Abstract
Motivation
Analysis of mutational signatures is a powerful approach for understanding the mutagenic processes that have shaped the evolution of a cancer genome. To evaluate the mutational signatures operative in a cancer genome, one first needs to quantify their activities by estimating the number of mutations imprinted by each signature.
Results
Here we present SigProfilerAssignment, a desktop and an online computational framework for assigning all types of mutational signatures to individual samples. SigProfilerAssignment is the first tool that allows both analysis of copy-number signatures and probabilistic assignment of signatures to individual somatic mutations. As its computational engine, the tool uses a custom implementation of the forward stagewise algorithm for sparse regression and nonnegative least squares for numerical optimization. Analysis of 2700 synthetic cancer genomes with and without noise demonstrates that SigProfilerAssignment outperforms four commonly used approaches for assigning mutational signatures.
Availability and implementation
SigProfilerAssignment is available under the BSD 2-clause license at https://github.com/AlexandrovLab/SigProfilerAssignment with a web implementation at https://cancer.sanger.ac.uk/signatures/assignment/.
1 Introduction
Somatic mutations accumulate in the genomes of all cells of the human body (Stratton et al. 2009, Martincorena and Campbell 2015). These mutations arise from different mutational processes, with each process generating a characteristic pattern of mutations, known as a mutational signature (Alexandrov et al. 2013). By leveraging the vast amounts of high-throughput DNA sequencing data generated over the past two decades, distinct mutational signatures have been elucidated from various cancer types (Alexandrov et al. 2020, Degasperi et al. 2022) and normal somatic tissues (Lee-Six et al. 2019, Lawson et al. 2020, Olafsson et al. 2020, Yoshida et al. 2020). Sets of mutation-type specific reference signatures have been developed and deposited in the Catalogue of Somatic Mutations in Cancer (COSMIC) database (Tate et al. 2019, Alexandrov et al. 2020), including signatures of single base substitutions (SBSs), doublet base substitutions (DBSs), small insertions and deletions (IDs), and copy number alterations (CNs).
There are at least two distinct approaches for analyzing mutational signatures. De novo extraction is an unsupervised machine learning approach that allows identifying the patterns of known and previously unknown mutational signatures (Islam et al. 2022). This type of analysis is predominately used for deriving reference signatures as it requires large cohorts of generally more than 100 samples. In contrast, refitting of mutational signatures is a numerical optimization approach that allows the assignment of known (in most cases, reference) signatures to an individual sample by quantifying the number of mutations attributed to each signature operative in that sample.
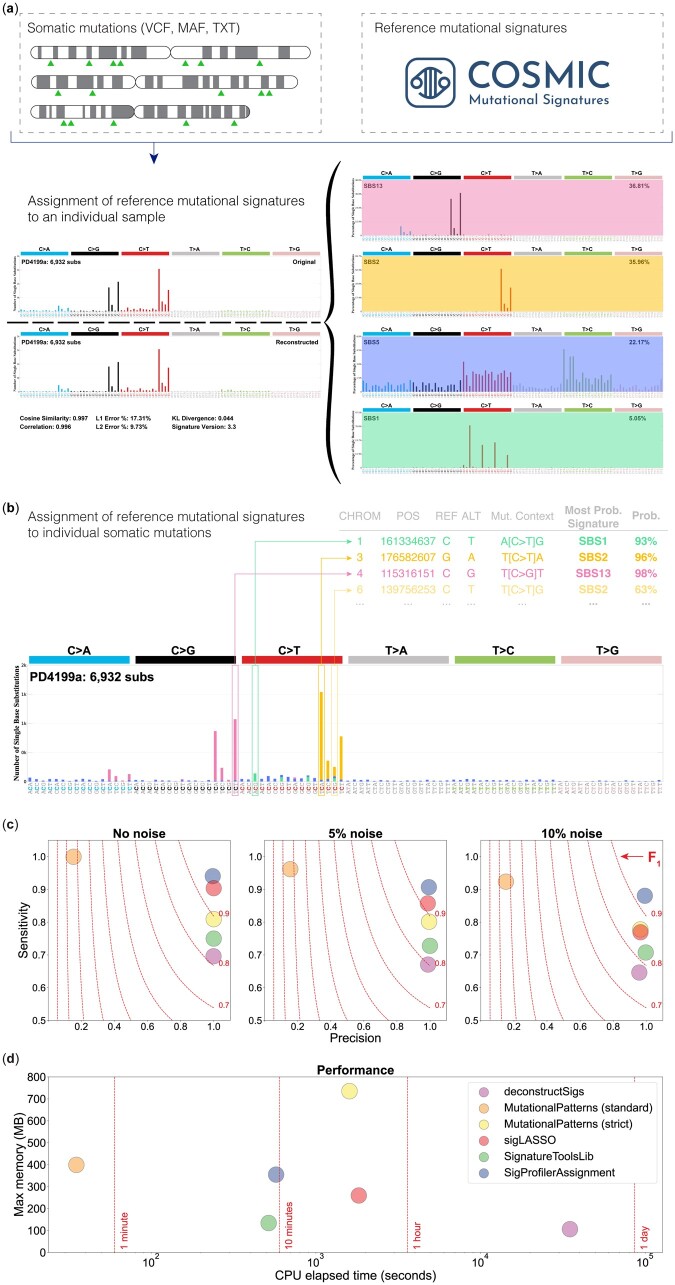
