Abstract
This study collected the stools of 10-km open-water swimmers after race and probiotic supplementation, and 16S rRNA sequencing and metabolomic analysis were performed to clarify their intestinal microbiota characteristics. The findings revealed a relatively high proportion of Firmicutes in all the athletes. Firmicutes in female athletes were significantly higher after probiotic supplementation. The intestinal microbiota of athletes was closely associated with the pathways of exercise against cancer, exercise against aging, exercise for improving cognition, sphingolipid metabolism and endocrine resistance. Future research should focus on the relationship between Firmicutes and Proteobacteria with super class metabolites in athletes. This report initially explored the changes in intestinal microbiota involved in metabolic pathways in athletes after race and after probiotic supplementation and provided a theoretical basis for the further improvement of the monitoring of their physical function after race and selection of nutritional strategies during exercise training.
Keywords: 10-km open-water swim, Athletes, Intestinal microbiota, Metabolomics
1. Introduction
The human gastrointestinal tract is home to numerous microbes that outnumber even the total number of cells in the body[1]. The intestinal microbiota and human beings have a symbiotic relationship, and this symbiotic system is mutually beneficial for both parties[2]; J [3]. Athletes are a particular category of people. Compared with populations with no exercise experience, athletically experienced individuals have a more relatively complex intestinal microbiota and a relatively more stable gut microbiota structure, which means that the intestinal microbiota of athletes is of more valuable to investigate[4]. The 10-km open-water swimming (OWS) is a typical endurance race that is held in rivers, lakes, oceans or water channels[5]. Unlike regular general training and mock races, during official races, athletes must ensure proper starting conditions, including neural excitability, warm-up level and psychological factors, which are related to the gut-brain axis[[6], [7], [8]]. Intestinal microbiota could stimulate serotonin, dopamine and other neurotransmitters which control the hypothalamic axis in athletes to modulate altered physical and emotional states[9]. It has also been shown that proposed interoceptomimetic molecules that stimulate the transmission of gut signals to the brain may enhance athletic performance in mice[10]. Having knowledge of the characteristics of and determining the changes in athletes’ intestinal microbiota during a race can provide targeted guidance for their post-competition recovery and help them achieve their best condition quickly. However, no detailed investigations have been conducted on the composition and characteristics of the intestinal microbiota of OWS athletes.
Probiotics are dietary supplements that can promote the health of the body[11]. In humans, the combination of Lactobacillus gasseri OLL2809 and α-lactalbumin can alleviate the fatiguing effects on university-student athletes after strenuous exercise while enhancing their immune function[12]. The incidence of upper respiratory tract infections decreased after 12 weeks of multi-species probiotic supplementation in 33 winter-trained athletes[13]. These observations indicate that probiotic supplementation is of great research value for the intestinal microbiota of athletes. However, current studies on the intestinal microbiota of OWS athletes after probiotic supplementation are a few, particularly in 10-km OWS, and there are no current studies related to the athletic performance benefits of probiotic supplementation [4,14]. Therefore, in this study, through 16S rRNA sequencing and metabolomics techniques, 15 swimmers from Shandong, China's swimming team were used to analyse the changes in intestinal microbiota before and after a 10-km OWS race and after probiotic supplementation. The results provide valuable clues for the monitoring of the physical function of OWS athletes and selection of their nutritional strategies after a race.
2. Materials and methods
2.1. Subjects
In this study, 15 elite athletes (males/females, 8/7) from Shandong, China's OWS team were selected as subjects (age: 18.32 ± 4.41 years for males and 18.04 ± 2.96 years for females, BMI: 20.91 ± 2.36 for males and 20.81 ± 1.07 for females). Faecal samples were collected on regular training days as a control group. Probiotic mixtures were supplemented on five consecutive training days, and faeces were collected as a probiotic supplementation group, with two servings per day and a practical volume of 2 g each (mixture probiotic formula: inulin, oligofructose, lactitol, sterilised fermented carrot (vegetable juice solid drink) added at 7 %, Bifidobacterium lactis HN019, Lactobacillus acidophilus NCFM, Lactobacillus plantarum Lp-115, Bifidobacterium longum Bl-05 and calcium silicate). Athlete meals during probiotic supplementation include carbohydrate intake: 50%–60 %, protein intake: 10%–20 % and fat intake: 20%–30 %. On the race day, the athletes maintained their regular diet and water intake. After the 10-km open-water swim, the faeces from the first defecation of the athletes were collected as the experimental group (within 8 h after the race) and stored at −80 °C immediately after collection. Participants have completed informed consent form to indicate that they are participating in the study of their own free will.
2.2. Faecal DNA extraction and specific quantitative real-time polymerase chain reaction
The total DNA of the microbiota in the faecal samples was extracted using OMEGA Soil DNA Kit (Omega Bio-Tek, Norcross, GA, USA), and the bacterial 16S rRNA gene V3–V4 region with a length of 468 bp was selected for amplification. The 16S rRNA V3–V4 region specific primers were as follows: 338F: 5′- + ACTCCTACGGGAGGCAGCA-3′, 806R: 5′-GGACTACHVGGGTWTCTAAT-3′. Library construction was performed using Illumina's TruSeq Nano DNA LT Library Prep Kit, and 2 × 250 bp double-end sequencing was performed using a NovaSeq 6000 SP Reagent Kit (500 cycles).
High-resolution mass spectrometry data acquisition was performed using a high-performance liquid chromatograph (Thermo Scientific™ Dionex™ ΜltiMate™ 3000 Rapid Separation LC). The ion source parameters were as follows: spray voltage: 2500 v, capillary temperature: 320 °C; ion source: HESI. Mobile phase A was acetonitrile/water (60/40), and mobile phase B was isopropanol/acetonitrile (90/10). Both A and B contained 0.1 % formic acid and 10 mmol/L ammonium, and the column was Waters ACQUITY UPLC BEH C8 (1.7 μm 2.1 mm*100 mm) for up sequencing.
2.3. Statistics
Sequence splicing was performed using FLASH[15]. Then, the spliced sequences were quality controlled using Mothur [16]; sequences with a sequence clustering similarity higher than 97 % were clustered into one operational taxonomic unit (OTU) using USEARCH based on sequence similarity using the SILVA [17] database as a reference, and systematic classification of OTU sequences was conducted based on Bergey's taxonomy; Metastats [18] and linear discriminant analysis effect size were used to test for differences in the relative abundance of species between groups, and PICRUST [19] was used to compare the OTUs' functional genes based on the Kyoto Encyclopedia of Genes and Genomes (KEGG) database.
The raw files (.uep) obtained from the mass spectrometry assay were imported into Progenesis QI 2.3 software, and data alignment was performed on different samples based on the retention time deviation of 0.2 min and mass deviation of 5 ppm, coefficient of variance of 30 %, signal-to-noise ratio = 3 and minimum signal intensity = 100000. Summed ions and other information for peak extraction anti-fold product were obtained while integrating target ions by molecular ion peaks and fragment ions for molecular formula prediction and comparison with the database. Normalisation and labelling matrix production were performed, and finally, the identification and quantitative results of the data were obtained.
3. Result
3.1. High-throughput sequencing results of intestinal microbiota samples from 10-km open-water swimmers
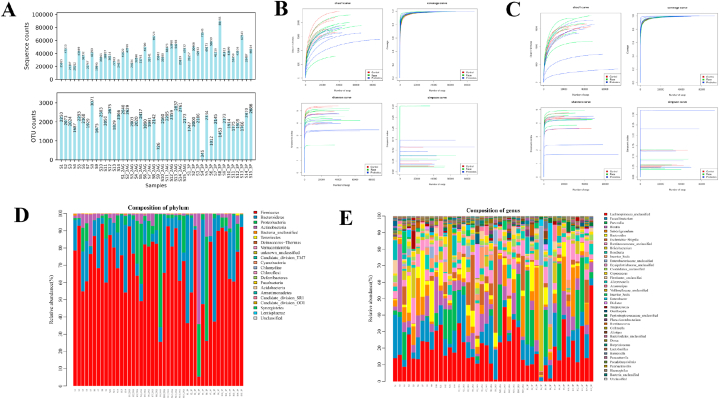
A total of 1772486 tags were available for all intestinal microbiota samples, and the sequences were clustered into OTUs based on sequence similarities with a similarity threshold of 97 %, i.e. sequences with a similarity higher than 97 % were clustered into one OTU. Fig. 1A shows the statistical plots of OTUs versus several sequences for each sample. Combined with the analysis of alpha diversity index values, no statistical difference was observed Shannon, Simpson and Chao indexes (P > 0.05) (Fig. 1B and C). And the compositions of the intestinal microbiota of each athlete after the race and probiotic supplementation in terms of phylum and genus were as follows (Fig. 1D and E).
Fig. 1.
Quality control of intestinal microbiota of all athletes. A: Distribution of the number of OTUs of samples before and after the 10-km open water swimming race; B: Alpha diversity sparsity curve of the intestinal microbiota of male athletes. The horizontal axis is the number of sequences, and the vertical axis is each diversity index; C: Alpha diversity sparsity curve of the intestinal microbiota of female athletes. The horizontal axis is the number of sequences, and the vertical axis is each diversity index; D: The horizontal distribution of the intestinal microbiota phylum of all athletes; E: The horizontal distribution of the intestinal microbiota genus of all athletes.
