Abstract
Purpose of Review
Enormous progress has been made in understanding the genetic architecture of obesity and the correlation of epigenetic marks with obesity and related traits. This review highlights current research and its challenges in genetics and epigenetics of obesity.
Recent Findings
Recent progress in genetics of polygenic traits, particularly represented by genome-wide association studies, led to the discovery of hundreds of genetic variants associated with obesity, which allows constructing polygenic risk scores (PGS). In addition, epigenome-wide association studies helped identifying novel targets and methylation sites being important in the pathophysiology of obesity and which are essential for the generation of methylation risk scores (MRS). Despite their great potential for predicting the individual risk for obesity, the use of PGS and MRS remains challenging.
Summary
Future research will likely discover more loci being involved in obesity, which will contribute to better understanding of the complex etiology of human obesity. The ultimate goal from a clinical perspective will be generating highly robust and accurate prediction scores allowing clinicians to predict obesity as well as individual responses to body weight loss-specific life-style interventions.
Supplementary Information
The online version contains supplementary material available at 10.1007/s13679-023-00526-z.
Keywords: Obesity, Genetic variants, Epigenetic marks, Polygenic risk scores, Methylation risk scores
Introduction
Obesity rates are steadily increasing [1] and represent a major public health thread worldwide. Being an important cause for concomitant metabolic co-morbidities such as type 2 diabetes, dyslipidemia, cardiometabolic diseases including coronary artery diseases, stroke and hypertension as well as for some types of cancers [2], obesity substantially reduces life expectancy [3]. As summarised by the World Obesity Atlas 2023 [4], about 988 million people (aged > 5 years) worldwide were affected with obesity (BMI ≥ 30 kg/m2) in 2020, which is estimated to dramatically increase by 2035 to 1.914 billion. This corresponds to a proportional increase of the population with obesity from 14% in 2020 to 24% in 2035, clearly illustrating the need to prevent and treat obesity.
Obesity is a multifactorial disease being governed by both genetics and environmental factors originating from a rather “obesogenic environment” such as sedentary lifestyle with reduced energy expenditure and high calorie diet intake. The existence of a genetic background in obesity is undisputable and first evidence was provided by family [5–8], twin [9–11] and adoption [12] studies that have clearly estimated heritability rates for BMI between 40 and 70%. Genome-wide association studies (GWAS) have to a large extent contributed to an improved understanding of the genetic architecture of common obesity and have provided hundreds of novel risk variants [13–15]. However, although significant advances have been made in describing the mechanistic circuitry for a least some of these genetic variants [16, 17], identifying novel risk variants in general precedes the biological and functional understanding of how these variants act in a certain target tissue in order to increase body weight. Furthermore, the variability of BMI attributed to genetic variation is still poorly explained [15]. The major challenge here is a combination of genetics with environmental factors such as energy intake, physical activity, smoking, but also gene–gene interactions. These interactions may introduce additional inter-individual variability, illustrating the highly dynamic and complex etiology underlying the pathophysiology of obesity.
Epigenetic analyses have therefore been largely accelerated during the last years with epigenome-wide association studies (EWAS) dominating the field. Numerous genes and novel CpG sites were identified conferring changes in methylation profiles in obesity [18]. However, causal interferences in obesity are still under debate, yet a few studies implicate a causal role of obesity in inducing changes in methylation levels [19•, 20].
To translate the bench-side generated knowledge into a clinical day life and to generate a useful tool helping to predict obesity (e.g. based on BMI changes), significant effort was put in designing polygenic risk scores and more recently, also methylation risk scores. These scores represent a weighted combination of several genetic variants or methylated CpG sites at many different positions across the human genome. However, so far, the use of such scores is rather limited as reliable prediction is not yet possible or to a substantial part inaccurate. Taken together, enormous progress has been made in understanding the genetic architecture of obesity and the correlation of epigenetic marks with obesity and related traits. This review aims at highlighting current research and its challenges in genetics and epigenetics of obesity.
Genetic Background of Common Polygenic Obesity
Lessons from monogenic obesity have significantly contributed to our general knowledge on genetics and physiology of body weight regulation. However, non-syndromic monogenic obesity affects only about 5% of the population with obesity [21]. About 95% of the individuals with obesity develop common polygenic obesity, which is multifactorial and assessing the heritability of polygenic obesity is still one of the major challenges, despite recent advances in genetics of obesity. Genome-wide strategies including linkage and genome-wide association studies (GWAS), which are hypothesis-free per se have been of paramount importance in discovering novel genes involved in the complex etiology of human obesity.
Identifying Novel Genetic Markers by Using Genome Wide Approaches—Linkage Analyses
Genome-wide linkage analyses allow testing for co-segregation of polymorphic genetic markers with phenotypic traits/disease in families, trios or sibling studies. The approach proved to be enormously efficient in discovering genetic variants in monogenic forms of obesity. However, when employed to discovery efforts for underlying genetic markers in polygenic forms of obesity, linkage analyses had only a marginal impact, as most of the identified susceptibility loci for obesity could not be replicated and confirmed in subsequent studies or fine mapped to identify the causal variants affecting the disease. This is most likely to be attributed to small sample sizes in the performed linkage analyses as well as to the poor coverage of genetic variation in tested genomes. One of the very few promising genes discovered in a linkage study was the ectonucleotide pyrophosphatase/phosphodiesterase 1 gene (ENPP1), located on chromosome 6q. The gene was initially discovered to be related to childhood obesity and associated traits by genome-wide linkage analyses [22] and one of its haplotypes further replicated in independent childhood cohorts as well as adults [23, 24]. It is of note however, that despite some inconsistencies in replication efforts, a large meta-analysis including 24,324 individuals clearly supported the potential role of the ENPP1 Q121 variant in the pathophysiology of obesity [25].
Identifying Novel Genetic Markers by Using Genome Wide Approaches—GWA Studies
Whilst the above-described approaches like candidate gene and genome-wide linkage studies showed only marginal success in discoveries of susceptibility genes for common polygenic obesity, prominent advances in molecular biology, including high-throughput genotyping techniques, have enabled researchers to use GWAS to identify novel genetic loci associated with human obesity. This has indeed led to a dramatic increase of until then unknown genetic variants associated with obesity. Started with the discovery of genetic variants in the fat mass and obesity-associated gene (FTO) reported in 2007 [26, 27], so far, more than 1000 loci carrying variants including single nucleotide polymorphisms (SNPs) significantly associated with measures of obesity like BMI have been identified in meta-analyses of large-scale GWAS. These efforts were mostly coordinated within international consortia such as GIANT (the Genetic Investigation of ANthropometric Traits) [13, 15], which predominantly included populations of European ancestry. However, a number of well-powered studies including populations of Asian [28, 29], Hispanic [30] and African [31] ancestries contributed to new discoveries or replication of already reported obesity susceptibility loci. These populations helped to increase the size of available cohorts and so the statistical power of the GWAS. Moreover, based on their specific demographic and evolutionary characteristics, they were particularly valuable in identifying genetic variants with larger effect sizes specific for the respective population. One of these ethnic groups is the Greenlandic population, which played a crucial role in identification of obesity-associated polymorphisms in ADCY3 [32, 33], a gene which may play a role in the regulation of human body weight [34].
The GWAS findings indicate that even with hundreds of obesity-associated loci identified to date, they only explain about 6% of the variation of BMI [15]. Although the remaining variability of BMI remains one of the major challenges of the future research efforts, genome-wide strategies have clearly demonstrated their enormous potential in discovering novel disease susceptibility loci (Fig. 1). In the context of obesity, they showed that most of the identified loci harbour genes involved in pathways affecting neuro-circuits of appetite and satiety regulation (BDNF, MC4R and NEGR) [35–37], energy and lipid metabolism (FTO, RPTOR and MAP2K5 [13, 27, 38], insulin secretion and action (TCF7L2, IRS1) [13, 38] as well as adipogenesis [14]. Furthermore, GWAS also suggested that many of the identified obesity-associated genes are common also for other metabolic diseases such as diabetes, hypertension, and coronary artery disease, which has been supported in gene ontology analyses (GO) highlighting gene clusters with common shared metabolic pathways for these diseases [39]. Another important takeaway from GWAS is the fact that numerous common polymorphisms associated with polygenic obesity in ethnically diverse population have been found in genes like PCSK1 [40–42], MC4R [43••] and POMC, known to carry rare loss of function variants leading to non-syndromic monogenic obesity. Although the GWAS are an excellent tool to uncover variants associated with complex non-Mendelian traits and diseases, understanding the underlying mechanisms behind these associations remains challenging. The majority of genetic variants associated with obesity map within non-coding regions without any obvious biological function, may however carry regulatory elements essential in molecular processes such as gene regulation. Finding the respective target gene of the associated variants appears often difficult since they may be located in distant chromosomal regions, which need to be assessed in subsequent follow up studies. For instance, despite the relatively large effect of the FTO SNPs on BMI with 0.35 kg/m2 per allele or 1 kg for a person who is 1.7 m tall reported in 2007 [27], it took until 2014 to explain the regulatory circuitry and mechanistic chains behind the associations between FTO variants and obesity. Claussnitzer et al. not only showed that the intronic BMI-associated FTO SNP maps within an enhancer element for ARID5B, but could also demonstrate that ARID5B regulates the expression of IRX3 and IRX5, which finally affect adipogenesis, lipid accumulation and thermogenesis [16].This study impressively demonstrated that comprehensive and well-designed functional studies are essential to elucidate molecular pathways underlying the observed associations of genetic loci with obesity.
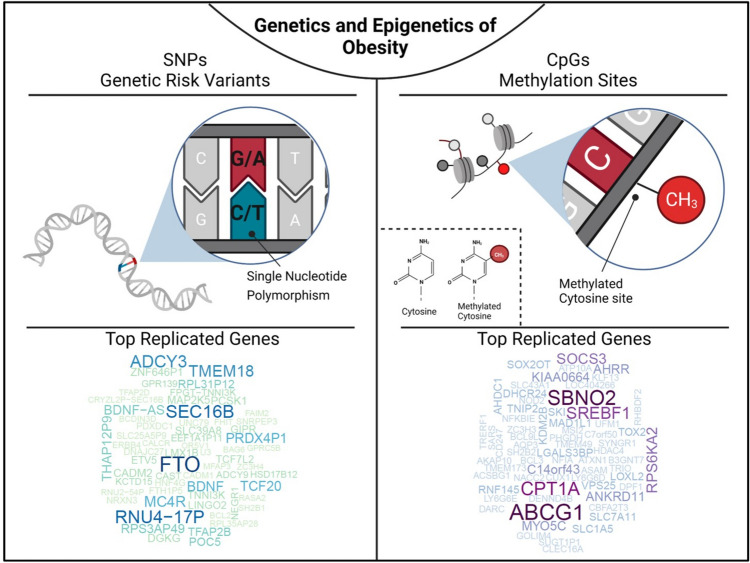
Fig. 1.
