Abstract
Several species of helicobacter have been isolated from laboratory mice, including H. bilis, H. hepaticus, H. muridarum, H. rodentium, and H. typhlonius, which appear to be the most common. The most widely used published method for molecular detection of these agents is PCR amplification of a conserved region of 16S rRNA, but differential speciation requires restriction enzyme digestion of the amplicons. This study was undertaken to determine PCR conditions that would simultaneously and specifically identify each of the five common species without restriction enzyme analyses. First, we designed novel and specific PCR primers for H. bilis, H. hepaticus, H. muridarum, H. rodentium, and H. typhlonius, using sequences from the heterologous regions of 16S rRNA. Because of comigration of amplified products, we next identified P17, an H. bilis-specific protein; P25, an H. hepaticus-specific protein; and P30, an H. muridarum-specific protein by screening genomic DNA expression libraries of each species. Primers were designed from these three genes, plus newly designed, species-specific 16S rRNA primers for H. rodentium and H. typhlonius that could be utilized for a five-plex PCR. The sizes of the amplicons from H. bilis, H. hepaticus, H. muridarum, H. rodentium, and H. typhlonius were 435, 705, 807, 191, and 122 bp, respectively, allowing simultaneous detection and effective discrimination among species.
Several species of helicobacter have been identified from naturally infected laboratory mice, most commonly H. bilis (10), H. hepaticus (9), H. muridarum (15), H. rodentium (25), and H. typhlonius (12). A recent study showed that infections with these bacteria are highly prevalent among research mouse colonies in the United States, Europe, and Asia (28). Among 88% of 34 sources surveyed, mice were infected with one or more helicobacter species. H. hepaticus was the most commonly isolated species, followed by H. typhlonius, and H. bilis, while only one case each of infection with H. rodentium and H. muridarum was found (28). A recent review described the significant effects of these bacteria upon research, including their association with hepatic neoplasia, intestinal neoplasia, and chronic proliferative enteritis in mice (30).
In addition to their impact upon rodent health and research, there is growing evidence that some of these helicobacter species may infect humans and may be associated with human disease. This was initially evident with PCR amplification of H. bilis DNA in bile and gall bladders of humans with cholecystitis (8). Subsequently, 24 of 29 patients with bile duct or gallbladder cancer tested PCR positive for H. bilis in their bile, compared to only 4 of 14 healthy subjects without biliary disease (17). In a recent study, 4 of 14 biliary tract cancer patients tested positive for H. bilis DNA by PCR (18). In addition to PCR, serologic studies have shown that human patients with chronic liver diseases, including autoimmune liver disease, developed antibodies to H. bilis and H. hepaticus (1, 20).
Because of the high prevalence of helicobacter species in research mouse colonies, the impact of infection on research, and the growing awareness of their zoonotic potential, it is important to have rapid and reliable diagnostic tests. The most sensitive and widely used method for detecting helicobacter infections is PCR targeting of a genus-specific, conserved region of 16S rRNA. For speciation, this has been followed by restriction enzyme digestion of the amplicons for identification of species-specific fragment lengths. For example, in one study, H. hepaticus, H. bilis, and H. muridarum were distinguished by digesting a 374-bp 16S rRNA fragment with three different restriction enzymes (21). Shen et al. analyzed 11 helicobacter species, including 4 murine species (H. bilis, H. hepaticus, H. muridarum, and H. rodentium). For the four murine species, a 1,219-bp 16S rRNA-amplified fragment was subjected to two different restriction enzymes, but H. bilis had to be further speciated using its own specific 16S rRNA primers (24). Most published diagnostic PCR tests for helicobacter utilize independent amplification of targets from each Helicobacter species (3, 21, 24, 27, 29). In addition to 16S rRNA targets, other genes have been targeted for PCR as well, including a urease gene (3, 26) and a cytolethal distending toxin B gene (14).
This study was undertaken to develop and optimize PCR conditions that would simultaneously detect and speciate five of the more common helicobacter species of the mouse (11) by multiplex PCR without the need for restriction enzyme analyses. We initially designed primers from species-specific heterologous regions of 16S rRNA, but some of the amplified products comigrated, precluding accurate discrimination. We therefore developed species-specific primers from other regions of the genomes, using genes that were cloned from H. bilis, H. hepaticus, and H. muridarum genomic expression libraries. Primers for these three targets and newly designed 16S rRNA primers for H. rodentium and H. typhlonius were utilized to develop a five-plex PCR.
MATERIALS AND METHODS
Bacterial culture and isolation.
Helicobacter bilis (ATCC 51630), H. hepaticus (ATCC 51448), and H. muridarum (ATCC 49282) were obtained from the American Type Culture Collection, and stock cultures of H. rodentium and H. typhlonius were provided by Lela K. Riley from the University of Missouri. These cultures were subjected to population cloning by 3× limiting dilution, as described previously (13). Helicobacters were cultured on moist Brucella agar, supplemented with 5% sheep blood. Plates were incubated for 3 to 7 days under microaerobic conditions at 37°C in high humidity in an anaerobic jar containing a GasPak with Campy-PakPlus system (Becton Dickinson, Cockeysville, Md.) (13). Borrelia burgdorferi cN40, a clonal isolate of B. burgdorferi sensu stricto, had been cloned by 3× limiting dilution and passage in mice, as described (2). B. burgdorferi was cultured in modified Barbour-Stoenner-Kelly (BSK II) medium at 33°C, as described (2). Campylobacter jejuni whole-cell lysate was prepared and provided as a gift from S. Jang, University of California, Davis, CA.
