Abstract
A unique aspect of meiosis is the segregation of homologous chromosomes at the meiosis I division. Homologs are physically connected prior to segregation by crossing over between nonsister chromatids. Crossovers arise from the repair of induced double-strand breaks (DSBs). In many organisms, more DSBs are formed than crossovers in a given nucleus. It has been previously suggested that repair of DSBs to noncrossover recombination products aids homolog alignment. Here we explore how two modes of the meiotic recombination pathway (crossover and noncrossover) and meiotic telomere reorganization contribute to the pairing and close juxtaposition of homologous chromosomes in budding yeast. We found that intermediates in the DSB repair pathway leading to both crossover and noncrossover recombination products contribute independently to close, stable homolog juxtaposition (CSHJ), a measurable state of homolog pairing. Analysis of the ndj1Δ mutant indicates that the effect of meiotic telomere reorganization on CSHJ is exerted through recombination intermediates at interstitial chromosomal loci, perhaps through the noncrossover branch of the DSB repair pathway. We suggest that transient, early DSB-initiated interactions, including those that give rise to noncrossovers, are important for homolog recognition and juxtaposition.
Keywords: Homolog, meiosis, pairing, recombination, telomeres
Meiosis is the process in which a parent diploid cell undergoes two rounds of chromosome segregation, after one round of replication, to yield four haploid gametes. A universal component of meiosis I is the reductional segregation of homologous chromosomes. Nondisjunction, or improper segregation of homologs, at this stage can lead to gamete aneuploidy. Three factors contribute to the tension required for the reductional segregation of homologs at anaphase I in most organisms: (1) crossing over between homologous chromosomes, (2) cohesion between sister chromatids, and (3) monopolar spindle attachment of sister chromatids at metaphase (Page and Hawley 2003).
Crossing over involves the reciprocal exchange of chromosome arms and can be visualized at late stages of meiotic prophase as chiasmata. Such exchange events are a manifestation of double-strand break (DSB)-promoted homologous recombination. Formation and repair of meiotically induced DSBs involve both DSB repair enzymes and meiosis-specific factors (for reviews, see Zickler and Kleckner 1999; Keeney 2001). Meiosis-specific factors bias recombination between homologous chromosomes instead of sister chromatids (Schwacha and Kleckner 1997) and ensure that at least one crossover occurs between every homolog pair (for review, see Bishop and Zickler 2004).
A key event in crossover formation is the recognition and pairing of homologous chromosomes. An outstanding question is what brings homologs together in meiosis. Is it early molecular events, late-forming molecules in which homologs are clearly linked by chemical or hydrogen-bonding interactions, or the assembly of specialized inter-homology chromosome structures? The emerging answer is that multiple mechanisms contribute to the recognition and stable pairing of homologous chromosomes (for review, see Zickler and Kleckner 1998; Burgess 2004). Interplay of homolog-pairing mechanisms can differ greatly; one mechanism may be dominant in one organism and missing in another. For example, Caenorhabditis elegans and female Drosophila melanogaster depend on the stabilizing force of the proteinaceous synaptonemal complex to juxtapose homologs until crossing over is achieved via recombination (Dernburg et al. 1998; McKim et al. 2002). Saccharomyces cerevisiae, Sordaria macrospora, Mus musculus, and Zea mays all rely primarily on recombination to stabilize pairing interactions between homologs (Mahadevaiah et al. 2001; Peoples et al. 2002; Storlazzi et al. 2003; Pawlowski et al. 2004). Although recombination remains necessary for juxtaposition, an important influence on pairing, recombination, and segregation of chromosomes in Schizosaccharomyces pombe is the association and movement of telomeres (Ding et al. 2004). Other organisms segregate chromosomes in the apparent absence of crossing over: In Drosophila melanogaster males, DSB-independent association may be sufficient to segregate nonrecombined homologs. In addition, backup mechanisms ensuring segregation of achiasmate chromosomes exist in Drosophila females and budding yeast (Dawson et al. 1986; Kramer and Hawley 2003). Together, these studies indicate that a unifying mechanism of homolog or DNA homology recognition either does not exist or remains to be discovered. For any of these systems, the extent of contribution from DNA base-pairing interactions remains a key question.
In budding yeast at least five known mechanisms that act either to bring chromosomes together or stabilize interaction can be monitored and dissected using genetic, molecular, and cytological techniques. These mechanisms of homolog association include DSB-independent pairing, meiotic recombination, synaptonemal complex formation, meiotic telomere reorganization, and an achiasmate segregation system (for review, see Roeder 1997; Zickler and Kleckner 1998, 1999; Scherthan 2001; Burgess 2002; Kemp et al. 2004). Elucidation of the relative contributions and interplay of these processes to accurate chromosome pairing and segregation has aided understanding of homolog associations in budding yeast. Genetic and cytological analyses have so far indicated that meiotic homologous recombination is a major determinant of close, stable homolog juxtaposition (CSHJ) during meiosis in yeast, while contributions from DSB-independent pairing and the central element of the synaptonemal complex are relatively weak (Weiner and Kleckner 1994; Nag et al. 1995; Peoples et al. 2002).
During meiosis in yeast an estimated 175–260 DSBs are created and repaired across the genome (Bishop 1994). A significant fraction (∼⅔) of DSB-induced homologous recombination events do not create a crossover (CR) product and instead yield a noncrossover (NCR) product. It has been proposed for some time that repair of DSBs to a NCR product may aid in facilitating and/or stabilizing interactions between homologous sequences at multiple sites along homologous chromosome pairs via DNA/DNA interactions (Smithies and Powers 1986; Carpenter 1987). One attractive mechanism by which such interactions could occur is through base-pairing a single-stranded 3′-end to complementary sequences within an intact homologous nonsister chromatid. Homology recognition may occur as part of the DSB repair mechanism, but other possibilities can also be imagined.
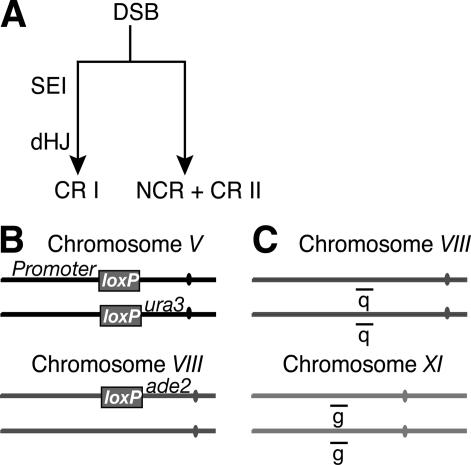
Clearly an important role for CR products is to ensure that paired homologs segregate accurately at meiosis I, but the process of their formation could also actively contribute to pairing (see Stahl et al. 2004 for a recent discussion of these models). At least two branches of the meiotic DSB repair pathway have been defined that yield CR products in yeast and Arabidopsis (Fig. 1A; de los Santos et al. 2003; Higgins et al. 2004). Class I CR products (CRI) are formed from one resected DSB end invading an intact homologous DNA sequence to form a single-end invasion (SEI) intermediate (Hunter and Kleckner 2001). The SEI is processed to a double-Holliday junction (dHJ) and resolved to form a crossover product (Schwacha and Kleckner 1995; Allers and Lichten 2001). In contrast, Class II CR products (CRII) and NCR products are thought to arise as the outcome of repair that does not involve SEI and dHJ formation, such as synthesis-dependent strand annealing (Paques and Haber 1999; Allers and Lichten 2001; de los Santos et al. 2003; Bishop and Zickler 2004). Only CRI products appear to be subject to CR interference (de los Santos et al. 2003; Higgins et al. 2004). Either one or both of these branches (CRI and/or NCR + CRII) of the DSB repair pathway may contribute to pairing interactions between homologs.
Figure 1.
Allelic and ectopic positions measure CSHJ and pairing in DSB repair mutants. (A) DSB repair pathway branches where SEI and dHJ intermediates lead to CRI products and NCR+CRII products are processed through a different mechanism (see text for details). (B) The Cre/loxP site-specific recombination assay measures collisions between chromosomal loci during meiosis, giving a readout on their relative spatial position in the nucleus. (C) Probes used for FISH experiments.
CRI and NCR + CRII products are thought to arise from a common DSB repair intermediate that follows resection of DSB 5′-ends and presumably the formation of an unstable D-loop structure by interaction of the resultant free 3′-end with a nonsister homolog (Allers and Lichten 2001; Hunter and Kleckner 2001; Borner et al. 2004). Formation of intermediates specific to the CRI branch of the DSB repair pathway depends on the ZMM class of proteins that includes Zip1, Zip2, Zip3, Mer3, and Msh5 (de los Santos et al. 2003; Borner et al. 2004). Mutations in the ZMM class of genes result in a similar qualitative phenotype: At high temperature (33°C), CRI levels are greatly decreased in the mutants relative to wild type, while NCRs are formed at wild-type levels. This temperature-dependent, CRI-specific defect occurs at the transition from DSB to SEI intermediates (Borner et al. 2004). The CRII pathway, on the other hand, represents ∼15% of total meiotic crossovers, arises independent of ZMM function, and requires Mms4/Mus81 (de los Santos et al. 2003). It is not clear how CRII is distinguished from NCR in this branch of the DSB repair pathway (for review, see Hollingsworth and Brill 2004).
