Abstract
Mutations in the five hrp and hrc genes in the hrpC operon of the phytopathogen Pseudomonas syringae pv. syringae 61 have different effects on bacterial interactions with host and nonhost plants. The hrcC gene within the hrpC operon encodes an outer membrane component of the Hrp secretion system that is conserved in all type III protein secretion systems and is required for most pathogenic phenotypes and for secretion of the HrpZ harpin to the bacterial milieu. The other four genes (in order), hrpF, hrpG, (hrcC), hrpT, and hrpV, appear to be unique to the group I hrp clusters found in certain phytopathogens (e.g., P. syringae and Erwinia amylovora) and are less well understood. We initiated an examination of their role in Hrp regulation and secretion by determining the effects of functionally nonpolar nptII cartridge insertions in each gene on the production and secretion of HrpZ, as determined by immunoblot analysis of cell fractions. P. syringae pv. syringae 61 hrpF, hrpG, and hrpT mutants were unable to secrete HrpZ, whereas the hrpV mutant overproduced and secreted the protein. This suggested that HrpV is a negative regulator of HrpZ production. Further immunoblot assays showed that the hrpV mutant produced higher levels of proteins encoded by all three of the major hrp operons tested—HrcJ (hrpZ operon), HrcC (hrpC operon), and HrcQB (hrpU operon)—and that constitutive expression of hrpV in trans abolished the production of each of these proteins. To determine the hierarchy of HrpV regulation in the P. syringae pv. syringae 61 positive regulatory cascade, which is composed of HrpRS (proteins homologous with ς54-dependent promoter-enhancer-binding proteins) and HrpL (alternate sigma factor), we tested the ability of constitutively expressed hrpV to repress the activation of HrcJ production that normally accompanies constitutive expression of hrpL or hrpRS. No repression was observed, indicating that HrpV acts upstream of HrpRS in the cascade. The effect of HrpV levels on transcription of the hrpZ operon was determined by monitoring the levels of β-glucuronidase produced by a hrpA′::uidA transcriptional fusion plasmid in different P. syringae pv. syringae 61 strains. The hrpV mutant produced higher levels of β-glucuronidase than the wild type, a hrcU (type III secretion) mutant produced the same level as the wild type, and the strain constitutively expressing hrpV in trans produced low levels equivalent to that of a hrpS mutant. These results suggest that HrpF, HrpG, and HrpT are all components of the type III protein secretion system whereas HrpV is a negative regulator of transcription of the Hrp regulon.
The characteristic ability of many phytopathogenic bacteria to elicit the hypersensitive response (HR) in nonhost plants or to be pathogenic in host plants is dependent on hrp and hrc genes (2). hrc genes represent a subset of the hrp genes that have been renamed to reflect their conservation among the type III protein secretion systems of both plant and animal pathogens (6). Among these, hrcC has been particularly well studied; it encodes an outer membrane protein that is essential for type III protein secretion and has a primary role in protein translocation across the outer membrane (2, 7, 32). The hrcC genes of Erwinia amylovora and Pseudomonas syringae are flanked by four small genes, which together form the hrpC operon. These four genes, hrpF, hrpG, hrpT, and hrpV, appear to be characteristic of group I hrp clusters, such as those of P. syringae and E. amylovora, and they are absent from the group II hrp clusters of Ralstonia solanacearum and Xanthomonas campestris pv. vesicatoria (9, 19). Group I and II hrp clusters also differ notably in their regulatory components, with group I hrp genes being activated by an alternate sigma factor and group II hrp genes being activated by an AraC homolog (2).
Essential activities in type III secretion can be ascribed to many of the Hrc proteins, such as HrcC, but less is known about the functions of the Hrp proteins. Notable exceptions are the HrpA, -L, -R, -S, and -Z proteins of P. syringae. HrpA is a Hrp-specific pilin (26). HrpR and -S show similarity with ς54-dependent promoter-enhancer-binding proteins, and both are required to activate the ς54-dependent production of HrpL, a sigma factor in the ECF (extracytoplasmic factor) family which activates the expression of other hrp genes and many avr genes (17). HrpZ is a harpin, a type of protein first reported from E. amylovora (31), which can elicit an apparent programmed cell death when infiltrated into the leaves of tobacco and several other plants (15). HrpZ is secreted in culture in a hrp-dependent manner from P. syringae (15), but the protein does not appear to be the physiological elicitor of the HR: mutations in hrmA, an avr-like gene, completely block the ability of a functional cluster of P. syringae hrp genes to function in Escherichia coli to elicit the HR, but they have no effect on HrpZ secretion (1, 3). Avr (avirulence) proteins appear to be the actual elicitors of the HR, and there is compelling evidence that many of these function inside plant cells following delivery by the P. syringae Hrp system (11, 22, 28, 30). Whether HrpZ has a primary role as an extracellular component of the Avr protein delivery system is unknown, but its secretion in culture provides an assay for the functioning of the Hrp secretion pathway in P. syringae.
In the accompanying paper, we have shown that mutations in the P. syringae pv. syringae 61 hrpF, hrpG, hrpT, and hrpV genes result in altered plant reaction phenotypes, with the effects of each mutation being quantitatively different (9). Unexpectedly, expression of hrpV in trans reduced the ability of wild-type P. syringae pv. syringae 61 to elicit the HR, suggesting that HrpV may be a negative regulator of the Hrp regulon. To test this hypothesis and to investigate further the functions of the other genes in the hrpC operon, we have determined the effects of mutations in these genes on the production of several Hrp marker proteins and on the secretion of the HrpZ harpin. Subsequently, we also investigated the place of HrpV inhibition in the HrpRS-HrpL regulatory cascade.
MATERIALS AND METHODS
Bacterial strains, plasmids, and media.