In the past decade, multiple tools for refitting known signatures were developed, including deconstructSigs (Rosenthal et al. 2016), MutationalPatterns (Blokzijl et al. 2018, Manders et al. 2022), sigLASSO (Li et al. 2020), and SignatureToolsLib (Degasperi et al. 2020, Degasperi et al. 2022). Most of these tools provide support almost exclusively for SBS signatures and lack an online interface, although a few web tools exist, including, MuSiCa (Díaz-Gay et al. 2018) and Mutalisk (Lee et al. 2018). Further, these tools have never been compared and no existing tool supports probabilistically assigning signatures to somatic mutations. To address these limitations, here, we present SigProfilerAssignment, a comprehensive bioinformatics tool for assigning mutational signatures to individual samples and individual somatic mutations (Fig. 1a and b). SigProfilerAssignment provides desktop and online support for all types of mutational signatures, including the COSMIC sets of reference SBS, DBS, ID, and CN signatures. Additionally, SigProfilerAssignment supports assignment of de novo extracted mutational signatures and of a user provided set of custom signatures. Our benchmarking based on 2700 simulated cancer genomes demonstrates that SigProfilerAssignment outperforms other commonly used tools on simulation data with and without noise.
Figure 1.
Assigning known mutational signatures to an individual sample and individual mutations with SigProfilerAssignment, and benchmarking with four other bioinformatics tools. SigProfilerAssignment supports input data in a standard format (VCF, MAF, or text) and it allows assigning a set of known signatures (e.g. ones from the COSMIC database) to an (a) individual sample and (b) probabilistically to an individual somatic mutation. Note that the probabilistic assignment of mutational signatures to an individual somatic mutation is only possible if a user provides a list of individual mutations (e.g. VCF file) for the examined sample instead of a mutational vector, as a mutational vector lacks information for individual mutations. (c) Accuracy benchmarking of SigProfilerAssignment and four other tools for assigning mutational signatures. Each tool was evaluated using 2700 synthetic cancer genomes generated using 21 different COSMIC reference mutational signatures. All COSMICv3.3 signatures were used as the input set of known mutational signatures. Three different levels of nonsystematic random noise (0%, 5%, and 10%) were used to evaluate the precision (x-axes), sensitivity (y-axes), and F1 scores (harmonic mean of precision and sensitivity; dotted lines) of each tool. (d) Computational benchmarking based on CPU elapsed time (x-axis; log-scaled) and maximum memory usage (y-axis) for each tool.
2 Materials and methods
Given a set of known mutational signatures and a set of mutations in a cancer genome, both classified under the same mutational schema (Alexandrov et al. 2013, Bergstrom et al. 2019), SigProfilerAssignment identifies the number of mutations caused by each signature in that cancer genome (Fig. 1a). To quantity the number of mutations imprinted by each signature, SigProfilerAssignment uses a custom implementation of the forward stagewise algorithm (Hastie et al. 2009) and it applies nonnegative least squares (NNLS), based on the Lawson-Hanson method (Ling et al. 1977). The tool’s algorithm is available in Algorithm 1, and it is described in Supplementary Data. In addition to quantifying the activity of each mutational signature, SigProfilerAssignment also assigns known signatures to individual mutations (Fig. 1b) based on their specific mutational context.