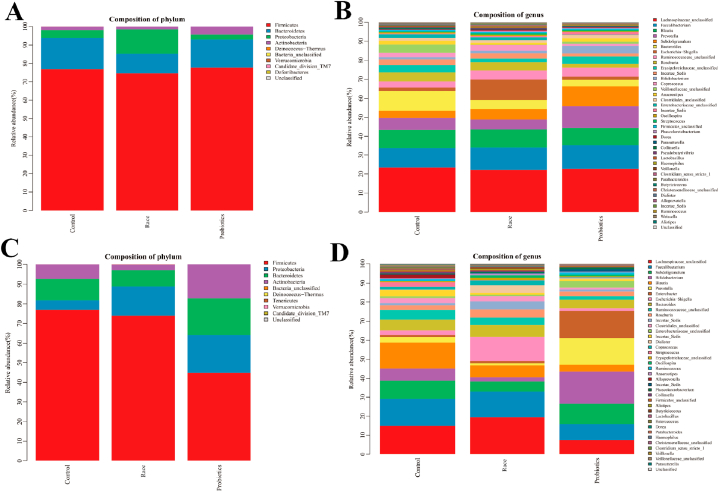
The intestinal microbiota of male athletes mainly included Firmicutes, Bacteroides, Proteobacteria and Actinobacteria, accounting for 99.93 % of the total microbiota. The dominant phyla of male athletes were Firmicutes and Bacteroides, which accounted for 93.84 % of the total intestinal microbiota. The sub-dominant phyla were Proteobacteria and Actinobacteria, which accounted for 6.09 % of the total intestinal microbiota, and the bacteria of other inferior phyla were less than 0.1 % of the total microbiota (Fig. 2A and Table 1).
Fig. 2.
Intestinal microbiota distribution of athletes. A&B: histogram of microbiota distribution after race and probiotic supplementation in male athletes (A: phylum level; B: genus level); C&D: Histogram of microbiota distribution after race and probiotic supplementation in female athletes (C: phylum level; D: genus level). The horizontal axis is the experimental grouping, and the vertical axis is the relative abundance of each taxonomic entry (taxon). The part of the histogram with less than 100 % is the taxa identified as unclassified at this taxonomic level.
Table 1.
Variation in the intestinal microbiota of male athletes after race and probiotic supplementation.
| Control | Race | Probiotics | Variation rate (C vs R) | Variation rate (C vs P) | |
|---|---|---|---|---|---|
| Firmicutes | 76.92 % | 74.62 % | 77.72 % | −2.99 % | 1.04 % |
| Bacteroidetes | 16.92 % | 10.55 % | 15.28 % | −37.66 % | −9.72 % |
| Proteobacteria | 4.17 % | 13.25 % | 2.65 % | 217.35 % | −36.52 % |
| Actinobacteria | 1.91 % | 1.52 % | 4.34 % | −20.33 % | 127.03 % |
| Others | 0.07 % | 0.06 % | 0.01 % | −15.19 % | −88.05 % |
Note: ‘Control’, ‘Race’, and ‘Probiotics’ represent the percentage of the corresponding phylum to the total microbiota in each group, respectively; ‘Variation rate (C vs R)’ represents the relative rate of change of the corresponding phylum to the total microbiota after the race; and ‘Variation rate (C vs P)’ represents the relative rate of change of the corresponding phylum to the total microbiota after probiotics supplementation.
The intestinal microbiota of female athletes mainly comprised Firmicutes, Bacteroides, Proteobacteria and Actinobacteria, accounting for 99.96 % of the total microbiota. The dominant phyla of female athletes were Firmicutes and Bacteroides, which accounted for 87.73 % of the total intestinal microbiota. The sub-dominant phyla were Proteobacteria and Actinobacteria, which accounted for 12.23 % of the total intestinal microbiota, and the bacteria of other inferior phyla were less than 0.1 % of the total microbiota (Fig. 2C and Table 2).
Table 2.
Variation in the intestinal microbiota of female athletes after race and probiotic supplementation.
| Control | Race | Probiotics | Variation rate (C vs R) | Variation rate (C vs P) | |
|---|---|---|---|---|---|
| Firmicutes | 76.91 % | 74.02 % | 44.69 % | −3.76 % | −41.89 % |
| Bacteroidetes | 10.83 % | 8.14 % | 18.76 % | −24.78 % | 73.29 % |
| Proteobacteria | 4.90 % | 14.82 % | 19.37 % | 202.27 % | 295.06 % |
| Actinobacteria | 7.32 % | 2.92 % | 17.16 % | −60.18 % | 134.41 % |
| Others | 0.04 % | 0.10 % | 0.01 % | 153.06 % | −69.10 % |
Note: ‘Control’, ‘Race’, and ‘Probiotics’ represent the percentage of the corresponding phylum to the total microbiota in each group, respectively; ‘Variation rate (C vs R)’ represents the relative rate of change of the corresponding phylum to the total microbiota after the race; and ‘Variation rate (C vs P)’ represents the relative rate of change of the corresponding phylum to the total microbiota after probiotics supplementation.
3.2. Distribution and changes in intestinal microbiota in 10-km open-water swimmers after race
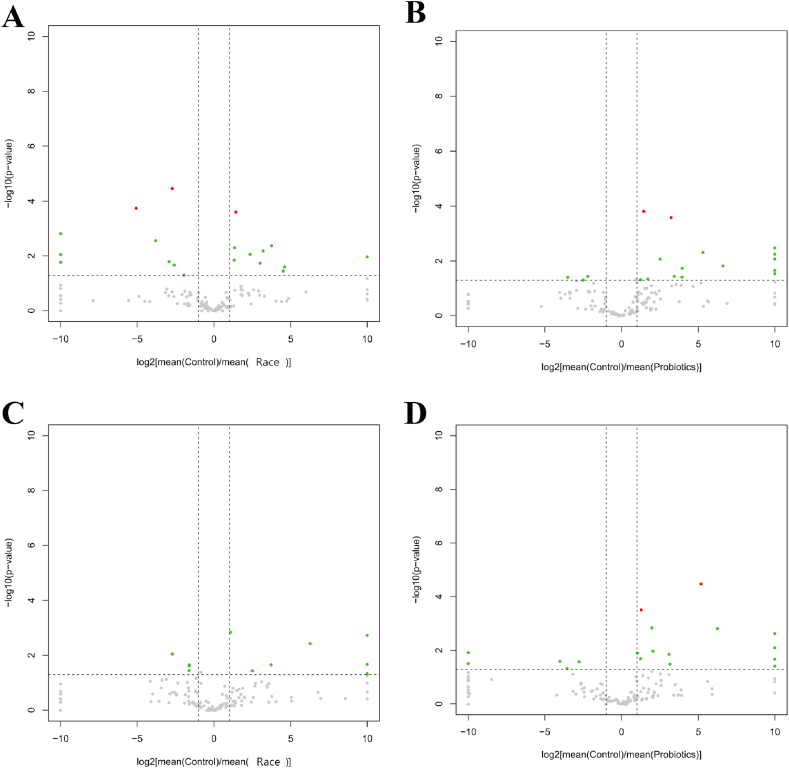
Compared with the control group, the variation rate of Proteobacteria was 217.35 %, and the variation rate of Bacteroides and Actinobacteria were −37.66 % and −20.33 % in male athletes. A total of 146 genera of bacteria were detected, with 19 genera presenting significant differences (Fig. 2, Fig. 3A and Table 3). Similarly, the female athletes showed the variation rate of Proteobacteria was 202.27 % and the variation rate of Bacteroides and Actinobacteria were −24.78 % and −60.18 %. A total of 162 bacterial genera were detected, with 15 genera presenting significant differences (Fig. 2, Fig. 3B and Table 3).
Fig. 3.
Differential microbiota volcano plots. A&B: Differential microbiota volcano plots for male athletes after race and probiotic supplementation. (A: [mean(C)/mean(AR)]; B: [mean(C)/mean(AP)]); C&D: Volcano plot of differential microbiota in female athletes after race and probiotic supplementation (A: [mean(C)/mean(AR)]; B: [mean(C)/mean(AP)]) Horizontal axis: log2[mean(A)/mean(B)] is the mean relative abundance of a taxon in group A divided by the mean relative abundance in group B, and then the logarithm of the base of 2 is taken. The vertical axis: log10(p-value) is the logarithm of the p-value with a base of 10 and a negative value; red dots: taxons that satisfy |log2[mean(A)/mean(B)]|>1 and p-value<0.05 and q-value<0.05 (advanced criteria); green dots: taxons that satisfy only |log2[mean(A)/mean(B)]|>1 and p-value<0.05 (primary criteria); grey dots: taxons that do not satisfy any of the above conditions.
Table 3.
Differential bacterial genes in male athletes after race and probiotic supplementation.
| Male |
C vs R |
Total: 146 | Male |
C vs P |
Total: 145 |
|---|---|---|---|---|---|
| Firmicutes |
2 |
Firmicutes |
7 |
||
| Bacteroidetes | 2 | Bacteroidetes | 2 | ||
| Proteobacteria | 4 | Diff: 19 | Proteobacteria | 2 | Diff: 19 |
| Actinobacteria | 8 | Actinobacteria | 5 | ||
| Others | 3 | Others | 3 |
Note: ‘C vs R’ or ‘C vs P’ represents the change in microbiota after race or probiotic supplementation, respectively; ‘Total’ means the number of genus detected in total, and ‘Diff’ means the number of differences in significant genus.
3.3. Distribution and changes in intestinal microbiota in 10-km open-water swimmers after probiotic supplementation
The most variation rate changes in the proportion of Actinobacteria were observed in male athletes. A total of 145 genera of bacteria were detected, with 19 genera presenting significant differences (Fig. 2, Fig. 3C and Table 4). Compared with male athletes, the range of phylum changes was more prominent in female athletes, with a substantial reduction in Firmicutes with a change rate of −41.89 % (P = 0.018). Meanwhile, the variation rates were 73.29 %, 295.06 % and 134.41 % for Bacteroidetes, Proteobacteria and Actinobacteria, respectively. A total of 163 genera of bacteria were detected, with 23 genera presenting significant differences (Fig. 2, Fig. 3D and Table 4).
