Abstract
HinP1I, a type II restriction endonuclease, recognizes and cleaves a palindromic tetranucleotide sequence (G↓CGC) in double-stranded DNA, producing 2 nt 5′ overhanging ends. Here, we report the structure of HinP1I crystallized as one protein monomer in the crystallographic asymmetric unit. HinP1I displays an elongated shape, with a conserved catalytic core domain containing an active-site motif of SDX18QXK and a putative DNA-binding domain. Without significant sequence homology, HinP1I displays striking structural similarity to MspI, an endonuclease that cleaves a similar palindromic DNA sequence (C↓CGG) and binds to that sequence crystallographically as a monomer. Almost all the structural elements of MspI can be matched in HinP1I, including both the DNA recognition and catalytic elements. Examining the protein–protein interactions in the crystal lattice, HinP1I could be dimerized through two helices located on the opposite side of the protein to the active site, generating a molecule with two active sites and two DNA-binding surfaces opposite one another on the outer surfaces of the dimer. A possible functional link between this unusual dimerization mode and the tetrameric restriction enzymes is discussed.
INTRODUCTION
Restriction endonucleases are ubiquitous among prokaryotes. Their principal biological function is to protect the host genome against invading foreign DNA (1). The type II class of restriction endonucleases forms the largest group with more than 3600 members characterized to date (2). The majority of type II enzymes recognize palindromic DNA sequences of 4–8 bp and, in the presence of Mg2+, cleave both strands of the DNA within or close to the recognition sequence, producing fragments with 5′ phosphate and 3′ hydroxyl ends (3).
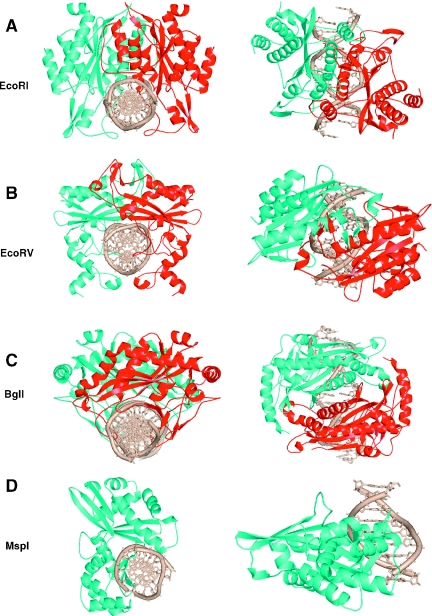
Seventeen crystal structures for type II restriction enzymes have been reported. All these structures share a catalytic architecture consisting of a β-sheet flanked by α helices (4). Consistent with their usually symmetrical DNA recognition and cleavage patterns, these enzymes were observed as dimers in the crystal lattices, with one exception, MspI, which occurs as a monomer (5). Dimerization results in a clamp-shaped molecule, allowing the enzyme to wrap around the DNA duplex and cleave the two DNA strands symmetrically and simultaneously. Mutation studies have revealed that dimerization is necessary for DNA cleavage (6). Interestingly, the dimerization mode appears to correlate with the DNA cleavage pattern (7). Restriction enzymes that cleave DNA giving 5′ overhangs, including EcoRI, BamHI, Bse634I, MunI, BstYI, NgoMIV, BglII, BsoBI, FokI and Cfr10I, share strikingly similar dimerization modes with four structurally equivalent α helices within the core domain dominating the contacts at the dimer interface (Figure 1A) (7–16). In contrast, the common dimerization feature of EcoRV, NaeI, PvuII and HincII (17–20), enzymes that produce blunt ended DNA fragments, is the intertwined contacts that are made by structural elements protruding away from the protein core and crossing over the neighboring subunit (Figure 1B). The BglI structure is the only example of an enzyme that generates DNA fragments with 3′ overhangs (21). Unlike the others, for the BglI–DNA complex, there was only one protein subunit and one DNA strand in the crystallographic asymmetric unit and thus the biologically active homodimer was generated by the 2-fold crystallographic symmetry (Figure 1C). The first exception to the rule of an endonuclease dimer came from the MspI structure (5). An MspI monomer, not a dimer, binds to a palindromic DNA sequence and makes specific (direct or water-mediated) contacts with all 4 bp in the CCGG recognition sequence (Figure 1D).
Figure 1.
Dimeric form of endonucleases: (A) EcoRI–DNA complex (PDB 1CKQ), (B) EcoRV–DNA complex (PDB 1RVA), (C) BglI–DNA complex (PDB 1DMU). (D) Monomeric form of endonuclease: MspI–DNA complex (PDB 1SA3).
Here, we present the structure of HinP1I, at 2.65 Å resolution, which contains a monomer in the crystallographic asymmetric unit. HinP1I is a type II restriction enzyme from Haemophilus infuenzae P1 (22) that recognizes the palindromic DNA sequence G↓CGC and cleaves between the first and second nucleotides, producing 2 nt 5′ overhangs. Although it displays no significant sequence homology to any known endonuclease, HinP1I shares high sequence identity with two hypothetical proteins from Haemophilus somnus 2336 (identity 71%) and Helicobacter pylori J99 (identity 54%). Here we show that, structurally, HinP1I is strikingly similar to MspI (5). The protein–protein interfaces in the HinP1I crystal lattice were examined, and a potential functional link between Hinp1I and the tetrameric restriction enzymes was discussed.
MATERIALS AND METHODS
Cloning
The genes for the HinP1I R-M system were cloned into Escherichia coli by the ‘methylase-selection’ technique (23,24). The DNA sequence of most of the larger PstI fragment (23,24) was determined by a thermocycling procedure using the Circumvent Sequencing Kit (New England Biolabs). A strategy of ‘primer-walking’ was employed, using the sequence obtained in one round to design the primer for the next. The primers, 14–16 bp in length and spaced at intervals of ∼250 nt, were obtained from New England Biolabs. The sequence of both DNA strands was determined.