Genetics and epigenetics of obesity. The figure illustrates that single nucleotide polymorphisms are genetic risk variants identified by GWAS and CpG sites being differentially methylated in obesity. To screen for the most frequently replicated genes close to identified SNPs and CpGs for associations with BMI, we accessed the GWAS (BMI in adults and children) and EWAS catalogue (BMI in adults), respectively. SNP and CpG associations with more than one annotation were handled as individual gene count. Only hits with a P < 1 × 10−8 were included. Associations were analysed for replication frequencies and blotted using the wordcloud package in R (version 4.2.0, https://blog.fellstat.com/?cat=11). Most replicated GWAS hits for BMI: Word cloud presenting the most often replicated gene hits for genome-wide association of SNPs with BMI in adults and children (GWAS catalogue accessed 20.03.2023 [44]). All genes are replicated at least fifteen times. Gene name size and colour intensity (light green to dark blue) are indicating the replication strength (from least [15 times] to most [56 times]). Long-non-coding RNAs were excluded. Most replicated EWAS hits for BMI: Word cloud presenting the most often replicated gene hits for epigenome-wide association studies with site-specific DNA methylation marks for BMI in adults (EWAS catalogue accessed (08.03.2023 [18]). All genes are replicated at least three times. Gene name size and colour intensity (light blue to dark purple) are indicating the replication strength (from least [three times] to most [nine times]). The upper panel of the figure was generated by using BioRender.com
Genome-Wide Association Studies and Polygenic Risk Scores in Children
The major part of research efforts in polygenic obesity has been focused on adult cohorts [45], whereas similar studies in childhood obesity are rather sparse [46] and are mainly concentrating on replication of findings achieved in adults. It is of note however, that most of the loci identified in adults also associate with obesity in children suggesting the impact of genetic variants across the entire lifespan [47, 48]. Exemplarily, polymorphisms in FTO and MC4R have been shown to be significantly associated with childhood and adolescent obesity in populations from diverse ethnic backgrounds [49–56]. Nevertheless, effects of some SNPs appear to be more pronounced in children and diminish later in life as has been shown for the associations of variants in TMEM18, GNDPA2, MC4R, NEGR1, BDNF and KTCD15 with early-onset obesity [57], and particularly for INSIG2 variants [58–60]. Interestingly, some studies reported that diabetes susceptibility alleles in the HHEX-IDE locus were associated with increased BMI in children, which may underpin the well-acknowledged association between childhood obesity and T2D later in adults [61].
In the context of childhood obesity, polygenic risk scores (PGS), which represent a simple model to determine genetic risk based on multiple genetic variants at different positions in the genome, may render an important tool in translation towards precision medicine. PGSs calculated in early life would enable detection and stratification of individuals with different degrees of obesity risk, and thus, the specific time windows for targeted individualised therapies could be developed [62]. Unfortunately, to date, PGSs are mainly calculated from GWAS performed in adults, which might cast doubts on their informative value for paediatrics. However, as shown by Khera et al. (2019), these doubts do not seem to be justified by the recently generated data [63••]. Here, the authors successfully demonstrated that a polygenic predictor based on 2.1 million known obesity variants is not only associated with a 13 kg increase in body weight in adulthood, but also at birth (+0.06 kg) and at 8 years of age (+3.5 kg) [63••]. Moreover, this study indicated that PGSs derived from adult data may have a comparable strong association with BMI in children. The predictive potential of PGSs in discriminating weight differences in this study was promising and could even be further refined by considering other non-genetic factors such as maternal BMI [64]. However, using this PGS in order to predict future obesity in the UK Biobank has been rather disappointing, showing a high proportion of unreliability making it less useful in clinical utility regarding disease prediction [45]. This clearly sheds light on the difficulties in using genetic information for common polygenic obesity and to translate it into a clinical prediction tool that can be a game changer in clinical day life and decision making. This is currently unrealistic although, it seems conceivable that the performance of PGSs can be increased by combining it with other factors such as environmental or epigenetic indicators to function more accurately [45]. Indeed, it is well-acknowledged that genetic profiling is gaining general popularity in extensive research endeavours, such as within large-scale biobanks linked to healthcare and clinical trials. As a result, it is more and more common for patients and their doctors to encounter PRS during clinical interactions, such as those related, e.g. to cardiovascular conditions [65], severe liver disease [66] and other human pathologies. Thus, refined and robust scores providing more accurate prediction of obesity in the future will undoubtedly become important measures in clinical settings potentially conferring also a predictive value for risk of developing obesity-related co-morbidities such as cardiovascular diseases, liver disease and several types of cancers.
The Importance of Epigenetic Mechanisms
In the context of the complex etiology of human obesity, epigenetic mechanisms based on, e.g. DNA methylation or histone modifications and gene-environment interactions are important to be considered in order to better understand the role of genetics in the development of this multifactorial disease. Despite large-scale GWAS and a simultaneously rising number of studies addressing gene-environment interactions, these studies remain challenging and their findings are often population-specific and not ubiquitously applicable and straightforward. Environmental factors such as physical activity, smoking and dietary components are acting as modifiers of the genetic predisposition to obesity manifestation. This clearly highlights obesity as a preventable disease and further indicates a highly beneficial potential of treatment strategies based on lifestyle interventions. A recent review reported the majority of SNP-environment interactions in association with alcohol consumption, smoking and physical activity [67]. However, among them were also robustly replicated associations as reported for the FTO locus which effects could be attenuated by increased physical activity, but exaggerated by non-healthy fried food consumption [68–70]. There is no doubt that the steadily increasing number of large-scale studies including cohorts such as the UK Biobank and similar large-scale efforts will lead to the discovery of new and more robust gene-environment interactions in the future, usable for more precise treatment opportunities. Nevertheless, the underlying causative mechanisms behind the observed associations remain unknown for most of the genes.
Epigenome-Wide Association Studies in Obesity
Epigenetic mechanisms such as DNA methylation or modification of histone core proteins are suggested to mediate gene-environment interactions and therefore may play a substantial role in susceptibility for obesity. DNA methylation is the most stable, easy to measure and best studied epigenetic mark and has been extensively studied over the last years in relation to obesity.
Genome wide DNA methylation patterns are widely used for EWAS aiming to uncover DNA methylation marks correlating with clinical variables of obesity or fat distribution. Thus, a rapid rise of well-powered EWAS (including multi-omics approaches) and partly large case–control studies in twins, family settings or independent subjects started almost one decade ago and discovered novel targets being involved in epigenetic dysregulation in obesity. In the present review, we summarised 45 genome-wide methylation studies including work mostly conducted in Caucasian subjects and performed in DNA samples originating from whole blood, isolated blood cells or adipose tissue, (Table 1). Importantly, although DNA methylation analyses truly identify novel candidate CpG sites and genes, additional information such as on genetic variation, gene expression, proteome/metabolome is warranted to understand the causative mechanistic circuitry underlying the correlation with disease relevant clinical traits. For instance, genetic variants may modulate the methylation at specific CpG sites potentially inducing co-methylation patterns at nearby sites, eventually translating into changes in clinical traits and suggesting a genotype–phenotype correlation. Therefore, a rising number of studies focus on multi-omics epigenetic associations with obesity or related traits, mainly promoted by the latest advances in high-throughput technologies and analytical approaches promoting (Table 1).
Table 1.
Epigenome wide Association Studies for BMI and related clinical traits of obesity
| Study reference | PMID | Clinical trait | Ethnicity | N/females; age in years | Tissue | Main findings | Multi-omics | Meta-analysis |
|---|---|---|---|---|---|---|---|---|
| Carless et al. (2013) [71] | 24,058,506 | WC, BMI | Mexican American |
183/101 19–75 |
Blood | Genetic effects on DNA methylation, no significant associations to obesity measures | ||
| Xu et al. (2013) [72] | 23,644,594 | BMI | African American |
96/48 14–20 |
Blood | Methylation variability in obesity | ||
| Dick et al. (2014) [73] | 24,630,777 | BMI | Caucasian |
459/84 ø55 |
Blood, adipose tissue | Methylation of HIF3A associated with BMI | ||
| Almén et al. (2014) [74] | 25,010,727 | BMI | Caucasian (Latvia) |
46/46 41–70 |
Blood | Age-associated epigenetic changes are influenced by obesity | ||
| Guenard et al. (2014) [75] | 24,495,915 | MetS | Caucasian (American) |
14/0 ø38 |
Adipose tissue | Multiple novel targets, pathways are related to cell membrane, inflammation; immunity; cell cycle | ||
| Aslibekyan et al. (2015) [76] | 26,110,892 | WC, BMI | Caucasian (European American) |
991/515 ø49 |
CD4 + T-cells | DNA methylation of CPT1A, PHGHD, CD38 and lncRNA00263 BMI/WC | ||
| Ollikainen et al. (2015) [77] | 25,866,590 | BMI | Caucasian |
80/46 ø27 |
Blood | Differences in DNA methylation due to excessive liver fat | ||
| Demerath et al. (2015) [78] | 25,935,004 | WC, BMI | African American |
2097/1334 47–70 |
Blood | 164 and 8 probes for WC and BMI respectively, including HIF3A, CPT1A and ABCG1 | ||
| Voisin et al. (2015) [79] | 26,449,484 | obesity | Caucasian |
355/141 14–34 |
Blood | 28 obesity SNPs associate with 107 proximal CpG site in obesity-related genes | Methylome, genotype | |
