Summary
Background
The pathology of keloid and especially the roles of bacteria on it were not well understood.
Methods
In this study, multi-omics analyses including microbiome, metaproteomics, metabolomic, single-cell transcriptome and cell-derived xenograft (CDX) mice model were used to explore the roles of bacteria on keloid disease.
Findings
We found that the types of bacteria are significantly different between keloid and healthy skin. The 16S rRNA sequencing and metaproteomics showed that more catalase (CAT) negative bacteria, Clostridium and Roseburia existed in keloid compared with the adjacent healthy skin. In addition, protein mass spectrometry shows that CAT is one of the differentially expressed proteins (DEPs). Overexpression of CAT inhibited the proliferation, migration and invasion of keloid fibroblasts, and these characteristics were opposite when CAT was knocked down. Furthermore, the CDX model showed that Clostridium butyricum promote the growth of patient's keloid fibroblasts in BALB/c female nude mice, while CAT positive bacteria Bacillus subtilis inhibited it. Single-cell RNA sequencing verified that oxidative stress was up-regulated and CAT was down-regulated in mesenchymal-like fibroblasts of keloid.
Interpretation
In conclusion, our findings suggest that bacteria and CAT contribute to keloid disease.
Funding
A full list of funding bodies that contributed to this study can be found in the Acknowledgements section.
Keywords: Keloid, Bacteria, Catalase, 16S rRNA, Metaproteomics
Research in context.
Evidence before this study
Previous studies have shown that bacterial diversity are associated with many skin diseases, such as seborrheic dermatitis, acne and psoriasis. The microenvironment changes and inflammation after bacteria infection have been verified to play important roles in these skin diseases. However, the roles of bacteria in keloid have not been reported so far.
Added value of this study
Keloid is a benign skin tumor that occurs as result of excessive scar formation. The global incidence rate of keloid increased significantly in the past decade. Currently, there is no effective treatment for keloid and the recurrence rate after surgery is extremely high. Bacteria may play important roles on the formation and growth of keloid. Therefore, characterizing the specific roles of bacteria in keloid may identify novel targets helpful for the new treatment strategies.
Implications of all the available evidence
This study aims to explore the function of bacteria in keloid with multi-omics analyses including microbiome, metaproteomics, metabolomic and single-cell transcriptome. We found that the types of bacteria are significantly different between keloid and healthy skin. More CAT-negative bacteria existed in keloid compared with the adjacent healthy skin. The expression level of CAT in fibroblasts was related with keloid disease. Furthermore, Clostridium butyricum can directly promote the formation and growth of keloid fibroblasts both in vitro and in vivo.
Introduction
Keloid is a benign skin tumor characterized by massive deposition of collagen fibers in the dermis and is often caused by inflammation after injury or infection, accompanied by symptoms such as pain and itching, which seriously affects the physical and mental health and life quality of patients.1,2 Its treatments remain a major challenge facing dermatologists and plastic surgeons due to the lack of effective drugs and the high recurrence rate after surgical excision or corticoid injection.3 The mechanisms underlying the disease are also unclear.
Keloids are developed from folliculitis sometimes.2,4,5 Histologically, the lesion area initially comprises follicular red papules, and the lesions gradually harden and merge into larger keloid-like indurations.2,4,5 Keloids often result from damage to the superficial reticular dermis layer followed by abnormal wound healing and persistent, localized inflammation.2 The microenvironment changes and inflammation after microorganism infection have been suspected to play an important role in keloid disease. Previous studies have shown that microorganisms are associated with inflammation in many skin diseases, such as seborrheic dermatitis, acne and psoriasis.6 However, to our knowledge, no study has reported the correlation between microorganisms and keloid. Thus, characterizing the roles of microorganisms in keloid may identify new targets for the treatment and prevention of keloid.
