Abstract
Deletions, translocations, or point mutations in the CREB-binding protein (CBP) gene have been associated with Rubinstein-Taybi Syndrome; a human developmental disorder characterized by retarded growth and reduced mental function. To examine the role of CBP in memory, transgenic mice were generated in which the CaMKIIα promoter drives expression of an inhibitory truncated CBP protein in forebrain neurons. Examination of hippocampal long-term potentiation (LTP), a form of synaptic plasticity thought to underlie memory storage, revealed significantly reduced late-phase LTP induced by dopamine-regulated potentiation in hippocampal slices from CBP transgenic mice. However, four-train induced late-phase LTP is normal. Behaviorally, CBP transgenic mice exhibited memory deficits in spatial learning in the Morris water maze and deficits in long-term memory for contextual fear conditioning, two hippocampus-dependent tasks. Together, these results demonstrate that CBP is involved in specific forms of hippocampal synaptic plasticity and hippocampus-dependent long-term memory formation.
Studies have demonstrated a pivotal role for transcription in learning and memory, from early experiments showing that RNA synthesis inhibitors block long-term memory without altering short-term memory, to studies in Drosophila, Aplysia, and mice demonstrating that transcription factor cAMP response element binding protein (CREB) is involved in long-term memory (Lonze and Ginty 2002; Kaplan and Abel 2003). Use-dependent changes in synaptic strength in the hippocampus and other brain regions are thought to underlie memory storage. One intensely studied form of synaptic plasticity is hippocampal long-term potentiation (LTP), a persistent, activity-dependent form of synaptic enhancement (Bliss and Richter-Levin 1993; Martin and Morris 2002). CREB has been shown to be involved in certain types of hippocampal LTP as well as long-term memory (for review, see Kaplan and Abel 2003). In most cases, CREB activates transcription of target genes by recruiting the coactivator CREB-binding protein (CBP) (Goodman and Smolik 2000), via the interaction between the Ser-133 phosphorylated kinase-inducible domain (KID) of CREB and the KIX domain of CBP (Chrivia et al. 1993). CBP stimulates transcription in two ways: via its intrinsic histone acetyltransferase (HAT) activity and via recruitment of components of the general transcriptional machinery (for review, see Vo and Goodman 2001).
A role for CBP in memory storage was first demonstrated in a mouse model of Rubinstein-Taybi Syndrome (Oike et al. 1999). RTS is a human developmental disorder characterized by retarded growth and reduced mental function (Rubinstein and Taybi 1963; Hennekam et al. 1992; Cantani and Gagliesi 1998), caused by breakpoints, microdeletions and point mutations in CBP (Petrij et al. 1995; Coupry et al. 2004). Mice heterozygous for an inhibitory truncated CBP allele, lacking the HAT domain and carboxyl terminus, showed many features of RTS such as growth retardation, cardiac anomalies, and skeletal abnormalities as well as deficient long-term memory (Oike et al. 1999; Bourtchouladze et al. 2003). These results suggest that CBP may have a role in long-term memory, but this interpretation needs to be made with caution due to the significant developmental abnormalities in this mouse model of RTS in which the alterations in CBP are not spatially or temporally restricted.
To examine the role of CBP in memory independent of its role in development or in peripheral tissues, we generated transgenic mice that express CBPΔ1, an inhibitory truncated CBP protein lacking the HAT domain and carboxyl terminus. CBPΔ1 is expressed from the calcium/calmodulin-dependent protein kinase IIα (CaMKIIα) promoter, which drives expression post-natally in neurons within the hippocampus, striatum, amygdala, and cortex (Mayford et al. 1996; Kojima et al. 1997). CaMKIIα-CBPΔ1 transgenic mice exhibit deficits in specific forms of hippocampal synaptic plasticity and impairments in spatial learning and long-term memory for contextual fear conditioning. Our analysis of CaMKIIα-CBPΔ1 transgenic mice suggests that this transcriptional coactivator has a role in synaptic plasticity and memory storage.
Results
Generation of transgenic mice expressing truncated CBP (CBPΔ1)
CBPΔ1 is based on the CBP truncation mutant generated by Oike et al. (1999) in their screen to identify genes associated with development in mice. Heterozygous CBP-deficient mice, which express the truncated CBP protein (lacking amino acids 1084-2441) thought to be acting as a dominant negative inhibitor, showed clinical features of Rubinstein-Taybi syndrome (RTS) (Oike et al. 1999). We deleted amino acids 1084-2441 to generate the same inhibitory form of CBP (CBPΔ1). CBPΔ1 contains the CREB-binding domain (KIX), which interacts with CREB and other factors, but lacks the BROMO domain and the histone acetyltransferase (HAT) domain in the carboxy-terminal half of CBP (Fig. 1A). Importantly, the CBPΔ1 truncation mutant is predicted to bind factors that interact with the amino terminus of CBP. Thus only a subset of CBP-interacting factors, including CREB, should be affected by this mutant.
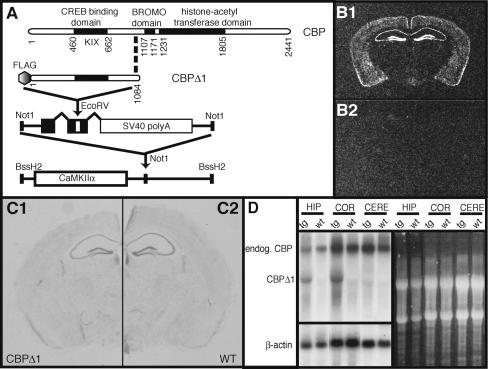
Figure 1.
Generation of CaMKIIα-CBPΔ1 transgenic animals. (A) A mouse CBP cDNA truncation mutant, including amino acids 1-1084, was FLAG-epitope tagged at the amino terminus and cloned into a vector containing intron and exon sequences with splice sites and the SV40 polyadenylation signal. This entire sequence was then cloned downstream of the 8.5 kb mouse CaMKIIα promoter. This construct was then used to generate CaMKIIα-CBPΔ1 transgenic mice via pronuclear injection. (B) Coronal sections from the brains of CBPΔ1 transgenic mice (B1) and a wild-type littermate (B2) were hybridized with a probe specific to the CBPΔ1 transgene. Expression of the transgene is observed in the hippocampus, cortex, striatum, and amygdala. (C) Coronal sections from a CBPΔ1 transgenic mouse (C1) and a wild-type littermate (C2) were Nissl stained to confirm integrity of the hippocampus and other structures. (D) Total RNA was isolated from the hippocampus, cortex, and cerebellum of CaMKIIα-CBPΔ1 transgenic mice and wild-type littermates and analyzed by Northern blot. The membrane was first hybridized to an aminoterminal CBP radiolabeled probe, stripped, and then subject to a β-actin radiolabeled probe. The ethidium bromide-stained RNA gel is also shown, revealing 18S and 28S rRNAs.
CBPΔ1 was FLAG-epitope tagged (Wood et al. 2000) and expressed from the CaMKIIα promoter (Mayford et al. 1996) to restrict transgene expression spatially to forebrain neurons. A transgenic approach was chosen because it allowed for expression of the CBP truncation product to be spatially restricted to neurons within the hippocampus, striatum, amygdala, and cortex as well as temporally restricted to post-natal development (Kojima et al. 1997) in order to avoid the developmental effects and lethality observed in more conventional CBP mutations (Tanaka et al. 1997; Oike et al. 1999). Additionally, CBP is thought to be present in the cell at limiting concentrations (for review, see Kamei et al. 1996; Vo and Goodman 2001) and due to transgene copy number and transgene insertion effects, different transgenic lines may have different levels of expression resulting in a range of impairments in CBP function. Three independent transgenic mouse lines expressing FLAG-CBPΔ1 were generated. The three lines had low (line 1352), medium (line 1353), and high (line 1364) levels of transgene expression as determined by semiquantitative in situ hybridization. Even though there is a wide range of expression from line 1352 to line 1364, all three lines have 25-30 transgene copies, as determined by Southern blot analysis (data not shown). As shown in Figure 1B, the transgene is expressed in the hippocampus, amygdala, striatum, and cortex in line 1364. All three lines were analyzed behaviorally, but as only line 1364 exhibited hippocampus-dependent memory deficits, we focused on line 1364 for further analysis.