| Arner et al. (2015) [80] | 26,351,548 | BMI | Caucasian |
29/29 ø46 |
Adipose tissue | 5529 differentially meth CpG sites correlate to 2223 diff expressed genes being related to metabolic function of fat cells | Methylome, transcriptome | |
| Rönn et al. (2015) [81] | 25,861,810 | BMI, HbA1c, age | Caucasian |
190/94 23–80 |
Blood, adipose tissue | DNA methylation and expression of 2825 genes correlated with BMI, of 1050 genes with age and 711 CpG site with HbA1c | Methylome, transcriptome | |
| Kirchner et al. (2016) [82] | 26,977,391 | BMI, T2D | Caucasian |
35/0 21–62 |
Liver | hypomethylated genes (ATF-motifs) in the liver of obese subjects independent of T2D | Methylome, transcriptome | |
| Ali et al. (2016) [83] | 27,564,309 | BMI, MetS | Caucasian |
192/106 8–90 |
Blood | Identification of SOCS3 methylation being associated with BMI, MetS and related lipid traits | ||
| Keller et al. (2016) [84] | 28,123,940 | BMI | Caucasian |
105/66 ø57 |
Adipose tissue | Identification of six robust adipose tissue depot-specific genes (HAND2, HOXC6, PPARG, SORBS2, CD36, and CLDN1) | Methylome, transcriptome | |
| Pietiläinen et al. (2016) [85] | 26,499,446 | BMI | Caucasian |
74/42 23–36 |
Adipose tissue | Diff methylated and expressed genes in monozygotic twins discordant for BMI, suggesting pathological adaptation of SAT to obesity is partly epigenetically regulated | Methylome, transcriptome | |
| Volkov et al. (2016) [86] | 27,322,064 | Caucasian |
119/ 22–80 |
Adipose tissue | Discovered mQTLs including ADCY3/POMC, APOA5, CETP, FADS2, GCKR, SORT1 and LEPR. 635 SNPs in mQTLs associated with expression of 86 genes | Methylome, genotype, transcriptome | ||
| Al Muftah et al. (2016) [87] | 26,823,690 | BMI, T2D | Arabs |
123/72 ø38 |
Blood | replicated eight CpG site associations in Arabs; suggest heterogeneity of effects underlies genetic variation | ||
| Sayols-Baixeras et al. (2017) [88] | 29,099,282 | WC, BMI | Caucasian |
641/325 35–79 |
Blood | Discovery of 70 and 30 novel CpGs associated with BMI and WC, respectively, explaining 25.94% and 29.22% of the variability of BMI and waist circumference | yes | |
| Crujeiras et al. (2017) [89] | 28,211,912 | obesity | Caucasian |
55/28 20–83 |
Blood, adipose tissue | FGFRL1, NCAPH2, PNKD and SMAD3 diff methylated between obese and non-obese subjects | ||
| Wahl et al. (2017) [20] | 28,002,404 | BMI | Caucasian, Indian Asian |
5387/2147 ø54 |
EWAS for BMI identified 187 genetic loci differentially methylated; the study demonstrated that epigenetic changes are likely the consequence of obesity rather that the cause | |||
| Meeks et al. (2017) [90] | 28,947,923 | WC, BMI, obesity | Sub-Saharan Africans |
547/316 ø50 |
Blood | The first EWAS for obesity in Africans identified three epigenome-wide significant loci (CPT1A, NLRC5 and BCAT1) | ||
| Wilson et al. (2017) [91] | 27,773,939 | BMI | Caucasian (North American) |
871/871 ø55 |
Blood | Identified CpG sites at ANGPT4, RORC, SOCS3, LGALS3BP associated with BMI | ||
| Guenard et al. (2017) [92] | 28,219,716 | MetS | Caucasian (North American) |
31/0 ø35 |
Blood, adipose tissue | Identified 2182 meQTLs regulating the methylation levels of 174 CpG sites; revealed COL11A2 meQTLs corr with fasting glucose levels | Methylome, genotype | |
| Mendelson et al. (2017) [93] | 28,095,459 | BMI | Caucasian |
3743/1947 ø72 |
Blood | The study discovered novel and replicated known BMI-associated at 83 CpGs loci | Methylome, transcriptome | |
| Kvaløy et al. (2018) [94] | 30,397,228 | BMI | Caucasian |
120/120 23–31 |
Blood | Identified 10 diff meth CpG sites: COX6A1P2/FGD2, SBNO2, TEX41, RPS6KA2, IGHE/IGHG1/IGHD, DMAP1, SOCS3 and SETBP1 | ||
| Dhana et al. (2018) [95] | 29,762,635 | WC, BMI | Caucasian |
1450/811 ø64 |
Blood | Discovered 12 and 13 CpGs associated with BMI and WC, respectively. The most significant CpGs were annotated to MSI2 and LARS2 | ||
| Campanella et al. (2018) [96] | 29,713,043 | WC, BMI, WHR, WHeR | Caucasian |
1941/1353 ø54 |
Blood | 40 CpG loci were associated with at least one adiposity measure. One CpG at ABCG1 was associated with all four traits other clinical variables | Methylome, transcriptome | Yes |
| Orozco et al. (2018) [97] | 29,566,149 | MetS | Caucasian |
201/0 45–73 |
Adipose tissue | Identification of 18 candidate genes, including known and novel genes. Methylation deconvolution demonstrated the loci were specific to adipocytes | Methylome, transcriptome | |
| Akinyemiju et al. (2018) [98] | 29,643,945 | MetS | African American |
614/411 ø49 |
Blood | Two differentially methylated CpGs annotated to IGF2BP1 and ABCG1 were identified, ABCG1 could be replicated | Yes | |
| Zaghlool et al. (2018) [99] | 29,325,019 | BMI | Arab Asians, Filipinos |
359/177 ø47 |
Blood | Reports potential causal effects of metabolite levels on methylation of obesity-associated CpG sites, such as on DHCR24, MYO5C and CPT1A | Methylome, proteome, metabolome | |
| Guo et al. (2018) [100] | 30,619,480 | BMI | Caucasian, Asian,African American, European American |
22,310/n.a n.a |
Blood, adipose tissue | 119 CpGs associated with obesity were identified, SOCS3 was part of enriched pathways and differentially expressed in adipose tissue | Methylome, transcriptome | Yes |
| Wang et al. (2018) [25] | 29,312,471 | BMI | African American |
700/n.a 14–36 |
Blood | The study identified 76 obesity-related CpG sites; 54 were replicated | Methylome, transcriptome | |
| Li et al. (2019) [101] | 31,152,155 | BMI | Asian |
60/30 39–72 |
Blood | The study identify 30 CpGs; however, none of the sites reached genome-wide significance. 11 differentially methylated regions were validated | Methylome, transcriptome | |
| Li et al. (2019) [102] | 31,480,455 | BMI | African American |
232/232 ø32 |
Saliva | The study suggests that high BMI accelerates DNA methylation age | ||
| Pan et al. (2019) [103] | 30,837,522 | BMI | African American, European American |
24/0 14–20 |
Blood | The group identified a novel neutrophil activation ALPL in obesity ALPL expression associates with CVD risk factors | Methylome, transcriptome, proteome | |
| Koh et al. (2020) [104] | 32,788,176 | BMI | Asian |
450/0 ø47 |
Blood, adipose tissue | The study discovered multiple diff meth CpG sites and altered gene expression, suggesting CPA3 as potential obesity-related gene | Methylome, transcriptome | |
| Giri et al. (2020) [105] | 32,363,570 | BMI | Indo-European |
1142/545 Ø50 |
Blood | The study discovered genetic markers in SLC22A11 and BAI3 that associated with DNA methylation at important cis-regulatory elements | Methylome, genotype | |
| Justice et al. (2020) [106] | 32,901,515 | BMI | African American |
2684/1702 45–64 |
Blood | Identified novel CpG sites near TXNIP, ADCY7, SREBF1 and RAP1GAP2 that were not previously described for obesity | ||
| Xie et al. (2021) [107] | 34,556,110 | WC, WHR | Caucasian |
210/105 ø28 |
Blood | This work idenfied a CpG site cg16170243 significantly associated with BMI adjusted WC | ||
| Chen et al. (2021) [108•] | 34,670,603 | WC, BMI | Asian (multi-ethnic: Chinese, Malay, Indian) |
409/338 ø51 |
Blood | Discovery of multiple CpG sites associated with BMI and WC. Analyses suggest high BMI is rather cause than consequence of changes in DNA methylation | ||
| Cao et al. (2021) [109] | 33,810,959 | WC, BMI, HC, WHR | Norfolk Island isolate |
47/24 ø42 |
Blood | Multi-trait analysis based PCA identifies two PC components explaining ~ 89% of the phenotypic variance; identified 5 CpGs at GOT2-CDH8, LYSMD3, HIBADH, ADGRD1 and EBF4 genes | ||
| Do et al. (2021) [110] | 34,278,703 | WC, BMI | Caucasian (European American) |
43,936/n.a 18–75 |
Blood and other tissues | The study discovered 52 CpG sites associated with BMI being potential mediators of obesity-associated chronic diseases | Yes | |
| Wu et al. (2022) [111] | 35,882,828 | WHR | Northern Han Chinease |
120/n.a ø52 |
Blood | The study identified numerous CpG sites whose methylation levels associate with WHR | ||
| Taylor et al. (2023) [112] | 36,479,596 | BMI | African American |
239/239 ø31 |
Saliva, blood | A combined sex and female-only meta-analyses discovered multiple CpG sites associated with BMI | Yes | |
| Do et al. (2023) [113•] | 36,649,705 | BMI | Caucasian/European, Asian, African |
17,034/n.a n.a |
Blood | This study identified ~ 700 novel CpG sites associated with BMI and that 397 CpG sites explained 32% of the variance in BMI | Yes |
This table summarises 45 studies identified by using the following criteria: We performed a PubMed search (dated 08/03/2023) for studies published during the last 10 years (2013–2023) using the following mesh-terms: DNA methylation AND obesity/EWAS AND obesity. We focused on studies analysing genome-wide DNA methylation by individual techniques (arrays, WGBS) associated with BMI or other obesity-related traits. We excluded longitudinal studies measuring DNA methylation changes induced by different types of interventions, studies mainly focusing on T2D, cancer, transgenerational effects or prediction models, studies only performed in vitro or other than human model organisms. In addition, we focused on DNA methylation only and excluded other epigenetic mechanisms such as histone modifications and non-coding RNAs. The table further provides information about the analysed trait(s) (BMI; WC waist circumference, WHR waist-to-hip ratio, WHeR waist-height-ratio, T2D type 2 diabetes, MetS metabolic syndrome etc.), ethnicity, sample size (discovery cohort), number of females included, age range or mean age (ø), tissue of origin, information about the reference (PMID), publication year and the major results. We further indicated studies that have analysed two or multi-omics levels (methylome, genotype, transcriptome, proteome and metabolome) and those using meta-analyses strategies. Here, we count a study to be “multi”-omics if the authors analysed more than one level of the omics cascade
Although the most powerful EWAS reported recognisable sample sizes with more than 5000 subjects in the discovery cohort [20], the effect sizes are highly variable ranging from 6 to 40 kg/m2 change in BMI per unit increase in blood DNA methylation. In general, sample sizes in EWAS studies are often much smaller than in GWAS analyses (Table 1), with the smaller cohorts mainly estimating methylation differences between individuals with and without obesity/metabolic syndrome. Studies with lower sample sizes also report smaller effect sizes such as 0.8–3.6% BMI increase per 0.1 increase in methylation ß-values [73]. Interestingly, Vehmeijer et al. demonstrated an increasing effect size with age by meta-analysing 187 methylation loci, previously reported to show cross-sectional association to BMI in adults, in children with an age between 2 and 18 years [114]. However, most studies are still of explorative nature focussing on the identification of novel candidate sites and genes rather than evaluating whether methylation changes are cause or consequence of obesity (Table 1).
