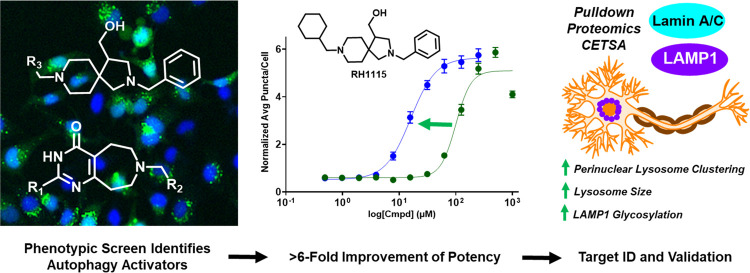
Abstract
Autophagy is a major catabolic degradation and recycling process that maintains homeostasis in cells and is especially important in postmitotic neurons. We implemented a high-content phenotypic assay to discover small molecules that promote autophagic flux and completed target identification and validation studies to identify protein targets that modulate the autophagy pathway and promote neuronal health and survival. Efficient syntheses of the prioritized compounds were developed to readily access analogues of the initial hits, enabling initial structure–activity relationship studies to improve potency and preparation of a biotin-tagged pulldown probe that retains activity. This probe facilitated target identification and validation studies through pulldown and competition experiments using both an unbiased proteomics approach and western blotting to reveal Lamin A/C and LAMP1 as the protein targets of compound RH1115. Evaluation of RH1115 in neurons revealed that this compound induces changes to LAMP1 vesicle properties and alters lysosome positioning. Dysfunction of the autophagy–lysosome pathway has been implicated in a variety of neurodegenerative diseases, including Alzheimer’s disease, highlighting the value of new strategies for therapeutic modulation and the importance of small-molecule probes to facilitate the study of autophagy regulation in cultured neurons and in vivo.
Keywords: small molecules, autophagy, phenotypic high-throughput screen, target identification, neurodegenerative diseases, medicinal chemistry
Introduction
Macroautophagy, hereafter referred to as autophagy, is an important cellular homeostasis pathway that has a critical role in the catabolic degradation and recycling of long-lived proteins and organelles.1 This intracellular process involves the engulfment of portions of the cytoplasm in a double-membraned structure known as an autophagosome, and upon fusion with a lysosome, the autophagic cargo is degraded.2 A functional autophagy pathway is especially important in neurons, which are postmitotic and do not replicate, so the need to remove cellular debris and toxins is paramount for neuron survival.3,4 In neurons, autophagosomes originate in distal axons and travel toward the soma, along the way fusing with lysosomes to create autolysosomes and facilitate degradation of the contents.5,6 Autophagy has been associated with neurodevelopment, neuronal homeostasis, and neuronal activity and plasticity.7−9 In addition, dysfunction of autophagy has also been related to many neurological disease states that involve protein misfolding and aggregation,10 including Huntington’s disease (HD), in which aggregation occurs from CAG repeats on the first exon of the huntingtin gene,11−13 Parkinson’s disease (PD), in which a variety of factors cause accumulation of α-synuclein and polyubiquinated proteins,14−16 and amyotrophic lateral sclerosis (ALS), in which genetic mutations lead to accumulation of ubiquitinated cytoplasmic inclusions like the protein TDP-43.17−19 Recent work has also linked autophagy and lysosome dysfunction to the progression of Alzheimer’s disease (AD).20−22 Studies in human and mouse models of the disease reveal the presence of numerous autophagic and lysosomal intermediates in brain tissue, suggestive of defects in fusion and/or transport and clearance of these organelles.23−25 Moreover, dysfunctional autophagy in neurons results in axonal organelle buildup similar to that observed in AD mouse models.26,27 Further studies have shown that the expression of the key autophagy regulator protein Beclin 1 is reduced in early AD, suggesting a connection between autophagy dysregulation and AD.22,28
Although a large body of literature indicates the importance of the autophagy–lysosome pathway (ALP) in AD and AD-related disorders, little emphasis is placed on neuronal ALP and the unique targeting of proteins for degradation from the axons and axon terminals, instead of just the degradative compartment in the cell body.23,24,29,30 In wild-type neurons, late endosomes and autophagosomes within distal regions of axons play important housekeeping functions by sequestering old or misfolded proteins and damaged organelles.29,30 The maturation process within axons is coupled with the retrograde axonal transport toward the neuronal cell body,30 and this retrograde movement of autophagosomes is dependent on fusion with the endolysosomal system. Therefore, any transport defect in endolysosomes or hybrid organelles would lead to autophagic cargo accumulation in axons. Indeed, perturbation of retrograde axonal lysosome transport has been shown to exacerbate amyloid plaque pathology in AD models.31 Taken together, these factors highlight autophagy activation and autophagosome maturation and clearance, including optimal lysosome transport, as a promising approach for therapeutic development to treat neurodegeneration. In addition, there is a growing appreciation for the intimate linkage of lysosome movement and positioning in cells to both lysosome function and the physiological state of the cell.32,33 For example, acidification of the cytoplasm causes dispersion of lysosomes toward the cell periphery, while its alkalinization causes their return to a perinuclear location. Furthermore, during dendritic cell maturation, lysosome tubulation and movement to the periphery aids in the delivery of MHC-II molecules to the cell surface.34,35 Therefore, a drug that modulates lysosome positioning could enable context-dependent control of lysosome functions in different cell types, creating additional therapeutic avenues beyond neurodegenerative diseases.
Recent therapeutic development efforts for AD have primarily focused on the reduction of Aβ aggregation by targeting secretase proteins responsible for Aβ formation from amyloid precursor protein (APP), including β-site APP cleavage enzyme (BACE1), a membrane-bound aspartyl protease responsible for the initiation of APP cleavage to generate Aβ, and γ-secretase, a complex of presenilin, nicastrin, Aph1, and Pen2 that performs the final cleavage of APP to generate Aβ.36−38 However, clinical trials revealed that despite a reduction in Aβ production, inhibition of APP cleavage resulted in limited cognitive improvement in AD patients, notable adverse side effects, and/or an inability to successfully clear tau and phosphorylated tau aggregates.39−43 These challenges highlight the importance of discovering new targets and strategies for the treatment of AD and other neurodegenerative diseases. Rapamycin, a well-established mTOR inhibitor and autophagy inducer, has been shown to reduce aggregates and increase protein degradation in HD and ALS mouse models and to delay the onset of behavioral abnormalities that arise in the mice.12,14,18 Alternative mTOR-independent activators of autophagy have shown similar success in clearing aggregation and preventing neuronal toxicity in HD and PD models.11,44−46 For example, the small-molecule trehalose has been shown to improve neuronal survival and clear tau aggregates in mouse models by restoring optimal autophagic flux in HD, PD, and ALS.47−50 Autophagy induction has also shown promise in AD models. For example, rapamycin treatment promotes clearance of Aβ aggregates and rescue of memory defects in AD mouse models.51,52 Other mTOR-independent autophagy inducers have also shown success in Aβ and tau clearance in addition to neuroprotective effects.53−55 Due to the important role of mTOR in immunity and the limited knowledge about targets and the mechanism of action of mTOR-independent autophagy inducers, we decided to capitalize on the major strength of phenotypic drug discovery to facilitate both hit discovery and optimization and target identification and validation.56−58 This approach led to successful identification of novel lead compounds and protein targets that modulate the autophagy pathway to support our efforts to develop novel therapeutic strategies for neurodegenerative diseases.
Results and Discussion
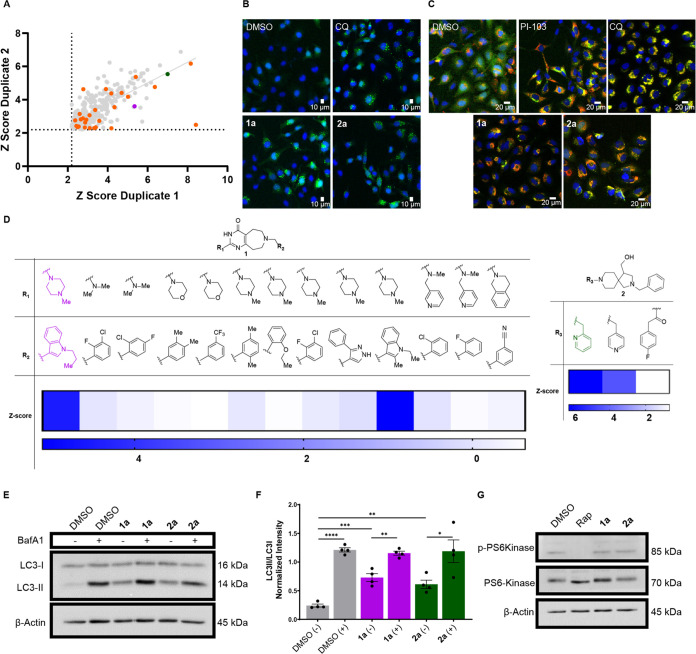
To measure autophagy modulation following compound treatment, we used the eGFP-LC3 puncta formation assay as our primary high-content screen (HCS) with the goal of identifying autophagy modulators with a variety of mechanisms of action to discover new cellular targets that improve disease-relevant phenotypes.59 Pro-LC3 is cleaved by the cysteine protease ATG4 to produce cytosolic LC3-I, and upon autophagy activation, ATG7 and ATG3 catalyze the conjugation of phosphatidylethanolamine (PE) to LC3-I by the ATG12-ATG5-ATG16L1 complex to form LC3-II, which is recruited to the autophagosome membrane and serves as a biomarker for autophagic flux.60−62 Compounds that modulate autophagy cause an increase in LC3-II, either by activating autophagy and increasing the number of autophagosomes formed, or by inhibiting autophagosome–lysosome fusion and causing an accumulation of autophagosomes. Both of these possibilities are identified by quantifying the number of green dots (puncta) per cell in the GFP-LC3 assay as an indication of autophagosome numbers.63 The antimalarial drug chloroquine (CQ) was used as a positive control in these experiments as it is a late-stage autophagy inhibitor that significantly increases LC3-II accumulation. The optimized HCS reliably performs with Z′ > 0.5 and % CV values <20%, supporting the robust nature of the screen (Table S1). We performed the HCS in duplicate with a library of 10,000 molecules that were obtained from the commercially available ChemDiv collection at a concentration of 20 μM to discover novel autophagy modulators. Of these, 312 hit molecules were identified based on a z-score of ≥2.2 for their ability to significantly increase the puncta per cell counts without cytotoxicity in HeLa cells (Figure 1A,B).
Figure 1.
Discovery of autophagy activators. (A) An eGFP-LC3 puncta formation assay in HeLa cells was performed in high throughput to screen 10,000 molecules, revealing 312 molecules that were able to increase the puncta/cell levels significantly (z-score >2.199 in both duplicate biological replicates). Out of the 312 compounds, 27 were found to be overtly cytotoxic (in orange). Compounds 1a (purple) and 2a (green) were prioritized. (B) Representative images from the eGFP-LC3 assay. The cells were plated at a density of 3000 cells/well and treated with dimethyl sulfoxide (DMSO), chloroquine (CQ) (20 μM), 1a (20 μM), or 2a (150 μM) for 4 h. The late-stage autophagy inhibitor CQ was used as a positive control for puncta accumulation. (C) mCherry-GFP-LC3 expressing HeLa cells were plated to a density of 3000 cells/well and compound-treated for 4 h with DMSO, CQ (20 μM), PI-103 (5 μM), 1a (20 μM), or 2a (40 μM). The PI3K inhibitor PI-103 and CQ were used as controls to show the effects of both early- and late-stage inhibition, respectively. (D) Structure–activity relationships (SAR) from the initial high-throughput screen revealed moieties critical for activity in the assay. Compounds 1a (left) and 2a (right) are highlighted. (E) Representative image of LC3 immunoblotting performed on HeLa cells treated with compounds for 4 h (DMSO, 1a (20 μM), or 2a (40 μM)). Co-treatment with autophagy inhibitor Bafilomycin A1 (BafA1, 100 nM) was used to confirm activation. (F) Quantification of LC3 immunoblots to measure activation and promotion of autophagic flux. Data are presented as mean ± SEM of four independent experiments (unpaired t test, *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001). (G) Immunoblot for phosphorylated P70S6Kinase (p-PS6Kinase) shows that compounds (1a (20 μM) and 2a (40 μM)) are activating autophagy in an mTOR-independent manner compared to mTOR inhibitor, rapamycin (Rap, 1 μM).
The 312 compounds were then obtained as a single plate for validation to differentiate activators from late-stage inhibitors using an mCherry-GFP-LC3 dual reporter assay. Upon fusion of the autophagosome to the lysosome, the green fluorescent protein (GFP) fluorescence is quenched due to the low pH, leaving the red color from the mCherry.64 By contrast, GFP fluorescence is not quenched by late-stage inhibitors that prevent autophagosome–lysosome fusion or lysosomal acidification, and thus red and green fluorescence overlap, resulting in a yellow color.64 The controls, late-stage inhibitors CQ and bafilomycin A1 (BafA1), cause robust accumulation of autophagosomes, observed by the yellow color, as expected. The 312 compounds were successfully classified as activators and late-stage inhibitors, and we focused on two activators, 1a and 2a, based on structure–activity relationships (SAR) observed in the HCS (Figure 1C,D). Compound 1a and analogues with the same core structure revealed that the presence of the indole moiety at R2 was critical for activity, as a lack of the indole resulted in compounds with no significant activity. The 4-methylpiperazine at R1 was present in both active analogues, so synthetic variation at this position was planned. For compound 2a, the tertiary amine in the R3 position was necessary for activity. Replacement of the tertiary amine with an amide resulted in a complete loss of activity, revealing the critical nature of the basic amine. Based on this observation, we designed synthetic analogues that modify the left side of compound 2a to incorporate other tertiary amines to improve potency in the eGFP-LC3 puncta formation assay.
To further confirm the conclusion from the dual reporter assay that 1a and 2a activate autophagy, LC3-II levels were also quantified with western blot flux assays (Figure 1E). If the compounds were late-stage inhibitors, we would expect to see no additional increase in LC3-II levels with co-treatment of the vATPase inhibitor, BafA1; however, we observed a significant increase in LC3-II levels following treatment with both compounds and a further increase when BafA1 was added to prevent LC3-II turnover through late-stage autophagy inhibition (Figure 1F).62,65 These results support the conclusion from the dual reporter assay that 1a and 2a are true autophagy activators and are enhancing autophagic flux. Next, we attempted to determine if our hits were activating autophagy independently of mTOR. The critical nature of mTOR in a variety of pathways means that mTOR inhibition may not be an ideal strategy for autophagy activation.66 One of the most well-known biomarkers for mTOR inhibition is the phosphorylation of the mTOR substrate, p70S6K.66 Treatment with our compounds did not affect the phosphorylation levels of p70S6K, unlike the mTOR inhibitor rapamycin (Figure 1G). This suggests our compounds activate autophagy through a different mechanism of action.
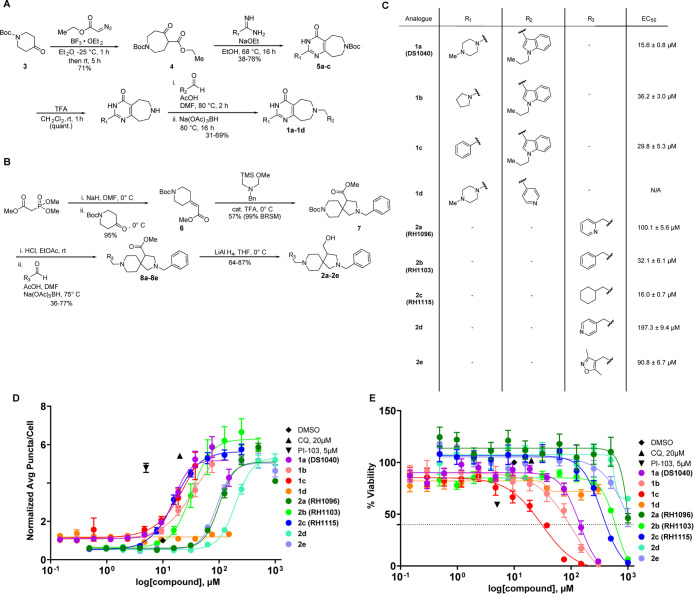
Two synthetic routes were developed to access 1a and 2a as well as a variety of analogues (Figure 2A,B). The synthesis of 1a began with a reaction of amidinopyrazole hydrochloride and methylpiperazine under basic conditions to afford the key amidine (Figure S1A). Subsequent analogues were prepared using alternative piperazine and aryl derivatives. Preparation of the indole reagent occurred by the straightforward alkylation of 1H-indole-3-carbaldehyde with propyl bromide and sodium hydride to afford the alkylated indole in good yield (Figure S1B).
Figure 2.
SAR studies provide analogues with improved potency. (A) Synthetic scheme for analogues of scaffold 1. The initial 1a (DS1040) hit and three additional analogues were prepared using a four-step method (longest linear sequence). (B) Synthetic scheme for analogues of scaffold 2. The initial 2a (RH1096) hit and four additional analogues were prepared using a four-step method (longest linear sequence). (C) Structures of the analogues and corresponding EC50 values in the eGFP-LC3 puncta formation assay. (D) Twelve-point dose–response curves were generated for each analogue in the eGFP-LC3 puncta formation assay (300–0.146 μM for 1 analogues, 1000–0.488 μM for 2 analogues). Data are presented as mean ± SEM of three independent experiments, each with duplicate biological replicates. Data points at the highest concentrations were omitted if they were found to be cytotoxic (% viability < 40%). (E) Percent viability was measured using nuclear count following treatment with compounds relative to DMSO controls. Data are presented as mean ± SEM of three independent experiments, each with duplicate biological replicates.
The synthetic strategy to generate pyrimidoazepine intermediates was adapted from Yang and co-workers (Figure 2A).67 The synthesis began with the Lewis acid-catalyzed ring expansion of 1-boc-4-piperidone with ethyl diazoacetate and BF3-etherate to access the oxoazepane ester 4 intermediate efficiently (71%).68 This intermediate then underwent a Pinner-type condensation with 4-methylpiperazine-1-carboximidamide (or relevant amidines for analogues) in the presence of sodium ethoxide base.67 This step enabled the generation of the key pyrimidoazepine intermediates 5a–5c in good yield (38–76%). Trifluoroacetic acid (TFA) was then used to deprotect the BOC-protecting group followed by reductive amination with the 1-propyl-1H-indole-3-carbaldehyde, catalytic acetic acid, and sodium triacetoxyborohydride to provide 1a (DS1040) in 35% over 2 steps.69 Two additional analogues were synthesized with pyrrolidine or phenyl moieties in the R1 position (1b, 1c) or a 4-pyridine moiety in the R2 position (1d).
To prepare 2a, trimethyl phosphonoacetate and 1-BOC-4-piperidinone underwent a Horner–Wadsworth–Emmons reaction using sodium hydride as a base to provide alkene 6 in excellent yield (95%) (Figure 2B). The alkene was then subjected to N-benzyl-1-methoxy-N-((trimethylsilyl)methyl)methanamine and catalytic TFA, to promote a [3 + 2] cycloaddition which provided spirocycle 7 in good yield (57% (99% BRSM)).70 Similar to the first synthetic route, a BOC deprotection and reductive amination using 2-pyridine carboxaldehyde provided the ester intermediate 8a in 32% over 2 steps. Analogues 8b–8e were formed using different aldehydes in the reductive amination step (36–77% over 2 steps). The final reduction of the ester to the primary alcohol using lithium aluminum hydride (LAH) provided 2a (RH1096) and its analogues in good to excellent yields (64–87%). In total, five analogues were created with different aryl or alkyl moieties in the R3 position.