Most of the bacterial strains and plasmids used in this study are described in the accompanying paper (9). Additional strains used were P. syringae pv. syringae 61–2074 (hrpL::TnphoA), P. syringae pv. syringae 61–2088 (hrcU::TnphoA), and P. syringae pv. syringae 61–2095 (hrpS::TnphoA) (16). E. coli was routinely grown in Luria broth or Terrific broth at 37°C (4). Pseudomonas strains were grown in King’s B (KB) medium (20) or LM medium (13) at 28 to 30°C, but for certain experiments the hrp-derepressing minimal medium of Huynh et al. (18), adjusted to pH 5.5, was used at 25°C. Antibiotics were used in selective media at the following concentrations (micrograms per milliliter): ampicillin, 100; kanamycin, 50; tetracycline, 10; spectinomycin 50; gentamicin 10; and nalidixic acid, 20.
Recombinant DNA techniques.
Restriction endonuclease digestion, agarose gel electrophoresis, DNA fragment preparation, plasmid extraction, DNA ligation, and transformation by the CaCl2 procedure were performed according to standard procedures (4). Plasmids were introduced into bacteria by transformation, electroporation (Gene Pulsar; Bio-Rad, Richmond, Calif.), or triparental mating (10).
Construction of pCPP2385, pCPP2389, and pCPP2383.
pCPP2385, which contains a constitutively expressed hrpL, was constructed by inserting an ΩSpr fragment (7) downstream of hrpL in pCPP2311. This provided a selectable marker that would be effective in P. syringae for pCPP2311, which contains hrpL subcloned downstream of the lac promoter in pUCP18 (27). pCPP2389 constitutively expresses hrpRS and was made by subcloning a 2.2-kb BamHI-BglII fragment derived from pHIR11 downstream of the lac promoter in pCPP30 (5). The 190-bp promoter-active SacI-HincII fragment upstream of hrpA was cloned into a promoterless uidA gene in pCPP45 (5) to create the hrpA′::uidA transcriptional fusion in plasmid pCPP2383.
Preparation of anti-HrcC and anti-HrcQB antibodies.
HrcC and HrcQB were purified from E. coli NovaBlue (λDE3) (Novagen, Madison, Wis.) carrying pNCHU316 and pNCHU366, respectively, according to previously described procedures (7). Before immunoblot analysis the antisera were preabsorbed with cell lysate mixtures of E. coli and P. syringae pv. syringae 61 mutants according to the following modifications of previously used procedures (15). Five-milliliter cultures of E. coli and P. syringae pv. syringae 61 mutants (P. syringae pv. syringae 61–N393 and P. syringae pv. syringae 61–N322 for incubation with HrcC and HrcQB antibodies, respectively) were grown overnight to stationary phase, washed several times with the same volume of 1× phosphate-buffered saline (0.058 M Na2HPO4, 0.017 M NaH2PO4 · H2O, 0.068 M NaCl), sonicated, incubated at 100°C for 10 min, and then allowed to cool to room temperature. Two hundred microliters of the corresponding antiserum and sodium azide at a final concentration of 0.02% were added into the cell lysate and incubated at room temperature for 2 h. The mixtures were centrifuged at 20,000 × g for 30 min, and the preabsorbed antisera were collected from the supernatant.
Immunoblot analysis of the expression of HrpZ, HrcC, HrcJ, and HrcQB proteins.
Wild-type P. syringae pv. syringae 61 and hrpF, hrpG, hrcC, hrpT, and hrpV mutants were grown in 5 ml of KB broth at 30°C to an optical density at 600 nm (OD600) of 0.5. The cells were collected by centrifugation, washed once in 5 ml of Hrp minimal medium, resuspended in 5 ml of the same medium, and incubated with shaking for 5 h. The cell and supernatant fractions were then separated by centrifugation. HrpZ expression was analyzed as described previously (24) or as outlined below. The supernatants were precipitated with trichloroacetic acid at a final concentration of 5%, washed with acetone, dissolved in 30 μl of 10 mM Tris buffer (pH 8.0), and boiled for 5 min after an equal volume of 2× loading buffer (0.625 M Tris [pH 6.8], 2% sodium dodecyl sulfate [SDS], 10% glycerol, 2% β-mercaptoethanol) was added. The cell pellets were washed with 10 mM Tris buffer (pH 8.0), resuspended in 125 μl of 10 mM Tris buffer, and boiled with an equal volume of 2× loading buffer for 5 min. A 20-μl sample of each fraction was subjected to SDS-10% polyacrylamide gel electrophoresis in a 0.75-mm-thick gel in a Mighty Small apparatus (Hoefer Scientific Instruments, San Francisco, Calif.). The prestained protein markers—ranging from 175.0 to 6.5 kDa—were from Bio-Rad. After separation, the protein bands were transferred to Immobilon-P membranes (Millipore Inc., Bedford, Mass.) in a TE70 semidry transfer unit (Hoefer Scientific Instruments) for 40 to 60 min. The membranes were probed with polyclonal anti-HrpZ, anti-HrcJ, anti-HrcC, anti-HrcQB, or anti-NPTII antibodies individually. Anti-NPTII antibodies (5 Prime→3 Prime, Inc., Boulder, Colo.) were used as an internal control to standardize the protein concentration of each lane. Immunodetection of the bands was performed by alkaline phosphatase-based chromogenic and chemiluminescent assays with Sigma Fast BCIP (5-bromo-4-chloro-3-indolylphosphate)-nitroblue tetrazolium tablets (0.15 mg of BCIP/ml and 0.3 mg of nitroblue tetrazolium/ml) or 0.25 mM disodium 2-chloro-5(4-methoxyspiro{1,2-dioxetane-3,2′-(5′-chloro)-tricyclo(3.3.1.13,7)decan}-4-yl)-1-phenyl phosphate (CDP-Star; Tropix Inc., Bedford, Mass.), respectively. Membranes were exposed for 2 to 15 min to OMAT X-ray film (Kodak, Rochester, N.Y.) for visualization of chemiluminescence.
