ABSTRACT
Multi-drug resistant (MDR) Pseudomonas aeruginosa harbor a complex array of β-lactamases and non-enzymatic resistance mechanisms. In this study, the activity of a β-lactam/β-lactam-enhancer, cefepime/zidebactam, and novel β-lactam/β-lactamase inhibitor combinations was determined against an MDR phenotype-enriched, challenge panel of P. aeruginosa (n = 108). Isolates were multi-clonal as they belonged to at least 29 distinct sequence types (STs) and harbored metallo-β-lactamases, serine β-lactamases, penicillin binding protein (PBP) mutations, and other non-enzymatic resistance mechanisms. Ceftazidime/avibactam, ceftolozane/tazobactam, imipenem/relebactam, and cefepime/taniborbactam demonstrated MIC90s of >128 mg/L, while cefepime/zidebactam MIC90 was 16 mg/L. In a neutropenic-murine lung infection model, a cefepime/zidebactam human epithelial-lining fluid-simulated regimen achieved or exceeded a translational end point of 1−log10 kill for the isolates with elevated cefepime/zidebactam MICs (16–32 mg/L), harboring VIM-2 or KPC-2 and alterations in PBP2 and PBP3. In the same model, to assess the impact of zidebactam on the pharmacodynamic (PD) requirement of cefepime, dose-fractionation studies were undertaken employing cefepime-susceptible P. aeruginosa isolates. Administered alone, cefepime required 47%–68% fT >MIC for stasis to ~1 log10 kill effect, while cefepime in the presence of zidebactam required just 8%–16% for >2 log10 kill effect, thus, providing the pharmacokinetic/PD basis for in vivo efficacy of cefepime/zidebactam against isolates with MICs up to 32 mg/L. Unlike β-lactam/β-lactamase inhibitors, β-lactam enhancer mechanism-based cefepime/zidebactam shows a potential to transcend the challenge of ever-evolving resistance mechanisms by targeting multiple PBPs and overcoming diverse β-lactamases including carbapenemases in P. aeruginosa.
IMPORTANCE
Compared to other genera of Gram-negative pathogens, Pseudomonas is adept in acquiring complex non-enzymatic and enzymatic resistance mechanisms thus remaining a challenge to even novel antibiotics including recently developed β-lactam and β-lactamase inhibitor combinations. This study shows that the novel β-lactam enhancer approach enables cefepime/zidebactam to overcome both non-enzymatic and enzymatic resistance mechanisms associated with a challenging panel of P. aeruginosa. This study highlights that the β-lactam enhancer mechanism is a promising alternative to the conventional β-lactam/β-lactamase inhibitor approach in combating ever-evolving MDR P. aeruginosa.
KEYWORDS: ß-lactamases, antibiotic resistance, Pseudomonas aeruginosa, cefepime, zidebactam, ß-lactam enhancer, ß-lactamase inhibitor
INTRODUCTION
Pseudomonas aeruginosa infections are often difficult to treat particularly in patients admitted to intensive care units with immunosuppression and other comorbidities. Often these patients have received inappropriate empiric therapy or are infected with multi-drug (MDR) or extreme-drug resistant (XDR) pathogens. The successful proliferation of high-risk MDR/XDR clones of P. aeruginosa is a consequence of the organism’s ability to manifest intrinsic and acquired resistance mechanisms, in turn, challenging the approaches traditionally employed for the discovery of novel antibiotics (1, 2).
Despite the introduction of several anti-pseudomonal antibiotics in the past decade, the conundrum of resistance mechanisms composed of hyper-efflux, impermeability, and β-lactamases in P. aeruginosa continues to pose therapeutic uncertainty during every treatment episode. Older anti-pseudomonal drugs (ceftazidime, cefepime, and piperacillin/tazobactam) are compromised by the hyper-production of pseudomonal-derived cephalosporinases (PDCs) (3), while OprD inactivation and/or hyper-efflux mechanisms impact carbapenems (4). Target modifications in the background of efflux compromise the activity of fluoroquinolones and aminoglycosides (5). As a result, in the United States, currently, 20%–30% of P. aeruginosa isolates display an MDR phenotype, which prompted the Centers for Disease Control and Prevention (CDC) to designate this pathogen as a “serious” threat. Likewise, the World Health Organization (WHO) designates P. aeruginosa as a “critical” pathogen for which new antibiotics are urgently needed (6–8). A recent CDC report describes a disturbing trend of a 32% rise in hospital-onset infections caused by MDR P. aeruginosa in 2020 highlighting the “collateral damage” of the COVID-19 pandemic (9).
Of late, much of the antibiotic discovery efforts have been directed towards finding novel β-lactam or β-lactamase inhibitor-based combinations to overcome diverse β-lactam-impacting resistance mechanisms in Gram-negatives including carbapenem-resistant-Enterobacterales, -P. aeruginosa and -Acinetobacter baumannii. Such efforts have led to the development of combinations such as ceftazidime/avibactam, ceftolozane/tazobactam, imipenem/relebactam, and cefepime/taniborbactam that show improved anti-pseudomonal activity compared to older therapies. However, a “coverage gap” continues to exist, as despite the chemical diversity of newer β-lactamase inhibitors, many are unable to inhibit the entire range of clinically significant β-lactamases in this pathogen (10). Also, reports of newer PDC variants continue to challenge the inhibitory activity of novel inhibitors paired with cephalosporins (11–13).
Recently, an unconventional discovery approach based on novel β-lactam enhancer action has been reported for phase 3-stage cefepime/zidebactam (WCK 5222) (14). The enhancer action of this combination is mediated by zidebactam, a novel bicyclo-acyl hydrazide (derived from diazabicyclooctane) possessing a potent penicillin-binding protein (PBP) 2 binding activity. When zidebactam is combined with PBP3-targeting cefepime, a mechanistic synergy is triggered resulting in the enhancement of cefepime’s bactericidal activity, both in vitro and in vivo against a broad spectrum of Gram-negative pathogens expressing diverse carbapenem-impacting resistance mechanisms (15, 16). Against A. baumannii, pharmacodynamic (PD) studies have established that zidebactam lowers cefepime’s exposure required for in vivo bactericidal activity (17) which forms the basis for the combination’s efficacy against isolates with cefepime/zidebactam minimum inhibitory concentrations (MICs) of up to 64 mg/L in translational animal infection models (18, 19).
For challenging P. aeruginosa infections involving MDR/XDR isolates, the potential clinical utility of novel agents developed through the aforementioned approaches would rely on their ability to overcome a multiplicity of resistance mechanisms. To investigate this aspect, a set of 108 whole genome sequenced heterogeneous P. aeruginosa isolates collected from the U.S. harboring diverse β-lactam-impacting resistance mechanisms was assembled. The in vitro activity of cefepime/zidebactam and novel anti-pseudomonal β-lactam/β-lactamase inhibitor combinations was determined against this panel. Furthermore, isolates with cefepime/zidebactam MICs > 8 mg/L (higher than the cefepime susceptible breakpoint) were employed in a translational neutropenic murine lung infection study to assess the in vivo efficacy of a human epithelial-lining fluid-simulated regimen (ELF-HSR) of cefepime/zidebactam. Finally, using the same model, the pharmacokinetic/pharmacodynamic (PK/PD) basis of in vivo efficacy of cefepime/zidebactam against P. aeruginosa was deciphered by studying the impact of zidebactam on cefepime’s % fT >MIC requirement. For this purpose, cefepime-susceptible P. aeruginosa were used, as such isolates enable identifying the standalone cefepime’s % fT >MIC requirement which then can be compared with cefepime’s requirement in the presence of zidebactam.
RESULTS
Genetic composition of challenge isolates
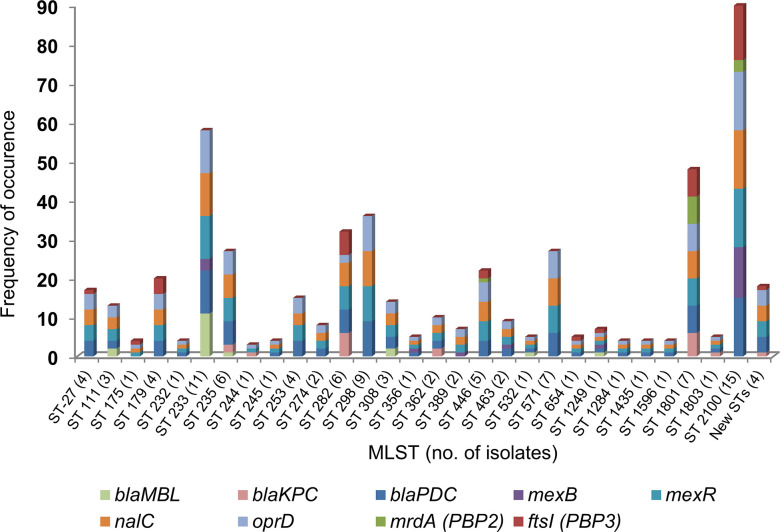
Analysis of the whole genome sequences revealed that the study isolates (n = 108) belonged to a diverse genetic background with at least 29 distinct, previously reported sequence types (STs). When analyzed in comparison to the genome of the reference isolate (P. aeruginosa PAO1), the study isolates demonstrated many nucleotide changes in the genes encoding several key functional proteins (PBP2, PBP3, PDC, AmpR, MexR, MexB, NalC, and OprD) known to be associated with β-lactam resistance in P. aeruginosa (Fig. 1; Table S1).