Table 4.
Differential bacterial genes in female athletes after race and probiotic supplementation.
| Female |
C vs R |
Total: 162 | Female |
C vs P |
Total: 163 |
|---|---|---|---|---|---|
| Firmicutes |
5 |
Firmicutes |
7 |
||
| Bacteroidetes | 1 | Bacteroidetes | 2 | ||
| Proteobacteria | 1 | Diff: 15 | Proteobacteria | 0 | Diff: 23 |
| Actinobacteria | 7 | Actinobacteria | 10 | ||
| Others | 1 | Others | 4 |
Note: ‘C vs R’ or ‘C vs P’ represents the change in microbiota after race or probiotic supplementation, respectively; ‘Total’ means the number of genus detected in total, and ‘Diff’ means the number of differences in significant genus.
3.4. Metabolite analysis of intestinal microbiota in 10-km open-water swimmers after race
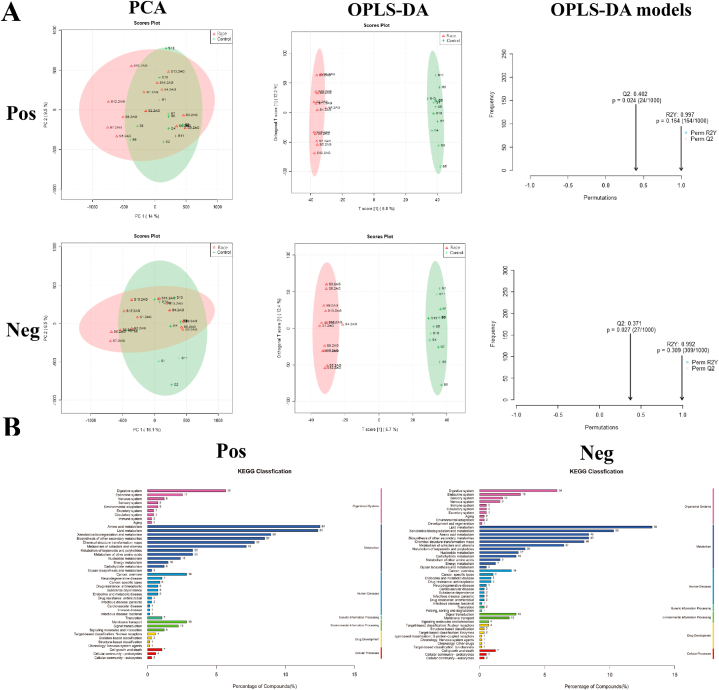
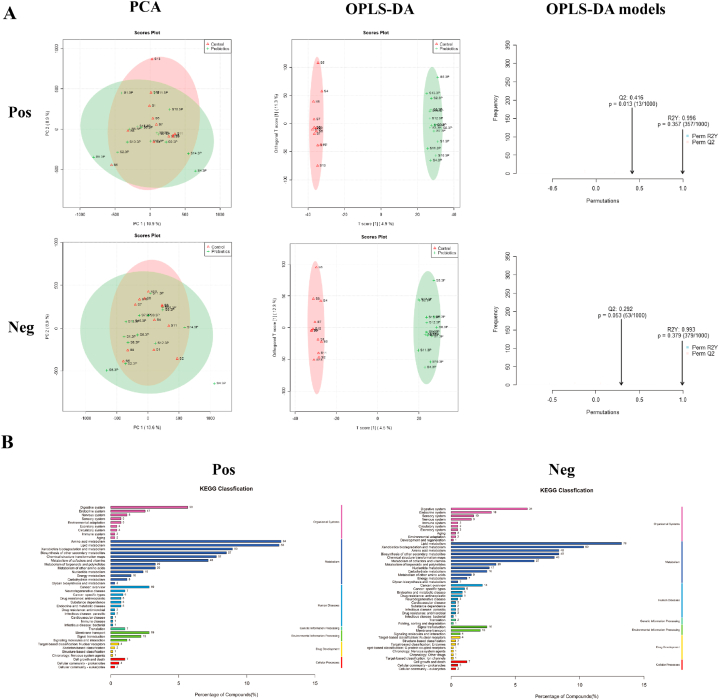
During the metabolomic analysis of the control and post-race athlete samples, the quality control samples showed central aggregation in principal component analysis (PCA), which indicates the good instrumental reproducibility of the samples during analysis. In addition, the results of PCA based on metabolomic data did not show considerable separation in positive and negative ion modes. Differences between groups were further distinguished using the orthogonal partial least squares discriminant analysis model, and the model fit was assessed by Q2 (cum) and R2Y (cum) (Fig. 4A). The post-race data indicated an R2Y value of 0.997 and a Q2 value of 0.402 in the positive ion mode and an R2Y value of 0.992 and a Q2 value of 0.371 in the negative ion mode. The positive and negative ion modes identified 8502 and 7228 annotated metabolites. After functional classification and statistical analysis through the KEGG database, the identified metabolites can be classified into organismal systems, metabolism, human diseases, genetic information processing, environmental information processing, drug development and cellular processes, among which metabolites related to the circulatory system and metabolism accounted for relatively high percentages (Fig. 4B).
Fig. 4.
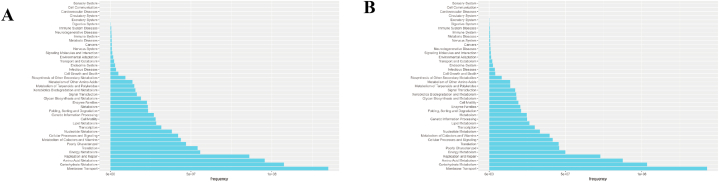
Assessment and statistical analysis of metabolomic identification results of the intestinal microbiota of athletes after the race. A: PCA and OPLS-DA scores in positive and negative ion mode and model prediction plots; (PCA: horizontal coordinate PC1 and vertical coordinate PC2 in the figure indicate the scores of the first and second-ranked principal components, respectively; different colored scatter points indicate samples from different experimental subgroups; ellipses are 95 % confidence intervals; OPLS-DA: R2Y and Q2 represent the variables explained by the model and the predictability of the model, respectively). B: Annotated and classified map of metabolite KEGG functions; (Note: horizontal coordinates are the number of each metabolite type as a percentage of the total number, left vertical coordinates are subclasses, right vertical coordinates are Class).
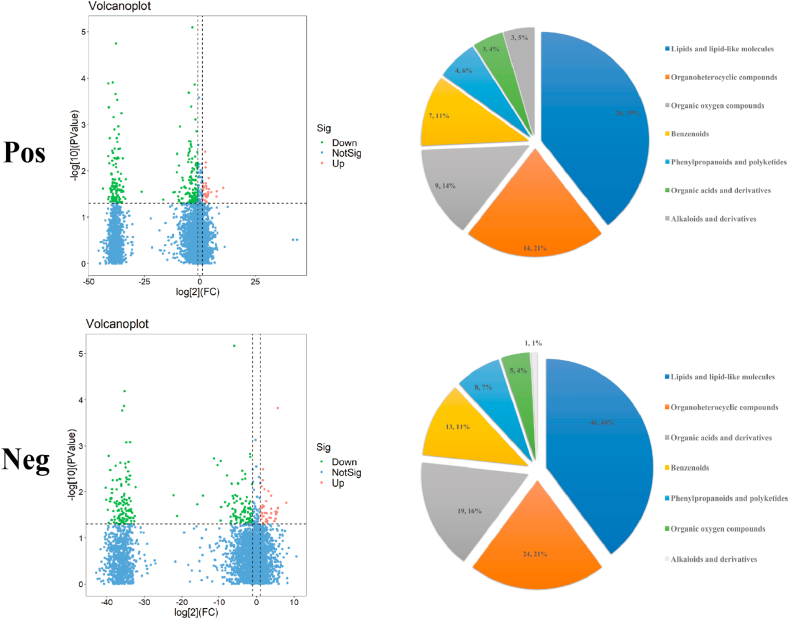
The metabolites identified after the race were screened using variable importance in projection (VIP) > 1 and P < 0.05 as criteria. Cluster analysis was performed on the screened differential metabolites, and 67 and 117 annotated differential metabolites were identified in positive and negative ion modes, respectively (Supplementary Data 1). According to the Human Metabolome Database, these metabolites were classified into lipids and lipid-like molecules, organoheterocyclic compounds, organic acids and derivatives; benzenoids, phenylpropanoids and polyketides; organic oxygen compounds, alkaloids and derivatives (Fig. 5).
Fig. 5.
Differential metabolites and differential metabolic pathways in the intestinal microbiota of athletes after race. Volcano plot and classification of differential metabolite changes in athletes' microbiota after race (Horizontal dashed line indicates the dashed line corresponding to P = 0.05 taken logarithmically, a vertical dashed line indicates the dashed line corresponding to FC threshold taken logarithmically. The dots meeting above the horizontal dashed line and on both sides of the vertical dashed line are also highlighted, where the red highlight on the right side indicates upward adjustment, and the green highlight on the left side indicates downward adjustment. (Blue dots indicate that the set threshold requirement is not met); Pie charts correspond horizontally to the categorical share of all differential metabolites in the positive/negative ion model (Categories include lipids and lipid-like molecules, organoheterocyclic compounds, organic acids and derivatives; benzenoids, phenylpropanoids and polyketides; organic oxygen compounds, alkaloids and derivatives).
