA fragment. Gene. 1984;29:303–313. doi: 10.1016/0378-1119(84)90059-3. [DOI] [PubMed] [Google Scholar]
- 47.Qian Y L, Tabita F R. A global signal transduction system regulates aerobic and anaerobic CO2 fixation in Rhodobacter sphaeroides. J Bacteriol. 1996;178:12–18. doi: 10.1128/jb.178.1.12-18.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Regensburger B, Hennecke H. RNA polymerase from Rhizobium japonicum. Arch Microbiol. 1983;135:103–109. doi: 10.1007/BF00408017. [DOI] [PubMed] [Google Scholar]
- 49.Rüther U. Construction and properties of a new cloning vehicle, allowing direct screening for recombinant plasmids. Mol Gen Genet. 1980;178:475–477. doi: 10.1007/BF00270503. [DOI] [PubMed] [Google Scholar]
- 50.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 51.Sanger F, Nicklen S, Coulson A R. Sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Schägger H, von Jagow G. Tricine-sodium dodecyl sulfate-polyacrylamide gel electrophoresis for the separation of proteins in the range from 1 to 100 kDa. Anal Biochem. 1987;166:368–379. doi: 10.1016/0003-2697(87)90587-2. [DOI] [PubMed] [Google Scholar]
- 53.Sganga M W, Bauer C E. Regulatory factors controlling photosynthetic reaction center and light harvesting gene expression in Rhodobacter capsulatus. Cell. 1992;68:945–954. doi: 10.1016/0092-8674(92)90037-d. [DOI] [PubMed] [Google Scholar]
- 54.Shingler V. Signal sensing by ς54-dependent regulators: derepression as a control mechanism. Mol Microbiol. 1996;19:409–416. doi: 10.1046/j.1365-2958.1996.388920.x. [DOI] [PubMed] [Google Scholar]
- 55.Simon R, Priefer U, Pühler A. Vector plasmids for in vivo and in vitro manipulation of gram-negative bacteria. In: Pühler A, editor. Molecular genetics of the bacteria-plant interaction. Heidelberg, Germany: Springer-Verlag; 1983. pp. 98–106. [Google Scholar]
- 56.Studer D, Gloudemans T, Franssen H J, Fischer H M, Bisseling T, Hennecke H. Involvement of the bacterial nitrogen fixation regulatory gene (nifA) in control of nodule-specific host-plant gene expression. Eur J Cell Biol. 1987;45:177–184. [Google Scholar]
- 57.Thöny B, Anthamatten D, Hennecke H. Dual control of the Bradyrhizobium japonicum symbiotic nitrogen fixation regulatory operon fixR nifA: analysis of cis- and trans-acting elements. J Bacteriol. 1989;171:4162–4169. doi: 10.1128/jb.171.8.4162-4169.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Thöny B, Fischer H M, Anthamatten D, Bruderer T, Hennecke H. The symbiotic nitrogen fixation regulatory operon (fixR-nifA) of Bradyrhizobium japonicum is expressed aerobically and is subject to a novel, nifA-independent type of activation. Nucleic Acids Res. 1987;15:8479–8499. doi: 10.1093/nar/15.20.8479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tiwari R P, Reeve W G, Dilworth M J, Glenn A R. Acid tolerance in Rhizobium meliloti strain WSM419 involves a two-component sensor-regulator system. Microbiology. 1996;142:1693–1704. doi: 10.1099/13500872-142-7-1693. [DOI] [PubMed] [Google Scholar]
- 60.Volz K. Structural and functional conservation in response regulators. In: Hoch J A, Silhavy T J, editors. Two-component signal transduction. Washington, D.C: American Society for Microbiology; 1995. pp. 53–64. [Google Scholar]
- 61.Wanner B L. Is cross regulation by phosphorylation of two-component response regulator proteins important in bacteria? J Bacteriol. 1992;174:2053–2058. doi: 10.1128/jb.174.7.2053-2058.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zeilstra-Ryalls J H, Kaplan S. Control of hemA expression in Rhodobacter sphaeroides 2.4.1: regulation through alterations in the cellular redox state. J Bacteriol. 1996;178:985–993. doi: 10.1128/jb.178.4.985-993.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]