Abstract
Mitochondria are critical cellular energy resources and are central to the life of the neuron. Mitophagy selectively clears damaged or dysfunctional mitochondria through autophagic machinery to maintain mitochondrial quality control and homeostasis. Mature neurons are postmitotic and consume substantial energy, thus require highly efficient mitophagy pathways to turn over damaged or dysfunctional mitochondria. Recent evidence indicates that mitophagy is pivotal to the pathogenesis of neurological diseases. However, more work is needed to study mitophagy pathway components as potential therapeutic targets. In this review, we briefly discuss the characteristics of nonselective autophagy and selective autophagy, including ERphagy, aggrephagy, and mitophagy. We then introduce the mechanisms of Parkin-dependent and Parkin-independent mitophagy pathways under physiological conditions. Next, we summarize the diverse repertoire of mitochondrial membrane receptors and phospholipids that mediate mitophagy. Importantly, we review the critical role of mitophagy in the pathogenesis of neurodegenerative diseases including Alzheimer’s disease, Parkinson’s disease, and amyotrophic lateral sclerosis. Last, we discuss recent studies considering mitophagy as a potential therapeutic target for treating neurodegenerative diseases. Together, our review may provide novel views to better understand the roles of mitophagy in neurodegenerative disease pathogenesis.
Keywords: Alzheimer’s disease, amyotrophic lateral sclerosis, autophagy, mitochondria, mitophagy, mitophagy receptor, Parkin, Parkinson’s disease, PINK1
Introduction
Neurons are the basic structural and functional cellular unit of the nervous system. The nervous system performs extremely complex functions using neurons to accept stimulation and transmit nerve impulses (Talifu et al., 2023). Both neurodevelopment and the long-term maintenance of neuronal health require effective removal of aggregated proteins and defective organelles. Autophagy (hereafter refers to as macroautophagy) is a major cellular system that degrades dysfunctional organelles and protein aggregates and is particularly critical for neurons (Tedesco et al., 2022). There are two main types of autophagy defined by their specificity. One is nonselective autophagy, which indiscriminately sequesters and degrades parts of the cytoplasm. The other is selective autophagy, which targets potentially dysfunctional cargo for degradation. Mitochondria play an essential role in cells and organisms. Mitochondrial homeostasis is a finely tuned process with pathways controlling mitochondrial size, biogenesis, and degradation (Huang et al., 2022). Mitophagy, which was first observed in electron micrographs of cultured cells, is an evolutionally conserved cellular process to selectively remove dysfunctional mitochondria (De Duve and Wattiaux, 1966). During mitophagy, transport receptors interact with mammalian light chain 3 (LC3) members to bridge targets to autophagosomes and lysosomes for degradation (Doblado et al., 2021). Recently, the basic biochemical steps of mitophagy in Parkin-dependent and Parkin-independent pathways have been elucidated. Mitophagy dysregulation connects with a variety of pathological conditions, especially neurological diseases, though the mechanisms of these connections are still unclear. More studies are required to determine if mitophagy is an effective therapeutic target for the potential intervention and treatment of neurological diseases.
In this review, we briefly discuss the characteristics of nonselective autophagy and selective autophagy, including ERphagy, aggrephagy, and mitophagy. We then focus on ubiquitin-dependent mitophagy with further descriptions of the Parkin-dependent and Parkin-independent pathways. We also discuss the mitochondrial membrane receptors and phospholipids that mediate mitophagy. We summarize the connections between mitophagy and pathological mechanisms of neurological diseases, including Alzheimer’s disease (AD), Parkinson’s disease (PD), and amyotrophic lateral sclerosis (ALS). Finally, we review recent studies of pharmacological drugs and natural compounds for which mitophagy is the therapeutic target.
Retrieval Strategy
We searched the PubMed database online to retrieve articles published through May 31, 2023. A combination of the following text words (MeSH terms) was used to maximize search specificity and sensitivity: “mitophagy”; “autophagy”; “mitochondria”; “PINK1”; “Parkin”; “mitophagy receptor”; “Alzheimer’s disease”; “Parkinson’s disease” and “amyotrophic lateral sclerosis”. The results were further screened by title and abstract, and only those studies were kept that explored the relationship between mitophagy and neurological disease pathogenesis, especially in AD, PD, and ALS. No language or study-type restrictions were applied. Articles were excluded that only discussed mitophagy in glial cells or chaperone-mediated autophagy and microautophagy in neurons.
Nonselective Autophagy
Nonselective autophagy is the major pathway to nonselectively target large cargo for degradation. Nonselective autophagy produces a bilayer vesicle structure in the cytoplasm that envelopes organelles and long-lived proteins to form autophagosomes that fuse with lysosomes and eventually degrade when stimulated (Figure 1A). Nonselective autophagy allows cells to survive through nutrient starvation until new nutrients become available. Decades of studies in yeast have defined the molecular participants in this pathway (Ohsumi et al., 1993; Ohsumi, 1994, 1997, 1999, 2001, 2014), and we gradually found that nonselective autophagy plays a critical role in the stability of cells. However, the importance of nonselective autophagy was not fully known until Yoshinori Ohsumi won the 2016 Nobel Prize in Physiology or Medicine. We now know that nonselective autophagy makes unique contributions to the nervous system, in addition to its global and basic functions. Nonselective autophagy modulates neuron physiology in diverse ways: (1) modulates the initial steps in neuronal development, differentiation, and generation (Iwata et al., 2023); (2) maintains neuronal homeostasis (Maday, 2016; Kulkarni et al., 2018); (3) regulates polarity establishment and axonal branching (Yang et al., 2017); (4) restricts synaptic transmission in dopamine neurons (Hernandez et al., 2012) and (5) impacts neuron longevity and life span (Bennetzen et al., 2012; Seah et al., 2016). However, an alternative pathway, selective autophagy, also prevents neurons from entering a pathological state (Fleming et al., 2022).
Figure 1.
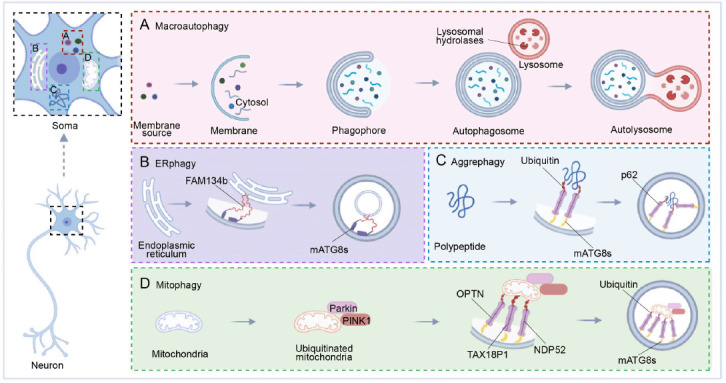
The spatial organization of autophagy in neurons.
Soma is depicted with four forms of autophagy in spatially distinct neuronal compartments: Macroautophagy (A), aggrephagy (B), ERphagy (C), and mitophagy (D). (A) substances to be cleared in neurons are gradually wrapped. First, a monolayer membrane and cytosol are formed, and with the formation of the bilayer, the phagophore is formed and then further wrapped to form the autophagosome. Finally, an autolysosome forms, and is cleared by lysosomal hydrolases in lysosomes. (B–D) Schematic diagrams for the subtypes of selective autophagy. For degradation of endoplasmic reticulum (ER), polypeptides, and mitochondria, the proteins involved are different from those for macroautophagy. (B) Family with sequence similarity 134, member B (FAM134b) connects to mammalian ATG8 proteins (mATG8s) on the membrane to participate in the degradation of damaged ER in ERphagy. (C) In aggrephagy, polypeptide-linked ubiquitin is linked to p62 on the membrane via mATG8s, and an autophagosome is then formed and degraded by lysosomes. (D) In mitophagy, Parkin and PINK1 are bound to ubiquitinated mitochondria. Using linkers, Optineurin (OPTN), T-cell leukemia virus type I binding protein 1 (TAX18P1), nuclear dot protein 52 (NDP52), ubiquitin, and mATG8s on mitochondria and membranes are linked, and the damaged mitochondria are eventually degraded. Created with BioRender.com.
Selective Autophagy
In contrast to starvation-induced autophagy, selective autophagy recognizes, sequesters, and destroys particular targets via autophagosomes. Specified soluble proteins, supramolecular complexes, droplets, abnormal or extra organelles, and invading pathogenic bacteria are all degraded by selective autophagy (Bourdenx et al., 2021). Though selective autophagy employs the same essential components as nonselective autophagy, it produces specialized autophagosomes that ingest specific cargos (Veljanovski and Batoko, 2014; Marshall and Vierstra, 2018). Receptors facilitate the selective formation of autophagosomes around target cargo via the mammalian ATG8 proteins (mATG8s) (Faruk et al., 2021), and these autophagosomes are designed to fuse with lysosomes to form autolysosomes. Finally, cargos are degraded by lysosomal hydrolase. Depending on the cargo type, selective autophagy pathways include ERphagy, aggrephagy, and mitophagy (Stavoe and Holzbaur, 2019), among which mitophagy is the most familiar pathway (Chu, 2019; Garcia-Macia et al., 2019). However, there is growing interest in the parallel pathways that mediate selective endoplasmic reticulum (ER) turnover and degradation of aggregated proteins.
ERphagy
The ER, which covers the entire cell, creates a vast and dynamic network of sheets, tubules, and cisternae. The ER effectively increases the membrane area for intracellular molecular transport (Lu et al., 2020). Notably, the neuronal ER must be renewed and remodeled, particularly under stress (Fernandes et al., 2016; Fowler and O’Sullivan, 2016), and ERphagy is the selective autophagic removal of ER segments (primarily ER membrane) to achieve this rapid renewal (Schuck et al., 2014; Grumati et al., 2018; Hubner and Dikic, 2020). Interestingly, neuropathies are accompanied by excess ER accumulation following failures in ERphagy, which may connect ERphagy to neurological disease pathogenesis (He et al., 2021; Reggiori and Molinari, 2022). Previous research reports that both ER stress and the unfolded protein response (UPR) increase ERphagy (Song et al., 2018; Zhao et al., 2020; Cherubini and Zito, 2022). The accumulation of unfolded and misfolded proteins harms the ER membrane, which then activates UPR stress-sensing proteins on the ER membrane that form links to mATG8s on the autophagic membrane to reduce or degrade unfolded proteins (Song et al., 2018; Wei et al., 2022; Figure 1B).
Aggrephagy
The ubiquitin-proteasome system clears misfolded or damaged proteins, but autophagy is indispensable for the clearance of aggregates of these misfolded proteins. Aggrephagy is the selective removal of aggregated proteins and is mediated by several receptors that work independently or in combination (Sarraf et al., 2022). Autophagic receptors such as Sequestosome 1 (p62/SQSTM1, hereafter p62), NBR1, TAX1BP1, TOLLIP, and CCT2 assist the aggrephagy system in recognizing and eliminating protein aggregates (Ma et al., 2022), and p62 is most central to the aggrephagy pathway (Danieli and Martens, 2018). p62 recruits autophagy machinery via its LIR motif and recruits cargo via its ubiquitin-binding domain, serving as an intermediary chain. To advance the degradation, p62 links mATG8s to the autophagic membrane with ubiquitinated cargo proteins (Yamada et al., 2018; Figure 1C). Although the relationship between protein aggregation and disease pathogenesis remains to be fully elucidated, several studies using mouse genetics, lentiviral delivery, and RNA interference have shown that aggregated protein clearance allows for symptomatic reversal in various neurological models (Watanabe et al., 2017; Suresh et al., 2018; Muscolino et al., 2020; Wetzel et al., 2020; Kumar et al., 2021).
Mitophagy
Mitophagy is another subtype of selective autophagy that clears damaged mitochondria in neurons (Eran and Ronit, 2022; Fang and Anisimov, 2023). Mitochondria use oxidative phosphorylation to generate energy in the form of ATP (Wallace et al., 1998; Wallace, 1999; Galluzzi et al., 2012; Palikaras and Tavernarakis, 2020), which generates harmful reactive oxygen species that damage mitochondria, which must then be efficiently removed. Neurons are particularly vulnerable to autophagic dysfunction as well as mitochondrial dysfunction because of their high energy dependence and post-mitotic state (Chinta et al., 2010; Cummins and Gotz, 2018; Ishikawa et al., 2018). Mitophagy uses an autophagy mechanism to selectively wrap and degrade damaged mitochondria, thus maintaining mitochondrial and neuronal homeostasis (Palikaras et al., 2015; Fivenson et al., 2017; Wu et al., 2019).