The newly synthesized molecules were tested in the eGFP-LC3 puncta formation assay in 12-point dose to generate EC50 values for each analogue (300–0.146 μM for 1 analogues, 1000–0.488 μM for 2 analogues) (Figure 2C,D). The analogues of DS1040 containing the indole had similar activity in this assay, highlighting the importance of this heterocycle. By contrast, replacement of the indole with a 4-pyridine completely eliminated autophagy activation activity. Variation of the R3 position in the 2 analogues resulted in improved activity compared to RH1096. Replacement of the 2-pyridine ring in RH1096 with benzene (2b, RH1103) or cyclohexane (2c, RH1115) resulted in a 3- and 6-fold increase in potency, respectively. Interestingly, modification of the nitrogen in the pyridine from the 2- to 4-position (2d) resulted in a 2-fold decrease in potency. Percent viability was also measured to ensure the activation response was not due to cytotoxicity (Figure 2E). All compounds were found to not be overtly cytotoxic at their EC50 values, indicating that the observed autophagy activation likely is not due to a cell death response. However, efforts to optimize DS1040 resulted in reduced cell viability and did not provide an improvement in activity. For example, analogue 1c caused an increase in cell death at concentrations that are close to the concentrations that induced autophagy. In contrast, optimization of RH1096 produced RH1115 which has an excellent, 22-fold selectivity window between autophagy activation and cytotoxicity. Active analogues were also confirmed to retain autophagy activation in the mCherry-GFP-LC3 dual reporter assay. Next, aqueous kinetic solubility was measured for molecules DS1040, RH1096, and RH1115.71 Even at 100 μM concentration, all three molecules were found to be highly soluble (Figure S2A), indicating that these molecules are excellent starting points for probe development. Based on these results, compound RH1115 was carried forward for target identification and validation experiments.
To determine the mechanism of action of the most potent hit, RH1115, a biotin-labeled probe was developed to use in a streptavidin bead pulldown assay. The modified synthetic route incorporated a terminal alkyne-containing precursor that was subjected to a copper-catalyzed azide–alkyne cycloaddition reaction with a biotin-tagged azide linker to link biotin to the molecule through formation of a triazole.72 The terminal alkyne was added to RH1115 in the late stages of the synthetic route after ester reduction to the primary alcohol. The primary alcohol underwent acylation using hex-5-ynoyl chloride and triethylamine to generate the alkyne in good yield (68%) (Figure S1C). The cycloaddition was performed using the alkyne and Biotin-PEG3-Azide with catalytic CuSO4 at 90 °C for 2 days to reach completion, which resulted in the desired product, Biotin-RH1115, with a yield of 36%. Biotin-RH1115 was evaluated in the eGFP-LC3 puncta formation assay and was able to significantly increase the puncta/cell count relative to DMSO with an EC50 of 46.2 μM (Figure S2B).
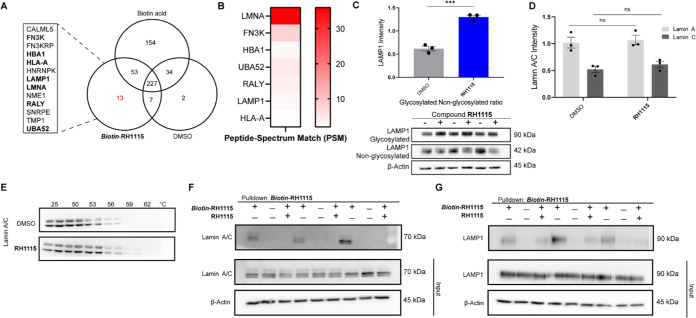
A pulldown experiment using Biotin-RH1115 was performed to identify the target of this molecule to obtain insight into the mechanism of action. Proteins bound to Biotin-RH1115 were eluted and prepared for mass spectrometry data acquisition and analysis.73 Through analysis of the resulting data, 13 proteins were identified as being pulled down by the biotinylated compound exclusively, i.e., they were not pulled down with the biotin acid or DMSO negative controls (Figure 3A). To confirm binding to the compounds, a competition assay was performed in which Biotin-RH1115 and excess RH1115 soluble competitor were incubated with the lysate prior to the pulldown. Proteins that were pulled down in the initial assay, but were not pulled down in the competition assay, likely have a specific interaction with RH1115. Of the initial 13 proteins pulled down, 7 proteins were no longer identified in the mass spectrometry analysis when treated with excess RH1115, providing putative targets for RH1115 (Figure 3A,B). Proteins with isoforms that were also pulled down by the controls or proteins that were not expressed in neurons were excluded (Table S2).
Figure 3.
Target identification and validation for RH1115. (A) Mass spectrometry-based identification of eluted proteins. Proteins captured by Biotin-RH1115 were subjected to digestion followed by mass spectrometry analysis. Eluted proteins identified were compared to DMSO and biotin acid as standards. Thirteen unique proteins (listed in alphabetical order) were identified to interact with the RH1115 probe and when treated with excess of RH1115, 7 proteins were no longer pulled down (bolded). (B) Number of peptide spectrum matches (PSMs) identified for each protein in pulldown sample with Biotin-RH1115 but not in the negative control samples. (C) Immunoblot for LAMP1 after 24 h compound treatment with RH1115 (50 μM) in HeLa cells (bottom). Proteins were quantified and represented as mean ± SEM of three biological replicates, normalized to β-actin (top). (D) Quantified immunoblots of Lamin A/C after 24 h compound treatment with RH1115 (50 μM) in HeLa cells. Proteins were quantified and represented as mean ± SEM of three biological replicates, normalized to β-actin. (E) Representative blots from three independent cellular thermal shift assays (CETSAs). A549 cells were treated with RH1115 (100 μM) or DMSO for 24 h and heated at each temperature in duplicate biological replicates for 3 min. (F) Immunoblot analysis of Lamin A/C capture by streptavidin pulldown protocol in the presence of Biotin-RH1115 (50 μM) (+/−), biotin acid (−/−), or Biotin-RH1115 (50 μM) with excess RH1115 (100 μM) following lysate addition to the beads (+/+). Data are presented as three independent experiments. β-actin was used as a loading control. (G) Immunoblot analysis of LAMP1 capture by streptavidin pulldown protocol in the presence of Biotin-RH1115 (50 μM) (+/−), biotin acid (−/−), or Biotin-RH1115 (50 μM) with excess RH1115 (100 μM) following lysate addition to the beads (+/+). Data are presented from three independent experiments. β-actin was used as a loading control. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001.
Validation experiments were performed with the remaining three proteins, Lamin A/C, ubiquitin A-52 residue ribosomal protein fusion (UBA-52), and lysosome-associated membrane glycoprotein 1 (LAMP1). Expression tests were performed for all 3 proteins to evaluate protein levels after 24 h RH1115 treatment (Figures 3C and S3A,B). A significant increase in the ratio of glycosylated/non-glycosylated LAMP1 was observed following treatment with RH1115 (Figure 3C) compared to control treatment. By contrast, expression levels of Lamin A/C were unchanged following treatment with compound RH1115 (Figures 3D and S3A). We then analyzed the interaction of RH1115 with Lamin A/C using a cellular thermal shift assay (CETSA) to measure changes in melting temperature following compound treatment to assess direct binding of the untagged compound to the protein(s) of interest. Both Lamin A and C were found to be stabilized by RH1115, further validating the direct interaction of Lamin A/C with unmodified RH1115 (Figures 3E and S3C).
Next, the mass spectrometry results were further validated through pulldown and competition studies followed by western blot analysis for Lamin A/C, LAMP1, and UBA-52 (Figures 3F,G, S3D,E, and S4A,B). UBA-52 was deemed a nonspecific target/false positive for RH1115 because western blot analysis of the pulldown eluent revealed an interaction with the negative control and the inability of the soluble RH1115 competitor to prevent binding of the Biotin-RH1115 probe (Figure S4A,B). By contrast, Lamin A/C was highly enriched in the eluent after treatment with Biotin-RH1115, and the interaction with the probe was almost completely prevented by the soluble RH1115 competitor, indicating a specific interaction between Lamin A/C and RH1115 (Figure 3F). Glycosylated LAMP1 was also pulled down and concentrated in the eluent after treatment with Biotin-RH1115, and this interaction was efficiently prevented by the addition of soluble RH1115 competitor (Figure 3G), thus confirming that this protein is also a target of compound RH1115.
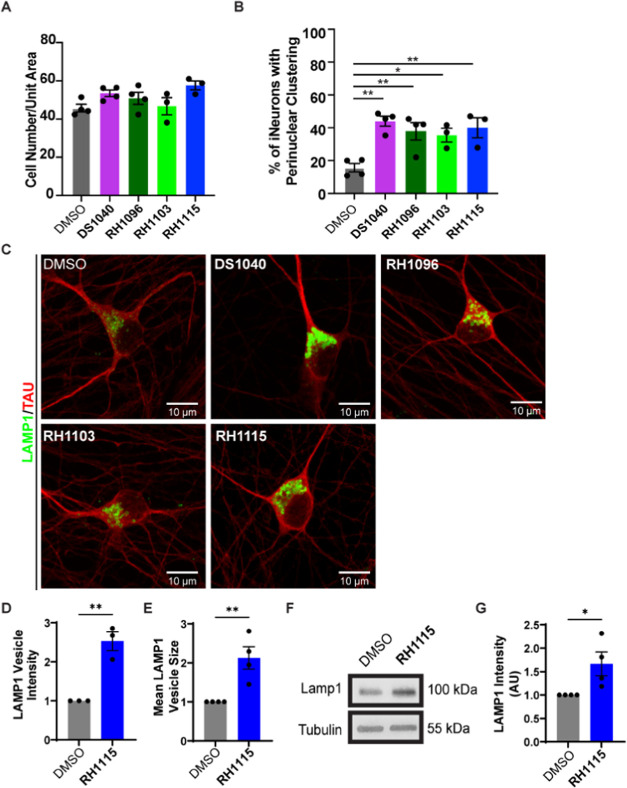
Given the potent effects of the compounds on autophagy, and the interest in modulation of autophagy as a therapeutic option for neurodegenerative diseases, such as AD, we next tested the compounds in human induced pluripotent stem cell (iPSC)-derived neurons (i3Neurons).74−77 DIV 10 i3Neurons treated with DS1040, RH1096, RH1103, and RH1115 for 72 h starting at DIV 7, showed no overt cytotoxicity as determined by their cell density at the end of the treatment (Figure 4A). Examination of LAMP1 staining in these neurons revealed that the compound treatment resulted in a profound effect on lysosome positioning in these i3Neurons (Figure 4B,C). While LAMP1 vesicles, which include a mixture of late endosomes and degradative lysosomes,78 are normally heterogeneous in their distribution in the neuronal cell body (soma), all of the compounds induced significant perinuclear clustering of these LAMP1 vesicles. In addition to the change in lysosome distribution, treatment with compound RH1115 resulted in increased intensity and mean size of the LAMP1 vesicles (Figures 4C–E and S5A), suggestive of increased LAMP1 levels on the endolysosomes. Indeed, the total levels of LAMP1 protein in lysates from neurons treated with compound RH1115 were also found to be increased by 1.5-fold when compared to control neurons (Figure 4F,G).
Figure 4.
Effects of autophagy activators on lysosome properties in neurons. (A) Quantification of the number of i3Neurons per unit area following treatment with DMSO (control), DS1040, RH1096, RH1103, and RH1115 as a read out of neuronal viability. Data are presented as mean ± SEM from three independent experiments, 150–200 neurons per treatment per experiment. (B) Quantification showing percentage of i3Neurons exhibiting enhanced perinuclear clustering of lysosomes after compound or DMSO treatment. Data are presented as mean ± SEM of four independent experiments, 100–150 neurons per treatment. (C) High-resolution confocal images of i3Neurons treated with compounds or DMSO and stained for LAMP1 (green) to label lysosomes and Tau (red) to label neurites, showing enlarged and brighter lysosomal vesicles. (D) Quantification of mean intensity of LAMP1-positive vesicles in DMSO and RH1115-treated (15 μM) i3Neurons from high-resolution confocal images. Data are presented as mean ± SEM of three independent experiments, 20–25 neurons per treatment. (E) Quantification of mean size of LAMP1-positive vesicles in DMSO and RH1115-treated (15 μM) i3Neurons from high-resolution confocal images. Data are presented as mean ± SEM of three independent experiments, 20–25 neurons per treatment. (F) Immunoblotting of LAMP1 in DIV20–21 i3Neurons treated with 15 μM RH1115 for 72 h vs 0.1% DMSO treatment. Tubulin is used as a loading control. (G) Quantification of LAMP1 immunoblot shows a significant increase in LAMP1 after RH1115 treatment (15 μM) compared to DMSO treatment from four independent experiments, mean ± SEM *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001.
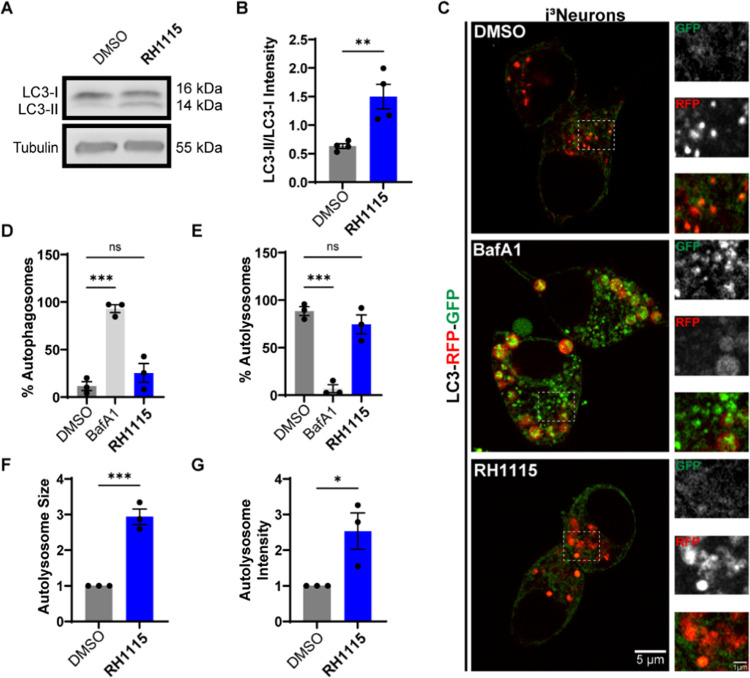
Given the effect of RH1115 on autophagic flux in HeLa cells and modulation of lysosome positioning, morphology, and potentially biogenesis (increased LAMP1 localization and recruitment to the perinuclear region), we next examined how the compound modulated autophagy in neurons. We found that i3Neurons treated with RH1115 did indeed exhibit a higher LC3-II/LC3-I ratio (Figure 5A,B) as determined by immunoblotting. To evaluate autophagic flux, we then examined the nature and distribution of autophagosomes and autolysosomes in i3Neurons derived from iPSCs generated to stably express LC3-RFP-GFP. Using Airyscan live imaging of i3Neurons treated with DMSO, BafA1, or RH1115, we determined the number and fraction of autophagosomes (GFP and RFP-positive vesicles) and autolysosomes (only RFP-positive vesicles) in each of these conditions (Figure 5C). We found that while BafA1 massively increased the number of autophagosomes per neuronal cell body (Figures 5C,D and S5B,C), RH1115 did not do the same. In contrast, the number of autophagosomes in DMSO and RH1115-treated i3Neurons was far lower. Consistent with BafA1 reducing fusion between autophagosomes and lysosomes, the percentage of autolysosomes in BafA1-treated neurons was far less than in RH1115 or DMSO-treated i3Neurons (Figures 5E and S5D,E). In these neurons, RH1115 did not inhibit autophagic flux (as seen by low numbers of autophagosomes), and in fact appeared to activate autophagic flux as evidenced by LC3-II/LC3-I ratio. Although a dramatic increase in the fraction of autolysosomes was not evident with RH1115 treatment when examining the absolute number of these vesicles, the autolysosomes were nearly 3 times as large as in the DMSO-treated condition (Figures 5F and S5F) and exhibited increased total RFP intensity per vesicle (Figure S5G), suggestive of potentially fused and larger autolysosomes. Thus, RH1115, in addition to affecting lysosome positioning and biogenesis, likely also increases autophagic flux.
Figure 5.
Confirmation of autophagy activation in neurons. (A) Immunoblotting for LC3 in DIV20–21 i3Neurons treated with RH1115 (15 μM) or DMSO (0.1%) for 72 h. (B) Quantification of LC3-II/LC3-I ratio shows that i3Neurons treated with RH1115 exhibit significantly increased LC3 lipidation in comparison to DMSO-treated i3Neurons. Mean ± SEM from four independent experiments. (C) Confocal images of DIV10 i3Neurons stably expressing LC3-RFP-GFP treated with DMSO, RH1115 (15 μM) for 72 h, or BafA1 (100 nM) for 24 h prior to live imaging using Airyscan. Higher-magnification images of region outlined by a dashed box are depicted to the right of each image. (D) Quantification of the percent of autophagosomes in i3Neurons treated with RH1115 (15 μM) or BafA1 (100 nM) compared to DMSO. (E) Quantification of the percent of autolysosomes in i3Neurons treated with RH1115 (15 μM) or BafA1 (100 nM) compared to DMSO. (F) Quantification of mean size of autolysosomes in i3Neurons treated with RH1115 (15 μM) compared to DMSO. (G) Quantification of the intensity of autolysosomes in i3Neurons treated with RH1115 (15 μM) compared to DMSO. In (D)–(G), data were collected from three independent experiments, 60–70 cells per treatment, mean ± SEM *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001.
Disease-modifying therapies for AD have remained a challenge in the field of drug discovery.79 As cases of AD rise globally, there is a critical need for alternative therapeutic strategies, and selective modulation of autophagy has emerged as a promising approach for the treatment of neurodegenerative and age-related diseases.80 Current clinical trials have evaluated BACE1 inhibitors, which prevent and clear Aβ protein aggregates but fail to improve cognitive function in AD patients and have significant side effects, minimizing their effectiveness.37 Moreover, some of these inhibitors lack the ability to clear tau aggregates in the somatodendritic compartment of neuronal brain cells, which can lead to the formation of tangles inside of neurons which promotes disease progression and eventual patient death.39 The drawbacks of BACE1 inhibitors demonstrate how target-based methods for drug discovery rely heavily on the modulation of candidate proteins, and even successful modulation of a promising target may not have the hypothesized impact in disease models. By contrast, phenotypic screening provides an unbiased approach to drug discovery.81 In recent years, phenotypic strategies for drug discovery have become increasingly popular because they can lead to the discovery of small molecules that function through unique mechanisms of action.82,83 Subsequent target identification and validation efforts can provide novel targets to affect disease-relevant phenotypes, which facilitates the development of highly effective, first-in-class therapeutics. Because autophagy has been implicated in a wide range of neurodegenerative diseases, we are particularly interested in developing optimized autophagy modulators with novel mechanisms to evaluate their ability to ameliorate autophagy defects observed in AD and to determine how the validated protein targets of these modulators are involved in disease pathology and progression.84
Lamin A/C was identified as a potential target of RH1115 using an unbiased proteomics approach (Figure 3B). Nuclear Lamins are divided into A- and B-type ligands depending on the structure and expression pattern. The LMNA gene encodes for multiple isoforms of the A-type Lamin proteins, including Lamin A and C, formed through alternative splicing that differ from each other by a modified C-terminus and absence of CAAX box in Lamin C.85,86 Subsequent validation experiments revealed a direct interaction between Lamin A/C and Biotin-RH1115, which was confirmed through a competition experiment with soluble RH1115 (Figure 3F). Expression levels of Lamin A/C did not significantly change following compound treatment (Figure 3D), but stabilization of Lamin A/C was observed by CETSA, which provided further support that a direct interaction occurs between RH1115 and Lamin A/C. Based on these observations, it is likely that modulation of Lamin A and/or C function or localization, but not expression, is important for the activity of RH1115. Lamin A/C are found in the nuclear envelope, where they contribute to several physiological processes, including the maintenance of cellular structure and stability, chromatin regulation, and telomere protection.86−88 Numerous diseases, known as Laminopathies, are caused by mutations in the LMNA gene,89 and recent work has attempted to clarify the effects of Lamins in neurodegeneration. Lamin abnormalities have been found to be present in both Drosophila and human tauopathy, leading to heterochromatin relaxation, DNA damage, and neuronal cell death.90 Interestingly, despite the importance of Lamin A/C in most cell types, healthy neurons notably have little to no Lamin A expression, which allows for improved flexibility and plasticity.91−93 While Méndez-López and co-workers identified Lamin A and C in both control and AD human hippocampal samples, they observed a significant increase of LMNA mRNA and Lamin A/C protein expression in AD samples characterized as high-severity cases.94 More recently, Gil and co-workers noted significant increases of hippocampal neuron expression of Lamin A and a lack of Lamin C in cases of early- and late-stage AD, while neurons from healthy elderly patients did not show Lamin A expression, suggesting a possible connection between abnormal Lamin A/C expression and AD progression.95 Although autophagy activation using rapamycin or its analogue temsirolimus has been shown to ameliorate various Laminopathic disease states, the connection of Lamin A/C to autophagy in neurons and neurodegeneration is much less explored.96−98 Future studies will investigate the role of Lamin A/C in the regulation of neuronal autophagy in AD and how the interaction of RH1115 with Lamin proteins may be modulating autophagy to assess the potential of Lamin A/C as a novel target for AD therapeutic development.