Previously, inflammatory cells, increased numbers of fibroblasts, newly formed blood vessels, and collagen deposits were reported in keloid tissue.7 Fibroblasts can ultimately lead to keloid formation by excessive extracellular matrix (ECM) deposition, a process driven by many growth factors, such as transforming growth factor-β (TGF-β), platelet-derived growth factor (PDGF), insulin-like growth factor, collagen Ⅰ and III, vimentin, chemokine receptor 7 (CCR7), and interleukin-4 (IL-4).8,9 Increased levels of reactive oxygen species (ROS) were also reported in keloid fibroblasts, which contribute to the inflammation and oxidative stress response of the disease.10 Catalase (CAT) is an important antioxidant enzyme in aerobic metabolism in cells and microorganisms. It plays an important role in removing ROS and maintaining the balance of the redox state.11 Some previous studies have found that some types of bacteria produce CAT, while others do not.12,13 In this study, we used multiomics to systemically explore specific typies of bacteria involved in keloid disease and used CDX model to study how CAT regulate the growth of keloid.
Methods
Clinical samples
Patients
This study was approved by the Medical Ethics Committee of Peking Union Medical College Hospital (JS-2907). All participants provided written informed consent. From August 2018 to March 2022, 132 patients diagnosed with keloid and 115 patients with cosmetic surgery were included in this study (Table 1 and Supplementary Table 1). The severity of keloids was assessed using the Modified Vancouver Scar Scale (mVSS).14 Keloid tissue was collected from the hypertrophic part of the mass, and normal skin was collected from the removed skin during operation 2 and 3 mm from the edge of the mass. Normal skin tissue from patients who underwent cosmetic surgery was collected from the removed skin during surgery, which included macromastia reduction, rhytidectomy, blepharoplasty, etc. More details of the samples for different experiments were shown in Supplementary Table 1. The incidence rate of keloid is higher in women than in men (Supplementary Table 2). To make the analysis more comprehensive, we kept the ratio of women to men no more than 2:1 when included the samples.
Table 1.
Characteristics of patients (n = 247).
| Characteristics | Patients with keloid | Cosmetic patients | P value | |
|---|---|---|---|---|
| Sex | 0.486a | |||
| Male | 91 | 46 (18.6%) | 45 (18.2%) | |
| Female | 156 | 86 (34.8%) | 70 (28.3%) | |
| Age | ||||
| 18–29 | 94 | 65 (26.3%) | 29 (11.7%) | <0.05a |
| 30–49 | 121 | 41 (16.6%) | 80 (32.4%) | |
| >50 | 32 | 26 (10.5%) | 6 (2.4%) | |
| Pathogenic site | <0.05b | |||
| Forebreast | 187 | 114 (46.2%) | 73 (29.6%) | |
| Ear | 5 | 5 (2%) | / | |
| Hypogastrium | 9 | 9 (3.6%) | / | |
| Perineum | 1 | 1 (0.4%) | / | |
| Head and neck | 44 | 2 (0.8%) | 42 (17%) | |
| Upper limb | 1 | 1 (0.4%) | / |
Pearman test was used to calculate P value.
Fisher's test was used to calculate P value.
Inclusion and exclusion criteria
The inclusion criteria were as follows: patients diagnosed with keloid who needed surgical treatment; patients undergoing cosmetic surgery but without keloid; patients aged over 18 years; and patients who signed an informed consent form. The exclusion criteria included patients with severe systemic disease, patients younger than 18 years old, and patients with mental disorders.