Ubiquitin-Dependent Mitophagy
Mitophagy is mediated through either ubiquitin-dependent or ubiquitin-independent pathways, where the primary molecular events involve the targeting of damaged mitochondria to the autophagy machinery (Iorio et al., 2021). In ubiquitin-dependent mitophagy, the damaged mitochondrial surface is ubiquitinated and core autophagy-related proteins are recruited, including OPTN, TAX18P1, and NDP52. When these proteins link with mATG8s, an isolating membrane is gradually formed around the mitochondria to generate the autophagic vesicle, which is eventually absorbed by lysosomes (Johansen and Lamark, 2020; Figure 1D). This ubiquitin-dependent mitophagy is further divided into Parkin-dependent and Parkin-independent pathways that differ in the initiating steps. Here, we focus on canonical mitophagy initiated by Parkin in a ubiquitin-dependent way, and we describe the diverse repertoire of receptor and adaptor molecules involved in canonical mitophagy (Figure 2A and B).
Figure 2.
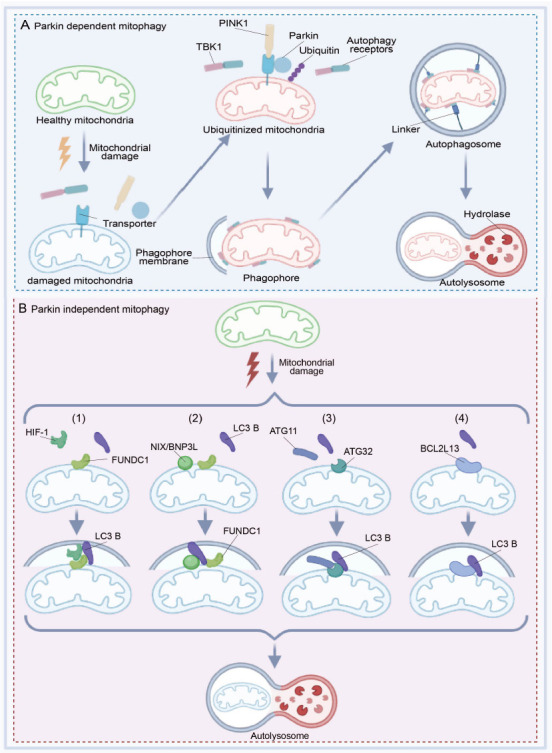
Schematic diagram of the Parkin-dependent and Parkin-independent pathways of mitophagy in neurons.
(A) When mitochondria are damaged by stimulating conditions, PINK1 is stabilized on the mitochondrial outer membrane (MOM), activating the Parkin. Parkin then assembles ubiquitin chains on numerous transporters, which can recruit ubiquitin-binding autophagy receptors to generate ubiquitinated mitochondria. Autophagy receptors and TBK1 bind to each other and bind to mitochondria, thus forming intermediates called phagophores. Using linkers, autophagosomes and autolysosomes are formed gradually, and the damaged mitochondria are eventually degraded. (B) Under different conditions, Parkin on damaged mitochondria is not activated and the damaged mitochondria bind to LC3 B via components other than ubiquitin. (B1) Hypoxia-inducible factor 1 (HIF-1) and LC3 B serve as linkers to connect FUN14 domain containing 1 (FUNDC1) on mitochondria, contributing to the further development of mitophagy. (B2) FUNDC1 is activated to mediate mitophagy, as the Bcl2/adenovirus E1B 19 kDa interacting protein 3 (BNIP3/NIX) interacts directly with LC3. (B3) Autophagy-related 11 (ATG11) binds to Autophagy-related 32 (ATG32) on the mitochondrial membrane and then binds to LC3B. (B4) LC3B binds to BCL2-like 13 (BCL2L13) on the mitochondrial membrane and then the damaged mitochondria are degraded by lysosomes. Created with BioRender.com.
Parkin-dependent mitophagy
Our knowledge of ubiquitin-dependent mitophagy is mostly obtained through the study of the E3 ubiquitin-protein ligase Parkin and its activating kinase PTEN-induced putative kinase 1 (PINK1). Ubiquitin-dependent mitophagy largely depends on Parkin, as the chain assembly process comprises a mitochondrial damage sensor (PINK1), a signal amplifier (Parkin), and a signal effector (ubiquitin chains) (Barazzuol et al., 2020; Figure 2A). Parkin functions as an E3 ubiquitin ligase, while the protein kinase PINK1 is induced by PTEN which is expressed at low levels in cells with healthy mitochondria (Kawajiri et al., 2010; Fivenson et al., 2017). When mitochondria generate energy, they store electrochemical potential energy on the mitochondrial membrane, and the asymmetric distribution of protons and other ions on both sides of the membrane forms the mitochondrial membrane potential (Tang et al., 2019). When mitochondria are damaged, the reduction in mitochondrial membrane potential induces the accumulation of PINK1 on the mitochondrial outer membrane (MOM), and activated PINK1 phosphorylates proximal ubiquitins, resulting in the enrichment of Parkin on the MOM and the accumulation of MOM proteins (Seabright et al., 2020).
Ubiquitin participates in this process by binding to a different cellular protein, designating it for degradation, and causing the release of amino acids. The ubiquitin ligase adds multiple ubiquitin molecules to the target protein to form a clustered ubiquitin protein chain that can be absorbed by the proteasome (Roverato et al., 2021; Sun et al., 2022). Damaged mitochondria are recognized by a group of ubiquitin-binding mitophagy receptors (e.g., TBK1, OPTN, NDP52, p62, NBR1, and TAX1BP1), which results in Parkin-dependent degradation of MOM proteins (Moore and Holzbaur, 2016b; Kumar and Reichert, 2021). The interaction between ubiquitin-conjugated mitophagy receptors and the autophagosomal membrane protein LC3B promotes autophagic clearance of damaged mitochondria (Figure 2A). Parkin-dependent mitophagy provides a framework for understanding the molecular mechanisms that link ubiquitin chain synthesis to the recruitment of autophagy receptors required for mitophagy.
There are several models to explain the activation of Parkin on mitochondria (Iorio et al., 2021). Here, we introduce a positive feedback loop model to summarize the mechanisms of ubiquitin and Parkin phosphorylation (Yamada et al., 2018). At first, phosphorylated PINK1 leads to the accumulation of pSer65-Ub on mitochondria, which then increases the recruitment of unphosphorylated Parkin. A minority of ubiquitin molecules on damaged mitochondria are phosphorylated in a PINK1-dependent manner when active Parkin is present. The Parkin-pSer65-Ub interaction leads to partial activation of the ubiquitin ligase activity of Parkin. The binding of pSer65-Ub to pSer65-Parkin is much stronger than binding to unphosphorylated Parkin. Finally, the accumulation of pSer65-Parkin facilitates ubiquitin chain assembly and provides more ubiquitins for phosphorylation by PINK1, thus forming a positive feedback loop for Parkin activation (Harper et al., 2018).
In canonical mitophagy, ubiquitin-binding autophagy receptors recruit the ATG8-positive phagophore, which holds the damaged mitochondria and permits fusion with lysosomes. The canonical autophagosome assembly pathway consists of three major arms. First, the phosphatidylinositol 3-kinase catalytic subunit type 3 arm (VPS34) produces the phosphatidylinositol-3-phosphate (PtdIns3P) on donor membranes. Second, the serine/threonine protein kinase ULK1 arm initiates the formation and expansion period for phagophores. Last, the ATG8 conjugation pathway involves ATG7 (E1), ATG3 (E2), and the ATG5/ATG12-ATG16 (E3) complex. Then, the ATG8 proteins are bound to phosphatidylethanolamine (PE) on the autophagosome membrane (Lamb et al., 2013; Hurley and Schulman, 2014). Tsuboyama et al. (2016) used live-cell imaging to show retardation of autophagosomal closure and inner membrane breakdown on lysosomal fusion in the absence of the ATG8 conjugating system, demonstrating a non-canonical mitophagy pathway in which the autophagosomes form independent of ATG8-conjugation. However, this non-canonical mitophagy pathway is less efficient than the canonical mitophagy pathway.
Parkin-independent mitophagy
Other forms of mitophagy that do not require Parkin and PINK1 generally involve the recruitment of receptor molecules for LC3 family members on the MOM, which then recognize and clear unwanted mitochondria (Khaminets et al., 2016). This pathway does not activate Parkin on damaged mitochondria. The damaged mitochondria bind to LC3B, mediated by other characteristic components. Owing to the demand of oxidative phosphorylation, mitochondrial function is directly related to oxygen content. When oxygen is scarce, cells will eliminate excess mitochondria via mitophagy to avoid mitochondrial stress caused by cellular hypoxia (Martinez-Vicente, 2017; Cen et al., 2021). Here, we introduce four critical receptor pathways for Parkin-independent mitophagy.
(1) HIF-1 and LC3B serve as linkers to connect FUNDC1 on mitochondria, contributing to the further development of mitophagy (Figure 2B1). FUNDC1 interacts with both fission and fusion machinery components to regulate mitochondrial dynamics. Mitochondrial phosphatase PGAM5 dephosphorylates FUNDC1, thereby disrupting its physical connection with OPA1 and inhibiting mitochondrial fusion under hypoxic conditions (Martinez-Vicente, 2017). Conversely, FUNDC1 shifts to the ER mitochondrial contact site, mediating dynamin-related protein 1 (DRP1) recruitment and mitochondrial cleavage. Therefore, FUNDC1 coordinates mitochondrial morphology and mitochondrial autophagy under stress (Chen et al., 2016; Wu et al., 2016b). Thus, FUNDC1 coordinates mitophagy under stress. ULK1 can also phosphorylate FUNDC1 to stimulate mitophagy (Wu et al., 2014).
(2) By binding to LC3B, FUNDC1 generates autophagic membranes. The FUNDC1-LC3 interaction and mitophagy are regulated by phosphorylation or dephosphorylation of the LIR domain (Zhou et al., 2018). FUNDC1 is activated to mediate mitophagy when mitochondria are depolarized and hypoxic. NIX/BNIP3L, a MOM protein that contains the Bcl-2 homology 3 (BH3) sequence, promotes cell death and autophagy (Moore and Holzbaur, 2016a; Figure 2B2).
(3) ATG32, an autophagy-related protein, was discovered in yeast as a mitophagy receptor on the MOM (Xia et al., 2018). ATG32 binds to ATG11, an adapter protein that promotes mitophagy, and this interaction mediates substrate selectivity which is required to form perimitochondrial autophagosomes (Li and Vierstra, 2014; Figure 2B3).
(4) BCL2L13 (Bcl-2-like protein 13) is a functional mammalian homologue of ATG32 that can independently drive mitophagy. BCL2L13 has a canonical LIR domain for binding LC3B to mediate mitochondrial clearance (Otsu et al., 2015; Murakawa et al., 2019; Figure 2B4). These pathways improve mitochondrial homeostasis in vivo by providing complementary options for Parkin-dependent mitophagy.
Protein Receptor and Phospholipid that Mediate Mitophagy
Mitophagy receptors, including B-cell lymphoma 2 nineteen kilodalton interacting protein 3 (BNIP3), NIX, BCL2L13, FUND1, and Prohibitin 2 (PHB2) are mitochondrial membrane protein receptors with a common LIR motif that promotes direct interaction of the LC3/GABARAP family members with mitochondria to recruit the autophagic machinery (Martinez-Vicente, 2017).
NIX (also called BNIP3L) is an analog of BNIP3, and both belong to the Bcl-2 family. They were originally reported to affect programmed cell death and were later found to be mitophagy receptors. NIX was revealed as a mitophagy receptor during reticulocyte maturation when mitochondria are eliminated from the erythrocyte (Peng et al., 2020). BCL2L13 is the mammalian homolog of ATG32 that mediates both mitophagy and mitochondrial fragmentation (Xia et al., 2018). FUND1 is an MOM protein activated by hypoxia, and FUND1 activity is regulated by several reversible phosphorylations mediated by various kinases and phosphatases (Wu et al., 2016a). AMBRA1 is a scaffold protein that stabilizes the mTOR complex 1 and the ULK1 complex. AMBRA1 localizes to the mitochondria as a mitophagy receptor that targets LC3 with its LIR motif (Strappazzon et al., 2015). Interestingly, PHB2 is a mitochondrial inner membrane (MIM) protein that also serves as a mitophagy receptor. This MIM protein receptor connects LC3-II via its LIR motif upon mitochondrial depolarization during the erasing of paternal mitochondria (Palikaras et al., 2018).