The CaMKIIα promoter does not become active until 10-21 d post-partum (Kojima et al. 1997) and expression is restricted spatially to forebrain neurons, which may be one reason why CBPΔ1 transgenic mice do not have the developmental impairments (growth retardation, significantly reduced weight, retarded osseous maturation and other skeletal abnormalities, cardiac abnormalities, and seizures) observed in the heterozygous CBP-deficient mice generated by Oike et al. (1999). CBPΔ1 transgenic mice are developmentally similar to their wild-type littermates. There was no observable weight difference between CBPΔ1 transgenic males (27.8 ± 1.2 g) and wild-type male littermates (25.4 ± 1.3 g) (t[7] = 1.11, p = NS) or between CBPΔ1 transgenic females (21.3 ± 0.4 g) and wild-type female littermates (20.8 ± 0.4 g) (t[7] = 0.83, p = NS). To observe locomotor activity and anxiety, mice were examined on the elevated zero maze. No differences were observed in the number of transitions between the open and closed quadrants (t[22] = 1.21, p = NS), head pokes into open quadrants (t[22] = 0.19, p = NS), or time spent in the open quadrant (t[22] = 1.47, p = NS). In Figure 1C, Nissl staining of coronal sections from CBPΔ1 transgenic (C1) and wild-type littermates (C2) showed no gross morphological differences. Northern blot analysis showed that the CBPΔ1 transcript is expressed in the hippocampus and cortex, but not the cerebellum, of CBPΔ1 transgenic mice (Fig. 1D). Quantification of the CBPΔ1 transcript and endogenous CBP transcript revealed that the transgene product is expressed at 95% of endogenous CBP levels in the hippocampus and 84% of endogenous CBP levels in the cortex. We used Affymetrix U74Av2 microarrays to examine changes in gene expression at basal conditions. Five CBPΔ1 transgenic mice and five wild-type littermates were taken from their homecages, hippocampi were dissected, and cRNA target was prepared according to the recommended protocol (Affymetrix). Data were processed using Robust Multiarray Average (RMA) (Bolstad et al. 2003). Statistical Analysis of Microarray (SAM) (Tusher et al. 2001) was used to estimate the significance of expression changes between transgenic and wild-type animals. In these basal-state conditions, we did not observe any significant changes in hippocampal gene expression between CBPΔ1 transgenic mice and wild-type littermates, including no differences in the following housekeeping genes: tubulin, β-actin, 28S rRNA, ribosomal proteins RPL23 and RPL13A, hprt, gapdh, and ubc (M.A. Wood, M.B. Keeley, C. Isiegas, K. Hellman, J. Stein, and T. Abel, unpubl.).
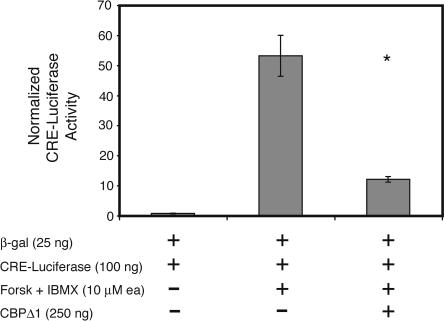
The truncated CBP mutant in CBPΔ1 transgenic mice was designed to interrupt transcription factors utilizing CBP as a co-activator for the expression of their target genes. One such transcription factor is CREB. To examine the effect of the truncated CBP mutant on CRE-mediated transcription, we expressed it in HEK293 cells along with a CRE-luciferase reporter gene. CRE-mediated transcription induced by forskolin/IBMX treatment was inhibited by CBPΔ1 by ∼75% (Fig. 2). Normalized CRE-luciferase levels reached 53.1 ± 6.8 relative light units (RLUs), but only 11.9 ± 0.9 RLUs in the presence of the truncated CBP mutant, indicating that this CBP mutant significantly affects CRE-dependent forskolin-stimulated transcription (t[10] = 6.01, p < 0.0001).
Figure 2.
CBPΔ1 inhibits CRE-mediated transcription in tissue culture. The truncated CBP mutant (CBPΔ1) inhibits CRE-mediated transcription by ∼75%. Normalized CRE-luciferase levels reached 53.1 ± 6.8 relative light units (RLUs), but only 11.9 ± 0.9 RLUs in the presence of the truncated CBP mutant, showing that this CBP mutant affects CRE-dependent transcription. * indicates p < 0.0001.
Reduced long-term potentiation in the hippocampus of CBPΔ1 transgenic mice
Long-term potentiation (LTP), the activity-dependent change in the strength of neuronal connections, is a form of long-lasting synaptic plasticity that has been proposed as a cellular mechanism of learning and memory (for review, see Martin and Morris 2002). To determine whether CBPΔ1 transgenic mice exhibit deficits in LTP, we characterized three forms of LTP in the CA1 region of hippocampal slices from mutant mice and their wild-type littermates: the transient early form of LTP resulting from a single 1 sec, 100 Hz train (E-LTP), and the longer-lasting forms of LTP resulting from four 1 sec, 100 Hz trains delivered 5 min apart (tetraburst L-LTP), or from a single 100 Hz train delivered in the presence of a D1 dopaminergic agonist, chloro-APB hydrobromide (Pittenger et al. 2002). The enhancement of LTP by D1 agonists is reduced by K-CREB, a dominant negative form of CREB (Pittenger et al. 2002), suggesting that it might also be affected by the inhibitory CBPΔ1 mutant. Conversely, E-LTP and tetraburst L-LTP are not affected by K-CREB and are predicted to be unaffected by the inhibitory CBPΔ1 mutant.
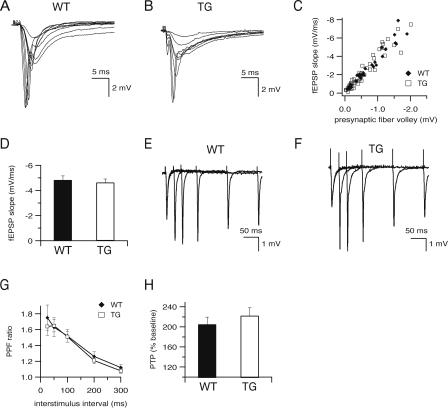
Before examining changes in LTP, it was necessary to determine whether hippocampal slices from CBPΔ1 transgenic mice showed any significant changes in baseline synaptic transmission. Field potentials in the CA1 region were elicited with bipolar stimulating electrodes placed in the Schaffer collaterals and characterized with respect to initial slope, peak amplitude, and input-output relationship. Hippocampal slices from CBPΔ1 transgenic and wild-type littermates were similar with respect to these baseline parameters (Fig. 3). Field potentials in slices from wild type and CBPΔ1 transgenic mice exhibit similar waveforms (Fig. 3A,B, respectively), and a scatter plot relating the initial slope of the fEPSP to the size of the presynaptic fiber volley (Fig. 3C) shows that expression of CBPΔ1 in transgenic animals (n = 7 slices from four mice) does not change the input-output characteristics in area CA1 as compared to wild-type littermates (n = 4 slices from three mice). There was also no difference in maximum responses, with average initial slopes of -4.8 ± 0.3 for wild type and -4.6 ± 0.3 mV/msec for transgenic littermates (Fig. 3D; t[19] = 0.26, p = NS; n = 26 slices from 16 wild-type mice and n = 32 slices from 15 CBPΔ1 transgenic mice).
Figure 3.
