Abstract
Like the translational elongation factor EF-Tu, RNase P interacts with a large number of substrates where RNase P with its RNA subunit generates tRNAs with matured 5′ termini by cleaving tRNA precursors immediately 5′ of the residue at +1, i.e. at the position that corresponds to the first residue in tRNA. Most tRNAs carry a G+1C+72 base pair at the end of the aminoacyl acceptor-stem whereas in tRNAGln G+1C+72 is replaced with U+1A+72. Here, we investigated RNase P RNA-mediated cleavage as a function of having G+1C+72 versus U+1A+72 in various substrate backgrounds, two full-size tRNA precursors (pre-tRNAGln and pre-tRNATyrSu3) and a model RNA hairpin substrate (pATSer). Our data showed that replacement of G+1C+72 with U+1A+72 influenced ground state binding, cleavage efficiency under multiple and single turnover conditions in a substrate-dependent manner. Interestingly, we observed differences both in ground state binding and rate of cleavage comparing two full-size tRNA precursors, pre-tRNAGln and pre-tRNATyrSu3. These findings provide evidence for substrate discrimination in RNase P RNA-mediated cleavage both at the level of binding, as previously observed for EF-Tu, as well as at the catalytic step. In our experiments where we used model substrate derivatives further indicated the importance of the +1/+72 base pair in substrate discrimination by RNase P RNA. Finally, we provide evidence that the structural architecture influences Mg2+ binding, most likely in its vicinity.
INTRODUCTION
The majority of tRNAs in bacteria and eukaryotic cells carry a guanosine at the 5′ termini (1). One of the exceptions is tRNAGln that harbors a U at this position that pairs with an A at position 72, and throughout this study we have inferred that this is the case. The U+1A+72 base pair in tRNAGln is an important determinant in the charging of tRNAGln [(2) and references therein]. In contrast to the various specific aminoacyl tRNA synthetases, elongation factor EF-Tu binds to all elongator aminoacyl-tRNAs. However, based on determination of the equilibrium dissociation constants differences in binding between EF-Tu and different correctly aminoacylated tRNAs have been detected (3–6). Like EF-Tu, the endoribonuclease RNase P interacts with a large number of substrates where tRNA precursor transcripts constitute the majority. The role of RNase P is to generate tRNA molecules with matured 5′ termini and as such RNase P is essential and found in all different kingdoms of life (7). Bacterial RNase P consists of the catalytic RNA moiety and a basic protein in a 1:1 ratio (8–10).
Available data suggests that it is the tRNA structure of the precursor that is recognized by RNase P and that the aminoacyl acceptor stem is a major determinant (7). More specifically, base substitutions of the residues at and in the vicinity of the cleavage site affect both cleavage efficiency and cleavage site recognition (7,11–16) indicating their importance for cleavage. The majority of these studies have been conducted using one specific substrate, i.e. different full-size tRNA precursor substrates [(7); see also the more recent Ref (17)]. However, to date very few studies have analyzed in detail the consequences of base and/or base pair replacement at different positions in different substrates. Consequently, the importance of the identity of, for example, the +1/+72 base pair in different substrate contexts with respect to cleavage rates, substrate binding and cleavage site recognition remain an open question. This together with the findings in the EF-Tu system (see above) and the fact that while most tRNAs carry G+1C+72 tRNAGln harbors a U+1A+72 base pair encouraged us to investigate RNase P RNA-mediated cleavage as a function of having G+1C+72 versus U+1A+72 in various substrate backgrounds, two full-size tRNA precursors (pre-tRNAGln and pre-tRNATyrSu3) and a model RNA hairpin substrate (pATSer). Considering that available data also suggest that the RNase P protein, the C5 protein, does not interact with the residues at positions +1 and +72 [(18) and references therein], we decided to perform our experiments in the absence of the C5 protein. As model system, we used Escherichia coli RNase P RNA, M1 RNA.
Our data showed that replacement of G+1C+72 with U+1A+72 influenced ground state binding, cleavage efficiency under multiple and single turnover (under conditions where chemistry is rate limiting) in a substrate-dependent manner. Interestingly, comparison of M1 RNA-mediated cleavage of ‘wild type’ precursors to tRNAGln and tRNATyrSu3 revealed differences both in ground state binding and rate under single turnover conditions. Moreover, based on our studies using various model RNA hairpin substrate derivatives, we consider it likely that the G+1C+72 to U+1A+72 influence metal ion binding in the vicinity of the cleavage site and that it is the presence of U at +1 which is the main reason to the observed effects. These findings provide evidence for substrate discrimination in RNase P RNA-mediated cleavage, and indicate the importance of the +1/+72 base pair in this process. These data will be discussed in relation to the findings in the EF-Tu system.
MATERIALS AND METHODS
Preparation of substrates and M1 RNA
The various pATSer derivatives were purchased from Dharmacon, USA, purified, labeled at the 5′ end and gel purified according to standard procedures as described elsewhere (12,13,19). The pGln and pSu3 variants and M1 RNA were generated as run-off transcripts using T7 DNA-dependent RNA polymerase (19–21).
Cleavage assay conditions
M1 RNA activity was monitored under single turnover conditions in buffer A [50 mM Tris–HCl (pH 7.2), 5% (w/v) PEG 6000, 100 mM NH4Cl] (22) or in buffer B [50 mM Bis-Tris Propane (pH 6.1 or as indicated), 5% (w/v) PEG 6000, 100 mM NH4Cl] in the presence of MgCl2 and/or MnCl2 as indicated. The pH was adjusted with HCl and the given pH values were measured at 37°C with all components added except M1 RNA and substrate. The concentrations of M1 RNA and substrates were ≥0.24 and ≤0.05 μM, respectively. All reactions were performed at 37°C and the reaction products were separated on denaturing 20–22% (w/v) polyacrylamide gels and cleavage was quantified on a Phosphorimager (Molecular Dynamics 400S) as described elsewhere [(13) and references therein].
Determination of the site of cleavage
The frequency of cleavage at different positions was quantified from the relative amounts of 5′ cleavage products as a result of cleavage at the different positions as described elsewhere (23). In the case of the pGln derivatives, the cleavage sites were also determined by primer extension analysis using an oligonucleotide complementary to positions A31 through C48 in tRNAGln (see Figure 1).
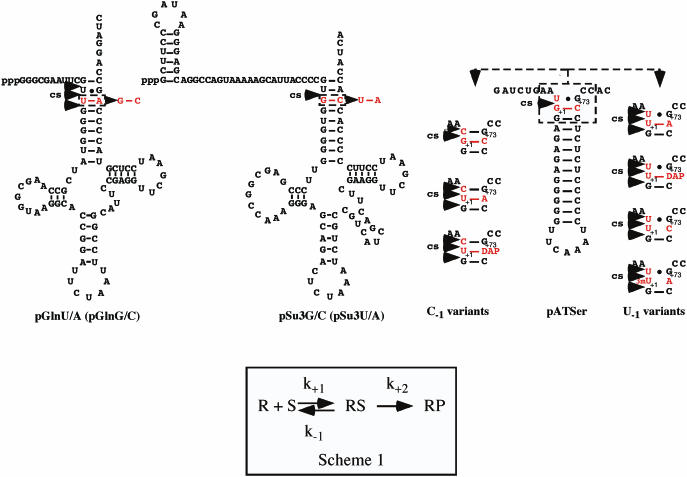
Figure 1.
Predicted secondary structure of the substrates used in this study, pGln, pSu3 and pATSer, as indicated. RNase P cleavage site(s), cs, are indicated with arrowheads and the residues that were substituted in red. Scheme 1: simplified reaction scheme where k+1 and k−1 represent rate constants for the formation and dissociation, respectively, of RNase P RNA–substrate (RS) complex, while k+2 is defined as the rate constant for cleavage of the scissile bond.