We set out to define the relative contribution of different branches of the DSB repair pathway to initial pairing interactions and close, stable homolog juxtaposition (CSHJ) in meiosis of budding yeast. Two different assays for detecting allelic and ectopic associations were applied. First, Cre/loxP site-specific recombination between pairs of loxP sites was used to measure relative collision levels between pairs of loci in living cells (Fig. 1B; Burgess and Kleckner 1999). Frequency of Cre-mediated loxP recombination events has been used to report on the relative spatial proximity of loci pairs located at allelic positions on homologous chromosomes and at ectopic positions on a nonhomologous pair of chromosomes during meiosis (Peoples et al. 2002). We define wild-type levels of allelic Cre/loxP recombination as representing close, stable homolog juxtaposition. The second method of analysis was the use of fluorescence in situ hybridization (FISH) to detect possibly evanescent interactions between homologs. These interactions would limit the degree to which allelic loci on homolog pairs spread from one another after hypotonic lysis of meiotic cells (Fig. 1C; Weiner and Kleckner 1994; Burgess et al. 1999). We refer to interactions detected by FISH as pairing interactions. DSB-independent pairing is detected by FISH in situations where it is not detected by the Cre/loxP assay (Peoples et al. 2002).
Our prior analysis of recombination defective mutants (e.g., spo11Δ, rec104Δ, dmc1Δ, hop2Δ, mnd1Δ) indicated that some early meiotic recombination intermediate or set of intermediates (DSBs to SEIs) was responsible for attaining wild-type levels of allelic Cre/loxP recombination interactions during meiosis (Peoples et al. 2002; T.L. Peoples-Holst and S.M. Burgess, unpubl.). All of these mutants gave similar allelic Cre/loxP recombination interaction levels that were about fivefold reduced from wild-type levels. That is, mutants defective in strand invasion have virtually the same CSHJ defect as mutants that fail to form DSBs. Ectopic levels in these mutants were no different from those in the wild-type strain.
We report here that CRI recombination (i.e., SEI-dHJ-CRI) is not the sole branch of the DSB repair pathway contributing to CSHJ. The effect of DSB repair on CSHJ can be divided into at least two branches of this pathway: One depends on a subset of ZMM genes acting in the CRI branch (MER3, ZIP3, ZIP2, and ZIP1); another depends on NDJ1, a gene encoding a telomere-binding protein involved in meiotic telomere reorganization; and at least one more is enacted by factors yet unknown. Results from epistasis analysis of single- and double-mutant phenotypes indicate that intermediates specific to both the CRI and NCR + CRII branches of the DSB repair pathway contribute independently to close, stable homolog juxtaposition. Additionally, through analysis of the ndj1Δ mutant, our data indicate the effect of meiotic telomere reorganization on both pairing and CSHJ at interstitial loci is exerted through the initiation of recombination and not through DSB-independent interactions (e.g., via recombination processing and not telomere clustering alone).
Results
Cre/loxP recombination indicates allelic interactions
Cre-mediated interactions between loxP sites located at allelic and ectopic positions in the genome were quantified as Ura+ or Ade+ prototrophs per colony forming unit (CFU), respectively (Fig. 1B). Data for allelic interactions in mutant and wild-type strains are presented in two ways. First, allelic interactions are compared between strains from time courses performed in triplicate on the same day. Qualitative comparisons of relative allelic interactions between strains in the same time course are highly repeatable, although absolute levels of allelic interactions may vary between time courses. Second, data are represented in Table 1 as an average allelic interaction value (±standard deviation) from the 8-h time point for all cultures of each strain monitored in this work. These averaged allelic interaction values are cited in the text as percent of wild-type values and were used in the t-test calculations to compare strains quantitatively.
Table 1.
Comparison of Cre/loxP allelic interaction levels
| Strains for allelic interaction comparison | Mean Ura+ prototrophs/CFU t = 8 h (n separate cultures) | Significant differencea | p value | |
|---|---|---|---|---|
| WT vs. single mutants | ||||
| A. | WT; zip1Δ | 0.14 ± 0.03 (45) vs. 0.099 ± 0.017 (15) | S | p < 0.001 |
| WT; zip2Δ | 0.14 ± 0.03 (45) vs. 0.10 ± 0.03 (9) | S | 0.001 < p < 0.005 | |
| WT; zip3Δ | 0.14 ± 0.03 (45) vs. 0.077 ± 0.018 (24) | S | p < 0.001 | |
| WT; mer3Δ | 0.14 ± 0.03 (45) vs. 0.082 ± 0.014 (12) | S | p < 0.001 | |
| WT; msh4Δb | 0.14 ± 0.03 (45) vs. 0.16 ± 0.01 (3) | S | 0.001 < p < 0.005 | |
| WT; msh5Δ | 0.14 ± 0.03 (45) vs. 0.14 ± 0.01 (3) | NS | 0.1 < p < 0.5 | |
| WT; mus81Δc | 0.14 ± 0.03 (45) vs. 0.16 ± 0.01 (6) | S | p < 0.001 | |
| WT; ndj1Δ | 0.14 ± 0.03 (45) vs. 0.087 ± 0.013 (15) | S | p < 0.001 | |
| WT; spo11Δ | 0.14 ± 0.03 (45) vs. 0.034 ± 0.006 (9) | S | p < 0.001 | |
| spo11Δ vs. single mutants | ||||
| B. | spo11Δ; zip1Δ | 0.034 ± 0.006 (9) vs. 0.099 ± 0.017 (15) | S | p < 0.001 |
| spo11Δ; zip2Δ | 0.034 ± 0.006 (9) vs. 0.10 ± 0.02 (9) | S | p < 0.001 | |
| spo11Δ; zip3Δ | 0.034 ± 0.006 (9) vs. 0.077 ± 0.018 (24) | S | p < 0.001 | |
| spo11Δ; mer3Δ | 0.034 ± 0.006 (9) vs. 0.082 ± 0.014 (12) | S | p < 0.001 | |
| spo11Δ; ndj1Δ | 0.034 ± 0.006 (9) vs. 0.087 ± 0.013 (15) | S | p < 0.001 | |
| Double vs. single mutants | ||||
| C. | zip1Δzip3Δ; zip1Δ | 0.065 ± 0.011 (9) vs. 0.099 ± 0.017 (15) | S | p < 0.001 |
| zip1Δzip3Δ; zip3Δ | 0.065 ± 0.011 (9) vs. 0.077 ± 0.018 (24) | B | 0.01 < p < 0.05 | |
| zip3Δmer3Δ; zip3Δ | 0.065 ± 0.009 (6) vs. 0.077 ± 0.018 (24) | B | 0.01 < p < 0.05 | |
| zip3Δmer3Δ; mer3Δ | 0.065 ± 0.009 (6) vs. 0.082 ± 0.014 (12) | S | 0.001 < p < 0.005 | |
| D. | ndj1Δspo11Δ; ndj1Δ | 0.028 ± 0.008 (9) vs. 0.087 ± 0.013 (15) | S | p < 0.001 |
| ndj1Δspo11Δ; spo11Δ | 0.028 ± 0.008 (9) vs. 0.034 ± 0.006 (9) | NS | 0.05 < p < 0.1 | |
| E. | ndj1Δzip3Δ; ndj1Δ | 0.046 ± 0.008 (9) vs. 0.087 ± 0.013 (15) | S | p < 0.001 |
| ndj1Δzip3Δ; zip3Δ | 0.046 ± 0.008 (9) vs. 0.077 ± 0.018 (24) | S | p < 0.001 | |
| ndj1Δzip3Δ; spo11Δ | 0.046 ± 0.008 (9) vs. 0.034 ± 0.006 (9) | S | 0.001 < p < 0.005 | |
| ndj1Δzip1Δ; ndj1Δ | 0.078 ± 0.027 (6) vs. 0.087 ± 0.013 (15) | NS | 0.1 < p < 0.5 | |
| ndj1Δzip1Δ; zip1Δ | 0.078 ± 0.027 (6) vs. 0.099 ± 0.017 (15) | NS | 0.05 < p < 0.1 | |
| ndj1Δzip1Δ; spo11Δ | 0.078 ± 0.027 (6) vs. 0.034 ± 0.006 (9) | S | 0.005 < p < 0.01 | |
| ndj1Δmer3Δ; ndj1Δ | 0.055 ± 0.009 (9) vs. 0.087 ± 0.013 (15) | S | p < 0.001 | |
| ndj1Δmer3Δ; mer3Δ | 0.055 ± 0.009 (9) vs. 0.082 ± 0.014 (12) | S | p < 0.001 | |
| ndj1Δmer3Δ; spo11Δ | 0.055 ± 0.009 (9) vs. 0.034 ± 0.006 (9) | S | p < 0.001 | |
| mus81Δ mutants | ||||
| F. | mus81Δmer3Δ; mus81Δ | 0.092 ± 0.013 (6) vs. 0.160 ± 0.012 (6) | S | p < 0.001 |
| mus81Δmer3Δ; mer3Δ | 0.092 ± 0.013 (6) vs. 0.082 ± 0.014 (12) | NS | 0.05 < p < 0.1 | |
| ndj1Δmus81Δ; ndj1Δ | 0.11 ± 0.01 (6) vs. 0.087 ± 0.013 (15) | S | p < 0.001 | |
| ndj1Δmus81Δ; mus81Δ | 0.11 ± 0.01 (6) vs. 0.16 ± 0.01 (6) | S | p < 0.001 | |
| mus81Δndj1Δmer3Δ; ndj1Δmer3Δ | 0.067 ± 0.006 (6) vs. 0.055 ± 0.009 (9) | S | 0.001 < p < 0.005 | |
(WT) Wild type; (S) significantly different, p < 0.01; (B) borderline, 0.01 < p < 0.05; (NS) not significantly different, 0.05 < p.