Notably, phospholipids are a component of the mitochondrial membrane and are also critical in mediating mitophagy. Cardiolipin is a mitochondrial phospholipid present mostly in the MIM. Cardiolipin regulates the stability of many mitochondrial membrane protein complexes, such as the respiratory chain, and also modulates mitochondrial dynamics by affecting the localization of cytochrome c at the MIM (Maguire et al., 2017).
Mitophagy in Neurodegenerative Diseases
Mitochondrial homeostasis is dynamically balanced between mitochondrial biogenesis and autophagic degradation of dysfunctional or excess mitochondria (Figure 3A). Mitophagy is induced upon gene deletions, lysosomal dysfunction, reactive oxygen species, and mitochondrial permeability transition pore formation (Tanida, 2011; Li et al., 2015; Yan and Finkel, 2017). Dysfunctional mitophagy disrupts the physiological homeostasis of mitochondria and neurons (Cen et al., 2021) and results in accelerated mitochondrial clearance with insufficient mitochondrial biogenesis, thus increasing the burden on remaining organelles and promoting mitophagy-mediated neuron death (Doxaki and Palikaras, 2020; Subramaniam, 2020). There is substantial evidence that disruption of mitophagy causes neurodegenerative disease pathogenesis (Morris and Hollenbeck, 1995; Wang et al., 2010; Zaninello et al., 2020).
Figure 3.
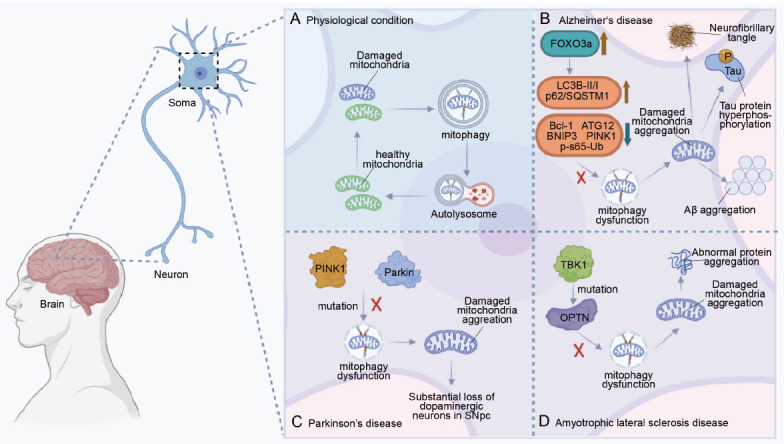
The schematic diagram of mitophagy in neurological disease pathogenesis.
Soma is depicted with mitophagy under physiological or pathological conditions: physiological condition (A), Alzheimer’s disease (B), Parkinson’s disease (C), Amyotrophic lateral sclerosis (D). (A) Under physiological conditions, neurons clear damaged mitochondria through mitophagy to maintain mitochondrial homeostasis. (B) In the neurons of Alzheimer’s patients, as the active form of FOXO3a in the cytosol increases, some mitophagy-related proteins decline, such as Bcl-1, autophagy-related 32 (ATG32), BNIP3, PINK1, and p-S65-Ub. Levels of LC3B-II/I and p62/SQSTM1 then increase, which leads to impaired mitophagy and ultimately the pathological features of Alzheimer’s, such as neurofibrillary tangles, Tau protein hyperphosphorylation, and Aβ aggregation. Brown arrows represent increases and blue arrows represent decreases. (C) In the neurons of Parkinson’s disease, mutations of Parkin and PINK1 directly lead to mitophagy dysfunction, resulting in substantial loss of dopaminergic neurons in the substantia nigra of Parkinson’s patients. (D) In the neurons of amyotrophic lateral sclerosis patients, mutations of TABK-binding kinase 1 (TBK1) affect the activity of OPTN, leading to mitophagy dysfunction. Thus, changes in OPTN cause the pathological protein aggregation. Created with BioRender.com.
AD
AD is the most common neurodegenerative disease worldwide (Royall et al., 2003). The main pathological manifestations are neurofibrillary tangles, Tau protein hyperphosphorylation, and amyloid-beta aggregation (Zhao et al., 2016; Reddy and Oliver, 2019; Figure 3B). Abnormal mitophagy prevents the proper clearance of damaged mitochondria, increases the accumulation of AD-related pathological proteins such as Aβ and hyperphosphorylated Tau, increases neuronal apoptosis, and hampers energy metabolism (Oliver and Reddy, 2019; Xie et al., 2019; Wang et al., 2020; Bell et al., 2021). Previous studies indicate that AD patients have brains with impaired mitophagy, as both mitochondrial proteins and the ratio of mitochondrial DNA/nuclear DNA change significantly (Onyango et al., 2017; Oliver and Reddy, 2019). In the hippocampus of AD patients, PINK1 increases in Braak II-III stage, while Parkin increases in Braak VI stage, and mitochondrial markers increase significantly in the early and late stages of AD (Araya et al., 2019), suggesting that mitophagy dysfunction may be related to defective initiation of PINK1/Parkin. Because of an increase in FOXO3a, other changes in proteins are reported in the brains of AD patients, such as decreased Bcl-1, ATG12, BNIP3, and p-S65-Ub, and an increased ratio of LC3-II/I to p62 (Khandelwal et al., 2011; Salminen et al., 2013; Liu et al., 2017; Von Schulze et al., 2018; Sohn et al., 2021; Yao et al., 2021; Figure 3B). Interestingly, ATG5 and Parkin are reduced in the peripheral blood of AD patients, and decreases in Parkin, PINK1, and LC3 are also observed in peripheral blood (Castellazzi et al., 2019).
Furthermore, studies with in vitro models indicate that AD is related to mitophagy dysfunction. Decreased mitophagy was found in neurons of a Caenorhabditis elegans model of AD (Shaerzadeh et al., 2014; Fang et al., 2019). In contrast, PARK2 (gene encoding Parkin) upregulation compensates for AD-related mitophagy alterations and the accumulation of ubiquitinated proteins (Martin-Maestro et al., 2016). Alzheimer’s pathology also affects mitophagy in turn; for instance, Tau accumulation impairs mitophagy by increasing mitochondrial membrane potential and reducing mitochondrial Parkin (Hu et al., 2016).
Therefore, an AD prevention strategy might consider stimulating mitophagy, and therapeutic strategies for mitophagy targets of AD include gene editing techniques, drug development, and other mitochondrial protective approaches. Lentiviral overexpression of Parkin in AD fibroblasts enhances mitochondrial function (Hong et al., 2014). In AD mouse models, treatment with Urolithin A rescues mitochondrial structure and improves cognitive function (Gimenez-Bastida et al., 2012; Cásedas et al., 2020). Studies have consistently proposed that physical exercise and caloric restriction attenuate AD pathology via the SIRT1/PINK1/Parkin signaling pathway (Anekonda, 2006; Zhou et al., 2022). However, the potency of a single mitophagy-stimulating molecule remains to be determined.
PD
PD is a chronic neurodegenerative disorder characterized by motor deficits resulting from the progressive loss of dopaminergic neurons in the substantia nigra (SN). The main pathological features of PD are tremors, increased muscle tone, bradykinesia, and postural balance disorders (Langston and Cookson, 2020). The loss of dopaminergic neurons is primarily caused by the high oxidative environment and low proteasome activity in the SN (Hashimoto et al., 2021; Figure 3C). However, the role of mitophagy in PD has gradually become clear.
There is growing evidence that mitophagy dysfunction occurs in the brains of PD patients. Mutations in Parkin and PINK1 have been found in autosomal recessive juvenile PD patients (Kitada et al., 1998; Marder et al., 2010; Lazarou et al., 2015; Figure 3C), and PD model mice have enlarged and swollen mitochondria. Similarly, mitophagy in the SN and amygdala of PD patients is compromised, eventually leading to the death of dopaminergic neurons (Lee and Trojanowski, 2006).
Parkin mutations lead to defects in mitophagy, while impaired mitophagy globally emerges in PD models (Youle and Narendra, 2011). Mitochondrial DNA loss and mitochondrial damage affect individual dopaminergic neurons in PD patients (Wager and Russell, 2013). XBP1 is a transcription factor activated by ER stress following unconventional splicing by the nuclease ERN1/IREα. The functional interaction between XBP1 and PINK1 controls mitophagy and may affect PD, and dexmedetomidine enhances PINK1/Parkin-mediated mitophagy in a neurotoxin-induced PD mouse model called 1-methyl-4-phenyl-1,2,3,6-tetrahydropyridine by activating AMPK (Chen et al., 2022). Recently, several drugs have focused on mitophagy in PD treatment. ROCK (a key regulator of mitophagy) inhibitors enhance the targeting of mitochondria to lysosomes by promoting the recruitment of HK2 (a positive regulator of Parkin) to mitochondria (Quadir et al., 2021). Artemisia leaf extract protects against neuron toxicity by promoting mitophagy clearance both in vitro and in vivo (Wu et al., 2022). Celastrol enhances mitophagy by reducing MPP (+)-induced dopaminergic neuronal death (Lin et al., 2019). Overall, molecular drugs for PD are based mainly on hub proteins for mitophagy.
ALS
ALS, also known as motor neuron disease, is characterized by abnormal protein aggregates called inclusion bodies in the cytoplasm of neurons, resulting in a vicious cycle that exacerbates oxidative damage (Figure 3D). These pathological changes ultimately lead to progressive muscle weakness and death (Kim et al., 2020). The pathogenesis of ALS is currently explained by several theories including a copper-zinc superoxide dismutase gene mutation theory, excitatory amino acid toxicity theory, autoimmune theory, and neurotrophic factor theory (Kim et al., 2020). Although there are recently available drugs and treatments for this complex multifactorial disease, the efficacy is limited. Fortunately, the targeting of mitophagy in ALS is being intensely researched.
Mitophagy dysfunction and ALS are connected. OPTN, a cause of ALS progress, plays an important role in the Parkin-mediated mitophagy pathway (Wong and Holzbaur, 2014, 2015; Zhang et al., 2015; Moore and Holzbaur, 2016b). Notably, there are about 100 different TBK1 mutations associated with OPTN (Harding et al., 2021) that eventually promote ALS-related pathology (Figure 3D). In addition to the abnormal expression of mitophagy-related proteins, inefficient turnover of damaged mitochondrial aggregates may contribute to the progression of ALS (Moore and Holzbaur, 2016a; Rogers et al., 2017). In turn, ALS-related genes interfere with the mitochondrial quality control system by disrupting the membrane potential of the mitochondrial network (Evans and Holzbaur, 2020).
Mitophagy dysfunction occurs in both patients and animal models of ALS. In an ALS mouse model with overexpressed recombinant superoxide dismutase 1, mitochondrial function and transport were disrupted (Shi et al., 2010). ALS mice have significantly increased translocator protein levels that are associated with decreased expression of ATG12 (Magri et al., 2023). In contrast, overexpression of TAR DNA binding protein-43, an ALS-associated protein, enhances mitophagy (Hong et al., 2012).
The connection between mitophagy and ALS provides a natural therapeutic target for ALS treatment. Human insulin-like growth factor-1, vitamin E, rilmenidine, and the combination of nicotinamide riboside and pterostilbene (PT), are protective of mitochondria against apoptosis and upregulate mitophagy in mouse and cell models of ALS (Perera et al., 2018; Chiricosta et al., 2019; Wen et al., 2019; Obrador et al., 2021). In addition, the mitochondria-protective drugs ketone bodies and Albrioza (sodium phenylbutyrate and taurursodiol) regulate mitophagy (Caplliure-Llopis et al., 2020). Mitochondrial protection is a valid option for novel ALS drug development.
Mitophagy as a Therapeutic Target
Mitophagy is critical to disease pathogenesis, and many pharmaceutical companies are attempting to regulate mitophagy to inhibit mitochondria-related pathologies (Doblado et al., 2021). Some pharmacological drugs and natural compounds with well-known metabolic activities have been tested in mitophagy-related studies, though most relevant studies have been limited to in vitro or preclinical trials (Aman et al., 2020).