LAMP1, well known for its role in the biogenesis and maintenance of lysosomes, was also identified as a target in our proteomics experiments, and validation experiments confirmed a direct interaction between Biotin-RH1115 and LAMP1 (Figure 3G). Lysosomes are a key contributor to neuronal protein and organelle homeostasis and the clearance of autophagic cargo,99 and lysosome function has been found to be altered in AD models.78,100 Studies in AD mouse neurons have shown accumulation of lysosome-like organelles in amyloid plaques found at swollen axon sites.24 Additionally, Aβ prevents autophagic flux by disrupting normal lysosome distribution in AD models.101 Treatment with RH1115 also resulted in the change of LAMP1 distribution in the soma of human iPSC neurons (Figure 4B,C) and increased LAMP1 intensity and vesicle size (Figure 4C,E). Retrograde movement of lysosomes to a perinuclear location has been suggested to facilitate autophagosome-lysosome fusion,102 and autophagy induction by compound treatment or transcription factor overexpression has been shown to increase LAMP1 protein levels and perinuclear clustering of lysosomes.103−105 Interestingly, we also noted a significant increase in the ratio of glycosylated to non-glycosylated LAMP1 following treatment with RH1115 (Figure 3C). Maturation of LAMP1 consists of glycosylation of the protein to form a stable glycoprotein layer that maintains the integrity of the lysosome and may indirectly modulate the fusion of lysosomes with phagosomes, autophagosomes, or the plasma membrane.106 While decreases in protein glycosylation have been observed in AD models, the results are not consistent across regions of the brain, and the glycosylation of LAMP1 specifically has not been extensively studied.107,108 Abnormal LAMP1 glycosylation has also been observed in another neurodegenerative disease, Niemann-Pick type C1 (NPC), which is a lysosomal storage disease that affects cholesterol trafficking due to mutations in the NPC1 gene.109 Both NPC and AD have common pathological features, including Aβ accumulation and neurofibrillary tangles, but one of the most significant similarities is the contribution of polymorphisms in the apolipoprotein E (ApoE) for the progression of both diseases.110,111 Along with the increase in glycosylation of LAMP1, direct interaction between RH1115 and LAMP1 was observed (Figure 3G). Taken together, these results suggest that treatment with RH1115 may rescue dysfunctional LAMP1 and restore autophagic flux by promoting lysosome movement and autophagosome–lysosome fusion in neurons. Changes in glycosylation pattern and the contribution of this increase in glycosylated LAMP1 to the observed phenotype will be further investigated.
Conclusions
In summary, we implemented a phenotypic assay to identify molecules that induce autophagy, and we confirmed that the prioritized molecules are mTOR-independent autophagy activators. Through synthetic optimization, we were able to access more potent analogues of our initial hits and to develop a biotinylated version of the RH1115 analogue that retained its biological activity and phenotypic properties to enable target identification studies that revealed two protein targets of interest with significant implications in neurodegeneration. Finally, we determined that this compound alters positioning of lysosomes and increases autophagic flux in human iPSC-derived neurons. Given the highly polarized and unique morphology of neurons, and the link between lysosome transport and maturation in these cells, a small molecule that mobilizes endolysosomes in neurons could be especially impactful in neurodegenerative diseases. Future studies will evaluate the effects of these small-molecule autophagy modulators on lysosomal pathology and Aβ generation in neuronal models of AD to provide additional insight into the therapeutic potential of the identified protein targets, which may reveal alternative biomarkers for clinical evaluation and enable the development of new therapeutic strategies to treat neurodegeneration.
Methods
Synthetic Methods
General Information
All chemicals for synthetic methods were purchased from Sigma-Aldrich, Alfa-Aesar, Acros Organics, TCI America, Oakwood Chemicals, or Chem Impex and were used without further purification unless otherwise noted. Reaction mixtures were purified on a Biotage Isolera One automated chromatography system with silica gel columns. Microwave reactions utilized the Biotage Initiator + microwave reactor. Reactions were monitored by TLC (Silica gel 60 F254 Glass Backed plates) and mass spectrometry using LCMS (Agilent 1260 Series automated chromatographic system outfitted with a Thermo Scientific Accucore column (2.1 mm × 50 mm, 2.6 μm particle size)) and an Agilent 6120 quadrupole MS, utilizing a gradient elution mobile phase of 25% ACN/H2O to 95% ACN/H2O over 3 min, then holding at 95% ACN/H2O for 2 min (0.200 mL/min flow rate, 30 °C column compartment, detection modes: wavelengths of 254 and 280 nm). NMR data were collected on a Bruker AV 500 MHz spectrometer outfitted with a Bruker 5 mm 1H19F/BBO S2 Z-gradient probe, and spectra were processed utilizing Mestrenova (Mestrelab Research). Data were recorded at ambient temperature and are reported as chemical shift (ppm) relative to solvent peak (1H NMR: CDCl3 = 7.26 ppm, MeOD = 3.31 ppm, D2O = 4.65 ppm; 13C NMR: CDCl3 = 77.16 ppm, MeOD = 49.00 ppm). IR data were collected on a Thermo Scientific Nicolet IS5 spectrometer outfitted with a Thermo Fisher Scientific iD5 ATR. HRMS data were collected by Dr. Furong Sun at the University of Illinois-Urbana-Champaign using Waters Q-TOF Ultima ESI.
General Procedures
General Procedure A
To a flame-dried flask equipped with a stir bar were added sodium ethoxide (2.0 equiv), 1-(tert-butyl) 4-ethyl 5-oxoazepane-1,4-dicarboxylate (1.0 equiv), the appropriate amidine (1.5 equiv), and ethanol (0.15 M). The mixture was heated to 75 °C for 3.5 h before being quenched with water and extracted with dichloromethane (DCM).
General Procedure B
To a reaction vessel equipped with a magnetic stir bar were added a 1:1 mixture of TFA and DCM (0.20 M) and the substituted BOC-azepine. The reaction was stirred for 1 h at room temperature (RT). The mixture was concentrated under vacuum and was used crude in the next step.
General Procedure C
To a flame-dried flask equipped with a stir bar were added crude azepine (1.0 equiv) and desired aldehyde (5.0 equiv) in 2% AcOH in dimethylformamide (DMF, 0.05 M). The solution was heated to 75 °C for 2 h before addition of sodium triacetoxyborohydride (5.0 equiv). The reaction stirred for 16 h before being removed from the heat. The reaction was quenched with H2O, and the aqueous layer was then washed in triplicate with DCM. The organic layer was concentrated under vacuum.
General Procedure D
To a flame-dried flask equipped with a stir bar were added crude methyl 2-benzyl-2,8-diazaspiro[4.5]decane-4-carboxylate (1.0 equiv) and desired aldehyde (5.0 equiv) in 2% AcOH in DMF (0.05 M). The solution was heated to 75 °C for 1 h before addition of sodium triacetoxyborohydride (5.0 equiv). The reaction stirred for 3 h before being removed from the heat and stirred for 16 h at RT. The reaction was quenched with H2O, and the aqueous layer was then washed in triplicate with EtOAc. The recovered aqueous layer was concentrated under vacuum.
General Procedure E
To a microwave vial equipped with a stir bar was added LiAlH4 (2.0 equiv) as a solution in anhydrous tetrahydrofuran (THF, 0.20 M). Desired carboxylate (1.0 equiv) was added dropwise in THF (0.20 M). The reaction vessel was sealed and stirred for 2 h at 0 °C. EtOAc (2 mL) and H2O (1 mL) were added to quench the reaction. The mixture was passed through a short-path column of Celite before being concentrated under vacuum.
Synthetic Protocols and Compound Characterization
4-Methylpiperazine-1-carboximidamide (S1)
To a flame-dried flask was added N-methylpiperazine (0.554 mL, 4.992 mmol) as a solution in DMF (0.50 M). Then, amidinopyrazole HCl (731.7 mg, 4.992 mmol) and N,N-diisopropylethylamine (0.869 mL, 4.992 mmol) were added before the solution was stirred at 80 °C for 10 h. The solution was concentrated under vacuum before crystallizing with ether to generate product as yellow crystals: 614.6 mg (87%). 1H NMR (500 MHz, D2O) δ 3.97 (s, 4H), 3.35 (s, 4H), 2.96 (s, 3H). 13C NMR (125 MHz, D2O) δ 156.64, 52.92, 44.45, 44.41. IR (neat) υmax = 3304, 3117, 2941, 2863, 2853, 2810, 1663, 1648, 1596, 1525, 1450, 1289, 1208, 1151, 1131, 1080, 1057, 996, 806. HRMS (ESI) C6H15N4. Calculated: [M + H]+ 143.1297, found: [M + H]+ 143.1293.
Pyrrolidine-1-carboximidamide (S2)
To a flame-dried microwave vial was added pyrrolidine (0.462 mL, 5.624 mmol) as a solution in DMF (0.67 M). Then, amidinopyrazole HCl (824 mg, 5.624 mmol) and N,N-diisopropylethylamine (0.980 mL, 5.624 mmol) were added, and the solution was heated using microwave irradiation for 35 min at 133 °C. The solution was concentrated under vacuum and taken without purification to the next step as a white powder: 710 mg (quant.). 1H NMR (500 MHz, MeOD) δ 7.06 (s, 1H), 3.48–3.39 (m, 4H), 2.09–1.97 (m, 4H). 13C NMR (125 MHz, MeOD) δ 154.73, 46.87, 24.83. IR (neat) υmax = 3315, 3159, 1651, 1621, 1558, 1480, 1467, 1457, 1364, 1223, 1157, 1064, 1038. HRMS (ESI) C5H12N3. Calculated: [M + H]+ 114.1031, found: [M + H]+ 114.1026.
Benzimidamide (S3)
To a flame-dried flask was added a solution of ammonium chloride (210.0 mg, 3.882 mmol) as a solution in toluene (2.0 M). Trimethylaluminum was added as a solution in toluene (2.0 M), and the resulting solution was stirred for 2 h. Benzonitrile (0.400 mL, 3.882 mmol) was added dropwise and heated at 83 °C for 16 h. The reaction was concentrated and poured over a silica slurry in DCM. The slurry was gravity-filtered and washed with MeOH before concentrating the pure product as a white powder: 410.5 mg (88%). 1H NMR (500 MHz, MeOD) δ 7.82 (dd, J = 8.44, 1.33 Hz, 2H), 7.78–7.72 (m, 1H), 7.65–7.59 (m, 2H), 7.57 (s, 1H), 1.46–1.32 (m, 1H). 13C NMR (125 MHz, MeOD) δ 167.14, 133.81, 129.06, 128.17, 127.51. IR (neat) υmax = 3160, 1660, 1624, 1346, 1224. HRMS (ESI) C7H9N2. Calculated: [M + H]+ 121.0766, found: [M + H]+, 121.0763.
1-Propyl-1H-indole-3-carbaldehyde (S4)
To a flame-dried flask were added 1H-indole-3-carbaldehyde (1.0023 g, 6.889 mmol), 1-bromopropane (12.50 mL, 13.78 mmol), and sodium hydride (0.3040 g, 8.267 mmol) as a solution in DMF (0.1 M). The reaction was stirred for 40 min before being quenched with H2O, and the aqueous layer was washed in triplicate with EtOAc. The organic layer was concentrated under vacuum. The crude product was purified by column chromatography (0–30% EtOAc in hexanes over 12 min) to afford the desired product as an orange solid: 1.0285 g (80%) 1H NMR (500 MHz, MeOD) δ 9.84 (s, 1H), 8.17 (d, J = 8.00 Hz, 1H), 8.09 (s, 1H), 7.51 (d, J = 8.21 Hz, 1H), 7.38–7.19 (m, 2H), 4.22 (t, J = 7.06 Hz, 2H), 1.91 (h, J = 7.31 Hz, 2H), 0.93 (t, J = 7.42 Hz, 3H). 13C NMR (125 MHz, MeOD) δ 185.40, 140.83, 137.58, 125.12, 123.59, 122.48, 121.33, 117.57, 110.30, 48.23, 22.75, 10.05. IR (neat) υmax = 2967, 2934, 2878, 2793, 2723, 2699, 1644, 1609, 1571, 1529, 1486, 1474, 1465, 1454, 1410, 1394, 1381, 1353, 1320, 1284, 1241, 1190, 1171. HRMS (ESI) C12H14NO. Calculated: [M + H]+ 188.1075, found: [M + H]+ 188.1071.
1-(tert-Butyl) 4-Ethyl 5-oxoazepane-1,4-dicarboxylate (3)
To a flame-dried flask equipped with a stir bar was added 1-Boc-4-piperidone (2.0000 g, 10.04 mmol) as a solution in diethyl ether (0.60 M). Then, ethyl diazoacetate (1.58 mL, 15.06 mmol) and BF3·OEt2 (1.24 mL, 10.04 mmol) were added dropwise, and the resulting mixture was stirred at −25 °C for 1 h. The reaction was neutralized with sodium bicarbonate, and the aqueous layer was extracted in triplicate with EtOAc. The combined organic extracts were washed with brine, dried with Na2SO4, and concentrated under vacuum. The crude product was purified by column chromatography (0–20% EtOAc in hexanes over 10 min) to afford the desired product as a yellow oil: 2.020 g (71%). Product was shown to exist as the keto- and enol-tautomers leading to the observed half-integrations in the proton NMR and splitting in the carbon NMR. 1H NMR (500 MHz, CDCl3) δ 4.11 (q, J = 7.13 Hz, 2H), 3.44 (s, 1H), 3.23 (t, J = 7.17 Hz, 1H), 2.54 (s, 1H), 2.27 (t, J = 7.47 Hz, 1H), 1.81 (p, J = 7.30 Hz, 1H), 1.43 (s, 6H), 1.24 (t, J = 7.16 Hz, 3H). 13C NMR (125 MHz, CDCl3) δ 173.07, 171.91, 155.27, 79.73, 60.43 (d, J = 16.50 Hz), 46.90 (d, J = 37.97 Hz), 43.31, 33.70 (d, J = 73.69 Hz), 28.36, 23.75 (d, J = 48.41 Hz), 21.79 (d, J = 3.24 Hz), 14.17 (d, J = 4.59 Hz). IR (neat) υmax = 2976, 2931, 1741, 1668, 1469, 1445, 1415, 1366, 1317, 1242, 1199, 1158, 1067, 1024, 897, 860 HRMS (ESI) C14H23NO5. Calculated: [M + Na]+ 308.1474, Found: [M + Na]+ 308.1464.
tert-Butyl 2-(4-Methylpiperazin-1-yl)-4-oxo-3,4,5,6,8,9-hexahydro-7H-pyrimido[4,5-d]azepine-7-carboxylate (4a)
Following General Procedure A, 1-(tert-butyl) 4-ethyl 5-oxoazepane-1,4-dicarboxylate (900.0 mg, 3.154 mmol), 4-methylpiperazine-1-carboximidamide (659 mg, 4.637 mmol), and sodium ethoxide as a 1 M solution in ethanol (3.2 mL) were used. Crude product was purified using column chromatography (0–100% MeOH in DCM for 5 min, holding at 100% for 4 min) to afford the desired product as an off-white solid: 800 mg (70%). 1H NMR (500 MHz, CDCl3) δ 3.73 (s, 4H), 3.51 (d, J = 44.71 Hz, 4H), 2.75 (d, J = 29.59 Hz, 4H), 2.47 (t, J = 4.90 Hz, 4H), 2.34 (s, 3H), 1.54–1.41 (m, 9H). 13C NMR (125 MHz, CDCl3) δ 167.87, 165.79, 155.02, 151.55, 111.19, 79.64, 54.54, 46.96, 46.00, 44.26, 43.71, 40.18, 28.48, 24.33. IR (neat) υmax = 2972, 2960, 2930, 2795, 1662, 1616, 1574, 1456, 1414, 1385, 1362, 1329, 1304, 1294, 1237, 1203, 1159, 1107, 1084, 1040, 1002, 935, 884, 862. HRMS (ESI) C18H30N5O3. Calculated: [M + H]+ 364.2349, found: [M + H]+ 364.2332.
tert-Butyl 4-Oxo-2-(Pyrrolidin-1-yl)-3,4,5,6,8,9-hexahydro-7H-pyrimido[4,5-d]azepine-7-carboxylate (4b)
Following General Procedure A, 1-(tert-butyl) 4-ethyl 5-oxoazepane-1,4-dicarboxylate (336.0 mg, 1.178 mmol), pyrrolidine-1-carboximidamide (200 mg, 1.767 mmol), and sodium ethoxide as a 0.15 M solution in ethanol (7.8 mL) were used. Crude product was purified using column chromatography (0–10% MeOH in DCM for 15 min) to afford the desired product as an off-white solid: 150 mg (38%). 1H NMR (500 MHz, CDCl3) δ 3.54 (t, J = 6.66 Hz, 6H), 3.45 (s, 2H), 2.74 (d, J = 20.18 Hz, 4H), 1.97 (d, J = 6.42 Hz, 4H), 1.46 (s, 9H). 13C NMR (125 MHz, CDCl3) δ 167.89, 165.35, 155.03, 150.37, 109.91, 79.53, 46.79 (d, J = 99.93 Hz), 46.66, 44.23 (d, J = 90.79 Hz), 40.13, 28.48, 25.37, 24.46 (d, J = 25.93 Hz). IR (neat) υmax = 2971, 1687, 1626, 1584, 1457, 1413, 1390, 1364, 1335, 1290, 1236, 1162, 1108, 1085, 949, 938, 868. HRMS (ESI) C17H27N4O3. Calculated: [M + H]+ 335.2083, found: [M + H]+ 335.2078.
tert-Butyl 4-Oxo-2-Phenyl-3,4,5,6,8,9-hexahydro-7H-pyrimido[4,5-d]azepine-7-carboxylate (4c)
Following General Procedure A, 1-(tert-butyl) 4-ethyl 5-oxoazepane-1,4-dicarboxylate (285 mg, 0.9987 mmol), benzimidamine (180 mg, 1.498 mmol), and sodium ethoxide as a 0.15 M solution in ethanol (6.7 mL) were used. Crude product was purified using column chromatography (0–10% MeOH in DCM for 10 min) to afford the desired product as an off-white solid: 260 mg (76%). 1H NMR (500 MHz, CDCl3) δ 8.23 (d, J = 6.89 Hz, 2H), 7.61–7.45 (m, 3H), 3.61 (d, J = 34.79 Hz, 4H), 3.00 (d, J = 49.63 Hz, 4H), 1.49 (s, 8H), 1.45–1.42 (m, 1H). 13C NMR (125 MHz, CDCl3) δ 166.09, 165.11, 154.98, 153.63, 131.99, 131.71, 128.81, 127.60, 122.67, 79.87, 45.68 (d, J = 98.61 Hz), 44.00 (d, J = 93.44 Hz), 39.70, 28.48, 24.81. IR (neat) υmax = 3359, 3140, 2973, 2929, 1661, 1630, 1542, 1456, 1417, 1366, 1330, 1313, 1282, 1263, 1250, 1234, 1168, 1113, 1087, 1062, 969, 940, 902, 857. HRMS (ESI) C19H24N3O3. Calculated: [M + H]+ 342.1818, found: [M + H]+ 342.1813.
2-(4-Methylpiperazin-1-yl)-7-((1-propyl-1H-indol-3-yl)methyl)-3,5,6,7,8,9-hexahydro-4H-pyrimido[4,5-d]azepin-4-one (1a, DS1040)
Following General Procedure B, tert-butyl 2-(4-methylpiperazin-1-yl)-4-oxo-3,4,5,6,8,9-hexahydro-7H-pyrimido[4,5-d]azepine-7-carboxylate (80.3 mg, 0.2209 mmol), and trifluoroacetic acid (1.1 mL) were added. Following conversion to the free amide, General Procedure C was followed. 1-Propyl-1H-indole-3-carbaldehyde (124.1 mg, 0.6630 mmol), sodium triacetoxyborohydride (234.2 mg, 1.105 mmol) in DMF (4.4 mL), and acetic acid (0.090 mL) were added. Crude product was purified using column chromatography (0–100% MeOH in DCM for 15 min) to afford the desired product as an orange solid: 33.7 mg (35%). 1H NMR (500 MHz, CDCl3) δ 7.73 (d, J = 7.91 Hz, 1H), 7.31 (d, J = 8.22 Hz, 1H), 7.20 (ddd, J = 8.15, 6.94, 1.19 Hz, 1H), 7.10 (ddd, J = 7.98, 6.97, 1.04 Hz, 1H), 7.05 (s, 1H), 4.05 (t, J = 7.10 Hz, 2H), 3.83 (s, 2H), 3.70 (t, J = 5.13 Hz, 4H), 2.82–2.55 (m, 8H), 2.44 (t, J = 5.01 Hz, 4H), 2.31 (s, 3H), 1.86 (h, J = 7.31 Hz, 2H), 0.93 (t, J = 7.37 Hz, 3H). 13C NMR (125 MHz, CDCl3) δ 169.22, 165.36, 151.92, 136.41, 128.73, 127.76, 121.47, 119.87, 119.02, 112.24, 109.47, 55.07, 54.73, 54.40, 52.91, 48.08, 46.21, 44.48, 39.28, 23.77, 23.65, 11.72. IR (neat) υmax = 2929, 2782, 1643, 1568, 1455, 1392, 1356, 1340, 1319, 1302, 1293, 1279, 1263, 1217, 1191, 1167, 1149, 1136, 1113, 1076, 1002, 972, 950, 853. HRMS (ESI) C25H35N6O. Calculated: [M + H]+ 435.2872, found: [M + H]+ 435.2857.
