Abstract
The secreted lipid transporter apolipoprotein E (APOE) plays important roles in atherosclerosis and Alzheimer’s disease and has been implicated as a suppressor of melanoma progression. The APOE germline genotype predicts human melanoma outcomes, with APOE4 and APOE2 allele carriers exhibiting prolonged and reduced survival, respectively, relative to APOE3 homozygotes. While the APOE4 variant was recently shown to suppress melanoma progression by enhancing antitumor immunity, further work is needed to fully characterize the melanoma cell-intrinsic effects of APOE variants on cancer progression. Using a genetically engineered mouse model, we showed that human germline APOE genetic variants differentially modulate melanoma growth and metastasis in an APOE2>APOE3>APOE4 manner. The low-density lipoprotein receptor-related protein 1 (LRP1) receptor mediated the cell-intrinsic effects of APOE variants on melanoma progression. Protein synthesis was a tumor cell-intrinsic process differentially modulated by APOE variants, with APOE2 promoting translation via LRP1. These findings reveal a gain-of-function role for the APOE2 variant in melanoma progression, which may aid in predicting melanoma patient outcomes and understanding the protective effect of APOE2 in Alzheimer’s disease.
Keywords: apolipoprotein E, melanoma, translation, LRP1, metastasis, genetic variants, genetically engineered mouse model
Introduction
Germline genetic variants are well-established regulators of cancer development. They underlie hereditary cancer predisposition syndromes, which account for 5–10% of all malignancies (1), and an estimated 33% of all cancer is thought to be heritable (2). In contrast, the impact of germline genetics on the progression of cancers once they are established is poorly understood. We previously identified genetic variation in the secreted glycoprotein apolipoprotein E (APOE) as a significant modulator of survival after melanoma development (3). Humans have three prevalent APOE alleles, termed APOE2, APOE3, and APOE4, that differ by variation at two amino acid residues. In contrast to Alzheimer’s disease, where APOE4 is the single greatest monogenetic risk factor for disease onset and APOE2 is protective, melanoma patients who carry an APOE4 allele exhibit improved survival whereas APOE2 carriers experience poorer outcomes in multiple large cohorts. The variant-dependent effects of APOE on melanoma progression are partly governed by differences in anti-tumor immunity, as stromal expression of APOE4 confers increased immune effector responses relative to APOE2. Moreover, APOE4 has been shown to suppress tumor angiogenesis and endothelial recruitment by cancer cells (3,4). In addition to tumor cell-extrinsic effects on the immune and vascular compartments, prior work also identified a cell-intrinsic effect of APOE in suppressing of melanoma cell invasiveness (4). This cell-intrinsic effect was also variant dependent, with APOE4 most potently suppressing invasion relative to APOE2 (3). Overall, the effects of APOE4 including enhanced anti-tumor immunity, reduced angiogenesis, and reduced invasiveness collectively contribute to its melanoma-suppressive effects relative to APOE2. These past studies revealed an APOE4>APOE3>APOE2 pattern of melanoma tumor suppression. While this past work led to the identification of multiple cellular and organismal cancer progression phenotypes that are differentially associated with APOE variants, the impact of APOE variants on intracellular processes that could contribute to cancer growth and metastasis outcomes has remained uncharacterized.
Previous studies investigating the role of APOE in melanoma progression have primarily utilized transplantable models, in which established cancer cells are injected directly into mice (3–5). Herein, we crossed the BrafV600E/Pten−/− conditional melanoma model (6) with mice in which the murine Apoe locus had been replaced with one of the three human APOE genes—thus generating an allelic series of genetically engineered mouse models (GEMMs) of melanoma harboring human APOE2, APOE3, or APOE4. These allelic strains vary from one another by just one or two amino acids. In contrast to transplantable models, GEMMs recapitulate all steps of the cancer progression cascade from tumor initiation through to metastatic colonization, and the genetics of the tumor match that of the host. We therefore reasoned that cancer cell exposure to allele-concordant host and tumoral APOE for the entire metastatic trajectory, as it occurs in patients, may enhance phenotypic expression of the variants’ cell-intrinsic effects. We found that genetically initiated melanoma growth and metastasis were regulated by APOE genotype in an APOE2>E3>E4 manner. Additionally, we identified mRNA translation as a melanoma cell-intrinsic process that is modulated by APOE, with APOE2 acting as a promoter of protein synthesis. These effects were dependent on melanoma cell expression of low-density lipoprotein receptor-related protein 1 (LRP1), a receptor for APOE. In the Alzheimer’s field, it has long been debated whether APOE2 is solely a loss-of-function allele with respect to its effects on neurodegeneration or whether it harbors gain-of-function properties (7). Our findings reveal clear gain-of-function effects for APOE2 in both protein translation and the promotion of melanoma progression. Through systematic interrogation of the effects of germline genetic variation of the APOE gene in mouse and patient tumors, our work uncovers a novel role for APOE in the regulation of protein synthesis via the LRP1 receptor and provides definitive evidence for hereditary regulation of cancer progression and metastasis by common human germline genetic variants.
Materials and Methods
Mice
Humanized APOE2 (#1547, C57BL/6NTac), APOE3 (#1548, C57BL/6), and APOE4 (#1549, C57BL/6NTac) knock-in mice were obtained from Taconic Biosciences. Braf V600E/+;Pten−/−;Tyr::CreER (BPC) mice (RRID:IMSR_JAX:013590, C57BL/6J) were obtained from The Jackson Laboratory. Lrp1flox/flox mice (C57BL/6J) were generously provided by David Hui (8). BPC mice were crossed with APOE knock-in mice to generate BPC/APOE2, BPC/APOE3, and BPC/APOE4 mice. BPC/APOE2 and BPC/APOE4 mice were crossed with Lrp1flox/flox mice to generate BPC/APOE2/LRP1Δ and BPC/APOE4/LRP1Δ mice. Crosses were maintained on a C57BL/6J background.
Mouse genotyping
Genotyping of Braf V600E/+;Pten−/−;Tyr::CreER mice was performed as instructed by The Jackson Laboratory. Genotyping for Lrp1flox/flox and discernment between mouse (200 bp) and human (~600 bp) APOE was performed using standard PCR protocols. To distinguish between human APOE alleles, restriction fragment length polymorphism (RFLP) genotyping was performed (9). Briefly, a 244 bp portion of APOE was amplified using standard PCR protocols and digested simultaneously with AflIII (R0541) and HaeII (R0107) restriction enzymes (New England Biolabs) for at least two hours at 37°C. Allele-specific banding was visualized on a 4% agarose gel. The following PCR primers were utilized:
Braf V600E/+;Pten−/−;Tyr::CreER genetic model
Cre transgene forward: 5’ – GCG GTC TGG CAG TAA AAA CTA TC – 3’
Cre transgene reverse: 5’ – GTG AAA CAG CAT TGC TGT CAC TT – 3’
Cre internal control forward: 5’ – CAC GTG GGC TCC AGC ATT – 3’
Cre internal control reverse: 5’ – TCA CCA GTC ATT TCT GCC TTT G – 3’
Braf forward: 5’ – TGA GTA TTT TTG TGG CAA CTG C – 3’
Braf reverse: 5’ – CTC TGC TGG GAA AGC GGC – 3’
Pten forward: 5’ – CAA GCA CTC TGC GAA CTG AG – 3’
Pten reverse: 5’ — AAG TTT TTG AAG GCA AGA TGC — 3’
Mouse versus human APOE
Common forward: 5’ – TAC CGG CTC AAC TAG GAA CCA T – 3’
Mouse Apoe reverse: 5’ – TTT AAT CGT CCT CCA TCC CTG C – 3’
Human APOE reverse: 5’ – GTT CCA TCT CAG TCC CAG TCTC – 3’
Human APOE allele RFLP
Human APOE forward: 5’ – ACA GAA TTC GCC CCG GCC TGG TAC AC – 3’
Human APOE reverse: 5’ – TAA GCT TGG CAC GGC TGT CCA AGG A – 3’
Lrp1flox/flox
Lrp1 forward: 5’ – CAT ACC CTC TTC AAA CCC CTT CCT G – 3’
Lrp1 reverse: 5’ – GCA AGC TCT CCT GCT CAG ACC TGG A – 3’
Cell lines
HEK293T cells were obtained from the American Tissue Type Collection (RRID:CVCL_0045). The B16F10 cell line transduced with a retroviral construct to express luciferase and GFP (10) and a short hairpin RNA (shRNA) targeting murine Apoe (Millipore Sigma, TRCN0000011799; B16F10-TR-shApoe) was previously described (3,5). B16F10-TR-shApoe and HEK293T cells were cultured in DMEM (Gibco, 11995) supplemented with 10% fetal bovine serum (FBS) (D10F). All cells were maintained in an incubator at 37°C and 5% CO2 and regularly tested for Mycoplasma contamination with the Universal Mycoplasma Detection Kit (ATCC, 30–1012K). Cells were discarded after the 15th passage.