Binding assay conditions
Spin columns were used to determine apparent equilibrium dissociation constants (appKd) in Buffer C: 50 mM MES at pH 6.0 at 37°C, 0.8 M NH4OAc, 0.05% (w/v) Nonidet P40, 0.1% (w/v) SDS in the presence of various concentrations of CaCl2 as described elsewhere (24,25) except that preincubation was 20 min and the time after mixing substrate with M1 RNA was 20 min. The substrate concentration was <10 nM and M1 RNA concentration was varied from 0.01 to 13.7 μM. appKd values were determined by non-linear regression analysis using Origin 7.0 software (Originlab) and the equation fc = ft × [M1 RNA]free/(Kd + [M1 RNA]free) where fc = fraction of precursor substrate in complex with M1 RNA and ft = maximum fraction of substrate able to bind M1 RNA, i.e. endpoint.
Determination of the kinetic constants under multiple turnover conditions
The kinetic constants kcat and Km were determined under multiple turnover conditions in buffer A at 160 mM MgCl2 and pH 7.2 as described elsewhere (12,13).
Determination of the kinetic constants, kobs/Ksto and/or kobs, under single turnover conditions
The kinetic constants kobs/Ksto(kcat/Km) and/or kobs (the rate constant of cleavage) were determined following two protocols.
Protocol A was used to determine kobs/Ksto and kobs for cleavage of the various pATSer derivatives using Buffer B (pH 6.1) in the presence of 40 mM MgCl2 as described elsewhere (20,22,26). In the case of cleavage of the different tRNAGln (pGlnG/C and pGlnU/A) and tRNATyrSu3 (pSu3G/C and pSu3U/A) precursors, we used Buffer C (without Nonidet P40 and SDS) in the presence of 40 mM Mg(OAc)2. The final concentration of substrate was 40 nM while for M1 RNA the concentration was varied between 0.040 and 5.16 μM. The kobs values were obtained by linear regression from Eadie–Hofstee plots.
Protocol B was used to determine the pseudo-first order rate constant of cleavage, kobs, at higher Mg2+ concentrations, 160 and 400 mM according to Warnecke et al. (24). The kobs was calculated from single turnover experiments with 40 nM substrate and 4.1 μM M1 RNA by non-linear regression analysis using Origin 7.0 software (Originlab) and the equation fcleaved = fendpoint × (1 − e−kobs×t) where fcleaved = fraction of cleaved substrate, t = time, fendpoint = maximum cleavable fraction of substrate.
Irrespective of protocol (A or B), the 5′ cleavage fragments were used in our calculations to determine the frequency of cleavage at each time point (and/or M1 RNA concentration).
RESULTS
To investigate the impact on cleavage site recognition, rate of cleavage (kobs) and ground state binding (appKd) in M1 RNA-mediated cleavage as a result of substituting the G+1C+72 with U+1A+72 (or vice versa in the case of tRNAGln), we decided to use two full-size tRNA precursors, a tRNAGln precursor (pGln) and the tRNATyrSu3 precursor (pSu3), and one model RNA hairpin substrate, pATSer (Figure 1). Model RNA hairpin substrates like pATSer have been used to obtain information about RNase P RNA substrate interaction, cleavage site recognition, chemical groups important for cleavage and role of metal ions (12,13,22,23,27–30). We will first describe the result obtained using full-size tRNA precursors, and this will be followed by our studies using various pATSer derivatives.
Replacement of U+1A+72 to G+1C+72 in a tRNAGln precursor affected both ground state binding and rate of cleavage
The tRNAGln precursor carries U+1A+72 while the majority of tRNAs harbor G+1C+72 (see above). Thus, to understand whether this difference in the identity of the +1/+72 base pair influenced M1 RNA-mediated cleavage we replaced U+1A+72 with G+1C+72 in a tRNAGln precursor generating pGlnG/C (Figure 1; underlined residues correspond to the +1/+72 residues throughout this study). The corresponding ‘wild type’ is referred to as pGlnU/A where the underlined residues indicate G+1C+72 and U+1A+72, respectively. In addition, we substituted G+1C+72 with U+1A+72 in the tRNATyrSu3 (pSu3) precursor that normally carries a GC-base pair at +1/+72. These two variants are referred to as pSu3G/C (‘wild type’) and pSu3U/A (Figure 1). These different tRNA precursor variants were analyzed with respect to ground state binding (appKd) and rate of cleavage (kobs) under saturating single turnover conditions at pH 6.0. At this pH, available data suggest that the chemistry of cleavage is rate-limiting (22,31–34; see also below). Note that RNase T1 structural probing revealed no apparent overall structural changes due to substituting the +1/+72 base pair in these tRNA precursors (data not shown).
As shown in Table 1 substituting U+1A+72 with G+1C+72 in pGln resulted in a significant reduction in appKd, while only a modest change was observed comparing appKd for pSu3G/C and pSu3U/A. Moreover, the U+1A+72 to G+1C+72 substitution in pGln resulted in a 5-fold increase in kobs, while kobs/Ksto (= kcat/Km) was changed almost 50-fold (Table 2) in keeping with the observed difference in appKd (65-fold). Likewise, the rate of cleavage (kobs) was reduced replacing G+1C+72 with U+1A+72 in the pSu3 context (Table 2). But, in this case we observed a dramatic reduction, >150-fold. Finally, while the pSu3 derivatives were cleaved only at the correct position +1 (data not shown) the pGln derivatives were cleaved at several positions (at −1, +1 and +2), irrespective of U+1A+72 or G+1C+72 and +/− C5 protein (Figure 2A, data not shown for cleavage of pGlnG/C or in the presence of C5). Taken together, these findings suggested that replacement of U+1A+72 with G+1C+72 in pGln influenced both ground state binding and rate of cleavage under conditions where chemistry is rate limiting. By contrast, in the case of pSu3 we only observed a dramatic effect on the rate of cleavage. Thus, the observed effects as a result of substituting the +1/+72 base pair appears to depend on tRNA/tRNA precursor identity. Moreover, comparison of the rate of cleavage and ground state binding for ‘wild type’ pGlnU/A and pSu3G/C, suggested substrate discrimination in M1 RNA-mediated cleavage both at the level of binding and at the catalytic step.
Table 1.
Apparent Kd values for the different substrate variants at different Ca2+ concentrations as indicated
| Substrate | appKd (μM) | ||
|---|---|---|---|
| 40 mM | 160 mM | 400 mM | |
| pATSerUGG/C | 0.11 ± 0.056 | 0.020 ± 0.0093 | 0.026 ± 0.018 |
| pATSerUGU/A | 1.3 ± 0.078 | 0.13 ± 0.023 | 0.030 ± 0.024 |
| pATSerUGU/DAP | 4.8 ± 1.2 | 0.73 ± 0.49 | 0.94 ± 0.39 |
| pATSerCGG/C | 11 ± 7 | 0.24 ± 0.06 | 0.057 ± 0.027 |
| pATSerCGU/A | n.d. | 3.2 ± 0.38 | 1.4 ± 0.8 |
| pATSerCGU/DAP | n.d. | 0.40 ± 0.077 | 0.21 ± 0.08 |
| pGlnU/A | 0.98 ± 0.18 | n.d. | n.d. |
| pGlnG/C | 0.015 ± 0.005 | n.d | n.d. |
| pSu3G/C | 0.079 ± 0.044 | n.d. | n.d. |
| pSu3U/A | 0.14 ± 0.043 | n.d. | n.d. |
The experiments were performed as described in Materials and Methods. Each value is an average of several independent experiments and n.d. = not determined. Underlined residues correspond to the identity of the +1/+72 base pair.
Table 2.