Measurement of β-glucuronidase activity.
Transcription of the hrpZ operon was determined by measuring the expression of β-glucuronidase from the hrpA′::uidA transcriptional fusion plasmid pCPP2383. P. syringae pv. syringae 61 derivatives carrying pCPP2383 were grown in KB medium to stationary phase and then transferred to Hrp minimal medium at an initial OD600 of 0.5. The bacteria were harvested 0, 4, and 10 h later for determination of β-glucuronidase activity and total protein concentration. β-Glucuronidase activity was estimated with 4-methylumbelliferyl-β-d-glucuronide and a TKO 100 minifluorometer (Hoefer Scientific Instruments) according to the procedure outlined in the operating instructions. Total protein concentration was determined with the Bio-Rad protein assay reagent (Bio-Rad Laboratories, Hercules, Calif.). Specific activity is presented in nanomoles of methylumbelliferone production per milligram of total protein per minute.
RESULTS
nptII cartridge insertions in hrpF, hrpG, hrpT, and hrpV result in three classes of mutants that are altered in their HrpZ production and HrpZ secretion phenotypes.
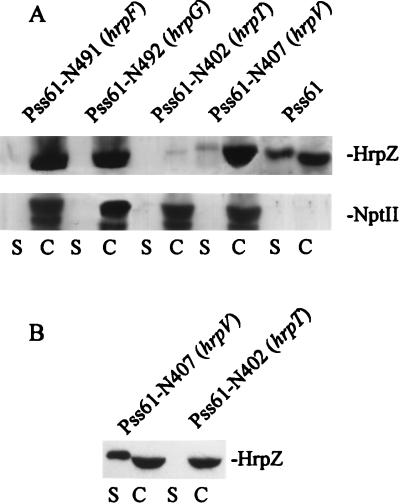
Functionally nonpolar nptII cartridge insertions were constructed previously in each gene in the P. syringae pv. syringae 61 hrpC operon (9). To determine the effects of mutations in hrpF, hrpG, hrpT, and hrpV on HrpZ production and secretion, we performed an immunoblot analysis of HrpZ levels in the cell and supernatant fractions of cultures that were in logarithmic growth in Hrp minimal medium 5 h after being shifted from logarithmic growth in complex KB medium (Fig. 1A). Three classes of mutants could be discerned: (i) hrpF and hrpG mutants were unable to secrete HrpZ, (ii) the hrpT mutant had strongly reduced HrpZ production, and (iii) the hrpV mutant overproduced HrpZ and secreted at least some of the protein. NptII expressed from its cognate promoter in the cartridge inserted in each mutant was used as a control for constitutive gene expression and for nonspecific leakage of cytoplasmic proteins to the medium.
FIG. 1.
Effects of nptII cartridge insertions in the P. syringae pv. syringae 61 (Pss61) hrpF, hrpG, hrpT, and hrpV genes on the production and secretion of HrpZ. (A) Bacteria in early-logarithmic-phase growth in KB medium were harvested by centrifugation and resuspended at an OD600 of 0.5 in Hrp minimal medium. After a further 5 h of incubation, the cultures were fractionated into cell (C) and supernatant (S) fractions by centrifugation and assayed for the accumulation of HrpZ and NptII by SDS-polyacrylamide gel electrophoresis followed by immunoblot analysis with appropriate antibodies and chemiluminescent immunodetection. (B) Bacteria were grown to stationary phase in KB medium and then handled as described above.
In repeated experiments we observed variability in the phenotypes of the hrpV, hrpT, and hrpG mutants that appeared to correlate with the duration of previous growth in complex media. When grown to higher culture densities, the hrpG mutant produced low levels of HrpZ whereas the hrpT and hrpV mutants produced and secreted more HrpZ, respectively. The phenotypes of hrpT and hrpV mutants grown to stationary phase in KB medium before being shifted to Hrp minimal medium are shown in Fig. 1B. The hrpG mutant produced no detectable HrpZ in this experiment (not shown), and we chose to employ the culture conditions described in the legend to Fig. 1A in subsequent experiments. In light of this variability, it is important to note that regardless of culture conditions, the hrpV mutant always secreted at least some HrpZ whereas the hrpT mutant did not. These data suggested that HrpT contributes to HrpZ secretion, whereas HrpV does not. The reduced production of HrpZ by the ΔhrpT::nptII mutant suggested that HrpV was a negative regulator that was being overexpressed from the proximal, upstream nptII promoter. The apparent overexpression of HrpZ that was observed in the hrpV mutant (Fig. 1A) further supported this hypothesis.
Effects on HrcC and HrcQB accumulation of nptII cartridge insertions in hrpF, hrpG, hrpT, and hrpV further indicate that HrpV is a negative regulator of hrp expression.