Fig 1.
Diversity and frequency in occurrence of resistance mechanisms (β-lactamases and mutations in proteins) in P. aeruginosa elucidated by whole genome sequencing (n = 108). blaMBL: 17 were VIM-2 and one was NDM-1, blaKPC: 18 were KPC-2 and one was KPC-5.
A total of 37 isolates (34.3%) were found to express carbapenemases (KPC, 19 [KPC-2, 18; KPC-5, 1]; MBL, 18 [VIM-2, 17; NDM-1, 1]). Out of 19 KPC-producing P. aeruginosa, 6 and 15 isolates were ceftazidime/avibactam (MICs > 8 mg/L) and imipenem/relebactam-non-susceptible (MICs > 2 mg/L), respectively. Among them, one isolate that produced KPC-5 (a KPC variant reported to possess increased hydrolytic activity against ceftazidime) (20) and harbored oprD mutations showed resistance to both ceftazidime/avibactam and imipenem/relebactam.
Three of the ceftazidime/avibactam-non-susceptible isolates belonged to ST 1801 and showed simultaneous substitutions in PBP3 (F533L) (21) and PBP2 (A174V, V517M) (22), while the other three (ST 235 and ST 244) isolates had substitutions in proteins involved in efflux regulation (e.g., the V126E substitution in MexR).
Among 15 imipenem/relebactam-non-susceptible isolates, 12 isolates had PBP3 substitutions (either F533L [n = 6] or T91A [n = 6]). Our analysis showed that F533L substitution in PBP3 was invariably associated with PBP2 substitutions (A174V, V517M).
The V126E substitution in MexR is frequently found in MDR P. aeruginosa isolates and has been associated with an increase in the MICs of imipenem in the background of impermeability (23–25). Thus, among KPC-producing isolates, the main reason for ceftazidime/avibactam and imipenem/relebactam non-susceptibilities seems to be linked with changes in PBP3 and efflux in the background of impermeability (OprD mutations). Though the OprD mutations were also observed in the ceftazidime/avibactam and imipenem/relebactam-susceptible isolates, the absence of high-level resistance in the majority of those isolates suggests that the major contributors in raising their MICs are PBP3 modifications and efflux.
Five out of nineteen KPC-producing isolates showed cefepime/zidebactam MICs of 16–32 mg/L (above cefepime’s susceptible breakpoint); they harbored mutations in PBP3 (F533L), PBP2 (A174V, V517M), and NalC (G71E, S209R). In contrast, 16/19 KPC-producing isolates showed cefepime/taniborbactam MICs in the range of 16–128 mg/L probably due to substitutions in multiple proteins including PBP3, OprD, and those related to efflux.
Among non-carbapenemase producing isolates (n = 71), in general, the mutations responsible for raising the MICs of different antibiotics were associated with both enzymatic (PDC) and non-enzymatic (efflux and target) mechanisms. Specific mutations that correlated with the increase in both the ceftazidime/avibactam and ceftolozane/tazobactam MICs were more often detected in PBP3 (R504C, F533L) (21, 26) and PDC (E219K, SANC numbering) (27). The non-susceptibility to imipenem/relebactam in non-carbapenemase producers seems to be multifactorial as mutations were observed in PBP3, efflux-related proteins, and OprD. Cefepime/zidebactam MICs of 16–32 mg/L were observed in isolates belonging to ST 356 (n = 1), ST 1801 (n = 1), and ST 2100 (n = 5). In this instance, the major detected mutations were in PBP3 (G63D, R504C, and F533L), PBP2 (A174V, V517M, and G591S), and PDC-537 (P153L, E219K, SANC numbering) (28).
Comparative in vitro activity
Table 1 shows the MIC distribution of antibiotics for the isolates categorized as MBL- or KPC-producers or non-carbapenemase producers. Fig. S2 shows the MICs of cefepime/zidebactam versus other β-lactam/β-lactamase inhibitor combinations for each carbapenemase-producing isolate.
TABLE 1.
MIC distributions of cefepime/zidebactam and other antibiotics for major resistance groupsa
| Organism group | Number of P. aeruginosa isolates with indicated MIC (mg/L) | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| ≤ 0.25 | 0.5 | 1 | 2 | 4 | 8 | 16 | 32 | 64 | ≥128 | |
| KPC producers (n = 19) | ||||||||||
| Cefepime | 19 | |||||||||
| Cefepime/zidebactam | 6 | 8 | 2 | 3 | ||||||
| Cefepime/taniborbactam | 1 | 2 | 7 | 1 | 8 | |||||
| Ceftolozane/tazobactam | 3 | 1 | 4 | 5 | 6 | |||||
| Ceftazidime/avibactam | 1 | 4 | 2 | 6 | 3 | 2 | 1 | |||
| Aztreonam/avibactam | 6 | 9 | 3 | 1 | ||||||
| Imipenem/relebactam | 1 | 1 | 2 | 2 | 3 | 3 | 2 | 1 | 4 | |
| Meropenem | 1 | 2 | 16 | |||||||
| Meropenem/vaborbactam | 2 | 1 | 2 | 3 | 11 | |||||
| MBL producers (n = 18) | ||||||||||
| Cefepime | 2 | 3 | 3 | 10 | ||||||
| Cefepime/zidebactam | 3 | 7 | 5 | 2 | 1 | |||||
| Cefepime/taniborbactam | 2 | 3 | 1 | 6 | 6 | |||||
| Ceftolozane/tazobactam | 18 | |||||||||
| Ceftazidime/avibactam | 4 | 5 | 9 | |||||||
| Aztreonam/avibactam | 1 | 1 | 6 | 3 | 2 | 4 | 1 | |||
| Imipenem/relebactam | 1 | 1 | 4 | 12 | ||||||
| Meropenem | 2 | 3 | 1 | 5 | 7 | |||||
| Meropenem/vaborbactam | 1 | 2 | 4 | 5 | 4 | 2 | ||||
| Non-carbapenemase producers (n = 71) | ||||||||||
| Cefepime | 2 | 2 | 10 | 14 | 10 | 11 | 12 | 10 | ||
| Cefepime/zidebactam | 1 | 1 | 14 | 23 | 25 | 4 | 3 | |||
| Cefepime/taniborbactam | 3 | 6 | 15 | 15 | 11 | 6 | 4 | 11 | ||
| Ceftolozane/tazobactam | 4 | 14 | 9 | 8 | 7 | 5 | 3 | 1 | 7 | 13 |
| Ceftazidime/avibactam | 1 | 5 | 7 | 8 | 14 | 11 | 2 | 7 | 16 | |
| Aztreonam/avibactam | 5 | 6 | 8 | 22 | 15 | 15 | ||||
| Imipenem/relebactam | 4 | 2 | 13 | 25 | 18 | 8 | 1 | |||
| Meropenem | 2 | 4 | 10 | 11 | 18 | 16 | 7 | 3 | ||
| Meropenem/vaborbactam | 1 | 3 | 4 | 10 | 13 | 25 | 9 | 4 | 2 | |
Susceptible range (FDA criteria) for each agent except for cefepime/zidebactam, cefepime/taniborbactam, aztreonam/avibactam and meropenem/vaborbactam is depicted by boldfaced numbers; for these approved antibiotics, FDA breakpoints are consistent with CLSI breakpoints. MIC of cefepime/zidebactam was determined at 1:1 ratio. A fixed 4 mg/L of inhibitor concentration was used for cefepime/taniborbactam, ceftolozane/tazobactam, ceftazidime/avibactam, and imipenem/relebactam. A fixed 8 mg/L of inhibitor concentration was used for meropenem/vaborbactam.