The most important of these drugs are SIRT1 activators such as resveratrol and polydatin, which have cardioprotective effects when administered before a heart ischemic episode. Post-infarction treatment with polydatin decreased myocardial IR injury and the size of myocardial infarct in mice via mitophagy activation, which reduces mitochondrial ROS production, cell death, and inflammation (Ling et al., 2016). Melatonin also activates mitophagy by activating the Parkin/SIRT3/FOXO3a pathway in an atherogenic mouse model (Ma et al., 2018), and melatonin treatment rescues mitophagy and PD phenotypes via the PINK1/Parkin/DJ-1/MUL1 network in a zebrafish model (Diaz-Casado et al., 2016). Metformin is also considered a therapeutic strategy for neurological diseases because it activates mitophagy by promoting the expression of vital mitophagy factors, such as LC3, PINK1, Parkin, and NIX. Metformin enhances mitophagy through AMPK activation and preserves mitochondrial health in mononuclear cells of type 2 diabetics (Bhansali et al., 2020). Metformin treatment benefits Parkin-mediated mitophagy by promoting the degradation of mitofusins and shutting down the inhibitory interaction of cytosolic p53 with Parkin in an obese mouse model (Song et al., 2016). Metformin can increase Parkin expression by inhibiting NF-κB activation and accelerating mitophagy in cultured cells exposed to high glucose levels (Zhao and Sun, 2020). Moreover, in primary cortical neuron culture, PINK1 is activated by the anthelmintic drug niclosamide and its analog AM85 (Dibromsalan), leading to further activation of the PINK1/Parkin mitophagy pathway. Thus, niclosamide and AM85 may have therapeutic potential in PD (Barini et al., 2018).
We have summarized the pharmacological drugs and natural compounds that target mitophagy as a therapeutic strategy (Table 1). The repurposing of mitophagy regulators as drugs for neurological disease is promising for future clinical applications (Doblado et al., 2021).
Table 1.
Mitophagic therapeutic targets in neurodegenerative diseases
| Neurodegenerative diseases | Drugs | Targets | References |
|---|---|---|---|
| AD | Overexpressing Parkin with lentivirus | Parkin-dependent pathway | Hong et al., 2014 |
| Urolithin A | A greater response to oxidative stress | Gimenez-Bastida et al., 2012; Cásedas et al., 2020 | |
| Physical exercise and caloric restriction | SIRT1/PINK1/Parkin signaling pathway | Anekonda, 2006; Zhou et al., 2022 | |
| Overexpression of transcription factor EB | Promotes degradation of fibrillar amyloid-beta via upregulation of lysosomal biogenesis in the microglia | Tan et al., 2019 | |
| Actinonin | Restores mitochondrial damage by activating BNIP3-mediated mitophagy | Fang et al., 2019 | |
| NAD+-precursor molecules | McWilliams et al., 2016; McLelland et al., 2018 | ||
| Ubisol-Q(10) | Increases expression of autophagy-related genes Bcl-1 and JNK1 | Vegh et al., 2019 | |
| AD-hepatocyte growth factor | Markedly increases the formation of Beclin1-Vps34-ATG14L complex | Liu et al., 2016 | |
| PD | Spermidine | Induces mitophagy and prevents memory loss | Wirth et al., 2018 |
| XBP1 (a transcription factor activated by ER stress) | Functional interaction between XBP1 and PINK1 | Fleming et al., 2022 | |
| Dexmedetomidine | Parkin-dependent pathway | Chen et al., 2022 | |
| ROCK (an inhibitor of mitophagy) | Enhances the targeting of mitochondria to lysosomes | Quadir et al., 2021 | |
| Artemisia leaf extract | Promotes mitophagy clearance | Wu et al., 2022 | |
| Cilostazol | Hampers both LC3-II and p62 levels | Hedya et al., 2018 | |
| ALS | Celastrol | Reduces MPP(+)-induced dopaminergic neuronal death | Lin et al., 2019 |
| Methionine sulfoxide reductase | Reduces patients in PD platelets and mutations in M192 | Aman et al., 2020 | |
| Human insulin-like growth factor-1 | Protects mitochondria from apoptosis and upregulates mitophagy | Chiricosta et al., 2019 | |
| Vitamin E | Obrador et al., 2021 | ||
| Rilmenidine | Perera et al., 2018 | ||
| Combination of nicotinamide | Wen et al., 2019 | ||
| Riboside and pterostilbene | |||
| Ketone bodies | Regulates mitophagy, reduces mitochondrial dysfunction | Caplliure-Llopis et al., 2020 | |
| ALBRIOZA | Caplliure-Llopis et al., 2020 | ||
| TAR DNA binding protein-43 | Interacts with voltage-dependent anion channel 1, Prohibitins 2, and pivotal mitophagy receptors | Davis et al., 2018 |
AD: Alzheimer's disease; ALS: amyotrophic lateral sclerosis; Aβ: amyloid-beta; ER: endoplasmic reticulum; LC3: light chain 3; MPP:1-methyl-4-phenylpyridinium; SIRT1: silent mating type information regulation 2 homolog-1; PD: Parkinson's disease; PINK1: PTEN induced putative kinase 1; ROCK: Rho-associated protein kinase/Rho-kinase; XBP1: mouse X-box binding protein 1.
Discussion
Mitochondria are the energy source of a cell and vital to survival, and the high energy demands of neurons ensure constant mitochondrial damage. Post-mitotic neurons no longer dilute damaged organelles via cell division, so the nervous system is uniquely vulnerable and largely dependent on selective autophagy. Mitophagy is an efficient and selective pathway that responds to this stress by degrading damaged mitochondria. Thus, mitophagy is critical to the homeostasis and physiological function of neurons (Harper et al., 2018).
Nonselective autophagy also targets mitochondria for degradation, though there is no specific recognition of characteristic proteins on the outer membrane of damaged mitochondria. Therefore, nonselective autophagy degrades some healthy mitochondria during early neuronal polarity establishment and axonal branching, and inhibition of nonselective autophagy leads to early neuronal axon development in vivo and in vitro (Yang et al., 2017). Mitophagy is distinct from nonselective autophagy in that specific proteins mark damaged mitochondria for degradation via mitophagy. Mitophagy is further classified into Parkin-dependent and Parkin-independent pathways that differ in their reliance on the PINK1-Parkin-Ub system. The Parkin-dependent pathway has been the more widely studied and reported mitophagy pathway (Vives-Bauza et al., 2010; Dorn, 2016).
In this review, we hypothesize that nonselective autophagy and selective autophagy contribute to neuron physiology and pathology, respectively. Neurons are post-mitotic and rapidly developing, with great demands for nutrition and energy. Early axon development depends upon the rapid recruitment of any available material to resist forthcoming crises (Schmidt, 2019). In this case, nonselective autophagy cycles rapidly to maintain mitochondrial homeostasis (Knowlton et al., 2017), but it is crucial to remove damaged mitochondria efficiently, especially for mature neurons that constantly resist environmental stressors. Selective mitophagy stabilizes the regulation of mitochondrial biogenesis, and mitophagy dysfunction leads to the collapse of neurons and various pathological states that result in further neuronal cell death and neurological disease. Therefore, the activation of mitophagy is a promising approach to the treatment of neurological diseases.
Notably, there is very little study on PINK1/Parkin-dependent mitophagy in neuronal systems with artificial overexpression of Parkin, which raises the concern that such conditions are not physiological and might not reflect the real state of mitophagy in neurons (Grenier et al., 2013). Utilizing the methods employed with non-neuronal cells, Parkin recruitment to mitochondria is barely detectable in neuronal systems, possibly because neurons have a different bioenergetic metabolism and largely depend on oxidative phosphorylation for ATP synthesis while non-neuronal systems depend on glycolytic metabolism (Van Laar et al., 2011). Alternatively, neurons may clear dysfunctional mitochondria using different mechanisms determined by the severity of mitochondrial damage, such that PINK1/Parkin-dependent mitophagy might be activated only in response to severe mitochondrial damage (Martinez-Vicente, 2017).
Limitations
The term autophagy describes various pathways that cells apply to deliver cytoplasmic material to lysosomes, including autophagy, chaperone-mediated autophagy, and microautophagy. The subclasses of selective autophagy include mitophagy, ERphagy, aggrephagy, pexophagy, ribophagy, and lysophagy. In this review, we primarily discuss mitophagy, including basal mitophagy and stress-induced mitophagy, though programmed mitophagy is not mentioned. In addition, we only emphasize neuronal mitophagy, while astrocytes, microglia, and oligodendrocytes also play a role in mitophagy and neuronal health that is worth discussing in-depth in a future review.
Conclusion
Here, we describe the molecular mechanisms of canonical mitophagy, which is largely dependent on the PINK1/Parkin/Ub system. We focus on the connection between mitophagy and AD, PD, and ALS, and introduced therapeutic targets and molecular drugs based on mitophagy. Overall, our review provides novel molecular clues to neuroscientists and neurologists in understanding the role of mitophagy in neurological disease pathogenesis.
Acknowledgments:
We thank Drs. Yi Li from the Center for Excellence in Brain Science and Intelligence Technology, Institute of Neuroscience, and Zhengrun Gao from Songjiang Research Institute, Shanghai Jiao Tong University School of Medicine, for valuable comments.
Funding Statement
Funding: This work was supported by the National Natural Science Foundation of China, Nos. 82001211 (to KY), 82101241 (to SW), and 82125032 (to FL).
Footnotes
Conflicts of interest: The authors declare no competing interests.
Data availability statement: Not applicable.