7-((1-Propyl-1H-indol-3-yl)methyl)-2-(pyrrolidin-1-yl)-3,5,6,7,8,9-hexahydro-4H-pyrimido[4,5-d]azepin-4-one (1b)
Following General Procedure B, tert-butyl 4-oxo-2-(pyrrolidin-1-yl)-3,4,5,6,8,9-hexahydro-7H-pyrimido[4,5-d]azepine-7-carboxylate (150 mg, 0.4500 mmol) and trifluoroacetic acid (2.2 mL) were added. Following conversion to the free amide, General Procedure C was followed. 1-Propyl-1H-indole-3-carbaldehyde (176.0 mg, 0.9390 mmol), sodium triacetoxyborohydride (497 mg, 2.347 mmol) in DMF (9.0 mL), and acetic acid (0.190 mL) were added. Crude product was purified using column chromatography (0–25% MeOH in DCM for 15 min) to afford the desired product as a yellow solid: 62.0 mg (33%). 1H NMR (500 MHz, MeOD) δ 7.71 (d, J = 7.94 Hz, 1H), 7.52 (s, 1H), 7.48 (d, J = 8.30 Hz, 1H), 7.25 (t, J = 7.65 Hz, 1H), 7.17 (t, J = 7.52 Hz, 1H), 4.50 (s, 2H), 4.19 (t, J = 7.02 Hz, 2H), 3.52–3.41 (m, 4H), 3.37 (s, 2H), 3.27 (s, 2H), 2.98 (t, J = 5.24 Hz, 2H), 2.91 (s, 2H), 2.06–1.93 (m, 4H), 1.88 (h, J = 6.46, 5.59 Hz, 2H), 0.92 (t, J = 7.38 Hz, 3H). 13C NMR (125 MHz, MeOD) δ 161.52, 151.21, 136.47, 131.12, 128.00, 122.00, 120.03, 118.15, 118.00, 109.91, 107.98, 102.37, 53.29, 52.54, 50.71, 46.46, 41.04, 33.49, 24.80, 23.14, 19.88, 10.22. IR (neat) υmax = 2957, 2875, 1632, 1588, 1456, 1393, 1333, 1262, 1237, 1197, 1175, 1125, 1014, 960, 898, 879. HRMS (ESI) C24H32N5O. Calculated: [M + H]+ 406.2607, found: [M + H]+ 406.2592.
2-Phenyl-7-((1-propyl-1H-indol-3-yl)methyl)-3,5,6,7,8,9-hexahydro-4H-pyrimido[4,5-d]azepin-4-one (1c)
Following General Procedure B, tert-butyl 4-oxo-2-phenyl-3,4,5,6,8,9-hexahydro-7H-pyrimido[4,5-d]azepine-7-carboxylate (260 mg, 0.7615 mmol) and trifluoroacetic acid (2.5 mL) were added. Following conversion to the free amide, General Procedure C was followed. 1-Propyl-1H-indole-3-carbaldehyde (250 mg, 1.335 mmol), sodium triacetoxyborohydride (707 mg, 3.336 mmol) in DMF (13 mL), and acetic acid (0.270 mL) were added. Crude product was purified using column chromatography (0–10% MeOH in DCM for 10 min) to afford the desired product as a yellow solid: 84.4 mg (31%). 1H NMR (500 MHz, MeOD) δ 7.98–7.92 (m, 2H), 7.70 (d, J = 7.89 Hz, 1H), 7.55 (t, J = 7.34 Hz, 1H), 7.49 (t, J = 7.46 Hz, 2H), 7.41 (d, J = 7.16 Hz, 2H), 7.20 (ddd, J = 8.21, 6.93, 1.04 Hz, 1H), 7.12 (ddd, J = 7.98, 7.04, 0.99 Hz, 1H), 4.30 (s, 2H), 4.14 (t, J = 7.04 Hz, 2H), 3.26–3.19 (m, 2H), 3.14 (q, J = 5.61 Hz, 4H), 3.01 (dd, J = 6.93, 3.61 Hz, 2H), 1.85 (h, J = 7.22 Hz, 2H), 0.90 (t, J = 7.37 Hz, 3H). 13C NMR (125 MHz, MeOD) δ 164.07, 163.92, 155.54, 136.42, 132.30, 131.45, 130.19, 128.49, 128.16, 127.34, 121.67, 121.52, 119.57, 118.36, 109.65, 104.61, 52.60, 52.29, 50.72, 47.48, 34.06, 23.15, 21.07, 10.25. IR (neat) υmax = 2930, 1635, 1603, 1542, 1505, 1466, 1398, 1327, 1195, 1128, 1013, 943, 898. HRMS (ESI) C26H29N4O. Calculated: [M + H]+ 413.2341, found: [M + H]+ 413.2324.
2-Phenyl-7-(pyridin-4-yl)methyl-3,5,6,7,8,9-hexahydro-4H-pyrimido[4,5-d]azepin-4-one (1d)
Following General Procedure B, tert-butyl 2-(4-methylpiperazin-1-yl)-4-oxo-3,4,5,6,8,9-hexahydro-7H-pyrimido[4,5-d]azepine-7-carboxylate (75.1 mg, 0.286 mmol) and trifluoroacetic acid (0.82 mL) were added. Following conversion to the free amide, General Procedure C was followed. 4-Pyridine carboxyaldehyde (0.67 mL, 0.572 mmol), sodium triacetoxyborohydride (302.5 mg, 1.430 mmol) in DMF (5.72 mL), and acetic acid (60.6 μL) were added. Crude product was purified using column chromatography (0–10% methanolic ammonia in DCM for 15 min, holding at 10% for 10 min) to afford the desired product as a yellow solid: 63.9 mg (69%). 1H NMR (500 MHz, MeOD) δ 8.47 (s, 1H), 8.41 (d, J = 5.28 Hz, 1H), 7.46 (d, J = 4.66 Hz, 1H), 7.32 (d, J = 5.03 Hz, 1H), 4.81 (s, 1H), 3.68 (s, 2H), 3.65 (s, 4H), 2.81 (d, J = 5.65 Hz, 2H), 2.74 (d, J = 5.36 Hz, 2H), 2.61 (d, J = 8.33 Hz, 2H), 2.54 (t, J = 4.85 Hz, 2H), 2.50 (s, 4H), 2.33 (s, 3H). 13C NMR (125 MHz, MeOD) δ 162.72, 152.55, 151.75, 149.38, 148.61, 147.98, 124.23, 122.80, 112.09, 75.67, 61.38, 54.86, 53.91, 52.66, 44.58, 43.77, 23.26. IR (neat) υmax = 2922, 2799, 1626, 1563, 1413, 1399, 1290, 1264, 1142, 1001, 950, 803, 790, 576. HRMS (ESI) C19H26N6O. Calculated: [M + H]+ 355.2246, found: [M + H]+ 355.2236.
tert-Butyl 4-(2-Methoxy-2-oxoethylidene)piperidine-1-carboxylate (6)
To a flame-dried flask equipped with a stir bar was added NaH (60% dispersion in mineral oil, 240.9 mg, 6.023 mmol) as a suspension in anhydrous DMF (0.10 M). The resulting mixture was cooled to 0 °C before dropwise addition of trimethyl phosphonoacetate (1.20 mL, 7.416 mmol). The reaction was stirred for 30 min before addition of 1-tert-butoxycarbonylpiperidin-4-one (0.9985 g, 5.019 mmol) dissolved in DMF (1 mL). The reaction proceeded for 6 h before quenching with NH4Cl. The aqueous layer was extracted in triplicate with EtOAc. The combined organic extracts were washed with brine, dried with Na2SO4, and concentrated under vacuum. The crude product was purified by column chromatography (0–100% EtOAc in hexanes over 15 min) to afford the desired product as a white solid: 1.058 g (95%). 1H NMR (500 MHz, CDCl3) δ 5.57 (s, 1H), 3.53 (s, 3H), 3.33 (dt, J = 14.87, 5.80 Hz, 4H), 2.78 (t, J = 5.86 Hz, 2H), 2.13 (t, J = 5.83 Hz, 2H), 1.32 (s, 9H). 13C NMR (125 MHz, CDCl3) δ 166.04, 157.83, 154.03, 114.52, 79.22, 50.46, 44.42, 36.09, 29.20, 28.05. IR (neat) υmax = 3014, 2968, 2871, 1680, 1652, 1422, 1885, 1478, 1421, 1385, 1364, 1340, 1314, 1255, 1236, 1213, 1139, 1114, 1009, 992, 980, 965, 864, 791, 767, 744, 726, 690, 634. HRMS (ESI) C13H21NO4. Calculated: [M + H-Boc]+ 156.1025, found: [M + H-Boc]+ 156.1029.
8-(tert-Butyl)-4-methyl-2-benzyl-2,8-diazaspiro[4.5]decane-4,8-dicarboxylate (7)
To a flame-dried flask that was purged with argon was equipped a magnetic stir bar. The flask was cooled to 0 °C before addition of tert-butyl 4-(2-methoxy-2-oxoethylidene)piperidine-1-carboxylate (1.005 g, 3.917 mmol) as a solution in anhydrous toluene (0.40 M) and N-(methoxymethyl)-N-(trimethylsilylmethyl)-benzylamine (1.3948 g, 5.875 mmol). The mixture was stirred for 15 min before dropwise addition of TFA (0.060 mL, 0.783 mmol) as a solution in DCM (1.0 M). The reaction was stirred for 2 h before being concentrated under vacuum. The crude product was purified by column chromatography (0–100% EtOAc in hexanes over 20 min) to afford the desired product as a clear oil: 861.6 mg (57%, 99% BRSM). 1H NMR (500 MHz, CDCl3) δ 7.28 (t, J = 7.05 Hz, 4H), 7.23–7.18 (m, 1H), 3.82 (s, 2H), 3.63 (s, 3H), 3.62 (d, J = 4.13 Hz, 2H), 2.94 (t, J = 8.53 Hz, 1H), 2.83 (d, J = 12.01 Hz, 1H), 2.78 (t, J = 8.93 Hz, 3H), 2.69 (t, J = 8.00 Hz, 1H), 2.31 (d, J = 9.18 Hz, 1H), 1.81–1.72 (m, 1H), 1.62 (d, J = 13.36 Hz, 1H), 1.51 (d, J = 13.10 Hz, 1H), 1.41 (s, 9H), 1.34 (t, J = 11.21 Hz, 1H). 13C NMR (125 MHz, CDCl3) δ 173.23, 154.77, 138.83, 128.42, 128.27, 126.98, 79.37, 61.93, 59.84, 55.27, 53.42, 51.46, 44.03, 41.32, 36.88, 32.48, 28.40. IR (neat) υmax = 2948, 1733, 1687, 1495, 1453, 1422, 1364, 1273, 1244, 1093, 1028, 978, 951, 911, 860, 738, 698. HRMS (ESI) C22H32N2O4. Calculated: [M + H]+ 389.2440, found: [M + H]+ 389.2444.
Methyl 2-Benzyl-2,8-diazaspiro[4.5]decane-4-carboxylate (S5)
To a reaction vessel equipped with a magnetic stir bar was added a 1:1 mixture of HCl/EtOAc (0.30 M) and 8-(tert-butyl)-4-methyl-2-benzyl-2,8-diazaspiro[4.5]decane-4,8-dicarboxylate (427.3 mg, 1.010 mmol). The reaction was stirred for 30 min at RT before being quenched with H2O. The aqueous layer was washed in triplicate with EtOAc. The recovered aqueous layer was concentrated under vacuum. The product was isolated as a yellow foaming solid (454.1 mg) and used crude in the next step.
Methyl 2-Benzyl-8-(pyridin-2-ylmethyl)-2,8-diazaspiro[4.5]decane-4-carboxylate (8a)
Following General Procedure D, 2-benzyl-2,8-diazaspiro[4.5]decane-4-carboxylate (227.7 mg, 0.7871 mmol), pyridine-2-carbaldehyde (421.6 mg, 3.936 mmol), and sodium triacetoxyborohydride (834.2 mg, 3.936 mmol) were used. Crude product was purified using column chromatography (0–10% MeOH in DCM over 10 min) to afford the desired product as a brown oil: 65.7 mg (62% over 2 steps). 1H NMR (500 MHz, CDCl3) δ 8.59–8.47 (m, 1H), 7.64 (t, J = 7.70 Hz, 1H), 7.39 (d, J = 7.64 Hz, 1H), 7.32 (q, J = 7.99, 7.44 Hz, 4H), 7.28–7.22 (m, 1H), 7.15 (t, J = 6.40 Hz, 1H), 3.67 (s, 5H), 3.61 (s, 2H), 2.99 (s, 1H), 2.86 (d, J = 9.26 Hz, 1H), 2.80–2.67 (m, 4H), 2.30 (d, J = 9.19 Hz, 1H), 2.17 (s, 1H), 2.08 (s, 1H), 1.96 (t, J = 12.73 Hz, 1H), 1.71 (d, J = 13.16 Hz, 1H), 1.56 (s, 2H). 13C NMR (125 MHz, MeOD) δ 172.58, 155.82, 148.49, 137.36, 136.64, 128.95, 128.18, 127.46, 123.98, 122.90, 62.60, 61.84, 59.57, 54.40, 52.58, 50.92, 50.83, 50.24, 42.95, 35.48, 20.96. IR (neat) υmax = 2947, 2804, 1683, 1580, 1570, 1495, 1475, 1434, 1364, 1260, 1168, 1028, 993, 911, 861, 757, 700. HRMS (ESI) C23H29N3O2. Calculated: [M + H]+ 380.2338, found: [M + H]+ 380.2338.
Methyl 2,8-Dibenzyl-2,8-diazaspiro[4.5]decane-4-carboxylate (8b)
Following General Procedure D, 2-benzyl-2,8-diazaspiro[4.5]decane-4-carboxylate (113.9 mg, 0.3953 mmol), benzaldehyde (209.6 mg, 1.976 mmol), and sodium triacetoxyborohydride (418.8 mg, 1.976 mmol) were used. Crude product was purified using column chromatography (0–25% MeOH in DCM over 25 min) to afford the desired product as a clear oil: 27.0 mg (36% over 2 steps). 1H NMR (500 MHz, CDCl3) δ 7.38–7.27 (m, 10H), 3.77 (s, 2H), 3.68 (s, 3H), 3.66 (s, 2H), 3.12 (s, 1H), 2.87 (q, J = 8.88, 6.44 Hz, 3H), 2.80 (t, J = 7.63 Hz, 2H), 2.40 (d, J = 9.62 Hz, 1H), 2.33 (s, 1H), 2.22 (s, 1H), 2.08 (s, 1H), 1.84 (d, J = 13.49 Hz, 1H), 1.71 (s, 1H), 1.63 (s, 1H). 13C NMR (125 MHz, CDCl3) δ 175.82, 172.72, 135.65, 132.84, 130.36, 129.34, 128.71, 128.62, 128.57, 128.00, 61.37, 59.57, 54.29, 52.17, 51.93, 49.90, 49.45, 43.21, 34.35, 30.61, 21.79 IR (neat) υmax = 2926, 2852, 2806, 1698, 1644, 1553, 1495, 1454, 1485, 1454, 1435, 1365, 1234, 1193, 1168, 1028, 738, 614, 603, 578, 568. HRMS (ESI) C24H30N2O2. Calculated: [M + H]+ 379.2386, found: [M + H]+ 379.2388.
Methyl 2-Benzyl-8-(cyclohexylmethyl)-2,8-diazaspiro[4.5]decane-4-carboxylate (8c)
Following General Procedure D, 2-benzyl-2,8-diazaspiro[4.5]decane-4-carboxylate (214.9 mg, 0.7450 mmol), cyclohexane carboxaldehyde (417.8 mg, 3.725 mmol), and sodium triacetoxyborohydride (789.5 mg, 3.725 mmol) were used. Crude product was purified using column chromatography (0–100% MeOH in DCM over 15 min, holding at 25% for 5 min) to afford the desired product as a yellow oil: 136.8 mg (77% over 2 steps). 1H NMR (500 MHz, CDCl3) δ 7.31 (q, J = 7.46 Hz, 4H), 7.28–7.22 (m, 1H), 3.82–3.72 (m, 2H), 3.71 (s, 3H), 3.44 (s, 1H), 3.14 (t, J = 8.96 Hz, 2H), 2.92 (t, J = 11.36 Hz, 2H), 2.83 (t, J = 7.63 Hz, 1H), 2.65 (s, 1H), 2.57 (d, J = 6.70 Hz, 2H), 2.47 (d, J = 9.69 Hz, 1H), 2.33 (t, J = 12.67 Hz, 1H), 2.00–1.91 (m, 4H), 1.81 (d, J = 11.84 Hz, 2H), 1.70 (d, J = 12.87 Hz, 3H), 1.62 (d, J = 12.63 Hz, 1H), 1.25–1.07 (m, 3H), 0.97 (q, J = 12.24, 11.05 Hz, 2H). 13C NMR (125 MHz, MeOD) δ 172.36, 137.41, 128.73, 128.14, 127.22, 62.96, 61.52, 59.42, 54.35, 52.24, 50.98, 50.65, 50.23, 42.22, 33.39, 32.94, 30.70, 29.42, 25.66, 25.23, 21.65. IR (neat) υmax = 2925, 2852, 1651, 1449, 1362, 1263, 1171, 1028, 945, 602, 568, 559. HRMS (ESI) C24H36N2O2. Calculated: [M + H]+ 385.2855, found: [M + H]+ 385.2861.
Methyl 2-Benzyl-8-(pyridin-4-ylmethyl)-2,8-diazaspiro[4.5]decane-4-carboxylate (8d)
Following General Procedure D, 2-benzyl-2,8-diazaspiro[4.5]decane-4-carboxylate (214.9 mg, 0.7450 mmol), 4-pyridine carboxaldehyde (399.1 mg, 3.726 mmol), and sodium triacetoxyborohydride (789.7 mg, 3.726 mmol) were used. Crude product was purified using column chromatography (0–8% MeOH in DCM over 10 min, holding at 8% for 5 min) to afford the desired product as a yellow oil: 137.2 mg (58% over 2 steps). 1H NMR (500 MHz, CDCl3) δ 8.53 (d, J = 5.01 Hz, 1H), 8.49 (d, J = 4.95 Hz, 2H), 7.31 (dd, J = 14.40, 7.15 Hz, 4H), 7.23 (d, J = 3.94 Hz, 2H), 3.68 (s, 5H), 3.42 (s, 2H), 3.02 (t, J = 8.29 Hz, 1H), 2.86 (d, J = 9.18 Hz, 1H), 2.77 (dt, J = 24.45, 8.14 Hz, 2H), 2.63 (dd, J = 28.93, 11.31 Hz, 2H), 2.31 (d, J = 9.30 Hz, 1H), 2.08 (t, J = 11.37 Hz, 1H), 1.99 (d, J = 9.16 Hz, 1H), 1.90 (td, J = 12.30, 11.75, 3.98 Hz, 1H), 1.72 (d, J = 13.19 Hz, 1H), 1.52 (d, J = 5.42 Hz, 2H). 13C NMR (125 MHz, MeOD) δ 172.98, 152.51, 148.60, 137.52, 128.76, 128.06, 127.14, 124.48, 121.37, 62.28, 61.92, 61.16, 59.78, 53.04, 51.08, 50.66, 50.35, 43.19, 36.53, 32.37. IR (neat) υmax = 3028, 2945, 2804, 1602, 1561, 1495, 1435, 1415, 1363, 1322, 1299, 1266, 1169, 1091, 1064, 1040, 1028, 993, 955, 912, 812, 794, 740, 700, 628, 616, 603, 593, 581, 568. HRMS (ESI) C23H29N3O2. Calculated: [M + H]+ 380.2338, found: [M + H]+ 380.2344.