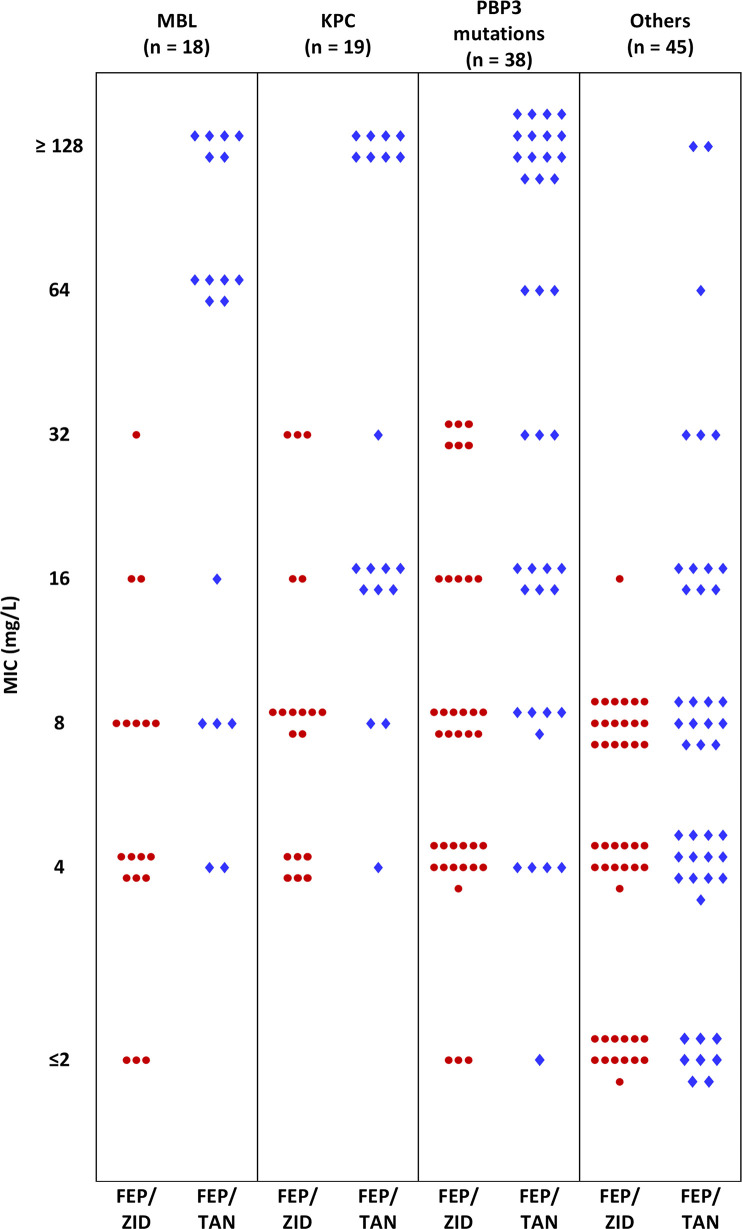
As anticipated, the MBL subset (n = 18) was resistant to ceftolozane/tazobactam (18/18), ceftazidime/avibactam (18/18), and imipenem/relebactam (17/18) using FDA breakpoints. Further, adding the MBL inhibitor, taniborbactam (4 mg/L) to cefepime, did not significantly improve the susceptibility to cefepime. While cefepime MICs were 16 to ≥128 mg/L for all MBL isolates, taniborbactam reduced the cefepime MICs to ≤8 mg/L for only 5 of 18 isolates (27.8%) with one additional isolate inhibited at 16 mg/L. For this MBL subset, cefepime/zidebactam was distinctly more active than cefepime/taniborbactam with 15 of 18 isolates (83.3%) inhibited at ≤8 mg/L and all except one were inhibited at ≤16 mg/L (94.4% inhibition) (Fig. 2). Despite the known stability of aztreonam towards MBL hydrolysis, aztreonam/avibactam inhibited just 33.3% of MBL isolates at 8 mg/L.
Fig 2.
Comparative distribution of MICs of two cefepime-based combinations categorized as per major resistance mechanisms identified through whole genome sequencing of 108 P. aeruginosa. Each symbol represents one isolate. FEP/ZID: cefepime/zidebactam; FEP/TAN: cefepime/taniborbactam.
As stated above, blaKPC was found in 19 isolates with KPC-2 in 18 isolates and KPC-5 in the remaining one isolate. All were non-susceptible to ceftolozane/tazobactam. Against this subset, despite avibactam, relebactam, and taniborbactam being known to inhibit KPC, their respective combinations showed limited activity at their corresponding susceptible breakpoints. On the other hand, cefepime/zidebactam MICs ranged from 4 to 32 mg/L; 16/19 were inhibited at ≤16 mg/L (Table 1).
No carbapenemase was detected in the remaining 71 isolates. Regardless, 91.5% of them were non-susceptible to meropenem which indicates enrichment of carbapenem-impacting non-enzymatic resistance mechanisms in this panel. Unexpectedly, 29/71 (40.8%) and 36/71 (50.7%) were non-susceptible to ceftolozane/tazobactam and ceftazidime/avibactam, respectively (Table 1). As described earlier, this non-susceptible population was enriched with substitutions in PBP3 and efflux proteins as well as in PDCs reported to be linked with a rise in ceftolozane/tazobactam and ceftazidime/avibactam MICs (Table S1). Imipenem/relebactam and cefepime/taniborbactam also showed sub-optimal activity; 27/71 (38%) and 32/71 (45.1%) of isolates were non-susceptible to these combinations, respectively.
With regards to non-β-lactam antibiotics, among the entire population, extreme resistance to ciprofloxacin, substantial resistance to amikacin, and potent activity of colistin were observed. The antibiotic panel was also inclusive of meropenem/vaborbactam but not discussed above as, predictably, the addition of vaborbactam did not improve the activity of meropenem (Table 2).
TABLE 2.
MIC range, MIC50 and MIC90 of cefepime/zidebactam and other antibiotics for all P. aeruginosa (n = 108)
| Antibiotic/combinations | MIC (mg/L) | % Susceptibilitya | ||
|---|---|---|---|---|
| MIC50 | MIC90 | Range | ||
| Cefepime | 32 | ≥128 | 1 - ≥ 128 | 25.9 |
| Cefepime/zidebactam | 4 | 16 | 0.5–32 | 86.1b / 100c |
| Cefepime/taniborbactam | 16 | ≥128 | 1 - ≥ 128 | 43.5b |
| Ceftolozane/tazobactam | 16 | ≥128 | 0.12 - ≥ 128 | 38.9 |
| Ceftazidime/avibactam | 16 | ≥128 | 0.5 - ≥ 128 | 44.4 |
| Aztreonam/avibactam | 32 | ≥128 | 0.25 - ≥ 128 | 23.1d |
| Imipenem | 32 | ≥128 | 0.12 - ≥ 128 | 9.3 |
| Imipenem/relebactam | 4 | ≥128 | 0.12 ≥ 128 | 45.4 |
| Meropenem | 32 | ≥128 | 0.5 - ≥ 128 | 5.6 |
| Meropenem/vaborbactam | 16 | ≥128 | 0.25 - ≥ 128 | 33.3e |
| Ciprofloxacin | 16 | ≥128 | 0.06 - ≥ 128 | 12 |
| Amikacin | 16 | ≥128 | 0.25 - ≥ 128 | 56.5 |
| Colistin | ≤0.25 | 1 | 0.03 - ≥ 128 | 94.4f |
Susceptibility interpreted against FDA criteria.
As per cefepime susceptible breakpoint.
As per cefepime/zidebactam’s proposed PK/PD breakpoint of ≤32 mg/L.
As per aztreonam standalone susceptibility breakpoint.
Based on meropenem susceptible breakpoint of ≤2 mg/L.
Based on CLSI intermediate breakpoint of ≤2 mg/L.
In vivo efficacy
Assessment of cefepime/zidebactam efficacy employing ELF-human-simulated regimen
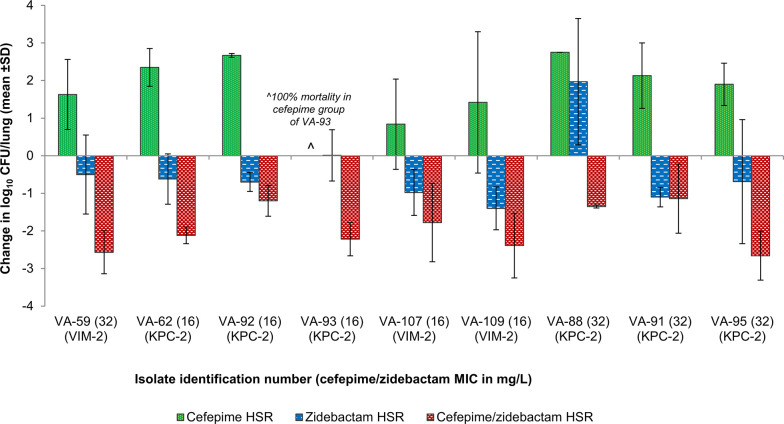
For the in vivo efficacy study, all the isolates with elevated cefepime/zidebactam MICs (>8 mg/L, n = 15) were chosen. However, only 9/15 isolates were able to successfully infect and grow in the lungs of neutropenic mice (the others were unfit) and were included in the efficacy assessment study (Table 3). The bacterial load in the lungs at 0 h ranged from 5.4 to 6.2 log10 CFU (mean 5.8 ± 0.2 log10 CFU). In the untreated groups, all the mice succumbed to infection by 24 h. Cefepime HSR was not efficacious; a net-growth of >1 log10 CFU/lung was noted in 7/9 isolates and in the remaining two isolates, 0.84 log10 net growth in VA107 and 100% mortality in VA93 were observed. Zidebactam HSR showed a bactericidal efficacy with a mean net-drop of 0.75 ± 0.42 log10 CFU/lung in 8/9 isolates and in a lone isolate, a net-growth of 1.97 log10 CFU/lung was observed. In contrast, the cefepime/zidebactam HSR demonstrated pronounced killing in all the studied isolates with a magnitude ranging from 1.1 log10 CFU/lung to 2.7 log10 CFU/lung (mean 1.9 ± 0.6 log10 CFU/lung), thus, exceeding the translational end point of 1-log10 kill (Fig. 3).