Baseline electrophysiological properties are normal in CBPΔ1 transgenic mice. (A-C) Input-output characteristics were examined by recording field potentials (fEPSPs) in area CA1 resulting from stimuli of increasing intensity delivered to the Schaeffer collaterals in 400 μm hippocampal slices from wild type (A) and CBPΔ1 transgenic mice (B). Plots of fEPSP initial slopes versus the corresponding presynaptic fiber volley amplitudes (C) are similar in slices from wild-type littermates (n = 4 slices from three mice) and CBPΔ1 transgenic mice (n = 7 slices from four mice). (D) Maximum fEPSP slopes are similar in CBPΔ1 transgenic mice (n = 32 slices from 15 mice) and wild-type littermates (n = 26 slices from 16 mice). Sample traces of paired pulse facilitation at 25, 50, 100, 200, and 300 msec are shown in slices from wild type (E) and CBPΔ1 transgenic mice (F). (G) Paired-pulse facilitation is not significantly different between wild type (n = 15 slices from eight mice) and CBPΔ1 transgenic mice (n = 21 slices from nine mice) at interstimulus intervals between 25 and 300 msec. (H) Post-tetanic potentiation is not altered in CBPΔ1 transgenic mice (n = 11 slices from eight transgenic mice and n = 10 slices from eight wild-type mice).
To determine whether short-term forms of synaptic plasticity are affected by expression of CBPΔ1 in transgenic mice, we examined post-tetanic potentiation and paired pulse facilitation, a form of short-term synaptic plasticity that is sensitive to changes in presynaptic probability of release (Zucker 1989), in hippocampal slices from CBPΔ1 transgenic and wild-type littermates. When pairs of stimuli were delivered at interstimulus intervals from 25 to 300 msec, both wild type (n = 15 slices from eight mice) and CBPΔ1 transgenic hippocampal slices (n = 21 slices from nine mice) display similar levels of facilitation (Fig. 3E-G). There were also no significant differences in post-tetanic potentiation following a single 1 sec, 100 Hz train, either in control saline (Fig. 3H; t[14] = 0.78, p = NS; n = 10 slices from eight wild-type mice and n = 11 slices from eight CBPΔ1 transgenic mice), or in the presence of chloro-APB (Fig. 4B; t[10] = 0.33, p = NS; n = 5 slices from five wild-type mice and n = 7 slices from seven CBPΔ1 transgenic mice).
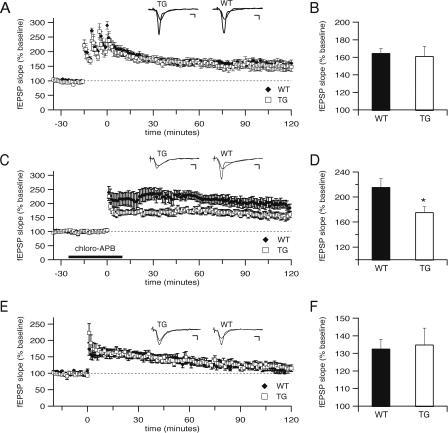
Figure 4.
Long-term potentiation deficits in CBPΔ1 transgenic mice. (A) Long-term potentiation (LTP) at Schaffer collateral synapses in response to four 1 sec, 100 Hz trains administered 5 min apart is normal in CBPΔ1 transgenic mice. Both CBPΔ1 transgenic mice (n = 7 slices from four mice) and wild-type littermates (n = 6 slices from five mice) show stable potentiation. Insets show superimposed sample sweeps from before and 60 min after tetanic stimulation for CBPΔ1 transgenic mice and wild-type littermates. (B) At 60 min following tetanic stimulation, wild-type mice showed potentiation to 165 ± 5% of control, and CBPΔ1 transgenic mice showed potentiation to 161 ± 11% of control. (C) LTP in response to a single 1 sec, 100 Hz train in the presence of a D1 dopaminergic agonist (10 μM chloro-APB hydrobromide) is impaired in CBPΔ1 transgenic mice. Both CBPΔ1 transgenic mice (n = 7 slices from seven mice) and wild-type littermates (n = 5 slices from five mice) show stable potentiation, but CBPΔ1 transgenic hippocampal slices show significantly reduced LTP. (D) At 60 min following tetanic stimulation, wild-type mice showed potentiation to 216 ± 13% of control, whereas CBPΔ1 transgenic mice showed significantly decreased potentiation to 176 ± 8% of control. (E) Long-term potentiation in response to a single 1 sec, 100 Hz train in control saline is not significantly different between slices from CBPΔ1 transgenic mice (n = 5 slices from four mice) and wild-type littermates (n = 4 slices from three mice). (F) At 60 min following tetanic stimulation by a single 1 sec, 100 Hz train in control saline, wild-type mice (133 ± 5% of control) show equivalent potentiation to CBPΔ1 transgenic mice (135 ± 9% of control). * indicates p < 0.05. Scale bars: 3 msec, 1 mV.
To assess the effects of the expression of CBPΔ1 on longterm potentiation, test pulses were delivered once per minute to the Schaffer collaterals and the resulting fEPSPs were recorded. After recording stable baseline fEPSP responses for at least 15 min, LTP was induced by one of three protocols. When tetraburst L-LTP was induced by four 1 sec, 100 Hz trains delivered 5 min apart, average responses for hippocampal slices from wild-type mice (n = 6 slices from five mice) and CBPΔ1 transgenic mice (n = 7 slices from four mice) show that this form of LTP is not significantly different between groups (Fig. 4A; F[1,1099] = 0.24, p = NS), with average potentiation at 60 min at 165 ± 5% of control for wild-type mice and 161 ± 11% for CBPΔ1 transgenic mice (Fig. 4B). To study a second form of LTP, sections were treated with 10 μM chloro-APB in 0.05% EtOH for 25 min, followed by a single 1 sec, 100 Hz train. Average responses for hippocampal slices from wild-type mice (n = 5 slices from five mice) and CBPΔ1 transgenic mice (n = 7 slices from seven mice) show that this form of LTP is significantly reduced in CBPΔ1 transgenic mice (Fig. 4C; F[1,1150] = 9.41, p < 0.05), with average potentiation at 60 min at 216 ± 13% of control for wild-type mice and 176 ± 8% for CBPΔ1 transgenic mice (Fig. 4D; t[10] = 2.68, p < 0.05). In contrast, there was no difference between hippocampal slices from wild-type mice (n = 4 slices from three mice) and CBPΔ1 transgenic mice (n = 5 slices from four mice) in the E-LTP resulting from a single 1 sec, 100 Hz train in control saline (Fig. 4E; F[1,426] = 0.23, p = NS), with no differences between wild type (133 ± 5% of control) and CBPΔ1 transgenic mice (135 ± 9% of control) in the average potentiation at 60 min (Fig. 4F; t[6] = 0.50, p = NS). These results indicate a role for CBP in long-lasting hippocampal synaptic plasticity induced by pairing tetanic electrical stimulation and D1 receptor activation.