Summary of cleavage rate constants in cleavage of different substrate variants at pH 6.1 at various Mg2+ concentrations
| Substrate | 40 mM | 160 mM | 400 mM | ||
|---|---|---|---|---|---|
| kobs | kobs/Ksto | kobs | kobs | ||
| pATSerUGG/C | |||||
| −1 | n.d. | n.d. | n.d. | ||
| +1 | 1.2 ± 0.2a | 3.8 ± 0.6 | 1.4 ± 0.18 | 0.46 ± 0.29 | |
| pATSerUGU/A | |||||
| −1 | n.d. | n.d. | n.d. | ||
| +1 | 0.0077 ± 0.0039 | 0.11 ± 0.05 | 0.10 ± 0.023 | 0.20 ± 0.10 | |
| +2 | n.d. | n.d. | n.d. | ||
| pATSerUGU/DAP | |||||
| −1 | n.d. | n.d. | n.d. | ||
| +1 | 0.0005 ± 0.00005 | 0.015 ± 0.004 | 0.016 ± 0.002 | 0.032 ± 0.010 | |
| +2 | n.d. | n.d. | n.d. | ||
| pATSerCGG/C | |||||
| −1 | 0.055 ± 0.0061 | 0.065 ± 0.0081 | |||
| +1 | n.d. | n.d. | 0.19 ± 0.026 | 0.15 ± 0.055 | |
| pATSerCGU/A | |||||
| −1 | 0.013 ± 0.001a | ||||
| +1 | n.d. | n.d. | n.d. | 0.016 ± 0.002a | |
| pATSerCGU/DAP | |||||
| −1 | 0.015 ± 0.001a | 0.0083 ± 0.0015 | |||
| +1 | n.d. | n.d. | 0.041 ± 0.002a | 0.045 ± 0.004 | |
| pGlnU/A | |||||
| −1 | n.d. | n.d. | |||
| +1 | 0.47 ± 0.12 | 0.20 ± 0.10 | n.d. | n.d. | |
| +2 | n.d. | n.d. | |||
| pGlnG/C | |||||
| −1 | n.d. | n.d. | |||
| +1 | 2.8 ± 0.50 | 9.4 ± 1.4 | n.d. | n.d. | |
| +2 | n.d. | n.d. | |||
| pSu3G/C | |||||
| +1 | 2.5 ± 0.77 | 4.3 ± 1.2 | n.d. | n.d. | |
| pSu3U/A | |||||
| +1 | 0.012 ± 0.0035 | 0.053 ± 0.027 | n.d. | n.d. | |
The experiments were performed as described in Materials and Methods. Each value is an average of several independent experiments and n.d. = not determined while −1, +1 and +2 refers to the cleavage site. kobs and kobs/Ksto (= kcat/Km) are expressed as min−1 and min−1 × μM−1, respectively.
aBased on two independent determinations. Underlined residues correspond to the identity of the +1/+72 base pair. The finding that the rates of cleavage for pATSerUGG/C, pATSerUGU/A and pATSerUGU/DAP were linear dependent on [OH−] up to at least pH 6.5 suggested that the rate of cleavage at pH 6.1 is rate limiting (data not shown).
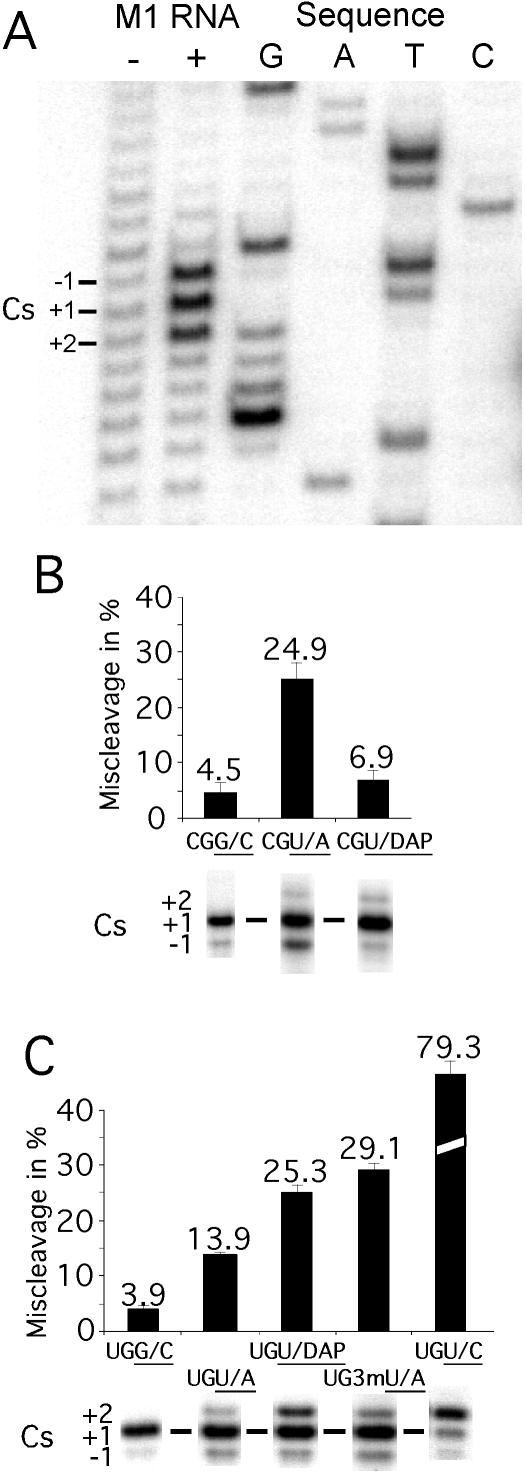
Figure 2.
Miscleavage of various substrates. (A) Identification of the cleavage site in pGlnU/A by primer extension analysis as described in Materials and Methods. In this experiment, ∼0.2 μM pGlnU/A was incubated for 8 min (after preincubation of M1 RNA for 7 min) in the presence or absence of 2 μM M1 RNA in 50 mM MES (pH 6.0) and 40 mM Mg(OAc)2. This was followed by primer extension analysis using an oligodeoxynucleotide complementary to residues A31 through C48 in tRNAGln (see Figure 1). Cs = cleavage site. (B and C) Miscleavage of various pATSer substrates [(B) with C at −1 and (C) with U at −1) represented as percentage of miscleavage and cleavage sites as indicated where for example UGG/C corresponds to the substrate pATSerUGG/C, for details see text. The 5′ cleavage fragments were separated on 22% denaturing PAGE as described in Materials and Methods.
Cleavage as a function of substituting G+1C+72 to U+1A+72 in a model hairpin substrate
To further study the consequences as a result of replacing G/C with U/A at +1/+72, we generated G+1C+72 and U+1A+72 variants of pATSer (Figure 1). The residue that pair with the +1 residue in these substrates corresponds to position +72 in tRNA precursors and will therefore be referred to as position +72. Likewise, the residue immediately 3′ of +72 corresponds to the discriminator base in tRNA and will be referred to as residue +73. In the experiments using pATSer model substrates, G+1C+72 variants were referred to as ‘wild type’ substrates. We also decided to study cleavage of a pATSer derivative carrying a U at +1 and a 2,6-diaminopurine (DAP) at position +72, where U and DAP has the potential to form a base pair with three H-bonds. In this context, we also used two substrates where U+1 cannot interact with the residue at +72 by conventional base-pairing: pATSerUG variants with U+1C+72 [but see Ref (35)] and 3mU+1A+72 (where 3mU = 3-methyl-U).
The residue at −1 plays an important role in cleavage by RNase P RNA (12–14,23). Therefore, we also studied pATSer-(+1/+72) variants with either C or U at −1 where position −1 corresponds to the position immediately 5′ of the canonical cleavage site at +1 (Figure 1).
These substrates were analyzed with respect to cleavage site recognition, cleavage rates (efficiency) under multiple as well as single turnover conditions, ground state binding (appKd = apparent Kd) and metal ion binding.
