ABSTRACT
The phytopathogenic fungus Rhizopus microsporus harbors a bacterial endosymbiont (Mycetohabitans rhizoxinica) that produces a crucial virulence factor responsible for the characteristic symptoms of rice seedling blight. The persistence of this unique microbial alliance is ensured by evasion of fungal defense mechanisms by the resident endosymbionts and their strict control of fungal reproduction. A functional bacterial Type 3 secretion system (T3SS) is essential for the establishment of the symbiosis. Yet, the nature of the effector(s) secreted through the T3SS, and their effects on fungal physiology, has remained elusive. Using bioinformatic analysis, we identified genes encoding potential T3SS effectors, namely an alanine-tryptophan-arginine triad (AWR) peptide and a set of transcription activator-like (TAL) effectors, in the genomes of eight endofungal Mycetohabitans strains. Co-culture experiments demonstrated that the sporulating phenotype of apo-symbiotic R. microsporus is unaffected upon reinfection with M. rhizoxinica Δawr compared to with wild-type M. rhizoxinica. In contrast, the ability of apo-symbiotic R. microsporus to produce mature sporangiophores when reinfected with M. rhizoxinica strains deficient in individual MTALs (M. rhizoxinica Δmtal1, M. rhizoxinica Δmtal2, and M. rhizoxinica Δmtal3) is significantly reduced. Trans-complementation experiments showed restoration of fungal sporulation, thus confirming that TAL effectors produced by M. rhizoxinica (MTAL1, MTAL2, and MTAL3) are needed for fungal sporulation. Using fluorescence microscopy, we show that AWR- and MTAL-deficient M. rhizoxinica strains successfully colonize apo-symbiotic R. microsporus, revealing the importance of bacterial MTALs in establishing a stable symbiosis after fungal colonization. Our findings attribute a new function to members of the MTAL family of T3SS-associated effectors and provide deeper insights into host control by prokaryotic symbionts.
IMPORTANCE
Interactions between fungi and bacteria are critically important in ecology, medicine, and biotechnology. In this study, we shed light on factors that promote the persistence of a toxin-producing, phytopathogenic Rhizopus-Mycetohabitans symbiosis that causes severe crop losses in Asia. We present an unprecedented case where bacterially produced transcription activator-like (TAL) effectors are key to maintaining a stable endosymbiosis. In their absence, fungal sporulation is abrogated, leading to collapse of the phytopathogenic alliance. The Mycetohabitans TAL (MTAL)-mediated mechanism of host control illustrates a unique role of bacterial effector molecules that has broader implications, potentially serving as a model to understand how prokaryotic symbionts interact with their eukaryotic hosts.
KEYWORDS: host control, Mycetohabitans, Rhizopus microsporus, sporulation, symbiosis
INTRODUCTION
Bacteria that live in close association with eukaryotic hosts may control and exploit their host via pathogenic or mutualistic interactions (1). To take control of their eukaryotic hosts, bacteria have evolved dedicated secretion systems (Types 1 to 9) that secrete a wide range of effectors into the extracellular space or directly into the target cell (2–4). These sophisticated molecules are central to some of the most devastating diseases (5). For example, the human and animal pathogen Burkholderia mallei, the causative agent of the highly lethal disease glanders, depends on Type 3 secretion system (T3SS) effectors to invade the cytoplasm of its host (6). Plant-pathogenic Xanthomonas and Ralstonia species colonize their hosts with the help of T3SS-associated transcription activator-like (TAL) effectors that imitate plant transcription factors (7–9). The fungal pathogen Janthinobacterium agaricidamnosum secretes degradative enzymes and putative effector proteins via the Type 2 secretion system (T2SS) and T3SS. These effectors aid bacterial invasion of the white button mushroom, leading to rapid tissue decay and soft rot disease (10).
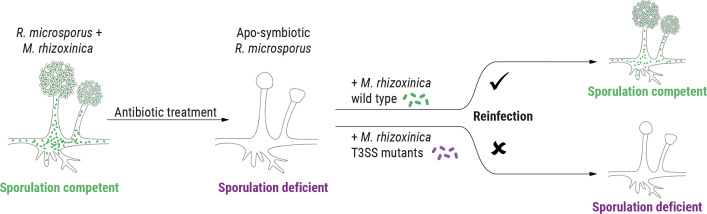
The effectors and associated secretion systems utilized by pathogenic bacteria to control their hosts have been disproportionately studied compared to those employed by symbiotic bacteria, which remain underexplored. The endosymbiosis between the Mucoromycota fungus Rhizopus microsporus and the bacterium Mycetohabitans rhizoxinica (formerly Burkholderia rhizoxinica) is an intriguing example of host control by a symbiont (11–13). Fungal reproduction through spores relies exclusively on the presence of endobacteria (Fig. 1). When R. microsporus is cured of its endosymbiont, the fungal host is unable to reproduce vegetatively (13). This strict control of sporulation ensures that the endosymbiosis persists over generations because the endosymbionts are translocated into the fungal spores during host reproduction (13). Maintaining its endosymbiont has clear advantages for the fungus, such as killing rice seedlings with the bacterial secondary metabolite rhizoxin (14) and protection from micropredators (15).
Fig 1.
Schematic representation of M. rhizoxinica-dependent sporulation of R. microsporus. Mature sporangia are absent in fungus cured of endosymbionts by antibiotic treatment. Co-cultivation of wild-type M. rhizoxinica with the apo-symbiotic fungus leads to reinfection, and the host’s ability to form sporangiophores is restored. T3SS mutants fail to reinfect hyphae, causing a lack of sporulation.
Effectors have been repeatedly associated with the establishment and maintenance of the R. microsporus–M. rhizoxinica symbiosis. For example, M. rhizoxinica invades the fungal host by secreting effector proteins via the T2SS (16), while a functional T3SS is required for the formation of a stable symbiosis (17). Gene expression analyses further hinted at the importance of effectors secreted via the T3SS since nearly 60 T3SS-associated effector genes are over-expressed during establishment of the symbiosis (18). However, Mycetohabitans TAL (MTAL1) is the only effector that has so far been recognized as essential for the maintenance of the R. microsporus–M. rhizoxinica symbiosis (19). Using a combination of microfluidics and fluorescence microscopy, we showed that MTAL1 is crucial for evading host immune response and thus facilitates the intracellular survival of M. rhizoxinica (19). While these findings indicate that effectors secreted through the T3SS are important symbiosis factors, there is a lack of knowledge about whether they could be involved in controlling fungal reproduction and development.
Here, we show that homologous genes encoding T3SS-associated effectors, namely alanine-tryptophan-arginine triad (AWR) peptides and Mycetohabitans TALs (MTAL1, MTAL2, and MTAL3), are widespread in the genomes of endofungal Mycetohabitans symbionts. Furthermore, we demonstrate that MTALs are instrumental in controlling fungal reproduction in the phytopathogenic R. microsporus–M. rhizoxinica bacterial–fungal alliance.
RESULTS
AWR peptides are universally conserved in endofungal Mycetohabitans symbionts
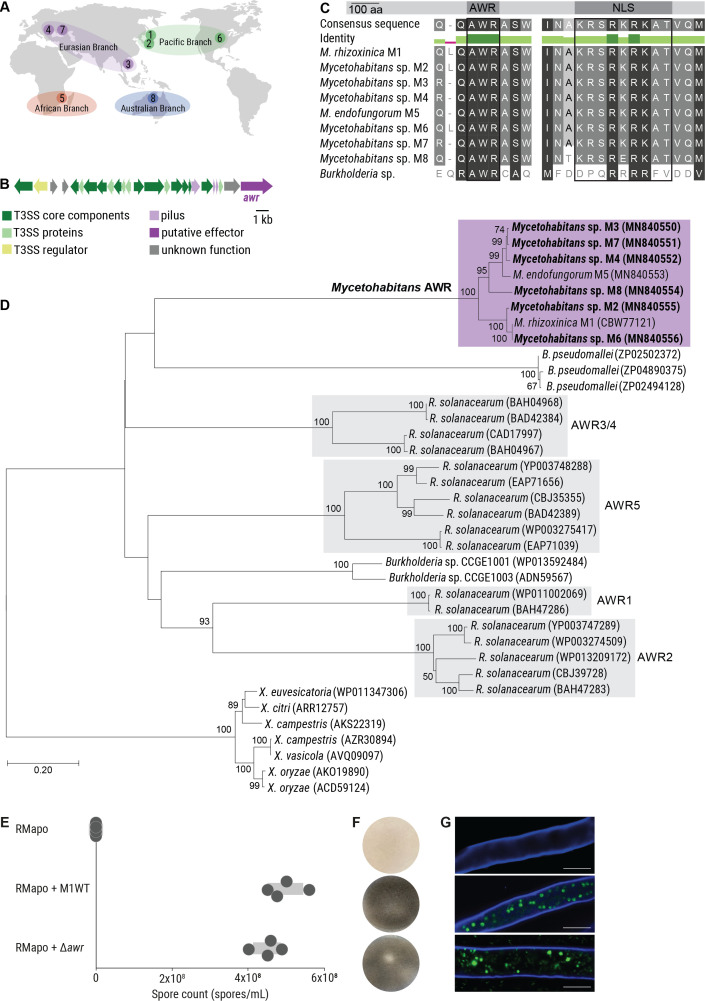
Initial physical contact of M. rhizoxinica with R. microsporus induces the expression of approximately 60 T3SS-associated effector genes in M. rhizoxinica (18), hinting that the cognate effectors act as mediators of this bacterial–fungal interaction. Therefore, we sought to identify genes coding for putative Type 3 effector proteins by searching publicly available genomes of eight Mycetohabitans species that are endosymbionts of globally distributed R. microsporus strains (see Table S1). The fungal strains and their corresponding endosymbionts were isolated from varying habitats (e.g., arid and forest soils, food products, and human tissue) and are grouped into the following four, geographically distant branches: (i) Pacific branch containing M. rhizoxinica HKI-454 (M1), Mycetohabitans sp. HKI-512 (M2), and Mycetohabitans sp. HKI-513 (M6); (ii) Eurasian branch containing Mycetohabitans sp. HKI-455 (M3), Mycetohabitans sp. HKI-402 (M4), and Mycetohabitans sp. HKI-403 (M7); (iii) African branch containing Mycetohabitans endofungorum HKI-456 (M5); and (iv) Australian branch containing Mycetohabitans sp. HKI-404 (M8) (Fig. 2A; see also Table S1) (20).