Purification
Aliquots containing 50 g E.coli RR1 carrying the cloned HinP1I R-M system on two plasmids (R gene on pUC19 and M gene on pACYC184) were resuspended in 200 ml Buffer A (10 mM potassium phosphate, pH 7.4, 1 mM DTT, 0.1 mM Na2EDTA and 5% glycerol) and disrupted by sonication for 4 × 5 min. Following centrifugation at 12 000 r.p.m. for 45 min, the supernatant was applied to a 70 ml Heparin Hyper D column, washed with 200 ml Buffer A, and a 600 ml gradient from 0 to 1.5 M NaCl in Buffer A was applied. HinP1I eluted between 0.75 and 0.9 M NaCl. The pooled fractions were dialyzed against Buffer A containing 50 mM KPO4, pH 7.4, and applied in two separate runs to an 8 ml Source S column. The column was washed with 16 ml Buffer A and then eluted with 100 ml of a gradient from 0 to 1.0 M NaCl in Buffer A. HinP1I eluted between 0.275 and 0.325 M NaCl. Pooled fractions (20 ml) were diluted with 62 ml Buffer B (10 mM Tris–HCl, pH 7.4, 1 mM DTT, 0.1 mM Na2EDTA and 5% glycerol) and applied to an 8 ml Source Q column. Following a wash with 16 ml Buffer B, the column was eluted with a gradient from 0 to 1.0 M NaCl in Buffer B. HinP1I eluted between 0.275 and 0.325 M NaCl, and the 7.5 ml pool was diluted with 15 ml Buffer A and applied in two separate runs to a 3 ml Heparin TSK column. The column was washed with 6 ml Buffer A and eluted with a 60 ml gradient from to 0 to 1.0 M NaCl. HinP1I eluted between 0.6 and 0.75 M NaCl. The final yield of HinP1I in the pooled fractions was 20 mg.
Crystallography
Native crystals of HinP1I were grown at 16°C by the hanging drop method. A 1 μl volume of 12 mg/ml protein in 0.5 M NaCl, 20 mM HEPES (pH 7.0) buffer was mixed with 1 μl of a well solution containing 6% PEG 4000, 0.2 M Ammonium acetate, 100 mM MES, pH 5.5 and 1.2 M NaCl. The mixture was then allowed to equilibrate over the well solution.
Diffraction data were collected at Brookhaven National Laboratory using beamlines X8C and X12C of the National Synchrotron Light Source. Native crystals were briefly soaked in 1 mM K2OsO4 solution, producing osmium (Os)-derivative crystals. One Os-containing crystal was used to collect data at peak wavelength. The native data were collected to a better resolution (Table 1). The data were subsequently reduced and scaled using the DENZO/SCALEPACK (25).
Table 1.
Crystallographic data and refinement statistics
| Parameters | Os-peak | Native |
|---|---|---|
| Wavelength (Å) | 1.1396 | 1.072 |
| Beamline (NSLS) | X12C | X8C |
| Space group | P6322 | |
| Unit cell dimensions (Å) | ||
| a = b | 117.0 | 117.5 |
| c | 113.6 | 115.1 |
| Resolution range (Å) | 26.0–3.30 | 22.0–2.60 |
| Completeness (%) | 93.2 | 99.9 |
| Rmerge = Σ|I − <I>|/ΣI | 0.089 | 0.075 |
| <I/σ> | 10.9 | 18.9 |
| Observed reflections | 56385 | 178050 |
| Unique reflections | 12314 | 14790 |
| Anomalous sites | 1 | — |
| Highest resolutions shell | ||
| Resolution range (Å) | 3.42–3.30 | 2.64–2.60 |
| Completeness (%) | 92.8 | 99.0 |
| Rmerge = Σ|I − <I>|/ΣI | 0.366 | 0.409 |
| <I/σ> | 3.1 | 5.1 |
| Refinement | ||
| Resolution range (Å) | 20.0–2.65 | |
| Molecules/asymmetric unit | 1 | |
| Rfactor = Σ|Fo − Fc|/Σ|Fo| | 0.221 | |
| Rfree (5% of total data) | 0.252 | |
| Root-mean-square deviation from ideal | ||
| Bond lengths (Å) | 0.007 | |
| Bond angels (°) | 1.5 | |
| Dihedrals (°) | 23.5 | |
| Improper (°) | 0.7 | |
Initial phases were obtained by single isomorphous replacement using the anomalous scattering phasing method. SOLVE (26) readily determined the single Os site. The phases were then greatly improved by maximum likelihood density modification using RESOLVE (27,28). Owing to the good quality of the electron density map, RESOLVE was able to auto-build 48% of the HinP1I model with side chains and an additional 28% as main chain atoms. All subsequent model rebuilding was performed using O (29). Crystallographic refinement employed the program CNS (30) against the native data set at 2.65 Å resolution (Table 1).
RESULTS
Sequence analysis
Analysis of the sequence revealed two complete open reading frames, ORF1 and ORF2, and the beginning of a third, all in the same orientation. ORF1 encoded a 322 residue protein belonging to the 5-methylcytosine family of DNA-methyltransferases (m5C-MTases) that was shown to encode the M.HinP1I methyltransferase. ORF2 encoded a 247 residue protein that seemed likely to encode HinP1I. This assignment was confirmed by N-terminal amino acid sequence analysis of the purified HinP1I endonuclease. The first 22 residue of HinP1I was found to be MNLVELGSKTAKDGFKNEKDIA…, exactly matching those coded at the start of ORF2.
The third, incomplete, ORF began 30 bp downstream of the end of the hinP1IR gene, and traversed the PstI site, beyond which no sequence was obtained. This ORF codes for the first 100 amino acids, or so, of the highly conserved enzyme valyl-tRNA synthetase, and so it is likely to be the beginning of the H.influenzae P1 valS gene. The HindIII R-M system of H.influenzae Rd also occurs immediately upstream of the valS gene (31) in the genome of that bacterium, and so the HinP1I and HindIII systems might be considered allelic. The two systems are entirely unrelated and so are unlikely to have diverged from a common ancestor. They differ in recognition sequence specificity (HindIII: AAGCTT; HinP1I: GCGC); in gene organization (HindIII: R gene then M; HinP1I: M gene then R); and in methyltransferase activity (M.HindIII: N6-adenine MTase; M.HinP1I: 5-cytosine MTase). The amino acid sequences of the HindIII and HinP1I endonucleases are completely dissimilar, and so too are those of the methyltransferases (32). The proximity of both systems to the valS gene would seem to be a coincidence, then, or a genetic convenience, rather than a sign of common origin.