Cleavage site recognition
At pH 6.1 in the presence of 160 mM Mg2+, the pATSerCGG/C substrate is cleaved at the correct site (+1) as well as with a low frequency at position −1 in keeping with our previous observations [Figure 2B; Refs (12,22)]. Replacement of G+1C+72 with U+1A+72 or U+1DAP+72 resulted in miscleavage both at the −1 and +2 positions. However, the total frequency of miscleavage (i.e. both at −1 and at +2) of the U+1DAP+72 variant was approximately the same compared to that for the G+1C+72 variant.
Replacing C−1 with U resulted in a similar miscleavage pattern for the U+1A+72 and U+1DAP+72 variants, i.e. miscleavage at −1 and at +2. But, here a significant increase in miscleavage was observed for the U+1DAP+72 variant mainly due to an increased frequency of cleavage at +2 (Figure 2C). Just replacing G+1 with U in pATSerUGG/C or U+1 in pATSerUGU/A with a 3-methyl U also resulted in miscleavage both at −1 and at +2 where pATSerUGU/C was cleaved preferentially at +2 (Figure 2C).
We conclude that the presence of U at +1 influence cleavage site recognition however, our data suggested that this depends on substrate context (see above). Moreover, in keeping with previous data, cleavage site recognition is also affected by the identity of the −1 residue (12–14,22). Interestingly, increasing the pH to 7.2 resulted in approximately a 2-fold increase in miscleavage for the C−1 derivatives, while for the U−1 variants no change was detected irrespective of modification at +1/+72 (data not shown) consistent with our previous findings (22).
Replacement of G+1C+72 influenced the efficiency of cleavage under multiple turnover conditions
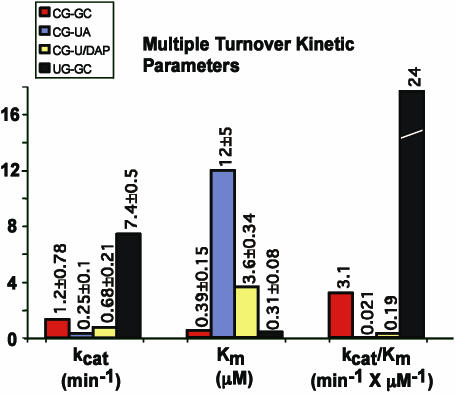
We previously reported the kinetic constants for cleavage of several pATSerCG variants under multiple turnover conditions at pH 7.2 (12,13,23,27). Hence, we first determined cleavage efficiency for the various +1/+72 derivatives by calculating kcat/Km values based on determination of kcat and Km under multiple turnover conditions at pH 7.2 and 160 mM Mg2+ (see Materials and Methods). As shown in Figure 3, reductions ranging between 16- and ∼150-fold in kcat/Km (for cleavage at +1) for pATSerCGU/DAP and pATSerCGU/A, respectively, were observed compared to cleavage of the corresponding G+1C+72 variant. This was mainly due to an increased Km (≈10-fold) for the former, while for pATSerCGU/A kcat was down 5-fold and Km was increased 30-fold. In keeping with our previous data (13), pATSerUGG/C was cleaved with a higher efficiency compared to cleavage of pATSerCGG/C indicating the importance of the identity of residue −1 (Figure 3). These data indicated that replacement of G+1C+72 with U+1A+72 (or U+1DAP+72) influence the efficiency of cleavage also in the context of the model substrate, pATSer.
Figure 3.
Kinetic constants of various pATSerCG derivatives cleaved by M1 RNA under multiple turnover conditions as indicated. The constants were determined at pH 7.2 as described in Materials and Methods. Given kcat and Km values are averages of several independent experiments and experimental errors are given as ‘±’. The kcat/Km values were calculated using the kcat and Km numbers.
Replacement of G+1C+72 influenced the rate of cleavage under conditions where chemistry is rate limiting also in the context of a model hairpin substrate
At pH 7.2, chemistry is not rate limiting, therefore these data do not provide information about the importance of the G+1C+72 to U+1A+72 change with respect to the chemistry of cleavage. We therefore determined the rate constant kobs for cleavage under single turnover conditions at pH 6.1 where chemistry of cleavage is suggested to be rate limiting [(22,31–34); see also Table 2 legend]. Furthermore, our initial experiments where we studied rate of cleavage under single turnover conditions as a function of increasing Mg2+-concentration indicated different Mg2+ requirements due to replacing G+1C+72 with U+1A+72 (Figure 4A). We therefore determined kobs at different Mg2+ concentrations. The results are summarized in Table 2.
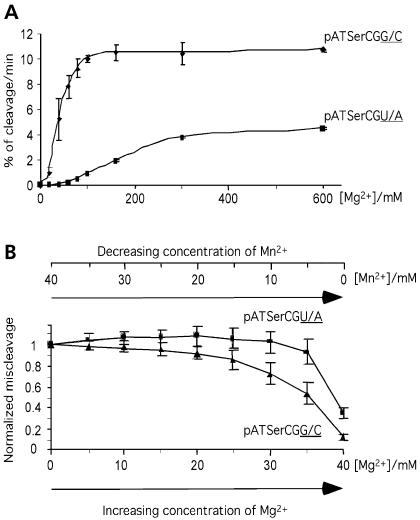
Figure 4.
Cleavage of pATSerCGG/C and pATSerCGU/A by M1 RNA at different concentrations of Mg2+ (A) and [Mg2+]/([Mg2+] + [Mn2+]) ratios (B). (A) Cleavage rates pATSerCGG/C and pATSerCGU/A expressed as percentage of cleavage per minute. (B) Normalized miscleavage at −1 of pATSerCGG/C and pATSerCGU/A as a function of [Mg2+]/([Mg2+] + [Mn2+]) as indicated. The total divalent metal ion concentration was kept constant at 40 mM and the concentrations of Mg2+ and Mn2+ were varied as indicated. The curves are averages of several independent experiments and the bars indicate experimental errors. The experiments were performed at 37°C as outlined in Materials and Methods under single turnover conditions at pH 6.1. pATSerCGG/C = triangles; pATSerCGU/A = squares.
In general, replacement of G+1C+72 to any of the other combinations tested resulted in a reduction in the rate of cleavage at the correct (+1) position irrespective of U or C at −1 with the exception of when kobs values for cleavage of the U+1DAP+72 derivatives were compared (see below). The extreme case was the 2500-fold decrease in rate of cleavage of pATSerUGU/DAP at 40 mM Mg2+ while kobs was reduced >150-fold for pATSerUGU/A at the same [Mg2+]. By increasing the Mg2+-concentration, a rescue in the rate of cleavage for the U−1-carrying variants was observed and at 400 mM Mg2+, the rates of cleavage for the U+1A+72 and U+1DAP+72 variants were only down 2.8- and 16-fold, respectively, compared to cleavage of the corresponding G+1C+72 variant, pATSerUGG/C.
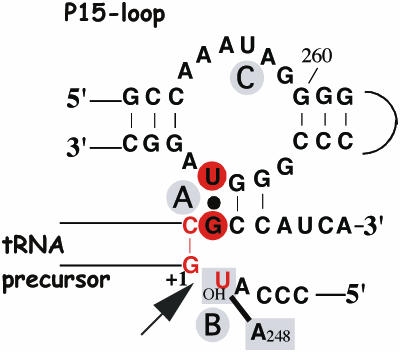
The C−1 derivatives pATSerCGG/C and pATSerCGU/A were cleaved with lower rates relative to the corresponding U−1 derivatives irrespective of [Mg2+] indicating the importance of the −1 residue (and/or +73, see below). Comparing kobs for cleavage of the U+1DAP+72 variants suggested an increased rate of cleavage with a C at position −1. Moreover, in contrast to what we observed for the U−1-variants, no rescue in the rate of cleavage was observed for the C−1 derivatives as a result of increasing [Mg2+] under the conditions tested. It is conceivable that this difference is influenced by the fact that C−1 is base-paired to G+73 in the substrate (see Figure 5).
Figure 5.