Experimental infection of mice with H. bilis, H. hepaticus, H. muridarum, H. rodentium, or H. typhlonius individually.
Virus antibody- and Helicobacter-free C3H/HeN (C3H) and C3H/Smn.CIcrHsd-scid (C3H-scid) mice were purchased at 3 to 5 weeks of age from the National Cancer Institute Animal Production Program, Frederick Cancer Research Center, Frederick, Md., (C3H) or Harlan Sprague-Dawley, Indianapolis, Ind. (C3H-scid). Upon arrival, fecal pellets from all mice were tested for Helicobacter by culture (below) and PCR (13, 23). Mice were maintained in a pathogen-free room with restricted access on a 12:12 light cycle. They were fed irradiated Pico Lab Mouse Diet 20 (PMI Nutrition International, Inc., Brentwood, MO). For mouse inoculation, bacteria were adjusted to 108 CFU per ml, and 0.1 ml was inoculated intraperitoneally into C3H-scid mice, as described (13). Once infection was established (4 to 8 weeks after inoculation) and confirmed by fecal PCR, the mice were killed and livers collected. Liver tissues containing host-adapted H. bilis or H. hepaticus or pools of liver and colon containing host-adapted H. muridarum, H. rodentium, or H. typhlonius were homogenized in 10 ml of brucella broth, and then 0.25 ml of each homogenate was inoculated by oral gavage into C3H mice. Infection status was monitored weekly by fecal PCR and culture. At 6 months after infection, blood was collected and serum harvested from positive mice. These immune sera were preabsorbed with phage/Escherichia coli lysate and stored at −20°C for screening the DNA expression libraries.
Mice that were naturally infected either with H. bilis or H. hepaticus.
Twenty retired SKH1 breeder mice that were naturally infected with H. bilis were purchased from Charles River Laboratory (Portage, MI). Twenty retired Hsd:ICR (CD-1) breeders that were infected with H. hepaticus were purchased from Harlan Sprague Dawley, Inc (San Diego, CA). Infection status of these colonies was determined by the vendors, but confirmed upon arrival. Upon arrival, all mice were euthanized and exsanguinated by cardiocentesis. Mucosal scrapings of cecum were collected and streaked onto brucella plates for culture. Mucosal scrapings of cecum, a section of cecum (25 to 30 mg), and livers from each mouse were processed for DNA using DNeasy Tissue kit (QIAGEN, Valencia, CA), according to the manufacturer's instruction. Extracted DNA was used for PCR amplification, using specific primers listed in Table 1.
TABLE 1.
Primer sets for amplification of five murine helicobacter species
| Species and primera | Sequence (5′ to 3′) |
|---|---|
| H. rodentium | |
| 1201f | TTGTGAAATGGAGCAAATCTTAAAAACT |
| 1375r | TAGCCAGTTTGGCATTCC |
| H. typhlonius | |
| 163f | AGGGACTCTTAAATATGCTCCTAGAGT |
| 262r | ATTCATCGTGTTTGAATGCGTCAA |
| H. bilis | |
| p17 f | ATGGAACAGATAAAGATTTTAAAGCAACTTCAG |
| p17 r | CTATGCAAGTTGTGCGTTAAGCAT |
| H. hepaticus | |
| p25 f | ATGGGTAAGAAAATAGCAAAAAGATTGCAA |
| p25 r | CTATTTCATATCCATAAGCTCTTGAGAATC |
| H. muridarum | |
| p30 f | ATGACAAAAAAATATTCTTTCACAAAACTATTCATTGGT |
| p30 r | TTTATTTTAGATTCCATTTAACTGCTAAATCATCAATAGT |
f, forward; r, reverse.
The University of California, Davis, laboratory animal care program is fully accredited by the Association for Assessment and Accreditation of Laboratory Animal Care, and this study was reviewed and approved by the Institutional Animal Care and Use Committee. All procedures and use of mice were in compliance with the PHS Guide for the Care and Use of Laboratory Animals.
DNA samples for PCR.
Genomic DNA was purified from H. bilis, H. hepaticus, H. muridarum, H. rodentium, and H. typhlonius, as previously described (5). DNA was also purified from fecal pellets from mice experimentally infected with H. bilis, H. hepaticus, or H. muridarum, as described (23). Briefly, 7 to 10 pellets were suspended in 1 ml of sterile phosphate-buffered saline, mixed with toothpick and vortexed thoroughly. The mixtures were centrifuged briefly at 6,000 rpm for 10 s in a benchtop Microfuge R (Beckman, Palo Alto, CA). The supernatants were collected using a sterile 1-cm3 syringe, and passed through a 0.8-μm filter attached to the syringe. The filtrates were centrifuged at 13,500 rpm for 5 min. The supernatants were discarded, and the remaining pellets were used to extract DNA with the QIAGEN DNeasy Tissue kit (QIAGEN Inc., Valencia, CA) for animal tissues. DNA was eluted from the columns with 200 μl of distilled water. Assays included negative controls from uninfected mice.
PCR.