Methyl 2-Benzyl-8-((3,5-dimethylisoxazol-4-yl)methyl)-2,8-diazaspiro[4.5]decane-4-carboxylate (8e)
Following General Procedure D, 2-benzyl-2,8-diazaspiro[4.5]decane-4-carboxylate (214.9 mg, 0.7450 mmol), 3,5-dimethyl-4-isoxazole carbaldehyde (399.1 mg, 3.726 mmol), and sodium triacetoxyborohydride (789.7 mg, 3.726 mmol) were used. Crude product was purified using column chromatography (0–100% MeOH in DCM over 10 min, holding at 10% for 5 min) to afford the desired product as a yellow oil: 137.2 mg (68% over 2 steps). 1H NMR (500 MHz, CDCl3) δ 7.37–7.28 (m, 4H), 7.26 (t, J = 7.12 Hz, 1H), 3.77 (s, 2H), 3.66 (s, 3H), 3.17 (s, 2H), 3.14–3.07 (m, 1H), 2.99–2.90 (m, 1H), 2.88–2.81 (m, 1H), 2.76 (t, J = 7.75 Hz, 1H), 2.62 (dd, J = 28.36, 10.62 Hz, 2H), 2.38 (d, J = 9.69 Hz, 1H), 2.29 (s, 3H), 2.20 (s, 3H), 2.00 (s, 1H), 1.94 (d, J = 11.05 Hz, 1H), 1.83 (td, J = 12.42, 11.50, 3.91 Hz, 1H), 1.70 (d, J = 13.40 Hz, 1H), 1.52–1.39 (m, 2H). 13C NMR (125 MHz, CDCl3) δ 173.43, 166.50, 160.39, 138.55, 128.61, 128.30, 127.06, 110.48, 62.49, 59.91, 55.21, 53.41, 51.44, 51.07, 50.58, 50.40, 43.88, 37.24, 32.89, 11.05, 10.28. IR (neat) υmax = 2921, 2804, 1730, 1679, 1640, 1583, 1495, 1452, 1434, 1362, 1298, 1260, 1194, 1069, 1028, 989, 956, 911, 886, 801, 742, 699. HRMS (ESI) C23H31N3O3. Calculated: [M + H]+ 398.2444, found: [M + H]+ 398.2452.
(2-Benzyl-8-(pyridin-2-ylmethyl)-2,8-diazaspiro[4.5]decan-4-yl)methanol (2a, RH1096)
Following the General Procedure E, LiAlH4 (3.9 mg, 0.1017 mmol) and methyl 2-benzyl-8-(pyridin-2-ylmethyl)-2,8-diazaspiro[4.5]decane-4-carboxylate (19.3 mg, 0.0509 mmol) were used. Crude product was purified using column chromatography (0–100% MeOH in DCM over 10 min) to afford the desired product as a clear oil: 11.4 mg (64%) 1H NMR (500 MHz, CDCl3) δ 8.55 (d, J = 4.70 Hz, 1H), 7.64 (t, J = 7.66 Hz, 1H), 7.39 (d, J = 7.80 Hz, 1H), 7.32 (t, J = 7.30 Hz, 2H), 7.28 (d, J = 7.13 Hz, 3H), 7.15 (t, 1H), 3.74 (s, 2H), 3.63 (d, J = 8.43 Hz, 2H), 3.60 (s, 2H), 2.87–2.73 (m, 3H), 2.67 (s, 2H), 2.28 (d, J = 9.62 Hz, 1H), 2.25–2.12 (m, 2H), 1.91–1.82 (m, 2H), 1.79 (d, J = 13.66 Hz, 1H), 1.69 (td, J = 11.67, 10.30, 3.68 Hz, 1H), 1.61 (d, J = 13.56 Hz, 1H). 13C NMR (125 MHz, CDCl3) δ 158.90, 149.22, 138.35, 136.35, 128.53, 128.45, 127.16, 123.13, 121.94, 64.98, 64.02, 63.64, 60.31, 57.59, 52.10, 51.34, 41.20, 39.14, 32.47. IR (neat) υmax = 3374, 2924, 1645, 1595, 1436, 1366, 1230, 1092, 1028, 743. HRMS (ESI) C22H29N3O. Calculated: [M + H]+ 352.2389, found: [M + H]+ 352.2393.
(2,8-Dibenzyl-2,8-diazaspiro[4.5]decan-4-yl)methanol (2b, RH1103)
Following the General Procedure E, LiAlH4 (8.3 mg, 0.2198 mmol) and methyl 2,8-dibenzyl-2,8-diazaspiro[4.5]decane-4-carboxylate (20.7 mg, 0.1099 mmol) were used. Crude product was purified using column chromatography (0–20% MeOH in DCM over 20 min) to afford the desired product as a clear oil: 26.1 mg (68%). 1H NMR (500 MHz, CDCl3) δ 7.43–7.29 (m, 10H), 5.80 (s, 1H), 3.88 (q, 2H), 3.75–3.64 (m, 4H), 3.11 (t, 1H), 3.00 (dd, J = 10.58, 4.47 Hz, 1H), 2.96–2.85 (m, 3H), 2.72 (d, J = 10.45 Hz, 1H), 2.44–2.29 (m, 2H), 2.07 (t, J = 9.57, 7.49 Hz, 1H), 2.00–1.90 (m, 2H), 1.81 (d, J = 14.04 Hz, 1H), 1.71 (d, J = 14.08 Hz, 1H). 13C NMR (125 MHz, MeOD) δ 132.72, 129.79, 129.17, 128.26, 128.17, 128.10, 127.79, 127.70, 62.84, 61.98, 60.45, 59.82, 56.27, 50.56, 50.07, 40.91, 35.71, 29.84, 29.34. IR (neat) υmax = 3369, 2921, 2851, 2802, 1557, 1494, 1452, 1378, 1261, 1074, 1028, 991, 913, 798, 740. HRMS (ESI) C23H30N2O. Calculated: [M + H]+ 351.2436, found: [M + H]+ 351.2440.
(2-Benzyl-8-(cyclohexylmethyl)-2,8-diazaspiro[4.5]decan-4-yl)methanol (2c, RH1115)
Following the General Procedure E, LiAlH4 (6.9 mg, 0.1815 mmol) and methyl 2-benzyl-8-(cyclohexylmethyl)-2,8-diazaspiro[4.5]decane-4-carboxylate (34.9 mg, 0.0908 mmol) were used. Crude product was purified using column chromatography (0–100% MeOH in DCM over 15 min) to afford the desired product as a yellow oil: 18.0 mg (74%). 1H NMR (500 MHz, CDCl3) δ 7.43–7.27 (m, 5H), 3.96–3.79 (m, 2H), 3.78–3.61 (m, 2H), 3.19 (s, 2H), 3.08 (s, 1H), 2.88 (s, 2H), 2.71 (d, J = 10.02 Hz, 1H), 2.58 (d, J = 6.79 Hz, 3H), 2.37–2.19 (m, 2H), 2.11 (s, 1H), 2.01 (s, 1H), 1.94 (d, J = 14.48 Hz, 1H), 1.83 (d, J = 13.31 Hz, 3H), 1.72 (d, J = 11.48 Hz, 3H), 1.68–1.61 (m, 1H), 1.23 (q, J = 12.58 Hz, 2H), 1.19–1.09 (m, 1H), 0.98 (q, J = 12.73, 11.89 Hz, 2H). 13C NMR (125 MHz, MeOD) δ 130.18, 129.06, 128.24, 127.56, 63.13, 62.69, 60.38, 59.68, 56.02, 50.88, 50.57, 40.34, 34.18, 33.00, 30.55, 28.33, 25.65, 25.18, 20.90. IR (neat) υmax = 3329, 2919, 2849, 2800, 1574, 1494, 1448, 1378, 1297, 1263, 1119, 1072, 1028, 994, 892, 844, 798, 739, 698, 652. HRMS (ESI) C23H36N2O. Calculated: [M + H]+ 357.2906, found: [M + H]+ 357.2904.
(2-Benzyl-8-(pyridin-4-ylmethyl)-2,8-diazaspiro[4.5]decan-4-yl)methanol (2d)
Following the General Procedure E, LiAlH4 (10.0 mg, 0.2635 mmol) and methyl 2-benzyl-8-(cyclohexylmethyl)-2,8-diazaspiro[4.5]decane-4-carboxylate (50.3 mg, 0.1325 mmol) were used. Crude product was purified using column chromatography (0–100% MeOH in DCM over 15 min) to afford the desired product as a yellow oil: 37.3 mg (81%). 1H NMR (500 MHz, CDCl3) δ 8.52 (d, J = 5.09 Hz, 2H), 7.38–7.27 (m, 5H), 7.25 (s, 2H), 6.55 (s, 1H), 3.86–3.75 (m, 2H), 3.73 (d, J = 3.62 Hz, 2H), 3.46 (s, 2H), 2.99 (d, J = 4.86 Hz, 2H), 2.90 (d, J = 10.27 Hz, 1H), 2.61 (s, 1H), 2.55 (d, J = 10.25 Hz, 1H), 2.18–2.09 (m, 1H), 2.04 (s, 2H), 1.97 (t, J = 4.75 Hz, 1H), 1.81 (t, J = 10.00 Hz, 1H), 1.73 (d, J = 16.20 Hz, 1H), 1.68 (d, J = 10.30 Hz, 1H), 1.60 (d, J = 13.51 Hz, 1H). 13C NMR (125 MHz, MeOD) δ 148.71, 148.58, 136.45, 129.12, 128.18, 127.50, 124.47, 63.07, 61.26, 60.69, 60.02, 56.54, 51.16, 50.51, 49.31, 40.98, 36.84, 30.97. IR (neat) υmax = 3396, 2913, 2803, 1604, 1560, 1417, 1362, 1221, 1131, 1090, 1028, 992, 813, 790, 744, 700, 663, 636, 611, 574, 567. HRMS (ESI) C22H29N3O. Calculated: [M + H]+ 352.2389, found: [M + H]+ 352.2394.
(2-Benzyl-8-((3,5-dimethylisoxazol-4-yl)methyl)-2,8-diazaspiro[4.5]decan-4-yl)methanol (2e)
Following the General Procedure E, LiAlH4 (9.5 mg, 0.2516 mmol) and methyl 2-benzyl-8-(cyclohexylmethyl)-2,8-diazaspiro[4.5]decane-4-carboxylate (50.0 mg, 0.1258 mmol) were used. Crude product was purified using column chromatography (0–100% MeOH in DCM over 20 min) to afford the desired product as a yellow oil: 40.3 mg (87%). 1H NMR (500 MHz, CDCl3) δ 7.49 (d, J = 5.42 Hz, 2H), 7.38 (d, J = 5.92 Hz, 3H), 4.09 (d, J = 13.63 Hz, 1H), 4.01 (d, J = 12.90 Hz, 1H), 3.72 (d, J = 4.65 Hz, 2H), 3.31 (t, J = 8.15 Hz, 1H), 3.22 (s, 3H), 3.06 (d, J = 11.03 Hz, 1H), 2.89 (d, J = 11.05 Hz, 1H), 2.60 (s, 2H), 2.32 (s, 3H), 2.22 (s, 3H), 2.12 (s, 1H), 2.02 (s, 2H), 1.69 (td, J = 33.01, 26.90, 12.77 Hz, 4H). 13C NMR (125 MHz, MeOD) δ 167.60, 160.36, 129.46, 128.41, 128.14, 110.02, 62.71, 60.24, 59.71, 56.16, 50.60, 50.12, 49.60, 48.70, 41.28, 36.36, 30.54, 9.62, 8.73. IR (neat) υmax = 3400, 2926, 2808, 1645, 1421, 1362, 1223, 1092, 1028, 989, 745, 701, 582, 580, 566. HRMS (ESI) C22H31N3O2. Calculated: [M + H]+ 370.2495, found: [M + H]+ 370.2497.
(2-Benzyl-8-(cyclohexylmethyl)-2,8-diazaspiro[4.5]decan-4-yl)methyl Hex-5-ynoate (S6)
To a flame-dried flask that was purged with argon was equipped a magnetic stir bar. The flask was cooled to 0 °C before addition of (2-benzyl-8-(cyclohexylmethyl)-2,8-diazaspiro[4.5]decan-4-yl)methanol (14.2 mg, 0.0398 mmol) and triethylamine (8.1 mg, 0.0797 mmol) as a solution in DCM (0.01 M). The reaction was stirred for 5 min before addition of hex-5-ynoyl chloride (10.4 mg, 0.0797 mmol). The mixture was stirred for 3 h with temperature increasing from 0 to 25 °C before the reaction was quenched with H2O. The aqueous layer was extracted in triplicate with DCM. The combined organic extracts were washed with brine, dried with Na2SO4, and concentrated under vacuum. The crude product was purified by column chromatography (0–100% MeOH in DCM over 10 min, holding at 15% MeOH for 5 min) to afford the desired product as a clear oil: 12.2 mg (68%). 1H NMR (500 MHz, MeOD) δ 7.36–7.29 (m, 4H), 7.27 (t, J = 6.76 Hz, 1H), 4.20 (dd, J = 11.21, 6.45 Hz, 1H), 4.07 (dd, J = 11.19, 7.38 Hz, 1H), 3.69 (d, J = 3.12 Hz, 2H), 2.99 (t, J = 8.71 Hz, 1H), 2.85 (d, J = 7.98 Hz, 1H), 2.76 (s, 3H), 2.53 (d, J = 9.75 Hz, 1H), 2.46 (t, J = 7.34 Hz, 2H), 2.41 (d, J = 8.90 Hz, 1H), 2.28–2.15 (m, 5H), 1.95 (t, J = 13.08 Hz, 1H), 1.85–1.67 (m, 13H), 1.33 (d, J = 12.83 Hz, 2H), 1.22 (d, J = 12.68 Hz, 1H), 1.01 (d, J = 11.76 Hz, 2H). 13C NMR (125 MHz, MeOD) δ 173.18, 137.86, 128.67, 128.03, 127.01, 68.95, 63.13, 62.58, 59.83, 55.96, 51.21, 50.64, 46.25, 40.62, 34.97, 33.64, 32.27, 31.02, 29.46, 25.86, 25.41, 23.50, 17.03. IR (neat) υmax = 2922, 2850, 2797, 1583, 1495, 1450, 1377, 1265, 1072, 1028, 984. HRMS (ESI) C29H42N2O2. Calculated: [M + H]+ 451.3325, found: [M + H]+ 451.3325.
(2-Benzyl-8-(cyclohexylmethyl)-2,8-diazaspiro[4.5]decan-4-yl)methyl 4-(1-(13-Oxo-17-(2-oxohexahydro-1H-thieno[3,4-d]imidazol-4-yl)-3,6,9-trioxa-12-azaheptadecyl)-1H-1,2,3-triazol-5-yl)butanoate (Biotin-RH1115)
To a flame-dried flask that was purged with argon was equipped a magnetic stir bar. To the flask were added (2-benzyl-8-(cyclohexylmethyl)-2,8-diazaspiro[4.5]decan-4-yl)methyl hex-5-ynoate (12.2 mg, 0.0271 mmol), azide-PEG3-biotin conjugate (14.4 mg, 0.0325 mmol), and CuSO4·5H2O (20 mol %) as a solution in 4:1 H2O/THF. The mixture was heated to 90 °C and stirred for 48 h. The mixture was concentrated and purified by column chromatography (0–100% MeOH in DCM over 35 min, holding at 18% MeOH for 10 min) to afford the desired product as a slightly yellow solid: 8.7 mg (36%). 1H NMR (500 MHz, MeOD) δ 7.80 (s, 1H), 7.31 (d, J = 6.59 Hz, 4H), 7.25 (d, J = 6.59 Hz, 1H), 5.23 (t, 1H), 4.54 (t, J = 5.04 Hz, 2H), 4.48 (t, J = 6.44 Hz, 1H), 4.36–4.27 (m, 3H), 4.23–4.13 (m, 3H), 4.03 (dd, J = 11.07, 7.88 Hz, 1H), 3.88 (t, J = 5.08 Hz, 2H), 3.63 (s, 1H), 3.60 (s, 3H), 3.58 (s, 3H), 3.52 (t, J = 5.41 Hz, 2H), 3.21–3.15 (m, 1H), 2.91 (dd, J = 11.69, 5.89 Hz, 2H), 2.80 (d, J = 9.67 Hz, 1H), 2.76–2.66 (m, 3H), 2.44–2.30 (m, 7H), 2.20 (t, J = 7.39 Hz, 2H), 2.04 (d, J = 2.65 Hz, 5H), 2.02 (s, 2H), 1.95 (t, J = 7.48 Hz, 2H), 1.78–1.70 (m, 6H), 1.66–1.58 (m, 8H), 1.53 (d, J = 6.40 Hz, 4H), 1.43 (q, J = 7.73, 7.00 Hz, 3H). 13C NMR (125 MHz, MeOD) δ 173.31, 166.78, 166.22, 128.84, 128.68, 128.03, 127.99, 126.94, 122.82, 76.84, 70.14, 70.08, 70.02, 69.86, 69.22, 69.02, 63.32, 62.82, 61.96, 60.23, 59.94, 56.06, 55.59, 54.85, 51.51, 50.77, 39.66, 38.94, 35.34, 32.81, 31.67, 31.36, 29.34, 29.06, 28.36, 28.11, 26.71, 26.05, 25.58, 25.45, 24.41, 24.21, 22.33, 18.66, 15.89, 13.03. IR (neat) υmax = 3137, 3079, 2922, 2854, 1698, 1607, 1538, 1501, 1435, 1402, 1347, 1331, 1309, 1276, 1254, 1219, 1114, 1086, 1048, 900, 879, 847, 814, 744, 733, 687, 607, 592, 585, 564. HRMS (ESI) C47H74N8O7S. Calculated: [M + H]+ 895.5479, found: [M + H]+ 895.5464.
Biological Assays
Cell Culture
HeLa and A549 Cells
HeLa cells stably expressing eGFP-LC3 and mCherry-GFP-LC3 were a gift from Ramnik Xavier at Massachusetts General Hospital, Boston, MA. HeLa and A549 cells were purchased from Sigma-Aldrich (90321013; 86012804). All cells were cultured in DMEM (Corning, 15-013-CV) with 10% FBS (Sigma-Aldrich 2442), 3.6 mM l-glutamine (Corning, 25-005-Cl), and 1× penicillin-streptomycin (Corning, 30-002-Cl). Cultured cells were maintained in a humidified incubator at 37 °C with 5% CO2.
iPSCs Culture and Neuronal Differentiation
WTC-11 human-induced pluripotent stem cells were differentiated into a glutamatergic cortical layer 2–3 neuronal fate (referred to as i3Neurons) using a doxycycline-inducible neurogenin2 transgene expressed from the AAVS1 safe harbor locus, as described previously.74 iPSCs were grown on Matrigel (Corning, 356230)-coated dishes, maintained in E8 Flex media (Life Technologies, A2858501), and passaged using accutase (Corning, 25058CI). i3Neuron differentiation was initiated by plating iPSCs (500,000 iPSCs per well) into a Matrigel-coated 6-well plate in induction media containing knockout DMEM F12 (Thermo Fisher Scientific Scientific, 12660012), 1× N2 supplement (Thermo Fisher Scientific Scientific, 17502048), 1× nonessential amino acids solution (Thermo Fisher Scientific Scientific, 11140050), 1× l-glutamine glutamax solution (Thermo Fisher Scientific Scientific, 35050061), 10 μM ROCK inhibitor (Tocris Biosciences, 1254/1), and 2 μg/mL doxycycline hydrochloride (Sigma-Aldrich, BP26531) for 3 days. Prior to plating, 35 mm acid-washed coverslips (Carolina Biological Supply Company, 633009) and 6-well plates were coated with 0.1 mg/mL poly-l-ornithine (Sigma-Aldrich, A-004-M) and glass coverslips were additionally coated with 10 μg/mL mouse Laminin (Gibco, 23017015). Induced i3Neurons at DIV3 were plated into either 35 mm glass-bottom dishes (MatTek Life Sciences, P35G-1.5-14-C) for live imaging (30,000 cells per MatTek dish), in 35 mm glass coverslips for immunofluorescence (30,000 cells per coverslip) or in a 6-well plate for immunoblotting (500,000 cells per well).