Wild-type and msh4Δ strains were not as different when cultures analyzed in parallel on the same day were compared [0.17 (3) vs. 0.16 (3), 0.1 < p < 0.5].
Wild-type and mus81Δ strains were not as different when cultures analyzed in parallel on the same day were compared [Exp. 1: 0.16 (3) vs. 0.17 (3), p > 0.1; and Exp. 2: 0.18 (3) vs. 0.15 (3), 0.01 < p < 0.1].
All mutant strains characterized in this study, including double- and triple-mutant strains, exhibited wild-type levels of survival after the return to growth procedure indicating that all cells plated, and not a subpopulation of viable cells, were assayed for allelic and ectopic interactions (data not shown). Additionally, no strain exhibited a difference in ectopic interactions compared with wild type (Fig. 2; data not shown). These results indicate that these mutations under analysis confer no general effect on Cre/loxP assay mechanics (e.g., via control of GAL1-Cre expression levels).
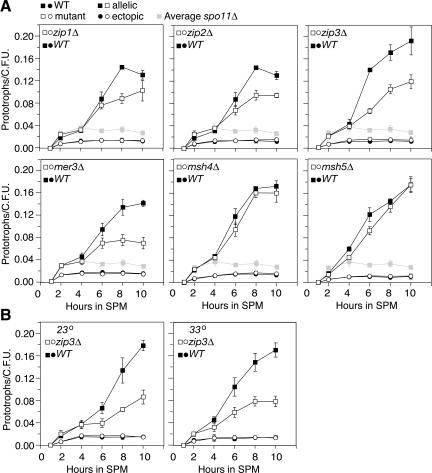
Figure 2.
Commitment to Cre-mediated recombination in wild-type and zmm mutant strains. (A) Synchronized meiotic cells were plated on selective media following Cre induction at t = 1 h after meiotic induction. Allelic interactions (squares) are reported by Ura+ prototrophs/CFU, and ectopic interactions (circles) are reported by Ade+ prototrophs/CFU. The wild-type control is indicated with filled black symbols; mutants are designated by open symbols. Measurements shown here are the average of three independent cultures (±SD) processed in parallel. Light-gray-filled symbols indicate averaged spo11Δ allelic interaction values from nine independent cultures. All cultures were maintained at 30°C. (B) Commitment to Cre-mediated recombination in wild-type and zip3Δ strains incubated during meiosis at 23°C and 33°C.
Allelic interactions in zip3Δ, zip2Δ, zip1Δ, and mer3Δ are reduced compared with wild-type levels but not to spo11Δ levels
zmm mutants specifically affect the CRI branch of the DSB repair pathway (de los Santos et al. 2003; Borner et al. 2004). We examined both allelic and ectopic interactions in each of these deletion mutant strain backgrounds during a meiotic time course to assess their contribution to CSHJ (Fig. 2A). If the CRI branch of the DSB repair pathway were solely responsible for CSHJ, we would expect these mutants to give allelic interaction levels similar to spo11Δ.
The zip3Δ, zip2Δ, zip1Δ, and mer3Δ mutants all exhibited allelic interactions that were significantly reduced compared to wild-type levels (p < 0.005) (Table 1A). Interestingly, allelic interactions in these zmm mutants were not reduced to the level exhibited by a mutant lacking meiotic recombination initiation (spo11Δ; p < 0.001) (Table 1B; Fig. 2A). Among this subset of zmm mutants, zip3Δ exhibited the most severe reduction compared to wild type (55%), followed by mer3Δ (60% of wild-type levels), and zip1Δ and zip2Δ (69% and 73% of wild type), respectively. In contrast, allelic interactions for spo11Δ mutants were found to be 22% of wild-type levels in this study (Table 1).
Previously, we had shown that an msh5Δ mutant exhibited allelic interactions indistinguishable from the wild-type control (Fig. 2A; Peoples et al. 2002). Msh4 has been reported to colocalize with Msh5 (Agarwal and Roeder 2000). Accordingly, we found that an msh4Δ single mutant also exhibited the same level of allelic interactions as msh5Δ and wild type (Fig. 2A). This result was somewhat surprising given that the zmm mutants have been shown to exhibit qualitatively similar phenotypes with respect to CR and NCR recombination, SEI, and dHJ formation (Borner et al. 2004). These results suggested to us that MSH5, and possibly SEI and dHJ formation, may play little or no role in CSHJ. Instead, CSHJ depends on only a subset of the ZMM group of genes, ZIP1, ZIP2, ZIP3, and MER3 but not MSH5 or MSH4.
Taken together, we interpret the effect of zip1Δ, zip2Δ, zip3Δ, and mer3Δ mutations on reducing allelic interactions to suggest that an early intermediate (i.e., preceding SEI formation) in the CRI branch of the DSB repair pathway plays a significant role, but is not the sole contributing determinant, of CSHJ during yeast meiotic prophase.
Allelic interactions are similar at low and high temperatures
zmm mutants exhibit a more severe CR defect at 33°C than at 23°C (Borner et al. 2004). One of the most extreme defects exhibited by these mutants is the reduction in SEI formation, to ≤15% of wild-type levels, seen in the zip3Δ mutant. In contrast, SEI levels in the zip3Δ mutant at 23°C are similar to those of wild type after a delay of ∼5 h (Borner et al. 2004). To relate the induction of CSHJ to the establishment of the first detectable physical interaction between homologous chromosomes in DSB repair (i.e., SEI formation), we assayed allelic and ectopic interactions in the zip3Δ mutant strain at 23°C and 33°C. Virtually no difference in allelic interaction levels was observed when the experiments were carried out in parallel at the two temperatures (Fig. 2B). Average allelic interactions in zip3Δ were 52% and 50% of wild-type levels at 23°C and 33°C, respectively. This is comparable to the 55% of wild-type levels observed for the zip3Δ mutant at 30°C (Table 1; see above). No difference was observed between wild-type and zip3Δ ectopic interactions at 23°C, 30°C, or 33°C (Fig. 2A,B).
For the cultures incubated at 23°C, allelic interactions were apparently still increasing at 10 h after transfer to sporulation medium. We considered the possibility that if incubated longer, levels would eventually increase to wild-type level and thus provide a correlation between Cre/loxP levels with SEI formation for the two extreme temperatures. We repeated the experiment with single cultures at 23°C, sampling wild type and zip3Δ up to 14 h after meiotic induction, and found no further increase in allelic interactions in the zip3Δ mutant following attainment of maximum levels at 10 h after transfer to sporulation medium (data not shown).
We interpret the lack of change in allelic interactions between 23°C, 30°C, and 33°C to indicate that SEIs play no detectable role in establishing CSHJ at the examined locus. In other words, the contribution of ZMM proteins to CSHJ is likely independent of SEIs.
Epistasis analysis of zip3Δ, zip1Δ, and mer3Δ alleles with respect to CSHJ
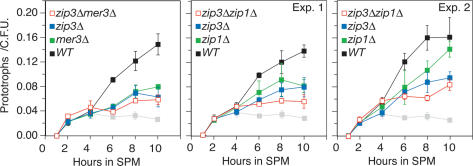
Comparison of single- and double-mutant phenotypes conferred by various zmm mutations showed that ZIP3, ZIP1, and MER3 act in the same branch of the meiotic recombination pathway (Borner et al. 2004). We analyzed zip3Δ zip1Δ and zip3Δ mer3Δ double mutants to determine whether these genes also act in the same pathway to facilitate CSHJ. Time-course analysis revealed that the double-mutant strains gave qualitatively similar levels of allelic interaction as the single mutants (Fig. 3). Upon closer inspection of the t = 8 time point for all experiments performed, there was a borderline to significant difference in interaction levels when comparing double and single mutants (Table 1C). This effect was repeatedly more pronounced for the zip3Δ zip1Δ double mutant compared with the two corresponding single mutants (Fig. 3, Exps. 1 and 2). This slight decrease of the zip3Δ zip1Δ double-mutant phenotype relative to the zip3Δ single mutant may indicate a very weak contribution of ZIP1 to CSHJ independent of ZIP3 function.