Generation of stable cell lines
APOE coding sequences from pCMV4-APOE2 (RRID:Addgene_87085), pCMV4-APOE3 (RRID:Addgene_87086), and pCMV4-APOE4 (RRID:Addgene_87087) plasmids were subcloned into the pBabe-hygro vector (RRID:Addgene_1765). Retrovirus was produced in HEK293T cells grown in 10 cm plates. Cells were transfected with retroviral Gag-pol (8 ug) and VSV-G (4 ug) packaging plasmids and pBabe vector (8 ug) using PEI Max transfection reagent (VWR, 75800–188). After 24 h, the medium was replaced with fresh DF10, and virus-containing supernatant was collected 48 and 72 h after transfection. The supernatant was filtered through a 0.45 μm filter, and viral supernatant was used with 8 μg/ml polybrene (Sigma, TR-1003-G) to transduce pre-plated B16F10-TR-shApoe cells for 8 hours. Following a second round of transduction, antibiotic selection was performed using 600 μg/ml Hygromycin B (Invitrogen, 10687010). Protein overexpression was validated by western blot.
Proliferation assay
2.5 × 104 B16F10-TR-shApoe cells stably expressing APOE2, APOE3, APOE4, or empty vector were plated in triplicate in a 12 well plate in DF10. Confluence was measured with an Incucyte S3 Live-Cell Analysis System (RRID:SCR_023147) with phase contrast at 10x magnification. Images were taken at 4-hour intervals for 72 hours.
Generation of CRISPR cell lines
Single guide RNA sequences targeting murine Lrp1 were obtained from the GeCKO v2 library (11) and cloned into the pSpCas(BB)-2A-Puro (PX459) V2.0 vector (RRID:Addgene_62988). B16F10-TR-shApoe cells were plated into a 6-well dish the day prior to transfection and transfected with 4 μg plasmid and TurboFect transfection reagent (Thermo Scientific, R0533) diluted in serum-free media, according to the manufacturer’s instructions. 24 hours after transfection, puromycin selection was initiated at 2 μg/mL concentration (Thermo Scientific, A1113803). Single clones were obtained by limiting dilution followed by Sanger sequencing of individual clones to confirm the presence of indels. Knockout clones were pooled to reconstitute heterogeneity, and LRP1 knockout was confirmed via western blot.
Guide RNA sequences:
sgCtrl: 5’ – GCGAGGTATTCGGCTCCGCG – 3’
sgLrp1-1: 5’ – CCCGTTGCAGAGACGAGACA – 3’
sgLrp1-2: 5’ – TTTGACGAGTGTTCCGTGTA – 3’
Tail vein metastasis assay
6–8-week-old male APOE2 and APOE4 knock-in mice were injected via lateral tail vein with 100 μl of PBS containing 1 × 105 B16F10-TR-shApoe cells. D-luciferin (GoldBio, 115144–35-9) was injected retro-orbitally, and bioluminescence was measured with an IVIS Lumina II (Caliper Life Sciences). Bioluminescence imaging was performed weekly, and signal was normalized to the signal obtained on day 0.
Histology
Mice were perfused via intracardiac injection with PBS followed by 4% paraformaldehyde (PFA). The lungs were resected, incubated in 4% PFA at 4°C overnight, and dehydrated in 70% ethanol at 4°C. Lungs were then embedded in paraffin, cut into 5 μm sections, and stained with hematoxylin and eosin (Histoserv, Inc.) or S100 (Brigham and Women’s Hospital Specialized Histopathology Service). Slides were digitally scanned with a PathScan Enabler (Meyer Instruments).
Genetic tumor initiation
Topical induction
10 mg/ml of 4-hydroxytamoxifen (4-OHT; Sigma, H6278) was dissolved in acetone with gentle heating. 6–8-week-old female mice were shaved on the back, and 5 μl of 4-OHT was applied to back skin and allowed to air dry. Mice were observed twice weekly for tumor formation, defined as a raised, pigmented lesion at the site of tamoxifen application. Tumor volume was measured as described previously (12) by assessing length, width and height using a digital caliper (V=l x w x h), as tumors tended to grow cuboidal rather than spherical. For survival analyses, mice were euthanized according to the humane endpoints outlined in the IACUC protocol.
Perinatal induction
Two-day-old female neonates were tail snipped as described previously (13) and genotyped. 10 μl of 4-OHT diluted in DMSO (50 mg/mL) was applied with a small paintbrush to the back skin of neonates on postnatal days 3, 5, and 7. Mice were euthanized on postnatal day 35 or when moribund, whichever occurred earlier.
Immunofluorescence of BPC tumor sections
Fresh tumors were excised, embedded in optimal cutting temperature (OCT) compound (Sakura Finetek, 4583), flash frozen in liquid nitrogen, and stored at −80°C. 20 μM tumor sections were obtained with a cryostat. Sections were fixed at −20°C with acetone/methanol and permeabilized with 0.1% Triton-X for 10 minutes at room temperature (RT). Blocking was performed for 30 minutes at RT with 5% goat serum in PBS with 0.1% Tween 20 (PBST). Sections were incubated at 4°C overnight with primary antibody diluted in blocking solution: LRP1 (1:100; abcam 92544, RRID:AB_2234877), CD8 (1:1000, abcam 217344, RRID:AB_2890649). Slides were washed with PBS and then incubated with Alexa Fluor 488 (Invitrogen A11008, RRID:AB_143165) or 555 (Invitrogen A21428, RRID:AB_141784) anti-rabbit secondary antibody diluted in PBST for 45 minutes (1:200). Slides were washed again with PBS, and nuclei were stained with 1 μg/mL of DAPI (Roche, 10236276001) followed by mounting with ProLong Gold Antifade Mountant (Invitrogen, P36930). Four independent fields per tumor section were imaged at random with a Nikon A1R MP confocal microscope (RRID:SCR_020319) with consistent instrument settings between samples. Sections stained with secondary antibody alone were used as negative controls.
Quantification of BPC lung metastases
Lungs were fixed and dehydrated as described above. Lungs were then visualized with an OMAX trinocular microscope (W43C1-L08-TP). The number of pigmented lesions on the surface of each lung was quantified in a blinded manner under high magnification.
Western blot
Cells were lysed in ice cold RIPA buffer (G-Biosciences, 786–490) supplemented with protease inhibitor cocktail (Roche, 11836153001). Samples were denatured, separated by SDS-PAGE with 4–12% Bis-tris gels (Sigma), and transferred to low fluorescence PVDF membranes with the Trans-Blot Turbo Transfer System according to manufacturer’s instructions (Bio-Rad, RRID:SCR_023156). Membranes were blocked for one hour with Intercept Blocking Buffer (LI-COR, 927–7000) and probed overnight at 4°C with the following primary antibodies diluted in blocking buffer containing 0.2% Tween-20: Puromycin (1:10000, Millipore MABE343, RRID:AB_2566826), APOE (1:1000, GeneTex GTX100053, RRID:AB_1949674), HSC70 (1:1000, Santa Cruz sc-7298, RRID:AB_627761), LRP1 (1:50000, abcam 92544, RRID:AB_2234877). Membranes were washed with PBST and incubated for an hour with IRDye 680RD goat anti-mouse (LI-COR 926–68070, RRID:AB_10956588) or 800CW goat anti-rabbit (LI-COR 926–32211, RRID:AB_621843) secondary antibodies diluted in blocking buffer containing 0.2% Tween 20 and 0.1% SDS (1:5000). Blots were imaged with a LI-COR Odyssey M Imager and analyzed with Image Studio Lite (RRID:SCR_013715) and Empiria Studio (RRID:SCR_022512) software.
SUnSET assay
In vivo
BPC tumors were topically induced in mice as described above. 35 days after induction, mice were weighed and injected intraperitoneally with 40 nmol/g of puromycin. Mice were placed back in their cage for 30 minutes, after which mice were anesthetized with isoflurane and sacrificed by cervical dislocation. Tumors were dissected and rinsed with PBS to remove blood. ~10mg of tumor was dissected from the center of tumors and homogenized with a Bead Ruptor Elite (Omni International) at 0°C in 200 μL of RIPA supplemented with protease inhibitor cocktail (Roche, 11836153001). To reduce viscosity, lysates were then treated with DNAse I (Norgen, 25710) according to manufacturer’s instructions. 40 μg of lysate was loaded and western blot was run and analyzed as described above. A mouse IgG2a-specific secondary antibody (1:5000; LI-COR 926–32351, RRID:AB_2782998) was used to eliminate background mouse IgG signal. Total protein was detected with the Revert 700 Total Protein Staining kit (LI-COR, 926–11010).
In vitro
5×104 B16F10-TR-shApoe cells stably expressing APOE2, APOE3, APOE4, or empty vector were plated in 8-well chamber slides (Nunc, 154941) the day before experiment. The next day, cells were serum starved for 6 hours in DMEM containing 0.2% FBS and then stimulated for 15 minutes with DF10. After stimulation, cells were pulsed with 10 μg/mL puromycin in DF10 for 30 minutes. As a positive control, one group of cells was treated with 100 μg/mL CHX for 10 minutes in DF10 prior to puromycin treatment. Cells were washed twice with PBS and then fixed with 4% PFA for 10 minutes. Cells were washed twice with PBS for 5 minutes each and then permeabilized with 0.5% Triton X-100 for 10 minutes. Two 5-minute PBS washes were performed, and cells were then incubated for 90 minutes at RT with 0.1% Triton X-100 containing Alexa Fluor 647-conjugated anti-puromycin antibody (1:5000; Millipore MABE343-AF647, RRID:AB_2736876). Cells were washed thrice with PBST for 5 minutes each. During the second wash, DAPI was added at a 1 μg/mL concentration. Slides were mounted with ProLong Gold and left to dry overnight. Four independent fields per condition were imaged at random with a Nikon A1R MP confocal microscope (RRID:SCR_020319) with consistent instrument settings between conditions. Mean fluorescence intensity was quantified with ImageJ (RRID:SCR_003070).