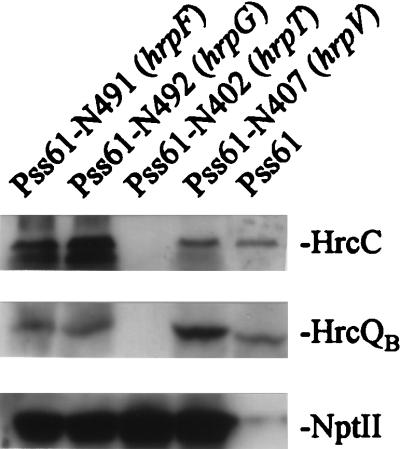
To determine the effects of nptII cartridge insertions within the hrpC operon on the production of other components of the Hrp system, immunoblot analysis of HrcC and HrcQB (encoded by the hrpU operon) was performed on the cell fractions of various mutants grown to early logarithmic phase in complex medium and then in minimal medium (Fig. 2). Again, three classes of mutants could be discerned: (i) hrpF and hrpG mutants (with nptII insertions upstream of hrcC) produced higher levels of HrcC but wild-type levels of HrpQB, (ii) the hrpT mutant produced no detectable HrcC or HrpQB, and (iii) the hrpV mutant produced higher levels of both HrcC and HrcQB than did wild-type P. syringae pv. syringae 61. NptII levels provided an internal control against protein loading variability that would account for these differences. These observations indicated that in early-logarithmic-phase growth in Hrp minimal medium, certain P. syringae pv. syringae 61 hrpC operon mutants—P. syringae pv. syringae 61–N491 (ΔhrpF::nptII) and P. syringae pv. syringae 61–N492 (hrpG::nptII)—accumulate more HrcC through expression of the nptII promoter than through expression of the native promoter, and they further support the notion that overproduction of HrpV represses hrp expression whereas disruption of hrpV increases hrp expression.
FIG. 2.
Effects of nptII cartridge insertions in the P. syringae pv. syringae 61 (Pss61) hrpF, hrpG, hrpT, and hrpV genes on hrcC and hrcQB expression. Cell fractions of cultures grown for 5 h in Hrp minimal medium, as described in the legend to Fig. 1A, were assayed for the accumulation of HrcC and HrcQB by SDS-polyacrylamide gel electrophoresis followed by immunoblot analysis with appropriate antibodies and chemiluminescent immunodetection.
Constitutive expression of hrpV in trans represses production of proteins encoded by three of the major hrp operons.
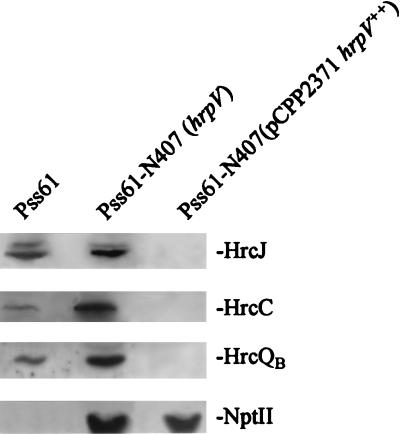
To directly test the effects of altered levels of hrpV expression on regulation of the hrp system, we examined the accumulation of the products of three of the major hrp secretion operons in P. syringae pv. syringae 61 cells that were either wild type, deficient in hrpV, or overexpressing hrpV (Fig. 3). Overexpression of hrpV was achieved by transforming ΔhrpV::nptII mutant P. syringae pv. syringae 61–N407 with pCPP2371, which has the hrpV gene expressed from the nptII promoter in vector pML122. HrcC, HrcQB, and HrcJ, which are products of the hrpC, hrpU, and hrpZ operons, respectively, were equally affected by these changes in hrpV expression. That is, accumulation of the three Hrc proteins was higher in P. syringae pv. syringae 61–N407 than in the wild type and almost undetectable in P. syringae pv. syringae 61–N407(pCPP2371).
FIG. 3.
Effects of mutating or overexpressing hrpV on the accumulation of Hrc proteins encoded by three different hrp operons. Bacteria grown for 5 h in Hrp minimal medium, as described in the legend to Fig. 1A, were assayed for the accumulation of HrcJ, HrcC, and HrcQB by SDS-polyacrylamide gel electrophoresis followed by immunoblot analysis with appropriate antibodies and chemiluminescent immunodetection. Pss61, P. syringae pv. syringae 61.
Constitutive expression of hrpL or hrpRS blocks the repressive effects of hrpV overexpression.
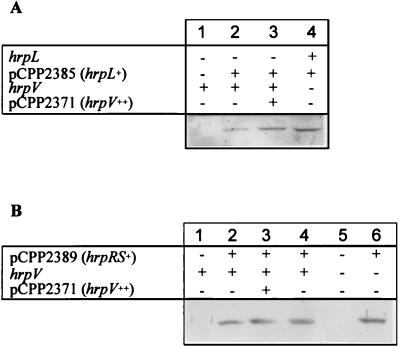
To determine at what level in the hrp regulatory cascade HrpV acts, hrpV was overexpressed in cells containing constitutively expressed hrpL or hrpRS, and the resulting levels of HrcJ production were determined (Fig. 4). HrcJ was analyzed because it is encoded by the hrpZ operon but its accumulation is not subject to apparent posttranscriptional regulation, as has been observed with HrpZ (7). Constitutive expression of hrpL or hrpRS results in constitutive expression of hrp genes under the normally repressive conditions of growth in complex media (34). To rigorously test the possibility that HrpV was an anti-sigma factor acting on HrpL we introduced plasmids into P. syringae pv. syringae 61–2074 (hrpL::TnphoA) that were designed to produce much higher levels of HrpV than HrpL. hrpL was expressed from the lac promoter in pUCP18, which is relatively weak in Pseudomonas spp., while hrpV was expressed from the nptII promoter in pML122, which is much stronger (21). This combination of copy number and promoter strength resulted in high production of HrpV, which was visible as a unique band of the predicted size on a Coomassie blue-stained SDS-polyacrylamide gel (data not shown). Nevertheless, HrpV did not reduce HrcJ production in the presence of HrpL, indicating that HrpV acts upstream of HrpL in the regulatory cascade (Fig. 4A). Similarly, hrpV failed to repress the accumulation of HrcJ that resulted from constitutive expression of hrpRS (Fig. 4B). Constitutive expression of hrpL or hrpRS also overcame the repressive effects of hrpV overexpression by pCPP2371 when the bacteria were grown in Hrp minimal medium rather than LM medium (data not shown). It is also important to note that ΔhrpV mutant P. syringae pv. syringae 61–N407 produced no detectable HrcJ in Hrp-repressive LM medium (Fig. 4B). These observations suggest that the step at which hrpV interferes with hrp gene expression lies upstream of the hrpRS and hrpL induction cascade.