Immunofluorescence Experiments
DIV 10 i3Neurons on 35 mm glass coverslips treated for 72 h with 0.1% DMSO (American Type Culture Collection, 4-X), RH1115 (15 μM), DS1040 (20 μM), RH1096 (40 μM), RH1103 (40 μM), or rapamycin (LC Laboratories, R-5000) (3 μM) or treated for 24 h with bafilomycin A1 (Sigma-Aldrich, B1793) (100 nM) and were fixed and immunostained as described previously.74 See Supporting Table S3 for antibody information.
Analysis of Lysosomal Vesicle Properties in i3Neurons
ImageJ “Analyze Particles” tool was used to measure lysosomal vesicle size, integrated density, and number within i3Neuron somas immunostained for LAMP1. Each soma was outlined using the “freehand selections” tool, and then using “Threshold” function, a threshold was set based on optimal coverage of most vesicles possible without compromising vesicle size and shape (avoiding false negatives or collapse/fusion of different vesicles). The “Analyze Particles” tool was then used to define the parameters for the measured vesicles, with a minimum size of 0.002 μm2 and no maximum size. Mean size, intensity of vesicles, and count per neuron were computed.
Evaluation of Lysosome Positioning in i3Neurons
DIV 10 i3Neurons on 35 mm glass coverslips treated with 0.1% DMSO, DS1040 (20 μM), RH1096 (40 μM), RH1103 (40 μM), or RH1115 (15 μM) for 72 h were fixed and immunostained for Tau or LAMP1 as described previously.74 Images were acquired using a high-magnification objective. Lysosome distribution was classified as “clustered”, “normal”, or “intermediate” based on proximity around nucleus (DAPI signal). Imaging and analysis were carried out in a double-blind fashion. Mean ± SEM of percent neurons exhibiting perinuclear clustering of lysosomes from three independent experiments were computed.
eGFP-LC3 Puncta Formation Assay
Methods have been adapted from previously published work.59 Compounds tested in the HCS were a curated ChemDiv library provided by the UICenter for Drug Discovery. HeLa cells expressing eGFP-LC3 were plated at a density of 3000 cells/well in a black 384-well plate (Corning, 3764) and incubated for 24 h at 37 °C. After 24 h, a Biomek NXP automated liquid handler (Beckman Coulter) transferred compounds (20 μM), DMSO (Corning, 259-50-CQC) (0.4%), chloroquine (CQ, Sigma-Aldrich, C6628) (20 μM), or PI-103 (LC Laboratories, P-9099) (5 μM). The plate was incubated for 4 h at 37 °C before the media was aspirated using a MultiFlo FX (BioTek SpA, MFXPW) and 25 μL of 4% paraformaldehyde (PFA) (Electron Microscopy Sciences, 15710) were added. The plate was incubated in the dark at room temperature (RT) for 12 min before PFA was aspirated and washed with 50 μL of 1× PBS (Corning, 21-040-CM). Following washing, 25 μL Hoechst 33342 nuclear stain (Thermo Scientific, H3570) was added at a concentration of 2 μg/mL and incubated in the dark at RT for 10 min. The solution was aspirated and 50 μL of 1× PBS was added before the plate was sealed using the PlateMax semiautomatic plate sealer (Axygen). The plate was imaged at 10× magnification using the DAPI and FITC filters on the ImageXpress Micro (IXM) XLS automated florescent microscope (Molecular Devices). Images were analyzed using Meta Xpress software in assays. Hits were selected based on assays with a Z′ (Z factor) above 0.4, a % CV of less than 20%, and a z-score greater than 2.199 in two replicates, yielding 312 molecules that meet these requirements. See Table S1 for further information. Subsequent eGFP-LC3 assays run on synthesized molecules were performed in 12-point dose (300 to 0.146 μM for 1 analogues, 1000 to 0.488 μM for 2 analogues) to generate EC50 values. Overtly cytotoxic concentrations (% viability <40) were excluded. Data are presented as mean ± SEM from three independent experiments, each with duplicate biological replicates.
Dual Reporter Assay
HeLa Cells
Cells stably expressing mCherry-GFP-LC3 were grown to a density of 3000 cells/well. The dual reported assay was performed following the same protocol as the eGFP-LC3 Puncta Formation assay. The plates were imaged using three filters: DAPI for nuclear stain, FITC for GFP, and Texas Red for mCherry. CellProfiler 3.1.9 was used to analyze these images to determine the number of autophagosomes and autolysosomes.
i3Neurons
i3Neurons derived from iPSCs stably expressing LC3-RFP-GFP were treated with 0.1% DMSO, RH1115 (15 μM) for 72 h or BafA1 (100 nM) for 24 h and imaged live at 37 °C in imaging media using Fast Airyscan on a Zeiss LSM880 microscope. A single scan image across the center of each cell (nucleus in full focus) was used to capture both GFP and RFP fluorescence using 488 and 560 nm lasers, respectively. Autophagosomes and autolysosomes were identified and analyzed in the following manner: brightness and contrast were adjusted equally for all images (green based on bafilomycin treatment and red based on DMSO treatment). Merged images, following adjustment of brightness and contrast, were used to identify “green and yellow” (autophagosomes) and the “red” (autolysosomes) vesicles, and their numbers were computed using the multipoint tool (ImageJ/Fiji). The percent autolysosomes were computed as the “red” vesicles divided by the “red” plus “green and yellow” vesicles. To evaluate the size and intensity of autolysosomes specifically, we first carried out particle analysis (“Analyze Particles” ImageJ) on the red channel images from DMSO and RH1115 treatments and filtered out the autophagosomes (using the merged image data as reference).
Immunoblotting
General Procedure
Cell lysate was centrifuged at 12,000 rpm for 30 min at 4 °C to remove cellular debris. The supernatant was combined with 4× NuPAGE LDS Sample Buffer (Thermo Fisher Scientific, NP0007) and 10x Bolt Sample Reducing Agent (Thermo Fisher Scientific, B0009) and separated by 10% SDS-PAGE (120 V, 1.5 h). Protein was transferred onto PVDF membrane (Millipore Sigma, IPFL00010) at 25 V for 1 h. The membrane was blocked with 5% Blotting-grade Blocker (Bio-Rad, 1706404) in TBS spiked with 0.1% Tween-20 (VWR, 97062) and incubated for 1 h at RT. Membranes were incubated overnight at 4 °C with the primary antibody. The blots were washed three times with TBS-T and incubated with secondary antibody for 1 h at RT. Membranes were washed with TBS-T and incubated with SuperSignal West Pico PLUS Chemiluminescent Substrate (Thermo Fisher Scientific, 34580) for 3 min at RT. Blots were visualized using the c Series Capture Software on the Azure Imaging System. Images were quantified using ImageJ. See Supporting Table S3 for antibody information. For the majority of blots, the samples were run on hand-cast Tris-Glycine Gel (10–12%) prior to transfer.
i3 Neurons
DIV 21 i3Neurons were washed with ice-cold phosphate-buffered saline and lysed in lysis buffer of 1% Triton in PBS with protease inhibitor (1×) (Thermo Fisher Scientific, A32961) and phosphatase inhibitor (1×) (PhosStop Roche, 4906845001) with benzonase (1 μL/75 μL lysate, Millipore Sigma, E1014) prior to centrifugation at 13,000 rpm for 10 min. The supernatant was collected and boiled at 95 °C for 3 min in 1× SDS sample buffer for subsequent SDS-PAGE.
LC3 Immunoblotting
HeLa cells were plated in at a density of 75,000 cells/well in a 24-well dish and left at RT for 1 h to adhere to the plate before being placed in the incubator at 37 °C for 24 h. 0.1% DMSO, compound 1a (20 μM), compound 2a (80 μM), and BafA1 (LC Laboratories, B-1080) (100 nM) were administered by hand, and the plate was returned to the incubator for 4 h. The cells were lysed using NP-40 Lysis Buffer containing 10 mL of 1× TBS (Corning, 46-012-CM), 1 Pierce protease and phosphate inhibitor tablet (Thermo Fisher Scientific, A32959), and 1% IGEPAL (Sigma-Aldrich, 56741). General procedures above were followed utilizing LC3 and β-actin primary antibodies and Anti-Rabbit IgG, HRP-linked secondary antibody in 5% Blotting-grade Blocker in TBS-T. Data are presented as mean ± SEM of four independent experiments.
P70S6K and Phospho-P70S6K Immunoblotting
Slight modifications were made to the procedure for LC3 immunoblotting. Compound 2a was treated at 40 μM. 1% bovine serum albumin (Sigma-Aldrich, A2153) was used for blocking steps. Phosphorylated p70S6K and β-Actin were used as primary antibodies and Anti-Rabbit IgG, HRP-linked was used as a secondary antibody before the blot was stripped using Restore Western Blot Stripping Buffer (Thermo Fisher Scientific, 21059). The blot was reprobed with p70S6K. Images were analyzed using ImageJ.
Lamin A/C, LAMP1, and UBA-52 Immunoblotting
Slight modifications were made to the procedure for LC3 immunoblotting. HeLa cells were plated at a density of 300,000 cells/mL in a 6-well dish. Treatment of 0.1% DMSO and RH1115 (50 μM) were administered. Lamin A/C and UBA-52 blots were run on hand-casted Tris-Glycine Gel (10–12%) prior to transfer. LAMP1 expression level samples were run on a NuPAGE 4 to 12% Bis-Tris Gel (Thermo, #NP0335BOX) following the manufacturer’s instructions for gel running buffer and transfer buffer. Data are presented as mean ± SEM of three biological replicates for each condition.
Cell Viability Assay
HeLa Cells
Following the procedure for dose-dependent eGFP-LC3 puncta formation, nuclear counts were determined using the DAPI filter. Nuclear counts were compared to the average DMSO nuclear count to generate a percent viability. Percent viability data were generated as mean ± SEM from three independent experiments, each with duplicate biological replicates.
i3Neurons
DIV 10 i3Neurons on 35 mm glass coverslips were treated with 0.1% DMSO, 1a (DS1040) (20 μM), 2a (RH1096) (40 μM), 2b (RH1103) (40 μM), or 2c (RH1115) (15 μM) for 72 h before being fixed and immunostained for Tau and LAMP1 as described previously.74 Images were acquired using a high-magnification objective (5 to 6 areas at random) using a Keyence BZ-X810 microscope (Osaka, Japan), and the number of neurons per unit area was computed. Tau staining was used to confirm neuronal viability (normal morphology and neurite integrity). Mean ± SEM of three independent experiments was computed.
Kinetic Aqueous Solubility Assay
Compounds were diluted with 1× PBS to a concentration of 100 μM in a 96-well clear assay plate (Corning, #3628). Diclofenac was used as our soluble control, while dipyridamole was used as our insoluble control. Optical density readings at 620 nm were taken on a SpectraMax i3x (Molecular Devices) using the Softmax Pro 6.5.1 software. Data are presented as the mean ± SEM of five independent experiments.
Biotinylated Compound Pulldown Assay
HeLa cells were plated at a density of 3.75 × 106 cells/mL in a T-225 flask (Thermo Fisher Scientific, 159934) in 50 mL of DMEM and then grown for 3 days to confluency. The cells were resuspended by trypsinization then pelleted and stored at −80 °C until ready for cell lysis. Prior to sonication, the pelleted cells were thawed and then washed with 1× PBS. The cells were then lysed using Pierce IP Lysis Buffer (Thermo Fisher Scientific, 87788) via sonication. Lysate was centrifuged at 18,000g for 30 min at 4 °C to remove debris and the supernatant was transferred to a clean microcentrifuge tube. Protein concentration was measured using the Pierce BCA Protein Assay Kit (Thermo Fisher Scientific, 23225). Thirty μL of Pierce Streptavidin Magnetic Beads (Thermo Fisher Scientific, 88816) were washed twice with 200 μL of 1× PBS. Biotinylated RH1115 (Biotin-RH1115) (50 μM), biotin acid (50 μM), and DMSO (2 μL) were incubated with the beads while rotating at 4 °C for 2 h. 500 μg of lysate was then added to the magnetic beads and diluted to 200 μL using the cell lysis buffer. The lysate was left with the beads and compounds for 16 h before washing twice with 200 μL of 1× PBS, then eluting with elution buffer (Thermo Fisher Scientific, 1858606). For competitor experiments, an additional sample containing the magnetic beads and Biotin-RH1115 (50 μM) was prepared. Upon addition of the 500 μg cell lysate, RH1115 (100 μM) was added and incubated with the other samples for 16 h at 4 °C. Samples were dried completely using a speed vacuum before proceeding onto sample digestion using S-Trap (ProfiTi, CO2-micro-10). The dried sample was solubilized in 25 μL of 5% SDS, 50 mM TEAB. Samples were reduced using 20 mM DTT and heated for 5 min at 95 °C. Disulfides were then alkylated using 40 mM iodoacetamide and incubated in the dark for 30 min. 12% aqueous phosphoric acid at a 1:10 ratio to yield a final concentration of 1.2% phosphoric acid. Then, 165 μL of S-Trap protein binding buffer (90% aqueous methanol, 100 mM TEAB, pH 7.1) was added to the acidified solution. The prepared solution was then added to the S-Trap microcolumn and centrifuged to capture protein. The column was washed with 150 μL of S-Trap protein binding buffer. Two μg of trypsin (Thermo Fisher Scientific, 90058) in 40 μL of digestion buffer (50 mM TEAB) was added to the top of the column and left for 16 h at 37 °C to digest. Peptides were eluted with 40 μL of 50 mM TEAB, then 35 μL of 50% ACN, and 0.2% formic acid. Samples were dried on speed vacuum and then resuspended in 0.1% formic acid for liquid chromatography-mass spectrometry (LC-MS) data collection and analysis.
Liquid Chromatography-Mass Spectrometry (LC-MS) Data Acquisition and Analysis
Resuspended samples were spun down at 14,000g for 30 min before being transferred to polypropylene vials. 0.5 μL of each sample was injected into an Agilent 1260 Infinity nanoLC system (Agilent Technologies) coupled with a Q Exactive mass spectrometer (Thermo Fisher Scientific). Samples were analyzed using the following method: First, digested peptides were loaded onto a Thermo NanoViper trap column (75 μm × 20 mm, 3 μm C18, 100 Å) (Thermo Fisher Scientific, 164946) and washed for 10 min with solvent A (0.1% FA in water) at 2 μL/min flow rate. Peptides were then loaded onto an Agilent Zorbax 300SB-C18 column (5065–9911) (0.075 mm × 150 mm, 3.5 μm 300 Å) at 5% B (0.1% FA in ACN). Separation was carried out using a 180 min gradient going from 5 to 30% B with a flow rate of 0.25 μL/min. The system was then increased from 30–60% B and from 60–90% B, with each increase in 10 min period. The system is then maintained at 90% B for 10 min prior to a 15 min re-equilibration segment at 5% B prior to the next run. Mass spectra were collected using data-dependent acquisition (DDA) with a capillary temperature of 250 °C and spray voltage of 1.7 kV. Full MS scans were collected at a mass resolution of 70,000 with a scan range of 375–2000 m/z. Automatic gain control (AGC) target was set at 1 × 106 for a maximum injection time (IT) of 100 ms. The top ten most intense 10 peaks were selected for MS/MS analysis, with an isolation window of 1.5 m/z. MS/MS spectra were acquired at a resolution of 35,000, ACG target 1 × 105, maximum IT of 50 ms. The first fixed mass was set at 100 m/z. Parent ions were fragmented at a normalized collision energy (NCE) of 27%. Dynamic exclusion was set for 20 s. Parent ions with charges of 1 and larger than 6 were excluded. All raw data were deposited on the Mass Spectrometry Interactive Virtual Environment (MassIVE: ftp://massive.ucsd.edu/MSV000091930/). Raw files were analyzed using Proteome Discoverer 2.3 (Thermo Fisher Scientific) using the Sequest HT search engine against the UniProt Homo Sapien database (42,368 gene sequence; downloaded June 12, 2019). Mass error tolerance was set to 10 ppm for precursors, cleaved by trypsin, allowing a maximum of two missed cleavages, with sequence lengths between 6 and 144 amino acids. Fragment masses were searched with a tolerance of ±0.02 Da. Dynamic modifications included oxidation (M), deamidation (N, Q), and acetylation (N-terminus). Carbamidomethylation was set as a static modification (C). Both peptides and PSMs were set to a target false discovery rate (FDR) of ≤0.01 for matches with high confidence. After database searching, protein list was filtered so that only proteins with at least two high-confidence peptides found in 3 biological replicates remain.
Pulldown Validation Experiments
HeLa cells were plated at a density of 3.75 × 106 cells/mL in a T-225 flask in 50 mL of DMEM and grown for 3 days to confluency. The cells were resuspended by trypsinization, then pelleted and stored at −80 °C until ready for cell lysis. Prior to sonication, the pelleted cells were thawed and then washed with 1× PBS. The cells were resuspended in Pierce IP Lysis Buffer (Thermo Fisher Scientific, 87788) supplemented with a Pierce protease and phosphatase inhibitor tablet (Thermo Fisher Scientific, A32959) and lysed via sonication. Lysate was centrifuged at 18,000g for 30 min at 4 °C to remove debris and the supernatant was transferred to a clean microcentrifuge tube. The supernatant was probed for the appropriate protein using immunoblotting. Protein concentration was measured using the Pierce BCA Protein Assay Kit (Thermo Fisher Scientific, 23225). 40 μL of Pierce Streptavidin Magnetic Beads (Thermo Fisher Scientific, 88816) was washed twice with 200 μL of 1× PBS. Biotin-RH1115 (50 μM) and biotin acid (50 μM) were incubated with the beads while rotating at 4 °C for 2 h. 500 μg of lysate was then added to the magnetic beads and diluted to 200 μL using the cell lysis buffer. The lysate was left with the beads and compounds for 16 h before washing twice with 200 μL of 1× PBS then eluting with 20 μL 1× SDS of loading buffer. For competitor experiments, an additional sample containing the magnetic beads and Biotin-RH1115 (50 μM) was prepared. Upon addition of the 500 μg cell lysate, RH1115 (100 μM) was added and incubated with the other samples for 16 h at 4 °C.
Cellular Thermal Shift Assay
A549 cells were plated at a density of 2.5 × 106 cells in a 10 cm dish in DMEM and grown for 16 h. The cells were then treated with RH1115 (100 μM) or DMSO (50 μL) and incubated for 24 h. Media was removed and cells were suspended by trypsinization, pelleted washed with PBS, and resuspended to a density of 1.0 × 107 cells/mL in PBS supplemented with Pierce protease inhibitor tablet. 50 μL of each cell suspension was then dispensed into PCR tubes (Bio-Rad, TBS0201), heated at 50–62 °C in 3° increments using a Bio-Rad T100 thermal cycler for 3 min, and immediately lysed by freeze–thaw. Debris was then removed by centrifugation at 18,000g for 30 min at 4 °C, and the supernatant was probed by immunoblot for the presence of Lamin A/C and β-actin.
Microscopy of i3Neurons
Live imaging was carried out using a Zeiss LSM880 Airyscan confocal microscope with a 63x oil immersion objective (1.4 NA). Live images were acquired at speeds of 1–2 frames per second. Zeiss Zen software further processed the Airyscan images and analysis was performed using FIJI/ImageJ software.112 Standard confocal imaging for immunostaining experiments was carried out using Zeiss LSM 710 or 880 with a 63x oil immersion objective (1.4 NA).
Statistical Analysis
Statistical analysis was performed using GraphPad Prism version 9.5.0. Data presented in bar graphs were analyzed using an ordinary one-way ANOVA for 3 or more groups followed by Dunnett’s multiple comparisons testing or unpaired t-test for comparison of two groups. EC50 values were determined using a nonlinear fit function comparing log(agnostic) vs response, variable slope (four parameters). Error bars are mean ± SEM unless otherwise noted. Significance stars correspond to the following P values: ns: P > 0.05; *: P ≤ 0.05; **: P ≤ 0.01; ***: P ≤ 0.001; ****: P ≤ 0.0001.
Acknowledgments
The authors thank Dr. Ramnik Xavier, Massachusetts General Hospital, for the gift of eGFP-LC3 and mCherry-GFP-LC3 HeLa cells. They acknowledge the University of Illinois Mass Spectrometry Core for collecting HRMS data, and Dr. Thomas Whitmarsh-Everiss and Adam Ruprecht for help with synthetic chemistry.
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acschemneuro.3c00573.