TABLE 3.
Major resistance mechanisms identified in P. aeruginosa isolates utilized in cefepime/zidebactam in vivo efficacy assessment study (cefepime/zidebactam MICs > 8 mg/L)
| Isolatesa | MLST | FEP/ZID MICs mg/L | Major resistance mechanisms |
|---|---|---|---|
| VA59 | 233 | 32 | VIM-2, OXA-4, OXA-486, PDC-3 |
| VA62 | 1801 | 16 | OXA-486, OXA-10, PDC-3; PBP2 (V517M A174V); PBP3 (F533L) |
| VA88 | 1801 | 32 | KPC-2, OXA-10, OXA-486, PDC-3; PBP2 (A174V V517M); PBP3 (F533L) |
| VA91 | 1801 | 32 | KPC-2, OXA-10, OXA-486, PDC-3; PBP2 (A174V V517M); PBP3 (F533L) |
| VA92 | 1801 | 16 | KPC-2, OXA-10, OXA-486, PDC-3; PBP2 (A174V V517M); PBP3 (F533L) |
| VA93 | 1801 | 16 | KPC-2, OXA-10, OXA-486, PDC-3; PBP2 (A174V V517M); PBP3 (F533L) |
| VA95 | 1801 | 32 | KPC-2, OXA-10, OXA-486, PDC-3; PBP2 (A174V V517M); PBP3 (F533L) |
| VA107 | 233 | 16 | VIM-2, OXA-4, OXA-486, PDC-3 |
| VA109 | 233 | 16 | VIM-2, OXA-4, OXA-486, PDC-3 |
The isolates were resistant to other BL/BLI combinations (imipenem/relebactam and ceftolozane/tazobactam) FEP/ZID: cefepime/zidebactam.
Fig 3.
In vivo efficacy of human-simulated regimen (HSR) of cefepime/zidebactam in neutropenic murine lung infection model against P. aeruginosa with cefepime/zidebactam MICs > 8 mg/L.
Effect on cefepime’s % fT >MIC requirement in the presence of zidebactam
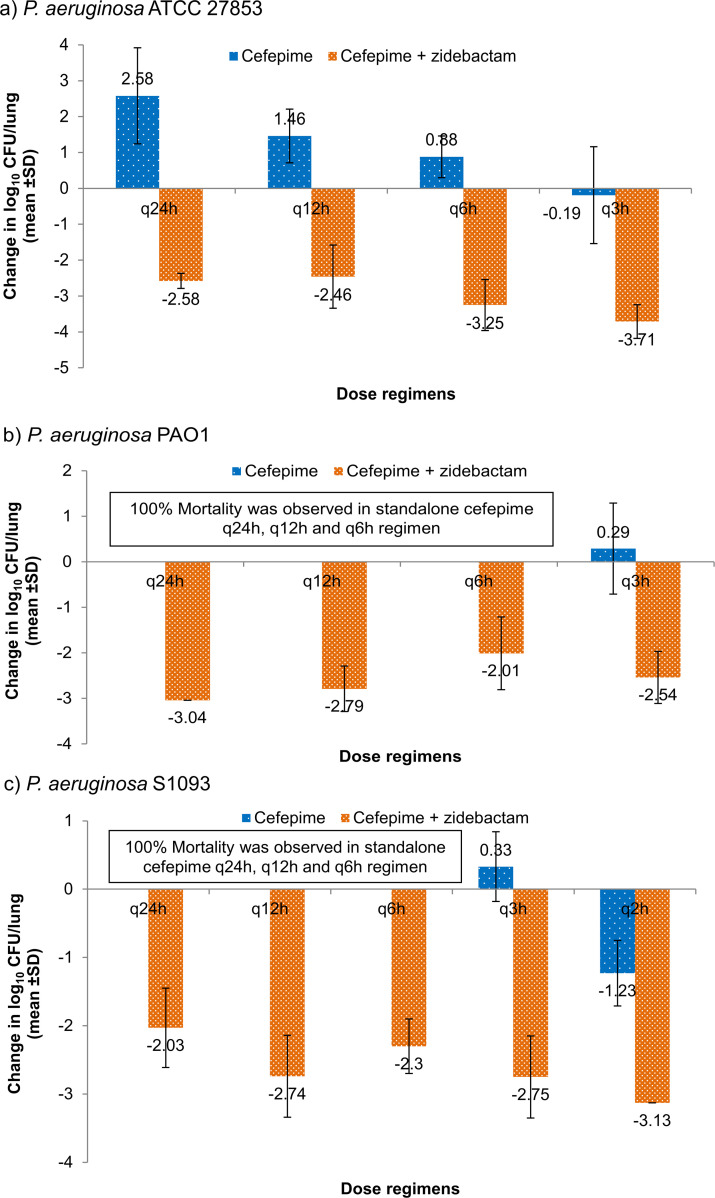
Zidebactam monotherapy at total daily dose (TDD) of 100 mg/kg, fractionated q2h showed mortality or no-efficacy. Infrequent regimens of cefepime monotherapy (q24h and q12h) resulted in net-growth or mortality showing lack of effectiveness of such regimens. However, as expected, more frequent regimens (q3h or q2h) resulted in net bacteriostatic effect to ~1 log10 kill with cefepime fT >MIC being 46.8%–68.2%. Interestingly, q24h and q12h cefepime regimens (ineffective as standalone) combined with zidebactam q2h regimen (ineffective as standalone) turned highly bactericidal (>2 log10 kill). At these regimens, cefepime’s fT >MIC was just 8%–16% (Fig. 4).
Fig 4.
Efficacy of cefepime as standalone and in the presence of zidebactam against cefepime-susceptible P. aeruginosa in neutropenic murine lung infection model. Cefepime total dose of 150 mg/kg was administered as single dose (q24h) and in various fractionated regimens [every 12 h (q12h), every 6 h (q6h), every 3 h (q3h), or every 2 h (q2h)] either standalone or in combination with a zidebactam regimen of total dose of 100 mg/kg, administered in q2h fraction (8.33 mg/kg). MICs of cefepime and cefepime/zidebactam were similar; 2, 1, and 2 mg/L against, P. aeruginosa ATCC 27853 (Fig. 4a), PAO1 (Fig. 4b), and S1093 (Fig. 4c), respectively.
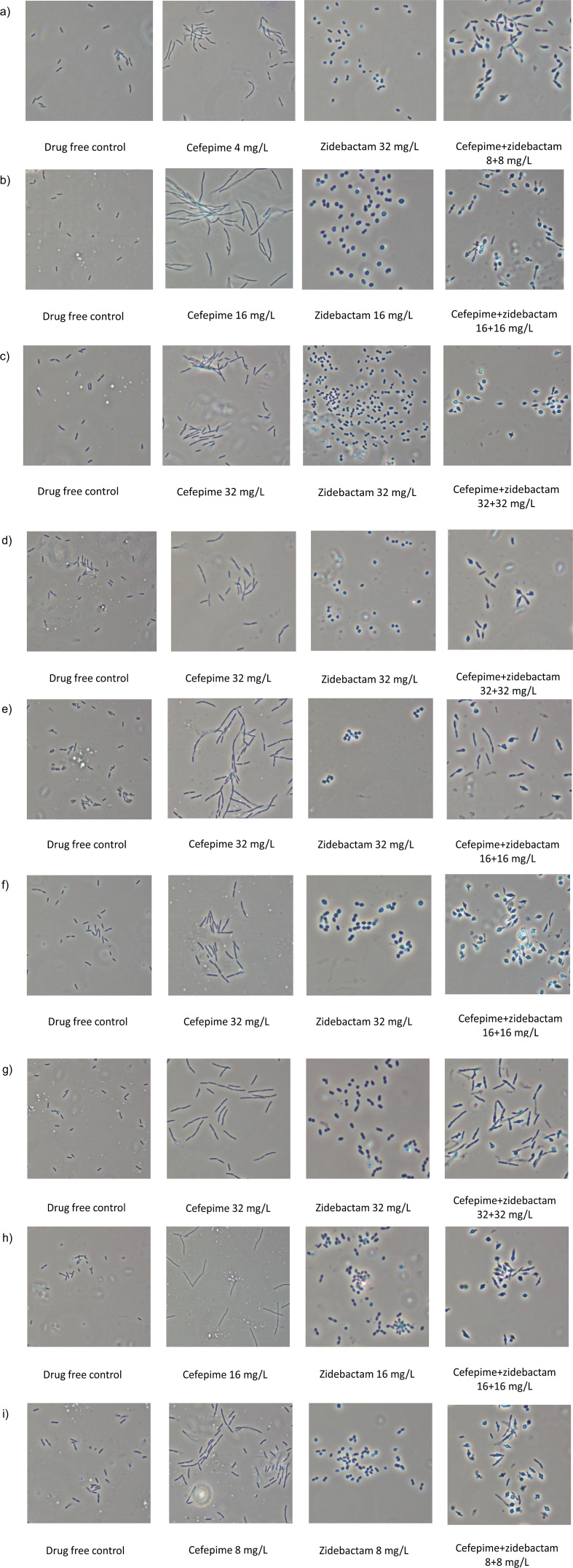
Morphology of cefepime, zidebactam, or cefepime/zidebactam treated cells
All nine isolates that were treated with zidebactam transformed into spherical forms consistent with an effect of PBP2 inactivation. Cefepime-treated cells showed elongation indicative of PBP3 binding. Cefepime/zidebactam-treated cells were pleomorphic and lysis-prone suggesting bactericidal action associated with synergistic effect of concurrent inactivation of multiple PBPs (Fig. 5).