FIG. 4.
Prevention of hrpV-dependent inhibition of HrcJ accumulation by constitutive expression of hrpL and hrpRS. All bacteria were grown in LM medium and assayed for the accumulation of HrcJ by immunoblot analysis, as described in the legend to Fig. 1, but with chromogenic immunodetection. Cultures denoted hrpL− carried a chromosomal hrpL::TnphoA mutation; those denoted hrpV− carried a ΔhrpV::nptII mutation. (A) Lanes: 1, P. syringae pv. syringae 61–2074; 2, P. syringae pv. syringae 61–2074(pCPP2385); 3, P. syringae pv. syringae 61–2074(pCPP2385/pCPP2371); 4, P. syringae pv. syringae 61–N407(pCPP2385). (B) Lanes: 1, P. syringae pv. syringae 61; 2, P. syringae pv. syringae 61(pCPP2389); 3, P. syringae pv. syringae 61(pCPP2389/pCPP2371); 4, P. syringae pv. syringae 61(pCPP2389/pML122); 5, P. syringae pv. syringae 61–N407; 6, P. syringae pv. syringae 61–N407(pCPP2389).
Constitutive expression of hrpV represses transcription of the hrpZ operon.
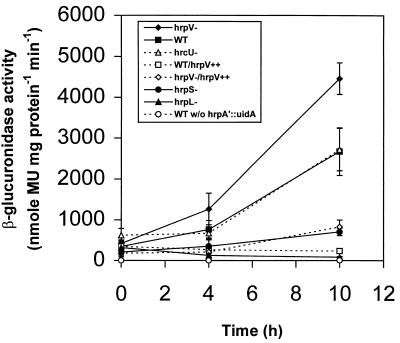
The previous experiments were based on the differential accumulation of HrpZ and three Hrc proteins, particularly HrcJ, which is in the hrpZ operon. To verify that overexpression of HrpV represses transcription of the hrpZ operon, as opposed to acting posttranscriptionally, we constructed plasmid pCPP2383 (hrpA′::uidA), in which the promoter of the hrpZ operon is transcriptionally fused to a uidA gene, and analyzed β-glucuronidase activity in P. syringae pv. syringae 61 derivatives that carried the plasmid and were altered in their production of HrpV and other components of the Hrp system (Fig. 5). Deletion of hrpV resulted in levels of hrpA′::uidA expression significantly higher than that in the wild type, whereas overexpression of hrpV resulted in a reduction in hrpA′::uidA expression to a level equivalent to that of hrpL and hrpS mutants. P. syringae pv. syringae 61–2088, a hrcU::TnphoA mutant that fails to export HrpZ out of the cytoplasm (7), expressed hrpA′::uidA at wild-type levels. Thus, the effects of hrpV on the accumulation of two products of the hrpZ operon, HrpZ and HrpJ, that were observed as described above can be attributed to the transcription of the operon.
FIG. 5.
Effects on hrpA′::uidA expression of altering the levels of expression of hrpV, hrpL, and hrpS. Bacteria were grown in KB medium to stationary phase and transferred to Hrp minimal medium at an initial OD600 of 0.5. β-Glucuronidase activity and total protein concentration were measured at the indicated times as described in the text. The values represent the means of three replicates. The vertical lines indicate standard errors, and where they are absent, the limits were within the symbol dimensions. Except where noted, all strains carried pCPP2383 (hrpA′::uidA). The strains tested were P. syringae pv. syringae 61–N407 (hrpV−), P. syringae pv. syringae 61–2088 (hrcU−), P. syringae pv. syringae 61 (WT), P. syringae pv. syringae 61–N407(pCPP2371) (hrpV−/hrpV++), P. syringae pv. syringae 61–2095 (hrpS−), P. syringae pv. syringae 61(pCPP2371) (hrpV++), P. syringae pv. syringae 61–2074 (hrpL−), and P. syringae pv. syringae 61 without pCPP2383 (WT w/o hrpA′::uidA). MU, methylumbelliferone.
DISCUSSION
By examining the effects of nptII cartridge insertions in the four small genes that flank hrcC in the hrpC operon, we have learned that hrpF, hrpG, and hrpT have roles in HrpZ secretion, whereas hrpV encodes a negative regulator of hrp gene expression. These observations raise several questions regarding the plant reaction and biochemical phenotypes of these genes, the functions of their products in Hrp secretion and regulation, and the global control of the hrp regulon.
To properly interpret the phenotypes of these mutants, it is necessary to consider the effects of the nptII cartridge that was used to construct them. The cartridge lacks a transcription terminator, thus permitting nptII and downstream genes to be driven by the cartridge promoter and enabling the construction of mutants in complex media that support robust bacterial growth but poor hrp expression. The efficacy of this approach for testing the functions of individual genes is supported by the observation that appropriate single-gene subclones can restore wild-type HR elicitation activity to nptII-marked hrpF, hrpG, hrpT, and hrcC mutants (9). These subclones can similarly restore the ability to secrete HrpZ (data not shown). However, constitutive expression of downstream genes can have unanticipated effects, as we have found here with hrpV and will discuss further below.
It is also noteworthy that the plant reaction phenotypes of some of these hrpC operon mutations, reported in the accompanying paper (9), are not as strong as their HrpZ secretion phenotypes. We anticipated that any mutation preventing HrpZ secretion would have a strong Hrp phenotype, as was observed with hrpF and hrcC. However, the hrpG and hrpT mutants retained some ability to elicit the HR in tobacco and disease in beans. Since the physiological elicitors of the HR are now thought to be Avr proteins that are transferred into plant cells by the Hrp system, one explanation is that these mutations do not completely inhibit that process. At present, this possibility is not testable because there is no assay for P. syringae Avr protein secretion in culture or for quantitative Avr protein transfer in planta.