Overall structure
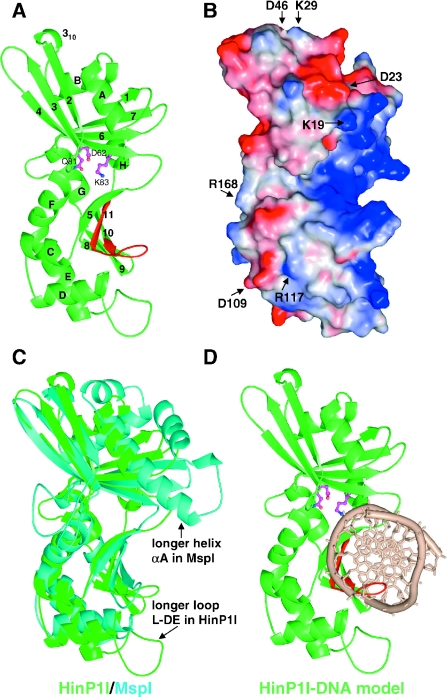
HinP1I comprises 247 amino acids, the first six N-terminal residues of which are invisible in the electron density map. In overall topology, HinP1I belongs to the α/β protein class, with elongated dimensions of 68 × 33 × 32 Å (Figure 2A), and contains two sets of β-sheets surrounded by eight α helices. The upper β-sheet is a mixed six-stranded (β1-7-6-4-3-2) sheet sandwiched by four α helices (αA and αB on one side, and αH and αG on the other). The lower sheet contains five anti-parallel strands (β5-11-10-9-8), with the left side of the sheet packing against five α helices (αC, αD, αE, αF and αG) and the right side forming a concave basic surface, which is the putative DNA-binding site (Figure 2B).
Figure 2.
Structure of HinP1I (A) Ribbon representation. Helices are labeled as letters A–H and strands labeled as numbers 1–11 from N- to C-termini. Putative catalytic residues are shown in magenta color. The C-terminal β-hairpin, colored in red, contains the invariant residues (presumably involved in DNA base specific interactions) between HinP1I and MspI. (B) Molecular surface representation, shown in the same orientation of (A). The surface is colored blue for positive, red for negative and white for neutral. The basic (blue) concave surface, shown on the right side of the molecule, represents a DNA-binding surface. Several conserved charged residues (labeled) are scattered throughout the surface. R168, shown on the left side of the molecule, represents a potential dimer interface (see Figure 5B). (C) Superimposition of HinP1I (green) and MspI (cyan). (D) A model of HinP1I monomer docked with DNA.
HinP1I shares striking structural similarity to MspI
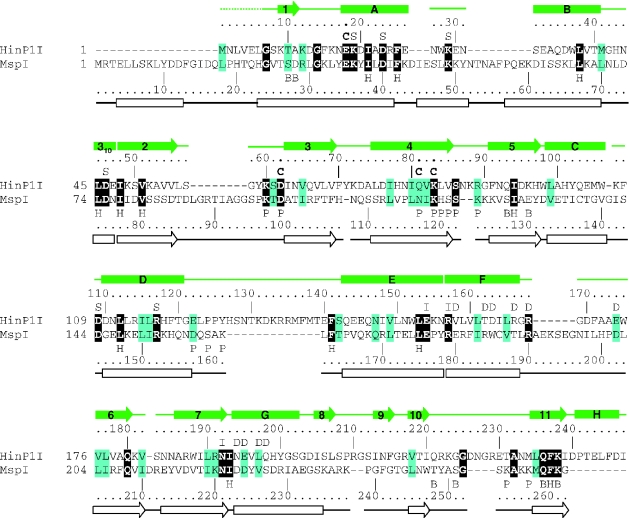
As revealed by a Dali structure comparison (33), a pairwise least squares superposition of HinP1I and MspI gives a root-mean-square deviation of 3.1 Å over 195 Cα pairs (Z-score of 15.2). This close structural similarity is not evident at the sequence level: guided by structural alignment, HinP1I shows only 14% amino acid sequence identity (34 out of 247 residues) to MspI (Figure 3). Among the 34 identical residues, 12 with hydrophobic side chains intercalate to form the hydrophobic core of the molecule; 3 are catalytic (E18, D62 and K83); 2 are potentially involved in specific DNA contacts (Q236 and K238), and another 2 in DNA phosphate contacts (K60 and S86); 3 glycines (G7, G14 and G225) might be structurally important for short turns; 2 charged residues form an intra-molecular salt bridge (E154 and R157); and 2 polar residues are involved in intra-molecular hydrogen bonds (Q180 and N192). Seven additional conserved charged residues of unknown function (K19, D23, K29, D46, D109, R117 and R168) are scattered upon the surface of the molecule (Figure 2B).
Figure 3.
Structure-based sequence alignment of HinP1I and MspI. The residue number and secondary structural elements of HinP1I and MspI are shown, respectively, above and below the aligned sequences. The amino acids highlighted are invariant (white letter against black background) and conserved (black against cyan). The letters immediately above or below the sequences indicate the structural and suggested functional roles of the corresponding residues: ‘B’ indicates DNA base interaction, ‘C’ indicates catalysis, ‘D’ indicates dimer interface, ‘H’ indicates hydrophobic core, ‘I’ indicates intra-molecule interaction, ‘P’ indicates DNA phosphate interaction and ‘S’ indicates conserved surface residues (whose locations are shown in Figure 2B). The protein–DNA interactions involving main chain atoms are not indicated.