Fig 5.
Morphological changes induced by cefepime, zidebactam, and cefepime+zidebactam in P. aeruginosa with cefepime/zidebactam MICs 16–32 mg/L (isolates employed in cefepime/zidebactam in vivo efficacy study). (a) P. aeruginosa VA 59; (b) P. aeruginosa VA 62; (c) P. aeruginosa VA 88; (d) P. aeruginosa VA 91; (e) P. aeruginosa VA 92; (f) P. aeruginosa VA 93; (g) P. aeruginosa VA 95; (h) P. aeruginosa VA 107; and (i) P. aeruginosa VA 109.
DISCUSSION
Contemporary MDR/XDR P. aeruginosa phenotypes are characterized by co-expression of various resistance mechanisms, mainly, increased efflux activity, OprD inactivation, PBP mutations and ever evolving β-lactamase variants. The daunting task has been to optimize a single antibiotic that overcomes all these resistance mechanisms. Unfortunately, older as well as newer anti-pseudomonal antibiotics are able to handle only a limited spectrum of resistance mechanisms, thus precluding their use as a reliable monotherapy for contemporary pseudomonal infections. Therefore, for seriously ill patients, combination therapies are required to improve clinical outcomes. However, they pose toxicity risks, adverse PK interactions, and dosing difficulties (29).
Challenged against a set of MDR P. aeruginosa isolates possessing diverse resistance mechanisms (PDC, KPC, MBLs, enhanced efflux, OprD inactivation, PBP3 and PBP2 substitutions), the present study revealed the limitations of novel anti-pseudomonal β-lactam/β-lactamase inhibitor combinations. Against this select collection, the susceptibilities to imipenem/relebactam, ceftazidime/avibactam, ceftolozane/tazobactam, and cefepime/taniborbactam were <60% even against a subset of isolates not producing any carbapenemase (overall <50%). This was somewhat unanticipated, since ceftazidime/avibactam, ceftolozane/tazobactam and cefepime/taniborbactam are expected to overcome PDC over-expression (avibactam and taniborbactam being potent β-lactamase inhibitors and ceftolozane being stable to PDC hydrolysis) (30, 31) and OprD truncations (cephalosporins are known to be less impacted than carbapenems) (32, 33). Thus, the modest activities of these β-lactam/β-lactamase inhibitors observed in this study could be attributed to hampered target binding due to the PBP3 substitutions, particularly, in the background of impermeability. Compromise in the activity of imipenem/relebactam could also be attributed to PBP changes, though contribution of concurrently operating additional resistance mechanisms cannot be ruled out.
The probable role of PBP substitutions in impacting the activity of these β-lactam/β-lactamase inhibitors stem from elevated MICs of ceftazidime/avibactam (8 to 32 mg/L) and imipenem/relebactam (16 to >128 mg/L) against ST 1801 isolates harboring KPC-2. Even though both avibactam and relebactam are potent inhibitors of KPC and mutations in genes encoding for hyper efflux were not identified in the ST 1801 isolates, ceftazidime/avibactam and imipenem/relebactam MICs remained high (Table S1). Thus, in conjunction with already well-established resistance mechanisms, proliferation of PBP substitutions in P. aeruginosa could further add to the challenge of optimizing novel antibiotics targeted towards this problematic pathogen.
In contrast, β-lactam-enhancer based cefepime/zidebactam demonstrated potent activity against the same set of P. aeruginosa isolates with 86.1% susceptibility at a cefepime (2 g, q8h) breakpoint of 8 mg/L which rose to 100% at 32 mg/L, an in vivo efficacy-supported cut-off being proposed as cefepime/zidebactam’s PK/PD breakpoint for P. aeruginosa. Earlier independent in vivo translational studies have established therapeutically relevant coverage of cefepime/zidebactam against P. aeruginosa isolates with MICs higher than cefepime’s susceptible breakpoint (up to 32 mg/L) (34, 35).
The consistent in vitro activity of cefepime/zidebactam against MDR P. aeruginosa isolates expressing a multitude of resistance mechanisms including PBP substitutions is a result of β-lactamase stable zidebactam’s PBP2 binding action that continues even in the isolates expressing enhanced efflux or impermeability as shown in this study. Likewise, cefepime’s ability to engage its high affinity PBP targets is facilitated by rapid cellular penetration and fast rate of PBP binding (14). As a result, in combination, a PBP level synergistic interaction enables cefepime/zidebactam to overcome multiple β-lactam-impacting non-enzymatic and enzymatic resistance mechanisms in P. aeruginosa.
Functional evidence of a multiple PBP binding driven synergistic interaction between cefepime and zidebactam was also manifested through changes in morphology of Pseudomonas cells. Upon exposure to the cefepime/zidebactam combination, we observed that the cell morphology changed from cocco-bacillary to lysis-prone spheroplasts (indication of multiple PBP engagement). Interestingly, such morphological change was noted at a significantly lower concentration of cefepime when combined with zidebactam, as compared to the concentration of standalone cefepime required to induce elongation (indication of PBP3 binding). Thus, we hypothesize that engagement of PBP3 and PBP2 (multiple target inactivation) by cefepime and zidebactam, respectively, has a synergetic effect by efficiently inhibiting or arresting cell wall synthesis allowing more rapid cell wall degradation (36).
A similar activity profile of cefepime/zidebactam was also proposed by Mullane et al., wherein, the in vitro activity of standalone cefepime/zidebactam was compared with several combinations against a panel of 30 carbapenem-resistant P. aeruginosa (37). While 97% isolates were inhibited by cefepime/zidebactam alone at ≤16 mg/L, the susceptibility rates to combination of other antibiotics (cefepime, ceftolozane-tazobactam, or meropenem combined with either amikacin or fosfomycin) were lower (<70% at established breakpoints). Pathogen coverage achieved with cefepime/zidebactam alone was even broader than that achieved with the most active combination of ceftolozane/tazobactam plus amikacin or fosfomycin (37).
We further investigated the impact of higher cefepime/zidebactam MICs of 16 and 32 mg/L obtained for nine isolates (Table 3) on its in vivo efficacy by employing a neutropenic murine pneumonia model. The results showed that, for all the isolates regardless of MICs, cefepime/zidebactam ELF-HSR caused a ≥ 1-log10 kill, thus exceeding the translational end point. Interestingly, for the majority of isolates, even zidebactam monotherapy showed considerable bactericidal effect. Notably, these isolates display several resistance mechanisms such as VIM-2, KPC-2, and PBP3 and PBP2 substitutions. To investigate the PK/PD basis of coverage of P. aeruginosa with higher cefepime/zidebactam MIC (higher than cefepime breakpoint of 8 mg/L), we assessed the impact of zidebactam on cefepime’s % fT >MIC requirement. This study showed that standalone cefepime fT >MIC of ≥46.8% provided merely bacteriostatic to ~1 log10 kill effect, while in combination with zidebactam, a lowered cefepime fT >MIC of 8%–16% imparted a substantially higher kill of >2 log10 CFU. Such modulation of the partner antibiotic’s PK/PD is an attribute associated with zidebactam’s β-lactam enhancer action, not reported with conventional β-lactamase inhibitors. Thus, a lowered requirement of cefepime’s % fT >MIC (linked with bactericidal effect) in the presence of zidebactam provided a rationale for observed in vivo bactericidal effect of cefepime/zidebactam against P. aeruginosa with higher cefepime/zidebactam MICs through its humanized regimen (cefepime 2g + zidebactam 1 g, TID). Moreover, adequacy of shorter % fT >MIC requirement of cefepime/zidebactam in rendering bactericidal effect is expected to be beneficial in critically-ill patient population often associated with reduced drug exposures (17).