Fig 2.
Identification of an AWR effector associated with the T3SS from endofungal Mycetohabitans species. (A) Map depicting global distribution of R. microsporus strains containing Mycetohabitans symbionts and their classification into four geographical branches. (B) Schematic representation of the T3SS gene cluster (sct) located on plasmid pBR01 that encodes the Type 3 secretion apparatus. The putative Type 3 effector-encoding gene (awr) is indicated in dark purple. (C) Multiple sequence alignment of the AWR peptides from endofungal Mycetohabitans strains, and a related Burkholderia species (CCGE1003). The functional domains of the protein (nuclear localization sequence, NLS) are labeled. (D) Phylogenetic tree of AWR homologs from plant pathogenic Ralstonia solanacearum and plant-associated Burkholderia species. The phylogenetic analyses were performed using MEGA7 (see Materials and Methods for details). Sequences newly identified in this study are highlighted in bold (see Table S3). GenBank accession numbers are given in brackets. (E) Spore count of apo-symbiotic R. microsporus incubated with solvent control (RMapo) or reinfected with M. rhizoxinica wild type (RMapo + M1WT), or AWR-deficient M. rhizoxinica (RMapo + Δawr). Dots represent four independent replicates (n = 4 biological replicates) and grey bars mark ± one standard error of the mean. (F) Representative photographs of fungal cultures depicting the formation of sporangiophores (black mycelium). Apo-symbiotic R. microsporus incubated with solvent control does not sporulate (white mycelium). Strain order as given in panel E. (G) Localization of bacteria (green) inside the fungal hyphae (blue) was confirmed by fluorescence microscopy. Strain order as given in panel E. Scale bars: 10 µm.
Using T3SS prediction tools (see Materials and Methods for details) (21, 22), we found that each Mycetohabitans genome sequence harbors a single gene encoding an AWR peptide (Fig. 2B and C), a potential T3SS effector. Homologs belonging to the awr gene family, present in the genomes of a number of bacterial pathogens including phytopathogenic Burkholderia and Ralstonia strains, collectively contribute to bacterial virulence (23). For example, deletion of all five awr genes in Ralstonia solanacearum severely impairs its capacity to multiply in its host plant (23, 24).
To confirm the identified sequences as genuine awr orthologs, a phylogeny was inferred from alignment of the predicted endofungal Mycetohabitans AWR peptide sequences. These sequences were aligned with homologous AWR sequences from Gram-negative plant and animal pathogens as well as a number of Burkholderia symbionts (Fig. S1) (23). The tree was rooted in the Xanthomonas (γ-proteobacteria) sequences, which are more distantly related to the Ralstonia and Burkholderia (β-proteobacteria) sequences. According to the inferred phylogeny, Mycetohabitans AWR peptides form a monophyletic group with the nearest relative being a protein of unknown function found in the mammalian pathogen Burkholderia pseudomallei (GenBank accession number: ZP02502372) (25). In addition, all eight endofungal Mycetohabitans AWR peptides possess sequences at their C termini that are consistent with the previously described monopartite nuclear localization sequence (NLS), K(K/R)X(K/R) (Fig. 2C; see also Table S2) (26).
AWR peptide plays no discernible role in R. microsporus sporulation
The association between R. microsporus ATCC62417 and M. rhizoxinica was used as a model system to probe whether the bacterial AWR plays a role in this bacterial–fungal symbiosis. To this end, we performed a targeted gene deletion of the M. rhizoxinica awr gene using a double-crossover strategy ( Fig. S2A and B) (17). Although M. rhizoxinica can be maintained in axenic cultures under laboratory conditions, genetic manipulations are challenging. The long doubling time and aggregation of cells (27) significantly impair the selection process. Despite these hurdles, we succeeded in deleting the awr gene in M. rhizoxinica to generate M. rhizoxinica Δawr::KanR (M. rhizoxinica Δawr; Fig. S2C) by continuous passaging of transformants on double-selection media. The AWR-deficient strain was examined for its ability to promote fungal reproduction using a previously described sporulation bioassay (17). First, R. microsporus is treated with antibiotics to eliminate its natural M. rhizoxinica endosymbionts (13) resulting in an apo-symbiotic (endosymbiont-free) fungal strain that is unable to sporulate (Fig. 1). Apo-symbiotic R. microsporus is then individually co-cultured with the M. rhizoxinica wild type and the M. rhizoxinica mutants of interest. An uninoculated medium control is also included. If spore formation occurs after 4–7 days, it indicates successful establishment of the symbiosis (13). Subsequently, sporulation efficiency can be calculated by comparing the sporulating phenotype conferred by the M. rhizoxinica mutants to that conferred by the M. rhizoxinica wild type.
When apo-symbiotic R. microsporus and wild-type M. rhizoxinica are co-cultured, mature sporangiophores can be seen after 4 days. Similar numbers of spores are observed upon co-cultivation with M. rhizoxinica Δawr, i.e., the wild-type phenotype is restored (Fig. 2E and F). In addition, fungal mycelium reinfected with M. rhizoxinica Δawr did not show any growth deficiencies and is characterized by a similar macroscopic phenotype as the wild type (i.e., fluffy mycelium with aerial hyphae) (Fig. 2F). Fluorescence microscopy was used to monitor whether bacteria (stained with SYTO9) are able to recolonize fungal hyphae (stained with calcofluor white). The level of intracellularly located M. rhizoxinica Δawr is comparable to that of wild-type M. rhizoxinica (Fig. 2G), suggesting that AWR is not essential for M. rhizoxinica to enter R. microsporus hyphae and to establish a stable symbiosis. Indeed, we observed no septa formation in R. microsporus reinfected with M. rhizoxinica Δawr (Fig. 2G), as expected for a stable endosymbiosis (19). These results suggest that AWR is not essential for M. rhizoxinica to enter R. microsporus hyphae and to establish a stable symbiosis. However, given that awr genes are universally conserved in the genomes of endofungal Mycetohabitans spp. despite their highly reduced genomes (22, 28), AWR peptides may still play as-yet-unknown physiological roles.
Genes encoding TALs are widespread in endofungal Mycetohabitans symbionts
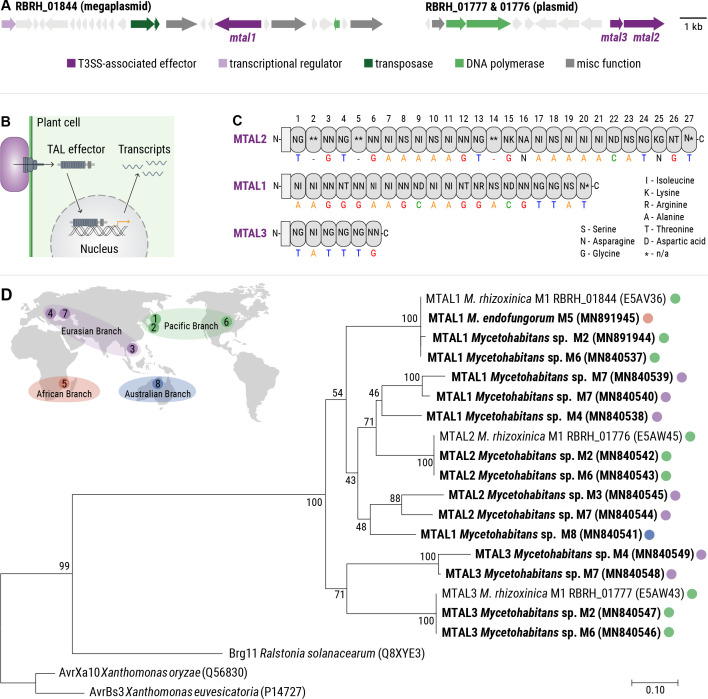
Since AWR deficiency had no discernible effect on fungal reproduction, we continued our search for putative Type 3 effectors in the genomes of the eight endofungal Mycetohabitans species (M1–M8). Genes encoding three putative T3SS effectors, with the locus tags RBRH_01844 (2,316bp), RBRH_01776 (2,994bp), and RBRH_01777 (936bp), were previously identified in the genome of M. rhizoxinica (Fig. 3A) (29–31). Since these genes are highly similar to those encoding TAL effectors, they were named MTAL (for Mycetohabitans transcription activator-like effectors; MTAL1: RBRH_01844; MTAL2: RBRH_01776; and MTAL3: RBRH_01777). TAL effectors are employed by bacterial pathogens such as phytopathogenic Burkholderia, Ralstonia, and Xanthomonas strains to modify the expression of host plant genes, which aids bacterial colonization and virulence (Fig. 3B) (29, 32, 33).