Each genomic DNA, fecal DNA, or tissue DNA sample was used as a template separately. Species-specific 16S rRNA primers for each helicobacter species were used (Table 1). Twenty-five μl of HotStarTaqMaster mix (QIAGEN Inc., Valencia, CA), 1 μl of DNA template, 1 μl of each primer at 100 μM, and 22 μl of distilled water were mixed for a 50 μl of reaction. The concentration of genomic DNA was 5 μg/ml, and the fecal DNA was 14 μg/ml. For single-plex PCR, DNA was denatured at 94°C for 1 min, annealed at 55°C for 1 min, and extended at 72°C for 1 min. This process was repeated for 30 cycles in a PTC-220 DNA Engine Dyad Cycler (MJ Research Inc., Waltham, MA). Amplicons were analyzed by 1% agarose gel electrophoresis. Five-plex PCR conditions were similar to single-plex PCR, except the annealing temperature was 53°C and 2.5% agarose gel electrophoresis was used.
H. bilis, H. hepaticus, and H. muridarum genomic expression libraries.
Genomic DNA was isolated from H. bilis, H. hepaticus, and H. muridarum. Two hundred μg of each genomic DNA was shipped to Stratagene, La Jolla, CA (for H. bilis) and BBI Biotech Research Laboratories, Inc., Gaithersburg, MD (for H. hepaticus and H. muridarum) to construct λ ZAP II genomic expression libraries. The λ ZAP II phage contains pBluescript that can be excised and cloned directly with ExAssist helper phage (Stratagene, La Jolla, Calif.). The respective libraries were screened with immune sera derived from mice that were naturally or experimentally infected with H. bilis, H. hepaticus, or H. muridarum. Immunoreactive clones were obtained by routine procedures, as described (4). DNA sequencing was performed at the W. M. Keck Foundation Biotechnology Resource Laboratory at Yale School of Medicine. DNA sequence was analyzed using the MacVector program (Accelrys, Madison, WI).
Expression and purification of recombinant proteins.
The primers for H. bilis p17 DNA corresponded to nucleotides 1 to 33 and 412 to 435 of the p17 gene. The primers for H. hepaticus p25 DNA corresponded to nucleotides 1 to 30 and 675 to 705 of the p25 gene. The primers for H. muridarum p30 DNA corresponded to nucleotides 1 to 39 and 768 to 807 of the p30. DNA from the original reactive clones for each gene was used as template. Amplified p17, p25, and p30 DNA fragments were cloned in frame with the glutathione S-transferase gene into pMX, a pGEX-2T vector (Pharmacia, Pistaway, N.J.) with a modified polylinker (22). The PCR-amplified DNA sequences of the recombinant DNA were confirmed by sequence comparison with the original inserts. Recombinant proteins were purified on glutathione columns and freed of their glutathione S-transferase fusion partner by thrombin cleavage, as described (4).
Antisera to H. bilis P17, H. hepaticus P25, and H. muridarum P30 recombinant proteins were generated by subcutaneous injection of 20 μg of recombinant protein emulsified in 0.1 ml of Freund's complete adjuvant, followed by two boosts of 10 μg of protein each in incomplete Freund's adjuvant at 2-week intervals. Sera were collected and tested by enzyme-linked immunosorbent assay (ELISA), and antibody reactivity of antisera was verified at a serum dilution of ≥1:100,000.
Native bacterial antigens.
To prepare whole-cell lysates, broth cultures of H. bilis, H. hepaticus, H. muridarum, B. burgdorferi, and C. jejuni were pelleted by centrifugation, washed with cold phosphate-buffered saline, and then sonicated to lyse cells. The lysates were stored at −20°C.
Immunoblots.
Four μg of whole-cell lysates or recombinant proteins was resolved in 15% sodium dodecyl sulfate-polyacrylamide gels by electrophoresis and transferred to nitrocellulose membranes. For dot blots, a Bio-Dot Microfiltration apparatus (Bio-Rad, Hercules, CA) was used to transfer proteins to nitrocellulose membranes. A sheet of Bio-Rad 9 × 12-cm Trans-Blot Transfer Medium nitrocellulose paper was soaked for 10 min in Tris-buffered saline (TBS) and then blotted with Whatman paper to dry. One hundred μl of TBS was applied to each well to rewet the membrane, and then a vacuum was applied to the apparatus to remove the TBS. Proteins were diluted in TBS at 10 μg/ml, and 100 μl was then added to each well (one well is equivalent to one dot). The TBS was allowed to pass through the nitrocellulose by gravity filtration (approximately 1.5 h). Once all the TBS had filtered through, the unit was disassembled and the nitrocellulose membranes were then processed as immunoblots. Membranes were probed with immune serum diluted 1:100, then labeled with alkaline phosphatase-conjugated goat anti-mouse immunoglobulin G secondary antibody, diluted at 1:4,500 (Sigma, St. Louis, MO).
ELISA.
One hundred μl of 1 μg/ml of whole-cell lysates or recombinant proteins in carbonate coating buffer (pH 9.6) was plated in 96-well plates, as described (7). Duplicate samples of each mouse serum, including uninfected normal mouse serum as a control, were diluted 1:200 for probing. Secondary antibody was alkaline phosphatase-conjugated goat anti-mouse immunoglobulin G (H+L) diluted at 1:5,000 (Jackson ImmunoResearch Lab. Inc., West Grove, PA). Optical density values were read on a Kinetic Microplate Reader (Molecular Devices, Sunnyvale, CA). Values were subtracted from background reactivity against normal mouse serum at optical density 405 nm.