Illustration of the ‘RCCA–RNase P RNA interaction’ (interacting residues underlined) and the interaction between A248 and residue −1 in the substrate, the ‘A248/N−1 interaction’ [for details see the text and (14,22), and references therein]. The residues indicated in red correspond to those that were replaced in this report. The residues in red circles show the ‘+73/294 interaction’ while the arrow mark the canonical RNase P cleavage site at +1. In the case of the C−1 variants, C−1 is inferred to base pair with G+73 in the absence of RNase P RNA. A and B (in gray circles) represent Mg2+ ions that have been suggested to be positioned at and in the vicinity of the cleavage site [(22) and references therein] while C is an additional ion positioned in the P15-loop. The 2′-OH at the −1 position has been suggested coordinating Mg2+ at the cleavage site, and/or interacting with RNase P RNA (14,15,22,28,29,31,34).
In conclusion, replacing G+1C+72 and/or the −1 residue resulted in changes in the rate of cleavage under conditions where the rate of cleavage is rate limiting in the context of the model RNA hairpin substrate. Our data also indicated that substituting G+1C+72 to U+1A+72 or U+1DAP+72 influenced Mg2+ binding (see also below), conceivably the Mg2+-ion(s) bound in the vicinity of the cleavage site. It also appears that the influence of Mg2+ was dependent on the identity of the −1 residue. In the case of U+1DAP+72, we have to consider that the introduction of DAP at this position also affects the electrostatics. Therefore, this might be part of the reason for our observations using the U+1DAP+72 variants.
The kobs values at 40 mM Mg2+ were determined from Eadie–Hofstee plots (Vi = f(Vi/[M1 RNA]). The intercept with the x-axis gives estimates of kobs/Ksto (= kcat/Km), which corresponds to the association rate constants k+1 (assuming that k−1<k+2, where k−1 represents the dissociation constant and k+2 the rate constant for cleavage, see scheme 1 in Figure 1), for the pATSerUG-(+1/+72) derivatives tested under these conditions. Based on this, our data suggested that replacing G+1C+72 with U+1A+72 and U+1DAP+72 resulted in a 35- and 250-fold decrease in k+1, respectively (kobs/Ksto; Table 2).
Replacing G+1C+72 and the −1 residue in the model substrate affected ground state binding
Next, we investigated whether substitution of G+1C+72 with U+1A+72 (or U+1DAP+72) and/or the −1 residue influenced ground state binding. As shown in Table 1, substituting G+1C+72 with U+1A+72 or U+1DAP+72 resulted in increased appKd values irrespective of Mg2+-concentration and U or C at the −1 position. But, comparison of appKd values for pATSerUGG/C and pATSerUGU/A showed a decreasing difference in appKd with increasing Mg2+. This was not observed in the case of the U+1DAP+72 variant. Moreover, comparing appKd values for the U+1DAP+72 derivatives with the respective ‘wild type’ values, i.e. appKd for pATSerUGG/C and pATSerCGG/C, respectively, showed a larger difference in the case where the substrate carried a U at −1 (≈40-fold versus 1.7-fold difference at 160 mM Ca2+). The appKd values were also found to be generally higher for the substrates carrying a C−1 compared to substrates with U−1. The reason to these observations might be that the C−1 and G+73 interact via base-pairing in the absence of M1 RNA (see also above). As a consequence, this would make these residues less accessible to interact with M1 RNA upon M1 RNA–substrate (RS) complex formation, e.g. establishing the +73/294 interaction [Figure 5; (13)]. In addition, U−1 would probably result in a more favorable interaction with A248 in M1 RNA than C−1 [(14); see also (12)]. To conclude, in addition to the influence on the rate of cleavage and possibly Mg2+ binding, replacing G+1C+72 with U+1A+72 (or U+1DAP+72) and the residue at −1 influenced ground state binding. This is similar to what was observed comparing binding of the pGln variants, but different in relation to when the pSu3 derivatives were used (see above).
Replacing G+1C+72 influenced metal ion binding in its vicinity
The data presented above suggested that replacement of G+1C+72 with U+1A+72 affects metal ion binding possibly in its vicinity (see Discussion). To study this in more detail, we took advantage of the fact that cleavage of pATSerC/GG/C results in substantial miscleavage at −1 in the presence of Mn2+. The addition of Mg2+ (without changing the total metal ion concentration, i.e. [Mg2+] + [Mn2+]) to the cleavage reaction suppresses miscleavage at −1 [Figure 4B; (12,13,23,27)]. We have used this approach elsewhere to obtain information about chemical groups important for Mg2+ binding in the M1 RNA cleavage reaction (12). Here, we studied the frequency of miscleavage as a function of [Mg2+]/([Mg2+] + [Mn2+]) for pATSerC/GG/C and pATSerC/GU/A. The data in Figure 4B showed that higher [Mg2+] was required to suppress the Mn2+-induced miscleavage as a result of replacement of G+1C+72 with U+1A+72. These findings supported the suggestion that the G+1C+72 to U+1A+72 change influences Mg2+ binding in the M1 RNA substrate complex.
DISCUSSION
Substrate discrimination in RNase P RNA-mediated cleavage
RNase P has to recognize and cleave tRNA precursors irrespective of whether it has a G/C or U/A base pair at position +1/+72 (or any other combination, e.g. C+1A+72 or C+1G+72 as in the precursors to tRNAfMet and tRNAPro, respectively). Here, we demonstrated that replacement of G+1C+72 with U+1A+72 in full-size tRNA precursor substrates and a model hairpin–loop substrate influenced cleavage by M1 RNA (RNase P RNA derived from E.coli) by affecting: ground state binding, rate of cleavage under conditions where chemistry is suggested to be rate limiting, cleavage site recognition and most likely Mg2+ binding in the vicinity of the cleavage site. These findings are of particular relevance in view of the presence of U+1A+72 in the wild-type tRNAGln precursor while most other tRNA precursors in bacterial cells carry G+1C+72 (1). Indeed, the tRNAGln precursor, ‘wild type’ pGlnU/A, used here was shown to bind to M1 RNA with a 10-fold lower binding affinity compared to the ‘wild type’ tRNATyrSu3 precursor pSu3G/C (this report). Hence, these findings provide evidence for substrate discrimination in RNase P RNA-mediated cleavage at the level of substrate binding. As RNase P, EF-Tu also interacts with a large number of different substrates, i.e. aminoacylated tRNAs, and a 10-fold difference in binding between EF-Tu and different correctly aminoacylated tRNA was reported [(6); see also (3–5)]. Thus, both RNase P RNA and EF-Tu discriminate at the level of substrate binding. It is conceivable that this is related to that RNase P RNA and EF-Tu bind the same tRNA regions: the aminoacyl acceptor stem, T-stem and T-loop (7,36,37). However, our kinetic data as determined under saturating single turnover conditions also suggested substrate discrimination at the catalytic step for RNase P RNA: 5-fold difference in kobs for pGlnU/A and pSu3G/C (Table 2).
Taken together, based on our studies where we replaced U+1A+72 with G+1C+72 in pGlnU/A (or vice versa in pSu3 and pATSer), we conclude that the +1/+72 plays an important role in substrate discrimination by RNase P RNA, and it will be of interest to investigate whether this is also the case for EF-Tu. Moreover, our findings provide evidence that the U+1A+72 base pair in tRNAGln plays a role not only in the interaction with glutaminyl-tRNA synthetase (38), but also in tRNA processing. This extends and supports the suggestion that different substrates interact differently with RNase P RNA (20,30,39,40). Note that changing the structural architecture at the cleavage site did not affect the overall structure of the substrate however, with respect to local structural consequences (see below).