Fig 3.
Identification of predicted MTAL effectors from endofungal Mycetohabitans species. (A) Schematic illustration of the gene clusters encoding MTAL1 (RBRH_01844), MTAL2 (RBRH_01776), and MTAL3 (RBRH_01777) indicated in dark purple. (B) Mode of action of TALs from Xanthomonas sp. TALs are secreted directly into plant cells via the T3SS, translocate to the nucleus, and induce expression of target genes. (C) Schematic representation of the overall domain structure of M. rhizoxinica MTALs and the amino acid tandem repeats (top) that specify the target nucleotide sequence (bottom). Cryptic repeats that have less than 45% amino acid identity with core repeats are marked with an asterisk (*, n/a: not annotated). Modified from Lange et al. (30). (D) Phylogenetic tree of TAL proteins from eight endofungal Mycetohabitans species, and plant pathogenic R. solanacearum and Xanthomonas sp. MTAL sequences identified in this study are highlighted in bold and GenBank accession numbers are given in brackets (Table S3). The distribution of MTALs across the four Mycetohabitans branches is indicated as follows: green, Pacific branch; purple, Eurasian branch; orange, African branch; and blue, Australian branch.
To investigate whether MTALs are a common feature in endofungal Mycetohabitans strains, we surveyed the publicly available genomes for mtal homologs. Using bioinformatic analysis, we discovered that endofungal Mycetohabitans strains commonly encode multiple mtal genes in their genomes (Table S3). Specifically, mtal1 was detected in the genomes of Mycetohabitans M1, M4, M6, M7, and M8, mtal2 in Mycetohabitans M1, M2, M3, M6, and M7, and mtal3 in Mycetohabitans M1, M2, M4, M6, and M7. The highly repetitive nature of mtal genes hinders prediction within the genome of a given organism. We attempted to amplify hidden mtal genes via polymerase chain reaction (PCR) (Table S4) but only obtained mtal1 sequences from Mycetohabitans strains M2 and M5. Details of all mtal genes, whether identified bioinformatically or by PCR amplification and Sanger sequencing, were deposited in GenBank under the accession numbers listed in Table S3.
All members of the TAL effector family, including Xanthomonas and Ralstonia TALs, are characterized by a central protein domain consisting of tandem-arranged repeats. These repeats mediate sequence-specific binding to host DNA, which alters gene expression to benefit the pathogen (Fig. 3C) (34). In addition, Xanthomonas and Ralstonia TAL proteins possess a Type 3 secretion signal, a nuclear localization sequence (NLS), and an activation domain, all of which are required for the transcriptional activation of host plant genes (32). As the short C termini of MTAL proteins do not contain NLSs, we searched for NLS motifs within the entire MTAL protein sequences using the NucPred prediction software (see Materials and Methods for details). Some MTAL1 and MTAL2 proteins are predicted to localize to the nucleus (52%), while the remainder localizes to the cytoplasm (48%). Localization of MTAL3 could not be predicted (Table S5). In addition, we identified the NLS-like sequence “RIRK” in the C termini of all MTAL1 and MTAL2 proteins (Table S5). This NLS-like sequence was previously shown to mediate translocation of MTAL2 from Mycetohabitans sp. B13 (M4) to the nucleus of Saccharomyces cerevisiae (35).
Phylogenetic analysis of predicted MTAL proteins was performed using the central protein domain consisting of tandem-arranged repeats (Fig. 3C) (34). The inferred phylogeny confirmed mtal sequences as encoding TAL orthologs, with R. solanacearum TALs being the closest relatives (Fig. 3D; see also Fig. S3). In addition, detection of mtal1, mtal2, and mtal3 in the genomes of Mycetohabitans species belonging to four geographically distant branches (Fig. 3D) (20) reveals a wide distribution of mtal genes in fungal endosymbionts, thus adding to the number of previously identified MTALs in Mycetohabitans strains (35). Based on the prevalence of mtal genes in the genomes of endofungal Mycetohabitans and their transcriptional upregulation during physical contact of M. rhizoxinica and R. microsporus (18), we deemed MTALs potentially important effectors in the Rhizopus-Mycetohabitans symbiosis.
Sporulation of R. microsporus containing MTAL-deficient M. rhizoxinica is greatly diminished
In order to investigate whether MTALs contribute to the Rhizopus-Mycetohabitans symbiosis, we performed targeted gene deletions using a double-crossover strategy (Fig. S2A and B) (17). The mtal genes were individually deleted in M. rhizoxinica to generate M. rhizoxinica Δmtal2::KanR (M. rhizoxinica Δmtal2) and M. rhizoxinica Δmtal3::KanR (M. rhizoxinica Δmtal3; Fig. S2D and E). The mtal1 gene was previously deleted in M. rhizoxinica, yielding M. rhizoxinica Δmtal1::ApraR (M. rhizoxinica Δmtal1) (19).
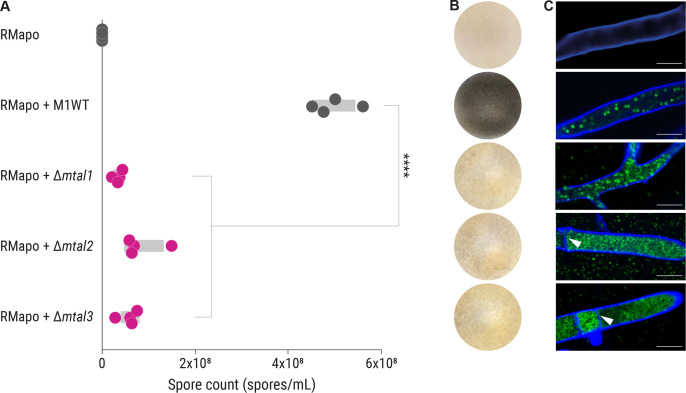
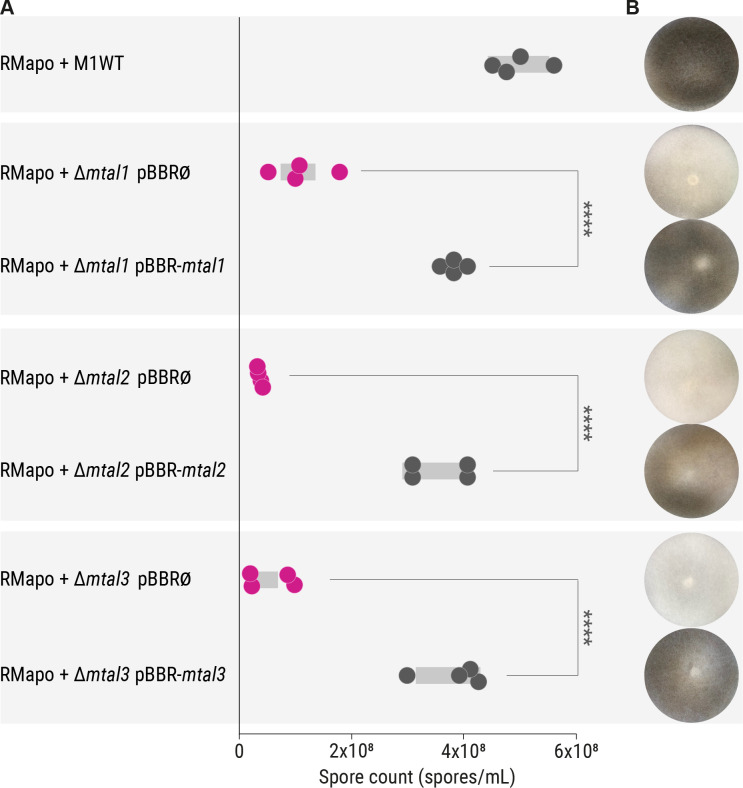
We probed whether MTAL-deficient M. rhizoxinica strains have an effect on host reproduction using the sporulation bioassay (17). Apo-symbiotic R. microsporus does not produce sporangiophores (Fig. 4A and B). When wild-type M. rhizoxinica and apo-symbiotic R. microsporus are co-cultured, sporulation can be seen after 4 days indicating the successful establishment of the symbiosis (Fig. 4A and B). In contrast, in the case of co-cultivation with M. rhizoxinica Δmtal1, M. rhizoxinica Δmtal2, and M. rhizoxinica Δmtal3, only a limited number of mature sporangia are formed (Fig. 4A and B). Indeed, the sporulation efficiency is significantly reduced (P < 0.001) when compared to wild-type M. rhizoxinica and M. rhizoxinica Δawr (Fig. 4A; see also Table S6).
Fig 4.