Multi-omics analysis
Microbiome
Bacteria 16S rRNA gene sequencing
Thirty-nine samples (20 keloid tissues and 19 normal skin tissues) were collected during the operation. The skins were in direct contact with the complex biological environment. To exclude the influence of microbes from the external environment, the epidermal layer was removed from the dermal layer of the skin. To prevent cross contamination, three 1-cm3 pieces of tissue from each sample were collected under sterile condition and prepared for sequencing. Total genomic DNA from the samples was extracted using the cetyl trimethyl ammonium bromide (CTAB) extraction protocol. The 16S rRNA gene of distinct regions (16S V3-V4) was amplified using specific primers: 341F (5′-CCTAYGGGRBGCASCAG-3′) and 806R (5′-GGACTACNNGGGTATCTAAT-3′).15,16 Sequencing libraries were generated using the TruSeq® DNA PCR-Free Sample Preparation Kit (Illumina, USA) following the manufacturer's instruction. The library quality was assessed on the Qubit@ 2.0 Fluorometer (Thermo Scientific). Finally, the library was sequenced on an Illumina NovaSeq platform and 250 bp paired-end reads were generated.17
Operational taxonomic units (OTUs) clustering and species annotation of amplicon sequencing
UPARSE software was used to cluster all clean reads of all samples. The sequences were clustered into OTUs with 97% identity, and representative sequences of OTUs were selected simultaneously. According to its algorithm principles, the sequence with the highest frequency in OTUs was selected as the representative sequence of OTUs. Species annotation was performed on OTU sequences, and species annotation analysis was performed. Taxonomic information was obtained at each taxonomic level, including the kingdom, phylum, class, order, family, genus, and species statistics of the community composition of each sample. A rapid multiple sequence alignment was performed using MUSCLE software to obtain the phylogenetic relationship of all OTU sequences. Finally, the data of each sample were normalized, and the sample with the least amount of data was used as the standard for normalization. The subsequent alpha diversity analysis and beta diversity analysis were based on the homogenized data.
Analysis of alpha and beta diversity of amplicon sequencing
The alpha diversity dilution curve (rarefaction curve) randomly selects a certain amount of data from the sample, counts the number of species or diversity indices they represent, and constructs a curve based on the amount of data and species diversity to illustrate whether the amount of sequencing data of samples is reasonable and indirectly reflects the abundance of species in the sample. The analysis of alpha diversity and beta diversity was mainly performed with the QIIME2 diversity plugin. The alpha diversity index is an analysis of species diversity in a sample, including two factors: the richness and evenness of the species composition in the sample. The Shannon index is usually used to evaluate the species diversity of a sample. This index shows that the diversity of the samples is more complex.
Phylogenetic tree analysis of amplicon sequencing
The evolutionary tree was drawn by the R language ggtree package. The representative sequences of OTUs of interest were selected for phylogenetic analysis (the top 30 OTUs in total abundance) to draw an evolutionary relationship tree diagram, and a heatmap was drawn in combination with the absolute abundance of OTUs in each group.
Metabolomics analysis
The metabolomic profiles of 20 keloid tissues and 20 normal skin tissues were analyzed. One hundred milligrams of the sample was weighed and and 1 mL of tissue extraction buffer (75% 9:1 methanol: chloroform, 25% H2O) was added. The samples were homogenized in a high-throughput tissue grinder by ultrasonication and this process was repeated twice. The samples were subsequently concentrated with a vacuum concentrator and dissolved in 200 μL of 2-chlorophenylalanine solution containing 50% acetonitrile. After filtration with 0.22 μm membrane, a total of 20 μL of each sample to be tested was mixed into a QC sample and analyzed by LC–MS with Thermo Q Exactive Plus and Thermo Ultimate 3000.
Metaproteomics
Eighteen keloid tissue and twelve normal skin tissue samples were analyzed for metaproteomics. 50 mg of tissue were rinsed first and then transferred to a magnetic bead-containing tubes with 500 μL of 2% SDS in 20 mM Tris pH added. The tissues were homogenized with a crusher, centrifuged at 14,000 × g at 4 °C for 10 min, and the protein-containing supernatant was subjected to SDS-PAGE followed by trypsin digestion. The digested protein was finally precipitated with acetone and analyzed by Orbitrap Exploris 480 mass spectrometer (Thermo Scientific).