RNA extraction from BPC tumors
Primary tumors were dissected 49 days after 4-OHT administration, flash frozen in liquid nitrogen, and stored at −80°C until RNA extraction. For RNA extraction, 10mg of tissue was dissected from the center of tumors on a ThermalTray (Corning, 432074) placed on dry ice. Tumor pieces were placed in homogenizer tubes containing ceramic beads along with lysis buffer from the Total RNA Purification Kit (Norgen, 37500) and RNAse inhibitors (10 μl/mL β-mercaptoethanol and 200 units/mL RNAsin Plus (Promega, N2615)), flash frozen in liquid nitrogen, and homogenized with a Bead Ruptor Elite (Omni International) at 0°C. RNA was then purified with the Total RNA Purification Kit with on-column DNAse treatment per manufacturer’s instructions.
Digestion, purification, and RNA extraction of tail vein lung metastases
6–10-week-old female APOE2 and APOE3 knock-in mice were injected via lateral tail vein with 100 μl of PBS containing 1 × 105 B16F10-TR-shApoe cells. Fifteen days after injection, mice were anesthetized via intraperitoneal injection of 2.5% Avertin in PBS (Sigma-Aldrich, T48402). Lungs were perfused with cold PBS intratracheally and via the left ventricle. Lungs were removed and placed in a 6-well plate on ice. Lungs were minced on ice with a scalpel and resuspended in 2mL of HBSS2+ (HBSS with calcium and magnesium (Gibco, 24020) supplemented with 2% FBS, 1 mM sodium pyruvate (Gibco, 11360), 25 mM HEPES (Gibco, 15630), 2 mg/mL collagenase IV (Worthington, LS004188), and 0.1 mg/ml DNAse I (Roche, 10104159001)) for 30 min at 37 °C on an orbital shaker at 80 rpm. 7 mL of HBSS2− (HBSS without calcium and magnesium (Gibco, 14170), 2% FBS, 1 mM sodium pyruvate and 25 mM HEPES) was added to the digested tissue, which was homogenized with a serological pipette. The mixture was mashed through a 70 μm strainer and centrifuged at 500g for 5 min at 4°C. The supernatant was aspirated, and the pelleted cells were incubated in ACK lysis buffer (Lonza, 10–548E) for 3 minutes at RT. The mixture was neutralized with HBSS2− and centrifuged. Cells were resuspended in 200uL of HBSS2− and transferred to microcentrifuge tubes for flow sorting.
Flow cytometry
Cells were centrifuged at 500g for 5 min at 4°C. Cells were then resuspended in 50 μl of flow buffer (1x PBS, 2% FBS, 10mM EDTA, 25mM HEPES) containing anti-CD16/CD32 antibody for Fc receptor blockade (1:200, Biolegend 101320, RRID:AB_1574975) and incubated on ice for 5 minutes. For surface marker staining, 50 μL of flow buffer containing anti-CD45-BV785 antibody (1:3000, Biolegend 103149, RRID:AB_2564590) was added, and samples were gently vortexed and incubated for another 20 minutes on ice under protection from light. Cells were then washed with flow buffer and resuspended in flow buffer containing TO-PRO-3 (Invitrogen, T3605). TO-PRO-3low/CD45−/GFP+ cells were sorted on a BD FACSAria II (RRID:SCR_018934). Sorted tumor cells were centrifuged at 200g for 20 min at 4°C, and RNA was purified with the Total RNA Purification Kit with on-column DNAse treatment per manufacturer’s instructions. RNA was then concentrated with an RNA Clean & Concentrator Kit (Zymo Research, R1015).
RNA-Seq
BPC tumors
RNA integrity numbers (RIN) were measured with an Agilent Bioanalyzer 2100 (RRID:SCR_018043), with an average RIN of 8.15. TruSeq RNA Library Prep Kit v2, Set A (Illumina, RS-122–2001) was used to generate RNA-seq libraries according to manufacturer’s instructions. Libraries were quantified with an Agilent TapeStation and pooled at equimolar concentrations. Pooled libraries were sequenced with an Illumina NextSeq 500 (RRID:SCR_014983; High Output, 75 SR). For analysis, FASTQ file quality was checked with FastQC (https://www.bioinformatics.babraham.ac.uk/projects/fastqc/). Kallisto v0.46.1 (14) was used to pseudoalign reads to the mm10 mouse transcriptome (version 101) downloaded from Ensembl. Quality data was aggregated with MultiQC (15). Counts were imported into R v4.1.3. with RStudio v2022.02.1 and tximport v1.18.0 (16). Differential expression analysis was performed with DESeq2 v1.28.1 (17) after prefiltering genes with less than 10 counts. Genes were annotated with AnnotationDbi v1.52.0 and the org.Mm.eg.db package. Genes were ranked based on Wald statistic, and GSEA was performed with the fgsea v1.20.0 (18). Mouse gene sets were downloaded from http://bioinf.wehi.edu.au/MSigDB/ based on MSigDB v7.1.
Tail vein lung metastases
RNA quality was assessed with an Agilent Bioanalyzer 2100, with an average RIN of 8.58. RNA-seq libraries were generated with a QuantSeq 3’ mRNA-Seq Library Prep Kit FWD (Lexogen) according to manufacturer’s instructions. Libraries were quantified, pooled, and sequenced as described above. FASTQ file quality was checked with FastQC. Adapter and poly A tail sequences were trimmed with bbduk v.38.31 (https://sourceforge.net/projects/bbmap/; options k=13, ktrim=r, forcetrimleft=11, useshortkmers=t, mink=5, qtrim=t, trimq=10, minlength=20). Reads were aligned to the GRCm38 mouse genome with STAR v.2.6.0 at default settings except for ‘outFilterMismatchNoverLmax’ set to 0.1 and ‘outFilterMultimapNmax’ set to 1. Mapped reads were counted with featureCounts (19). Differential expression analysis, gene annotation, and GSEA were performed as described above.
TCGA analysis
Harmonized raw counts of tumor transcriptomes from the TCGA-SKCM study were downloaded from the Genomic Data Commons API and imported into R with TCGAbiolinks v2.18.0 (20). APOE genotype information from whole exome sequencing was utilized as determined previously (3). Differential expression analysis was performed as described for BPC tumors, with tumor stage included as a covariate for primary tumor analysis to account for differences in tumor progression between genotypes. Genes were annotated with AnnotationDbi v1.52.0 and the org.Hs.eg.db package. The Reactome gene set was downloaded from MSigDB v7.1, and GSEA was performed as described above.
Statistical analysis
All data are expressed as mean ± SEM, unless indicated otherwise. Groups were compared using statistical tests for significance as described in the figure legends. A P value less than 0.05 was considered statistically significant. Statistical tests were performed with GraphPad Prism 9 (RRID:SCR_002798).
Study approval
All animal experiments were conducted in accordance with a protocol (#20010) approved by the Institutional Animal Care and Use Committee at The Rockefeller University.
Data availability
RNA-Seq data generated for this study have been deposited at the Gene Expression Omnibus under accession numbers GSE208718 and GSE209873. Data from the TCGA-SKCM study were obtained from the Genomic Data Commons: https://portal.gdc.cancer.gov/. All other raw data generated in this study are available upon request from the corresponding author.