Sporulation ability of R. microsporus containing MTAL-deficient M. rhizoxinica is reduced. (A) Spore count of apo-symbiotic R. microsporus incubated with solvent control (RMapo) or reinfected with M. rhizoxinica wild type (RMapo + M1WT) or MTAL-deficient M. rhizoxinica (RMapo + Δmtal1, RMapo + Δmtal2, and RMapo + Δmtal3). Dots represent four independent replicates (n = 4 biological replicates) and grey bars mark ± one standard error of the mean. One-way analysis of variance with Tukey’s multiple comparison test (****P < 0.0001; Table S6). (B) Representative photographs of fungal cultures. Apo-symbiotic R. microsporus reinfected with M. rhizoxinica wild type sporulates after 4 days of incubation (black mycelium). The apo-symbiotic fungus shows no sporulation. Strain order as given in panel A. (C) Localization of bacteria (green) inside the fungal hyphae (blue) was confirmed by fluorescence microscopy. Strain order as given in panel A. Scale bars: 10 µm. White arrowheads point to septa. Data points, sporulation image, and fluorescence microscopic image of RMapo and RMapo + M1WT are the same as depicted in Fig. 2E through G, respectively.
We considered the possibility that a lack of sporulation could be due to the inability of the MTAL-deficient M. rhizoxinica to recolonize the fungal host, as was reported for strains lacking a functional T2SS and T3SS (16, 17). We therefore used fluorescence microscopy to monitor bacteria (stained with SYTO9) inside the fungal hyphae (stained with calcofluor white) following reinfection. We noted that M. rhizoxinica Δmtal1, M. rhizoxinica Δmtal2, and M. rhizoxinica Δmtal3 successfully recolonize apo-symbiotic R. microsporus (Fig. 4C). In addition, MTAL-deficient M. rhizoxinica reaches higher cell densities within the host cytosol compared to wild-type M. rhizoxinica and M. rhizoxinica Δawr (Fig. 2G and 4C). In addition, we observed formation of septa in hyphae containing M. rhizoxinica Δmtal2 and M. rhizoxinica Δmtal3, as was reported for R. microsporus hyphae containing M. rhizoxinica Δmtal1 (19).
In order to confirm that the inability of the MTAL-deficient M. rhizoxinica strains to restore fungal sporulation is solely due to disruption of the mtal genes, we performed an in vivo trans-complementation experiment. We constructed the expression vectors, pBBR_Ps12_mtal2 and pBBR_Ps12_mtal3, in which the mtal gene is under the control of a constitutive promoter. We transformed M. rhizoxinica Δmtal2 and M. rhizoxinica Δmtal3 with pBBR_Ps12_mtal2 and pBBR_Ps12_mtal3, respectively (19), yielding M. rhizoxinica Δmtal2 pBBR-mtal2 and M. rhizoxinica Δmtal3 pBBR-mtal3 (complemented M. rhizoxinica mtal mutants; Fig. S2F). M. rhizoxinica Δmtal2 and M. rhizoxinica Δmtal3 were also transformed with the empty vector pBBR_Ps12 to generate the control strains M. rhizoxinica Δmtal2 pBBR∅ and M. rhizoxinica Δmtal3 pBBR∅ (empty vector controls; Fig. S2F). M. rhizoxinica Δmtal1 pBBR-mtal1 (complemented M. rhizoxinica mtal1 mutant) and M. rhizoxinica Δmtal1 pBBR∅ (empty vector control) were generated previously (19).
The ability of the complemented M. rhizoxinica mtal mutants to restore sporulation in the fungal host was assessed in the sporulation bioassay. Reinfection of apo-symbiotic R. microsporus with each of the three empty vector control strains (M. rhizoxinica Δmtal1 pBBR∅, M. rhizoxinica Δmtal2 pBBR∅, and M. rhizoxinica Δmtal3 pBBR∅) does not restore sporulation of the fungal host, whereas the complemented M. rhizoxinica mtal mutants (M. rhizoxinica Δmtal1 pBBR-mtal1, M. rhizoxinica Δmtal2 pBBR-mtal2, and M. rhizoxinica Δmtal3 pBBR-mtal3) readily trigger sporulation, in all cases restoring the wild-type phenotype (Fig. 5A and B). The sporulation efficiency is significantly increased in the complemented M. rhizoxinica mtal mutants (P < 0.001) when compared to the appropriate MTAL-deficient M. rhizoxinica strains (Fig. 5A; see also Table S7). The impaired ability of the MTAL-deficient strains to induce sporulation and the phenotypic complementation by trans expression of the relevant mtaI genes supports the proposal that T3SS-associated MTALs are essential effectors in the sporulation process of R. microsporus.
Fig 5.
Sporulation ability of R. microsporus containing complemented M. rhizoxinica mtal mutants is restored. (A) Spore count of apo-symbiotic R. microsporus reinfected with M. rhizoxinica wild type (RMapo + M1WT), M. rhizoxinica mtal mutants containing the relevant empty vector (RMapo + M. rhizoxinica Δmtal1 pBBR∅, RMapo + M. rhizoxinica Δmtal2 pBBR∅, and RMapo + M. rhizoxinica Δmtal3 pBBR∅), or M. rhizoxinica mtal mutants constitutively expressing a plasmid-borne copy of the relevant mtal gene (RMapo + M. rhizoxinica Δmtal1 pBBR-mtal1, RMapo + M. rhizoxinica Δmtal2 pBBR-mtal2, and RMapo + M. rhizoxinica Δmtal3 pBBR-mtal3). Dots represent four independent replicates (n = 4 biological replicates) and grey bars mark ± one standard error of the mean. One-way analysis of variance with Tukey’s multiple comparison test (****P < 0.0001; see Table S7). (B) Representative photographs of apo-symbiotic R. microsporus reinfected with M. rhizoxinica strains after 1 week of co-cultivation. Strain order as given in panel A. Data points and sporulation image of RMapo + M1WT are the same as depicted in Fig. 2E and F , respectively.
DISCUSSION
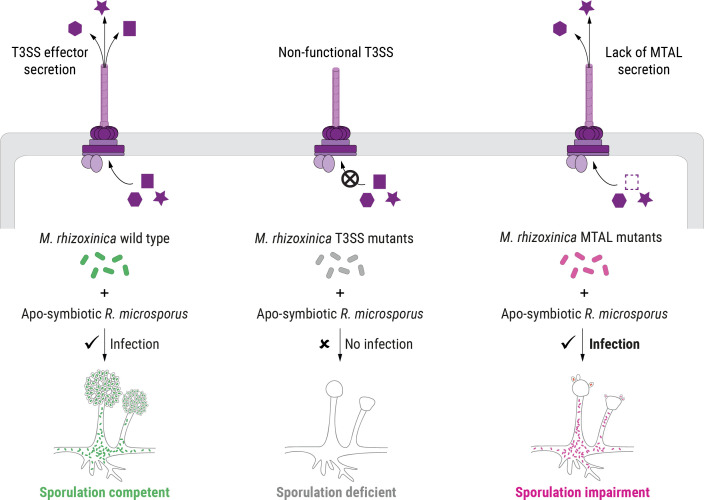
Bacteria that live in close association with eukaryotes can control their host by employing specialized effectors that are released through dedicated secretion systems (36). Among them, effector proteins delivered through the T3SS can be essential to host cell entry of Gram-negative bacterial pathogens, mutualists and, in rare cases, endosymbionts (17, 37, 38). TAL effectors are a prominent example of T3SS-associated proteins that allow plant-pathogenic bacteria to enter and control their hosts (8, 39, 40). Over the last two decades, TAL effectors have been engineered for application as genome editing tools due to their programmable DNA-binding properties (41). This has reinvigorated interest in TAL effectors leading to the discovery of TAL-related proteins in a wide range of bacterial species, including M. rhizoxinica (MTAL1, MTAL2, and MTAL3) (31). Here, we used a combination of genomic and functional studies to characterize T3SS-associated proteins in the fungal endosymbiont M. rhizoxinica. We found that fungal sporulation is independent of the T3SS-associated AWR protein, whereas MTAL proteins are instrumental in controlling host reproduction (Fig. 6).
Fig 6.
Schematic model of host control by endofungal bacteria. The recolonization of apo-symbiotic R. microsporus by the isolated wild-type endosymbiont, M. rhizoxinica, restores the host’s ability to form sporangiospores (left panel). M. rhizoxinica strains lacking a functional T3SS are unable to infect fungal hyphae and, as a consequence, fail to cause sporulation (middle panel). M. rhizoxinica that lack MTALs successfully reinfect the fungal host; however, in the absence of the T3SS-associated MTALs, the sporulating phenotype indicative of a stable symbiosis is profoundly impaired (right panel).