To generate an overview about genes reported to show associations between methylation level of specific CpG sites and BMI, we used data from the EWAS catalogue (all P < 1 × 10−8; EWAS catalogue accessed 08.03.2023, [18]) and performed a word-cloud analysis using gene IDs in R (wordcloud package, R version 4.2.0). Based on this analysis, we estimated the most replicated genes originating from EWAS for BMI (Fig. 1). All genes included were replicated at least in three studies with ABCG1 (ATP-binding Cassette Sub-family G Member 1) being the mostly replicated gene locus followed by CPT1A (Carnitin Palmitoyltransferase 1), SREBF1 (Sterol Regulatory Element Binding Transcription Factor 1), SBNO2 (Strawberry Notch Homolog 2) and SOCS3 (Suppressor of Cytokine Signaling 3). Among them ABCG1 [78, 91, 96, 98] and CPT1A [76, 78, 87, 90, 99] were described across different ethnicities such as Caucasian, African American, Africans and Asians (Table 1). Of note, some larger cohorts such as LOLIPOP (London Life Sciences Prospective Population [115]) or KORA (Cooperative Health Research in the Region of Augsburg [116]) are more frequently used as replication cohorts.
Several studies support the functional role of, e.g. ABCG1 and CPT1A in obesity. Wahl et al. [20] for instance reported an association of the BMI genetic risk score with the ABCG1 methylation being consistent with other studies reporting effects of overweight and weight-loss on methylation, expression or protein activity [20, 117, 118]. In general, ABCG1 is involved in mitochondrial cholesterol efflux, thus promoting cellular efflux to HDL. Its silencing leads to massive lipid accumulation in tissues of high fat and high cholesterol fed mice and in 3T3L1 adipocytes [119–121]. In line with this, ABCG1 and, e.g. SREBF1 methylation levels are also known to correlate with T2D [115, 122–124], postulating direct or indirect effects on metabolic consequences of obesity. Similar to ABCG1, CPT1A is involved in mitochondrial fatty acid oxidation and ROS production by regulating the entry of long-chain fatty acids into the mitochondrial matrix [125] and thereby also contributing to the activation of inflammasomes [126]. Furthermore, high-fat diet (+/− fructose) fed mice revealed a decreased CTP1a activity and thus a decreased fat metabolism, whereas knockdown of the fructose metabolism enhanced CPT1a activity [127]. Taken together, EWAS studies have extensively helped to discover CpG sites whose differential methylation levels correlate with important clinical traits of obesity and fat distribution, thus clearly illustrating the importance of epigenetic marks in obesity and its potential dysregulation in disease. However, despite these efforts and multiple novel candidate genes identified during the last years, the precise mechanistic circuitry of those genes in the human pathophysiology of obesity and relevant metabolic traits is still not well understood. Furthermore, although recent studies support the role of methylation changes in obesity, to what extent whole blood methylation profiles can mirror their patterns in target tissues remains under discussion. In addition, the majority of studies included in this review used array based approaches for genome-wide association studies, providing a limited overview of 1.7–3% of all CpG positions in the genome, illustrating that a large part of the remaining sites is undiscovered among these studies [128].
Ethnicity Specific Findings and Meta-analyses
Although most genome wide DNA methylation analyses were conducted in cohorts with Caucasian ancestry (Table 1), recent studies focussed more on the homo- or heterogeneity between the ethnic groups. For instance, a EWAS performed in an Arab population confirmed seven previously identified BMI loci but reported higher effect sizes compared to their replication cohort from the UK [87]. However, it has to be acknowledged that the reported association of these loci did not reach genome-wide significance level in the Arab discovery cohort. In line with this, previously reported associations between, e.g. CPT1A methylation and BMI have been confirmed across multiple ethnicities such as Caucasian, African American, African, Asian and Arab [76, 78, 87, 90, 99]. Moreover, a recently published multi-ethnic study in Asians was able to replicate 110 BMI-associated loci, which were previously reported for Europeans, South Asians and African Americans with a high consistency of the effect directions. Although they reported a great homogeneity across the different Asian ethnicities, they also demonstrated heterogeneity across several loci, where for instance the effects are mainly driven by the Chinese subjects [108•]. Another study, taking into account a longitudinal setting, discovered a total of 287 novel CpG sites correlating with BMI (266 in white participants, 21 in black individuals). Importantly, a major take home message from this report is that, based on the longitudinal design, the authors concluded that obesity seems to precede changes in methylation, underlining, in line with Wahl et al. [20] that obesity may rather be cause than consequence of epigenetic changes [19•].
In a very recent study, representing the largest meta-analysis so far, Do et al. [113•] performed an EWAS in more than 17,000 individuals to detect CpG sites associated with BMI of European (Caucasian), African and Asian subjects. Following this approach, the study confirmed 553 previously reported loci but also identified 685 novel sites, which were successfully replicated. Interestingly, only five CpG sites were reported showing an interaction with BMI by race/ethnicity among individuals with a European or African ancestry. Importantly, in an attempt to assess the value of such CpG sites in predicting BMI, the study demonstrated that 397 of those identified CpG positions explained 32% of BMI variability illustrating that a methylome-based prediction of BMI in this study performed relatively good [113•].
The Utility of Methylation Risk Scores in Predicting Disease Risk
Similar to genetic analyses, the concept of polygenic risks scores can be transferred to CpG methylation data and can be used to construct methylation risk scores (MRS). Such MRSs may prove useful tools in predicting disease risk or assessing exposure to specific environmental factors in the future. It is noteworthy, however, that in addition to methodological challenges in constructing weighted MRSs, all EWAS findings, and thereby MRSs are highly sensitive to potential confounders such as age, gender, ethnicity and also technological differences in assessing DNA methylation [129]. In line with Do et al. [113•], who reported 32% of the variability in BMI accounted for by an MRS, also others, such as Hamilton and colleagues [130], observed that an MRS correlates with adverse health outcomes and accounts for 10% of the variance in BMI. This is similar to observations in adult women where DNA methylation scores roughly explained 10% BMI variance in the population, whilst much less variance was explained in children (1–2%) and young adolescents (3%) [131]. Furthermore, the same study concluded that MRS is a poor marker for future BMI prediction, illustrating the challenges in using MRSs as a meaningful prediction tool in clinical day life so far. However, it has been shown that epigenetic predictors based on DNA methylation at CpG sites are valuable tools in predicting mortality and exposure to certain environmental factors such as to smoking [132]. This is largely corroborated by a recent study showing that MRSs performed better in adult individuals than polygenic risk scores in explaining variance in smoking and BMI [133]. Taken together, there is a potential that MRSs can evolve into useful tools for clinical decision making in the future, although until now there are still conflicting results published. Of note, by increasing the sample sizes, taking into account potential confounders and combining MRSs with polygenic risk scores in the future might help to overcome current obstacles.
Conclusion
Enormous advances have been made during the last years in identifying genetic and epigenetic loci being involved in the pathophysiology of obesity and related clinical traits. GWAS and EWAS approaches are both by nature hypothesis-free strategies that have proven excellent tools in discovering such novel susceptibility loci and sites. Although for most of the genetic risk variants still the mechanistic circuitry needs to be investigated, important progress has been made for a number of important players.. The use of polygenic risk scores in predicting future BMI or obesity is still in its infancy as a relatively frequent mis-prediction is complicating effective use in clinical settings. Likewise, numerous epigenetic studies have identified novel candidate CpGs and genes conferring changes in DNA methylation. Multiple genes also provide a plausible functional implication in related clinical traits. Construction of methylation risk scores has proven successful for predicting exposure to specific environmental factors such as smoking, but again, its utility in clinical day life is limited for prediction of disease risk. At this stage of research, it seems unlikely so far to use either polygenic or methylation risk scores as a valid clinical prediction tool in the near future. However, by further refining the scores, increasing sample sizes and improving weighting statistic, taking into account typical confounders and combining potentially polygenic risk scores with methylation risk scores may prove successful instruments useful in clinical settings such as predicting future BMI and obesity or predicting successful weight loss in the future.
Supplementary Information
Below is the link to the electronic supplementary material.
Author Contributions
All authors were responsible for the conception and design of the manuscript, drafting the manuscript, revising it critically for intellectual content and approving the final version.
Funding
Open access funding provided by University of Oslo (incl Oslo University Hospital)
Compliance with Ethical Standards
Competing Interests
The authors have no competing interests to declare that are relevant to the content of this article.
Human and Animal Rights and Informed Consent
This article does not contain any studies with human or animal subjects performed by any of the authors.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Maria Keller and Stina Ingrid Alice Svensson contributed equally to this work.
References
Papers of particular interest, published recently, have been highlighted as: • Of importance •• Of major importance
- 1.Ng M, Fleming T, Robinson M, Thomson B, Graetz N, Margono C, Mullany EC, Biryukov S, Abbafati C, Abera SF, et al. Global, regional, and national prevalence of overweight and obesity in children and adults during 1980–2013: a systematic analysis for the Global Burden of Disease Study 2013. Lancet. 2014;384(9945):766–781. doi: 10.1016/S0140-6736(14)60460-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bluher M. Adipose tissue inflammation: a cause or consequence of obesity-related insulin resistance. Clin Sci (London, England : 1979) 2016;130(18):1603–1614. doi: 10.1042/CS20160005. [DOI] [PubMed] [Google Scholar]
- 3.Fontaine KR, Redden DT, Wang C, Westfall AO, Allison DB. Years of life lost due to obesity. JAMA. 2003;289(2):187–193. doi: 10.1001/jama.289.2.187. [DOI] [PubMed] [Google Scholar]
- 4.World Obesity Atlas 2023. https://www.worldobesityday.org/resources/entry/world-obesity-atlas-2023.