Results
Human APOE variants differentially modulate melanoma progression in a genetically engineered mouse model
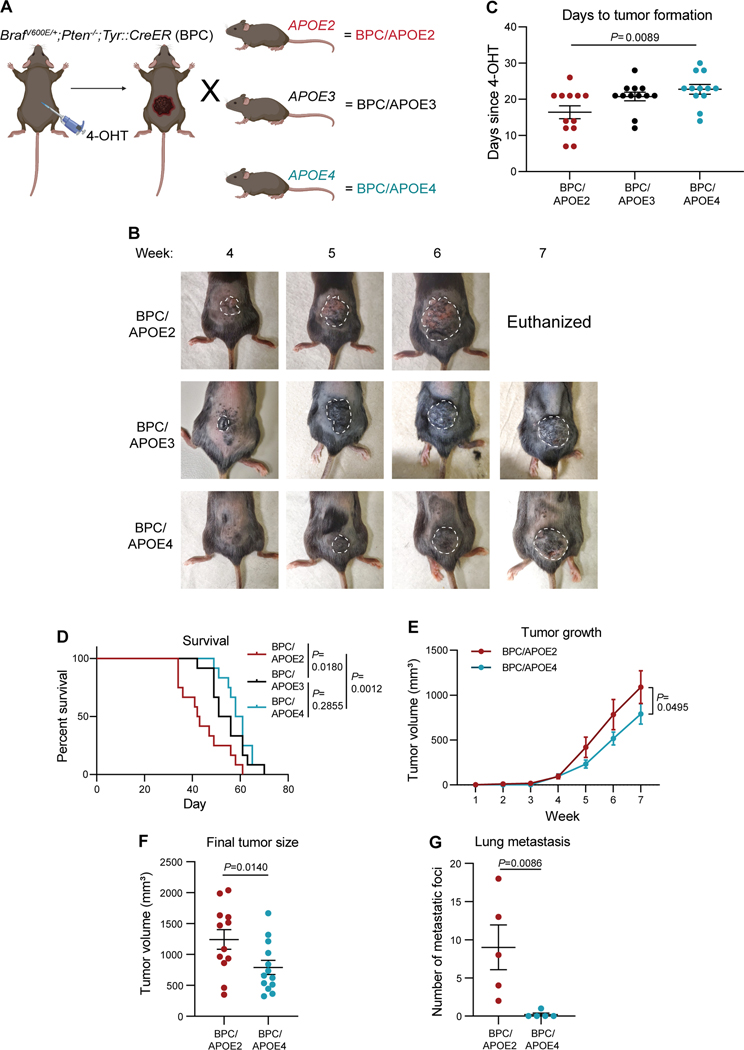
To model the impact of human APOE variants on all stages of melanoma progression, we crossed the well-established BrafV600E/+;Pten−/−;Tyr::CreER (BPC) GEMM with APOE-targeted replacement (knock-in) mice, in which the endogenous murine Apoe locus has been replaced with one of the three human APOE genes (21–23) (Fig. 1A). The BPC model enables tamoxifen-inducible deletion of the Pten tumor suppressor and activation of the BrafV600E oncogene specifically in melanocytes, resulting in melanoma formation in 3–4 weeks with 100% penetrance and recapitulation of the entirety of the metastatic cascade (6). To initiate primary tumor growth, 4-hydroxytamoxifen (4-OHT) was applied topically to the lower backs of adult BPC;APOE2 (BPC/APOE2), BPC;APOE3 (BPC/APOE3), and BPC;APOE4 (BPC/APOE4) mice. Western blot confirmed tumoral APOE expression, which was most prominent in the APOE2 background (Supplementary Fig. S1A). This is consistent with human expression patterns, where APOE2 carriers exhibit higher plasma APOE levels than APOE3 homozygotes and APOE4 carriers, and is likely due to feedback compensation for reduced binding of APOE2 to APOE receptors (24,25). We next evaluated genetically initiated tumor progression amongst the APOE genotypes. Melanoma onset occurred with shortest latency in BPC/APOE2 mice, followed by BPC/APOE3 mice and then BPC/APOE4 mice (Figs. 1B, 1C). Mouse survival followed a similar pattern, with BPC/APOE2 mice having the shortest median survival at 42.5 days, BPC/APOE3 mice intermediate at 53.5 days, and BPC/APOE4 mice having the longest survival at 59.5 days (Fig. 1D). We thus focused mainly on the APOE2 and APOE4 genotypes for the remainder of the study, as they produced the most divergent tumor phenotypes in the GEMM. In an independent cohort of mice whose tumor growth rate was tracked, melanomas grew faster (Fig. 1E) and were significantly larger at the day 49 endpoint (Fig. 1F) in BPC/APOE2 mice relative to BPC/APOE4 mice—consistent with the aforementioned findings.
Figure 1. Common human APOE variants differentially impact melanoma growth and metastasis in a genetically engineered mouse model.
A, Schematic depicting generation of, and tumor induction in, the Braf V600E/+;Pten−/−;Tyr::CreER;APOE2 (BPC/APOE2), ;APOE3 (BPC/APOE3), and; APOE4 (BPC/APOE4) mouse models. Created with BioRender.com. B, Representative images of tumor growth in BPC/APOE2, BPC/APOE3, and BPC/APOE4 mice 4 to 7 weeks after topical administration of 4-OHT. C, Number of days after topical 4-OHT administration until tumors were palpated and visualized in BPC/APOE2, BPC/APOE3, and BPC/APOE4 mice (n=12 per group). One-way ANOVA. D, Kaplan-Meier survival curves of BPC/APOE2, BPC/APOE3, and BPC/APOE4 mice after topical 4-OHT administration (n=12 per group). Log-rank test. E, Tumor growth curve of BPC/APOE2 (n=12) and BPC/APOE4 (n=13) mice after topical 4-OHT administration. Two-way ANOVA. F, Final tumor volumes of BPC/APOE2 (n=12) and BPC/APOE4 (n=13) mice from E at the experimental endpoint of 49 days after topical 4-OHT administration. Unpaired t-test. G, Quantification of lung metastatic foci in BPC/APOE2 (n=5) and BPC/APOE4 (n=5) mice after neonatal tumor induction. Unpaired t-test.
As APOE4 has been shown to enhance anti-tumor effector immune cell activity in melanoma (3), we evaluated the contribution of anti-tumor immunity to the observed growth differences between BPC/APOE2 and BPC/APOE4 tumors. Histological staining revealed no significant difference in CD8+ T cell infiltration between BPC/APOE2 and BPC/APOE4 tumors at the day 49 endpoint (Supplementary Figs. S1B and S1C). This is likely because the BPC GEMM is a poorly immunogenic mouse model (26), which may preclude immune differences from developing by this time point. Nevertheless, this result suggests a mechanism distinct from anti-tumor immunity by which APOE variants can differentially modulate tumor growth in the BPC GEMM.
Because APOE is a potent suppressor of melanoma metastasis (4), we next evaluated whether genetic variation in APOE could modulate metastatic capacity in the BPC GEMM. Metastatic burden in the BPC model is determined by quantifying pigmented foci on the surface of the lung after neonatal administration of 4-OHT (27). This contrasts with analysis of localized primary tumor growth, where 4-OHT is administered to adult mice. Accordingly, BPC/APOE2 and BPC/APOE4 neonates received 4-OHT topically and were euthanized after weaning age. Pigmented foci were visible on the lungs of mice (Supplementary Fig. S1D), and BPC/APOE2 mice exhibited substantially more lung metastatic foci compared to BPC/APOE4 mice (Fig. 1G). Histological analysis of a representative pigmented lesion revealed features consistent with metastatic melanoma on hematoxylin and eosin (H&E) staining as well as positive staining for S100, a sensitive marker for melanoma (Supplementary Fig. S1E). These results in the APOE variant GEMM revealed a potent impact of hereditary genetics on tumor growth and metastatic colony formation in an autochthonous model of melanoma progression.
Genetically engineered mouse model reveals protein translation upregulation in APOE2 tumors
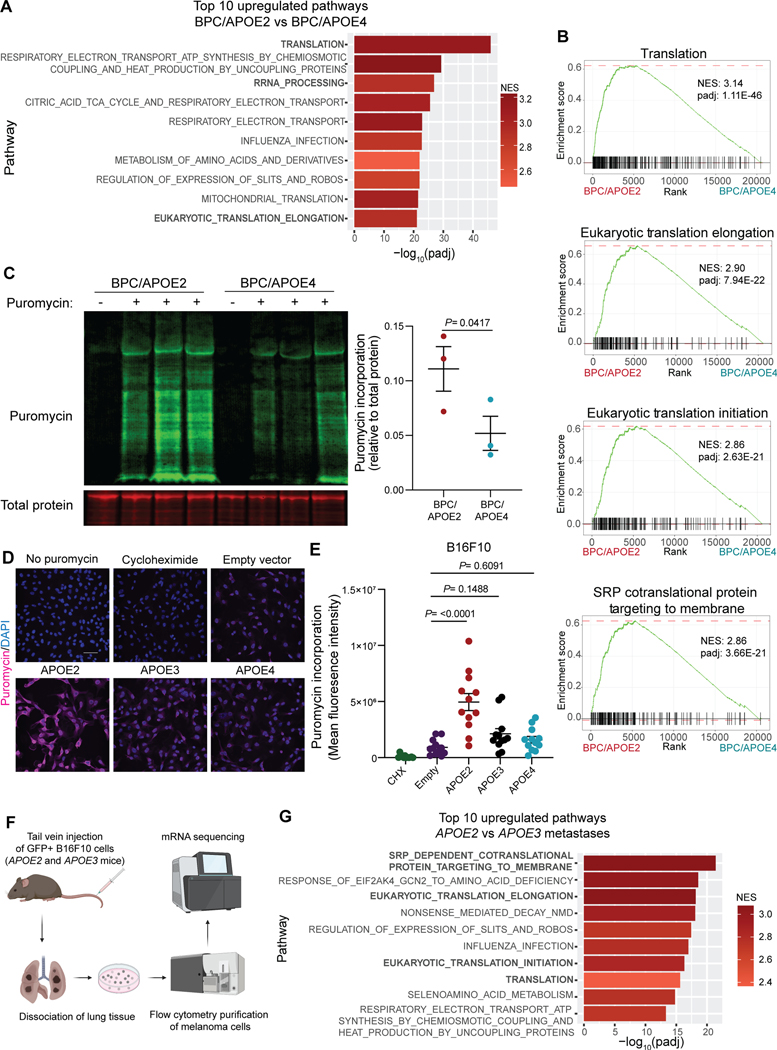
We next utilized the APOE allelic GEMM series as a tool to search for cellular processes that might be altered in an APOE variant-dependent manner and that could influence cancer progression. To this end, we performed bulk RNA-Seq of time-matched BPC/APOE2 and BPC/APOE4 tumors. Gene set enrichment analysis (GSEA) (28) revealed that translation was the most upregulated pathway in BPC/APOE2 tumors relative to BPC/APOE4 tumors (Fig. 2A; Supplementary Table 1). The additional protein synthesis-related pathways ‘Ribosomal RNA processing’ and ‘Eukaryotic translation elongation’ were also among the top ten upregulated pathways. BPC/APOE2 tumors displayed significant upregulation in all major steps of translation present in the Reactome (29) gene set relative to BPC/APOE4 tumors (Fig. 2B).
Figure 2. Common APOE germline genetic variants modulate protein synthesis in melanoma.