Algorithm 1: Assigning mutational signatures to samples with SigProfilerAssignment
Input: (a vector corresponding to a set of mutations in a sample) and
(a matrix corresponding to a set of known mutational signatures)
Output: (the vector reflecting the activities of the known signatures in sample
1: calcNNLS ( )
2: While FLAG = True:
3: = removeSignatures (, )
4: = addSignatures (, )
5: Set FLAG = False if remains constant and there is no addition or removal of signatures
END While
6: calcNNLS( )
7: Return
8: FUNCTION removeSignatures (, )
9: While FLAG = True:
10: Forin 1 to size(, 2) do //loop from 1 to the total number of signatures in
11: //remove the jth signature from
12: = calcNNLS()
END For
13: //find the signature set with least relative error
14: If (
15:
else
16: Return
END If
END While
END removeSignatures
17: FUNCTION addSignatures (, )
18: While FLAG = True:
19: Forin 1 to size(, 2) do //loop from 1 to the total number of signatures in
20: //add the pth signature from
21: = calcNNLS()
END For
22: //find the signature set with least relative error
23: If (
24:
else
25: Return
END If
END While
END addSignatures
26: FUNCTION calcNNLS( )
27: //Calculating NNLS with the Lawson-Hanson method
28: //Computing relative error
29: Return
END calcNNLS
3 Results
To evaluate the performance of SigProfilerAssignment and another four commonly used tools for refitting mutational signatures (Rosenthal et al. 2016, Blokzijl et al. 2018, Degasperi et al. 2020, Li et al. 2020, Degasperi et al. 2022, Manders et al. 2022), we performed a comparative benchmarking using a previously generated independent synthetic dataset (Islam et al. 2022) (Fig. 1c and d). The dataset encompasses the SBS patterns of 2700 simulated cancer genomes, corresponding to 300 tumors from 9 different cancer types, generated using 21 different COSMIC reference signatures. To emulate a typical refitting of mutational signatures, the complete set of 79 COSMICv3.3 SBS signatures was used as input. The mutational signatures’ activities obtained by each tool were compared against the ground truth activities used to synthetically generate these samples. Three different levels of random noise (0%, 5%, and 10%) were tested to assess the robustness of the different algorithms in a real biological context. To evaluate the accuracy of the signature refitting, we calculated sensitivity, specificity, and F1 score (Supplementary Data). In addition, we also examined the runtime and memory utilization of each tool.
Our synthetic benchmarking revealed that SigProfilerAssignment outperforms all other approaches for the examined noise levels (Fig. 1c). For 10% random noise, only SigProfilerAssignment obtained an F1 score >0.90. In all cases, SigProfilerAssignment exhibited a high precision while showing an improved sensitivity compared to other approaches (Fig. 1c), with consistent top performance across cancer types (Supplementary Fig. S1) and most mutational signatures (Supplementary Fig. S2). In terms of computational performance, SigProfilerAssignment processed the 2700 samples within 9.6 min (0.21 s per sample; Fig. 1d). Only the standard mode of MutationalPatterns generated results substantially faster. However, MutationalPatterns’ standard mode exhibited sub-optimal performance, with a significant drop in precision for all noise levels, likely due to overfitting of the input data (Fig. 1c) (Blokzijl et al. 2018). This issue has been addressed in the most recent version of MutationalPatterns with the addition of a strict mode (Manders et al. 2022), albeit with a significant computational performance cost (Fig. 1d). Other approaches limit overfitting by implementing different penalties based on the L1 error (viz. sigLASSO) (Li et al. 2020) or the sum-squared error (viz. deconstructSigs) (Rosenthal et al. 2016), and post-hoc filters based on the percentage of the total number of mutations attributed to a given signature (viz. deconstructSigs and SignatureToolsLib) (Rosenthal et al. 2016, Degasperi et al. 2022) (Supplementary Table S1). No significant memory requirements were observed for any of the tools (Fig. 1d). Analysis of similar benchmarking datasets for DBS, ID, and CN mutational signatures (Supplementary Data) revealed that SigProfilerAssignment exhibits high precision and sensitivity, with F1 scores >0.85 for all assessed noise levels (Supplementary Fig. S3).
4 Conclusion
Assigning mutational signatures to individual samples provides an opportunity to identify the processes responsible for somatic mutations on a sample-by-sample basis. Considering our synthetic benchmarking, SigProfilerAssignment stands out as the most precise and sensitive tool while maintaining high computational performance and bringing novel capabilities. To the best of our knowledge, SigProfilerAssignment represents the first computational tool for assigning signature probabilities to individual mutations, which can allow uncovering the mutational processes responsible for specific driver genomic alterations leading to tumor evolution. SigProfilerAssignment is also the first tool that supports assignment of the recently developed copy number signatures (Steele et al. 2022), which are good predictors of clinical survival (Drews et al. 2022, Steele et al. 2022).