3.5. Metabolite analysis of intestinal microbiota in 10-km open-water swimmers after probiotic supplementation
Post-probiotic supplementation indicated an R2Y value of 0.996 and a Q2 value of 0.416 in the positive ion mode and an R2Y value of 0.993 and a Q2 value of 0.292 in the negative ion mode. Thus, the established model had a relatively good fit and reliable prediction for subsequent screening of differential metabolites (Fig. 6A). The positive and negative ion modes identified 8501 and 7229 annotated metabolites, respectively. After functional classification and statistical analysis through the KEGG database, the identified metabolites can be classified into organismal systems, metabolism, human diseases, genetic information processing, environmental information processing, drug development and cellular processes, among which metabolites related to the circulatory system and metabolism accounted for relatively high percentages (Fig. 6B).
Fig. 6.
Assessment and statistical analysis of metabolomic identification results of intestinal microbiota in athletes after probiotic supplementation. A: PCA and OPLS-DA scores and model prediction plots in positive and negative ion mode; B: Annotated and classified map of metabolite KEGG functions.
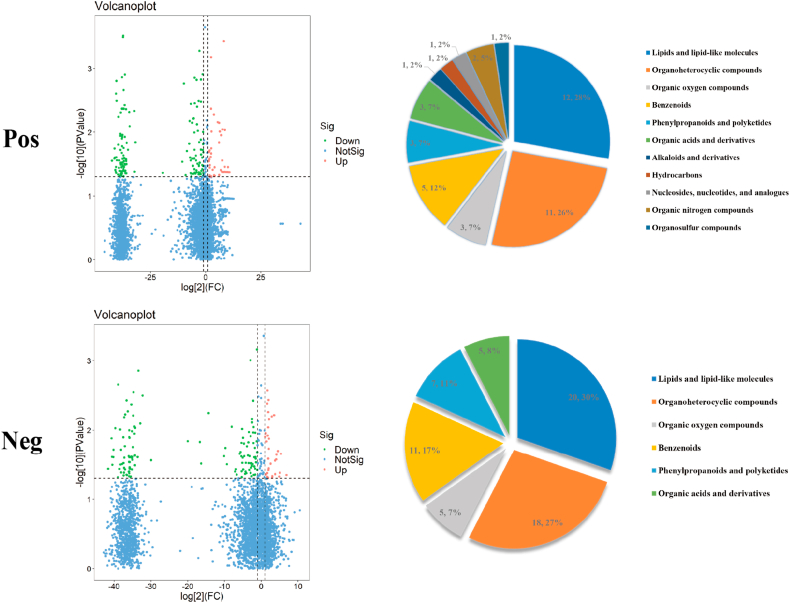
The metabolites identified after probiotic supplementation were screened using VIP >1 and P < 0.05 as criteria. The screened differential metabolites were subjected to cluster analysis, and 43 and 87 annotated differential metabolites were identified in the positive and negative ion modes, respectively (Supplementary Data 1). The positive metabolites were classified as hydrocarbons, nucleosides, nucleotides and analogues; organic nitrogen compounds, organosulfur compounds, lipids and lipid-like molecules; organoheterocyclic compounds, organic acids and derivatives; benzenoids, phenylpropanoids and polyketides; organic oxygen compounds, alkaloids and derivatives. By comparison, the negative metabolites were classified as lipids and lipid-like molecules, organoheterocyclic compounds, organic acids and derivatives; benzenoids, phenylpropanoids and polyketides; organic oxygen compounds (Fig. 7).
Fig. 7.
Differential metabolites and differential metabolic pathways of the intestinal microbiota of athletes after probiotic supplementation. Volcano diagram and classification of differential metabolite changes in athletes' microbiota after probiotic supplementation; Pie charts correspond horizontally to the categorical share of all differential metabolites in the positive/negative ion model.
3.6. Prediction of gut microbiota function and changes in metabolic pathways in male and female athletes in the same conditions
3.6.1. Differences in metabolic pathways between male and female athletes
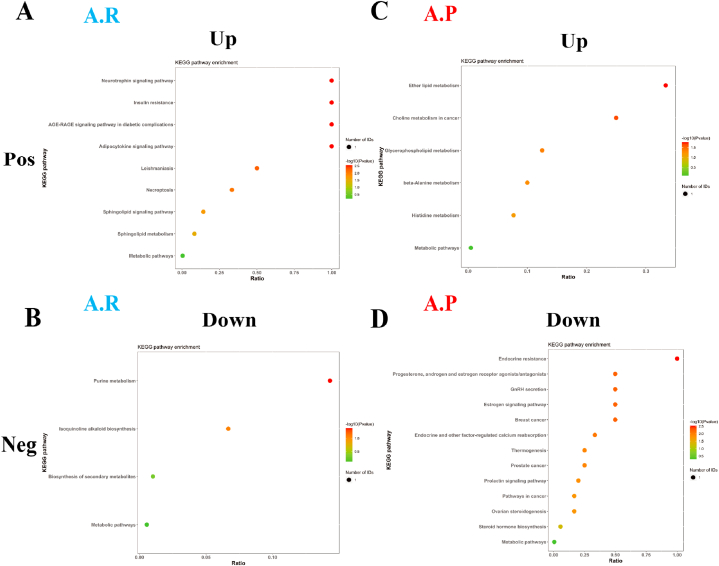
Metabolic pathway enrichment analysis of differential metabolites was performed through the KEGG database to analyse further the differences in the metabolic pathways of the intestinal microbiota of male and female athletes. After the race, enrichment was upregulated in nine metabolic pathways in the positive ion mode, including neurotrophin signalling pathway, adipocytokine signalling pathway, AGE-RAGE signalling pathway in diabetic complications and insulin resistance. Four metabolic pathways, including purine metabolism, biosynthesis of secondary metabolites, etc. (Fig. 8A and B and Supplementary Data 2), were downregulated in negative ion mode.
Fig. 8.
Differential metabolic pathways of intestinal microbiota in male and female athletes after race and probiotic supplementation. A&B: bubble diagram of metabolic pathways of intestinal microbiota differences between male and female athletes after race; C&D: bubble diagram of metabolic pathways of intestinal microbiota differences between male and female athletes after probiotic supplementation. (Note: horizontal coordinates are the number of each metabolite type as a percentage of the total number, left vertical coordinates are subclasses, correct vertical coordinates are Class.
After supplementation with probiotics, enrichment was upregulated in 6 metabolic pathways in positive ion mode, including ether lipid metabolism, glycerophospholipid metabolism, histidine metabolism, etc. 13 metabolic pathways were upregulated, including endocrine resistance, oestrogen signalling pathway, gonadotropin-releasing hormone secretion, etc., in negative ion mode (Fig. 8C and D and Supplementary Data 2).
3.6.2. Prediction of gut microbiota function in male and female athletes
The predicted KEEG metabolic pathways involving 41 functional genes were obtained by calculating the 16S rRNA gene abundance values for each group of samples based on the PCRUSI abundance table and their comparison with the KEGG database. The intestinal microbiota of male and female athletes had high proportions of genes involved in membrane transport, carbohydrate metabolism, amino acid metabolism and energy metabolism (Fig. 9A and B).
Fig. 9.
Functional enrichment of intestinal microbiota and correlation of differential microbiota with metabolite species in male athletes after race and probiotic supplementation. A: Gene functional profile of the intestinal microbiota in male athletes (vertical coordinates are gene functions, horizontal coordinates are functional abundance.); B: Gene functional profile of the intestinal microbiota in female athletes.
3.6.3. Correlation of differential microbiota with metabolite species in male and female athletes
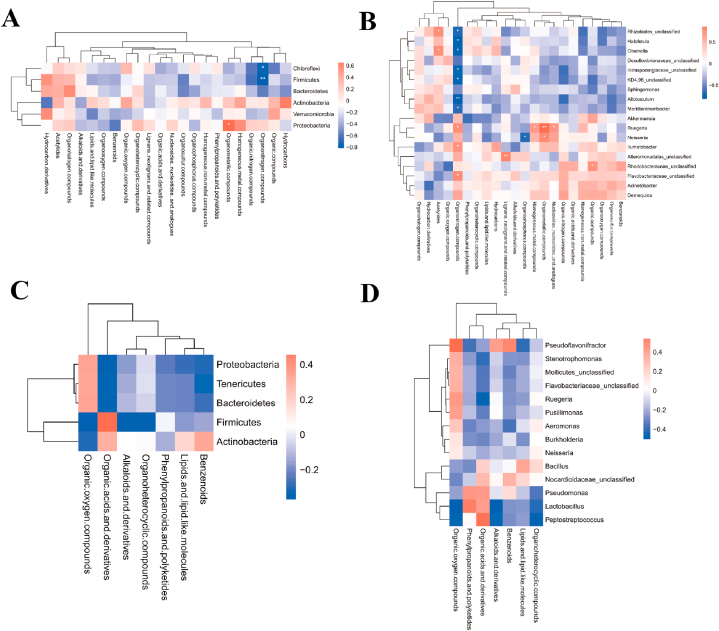
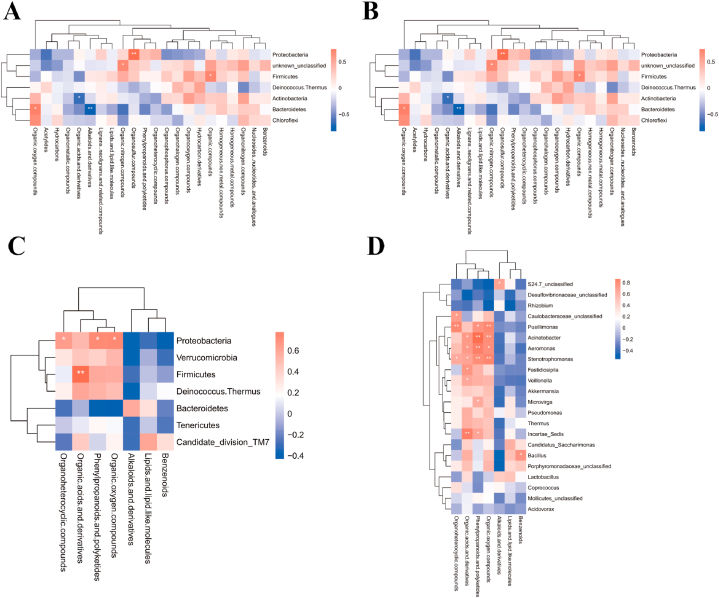
After the race, a joint analysis of differential genera and metabolites revealed a highly positive correlation between Proteobacteria and Organometallic compounds (P = 0.0217) in male athletes and a highly negative correlation between Firmicutes and Organonitrogen compounds (P = 0.0011) (Fig. 10A and B and Supplementary data 3). No significant correlation was found between the phylum and genus of female athletes and super class after the race (Fig. 10C and D and Supplementary data 3).