By means of a sporulation bioassay, we show that each individual M. rhizoxinica mtal mutant (M. rhizoxinica Δmtal1, M. rhizoxinica Δmtal2, and M. rhizoxinica Δmtal3) is unable to completely restore fungal sporulation, suggesting that MTALs are not functionally redundant in regulating sporulation. It is conceivable that distinct MTAL functions could arise from the varying number of the tandem-arranged repeats, which mediate sequence-specific DNA binding (34). In fact, MTAL proteins contain so-called core repeats (>45% identity to each other), which form the central section of the proteins with cryptic repeats (<45% identity to each other) at both the C-terminal and N-terminal ends (30). The number of these core repeats is highly variable between individual MTALs (M. rhizoxinica MTAL1: 20 repeats; M. rhizoxinica MTAL2: 27 repeats; and M. rhizoxinica MTAL3: 6 repeats; Fig. 2D) (30). Within each conserved repeat, only amino acids at positions 12 and 13 are highly variable, which are termed repeat variable di-residue (RVD). The RVD of each repeat determines the DNA base to which each repeat binds, referred to as the canonical TAL code (30, 39). The hypothesis that individual M. rhizoxinica MTAL proteins may have distinct functions in controlling host sporulation is supported by the two observations: (i) slight changes in the RVD sequence or in the number of repeats can change the DNA-binding specificity (30), and (ii) the RVD repeats of MTAL proteins are less conserved compared to Xanthomonas and Ralstonia TALs (29, 42). This functional diversity of individual MTALs is exemplified by MTAL2 from Mycetohabitans strain B13, consisting of 19 repeats, which is not involved in controlling fungal sporulation but instead contributes to the ability of R. microsporus to tolerate cell membrane stress (35). In addition, the reduced ability to tolerate membrane stress in R. microsporus containing MTAL2-deficient Mycetohabitans strain B13 could not be rescued by expressing MTAL2 from Mycetohabitans strain B14 (35), suggesting that MTAL proteins may target host genes with different functions.
In an effort to gain a glimpse at the possible means by which MTALs modulate host sporulation, we performed a bioinformatic search for possible target sequences in the R. microsporus genome. The analysis returned only partial matches for MTAL1 and MTAL2, and none for MTAL3. While the majority of matching sequences fall into the 5′ untranslated region of genes coding for DNA-associated proteins or within assembly gaps, the predicted MTAL1 target is located within the coding sequence of a membrane-bound transporter. MTAL2 was predicted to bind to DNA in the vicinity of genes coding for a histone binding protein (RBBP4), a fungal transcription activator, and a molecular chaperon protein (DnaJ). While these preliminary findings suggest that MTAL2 may control host sporulation through epigenetic regulation, experimental studies are needed to provide evidence of promoter binding.
We discovered a role for MTAL3 in controlling fungal reproduction. This is somewhat surprising as MTAL3 is predicted to contain only six tandem-arranged repeats and, consequently, is unable to bind DNA efficiently (30). However, it was previously reported that TAL effectors from various Xanthomonas strains can act through protein-protein interactions instead of binding to DNA (43, 44). For example, a class of structurally degenerate TAL effector-like proteins (TruncTALs) from rice pathogenic X. oryzae strains suppress certain plant disease resistance genes despite the inability to bind to DNA (43). In fact, some X. oryzae TAL effectors consisting of only 3.5 tandem-arranged repeats are able to mediate control of their host independent of a specific RVD sequence (45). Thus, it is conceivable that MTAL3 might function in a DNA-independent manner in the Rhizopus-Mycetohabitans symbiosis.
M. rhizoxinica that lack a functional T3SS are incapable of triggering visible spore formation, presumably since they are unable to consistently reinfect the apo-symbiotic host (17). Using fluorescence microscopy, we show that MTAL-deficient M. rhizoxinica strains are able to reinfect apo-symbiotic R. microsporus efficiently, yet their ability to induce fungal sporulation is profoundly impaired. Furthermore, the levels of apo-symbiotic R. microsporus reinfection by wild-type M. rhizoxinica are comparable. It follows that MTALs are T3SS-associated effectors that are not responsible for the absence of colonization by T3SS-deficient M. rhizoxinica. This is in stark contrast to Xanthomonas and Ralstonia TALs, which induce the expression of host plant genes that aid bacterial colonization and virulence (46). The nature of the T3 effectors responsible for establishment of the Rhizopus-Mycetohabitans symbiosis before and during invasion remains to be discovered. A recent transcriptomics study revealed upregulation of nearly 60 T3SS-associated effector genes in M. rhizoxinica upon initial physical contact with R. microsporus (18). Two candidate effectors were identified that may play a role during infection because they contain F-box-like and leucine-rich repeats (18), both features of known virulence effectors in pathogenic bacteria (47, 48).
Notably, this study provides the first functional characterization of T3SS-associated effector(s) in the maintenance of the Rhizopus-Mycetohabitans symbiosis. We demonstrate that MTAL proteins are crucial symbiosis factors that control fungal reproduction after colonization of the host. Reflecting their fundamental role in the maintenance of a stable Rhizopus-Mycetohabitans symbiosis, we show that mtal genes are prevalent in the genomes of eight endosymbiotic Mycetohabitans strains that were isolated from globally distributed R. microsporus strains inhabiting diverse habitats (20). Indeed, despite their heavily reduced genomes (22, 28), every endofungal Mycetohabitans strain analyzed so far contains at least one mtal gene (35). In addition to impaired fungal sporulation, we observed physiological changes in R. microsporus containing MTAL2- and MTAL3-deficient M. rhizoxinica. Fluorescence microscopy revealed septa formation by R. microsporus, an unusual phenomenon previously reported in R. microsporus reinfected with MTAL1-deficient M. rhizoxinica (19). Based on high-resolution live imaging, it was reported that absence of MTAL1 induces biogenesis of septa in R. microsporus leading to hyphal trapping of endobacteria and subsequent death of MTAL1-deficient M. rhizoxinica (19). It follows that the impaired sporulation of R. microsporus containing MTAL-deficient M. rhizoxinica may be an indirect consequence of the protective host response. The vital role of MTALs in the Rhizopus-Mycetohabitans endosymbiotic relationship is reinforced when one considers that the persistence of the symbiosis relies on spores containing healthy endobacteria (13).
In summary, we show that M. rhizoxinica MTALs do not promote colonization of R. microsporus but are essential factors in fungal sporulation (Fig. 6), representing an unprecedented case of bacterially produced T3SS effectors controlling host reproduction. The revelation that endobacteria affect the physiology of a fungal host by way of T3SS effectors offers a deeper insight into the dynamic interactions between bacteria and fungi. Our results illuminate a possible research avenue into the development of secretion system inhibitors to impede the Rhizopus-Mycetohabitans pathogenic alliance, which could potentially alleviate the economic damage caused by rice seedling blight.
MATERIALS AND METHODS
Strains and growth conditions
Eight R. microsporus strains harboring Mycetohabitans sp. endobacteria were used in this study (Table S1) (20). Endobacteria from R. microsporus ATCC62417 were eliminated by continuous antibiotic treatment (49), and the endosymbiont-free fungal strain was named apo-symbiotic R. microsporus (ATCC62417/S). Absence of endobacteria was confirmed by fluorescence microscopy and an absence of rhizoxin in extracts of the fungal mycelium (14). Both R. microsporus strains (ATCC62417 and ATCC62417/S) were cultivated on potato dextrose agar (PDA; Becton, Dickinson & Company, Sparks, MD, USA) at 30°C. Bacterial endosymbionts (M1-M8) were isolated from the mycelium of eight fungal strains as previously reported (50). Pure cultures of M. rhizoxinica were grown at 30°C in MGY M9 medium (10g/L glycerol, 1.25 g/L yeast extract, M9 salts: 40 mM K2HPO4, 14 mM KH2PO4, 2.2 mM C6H7NaO7, 7.5 mM (NH4)2SO4, and 0.8 mM Mg2SO4) or Standard Nutrient Agar I (Merck, Darmstadt, Germany) supplemented with 1% glycerol.
In silico predictions and characterization of Type 3 effectors
Potential Type 3-secreted effector proteins were predicted using the T3SS PREDICTION server (51) and the EFFECTIVE Type 3 prediction tool (52). Nuclear localization sequences (NLSs) were predicted using the cNLS domain prediction tool (53) and NucPred (54). The nucleotide sequences of potential Mycetohabitans Type 3 effector genes have been deposited in GenBank under the accession numbers provided in Table S3.
Amplification and Sanger sequencing of Mycetohabitans sp. mtal genes
Genomic DNA was isolated from eight axenic Mycetohabitans sp. cultures (Table S1) and quantified using a NanoDrop (Thermo Fisher Scientific, Waltham, MA, USA). PCR primers were designed to amplify partial coding sequences of mtal1 (GenBank accession number: RBRH_01844; Table S4). PCRs were performed in 25.0 µL volumes containing 12.5 µL of high-fidelity Taq DNA polymerase (Phusion Master Mix, New England Biolabs, Ipswich, MA, USA), forward and reverse primers (both 0.4 µM), and 100 ng of template gDNA. The following thermocycling conditions were used for amplification: 98°C/30 s, 1 cycle; 98°C/10 s, 65°C/30 s, 72°C/3 min, 30 cycles; 72°C/7 min, 1 cycle; 16°C/hold.
The PCR products were visualized on a 1.5% agarose gel stained with ethidium bromide before gel extraction (Zymoclean Gel DNA Recovery Kit, Zymo Research, Irvine, CA, USA). The purified amplicons were ligated into pCR-Blunt II-TOPO (Invitrogen, Carlsbad, CA, USA), followed by transformation into chemically competent Escherichia coli TOP10 cells (Invitrogen, One Shot). The plasmids were purified (Monarch Plasmid Miniprep Kit, New England Biolabs), and plasmid inserts were bi-directionally sequenced by an external contractor (Eurofins Genomics, Ebersberg, Germany). Sequences were deposited in GenBank under the accession numbers provided in Table S3.