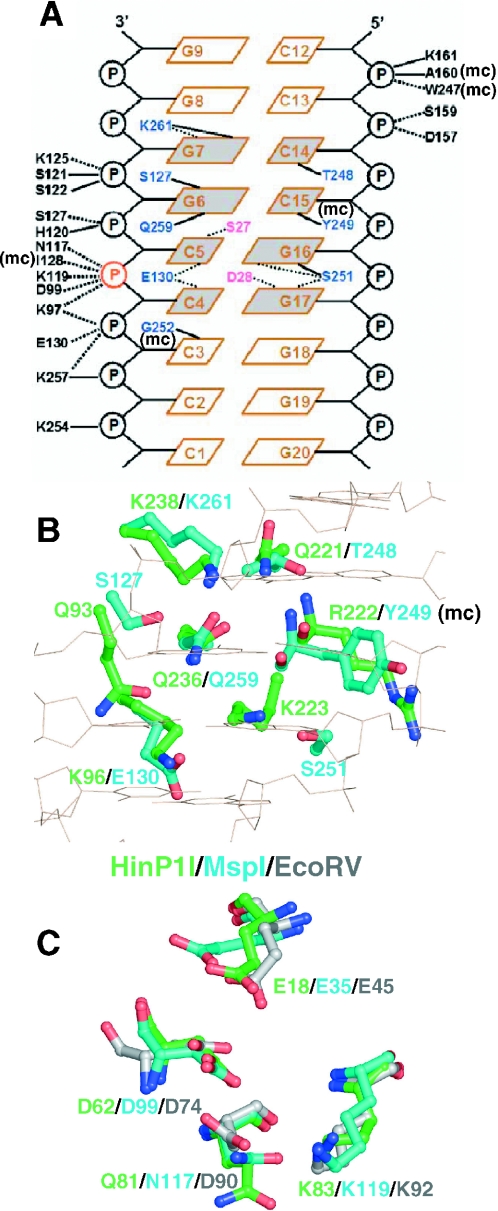
Almost all the structural elements of HinP1I can be matched in MspI (Figure 2C), including both the DNA recognition elements and the catalytic elements. This near-perfect match allowed us to create a model of HinP1I bound to DNA (Figure 2D). Using the coordinates for the complex of MspI–DNA (see Figure 1D), we superimposed the protein components and then positioned the DNA over the basic concave surface. The resulting model showed that HinP1I could contact the DNA without physical distortion of either the protein or the DNA component. Besides an extra N-terminal helix in MspI and an extra C-terminal helix in HinP1I (Figure 3), the most obvious differences between the two structures are helix αA, which is three helical turns longer in MspI and inserts into the minor groove of DNA to make water-mediated hydrogen bonds with the recognition sequence, and the loop between helices αD and αE, which is 13 residue longer in HinP1I and could adopt a closed conformation upon association with DNA (Figure 2C). The shorter corresponding loop in MspI (residues D157–K161) is involved in phosphate contacts outside of the recognition sequence (Figure 4A).
Figure 4.
Structural similarity between HinP1I and MspI (A) Schematic diagram of MspI–DNA interactions, reproduced and modified from Xu et al. (5). Solid lines indicate direct hydrogen bonds, dotted lines indicate water-mediated hydrogen bonds and ‘mc’ indicates interaction involving main chain atom. (B) Residues potentially important for DNA base specific recognition: superimposition of residues of HinP1I (green) and MspI (cyan). (C) Residues potentially important for catalysis: superimposition of active site residues of HinP1I (green), MspI (cyan) and EcoRV (grey). Four residues belong to a common motif of E……PDX15DXK (EcoRV), E……SDX18QXK (HinP1I) and E……TDX17NXK (MspI).
HinP1I and MspI recognize similar 4 bp double-stranded DNA sequences (G↓CGC and C↓CGG, respectively) and cleave them in the same positions. MspI makes specific contacts with all 8 nt within its recognition sequence (5), one-half of the sequence being recognized mainly by direct hydrogen bonds (5/6 interactions), and the other half mainly by water-mediated hydrogen bonds (6/7 interactions) (Figure 4A). Of the six residues in MspI that make direct hydrogen bonds (S127, T248, S251, Q259, K261 and the main chain carbonyl oxygen of Y249), four have equivalents in HinP1I: Q221, Q236, K238 and the main chain carbonyl of R222 (Figure 4B). These four residues are located in the C-terminal hairpin strands β10 and β11. The loop between these two strands, which is four residues longer in HinP1I than in MspI, is flexible and characterized by high thermal factors, but it might become ordered upon DNA binding, and positioned to make DNA contacts. Residues Q236 and K238 flank the most highly conserved segment between HinP1I and MspI, three consecutive invariant residues, QFK, present at the extreme C-terminus of MspI (Figure 3). Two residues that make phosphate contacts are also invariant: K97 in MspI (K60 in HinP1I) that contacts the scissile phosphate and the succeeding 5′ phosphate; and S122 in MspI (S86 in HinP1I) that contacts the second phosphate 3′ to the scissile phosphate (Figure 4A).
A common catalytic site motif among restriction enzymes is characterized as PDXn(D/E)XK, the consensus residues of which cluster around the scissile phosphate. The catalytic residues of EcoRV, D74, D90 and K92 align spatially with HinP1I residues D62, Q81 and K83, in which glutamine occurs in place of the second acidic residue (Figure 4C). The catalytic motif of HinP1I appears to be SDX18QXK, an unusual unique feature that is also represented in MspI as TDX17NXK, in which enzyme asparagine takes the place of the second acidic residue (5). In addition, E18 of helix αA in HinP1I and E35 in MspI are superimposable on E45 in EcoRV, and represent the third acidic residue important for catalysis (34).
Unexpected dimerization mode in crystal lattice
Because HinP1I crystallized as a single protomer in the crystallographic asymmetric unit and restriction endonucleases that recognize symmetric DNA sequences generally are thought to do so as homodimers, we examined all possible protein–protein interfaces in the crystal lattice based on the change in solvent accessible surface area (ΔASA) when going from a monomeric to a dimeric state. The largest change in ΔASA was found for a pair of the protomers related by a 2-fold crystallographic axis of rotation through two helices (αF and αG) located on the opposite side of the concave basic surface (Figure 5A). These helices are arranged as a parallel four-helix bundle at the dimer interface and participate in network of hydrogen bonds, van der Waals interactions and hydrophobic contacts, and electrostatic interactions (Figure 5B). Involved in this network are two arginines: R166 and R168. R168 interacts with E195; the corresponding residues in MspI are R190 and D224, a conservative change. R166 interacts with two residues, E174 in the loop following helix αF and N194 in helix αG. Although the set of three residues in the E174–R166–N194 interaction (E–R–N) is not present in MspI, there is covariation that suggests a conserved interaction: the side chains of T162 and R166 point out from the same surface of helix αF (Figure 5B); the corresponding positions in MspI are R184 and T188. Therefore, an arginine is present in MspI at a position probably still maintaining the ion-pair interaction (D202–R184–D223).