Nucleotide sequence accession number.
The GenBank accession numbers for H. bilis p17, H. hepaticus p25, and H. muridarum p30 are AF444005, AF444004, and AY734881, respectively.
RESULTS
Identification of three species-specific genes for H. bilis, H. hepaticus, and H. muridarum.
Because it was apparent that species-specific multiplexing could not be accomplished with 16S rRNA targets alone, additional species-specific gene targets were sought. Genomic DNA expression libraries were generated from the clonal populations of H. bilis, H. hepaticus, and H. muridarum. Each library was screened with serum collected from mice infected with the homologous helicobacter species, 6 months after inoculation.
The H. bilis DNA library yielded a 435-bp immunoreactive clone encoding p17. P17 recombinant protein was generated, and it was shown to be reactive with serum from H. bilis-infected mice with an ELISA titer of ≥100,000. When P17 recombinant protein was probed with immune sera from mice infected with H. bilis, H. hepaticus, H. muridarum, H. rodentium, or H. typhlonius, only H. bilis immune serum was reactive. Hyperimmune antiserum to P17 recombinant protein was generated, and it was reactive with a 17-kDa protein on H. bilis whole-cell lysate immunoblots, but not against lysates of H. hepaticus, H. muridarum, H. rodentium, or H. typhlonius (data not shown), suggesting that it was an H. bilis-specific protein.
GenBank BLAST revealed that H. bilis P17 shares 59% identity and 75% similarity with H. hepaticus starvation-inducible DNA-binding protein, Dps (GenBank accession number AAP76807.1). The calculated molecular mass of Dps is 17.8 kDa, whereas the calculated molecular mass of P17 is 16.6 kDa. P17 and Dps do not share identity on the DNA level in their 5′ regions; therefore, p17 primers did not amplify Dps (or p17 homologs) from H. hepaticus genomic DNA. Only the H. hepaticus genome sequence is currently available in published form. However, based on the relatively high similarity between P17 and Dps, these proteins may be conserved among helicobacter species.
The H. hepaticus DNA library yielded a 705-bp immunoreactive clone that encoded p25, which was likewise sequenced and expressed as a recombinant protein. P25 recombinant protein was reactive at low titer (1:100) on dot blots with serum from H. hepaticus-infected mice. Antiserum to P25 reacted with H. hepaticus lysates as well as H. bilis, H. muridarum, and H. typhlonius whole-cell lysates, but not with H. rodentium whole-cell lysate; suggesting there was cross-reactivity among the former four species. However, p25 primers could only amplify a DNA fragment corresponding to the p25 gene from H. hepaticus genomic DNA, but not from H. bilis, H. muridarum, H. rodentium, or H. typhlonius genomic DNA, indicating that p25 primers were H. hepaticus specific.
GenBank BLAST revealed that P25 is identical to H. hepaticus 50S ribosomal protein L1 (GenBank accession number AAP76961.1) and shares 77% identity and 87% similarity with H. pylori L1 and 65% identity and 82% similarity with C. jejuni L1. P25 primers did not amplify L1 from C. jejuni genomic DNA.
The H. muridarum DNA library yielded an 807-bp immunoreactive clone containing p30. Sequence analysis suggested that the gene encodes a putative lipoprotein with a hydrophobic leader sequence (amino acids 1 to 26) plus a signal peptidase II consensus sequence Leu-x-y-z-Cys (amino acids 22 to 26). P30 recombinant protein was generated; and ELISA and Western blot results indicated that P30 was specific to H. muridarum. When P30 recombinant protein was probed with immune sera from H. bilis-, H. hepaticus-, H. muridarum-, H. rodentium-, or H. typhlonius-infected mice, only H. muridarum immune serum was reactive (data not shown). Serum from H. muridarum-infected mice had antibody to P30 with a reciprocal titer of ≥2,700. Also, mouse hyperimmune serum to P30 was generated and when it was immunoblotted against H. bilis, H. hepaticus, H. muridarum, H. rodentium, or H. typhlonius whole-cell lysates, it reacted against a native 30-kDa protein from H. muridarum whole-cell lysates but not other helicobacter species (data not shown).
GenBank BLAST revealed that P30 shares 63% identity and 78% similarity with Campylobacter coli amino acid-binding protein. In addition, P30 shares 61% identity and 79% similarity with a probable H. hepaticus ABC-type amino acid transporter periplasmic solute-binding protein, and 59% identity and 75% similarity with C. jejuni amino acid ABC transporter. P30 primers did not amplify any DNA fragments from either H. hepaticus or C. jejuni genomic DNA.
P17, P25, and P30 primers for H. bilis, H. hepaticus, and H. muridarum PCR.
Primers for H. bilis p17, H. hepaticus p25, and H. muridarum p30 were designed from the sequences generated for each clone (Table 1). Single-plex PCR with each set of primers including newly designed 16S rRNA primers for H. rodentium and H. typhlonius (Table 1) was first investigated. Each set of primers amplified DNA from only the homologous species, but not from the heterologous helicobacter species, and no DNA could be amplified from B. burgdorferi or C. jejuni by any of these five sets of primers. Each genomic DNA was also serially diluted from 50, 5, 0.5, and 0.05 ng to determine the sensitivity of PCR with each set of these primers. The results were consistently positive at 0.5 ng of the genomic DNA with the corresponding primers for all five species, and the results were positive at 0.05 ng for H. bilis, H. hepaticus, H. muridarum, and H. typhlonius.