Single-cell sequencing
Fifty to one hundred milligrams of tissue was minced and dissociated with 2 mL of GEXSCOPE™ tissue dissociation solution (Singleron, Nanjing, China) at 37 °C for 15 min. After dissociation into a single cell suspension, the number of cells was counted using trypan blue staining. The cell concentration was adjusted to 1 × 105 cells/mL; then cells were added to the microfluidic chip and sequenced using the Illumina HiSeq X sequencing platform. The sequencing mode was 150 bp paired-end sequencing. Cell type identification and cluster analysis were performed on RNA-seq data using the Seurat program (http://satijalab.org/seurat/, R package, v.3.0.1).
Experimental verification
Keloid primary fibroblasts culture
The subcutaneous fat and epidermis were removed after repeated rinsing 5–6 times in PBS buffer containing 100 U/ml penicillin and 100 U/ml streptomycin. The skin sample was then cut into small pieces with ophthalmic scissors in a Petri dish and digested with 0.2% type II collagenase at 37 °C in a water bath shaker for 45 min. The digestion was repeated for a second time for 20 min. The cells were then pooled from the first and second digestion, centrifuged and counted. A total of 105 cells/ml were seeded in petri dishes and cultured with DMEM containing 10% FBS, 100 U/ml penicillin and 100 U/ml streptomycin for 3 passages. After the cells grew into dense monolayers, the cells were digested with 0.25% trypsin and the detached cells were collected to obtain purified fibroblasts.
CAT knockdown and overexpression in keloid fibroblasts
CAT knockdown (KD-CAT) constructs were established by Shanghai Jikein Medical Technology Company. A lentivirus vector (hU6-MCS-CBh-gcGFP-IRES-puromycin, Item No.: GV493) was used for the gene transfer. The target sequence for CAT knockdown was CGGAGATTCAACACTGCCAAT. The target sequence for negative control (KD-Control) was TTCTCCGAACGTGTCACGT. CAT overexpression (OV-CAT) and control (OV-Control) constructs were also established by Shanghai Jikein Medical Technology Company. The gene transfer vector was Ubi-MCS-3FLAG-CBh-gcGFP-IRES-puromycin (Item No.: GV492). Full-length human CAT gene sequence (Sequence IDs included in CCDS 7891.1, NCBI) was inserted in the vector and used for overexpression, while control transfer with the empty vector (GV492).
Fibroblasts of logarithmic growth stage were inoculated into 12-well plates with 5000 cells per well. The cells were divided into four groups: KD-CAT, KD-Control, OV-CAT, OV-Control. When the cells were 70% full, 12 μL of lentivirus (titer: 2E+9TU/mL) was added. After 12H, the cell status was observed and if there was no obvious cytotoxic effect, the culture medium was changed after 24H. If there is obvious cytotoxic effect, replace the medium immediately. Puromycin (2 μg/mL) was added to screen out successful infected cells for subsequent experiments.
Catalase activity detection
The catalase activity in cells, tissue or other samples was analyzed by colorimetric detection with Catalase Assay Kit (Beyotime, S0051, Shanghai, China) using a UV spectrophotometer at A240. The concentration calculation formula was c = A/(ε × b). In this formula, c is the sample concentration (unit is mol/L or M); A is the absorbance value; ε is the wavelength-dependent molar extinction coefficient (unit is L × mol−1 × cm−1 or M−1 × cm−1), the molar extinction coefficient of hydrogen peroxide is 43.6 M−1cm−1; and b is the light path (unit is cm). Therefore, the hydrogen peroxide concentration (M) = A240/(43.6 × b).
H2O2 and ROS detection
A hydrogen peroxide detection kit (Beyotime, S0038, Shanghai, China) was used for the quantitative detection of H2O2. One hundred microliters of hydrogen peroxide detection reagent was added to 50 μL of sample or standard. The mixture was gently mixed and incubated at room temperature for 30 min before absorbance at 560 nm was determined. The concentration of H2O2 in the sample was calculated from a standard curve. Amount of ROS was determined by flow cytometry using a reactive oxygen species fluorescence assay kit (Elabscience, E-BC-K138-F, Wuhan, China).