CBPΔ1 transgenic mice exhibit deficit in spatial learning as observed in the hidden-platform version of the Morris water maze
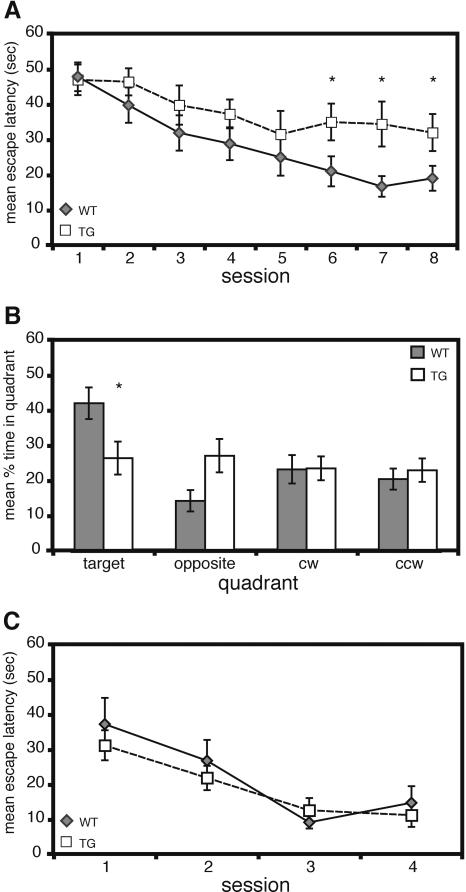
The hidden-platform version of the Morris water maze, a hippocampus-dependent task, requires an animal to learn and remember the relationships between multiple distal cues and the platform location to escape the water (for review, see Lipp and Wolfer 1998; D'Hooge and De Deyn 2001). Acquisition of this task normally requires between six and 10 days of training depending on the genetic background of the mouse and the number of trials per day (Kogan et al. 1997; Lipp and Wolfer 1998). As shown in Figure 5A, CBPΔ1 transgenic mice have a significant overall impairment in acquisition as compared to wild-type littermates (main effect of genotype: F[1,133] = 7.07, p < 0.05; main effect of day: F[7,133] = 6.89, p < 0.0001; genotype X day interaction: F[7,133] = 0.82, p = NS). CBPΔ1 transgenic mice (35.0 ± 5.2 sec) reached an acquisition plateau on the fifth day of training whereas wild-type littermates (21.1 ± 4.3 sec) reached a significantly lower average escape latency. There was no difference in swim speed (main effect of genotype: F[1,133] = 1.87, p = NS) or percent time spent floating (defined as swimming less than 5 cm/sec) between the two groups (main effect of genotype: F[1,133] = 0.24, p = NS). CBPΔ1 transgenic mice did demonstrate significantly more thigmotaxis (swimming in the outer 10% of the pool) during training than did wild-type littermates (main effect of genotype: F[1,133] = 5.92, p < 0.05). To assess spatial memory more directly, the mice were subject to probe trials, in which the platform was removed and the mice were allowed 60 sec to search the pool. The search pattern of an animal reveals spatial bias, which is thought to represent spatial long-term memory (Schenk and Morris 1985). There were no significant differences observed between groups on the probe trial the day after the fourth session (data not shown; main effect of quadrant: F[3,57] = 1.94, p = NS; main effect of genotype: F[1,57] = 2.35, p = NS; genotype X quadrant interaction: F[3,57] = 0.62, p = NS). However, during the probe trial at the end of training on the tenth day, CBPΔ1 transgenic mice spent significantly less time in the target quadrant, where the platform was located during training, than wild-type littermates (Fig. 5B; main effect of quadrant: F[3,57] = 3.78, p < 0.05; main effect of genotype: F[1,57] = 0.97, p = NS; genotype X quadrant interaction: F[3,57] = 3.36, p < 0.05). Wild-type mice show a preference for the target quadrant relative to other quadrants whereas CBPΔ1 transgenic mice do not show a spatial preference (Fig. 5B). No differences were observed in swim speed, floating, or thigmotaxis between the two groups during the probe trial (swim speed: t[19] = 0.89, p = NS; floating: t[19] = 0.48, p = NS; thigmotaxis: t[19] = 1.67, p = NS). We have also observed significant impairments in a recently derived CBPΔ1 transgenic mouse line (line 1611) for the hidden platform version of the water maze (data not shown), supporting our findings in the CBPΔ1 transgenic mouse line 1364 presented here.
Figure 5.
CBPΔ1 trangenic mice exhibit impaired spatial learning. (A) Mean escape latency during acquisition of the hidden-platform version of the Morris water maze. CBPΔ1 transgenic mice (n = 9) have a significant overall impairment in acquisition as compared to wild-type littermates (n = 12). (B) Mean percent time spent in each quadrant during the post-training probe trial on day ten. CBPΔ1 transgenic mice (n = 9) spent significantly less time in the target quadrant, where the platform was located during training, than did wild-type littermates (n = 12). (C) Mean escape latency during acquisition of the visible-platform version of the Morris water maze. A different group of mice were tested in the visible-platform experiment than those tested in the hidden-platform experiment. There was no difference observed between CBPΔ1 transgenic mice (n = 8) and wild-type littermates (n = 5) in the visible platform version of the Morris water maze. * indicates p < 0.05.
There was no difference observed between CBPΔ1 transgenic mice and wild-type littermates in escape latencies during training in the visible platform version of the water maze (Fig. 5C; main effect of genotype: F[1,33] = 0.58, p = NS). Both groups did improve across days demonstrating that the task was acquired (main effect of day: F[3,33] = 13.45, p < 0.0001; genotype X day interaction: F[3,33] = 0.53, p = NS). A different set of mice were used for the visible platform version of the water maze than for the hidden platform version. Overall, these results indicate that CBPΔ1 transgenic mice have impaired spatial learning.
CBPΔ1 transgenic mice exhibit impaired contextual fear conditioning but normal cued fear conditioning
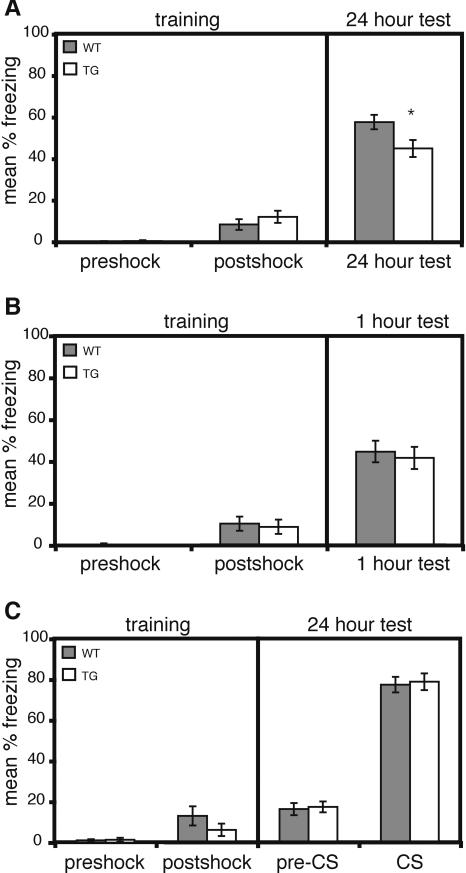
Contextual and cued fear conditioning are two forms of associative learning that induce effective memory for either the context or the cue after a single training session (e.g., Abel et al. 1997; LeDoux 2000). These two types of fear conditioning are thought to require partially distinct, but overlapping, neural systems. Lesions of the hippocampus affect contextual conditioning whereas lesions of the amygdala affect both contextual and cued conditioning (for review, see Maren 2001). Memory for the conditioned stimulus was measured as freezing, an absence of visible movement, when presented with the conditioned context or cue in a 24 h retention test. CBPΔ1 transgenic mice and wild-type littermates showed the same level of freezing before and after the shock on training day (Fig. 6A; preshock: wild-type mice exhibited 0.2 ± 0.2% freezing; transgenic mice exhibited 0.4 ± 0.2% freezing; t[36] = 0.52, p = NS; post-shock: wild-type mice exhibited 8.3 ± 2.5% freezing; transgenic mice exhibited 12.1 ± 2.9% freezing; t[36] = 0.95, p = NS). However, when re-exposed to the conditioned context 24 h later, CBPΔ1 transgenic mice show a significant decrease in freezing compared to their wild-type littermates (Fig. 6A; 24 h test: wild-type mice exhibited 57.6 ± 3.4% freezing; transgenic mice exhibited 44.9 ± 4.1% freezing; t[36] = 2.37, p < 0.05). To assess whether CBPΔ1 transgenic mice have normal short-term memory for contextual fear, a separate set of CBPΔ1 transgenic mice and wild-type littermates were tested in the conditioned context 1 h after training (Fig. 6B). CBPΔ1 transgenic mice and wild-type littermates showed similar levels of freezing 1 h after training (Fig. 6B; wild-type mice exhibited 45.0 ± 5.2% freezing; transgenic mice exhibited 41.9 ± 5.3% freezing; t[20] = 0.69, p = NS). This indicates that CBPΔ1 transgenic mice have normal short-term memory for the conditioned context.
Figure 6.
CBPΔ1 transgenic mice exhibit impaired long-term memory for contextual fear conditioning but normal cued fear conditioning. (A) Training and 24 h long-term memory test during contextual fear conditioning. There was no difference in freezing behavior between CBPΔ1 transgenic mice (n = 18) and wild-type littermates (n = 20) during the first 2.5 min before the shock is presented (preshock) or the 0.5 min after the shock (post-shock). However, CBPΔ1 transgenic mice exhibited a significant decrease in freezing in a 24 h retention test, performed in the same conditioned context, as compared to wild-type littermates. (B) Training and 1 h short-term memory test during contextual fear conditioning. CBPΔ1 transgenic mice (n = 11) and wild-type littermates (n = 11) showed no differences in freezing in a 1 h retention test, performed in the same conditioned context. (C) Training and 24 h long-term memory test during cued fear conditioning. No differences in freezing behavior were observed between CBPΔ1 transgenic mice (n = 14) and wild-type littermates (n = 8) during training or the 24 h retention test. The 24 h test was performed in a novel context for cued fear conditioning. * indicates p < 0.05.