Figure 5.
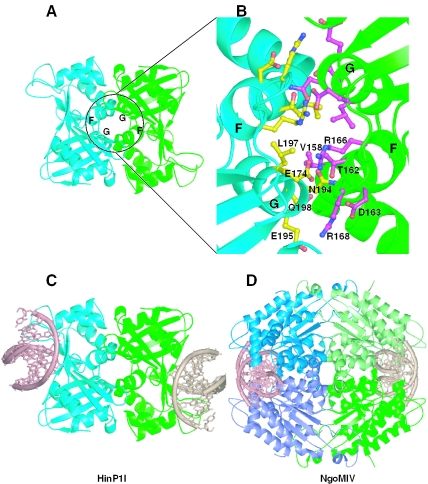
Potential link between the dimeric form of HinP1I and the tetrameric restriction enzymes (A) The HinP1I dimer interface mediated by a crystallographic 2-fold symmetry. (B) The enlarged dimer interface of HinP1I consists of residues from helices αG and αF. (C) A model of HinP1I dimer docked with two DNA molecules. (D) Structure of a tetramer of the NgoMIV restriction endonuclease in complex with two DNA molecules (PDB 1FIU). Two primary dimers (blue and green) are positioned back-to-back to each other.
We consider this potential dimer interface to be significant; approximately 1900 Å2 of solvent-accessible surface is buried by it (35). This area is comparable with those in other restriction enzymes, which range from ∼800 to 4000 Å2. If such a dimer interaction does exist in solution, it positions the catalytic sites and DNA-binding sites on opposite surfaces of the dimer facing away from one another (Figure 5C), rather than toward one another as occurs in all other endonuclease homodimers that have characterized.
The functional significance of the crystal symmetry (CS)-related HinP1I dimer (Figure 5C) is unclear, but it might enable HinP1I to link in a back-to-back fashion similar to that observed in tetrameric restriction enzymes, such as Cfr10I, NgoMIV and Bse634I (Figure 5D) (10,12,16,36). The tetrameric arrangement of these enzymes can be considered to be dimer–dimer interactions. The primary, face-to-face, dimerization is similar to that of type II restriction enzymes, such as EcoRI, enabling recognition of a symmetric sequence and simultaneous double-strand DNA cleavage. These primary dimers then position back-to-back to form a tetramer, which binds to two copies of DNA. Kinetic studies have shown that the tetrameric architecture is of functional importance, and back-to-back dimer–dimer interaction is required for effective DNA cleavage (10,36). Similar to these tetrameric enzymes, HinP1I could also interact with two recognition sites facilitated by its unique dimerization mode. In addition, processing at these sites may be cooperative, and HinP1I may also require two recognition sites to be present for effective catalysis.
Another possibility is that the dimeric form of HinP1I could allow the formation of a large enzyme–substrate network with high molecular weight DNA, such as demonstrated for Serratia nuclease (37). Although the two subunits of Serratia nuclease function independently of each other, they bind simultaneously to a single macromolecular DNA, forming a large network between the enzyme and the substrate. The CS-mediated homodimeric HinP1I may serve the same function.
HinP1I occurs as a monomer in solution
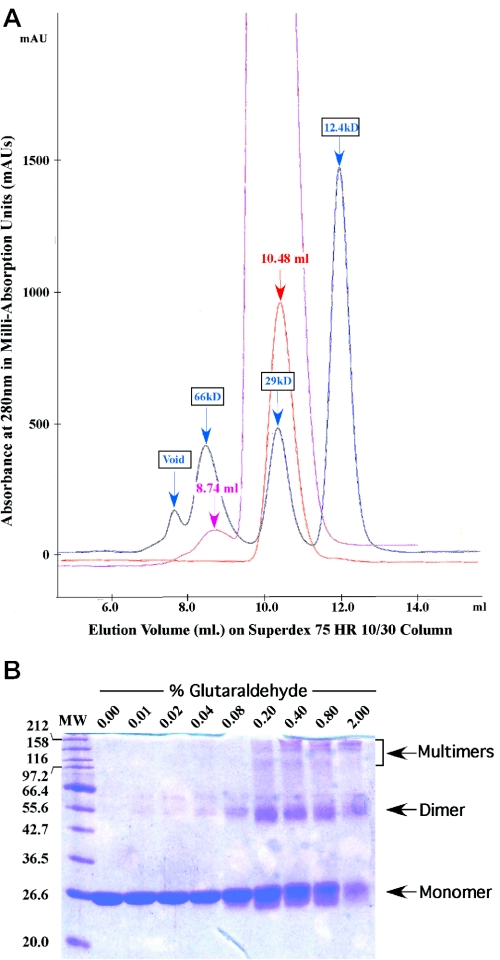
While a monomer/dimer equilibrium was observed for MspI in analytical ultracentrifuge experiments (5), we used several alternative ways to establish whether HinP1I exists as a monomer, dimer or tetramer in solution. First, gel filtration suggests that HinP1I has an apparent molecular weight of a monomer. On a Superdex75 column (Pharmacia), Hinp1I was loaded onto the column at an initial concentration of 3.7 mg/ml and eluted at an elution volume of 10.48, equivalent to a 28 kDa globular protein (Figure 6A). This is similar to the calculated molecular weight of 28.75 kDa. Only at a loading concentration of ∼75 mg/ml was a small fraction of HinP1I eluted at a size close to that of a dimer at ∼59 kDa (Figure 6A).
Figure 6.
HinP1I solution properties (A) Overlay of chromatographs from elution experiments on a Superdex 75 HR 10/30 sizing column. Blue: elution of three molecular weight standard proteins, cytochrome c (12.4 kDa), carbonic anhydrase (29.0 kDa) and BSA (66.0 kDa). Red: elution of HinPI1 after loading 100 μl at ∼3.7 mg/ml concentration. Magenta: elution of HinPI1 after loading 100 μl at ∼75 mg/ml concentration. (B) HinPI1 in the presence of increasing amounts of glutaraldehyde. HinPI1 (∼1.9 mg/ml) was treated with glutaraldehyde and incubated at room temperature for 1 h (total volume 20 μl). The reaction was stopped by the addition of 10 μl of 1 M glycine. An aliquot of 15 μl of 3× loading buffer was added to these samples and subsequently 20 μl of this final solution was loaded onto a 13% SDS–PAGE gel (Coomassie stain).