Isolation and culture of bacteria via microfluidic cultivation
To isolate bacteria, dermal tissue from dissected skin (approximately 200–500 mg) was grounded in 1 mL of brain-heart infusion broth using a glass homogenizer. At the same time, the control group was washed with sterile PBS to exclude the environmental pollution caused by the experiment. Ten keloid tissues and ten adjunct normal skin tissues were used for bacteria isolation. Bacteria in the skin were isolated and cultivated by picodroplet microfluidic cultivation.18 Aerobic culture: A 150 μL sample homogenate was plated on Columbia blood agar media (Thermo Fisher Scientific, PB0123A, Waltham, USA), chocolate agar media (no antibiotic) (Thermo Fisher Scientific, PB8124A, Waltham, USA), and China blue agar media (Sigma, 1023480500, USA) at 37 °C aerobically with 5% CO2 for 3 days. Anaerobic culture: A total of 150 μL sample homogenate was plated on anaerobic blood culture medium. Columbia agar (Thermo Fisher Scientific, CM331, Waltham, USA), Sterile Hemin and Microbial K1 Mixture were used to prepare anaerobic blood culture medium. The samples were placed in a GENbag (Marcel Mérieux, 45534, French) at 37 °C for 5–7 days. A fully automatic microbial nucleic acid detection system (Antu Biology, Zhengzhou, China) was used to identify cultured bacterial species.
Infection of keloid fibroblasts with target bacteria
The target bacteria was cultured overnight and harvested in their exponential growth phase. To determine the optimal multiplicity of infection (MOI), three following ratios (10:1, 50:1, 250:1) were first examined and a MOI (bacteria: cells) of 10:1 was chosen. Briefly, fibroblasts were plated in 96-well cell culture plates (5000 cells/well). Then 50,000 bacteria were added to each well with the density of bacteria as 1.5 × 108 CFU/mL. For Clostridium butyricum cultured with fibroblasts in a low oxygen environment, AnaeroPack™ 2.5L Rectangular Jar and AnaeroPack-Anaero 5% (C-04) were used. LPS (10 ng/mL, AbMole; cat. M9524) mixed with fibroblasts was used as the negative control.
Immunocytochemistry (IHC)
Skin samples were incubated with 3% H2O2 for 10 min at room temperature to eliminate endogenous peroxidase activity and the sections were prepared according to a routine protocol. For immunodetection, the cells were blocked with 10% serum and incubated at room temperature for 10 min. Primary antibody was added to the cells and incubated at 37 °C for 2 h. After rinsing with PBS, biotin-labeled secondary antibody was added and incubated at 37 °C for 30 min, followed by the addition of alkaline phosphatase-labeled streptavidin and incubation at 37 °C for 30 min. Finally, the immunolabled protein was detected by incubation for 15 min with chromogenic reagent, counterstained and the section was dehydrated and mounted.