Proteomics data (XLSX)
NMR spectra and uncropped western blots (Tables S1–S3) (Figures S1–S5). All raw data were deposited on the Mass Spectrometry Interactive Virtual Environment (MassIVE: ftp://massive.ucsd.edu/MSV000091930/) (PDF)
Author Contributions
§ R.S.H., A.M.S., and Z.A.P. equal contributions. R.S.H., A.M.S., and Z.A.P. designed and performed experiments, analyzed data, prepared figures, and edited drafts of the manuscript. R.S.H. performed synthesis and medicinal chemistry experiments, A.M.S. performed experiments in neurons, and Z.A.P. performed target identification and validation experiments. R.S.H. prepared the initial draft of the manuscript with help from A.M.S. and Z.A.P. M.A.K.-V. performed the high-content screen with help from M.S. S.P. and R.L. performed and analyzed several experiments in neurons. D.S. and A.D. contributed to the synthesis of hits and analogues. T.T.A.N. contributed to mass spectrometry experiments and data analysis. S.M.C. directed mass spectrometry experiments, analyzed results, and contributed to writing the manuscript. S.G. and L.N.A. directed the project, designed experiments, analyzed experimental results, and led the efforts to prepare the manuscript.
Research reported in this publication was supported by the Wolverine Foundation, The Ralph and Marian Falk Medical Trust, the UIC Center for Clinical and Translational Science (NIH UL1TR002003), and the National Institute on Aging and National Institute of Neurological Disorders and Stroke of the National Institutes of Health (RF1AG076653 (L.N.A., S.G.), R01AG074248 (S.G.), R01NS124784 (S.M.C.), R01NS114413 (L.N.A., S.M.C.), T32AG057468 (A.M.S., D.S.), and F30AG081091 (A.S.)). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
The authors declare no competing financial interest.
Supplementary Material
References
- Levine B.; Klionsky D. J. Development by Self-Digestion: Molecular Mechanisms and Biological Functions of Autophagy. Dev. Cell 2004, 6 (4), 463–477. 10.1016/S1534-5807(04)00099-1. [DOI] [PubMed] [Google Scholar]
- Menzies F. M.; Moreau K.; Rubinsztein D. C. Protein Misfolding Disorders and Macroautophagy. Curr. Opin. Cell Biol. 2011, 23 (2), 190–197. 10.1016/j.ceb.2010.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Son J. H.; Shim J. H.; Kim K.-H.; Ha J.-Y.; Han J. Y. Neuronal Autophagy and Neurodegenerative Diseases. Exp Mol. Med. 2012, 44 (2), 89–98. 10.3858/emm.2012.44.2.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cajigas I. J.; Will T.; Schuman E. M. Protein Homeostasis and Synaptic Plasticity. EMBO J. 2010, 29 (16), 2746–2752. 10.1038/emboj.2010.173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill S. E.; Colón-Ramos D. A. The Journey of the Synaptic Autophagosome: A Cell Biological Perspective. Neuron 2020, 105 (6), 961–973. 10.1016/j.neuron.2020.01.018. [DOI] [PubMed] [Google Scholar]
- Kulkarni A.; Chen J.; Maday S. Neuronal Autophagy and Intercellular Regulation of Homeostasis in the Brain. Curr. Opin. Neurobiol. 2018, 51, 29–36. 10.1016/j.conb.2018.02.008. [DOI] [PubMed] [Google Scholar]
- Andres-Alonso M.; Ammar M. R.; Butnaru I.; Gomes G. M.; Acuña Sanhueza G.; Raman R.; Yuanxiang P.; Borgmeyer M.; Lopez-Rojas J.; Raza S. A.; Brice N.; Hausrat T. J.; Macharadze T.; Diaz-Gonzalez S.; Carlton M.; Failla A. V.; Stork O.; Schweizer M.; Gundelfinger E. D.; Kneussel M.; Spilker C.; Karpova A.; Kreutz M. R. SIPA1L2 Controls Trafficking and Local Signaling of TrkB-Containing Amphisomes at Presynaptic Terminals. Nat. Commun. 2019, 10 (1), 5448 10.1038/s41467-019-13224-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu W. H.; Dorado B.; Figueroa H. Y.; Wang L.; Planel E.; Cookson M. R.; Clark L. N.; Duff K. E. Metabolic Activity Determines Efficacy of Macroautophagic Clearance of Pathological Oligomeric α-Synuclein. Am. J. Pathol. 2009, 175 (2), 736–747. 10.2353/ajpath.2009.080928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katsumata K.; Nishiyama J.; Inoue T.; Mizushima N.; Takeda J.; Yuzaki M. Dynein- and Activity-Dependent Retrograde Transport of Autophagosomes in Neuronal Axons. Autophagy 2010, 6 (3), 378–385. 10.4161/auto.6.3.11262. [DOI] [PubMed] [Google Scholar]
- Aguzzi A.; O’Connor T. Protein Aggregation Diseases: Pathogenicity and Therapeutic Perspectives. Nat. Rev. Drug Discovery 2010, 9 (3), 237–248. 10.1038/nrd3050. [DOI] [PubMed] [Google Scholar]
- Williams A.; Sarkar S.; Cuddon P.; Ttofi E. K.; Saiki S.; Siddiqi F. H.; Jahreiss L.; Fleming A.; Pask D.; Goldsmith P.; O’Kane C. J.; Floto R. A.; Rubinsztein D. C. Novel Targets for Huntington’s Disease in an mTOR-Independent Autophagy Pathway. Nat. Chem. Biol. 2008, 4 (5), 295–305. 10.1038/nchembio.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravikumar B.; Vacher C.; Berger Z.; Davies J. E.; Luo S.; Oroz L. G.; Scaravilli F.; Easton D. F.; Duden R.; O’Kane C. J.; Rubinsztein D. C. Inhibition of mTOR Induces Autophagy and Reduces Toxicity of Polyglutamine Expansions in Fly and Mouse Models of Huntington Disease. Nat. Genet. 2004, 36 (6), 585–595. 10.1038/ng1362. [DOI] [PubMed] [Google Scholar]
- Sarkar S.; Perlstein E. O.; Imarisio S.; Pineau S.; Cordenier A.; Maglathlin R. L.; Webster J. A.; Lewis T. A.; O’Kane C. J.; Schreiber S. L.; Rubinsztein D. C. Small Molecules Enhance Autophagy and Reduce Toxicity in Huntington’s Disease Models. Nat. Chem. Biol. 2007, 3 (6), 331–338. 10.1038/nchembio883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarkar S.; Ravikumar B.; Floto R. A.; Rubinsztein D. C. Rapamycin and mTOR-Independent Autophagy Inducers Ameliorate Toxicity of Polyglutamine-Expanded Huntingtin and Related Proteinopathies. Cell Death Differ. 2009, 16 (1), 46–56. 10.1038/cdd.2008.110. [DOI] [PubMed] [Google Scholar]
- Murphy K. E.; Gysbers A. M.; Abbott S. K.; Spiro A. S.; Furuta A.; Cooper A.; Garner B.; Kabuta T.; Halliday G. M. Lysosomal-Associated Membrane Protein 2 Isoforms Are Differentially Affected in Early Parkinson’s Disease. Mov. Disord. 2015, 30 (12), 1639–1647. 10.1002/mds.26141. [DOI] [PubMed] [Google Scholar]
- Dehay B.; Bové J.; Rodríguez-Muela N.; Perier C.; Recasens A.; Boya P.; Vila M. Pathogenic Lysosomal Depletion in Parkinson’s Disease. J. Neurosci. 2010, 30 (37), 12535–12544. 10.1523/JNEUROSCI.1920-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morimoto N.; Nagai M.; Ohta Y.; Miyazaki K.; Kurata T.; Morimoto M.; Murakami T.; Takehisa Y.; Ikeda Y.; Kamiya T.; Abe K. Increased Autophagy in Transgenic Mice with a G93A Mutant SOD1 Gene. Brain Res. 2007, 1167, 112–117. 10.1016/j.brainres.2007.06.045. [DOI] [PubMed] [Google Scholar]
- Zhang X.; Li L.; Chen S.; Yang D.; Wang Y.; Zhang X.; Wang Z.; Le W. Rapamycin Treatment Augments Motor Neuron Degeneration in SOD1G93A Mouse Model of Amyotrophic Lateral Sclerosis. Autophagy 2011, 7 (4), 412–425. 10.4161/auto.7.4.14541. [DOI] [PubMed] [Google Scholar]
- Sasaki S. Autophagy in Spinal Cord Motor Neurons in Sporadic Amyotrophic Lateral Sclerosis. J. Neuropathol. Exp. Neurol. 2011, 70 (5), 349–359. 10.1097/NEN.0b013e3182160690. [DOI] [PubMed] [Google Scholar]
- Piras A.; Collin L.; Grüninger F.; Graff C.; Rönnbäck A. Autophagic and Lysosomal Defects in Human Tauopathies: Analysis of Post-Mortem Brain from Patients with Familial Alzheimer Disease, Corticobasal Degeneration and Progressive Supranuclear Palsy. Acta Neuropathol. Commun. 2016, 4 (1), 22 10.1186/s40478-016-0292-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahman M. A.; Rahman M. H.; Mamun-Or-Rashid A. N. M.; Hwang H.; Chung S.; Kim B.; Rhim H. Autophagy Modulation in Aggresome Formation: Emerging Implications and Treatments of Alzheimer’s Disease. Biomedicines 2022, 10 (5), 1027. 10.3390/biomedicines10051027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pickford F.; Masliah E.; Britschgi M.; Lucin K.; Narasimhan R.; Jaeger P. A.; Small S.; Spencer B.; Rockenstein E.; Levine B.; Wyss-Coray T. The Autophagy-Related Protein Beclin 1 Shows Reduced Expression in Early Alzheimer Disease and Regulates Amyloid β Accumulation in Mice. J. Clin. Invest. 2008, 118 (6), 2190–2199. 10.1172/JCI33585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nixon R. A.; Wegiel J.; Kumar A.; Yu W. H.; Peterhoff C.; Cataldo A.; Cuervo A. M. Extensive Involvement of Autophagy in Alzheimer Disease: An Immuno-Electron Microscopy Study. J. Neuropathol. Exp. Neurol. 2005, 64 (2), 113–122. 10.1093/jnen/64.2.113. [DOI] [PubMed] [Google Scholar]
- Gowrishankar S.; Yuan P.; Wu Y.; Schrag M.; Paradise S.; Grutzendler J.; De Camilli P.; Ferguson S. M. Massive Accumulation of Luminal Protease-Deficient Axonal Lysosomes at Alzheimer’s Disease Amyloid Plaques. Proc. Natl. Acad. Sci. U.S.A. 2015, 112 (28), E3699–E3708. 10.1073/pnas.1510329112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Condello C.; Schain A.; Grutzendler J. Multicolor Time-Stamp Reveals the Dynamics and Toxicity of Amyloid Deposition. Sci. Rep. 2011, 1 (1), 19 10.1038/srep00019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boland B.; Kumar A.; Lee S.; Platt F. M.; Wegiel J.; Yu W. H.; Nixon R. A. Autophagy Induction and Autophagosome Clearance in Neurons: Relationship to Autophagic Pathology in Alzheimer’s Disease. J. Neurosci. 2008, 28 (27), 6926–6937. 10.1523/JNEUROSCI.0800-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hara T.; Nakamura K.; Matsui M.; Yamamoto A.; Nakahara Y.; Suzuki-Migishima R.; Yokoyama M.; Mishima K.; Saito I.; Okano H.; Mizushima N. Suppression of Basal Autophagy in Neural Cells Causes Neurodegenerative Disease in Mice. Nature 2006, 441 (7095), 885–889. 10.1038/nature04724. [DOI] [PubMed] [Google Scholar]
- Lucin K. M.; O’Brien C. E.; Bieri G.; Czirr E.; Mosher K. I.; Abbey R. J.; Mastroeni D. F.; Rogers J.; Spencer B.; Masliah E.; Wyss-Coray T. Microglial Beclin 1 Regulates Retromer Trafficking and Phagocytosis and Is Impaired in Alzheimer’s Disease. Neuron 2013, 79 (5), 873–886. 10.1016/j.neuron.2013.06.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maday S.; Holzbaur E. L. F. Autophagosome Biogenesis in Primary Neurons Follows an Ordered and Spatially Regulated Pathway. Dev. Cell 2014, 30 (1), 71–85. 10.1016/j.devcel.2014.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maday S.; Wallace K. E.; Holzbaur E. L. F. Autophagosomes Initiate Distally and Mature during Transport toward the Cell Soma in Primary Neurons. J. Cell Biol. 2012, 196 (4), 407–417. 10.1083/jcb.201106120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tammineni P.; Ye X.; Feng T.; Aikal D.; Cai Q. Impaired Retrograde Transport of Axonal Autophagosomes Contributes to Autophagic Stress in Alzheimer’s Disease Neurons. eLife 2017, 6, e21776 10.7554/eLife.21776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pu J.; Guardia C. M.; Keren-Kaplan T.; Bonifacino J. S. Mechanisms and Functions of Lysosome Positioning. J. Cell Sci. 2016, 129 (23), 4329–4339. 10.1242/jcs.196287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ballabio A.; Bonifacino J. S. Lysosomes as Dynamic Regulators of Cell and Organismal Homeostasis. Nat. Rev. Mol. Cell Biol. 2020, 21 (2), 101–118. 10.1038/s41580-019-0185-4. [DOI] [PubMed] [Google Scholar]
- Chow A.; Toomre D.; Garrett W.; Mellman I. Dendritic Cell Maturation Triggers Retrograde MHC Class II Transport from Lysosomes to the Plasma Membrane. Nature 2002, 418 (6901), 988–994. 10.1038/nature01006. [DOI] [PubMed] [Google Scholar]
- Vyas J. M.; Kim Y.-M.; Artavanis-Tsakonas K.; Love J. C.; Van der Veen A. G.; Ploegh H. L. Tubulation of Class II MHC Compartments Is Microtubule Dependent and Involves Multiple Endolysosomal Membrane Proteins in Primary Dendritic Cells1. J. Immunol. 2007, 178 (11), 7199–7210. 10.4049/jimmunol.178.11.7199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole S. L.; Vassar R. The Alzheimer’s Disease β-Secretase Enzyme, BACE1. Mol. Neurodegener. 2007, 2 (1), 22 10.1186/1750-1326-2-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das B.; Yan R. A Close Look at BACE1 Inhibitors for Alzheimer’s Disease Treatment. CNS Drugs 2019, 33 (3), 251–263. 10.1007/s40263-019-00613-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hur J.-Y. γ-Secretase in Alzheimer’s Disease. Exp. Mol. Med. 2022, 54 (4), 433–446. 10.1038/s12276-022-00754-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egan M. F.; Kost J.; Tariot P. N.; Aisen P. S.; Cummings J. L.; Vellas B.; Sur C.; Mukai Y.; Voss T.; Furtek C.; Mahoney E.; Harper Mozley L.; Vandenberghe R.; Mo Y.; Michelson D. Randomized Trial of Verubecestat for Mild-to-Moderate Alzheimer’s Disease. N. Engl. J. Med. 2018, 378 (18), 1691–1703. 10.1056/NEJMoa1706441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cebers G.; Alexander R. C.; Haeberlein S. B.; Han D.; Goldwater R.; Ereshefsky L.; Olsson T.; Ye N.; Rosen L.; Russell M.; Maltby J.; Eketjäll S.; Kugler A. R. AZD3293: Pharmacokinetic and Pharmacodynamic Effects in Healthy Subjects and Patients with Alzheimer’s Disease. J. Alzheimer’s Dis. 2016, 55 (3), 1039–1053. 10.3233/JAD-160701. [DOI] [PubMed] [Google Scholar]
- Lahiri D. K.; Maloney B.; Long J. M.; Greig N. H. Lessons from a BACE Inhibitor Trial: Off-Site but Not off Base. Alzheimer’s Dementia 2014, 10 (0), S411–S419. 10.1016/j.jalz.2013.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doody R. S.; Raman R.; Farlow M.; Iwatsubo T.; Vellas B.; Joffe S.; Kieburtz K.; He F.; Sun X.; Thomas R. G.; Aisen P. S.; Siemers E.; Sethuraman G.; Mohs R. A Phase 3 Trial of Semagacestat for Treatment of Alzheimer’s Disease. N. Engl. J. Med. 2013, 369 (4), 341–350. 10.1056/NEJMoa1210951. [DOI] [PubMed] [Google Scholar]
- Lanz T. A.; Karmilowicz M. J.; Wood K. M.; Pozdnyakov N.; Du P.; Piotrowski M. A.; Brown T. M.; Nolan C. E.; Richter K. E. G.; Finley J. E.; Fei Q.; Ebbinghaus C. F.; Chen Y. L.; Spracklin D. K.; Tate B.; Geoghegan K. F.; Lau L.-F.; Auperin D. D.; Schachter J. B. Concentration-Dependent Modulation of Amyloid-β in Vivo and in Vitro Using the γ-Secretase Inhibitor, LY-450139. J. Pharmacol. Exp. Ther. 2006, 319 (2), 924–933. 10.1124/jpet.106.110700. [DOI] [PubMed] [Google Scholar]
- Hebron M. L.; Lonskaya I.; Moussa C. E.-H. Nilotinib Reverses Loss of Dopamine Neurons and Improves Motor Behavior via Autophagic Degradation of α-Synuclein in Parkinson’s Disease Models. Hum. Mol. Genet. 2013, 22 (16), 3315–3328. 10.1093/hmg/ddt192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menzies F. M.; Garcia-Arencibia M.; Imarisio S.; O’Sullivan N. C.; Ricketts T.; Kent B. A.; Rao M. V.; Lam W.; Green-Thompson Z. W.; Nixon R. A.; Saksida L. M.; Bussey T. J.; O’Kane C. J.; Rubinsztein D. C. Calpain Inhibition Mediates Autophagy-Dependent Protection against Polyglutamine Toxicity. Cell Death Differ. 2015, 22 (3), 433–444. 10.1038/cdd.2014.151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose C.; Menzies F. M.; Renna M.; Acevedo-Arozena A.; Corrochano S.; Sadiq O.; Brown S. D.; Rubinsztein D. C. Rilmenidine Attenuates Toxicity of Polyglutamine Expansions in a Mouse Model of Huntington’s Disease. Hum. Mol. Genet. 2010, 19 (11), 2144–2153. 10.1093/hmg/ddq093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarkar S.; Davies J. E.; Huang Z.; Tunnacliffe A.; Rubinsztein D. C. Trehalose, a Novel mTOR-Independent Autophagy Enhancer, Accelerates the Clearance of Mutant Huntingtin and α-Synuclein*. J. Biol. Chem. 2007, 282 (8), 5641–5652. 10.1074/jbc.M609532200. [DOI] [PubMed] [Google Scholar]
- Rodríguez-Navarro J. A.; Rodríguez L.; Casarejos M. J.; Solano R. M.; Gómez A.; Perucho J.; Cuervo A. M.; García de Yébenes J.; Mena M. A. Trehalose Ameliorates Dopaminergic and Tau Pathology in Parkin Deleted/Tau Overexpressing Mice through Autophagy Activation. Neurobiol. Dis. 2010, 39 (3), 423–438. 10.1016/j.nbd.2010.05.014. [DOI] [PubMed] [Google Scholar]
- Castillo K.; Nassif M.; Valenzuela V.; Rojas F.; Matus S.; Mercado G.; Court F. A.; van Zundert B.; Hetz C. Trehalose Delays the Progression of Amyotrophic Lateral Sclerosis by Enhancing Autophagy in Motoneurons. Autophagy 2013, 9 (9), 1308–1320. 10.4161/auto.25188. [DOI] [PubMed] [Google Scholar]