The +1/+72 base pair and substrate context
Previous studies using P RNA from Bacillus subtilis indicated that substitution of G+1C+72 with U+1A+72 (A+1U+72 or C+1G+72) resulted only in modest effects on binding and chemistry of cleavage of a yeast tRNAPhe precursor in the context when the 2′OH at −1 had been replaced with a 2′H (15). But, in the presence of the B.subtilis RNase P protein, changing of G+1C+72 to U+1A+72 (A+1U+72 or C+1G+72) resulted in a significantly larger effect on both binding and rate of cleavage [(15); see also below]. Moreover, certain tRNA-like structures carrying a U/A base pair at the position corresponding to +1/+72 were cleaved at more than one position (41), similar to our findings using pATSer and pGln. However, these studies provided very little information about the consequences of replacing the +1/+72 base pair in different substrate contexts with respect to ground state binding, rate of cleavage and cleavage site recognition. As demonstrated here, substrate context is an important aspect to elucidate the impact on RNase P RNA-mediated cleavage caused by changes at the cleavage site, i.e. the +1/+72 base pair. The importance of context is further emphasized by our data where we used the model substrate variants with U and C at position −1. Here, we observed that the presence of an amino group, i.e. the presence of a H-donor, in the shallow groove in the substrate (due to the presence of DAP, at +72) influenced binding to a larger extent in the presence of U−1 compared to C−1 (Table 1). The significance of this is presently not clear, but it might be associated by having U at both −1 and +1 that influence the interaction with A248 and/or point to the interplay between the different components of the active site (see also below). It is also conceivable that the effects we observed are caused by a change in the electrostatics due to the introduction of DAP at position 72.
To conclude, these data indicate the importance of substrate context and structural architecture (in particular the +1/+72 base pair) of the cleavage site in RNase P RNA-mediated cleavage. Moreover, based on our studies, it is likely that the presence of U at +1 (and not A at +72) is the reason for the effects observed in the present study and in previous in vitro studies. Together this rationalizes earlier in vivo findings where it was suggested that RNase P processing of the dimeric Pro-Ser tRNA precursor was blocked due to replacement of G+1 with U+1 in tRNASer (42); the suppression efficiency for SupE (Su2/glnV) encoding tRNAGln was significantly reduced in a mutant RNase P protein (rnpA49) background (43). We also note the importance of the identity of the +1/+72 base irrespective of type of RNase P RNA, i.e. type A as represented by M1 RNA and B.subtilis P RNA that belongs to type B [(44); see also (20)].
Differences in cleavage and ground state binding of different substrates and residue −1
Recent data suggested that the residue at the −1 position immediately 5′ of the scissile bond interacts with residue A248 in M1 RNA [(11); see also (9)]. In E.coli as well as in other bacteria, a U at this position is the preferred residue (12–14,39). Substitution of G+1 with U+1 might therefore affect correct interaction with A248 and thereby influence the cleavage site recognition process resulting in miscleavage as observed in the present study for the model substrate pATSer. This is corroborated by our findings using the U+1C+72 and 3mU+1A+72 variants. In the case of the former, the U at +1 would become accessible to interact with A248 (Figure 5) resulting in cleavage preferentially at +2 whereas the presence of the methyl at position 3 would influence the potential interaction between A248 and 3mU+1. It is also likely, but not mutually exclusive, that Mg2+ binding in the vicinity of the cleavage site is affected as a result of substituting G+1C+72 (see also below) and as a consequence this results in miscleavage. Thus, our data are in keeping with the establishment of a functionally important interaction between the −1 residue in the 5′ leader and A248 [Figure 5; (14); see also (12)].
A U at +1 in pSu3 did not result in miscleavage. This can be rationalized by our previous findings that cleavage site recognition of pSu3 is dictated by several determinants (11) and changing one in pSu3 is not enough to result in cleavage at other positions. Moreover, pGln that naturally carries U+1A+72 was miscleaved at several positions irrespective of the identity of +1/+72, i.e. U+1A+72 or G+1C+72, or absence/presence of the RNase P protein C5. Thus, other factors must play a role in cleavage site recognition of this substrate. For example, the presence of the 6 nt long 3′ trailer and/or that the tRNAGln are not naturally transcribed as a monomeric precursor in E.coli, but as part of a multimeric tRNA gene transcript. Also, the length and structure of the 5′ leader might play a role. Indeed, earlier data suggested that the 43 nt long 5′ leader of pSu3 is involved in stabilizing the M1 RNA substrate interaction (45). This would be in keeping with the fact that no significant change in ground state binding was observed as a result of replacing G+1C+72 with U+1A+72 in pSu3.
We also observed that replacing U−1 with C−1 in the model pATSer substrate resulted in a significant increase in ground state binding. This is in keeping with Zahler et al. (14) who used a tRNA precursor substrate. As discussed in detail elsewhere, one likely reason for this is that C−1 in these substrates are engaged in base pairing with G73, and this results in G73 being less accessible for interaction with U294 in M1 RNA [Figure 5; (12–14,22,46)]. In addition, we observed a significant difference in the rate of cleavage (kobs) as a result of introduction of C at −1 (Table 2). Harris and coworkers using a tRNA precursor substrate did not detect any difference in the rate of cleavage comparing cleavage with U−1 versus C−1 (14). But, Loria and Pan (15) did observe that changing the 5′ leader, where one carried U−1 and the other C−1, resulted in different rates of cleavage. Together, this again indicates differences in cleavage of different substrates and corroborates the importance of the −1 residue. It is conceivable that binding of Mg2+ does play a role in this, given the difference in response to the Mg2+ concentration where the C−1 variants required higher Mg2+ concentration than the U−1 derivatives. Moreover, these differences again emphasize the importance to study different substrates [see also Discussion and ref. 22].
Structural architecture of the cleavage site and possible role of Mg2+
Although the overall structure did not change, as a result of replacing G+1C+72 with U+1A+72 (or vice versa) and/or substitution of the residue at −1 (see above), the local structural topography has been changed by definition. From the above and previous data [Ref (22) and references therein; see also (47)], it is clear that different chemical groups at and in the vicinity of the cleavage site are involved in aligning the scissile bond to ensure efficient and correct cleavage. At present, we do not have access to a high-resolution structure of the RNase P cleavage site in the RS-complex. Nonetheless, a picture is emerging based on genetic and biochemical data from several laboratories, and our current understanding of RNase P RNA substrate interactions are modeled in Figure 5 (for references see legend in Figure 5). Moreover, based on our present results we can infer the following: Hexahydrated Mg2+ ions bind to RNA and structural studies suggested binding in the deep groove of RNA [(48); see also for example (49)]. The structural studies further suggested that in one of the binding modes O6 and N7 of the guanosine of GpN in the deep groove are involved in hydrogen bonding with the hexahydrated ion. Several studies indicate that Mg2+ is bound in the vicinity of the cleavage site [(22,29,31,34,50,51) and unpublished data]. In this report, where we studied cleavage/ground state binding as a function of replacing G+1C+72 with U+1A+72, our data suggested an influence on Mg2+ binding. Since chemical groups of the +1/+72 are positioned very close to the scissile bond, these are likely to affect the structural architecture of the center of action. Given the importance of Mg2+ for cleavage, it is not unlikely therefore that changing the structure of the +1/+72 base pair results in modulation of Mg2+ binding in its vicinity and as a consequence affects RNase P RNA-mediated cleavage as we observed in the present report. This would be consistent with the importance of O6 and N7 in binding of hexahydrated Mg2+ (see above) and that a G at the +1 position is ≈80% conserved in tRNA precursors, and as such makes a positive contribution to catalysis [(7) and references therein]. This would also rationalize the finding in the B.subtilis RNase P system that an influence on both substrate binding and rate of cleavage was observed at lower Mg2+-concentrations when G+1C+72 was replaced with, for example, U+1A+72 (15,16).
To conclude, RNase P and elongation factor EF-Tu bind all tRNA precursors and elongator aminoacyl-tRNAs, respectively, in the cell (see above). In the RNase P case, the +1/+72 base pair and most likely Mg2+ is suggested to play an important role in this process. It will therefore be of interest to understand whether the +1/+72 base pair is also a determinant for EF-Tu. If the +1/+72 base pair indeed plays a role, then the question arises whether potential Mg2+ binding in the vicinity of this base pair also influences the discrimination in the interaction between different aminoacylated tRNAs and EF-Tu.