A, Top ten pathways upregulated in melanomas of BPC/APOE2 mice relative to BPC/APOE4 mice as determined by GSEA and ranked by adjusted p-value (n=4 per group; NES, normalized enrichment score; padj, adjusted p-value). B, Enrichment plots of translation-related pathways within the Reactome gene set. C, Western blot of puromycin incorporation into BPC/APOE2 and BPC/APOE4 tumors 35 days after 4-OHT administration (n=3 per group). Non-puromycin-pulsed mice were included as an antibody control. Unpaired t-test. D, Representative immunofluorescence images of puromycin incorporation into B16F10-shApoe pBabe Empty, APOE2, APOE3, and APOE4 cells. Non-puromycin-pulsed and cycloheximide (CHX)-treated cells were included as negative and positive controls, respectively (scale bar = 50μM). E, Quantification of mean fluorescence intensity from SUnSET assay in D. One-way ANOVA (n=3 independent experiments). F, Schematic depicting the workflow utilized to analyze transcriptomes of B16F10 lung metastases derived from APOE2 and APOE3 knock-in mice. Created with BioRender.com. G, Top ten pathways upregulated in APOE2 (n=8) lung metastases relative to APOE3 (n=5) as determined by GSEA and ranked by adjusted p-value (NES, normalized enrichment score; padj, adjusted p-value)
Translational control plays a critical role in all steps of cancer progression, and most oncogenic signaling pathways converge to enhance the translational capacity of tumor cells (30–32). We therefore sought to experimentally validate whether there were differences in translational efficiency between APOE2 and APOE4 melanomas. We utilized the surface sensing of translation (SUnSET) assay, a well-established method for measuring protein synthesis (33). In this assay puromycin can be administered to live mice, which then incorporates into nascent polypeptide chains synthesized in mouse tissues (34). Puromycin incorporation can then be quantified with an anti-puromycin antibody, providing a readout of global cellular translation. To control for tumor size differences, BPC/APOE2 and BPC/APOE4 mice were injected with puromycin 35 days after 4-OHT administration, an early time point at which a significant difference in tumor volumes was not yet detectable (Supplementary Fig. S2A). Consistent with our RNA-Seq results, BPC/APOE2 tumors exhibited significantly higher puromycin incorporation than BPC/APOE4 tumors, indicative of either a slower translation rate in the APOE4 background or enhanced translation rate in the APOE2 background (Fig. 2C). To further control for differences in tumor proliferation and the presence of stromal cells, we performed the SUnSET assay in vitro with melanoma cells stably overexpressing APOE2, APOE3, APOE4, or an empty control vector. Importantly, APOE was expressed at equivalent levels amongst the APOE2, APOE3, and APOE4 cell lines, thus controlling for differences in APOE expression levels present in the genetic model (Supplementary Fig. S2B). Additionally, under normal growth conditions there was no difference in proliferation rates among the cell lines, thus enabling us to control for the tumor growth differences observed in vivo (Supplementary Fig. S2C). Cells were serum starved and then pulsed with puromycin following growth factor stimulation. We observed no difference in puromycin incorporation between control, APOE3, and APOE4 cells, whereas APOE2 cells exhibited substantially increased puromycin signal (Figs. 2D and 2E). These findings are consistent with a model whereby APOE2 promotes translation rather than APOE4 inhibiting translation.
We next sought orthogonal support for a role of APOE2 in protein translation regulation and investigated whether APOE-dependent effects on translation were also present in metastatic disease. APOE protein is known to act extracellularly on tumoral APOE receptors, and both tumor-derived and stroma-derived APOE have been found to regulate melanoma metastatic capacity in this manner (4,5). We performed tail vein metastasis assays with syngeneic, GFP-expressing B16F10 melanoma cells depleted of murine APOE. Cells were injected into APOE2 or APOE3 knock-in mice, thereby assessing the impact of APOE variants secreted by the stromal compartment on cancer cells. APOE3 served as an ideal isogenic control for determining whether APOE2 mediates translational enhancement because it exhibited an intermediate phenotype in our GEMM studies and is the most common APOE variant in the human population (7). The injections were followed by bulk RNA-Seq of metastatic melanoma cells that were purified from lungs by flow cytometry (Fig. 2F), thus allowing us to examine the tumor cell-intrinsic effects of stroma-derived APOE2 and APOE3. APOE2 mice had a significantly higher fraction of GFP+ tumor cells within their dissociated lungs relative to APOE3 mice, indicative of higher metastatic burden (Supplementary Fig. S2D). Importantly, translation-related processes again dominated the top ten upregulated pathways in APOE2 metastases relative to APOE3 (Fig. 2G; Supplementary Table 2). These results indicate that the APOE variants differentially impact melanoma protein synthesis, with APOE2 stimulating translation in both localized and metastatic melanoma.
LRP1 mediates cell-intrinsic effects of APOE variants on melanoma progression
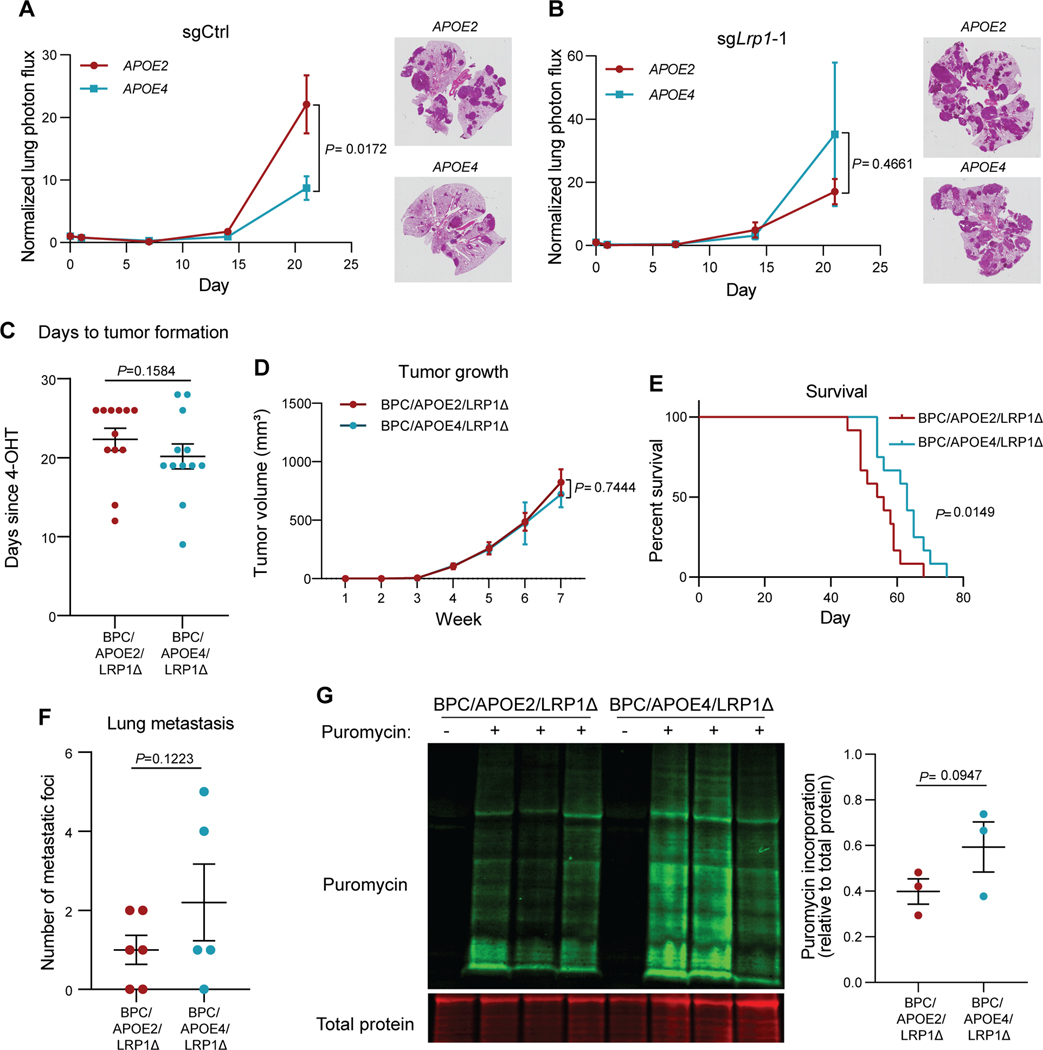
We previously found that the APOE receptor LRP1 mediates the cell-intrinsic effects of APOE on melanoma cells (4). We thus investigated whether tumoral LRP1 is necessary for the differential impact of APOE variants on melanoma progression. We first sought to determine whether melanoma LRP1 mediates differences between APOE2 and APOE4 mice in tail vein lung metastatic colonization assays. After tail vein injection, control B16F10 melanoma cells depleted of murine APOE metastasized to the lung more efficiently in syngeneic APOE2 knock-in mice relative to APOE4 mice (Fig. 3A). However, after CRISPR/Cas9-mediated deletion of melanoma Lrp1 via two independent guide RNAs (Supplementary Fig. S3A), the difference in metastatic burden between APOE2 and APOE4 mice was abrogated (Fig. 3B, Supplementary Fig. S3B). These findings reveal that the effects of APOE variants on melanoma metastatic capacity are mediated via the APOE receptor LRP1.
Figure 3. Differential effects of APOE variants on melanoma progression and protein translation are abrogated upon tumoral Lrp1 genetic deletion.