A different set of CBPΔ1 transgenic and wild-type mice that were naive to fear conditioning were used for the cued fear conditioning experiment. CBPΔ1 transgenic mice and wild-type littermates showed the same level of freezing before and after the shock on training day (Fig. 6C; preshock: wild-type mice exhibited 1.1 ± 0.6% freezing; transgenic mice exhibited 1.4 ± 0.9% freezing; t[20] = 0.31, p = NS; post-shock: wild-type mice exhibited 13.1 ± 4.7% freezing; transgenic mice exhibited 6.3 ± 3.1% freezing; t[20] = 1.03, p = NS). No differences were observed in the 24 h retention test, performed in a novel context, between groups either before or after the cue (Fig. 6C; pre-CS: wild-type mice exhibited 16.5 ± 2.9% freezing; transgenic mice exhibited 17.5 ± 2.6% freezing; t[20] = 0.26, p = NS; CS: wild-type mice exhibited 77.6 ± 3.8% freezing; transgenic mice exhibited 78.9 ± 4.1% freezing; t[20] = 0.23, p = NS).
Discussion
We have shown that CBP is involved in a specific form of hippocampal synaptic plasticity and spatial and contextual hippocampus-dependent memory. In CBPΔ1 transgenic mice, we observed a reduction in LTP induced by a single tetanus paired with a D1 dopaminergic agonist, the first demonstration of a role for CBP in this form of LTP. Behaviorally, we found that CBPΔ1 transgenic mice have spatial memory deficits in the hidden-platform version of the Morris water maze, which were not due to visual, motivational, or motor coordination difficulties. Additionally, long-term memory for contextual fear conditioning was impaired in CBPΔ1 transgenic mice, supporting the idea that CBP may have a specific role in hippocampus-dependent learning and memory.
Role for CBP in hippocampus-dependent memory
We generated CBPΔ1 transgenic mice that express an inhibitory truncated CBP mutant because this mutant CBP protein appears to act in a dominant negative fashion (Oike et al. 1999). Indeed, we found CBPΔ1 to significantly impair CRE-dependent transcription. The CBPΔ1 truncation mutant is predicted to bind only to factors that interact with the amino terminus of this transcriptional coactivator. Factors that bind to CBPΔ1 do not recruit HAT activity because of the removal of this domain in CBPΔ1. Thus, only a subset of CBP-interacting factors, including CREB, should be affected by this mutant. This same inhibitory truncated CBP mutant is expressed by mice generated by Oike et al. (1999), which have one wild-type CBP allele and one truncated CBP allele, and these mice phenocopy Rubinstein-Taybi Syndrome (RTS) in humans (we refer to these mice here as RTS mice). Two important differences between our CBPΔ1 transgenic mice and the RTS mice are that CBPΔ1 transgenic mice express the truncated CBP mutant in the presence of two wild-type CBP alleles, and that expression is spatially and temporally restricted in CBPΔ1 transgenic mice to post-natal neurons in specific regions of the brain. As a result, the CBPΔ1 mutant is less deleterious in our transgenic mice, and we were able to study the role of CBP in memory storage independent of the developmental abnormalities observed in RTS mice.
RTS mice exhibit impaired long-term memory in a stepthrough-avoidance test and cued fear conditioning, with no observable deficits in contextual fear conditioning or the Morris water maze (Oike et al. 1999). RTS mice have also been found to have long-term memory deficits for object recognition (Bourt-chouladze et al. 2003). The discrepancy between our results and those of Oike et al. (1999) may be due to the aforementioned differences in the expression of the truncated CBP protein between RTS mice (throughout the organism) and CBPΔ1 transgenic mice (specific areas of the brain), or to the significant developmental abnormalities seen in RTS mice. Additionally, differences in behavioral training protocols and genetic background can contribute significantly to observed differences (Bucan and Abel 2002).
Interestingly, although we observe transgene expression in the amygdala, CBPΔ1 transgenic mice do not exhibit cued fear conditioning deficits. One possibility for this observation is that the CaMKIIα promoter drives expression of a transgene to a lesser extent in the amygdala than the hippocampus. Indeed, transgenic mice expressing a dominant negative mutation of PKA from the CaMKIIα promoter express more highly in the hippocampus than the amygdala (Bourtchouladze et al. 2000) and exhibit impaired long-term memory for contextual fear conditioning, but normal long-term memory for cued fear conditioning as well as conditioned taste aversion, two amygdala-dependent tasks (Abel et al. 1997). Another possibility is that different memory systems have differential molecular requirements. Thus, long-term memory for hippocampus-dependent contextual fear conditioning may be more sensitive to alterations in CBP activity, whereas memory for amygdala-dependent cued fear conditioning may not. In support of this last possibility, we have observed that knock-in mice homozygous for point mutations in the CREB-binding domain (KIX) of CBP exhibit significantly impaired long-term memory for contextual fear conditioning, but normal memory for cued fear conditioning (M. Wood, unpubl.). These results are similar to what we have observed in the CBPΔ1 transgenic mice, suggesting that CBP function is particularly important for hippocampus-dependent forms of memory.
Role for CBP in hippocampal synaptic plasticity
CBPΔ1 transgenic mice showed normal tetraburst L-LTP induced by four 100 Hz, 1 sec trains and normal E-LTP induced by a single 1 sec, 100 Hz train but showed a significant reduction in L-LTP induced by the same 100 Hz train paired with the dopamine D1 agonist, chloro-APB. This form of LTP was also impaired in transgenic mice expressing KCREB, a dominant negative form of CREB (Pittenger et al. 2002). Like our CBPΔ1 transgenic mice, transgenic mice expressing KCREB showed normal tetraburst L-LTP and normal E-LTP in response to a single train of stimuli. It has become increasingly clear that there are multiple forms of long-lasting hippocampal synaptic plasticity, including LTP induced by the pairing of one train with D1 activation (Pittenger et al. 2002), theta-burst stimulation (Woo et al. 2000) and massed or spaced delivery of three or four 100 Hz tetani (Scharf et al. 2002). These forms of LTP have been shown to differ in their molecular mechanisms. For example, transgenic mice expressing KCREB, show normal L-LTP resulting from theta burst or tetraburst protocols (Pittenger et al. 2002). Further, other studies using different genetic manipulations of CREB in mice have also not found impairments in LTP induced by induction protocols using multiple tetani (Gass et al. 1998; Balschun et al. 2003). The similar phenotypes of reduced LTP induced by pairing of synaptic stimulation with chloro-APB in KCREB and CBPΔ1 transgenic mice suggests that CBP may be functioning as a transcriptional coactivator for a CREB family member in the Schaffer collateral path-way of the hippocampus to mediate the transcription of genes required for this form of synaptic plasticity. It also raises the intriguing question of what transcriptional mechanisms mediate LTP in response to multiple tetanic trains.