Figure 3.
Commitment to Cre-mediated prototroph formation in wild-type, single-mutant and double-mutant zmm strains. Graph parameters are the same as in Figure 2 except that ectopic interactions have been omitted for simplicity (no changes compared with wild-type or single mutants were observed). Single-mutant graphs have filled blue or green symbols and double-mutant graphs have open red symbols as indicated. Experiment 1 and 2 graphs indicate two separate time courses comparing zip3Δ zip1Δ and zip3Δ mutants.
A mutant defective in DSB formation and meiotic telomere reorganization (spo11Δ ndj1Δ) exhibits allelic interactions similar to the spo11Δ mutant
In premeiotic yeast cells, centromeres are clustered near the spindle pole body, and telomeres are localized at the nuclear envelope in eight to nine clusters (Jin et al. 1998; Trelles-Sticken et al. 1999). Concomitant with DNA replication, telomeres are released from the clusters and, by late leptotene, are dispersed around the nuclear envelope. At the leptotene–zygotene transition, all telomeres cluster tightly near the spindle pole body in the bouquet configuration and centromeres are no longer clustered. By pachytene, when synaptonemal complex extension is complete, the bouquet is no longer present, and telomeres again appear dispersed around the nuclear envelope and within the nucleus (Trelles-Sticken et al. 2000). We refer to these events, collectively, as meiotic telomere reorganization.
Ndj1 is a telomere-binding protein required for telomere attachment to the nuclear envelope and subsequent bouquet formation (Chua and Roeder 1997; Conrad et al. 1997; Trelles-Sticken et al. 2000). Previously, we reported the ndj1Δ single mutant to exhibit allelic Cre-mediated recombination levels intermediate to wild-type and spo11Δ levels and suggested that meiotic telomere reorganization contributes to CSHJ (Peoples et al. 2002).
We reasoned that there were two possible ways in which the ndj1Δ mutation could elicit a reduction in allelic interactions at an interstitial locus in the genome. First, the role of NDJ1 in meiotic telomere reorganization could contribute directly to interactions between homologs, independent of interactions mediated through DSB repair, through the role of Ndj1 in tethering telomeres to the nuclear envelope (Rockmill and Roeder 1998). Alternatively, the ndj1Δ mutation could elicit a defect in CSHJ through a negative effect on recombination. Indeed, DSB repair recombination intermediates and products are delayed in an ndj1Δ mutant (H.Y. Wu and S.M. Burgess, in prep.). Analysis of allelic interactions in spo11Δ ndj1Δ distinguishes between these two models. Allelic interactions in the spo11Δ ndj1Δ double mutant closely resemble those seen in the spo11Δ single mutant (Table 1D; Fig. 4A). Average allelic interactions were 18% of wild-type levels in the spo11Δ ndj1Δ double mutant and 22% of wild-type levels in the spo11Δ single mutant. No change from wild-type ectopic interaction levels was seen in the spo11Δ ndj1Δ, spo11Δ, or ndj1Δ mutants (data not shown).
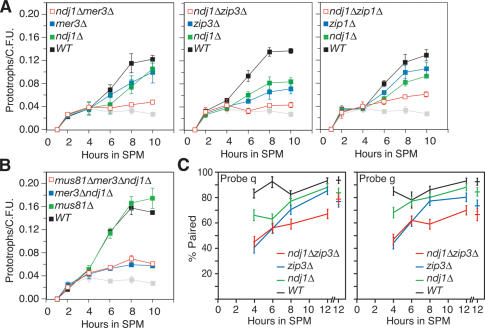
Figure 4.
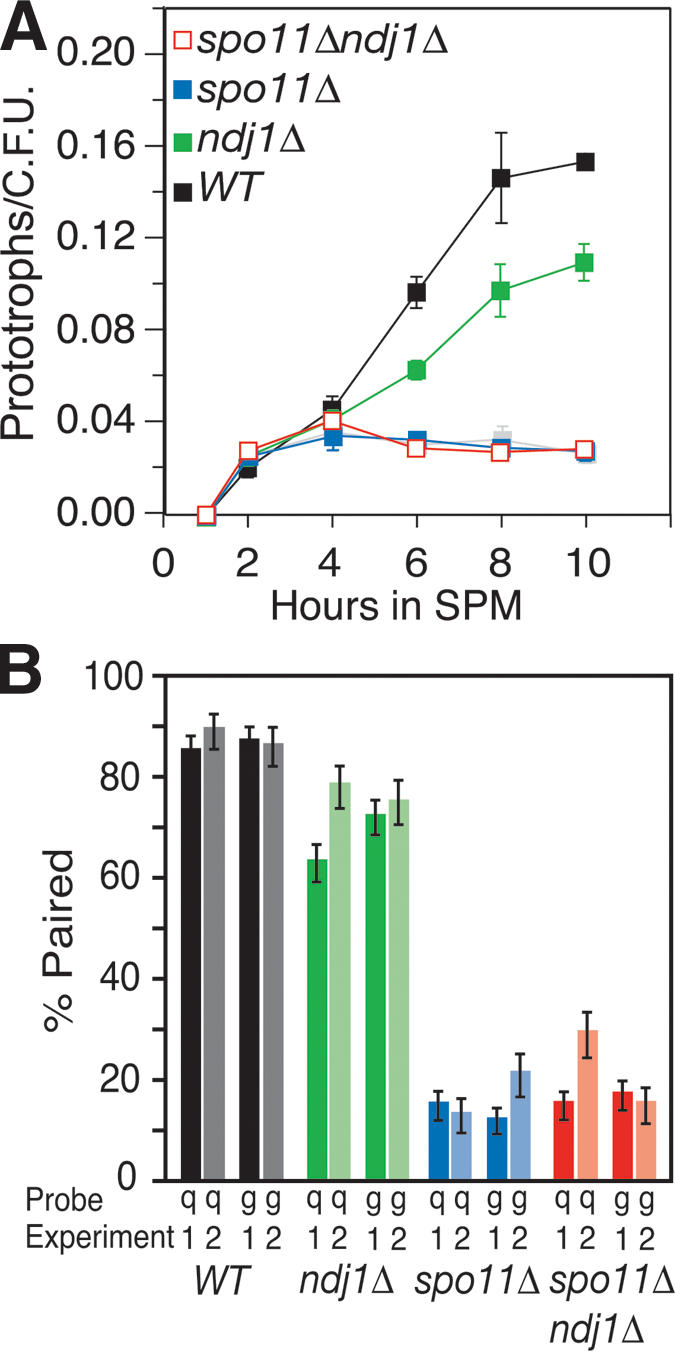
Epistasis analysis of ndj1Δ and spo11Δ mutant Cre/loxP and FISH phenotypes. (A) Commitment to Cre-mediated prototroph formation in wild-type, ndj1Δ, spo11Δ, and spo11Δ ndj1Δ strains. Graph parameters are the same as in Figure 3. Single-mutant graphs have filled blue or green symbols, and double-mutant graphs have open red symbols as indicated. (B) Two independent FISH experiments assess pairing at t = 6 h in sporulation medium (SPM) with fluorescently labeled cosmid probes hybridized to spread nuclei preparations. Pairing is defined as a distance between foci of 0.7 μm or less. Background pairing levels for each hybridization were determined as the fraction of nuclei scored with ectopic pairs of loci that were <0.7 μm apart. (Dark bars) Data collected in Experiment 1; (light bars) Experiment 2. Error bars indicate standard error based on the number of nuclei scored (n = #1, #2). (Black) Wild type (n = 139, 86); (green) ndj1Δ (n = 175, 101); (blue) spo11Δ (n = 151, 89); (red) spo11Δ ndj1Δ (n = 158, 98). Background pairing of heterologous FISH probes has been subtracted from pairing values shown (#1, #2): wild type (0.07, 0.10), ndj1Δ (0.14, 0.05), spo11Δ (0.14, 0.07), and spo11Δ ndj1Δ (0.06, 0.08).
Ectopic interactions are about twofold lower than the lowest detected allelic interactions observed using the Cre/loxP assay for any mutant analyzed to date. The question then arises as to whether the Cre/loxP assay has reached a minimum threshold for gauging allelic interactions. To address this issue, we used FISH to compare chromosome colocalization levels in wild type and the spo11Δ ndj1Δ, spo11Δ, and ndj1Δ mutants 6 h after entry into meiosis. For one experiment, we found the percent pairing for chromosomes VIII and XI to be 84% and 86% in wild type, 62% and 71% in ndj1Δ, 14% and 11% in spo11Δ, and 14% and 16% in spo11Δ ndj1Δ (Fig. 4B, Exp. 1, dark bars). An independent experiment showed qualitatively similar results (Fig. 4B, Exp. 2, light bars). Thus, there remains no qualitative difference in phenotype between the Cre/loxP assay and FISH for spo11Δ ndj1Δ and spo11Δ. The similarity between the spo11Δ and spo11Δ ndj1Δ mutants indicates that meiotic telomere reorganization is not a component of DSB-independent pairing and exerts its effect on CSHJ through its role, either directly or indirectly, in meiotic recombination at the interstitial sites we monitored.