Acknowledgments
We thank Dr C. Guerrier-Takada for the gift of the plasmid encoding ‘wild type’ pGln. Drs S. Dasgupta and S.G. Svärd are acknowledged for critical reading of the manuscript, and all our colleagues for discussions. This work was supported by grants from the Swedish Natural Research Council and the Foundation for Strategic Research Nucleic Acid Strategic Network to L.A.K. Funding to pay the Open Access publication charges for this article was provided by grants to L.A.K. from the Swedish Research Council and the Foundation for Strategic Research Nucleic Acid Strategic Network.
Conflict of interest statement. None declared.
REFERENCES
- 1.Sprinzl M., Horn C., Brown M., Ioudovitch A., Steinberg S. Compilation of tRNA sequences and sequences of tRNA genes. Nucleic Acids Res. 1998;26:148–153. doi: 10.1093/nar/26.1.148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Giégè R., Sissler M., Florentz C. Universal rules and idiosyncratic features in tRNA identity. Nucleic Acids Res. 1998;26:5017–5035. doi: 10.1093/nar/26.22.5017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Louie A., Ribeiro N.S., Reid B.R., Jurnak F. Relative affinities of all Escherichia coli aminoacyl-tRNAs for elongation factor Tu-GTP. J. Biol. Chem. 1984;259:5010–5016. [PubMed] [Google Scholar]
- 4.Abrahamson J.K., Laue T.M., Miller D.L., Johnson A.E. Direct determination of the association constant between elongation factor Tu-GTP and aminoacyl-tRNA using fluorescence. Biochemistry. 1985;24:692–700. doi: 10.1021/bi00324a023. [DOI] [PubMed] [Google Scholar]
- 5.Louie A., Jurnak F. Kinetic studies of Escherichia coli elongation factor Tu-guanosine 5′-triphosphate-aminoacyl-tRNA complexes. Biochemistry. 1985;24:6433–6439. doi: 10.1021/bi00344a019. [DOI] [PubMed] [Google Scholar]
- 6.LaRiviere F.J., Wolfson A.D., Uhlenbeck O.C. Uniform binding of aminoacyl-tRNAs to elongation factor Tu by thermodynamic compensation. Science. 2001;294:165–168. doi: 10.1126/science.1064242. [DOI] [PubMed] [Google Scholar]
- 7.Altman S., Kirsebom L.A. Ribonuclease P. In: Gesteland R.F., Cech T.R., Atkins J.F., editors. The RNA World, 2nd edn. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1999. pp. 351–380. [Google Scholar]
- 8.Guerrier-Takada C., Gardiner K., Marsh T., Pace N., Altman S. The RNA moiety of ribonuclease P is the catalytic subunit of the enzyme. Cell. 1983;35:849–857. doi: 10.1016/0092-8674(83)90117-4. [DOI] [PubMed] [Google Scholar]
- 9.Vioque A., Arnez J., Altman S. Protein–RNA interactions in the RNase P holoenzyme from Escherichia coli. J. Mol. Biol. 1988;202:835–848. doi: 10.1016/0022-2836(88)90562-1. [DOI] [PubMed] [Google Scholar]
- 10.Fang X.W., Yang X.J., Littrell K., Niranjanakumari S., Thiyagarajan P., Fierke C.A., Sosnick T.R., Pan T. The Bacillus subtilis RNase P holoenzyme contains two RNase RNA and two RNase P protein subunits. RNA. 2001;7:233–241. doi: 10.1017/s1355838201001352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kirsebom L.A. RNase P—a ‘Scarlet Pimpernel’. Mol. Microbiol. 1995;17:411–420. doi: 10.1111/j.1365-2958.1995.mmi_17030411.x. [DOI] [PubMed] [Google Scholar]
- 12.Brännvall M., Pettersson B.M.F., Kirsebom L.A. The residue immediately upstream of the RNase P cleavage site is a positive determinant. Biochimie. 2002;84:693–703. doi: 10.1016/s0300-9084(02)01462-1. [DOI] [PubMed] [Google Scholar]
- 13.Brännvall M., Pettersson B.M.F., Kirsebom L.A. Importance of the +73/294 interaction in Escherichia coli RNase P RNA substrate complexes for cleavage and metal ion coordination. J. Mol. Biol. 2003;325:697–709. doi: 10.1016/s0022-2836(02)01195-6. [DOI] [PubMed] [Google Scholar]
- 14.Zahler N.H., Christian E.L., Harris M.E. Recognition of the 5′ leader of pre-tRNA substrates by the active site of ribonuclease P. RNA. 2003;9:734–745. doi: 10.1261/rna.5220703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Loria A., Pan T. Recognition of the 5′ leader and the acceptor stem of a pre-tRNA substrate by the ribozyme from Bacillus subtilis RNase P. Biochemistry. 1998;37:10126–10133. doi: 10.1021/bi980220d. [DOI] [PubMed] [Google Scholar]
- 16.Loria A., Niranjanakumari S., Fierke C.A., Pan T. Recognition of a pre-tRNA substrate by the Bacillus subtilis RNase P holoenzyme. Biochemistry. 1998;37:15466–15473. doi: 10.1021/bi9816507. [DOI] [PubMed] [Google Scholar]
- 17.Harris M.E., Christian E.L. Recent insights into the structure and function of the ribonucleoprotein enzyme ribonuclease P. Curr. Opin. Struct. Biol. 2003;13:325–333. doi: 10.1016/s0959-440x(03)00069-1. [DOI] [PubMed] [Google Scholar]
- 18.Tsai H.-Y., Masquida B., Biswas R., Westhof E., Gopalan V. Molecular modeling of the three-dimensional structure of the bacterial RNase P holoenzyme. J. Mol. Biol. 2003;325:661–675. doi: 10.1016/s0022-2836(02)01267-6. [DOI] [PubMed] [Google Scholar]
- 19.Kufel J., Kirsebom L.A. The P15-loop of Escherichia coli RNase P RNA is an autonomous divalent metal ion binding domain. RNA. 1998;4:777–788. doi: 10.1017/s1355838298970923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Brännvall M., Mattsson J.G., Svärd S.G., Kirsebom L.A. RNase P RNA structure and cleavage reflect the primary structure of tRNA genes. J. Mol. Biol. 1998;283:771–783. doi: 10.1006/jmbi.1998.2135. [DOI] [PubMed] [Google Scholar]
- 21.Milligan J.F., Groebe D.R., Whiterell G.W., Uhlenbeck O.C. Oligoribonucleotide synthesis using T7 RNA polymerase and DNA templates. Nucleic Acids Res. 1987;15:8783–8798. doi: 10.1093/nar/15.21.8783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Brännvall M., Kikovska E., Kirsebom L.A. Cross talk between the +73/294 interaction and the cleavage site in RNase P RNA mediated cleavage. Nucleic Acids Res. 2004;32:5418–5429. doi: 10.1093/nar/gkh883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Brännvall M., Kirsebom L.A. Manganese ions induce miscleavage in the Escherichia coli RNase P RNA-catalyzed reaction. J. Mol. Biol. 1999;292:53–63. doi: 10.1006/jmbi.1999.3048. [DOI] [PubMed] [Google Scholar]
- 24.Warnecke J.M., Held R., Busch S., Hartmann R.K. Role of metal ions in the hydrolysis reaction catalyzed by RNase P RNA from Bacillus subtilis. J. Mol. Biol. 1999;290:433–445. doi: 10.1006/jmbi.1999.2890. [DOI] [PubMed] [Google Scholar]
- 25.Beebe J.A., Fierke C.A. A kinetic mechanism for cleavage of precursor tRNAAsp catalyzed by the RNA component of Bacillus subtilis ribonuclease P. Biochemistry. 1994;33:10294–10304. doi: 10.1021/bi00200a009. [DOI] [PubMed] [Google Scholar]