Second, dynamic light scattering measurements at a concentration of 3.7 mg/ml, at temperatures of 4, 16 and 25°C, suggested that the hydrodynamic radius of HinP1I is 2.7–2.8 nm, which would correspond to a 31–36 kDa globular protein (Table 2). This dimension (2 × 2.8 = 5.6 nm) is between the shorter (3.3 nm) and the longer (6.8 nm) dimensions of the monomer (Figure 2). Interestingly, the size increased as a function of protein concentration to 56 kDa (close to a dimer at 8.0 mg/ml) and 183 kDa (close to a tetramer at ∼75 mg/ml).
Table 2.
Summary of the dynamic light scattering experiments
| [c] (mg/ml) | DTa | RH (nm) | Mw (kDa) | Polydispersity (%) | Temperature (°C) |
|---|---|---|---|---|---|
| 3.6 | 411 | 2.8 | 36 | 48.5 | 4 |
| 3.6 | 644 | 2.7 | 31 | 48.9 | 16 |
| 3.6 | 777 | 2.8 | 36 | 42.9 | 25 |
| 8.0 | 506 | 3.4 | 56 | 24.6 | 16 |
| ∼75 | 301 | 5.5 | 183 | 31.5 | 15 |
aThe diffusion constant, expressed in units of 10−9 cm2/s, DT = KB T/6 π η RH where KB is the Boltzmann constant, T is the absolute temperature in Kelvin, η is the solvent viscosity (5% glycerol used in the experiment) and RH is the hydrodynamic radius of the average scattering particle.
Third, a portion of HinP1I formed a covalent dimer after cross-linking by glutaraldehyde (Figure 6B). At higher concentrations of glutaraldehyde, a second band appeared at a molecular weight close to tetramer. Taken together, these data suggest that HinP1I occurs predominantly as a monomer in solution, but can form a dimer or a higher order molecule weight complex at high protein concentrations.
DISCUSSION
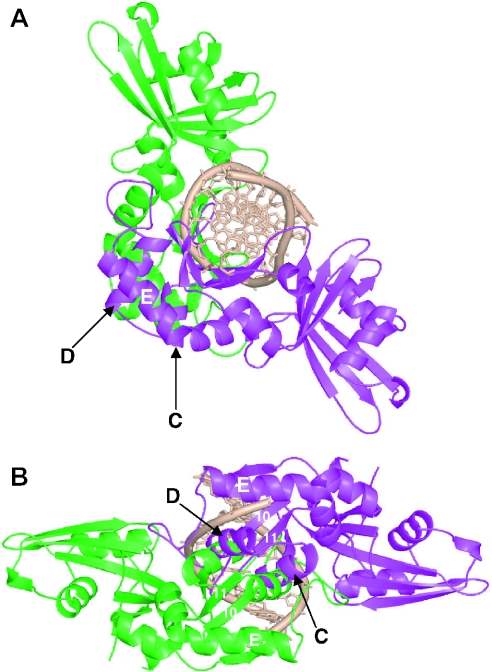
We do not know whether the solution properties of HinP1I are dependent on the presence of substrate DNA, although, most of the structurally characterized type II endonuclease–DNA complexes comprise a protein dimer of identical subunits and a single copy of a double-stranded oligonucleotide, with one active site for each DNA strand. Certain type IIS enzymes, such as FokI, dimerize transiently during catalysis (6,15,38) and it seems reasonable to assume that HinP1I would, too. We attempted to create a model of the HinP1I dimer bound to a single copy of DNA. To prepare the model, we rotated the HinP1I monomer (green) along the 2-fold symmetry of the palindromic DNA and generated the second monomer (magenta) with its active site for the other strand (Figure 7). The most serious steric clash occurs between helices αC and αD of one monomer and their 2-fold symmetry-related counterparts of the other monomer. We imagine that upon associating with DNA, helices αC and αD could adopt a different non-clashing conformation that would allow the formation of an anti-parallel four-helix bundle in the dimer interface. Interestingly, helix αD contains two invariant residues between HinP1I (D109 and R117) and MspI (D144 and R152) (see Figures 3), whose positions are exposed on the monomer surface (see Figure 2B), and which might therefore participate in face-to-face dimerization much as such residues do in the back-to-back dimerization discussed earlier.
Figure 7.
A hypothetical model of a HinP1I dimer bound with a single copy of DNA. The two monomers of Hinp1I are shown in green and magenta. (A) The presentation of green monomer of Hinp1I is similar to that shown in Figure 2D. (B) A view looking into the DNA major groove.
Like FokI, BfiI is a type IIS endonuclease that forms a homodimer as it makes double-strand breaks in DNA (39). However, the BfiI homodimer, and its related Nuc enzyme from Salmonella typhimurium (40), has only one active site present at the dimer interface, and this single active site acts sequentially on two DNA strands (41). Hence, it was proposed that, after cutting one strand, a rearrangement of either the protein and/or the DNA in the BfiI–DNA complex must switch the active site to the other strand (41). HinP1I (and possibly its structural homolog MspI) could in principle operate in the same sequential way, binding to palindromic recognition sequence as a monomer and cleaving first one DNA strand, and then the other. If so, it could likely produce considerable nicked intermediate, such as the monomeric mismatch repair endonuclease MutH (42), for the introduction of a nick into the target strand; however, we found it does not (G. G. Wilson, unpublished data). On the other hand, HinP1I could cleave DNA processively, i.e. repetitively continuing catalytic function without dissociating from the DNA. Clearly, further biochemical and structural studies of HinP1I are warranted.
Acknowledgments
The study was partly supported by US Public Health Service grant GM49245 (X.C., Z.Y. and J.R.H.) and New England Biolabs (R.J.R., R.M. and G.G.W.). Data for this study were measured at beamlines X12C and X8C of the National Synchrotron Light Source. Financial support for the beamlines comes principally from the Offices of Biological and Environmental Research and of Basic Energy Sciences of the US Department of Energy, and from the National Center for Research Resources of the National Institutes of Health. Figures were drawn using the program Pymol, a user-sponsored molecular modeling system with an OPEN-SOURCE foundation (http://pymol.sourceforge.net). Atomic coordinates have been deposited in the PDB with accession no. 1YNM. The GenBank sequence accession no. is AY849924. Funding to pay the Open Access publication charges for this article was provided by New England Biolabs.