Fig. 10.
Correlation of differential microbiota with metabolite species in male and female athletes after race. A&B: Heat map of correlation between differential microbiota and metabolite super class in male athletes after race (A: phylum level; B: genus level); C&D: Heat map of correlation between differential microbiota and metabolite super class in female athletes after race (C: phylum level; D: genus level (The results of correlation analyses are generally expressed as correlation coefficients, and the values of correlation coefficients are generally between −1 and 1. The closer the absolute value of correlation coefficients is to 1, the stronger the correlation is; the closer the absolute value is to 0, the weaker the correlation is). The vertical coordinates are the names of microorganisms in the "phylum" or "genus," and the horizontal coordinates are the names of metabolites. The red color represents a positive correlation, the blue color represents a negative correlation, and the heat map color in the upper right corner represents a strong (dark) and weak (light) correlation).
After probiotic supplementation, Proteobacteria was highly positively correlated with organosulfur compounds (P = 0.0059), and Bacteroidetes was highly negatively correlated with alkaloids and derivatives (P = 0.0010) in male athletes. Firmicutes was highly positively correlated with organic acids and derivatives (P = 0.0030) in female athletes (Fig. 11A–D and Supplementary data 3).
Fig. 11.
Correlation of differential microbiota with metabolite species in male and female athletes after probiotics supplementation. A&B: Heat map of correlation between differential microbiota and metabolite super class in male athletes after probiotics supplementation (A: phylum level; B: genus level); C&D: Heat map of correlation between differential microbiota and metabolite super class in female athletes after probiotics supplementation (C: phylum level; D: genus level).
4. Discussion
4.1. Intestinal microbiota characteristics of 10-km open-water swimmers
The 10-km OWS, also known as the ‘marathon’ swim, is a typical endurance sport[20]. Intestinal microbiota can improve endurance exercise performance by breaking down polysaccharides in food into short-chain fatty acids (SCFAs), which can be used as a source of energy for the liver and skeletal muscle to maintain blood glucose stability[21]. However, current studies have not provided an in-depth understanding of human intestinal microbiota. Especially, the characteristics of microbiota in athletes of different sports have not been systematically summarised.
According to Fig. 2A and Table 1, Firmicutes was the absolute dominant phylum in male athletes in daily training state, and it accounted for 76.92 % of the total bacterial microbiota, followed Bacteroidetes (16.92 %). The main inferior phyla were Proteobacteria and Actinobacteria, and they accounted for 4.17 % and 1.91 % of the total bacterial microbiota, respectively. The main disadvantageous phyla were Proteobacteria and Actinobacteria (4.17 % and 1.91 % of the total population, respectively). Firmicutes was the absolute dominant phyla in female athletes, and it accounted for 76.91 % of the total microbiota, followed by Bacteroidetes at 10.83 %. The main inferior phyla were Actinobacteria and Proteobacteria, and they accounted for 7.32 % and 4.90 % of the total microbiota, respectively. Male and female distributions in swimming sports were similar, but female athletes had more Actinobacteria microbiota than male athletes. In the intestinal microbiota system of the general population, Firmicutes is the dominant phylum (about 60 % of the total microbiota), followed by Bacteroidetes (20 %), whereas the abundances of Actinobacteria, Proteobacteria and Verrucomicrobia are relatively low[15]. Our data suggest that the characteristics of the intestinal microbiota composition in this type of movement differ significantly from those in the general population.
4.2. Alterations in the intestinal microbiota of athletes after race
According to Fig. 2C and Table 1, the variation rate of Firmicutes abundance were −2.99 % (P > 0.05) and −3.76 % (P > 0.05) in male and female athletes after the race, respectively. As the main dominant phylum, Firmicutes is the basis for athletes to train and maintain metabolic homeostasis continuously[22,23]. The results of this study suggest that the abundance of Firmicutes in this type of athletes almost did not change after the race, which suggests that the stable structure of the microbiota comprised by the main dominant phylum in endurance athletes is a vital indicator reflecting their athletic quality. Firmicutes is a part of the immune response and attenuates the inflammatory response of an organism[24]. At the phylum level, in the male athletes, after a race, a highly negative correlation between Firmicutes and Organonitrogen compounds (P = 0.0011) (Supplementary data 3). Organonitrogen chemicals are essential in many aspects of modern life[25], we have not found any relevant research on intestinal microbiota and Organonitrogen compounds, but there have been relevant studies indicating that biomass has begun to gradually replace chemical energy sources, and that joint research on intestinal microbiota and Organonitrogen compounds may be a future research direction. In addition, the variation rate in the abundance of Bacteroidetes in male and female athletes after the race were −37.66 % (P > 0.05) and −24.78 % (P > 0.05), respectively. Similar to Firmicutes, Bacteroidetes includes beneficial bacteria in humans and plays a crucial role in the stability of the intestinal microbiota structure[26,27].
The present study confirmed that the variation rate in the abundance of Proteobacteria reached 217.35 % (P > 0.05) and 202.27 % (P > 0.05) in male and female athletes after the race, respectively. In addition, we found a high positive correlation between Proteobacteria and organometallic compounds (Fig. 10A), a specific class of metal complexes, in male athletes after race[28]. Organometallic compounds have outstanding physicochemical properties. These compounds are usually associated with catalysis and are critical components of numerous drugs, such as anticancer drugs, antimalarials and enzyme inhibitors in biology[28,29]. Proteobacteria are often considered a representative of an imbalance in the intestinal microbiota, which is triggered by inflammation, metabolic dysregulation and other abnormalities[30]. We also observed that the rate of change in Actinobacteria abundance reached −20.33 % (P > 0.05) and −60.18 % (P > 0.05) in male and female athletes after the race, respectively. Actinobacteria accumulate triglyceride-based neutral lipids[15], and their abundance in mice decreased sharply after exercise intervention [31]. The present study confirmed that the abundance of Actinobacteria in athletes decreased after the race. We assessed the training and fatigue of athletes based on the change in the ratio of Proteobacteria to Actinobacteria abundance before and after the race (male athletes: pre-race: 2.18, post-race: 8.69; female athletes: pre-race: 0.67, post-race: 5.08).
Our data indicate the significant downregulation of the prolactin signalling pathway, cancer and prostate cancer signalling pathways in athletes and significant upregulation of the phosphatidylinositol signalling system after the race (Fig. 8A and B and Supplementary data 2). Prolactin is a typically multifunctional hormone; circulating prolactin concentrations increase with exercise, and the increase is about proportional to the intensity of exercise[32]. The prolactin response to sub-maximal exercise intensity in men is diminished after training but enhanced after performing maximal exercise intensity training; prolactin is one of the critical factors of the hypothalamic–pituitary–thyroid axis. Growth hormone–insulin-like growth factor (IGF), thyroid and prolactin are interconnected to regulate the body's immune response[33]. In the present study, prolactin signalling was downregulated after the race, which suggests that intensity of 10-km OWS may affect the athletes' intestinal microbiota involved in immune response. The phosphatidylinositol signalling pathway is commonly associated with anti-aging effects[34]. Swimming stimulates the IGF1–phosphatidylinositol-3 kinase–Akt signalling pathway in the rat hippocampus and thereby achieves an anti-aging effect(J. Y [35]. The present study detected an upregulation of the phosphatidylinositol signalling pathway, which suggests that intestinal microbiota may be involved in the metabolic process of exercise anti-aging. In addition, the results of this study imply that metabolic pathways associated with cancer are significantly downregulated after race, which also provides research evidence for the involvement of metabolic processes of intestinal microbiota in exercise against cancer. The neurotrophin signalling pathway was one of the most pronounced pathways with differential metabolic pathway changes in male and female athletes after the race. Exercise can effectively improve human cognitive function, especially endurance exercise, which can increase the hippocampal size and blood flow and enhance synaptic plasticity in humans[36,37]. The expression of brain-derived neurotrophic factor (BDNF), which interacts with Fibronectin type III domain-containing protein 5 (FNDC5) and Peroxisome proliferator-activated receptor gamma coactivator 1-alpha (PGC-1α) in the periphery, can be induced during endurance exercise [38]. In addition, the neurotrophin signalling pathway is an essential pathway in the intestinal–brain axis that regulates human emotions[39]. Our data shows that athletes need not only physical function monitoring after significant races but also targeted adjustment of their psychological state based on gender differences as an essential indicator of excellent performance.
4.3. Alterations in the intestinal microbiota of athletes after probiotic supplementation
After probiotic supplementation, the change rates of phylum abundance in male and female athletes were significantly different, and male athletes retained the original distribution ratio of intestinal phylum and relatively optimised original microbiota structure. We observed that Proteobacteria, which are often found as dietary fibre in several natural vegetables or food additives [40], in male athletes were highly positively correlated with organosulfur compounds, which are essential compounds for antibacterial, antiparasitic, antiviral, immunomodulatory and anticancer purposes [41]. Thus, probiotic supplementation is a fundamental cause of the relationship between Proteobacteria and organosulfur compounds in male athletes; organosulfur compounds benefit intestinal health in mice, and ingestion of these compounds can effectively regulate the intestinal imbalance of intestinal microbiota and treat colitis in mice[42]. However, current research on humans has not directly complemented organosulfur compounds.