C-Editors: Wang J, Zhao M; S-Editor: Li CH; L-Editors: Li CH, Song LP; T-Editor: Jia Y
References
- 1.Aman Y, Ryan B, Torsetnes SB, Knapskog AB, Watne LO, McEwan WA, Fang EF. Enhancing mitophagy as a therapeutic approach for neurodegenerative diseases. Int Rev Neurobiol. (2020);155:169–202. doi: 10.1016/bs.irn.2020.02.008. [DOI] [PubMed] [Google Scholar]
- 2.Anekonda TS. Resveratrol--a boon for treating Alzheimer's disease? Brain Res Rev. (2006);52:316–326. doi: 10.1016/j.brainresrev.2006.04.004. [DOI] [PubMed] [Google Scholar]
- 3.Araya J, Tsubouchi K, Sato N, Ito S, Minagawa S, Hara H, Hosaka Y, Ichikawa A, Saito N, Kadota T, Yoshida M, Fujita Y, Utsumi H, Kobayashi K, Yanagisawa H, Hashimoto M, Wakui H, Ishikawa T, Numata T, Kaneko Y, et al. PRKN-regulated mitophagy and cellular senescence during COPD pathogenesis. Autophagy. (2019);15:510–526. doi: 10.1080/15548627.2018.1532259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barazzuol L, Giamogante F, Brini M, Calì T. PINK1/Parkin mediated mitophagy. Ca2+signalling and ER-mitochondria contacts in Parkinson's disease. Int J Mol Sci. (2020);21:1772. doi: 10.3390/ijms21051772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Barini E, Miccoli A, Tinarelli F, Mulholland K, Kadri H, Khanim F, Stojanovski L, Read KD, Burness K, Blow JJ, Mehellou Y, Muqit MMK. The anthelmintic drug niclosamide and its analogues activate the Parkinson's disease associated protein kinase PINK1. Chembiochem. (2018);19:425–429. doi: 10.1002/cbic.201700500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bell SM, Barnes K, De Marco M, Shaw PJ, Ferraiuolo L, Blackburn DJ, Venneri A, Mortiboys H. Mitochondrial dysfunction in Alzheimer's disease:a biomarker of the future? Biomedicines. (2021);9:63. doi: 10.3390/biomedicines9010063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bennetzen MV, Marino G, Pultz D, Morselli E, Faergeman NJ, Kroemer G, Andersen JS. Phosphoproteomic analysis of cells treated with longevity-related autophagy inducers. Cell Cycle. (2012);11:1827–1840. doi: 10.4161/cc.20233. [DOI] [PubMed] [Google Scholar]
- 8.Bhansali S, Bhansali A, Dhawan V. Metformin promotes mitophagy in mononuclear cells:a potential in vitro model for unraveling metformin's mechanism of action. Ann N Y Acad Sci. (2020);1463:23–36. doi: 10.1111/nyas.14141. [DOI] [PubMed] [Google Scholar]
- 9.Bourdenx M, Martín-Segura A, Scrivo A, Rodriguez-Navarro JA, Kaushik S, Tasset I, Diaz A, Storm NJ, Xin Q, Juste YR, Stevenson E, Luengo E, Clement CC, Choi SJ, Krogan NJ, Mosharov EV, Santambrogio L, Grueninger F, Collin L, Swaney DL, et al. Chaperone-mediated autophagy prevents collapse of the neuronal metastable proteome. Cell. (2021);184:2696–2714.e2625. doi: 10.1016/j.cell.2021.03.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Caplliure-Llopis J, Peralta-Chamba T, Carrera-Julia S, Cuerda-Ballester M, Drehmer-Rieger E, Lopez-Rodriguez MM, de la Rubia Orti JE. Therapeutic alternative of the ketogenic mediterranean diet to improve mitochondrial activity in amyotrophic lateral sclerosis (ALS):A comprehensive review. Food Sci Nutr. (2020);8:23–35. doi: 10.1002/fsn3.1324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cásedas G, Les F, Choya-Foces C, Hugo M, López V. The metabolite urolithin-A ameliorates oxidative stress in Neuro-2a cells becoming a potential neuroprotective agent. Antioxidants (Basel) (2020);9:177. doi: 10.3390/antiox9020177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Castellazzi M, Patergnani S, Donadio M, Giorgi C, Bonora M, Bosi C, Brombo G, Pugliatti M, Seripa D, Zuliani G, Pinton P. Autophagy and mitophagy biomarkers are reduced in sera of patients with Alzheimer's disease and mild cognitive impairment. Sci Rep. (2019);9:20009. doi: 10.1038/s41598-019-56614-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cen X, Zhang M, Zhou M, Ye L, Xia H. Mitophagy regulates neurodegenerative diseases. Cells. (2021);(1876);10 doi: 10.3390/cells10081876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen C, Chen Y, Liu T, Song D, Ma D, Cheng O. Dexmedetomidine can enhance PINK1/Parkin-mediated mitophagy in MPTP-induced PD mice model by activating AMPK. Oxid Med Cell Longev. (2022);2022:7511393. doi: 10.1155/2022/7511393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen M, Chen Z, Wang Y, Tan Z, Zhu C, Li Y, Han Z, Chen L, Gao R, Liu L, Chen Q. Mitophagy receptor FUNDC1 regulates mitochondrial dynamics and mitophagy. Autophagy. (2016);12:689–702. doi: 10.1080/15548627.2016.1151580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cherubini A, Zito E. ER stress as a trigger of UPR and ER-phagy in cancer growth and spread. Front Oncol. (2022);12:997235. doi: 10.3389/fonc.2022.997235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chinta SJ, Mallajosyula JK, Rane A, Andersen JK. Mitochondrial alpha-synuclein accumulation impairs complex I function in dopaminergic neurons and results in increased mitophagy in vivo. Neurosci Lett. (2010);486:235–239. doi: 10.1016/j.neulet.2010.09.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chiricosta L, Gugliandolo A, Tardiolo G, Bramanti P, Mazzon E. Transcriptomic analysis of MAPK signaling in NSC-34 motor neurons treated with vitamin E. Nutrients. (2019);(1081);11 doi: 10.3390/nu11051081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chu CT. Mechanisms of selective autophagy and mitophagy:Implications for neurodegenerative diseases. Neurobiol Dis. (2019);122:23–34. doi: 10.1016/j.nbd.2018.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cummins N, Gotz J. Shedding light on mitophagy in neurons:what is the evidence for PINK1/Parkin mitophagy in vivo? Cell Mol Life Sci. (2018);75:1151–1162. doi: 10.1007/s00018-017-2692-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Danieli A, Martens S. p62-mediated phase separation at the intersection of the ubiquitin-proteasome system and autophagy. J Cell Sci. (2018);131:jcs214304. doi: 10.1242/jcs.214304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Davis SA, Itaman S, Khalid-Janney CM, Sherard JA, Dowell JA, Cairns NJ, Gitcho MA. TDP-43 interacts with mitochondrial proteins critical for mitophagy and mitochondrial dynamics. Neurosci Lett. (2018);678:8–15. doi: 10.1016/j.neulet.2018.04.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.De Duve C, Wattiaux R. Functions of lysosomes. Annu Rev Physiol. (1966);28:435–492. doi: 10.1146/annurev.ph.28.030166.002251. [DOI] [PubMed] [Google Scholar]
- 24.Diaz-Casado ME, Lima E, Garcia JA, Doerrier C, Aranda P, Sayed RK, Guerra-Librero A, Escames G, Lopez LC, Acuna-Castroviejo D. Melatonin rescues zebrafish embryos from the parkinsonian phenotype restoring the parkin/PINK1/DJ-1/MUL1 network. J Pineal Res. (2016);61:96–107. doi: 10.1111/jpi.12332. [DOI] [PubMed] [Google Scholar]
- 25.Doblado L, Lueck C, Rey C, Samhan-Arias AK, Prieto I, Stacchiotti A, Monsalve M. Mitophagy in human diseases. Int J Mol Sci. (2021);(3903);22 doi: 10.3390/ijms22083903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dorn GW., 2nd Parkin-dependent mitophagy in the heart. J Mol Cell Cardiol. (2016);95:42–49. doi: 10.1016/j.yjmcc.2015.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Doxaki C, Palikaras K. Neuronal mitophagy:friend or foe? Front Cell Dev Biol. (2020);8:611938. doi: 10.3389/fcell.2020.611938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Eran S, Ronit PK. APOE4 expression is associated with impaired autophagy and mitophagy in astrocytes. Neural Regen Res. (2022);17:777–778. doi: 10.4103/1673-5374.322452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Evans CS, Holzbaur ELF. Lysosomal degradation of depolarized mitochondria is rate-limiting in OPTN-dependent neuronal mitophagy. Autophagy. (2020);16:962–964. doi: 10.1080/15548627.2020.1734330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fang EF, Hou Y, Palikaras K, Adriaanse BA, Kerr JS, Yang B, Lautrup S, Hasan-Olive MM, Caponio D, Dan X, Rocktaschel P, Croteau DL, Akbari M, Greig NH, Fladby T, Nilsen H, Cader MZ, Mattson MP, Tavernarakis N, Bohr VA. Mitophagy inhibits amyloid-beta and tau pathology and reverses cognitive deficits in models of Alzheimer's disease. Nat Neurosci. (2019);22:401–412. doi: 10.1038/s41593-018-0332-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fang EF, Anisimov A. Turning up the NAD+-mitophagy axis to treat Alzheimer's disease. Neural Regen Res. (2023);18:319. doi: 10.4103/1673-5374.346472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Faruk MO, Ichimura Y, Komatsu M. Selective autophagy. Cancer Sci. (2021);112:3972–3978. doi: 10.1111/cas.15112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fernandes HJ, Hartfield EM, Christian HC, Emmanoulidou E, Zheng Y, Booth H, Bogetofte H, Lang C, Ryan BJ, Sardi SP, Badger J, Vowles J, Evetts S, Tofaris GK, Vekrellis K, Talbot K, Hu MT, James W, Cowley SA, Wade-Martins R. ER stress and autophagic perturbations lead to elevated extracellular alpha-synuclein in GBA-N370S Parkinson's iPSC-derived dopamine neurons. Stem Cell Reports. (2016);6:342–356. doi: 10.1016/j.stemcr.2016.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fivenson EM, Lautrup S, Sun N, Scheibye-Knudsen M, Stevnsner T, Nilsen H, Bohr VA, Fang EF. Mitophagy in neurodegeneration and aging. Neurochem Int. (2017);109:202–209. doi: 10.1016/j.neuint.2017.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fleming A, Bourdenx M, Fujimaki M, Karabiyik C, Krause GJ, Lopez A, Martin-Segura A, Puri C, Scrivo A, Skidmore J, Son SM, Stamatakou E, Wrobel L, Zhu Y, Cuervo AM, Rubinsztein DC. The different autophagy degradation pathways and neurodegeneration. Neuron. (2022);110:935–966. doi: 10.1016/j.neuron.2022.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fowler PC, O'Sullivan NC. ER-shaping proteins are required for ER and mitochondrial network organization in motor neurons. Hum Mol Genet. (2016);25:2827–2837. doi: 10.1093/hmg/ddw139. [DOI] [PubMed] [Google Scholar]
- 37.Galluzzi L, Kepp O, Trojel-Hansen C, Kroemer G. Mitochondrial control of cellular life stress and death. Circ Res. (2012);111:1198–1207. doi: 10.1161/CIRCRESAHA.112.268946. [DOI] [PubMed] [Google Scholar]
- 38.Garcia-Macia M, Santos-Ledo A, Caballero B, Rubio-Gonzalez A, de Luxan-Delgado B, Potes Y, Rodriguez-Gonzalez SM, Boga JA, Coto-Montes A. Selective autophagy lipophagy and mitophagy in the Harderian gland along the oestrous cycle:a potential retrieval effect of melatonin. Sci Rep. (2019);9:18597. doi: 10.1038/s41598-019-54743-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gimenez-Bastida JA, Truchado P, Larrosa M, Espin JC, Tomas-Barberan FA, Allende A, Garcia-Conesa MT. Urolithins ellagitannin metabolites produced by colon microbiota inhibit Quorum Sensing in Yersinia enterocolitica:Phenotypic response and associated molecular changes. Food Chem. (2012);132:1465–1474. doi: 10.1016/j.foodchem.2011.12.003. [DOI] [PubMed] [Google Scholar]
- 40.Grenier K, McLelland GL, Fon EA. Parkin- and PINK1-dependent mitophagy in neurons:will the real pathway please stand up? Front Neurol. (2013);4:100. doi: 10.3389/fneur.2013.00100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Grumati P, Dikic I, Stolz A. ER-phagy at a glance. J Cell Sci. (2018);131:jcs217364. doi: 10.1242/jcs.217364. [DOI] [PubMed] [Google Scholar]
- 42.Harding O, Evans CS, Ye J, Cheung J, Maniatis T, Holzbaur ELF. ALS- and FTD-associated missense mutations in TBK1 differentially disrupt mitophagy. Proc Natl Acad Sci U S A. (2021);118:e2025053118. doi: 10.1073/pnas.2025053118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Harper JW, Ordureau A, Heo JM. Building and decoding ubiquitin chains for mitophagy. Nat Rev Mol Cell Biol. (2018);19:93–108. doi: 10.1038/nrm.2017.129. [DOI] [PubMed] [Google Scholar]