The functions of HrpF, HrpG, and HrpT in the Hrp secretion pathway are unknown. Although they are not present in animal-associated bacteria, they are conserved among different pathovars of P. syringae (9) and are found in other phytopathogenic bacteria, such as E. amylovora (19), suggesting that they have specific functions in plant pathogenesis. The amino acid sequence of HrpT reveals that it is a putative lipoprotein, and it may have a role as a HrcC chaperone. This notion is based on two observations: (i) homologs of HrcC that are involved in type II secretion require a similarly small lipoprotein chaperone for insertion into the outer membrane (14) and (ii) group II hrp clusters lack a lipoprotein like HrpT, and their HrcC proteins lack the C-terminal region that is thought to interact with these chaperones, whereas the HrcC proteins of group I hrp systems possess this region (8). However, the biological function of HrpT is yet to be determined in these bacteria.
HrpV appears to be a negative regulator of the HrpR/S-HrpL activator cascade and is the first negative regulator reported for the P. syringae Hrp system. However, in assessing its role it is important to note that overexpression of hrpV eliminates expression of the Hrp regulon, whereas deletion of hrpV results only in moderate increases in hrp expression. One explanation for this would be the presence of a functionally equivalent copy of hrpV elsewhere in the genome. This would be analogous to the two recently reported negative regulators, YscM1 and YscM2, of the Yop virulon (which includes the type III secretion system) in Yersinia enterocolitica (29). To seek a second copy of hrpV, we probed P. syringae pv. syringae 61 total DNA with PCR-amplified hrpV genes in a DNA gel blot at moderate stringency, but we observed no extra hybridizing bands (data not shown).
We can postulate three potential functions for HrpV as a negative regulator. First, HrpV may be a negative-feedback regulator preventing overproduction of HrcC. The location of hrpV at the end of the hrpC operon is consistent with this hypothesis, as is the observation that the X. campestris pv. vesicatoria HrcC (HrpA1) protein induces the phage shock protein operon when expressed in E. coli (32).
A second potential function for HrpV is to delay the expression of other hrp operons until the channel-forming HrcC multimers have formed in the outer membrane. This model is based on the concept that the type III protein secretion system has evolved through the recruitment of two separate translocators: a flagellum export-derived system for translocation across the inner membrane (encoded by the hrpJ and hrpU operons) and HrcC for translocation across the outer membrane. We have previously shown that P. syringae pv. syringae 61 hrcC mutants accumulate some HrpZ in the periplasm, whereas mutants affected in the hrpJ and hrpU operons accumulate HrpZ only in the cytoplasm (7). This suggests that premature expression of the hrpJ and hrpU operons could result in deleterious localization of some Hrp and Avr proteins in the periplasm. Supporting this hypothesis is the observation that hrcC (hrpA1) in X. campestris pv. vesicatoria is activated by HrpG, which is higher in the regulatory cascade than HrpX, which in turn activates the other hrp genes (33). If HrpV preferentially represses either the hrpC operon or the hrpJ and hrpU operons, this must be determined by factors in addition to HrpL that affect the expression of these operons.
A third potential function for HrpV is to increase expression of the Hrp regulon upon contact with host cells through removal of the protein from the bacterial cell. This would be analogous to the host contact-dependent secretion of LcrQ, a negative regulator of yop and ysc expression, by Yersinia spp. (23). If HrpV can be secreted, this does not appear to happen in culture, because mutations blocking Hrp secretion do not inhibit hrp expression, as would be expected with accumulation of a normally secreted negative regulator. Furthermore, there is no consensus that P. syringae hrp expression is higher in planta than in minimal media that mimic plant intercellular fluids (25, 35). However, any increase in hrp expression in planta may be hard to detect if it is transient or of moderate magnitude. Support for the hypothesis that HrpV can be secreted is found in the puzzling observation that HrpV overexpression has a relatively minor effect on elicitation of the HR in tobacco leaves (9) whereas it virtually abolishes hrp expression in culture. This would be explainable if the HrpV pool could be secreted in planta but not in culture. Regardless of which hypothesis is correct, it is important to note that the hrpV mutation does have an effect on the pathogenesis of P. syringae pv. syringae 61 in beans (9).
Our data indicate that HrpV acts upstream of HrpRS. Several aspects of HrpRS remain puzzling. There is controversy regarding whether HrpS alone or both HrpR and HrpS are required to activate hrp expression (12, 34) and whether the hrpRS operon is expressed at a higher level in planta than in culture under Hrp-derepressing conditions (25, 35). The observation that the N-terminal domains associated with regulation in related proteins are missing from HrpR and HrpS suggests that the levels of active HrpRS may be key in regulating hrp expression, and our data provide indirect evidence that high levels of HrpV inhibit HrpRS production. Key questions for the future are whether HrpV directly controls expression of the hrpR promoter, whether bacteria in planta secrete HrpV and thereby increase hrp gene expression, and why hrpV mutants overexpressing the Hrp system are impaired in pathogenesis. Given the complex functions of the Hrp pathway in protein secretion and pathogenesis, it is not surprising that there is at least one negative regulator controlling the system.
ACKNOWLEDGMENTS
We thank Kent Loeffler for photography, David W. Bauer for constructing pCPP2311, Jihyun F. Kim for advice regarding potential chaperone interaction domains in HrcC, and C.-J. Chang for technical assistance.
This work was supported by NSF grant MCB-9631530 from the National Science Foundation and NSC grant 86-2321-B-005-020 from Taiwan.