The in vivo efficacy results obtained in this study are in agreement with the Kidd et al. study that showed a pronounced in vivo efficacy (static to 2 log10 kill) of cefepime/zidebactam ELF-HSR against several carbapenem and ceftolozane/tazobactam-resistant P. aeruginosa including isolates with cefepime/zidebactam MICs up to 32 mg/L (34). Taking into account the MIC90 of cefepime/zidebactam against global isolates (4 mg/L, n = 4808) and MIC90 against the subset of meropenem-non-susceptible isolates (8 mg/L, n = 1147) (38), a translational efficacy of cefepime/zidebactam against P. aeruginosa isolates with MICs up to 32 mg/L, as demonstrated in this study, potentially suggests a near-total coverage of MDR/XDR P. aeruginosa. If 108 P. aeruginosa clinical isolates included in this study were to represent pathogens causing infections in high-resistance regions or in ICU setting, cefepime/zidebactam is expected to be an important future arsenal for the treatment of MDR P. aeruginosa infections.
In summary, in the present study, β-lactam enhancer based approach showed promise in overcoming MDR P. aeruginosa regardless of resistance mechanisms expressed. While a multitude of resistance mechanisms expressed by MDR P. aeruginosa pose severe impediments to newer anti-pseudomonal drugs, the novel β-lactam enhancer approach, as exhibited by zidebactam, shows potential to transcend this challenge.
MATERIALS AND METHODS
Bacterial isolates
A collection of 108 well-characterized clinical P. aeruginosa isolates was used in this study. These isolates have been collected from northeast Ohio and the Mid-Atlantic states and were stored in the investigator’s laboratory. They were previously determined by phenotypic testing to be carbapenem resistant (>90% of isolates), and most were previously described (39–42).
Antibiotics and minimum inhibitory concentrations
Zidebactam, avibactam, relebactam, vaborbactam, taniborbactam and ceftolozane were synthesized at Wockhardt Research Centre, Aurangabad, India (>90% HPLC purity). Commercial formulation or >90% pure active pharmaceutical ingredient was used for tazobactam, cefepime, imipenem, meropenem, ciprofloxacin, amikacin, and colistin.
MICs of antibiotics were determined by broth microdilution method as recommended by Clinical & Laboratory Standards Institute, M100 guideline (43). Cefepime/zidebactam MICs were determined at 1:1 ratio. The fixed inhibitor concentration of 4 mg/L was used for avibactam, relebactam, tazobactam, and taniborbactam while 8 mg/L was used for vaborbactam. FDA breakpoints were employed for determining the susceptibility rates of isolates to the comparator antibiotics.
Whole genome sequencing (WGS)
WGS of all the study isolates (n = 108) was performed. Briefly, the sequencing libraries were prepared using the Nextra DNA Flex library preparation kit (Illumina, San Diego, CA) as per the manufacturer’s instructions. Subsequently, the paired-end library was subjected to sequencing on a HiSeq 2500 platform (Illumina, USA) generating 2 × 150 bp reads. Sequencing reads with a PHRED quality score below 20 were discarded and adapters were trimmed using cutadapt v1.8.1 and assessed with FastQC v0.11.4. Draft genome sequence data generated using Illumina were assembled using SPAdes (v.3.13.0) (44). Genome assemblies were annotated using the Prokaryotic Genome Annotation Pipeline (PGAP v.4.1) from NCBI (45).
Core genome single-nucleotide polymorphisms (SNPs) were identified using Snippy v.0.2.6 (https://github.com/tseemann/snippy) with P. aeruginosa PAO1 (accession no. CP053028.1) as the reference.
In vivo efficacy
Assessment of cefepime/zidebactam efficacy employing ELF-HSR
In vivo efficacy employing HSR of cefepime alone, or zidebactam alone, or cefepime/zidebactam was evaluated in a murine neutropenic infection model as described previously (34) against isolates with cefepime/zidebactam MICs of >8 mg/L (Table 3). These dosing regimens (designed at Wockhardt) produced cefepime, or zidebactam, or cefepime plus zidebactam exposures in mice epithelial lining fluid (ELF) comparable to that of respective exposures obtained in human ELF after 2 + 1 g, q8h administration of cefepime/zidebactam. The comparability between mice and human exposures in the ELF was in terms of the proportion of time during which cefepime and zidebactam concentrations remained above cefepime/zidebactam MICs (Table S2A and B; Fig. S1). Male/female Swiss Albino mice were rendered neutropenic by intra-peritoneal injections of cyclophosphamide 150 and 100 mg/kg on 4 days and 1 day prior to the infection, respectively. The humanized regimen of cefepime/zidebactam in mice was rendered feasible by slowing the renal elimination with the help of uranyl nitrate 5 mg/kg, intra-peritoneal injection administered 3 days before the infection (46).
Animals were infected with 0.05 mL of normal saline containing ~107 CFU/ml of P. aeruginosa through nostrils under isofluorane-induced transient anesthesia. Nine P. aeruginosa with cefepime/zidebactam MICs of 16–32 mg/L with resistance to imipenem/relebactam and ceftolozane/tazobactam were employed in this study (Table 3). Treatment (subcutaneous injections) was initiated 2 h post-infection with cefepime HSR or zidebactam HSR or cefepime/zidebactam combination HSR. A group of animals was administered with vehicle control. After 24-h treatment duration, animals were humanely sacrificed, and the lung bacterial load was estimated. Earlier at the time of initiation of treatment (0 h), a group of infected mice was sacrificed to determine the lung bacterial burden at time 0 h. Efficacy was defined as change in the bacterial load at 24 h as compared to 0 h. All the groups consisted of six animals.
Effect on cefepime’s % fT>MIC requirement in the presence of zidebactam
The above-described murine neutropenic infection model was employed with the exception that animals were not administered with uranyl nitrate. Cefepime was administered as a single dose (q24h) or in fractionated regimens over 24 h; every 12 h (q12h), every 6 h (q6h), every 3 h (q3h), or every 2 h (q2h) and the same regimen was combined with zidebactam given 8.33 mg/kg, every 2 h (q2h). The infecting isolates were cefepime-susceptible (n = 3) which enabled determining the efficacy-linked magnitude of % fT >MIC for cefepime monotherapy, which could then be compared with that of cefepime in the presence of zidebactam. The % fT >MIC of cefepime in the various fractionated regimens was determined using non-linear sigmoidal Emax model (GraphPad Prism version 7) and previously reported mouse plasma PK of cefepime (18).
Morphology of cefepime, zidebactam, or cefepime/zidebactam-treated cells
Morphological changes were studied for isolates with cefepime/zidebactam MICs of 16–32 mg/L by exposing sub-minimum inhibitory concentrations or at inhibitory concentrations of zidebactam or cefepime or cefepime/zidebactam to 106 CFU/mL bacterial density in cation-adjusted Mueller-Hinton broth under shaking condition. The treated cells were visualized after 3 h of exposure using a phase contrast microscope.
ACKNOWLEDGMENTS
The study was carried out as part of routine research activities at the Veterans Affairs Medical Center, Christian Medical College, and Wockhardt Research Centre.
AFTER EPUB
[This article was published on 27 October 2023 with text missing from the Introduction, 4th paragraph. The Introduction was corrected in the current version, posted on 1 November 2023.]
Contributor Information
Balaji Veeraraghavan, Email: vbalaji@cmcvellore.ac.in.
Robert A. Bonomo, Email: Robert.Bonomo@va.gov.
Steven J. Projan, MedImmune, Gaithersburg, Maryland, USA
ETHICS APPROVAL
Animal study protocols were reviewed and approved by Wockhardt's Institutional Animal Ethics Committee.
DATA AVAILABILITY
Genome assemblies were submitted to the National Center for Biotechnology Information (NCBI) GenBank database (BioProject no. PRJNA869185).
SUPPLEMENTAL MATERIAL
The following material is available online at https://doi.org/10.1128/mbio.01118-23.
ASM does not own the copyrights to Supplemental Material that may be linked to, or accessed through, an article. The authors have granted ASM a non-exclusive, world-wide license to publish the Supplemental Material files. Please contact the corresponding author directly for reuse.