Allelic interactions are reduced significantly in ndj1Δ zip3Δ, ndj1Δ mer3Δ, and somewhat in ndj1Δ zip1Δ double mutants compared to the corresponding single-mutant strains
Since SPO11 function is necessary for NDJ1 to exert its effect on CSHJ, we were interested in knowing whether NDJ1 acts through the same branch of the DSB repair pathway as MER3, ZIP3, and ZIP1 in establishment of CSHJ. Wild-type, double-mutant, and corresponding single-mutant strains were analyzed in parallel, meiotic time-course experiments. The ndj1Δ zmmΔ double mutants gave a more severe reduction in allelic interactions than the corresponding single mutants (Fig. 5A). Analysis indicated that the ndj1Δ zip3Δ double mutant exhibited allelic interactions that were on average 30% of wild-type levels, while the ndj1Δ and zip3Δ single mutants showed 63% and 55% of wild-type allelic interaction levels, respectively (Table 1E). Likewise, in the ndj1Δ mer3Δ double mutant, allelic interactions were 36% of wild-type levels, while ndj1Δ and mer3Δ single mutants gave 63% and 60% of wild-type levels, respectively (Fig. 5A; Table 1E). The ndj1Δ zip1Δ double mutant was on average 53% of wild-type levels, while ndj1Δ and zip1Δ allelic interactions were 63% and 69% of wild-type levels, respectively (Fig. 5A). While this decrease in the ndj1Δ zip1Δ double mutant appears significant when comparing strains within each time course, t-test analysis of average allelic interactions from the 8-h time point indicates this is not a significant difference (Table 1E). In each case, the defect exhibited by the double mutant was comparable to the product of the defect exhibited by each single mutant compared to wild type.
Figure 5.
Epistasis analysis of zmm, mus81Δ, and ndj1Δ mutant Cre/loxP and FISH phenotypes. (A) Graph parameters are the same as in Figure 3. Graphs plotting Cre/loxP data for single mutants have filled blue or green symbols, and data for double-mutant levels are plotted as open red symbols as indicated. (B) Graph parameters are the same as in Figure 3. Graphs plotting Cre/loxP data for single and double mutants have filled green or blue symbols and triple-mutant data is indicated by open red symbols. (C) FISH parameters are the same as stated in Figure 4B except that chromosome pairing was monitored at 4, 6, 8, and 12 h after meiotic induction. Percent pairing levels (with background subtracted) for FISH probes q and g are reported at 4, 6, 8, and 12 h. Error bars indicate standard error based on number of nuclei scored, n. (Black) Wild type (n = 124, 42, 149, 128); (green) ndj1Δ (n = 129, 142, 145, 138); (blue) zip3Δ (n = 121, 164, 153, 142); and (red) ndj1Δ zip3Δ (n = 146, 189, 165, 163). Data points after break in graph axis indicate an independent FISH experiment that assessed percent pairing levels (with background subtracted) for FISH probes q and g at 12 h for wild type (n= 84), ndj1Δ (n = 108), zip3Δ (n = 94), and ndj1Δ zip3Δ (n = 94). Background heterologous pairing subtracted from pairing values shown (t = 4, 6, 8, 12, 12): wild type (0.05, 0.08, 0.07, 0.02, 0.05), ndj1Δ (0.05, 0.13, 0.11, 0.07, 0.10), zip3Δ (0.11, 0.13, 0.04, 0.03, 0.08), and ndj1Δ zip3Δ (0.05, 0.11, 0.09, 0.04, 0.06).
We also measured allelic pairing levels in spread nuclei preparations using FISH. The ndj1Δ zip3Δ mutant exhibited decreased levels of pairing by FISH that mirror Cre/loxP assay results (Fig. 5C). Differences between double and single mutants, however, are not as notable as for Cre/loxP results, likely reflecting the ability of the FISH assay to detect DSB-independent homolog interactions that are not detected using the Cre/loxP assay (Peoples et al. 2002).
Interestingly, while allelic interactions are reduced in double-mutant combinations tested relative to the corresponding single mutants, all are greater than allelic interactions measured in the spo11Δ mutant (Table 1E). Together these results suggest that the effect of Ndj1 on CSHJ is exacted through meiotic recombination independently of Zip3, Zip1, and Mer3 and that these two pathways are not exclusively responsible for CSHJ. Indeed, there may be a third branch of the DSB repair pathway contributing to CSHJ.
Allelic interactions are not reduced in mus81Δ strains
The results presented above suggest that at least one more additional recombination pathway is playing a role in CSHJ. Since Mus81 and Mms4 are known to promote the CRII pathway that was recently shown to be distinct from the CRI pathway (de los Santos et al. 2003; Argueso et al. 2004; Borner et al. 2004), we tested the involvement of Mus81 in CSHJ. Allelic and ectopic interactions in wild-type, mus81Δ, mus81Δ ndj1Δ, mus81Δ mer3Δ, mer3Δ ndj1Δ, and mus81Δ mer3Δ ndj1Δ strains assayed in parallel indicated no negative effect of the mus81Δ mutation on either allelic or ectopic interactions (Fig. 5B; Table 1F; data not shown). Student's t-test analysis indicates that allelic interactions in mus81Δ ndj1Δ are significantly greater than in ndj1Δ and allelic interactions in mus81Δ ndj1Δ mer3Δ are significantly greater than in ndj1Δ mer3Δ. Although we cannot rule out some role for Mus81 as a negative regulator of allelic interactions, we did not observe consistent increases in allelic interactions in the presence of mus81Δ within each time course. Data from this experiment indicate that Mus81 does not play a direct role in facilitating CSHJ. These results, however, do not rule out the possibility that earlier intermediates in the CRII branch of the DSB repair pathway are promoting CSHJ.
Discussion
Early DSB-initiated interactions between homologs, including those giving rise to noncrossovers, promote CSHJ
Our previous analysis of single-mutant strains defective for meiotic chromosome dynamics revealed that the process of forming and repairing DSBs plays a major role in achieving CSHJ during meiosis of budding yeast. In this study, we found that mutants defective for both synapsis initiation and the formation of physical intermediates in a branch of the DSB repair pathway leading to crossovers (CRI; i.e., SEI and dHJ) exhibit significant and substantial levels of allelic interaction, albeit not at wild-type levels. Since the branch of the DSB repair pathway responsible for NCR and CRII formation is presumably still intact in zip3Δ, zip2Δ, zip1Δ, and mer3Δ mutants, we interpret this result to mean NCR (and probably also CRII)-specific recombination intermediates contribute to CSHJ during meiosis in yeast.
The first detectable physical recombination intermediate involving homologous nonsister chromatids is the SEI specific to the CRI pathway (Hunter and Kleckner 2001). The zip3Δ mutant exhibits similar allelic interactions when analyzed under conditions in which the SEI is presumably present or absent (i.e., at 23°C and 33°C, respectively). It is formally possible, however, that SEIs do not form in the region examined in the zip3Δ at any temperature. Yet zmm mutations vary in effect on CSHJ at the measured locus. For example, the msh5Δ mutant exhibits varying temperature-dependent levels of SEI formation at the HIS4LEU2 hotspot, as does zip3Δ, yet exhibits a nearly wild-type level of allelic interactions in our analysis. Taken together, we argue that the contribution of ZMM proteins to CSHJ is independent of their role in SEI formation.
If SEIs are not promoting CSHJ, then what type of interaction between homologs is involved? Nascent interactions have been proposed to exist prior to the establishment of detectable SEIs (Hunter and Kleckner 2001). These nascent interactions are postulated to be either unstable paranemic contacts between homologs or extremely short plectonemic joints located with axial association bridges (Hunter and Kleckner 2001). They may be analogous to cytologically detectable intermediates of homolog interaction in mammals (Moens et al. 1997). Moreover, the interactions could be mediated by components of the synapsis initiation complex (see below). We propose that homolog pairing is initially mediated by premeiotic DSB-independent contributions—the nature of which is unknown—which are then stabilized through both CRI and NCR + CRII (discussed below) recombination intermediates to bring about CSHJ, the final outcome of the collision rate assay in wild-type cells.
Components of the synapsis initiation complex contribute to CSHJ
We looked at mutants under conditions lacking both synapsis initiation complex formation and all known structures (SEI and dHJ) in which homologs are linked by chemical or hydrogen bonds and still saw significant and substantial levels of homolog interaction. In the CRI pathway, components acting in synapsis initiation complex formation facilitate CSHJ, while MSH4, MSH5, and the SEI recombination intermediate play no detectable role (see discussion below). Although recent work has led to the conclusion that Msh4 and Msh5 act early in the pathway destined for CRI formation (Colaiacovo et al. 2003; Borner et al. 2004; Higgins et al. 2004), we find that in terms of contribution to CSHJ, MSH5 is genetically distinguished from ZIP1, ZIP2, ZIP3, and MER3.