- Zhang X.; Chen S.; Song L.; Tang Y.; Shen Y.; Jia L.; Le W. MTOR-Independent, Autophagic Enhancer Trehalose Prolongs Motor Neuron Survival and Ameliorates the Autophagic Flux Defect in a Mouse Model of Amyotrophic Lateral Sclerosis. Autophagy 2014, 10 (4), 588–602. 10.4161/auto.27710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caccamo A.; Majumder S.; Richardson A.; Strong R.; Oddo S. Molecular Interplay between Mammalian Target of Rapamycin (mTOR), Amyloid-β, and Tau: EFFECTS ON COGNITIVE IMPAIRMENTS*. J. Biol. Chem. 2010, 285 (17), 13107–13120. 10.1074/jbc.M110.100420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X.; Gao F.; Lin C.; Chen A.; Deng J.; Chen P.; Lin M.; Xie B.; Liao Y.; Gong C.; Zheng X. mTOR-Mediated Autophagy in the Hippocampus Is Involved in Perioperative Neurocognitive Disorders in Diabetic Rats. CNS Neurosci. Ther. 2022, 28 (4), 540–553. 10.1111/cns.13762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirchenwitz M.; Stahnke S.; Grunau K.; Melcher L.; van Ham M.; Rottner K.; Steffen A.; Stradal T. E. B. The Autophagy Inducer SMER28 Attenuates Microtubule Dynamics Mediating Neuroprotection. Sci. Rep. 2022, 12 (1), 17805 10.1038/s41598-022-20563-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lonskaya I.; Hebron M. L.; Desforges N. M.; Schachter J. B.; Moussa C. E.-H. Nilotinib-Induced Autophagic Changes Increase Endogenous Parkin Level and Ubiquitination, Leading to Amyloid Clearance. J. Mol. Med. 2014, 92 (4), 373–386. 10.1007/s00109-013-1112-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Z.; Yan J.; Jiang W.; Yao X.; Chen J.; Chen L.; Li C.; Hu L.; Jiang H.; Shen X. Arctigenin Effectively Ameliorates Memory Impairment in Alzheimer’s Disease Model Mice Targeting Both β-Amyloid Production and Clearance. J. Neurosci. 2013, 33 (32), 13138–13149. 10.1523/JNEUROSCI.4790-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarkar S. Chemical Screening Platforms for Autophagy Drug Discovery to Identify Therapeutic Candidates for Huntington’s Disease and Other Neurodegenerative Disorders. Drug Discovery Today: Technol. 2013, 10 (1), e137–e144. 10.1016/j.ddtec.2012.09.010. [DOI] [PubMed] [Google Scholar]
- Lesire L.; Chaput L.; Cruz De Casas P.; Rousseau F.; Piveteau C.; Dumont J.; Pointu D.; Déprez B.; Leroux F. High-Throughput Image-Based Aggresome Quantification. SLAS Discovery 2020, 25 (7), 783–791. 10.1177/2472555220919708. [DOI] [PubMed] [Google Scholar]
- Chu C.; Zhang X.; Ma W.; Li L.; Wang W.; Shang L.; Fu P. Induction of Autophagy by a Novel Small Molecule Improves Aβ Pathology and Ameliorates Cognitive Deficits. PLoS One 2013, 8 (6), e65367 10.1371/journal.pone.0065367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klionsky D. J.; Abdelmohsen K.; Abe A.; et al. Guidelines for the Use and Interpretation of Assays for Monitoring Autophagy (3rd Edition). Autophagy 2016, 12 (1), 1–222. 10.1080/15548627.2015.1100356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lystad A. H.; Simonsen A. Mechanisms and Pathophysiological Roles of the ATG8 Conjugation Machinery. Cells 2019, 8 (9), 973. 10.3390/cells8090973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabeya Y.; Mizushima N.; Ueno T.; Yamamoto A.; Kirisako T.; Noda T.; Kominami E.; Ohsumi Y.; Yoshimori T. LC3, a Mammalian Homologue of Yeast Apg8p, Is Localized in Autophagosome Membranes after Processing. EMBO J. 2000, 19 (21), 5720–5728. 10.1093/emboj/19.21.5720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Streeter A.; Menzies F. M.; Rubinsztein D. C.. LC3-II Tagging and Western Blotting for Monitoring Autophagic Activity in Mammalian Cells. In Systems Biology of Alzheimer’s Disease; Castrillo J. I.; Oliver S. G., Eds.; Methods in Molecular Biology; Springer: New York, NY, 2016; pp 161–170. [DOI] [PubMed] [Google Scholar]
- Mizushima N.; Yoshimori T.; Levine B. Methods in Mammalian Autophagy Research. Cell 2010, 140 (3), 313–326. 10.1016/j.cell.2010.01.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizushima N.; Murphy L. O. Autophagy Assays for Biological Discovery and Therapeutic Development. Trends Biochem. Sci. 2020, 45 (12), 1080–1093. 10.1016/j.tibs.2020.07.006. [DOI] [PubMed] [Google Scholar]
- Liebl M. P.; Meister S. C.; Frey L.; Hendrich K.; Klemmer A.; Wohlfart B.; Untucht C.; Nuber J.; Pohl C.; Lakics V. Robust LC3B Lipidation Analysis by Precisely Adjusting Autophagic Flux. Sci. Rep. 2022, 12 (1), 79 10.1038/s41598-021-03875-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartmann B. p70S6 Kinase Phosphorylation for Pharmacodynamic Monitoring. Clin. Chim. Acta 2012, 413 (17), 1387–1390. 10.1016/j.cca.2012.03.023. [DOI] [PubMed] [Google Scholar]
- Yang H. Y.; Tae J.; Seo Y. W.; Kim Y. J.; Im H. Y.; Choi G. D.; Cho H.; Park W.-K.; Kwon O. S.; Cho Y. S.; Ko M.; Jang H.; Lee J.; Choi K.; Kim C.-H.; Lee J.; Pae A. N. Novel Pyrimidoazepine Analogs as Serotonin 5-HT2A and 5-HT2C Receptor Ligands for the Treatment of Obesity. Eur. J. Med. Chem. 2013, 63, 558–569. 10.1016/j.ejmech.2013.02.020. [DOI] [PubMed] [Google Scholar]
- Kantorowski E. J.; Kurth M. J. Expansion to Seven-Membered Rings. Tetrahedron 2000, 56 (26), 4317–4353. 10.1016/S0040-4020(00)00218-0. [DOI] [Google Scholar]
- Abdel-Magid A. F.; Carson K. G.; Harris B. D.; Maryanoff C. A.; Shah R. D. Reductive Amination of Aldehydes and Ketones with Sodium Triacetoxyborohydride. Studies on Direct and Indirect Reductive Amination Procedures1. J. Org. Chem. 1996, 61 (11), 3849–3862. 10.1021/jo960057x. [DOI] [PubMed] [Google Scholar]
- Chalyk B. A.; Butko M. V.; Yanshyna O. O.; Gavrilenko K. S.; Druzhenko T. V.; Mykhailiuk P. K. Synthesis of Spirocyclic Pyrrolidines: Advanced Building Blocks for Drug Discovery. Chem. - Eur. J. 2017, 23 (66), 16782–16786. 10.1002/chem.201702362. [DOI] [PubMed] [Google Scholar]
- Chung T. D. Y.; Terry D. B.; Smith L. H.. In Vitro and In Vivo Assessment of ADME and PK Properties During Lead Selection and Lead Optimization – Guidelines, Benchmarks and Rules of Thumb. In Assay Guidance Manual; Markossian S.; Grossman A.; Brimacombe K.; Arkin M.; Auld D.; Austin C.; Baell J.; Chung T. D. Y.; Coussens N. P.; Dahlin J. L.; Devanarayan V.; Foley T. L.; Glicksman M.; Haas J. V.; Hall M. D.; Hoare S.; Inglese J.; Iversen P. W.; Kales S. C.; Lal-Nag M.; Li Z.; McGee J.; McManus O.; Riss T.; Saradjian P.; Sittampalam G. S.; Tarselli M.; Trask O. J.; Wang Y.; Weidner J. R.; Wildey M. J.; Wilson K.; Xia M.; Xu X., Eds.; Eli Lilly & Company and the National Center for Advancing Translational Sciences: Bethesda (MD), 2004. [PubMed] [Google Scholar]
- Spiteri C.; Moses J. E. Copper-Catalyzed Azide–Alkyne Cycloaddition: Regioselective Synthesis of 1,4,5-Trisubstituted 1,2,3-Triazoles. Angew. Chem., Int. Ed. 2010, 49 (1), 31–33. 10.1002/anie.200905322. [DOI] [PubMed] [Google Scholar]
- Xu T.-H.; Yan Y.; Harikumar K. G.; Miller L. J.; Melcher K.; Xu E. H. Streptavidin Bead Pulldown Assay to Determine Protein Homooligomerization. Bio-protocol 2017, 7 (22), e2901 10.21769/BioProtoc.2901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gowrishankar S.; Lyons L.; Rafiq N. M.; Roczniak-Ferguson A.; De Camilli P.; Ferguson S. M. Overlapping Roles of JIP3 and JIP4 in Promoting Axonal Transport of Lysosomes in Human iPSC-Derived Neurons. MBoC 2021, 32 (11), 1094–1103. 10.1091/mbc.E20-06-0382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandopulle M. S.; Prestil R.; Grunseich C.; Wang C.; Gan L.; Ward M. E. Transcription Factor-Mediated Differentiation of Human iPSCs into Neurons. Curr. Protoc. Cell Biol. 2018, 79 (1), e51 10.1002/cpcb.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C.; Ward M. E.; Chen R.; Liu K.; Tracy T. E.; Chen X.; Xie M.; Sohn P. D.; Ludwig C.; Meyer-Franke A.; Karch C. M.; Ding S.; Gan L. Scalable Production of iPSC-Derived Human Neurons to Identify Tau-Lowering Compounds by High-Content Screening. Stem Cell Rep. 2017, 9 (4), 1221–1233. 10.1016/j.stemcr.2017.08.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snead A. M.; Gowrishankar S. Loss of MAPK8IP3 Affects Endocytosis in Neurons. Front. Cell. Neurosci. 2022, 16, 828071 10.3389/fncel.2022.828071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng X.-T.; Xie Y.-X.; Zhou B.; Huang N.; Farfel-Becker T.; Sheng Z.-H. Characterization of LAMP1-Labeled Nondegradative Lysosomal and Endocytic Compartments in Neurons. J. Cell Biol. 2018, 217 (9), 3127–3139. 10.1083/jcb.201711083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mangialasche F.; Solomon A.; Winblad B.; Mecocci P.; Kivipelto M. Alzheimer’s Disease: Clinical Trials and Drug Development. Lancet Neurol. 2010, 9 (7), 702–716. 10.1016/S1474-4422(10)70119-8. [DOI] [PubMed] [Google Scholar]
- Cummings J.; Lee G.; Nahed P.; Kambar M. E. Z. N.; Zhong K.; Fonseca J.; Taghva K. Alzheimer’s Disease Drug Development Pipeline: 2022. Alzheimer’s Dementia 2022, 8 (1), e12295 10.1002/trc2.12295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blay V.; Tolani B.; Ho S. P.; Arkin M. R. High-Throughput Screening: Today’s Biochemical and Cell-Based Approaches. Drug Discovery Today 2020, 25 (10), 1807–1821. 10.1016/j.drudis.2020.07.024. [DOI] [PubMed] [Google Scholar]
- Swinney D. C.; Lee J. A.. Recent Advances in Phenotypic Drug Discovery. F1000Research August 7, 2020. 10.12688/f1000research.25813.1. [DOI] [PMC free article] [PubMed]
- Moffat J. G.; Vincent F.; Lee J. A.; Eder J.; Prunotto M. Opportunities and Challenges in Phenotypic Drug Discovery: An Industry Perspective. Nat. Rev. Drug Discovery 2017, 16 (8), 531–543. 10.1038/nrd.2017.111. [DOI] [PubMed] [Google Scholar]
- Guo F.; Liu X.; Cai H.; Le W. Autophagy in Neurodegenerative Diseases: Pathogenesis and Therapy. Brain Pathol. 2018, 28 (1), 3–13. 10.1111/bpa.12545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo Y.-B.; Mastaglia F. L.; Wilton S. D. Normal and Aberrant Splicing of LMNA. J. Med. Genet. 2014, 51 (4), 215–223. 10.1136/jmedgenet-2013-102119. [DOI] [PubMed] [Google Scholar]
- Dittmer T. A.; Misteli T. The Lamin Protein Family. Genome Biol. 2011, 12 (5), 222 10.1186/gb-2011-12-5-222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casasola A.; Scalzo D.; Nandakumar V.; Halow J.; Recillas-Targa F.; Groudine M.; Rincón-Arano H. Prelamin A Processing, Accumulation and Distribution in Normal Cells and Laminopathy Disorders. Nucleus 2016, 7 (1), 84–102. 10.1080/19491034.2016.1150397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burla R.; La Torre M.; Saggio I. Mammalian Telomeres and Their Partnership with Lamins. Nucleus 2016, 7 (2), 187–202. 10.1080/19491034.2016.1179409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rankin J.; Ellard S. The Laminopathies: A Clinical Review. Clin. Genet. 2006, 70 (4), 261–274. 10.1111/j.1399-0004.2006.00677.x. [DOI] [PubMed] [Google Scholar]
- Frost B.; Bardai F. H.; Feany M. B. Lamin Dysfunction Mediates Neurodegeneration in Tauopathies. Curr. Biol. 2016, 26 (1), 129–136. 10.1016/j.cub.2015.11.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon D. N.; Wilson K. L. The Nucleoskeleton as a Genome-Associated Dynamic “Network of Networks. Nat. Rev. Mol. Cell Biol. 2011, 12 (11), 695–708. 10.1038/nrm3207. [DOI] [PubMed] [Google Scholar]
- Lammerding J.; Fong L. G.; Ji J. Y.; Reue K.; Stewart C. L.; Young S. G.; Lee R. T. Lamins A and C but Not Lamin B1 Regulate Nuclear Mechanics*. J. Biol. Chem. 2006, 281 (35), 25768–25780. 10.1074/jbc.M513511200. [DOI] [PubMed] [Google Scholar]
- Jung H.-J.; Coffinier C.; Choe Y.; Beigneux A. P.; Davies B. S. J.; Yang S. H.; Barnes R. H.; Hong J.; Sun T.; Pleasure S. J.; Young S. G.; Fong L. G. Regulation of Prelamin A but Not Lamin C by miR-9, a Brain-Specific microRNA. Proc. Natl. Acad. Sci. U.S.A. 2012, 109 (7), E423–E431. 10.1073/pnas.1111780109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Méndez-López I.; Blanco-Luquin I.; Sánchez-Ruiz de Gordoa J.; Urdánoz-Casado A.; Roldán M.; Acha B.; Echavarri C.; Zelaya V.; Jericó I.; Mendioroz M. Hippocampal LMNA Gene Expression Is Increased in Late-Stage Alzheimer’s Disease. Int. J. Mol. Sci. 2019, 20 (4), 878 10.3390/ijms20040878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gil L.; Niño S. A.; Chi-Ahumada E.; Rodríguez-Leyva I.; Guerrero C.; Rebolledo A. B.; Arias J. A.; Jiménez-Capdeville M. E. Perinuclear Lamin A and Nucleoplasmic Lamin B2 Characterize Two Types of Hippocampal Neurons through Alzheimer’s Disease Progression. Int. J. Mol. Sci. 2020, 21 (5), 1841 10.3390/ijms21051841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi J. C.; Muchir A.; Wu W.; Iwata S.; Homma S.; Morrow J. P.; Worman H. J. Temsirolimus Activates Autophagy and Ameliorates Cardiomyopathy Caused by Lamin A/C Gene Mutation. Sci. Transl. Med. 2012, 4 (144), 144ra102 10.1126/scitranslmed.3003875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cenni V.; Capanni C.; Columbaro M.; Ortolani M.; D’Apice M. R.; Novelli G.; Fini M.; Marmiroli S.; Scarano E.; Maraldi N. M.; Squarzoni S.; Prencipe S.; Lattanzi G. Autophagic Degradation of Farnesylated Prelamin A as a Therapeutic Approach to Lamin-Linked Progeria. Eur. J. Histochem. 2011, 55 (4), e36 10.4081/ejh.2011.e36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akinci B.; Sankella S.; Gilpin C.; Ozono K.; Garg A.; Agarwal A. K. Progeroid Syndrome Patients with ZMPSTE24 Deficiency Could Benefit When Treated with Rapamycin and Dimethylsulfoxide. Cold Spring Harbor Mol. Case Stud. 2017, 3 (1), a001339 10.1101/mcs.a001339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eskelinen E.-L. Roles of LAMP-1 and LAMP-2 in Lysosome Biogenesis and Autophagy. Mol. Aspects Med. 2006, 27 (5), 495–502. 10.1016/j.mam.2006.08.005. [DOI] [PubMed] [Google Scholar]
- Orr M. E.; Oddo S. Autophagic/Lysosomal Dysfunction in Alzheimer’s Disease. Alzheimer’s Res. Ther. 2013, 5 (5), 53. 10.1186/alzrt217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monaco A.; Fraldi A. Protein Aggregation and Dysfunction of Autophagy-Lysosomal Pathway: A Vicious Cycle in Lysosomal Storage Diseases. Front. Mol. Neurosci. 2020, 13, 37 10.3389/fnmol.2020.00037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korolchuk V. I.; Saiki S.; Lichtenberg M.; Siddiqi F. H.; Roberts E. A.; Imarisio S.; Jahreiss L.; Sarkar S.; Futter M.; Menzies F. M.; O’Kane C. J.; Deretic V.; Rubinsztein D. C. Lysosomal Positioning Coordinates Cellular Nutrient Responses. Nat. Cell Biol. 2011, 13 (4), 453–460. 10.1038/ncb2204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ba Q.; Raghavan G.; Kiselyov K.; Yang G. Whole-Cell Scale Dynamic Organization of Lysosomes Revealed by Spatial Statistical Analysis. Cell Rep. 2018, 23 (12), 3591–3606. 10.1016/j.celrep.2018.05.079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armstrong A.; Mattsson N.; Appelqvist H.; Janefjord C.; Sandin L.; Agholme L.; Olsson B.; Svensson S.; Blennow K.; Zetterberg H.; Kågedal K. Lysosomal Network Proteins as Potential Novel CSF Biomarkers for Alzheimer’s Disease. Neuromol. Med. 2014, 16 (1), 150–160. 10.1007/s12017-013-8269-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pi H.; Li M.; Tian L.; Yang Z.; Yu Z.; Zhou Z. Enhancing Lysosomal Biogenesis and Autophagic Flux by Activating the Transcription Factor EB Protects against Cadmium-Induced Neurotoxicity. Sci. Rep. 2017, 7 (1), 43466 10.1038/srep43466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwake M.; Schröder B.; Saftig P. Lysosomal Membrane Proteins and Their Central Role in Physiology. Traffic 2013, 14 (7), 739–748. 10.1111/tra.12056. [DOI] [PubMed] [Google Scholar]
- Frenkel-Pinter M.; Shmueli M. D.; Raz C.; Yanku M.; Zilberzwige S.; Gazit E.; Segal D. Interplay between Protein Glycosylation Pathways in Alzheimer’s Disease. Sci. Adv. 2017, 3 (9), e1601576 10.1126/sciadv.1601576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gizaw S. T.; Ohashi T.; Tanaka M.; Hinou H.; Nishimura S.-I. Glycoblotting Method Allows for Rapid and Efficient Glycome Profiling of Human Alzheimer’s Disease Brain, Serum and Cerebrospinal Fluid towards Potential Biomarker Discovery. Biochim. Biophys. Acta, Gen. Subj. 2016, 1860 (8), 1716–1727. 10.1016/j.bbagen.2016.03.009. [DOI] [PubMed] [Google Scholar]
- Cawley N. X.; Sojka C.; Cougnoux A.; Lyons A. T.; Nicoli E.; Wassif C. A.; Porter F. D. Abnormal LAMP1 Glycosylation May Play a Role in Niemann-Pick Disease, Type C Pathology. PLoS One 2020, 15 (1), e0227829 10.1371/journal.pone.0227829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu R.; Yanjanin N. M.; Elrick M. J.; Ware C.; Lieberman A. P.; Porter F. D. Apolipoprotein E Genotype and Neurological Disease Onset in Niemann–Pick Disease, Type C1. Am. J. Med. Genet., Part A 2012, 158A (11), 2775–2780. 10.1002/ajmg.a.35395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sienski G.; Narayan P.; Bonner J. M.; Kory N.; Boland S.; Arczewska A. A.; Ralvenius W. T.; Akay L.; Lockshin E.; He L.; Milo B.; Graziosi A.; Baru V.; Lewis C. A.; Kellis M.; Sabatini D. M.; Tsai L.-H.; Lindquist S. APOE4 Disrupts Intracellular Lipid Homeostasis in Human iPSC-Derived Glia. Sci. Transl. Med. 2021, 13 (583), eaaz4564 10.1126/scitranslmed.aaz4564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindelin J.; Arganda-Carreras I.; Frise E.; Kaynig V.; Longair M.; Pietzsch T.; Preibisch S.; Rueden C.; Saalfeld S.; Schmid B.; Tinevez J.-Y.; White D. J.; Hartenstein V.; Eliceiri K.; Tomancak P.; Cardona A. Fiji: An Open-Source Platform for Biological-Image Analysis. Nat. Methods 2012, 9 (7), 676–682. 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.