REFERENCES
- 1. Horcajada JP, Montero M, Oliver A, Sorlí L, Luque S, Gómez-Zorrilla S, Benito N, Grau S. 2019. Epidemiology and treatment of multidrug-resistant and extensively drug-resistant Pseudomonas aeruginosa infections. Clin Microbiol Rev 32:e00031-19. doi: 10.1128/CMR.00031-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Del Barrio-Tofiño E, López-Causapé C, Oliver A. 2020. Pseudomonas aeruginosa epidemic high-risk clones and their association with horizontally-acquired β-lactamases: 2020 update. Int J Antimicrob Agents 56:106196. doi: 10.1016/j.ijantimicag.2020.106196 [DOI] [PubMed] [Google Scholar]
- 3. Tam VH, Schilling AN, LaRocco MT, Gentry LO, Lolans K, Quinn JP, Garey KW. 2007. Prevalence of AmpC over-expression in bloodstream isolates of Pseudomonas aeruginosa. Clin Microbiol Infect 13:413–418. doi: 10.1111/j.1469-0691.2006.01674.x [DOI] [PubMed] [Google Scholar]
- 4. Bonomo RA, Szabo D. 2006. Mechanisms of multidrug resistance in Acinetobacter species and Pseudomonas aeruginosa. Clin Infect Dis 43 Suppl 2:S49–56. doi: 10.1086/504477 [DOI] [PubMed] [Google Scholar]
- 5. Nakajima A, Sugimoto Y, Yoneyama H, Nakae T. 2002. High-level fluoroquinolone resistance in Pseudomonas aeruginosa due to interplay of the MexAB-OprM efflux pump and the DNA gyrase mutation. Microbiol Immunol 46:391–395. doi: 10.1111/j.1348-0421.2002.tb02711.x [DOI] [PubMed] [Google Scholar]
- 6. Lob SH, DePestel DD, DeRyke CA, Kazmierczak KM, Young K, Motyl MR, Sahm DF. 2021. Ceftolozane/tazobactam and imipenem/relebactam cross-susceptibility among clinical isolates of Pseudomonas aeruginosa from patients with respiratory tract infections in ICU and non-ICU wards-SMART United States 2017-2019. Open Forum Infect Dis 8:fab320. doi: 10.1093/ofid/ofab320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. CDC . 2019. Antibiotic resistance threats in the United States. [Google Scholar]
- 8. Mancuso G, Midiri A, Gerace E, Biondo C. 2021. Bacterial antibiotic resistance: the most critical pathogens. Pathogens 10:1310. doi: 10.3390/pathogens10101310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. CDC. COVID-19: USA. Impact on antimicrobial resistance, special report. 2022. Atlanta, GA: U.S. Department of Health and Human Services; AMR report. https://www.cdc.gov/drugresistance/covid19.html. [Google Scholar]
- 10. Yusuf E, Bax HI, Verkaik NJ, van Westreenen M. 2021. An update on eight "new" antibiotics against multidrug-resistant Gram-negative bacteria. J Clin Med 10:1068. doi: 10.3390/jcm10051068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Ruedas-López A, Alonso-García I, Lasarte-Monterrubio C, Guijarro-Sánchez P, Gato E, Vázquez-Ucha JC, Vallejo JA, Fraile-Ribot PA, Fernández-Pérez B, Velasco D, Gutiérrez-Urbón JM, Oviaño M, Beceiro A, González-Bello C, Oliver A, Arca-Suárez J, Bou G. 2022. Selection of AmpC β-lactamase variants and metallo-β-lactamases leading to ceftolozane/tazobactam and ceftazidime/avibactam resistance during treatment of MDR/XDR Pseudomonas aeruginosa infections. Antimicrob Agents Chemother 66:e0206721. doi: 10.1128/AAC.02067-21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. MacVane SH, Pandey R, Steed LL, Kreiswirth BN, Chen L. 2017. Emergence of ceftolozane-tazobactam-resistant Pseudomonas aeruginosa during treatment is mediated by a single AmpC structural mutation. Antimicrob Agents Chemother 61:e01183-17. doi: 10.1128/AAC.01183-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Hujer AM, Bethel CR, Taracila MA, Marshall SH, Rojas LJ, Winkler ML, Painter RE, Domitrovic TN, Watkins RR, Abdelhamed AM, D’Souza R, Mack AR, White RC, Clarke T, Fouts DE, Jacobs MR, Young K, Bonomo RA. 2022. Imipenem/relebactam resistance in clinical isolates of extensively drug resistant Pseudomonas aeruginosa: inhibitor-resistant β-lactamases and their increasing importance. Antimicrob Agents Chemother 66:e0179021. doi: 10.1128/aac.01790-21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Moya B, Bhagwat S, Cabot G, Bou G, Patel M, Oliver A. 2020. Effective inhibition of PBPs by cefepime and zidebactam in the presence of VIM-1 drives potent bactericidal activity against MBL-expressing Pseudomonas aeruginosa. J Antimicrob Chemother 75:1474–1478. doi: 10.1093/jac/dkaa036 [DOI] [PubMed] [Google Scholar]
- 15. Almarzoky Abuhussain SS, Avery LM, Abdelraouf K, Nicolau DP. 2019. In vivo efficacy of humanized WCK 5222 (cefepime-zidebactam) exposures against carbapenem-resistant Acinetobacter baumannii in the neutropenic thigh model. Antimicrob Agents Chemother 63:e01931-18. doi: 10.1128/AAC.01931-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Moya B, Barcelo IM, Bhagwat S, Patel M, Bou G, Papp-Wallace KM, Bonomo RA, Oliver A. 2017. WCK 5107 (Zidebactam) and WCK 5153 are novel inhibitors of PBP2 showing potent "β-lactam enhancer" activity against Pseudomonas aeruginosa, including multidrug-resistant metallo-β-lactamase-producing high-risk clones. Antimicrob Agents Chemother 61:e02529-16. doi: 10.1128/AAC.02529-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Bhagwat S, Chavan R, Friedland D, Patel M. 2017. WCK 5222: Cefepime (FEP) + WCK 5107: in vivo demonstration of ZID-mediated Β-lactam enhancer effect leading to lowering of FEP % f T &Amp;Amp;Gt;MIC against P. Aeruginosa (PA) and A. Baumannii (AB), Abstr A4665 ASM MICROBE, New Orleans, USA
- 18. Bhagwat SS, Periasamy H, Takalkar SS, Palwe SR, Khande HN, Patel MV. 2019. The novel β-lactam enhancer zidebactam augments the in vivo pharmacodynamic activity of cefepime in a neutropenic mouse lung Acinetobacter baumannii infection model. Antimicrob Agents Chemother 63:e02146-18. doi: 10.1128/AAC.02146-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Avery LM, Abdelraouf K, Nicolau DP. 2018. Assessment of the in vivo efficacy of WCK 5222 (cefepime-zidebactam) against carbapenem-resistant Acinetobacter baumannii in the neutropenic murine lung infection model. Antimicrob Agents Chemother 62:e00948-18. doi: 10.1128/AAC.00948-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Wolter DJ, Kurpiel PM, Woodford N, Palepou M-FI, Goering RV, Hanson ND. 2009. Phenotypic and enzymatic comparative analysis of the novel KPC variant KPC-5 and its evolutionary variants, KPC-2 and KPC-4. Antimicrob Agents Chemother 53:557–562. doi: 10.1128/AAC.00734-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Lasarte-Monterrubio C, Fraile-Ribot PA, Vázquez-Ucha JC, Cabot G, Guijarro-Sánchez P, Alonso-García I, Rumbo-Feal S, Galán-Sánchez F, Beceiro A, Arca-Suárez J, Oliver A, Bou G. 2022. Activity of cefiderocol, imipenem/relebactam, cefepime/taniborbactam and cefepime/zidebactam against ceftolozane/tazobactam- and ceftazidime/avibactam-resistant Pseudomonas aeruginosa. J Antimicrob Chemother 77:2809–2815. doi: 10.1093/jac/dkac241 [DOI] [PubMed] [Google Scholar]
- 22. Pan X, Zhao X, Song Y, Ren H, Tian Z, Liang Q, Jin Y, Bai F, Cheng Z, Feng J, Wu W. 2022. Molecular characterization of WCK 5222 (Cefepime/Zidebactam)-resistant mutants developed from a carbapenem-resistant Pseudomonas aeruginosa clinical isolate. Microbiol Spectr 10:e0267821. doi: 10.1128/spectrum.02678-21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Aguilar-Rodea P, Zúñiga G, Cerritos R, Rodríguez-Espino BA, Gomez-Ramirez U, Nolasco-Romero CG, López-Marceliano B, Rodea GE, Mendoza-Elizalde S, Reyes-López A, Olivares Clavijo H, Vigueras Galindo JC, Velázquez-Guadarrama N, Rosas-Pérez I. 2022. Nucleotide substitutions in the mexR, nalC and nalD regulator genes of the MexAB-OprM efflux pump are maintained in Pseudomonas aeruginosa genetic lineages. PLoS One 17:e0266742. doi: 10.1371/journal.pone.0266742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Horna G, López M, Guerra H, Saénz Y, Ruiz J. 2018. Interplay between MexAB-OprM and MexEF-OprN in clinical isolates of Pseudomonas aeruginosa. Sci Rep 8:16463. doi: 10.1038/s41598-018-34694-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Pai H, Kim J, Kim J, Lee JH, Choe KW, Gotoh N. 2001a. Carbapenem resistance mechanisms in Pseudomonas aeruginosa clinical isolates. Antimicrob Agents Chemother 45:480–484. doi: 10.1128/AAC.45.2.480-484.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Mojica MF, De La Cadena E, Ríos R, García-Betancur JC, Díaz L, Reyes J, Hernández-Gómez C, Radice M, Gales AC, Castañeda Méndez P, Munita JM, Pallares CJ, Martínez JRW, Villegas MV. 2022. Molecular mechanisms leading to ceftolozane/tazobactam resistance in clinical isolates of Pseudomonas aeruginosa from five Latin American countries. Front Microbiol 13:1035609. doi: 10.3389/fmicb.2022.1035609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Fernández-Esgueva M, López-Calleja AI, Mulet X, Fraile-Ribot PA, Cabot G, Huarte R, Rezusta A, Oliver A. 2020. Characterization of AmpC β-lactamase mutations of extensively drug-resistant Pseudomonas aeruginosa isolates that develop resistance to ceftolozane/tazobactam during therapy. Enferm Infecc Microbiol Clin (Engl Ed) 38:474–478. doi: 10.1016/j.eimc.2020.01.017 [DOI] [PubMed] [Google Scholar]