- 5.Katzmarzyk PT, Perusse L, Rao DC, Bouchard C. Familial risk of overweight and obesity in the Canadian population using the WHO/NIH criteria. Obes Res. 2000;8(2):194–197. doi: 10.1038/oby.2000.21. [DOI] [PubMed] [Google Scholar]
- 6.Koeppen-Schomerus G, Wardle J, Plomin R. A genetic analysis of weight and overweight in 4-year-old twin pairs. International journal of obesity and related metabolic disorders : journal of the International Association for the Study of Obesity. 2001;25(6):838–844. doi: 10.1038/sj.ijo.0801589. [DOI] [PubMed] [Google Scholar]
- 7.Pietilainen KH, Kaprio J, Rissanen A, Winter T, Rimpela A, Viken RJ, Rose RJ. Distribution and heritability of BMI in Finnish adolescents aged 16y and 17y: a study of 4884 twins and 2509 singletons. International journal of obesity and related metabolic disorders : journal of the International Association for the Study of Obesity. 1999;23(2):107–115. doi: 10.1038/sj.ijo.0800767. [DOI] [PubMed] [Google Scholar]
- 8.Allison DB, Kaprio J, Korkeila M, Koskenvuo M, Neale MC, Hayakawa K. The heritability of body mass index among an international sample of monozygotic twins reared apart. International journal of obesity and related metabolic disorders : journal of the International Association for the Study of Obesity. 1996;20(6):501–506. [PubMed] [Google Scholar]
- 9.Feinleib M, Garrison RJ, Fabsitz R, Christian JC, Hrubec Z, Borhani NO, Kannel WB, Rosenman R, Schwartz JT, Wagner JO. The NHLBI twin study of cardiovascular disease risk factors: methodology and summary of results. Am J Epidemiol. 1977;106(4):284–285. doi: 10.1093/oxfordjournals.aje.a112464. [DOI] [PubMed] [Google Scholar]
- 10.Stunkard AJ, Foch TT, Hrubec Z. A twin study of human obesity. JAMA. 1986;256(1):51–54. doi: 10.1001/jama.1986.03380010055024. [DOI] [PubMed] [Google Scholar]
- 11.Stunkard AJ, Harris JR, Pedersen NL, McClearn GE. The body-mass index of twins who have been reared apart. N Engl J Med. 1990;322(21):1483–1487. doi: 10.1056/NEJM199005243222102. [DOI] [PubMed] [Google Scholar]
- 12.Stunkard AJ, Sorensen TI, Hanis C, Teasdale TW, Chakraborty R, Schull WJ, Schulsinger F. An adoption study of human obesity. N Engl J Med. 1986;314(4):193–198. doi: 10.1056/NEJM198601233140401. [DOI] [PubMed] [Google Scholar]
- 13.Locke AE, Kahali B, Berndt SI, Justice AE, Pers TH, Day FR, Powell C, Vedantam S, Buchkovich ML, Yang J, et al. Genetic studies of body mass index yield new insights for obesity biology. Nature. 2015;518(7538):197–206. doi: 10.1038/nature14177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shungin D, Winkler TW, Croteau-Chonka DC, Ferreira T, Locke AE, Mägi R, Strawbridge RJ, Pers TH, Fischer K, Justice AE, et al. New genetic loci link adipose and insulin biology to body fat distribution. Nature. 2015;518(7538):187–196. doi: 10.1038/nature14132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yengo L, Sidorenko J, Kemper KE, Zheng Z, Wood AR, Weedon MN, Frayling TM, Hirschhorn J, Yang J, Visscher PM. Meta-analysis of genome-wide association studies for height and body mass index in ∼700000 individuals of European ancestry. Hum Mol Genet. 2018;27(20):3641–3649. doi: 10.1093/hmg/ddy271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Claussnitzer M, Dankel SN, Kim KH, Quon G, Meuleman W, Haugen C, Glunk V, Sousa IS, Beaudry JL, Puviindran V, et al. FTO obesity variant circuitry and adipocyte browning in humans. N Engl J Med. 2015;373(10):895–907. doi: 10.1056/NEJMoa1502214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Glunk V, Laber S, Sinnott-Armstrong N, Sobreira DR, Strobel SM, Batista TM, Kubitz P, Moud BN, Ebert H, Huang Y, et al. A non-coding variant linked to metabolic obesity with normal weight affects actin remodelling in subcutaneous adipocytes. Nat Metab. 2023;5(5):861–879. doi: 10.1038/s42255-023-00807-w. [DOI] [PubMed] [Google Scholar]
- 18.Battram T, Yousefi P, Crawford G, Prince C, Sheikhali Babaei M, Sharp G, Hatcher C, Vega-Salas MJ, Khodabakhsh S, Whitehurst O, et al. The EWAS Catalog: a database of epigenome-wide association studies. Wellcome Open Res. 2022;7:41. doi: 10.12688/wellcomeopenres.17598.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.• Sun D, Zhang T, Su S, Hao G, Chen T, Li QZ, Bazzano L, He J, Wang X, Li S, et al. Body mass index drives changes in DNA methylation: a longitudinal study. Circ Res. 2019;125(9):824–33. The study perform ethnicity-specific EWASs and suggests that change in DNA methylation follow rather than precede obesity. [DOI] [PMC free article] [PubMed]
- 20.Wahl S, Drong A, Lehne B, Loh M, Scott WR, Kunze S, Tsai PC, Ried JS, Zhang W, Yang Y, et al. Epigenome-wide association study of body mass index, and the adverse outcomes of adiposity. Nature. 2017;541(7635):81–86. doi: 10.1038/nature20784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Salum KCR, Rolando JM, Zembrzuski VM, Carneiro JRI, Mello CB, Maya-Monteiro CM, Bozza PT, Kohlrausch FB, da Fonseca ACP. When leptin is not there: a review of what nonsyndromic monogenic obesity cases tell us and the benefits of exogenous leptin. Front Endocrinol (Lausanne) 2021;12:722441. doi: 10.3389/fendo.2021.722441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Meyre D, Lecoeur C, Delplanque J, Francke S, Vatin V, Durand E, Weill J, Dina C, Froguel P. A genome-wide scan for childhood obesity-associated traits in French families shows significant linkage on chromosome 6q22.31-q23.2. Diabetes. 2004;53(3):803–811. doi: 10.2337/diabetes.53.3.803. [DOI] [PubMed] [Google Scholar]
- 23.Böttcher Y, Körner A, Reinehr T, Enigk B, Kiess W, Stumvoll M, Kovacs P. ENPP1 variants and haplotypes predispose to early onset obesity and impaired glucose and insulin metabolism in German obese children. J Clin Endocrinol Metab. 2006;91(12):4948–4952. doi: 10.1210/jc.2006-0540. [DOI] [PubMed] [Google Scholar]
- 24.Meyre D, Bouatia-Naji N, Tounian A, Samson C, Lecoeur C, Vatin V, Ghoussaini M, Wachter C, Hercberg S, Charpentier G, et al. Variants of ENPP1 are associated with childhood and adult obesity and increase the risk of glucose intolerance and type 2 diabetes. Nat Genet. 2005;37(8):863–867. doi: 10.1038/ng1604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang R, Zhou D, Xi B, Ge X, Zhu P, Wang B, Zhou M, Huang Y, Liu J, Yu Y, et al. ENPP1/PC-1 gene K121Q polymorphism is associated with obesity in European adult populations: evidence from a meta-analysis involving 24,324 subjects. Biomed Environ Sci. 2011;24(2):200–206. doi: 10.3967/0895-3988.2011.02.015. [DOI] [PubMed] [Google Scholar]
- 26.Dina C, Meyre D, Gallina S, Durand E, Körner A, Jacobson P, Carlsson LM, Kiess W, Vatin V, Lecoeur C, et al. Variation in FTO contributes to childhood obesity and severe adult obesity. Nat Genet. 2007;39(6):724–726. doi: 10.1038/ng2048. [DOI] [PubMed] [Google Scholar]
- 27.Frayling TM, Timpson NJ, Weedon MN, Zeggini E, Freathy RM, Lindgren CM, Perry JR, Elliott KS, Lango H, Rayner NW, et al. A common variant in the FTO gene is associated with body mass index and predisposes to childhood and adult obesity. Science. 2007;316(5826):889–894. doi: 10.1126/science.1141634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Akiyama M, Okada Y, Kanai M, Takahashi A, Momozawa Y, Ikeda M, Iwata N, Ikegawa S, Hirata M, Matsuda K, et al. Genome-wide association study identifies 112 new loci for body mass index in the Japanese population. Nat Genet. 2017;49(10):1458–1467. doi: 10.1038/ng.3951. [DOI] [PubMed] [Google Scholar]
- 29.Buniello A, MacArthur JAL, Cerezo M, Harris LW, Hayhurst J, Malangone C, McMahon A, Morales J, Mountjoy E, Sollis E, et al. The NHGRI-EBI GWAS Catalog of published genome-wide association studies, targeted arrays and summary statistics 2019. Nucleic Acids Res. 2019;47(D1):D1005–d1012. doi: 10.1093/nar/gky1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wojcik GL, Graff M, Nishimura KK, Tao R, Haessler J, Gignoux CR, Highland HM, Patel YM, Sorokin EP, Avery CL, et al. Genetic analyses of diverse populations improves discovery for complex traits. Nature. 2019;570(7762):514–518. doi: 10.1038/s41586-019-1310-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ng MCY, Graff M, Lu Y, Justice AE, Mudgal P, Liu CT, Young K, Yanek LR, Feitosa MF, Wojczynski MK, et al. Discovery and fine-mapping of adiposity loci using high density imputation of genome-wide association studies in individuals of African ancestry: African Ancestry Anthropometry Genetics Consortium. PLoS Genet. 2017;13(4):e1006719. doi: 10.1371/journal.pgen.1006719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Grarup N, Moltke I, Andersen MK, Dalby M, Vitting-Seerup K, Kern T, Mahendran Y, Jørsboe E, Larsen CVL, Dahl-Petersen IK, et al. Loss-of-function variants in ADCY3 increase risk of obesity and type 2 diabetes. Nat Genet. 2018;50(2):172–174. doi: 10.1038/s41588-017-0022-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Saeed S, Bonnefond A, Tamanini F, Mirza MU, Manzoor J, Janjua QM, Din SM, Gaitan J, Milochau A, Durand E, et al. Loss-of-function mutations in ADCY3 cause monogenic severe obesity. Nat Genet. 2018;50(2):175–179. doi: 10.1038/s41588-017-0023-6. [DOI] [PubMed] [Google Scholar]
- 34.Siljee JE, Wang Y, Bernard AA, Ersoy BA, Zhang S, Marley A, Von Zastrow M, Reiter JF, Vaisse C. Subcellular localization of MC4R with ADCY3 at neuronal primary cilia underlies a common pathway for genetic predisposition to obesity. Nat Genet. 2018;50(2):180–185. doi: 10.1038/s41588-017-0020-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Boender AJ, van Rozen AJ, Adan RA. Nutritional state affects the expression of the obesity-associated genes Etv5, Faim2, Fto, and Negr1. Obesity (Silver Spring) 2012;20(12):2420–2425. doi: 10.1038/oby.2012.128. [DOI] [PubMed] [Google Scholar]