Specificity of PCR primer sets for amplification of DNA from feces of mice experimentally infected with H. bilis, H. hepaticus, H. muridarum, H. rodentium, or H. typhlonius.
The newly designed 16S rRNA primers for H. rodentium and H. typhlonius amplified homologous, but not heterologous, species-specific amplicons from fecal DNA of mice experimentally infected with each helicobacter species. In addition, H. bilis p17, H. hepaticus p25, and H. muridarum p30 primers specifically amplified DNA from feces of H. bilis-, H. hepaticus-, or H. muridarum-infected mice, respectively.
Specificity of PCR primer sets for amplification of DNA from mice that were naturally infected either with H. bilis or H. hepaticus.
Liver, colon, and colonic mucosal scrapings were collected from mice obtained from commercial colonies infected with H. bilis or H. hepaticus. As shown in Table 2, all mice that were infected with H. bilis were PCR negative for H. hepaticus, H. muridarum, H. rodentium, and H. typhlonius. All 20 H. bilis naturally infected mice were PCR positive with p17 primers. Amplicons generated by p17 primers from mouse 2 and mouse 3 were subcloned and sequenced, and the DNA sequences confirmed they represented the corresponding sequences for each DNA fragment of H. bilis.
TABLE 2.
Results of PCR amplification from target DNA derived from liver, cecum, or cecal mucosal scrapings of mice naturally infected with either H. bilis or H. hepaticus, using different helicobacter primer sets
| Infecting organism | No. of positive mice/no. testeda
|
||||
|---|---|---|---|---|---|
| H. bilis P17 | H. hepaticus P25 | H. muridarum P30 | H. rodentium Hr1201f/1357r | H. typhlonius Ht163f/262r | |
| H. bilis | 20/20 | 0/20 | 0/20 | 0/20 | 0/20 |
| H. hepaticus | 0/20 | 20/20 | 0/20 | 0/20 | 0/20 |
PCR was performed using DNA from three tissues: liver, cecum, and cecal mucosal scraping. A mouse was considered PCR positive if any one or all of the tissues were positive.
Using the same sets of primers for samples derived from naturally infected mice with H. hepaticus, all 20 H. hepaticus-infected mice were PCR negative with H. bilis, H. muridarum, H. rodentium, and H. typhlonius (Table 2). When primers for H. hepaticus p25 were used, all 20 mice were PCR positive, and sequences of the amplicons were identical to p25 gene sequence. These results indicate that these 20 mice were only infected with H. hepaticus, as the mouse vendor suggested.
Multiplex PCR for helicobacter species differentiation.
The initial attempts to develop a five-plex PCR with primer sets targeting only 16S rRNA sequences were unsuccessful due to the inability to generate distinctively sized amplicons. The above findings allowed the development of a five-plex PCR with sufficient sensitivity for detection of each helicobacter species and with sufficient specificity for differential speciation based on the size differences among the five chosen targets. A five-plex PCR was performed with a mixture of primers for p17, p25, p30, H. rodentium 16S rRNA (Hr1021f/1375r), and H. typhlonius 16S rRNA (Ht163f/262r) (Table 1). The multiplexed primers were added to genomic DNA of each helicobacter species and to DNA from B. burgdorferi and C. jejuni as negative controls. As shown in Fig. 1, only the homologous targeted gene was amplified from each species. No DNA was amplified from B. burgdorferi or C. jejuni (data not shown). Furthermore, amplification products were sufficiently diverse in size to allow differential speciation (Fig. 1).
FIG. 1.
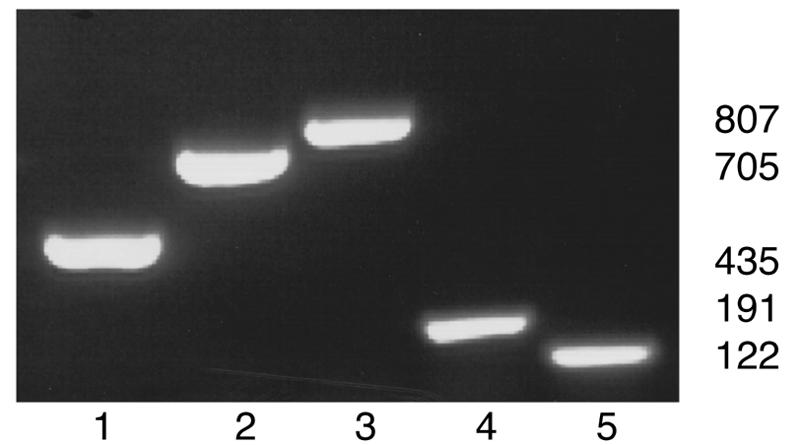
Five-plex PCR for amplification of DNA from five murine helicobacter species. Five sets of primers—p17, p25, p30, and 16S rRNA primers for H. rodentium (Hr1201f/1375r) and H. typhlonius (Hr163f/262r)—were added to each genomic DNA for PCR. Genomic DNA: lane 1, H. bilis; lane 2, H. hepaticus; lane 3, H. muridarum; lane 4, H. rodentium; and lane 5, H. typhlonius.