Synapsis initiation complexes are at least partially composed of Zip3 and Zip2 and form at the sites of axial associations, discrete locations where homologous chromosomes are closely connected (Chua and Roeder 1998; Fung et al. 2004). From visualization of Zip2 foci, Fung et al. (2004) has reported that msh4Δ does not affect interference between synapsis initiation complex sites. In this mutant situation, the interference of synapsis initiation complexes remains intact while CRs are reduced two- to threefold, and genetic interference is reduced dramatically (Ross-Macdonald and Roeder 1994; Novak et al. 2001; Fung et al. 2004). This uncoupling of synapsis initiation complex interference from genetic interference suggested a functional distinction between members of the ZMM class. A recent study from Snowden et al. (2004) has shown that the hMsh4–hMsh5 complex recognizes Holliday junctions and forms a meiosis-specific sliding clamp that embraces homologous chromosomes; they suggest this sliding clamp complex is involved in recombination intermediate stabilization. One possibility is that formation of synapsis initiation complexes at recombination sites leads to the stabilization of CRI intermediates at those sites, and hence mediates CR interference through the stabilization of these intermediates. Although this suggests a temporal relationship for the realization of synapsis initiation complex interference and genetic interference, the initial selection of synapsis initiation complex sites remains a mystery. In interpretation of our data, however, we suggest that it is the formation of synapsis initiation complexes, and not any subsequent steps involving Msh4 and Msh5, that facilitates CSHJ.
Ndj1 contributes to CSHJ through the DSB repair pathway
It has been shown in the fungus Sordaria that presynaptic alignment of homologous chromosomes occurs prior to bouquet formation (Storlazzi et al. 2003), suggesting that the bouquet may augment pre-existing pairing interactions. Even if a function in early pairing is not a manifestation of the bouquet per se, substantial data exist to postulate a role for telomere dynamics in homolog juxtaposition. For example, attachment of telomeres to the nuclear envelope alone could provide a basis for pairing by constraining chromosome mobility or topology (Zickler and Kleckner 1998). Indeed, a role for meiotic telomere reorganization in homology searching through limitation of the available search region has been proposed based on the observation that homologous recombination between heteroalleles placed at ectopic locations is apparently restricted to a lesser extent in an ndj1Δ mutant than in wild type (Goldman and Lichten 2000). Moreover, a role for Ndj1 in telomere-mediated interactions has been proposed by Rockmill and Roeder (1998).
Our analysis, using both Cre/loxP and FISH, indicates that spo11Δ is epistatic to ndj1Δ with respect to CSHJ and pairing. Thus, the majority of the effect of Ndj1 on CSHJ at interstitial loci depends on the initiation of meiotic recombination. The outcome of these experiments may have been influenced by our choice in FISH probes and location of loxP sites. In our analyses to date, we have used FISH probes and loxP constructs that are approximately equidistant from centromere and telomere on similarly sized chromosomes to minimize genome position effects and maximize homology-driven events. In the spo11Δ ndj1Δ double mutant, it may be that pairing defects would be exacerbated, compared to the two single mutants, if homolog pairing were monitored close to telomeres. In fact, greater pairing at subtelomeric loci than interstitial loci in a spo11Δ mutant has been measured using FISH (Weiner and Kleckner 1994; Bhuiyan and Schmekel 2004). If CSHJ at telomeres is, in fact, more affected in the spo11Δ ndj1Δ double mutant than in the spo11Δ single mutant, the possibility then arises that Ndj1 may actually facilitate CSHJ via two effects, first through DSB-independent pairing interactions at telomeres and second by aiding repair of recombination intermediates at interstitial locations. In this interpretation, meiotic telomere reorganization might play roles both in facilitating early homology searches and in stabilizing later recombination intermediates. We propose that Ndj1, via meiotic telomere reorganization, kinetically facilitates the homology search step of early meiotic recombination to contribute to the coalignment of homologs prior to synapsis, either by simple bulk mixing of the chromosomes or otherwise reducing the effective volume between two homologous sites in the nucleus.
Multiple intermediates in the DSB repair pathway contribute independently to CSHJ
In the absence of Ndj1 and the CRI pathway, CSHJ is not fully achieved, indicating that a pathway including Ndj1 acts independently of the CRI pathway to promote CSHJ. Supporting this idea, the reduction in measurable CSHJ caused by addition of a zmm mutant is very similar in wild-type and ndj1Δ backgrounds. Recent results from our lab have shown by physical analysis of recombinant chromosomes that the ndj1Δ delays both CR and NCR formation and reduces the levels of NCR products approximately twofold (H.Y. Wu and S. Burgess, in prep.). At this point, we cannot distinguish between NDJ1 predominantly influencing the NCR + CRII pathway versus affecting both CRI and NCR + CRII pathways. Taken together, our data have led to the conclusion that both CRI and NCR + CRII pathways contribute to CSHJ. In the NCR+CRII pathway, NDJ1 facilitates CSHJ likely via meiotic telomere reorganization.
Double mutants involving ndj1Δ and zip3Δ, zip1Δ, or mer3Δ exhibit allelic interaction levels greater than the levels observed for the spo11Δ single mutant. This result indicates that at least one more branch of the DSB repair pathway may be acting to facilitate CSHJ. While we have ruled out a role for Mus81 in promoting CSHJ (this work), the possibility exists that some unknown earlier intermediate in the Mus81/Mms4 CRII pathway is playing a role in CSHJ. An additional recombination pathway could also be the extra factor contributing to CSHJ. In support of this, ∼17% of the wild-type crossover level is still detected in an mms4Δ msh5Δ double mutant (de los Santos et al. 2003). A recent study also presents evidence for a branch of the DSB repair pathway separate from those branches dependent on MMS4 and MSH5 function (Argueso et al. 2004).
Alternatively, a more simplistic CSHJ mechanism is also suggested by our data. Analysis of the zip3Δ zip1Δ double mutant shows a reduction in CSHJ at the 8-h time point, indicating that Zip3 and Zip1 may not act entirely in the same pathway to facilitate CSHJ. It may well be that an ndj1Δ zip3Δ zip1Δ triple mutant would exhibit an allelic interaction level equal to that of spo11Δ, indicating, perhaps, separate CR pathway, NCR pathway, and Zip1-specific contributions to CSHJ.
It should be noted that effects of the ndt80Δ on crossover formation could influence our conclusions concerning the relative importance of different intermediates in facilitating CSHJ. NDT80, encoding a mid-prophase meiotic transcription factor, has been shown to directly affect crossover formation as ndt80Δ mutants arrest with unresolved Holliday-junction intermediates and reduced crossovers (Allers and Lichten 2001). While likely acting later than achievement of CSHJ, we cannot rule out the idea that ndt80Δ is differentially affecting the mutants we analyzed.
In terms of physical distance
How do different measurable levels of interaction between homologs in mutant situations translate to physical and/or spatial differences within the nucleus of a meiotic cell? It is likely that the Cre/loxP assay is measuring a combination of different “pairing” states. In physical terms, mutants that give similar intermediate levels of allelic interactions do not necessarily exhibit the same chromosome state. For example, proximity of two allelic loci to one another may be influenced by both segmental pairing and chromosome condensation. If loxP sites are located in chromosome loops, then Cre-mediated interactions will be affected both by how close the axes are to each other and by the relative distance the loxP sites are from the axis; the more compact the loops, the more likely it will be that loxP sites encounter one another. It is thought that DSBs form in chromosome loops and are later brought to the chromosomal axis for repair (Blat et al. 2002). At this time, we cannot distinguish whether we are detecting loop–loop, axis–loop, or axis–axis interactions. It is possible that two mutations that actually act in the same molecular “pathway” could show additive effects when combined because each mutation only provides a small increase in the physical space within which homologous chromosomes are constrained.
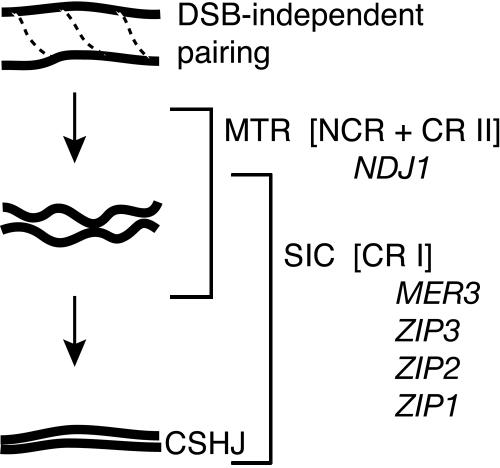
We favor the interpretation that CSHJ is achieved through several different meiotic processes coordinated to bring homologous chromosomes into a closer proximity that aids in the stabilization of progressively more robust recombination interactions. Support for this type of stepwise pairing model comes from a recent study in Sordaria that clearly describes visual distinction of progressive homolog recognition and pairing stages (Tesse et al. 2003). In our model, homologs involved in DSB-independent pairing are brought into a closer and increasingly more stable configuration by recombination events influenced by meiotic telomere reorganization and also by the formation of synapsis initiation complexes (Fig. 6). It is these dynamic stages (DSB-independent pairing + NCR intermediate + CR intermediate) that all contribute to CSHJ.