The abundance of Firmicutes decreased substantially after probiotic supplementation in female athletes, with a change rate of −41.89 % (P = 0.018). Our data show that after probiotic supplementation, Firmicutes in female athletes were highly positively correlated with organic acids and derivatives, which are commonly used as essential food additives or preservatives[43]. At the genus level, in the male athletes, Lactobacillus from Firmicutes was highly positively correlated with organophosphorus compounds after probiotic supplementation. Lactobacillus can promote intestinal mucosal homeostasis and the barrier function of an organism. In addition, the products secreted by Lactobacillus are closely related to immune function and intestinal barrier function (X. B [44]. During exercise, human intestinal microbiota usually produces several beneficial SCFAs, which are an essential source of energy for intestinal epithelial cells (IECs) and regulate the proliferation and differentiation of IECs and the function of subpopulations, such as enteroendocrine cells through different mechanisms; thus, intestinal microbiota affects intestinal motility and enhances intestinal barrier function and metabolism[45]. Current studies on organic acids and intestinal microbiota have mainly focused on the level of SCFA and the whole phylum. Exercise can increase SCFA content by altering the composition of microbiota, for endurance athletes, SCFA could be an ergogenic resource to promote energy metabolism and boost exercise performance so that exercise and SCFA complement each other[46,47]. Our data narrowed the relationship between organic acids and Firmicutes as intestinal microbiota while demonstrating that SCFAs are more closely related to female athletes with OWS.
Meanwhile, after probiotic supplementation, a significant downregulation of differential pathways, such as endocrine resistance, oestrogen signalling pathways and breast cancer, occurred in athletes. These pathways were relatively more relevant in female athletes. Endocrine resistance usually accompanies the treatment process of breast cancer[48,49]. Combined with the relatively significant changes in the intestinal microbiota that occurred in female athletes after probiotic supplementation, the downregulation of these associated pathways further emphasised the effect of probiotics on female athletes. The downregulation of pathways associated with breast cancer following probiotic supplementation suggests that probiotic supplementation is beneficial to a certain extent, especially for female athletes. In addition, after probiotic supplementation, Pusillimonas, Acinetobacter, Aeromonas and Stenotrophomona (Proteobacteria) in female athletes were associated with phenylpropanoids, polyketides and organic oxygen compounds, and organic acids and derivatives were highly correlated. Aeromonas is mainly associated with gastroenteritis and wound infections[50], and Acinetobacter is a common cause of acquired pneumonia, especially late-onset ventilator-associated pneumonia. This genus can cause other infections, including skin and wound infections, bacteraemia and meningitis[51]. Bacillus from Firmicutes, a common spoilage microorganism, is significantly and positively correlated with benzenoids[52]. Bacillus clausii is closely associated with the intestinal barrier and immune system[53]. However, the abundance of Actinobacteria related to energy supply during exercise and the inflammation-related Proteobacteria abundance increased significantly. Probiotics enhance the overall health and immunity, and probiotic supplementation can enhance the athletic performance and exercise adaptation of athletes[54]. The intestinal microbiota of female athletes is more likely to respond to exogenous probiotics and thus change their microbiota structure than male athletes[55]. This study revealed that the change in the microbiota structure after probiotic supplementation showed gender differences, and the results determining the more optimal structure between male and female athletes are inconclusive.
The relationship between the intestinal microbiota of athletes and gender differences needs further exploration. There are significant differences between males and females in the diversity, composition and metabolic profiles of intestinal microbiota. From birth, changes in sex hormones, daily activities and many other factors are associated with sex differences in intestinal microbiota[56]. A cohort study indicated that the composition of intestinal microbiota in athletes of different sport types also showed gender dependence, with female athletes typically having higher shannon diversity than non-athletes. Meanwhile the gender of the athlete is also significant in the potential risk of inflammation due to microbiological changes (Y [57]. Another study examined the composition of intestinal microbiota based on differences in skeletal muscle mass index (SMI) between men and women, and showed that males' SMI was more closely related to the compositional diversity of the intestinal microbiota[58]. Our data found significant changes in Firmicutes after probiotic supplementation in 10-km OWS female athletes, but not in male athletes. This relationship between intestinal microbiota and gender differences needs to be further explored. We also discovered insulin resistance-related pathways upregulated after race, and downregulation of sphingolipid metabolism occurred after probiotic supplementation. This finding suggests that we can reduce the adverse effects after a primary race by appropriately supplementing athletes with probiotics. Therefore, targeted interventions, personalized dietary treatment and recovery tools based on gender can be applied during the training and monitoring of athletes.
In order to reduce the bias caused by artificial control of the diet, we did not make additional restrictions on the athlete's diet and which caused limitations in our research process. Therefore, gender-based targeted interventions, personalized dietary treatments and rehabilitation tools could be used to regulate and optimize the intestinal microbiota during future training and monitoring of athletes.
5. Conclusion
The present work used faecal metabolomics combined with 16S rRNA gene sequencing to analyse the intestinal microbiota characteristics and metabolic features of 10-km open-water swimmers after race and probiotic supplementation. This type of athlete had unique proportions of intestinal microbiota phylum distribution, with Firmicutes in female athletes more likely to respond to exogenous probiotics. The intestinal microbiota of athletes was closely associated with pathways, such as exercise against cancer, exercise against aging, exercise for improving cognition, sphingolipid metabolism and endocrine resistance. In addition, Firmicutes and Proteobacteria were relatively closely associated with metabolite species. Overall, our findings present the intestinal microbiota characteristics of 10-km open-water swimmers and preliminarily explored the athletes' post-race and changes in intestinal microbiota involved in metabolic pathways after probiotic supplementation to provide a theoretical basis for further refining the relationship between athletes’ intestinal microbiota and exercise performance.
Funding
This work was supported by the National Natural Science Foundation of China (No. 32071173), the Central University's Special Fund for Basic Scientific Research (No.2022YB013) and the National Key Research and Development Program of China (No.2019YFF0301702-02-04).
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Data availability
The data are not stored in publicly available repositories and data that support the findings of this study are available from the corresponding author, upon reasonable request.
Ethics statement
The study was approved by the Ethics Committee, Institute of Sports Medicine, General Administration of Sport, China (No.202107). All participants were informed.
CRediT authorship contribution statement
Xuehan Li: Writing – original draft, Methodology. Yihsuan Lin: Writing – review & editing. Yue Chen: Data curation. Hongtao Sui: Data curation. Jianhao Chen: Formal analysis. Jiaqi Li: Formal analysis. Guoqing Zhang: Writing – review & editing, Validation, Resources, Conceptualization. Yi Yan: Writing – review & editing, Validation, Supervision, Resources, Methodology, Investigation, Funding acquisition, Conceptualization.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.heliyon.2023.e22735.
Contributor Information
Guoqing Zhang, Email: 13964000871@139.com.
Yi Yan, Email: yanyi@bsu.edu.cn.
Appendix A. Supplementary data
The following are the Supplementary data to this article.