A, B, Quantification of lung metastatic progression via bioluminescence imaging of B16F10-TR-shApoe sgCtrl (A) or sgLrp1-1 (B) cells injected via lateral tail vein into APOE2 and APOE4 mice. Representative images of H&E-stained lungs taken from mice at the day 21 endpoint (n = 9–10 mice per group; representative of two independent experiments; two-way ANOVA). C, Number of days after topical 4-OHT administration until tumors were palpated and visualized in BPC/APOE2/LRP1Δ and BPC/APOE4/LRP1Δ mice (n=12 per group). Unpaired t-test. D, Melanoma tumor growth curves of BPC/APOE2/LRP1Δ (n=12) and BPC/APOE4/LRP1Δ (n=10) mice after topical 4-OHT administration. Two-way ANOVA. E, Kaplan-Meier survival curves of BPC/APOE2/LRP1Δ and BPC/APOE4/LRP1Δ mice after topical 4-OHT administration (n=12 per group). Log-rank test. F, Quantification of lung metastatic foci in BPC/APOE2/LRP1Δ (n=6) and BPC/APOE4/LRP1Δ (n=5) mice after neonatal tumor induction. Unpaired t-test. G, Western blot of puromycin incorporation into BPC/APOE2/LRP1Δ and BPC/APOE4/LRP1Δ tumors 35 days after 4-OHT administration (n=3 per group). Non-puromycin-pulsed mice were included as an antibody control. Unpaired t-test.
As described earlier, transplantable mouse models are limited in their ability to recapitulate all steps of the metastatic cascade. Specifically, tail vein injections bypass the initial steps of tumor formation, stromal invasion, and intravasation because cancer cells are injected directly into the bloodstream. To better interrogate the ability of tumoral LRP1 to mediate differences in melanoma progression between APOE2 and APOE4 genotypes, we crossed BPC/APOE2 and BPC/APOE4 mice with Lrp1flox/flox mice. This enabled the deletion of Lrp1 specifically in the melanocytes that form melanomas after 4-OHT administration. Immunofluorescence staining of BPC/APOE2;Lrp1flox/flox (BPC/APOE2/LRP1Δ) and BPC/APOE4;Lrp1flox/flox (BPC/APOE4/LRP1Δ) tumors showed substantial loss of LRP1 signal compared to Lrp1 wild-type tumors, confirming successful Cre-mediated deletion (Supplementary Fig. S4A). We again monitored tumor growth in this model after 4-OHT administration to back skin of adult mice. In contrast to the differential effects of APOE variants observed in Lrp1 wild-type mice, there was no significant difference in tumor latency (Fig. 3C), tumor growth rate (Fig. 3D), or tumor volume at the experimental endpoint (Supplementary Fig. S4B) between BPC/APOE2/LRP1Δ and BPC/APOE4/LRP1Δ mice. In a separate survival experiment, there was a significant difference with BPC/APOE2/LRP1Δ mice exhibiting a median survival of 55 days compared to 63 days in BPC/APOE4/LRP1Δ mice (Fig. 3E). However, this difference was diminished compared to Lrp1 wild-type BPC/APOE2 and BPC/APOE4 mice (Fig. 1D). This result is consistent with our prior findings revealing that the APOE4 background confers potent anti-tumor immunity (3). BPC/APOE4/LRP1Δ mice are likely provided an advantage over BPC/APOE2/LRP1Δ mice in a survival experiment that allows more time for anti-tumor immunity to develop in this poorly immunogenic genetic model. Indeed, immunofluorescence staining of BPC/APOE2/LRP1Δ and BPC/APOE4/LRP1Δ tumors at the mouse survival endpoint revealed enhanced CD8+ T cell infiltration into APOE4 tumors (Supplementary Fig. S4C and S4D). There was no significant difference in lung metastasis between BPC/APOE2/LRP1Δ and BPC/APOE4/LRP1Δ mice after neonatal 4-OHT administration (Fig. 3F), consistent with melanoma cell-intrinsic effects of APOE dominating at the metastatic site. These results indicate that Lrp1 deletion significantly abrogates the impact of APOE variants on melanoma progression, with differences only emerging when the cell-extrinsic effects of APOE become impactful at later primary tumor stages in the genetic model.
Of note, equalization of melanoma progression between BPC/APOE2 and BPC/APOE4 mice upon Lrp1 deletion was largely driven by decreased tumor growth and metastasis in the APOE2 background. Tumor progression and metastasis between BPC/APOE4 and BPC/APOE4/LRP1Δ mice was unchanged (Supplementary Figs. S4E–G), again supporting the notion that APOE2 is acting in a gain-of-function manner to promote melanoma progression in this context. Having determined that LRP1 mediates the cell-intrinsic effects of the APOE variants, we next investigated whether the effects of APOE variants on mRNA translation are LRP1-dependent. The SUnSET assay was performed with day 35 BPC/APOE2/LRP1Δ and BPC/APOE4/LRP1Δ tumors and showed equalization of puromycin incorporation between the genotypes upon Lrp1 deletion (Fig. 3G). These findings reveal that the APOE receptor LRP1 is a required mediator of the cell-intrinsic effect of APOE2 on melanoma progression at the early primary tumor and metastasis stages. Moreover, LRP1 is required for the enhanced protein synthesis effect mediated by APOE2 in melanoma. We finally investigated the potential for the existence of a feedback loop between LRP1 and the APOE variants by evaluating LRP1 expression among the different APOE genotypes. We observed no difference in LRP1 expression among the APOE2, APOE3, and APOE4 backgrounds in vivo or in vitro (Supplementary Figs. S4H and S4I). LRP1 thus likely mediates downstream molecular events that are preferentially activated by APOE2 and remain to be identified.
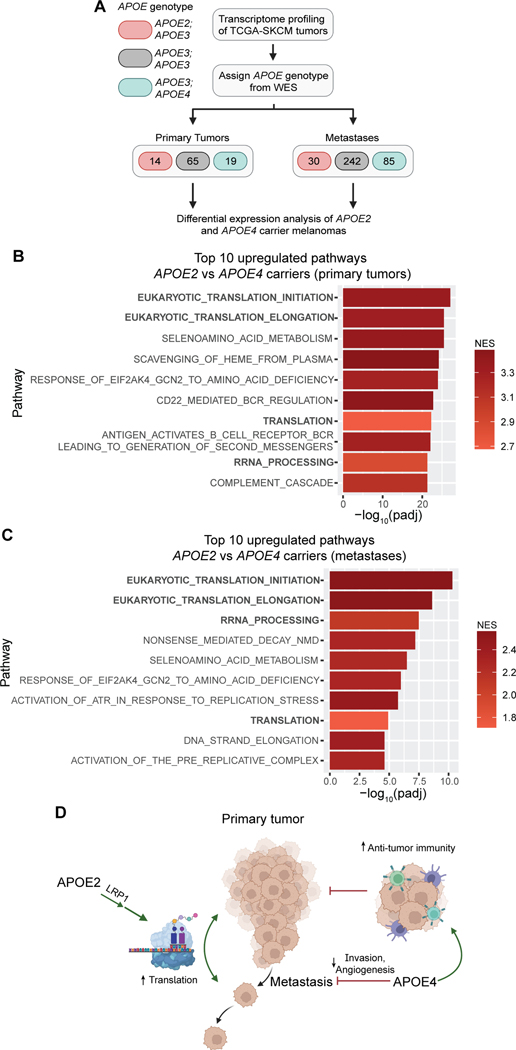
APOE genetics impact translation regulation in human melanomas
We next sought to determine whether the APOE variant-dependent translation phenotype could also be observed in human melanomas. Given that the worldwide allele frequency of APOE2 is less than ten percent (35), which limits statistical power, we turned to large public datasets. The Cancer Genome Atlas skin cutaneous melanoma (TCGA-SKCM) cohort contains over 400 patients with both RNA sequencing and whole exome sequencing data, enabling differential gene expression analysis of human tumors based on the germline APOE genotype (36) (Fig. 4A). The TCGA-SKCM dataset was previously used to determine that APOE4 carriers have a survival advantage relative to APOE3 patients while APOE2 carriers experience worse survival outcomes (3). Consistent with our findings in BPC/APOE2 and BPC/APOE4 mice, primary tumors from APOE2 carrier patients exhibited upregulation of translation pathways relative to tumors from APOE4 carriers (Fig. 4B; Supplementary Table 3). Also consistent with the notion that APOE2 is an active promoter of protein synthesis, translation pathways were also the top upregulated processes in APOE2 carrier primary tumors compared to APOE3 homozygotes (Supplementary Figs. S5A and 5B; Supplementary Table 3). Protein translation upregulation in the APOE2 background relative to APOE4 was also maintained in metastatic tumors, suggesting that APOE2 promotes protein synthesis in the melanomas of APOE2 carriers throughout the metastatic cascade (Fig. 4C; Supplementary Table 3). Taken together with our findings from mouse modeling, we propose that APOE2-mediated enhancement of translation—a key driver of tumor growth and metastasis—supports melanoma growth and metastasis in APOE2 carriers, consistent with their poor survival compared to both APOE3 homozygotes and APOE4 carriers.
Figure 4. Protein translation pathways are upregulated in melanomas of APOE2 carrier patients.