Figure 6.
CSHJ facilitated by multiple recombination pathways. CSHJ is facilitated through DSB-independent pairing interactions that are enhanced by both meiotic telomere reorganization-aided recombination interactions and intermediates in the CRI pathway that also act at the point of synapsis initiation complex formation. See text for details.
Materials and methods
Yeast strains
All yeast strains are isogenic derivatives of SK1 (Kane and Roth 1974). Parental haploid strains (SBY1338 and SBY1438) containing Cre/loxP constructs used in this analysis have been described previously; all strains used in this study also contain the ndt80Δ (Xu et al. 1995; Peoples et al. 2002). For ndj1Δ::kanMX4 and zip1Δ::kanMX4, knockout mutations in SBY1338 and SBY1438 were generated by transformation using PCR-based disruption that replaced the entire open reading frame with the kanMX4 marker (Wach et al. 1994). For mer3Δ::kanMX4, zip3Δ::kanMX4, and zip2Δ::kanMX4 strains, knockouts were made similarly except that previously constructed knockout strains were purchased from Research Genetics and PCR primers were designed to amplify regions ∼200 bp upstream and downstream of the disrupted open reading frames for use in transformation. SK1 strains containing spo11Δ::kanMX4 were kindly provided by Scott Keeney (Sloan-Kettering Cancer Center, New York) and described in Kee and Keeney (2002). Strains used for double- and triple-mutant analysis were constructed by transformation using PCR-based disruption that replaced the entire open reading frame with either natMX4 or hphMX4 (Wach et al. 1994). The markers used in these strain constructions are as follows: spo11Δ::hphMX4 ndj1Δ::kanMX4, ndj1Δ::hphMX4 mer3Δ::kanMX4, ndj1Δ::kanMX4 zip3Δ::hphMX4, ndj1Δ::hphMX4 zip1Δ::kanMX4, zip3Δ::hphMX4 zip1Δ::kanMX4, zip3Δ::hphMX4 mer3Δ::kanMX4, mus81Δ::natMX4, mus81Δ::natMX4 mer3Δ::kanMX4, mus81Δ::natMX4 ndj1Δ::kanMX4, mus81Δ::natMX4 mer3Δ::kanMX4 ndj1Δ::hphMX4. Diploids used in this study were constructed by crossing the SBY1338- and SBY1438-derived knockout strains.
Synchronous meiotic time courses
Media used for meiotic time courses were described previously (Peoples et al. 2002). The location of Cre/loxP constructs and synchronous meiotic time courses at 30°C were the same as described previously (Peoples et al. 2002). Orientation and chromosome location of promoter (pGPD1-loxP) and two reporter (loxP-ura3 and loxP-ade2) constructs is shown in Figure 1. Cre-mediated recombination between loxP sites located at allelic positions at the FLO8 locus on chromosome V result in Ura+ prototrophs while ectopic interactions between flo8::pGPD1-loxP and a loxP-ade2 site located at the NDT80 locus on chromosome VIII give Ade+ prototrophs upon return to mitotic growth. All strains contain the ndt80Δ and arrest in prophase I, prior to the first meiotic division. For time courses carried out at temperatures other than 30°C, cultures were shifted to either 23°C or 33°C upon resuspension in sporulation medium (SPM). The time course comparing wild type (WT) and the zip3Δ mutantat23°Cfor t = 1 through t = 14 was carried out in a 100-mL meiotic culture.
FISH analysis
Meiotic nuclear spreads, labeling of probes, and nuclei visualization was the same as reported in Peoples et al. (2002). Alexa-488-labeled cosmid probe q (ATCC70891, American Type Culture Collection, Manassas, VA), hybridizes to chromosome VIII, while Cy3-labeled cosmid probe g (pUKG141, B. Dujon, Institut Pasteur, Paris, France) identifies chromosome XI. Hybridization and washing of probes were the same as that described in Weiner and Kleckner (1994) except that no detection antibodies were used.
Acknowledgments
We thank Michael Lichten, JoAnne Engebrecht, Joshua Chang Mell, and Hsin-Yen Wu for critical review of this manuscript, and the Kleckner lab for sharing ZMM data prior to publication. Special thanks to Joshua Chang Mell, Hsin-Yen Wu, JoAnne Engebrecht, and Neil Hunter for discussion of this work. Work supported by NIH Molecular and Cellular Biology Training Grant NIH T 32 GM007377-26 award to T.L.P.H. and American Cancer Society RSG-01-053-01-CCG award to S.M.B.
Article and publication are at http://www.genesdev.org/cgi/doi/10.1101/gad.1293605.
References
- Agarwal S. and Roeder, G.S. 2000. Zip3 provides a link between recombination enzymes and synaptonemal complex proteins. Cell 102: 245–255. [DOI] [PubMed] [Google Scholar]
- Allers T. and Lichten, M. 2001. Differential timing and control of noncrossover and crossover recombination during meiosis. Cell 106: 47–57. [DOI] [PubMed] [Google Scholar]
- Argueso J.L., Wanat, J., Gemici, Z., and Alani, E. 2004. Competing crossover pathways act during meiosis in Saccharomyces cerevisiae. Genetics 168: 1805–1816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhuiyan H. and Schmekel, K. 2004. Meiotic chromosome synapsis in yeast can occur without Spo11-induced DNA double-strand breaks. Genetics 168: 775–783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bishop D.K. 1994. RecA homologs Dmc1 and Rad51 interact to form multiple nuclear complexes prior to meiotic chromosome synapsis. Cell 79: 1081–1092. [DOI] [PubMed] [Google Scholar]
- Bishop D.K. and Zickler, D. 2004. Early decision: Meiotic crossover interference prior to stable strand exchange and synapsis. Cell 117: 9–15. [DOI] [PubMed] [Google Scholar]
- Blat Y., Protacio, R.U., Hunter, N., and Kleckner, N. 2002. Physical and functional interactions among basic chromosome organizational features govern early steps of meiotic chiasma formation. Cell 111: 791–802. [DOI] [PubMed] [Google Scholar]
- Borner G.V., Kleckner, N., and Hunter, N. 2004. Crossover/noncrossover differentiation, synaptonemal complex formation, and regulatory surveillance at the leptotene/zygotene transition of meiosis. Cell 117: 29–45. [DOI] [PubMed] [Google Scholar]
- Burgess S.M. 2002. Homologous chromosome associations and nuclear order in meiotic and mitotically dividing cells of budding yeast. Adv. Genet. 46: 49–90. [DOI] [PubMed] [Google Scholar]
- ____. 2004. Homolog pairing in S. pombe: The ends are the means. Mol. Cell 13: 766–768. [DOI] [PubMed] [Google Scholar]
- Burgess S.M. and Kleckner, N. 1999. Collisions between yeast chromosomal loci in vivo are governed by three layers of organization. Genes & Dev. 13: 1871–1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgess S.M., Kleckner, N., and Weiner, B.M. 1999. Somatic pairing of homologs in budding yeast: Existence and modulation. Genes & Dev. 13: 1627–1641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carpenter A.T. 1987. Gene conversion, recombination nodules, and the initiation of meiotic synapsis. Bioessays 6: 232–236. [DOI] [PubMed] [Google Scholar]
- Chua P.R. and Roeder, G.S. 1997. Tam1, a telomere-associated meiotic protein, functions in chromosome synapsis and crossover interference. Genes & Dev. 11: 1786–1800. [DOI] [PubMed] [Google Scholar]
- ____. 1998. Zip2, a meiosis-specific protein required for the initiation of chromosome synapsis. Cell 93: 349–359. [DOI] [PubMed] [Google Scholar]
- Colaiacovo M.P., MacQueen, A.J., Martinez-Perez, E., McDonald, K., Adamo, A., La Volpe, A., and Villeneuve, A.M. 2003. Synaptonemal complex assembly in C. elegans is dispensable for loading strand-exchange proteins but critical for proper completion of recombination. Dev. Cell 5: 463–474. [DOI] [PubMed] [Google Scholar]
- Conrad M.N., Dominguez, A.M., and Dresser, M.E. 1997. Ndj1p, a meiotic telomere protein required for normal chromosome synapsis and segregation in yeast. Science 276: 1252–1255. [DOI] [PubMed] [Google Scholar]
- Dawson D.S., Murray, A.W., and Szostak, J.W. 1986. An alternative pathway for meiotic chromosome segregation in yeast. Science 234: 713–717. [DOI] [PubMed] [Google Scholar]
- de los Santos T., Hunter, N., Lee, C., Larkin, B., Loidl, J., and Hollingsworth, N.M. 2003. The Mus81/Mms4 endonuclease acts independently of double-Holliday junction resolution to promote a distinct subset of crossovers during meiosis in budding yeast. Genetics 164: 81–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dernburg A.F., McDonald, K., Moulder, G., Barstead, R., Dresser, M., and Villeneuve, A.M. 1998. Meiotic recombination in C. elegans initiates by a conserved mechanism and is dispensable for homologous chromosome synapsis. Cell 94: 387–398. [DOI] [PubMed] [Google Scholar]