Phylogenetic analysis
For phylogenetic analysis, predicted T3 effector protein sequences were aligned using ClustalW (55). Alignments were generated using a gap open penalty of 10 and a gap extension penalty of 0.1 as implemented in the MEGA7 package (Molecular Evolutionary Genetics Analysis software, version 5.0) (56). All positions containing gaps and missing data were eliminated. The evolutionary history was inferred using the Neighbor-Joining method with maximum composite likelihood distances and 10,000 bootstrap repetitions (57, 58). The alignments of sequences used in this study are shown in Fig. S1 and S3.
Generation of M. rhizoxinica mtal mutant strains
To investigate the role of both AWR and MTAL proteins in the symbiosis, three genes (awr: RBRH_03012; mtal2: RBRH_01776; and mtal3: RBRH_01777) were deleted using a double crossover strategy as previously described (17). The gene mtal1 (RBRH_01844) was previously deleted in M. rhizoxinica, yielding M. rhizoxinica Δmtal1::ApraR (M. rhizoxinica Δmtal1) (19).
Using a proofreading polymerase, the upstream and downstream regions of the genes of interest were amplified. Primers were designed to contain 20 bp overlap with the gene of interest as well as a 20 bp overlap with an antibiotic resistance cassette (kanamycin). The kanamycin cassette was amplified from pK19, using primers carrying the same 20 bp overlaps (Fig. S2B).
The gene disruption vector pGL42a was used to generate M. rhizoxinica Δawr::KanR, Δmtal2::KanR, and Δmtal3::KanR mutants. pGL42a was double-digested with the restriction enzymes SpeI and KpnI (New England Biolabs). The linear vector was gel-purified (Monarch DNA Gel Extraction Kit, New England Biolabs) and quantified on a NanoDrop (Thermo Fisher Scientific).
For each target gene, equimolar amounts of the three relevant PCR products were mixed with linear pGL42a in 2× Master Mix (NEBuilder HiFi DNA Assembly Cloning Kit, New England Biolabs) and incubated at 60°C for 1h following the manufacturer’s recommendations. The new plasmids pZU52, pZU21, and pZU19 (targeting awr, mtal2, and mtal3 for disruption, respectively) were introduced into E. coli by chemical transformation. Transformants were selected on Standard Nutrient Agar I supplemented with 50 µg/mL kanamycin.
Competent M. rhizoxinica was transformed with vectors pZU52, pZU21, or pZU19 via electroporation (17). Transformants were grown on Standard Nutrient Agar I containing 50 µg/mL kanamycin. Colonies were subsequently passaged onto agar plates containing double selection medium (17) until the correct gene disruption vectors were observed using colony PCR. Colony PCRs were carried out in 12.0µL volumes containing 5µL of high-fidelity OneTaq Quick-Load 2× Master Mix (New England Biolabs), appropriate forward and reverse primers (both 0.4 µM; Table S4), and 5µL colony suspension. The following thermocycling conditions were used for amplification: 96°C/3min, 1 cycle; 96°C/10s, 58°C/15s, 68°C/1min, 30 cycles; 68°C/5min, 1 cycle; 16°C/hold. The resulting PCR products were visualized on an agarose gel. Primers were designed to span the two recombination sites, yielding amplicons A and B in mutant strains and amplicons C and D in M. rhizoxinica wild-type strains (Fig. S2A and B).
Generation of genetically complemented M. rhizoxinica Δmtal1 strains
In order to genetically complement the MTAL-deficient strains M. rhizoxinica ∆mtal2 and M. rhizoxinica ∆mtal3, genomic DNA from M. rhizoxinica was isolated using the MasterPure DNA Purification Kit (Biozym Scientific, Hessisch Oldendorf, Germany) following the manufacturer’s recommendations. The mtal2 and mtal3 genes were amplified by PCR with the primer pairs listed in Table S4 using Phusion High-Fidelity PCR Master Mix with HF Buffer (New England Biolabs). The PCR products were gel-purified with the Monarch DNA Gel Extraction Kit (New England Biolabs). The purified amplicons were cloned into the NdeI/AflII restricted pBBR_Ps12_gfp downstream of the constitutive promoter Ps12 with the 2× Master Mix (NEBuilder HiFi DNA Assembly Cloning Kit, New England Biolabs), yielding pBBR-mtal2 and pBBR-mtal3. The reaction mixture was subsequently used to transform chemically competent E. coli TOP10 cells (Invitrogen, One Shot).
To generate an empty vector control, pBBR_Ps12_gfp was digested with the restriction enzyme BstBI. The resulting linear vector lacking gfp was self-circularized using T4 DNA Ligase (New England Biolabs) to yield pBBR∅ and used to transform chemically competent E. coli TOP10 cells (Invitrogen, One Shot). All plasmids were purified from E. coli TOP10 overnight cultures using the Monarch Plasmid Miniprep Kit (New England Biolabs) and verified by restriction digest and Sanger sequencing using the primers cmr_seq_fw and BBR_seq_rv (Table S4).
The new plasmids (pBBR-mtal2, pBBR-mtal3, or pBBR∅) were introduced into competent M. rhizoxinica ∆mtal2 or M. rhizoxinica ∆mtal3 cells as appropriate via electroporation. Transformants were grown on Standard Nutrient Agar I containing 50µg/mL chloramphenicol and 50 µg/mL kanamycin. Colonies containing the respective plasmids were verified using colony PCR (Fig. S2D and E; see also Table S4). The complemented M. rhizoxinica Δmtal1 mutant (M. rhizoxinica Δmtal1 pBBR-mtal1) and M. rhizoxinica Δmtal1 containing the relevant empty vector (M. rhizoxinica Δmtal1 pBBR∅) were generated previously (19).
Sporulation bioassay
In a liquid sporulation bioassay, apo-symbiotic R. microsporus aerial hypha (∼0.1 cm3) was grown in 24-well plates containing 750 µL Vorkultur medium (5 g/L glycerol, 10 g/L yeast extract, 10 g/L corn starch, 10 g/L corn step solids, 10 g/L CaCO3; pH 6.5). After 15 h of incubation, 100 µL of overnight cultures of M. rhizoxinica wild type (M1WT), M. rhizoxinica mtal mutants (M. rhizoxinica Δmtal1, M. rhizoxinica Δmtal2, and M. rhizoxinica Δmtal3), M. rhizoxinica mtal mutants containing the relevant empty vector (M. rhizoxinica Δmtal1 pBBR∅, M. rhizoxinica Δmtal2 pBBR∅, and M. rhizoxinica Δmtal3 pBBR∅), or M. rhizoxinica mtal mutants expressing a plasmid-borne copy of the relevant mtal gene (M. rhizoxinica Δmtal1 pBBR-mtal1, M. rhizoxinica Δmtal2 pBBR-mtal2, and M. rhizoxinica Δmtal3 pBBR-mtal3) were added to individual wells. Co-culture plates were incubated at 30°C for 5–14 days. Fungal mycelium was transferred from co-culture plates to PDA petri dishes, which were incubated at 30°C for 5 days. Spores were harvested from PDA plate using 10 mL NaCl (0.15 M) and counted using a Thoma Chamber.
Experiments were performed four times independently (n = 4 biological replicates) with six technical replicates on each plate. Data are presented as means with grey bars marking ±1 standard error of the mean. Raw data from sporulation experiments were processed with MS Excel. GraphPad Prism 5.03 (GraphPad Software, La Jolla, CA, USA; https://www.graphpad.com/) was used for statistical analysis and graphing. Data from spore counts were compared between M. rhizoxinica strains using one-way analysis of variance and Tukey HSD test function in GraphPad. P values < 0.05 were considered statistically significant. The Brown-Forsythe test was used to test for equal variance and a P < 0.05 was considered significant.
Fluorescence microscopy of co-cultures
One-week-old fungal-bacterial co-cultures were used to visualize the localization of the M. rhizoxinica strains. The bacterial cells were stained with 5 µM Syto 9 (Invitrogen), and fungal cells were counter-stained with 2 µg/mL calcofluor white (Fluka, Germany) for 5–10 min. Fluorescence microscopy was carried out using a Zeiss LSM 710 confocal laser-scanning microscope (Zeiss, Oberkochen, Germany), and images were captured using the Zeiss-Zen software.
ACKNOWLEDGMENTS
I.R. is grateful for financial support from the European Union’s Horizon 2020 Research and Innovation Program under the Marie Skłodowska-Curie grant agreement no. 794343. Financial support by the Deutsche Forschungsgemeinschaft (DFG, German Research Foundation) under Germany’s Excellence Strategy—EXC 2051 (Cluster of Excellence "Balance of the Microverse") Project-ID 390713860 and the SFB 1127 ChemBioSys, Project-ID 239748522, and the Leibniz Award (to C.H.) and the JSMC to Z.U. is gratefully acknowledged.
I.R. conceived the idea, developed the study design, interpreted the data, and wrote the manuscript. Z.U. conceived the idea, generated M. rhizoxinica mtal mutants, and performed fluorescence microscopy. P.W. designed plasmids and generated complemented strains. E.M.M. assisted in data interpretation and manuscript revision. N.M. conceived the idea. S.J.P. performed bioinformatic analysis and revised the manuscript. T.P.S. assisted in data interpretation and manuscript revision. C.H. conceived the idea, drafted, and revised the manuscript.