While this manuscript was in preparation, two papers appeared showing that conditional transgenic mice expressing histone acetyltransferase (HAT)-deficient CBP exhibit impaired memory in the Morris water maze and object recognition tasks (Korzus et al. 2004) and that mice heterozygous for a null mutation of CBP exhibit impaired memory for object recognition and contextual fear conditioning (Alarcon et al. 2004). Korzus et al. (2004) were able to ameliorate the object recognition memory deficit by intraperitioneal administration of the histone deacetylase (HDAC) inhibitor trichostatin A (TSA). Alarcon et al. (2004) were able to ameliorate the contextual fear conditioning memory deficit by intracerebroventricular injection of suberoylanilide hydroxamic acid (SAHA). These HDAC inhibitors induce a state of increased histone acetylation that presumably compensates for the lack of CBP histone acetyltransferase activity in the CBP mutant mice. Levenson et al. (2004) demonstrated that HDAC inhibitors enhance induction of LTP at Schaffer-collateral synapses in area CA1 and enhance long-term memory formation for contextual fear conditioning. Additionally, in RTS mice, inhibitors of phosphodiesterase 4 enhance CREB-dependent gene expression and ameliorate memory deficits for object recognition in a dose dependent manner (Bourtchouladze et al. 2003). Interestingly, these authors observed a shift in the dose response curve in the RTS mice as compared to wild-type mice, suggesting that an increased upstream enhancement of the cAMP signaling cascade is necessary to compensate for the downstream disruptions in this pathway caused by the inhibitory truncated CBP mutant expressed in RTS mice. These results agree with our own in pointing to a role for CBP and its histone acetylation activity in memory storage. However, mice heterozygous for a null mutation of CBP exhibit impaired L-LTP induced by four 100 Hz trains stimulation (Alarcon et al. 2004), a form of LTP that we found to be normal in our CBPΔ1 transgenic mice. Results from mice heterozygous for a null mutation of CBP are difficult to interpret due to the developmental abnormalities and embryonic lethality present in these mice (Tanaka et al. 1997; Kung et al. 2000). With our transgenic approach, we are able to attribute LTP deficits in CBPΔ1 transgenic mice to the specific expression of CBPΔ1 in hippocampal neurons.
Our results demonstrate a role for CBP in hippocampal synaptic plasticity and hippocampus-dependent long-term memory formation. The involvement of CBP, a transcriptional coactivator with histone acetyltransferase activity, in memory storage further defines the complex nature of transcriptional activation required for memory. CBP is recruited to activated transcription factors and then CBP itself is activated to initiate full transcriptional activation. Thus, CBP may provide a mechanism by which multiple signaling cascades are integrated to coordinately regulate gene expression. The future identification of genes regulated by CBP and histone acetylation will be critical to our understanding of the cellular processes activated by CBP during memory storage.
Materials and Methods
Generation of CBPΔ1 transgenic mice
The full-length wild-type cDNA (pRc/RSV-mCBP.HA.RK; Chrivia et al. 1993) for mouse CBP was kindly provided by Dr. Marc Montminy (The Salk Institute for Biological Studies). Amino acids 1-1084 (CBPΔ1) were cloned into pBluescript II KS (+/-) using BamH1 and Xba1 sites. The truncated CBP cDNA was then FLAG-epitope tagged by Muta-Gene Phagemid In Vitro Mutagenesis (BioRad). FLAG-CBPΔ1 was cloned into the EcoRV site of MM400, a vector containing the untranslated leader with a hybrid intron (Choi et al. 1991) and SV40 polyA sequences. Then, FLAG-CBPΔ1 with an untranslated leader and SV40 sequences were cloned into the Not1 site of MM403, which contains the 8.5 kb mouse CaMKIIα promoter (Mayford et al. 1996). All cloning junctions were verified by DNA sequencing.
The FLAG-CBPΔ1 transgene construct was excised from MM403 using BssHII sites and purified by CsCl gradient centrifugation. The 13 kb transgene was isolated by agarose gel electrophoresis, electroeluted, and purified using Elutip (Schleicher and Schuell). Transgenic mice were generated by injecting purified CaMKIIα-FLAG-CBPΔ1 transgene into pronuclei of B6-SJL/F1 zygotes (Transgenic and Chimeric Mouse Facility at the University of Pennsylvania). Founders were backcrossed to C57BL/6J mice. Mice were maintained and bred under standard conditions, consistent with National Institute of Health guidelines and approved by the Institutional Animal Care and Use Committee. Mice were maintained on a 12-h light/12-h dark cycle with behavioral testing occurring during the light phase. For experiments, subjects were N2-N4s and 8-12 wk old at the time of testing and had free access to food and water. Littermate mice were used for controls in all experiments. For genotyping, tail DNA was prepared and analyzed by Southern blotting using a transgene-specific probe as previously described (Abel et al. 1997).
In situ hybridization and Nissl staining
Mouse brains were dissected and flash frozen in 2-methylbutane. Sagittal cryostat sections (20 μm) were fixed and hybridized as previously described (Abel et al. 1997). An [α-35S]-dATP-labeled, transgene-specific oligonucleotide (5′-GCTTGTCATCATCGTCCTTGTAGTCCATCCCATCCGCAGG-3′) was used that hybridizes to the FLAG-epitope and the beginning of the CBP cDNA. Slides were exposed to Kodak Biomax autoradiographic film. For Nissl staining, brains were dissected and flash frozen in liquid nitrogen. Cryostat sections (20 μm) were fixed in 4% paraformaldehyde in PBS.
Northern blot analysis
Hippocampus, cortex, and cerebellum were dissected and placed in RNAlater (Ambion). Total RNA was then isolated from tissue homogenized in TRIzol reagent (Gibco). Eight micrograms of total RNA for each sample was separated on an RNA borate/formaldehyde gel and then transferred onto Hybond-XL membrane (Amersham Biosciences). The membrane was then probed with a 32P-radiolabeled CBP amino-terminal cDNA fragment, washed and then exposed to XAR-5 film (Kodak). The membrane was then stripped using 0.1× SSC and 0.1% SDS at 95°C and re-probed with a 32P-radiolabeled β-actin cDNA fragment to normalize loading and transfer.
Tissue culture
HEK 293 cells (ATCC) were maintained in Minimum Essential Media with Earle's Salts (Gibco) supplemented with 10% horse serum (ATCC) and 1% penicillin/streptomycin (LTI) and grown at 37°C, 5% CO2. Before transfection, cells at 60%-70% confluence were incubated in serum free media with 1% penicillin/streptomycin for 3 h. Cells were transfected with a total of 375 ng plasmid DNA using FuGene 6 Transfection Reagent (Roche) according to manufacturer's protocol. The following plasmids were used: pβgal-Crontrol (BD Biosciences), CRE-Luciferase (Ohda et al. 2003), and CBPΔ1-MM400. pβgal-Control contains the SV40 early promoter and enhancer sequences inserted upstream and downstream, respectively, of the lacZ gene and was used to normalize the transfection efficiency. CRE-luciferase expresses firefly luciferase under the control of cAMP-responsive element (CRE) and functioned as a reporter. CBPΔ1-MM400 expresses CBPΔ1 (residues 1-1084) from the CMV promoter. MM400 was added to keep the amount of DNA in each transfection equal. One day after transfection, the cells were treated with 10 μM of forskolin (Sigma) and 10 μM of IBMX (Sigma) and incubated for 6 h. At the end of incubation, cells were harvested and assayed for luciferase activity with the Luciferase Assay System with Reporter Lysis Buffer (Promega) and β-Galactosidase activity with β-Gal Reporter Gene Assay (Roche).
Electrophysiological analysis
Acute, transverse hippocampal slices (400 μm thick) were prepared from 8- to 12-wk old transgenics and wild-type controls and maintained in an interface chamber (Abel et al. 1997). Sections were continuously perfused with oxygenated artificial cerebrospinal fluid (ACSF) at 31°C, pH 7.4 and allowed to recover for at least 2 h after dissection before starting electrophysiological experiments. Action potentials were elicited once per minute in CA3 axons with a bipolar stimulating electrode (A-M systems, Inc.; 0.002 inch diameter nichrome wire) placed in the stratum radiatum, with the stimulus intensity set to produce 40% of the maximum response. The resulting field potentials (fEPSPs) were recorded using a glass microelectrode (A-M systems, Inc.; 1.5 mm × 0.85 mm) filled with ACSF with resistance between 1 and 2 MΩ, placed in the stratum radiatum of CA1. Data were acquired using Clampex 7 (Axon Instruments, Inc.) and exported to Igor 4.05A (Wavemetrics, Inc.), where both peak amplitude and initial slope of fEPSPs were calculated. Slices with peak amplitudes less than 2.5 mV were not utilized. Paired-pulse facilitation was assessed at interstimulus intervals ranging from 25 to 300 msec. For slices in which the presynaptic fiber volley was distinguishable, input-output relations were characterized by plotting the initial slope of the fEPSP against the amplitude of the presynaptic fiber volley. For E-LTP experiments, baseline responses were monitored for at least 15 min before applying one train of stimuli at 100 Hz for 1 sec. L-LTP was induced using two stimulus protocols: a tetraburst protocol consisting of four 100 Hz, 1 sec trains of stimuli administered 5 min apart, or a single 100 Hz, 1 sec train delivered after 25 min in the presence of 10 μM chloro-APB (Sigma), a D1 agonist, delivered in 0.05% EtOH. For both types of L-LTP experiments, baseline responses were monitored for at least 15 min, and recordings were continued for at least 2 h following LTP induction. Initial fEPSP slopes were normalized against the average of the last 20 responses before LTP induction and expressed as percent. Post-tetanic potentiation was assessed relative to baseline responses, 1 min after the 100 Hz train.