- 36.Ho EV, Klenotich SJ, McMurray MS, Dulawa SC. Activity-based anorexia alters the expression of BDNF transcripts in the mesocorticolimbic reward circuit. PLoS ONE. 2016;11(11):e0166756. doi: 10.1371/journal.pone.0166756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Horstmann A, Kovacs P, Kabisch S, Boettcher Y, Schloegl H, Tönjes A, Stumvoll M, Pleger B, Villringer A. Common genetic variation near MC4R has a sex-specific impact on human brain structure and eating behavior. PLoS ONE. 2013;8(9):e74362. doi: 10.1371/journal.pone.0074362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kilpeläinen TO, Zillikens MC, Stančákova A, Finucane FM, Ried JS, Langenberg C, Zhang W, Beckmann JS, Luan J, Vandenput L, et al. Genetic variation near IRS1 associates with reduced adiposity and an impaired metabolic profile. Nat Genet. 2011;43(8):753–760. doi: 10.1038/ng.866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Su LN, Wang YB, Wnag CG, Wei HP. Network analysis identifies common genes associated with obesity in six obesity-related diseases. J Zhejiang Univ Sci B. 2017;18(8):727–732. doi: 10.1631/jzus.B1600454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Benzinou M, Creemers JW, Choquet H, Lobbens S, Dina C, Durand E, Guerardel A, Boutin P, Jouret B, Heude B, et al. Common nonsynonymous variants in PCSK1 confer risk of obesity. Nat Genet. 2008;40(8):943–945. doi: 10.1038/ng.177. [DOI] [PubMed] [Google Scholar]
- 41.Choquet H, Kasberger J, Hamidovic A, Jorgenson E. Contribution of common PCSK1 genetic variants to obesity in 8,359 subjects from multi-ethnic American population. PLoS ONE. 2013;8(2):e57857. doi: 10.1371/journal.pone.0057857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rouskas K, Kouvatsi A, Paletas K, Papazoglou D, Tsapas A, Lobbens S, Vatin V, Durand E, Labrune Y, Delplanque J, et al. Common variants in FTO, MC4R, TMEM18, PRL, AIF1, and PCSK1 show evidence of association with adult obesity in the Greek population. Obesity (Silver Spring) 2012;20(2):389–395. doi: 10.1038/oby.2011.177. [DOI] [PubMed] [Google Scholar]
- 43.•• Loos RJ, Lindgren CM, Li S, Wheeler E, Zhao JH, Prokopenko I, Inouye M, Freathy RM, Attwood AP, Beckmann JS, et al. Common variants near MC4R are associated with fat mass, weight and risk of obesity. Nat Genet. 2008;40(6):768–75. In addition to being an excellent review summarize the authors the predictive value of polygenic risk scores in predicting future BMI and obesity. [DOI] [PMC free article] [PubMed]
- 44.Sollis E, Mosaku A, Abid A, Buniello A, Cerezo M, Gil L, Groza T, Güneş O, Hall P, Hayhurst J, et al. The NHGRI-EBI GWAS Catalog: knowledgebase and deposition resource. Nucleic Acids Res. 2023;51(D1):D977–d985. doi: 10.1093/nar/gkac1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Loos RJF, Yeo GSH. The genetics of obesity: from discovery to biology. Nat Rev Genet. 2022;23(2):120–133. doi: 10.1038/s41576-021-00414-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Warner ET, Jiang L, Adjei DN, Turman C, Gordon W, Wang L, Tamimi R, Kraft P, Lindström S. A genome-wide association study of childhood body fatness. Obesity (Silver Spring) 2021;29(2):446–453. doi: 10.1002/oby.23070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bradfield JP, Taal HR, Timpson NJ, Scherag A, Lecoeur C, Warrington NM, Hypponen E, Holst C, Valcarcel B, Thiering E, et al. A genome-wide association meta-analysis identifies new childhood obesity loci. Nat Genet. 2012;44(5):526–531. doi: 10.1038/ng.2247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Scherag A, Dina C, Hinney A, Vatin V, Scherag S, Vogel CI, Müller TD, Grallert H, Wichmann HE, Balkau B, et al. Two new loci for body-weight regulation identified in a joint analysis of genome-wide association studies for early-onset extreme obesity in French and german study groups. PLoS Genet. 2010;6(4):e1000916. doi: 10.1371/journal.pgen.1000916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Costa-Urrutia P, Colistro V, Jiménez-Osorio AS, Cárdenas-Hernández H, Solares-Tlapechco J, Ramirez-Alcántara M, Granados J, Ascencio-Montiel IJ, Rodríguez-Arellano ME. Genome-wide association study of body mass index and body fat in Mexican-Mestizo Children. Genes (Basel) 2019;10(11):945. doi: 10.3390/genes10110945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Couto Alves A, De Silva NMG, Karhunen V, Sovio U, Das S, Taal HR, Warrington NM, Lewin AM, Kaakinen M, Cousminer DL, et al. GWAS on longitudinal growth traits reveals different genetic factors influencing infant, child, and adult BMI. Sci Adv. 2019;5(9):eaaw3095. doi: 10.1126/sciadv.aaw3095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.den Hoed M, Ekelund U, Brage S, Grontved A, Zhao JH, Sharp SJ, Ong KK, Wareham NJ, Loos RJ. Genetic susceptibility to obesity and related traits in childhood and adolescence: influence of loci identified by genome-wide association studies. Diabetes. 2010;59(11):2980–2988. doi: 10.2337/db10-0370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Liu G, Zhu H, Lagou V, Gutin B, Stallmann-Jorgensen IS, Treiber FA, Dong Y, Snieder H. FTO variant rs9939609 is associated with body mass index and waist circumference, but not with energy intake or physical activity in European- and African-American youth. BMC Med Genet. 2010;11:57. doi: 10.1186/1471-2350-11-57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Liu HY, Alyass A, Abadi A, Peralta-Romero J, Suarez F, Gomez-Zamudio J, Audirac A, Parra EJ, Cruz M, Meyre D. Fine-mapping of 98 obesity loci in Mexican children. Int J Obes (Lond) 2019;43(1):23–32. doi: 10.1038/s41366-018-0056-7. [DOI] [PubMed] [Google Scholar]
- 54.Wheeler E, Huang N, Bochukova EG, Keogh JM, Lindsay S, Garg S, Henning E, Blackburn H, Loos RJ, Wareham NJ, et al. Genome-wide SNP and CNV analysis identifies common and low-frequency variants associated with severe early-onset obesity. Nat Genet. 2013;45(5):513–517. doi: 10.1038/ng.2607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Xi B, Shen Y, Zhang M, Liu X, Zhao X, Wu L, Cheng H, Hou D, Lindpaintner K, Liu L, et al. The common rs9939609 variant of the fat mass and obesity-associated gene is associated with obesity risk in children and adolescents of Beijing. China BMC Med Genet. 2010;11:107. doi: 10.1186/1471-2350-11-107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yao S, Wu H, Ding JM, Wang ZX, Ullah T, Dong SS, Chen H, Guo Y. Transcriptome-wide association study identifies multiple genes associated with childhood body mass index. Int J Obes (Lond) 2021;45(5):1105–1113. doi: 10.1038/s41366-021-00780-y. [DOI] [PubMed] [Google Scholar]
- 57.Zhao J, Bradfield JP, Li M, Wang K, Zhang H, Kim CE, Annaiah K, Glessner JT, Thomas K, Garris M, et al. The role of obesity-associated loci identified in genome-wide association studies in the determination of pediatric BMI. Obesity (Silver Spring) 2009;17(12):2254–2257. doi: 10.1038/oby.2009.159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bressler J, Fornage M, Hanis CL, Kao WH, Lewis CE, McPherson R, Dent R, Mosley TH, Pennacchio LA, Boerwinkle E. The INSIG2 rs7566605 genetic variant does not play a major role in obesity in a sample of 24,722 individuals from four cohorts. BMC Med Genet. 2009;10:56. doi: 10.1186/1471-2350-10-56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Campa D, Hüsing A, McKay JD, Sinilnikova O, Vogel U, Tjønneland A, Overvad K, Stegger J, Clavel-Chapelon F, Chabbert-Buffet N, et al. The INSIG2 rs7566605 polymorphism is not associated with body mass index and breast cancer risk. BMC Cancer. 2010;10:563. doi: 10.1186/1471-2407-10-563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Herbert A, Gerry NP, McQueen MB, Heid IM, Pfeufer A, Illig T, Wichmann HE, Meitinger T, Hunter D, Hu FB, et al. A common genetic variant is associated with adult and childhood obesity. Science. 2006;312(5771):279–283. doi: 10.1126/science.1124779. [DOI] [PubMed] [Google Scholar]
- 61.Zhao J, Bradfield JP, Zhang H, Annaiah K, Wang K, Kim CE, Glessner JT, Frackelton EC, Otieno FG, Doran J, et al. Examination of all type 2 diabetes GWAS loci reveals HHEX-IDE as a locus influencing pediatric BMI. Diabetes. 2010;59(3):751–755. doi: 10.2337/db09-0972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Maher BS. Polygenic scores in epidemiology: risk prediction, etiology, and clinical utility. Curr Epidemiol Rep. 2015;2(4):239–244. doi: 10.1007/s40471-015-0055-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.•• Khera AV, Chaffin M, Wade KH, Zahid S, Brancale J, Xia R, Distefano M, Senol-Cosar O, Haas ME, Bick A, et al. Polygenic prediction of weight and obesity trajectories from birth to adulthood. Cell. 2019;177(3):587–96.e589. The study describes a new polygenic risk score that has ability to discriminate differences in weight, obesity, cardiometabolic disease and mortality in adults. [DOI] [PMC free article] [PubMed]
- 64.Lange K, Kerr JA, Mansell T, O’Sullivan JM, Burgner DP, Clifford SA, Olds T, Dwyer T, Wake M, Saffery R. Can adult polygenic scores improve prediction of body mass index in childhood? Int J Obes (Lond) 2022;46(7):1375–1383. doi: 10.1038/s41366-022-01130-2. [DOI] [PubMed] [Google Scholar]
- 65.O'Sullivan JW, Raghavan S, Marquez-Luna C, Luzum JA, Damrauer SM, Ashley EA, O'Donnell CJ, Willer CJ, Natarajan P. Polygenic risk scores for cardiovascular disease: a scientific statement from the American Heart Association. Circulation. 2022;146(8):e93–e118. doi: 10.1161/CIR.0000000000001077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.De Vincentis A, Tavaglione F, Jamialahmadi O, Picardi A, Antonelli Incalzi R, Valenti L, Romeo S, Vespasiani-Gentilucci U. A polygenic risk score to refine risk stratification and prediction for severe liver disease by clinical fibrosis scores. Clin Gastroenterol Hepatol. 2022;20(3):658–673. doi: 10.1016/j.cgh.2021.05.056. [DOI] [PubMed] [Google Scholar]