It was not possible to obtain mice that were coinfected with all five species of helicobacter. In order to evaluate the efficacy of multiplex PCR on feces from mice coinfected with multiple helicobacter species, fecal DNA from a normal mouse was spiked with genomic DNA of each helicobacter species. In the presence of 1 μl of normal mouse fecal DNA per 50 μl of PCR, the sensitivity of the five-plex PCR primer amplification of each individual target was not affected by the presence of fecal DNA. Results were consistently positive at 0.5 ng of genomic DNA for the corresponding primers of each of the five species, and the results were positive at 0.05 ng for H. bilis, H. rodentium, and H. typhlonius (data not shown). Next, 1 μl each of the five Helicobacter genomic DNAs was mixed and then serially diluted ten-fold. One μl from each dilution plus 1 μl of normal mouse fecal DNA were added to a 50-μl five-plex PCR. All five targeted DNA fragments were amplified at 0.5 ng (Fig. 2), but not at 0.05 ng. These results suggested that fecal DNA did not affect sensitivity of the five-plex PCR; however, multiple templates did reduce the sensitivity of the five-plex PCR about 10-fold.
FIG. 2.
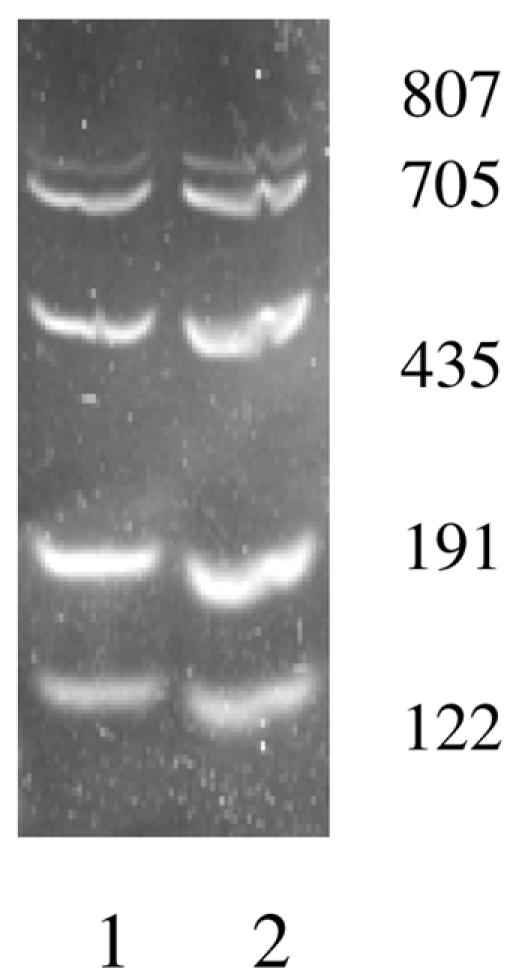
Five-plex PCR for amplification of five targets in single reaction. Five sets of primers—p17, p25, p30, and 16S rRNA primers for H. rodentium (Hr1201f/1375r) and H. typhlonius (Hr163f/262r)—and five genomic DNAs each at concentrations of 5 ng (lane 1) and 0.5 ng (lane 2) plus 1 μl of fecal DNA were mixed in a single PCR.
DISCUSSION
The current study successfully developed five primer sets for differential amplification of DNA targets from H. bilis, H. hepaticus, H. muridarum, H. rodentium, and H. typhlonius. Development of primers from heterologous regions of 16S rRNA was effective, but several of the amplification products comigrated, thereby precluding accurate discrimination in a multiplex format. In a series of experiments, we developed a set of five primer sets with sufficient sensitivity, specificity, and diversity in amplicon size to allow accurate and simultaneous detection of the five named species of mouse helicobacters. Ultimately, it was deemed appropriate to target three novel genes, H. bilis p17, H. hepaticus p25, and H. muridarum p30, in combination with 16S rRNA sequences that were specific for H. rodentium and H. typhlonius. Because of the difficulty in obtaining DNA samples from mice naturally coinfected with all five murine species, we tested fecal DNA spiked with mixtures of all five genomic DNAs in a five-plex format. Under these conditions, all five targeted DNAs could be amplified at 0.5 ng in a single five-plex PCR. For better resolution, the annealing temperature had to be lowered to 53°C following 2.5% agarose gel analysis instead of 55°C and 1% gel analysis for single target DNA five-plex PCR. Thus, a single five-plex PCR, followed by one agarose gel electrophoresis, was enough to differentiate among the five species. Recently, Nilsson et al. (19) also developed a multiplex PCR assay for helicobacters, but two sets of PCR had to be performed, targeting V3 and V6 to -7 regions of 16S rRNA, followed by 9% polyacrylamide gel electrophoresis running for 4 h, which required more work and expense.