References
- 1.Rajilic-Stojanovic M., de Vos W.M. The first 1000 cultured species of the human gastrointestinal microbiota. FEMS Microbiol. Rev. 2014;38(5):996–1047. doi: 10.1111/1574-6976.12075. https://www.ncbi.nlm.nih.gov/pubmed/24861948 Retrieved from. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hooper L.V., Macpherson A.J. Immune adaptations that maintain homeostasis with the intestinal microbiota. Nat. Rev. Immunol. 2010;10(3):159–169. doi: 10.1038/nri2710. https://www.ncbi.nlm.nih.gov/pubmed/20182457 Retrieved from. [DOI] [PubMed] [Google Scholar]
- 3.Li J., Jia H., Cai X., Zhong H., Feng Q., Sunagawa S.…Meta H.I.T.C. An integrated catalog of reference genes in the human gut microbiome. Nat. Biotechnol. 2014;32(8):834–841. doi: 10.1038/nbt.2942. https://www.ncbi.nlm.nih.gov/pubmed/24997786 Retrieved from. [DOI] [PubMed] [Google Scholar]
- 4.Marttinen M., Ala-Jaakkola R., Laitila A., Lehtinen M.J. Gut microbiota, probiotics and physical performance in athletes and physically active individuals. Nutrients. 2020;12(10) doi: 10.3390/nu12102936. https://www.ncbi.nlm.nih.gov/pubmed/32992765 Retrieved from. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Baldassarre R., Ieno C., Bonifazi M., Di Castro A., Gianfelici A., Piacentini M.F. Carbohydrate supplementation during a simulated 10-km open water swimming race: effects on physiological, perceptual parameters and performance. Eur. J. Sport Sci. 2022;22(3):390–398. doi: 10.1080/17461391.2021.1880644. https://www.ncbi.nlm.nih.gov/pubmed/33487101 Retrieved from. [DOI] [PubMed] [Google Scholar]
- 6.Baumler A.J., Sperandio V. Interactions between the microbiota and pathogenic bacteria in the gut. Nature. 2016;535(7610):85–93. doi: 10.1038/nature18849. https://www.ncbi.nlm.nih.gov/pubmed/27383983 Retrieved from. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bermon S., Petriz B., Kajeniene A., Prestes J., Castell L., Franco O.L. The microbiota: an exercise immunology perspective. Exerc. Immunol. Rev. 2015;21:70–79. https://www.ncbi.nlm.nih.gov/pubmed/25825908 Retrieved from. [PubMed] [Google Scholar]
- 8.Clancy R.L., Gleeson M., Cox A., Callister R., Dorrington M., D'Este C., Henriksson A. Reversal in fatigued athletes of a defect in interferon gamma secretion after administration of Lactobacillus acidophilus. Br. J. Sports Med. 2006;40(4):351–354. doi: 10.1136/bjsm.2005.024364. https://www.ncbi.nlm.nih.gov/pubmed/16556792 Retrieved from. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Clark A., Mach N. Exercise-induced stress behavior, gut-microbiota-brain axis and diet: a systematic review for athletes. J Int Soc Sports Nutr. 2016;13:43. doi: 10.1186/s12970-016-0155-6. https://www.ncbi.nlm.nih.gov/pubmed/27924137 Retrieved from. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dohnalova L., Lundgren P., Carty J.R.E., Goldstein N., Wenski S.L., Nanudorn P.…Thaiss C.A. A microbiome-dependent gut-brain pathway regulates motivation for exercise. Nature. 2022;612(7941):739–747. doi: 10.1038/s41586-022-05525-z. https://www.ncbi.nlm.nih.gov/pubmed/36517598 Retrieved from. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jager R., Mohr A.E., Carpenter K.C., Kerksick C.M., Purpura M., Moussa A., Antonio J. International society of sports nutrition position stand: probiotics. J Int Soc Sports Nutr. 2019;16(1):62. doi: 10.1186/s12970-019-0329-0. https://www.ncbi.nlm.nih.gov/pubmed/31864419 Retrieved from. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sashihara T., Nagata M., Mori T., Ikegami S., Gotoh M., Okubo K., Itoh H. Effects of Lactobacillus gasseri OLL2809 and alpha-lactalbumin on university-student athletes: a randomized, double-blind, placebo-controlled clinical trial. Appl. Physiol. Nutr. Metabol. 2013;38(12):1228–1235. doi: 10.1139/apnm-2012-0490. https://www.ncbi.nlm.nih.gov/pubmed/24195623 Retrieved from. [DOI] [PubMed] [Google Scholar]
- 13.Strasser B., Geiger D., Schauer M., Gostner J.M., Gatterer H., Burtscher M., Fuchs D. Probiotic supplements beneficially affect tryptophan-kynurenine metabolism and reduce the incidence of upper respiratory tract infections in trained athletes: a randomized, double-blinded, placebo-controlled trial. Nutrients. 2016;8(11) doi: 10.3390/nu8110752. https://www.ncbi.nlm.nih.gov/pubmed/27886064 Retrieved from. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shaw G., Koivisto A., Gerrard D., Burke L.M. Nutrition considerations for open-water swimming. Int. J. Sport Nutr. Exerc. Metabol. 2014;24(4):373–381. doi: 10.1123/ijsnem.2014-0018. https://www.ncbi.nlm.nih.gov/pubmed/24667305 Retrieved from. [DOI] [PubMed] [Google Scholar]
- 15.Magoc T., Salzberg S.L. FLASH: fast length adjustment of short reads to improve genome assemblies. Bioinformatics. 2011;27(21):2957–2963. doi: 10.1093/bioinformatics/btr507. https://www.ncbi.nlm.nih.gov/pubmed/21903629 Retrieved from. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schloss P.D., Westcott S.L., Ryabin T., Hall J.R., Hartmann M., Hollister E.B., Weber C.F. Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl. Environ. Microbiol. 2009;75(23):7537–7541. doi: 10.1128/AEM.01541-09. https://www.ncbi.nlm.nih.gov/pubmed/19801464 Retrieved from. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pruesse E., Quast C., Knittel K., Fuchs B.M., Ludwig W., Peplies J., Glockner F.O. SILVA: a comprehensive online resource for quality checked and aligned ribosomal RNA sequence data compatible with ARB. Nucleic Acids Res. 2007;35(21):7188–7196. doi: 10.1093/nar/gkm864. https://www.ncbi.nlm.nih.gov/pubmed/17947321 Retrieved from. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.White J.R., Nagarajan N., Pop M. Statistical methods for detecting differentially abundant features in clinical metagenomic samples. PLoS Comput. Biol. 2009;5(4) doi: 10.1371/journal.pcbi.1000352. https://www.ncbi.nlm.nih.gov/pubmed/19360128 Retrieved from. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Langille M.G., Zaneveld J., Caporaso J.G., McDonald D., Knights D., Reyes J.A., Huttenhower C. Predictive functional profiling of microbial communities using 16S rRNA marker gene sequences. Nat. Biotechnol. 2013;31(9):814–821. doi: 10.1038/nbt.2676. https://www.ncbi.nlm.nih.gov/pubmed/23975157 Retrieved from. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rodriguez L., Veiga S. Effect of the pacing strategies on the open-water 10-km world swimming championships performances. Int. J. Sports Physiol. Perform. 2018;13(6):694–700. doi: 10.1123/ijspp.2017-0274. https://www.ncbi.nlm.nih.gov/pubmed/29035600 Retrieved from. [DOI] [PubMed] [Google Scholar]
- 21.Hou H., Chen D., Zhang K., Zhang W., Liu T., Wang S., Cao H. Gut microbiota-derived short-chain fatty acids and colorectal cancer: ready for clinical translation? Cancer Lett. 2022;526:225–235. doi: 10.1016/j.canlet.2021.11.027. https://www.ncbi.nlm.nih.gov/pubmed/34843863 Retrieved from. [DOI] [PubMed] [Google Scholar]
- 22.Aragon-Vela J., Solis-Urra P., Ruiz-Ojeda F.J., Alvarez-Mercado A.I., Olivares-Arancibia J., Plaza-Diaz J. Impact of exercise on gut microbiota in obesity. Nutrients. 2021;13(11) doi: 10.3390/nu13113999. https://www.ncbi.nlm.nih.gov/pubmed/34836254 Retrieved from. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mankowska K., Marchelek-Mysliwiec M., Kochan P., Kosik-Bogacka D., Konopka T., Grygorcewicz B., Dolegowska B. Microbiota in sports. Arch. Microbiol. 2022;204(8):485. doi: 10.1007/s00203-022-03111-5. https://www.ncbi.nlm.nih.gov/pubmed/35834007 Retrieved from. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Verdam F.J., Fuentes S., de Jonge C., Zoetendal E.G., Erbil R., Greve J.W.…Rensen S.S. Human intestinal microbiota composition is associated with local and systemic inflammation in obesity. Obesity. 2013;21(12):E607–E615. doi: 10.1002/oby.20466. https://www.ncbi.nlm.nih.gov/pubmed/23526699 Retrieved from. [DOI] [PubMed] [Google Scholar]
- 25.Chen X., Song S., Li H., Gozaydin G., Yan N. Expanding the boundary of biorefinery: organonitrogen chemicals from biomass. Acc. Chem. Res. 2021;54(7):1711–1722. doi: 10.1021/acs.accounts.0c00842. https://www.ncbi.nlm.nih.gov/pubmed/33576600 Retrieved from. [DOI] [PubMed] [Google Scholar]
- 26.Ley R.E., Turnbaugh P.J., Klein S., Gordon J.I. Microbial ecology: human gut microbes associated with obesity. Nature. 2006;444(7122):1022–1023. doi: 10.1038/4441022a. https://www.ncbi.nlm.nih.gov/pubmed/17183309 Retrieved from. [DOI] [PubMed] [Google Scholar]
- 27.Mariat D., Firmesse O., Levenez F., Guimaraes V., Sokol H., Dore J., Furet J.P. The Firmicutes/Bacteroidetes ratio of the human microbiota changes with age. BMC Microbiol. 2009;9:123. doi: 10.1186/1471-2180-9-123. https://www.ncbi.nlm.nih.gov/pubmed/19508720 Retrieved from. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Patra M., Gasser G. Organometallic compounds: an opportunity for chemical biology? Chembiochem. 2012;13(9):1232–1252. doi: 10.1002/cbic.201200159. https://www.ncbi.nlm.nih.gov/pubmed/22619182 Retrieved from. [DOI] [PubMed] [Google Scholar]
- 29.Babak M.V., Ang W.H. Multinuclear organometallic ruthenium-arene complexes for cancer therapy. Met Ions Life Sci. 2018;18 doi: 10.1515/9783110470734-012. https://www.ncbi.nlm.nih.gov/pubmed/29394025 Retrieved from. [DOI] [PubMed] [Google Scholar]
- 30.Shin N.R., Whon T.W., Bae J.W. Proteobacteria: microbial signature of dysbiosis in gut microbiota. Trends Biotechnol. 2015;33(9):496–503. doi: 10.1016/j.tibtech.2015.06.011. https://www.ncbi.nlm.nih.gov/pubmed/26210164 Retrieved from. [DOI] [PubMed] [Google Scholar]