- 44.Hashimoto K, Nishimura S, Sakata N, Inoue M, Sawada A, Akagi M. Characterization of PD-1/PD-L1 immune checkpoint expression in the pathogenesis of musculoskeletal Langerhans cell histiocytosis:A retrospective study. Medicine (Baltimore) (2021);100:e27650. doi: 10.1097/MD.0000000000027650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.He L, Qian X, Cui Y. Advances in ER-phagy and its diseases relevance. Cells. (2021);(2328);10 doi: 10.3390/cells10092328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hedya SA, Safar MM, Bahgat AK. Cilostazol mediated Nurr1 and autophagy enhancement:neuroprotective activity in rat rotenone PD model. Mol Neurobiol. (2018);55:7579–7587. doi: 10.1007/s12035-018-0923-1. [DOI] [PubMed] [Google Scholar]
- 47.Hernandez D, Torres CA, Setlik W, Cebrian C, Mosharov EV, Tang G, Cheng HC, Kholodilov N, Yarygina O, Burke RE, Gershon M, Sulzer D. Regulation of presynaptic neurotransmission by macroautophagy. Neuron. (2012);74:277–284. doi: 10.1016/j.neuron.2012.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hong K, Li Y, Duan W, Guo Y, Jiang H, Li W, Li C. Full-length TDP-43 and its C-terminal fragments activate mitophagy in NSC34 cell line. Neurosci Lett. (2012);530:144–149. doi: 10.1016/j.neulet.2012.10.003. [DOI] [PubMed] [Google Scholar]
- 49.Hong X, Liu J, Zhu G, Zhuang Y, Suo H, Wang P, Huang D, Xu J, Huang Y, Yu M, Bian M, Sheng Z, Fei J, Song H, Behnisch T, Huang F. Parkin overexpression ameliorates hippocampal long-term potentiation and beta-amyloid load in an Alzheimer's disease mouse model. Hum Mol Genet. (2014);23:1056–1072. doi: 10.1093/hmg/ddt501. [DOI] [PubMed] [Google Scholar]
- 50.Hu Y, Li XC, Wang ZH, Luo Y, Zhang X, Liu XP, Feng Q, Wang Q, Yue Z, Chen Z, Ye K, Wang JZ, Liu GP. Tau accumulation impairs mitophagy via increasing mitochondrial membrane potential and reducing mitochondrial Parkin. Oncotarget. (2016);7:17356–17368. doi: 10.18632/oncotarget.7861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Huang Y, Ji J, Zhao Q, Song J. Editorial:Regulation of endoplasmic reticulum and mitochondria in cellular homeostasis. Front Cell Dev Biol. (2022);10:1004376. doi: 10.3389/fcell.2022.1004376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hubner CA, Dikic I. ER-phagy and human diseases. Cell Death Differ. (2020);27:833–842. doi: 10.1038/s41418-019-0444-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hurley JH, Schulman BA. Atomistic autophagy:the structures of cellular self-digestion. Cell. (2014);157:300–311. doi: 10.1016/j.cell.2014.01.070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Iorio R, Celenza G, Petricca S. Mitophagy:molecular mechanisms new concepts on Parkin activation and the emerging role of AMPK/ULK1 axis. Cells. (2021);11:30. doi: 10.3390/cells11010030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ishikawa KI, Yamaguchi A, Okano H, Akamatsu W. Assessment of mitophagy in iPS cell-derived neurons. Methods Mol Biol. (2018);1759:59–67. doi: 10.1007/7651_2017_10. [DOI] [PubMed] [Google Scholar]
- 56.Iwata R, Casimir P, Erkol E, Boubakar L, Planque M, Gallego López IM, Ditkowska M, Gaspariunaite V, Beckers S, Remans D, Vints K, Vandekeere A, Poovathingal S, Bird M, Vlaeminck I, Creemers E, Wierda K, Corthout N, Vermeersch P, Carpentier S, et al. Mitochondria metabolism sets the species-specific tempo of neuronal development. Science. (2023);379:eabn4705. doi: 10.1126/science.abn4705. [DOI] [PubMed] [Google Scholar]
- 57.Johansen T, Lamark T. Selective autophagy:ATG8 family proteins. LIR motifs and cargo receptors. J Mol Biol. (2020);432:80–103. doi: 10.1016/j.jmb.2019.07.016. [DOI] [PubMed] [Google Scholar]
- 58.Kawajiri S, Saiki S, Sato S, Sato F, Hatano T, Eguchi H, Hattori N. PINK1 is recruited to mitochondria with parkin and associates with LC3 in mitophagy. FEBS Lett. (2010);584:1073–1079. doi: 10.1016/j.febslet.2010.02.016. [DOI] [PubMed] [Google Scholar]
- 59.Khaminets A, Behl C, Dikic I. Ubiquitin-dependent and independent signals in selective autophagy. Trends Cell Biol. (2016);26:6–16. doi: 10.1016/j.tcb.2015.08.010. [DOI] [PubMed] [Google Scholar]
- 60.Khandelwal PJ, Herman AM, Hoe HS, Rebeck GW, Moussa CE. Parkin mediates beclin-dependent autophagic clearance of defective mitochondria and ubiquitinated Abeta in AD models. Hum Mol Genet. (2011);20:2091–2102. doi: 10.1093/hmg/ddr091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kim G, Gautier O, Tassoni-Tsuchida E, Ma XR, Gitler AD. ALS genetics:gains losses and implications for future therapies. Neuron. (2020);108:822–842. doi: 10.1016/j.neuron.2020.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kitada T, Asakawa S, Hattori N, Matsumine H, Yamamura Y, Minoshima S, Yokochi M, Mizuno Y, Shimizu N. Mutations in the parkin gene cause autosomal recessive juvenile parkinsonism. Nature. (1998);392:605–608. doi: 10.1038/33416. [DOI] [PubMed] [Google Scholar]
- 63.Knowlton WM, Hubert T, Wu Z, Chisholm AD, Jin Y. A select subset of electron transport chain genes associated with optic atrophy link mitochondria to axon regeneration in Caenorhabditis elegans. Front Neurosci. (2017);11:263. doi: 10.3389/fnins.2017.00263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kulkarni A, Chen J, Maday S. Neuronal autophagy and intercellular regulation of homeostasis in the brain. Curr Opin Neurobiol. (2018);51:29–36. doi: 10.1016/j.conb.2018.02.008. [DOI] [PubMed] [Google Scholar]
- 65.Kumar MJV, Shah D, Giridharan M, Yadav N, Manjithaya R, Clement JP. Spatiotemporal analysis of soluble aggregates and autophagy markers in the R6/2 mouse model. Sci Rep. (2021);11:96. doi: 10.1038/s41598-020-78850-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kumar R, Reichert AS. Common Principles and Specific Mechanisms of Mitophagy from Yeast to Humans. Int J Mol Sci. (2021);22:4363. doi: 10.3390/ijms22094363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Lamb CA, Yoshimori T, Tooze SA. The autophagosome:origins unknown biogenesis complex. Nat Rev Mol Cell Biol. (2013);14:759–774. doi: 10.1038/nrm3696. [DOI] [PubMed] [Google Scholar]
- 68.Langston RG, Cookson MR. Pathways of protein synthesis and degradation in PD pathogenesis. Prog Brain Res. (2020);252:217–270. doi: 10.1016/bs.pbr.2020.01.002. [DOI] [PubMed] [Google Scholar]
- 69.Lazarou M, Sliter DA, Kane LA, Sarraf SA, Wang C, Burman JL, Sideris DP, Fogel AI, Youle RJ. The ubiquitin kinase PINK1 recruits autophagy receptors to induce mitophagy. Nature. (2015);524:309–314. doi: 10.1038/nature14893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Lee VM, Trojanowski JQ. Mechanisms of Parkinson's disease linked to pathological alpha-synuclein:new targets for drug discovery. Neuron. (2006);52:33–38. doi: 10.1016/j.neuron.2006.09.026. [DOI] [PubMed] [Google Scholar]
- 71.Li F, Vierstra RD. Arabidopsis ATG11 a scaffold that links the ATG1-ATG13 kinase complex to general autophagy and selective mitophagy. Autophagy. (2014);10:1466–1467. doi: 10.4161/auto.29320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Li L, Tan J, Miao Y, Lei P, Zhang Q. ROS and autophagy:interactions and molecular regulatory mechanisms. Cell Mol Neurobiol. (2015);35:615–621. doi: 10.1007/s10571-015-0166-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Lin MW, Lin CC, Chen YH, Yang HB, Hung SY. Celastrol inhibits dopaminergic neuronal death of Parkinson's disease through activating mitophagy. Antioxidants (Basel) (2019);9:37. doi: 10.3390/antiox9010037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Ling Y, Chen G, Deng Y, Tang H, Ling L, Zhou X, Song X, Yang P, Liu Y, Li Z, Zhao C, Yang Y, Wang X, Kitakaze M, Liao Y, Chen A. Polydatin post-treatment alleviates myocardial ischaemia/reperfusion injury by promoting autophagic flux. Clin Sci (Lond) (2016);130:1641–1653. doi: 10.1042/CS20160082. [DOI] [PubMed] [Google Scholar]
- 75.Liu H, Dai C, Fan Y, Guo B, Ren K, Sun T, Wang W. From autophagy to mitophagy:the roles of P62 in neurodegenerative diseases. J Bioenerg Biomembr. (2017);49:413–422. doi: 10.1007/s10863-017-9727-7. [DOI] [PubMed] [Google Scholar]
- 76.Liu J, Wu P, Wang Y, Du Y, A N, Liu S, Zhang Y, Zhou N, Xu Z, Yang Z. Ad-HGF improves the cardiac remodeling of rat following myocardial infarction by upregulating autophagy and necroptosis and inhibiting apoptosis. Am J Transl Res. (2016);8:4605–4627. [PMC free article] [PubMed] [Google Scholar]
- 77.Lu M, van Tartwijk FW, Lin JQ, Nijenhuis W, Parutto P, Fantham M, Christensen CN, Avezov E, Holt CE, Tunnacliffe A, Holcman D, Kapitein L, Schierle GSK, Kaminski CF. The structure and global distribution of the endoplasmic reticulum network are actively regulated by lysosomes. Sci Adv. (2020);6:eabc7209. doi: 10.1126/sciadv.abc7209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Ma S, Chen J, Feng J, Zhang R, Fan M, Han D, Li X, Li C, Ren J, Wang Y, Cao F. Melatonin ameliorates the progression of atherosclerosis via mitophagy activation and NLRP3 inflammasome inhibition. Oxid Med Cell Longev. (2018);2018:9286458. doi: 10.1155/2018/9286458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Ma X, Zhang W, Deng H, Zhang M, Ge L. A biochemical reconstitution approach to identify autophagy receptors for aggrephagy in mammalian cells. STAR Protoc. (2022);3:101662. doi: 10.1016/j.xpro.2022.101662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Maday S. Mechanisms of neuronal homeostasis:Autophagy in the axon. Brain Res. (2016);1649:143–150. doi: 10.1016/j.brainres.2016.03.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Magri A, Lipari CLR, Risiglione P, Zimbone S, Guarino F, Caccamo A, Messina A. ERK1/2-dependent TSPO overactivation associates with the loss of mitophagy and mitochondrial respiration in ALS. Cell Death Dis. (2023);14:122. doi: 10.1038/s41419-023-05643-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Maguire JJ, Tyurina YY, Mohammadyani D, Kapralov AA, Anthonymuthu TS, Qu F, Amoscato AA, Sparvero LJ, Tyurin VA, Planas-Iglesias J, He RR, Klein-Seetharaman J, Bayir H, Kagan VE. Known unknowns of cardiolipin signaling:The best is yet to come. Biochim Biophys Acta Mol Cell Biol Lipids. (2017);1862:8–24. doi: 10.1016/j.bbalip.2016.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Marder KS, Tang MX, Mejia-Santana H, Rosado L, Louis ED, Comella CL, Colcher A, Siderowf AD, Jennings D, Nance MA, Bressman S, Scott WK, Tanner CM, Mickel SF, Andrews HF, Waters C, Fahn S, Ross BM, Cote LJ, Frucht S, et al. Predictors of parkin mutations in early-onset Parkinson disease:the consortium on risk for early-onset Parkinson disease study. Arch Neurol. (2010);67:731–738. doi: 10.1001/archneurol.2010.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Marshall RS, Vierstra RD. Autophagy:The master of bulk and selective recycling. Annu Rev Plant Biol. (2018);69:173–208. doi: 10.1146/annurev-arplant-042817-040606. [DOI] [PubMed] [Google Scholar]