Water maze
The hidden and visible platform water maze experiments were performed in a circular pool using the methods previously described (Lattal and Abel 2001a), except that two trials per day (4-6 min intertrial interval) were given during the eight training days and mice were handled for 1 min every day for three days before training began. Briefly, on the first day, mice were trained to sit on a submerged platform in a bucket for two 30 sec trials. Then, mice were trained for four days, a probe trial was given on the fifth day, then four more days of training occurred (to the same platform location) followed by a second probe trial on the tenth day. The platform was removed during probe trials on the fifth and tenth days, which lasted for 1 min and were followed by a 20 sec platform sit. The nonspatial version of the water maze was performed by attaching a visible cue to the platform, which was placed in different locations during each of the two trials per day, for four consecutive days. Naive animals were used for the visible platform task. The path of the mouse was recorded using a video tracking system (HVS Image, Water2020 version 1/2001).
Fear conditioning
Fear conditioning experiments were performed in chambers using the methods previously described (Lattal and Abel 2001b). Mice were handled for three consecutive days for 1 min each day. For contextual fear conditioning, mice were placed into the conditioning chamber and received a 2 sec 1.5 mA scrambled footshock 2.5 min after placement into the chamber. Mice were removed from the chamber after 3 min. During testing, mice received one 5 min exposure to the same conditioned context in the absence of shock 1 or 24 h after conditioning. Different sets of mice were used for the 1 h and 24 h retention tests. For cued fear conditioning, mice were placed into the chamber and the cue (white noise) was activated from 2-2.5 min after placement into the chamber with a 2 sec 1.5 mA footshock 2.5 min after placement into the chamber. Mice were removed from the chamber after a total of 3 min. On testing day, mice in the cued group received one 5 min exposure to a novel context (another conditioning chamber with smooth flat floor, altered dimensions, and a novel odorant) for 0-2 min (pre-CS) followed by exposure to the cue from 2-5 min (CS), 24 h after conditioning. Conditioning was assayed by measuring freezing behavior, the complete absence of movement (Fanselow 1980). Freezing was scored during conditioning as well as testing. The behavior of each mouse was sampled at 5 sec intervals and the percentage of those intervals in which the mouse froze was calculated. Different sets of mice were used for contextual and cued conditioning experiments.
Data analysis
Analyses of variance (ANOVAs) were performed in all experiments using SigmaStat (version 2.03) and Systat (version 7.0.1). Repeated measures ANOVA were used where appropriate. Simple planned comparisons were made using Student's t-test. There were no overall significant differences between males and females for either the wild type or transgenic groups, therefore results were collapsed for final data analysis. Experimenters were blind to genotype and genotypes were confirmed by Southern blot analysis or PCR following behavioral and electrophysiological tests.
Acknowledgments
We thank Dr. Jean Richa at the Transgenic and Chimeric Mouse Facility at the University of Pennsylvania for generating CBPΔ1 transgenic animals, Dr. Marc Montminy for the mouse CBP cDNA, and Dr. Michele P. Kelly for help with statistical analysis. We thank Dr. Tom Gould, Dr. Matt Lattal, and Conor McDonough for comments on the manuscript. This research was supported by a SFN MNFP post-doctoral fellowship (to M.A.W.), post-doctoral Training Program fellowship in Neuropsychopharmacology (to M.K.), and by grants from the Merck Foundation, the National Institutes of Health, the Packard Foundation, the University of Pennsylvania Research Foundation, and the White-hall Foundation (to T.A.).
Article and publication are at http://www.learnmem.org/cgi/doi/10.1101/lm.86605.
References
- Abel, T., Nguyen, P.V., Barad, M., Deuel, T.A., Kandel, E.R., and Bourtchouladze, R. 1997. Genetic demonstration of a role for PKA in the late phase of LTP and in hippocampus-based long-term memory. Cell 88: 615-626. [DOI] [PubMed] [Google Scholar]
- Alarcon, J.M., Malleret, G., Touzani, K., Vronskaya, S., Ishii, S., Kandel, E.R., and Barco, A. 2004. Chromatin acetylation, memory, and LTP are impaired in CBP(+/-) mice: A model for the cognitive deficit in Rubinstein-Taybi Syndrome and its amelioration. Neuron 42: 947-959. [DOI] [PubMed] [Google Scholar]
- Balschun, D., Wolfer, D.P., Gass, P., Mantamadiotis, T., Welzl, H., Schutz, G., Frey, J.U., and Lipp, H.P. 2003. Does cAMP response element-binding protein have a pivotal role in hippocampal synaptic plasticity and hippocampus-dependent memory? J. Neurosci. 23: 6304-6314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bliss, T.V. and Richter-Levin, G. 1993. Spatial learning and the saturation of long-term potentiation. Hippocampus 3: 123-125. [DOI] [PubMed] [Google Scholar]
- Bolstad, B.M., Irizarry, R.A., Astrand, M., and Speed, T.P. 2003. A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics 19: 185-193. [DOI] [PubMed] [Google Scholar]
- Bourtchouladze, R., Abel, T., Bailey, C.H., and Kandel, E.R. 2000. A common requirement for PKA signaling in hippocampus suggests a similarity in the molecular storage mechanism between trace and contextual conditioning. Society for Neuroscience Abstract 26: 1509. [Google Scholar]
- Bourtchouladze, R., Lidge, R., Catapano, R., Stanley, J., Gossweiler, S., Romashko, D., Scott, R., and Tully, T. 2003. A mouse model of Rubinstein-Taybi syndrome: Defective long-term memory is ameliorated by inhibitors of phosphodiesterase 4. Proc. Natl. Acad. Sci. 100: 10518-10522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bucan, M. and Abel, T. 2002. The mouse: Genetics meets behaviour. Nat. Rev. Genet. 3: 114-123. [DOI] [PubMed] [Google Scholar]
- Cantani, A. and Gagliesi, D. 1998. Rubinstein-Taybi syndrome. Review of 732 cases and analysis of the typical traits. Eur. Rev. Med. Pharmacol. Sci. 2: 81-87. [PubMed] [Google Scholar]
- Choi, T., Huang, M., Gorman, C., and Jaenisch, R. 1991. A generic intron increases gene expression in transgenic mice. Mol. Cell. Biol. 11: 3070-3074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chrivia, J.C., Kwok, R.P., Lamb, N., Hagiwara, M., Montminy, M.R., and Goodman, R.H. 1993. Phosphorylated CREB binds specifically to the nuclear protein CBP. Nature 365: 855-859. [DOI] [PubMed] [Google Scholar]
- Coupry, I., Monnet, L., Attia, A.A., Taine, L., Lacombe, D., and Arveiler, B. 2004. Analysis of CBP (CREBBP) gene deletions in Rubinstein-Taybi syndrome patients using real-time quantitative PCR. Hum. Mutat. 23: 278-284. [DOI] [PubMed] [Google Scholar]
- D'Hooge, R. and De Deyn, P.P. 2001. Applications of the Morris water maze in the study of learning and memory. Brain Res. Rev. 36: 60-90. [DOI] [PubMed] [Google Scholar]