- 26.Tallsjö A., Kirsebom L.A. Product release is a rate-limiting step during cleavage by the catalytic RNA subunit of Escherichia coli RNase P. Nucleic Acids Res. 1993;21:51–57. doi: 10.1093/nar/21.1.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Brännvall M., Kirsebom L.A. Metal ion cooperativity in ribozyme cleavage of RNA. Proc. Natl Acad. Sci. USA. 2001;98:12943–12947. doi: 10.1073/pnas.221456598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Perreault J-P., Altman S. Important 2′-hydroxyl groups in model substrates for M1 RNA, the catalytic RNA subunit of RNase P from Escherichia coli. J. Mol. Biol. 1992;226:399–409. doi: 10.1016/0022-2836(92)90955-j. [DOI] [PubMed] [Google Scholar]
- 29.Perreault J-P., Altman S. Pathway of activation by magnesium ions of substrates for the catalytic subunit of RNase P RNA from Escherichia coli. J. Mol. Biol. 1993;230:750–756. doi: 10.1006/jmbi.1993.1197. [DOI] [PubMed] [Google Scholar]
- 30.Pan T., Jakacka M. Multiple substrate binding sites in the ribozyme from Bacillus subtilis RNase P. EMBO J. 1996;15:2249–2255. [PMC free article] [PubMed] [Google Scholar]
- 31.Smith D., Pace N.R. Multiple magnesium ions in the ribonuclease P reaction mechanism. Biochemistry. 1993;32:5273–5281. doi: 10.1021/bi00071a001. [DOI] [PubMed] [Google Scholar]
- 32.Warnecke J.M., Fürste J.P., Hardt W.-D., Erdmann V.A., Hartmann R.K. Ribonuclease P (RNase P) RNA is converted to a Cd2+-ribozyme by a single Rp-phosphorothioate modification in the precursor tRNA at the RNase P cleavage site. Proc. Natl Acad. Sci. USA. 1996;93:8924–8928. doi: 10.1073/pnas.93.17.8924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Loria A., Pan T. Recognition of the T stem–loop of a pre-tRNA substrate by the ribozyme from Bacillus subtilis ribonuclease P. Biochemistry. 1997;36:6317–6325. doi: 10.1021/bi970115o. [DOI] [PubMed] [Google Scholar]
- 34.Persson T., Cuzic S., Siedler S., Hartmann R.K. Catalysis by RNase P RNA: unique features and unprecedented active site plasticity. J. Biol. Chem. 2003;278:43394–43401. doi: 10.1074/jbc.M305939200. [DOI] [PubMed] [Google Scholar]
- 35.Holbrook S.R., Cheong C., Tinoco I., Jr, Kim S-H. Crystal structure of an RNA double helix incorporating a track of non-Watson–Crick base pairs. Nature. 1991;353:579–581. doi: 10.1038/353579a0. [DOI] [PubMed] [Google Scholar]
- 36.Nissen P., Kjeldgaard M., Thirup S., Polekhina G., Reshetnikova L., Clark B.F., Nyborg J. Crystal structure of the ternary complex of Phe-tRNA(Phe), EF-Tu, and a GTP analogue. Science. 1995;270:1464–1472. doi: 10.1126/science.270.5241.1464. [DOI] [PubMed] [Google Scholar]
- 37.Nissen P., Thirup S., Kjeldgaard M., Nyborg J. The crystal structure of Cys-tRNA(Cys)-EF-Tu-GDPNP reveals general and specific features in the ternary complex and in tRNA. Structure. 1999;7:143–156. doi: 10.1016/s0969-2126(99)80021-5. [DOI] [PubMed] [Google Scholar]
- 38.Rath V.L., Silvianm L.F., Beijer B., Sproat B.S., Steitz T.A. How glutaminyl-tRNA synthetase selects glutamine. Structure. 1998;6:439–449. doi: 10.1016/s0969-2126(98)00046-x. [DOI] [PubMed] [Google Scholar]
- 39.Kufel J., Kirsebom L.A. Different cleavage sites are aligned differently in the active site of M1 RNA, the catalytic subunit of Escherichia coli RNase P. Proc. Natl Acad. Sci. USA. 1996;93:6085–6090. doi: 10.1073/pnas.93.12.6085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gaur R.K., Hanne A., Conrad F., Kahle D., Krupp G. Differences in the interaction of Escherichia coli RNase P RNA with tRNAs containing a short or a long extra arm. RNA. 1996;2:674–681. [PMC free article] [PubMed] [Google Scholar]
- 41.Mans R.M.W., Guerrier-Takada C., Altman S., Pleij C.W.A. Interaction of RNase P from Escherichia coli with pseudoknotted structures in viral RNAs. Nucleic Acids Res. 1990;18:3479–3487. doi: 10.1093/nar/18.12.3479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.McClain W.H., Wilson J.H., Seidman J.G. Genetic analysis of structure and function in phage T4 tRNASer. J. Mol. Biol. 1988;203:549–553. doi: 10.1016/0022-2836(88)90191-x. [DOI] [PubMed] [Google Scholar]
- 43.Kirsebom L.A., Baer M.F., Altman S. Differential effects of mutations in the protein and RNA moieties of RNase P on the efficiency of suppression by various tRNA suppressors. J. Mol. Biol. 1988;204:879–888. doi: 10.1016/0022-2836(88)90048-4. [DOI] [PubMed] [Google Scholar]
- 44.Haas E.S., Banta A.B., Harris J.K., Pace N.R., Brown J.W. Structure and evolution of ribonuclease P RNA in gram-positive. Nucleic Acids Res. 1996;24:4775–4782. doi: 10.1093/nar/24.23.4775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Guerrier-Takada C., Altman S. A physical assay for and kinetic analysis of the interactions between M1 RNA and tRNA precursor substrates. Biochemistry. 1993;32:7152–7161. doi: 10.1021/bi00079a012. [DOI] [PubMed] [Google Scholar]
- 46.Tallsjö A., Kufel J., Kirsebom L.A. Interaction between Escherichia coli RNase P RNA and the discriminator base results in slow product release. RNA. 1996;2:299–307. [PMC free article] [PubMed] [Google Scholar]
- 47.Zahler N.H., Sun L., Christian E.L., Harris M.E. The pre-tRNA nucleotide base and 2′-hydroxyl at N(−1) contribute to fidelity in tRNA processing by RNase P. J. Mol. Biol. 2005;345:969–985. doi: 10.1016/j.jmb.2004.10.080. [DOI] [PubMed] [Google Scholar]
- 48.Misra V.K., Draper D.E. A thermodynamic framework for Mg2+ binding to RNA. Proc. Natl Acad. Sci. USA. 2001;98:12456–12461. doi: 10.1073/pnas.221234598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Robinson H., Gao Y-G., Sanishvili R., Joachimiak A., Wang A.H.-J. Hexahydrated magnesium ions bind in the deep major groove and at the outer mouth of A-form nucleic acid duplexes. Nucleic Acids Res. 2000;28:1760–1766. doi: 10.1093/nar/28.8.1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zuleeg T., Hartmann R.K., Kreutzer R., Limmer S. NMR spectroscopic evidence for Mn2+ (Mg2+) binding to a precursor-tRNA microhelix near the potential RNase P cleavage site. J. Mol. Biol. 2001;305:181–189. doi: 10.1006/jmbi.2000.4299. [DOI] [PubMed] [Google Scholar]
- 51.Brännvall M., Mikkelsen N.E., Kirsebom L.A. Monitoring the structure of Escherichia coli RNase P RNA in the presence of various metal ions. Nucleic Acids Res. 2001;29:1426–1432. doi: 10.1093/nar/29.7.1426. [DOI] [PMC free article] [PubMed] [Google Scholar]