- 67.San-Cristobal R, de Toro-Martín J, Vohl MC. Appraisal of gene-environment interactions in GWAS for evidence-based precision nutrition implementation. Curr Nutr Rep. 2022;11(4):563–573. doi: 10.1007/s13668-022-00430-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Feitosa MF, Kraja AT, Chasman DI, Sung YJ, Winkler TW, Ntalla I, Guo X, Franceschini N, Cheng CY, Sim X, et al. Novel genetic associations for blood pressure identified via gene-alcohol interaction in up to 570K individuals across multiple ancestries. PLoS ONE. 2018;13(6):e0198166. doi: 10.1371/journal.pone.0198166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Graff M, Scott RA, Justice AE, Young KL, Feitosa MF, Barata L, Winkler TW, Chu AY, Mahajan A, Hadley D, et al. Genome-wide physical activity interactions in adiposity - A meta-analysis of 200,452 adults. PLoS Genet. 2017;13(4):e1006528. doi: 10.1371/journal.pgen.1006528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kilpeläinen TO, Qi L, Brage S, Sharp SJ, Sonestedt E, Demerath E, Ahmad T, Mora S, Kaakinen M, Sandholt CH, et al. Physical activity attenuates the influence of FTO variants on obesity risk: a meta-analysis of 218,166 adults and 19,268 children. PLoS Med. 2011;8(11):e1001116. doi: 10.1371/journal.pmed.1001116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Carless MA, Kulkarni H, Kos MZ, Charlesworth J, Peralta JM, Göring HH, Blangero J. Genetic effects on DNA methylation and its potential relevance for obesity in Mexican Americans. PloS one. 2013;8(9):e73950. doi: 10.1371/journal.pone.0073950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Xu X, Su S, Barnes VA, De Miguel C, Pollock J, Ownby D, Wang X. A genome-wide methylation study on obesity: differential variability and differential methylation. Epigenetics. 2013;8(5):522–533. doi: 10.4161/epi.24506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Dick KJ, Nelson CP, Tsaprouni L, Sandling JK, Aïssi D, Wahl S, Meduri E, Morange PE, Gagnon F, Grallert H, et al. DNA methylation and body-mass index: a genome-wide analysis. Lancet. 2014;383(9933):1990–1998. doi: 10.1016/S0140-6736(13)62674-4. [DOI] [PubMed] [Google Scholar]
- 74.Almén MS, Nilsson EK, Jacobsson JA, Kalnina I, Klovins J, Fredriksson R, Schiöth HB. Genome-wide analysis reveals DNA methylation markers that vary with both age and obesity. Gene. 2014;548(1):61–67. doi: 10.1016/j.gene.2014.07.009. [DOI] [PubMed] [Google Scholar]
- 75.Guénard F, Tchernof A, Deshaies Y, Pérusse L, Biron S, Lescelleur O, Vohl MC. Differential methylation in visceral adipose tissue of obese men discordant for metabolic disturbances. Physiol Genomics. 2014;46(6):216–222. doi: 10.1152/physiolgenomics.00160.2013. [DOI] [PubMed] [Google Scholar]
- 76.Aslibekyan S, Demerath EW, Mendelson M, Zhi D, Guan W, Liang L, Sha J, Pankow JS, Liu C, Irvin MR, et al. Epigenome-wide study identifies novel methylation loci associated with body mass index and waist circumference. Obesity (Silver Spring) 2015;23(7):1493–1501. doi: 10.1002/oby.21111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Ollikainen M, Ismail K, Gervin K, Kyllönen A, Hakkarainen A, Lundbom J, Kaprio J. Genome-wide blood DNA methylation alterations at regulatory elements and heterochromatic regions in monozygotic twins discordant for obesity and liver fat. Clin Epigenetics. 2015;7(1):39. doi: 10.1186/s13148-015-0073-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Demerath EW, Guan W, Grove ML, Aslibekyan S, Mendelson M, Zhou YH, Hedman ÅK, Sandling JK, Li LA, Irvin MR, et al. Epigenome-wide association study (EWAS) of BMI, BMI change and waist circumference in African American adults identifies multiple replicated loci. Hum Mol Genet. 2015;24(15):4464–4479. doi: 10.1093/hmg/ddv161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Voisin S, Almén MS, Zheleznyakova GY, Lundberg L, Zarei S, Castillo S, Schiöth HB. Many obesity-associated SNPs strongly associate with DNA methylation changes at proximal promoters and enhancers. Genome Med. 2015;7:103. doi: 10.1186/s13073-015-0225-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Arner P, Sinha I, Thorell A, Rydén M, Dahlman-Wright K, Dahlman I. The epigenetic signature of subcutaneous fat cells is linked to altered expression of genes implicated in lipid metabolism in obese women. Clin Epigenetics. 2015;7(1):93. doi: 10.1186/s13148-015-0126-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Rönn T, Volkov P, Gillberg L, Kokosar M, Perfilyev A, Jacobsen AL, Ling C. Impact of age, BMI and HbA1c levels on the genome-wide DNA methylation and mRNA expression patterns in human adipose tissue and identification of epigenetic biomarkers in blood. Hum Mol Genet. 2015;24(13):3792–3813. doi: 10.1093/hmg/ddv124. [DOI] [PubMed] [Google Scholar]
- 82.Kirchner H, Sinha I, Gao H, Ruby MA, Schönke M, Lindvall JM, Zierath JR. Altered DNA methylation of glycolytic and lipogenic genes in liver from obese and type 2 diabetic patients. Mol Metab. 2016;5(3):171–183. doi: 10.1016/j.molmet.2015.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Ali O, Cerjak D, Kent JW, Jr, James R, Blangero J, Carless MA, Zhang Y. Methylation of SOCS3 is inversely associated with metabolic syndrome in an epigenome-wide association study of obesity. Epigenetics. 2016;11(9):699–707. doi: 10.1080/15592294.2016.1216284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Keller M, Hopp L, Liu X, Wohland T, Rohde K, Cancello R, Böttcher Y. Genome-wide DNA promoter methylation and transcriptome analysis in human adipose tissue unravels novel candidate genes for obesity. Mol Metab. 2017;6(1):86–100. doi: 10.1016/j.molmet.2016.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Pietiläinen KH, Ismail K, Järvinen E, Heinonen S, Tummers M, Bollepalli S, Ollikainen M. DNA methylation and gene expression patterns in adipose tissue differ significantly within young adult monozygotic BMI-discordant twin pairs. Int J Obes. 2016;40(4):654–661. doi: 10.1038/ijo.2015.221. [DOI] [PubMed] [Google Scholar]
- 86.Volkov P, Olsson AH, Gillberg L, Jørgensen SW, Brøns C, Eriksson KF, Ling C. A genome-wide mQTL analysis in human adipose tissue identifies genetic variants associated with DNA methylation, gene expression and metabolic traits. PloS One. 2016;11(6):e0157776. doi: 10.1371/journal.pone.0157776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Al Muftah WA, Al-Shafai M, Zaghlool SB, Visconti A, Tsai PC, Kumar P, Spector T, Bell J, Falchi M, Suhre K. Epigenetic associations of type 2 diabetes and BMI in an Arab population. Clin Epigenetics. 2016;8:13. doi: 10.1186/s13148-016-0177-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Sayols-Baixeras S, Subirana I, Fernández-Sanlés A, Sentí M, Lluís-Ganella C, Marrugat J, Elosua R. DNA methylation and obesity traits: An epigenome-wide association study. The REGICOR study. Epigenetics. 2017;12(10):909–916. doi: 10.1080/15592294.2017.1363951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Crujeiras AB, Diaz-Lagares A, Sandoval J, Milagro FI, Navas-Carretero S, Carreira MC, Martinez JA. DNA methylation map in circulating leukocytes mirrors subcutaneous adipose tissue methylation pattern: a genome-wide analysis from non-obese and obese patients. Sci Rep. 2017;7(1):41903. doi: 10.1038/srep41903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Meeks KAC, Henneman P, Venema A, Burr T, Galbete C, Danquah I, Schulze MB, Mockenhaupt FP, Owusu-Dabo E, Rotimi CN, et al. An epigenome-wide association study in whole blood of measures of adiposity among Ghanaians: the RODAM study. Clin Epigenetics. 2017;9:103. doi: 10.1186/s13148-017-0403-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Wilson LE, Harlid S, Xu Z, Sandler DP, Taylor JA. An epigenome-wide study of body mass index and DNA methylation in blood using participants from the Sister Study cohort. Int J Obes (Lond) 2017;41(1):194–199. doi: 10.1038/ijo.2016.184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Guénard F, Tchernof A, Deshaies Y, Biron S, Lescelleur O, Biertho L, Vohl MC. Genetic regulation of differentially methylated genes in visceral adipose tissue of severely obese men discordant for the metabolic syndrome. Transl Res. 2017;184:1–11. doi: 10.1016/j.trsl.2017.01.002. [DOI] [PubMed] [Google Scholar]
- 93.Mendelson MM, Marioni RE, Joehanes R, Liu C, Hedman ÅK, Aslibekyan S, Deary IJ. Association of body mass index with DNA methylation and gene expression in blood cells and relations to cardiometabolic disease: a Mendelian randomization approach. PLoS Med. 2017;14(1):e1002215. doi: 10.1371/journal.pmed.1002215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Kvaløy K, Page CM, Holmen TL. Epigenome-wide methylation differences in a group of lean and obese women–A HUNT Study. Sci Rep. 2018;8(1):16330. doi: 10.1038/s41598-018-34003-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Dhana K, Braun KV, Nano J, Voortman T, Demerath EW, Guan W, Dehghan A. An epigenome-wide association study of obesity-related traits. Am J Epidemiol. 2018;187(8):1662–1669. doi: 10.1093/aje/kwy025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Campanella G, Gunter MJ, Polidoro S, Krogh V, Palli D, Panico S, Sacerdote C, Tumino R, Fiorito G, Guarrera S, et al. Epigenome-wide association study of adiposity and future risk of obesity-related diseases. Int J Obes (Lond) 2018;42(12):2022–2035. doi: 10.1038/s41366-018-0064-7. [DOI] [PubMed] [Google Scholar]
- 97.Orozco LD, Farrell C, Hale C, Rubbi L, Rinaldi A, Civelek M, Pellegrini M. Epigenome-wide association in adipose tissue from the METSIM cohort. Hum Mol Genet. 2018;27(10):1830–1846. doi: 10.1093/hmg/ddy093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Akinyemiju T, Do AN, Patki A, Aslibekyan S, Zhi D, Hidalgo B, Irvin MR. Epigenome-wide association study of metabolic syndrome in African-American adults. Clin Epigenetics. 2018;10(1):49. doi: 10.1186/s13148-018-0483-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Zaghlool SB, Mook-Kanamori DO, Kader S, Stephan N, Halama A, Engelke R, Sarwath H, Al-Dous EK, Mohamoud YA, Roemisch-Margl W, et al. Deep molecular phenotypes link complex disorders and physiological insult to CpG methylation. Hum Mol Genet. 2018;27(6):1106–1121. doi: 10.1093/hmg/ddy006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Guo Q, Zheng R, Huang J, He M, Wang Y, Guo Z, Chen P. Using integrative analysis of DNA methylation and gene expression data in multiple tissue types to prioritize candidate genes for drug development in obesity. Front Genet. 2018;9:663. doi: 10.3389/fgene.2018.00663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Li W, Zhang D, Wang W, Wu Y, Mohammadnejad A, Lund J, Tan Q. DNA methylome profiling in identical twin pairs discordant for body mass index. Int J Obes. 2019;43(12):2491–2499. doi: 10.1038/s41366-019-0382-4. [DOI] [PubMed] [Google Scholar]