- Fanselow, M.S. 1980. Conditional and unconditional components of post-shock freezing. Pavlovian J. of Biol. Sci. 95: 177-182. [DOI] [PubMed] [Google Scholar]
- Gass, P., Wolfer, D.P., Balschun, D., Rudolph, D., Frey, U., Lipp, H., and Schutz, G. 1998. Deficits in memory tasks of mice with CREB mutations depend on gene dosage. Learn. Mem. 5: 274-288. [PMC free article] [PubMed] [Google Scholar]
- Goodman, R.H. and Smolik, S. 2000. CBP/p300 in cell growth, transformation, and development. Genes & Dev. 14: 1553-1577. [PubMed] [Google Scholar]
- Hennekam, R.C., Baselier, A.C., Beyaert, E., Bos, A., Blok, J.B., Jansma, H.B., Thorbecke-Nilsen, V.V., and Veerman, H. 1992. Psychological and speech studies in Rubinstein-Taybi syndrome. Am. J. Ment. Retard. 96: 645-660. [PubMed] [Google Scholar]
- Kamei, Y., Xu, L., Heinzel, T., Torchia, J., Kurokawa, R., Gloss, B., Lin, S.C., Heyman, R.A., Rose, D.W., Glass, C.K., et al. 1996. A CBP integrator complex mediates transcriptional activation and AP-1 inhibition by nuclear receptors. Cell 85: 403-414. [DOI] [PubMed] [Google Scholar]
- Kaplan, M.P. and Abel, T. 2003. Genetic approaches to the study of synaptic plasticity and memory storage. CNS Spectr. 8: 597-610. [DOI] [PubMed] [Google Scholar]
- Kogan, J.H., Frankland, P.W., Blendy, J.A., Coblentz, J., Marowitz, Z., Schütz, G., and Silva, A. 1997. Spaced training induces normal long-term memory in CREB mutant mice. Curr. Biol. 7: 1-11. [DOI] [PubMed] [Google Scholar]
- Kojima, N., Wang, J., Mansuy, I.M., Grant, S.G., Mayford, M., and Kandel, E.R. 1997. Rescuing impairment of long-term potentiation in fyn-deficient mice by introducing Fyn transgene. Proc. Natl. Acad. Sci. 94: 4761-4765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korzus, E., Rosenfeld, M.G., and Mayford, M. 2004. CBP histone acetyltransferase activity is a critical component of memory consolidation. Neuron 42: 961-972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kung, A.L., Rebel, V.I., Bronson, R.T., Ch'ng, L.E., Sieff, C.A., Livingston, D.M., and Yao, T.P. 2000. Gene dose-dependent control of hematopoiesis and hematologic tumor suppression by CBP. Genes & Dev. 14: 272-277. [PMC free article] [PubMed] [Google Scholar]
- Lattal, K.M. and Abel, T. 2001a. Different requirements for protein synthesis in acquisition and extinction of spatial preferences and context-evoked fear. J. Neurosci. 21: 5773-5780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ———. 2001b. An immediate-shock freezing deficit with discrete cues: A possible role for unconditioned stimulus processing mechanisms. J. Exp. Psychol. Anim. Behav. Process. 27: 394-406. [PubMed] [Google Scholar]
- LeDoux, J.E. 2000. Emotion circuits in the brain. Annu. Rev. Neurosci. 23: 155-184. [DOI] [PubMed] [Google Scholar]
- Levenson, J.M., O'Riordan, K.J., Brown, K.D., Trinh, M.A., Molfese, D.L., and Sweatt, J.D. 2004. Regulation of histone acetylation during memory formation in the hippocampus. J. Biol. Chem. 39: 40545-40559. [DOI] [PubMed] [Google Scholar]
- Lipp, H.P. and Wolfer, D.P. 1998. Genetically modified mice and cognition. Curr. Opin. Neurobiol. 8: 272-280. [DOI] [PubMed] [Google Scholar]
- Lonze, B. and Ginty, D. 2002. Function and regulation of CREB family transcription factors in the nervous system. Neuron 35: 605-623. [DOI] [PubMed] [Google Scholar]
- Maren, S. 2001. Neurobiology of Pavlovian fear conditioning. Annu. Rev. Neurosci. 24: 897-931. [DOI] [PubMed] [Google Scholar]
- Martin, S.J. and Morris, R.G. 2002. New life in an old idea: The synaptic plasticity and memory hypothesis revisited. Hippocampus 12: 609-636. [DOI] [PubMed] [Google Scholar]
- Mayford, M., Bach, M.E., Huang, Y.-Y., Wang, L., Hawkins, R.D., and Kandel, E.R. 1996. Control of memory formation through regulated expression of a CaMKII transgene. Science 274: 1678-1683. [DOI] [PubMed] [Google Scholar]
- Ohda, Y., Wang, L., Ahn, R.S., Park, J.Y., Seong, J.Y., and Kwon, H.B. 2003. Differential G protein coupling preference of mammalian and nonmammalian gonadotropin-releasing hormone receptors. Mol. Cell. Endocrinol. 205: 89-98. [DOI] [PubMed] [Google Scholar]
- Oike, Y., Hata, A., Mamiya, T., Kaname, T., Noda, Y., Suzuki, M., Yasue, H., Nabeshima, T., Araki, K., and Yamamura, K. 1999. Truncated CBP protein leads to classical Rubinstein-Taybi syndrome phenotypes in mice: implications for a dominant-negative mechanism. Hum. Mol. Genet. 8: 387-396. [DOI] [PubMed] [Google Scholar]
- Petrij, F., Giles, R.H., Dauwerse, H.G., Saris, J.J., Hennekam, R.C.M., Masuno, M., Tommerup, N., van Ommen, G.-J.B., Goodman, R.H., Peters, D.J.M., et al. 1995. Rubinstein-Taybi syndrome caused by mutations in the transcriptional co-activator CBP. Nature 376: 348-351. [DOI] [PubMed] [Google Scholar]
- Pittenger, C., Huang, Y.Y., Paletzki, R.F., Bourtchouladze, R., Scanlin, H., Vronskaya, S., and Kandel, E.R. 2002. Reversible inhibition of CREB/ATF transcription factors in region CA1 of the dorsal hippocampus disrupts hippocampus-dependent spatial memory. Neuron 34: 447-462. [DOI] [PubMed] [Google Scholar]
- Rubinstein, J.H. and Taybi, H. 1963. Broad thumbs and toes and facial abnormalities. A possible mental retardation syndrome. Am. J. Dis. Child 105: 588-608. [DOI] [PubMed] [Google Scholar]
- Scharf, M.T., Woo, N.H., Lattal, K.M., Young, J.Z., Nguyen, P.V., and Abel, T. 2002. Protein synthesis is required for the enhancement of long-term potentiation and long-term memory by spaced training. J. Neurophysiol. 87: 2770-2777. [DOI] [PubMed] [Google Scholar]
- Schenk, F. and Morris, R.G. 1985. Dissociation between components of spatial memory in rats after recovery from the effects of retrohippocampal lesions. Exp. Brain Res. 58: 11-28. [DOI] [PubMed] [Google Scholar]
- Tanaka, Y., Naruse, I., Maekawa, T., Masuya, H., Shiroishi, T., and Ishii, S. 1997. Abnormal skeletal patterning in embryos lacking a single Cbp allele: A partial similarity with Rubinstein-Taybi syndrome. Proc. Natl. Acad. Sci. 94: 10215-10220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tusher, V.G., Tibshirani, R., and Chu, G. 2001. Significance analysis of microarrays applied to the ionizing radiation response. Proc. Natl. Acad. Sci. 98: 5116-5121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vo, N., and Goodman, R.H. 2001. CREB-binding protein and p300 in transcriptional regulation. J. Biol. Chem. 276: 13505-13508. [DOI] [PubMed] [Google Scholar]
- Woo, N.H., Duffy, S.N., Abel, T., and Nguyen, P.V. 2000. Genetic and pharmacological demonstration of differential recruitment of cAMP-dependent protein kinases by synaptic activity. J. Neurophysiol. 84: 2739-2745. [DOI] [PubMed] [Google Scholar]
- Wood, M.A., McMahon, S.B., and Cole, M.D. 2000. An ATPase/helicase complex is an essential cofactor for oncogenic transformation by c-Myc. Mol. Cell 5: 321-330. [DOI] [PubMed] [Google Scholar]
- Zucker, R.S. 1989. Short-term synaptic plasticity. Annu. Rev. Neurosci. 12: 13-31. [DOI] [PubMed] [Google Scholar]