REFERENCES
- 1.Alfano J R, Bauer D W, Milos T M, Collmer A. Analysis of the role of the Pseudomonas syringae pv. syringae HrpZ harpin in elicitation of the hypersensitive response in tobacco using functionally nonpolar deletion mutations, truncated HrpZ fragments, and hrmA mutations. Mol Microbiol. 1996;19:715–728. doi: 10.1046/j.1365-2958.1996.415946.x. [DOI] [PubMed] [Google Scholar]
- 2.Alfano J R, Collmer A. The type III (Hrp) secretion pathway of plant pathogenic bacteria: trafficking harpins, Avr proteins, and death. J Bacteriol. 1997;179:5655–5662. doi: 10.1128/jb.179.18.5655-5662.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Alfano J R, Kim H-S, Delaney T P, Collmer A. Evidence that the Pseudomonas syringae pv. syringae hrp-linked hrmA gene encodes an Avr-like protein that acts in a hrp-dependent manner within tobacco cells. Mol Plant-Microbe Interact. 1997;10:580–588. doi: 10.1094/MPMI.1997.10.5.580. [DOI] [PubMed] [Google Scholar]
- 4.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K E. Short protocols in molecular biology. 3rd ed. New York, N.Y: John Wiley & Sons; 1995. [Google Scholar]
- 5.Bauer D W, Collmer A. Molecular cloning, characterization, and mutagenesis of a pel gene from Pseudomonas syringae pv. lachrymans encoding a member of the Erwinia chrysanthemi PelADE family of pectate lyases. Mol Plant-Microbe Interact. 1997;10:369–379. doi: 10.1094/MPMI.1997.10.3.369. [DOI] [PubMed] [Google Scholar]
- 6.Bogdanove A J, Beer S V, Bonas U, Boucher C A, Collmer A, Coplin D L, Cornelis G R, Huang H-C, Hutcheson S W, Panopoulos N J, Van Gijsegem F. Unified nomenclature for broadly conserved hrp genes of phytopathogenic bacteria. Mol Microbiol. 1996;20:681–683. doi: 10.1046/j.1365-2958.1996.5731077.x. [DOI] [PubMed] [Google Scholar]
- 7.Charkowski A O, Huang H-C, Collmer A. Altered localization of HrpZ in Pseudomonas syringae pv. syringae hrp mutants suggests that different components of the type III secretion pathway control protein translocation across the inner and outer membranes of gram-negative bacteria. J Bacteriol. 1997;179:3866–3874. doi: 10.1128/jb.179.12.3866-3874.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Daefler S, Guilvout I, Hardie K R, Pugsley A P, Russel M. The C-terminal domain of the secretin PulD contains the binding site for its cognate chaperone, PulS, and confers PulS dependence on pIVf1 function. Mol Microbiol. 1997;24:465–475. doi: 10.1046/j.1365-2958.1997.3531727.x. [DOI] [PubMed] [Google Scholar]
- 9.Deng W-L, Preston G, Collmer A, Chang C-J, Huang H-C. Characterization of the hrpC and hrpRS operons of Pseudomonas syringae pathovars syringae, tomato, and glycinea and analysis of the ability of hrpF, hrpG, hrcC, hrpT, and hrpV mutants to elicit the hypersensitive response and disease in plants. J Bacteriol. 1998;180:4523–4531. doi: 10.1128/jb.180.17.4523-4531.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ditta G, Stanfield S, Corbin D, Helinski D R. Broad host range DNA cloning system for Gram-negative bacteria: construction of a gene bank of Rhizobium meliloti. Proc Natl Acad Sci USA. 1980;77:7347–7351. doi: 10.1073/pnas.77.12.7347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gopalan S, Bauer D W, Alfano J R, Loniello A O, He S Y, Collmer A. Expression of the Pseudomonas syringae avirulence protein AvrB in plant cells alleviates its dependence on the hypersensitive response and pathogenicity (Hrp) secretion system in eliciting genotype-specific hypersensitive cell death. Plant Cell. 1996;8:1095–1105. doi: 10.1105/tpc.8.7.1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Grimm C, Aufsatz W, Panopoulos N J. The hrpRS locus of Pseudomonas syringae pv. phaseolicola constitutes a complex regulatory unit. Mol Microbiol. 1995;15:155–165. doi: 10.1111/j.1365-2958.1995.tb02230.x. [DOI] [PubMed] [Google Scholar]
- 13.Hanahan D. Techniques for transformation of E. coli. In: Glover D M, editor. DNA cloning: a practical approach. Oxford, United Kingdom: IRL Press; 1985. pp. 109–135. [Google Scholar]
- 14.Hardie K R, Lory S, Pugsley A P. Insertion of an outer membrane protein in Escherichia coli requires a chaperone-like protein. EMBO J. 1996;15:978–988. [PMC free article] [PubMed] [Google Scholar]
- 15.He S Y, Huang H-C, Collmer A. Pseudomonas syringae pv. syringae harpinPss: a protein that is secreted via the Hrp pathway and elicits the hypersensitive response in plants. Cell. 1993;73:1255–1266. doi: 10.1016/0092-8674(93)90354-s. [DOI] [PubMed] [Google Scholar]
- 16.Huang H-C, Hutcheson S W, Collmer A. Characterization of the hrp cluster from Pseudomonas syringae pv. syringae 61 and TnphoA tagging of genes encoding exported or membrane-spanning Hrp proteins. Mol Plant-Microbe Interact. 1991;4:469–476. [Google Scholar]
- 17.Hutcheson S W, Heu S, Hin S, Lidell M C, Pirhonen M U, Rowley D L. Function and regulation of Pseudomonas syringae hrp genes. In: Nakazawa T, Rurukawa K, Haas D, Silver S, editors. Molecular biology of pseudomonads. Washington, D.C: ASM Press; 1995. pp. 512–521. [Google Scholar]