- 85.Martin-Maestro P, Gargini R, Perry G, Avila J, Garcia-Escudero V. PARK2 enhancement is able to compensate mitophagy alterations found in sporadic Alzheimer's disease. Hum Mol Genet. (2016);25:792–806. doi: 10.1093/hmg/ddv616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Martinez-Vicente M. Neuronal mitophagy in neurodegenerative diseases. Front Mol Neurosci. (2017);10:64. doi: 10.3389/fnmol.2017.00064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.McLelland GL, Goiran T, Yi W, Dorval G, Chen CX, Lauinger ND, Krahn AI, Valimehr S, Rakovic A, Rouiller I, Durcan TM, Trempe JF, Fon EA. Mfn2 ubiquitination by PINK1/parkin gates the p97-dependent release of ER from mitochondria to drive mitophagy. Elife. (2018);7:e32866. doi: 10.7554/eLife.32866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.McWilliams TG, Prescott AR, Allen GF, Tamjar J, Munson MJ, Thomson C, Muqit MM, Ganley IG. mito-QC illuminates mitophagy and mitochondrial architecture in vivo. J Cell Biol. (2016);214:333–345. doi: 10.1083/jcb.201603039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Moore AS, Holzbaur EL. Dynamic recruitment and activation of ALS-associated TBK1 with its target optineurin are required for efficient mitophagy. Proc Natl Acad Sci U S A. (2016a);113:E3349–3358. doi: 10.1073/pnas.1523810113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Moore AS, Holzbaur EL. Spatiotemporal dynamics of autophagy receptors in selective mitophagy. Autophagy. (2016b);12:1956–1957. doi: 10.1080/15548627.2016.1212788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Morris RL, Hollenbeck PJ. Axonal transport of mitochondria along microtubules and F-actin in living vertebrate neurons. J Cell Biol. (1995);131:1315–1326. doi: 10.1083/jcb.131.5.1315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Murakawa T, Okamoto K, Omiya S, Taneike M, Yamaguchi O, Otsu K. A mammalian mitophagy receptor. Bcl2-L-13 recruits the ULK1 complex to induce mitophagy. Cell Rep. (2019);26:338–345.e336. doi: 10.1016/j.celrep.2018.12.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Muscolino E, Schmitz R, Loroch S, Caragliano E, Schneider C, Rizzato M, Kim YH, Krause E, Juranic Lisnic V, Sickmann A, Reimer R, Ostermann E, Brune W. Herpesviruses induce aggregation and selective autophagy of host signalling proteins NEMO and RIPK1 as an immune-evasion mechanism. Nat Microbiol. (2020);5:331–342. doi: 10.1038/s41564-019-0624-1. [DOI] [PubMed] [Google Scholar]
- 94.Obrador E, Salvador R, Marchio P, Lopez-Blanch R, Jihad-Jebbar A, Rivera P, Valles SL, Banacloche S, Alcacer J, Colomer N, Coronado JA, Alandes S, Drehmer E, Benlloch M, Estrela JM. Nicotinamide riboside and pterostilbene cooperatively delay motor neuron failure in ALS SOD1(G93A) mice. Mol Neurobiol. (2021);58:1345–1371. doi: 10.1007/s12035-020-02188-7. [DOI] [PubMed] [Google Scholar]
- 95.Ohsumi Y. Autophagy in yeast, Saccharomyces cerevisiae. Tanpakushitsu Kakusan Koso. (1994);39:632–639. [PubMed] [Google Scholar]
- 96.Ohsumi Y. Autophagy in yeast, bulk protein degradation in the vacuole. Seikagaku. (1997);69:39–44. [PubMed] [Google Scholar]
- 97.Ohsumi Y. Molecular mechanism of autophagy in yeast, Saccharomyces cerevisiae. Philos Trans R Soc Lond B Biol Sci. (1999);354:1577–1580. doi: 10.1098/rstb.1999.0501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Ohsumi Y. Molecular dissection of autophagy:two ubiquitin-like systems. Nat Rev Mol Cell Biol. (2001);2:211–216. doi: 10.1038/35056522. [DOI] [PubMed] [Google Scholar]
- 99.Ohsumi Y. Historical landmarks of autophagy research. Cell Res. (2014);24:9–23. doi: 10.1038/cr.2013.169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Ohsumi Y, Ohsumi M, Baba M. Autophagy in yeast. Tanpakushitsu Kakusan Koso. (1993);38:46–52. [PubMed] [Google Scholar]
- 101.Okamoto K, Kondo-Okamoto N, Ohsumi Y. Mitochondria-anchored receptor Atg32 mediates degradation of mitochondria via selective autophagy. Dev Cell. (2009);17:87–97. doi: 10.1016/j.devcel.2009.06.013. [DOI] [PubMed] [Google Scholar]
- 102.Okatsu K, Kimura M, Oka T, Tanaka K, Matsuda N. Unconventional PINK1 localization to the outer membrane of depolarized mitochondria drives Parkin recruitment. J Cell Sci. (2015);128:964–978. doi: 10.1242/jcs.161000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Oliver DMA, Reddy PH. Molecular basis of Alzheimer's disease:focus on mitochondria. J Alzheimers Dis. (2019);72:S95–116. doi: 10.3233/JAD-190048. [DOI] [PubMed] [Google Scholar]
- 104.Onyango IG, Khan SM, Bennett JP., Jr Mitochondria in the pathophysiology of Alzheimer's and Parkinson's diseases. Front Biosci (Landmark Ed) (2017);22:854–872. doi: 10.2741/4521. [DOI] [PubMed] [Google Scholar]
- 105.Otsu K, Murakawa T, Yamaguchi O. BCL2L13 is a mammalian homolog of the yeast mitophagy receptor Atg32. Autophagy. (2015);11:1932–1933. doi: 10.1080/15548627.2015.1084459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Palikaras K, Lionaki E, Tavernarakis N. Balancing mitochondrial biogenesis and mitophagy to maintain energy metabolism homeostasis. Cell Death Differ. (2015);22:1399–1401. doi: 10.1038/cdd.2015.86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Palikaras K, Lionaki E, Tavernarakis N. Mechanisms of mitophagy in cellular homeostasis physiology and pathology. Nat Cell Biol. (2018);20:1013–1022. doi: 10.1038/s41556-018-0176-2. [DOI] [PubMed] [Google Scholar]
- 108.Palikaras K, Tavernarakis N. Regulation and roles of mitophagy at synapses. Mech Ageing Dev. (2020);187:111216. doi: 10.1016/j.mad.2020.111216. [DOI] [PubMed] [Google Scholar]
- 109.Peng X, Chen H, Li Y, Huang D, Huang B, Sun D. Effects of NIX-mediated mitophagy on ox-LDL-induced macrophage pyroptosis in atherosclerosis. Cell Biol Int. (2020);44:1481–1490. doi: 10.1002/cbin.11343. [DOI] [PubMed] [Google Scholar]
- 110.Perera ND, Sheean RK, Lau CL, Shin YS, Beart PM, Horne MK, Turner BJ. Rilmenidine promotes MTOR-independent autophagy in the mutant SOD1 mouse model of amyotrophic lateral sclerosis without slowing disease progression. Autophagy. (2018);14:534–551. doi: 10.1080/15548627.2017.1385674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Quadir H, Hakobyan K, Gaddam M, Ojinnaka U, Ahmed Z, Kannan A, Mostafa JA. Role of Rho-associated protein kinase inhibition as therapeutic strategy for Parkinson's disease:dopaminergic survival and enhanced mitophagy. Cureus. (2021);13:e16973. doi: 10.7759/cureus.16973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Reddy PH, Oliver DM. Amyloid beta and phosphorylated Tau-induced defective autophagy and mitophagy in Alzheimer's disease. Cells. (2019);8:488. doi: 10.3390/cells8050488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Reggiori F, Molinari M. ER-phagy:mechanisms regulation and diseases connected to the lysosomal clearance of the endoplasmic reticulum. Physiol Rev. (2022);102:1393–1448. doi: 10.1152/physrev.00038.2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Rogers RS, Tungtur S, Tanaka T, Nadeau LL, Badawi Y, Wang H, Ni HM, Ding WX, Nishimune H. Impaired mitophagy plays a role in denervation of neuromuscular junctions in ALS mice. Front Neurosci. (2017);11:473. doi: 10.3389/fnins.2017.00473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Roverato ND, Sailer C, Catone N, Aichem A, Stengel F, Groettrup M. Parkin is an E3 ligase for the ubiquitin-like modifier FAT10 which inhibits Parkin activation and mitophagy. Cell Rep. (2021);34:108857. doi: 10.1016/j.celrep.2021.108857. [DOI] [PubMed] [Google Scholar]
- 116.Royall DR, Roman GC, Delacourte A. Pathological determinants of dementia in Alzheimer's disease (AD) Exp Aging Res. (2003);29:107–110. doi: 10.1080/03610730303707. [DOI] [PubMed] [Google Scholar]
- 117.Salminen A, Kaarniranta K, Kauppinen A, Ojala J, Haapasalo A, Soininen H, Hiltunen M. Impaired autophagy and APP processing in Alzheimer's disease:The potential role of Beclin 1 interactome. Prog Neurobiol. (2013);106-107:33–54. doi: 10.1016/j.pneurobio.2013.06.002. [DOI] [PubMed] [Google Scholar]
- 118.Sarraf SA, Shah HV, Kanfer G, Pickrell AM, Holtzclaw LA, Ward ME, Youle RJ. Loss of TAX1BP1-directed autophagy results in protein aggregate accumulation in the brain. Mol Cell. (2022);82:1383–1385. doi: 10.1016/j.molcel.2022.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Schmidt RE. Mitochondriopathy:The unifying concept in distal neuropathies? Int Rev Neurobiol. (2019);145:1–12. doi: 10.1016/bs.irn.2019.05.004. [DOI] [PubMed] [Google Scholar]
- 120.Schuck S, Gallagher CM, Walter P. ER-phagy mediates selective degradation of endoplasmic reticulum independently of the core autophagy machinery. J Cell Sci. (2014);127:4078–4088. doi: 10.1242/jcs.154716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Seabright AP, Fine NHF, Barlow JP, Lord SO, Musa I, Gray A, Bryant JA, Banzhaf M, Lavery GG, Hardie DG, Hodson DJ, Philp A, Lai YC. AMPK activation induces mitophagy and promotes mitochondrial fission while activating TBK1 in a PINK1-Parkin independent manner. FASEB J. (2020);34:6284–6301. doi: 10.1096/fj.201903051R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Seah NE, de Magalhaes Filho CD, Petrashen AP, Henderson HR, Laguer J, Gonzalez J, Dillin A, Hansen M, Lapierre LR. Autophagy-mediated longevity is modulated by lipoprotein biogenesis. Autophagy. (2016);12:261–272. doi: 10.1080/15548627.2015.1127464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Shaerzadeh F, Motamedi F, Minai-Tehrani D, Khodagholi F. Monitoring of neuronal loss in the hippocampus of Abeta-injected rat:autophagy mitophagy and mitochondrial biogenesis stand against apoptosis. Neuromolecular Med. (2014);16:175–190. doi: 10.1007/s12017-013-8272-8. [DOI] [PubMed] [Google Scholar]
- 124.Shi P, Wei Y, Zhang J, Gal J, Zhu H. Mitochondrial dysfunction is a converging point of multiple pathological pathways in amyotrophic lateral sclerosis. J Alzheimers Dis. (2010);2(20 Suppl):S311–324. doi: 10.3233/JAD-2010-100366. [DOI] [PubMed] [Google Scholar]
- 125.Sohn HY, Kim SI, Park JY, Park SH, Koh YH, Kim J, Jo C. ApoE4 attenuates autophagy via FoxO3a repression in the brain. Sci Rep. (2021);11:17604. doi: 10.1038/s41598-021-97117-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Song S, Tan J, Miao Y, Zhang Q. Crosstalk of ER stress-mediated autophagy and ER-phagy:Involvement of UPR and the core autophagy machinery. J Cell Physiol. (2018);233:3867–3874. doi: 10.1002/jcp.26137. [DOI] [PubMed] [Google Scholar]
- 127.Song YM, Lee WK, Lee YH, Kang ES, Cha BS, Lee BW. Metformin restores Parkin-mediated mitophagy, suppressed by cytosolic p53. Int J Mol Sci. (2016);17:122. doi: 10.3390/ijms17010122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Stavoe AKH, Holzbaur ELF. Autophagy in neurons. Annu Rev Cell Dev Biol. (2019);35:477–500. doi: 10.1146/annurev-cellbio-100818-125242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Strappazzon F, Nazio F, Corrado M, Cianfanelli V, Romagnoli A, Fimia GM, Campello S, Nardacci R, Piacentini M, Campanella M, Cecconi F. AMBRA1 is able to induce mitophagy via LC3 binding, regardless of PARKIN and p62/SQSTM1. Cell Death Differ. (2015);22:517. doi: 10.1038/cdd.2014.190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Subramaniam S. Exaggerated mitophagy:a weapon of striatal destruction in the brain? Biochem Soc Trans. (2020);48:709–717. doi: 10.1042/BST20191283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Sun S, Hou H, Ma G, Ma Q, Li N, Zhang L, Dong C, Cao M, Tam KY, Ying Z, Wang H. The interaction between E3 ubiquitin ligase Parkin and mitophagy receptor PHB2 links inner mitochondrial membrane ubiquitination to efficient mitophagy. J Biol Chem. (2022);298:102704. doi: 10.1016/j.jbc.2022.102704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Suresh SN, Chavalmane AK, Pillai M, Ammanathan V, Vidyadhara DJ, Yarreiphang H, Rai S, Paul A, Clement JP, Alladi PA, Manjithaya R. Modulation of autophagy by a small molecule inverse agonist of ERRalpha is neuroprotective. Front Mol Neurosci. (2018);11:109. doi: 10.3389/fnmol.2018.00109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Talifu Z, Liu JY, Pan YZ, Ke H, Zhang CJ, Xu X, Gao F, Yu Y, Du LJ, Li JJ. In vivo astrocyte-to-neuron reprogramming for central nervous system regeneration:a narrative review. Neural Regen Res. (2023);18:750–755. doi: 10.4103/1673-5374.353482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Tan S, Yu CY, Sim ZW, Low ZS, Lee B, See F, Min N, Gautam A, Chu JJH, Ng KW, Wong E. Pomegranate activates TFEB to promote autophagy-lysosomal fitness and mitophagy. Sci Rep. (2019);9:727. doi: 10.1038/s41598-018-37400-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Tang C, Han H, Liu Z, Liu Y, Yin L, Cai J, He L, Liu Y, Chen G, Zhang Z, Yin XM, Dong Z. Activation of BNIP3-mediated mitophagy protects against renal ischemia-reperfusion injury. Cell Death Dis. (2019);10:677. doi: 10.1038/s41419-019-1899-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Tanida I. Autophagosome formation and molecular mechanism of autophagy. Antioxid Redox Signal. (2011);14:2201–2214. doi: 10.1089/ars.2010.3482. [DOI] [PubMed] [Google Scholar]