Conflict of interest statement. None declared.
REFERENCES
- 1.Wilson G.G., Murray N.E. Restriction and modification systems. Annu. Rev. Genet. 1991;25:585–627. doi: 10.1146/annurev.ge.25.120191.003101. [DOI] [PubMed] [Google Scholar]
- 2.Roberts R.J., Vincze T., Posfai J., Macelis D. REBASE: restriction enzymes and DNA methyltransferases. Nucleic Acids Res. 2005;33:D230–D232. doi: 10.1093/nar/gki029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Roberts R.J., Belfort M., Bestor T., Bhagwat A.S., Bickle T.A., Bitinaite J., Blumenthal R.M., Degtyarev S.K., Dryden D.T.F., Dybvig K., et al. A nomenclature for restriction enzymes, DNA methyltransferases, homing endonucleases and their genes. Nucleic Acids Res. 2003;31:1805–1812. doi: 10.1093/nar/gkg274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pingoud A., Jeltsch A. Structure and function of type II restriction endonucleases. Nucleic Acids Res. 2001;29:3705–3727. doi: 10.1093/nar/29.18.3705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Xu Q.S., Kucera R.B., Roberts R.J., Guo H.C. An asymmetric complex of restriction endonuclease MspI on its palindromic DNA recognition site. Structure. 2004;12:1741–1747. doi: 10.1016/j.str.2004.07.014. [DOI] [PubMed] [Google Scholar]
- 6.Bitinaite J., Wah D.A., Aggarwal A.K., Schildkraut I. FokI dimerization is required for DNA cleavage. Proc. Natl Acad. Sci. USA. 1998;95:10570–10575. doi: 10.1073/pnas.95.18.10570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Deibert M., Grazulis S., Janulaitis A., Siksnys V., Huber R. Crystal structure of MunI restriction endonuclease in complex with cognate DNA at 1.7 A resolution. EMBO J. 1999;18:5805–5816. doi: 10.1093/emboj/18.21.5805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kim Y.C., Grable J.C., Love R., Greene P.J., Rosenberg J.M. Refinement of Eco RI endonuclease crystal structure: a revised protein chain tracing. Science. 1990;249:1307–1309. doi: 10.1126/science.2399465. [DOI] [PubMed] [Google Scholar]
- 9.Newman M., Strzelecka T., Dorner L.F., Schildkraut I., Aggarwal A.K. Structure of Bam HI endonuclease bound to DNA: partial folding and unfolding on DNA binding. Science. 1995;269:656–663. doi: 10.1126/science.7624794. [DOI] [PubMed] [Google Scholar]
- 10.Grazulis S., Deibert M., Rimseliene R., Skirgaila R., Sasnauskas G., Lagunavicius A., Repin V., Urbanke C., Huber R., Siksnys V. Crystal structure of the Bse634I restriction endonuclease: comparison of two enzymes recognizing the same DNA sequence. Nucleic Acids Res. 2002;30:876–885. doi: 10.1093/nar/30.4.876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Townson S.A., Samuelson J.C., Vanamee E.S., Edwards T.A., Escalante C.R., Xu S.Y., Aggarwal A.K. Crystal structure of BstYI at 1.85 A resolution: a thermophilic restriction endonuclease with overlapping specificities to BamHI and BglII. J. Mol. Biol. 2004;338:725–733. doi: 10.1016/j.jmb.2004.02.074. [DOI] [PubMed] [Google Scholar]
- 12.Deibert M., Grazulis S., Sasnauskas G., Siksnys V., Huber R. Structure of the tetrameric restriction endonuclease NgoMIV in complex with cleaved DNA. Nature Struct. Biol. 2000;7:792–799. doi: 10.1038/79032. [DOI] [PubMed] [Google Scholar]
- 13.Lukacs C.M., Kucera R., Schildkraut I., Aggarwal A.K. Understanding the immutability of restriction enzymes: crystal structure of BglII and its DNA substrate at 1.5 A resolution. Nature Struct. Biol. 2000;7:134–140. doi: 10.1038/72405. [DOI] [PubMed] [Google Scholar]
- 14.van der Woerd M.J., Pelletier J.J., Xu S., Friedman A.M. Restriction enzyme BsoBI–DNA complex: a tunnel for recognition of degenerate DNA sequences and potential histidine catalysis. Structure. 2001;9:133–144. doi: 10.1016/s0969-2126(01)00564-0. [DOI] [PubMed] [Google Scholar]
- 15.Wah D.A., Bitinaite J., Schildkraut I., Aggarwal A.K. Structure of FokI has implications for DNA cleavage. Proc. Natl Acad. Sci. USA. 1998;95:10564–10569. doi: 10.1073/pnas.95.18.10564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bozic D., Grazulis S., Siksnys V., Huber R. Crystal structure of Citrobacter freundii restriction endonuclease Cfr10I at 2.15 A resolution. J. Mol. Biol. 1996;255:176–186. doi: 10.1006/jmbi.1996.0015. [DOI] [PubMed] [Google Scholar]
- 17.Winkler F.K., Banner D.W., Oefner C., Tsernoglou D., Brown R.S., Heathman S.P., Bryan R.K., Martin P.D., Petratos K., Wilson K.S. The crystal structure of EcoRV endonuclease and of its complexes with cognate and non-cognate DNA fragments. EMBO J. 1993;12:1781–1795. doi: 10.2210/pdb4rve/pdb. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Huai Q., Colandene J.D., Topal M.D., Ke H. Structure of NaeI–DNA complex reveals dual-mode DNA recognition and complete dimer rearrangement. Nature Struct. Biol. 2001;8:665–669. doi: 10.1038/90366. [DOI] [PubMed] [Google Scholar]
- 19.Horton J.R., Cheng X. PvuII endonuclease contains two calcium ions in active sites. J. Mol. Biol. 2000;300:1049–1056. doi: 10.1006/jmbi.2000.3938. [DOI] [PubMed] [Google Scholar]
- 20.Horton N.C., Dorner L.F., Perona J.J. Sequence selectivity and degeneracy of a restriction endonuclease mediated by DNA intercalation. Nature Struct. Biol. 2002;9:42–47. doi: 10.1038/nsb741. [DOI] [PubMed] [Google Scholar]