In summary, SigProfilerAssignment provides a novel computational package and an accessible online interface to accurately assign known mutational signatures to an individual cancer and individual somatic mutations, thus, enabling users to ascertain the mutational processes operative in a cancer genome.
Supplementary Material
Contributor Information
Marcos Díaz-Gay, Department of Cellular and Molecular Medicine, UC San Diego, La Jolla, CA 92093, United States; Department of Bioengineering, UC San Diego, La Jolla, CA 92093, United States; Moores Cancer Center, UC San Diego, La Jolla, CA 92037, United States.
Raviteja Vangara, Department of Cellular and Molecular Medicine, UC San Diego, La Jolla, CA 92093, United States; Department of Bioengineering, UC San Diego, La Jolla, CA 92093, United States; Moores Cancer Center, UC San Diego, La Jolla, CA 92037, United States.
Mark Barnes, Department of Cellular and Molecular Medicine, UC San Diego, La Jolla, CA 92093, United States; Department of Bioengineering, UC San Diego, La Jolla, CA 92093, United States; Moores Cancer Center, UC San Diego, La Jolla, CA 92037, United States.
Xi Wang, Department of Cellular and Molecular Medicine, UC San Diego, La Jolla, CA 92093, United States; Department of Bioengineering, UC San Diego, La Jolla, CA 92093, United States; Moores Cancer Center, UC San Diego, La Jolla, CA 92037, United States.
S M Ashiqul Islam, Department of Cellular and Molecular Medicine, UC San Diego, La Jolla, CA 92093, United States; Department of Bioengineering, UC San Diego, La Jolla, CA 92093, United States; Moores Cancer Center, UC San Diego, La Jolla, CA 92037, United States.
Ian Vermes, COSMIC, Wellcome Sanger Institute, Hinxton, Cambridgeshire CB10 1SA, United Kingdom.
Stephen Duke, COSMIC, Wellcome Sanger Institute, Hinxton, Cambridgeshire CB10 1SA, United Kingdom.
Nithish Bharadhwaj Narasimman, Department of Cellular and Molecular Medicine, UC San Diego, La Jolla, CA 92093, United States; Department of Bioengineering, UC San Diego, La Jolla, CA 92093, United States; Moores Cancer Center, UC San Diego, La Jolla, CA 92037, United States.
Ting Yang, Department of Cellular and Molecular Medicine, UC San Diego, La Jolla, CA 92093, United States; Department of Bioengineering, UC San Diego, La Jolla, CA 92093, United States; Moores Cancer Center, UC San Diego, La Jolla, CA 92037, United States.
Zichen Jiang, Department of Cellular and Molecular Medicine, UC San Diego, La Jolla, CA 92093, United States; Department of Bioengineering, UC San Diego, La Jolla, CA 92093, United States; Moores Cancer Center, UC San Diego, La Jolla, CA 92037, United States.
Sarah Moody, Cancer, Ageing and Somatic Mutation, Wellcome Sanger Institute, Wellcome Genome Campus, Cambridge CB10 1SA, United Kingdom.
Sergey Senkin, Genetic Epidemiology Group, International Agency for Research on Cancer, 69372 Lyon, France.
Paul Brennan, Genetic Epidemiology Group, International Agency for Research on Cancer, 69372 Lyon, France.
Michael R Stratton, Cancer, Ageing and Somatic Mutation, Wellcome Sanger Institute, Wellcome Genome Campus, Cambridge CB10 1SA, United Kingdom.
Ludmil B Alexandrov, Department of Cellular and Molecular Medicine, UC San Diego, La Jolla, CA 92093, United States; Department of Bioengineering, UC San Diego, La Jolla, CA 92093, United States; Moores Cancer Center, UC San Diego, La Jolla, CA 92037, United States.
Supplementary data
Supplementary data are available at Bioinformatics online.
Conflict of interest
L.B.A. is a co-founder, CSO, scientific advisory member, and consultant for io9, has equity, and receives income. The terms of this arrangement have been reviewed and approved by the University of California, San Diego in accordance with its conflict of interest policies. L.B.A.’s spouse is an employee of Biotheranostics, Inc. L.B.A. is also an inventor of a US Patent 10776718 for source identification by non-negative matrix factorization. L.B.A. declares U.S. provisional applications with serial numbers: 63/289601; 63/269033; 63/366392; 63/367846; 63/412835. All other authors declare no competing interests.