- Ding D.Q., Yamamoto, A., Haraguchi, T., and Hiraoka, Y. 2004. Dynamics of homologous chromosome pairing during meiotic prophase in fission yeast. Dev. Cell 6: 329–341. [DOI] [PubMed] [Google Scholar]
- Fung J.C., Rockmill, B., Odell, M., and Roeder, G.S. 2004. Imposition of crossover interference through the nonrandom distribution of synapsis initiation complexes. Cell 116: 795–802. [DOI] [PubMed] [Google Scholar]
- Goldman A.S. and Lichten, M. 2000. Restriction of ectopic recombination by interhomolog interactions during Saccharomyces cerevisiae meiosis. Proc. Natl. Acad. Sci. 97: 9537–9542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higgins J.D., Armstrong, S.J., Franklin, F.C., and Jones, G.H. 2004. The Arabidopsis MutS homolog AtMSH4 functions at an early step in recombination: Evidence for two classes of recombination in Arabidopsis. Genes & Dev. 18: 2557–2570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollingsworth N.M. and Brill, S.J. 2004. The Mus81 solution to resolution: Generating meiotic crossovers without Holliday junctions. Genes & Dev. 18: 117–125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter N. and Kleckner, N. 2001. The single-end invasion: An asymmetric intermediate at the double-strand break to double-Holliday junction transition of meiotic recombination. Cell 106: 59–70. [DOI] [PubMed] [Google Scholar]
- Jin Q., Trelles-Sticken, E., Scherthan, H., and Loidl, J. 1998. Yeast nuclei display prominent centromere clustering that is reduced in nondividing cells and in meiotic prophase. J. Cell Biol. 141: 21–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kane S.M. and Roth, R. 1974. Carbohydrate metabolism during ascospore development in yeast. J. Bacteriol. 18: 8–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kee K. and Keeney, S. 2002. Functional interactions between SPO11 and REC102 during initiation of meiotic recombination in Saccharomyces cerevisiae. Genetics 160: 111–122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keeney S. 2001. Mechanism and control of meiotic recombination initiation. Curr. Top. Dev. Biol. 52: 1–53. [DOI] [PubMed] [Google Scholar]
- Kemp B., Boumil, R.M., Stewart, M.N., and Dawson, D.S. 2004. A role for centromere pairing in meiotic chromosome segregation. Genes & Dev. 18: 1946–1951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kramer J. and Hawley, R.S. 2003. The spindle-associated transmembrane protein Axs identifies a membranous structure ensheathing the meiotic spindle. Nat. Cell Biol. 5: 261–263. [DOI] [PubMed] [Google Scholar]
- Mahadevaiah S.K., Turner, J.M., Baudat, F., Rogakou, E.P., de Boer, P., Blanco-Rodriguez, J., Jasin, M., Keeney, S., Bonner, W.M., and Burgoyne, P.S. 2001. Recombinational DNA double-strand breaks in mice precede synapsis. Nat. Genet. 27: 271–276. [DOI] [PubMed] [Google Scholar]
- McKim K.S., Jang, J.K., and Manheim, E.A. 2002. Meiotic recombination and chromosome segregation in Drosophila females. Annu. Rev. Genet. 36: 205–232. [DOI] [PubMed] [Google Scholar]
- Moens P.B., Chen, D.J., Shen, Z., Kolas, N., Tarsounas, M., Heng, H.H., and Spyropoulos, B. 1997. Rad51 immunocytology in rat and mouse spermatocytes and oocytes. Chromosoma 106: 207–215. [DOI] [PubMed] [Google Scholar]
- Nag D.K., Scherthan, H., Rockmill, B., Bhargava, J., and Roeder, G.S. 1995. Heteroduplex DNA formation and homolog pairing in yeast meiotic mutants. Genetics 141: 75–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novak J.E., Ross-Macdonald, P.B., and Roeder, G.S. 2001. The budding yeast Msh4 protein functions in chromosome synapsis and the regulation of crossover distribution. Genetics 158: 1013–1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Page S.L. and Hawley, R.S. 2003. Chromosome choreography: The meiotic ballet. Science 301: 785–789. [DOI] [PubMed] [Google Scholar]
- Paques F. and Haber, J.E. 1999. Multiple pathways of recombination induced by double-strand breaks in Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev. 63: 349–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pawlowski W.P., Golubovskaya, I.N., Timofejeva, L., Meeley, R.B., Sheridan, W.F., and Cande, W.Z. 2004. Coordination of meiotic recombination, pairing, and synapsis by PHS1. Science 303: 89–92. [DOI] [PubMed] [Google Scholar]
- Peoples T.L., Dean, E., Gonzalez, O., Lambourne, L., and Burgess, S.M. 2002. Close, stable homolog juxtaposition during meiosis in budding yeast is dependent on meiotic recombination, occurs independently of synapsis, and is distinct from DSB-independent pairing contacts. Genes & Dev. 16: 1682–1695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rockmill B. and Roeder, G.S. 1998. Telomere-mediated chromosome pairing during meiosis in budding yeast. Genes & Dev. 12: 2574–2586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roeder G.S. 1997. Meiotic chromosomes: It takes two to tango. Genes & Dev. 11: 2600–2621. [DOI] [PubMed] [Google Scholar]
- Ross-Macdonald P. and Roeder, G.S. 1994. Mutation of a meiosis-specific MutS homolog decreases crossing over but not mismatch correction. Cell 79: 1069–1080. [DOI] [PubMed] [Google Scholar]
- Scherthan H. 2001. A bouquet makes ends meet. Nat. Rev. Mol. Cell Biol. 2: 621–627. [DOI] [PubMed] [Google Scholar]
- Schwacha A. and Kleckner, N. 1995. Identification of double Holliday junctions as intermediates in meiotic recombination. Cell 83: 783–791. [DOI] [PubMed] [Google Scholar]
- ____. 1997. Interhomolog bias during meiotic recombination: Meiotic functions promote a highly differentiated interhomolog-only pathway. Cell 90: 1123–1135. [DOI] [PubMed] [Google Scholar]
- Smithies O. and Powers, P.A. 1986. Gene conversions and their relation to homologous chromosome pairing. Philos. Trans. R Soc. Lond. B Biol. Sci. 312: 291–302. [DOI] [PubMed] [Google Scholar]
- Snowden T., Acharya, S., Butz, C., Berardini, M., and Fishel, R. 2004. hMSH4–hMSH5 recognizes Holliday junctions and forms a meiosis-specific sliding clamp that embraces homologous chromosomes. Mol. Cell 15: 437–451. [DOI] [PubMed] [Google Scholar]
- Stahl F.W., Foss, H.M., Young, L.S., Borts, R.H., Abdullah, M.F., and Copenhaver, G.P. 2004. Does crossover interference count in Saccharomyces cerevisiae? Genetics 168: 35–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storlazzi A., Tesse, S., Gargano, S., James, F., Kleckner, N., and Zickler, D. 2003. Meiotic double-strand breaks at the interface of chromosome movement, chromosome remodeling, and reductional division. Genes & Dev. 17: 2675–2687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tesse S., Storlazzi, A., Kleckner, N., Gargano, S., and Zickler, D. 2003. Localization and roles of Ski8p protein in Sordaria meiosis and delineation of three mechanistically distinct steps of meiotic homolog juxtaposition. Proc. Natl. Acad. Sci. 100: 12865–12870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trelles-Sticken E., Loidl, J., and Scherthan, H. 1999. Bouquet formation in budding yeast: Initiation of recombination is not required for meiotic telomere clustering. J. Cell Sci. 112: 651–658. [DOI] [PubMed] [Google Scholar]
- Trelles-Sticken E., Dresser, M.E., and Scherthan, H. 2000. Meiotic telomere protein Ndj1p is required for meiosis-specific telomere distribution, bouquet formation and efficient homologue pairing. J. Cell Biol. 151: 95–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wach A., Brachat, A., Pohlmann, R., and Philippsen, P. 1994. New heterologous modules for classical or PCR-based gene disruptions in Saccharomyces cerevisiae. Yeast 10: 1793–1808. [DOI] [PubMed] [Google Scholar]
- Weiner B.M. and Kleckner, N. 1994. Chromosome pairing via multiple interstitial interactions before and during meiosis in yeast. Cell 77: 977–991. [DOI] [PubMed] [Google Scholar]
- Xu L., Ajimura, M., Padmore, R., Klein, C., and Kleckner, N. 1995. NDT80, a meiosis-specific gene required for exit from pachytene in Saccharomyces cerevisiae. Mol. Cell. Biol. 15: 6572–6581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zickler D. and Kleckner, N. 1998. The leptotene–zygotene transition of meiosis. Annu. Rev. Genet. 32: 619–697. [DOI] [PubMed] [Google Scholar]
- ____. 1999. Meiotic chromosomes: Integrating structure and function. Annu. Rev. Genet. 33: 603–754. [DOI] [PubMed] [Google Scholar]