- 31.Evans C.C., LePard K.J., Kwak J.W., Stancukas M.C., Laskowski S., Dougherty J., Ciancio M.J. Exercise prevents weight gain and alters the gut microbiota in a mouse model of high fat diet-induced obesity. PLoS One. 2014;9(3) doi: 10.1371/journal.pone.0092193. https://www.ncbi.nlm.nih.gov/pubmed/24670791 Retrieved from. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.McMurray R.G., Hackney A.C. Interactions of metabolic hormones, adipose tissue and exercise. Sports Med. 2005;35(5):393–412. doi: 10.2165/00007256-200535050-00003. https://www.ncbi.nlm.nih.gov/pubmed/15896089 Retrieved from. [DOI] [PubMed] [Google Scholar]
- 33.Hackney A.C., Davis H.C., Lane A.R. Growth hormone-insulin-like growth factor Axis, thyroid Axis, prolactin, and exercise. Front. Horm. Res. 2016;47:1–11. doi: 10.1159/000445147. https://www.ncbi.nlm.nih.gov/pubmed/27348437 Retrieved from. [DOI] [PubMed] [Google Scholar]
- 34.Yang L., Xu H., Chen Y., Miao C., Zhao Y., Xing Y., Zhang Q. Melatonin: multi-target mechanism against diminished ovarian reserve based on network pharmacology. Front. Endocrinol. 2021;12 doi: 10.3389/fendo.2021.630504. https://www.ncbi.nlm.nih.gov/pubmed/33959095 Retrieved from. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lin J.Y., Kuo W.W., Baskaran R., Kuo C.H., Chen Y.A., Chen W.S., Huang C.Y. Swimming exercise stimulates IGF1/PI3K/Akt and AMPK/SIRT1/PGC1alpha survival signaling to suppress apoptosis and inflammation in aging hippocampus. Aging (Albany NY) 2020;12(8):6852–6864. doi: 10.18632/aging.103046. https://www.ncbi.nlm.nih.gov/pubmed/32320382 Retrieved from. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cotman C.W., Berchtold N.C., Christie L.A. Exercise builds brain health: key roles of growth factor cascades and inflammation. Trends Neurosci. 2007;30(9):464–472. doi: 10.1016/j.tins.2007.06.011. https://www.ncbi.nlm.nih.gov/pubmed/17765329 Retrieved from. [DOI] [PubMed] [Google Scholar]
- 37.Mattson M.P. Energy intake and exercise as determinants of brain health and vulnerability to injury and disease. Cell Metabol. 2012;16(6):706–722. doi: 10.1016/j.cmet.2012.08.012. https://www.ncbi.nlm.nih.gov/pubmed/23168220 Retrieved from. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wrann C.D., White J.P., Salogiannnis J., Laznik-Bogoslavski D., Wu J., Ma D.…Spiegelman B.M. Exercise induces hippocampal BDNF through a PGC-1alpha/FNDC5 pathway. Cell Metabol. 2013;18(5):649–659. doi: 10.1016/j.cmet.2013.09.008. https://www.ncbi.nlm.nih.gov/pubmed/24120943 Retrieved from. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Du Y., Gao X.R., Peng L., Ge J.F. Crosstalk between the microbiota-gut-brain axis and depression. Heliyon. 2020;6(6) doi: 10.1016/j.heliyon.2020.e04097. https://www.ncbi.nlm.nih.gov/pubmed/32529075 Retrieved from. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Miekus N., Marszalek K., Podlacha M., Iqbal A., Puchalski C., Swiergiel A.H. Health benefits of plant-derived sulfur compounds, glucosinolates, and organosulfur compounds. Molecules. 2020;25(17) doi: 10.3390/molecules25173804. https://www.ncbi.nlm.nih.gov/pubmed/32825600 Retrieved from. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Costea T., Hudita A., Ciolac O.A., Galateanu B., Ginghina O., Costache M.…Mocanu M.M. Chemoprevention of colorectal cancer by dietary compounds. Int. J. Mol. Sci. 2018;19(12) doi: 10.3390/ijms19123787. https://www.ncbi.nlm.nih.gov/pubmed/30487390 Retrieved from. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Guillamon E., Andreo-Martinez P., Mut-Salud N., Fonolla J., Banos A. Beneficial effects of organosulfur compounds from Allium cepa on gut health: a systematic review. Foods. 2021;10(8) doi: 10.3390/foods10081680. https://www.ncbi.nlm.nih.gov/pubmed/34441457 Retrieved from. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Qiu X., Zhang Y., Zhou Y., Li G.H., Feng X.S. Progress in pretreatment and analysis of organic Acids: an update since 2010. Food Chem. 2021;360 doi: 10.1016/j.foodchem.2021.129977. https://www.ncbi.nlm.nih.gov/pubmed/34023712 Retrieved from. [DOI] [PubMed] [Google Scholar]
- 44.Lin X.B., Wang T., Stothard P., Corander J., Wang J., Baines J.F.…Walter J. The evolution of ecological facilitation within mixed-species biofilms in the mouse gastrointestinal tract. ISME J. 2018;12(11):2770–2784. doi: 10.1038/s41396-018-0211-0. https://www.ncbi.nlm.nih.gov/pubmed/30013162 Retrieved from. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Martin-Gallausiaux C., Marinelli L., Blottiere H.M., Larraufie P., Lapaque N. SCFA: mechanisms and functional importance in the gut. Proc. Nutr. Soc. 2021;80(1):37–49. doi: 10.1017/S0029665120006916. https://www.ncbi.nlm.nih.gov/pubmed/32238208 Retrieved from. [DOI] [PubMed] [Google Scholar]
- 46.Bongiovanni T., Yin M.O.L., Heaney L.M. Correction: the athlete and gut microbiome: short-chain fatty acids as potential ergogenic aids for exercise and training. Int. J. Sports Med. 2021;42(13):e1. doi: 10.1055/a-1524-2095. https://www.ncbi.nlm.nih.gov/pubmed/35176789 Retrieved from. [DOI] [PubMed] [Google Scholar]
- 47.Tang Q., Jin G., Wang G., Liu T., Liu X., Wang B., Cao H. Current sampling methods for gut microbiota: a call for more precise devices. Front. Cell. Infect. Microbiol. 2020;10:151. doi: 10.3389/fcimb.2020.00151. https://www.ncbi.nlm.nih.gov/pubmed/32328469 Retrieved from. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Alataki A., Dowsett M. Human epidermal growth factor receptor-2 and endocrine resistance in hormone-dependent breast cancer. Endocr. Relat. Cancer. 2022;29(8):R105–R122. doi: 10.1530/ERC-21-0293. https://www.ncbi.nlm.nih.gov/pubmed/35613334 Retrieved from. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.D'Souza A., Spicer D., Lu J. Overcoming endocrine resistance in metastatic hormone receptor-positive breast cancer. J. Hematol. Oncol. 2018;11(1):80. doi: 10.1186/s13045-018-0620-6. https://www.ncbi.nlm.nih.gov/pubmed/29891002 Retrieved from. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Litwinowicz A., Blaszkowska J. Preventing infective complications following leech therapy: elimination of symbiotic Aeromonas spp. from the intestine of Hirudo verbana using antibiotic feeding. Surg. Infect. 2014;15(6):757–762. doi: 10.1089/sur.2014.036. https://www.ncbi.nlm.nih.gov/pubmed/24897173 Retrieved from. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hu Q., Hu Z., Li J., Tian B., Xu H., Li J. Detection of OXA-type carbapenemases and integrons among carbapenem-resistant Acinetobactor baumannii in a teaching hospital in China. J. Basic Microbiol. 2011;51(5):467–472. doi: 10.1002/jobm.201000402. https://www.ncbi.nlm.nih.gov/pubmed/21656808 Retrieved from. [DOI] [PubMed] [Google Scholar]
- 52.Cairo J., Gherman I., Day A., Cook P.E. Bacillus cytotoxicus-A potentially virulent food-associated microbe. J. Appl. Microbiol. 2022;132(1):31–40. doi: 10.1111/jam.15214. https://www.ncbi.nlm.nih.gov/pubmed/34260791 Retrieved from. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lopetuso L.R., Scaldaferri F., Franceschi F., Gasbarrini A. Bacillus clausii and gut homeostasis: state of the art and future perspectives. Expet Rev. Gastroenterol. Hepatol. 2016;10(8):943–948. doi: 10.1080/17474124.2016.1200465. https://www.ncbi.nlm.nih.gov/pubmed/27291780 Retrieved from. [DOI] [PubMed] [Google Scholar]
- 54.Shing C.M., Peake J.M., Lim C.L., Briskey D., Walsh N.P., Fortes M.B., Vitetta L. Effects of probiotics supplementation on gastrointestinal permeability, inflammation and exercise performance in the heat. Eur. J. Appl. Physiol. 2014;114(1):93–103. doi: 10.1007/s00421-013-2748-y. https://www.ncbi.nlm.nih.gov/pubmed/24150782 Retrieved from. [DOI] [PubMed] [Google Scholar]
- 55.Yadav M.K., Kumari I., Singh B., Sharma K.K., Tiwari S.K. Probiotics, prebiotics and synbiotics: safe options for next-generation therapeutics. Appl. Microbiol. Biotechnol. 2022;106(2):505–521. doi: 10.1007/s00253-021-11646-8. https://www.ncbi.nlm.nih.gov/pubmed/35015145 Retrieved from. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Chen J., Li H., Hird S.M., Chen M.H., Xu W., Maas K., Cong X. Sex differences in gut microbial development of preterm infant twins in early life: a longitudinal analysis. Front. Cell. Infect. Microbiol. 2021;11 doi: 10.3389/fcimb.2021.671074. https://www.ncbi.nlm.nih.gov/pubmed/34458157 Retrieved from. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Li Y., Cheng M., Zha Y., Yang K., Tong Y., Wang S., Ning K. Gut microbiota and inflammation patterns for specialized athletes: a multi-cohort study across different types of sports. mSystems. 2023;8(4) doi: 10.1128/msystems.00259-23. https://www.ncbi.nlm.nih.gov/pubmed/37498086 Retrieved from. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Park C.H., Lee E.J., Kim H.L., Lee Y.T., Yoon K.J., Kim H.N. Sex-specific associations between gut microbiota and skeletal muscle mass in a population-based study. J Cachexia Sarcopenia Muscle. 2022;13(6):2908–2919. doi: 10.1002/jcsm.13096. https://www.ncbi.nlm.nih.gov/pubmed/36218092 Retrieved from. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data are not stored in publicly available repositories and data that support the findings of this study are available from the corresponding author, upon reasonable request.