A, Schematic depicting the workflow utilized to analyze transcriptomes of APOE2 and APOE4 carrier melanomas in the TCGA-SKCM cohort (WES, whole exome sequencing). Created with BioRender.com. B, Top ten pathways upregulated in primary tumors of APOE2 carrier patients (n=14) relative to APOE4 carrier patients (n=19) as determined by GSEA and ranked by adjusted p-value (NES, normalized enrichment score; padj, adjusted p-value). C, Top ten pathways upregulated in metastases of APOE2 carrier patients (n=30) relative to APOE4 carrier patients (n=85) as determined by GSEA and ranked by adjusted p-value. D, Model depicting our current understanding of the role of common APOE genetic variants in melanoma progression. The model depicts APOE4 acting as a suppressor of melanoma progression by enhancing anti-tumor immunity as well as repressing angiogenesis and invasion. In contrast, APOE2 is shown as a driver of melanoma progression through its stimulation of protein synthesis via the LRP1 receptor. Created with BioRender.com.
In sum, our findings uncover a new, surprising role of APOE and its genetic variants as differential modulators of mRNA translation in melanoma. We moreover provide evidence from a genetically initiated model that common germline variation in a human gene —APOE— differentially regulates tumor and metastatic progression. We propose a model in which APOE2 and APOE4 carry out opposing functions in melanoma, with APOE2 promoting growth, metastasis, and protein translation via the LRP1 receptor and APOE4 repressing progression via its suppression of multiple cancer phenotypes including invasion, angiogenesis and immune evasion, as demonstrated in previous work (3) (Fig. 4D).
Discussion
Despite nearly 30 years of research following the discovery that APOE genotype impacts Alzheimer’s disease risk, the molecular mechanisms underlying this link remain elusive (7). This is a product of the pronounced complexity of APOE, which has diverse roles in numerous biological processes including lipid metabolism, immunity, mitochondrial function, and neuronal repair (7,37). It has also been unclear whether the APOE alleles represent a progressive gain or loss of function of the APOE protein, as the risk of APOE-influenced diseases other than Alzheimer’s does not always follow an APOE4>APOE3>APOE2 order (7). Our study adds to an increasing body of evidence that the pleiotropic behavior of APOE also applies to melanoma (3,4) and supports the notion that APOE2 can act as a gain-of-function allele in a disease context. We reveal that the APOE variants exert cell-intrinsic effects on melanoma that impact primary tumor growth and metastatic capacity in a BrafV600E-driven GEMM, a model that has provided numerous important insights into melanoma biology (12,38–40). Tumors in this melanoma GEMM progressed in an APOE2>APOE3>APOE4 manner, consistent with melanoma patient survival outcomes. We expand the mechanism of this differential modulation beyond anti-tumor immunity, angiogenesis, and cellular invasion by identifying mRNA translation as an oncogenic process that is actively enhanced by APOE2.
Supporting the idea that our findings may have broader disease implications, our examination of previously published transcriptomic and proteomic Alzheimer’s datasets revealed that mRNA translation is a pathway that is upregulated in the brains of APOE2 carriers relative to those of APOE3 homozygotes and APOE4 carriers (41,42). Neuronal protein synthesis has been well established as crucial for synaptic function and memory formation, and it is dysregulated in Alzheimer’s (43). Thus, enhancement of translation may contribute to the protective effect of APOE2 in Alzheimer’s and may partly explain the inverse impact of APOE variants in melanoma versus Alzheimer’s disease. Indeed, additional mechanistic connections between melanoma and Alzheimer’s disease are being increasingly uncovered (44).
We identified the APOE receptor LRP1 as necessary for the pro-tumorigenic and pro-metastatic activity of APOE2. Deletion of Lrp1 abrogated differences in tumor growth, metastasis, and protein synthesis between APOE2 and APOE4 mice. LRP1 is a well-established regulator of intracellular signaling (45), thus making it suitable for mediating signals that could impact protein synthesis. Future studies are needed to elucidate the molecular mechanism downstream of this APOE2/LRP1 axis, as it may reveal new therapeutic targets for the treatment of melanoma and perhaps Alzheimer’s disease. Taken together, our work further highlights how germline genetic variation in the APOE gene impacts melanoma progression through disparate mechanisms. Although long speculated to exist, causal somatic mutational drivers of metastasis have not been identified despite extensive sequencing efforts, suggesting that alternative mechanisms may underlie propensity for metastatic progression (46). Our findings provide support for germline genetic variants as causal contributors to metastatic outcomes—revealing hereditary genetics as a predictor of cancer progression and providing new avenues for mechanistic and therapeutic studies.
Supplementary Material
Significance.
APOE germline variants impact melanoma progression through disparate mechanisms, such as the protein synthesis–promoting function of the APOE2 variant, indicating that germline genetic variants are causal contributors to metastatic outcomes.
Acknowledgements
We are grateful to members of the Tavazoie laboratory at Rockefeller University for feedback on the manuscript. We thank J. Posada for her pathology advice. We also thank the following Rockefeller University resource centers: V. Francis and other veterinary staff of the Comparative Bioscience Center, S. Mazel and staff of the Flow Cytometry Resource Center, C. Zhao and staff of the Genomics Resource Center, and staff of the Fisher Drug Discovery Resource Center. N. Adaku was supported by a Medical Scientist Training Program grant from the National Institute of General Medical Sciences of the National Institutes of Health under award number T32GM007739 to the Weill Cornell/Rockefeller/Sloan Kettering Tri-Institutional MD-PhD Program, an F30 Predoctoral Fellowship from the National Cancer Institute of the National Institutes of Health under award number F30CA257226, a medical student grant from the Melanoma Research Foundation, and the Shapiro-Silverberg Fund for the Advancement of Translational Research. S.F. Tavazoie was supported by grants from the National Cancer Institute of the National Institutes of Health under award numbers R01CA184804, R35CA274446, and U54CA261701 as well as the Black Family Metastasis Center.
Financial Support:
National Institutes of Health grant T32GM007739 (NA)
National Institutes of Health grant F30CA257226 (NA)
Melanoma Research Foundation (NA)
Shapiro-Silverberg Fund for the Advancement of Translational Research (NA)
National Institutes of Health grant R01CA184804 (SFT)
National Institutes of Health grant R35CA274446 (SFT)
National Institutes of Health grant U54CA261701 (SFT)
Black Family Metastasis Center (SFT)
Footnotes
Conflicts of Interest:
S.F.T. is a cofounder, shareholder, and member of the scientific advisory board of Inspirna. The remaining authors declare no competing interests.
References
- 1.Nagy R, Sweet K, Eng C. Highly penetrant hereditary cancer syndromes. Oncogene 2004;23(38):6445–70 doi 10.1038/sj.onc.1207714. [DOI] [PubMed] [Google Scholar]
- 2.Mucci LA, Hjelmborg JB, Harris JR, Czene K, Havelick DJ, Scheike T, et al. Familial Risk and Heritability of Cancer Among Twins in Nordic Countries. JAMA 2016;315(1):68–76 doi 10.1001/jama.2015.17703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ostendorf BN, Bilanovic J, Adaku N, Tafreshian KN, Tavora B, Vaughan RD, et al. Common germline variants of the human APOE gene modulate melanoma progression and survival. Nat Med 2020;26(7):1048–53 doi 10.1038/s41591-020-0879-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pencheva N, Tran H, Buss C, Huh D, Drobnjak M, Busam K, et al. Convergent multi-miRNA targeting of ApoE drives LRP1/LRP8-dependent melanoma metastasis and angiogenesis. Cell 2012;151(5):1068–82 doi 10.1016/j.cell.2012.10.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pencheva N, Buss CG, Posada J, Merghoub T, Tavazoie SF. Broad-spectrum therapeutic suppression of metastatic melanoma through nuclear hormone receptor activation. Cell 2014;156(5):986–1001 doi 10.1016/j.cell.2014.01.038. [DOI] [PubMed] [Google Scholar]
- 6.Dankort D, Curley DP, Cartlidge RA, Nelson B, Karnezis AN, Damsky WE, Jr., et al. Braf(V600E) cooperates with Pten loss to induce metastatic melanoma. Nat Genet 2009;41(5):544–52 doi 10.1038/ng.356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Belloy ME, Napolioni V, Greicius MD. A Quarter Century of APOE and Alzheimer’s Disease: Progress to Date and the Path Forward. Neuron 2019;101(5):820–38 doi 10.1016/j.neuron.2019.01.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Basford JE, Wancata L, Hofmann SM, Silva RA, Davidson WS, Howles PN, et al. Hepatic deficiency of low density lipoprotein receptor-related protein-1 reduces high density lipoprotein secretion and plasma levels in mice. J Biol Chem 2011;286(15):13079–87 doi 10.1074/jbc.M111.229369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zivelin A, Rosenberg N, Peretz H, Amit Y, Kornbrot N, Seligsohn U. Improved Method for Genotyping Apolipoprotein E Polymorphisms by a PCR-Based Assay Simultaneously Utilizing Two Distinct Restriction Enzymes. Clinical Chemistry 1997;43(9):1657–9 doi 10.1093/clinchem/43.9.1657. [DOI] [PubMed] [Google Scholar]