Contributor Information
Christian Hertweck, Email: christian.hertweck@leibniz-hki.de.
Reinhard Fischer, Karlsruhe Institute of Technology, Karlsruhe, Germany.
DATA AVAILABILITY
All data generated or analyzed during this study are included in the article and in the supporting files.
SUPPLEMENTAL MATERIAL
The following material is available online at https://doi.org/10.1128/mbio.01824-23.
Supplemental figures and tables.
ASM does not own the copyrights to Supplemental Material that may be linked to, or accessed through, an article. The authors have granted ASM a non-exclusive, world-wide license to publish the Supplemental Material files. Please contact the corresponding author directly for reuse.
REFERENCES
- 1. Dale C, Moran NA. 2006. Molecular interactions between bacterial symbionts and their hosts. Cell 126:453–465. doi: 10.1016/j.cell.2006.07.014 [DOI] [PubMed] [Google Scholar]
- 2. Costa TRD, Felisberto-Rodrigues C, Meir A, Prevost MS, Redzej A, Trokter M, Waksman G. 2015. Secretion systems in Gram-negative bacteria: structural and mechanistic insights. Nat Rev Microbiol 13:343–359. doi: 10.1038/nrmicro3456 [DOI] [PubMed] [Google Scholar]
- 3. Phan TH, Houben ENG. 2018. Bacterial secretion chaperones: the mycobacterial type VII case. FEMS Microbiol Lett 365:fny197. doi: 10.1093/femsle/fny197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Lasica AM, Ksiazek M, Madej M, Potempa J. 2017. The type IX secretion system (T9SS): highlights and recent insights into its structure and function. Front Cell Infect Microbiol 7:215. doi: 10.3389/fcimb.2017.00215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Dean P. 2011. Functional domains and motifs of bacterial type III effector proteins and their roles in infection. FEMS Microbiol Rev 35:1100–1125. doi: 10.1111/j.1574-6976.2011.00271.x [DOI] [PubMed] [Google Scholar]
- 6. Ribot WJ, Ulrich RL. 2006. The animal pathogen-like type III secretion system is required for the intracellular survival of Burkholderia mallei within J774.2 macrophages. Infect Immun 74:4349–4353. doi: 10.1128/IAI.01939-05 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Moscou MJ, Bogdanove AJ. 2009. A simple cipher governs DNA recognition by TAL effectors. Science 326:1501. doi: 10.1126/science.1178817 [DOI] [PubMed] [Google Scholar]
- 8. de Lange O, Schreiber T, Schandry N, Radeck J, Braun KH, Koszinowski J, Heuer H, Strauß A, Lahaye T. 2013. Breaking the DNA-binding code of Ralstonia solanacearum TAL effectors provides new possibilities to generate plant resistance genes against bacterial wilt disease. New Phytol 199:773–786. doi: 10.1111/nph.12324 [DOI] [PubMed] [Google Scholar]
- 9. Verdier V, Triplett LR, Hummel AW, Corral R, Cernadas RA, Schmidt CL, Bogdanove AJ, Leach JE. 2012. Transcription activator-like (TAL) effectors targeting OsSWEET genes enhance virulence on diverse rice (Oryza sativa) varieties when expressed individually in a TAL effector-deficient strain of Xanthomonas oryzae. New Phytol 196:1197–1207. doi: 10.1111/j.1469-8137.2012.04367.x [DOI] [PubMed] [Google Scholar]
- 10. Wein P, Dornblut K, Herkersdorf S, Krüger T, Molloy EM, Brakhage AA, Hoffmeister D, Hertweck C. 2023. Bacterial secretion systems contribute to rapid tissue decay in button mushroom soft rot disease. mBio 14:e0078723. doi: 10.1128/mbio.00787-23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Partida-Martinez LP, Hertweck C. 2005. Pathogenic fungus harbours endosymbiotic bacteria for toxin production. Nature 437:884–888. doi: 10.1038/nature03997 [DOI] [PubMed] [Google Scholar]
- 12. Estrada-de Los Santos P, Palmer M, Chávez-Ramírez B, Beukes C, Steenkamp ET, Briscoe L, Khan N, Maluk M, Lafos M, Humm E, Arrabit M, Crook M, Gross E, Simon MF, Dos Reis Junior FB, Whitman WB, Shapiro N, Poole PS, Hirsch AM, Venter SN, James EK. 2018. Whole genome analyses suggests that Burkholderia sensu lato contains two additional novel genera (Mycetohabitans gen. nov., and Trinickia gen. nov.): implications for the evolution of diazotrophy and nodulation in the Burkholderiaceae. Genes (Basel) 9:389. doi: 10.3390/genes9080389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Partida-Martinez LP, Monajembashi S, Greulich KO, Hertweck C. 2007. Endosymbiont-dependent host reproduction maintains bacterial-fungal mutualism. Curr Biol 17:773–777. doi: 10.1016/j.cub.2007.03.039 [DOI] [PubMed] [Google Scholar]
- 14. Scherlach K, Partida-Martinez LP, Dahse HM, Hertweck C. 2006. Antimitotic rhizoxin derivatives from a cultured bacterial endosymbiont of the rice pathogenic fungus Rhizopus microsporus. J Am Chem Soc 128:11529–11536. doi: 10.1021/ja062953o [DOI] [PubMed] [Google Scholar]
- 15. Richter I, Radosa S, Cseresnyés Z, Ferling I, Büttner H, Niehs SP, Gerst R, Scherlach K, Figge MT, Hillmann F, Hertweck C. 2022. Toxin-producing endosymbionts shield pathogenic fungus against micropredators. mBio 13:e0144022. doi: 10.1128/mbio.01440-22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Moebius N, Üzüm Z, Dijksterhuis J, Lackner G, Hertweck C. 2014. Active invasion of bacteria into living fungal cells. Elife 3:e03007. doi: 10.7554/eLife.03007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Lackner G, Moebius N, Hertweck C. 2011. Endofungal bacterium controls its host by an hrp type III secretion system. ISME J 5:252–261. doi: 10.1038/ismej.2010.126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Lastovetsky OA, Krasnovsky LD, Qin X, Gaspar ML, Gryganskyi AP, Huntemann M, Clum A, Pillay M, Palaniappan K, Varghese N, Mikhailova N, Stamatis D, Reddy TBK, Daum C, Shapiro N, Ivanova N, Kyrpides N, Woyke T, Pawlowska TE. 2020. Molecular dialogues between early divergent fungi and bacteria in an antagonism versus a mutualism. mBio 11:e02088-20. doi: 10.1128/mBio.02088-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Richter I, Wein P, Uzum Z, Stanley CE, Krabbe J, Molloy EM, Moebius N, Ferling I, Hillmann F, Hertweck C. 2023. Transcription activator-like effector protects bacterial endosymbionts from entrapment within fungal hyphae. Curr Biol 33:2646–2656. doi: 10.1016/j.cub.2023.05.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Lackner Gerald, Möbius N, Scherlach K, Partida-Martinez LP, Winkler R, Schmitt I, Hertweck C. 2009. Global distribution and evolution of a toxinogenic Burkholderia-rhizopus symbiosis. Appl Environ Microbiol 75:2982–2986. doi: 10.1128/AEM.01765-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Lackner G, Moebius N, Partida-Martinez L, Hertweck C. 2011. Complete genome sequence of Burkholderia rhizoxinica, an endosymbiont of Rhizopus microsporus. J Bacteriol 193:783–784. doi: 10.1128/JB.01318-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Niehs SP, Scherlach K, Dose B, Uzum Z, Stinear TP, Pidot SJ, Hertweck C. 2022. A highly conserved gene locus in endofungal bacteria codes for the biosynthesis of symbiosis-specific cyclopeptides. PNAS Nexus 1:gac152. doi: 10.1093/pnasnexus/pgac152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Solé M, Popa C, Mith O, Sohn KH, Jones JDG, Deslandes L, Valls M. 2012. The awr gene family encodes a novel class of Ralstonia solanacearum type III effectors displaying virulence and avirulence activities. Mol Plant Microbe Interact 25:941–953. doi: 10.1094/MPMI-12-11-0321 [DOI] [PubMed] [Google Scholar]
- 24. Uematsu T, Yoshimura D, Nishiyama K, Ibaragi T, Fujii H. 1976. Pathogenic bacterium causing seedling rot of rice. Jpn J Phytopathol 42:464–471. doi: 10.3186/jjphytopath.42.464 [DOI] [Google Scholar]
- 25. Van Zandt KE, Greer MT, Gelhaus HC. 2013. Glanders: an overview of infection in humans. Orphanet J Rare Dis 8:131. doi: 10.1186/1750-1172-8-131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Chelsky D, Ralph R, Jonak G. 1989. Sequence requirements for synthetic peptide-mediated translocation to the nucleus. Mol Cell Biol 9:2487–2492. doi: 10.1128/mcb.9.6.2487-2492.1989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Uzum Z, Silipo A, Lackner G, De Felice A, Molinaro A, Hertweck C. 2015. Structure, genetics and function of an exopolysaccharide produced by a bacterium living within fungal hyphae. Chembiochem 16:387–392. doi: 10.1002/cbic.201402488 [DOI] [PubMed] [Google Scholar]