- 18.Huynh T V, Dahlbeck D, Staskawicz B J. Bacterial blight of soybean: regulation of a pathogen gene determining host cultivar specificity. Science. 1989;245:1374–1377. doi: 10.1126/science.2781284. [DOI] [PubMed] [Google Scholar]
- 19.Kim J F, Wei Z-M, Beer S V. The hrpA and hrpC operons of Erwinia amylovora encode components of a type III pathway that secretes harpin. J Bacteriol. 1997;179:1690–1697. doi: 10.1128/jb.179.5.1690-1697.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.King E O, Ward M K, Raney D E. Two simple media for the demonstration of pyocyanine and fluorescein. J Lab Clin Med. 1954;44:301–307. [PubMed] [Google Scholar]
- 21.Labes M, Puhler A, Simon R. A new family of RSF1010-derived expression and lac-fusion broad-host-range vectors for gram-negative bacteria. Gene. 1990;89:37–46. doi: 10.1016/0378-1119(90)90203-4. [DOI] [PubMed] [Google Scholar]
- 22.Leister R T, Ausubel F M, Katagiri F. Molecular recognition of pathogen attack occurs inside of plant cells in plant disease resistance specified by the Arabidopsis genes RPS2 and RPM1. Proc Natl Acad Sci USA. 1996;93:15497–15502. doi: 10.1073/pnas.93.26.15497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pettersson J, Nordfelth R, Dubinina E, Bergman T, Gustafsson M, Magnusson K E, Wolf-Watz H. Modulation of virulence factor expression by pathogen target cell contact. Science. 1996;273:1231–1233. doi: 10.1126/science.273.5279.1231. [DOI] [PubMed] [Google Scholar]
- 24.Preston G, Huang H-C, He S Y, Collmer A. The HrpZ proteins of Pseudomonas syringae pvs. syringae, glycinea, and tomato are encoded by an operon containing Yersinia ysc homologs and elicit the hypersensitive response in tomato but not soybean. Mol Plant-Microbe Interact. 1995;8:717–732. doi: 10.1094/mpmi-8-0717. [DOI] [PubMed] [Google Scholar]
- 25.Rahme L G, Mindrinos M N, Panopoulos N J. Plant and environmental sensory signals control the expression of hrp genes in Pseudomonas syringae pv. phaseolicola. J Bacteriol. 1992;174:3499–3507. doi: 10.1128/jb.174.11.3499-3507.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Roine E, Wei W, Yuan J, Nurmiaho-Lassila E-L, Kalkkinen N, Romantschuk M, He S Y. Hrp pilus: an hrp-dependent bacterial surface appendage produced by Pseudomonas syringae pv. tomato DC3000. Proc Natl Acad Sci USA. 1997;94:3459–3464. doi: 10.1073/pnas.94.7.3459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schweizer H P. Escherichia-Pseudomonas shuttle vectors derived from pUC18/19. Gene. 1991;97:109–112. doi: 10.1016/0378-1119(91)90016-5. [DOI] [PubMed] [Google Scholar]
- 28.Scofield S R, Tobias C M, Rathjen J P, Chang J H, Lavelle D T, Michelmore R W, Staskawicz B J. Molecular basis of gene-for-gene specificity in bacterial speck disease of tomato. Science. 1996;274:2063–2065. doi: 10.1126/science.274.5295.2063. [DOI] [PubMed] [Google Scholar]
- 29.Stanier I, Iriarte M, Cornelis G R. YscM1 and YscM2, two Yersinia enterocolitica proteins causing downregulation of yop transcription. Mol Microbiol. 1997;26:833–843. doi: 10.1046/j.1365-2958.1997.6281995.x. [DOI] [PubMed] [Google Scholar]
- 30.Tang X, Frederick R D, Zhou J, Halterman D A, Jia Y, Martin G B. Initiation of plant disease resistance by physical interaction of AvrPto and Pto kinase. Science. 1996;274:2060–2062. doi: 10.1126/science.274.5295.2060. [DOI] [PubMed] [Google Scholar]
- 31.Wei Z-M, Laby R J, Zumoff C H, Bauer D W, He S Y, Collmer A, Beer S V. Harpin, elicitor of the hypersensitive response produced by the plant pathogen Erwinia amylovora. Science. 1992;257:85–88. doi: 10.1126/science.1621099. [DOI] [PubMed] [Google Scholar]
- 32.Wengelnik K, Marie C, Russel M, Bonas U. Expression and localization of HrpA1, a protein of Xanthomonas campestris pv. vesicatoria essential for pathogenicity and induction of the hypersensitive response. J Bacteriol. 1996;178:1061–1069. doi: 10.1128/jb.178.4.1061-1069.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wengelnik K, Van den Ackerveken G, Bonas U. HrpG, a key hrp regulatory protein of Xanthomonas campestris pv. vesicatoria, is homologous to two-component response regulators. Mol Plant-Microbe Interact. 1996;9:704–712. doi: 10.1094/mpmi-9-0704. [DOI] [PubMed] [Google Scholar]
- 34.Xiao Y, Heu S, Yi J, Lu Y, Hutcheson S W. Identification of a putative alternate sigma factor and characterization of a multicomponent regulatory cascade controlling the expression of Pseudomonas syringae pv. syringae Pss61 hrp and hrmA genes. J Bacteriol. 1994;176:1025–1036. doi: 10.1128/jb.176.4.1025-1036.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Xiao Y, Lu Y, Heu S, Hutcheson S W. Organization and environmental regulation of the Pseudomonas syringae pv. syringae 61 hrp cluster. J Bacteriol. 1992;174:1734–1741. doi: 10.1128/jb.174.6.1734-1741.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]