- 10.Ponomarev V, Doubrovin M, Serganova I, Vider J, Shavrin A, Beresten T, et al. A novel triple-modality reporter gene for whole-body fluorescent, bioluminescent, and nuclear noninvasive imaging. Eur J Nucl Med Mol Imaging 2004;31(5):740–51 doi 10.1007/s00259-003-1441-5. [DOI] [PubMed] [Google Scholar]
- 11.Sanjana NE, Shalem O, Zhang F. Improved vectors and genome-wide libraries for CRISPR screening. Nat Methods 2014;11(8):783–4 doi 10.1038/nmeth.3047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Spranger S, Bao R, Gajewski TF. Melanoma-intrinsic beta-catenin signalling prevents anti-tumour immunity. Nature 2015;523(7559):231–5 doi 10.1038/nature14404. [DOI] [PubMed] [Google Scholar]
- 13.Westmark PR, Gutierrez A, Westmark CJ. A Simple, Reliable and Inexpensive Method to Individually Identify Neonate Mice. Lab Animal Sci Prof 2021;9(1):46–8. [PMC free article] [PubMed] [Google Scholar]
- 14.Bray NL, Pimentel H, Melsted P, Pachter L. Near-optimal probabilistic RNA-seq quantification. Nat Biotechnol 2016;34(5):525–7 doi 10.1038/nbt.3519. [DOI] [PubMed] [Google Scholar]
- 15.Ewels P, Magnusson M, Lundin S, Kaller M. MultiQC: summarize analysis results for multiple tools and samples in a single report. Bioinformatics 2016;32(19):3047–8 doi 10.1093/bioinformatics/btw354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Soneson C, Love MI, Robinson MD. Differential analyses for RNA-seq: transcript-level estimates improve gene-level inferences. F1000Res 2015;4:1521 doi 10.12688/f1000research.7563.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 2014;15(12):550 doi 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Korotkevich G, Sukhov V, Budin N, Shpak B, Artyomov MN, Sergushichev A. Fast gene set enrichment analysis. bioRxiv 2021:060012 doi 10.1101/060012. [DOI] [Google Scholar]
- 19.Liao Y, Smyth GK, Shi W. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 2014;30(7):923–30 doi 10.1093/bioinformatics/btt656. [DOI] [PubMed] [Google Scholar]
- 20.Colaprico A, Silva TC, Olsen C, Garofano L, Cava C, Garolini D, et al. TCGAbiolinks: an R/Bioconductor package for integrative analysis of TCGA data. Nucleic Acids Res 2016;44(8):e71 doi 10.1093/nar/gkv1507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sullivan PM, Mezdour H, Quarfordt SH, Maeda N. Type III hyperlipoproteinemia and spontaneous atherosclerosis in mice resulting from gene replacement of mouse Apoe with human Apoe*2. J Clin Invest 1998;102(1):130–5 doi 10.1172/JCI2673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sullivan PM, Mezdour H, Aratani Y, Knouff C, Najib J, Reddick RL, et al. Targeted replacement of the mouse apolipoprotein E gene with the common human APOE3 allele enhances diet-induced hypercholesterolemia and atherosclerosis. J Biol Chem 1997;272(29):17972–80 doi 10.1074/jbc.272.29.17972. [DOI] [PubMed] [Google Scholar]
- 23.Knouff C, Hinsdale ME, Mezdour H, Altenburg MK, Watanabe M, Quarfordt SH, et al. Apo E structure determines VLDL clearance and atherosclerosis risk in mice. J Clin Invest 1999;103(11):1579–86 doi 10.1172/JCI6172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rasmussen KL, Tybjaerg-Hansen A, Nordestgaard BG, Frikke-Schmidt R. Plasma levels of apolipoprotein E and risk of dementia in the general population. Ann Neurol 2015;77(2):301–11 doi 10.1002/ana.24326. [DOI] [PubMed] [Google Scholar]
- 25.Kowal RC, Herz J, Weisgraber KH, Mahley RW, Brown MS, Goldstein JL. Opposing effects of apolipoproteins E and C on lipoprotein binding to low density lipoprotein receptor-related protein. J Biol Chem 1990;265(18):10771–9. [PubMed] [Google Scholar]
- 26.Wang J, Perry CJ, Meeth K, Thakral D, Damsky W, Micevic G, et al. UV-induced somatic mutations elicit a functional T cell response in the YUMMER1.7 mouse melanoma model. Pigment Cell Melanoma Res 2017;30(4):428–35 doi 10.1111/pcmr.12591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Damsky WE, Curley DP, Santhanakrishnan M, Rosenbaum LE, Platt JT, Gould Rothberg BE, et al. beta-catenin signaling controls metastasis in Braf-activated Pten-deficient melanomas. Cancer Cell 2011;20(6):741–54 doi 10.1016/j.ccr.2011.10.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A 2005;102(43):15545–50 doi 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jassal B, Matthews L, Viteri G, Gong C, Lorente P, Fabregat A, et al. The reactome pathway knowledgebase. Nucleic Acids Research 2019;48(D1):D498–D503 doi 10.1093/nar/gkz1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Truitt ML, Ruggero D. New frontiers in translational control of the cancer genome. Nat Rev Cancer 2016;16(5):288–304 doi 10.1038/nrc.2016.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Knight JRP, Alexandrou C, Skalka GL, Vlahov N, Pennel K, Officer L, et al. MNK Inhibition Sensitizes KRAS-Mutant Colorectal Cancer to mTORC1 Inhibition by Reducing eIF4E Phosphorylation and c-MYC Expression. Cancer Discov 2021;11(5):1228–47 doi 10.1158/2159-8290.CD-20-0652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Krishnamoorthy GP, Davidson NR, Leach SD, Zhao Z, Lowe SW, Lee G, et al. EIF1AX and RAS Mutations Cooperate to Drive Thyroid Tumorigenesis through ATF4 and c-MYC. Cancer Discov 2019;9(2):264–81 doi 10.1158/2159-8290.CD-18-0606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schmidt EK, Clavarino G, Ceppi M, Pierre P. SUnSET, a nonradioactive method to monitor protein synthesis. Nat Methods 2009;6(4):275–7 doi 10.1038/nmeth.1314. [DOI] [PubMed] [Google Scholar]
- 34.Goodman CA, Hornberger TA. Measuring protein synthesis with SUnSET: a valid alternative to traditional techniques? Exerc Sport Sci Rev 2013;41(2):107–15 doi 10.1097/JES.0b013e3182798a95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Singh PP, Singh M, Mastana SS. APOE distribution in world populations with new data from India and the UK. Ann Hum Biol 2006;33(3):279–308 doi 10.1080/03014460600594513. [DOI] [PubMed] [Google Scholar]
- 36.Cancer Genome Atlas Network. Genomic Classification of Cutaneous Melanoma. Cell 2015;161(7):1681–96 doi 10.1016/j.cell.2015.05.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Huang Y, Mahley RW. Apolipoprotein E: structure and function in lipid metabolism, neurobiology, and Alzheimer’s diseases. Neurobiol Dis 2014;72 Pt A:3–12 doi 10.1016/j.nbd.2014.08.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Deuker MM, Marsh Durban V, Phillips WA, McMahon M. PI3’-kinase inhibition forestalls the onset of MEK1/2 inhibitor resistance in BRAF-mutated melanoma. Cancer Discov 2015;5(2):143–53 doi 10.1158/2159-8290.CD-14-0856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Maertens O, Johnson B, Hollstein P, Frederick DT, Cooper ZA, Messiaen L, et al. Elucidating distinct roles for NF1 in melanomagenesis. Cancer Discov 2013;3(3):338–49 doi 10.1158/2159-8290.CD-12-0313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Xie X, Koh JY, Price S, White E, Mehnert JM. Atg7 Overcomes Senescence and Promotes Growth of BrafV600E-Driven Melanoma. Cancer Discov 2015;5(4):410–23 doi 10.1158/2159-8290.CD-14-1473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lefterov I, Wolfe CM, Fitz NF, Nam KN, Letronne F, Biedrzycki RJ, et al. APOE2 orchestrated differences in transcriptomic and lipidomic profiles of postmortem AD brain. Alzheimers Res Ther 2019;11(1):113 doi 10.1186/s13195-019-0558-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dai J, Johnson ECB, Dammer EB, Duong DM, Gearing M, Lah JJ, et al. Effects of APOE Genotype on Brain Proteomic Network and Cell Type Changes in Alzheimer’s Disease. Front Mol Neurosci 2018;11:454 doi 10.3389/fnmol.2018.00454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Buffington SA, Huang W, Costa-Mattioli M. Translational control in synaptic plasticity and cognitive dysfunction. Annu Rev Neurosci 2014;37:17–38 doi 10.1146/annurev-neuro-071013-014100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kleffman K, Levinson G, Rose IVL, Blumenberg LM, Shadaloey SAA, Dhabaria A, et al. Melanoma-Secreted Amyloid Beta Suppresses Neuroinflammation and Promotes Brain Metastasis. Cancer Discov 2022;12(5):1314–35 doi 10.1158/2159-8290.CD-21-1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Boucher P, Herz J. Signaling through LRP1: Protection from atherosclerosis and beyond. Biochem Pharmacol 2011;81(1):1–5 doi 10.1016/j.bcp.2010.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Vogelstein B, Papadopoulos N, Velculescu VE, Zhou S, Diaz LA, Jr., Kinzler KW. Cancer genome landscapes. Science 2013;339(6127):1546–58 doi 10.1126/science.1235122. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
RNA-Seq data generated for this study have been deposited at the Gene Expression Omnibus under accession numbers GSE208718 and GSE209873. Data from the TCGA-SKCM study were obtained from the Genomic Data Commons: https://portal.gdc.cancer.gov/. All other raw data generated in this study are available upon request from the corresponding author.