Abstract
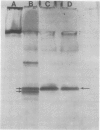
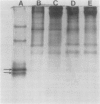
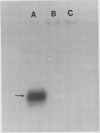
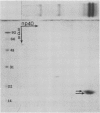
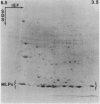
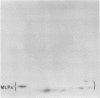
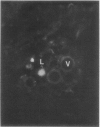
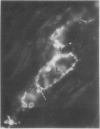
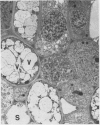
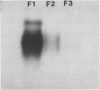
Latex from the opium poppy, Papaver somniferum L., was analyzed by polyacrylamide gel electrophoresis (PAGE). Two latex-specific bands were identified in protein samples of poppy latex using one-dimensional native PAGE. Second dimension analysis with SDS-PAGE indicates that these proteins have a relative molecular weight of approximately 20 kilodaltons. We have termed these polypeptides the major latex proteins (MLPs). Polyclonal antibodies prepared against the MLPs were used to probe protein gel blots of latex and poppy tissues known to lack laticifers. Laticifer-free tissues showed no reaction with anti-MLP immunoglobulin G indicating that MLPs are found only in poppy latex. MLP distribution was also examined in mature opium poppy tissues by immunocytochemistry. Laticifers were differentially labeled by fluorescein isothiocyanate secondary labeling of anti-MLP immunoglobulin G and could easily be identified in both transverse and longitudinal section. Fractionation studies of isolated latex showed that MLPs are concentrated in the latex cytosol and not in alkaloidal vesicles. Analysis of latex proteins by conventional two-dimensional electrophoresis indicates that the two MLP bands are composed of several distinct polypeptides with similar relative molecular weights. The pIs of these molecules range from 6.0 to 3.5. The role(s) of MLPs in laticifer metabolism has not been determined.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bradford M. M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976 May 7;72:248–254. doi: 10.1006/abio.1976.9999. [DOI] [PubMed] [Google Scholar]
- Fairbanks G., Steck T. L., Wallach D. F. Electrophoretic analysis of the major polypeptides of the human erythrocyte membrane. Biochemistry. 1971 Jun 22;10(13):2606–2617. doi: 10.1021/bi00789a030. [DOI] [PubMed] [Google Scholar]
- Friedman R. D. Comparison of four different silver-staining techniques for salivary protein detection in alkaline polyacrylamide gels. Anal Biochem. 1982 Nov 1;126(2):346–349. doi: 10.1016/0003-2697(82)90525-5. [DOI] [PubMed] [Google Scholar]
- KLEINSCHMIDT G., MOTHES K. Zur Physiologie und Biosynthese der Alkaloide von Papaver somniferum. Z Naturforsch B. 1959 Jan;14B(1):52–56. [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- O'Farrell P. H. High resolution two-dimensional electrophoresis of proteins. J Biol Chem. 1975 May 25;250(10):4007–4021. [PMC free article] [PubMed] [Google Scholar]
- Roberts M. F., McCarthy D., Kutchan T. M., Coscia C. J. Localization of enzymes and alkaloidal metabolites in Papaver latex. Arch Biochem Biophys. 1983 Apr 15;222(2):599–609. doi: 10.1016/0003-9861(83)90558-1. [DOI] [PubMed] [Google Scholar]
- Towbin H., Staehelin T., Gordon J. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc Natl Acad Sci U S A. 1979 Sep;76(9):4350–4354. doi: 10.1073/pnas.76.9.4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wray W., Boulikas T., Wray V. P., Hancock R. Silver staining of proteins in polyacrylamide gels. Anal Biochem. 1981 Nov 15;118(1):197–203. doi: 10.1016/0003-2697(81)90179-2. [DOI] [PubMed] [Google Scholar]