- 28. Lackner G, Moebius N, Partida-Martinez LP, Boland S, Hertweck C. 2011. Evolution of an endofungal lifestyle: deductions from the Burkholderia rhizoxinica genome. BMC Genomics 12:210. doi: 10.1186/1471-2164-12-210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Juillerat A, Bertonati C, Dubois G, Guyot V, Thomas S, Valton J, Beurdeley M, Silva GH, Daboussi F, Duchateau P. 2014. BurrH: a new modular DNA binding protein for genome engineering. Sci Rep 4:3831. doi: 10.1038/srep03831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. de Lange O, Wolf C, Dietze J, Elsaesser J, Morbitzer R, Lahaye T. 2014. Programmable DNA-binding proteins from Burkholderia provide a fresh perspective on the TALE-like repeat domain. Nucleic Acids Res 42:7436–7449. doi: 10.1093/nar/gku329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Stella S, Molina R, Bertonatti C, Juillerrat A, Montoya G. 2014. Expression, purification, crystallization and preliminary X-ray diffraction analysis of the novel modular DNA-binding protein BurrH in its apo form and in complex with its target DNA. Acta Crystallogr F Struct Biol Commun 70:87–91. doi: 10.1107/S2053230X13033037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Boch J, Bonas U. 2010. Xanthomonas AvrBs3 family-type III effectors: discovery and function. Annu Rev Phytopathol 48:419–436. doi: 10.1146/annurev-phyto-080508-081936 [DOI] [PubMed] [Google Scholar]
- 33. Cunnac S, Occhialini A, Barberis P, Boucher C, Genin S. 2004. Inventory and functional analysis of the large Hrp regulon in Ralstonia solanacearum: identification of novel effector proteins translocated to plant host cells through the type III secretion system. Mol Microbiol 53:115–128. doi: 10.1111/j.1365-2958.2004.04118.x [DOI] [PubMed] [Google Scholar]
- 34. Deng D, Yan C, Pan X, Mahfouz M, Wang J, Zhu JK, Shi Y, Yan N. 2012. Structural basis for sequence-specific recognition of DNA by TAL effectors. Science 335:720–723. doi: 10.1126/science.1215670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Carter ME, Carpenter SCD, Dubrow ZE, Sabol MR, Rinaldi FC, Lastovetsky OA, Mondo SJ, Pawlowska TE, Bogdanove AJ. 2020. A TAL effector-like protein of an endofungal bacterium increases the stress tolerance and alters the transcriptome of the host. Proc Natl Acad Sci U S A 117:17122–17129. doi: 10.1073/pnas.2003857117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Rapisarda C, Fronzes R. 2018. Secretion systems used by bacteria to subvert host functions. Curr Issues Mol Biol 25:1–42. doi: 10.21775/cimb.025.001 [DOI] [PubMed] [Google Scholar]
- 37. Galán JE, Collmer A. 1999. Type III secretion machines: bacterial devices for protein delivery into host cells. Science 284:1322–1328. doi: 10.1126/science.284.5418.1322 [DOI] [PubMed] [Google Scholar]
- 38. Dale C, Plague GR, Wang B, Ochman H, Moran NA. 2002. Type III secretion systems and the evolution of mutualistic endosymbiosis. Proc Natl Acad Sci U S A 99:12397–12402. doi: 10.1073/pnas.182213299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Boch J, Scholze H, Schornack S, Landgraf A, Hahn S, Kay S, Lahaye T, Nickstadt A, Bonas U. 2009. Breaking the code of DNA binding specificity of TAL-type III effectors. Science 326:1509–1512. doi: 10.1126/science.1178811 [DOI] [PubMed] [Google Scholar]
- 40. Kay S, Bonas U. 2009. How Xanthomonas type III effectors manipulate the host plant. Curr Opin Microbiol 12:37–43. doi: 10.1016/j.mib.2008.12.006 [DOI] [PubMed] [Google Scholar]
- 41. Perez-Quintero AL, Szurek B. 2019. A decade decoded: spies and hackers in the history of TAL effectors research. Annu Rev Phytopathol 57:459–481. doi: 10.1146/annurev-phyto-082718-100026 [DOI] [PubMed] [Google Scholar]
- 42. Stella S, Molina R, López-Méndez B, Juillerat A, Bertonati C, Daboussi F, Campos-Olivas R, Duchateau P, Montoya G. 2014. BuD, a helix-loop-helix DNA-binding domain for genome modification. Acta Crystallogr D Biol Crystallogr 70:2042–2052. doi: 10.1107/S1399004714011183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Read AC, Rinaldi FC, Hutin M, He Y-Q, Triplett LR, Bogdanove AJ. 2016. Suppression of Xo1-mediated disease resistance in rice by a truncated, non-DNA-binding TAL effector of Xanthomonas oryzae. Front Plant Sci 7:1516. doi: 10.3389/fpls.2016.01516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Schornack S, Ballvora A, Gürlebeck D, Peart J, Baulcombe D, Ganal M, Baker B, Bonas U, Lahaye T. 2004. The tomato resistance protein Bs4 is a predicted non-nuclear TIR-NB-LRR protein that mediates defense responses to severely truncated derivatives of AvrBs4 and overexpressed AvrBs3. Plant J 37:46–60. doi: 10.1046/j.1365-313x.2003.01937.x [DOI] [PubMed] [Google Scholar]
- 45. Triplett LR, Cohen SP, Heffelfinger C, Schmidt CL, Huerta AI, Tekete C, Verdier V, Bogdanove AJ, Leach JE. 2016. A resistance locus in the American heirloom rice variety Carolina gold select is triggered by TAL effectors with diverse predicted targets and is effective against African strains of Xanthomonas oryzae Pv. oryzicola. Plant J 87:472–483. doi: 10.1111/tpj.13212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Scholze H, Boch J. 2011. TAL effectors are remote controls for gene activation. Curr Opin Microbiol 14:47–53. doi: 10.1016/j.mib.2010.12.001 [DOI] [PubMed] [Google Scholar]
- 47. Rohde JR, Breitkreutz A, Chenal A, Sansonetti PJ, Parsot C. 2007. Type III secretion effectors of the IpaH family are E3 ubiquitin ligases. Cell Host Microbe 1:77–83. doi: 10.1016/j.chom.2007.02.002 [DOI] [PubMed] [Google Scholar]
- 48. Price CT, Al-Khodor S, Al-Quadan T, Santic M, Habyarimana F, Kalia A, Kwaik YA. 2009. Molecular mimicry by an F-box effector of Legionella pneumophila hijacks a conserved polyubiquitination machinery within macrophages and protozoa. PLoS Pathog 5:e1000704. doi: 10.1371/journal.ppat.1000704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Partida-Martinez LP, Hertweck C. 2007. A gene cluster encoding rhizoxin biosynthesis in Burkholderia rhizoxina, the bacterial endosymbiont of the fungus Rhizopus microsporus. Chembiochem 8:41–45. doi: 10.1002/cbic.200600393 [DOI] [PubMed] [Google Scholar]
- 50. Partida-Martinez LP, Groth I, Schmitt I, Richter W, Roth M, Hertweck C. 2007. Burkholderia rhizoxinica sp. nov. and Burkholderia endofungorum sp. nov., bacterial endosymbionts of the plant-pathogenic fungus Rhizopus microsporus. Int J Syst Evol Microbiol 57:2583–2590. doi: 10.1099/ijs.0.64660-0 [DOI] [PubMed] [Google Scholar]
- 51. Löwer M, Schneider G. 2009. Prediction of type III secretion signals in genomes of Gram-negative bacteria. PLoS One 4:e5917. doi: 10.1371/journal.pone.0005917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Arnold R, Brandmaier S, Kleine F, Tischler P, Heinz E, Behrens S, Niinikoski A, Mewes H-W, Horn M, Rattei T, Stebbins CE. 2009. Sequence-based prediction of type III secreted proteins. PLoS Pathog 5:e1000376. doi: 10.1371/journal.ppat.1000376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Kosugi S, Hasebe M, Tomita M, Yanagawa H. 2009. Systematic identification of cell cycle-dependent yeast nucleocytoplasmic shuttling proteins by prediction of composite motifs. Proc Natl Acad Sci U S A 106:10171–10176. doi: 10.1073/pnas.0900604106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Brameier M, Krings A, MacCallum RM. 2007. NucPred-predicting nuclear localization of proteins. J Bioinform 23:1159–1160. doi: 10.1093/bioinformatics/btm066 [DOI] [PubMed] [Google Scholar]
- 55. Thompson JD, Higgins DG, Gibson TJ. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res 22:4673–4680. doi: 10.1093/nar/22.22.4673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Kumar S, Stecher G, Tamura K. 2016. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol 33:1870–1874. doi: 10.1093/molbev/msw054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Saitou N, Nei M. 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454 [DOI] [PubMed] [Google Scholar]
- 58. Felsenstein J. 1985. Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39:783–791. doi: 10.1111/j.1558-5646.1985.tb00420.x [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental figures and tables.
Data Availability Statement
All data generated or analyzed during this study are included in the article and in the supporting files.