- 102.Li C, Wang Z, Hardy T, Huang Y, Hui Q, Crusto CA, Sun YV. Association of obesity with DNA methylation age acceleration in African American mothers from the InterGEN study. Int J Mol Sci. 2019;20(17):4273. doi: 10.3390/ijms20174273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Pan Y, Choi JH, Shi H, Zhang L, Su S, Wang X. Discovery and validation of a novel neutrophil activation marker associated with obesity. Sci Rep. 2019;9(1):3433. doi: 10.1038/s41598-019-39764-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Koh IU, Choi NH, Lee K, Yu HY, Yun JH, Kong JH, Moon S. Obesity susceptible novel DNA methylation marker on regulatory region of inflammation gene: results from the Korea Epigenome Study (KES) BMJ Open Diabetes Res Care. 2020;8(1):e001338. doi: 10.1136/bmjdrc-2020-001338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Giri AK, Prasad G, Bandesh K, Parekatt V, Mahajan A, Banerjee P, Bharadwaj D. Multifaceted genome-wide study identifies novel regulatory loci in SLC22A11 and ZNF45 for body mass index in Indians. Mol Genet Genom. 2020;295:1013–1026. doi: 10.1007/s00438-020-01678-6. [DOI] [PubMed] [Google Scholar]
- 106.Justice AE, Chittoor G, Gondalia R, Melton PE, Lim E, Grove ML, North KE. Methylome-wide association study of central adiposity implicates genes involved in immune and endocrine systems. Epigenomics. 2020;12(17):1483–1499. doi: 10.2217/epi-2019-0276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Xie T, Gorenjak V, Stathopoulou MG, Dadé S, Marouli E, Masson C, Visvikis-Siest S. Epigenome-wide association study detects a novel loci associated with central obesity in healthy subjects. BMC Med Genom. 2021;14(1):233. doi: 10.1186/s12920-021-01077-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.• Chen Y, Kassam I, Lau SH, Kooner JS, Wilson R, Peters A, Sim X. Impact of BMI and waist circumference on epigenome-wide DNA methylation and identification of epigenetic biomarkers in blood: an EWAS in multi-ethnic Asian individuals. Clin Epigenetics. 2021;13(1):195. 10.1186/s13148-021-01162-x. The study suggest that obesity is preceding methylation changes being likely cause rather than consequence. [DOI] [PMC free article] [PubMed]
- 109.Cao VT, Lea RA, Sutherland HG, Benton MC, Pishva RS, Haupt LM, Griffiths LR. A genome-wide methylation study of body fat traits in the Norfolk Island isolate. Nutr Metab Cardiovasc Dis. 2021;31(5):1556–1563. doi: 10.1016/j.numecd.2021.01.027. [DOI] [PubMed] [Google Scholar]
- 110.Do WL, Gohar J, McCullough LE, Galaviz KI, Conneely KN, Narayan KV. Examining the association between adiposity and DNA methylation: a systematic review and meta-analysis. Obes Rev. 2021;22(10):e13319. doi: 10.1111/obr.13319. [DOI] [PubMed] [Google Scholar]
- 111.Wu Y, Tian H, Wang W, Li W, Duan H, Zhang D. DNA methylation and waist-to-hip ratio: an epigenome-wide association study in Chinese monozygotic twins. J Endocrinol Invest. 2022;45(12):2365–2376. doi: 10.1007/s40618-022-01878-4. [DOI] [PubMed] [Google Scholar]
- 112.Taylor JY, Huang Y, Zhao W, Wright ML, Wang Z, Hui Q, Sun YV. Epigenome-wide association study of BMI in Black populations from InterGEN and GENOA. Obesity. 2023;31(1):243–255. doi: 10.1002/oby.23589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.• Do WL, Sun D, Meeks K, Dugué PA, Demerath E, Guan W, Li S, Chen W, Milne R, Adeyemo A, et al. Epigenome-wide meta-analysis of BMI in nine cohorts: examining the utility of epigenetically predicted BMI. Am J Hum Genet. 2023;110(2):273–83. This work suggests that ~400 CpG sites can be attributed to 32% BMI variance in predicting BMI in a test set. [DOI] [PMC free article] [PubMed]
- 114.Vehmeijer FOL, Küpers LK, Sharp GC, Salas LA, Lent S, Jima DD, Tindula G, Reese S, Qi C, Gruzieva O, et al. DNA methylation and body mass index from birth to adolescence: meta-analyses of epigenome-wide association studies. Genome Med. 2020;12(1):105. doi: 10.1186/s13073-020-00810-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Chambers JC, Loh M, Lehne B, Drong A, Kriebel J, Motta V, Wahl S, Elliott HR, Rota F, Scott WR, et al. Epigenome-wide association of DNA methylation markers in peripheral blood from Indian Asians and Europeans with incident type 2 diabetes: a nested case-control study. Lancet Diabetes Endocrinol. 2015;3(7):526–534. doi: 10.1016/S2213-8587(15)00127-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Holle R, Happich M, Löwel H, Wichmann HE. KORA–a research platform for population based health research. Gesundheitswesen. 2005;67(Suppl 1):S19–25. doi: 10.1055/s-2005-858235. [DOI] [PubMed] [Google Scholar]
- 117.Aron-Wisnewsky J, Julia Z, Poitou C, Bouillot JL, Basdevant A, Chapman MJ, Clement K, Guerin M. Effect of bariatric surgery-induced weight loss on SR-BI-, ABCG1-, and ABCA1-mediated cellular cholesterol efflux in obese women. J Clin Endocrinol Metab. 2011;96(4):1151–1159. doi: 10.1210/jc.2010-2378. [DOI] [PubMed] [Google Scholar]
- 118.Edgel KA, McMillen TS, Wei H, Pamir N, Houston BA, Caldwell MT, Mai PO, Oram JF, Tang C, Leboeuf RC. Obesity and weight loss result in increased adipose tissue ABCG1 expression in db/db mice. Biochim Biophys Acta. 2012;1821(3):425–434. doi: 10.1016/j.bbalip.2011.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Frisdal E, Le Lay S, Hooton H, Poupel L, Olivier M, Alili R, Plengpanich W, Villard EF, Gilibert S, Lhomme M, et al. Adipocyte ATP-binding cassette G1 promotes triglyceride storage, fat mass growth, and human obesity. Diabetes. 2015;64(3):840–855. doi: 10.2337/db14-0245. [DOI] [PubMed] [Google Scholar]
- 120.Kennedy MA, Barrera GC, Nakamura K, Baldán A, Tarr P, Fishbein MC, Frank J, Francone OL, Edwards PA. ABCG1 has a critical role in mediating cholesterol efflux to HDL and preventing cellular lipid accumulation. Cell Metab. 2005;1(2):121–131. doi: 10.1016/j.cmet.2005.01.002. [DOI] [PubMed] [Google Scholar]
- 121.Wang N, Lan D, Chen W, Matsuura F, Tall AR. ATP-binding cassette transporters G1 and G4 mediate cellular cholesterol efflux to high-density lipoproteins. Proc Natl Acad Sci U S A. 2004;101(26):9774–9779. doi: 10.1073/pnas.0403506101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Dayeh T, Tuomi T, Almgren P, Perfilyev A, Jansson PA, de Mello VD, Pihlajamäki J, Vaag A, Groop L, Nilsson E, et al. DNA methylation of loci within ABCG1 and PHOSPHO1 in blood DNA is associated with future type 2 diabetes risk. Epigenetics. 2016;11(7):482–488. doi: 10.1080/15592294.2016.1178418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Hidalgo B, Irvin MR, Sha J, Zhi D, Aslibekyan S, Absher D, Tiwari HK, Kabagambe EK, Ordovas JM, Arnett DK. Epigenome-wide association study of fasting measures of glucose, insulin, and HOMA-IR in the Genetics of Lipid Lowering Drugs and Diet Network study. Diabetes. 2014;63(2):801–807. doi: 10.2337/db13-1100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Kulkarni H, Kos MZ, Neary J, Dyer TD, Kent JW, Jr, Göring HH, Cole SA, Comuzzie AG, Almasy L, Mahaney MC, et al. Novel epigenetic determinants of type 2 diabetes in Mexican-American families. Hum Mol Genet. 2015;24(18):5330–5344. doi: 10.1093/hmg/ddv232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Moon JS, Nakahira K, Chung KP, DeNicola GM, Koo MJ, Pabón MA, Rooney KT, Yoon JH, Ryter SW, Stout-Delgado H, et al. NOX4-dependent fatty acid oxidation promotes NLRP3 inflammasome activation in macrophages. Nat Med. 2016;22(9):1002–1012. doi: 10.1038/nm.4153. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 126.Hall CJ, Sanderson LE, Lawrence LM, Pool B, van der Kroef M, Ashimbayeva E, Britto D, Harper JL, Lieschke GJ, Astin JW, et al. Blocking fatty acid-fueled mROS production within macrophages alleviates acute gouty inflammation. J Clin Invest. 2018;128(5):1752–1771. doi: 10.1172/JCI94584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Softic S, Meyer JG, Wang GX, Gupta MK, Batista TM, Lauritzen H, Fujisaka S, Serra D, Herrero L, Willoughby J, et al. Dietary sugars alter hepatic fatty acid oxidation via transcriptional and post-translational modifications of mitochondrial proteins. Cell Metab. 2019;30(4):735–753.e734. doi: 10.1016/j.cmet.2019.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Villicaña S, Bell JT. Genetic impacts on DNA methylation: research findings and future perspectives. Genome Biol. 2021;22(1):127. doi: 10.1186/s13059-021-02347-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Hüls A, Czamara D. Methodological challenges in constructing DNA methylation risk scores. Epigenetics. 2020;15(1–2):1–11. doi: 10.1080/15592294.2019.1644879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Hamilton OKL, Zhang Q, McRae AF, Walker RM, Morris SW, Redmond P, Campbell A, Murray AD, Porteous DJ, Evans KL, et al. An epigenetic score for BMI based on DNA methylation correlates with poor physical health and major disease in the Lothian Birth Cohort. Int J Obes (Lond) 2019;43(9):1795–1802. doi: 10.1038/s41366-018-0262-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Reed ZE, Suderman MJ, Relton CL, Davis OSP, Hemani G. The association of DNA methylation with body mass index: distinguishing between predictors and biomarkers. Clin Epigenetics. 2020;12(1):50. doi: 10.1186/s13148-020-00841-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.McCartney DL, Hillary RF, Stevenson AJ, Ritchie SJ, Walker RM, Zhang Q, Morris SW, Bermingham ML, Campbell A, Murray AD, et al. Epigenetic prediction of complex traits and death. Genome Biol. 2018;19(1):136. doi: 10.1186/s13059-018-1514-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Odintsova VV, Rebattu V, Hagenbeek FA, Pool R, Beck JJ, Ehli EA, van Beijsterveldt CEM, Ligthart L, Willemsen G, de Geus EJC, et al. Predicting complex traits and exposures from polygenic scores and blood and buccal DNA methylation profiles. Front Psychiatry. 2021;12:688464. doi: 10.3389/fpsyt.2021.688464. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.