- 137.Tedesco B, Ferrari V, Cozzi M, Chierichetti M, Casarotto E, Pramaggiore P, Mina F, Piccolella M, Cristofani R, Crippa V, Rusmini P, Galbiati M, Poletti A. The role of autophagy-lysosomal pathway in motor neuron diseases. Biochem Soc Trans. (2022);50:1489–1503. doi: 10.1042/BST20220778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Tsuboyama K, Koyama-Honda I, Sakamaki Y, Koike M, Morishita H, Mizushima N. The ATG conjugation systems are important for degradation of the inner autophagosomal membrane. Science. (2016);354:1036–1041. doi: 10.1126/science.aaf6136. [DOI] [PubMed] [Google Scholar]
- 139.Van Laar VS, Arnold B, Cassady SJ, Chu CT, Burton EA, Berman SB. Bioenergetics of neurons inhibit the translocation response of Parkin following rapid mitochondrial depolarization. Hum Mol Genet. (2011);20:927–940. doi: 10.1093/hmg/ddq531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Vegh C, Pupulin S, Wear D, Culmone L, Huggard R, Ma D, Pandey S. Resumption of autophagy by Ubisol-Q(10) in Presenilin-1 mutated fibroblasts and transgenic AD mice:implications for inhibition of senescence and neuroprotection. Oxid Med Cell Longev. (2019);2019:7404815. doi: 10.1155/2019/7404815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Veljanovski V, Batoko H. Selective autophagy of non-ubiquitylated targets in plants:looking for cognate receptor/adaptor proteins. Front Plant Sci. (2014);5:308. doi: 10.3389/fpls.2014.00308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Vives-Bauza C, Zhou C, Huang Y, Cui M, de Vries RL, Kim J, May J, Tocilescu MA, Liu W, Ko HS, Magrane J, Moore DJ, Dawson VL, Grailhe R, Dawson TM, Li C, Tieu K, Przedborski S. PINK1-dependent recruitment of Parkin to mitochondria in mitophagy. Proc Natl Acad Sci U S A. (2010);107:378–383. doi: 10.1073/pnas.0911187107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Von Schulze A, McCoin CS, Onyekere C, Allen J, Geiger P, Dorn GW, 2nd, Morris EM, Thyfault JP. Hepatic mitochondrial adaptations to physical activity:impact of sexual dimorphism. PGC1alpha and BNIP3-mediated mitophagy. J Physiol. (2018);596:6157–6171. doi: 10.1113/JP276539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Wager K, Russell C. Mitophagy and neurodegeneration:the zebrafish model system. Autophagy. (2013);9:1693–1709. doi: 10.4161/auto.25082. [DOI] [PubMed] [Google Scholar]
- 145.Wallace DC, Brown MD, Melov S, Graham B, Lott M. Mitochondrial biology, degenerative diseases and aging. Biofactors. (1998);7:187–190. doi: 10.1002/biof.5520070303. [DOI] [PubMed] [Google Scholar]
- 146.Wallace DC. Mitochondrial diseases in man and mouse. Science. (1999);283:1482–1488. doi: 10.1126/science.283.5407.1482. [DOI] [PubMed] [Google Scholar]
- 147.Wang W, Zhao F, Ma X, Perry G, Zhu X. Mitochondria dysfunction in the pathogenesis of Alzheimer's disease:recent advances. Mol Neurodegener. (2020);15:30. doi: 10.1186/s13024-020-00376-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Wang X, Perry G, Smith MA, Zhu X. Amyloid-beta-derived diffusible ligands cause impaired axonal transport of mitochondria in neurons. Neurodegener Dis. (2010);7:56–59. doi: 10.1159/000283484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Watanabe Y, Tsujimura A, Taguchi K, Tanaka M. HSF1 stress response pathway regulates autophagy receptor SQSTM1/p62-associated proteostasis. Autophagy. (2017);13:133–148. doi: 10.1080/15548627.2016.1248018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Wei F, Yang A, Zhao Z, An H, Li Y, Duan Y. Mechanism of ER stress-mediated ER-phagy by CdTe-QDs in yeast cells. Toxicol Lett. (2022);365:36–45. doi: 10.1016/j.toxlet.2022.05.010. [DOI] [PubMed] [Google Scholar]
- 151.Wen D, Cui C, Duan W, Wang W, Wang Y, Liu Y, Li Z, Li C. The role of insulin-like growth factor 1 in ALS cell and mouse models:A mitochondrial protector. Brain Res Bull. (2019);144:1–13. doi: 10.1016/j.brainresbull.2018.09.015. [DOI] [PubMed] [Google Scholar]
- 152.Wetzel L, Blanchard S, Rama S, Beier V, Kaufmann A, Wollert T. TECPR1 promotes aggrephagy by direct recruitment of LC3C autophagosomes to lysosomes. Nat Commun. (2020);(2993);11 doi: 10.1038/s41467-020-16689-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Wirth M, Benson G, Schwarz C, Kobe T, Grittner U, Schmitz D, Sigrist SJ, Bohlken J, Stekovic S, Madeo F, Floel A. The effect of spermidine on memory performance in older adults at risk for dementia:A randomized controlled trial. Cortex. (2018);109:181–188. doi: 10.1016/j.cortex.2018.09.014. [DOI] [PubMed] [Google Scholar]
- 154.Wong YC, Holzbaur EL. Optineurin is an autophagy receptor for damaged mitochondria in parkin-mediated mitophagy that is disrupted by an ALS-linked mutation. Proc Natl Acad Sci U S A. (2014);111:E4439–4448. doi: 10.1073/pnas.1405752111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Wong YC, Holzbaur EL. Temporal dynamics of PARK2/parkin and OPTN/optineurin recruitment during the mitophagy of damaged mitochondria. Autophagy. (2015);11:422–424. doi: 10.1080/15548627.2015.1009792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Wu H, Wei H, Sehgal SA, Liu L, Chen Q. Mitophagy receptors sense stress signals and couple mitochondrial dynamic machinery for mitochondrial quality control. Free Radic Biol Med. (2016a);100:199–209. doi: 10.1016/j.freeradbiomed.2016.03.030. [DOI] [PubMed] [Google Scholar]
- 157.Wu LK, Agarwal S, Kuo CH, Kung YL, Day CH, Lin PY, Lin SZ, Hsieh DJ, Huang CY, Chiang CY. Artemisia Leaf Extract protects against neuron toxicity by TRPML1 activation and promoting autophagy/mitophagy clearance in both in vitro and in vivo models of MPP+/MPTP-induced Parkinson's disease. Phytomedicine. (2022);104:154250. doi: 10.1016/j.phymed.2022.154250. [DOI] [PubMed] [Google Scholar]
- 158.Wu NN, Zhang Y, Ren J. Mitophagy mitochondrial dynamics and homeostasis in cardiovascular aging. Oxid Med Cell Longev. (2019);2019:9825061. doi: 10.1155/2019/9825061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Wu W, Tian W, Hu Z, Chen G, Huang L, Li W, Zhang X, Xue P, Zhou C, Liu L, Zhu Y, Zhang X, Li L, Zhang L, Sui S, Zhao B, Feng D. ULK1 translocates to mitochondria and phosphorylates FUNDC1 to regulate mitophagy. EMBO Rep. (2014);15:566–575. doi: 10.1002/embr.201438501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Wu W, Lin C, Wu K, Jiang L, Wang X, Li W, Zhuang H, Zhang X, Chen H, Li S, Yang Y, Lu Y, Wang J, Zhu R, Zhang L, Sui S, Tan N, Zhao B, Zhang J, Li L, et al. FUNDC1 regulates mitochondrial dynamics at the ER-mitochondrial contact site under hypoxic conditions. EMBO J. (2016b);35:1368–1384. doi: 10.15252/embj.201593102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Xia X, Katzenell S, Reinhart EF, Bauer KM, Pellegrini M, Ragusa MJ. A pseudo-receiver domain in Atg32 is required for mitophagy. Autophagy. (2018);14:1620–1628. doi: 10.1080/15548627.2018.1472838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Xie C, Aman Y, Adriaanse BA, Cader MZ, Plun-Favreau H, Xiao J, Fang EF. Culprit or bystander:defective mitophagy in Alzheimer's disease. Front Cell Dev Biol. (2019);7:391. doi: 10.3389/fcell.2019.00391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Yamada T, Murata D, Adachi Y, Itoh K, Kameoka S, Igarashi A, Kato T, Araki Y, Huganir RL, Dawson TM, Yanagawa T, Okamoto K, Iijima M, Sesaki H. Mitochondrial stasis reveals p62-mediated ubiquitination in parkin-independent mitophagy and mitigates nonalcoholic fatty liver disease. Cell Metab. (2018);28:588–604.e585. doi: 10.1016/j.cmet.2018.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Yan Y, Finkel T. Autophagy as a regulator of cardiovascular redox homeostasis. Free Radic Biol Med. (2017);109:108–113. doi: 10.1016/j.freeradbiomed.2016.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Yang K, Yu B, Cheng C, Cheng T, Yuan B, Li K, Xiao J, Qiu Z, Zhou Y. Mir505-3p regulates axonal development via inhibiting the autophagy pathway by targeting Atg12. Autophagy. (2017);13:1679–1696. doi: 10.1080/15548627.2017.1353841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Yao RQ, Ren C, Xia ZF, Yao YM. Organelle-specific autophagy in inflammatory diseases:a potential therapeutic target underlying the quality control of multiple organelles. Autophagy. (2021);17:385–401. doi: 10.1080/15548627.2020.1725377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Youle RJ, Narendra DP. Mechanisms of mitophagy. Nat Rev Mol Cell Biol. (2011);12:9–14. doi: 10.1038/nrm3028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Zaninello M, Palikaras K, Naon D, Iwata K, Herkenne S, Quintana-Cabrera R, Semenzato M, Grespi F, Ross-Cisneros FN, Carelli V, Sadun AA, Tavernarakis N, Scorrano L. Inhibition of autophagy curtails visual loss in a model of autosomal dominant optic atrophy. Nat Commun. (2020);(4029);11 doi: 10.1038/s41467-020-17821-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Zhang CW, Hang L, Yao TP, Lim KL. Parkin regulation and neurodegenerative disorders. Front Aging Neurosci. (2015);7:248. doi: 10.3389/fnagi.2015.00248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Zhao D, Zou CX, Liu XM, Jiang ZD, Yu ZQ, Suo F, Du TY, Dong MQ, He W, Du LL. A UPR-induced soluble ER-Phagy receptor acts with VAPs to confer ER stress resistance. Mol Cell. (2020);79:963–977.e963. doi: 10.1016/j.molcel.2020.07.019. [DOI] [PubMed] [Google Scholar]
- 171.Zhao S, Zhao J, Zhang T, Guo C. Increased apoptosis in the platelets of patients with Alzheimer's disease and amnestic mild cognitive impairment. Clin Neurol Neurosurg. (2016);143:46–50. doi: 10.1016/j.clineuro.2016.02.015. [DOI] [PubMed] [Google Scholar]
- 172.Zhao Y, Sun M. Metformin rescues Parkin protein expression and mitophagy in high glucose-challenged human renal epithelial cells by inhibiting NF-kappaB via PP2A activation. Life Sci. (2020);246:117382. doi: 10.1016/j.lfs.2020.117382. [DOI] [PubMed] [Google Scholar]
- 173.Zhou H, Zhu P, Wang J, Zhu H, Ren J, Chen Y. Pathogenesis of cardiac ischemia reperfusion injury is associated with CK2alpha-disturbed mitochondrial homeostasis via suppression of FUNDC1-related mitophagy. Cell Death Differ. (2018);25:1080–1093. doi: 10.1038/s41418-018-0086-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Zhou S, Liu L, Zhang Y, Zhang Z, Li H, Fan F, He J, Kang J, Zuo L. Integrated untargeted and targeted metabolomics to reveal therapeutic effect and mechanism of Alpiniae oxyphyllae fructus on Alzheimer's disease in APP/PS1 mice. Front Pharmacol. (2022);13:1104954. doi: 10.3389/fphar.2022.1104954. [DOI] [PMC free article] [PubMed] [Google Scholar]