Funding
This work was supported by Cancer Research UK [Grand Challenge Award C98/A24032]; the US National Institute of Health [grants R01ES030993-01A, R01ES032547, R01CA269919, and U01CA290479]; and a Packard Fellowship for Science and Engineering to L.B.A. The computational analyses reported in this manuscript have utilized the Triton Shared Computing Cluster at the San Diego Supercomputer Center of UC San Diego. The funders had no roles in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Data availability
The data Data availability statement can be found in the supplementary materials.
References
- Alexandrov LB, Kim J, Haradhvala NJ. et al. ; PCAWG Consortium. The repertoire of mutational signatures in human cancer. Nature 2020;578:94–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexandrov LB, Nik-Zainal S, Wedge DC. et al. Deciphering signatures of mutational processes operative in human cancer. Cell Rep 2013;3:246–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergstrom EN, Huang MN, Mahto U. et al. SigProfilerMatrixGenerator: a tool for visualizing and exploring patterns of small mutational events. BMC Genomics 2019;20:685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blokzijl F, Janssen R, van Boxtel R. et al. MutationalPatterns: comprehensive genome-wide analysis of mutational processes. Genome Med 2018;10:33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Degasperi A, Amarante TD, Czarnecki J. et al. A practical framework and online tool for mutational signature analyses show inter-tissue variation and driver dependencies. Nat Cancer 2020;1:249–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Degasperi A, Zou X, Amarante TD. et al. ; Genomics England Research Consortium. Substitution mutational signatures in whole-genome–sequenced cancers in the UK population. Science 2022;376:abl9283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Díaz-Gay M, Vila-Casadesús M, Franch-Expósito S. et al. Mutational signatures in cancer (MuSiCa): a web application to implement mutational signatures analysis in cancer samples. BMC Bioinformatics 2018;19:224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drews RM, Hernando B, Tarabichi M. et al. A pan-cancer compendium of chromosomal instability. Nature 2022;606:976–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hastie T, Tibshirani R, Friedman JH.. The Elements of Statistical Learning: Data Mining, Inference, and Prediction. New York City, New York, USA: Springer, 2009. [Google Scholar]
- Islam SMA, Díaz-Gay M, Wu Y. et al. Uncovering novel mutational signatures by de novo extraction with SigProfilerExtractor. Cell Genom 2022;2:100179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawson ARJ, Abascal F, Coorens THH. et al. Extensive heterogeneity in somatic mutation and selection in the human bladder. Science 2020;370:75–82. [DOI] [PubMed] [Google Scholar]
- Ling RF, Lawson CL, Hanson RJ.. Solving least squares problems. J Am Stat Assoc 1977;72:930–1. [Google Scholar]
- Lee J, Lee AJ, Lee J-K. et al. Mutalisk: a web-based somatic MUTation AnaLyIS toolKit for genomic, transcriptional and epigenomic signatures. Nucleic Acids Res 2018;46:W102–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee-Six H, Olafsson S, Ellis P. et al. The landscape of somatic mutation in normal colorectal epithelial cells. Nature 2019;574:532–7. [DOI] [PubMed] [Google Scholar]
- Li S, Crawford FW, Gerstein MB.. Using sigLASSO to optimize cancer mutation signatures jointly with sampling likelihood. Nat Commun 2020;11:3575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manders F, Brandsma AM, de Kanter J. et al. MutationalPatterns: the one stop shop for the analysis of mutational processes. BMC Genomics 2022;23:134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martincorena I, Campbell PJ.. Somatic mutation in cancer and normal cells. Science 2015;349:1483–9. [DOI] [PubMed] [Google Scholar]
- Olafsson S, McIntyre RE, Coorens T. et al. Somatic evolution in non-neoplastic IBD-affected colon. Cell 2020;182:672–84.e11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenthal R, McGranahan N, Herrero J. et al. DeconstructSigs: delineating mutational processes in single tumors distinguishes DNA repair deficiencies and patterns of carcinoma evolution. Genome Biol 2016;17:31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steele CD, Abbasi A, Islam SMA. et al. Signatures of copy number alterations in human cancer. Nature 2022;606:984–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stratton MR, Campbell PJ, Futreal PA.. The cancer genome. Nature 2009;458:719–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tate JG, Bamford S, Jubb HC. et al. COSMIC: the catalogue of somatic mutations in cancer. Nucleic Acids Res 2019;47:D941–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida K, Gowers KHC, Lee-Six H. et al. Tobacco smoking and somatic mutations in human bronchial epithelium. Nature 2020;578:266–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data Data availability statement can be found in the supplementary materials.