- 28. Glen KA, Lamont IL. 2021. β-lactam resistance in Pseudomonas aeruginosa: current status, future prospects. Pathogens 10:1638. doi: 10.3390/pathogens10121638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Kunz Coyne AJ, El Ghali A, Holger D, Rebold N, Rybak MJ. 2022. Therapeutic strategies for emerging multidrug-resistant Pseudomonas aeruginosa. Infect Dis Ther 11:661–682. doi: 10.1007/s40121-022-00591-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Liu B, Trout REL, Chu G-H, McGarry D, Jackson RW, Hamrick JC, Daigle DM, Cusick SM, Pozzi C, De Luca F, Benvenuti M, Mangani S, Docquier J-D, Weiss WJ, Pevear DC, Xerri L, Burns CJ. 2020. Discovery of taniborbactam (VNRX-5133): a broad-spectrum serine- and metallo-β-lactamase inhibitor for carbapenem-resistant bacterial infections. J Med Chem 63:2789–2801. doi: 10.1021/acs.jmedchem.9b01518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Barnes MD, Taracila MA, Rutter JD, Bethel CR, Galdadas I, Hujer AM, Caselli E, Prati F, Dekker JP, Papp-Wallace KM, Haider S, Bonomo RA. 2018. Deciphering the evolution of cephalosporin resistance to ceftolozane-tazobactam in Pseudomonas aeruginosa. mBio 9:e02085-18. doi: 10.1128/mBio.02085-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Campana EH, Xavier DE, Petrolini F-B, Cordeiro-Moura JR, Araujo M de, Gales AC. 2017. Carbapenem-resistant and cephalosporin-susceptible: a worrisome phenotype among Pseudomonas aeruginosa clinical isolates in Brazil. Braz J Infect Dis 21:57–62. doi: 10.1016/j.bjid.2016.10.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Zeng ZR, Wang WP, Huang M, Shi LN, Wang Y, Shao HF. 2014. Mechanisms of carbapenem resistance in cephalosporin-susceptible Pseudomonas aeruginosa in China. Diagn Microbiol Infect Dis 78:268–270. doi: 10.1016/j.diagmicrobio.2013.11.014 [DOI] [PubMed] [Google Scholar]
- 34. Kidd JM, Abdelraouf K, Nicolau DP. 2020. Efficacy of human-simulated bronchopulmonary exposures of cefepime, zidebactam and the combination (WCK 5222) against MDR Pseudomonas aeruginosa in a neutropenic murine pneumonia model. J Antimicrob Chemother 75:149–155. doi: 10.1093/jac/dkz414 [DOI] [PubMed] [Google Scholar]
- 35. Monogue ML, Tabor-Rennie J, Abdelraouf K, Nicolau DP. 2019. In vivo efficacy of WCK 5222 (cefepime-zidebactam) against multidrug-resistant Pseudomonas aeruginosa in the neutropenic murine thigh infection model. Antimicrob Agents Chemother 63:e00233-19. doi: 10.1128/AAC.00233-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Papp-Wallace K, Nguyen N, Jacobs M, Bethel C, Barnes M, Kumar V, Bajaksouzian S, Rudin S, Rather PN, Bhavsar S, Ravikumar T, Deshpande P, Patil V, Yeole R, Bhagwat S, Patel M, Akker F, Bonomo R. 2018. Strategic approaches to overcome resistance against Gram-negative pathogens using β-lactamase inhibitors and β-lactam enhancers. doi: 10.1021/acs.jmedchem.8b00091 [DOI] [PMC free article] [PubMed]
- 37. Mullane EM, Avery LM, Nicolau DP. 2020. Comparative evaluation of the in vitro activities of WCK 5222 (cefepime-zidebactam) and combination antibiotic therapies against carbapenem-resistant Pseudomonas aeruginosa. Antimicrob Agents Chemother 64:e01669-19. doi: 10.1128/AAC.01669-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Sader HS, Mendes RE, Duncan LR, Carvalhaes CG, Castanheria M. 2022. Antimicrobial activity of Cefepime/Zidebactam (WCK 5222), a β-lactam/β-lactam enhancer combination, against clinical isolates of Gram-negative bacteria collected worldwide (2018-19). J Antimicrob Chemother 77:2642–2649. doi: 10.1093/jac/dkac233 [DOI] [PubMed] [Google Scholar]
- 39. Evans SR, Tran TTT, Hujer AM, Hill CB, Hujer KM, Mediavilla JR, Manca C, Domitrovic TN, Perez F, Farmer M, Pitzer KM, Wilson BM, Kreiswirth BN, Patel R, Jacobs MR, Chen L, Fowler VG, Chambers HF, Bonomo RA, Antibacterial Resistance Leadership Group (ARLG) . 2019. Rapid molecular diagnostics to inform empiric use of ceftazidime/avibactam and ceftolozane/tazobactam against Pseudomonas aeruginosa: PRIMERS IV. Clin Infect Dis 68:1823–1830. doi: 10.1093/cid/ciy801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Goldberg JA, Kumar V, Spencer EJ, Hoyer D, Marshall SH, Hujer AM, Hujer KM, Bethel CR, Papp-Wallace KM, Perez F, Jacobs MR, van Duin D, Kreiswirth BN, van den Akker F, Plummer MS, Bonomo RA. 2021. A γ-lactam siderophore antibiotic effective against multidrug-resistant Pseudomonas aeruginosa, Klebsiella pneumoniae, and Acinetobacter spp. Eur J Med Chem 220:113436. doi: 10.1016/j.ejmech.2021.113436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Goldberg JA, Nguyen H, Kumar V, Spencer EJ, Hoyer D, Marshall EK, Cmolik A, O’Shea M, Marshall SH, Hujer AM, Hujer KM, Rudin SD, Domitrovic TN, Bethel CR, Papp-Wallace KM, Logan LK, Perez F, Jacobs MR, van Duin D, Kreiswirth BM, Bonomo RA, Plummer MS, van den Akker F. 2020. A β-lactam siderophore antibiotic effective against multidrug-resistant Gram-negative bacilli. J Med Chem 63:5990–6002. doi: 10.1021/acs.jmedchem.0c00255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Perez F, Hujer AM, Marshall SH, Ray AJ, Rather PN, Suwantarat N, Dumford D III, O’Shea P, Domitrovic TNJ, Salata RA, Chavda KD, Chen L, Kreiswirth BN, Vila AJ, Haussler S, Jacobs MR, Bonomo RA. 2014. Extensively drug-resistant Pseudomonas aeruginosa isolates containing blaVIM-2 and elements of Salmonella genomic Island 2: a new genetic resistance determinant in northeast Ohio . Antimicrob Agents Chemother 58:5929–5935. doi: 10.1128/AAC.02372-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Clinical and Laboratory Standards Institute . 2022. M100-S32. Performance standards for antimicrobial susceptibility testing. 32nd Edition. Clinical and Laboratory Standards Institute, Malvern, PA. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, Pyshkin AV, Sirotkin AV, Vyahhi N, Tesler G, Alekseyev MA, Pevzner PA. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol 19:455–477. doi: 10.1089/cmb.2012.0021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Tatusova T, DiCuccio M, Badretdin A, Chetvernin V, Nawrocki EP, Zaslavsky L, Lomsadze A, Pruitt KD, Borodovsky M, Ostell J. 2016. NCBI prokaryotic genome annotation pipeline. Nucleic Acids Res. 44:6614–6624. doi: 10.1093/nar/gkw569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Fratoni AJ, Nicolau DP, Kuti JL. 2021. 1103. Minocycline (MIN) pharmacodynamics (PD) against Stenotrophomonas maltophilia (STM) in a neutropenic murine thigh infection model. Open Forum Infectious Diseases 8:S643–S643. doi: 10.1093/ofid/ofab466.1297 [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Citations
- Bhagwat S, Chavan R, Friedland D, Patel M. 2017. WCK 5222: Cefepime (FEP) + WCK 5107: in vivo demonstration of ZID-mediated Β-lactam enhancer effect leading to lowering of FEP % f T &Amp;Amp;Gt;MIC against P. Aeruginosa (PA) and A. Baumannii (AB), Abstr A4665 ASM MICROBE, New Orleans, USA
Supplementary Materials
Data Availability Statement
Genome assemblies were submitted to the National Center for Biotechnology Information (NCBI) GenBank database (BioProject no. PRJNA869185).