- 21.Newman M., Lunnen K., Wilson G., Greci J., Schildkraut I., Phillips S.E. Crystal structure of restriction endonuclease BglI bound to its interrupted DNA recognition sequence. EMBO J. 1998;17:5466–5476. doi: 10.1093/emboj/17.18.5466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shen S., Li Q., Yan P., Zhou B., Ye S., Lu Y., Wang D. Restriction endonucleases from three strains of Haemophilus influenzae. Sci. Sin. 1980;23:1435–1442. [PubMed] [Google Scholar]
- 23.Barsomian J.M., Card C.O., Wilson G.G. Cloning of the HhaI and HinPI restriction–modification systems. Gene. 1988;74:5–7. doi: 10.1016/0378-1119(88)90237-5. [DOI] [PubMed] [Google Scholar]
- 24.Lunnen K.D., Barsomian J.M., Camp R.R., Card C.O., Chen S.Z., Croft R., Looney M.C., Meda M.M., Moran L.S., Nwankwo D.O., Slatko B.E., Van Cott E.M., Wilson G.G. Cloning type II restriction and modification genes. Gene. 1988;74:25–32. doi: 10.1016/0378-1119(88)90242-9. [DOI] [PubMed] [Google Scholar]
- 25.Otwinowski Z., Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 26.Terwilliger T.C., Berendzen J. Automated MAD and MIR structure solution. Acta Crystallogr. Sect. D. 1999;55:849–861. doi: 10.1107/S0907444999000839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Terwilliger T.C. Maximum likelihood density modification. Acta Crystallogr. Sect. D. 2000;56:965–972. doi: 10.1107/S0907444900005072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Terwilliger T.C., Berendzen J. Automated main-chain model-building by template-matching and iterative fragment extension. Acta Crystallogr. Sect. D. 2002;59:34–44. doi: 10.1107/S0907444902018036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jones T.A., Zou J.Y., Cowan S.W., Kjeldgaard Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr. Sect. A. 1991;47:110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- 30.Brunger A.T., Adams P.D., Clore G.M., DeLano W.L., Gros P., Grosse-Kunstleve R.W., Jiang J.S., Kuszewski J., Nilges M., Pannu N.S., et al. Crystallography & NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr. D Biol. Crystallogr. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 31.Fleischmann R.D., Adams M.D., White O., Clayton R.A., Kirkness E.F., Kerlavage A.R., Bult C.J., Tomb J.-F., Dougherty B.A., Merrick J.M., et al. Whole-genome random sequencing and assembly of Haemophilus influenzae Rd. Science. 1995;269:496–512. doi: 10.1126/science.7542800. [DOI] [PubMed] [Google Scholar]
- 32.Nwankwo D.O., Moran L.S., Slatko B.E., Waite-Rees P.A., Dorner L.F., Benner J.S., Wilson G.G. Cloning, analysis and expression of the HindIII R-M-encoding genes. Gene. 1994;150:75–80. doi: 10.1016/0378-1119(94)90861-3. [DOI] [PubMed] [Google Scholar]
- 33.Holm L., Sander C. Protein structure comparison by alignment of distance matrices. J. Mol. Biol. 1993;233:123–138. doi: 10.1006/jmbi.1993.1489. [DOI] [PubMed] [Google Scholar]
- 34.Groll D.H., Jeltsch A., Selent U., Pingoud A. Does the restriction endonuclease EcoRV employ a two-metal-ion mechanism for DNA cleavage? Biochemistry. 1997;36:11389–11401. doi: 10.1021/bi9705826. [DOI] [PubMed] [Google Scholar]
- 35.Lee B., Richards F.M. The interpretation of protein structures: estimation of static accessibility. J. Mol. Biol. 1971;55:379–400. doi: 10.1016/0022-2836(71)90324-x. [DOI] [PubMed] [Google Scholar]
- 36.Siksnys V., Skirgaila R., Sasnauskas G., Urbanke C., Cherny D., Grazulis S., Huber R. The Cfr10I restriction enzyme is functional as a tetramer. J. Mol. Biol. 1999;291:1105–1118. doi: 10.1006/jmbi.1999.2977. [DOI] [PubMed] [Google Scholar]
- 37.Franke I., Meiss G., Pingoud A. On the advantage of being a dimer, a case study using the dimeric Serratia nuclease and the monomeric nuclease from Anabaena sp. strain PCC 7120. J. Biol. Chem. 1999;274:825–832. doi: 10.1074/jbc.274.2.825. [DOI] [PubMed] [Google Scholar]
- 38.Vanamee E.S., Santagata S., Aggarwal A.K. FokI requires two specific DNA sites for cleavage. J. Mol. Biol. 2001;309:69–78. doi: 10.1006/jmbi.2001.4635. [DOI] [PubMed] [Google Scholar]
- 39.Lagunavicius A., Sasnauskas G., Halford S.E., Siksnys V. The metal-independent type IIs restriction enzyme BfiI is a dimer that binds two DNA sites but has only one catalytic centre. J. Mol. Biol. 2003;326:1051–1064. doi: 10.1016/s0022-2836(03)00020-2. [DOI] [PubMed] [Google Scholar]
- 40.Stuckey J.A., Dixon J.E. Crystal structure of a phospholipase D family member. Nature Struct. Biol. 1999;6:278–284. doi: 10.1038/6716. [DOI] [PubMed] [Google Scholar]
- 41.Sasnauskas G., Halford S.E., Siksnys V. How the BfiI restriction enzyme uses one active site to cut two DNA strands. Proc. Natl Acad. Sci. USA. 2003;100:6410–6415. doi: 10.1073/pnas.1131003100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ban C., Yang W. Structural basis for MutH activation in E.coli mismatch repair and relationship of MutH to restriction endonucleases. EMBO J. 1998;17:1526–1534. doi: 10.1093/emboj/17.5.1526. [DOI] [PMC free article] [PubMed] [Google Scholar]