Although there is growing evidence that different murine helicobacters may be associated with human infection and disease, including H. bilis and H. hepaticus (1, 8, 17, 18, 20), other murine helicobacter species have yet to be incriminated as potential zoonotic agents. Mouse helicobacter infections are particularly insidious, as they seldom produce clinical signs or lesions in immunocompetent mice. Nevertheless, it is incumbent upon laboratory mouse programs to effectively test for the presence of infected mice, which may threaten disease-susceptible, immunodeficient, or genetically altered mice and compromise research derived from these infected mice (30). Serologic assays for testing mouse populations are problematic, in that immunocompetent mice may not seroconvert, or do so at low titer during late infection, and immunodeficient mice may not mount detectable antibody responses. Furthermore, antigens that are used for serodiagnosis of helicobacters are relatively insensitive. Recombinant proteins are being analyzed, but there are still no strongly immunogenic antigens for all of the mouse helicobacters. Two recombinant proteins have been described for serodiagnostic purposes, including P167 of H. bilis (6) and Map18 of H. hepaticus (16). P167 recombinant protein proved to be H. bilis specific (6), while Map18 recombinant protein proved to be H. hepaticus specific, but less sensitive than membrane extract antigen (16). For these reasons, surveillance of mice for helicobacter infections is generally done with fecal PCR, using universal Helicobacter genus-specific primers (21). However, the growing interest in these agents will increasingly warrant differential speciation. Thus, a multiplex PCR such as that described in this study will allow efficient screening of mouse populations.
Acknowledgments
This work was supported by Public Health Service grant RR14034 to S. W. Barthold from the National Center for Research Resources.
REFERENCES
- 1.Ananieva, O., I. Nilsson, T. Vorobjova, R. Uibo, and T. Wadstrom. 2002. Immune responses to bile-tolerant Helicobacter species in patients with chronic liver diseases, a randomized population group, and healthy blood donors. Clin. Diagn. Lab. Immunol. 9:1160-1164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barthold, S. W., M. S. de Souza, J. L. Janotka, A. L. Smith, and D. H. Persing. 1993. Chronic Lyme borreliosis in the laboratory mouse. Am. J. Pathol. 143:951-971. [PMC free article] [PubMed] [Google Scholar]
- 3.Drazenovich, N. L., C. L. Franklin, R. S. Livingston, and D. G. Besselsen. 2002. Detection of rodent Helicobacter spp. by use of fluorogenic nuclease polymerase chain reaction assays. Comp. Med. 52:347-353. [PubMed] [Google Scholar]
- 4.Feng, S., S. Das, T. Lam, R. A. Flavell, and E. Fikrig. 1995. A 55-kilodalton antigen encoded by a gene on a Borrelia burgdorferi 49-kilobase plasmid is recognized by antibodies in sera from patients with Lyme disease. Infect. Immun. 63:3459-3466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Feng, S., E. Hodzic, and S. W. Barthold. 2000. Lyme arthritis resolution with antiserum to a 37-kilodalton Borrelia burgdorferi protein. Infect. Immun. 68:4169-4173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Feng, S., E. Hodzic, L. V. Kendall, A. Smith, K. Freet, and S. W. Barthold. 2002. Cloning and expression of a Helicobacter bilis immunoreactive protein. Clin. Diagn. Lab. Immunol. 9:627-632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Feng, S., E. Hodzic, B. Stevenson, and S. W. Barthold. 1998. Humoral immunity to Borrelia burgdorferi N40 decorin binding proteins during infection of laboratory mice. Infect. Immun. 66:2827-2835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fox, J. G., F. E. Dewhirst, Z. Shen, Y. Feng, N. S. Taylor, B. J. Paster, R. L. Ericson, C. N. Lau, P. Correa, J. C. Araya, and I. Roa. 1998. Hepatic Helicobacter species identified in bile and gallbladder tissue from Chileans with chronic cholecystitis. Gastroenterology 114:755-763. [DOI] [PubMed] [Google Scholar]
- 9.Fox, J. G., F. E. Dewhirst, J. G. Tully, B. J. Paster, L. Yan, N. S. Taylor, M. J. Collins, Jr., P. L. Gorelick, and J. M. Ward. 1994. Helicobacter hepaticus sp. nov., a microaerophilic bacterium isolated from livers and intestinal mucosal scrapings from mice. J. Clin. Microbiol. 32:1238-1245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fox, J. G., L. L. Yan, F. E. Dewhirst, B. J. Paster, B. Shames, J. C. Murphy, A. Hayward, J. C. Belcher, and E. N. Mendes. 1995. Helicobacter bilis sp. nov., a novel Helicobacter species isolated from bile, livers, and intestines of aged, inbred mice. J. Clin. Microbiol. 33:445-454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Franklin, C. L., P. L. Gorelick, L. K. Riley, F. E. Dewhirst, R. S. Livingston, J. M. Ward, C. S. Beckwith, and J. G. Fox. 2001. Helicobacter typhlonius sp. nov., a novel murine urease-negative Helicobacter species. J. Clin. Microbiol. 39:3920-3926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Franklin, C. L., L. K. Riley, R. S. Livingston, C. S. Beckwith, R. R. Hook, Jr., C. L. Besch-Williford, R. Hunziker, and P. L. Gorelick. 1999. Enteric lesions in SCID mice infected with “Helicobacter typhlonicus,” a novel urease-negative Helicobacter species. Lab. Anim. Sci. 49:496-505. [PubMed] [Google Scholar]
- 13.Hodzic, E., M. McKisic, S. Feng, and S. W. Barthold. 2001. Evaluation of diagnostic methods for Helicobacter bilis infection in laboratory mice. Comp. Med. 51:406-412. [PubMed] [Google Scholar]
- 14.Kostia, S., P. Veijalainen, U. Hirvi, and M. L. Hanninen. 2003. Cytolethal distending toxin B gene (cdtB) homologues in taxa 2, 3 and 8 and in six canine isolates of Helicobacter sp. flexispira. J. Med. Microbiol. 52:103-108. [DOI] [PubMed] [Google Scholar]
- 15.Lee, A., M. W. Phillips, J. L. O'Rourke, B. J. Paster, F. E. Dewhirst, G. J. Fraser, J. G. Fox, L. I. Sly, P. J. Romaniuk, T. J. Trust et al. 1992. Helicobacter muridarum sp. nov., a microaerophilic helical bacterium with a novel ultrastructure isolated from the intestinal mucosa of rodents. Int. J. Syst. Bacteriol. 42:27-36. [DOI] [PubMed] [Google Scholar]
- 16.Livingston, R. S., L. K. Riley, R. R. Hook, Jr., C. L. Besch-Williford, and C. L. Franklin. 1999. Cloning and expression of an immunogenic membrane-associated protein of Helicobacter hepaticus for use in an enzyme-linked immunosorbent assay. Clin. Diagn. Lab. Immunol. 6:745-750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Matsukura, N., S. Yokomuro, S. Yamada, T. Tajiri, T. Sundo, T. Hadama, S. Kamiya, Z. Naito, and J. G. Fox. 2002. Association between Helicobacter bilis in bile and biliary tract malignancies: H. bilis in bile from Japanese and Thai patients with benign and malignant diseases in the biliary tract. Jpn J. Cancer Res. 93:842-847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Murata, H., S. Tsuji, M. Tsujii, H. Y. Fu, H. Tanimura, M. Tsujimoto, N. Matsuura, S. Kawano, and M. Hori. 2004. Helicobacter bilis infection in biliary tract cancer. Aliment. Pharmacol. Ther. 20(Suppl. 1):90-94. [DOI] [PubMed] [Google Scholar]
- 19.Nilsson, H.-O., I.-S. Ouis, U. Stenram, Å. Ljungh, A. P. Moran, T. Wadström, and W. A. Al-Soud. 2004. High prevalence of Helicobacter species detected in laboratory mouse strains by multiplex PCR-denaturing gradient gel electrophoresis and pyrosequencing. J. Clin. Microbiol. 42:3781-3788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nilsson, I., I. Kornilovs'ka, S. Lindgren, A. Ljungh, and T. Wadstrom. 2003. Increased prevalence of seropositivity for non-gastric Helicobacter species in patients with autoimmune liver disease. J. Med. Microbiol. 52:949-953. [DOI] [PubMed] [Google Scholar]
- 21.Riley, L. K., C. L. Franklin, R. R. Hook, Jr., and C. Besch-Williford. 1996. Identification of murine helicobacters by PCR and restriction enzyme analyses. J. Clin. Microbiol. 34:942-946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sears, J. E., E. Fikrig, T. Y. Nakagawa, K. Deponte, N. Marcantonio, F. S. Kantor, and R. A. Flavell. 1991. Molecular mapping of OspA-mediated immunity against Borrelia burgdorferi, the agent of Lyme disease. J. Immunol. 147:1995-2001. [PubMed] [Google Scholar]
- 23.Shames, B., J. G. Fox, F. Dewhirst, L. Yan, Z. Shen, and N. S. Taylor. 1995. Identification of widespread Helicobacter hepaticus infection in feces in commercial mouse colonies by culture and PCR assay. J. Clin. Microbiol. 33:2968-2972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shen, Z., Y. Feng, and J. G. Fox. 2000. Identification of enterohepatic Helicobacter species by restriction fragment-length polymorphism analysis of the 16S rRNA gene. Helicobacter 5:121-128. [DOI] [PubMed] [Google Scholar]
- 25.Shen, Z., J. G. Fox, F. E. Dewhirst, B. J. Paster, C. J. Foltz, L. Yan, B. Shames, and L. Perry. 1997. Helicobacter rodentium sp. nov., a urease-negative Helicobacter species isolated from laboratory mice. Int. J. Syst. Bacteriol. 47:627-634. [DOI] [PubMed] [Google Scholar]
- 26.Shen, Z., D. B. Schauer, H. L. T. Mobley, and J. G. Fox. 1998. Development of a PCR-restriction fragment length polymorphism assay using the nucleotide sequence of the Helicobacter hepaticus urease structural genes ureAB. J. Clin. Microbiol. 36:2447-2453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shomer, N. H., C. A. Dangler, R. P. Marini, and J. G. Fox. 1998. Helicobacter bilis/Helicobacter rodentium co-infection associated with diarrhea in a colony of scid mice. Lab. Anim. Sci. 48:455-459. [PubMed] [Google Scholar]
- 28.Taylor, N. S., S. Xu, V. Ng, F. E. Dewhirst, and J. G. Fox. 2004. High prevalence of Helicobacter spp. in mouse research colonies in the United States, Europe and Asia, abstr. P003. 55th AALAS National Meeting, 17 to 21 October, 2004, Tampa, Fla.
- 29.Whary, M. T., J. H. Cline, A. E. King, K. M. Hewes, D. Chojnacky, A. Salvarrey, and J. G. Fox. 2000. Monitoring sentinel mice for Helicobacter hepaticus, H rodentium, and H bilis infection by use of polymerase chain reaction analysis and serologic testing. Comp. Med. 50:436-443. [PubMed] [Google Scholar]
- 30.Whary, M. T., and J. G. Fox. 2004. Natural and experimental Helicobacter infections. Comp. Med. 54:125-158. [PubMed] [Google Scholar]