Mouse xenograft
BALB/c female nude mice were purchased from the Animal Husbandry Centre of the Beijing Institute of Cell Biology, Academia Sinica, Beijing, China (#110324231101542165, 2022). All experiments were approved by and conducted in accordance with the guidelines of the Institutional Animal Care and Use Committee at Peking Union Medical College (Approval No.: 2022096) and the guidelines of ARRIVE (Animal Research: Reporting of In Vivo Experiments). Mice were housed 3/cage at 24 ± 1 °C for 12 h/day light cycle with free access to food and water. For studying the roles of CAT on keloid mass growth, 1 × 107 cells/mL keloid fibroblasts were injected into the subcutaneous layer of left lower abdomen of BALB/c nude mice (4 weeks, n = 6 per group) at a volume of 0.1 mL. For studying the roles of bacterial on keloid mass growth, 1 × 107 cells/mL keloid fibroblasts were mixed with 1.5 × 108 CFU/mL Clostridium butyricum or Bacillus subtilis, and then immediately injected into the subcutaneous layer of right abdomen of BALB/c nude mice (4 weeks, n = 6 per group) at a volume of 0.1 mL. Sterilized PBS mixed with keloid fibroblasts were injected as a negative control. The growth of mass in the abdomen area was monitored for 1 month. Mass size was measured after 2 weeks in nude mice and every 5 days afterwards. Mass volume = ab2/2 (a: long axis of the mass; b: short axis of the mass). Zoletil 50 (50 mg/mL, 3 mg/kg animal weight, Virbac, French) was used for anesthesia in nude mice.
Western blot
Approximately 300 μL RIPA buffer (Sigma–Aldrich, R0278, Missouri, USA) and 3 μL of PMSF (Solarbio, P0100, Beijing, China) were added to 100 μg of skin tissues or cells. The samples were centrifuged at 16,000 × g in a precooled centrifuge at 4 °C for 20 min, and the supernatant was harvested. The proteins were separated by SDS–PAGE and transferred to polyvinylidene fluoride membranes. Immunodetection was carried out following a standard protocol. Detailed information of the source of the antibodies used for immunoblotting in this study is as follows: GAPDH (Proteintech Cat# 60004-1-Ig, RRID:AB_2107436). Catalase (Proteintech Cat# 21260-1-AP, RRID:AB_10733099), Collagen type III (Proteintech Cat# 22734-1-AP, RRID:AB_2879158), NF-κB P65 (Proteintech Cat# 10745-1-AP, RRID:AB_2178878), and AKT (Proteintech Cat# 10176-2-AP, RRID:AB_2224574). Phospho-NF-κB p65 (Ser468) recombinant antibody (Proteintech Cat# 82335-1-RR, Proteintech, USA), Phospho-AKT (Ser473) monoclonal antibody (Proteintech Cat# 66444-1-Ig, RRID:AB_2782958).
Hematoxylin-eosin staining
First, the tissue samples were fixed with formalin, embedded in paraffin and sectioned with a paraffin microtome to a thickness of 4–8 μm. The tissue sections placed on glass slides were washed multiple times with xylene to solubilize and remove the paraffin, with xylene subsequently removed by graded washes with xylene and ethanol. Finally, the tissue was rehydrated through graded ethanol in descending order, with the final rinse in distilled water. Next, 100 μL of hematoxylin or eosin solution was added dropwise to the tissue section for staining. The tissue sections were then dehydrated with graded alcohol in ascending order with the final rinse in absolute alcohol.
Electron microscopy
Full-thickness keloid samples from patient No. 75 were obtained, fixed with 2% glutaraldehyde for 2 h, washed with phosphate buffer, fixed with 1% osmic acid for 2 h, dehydrated with an ethanol gradient, and embedded in acetone after immersion. The samples were polymerized for 48 h at 60 °C, sliced and observed with TEM-1400 pulses and a JEM-1400 transmission electron microscope.
Fluorescence in situ hybridization (FISH)
Immediately after harvesting, the tissue was rinsed and fixed with a formalin solution prepared with DEPC water for more than 12 h. After the tissue was fixed, it was embedded in paraffin and sectioned by a microtome. After deparafinnation in xylene and rehydration in a graded series dilution of alcohol, the tissues were first treated with Proteinase K (20 μg/mL) (Servicebio, G1205, Wuhan, China) at 37 °C for 20 min, then incubated with prehybridization solution at 37 °C for 1 h, and finally incubated with the hybridization solution containing probe EUB338 at 1 μM overnight at 42 °C. After wash with 2× SSC (Servicebio, G3016-4, Wuhan, China) at 37 °C for 10 min, 1× SSC at 37 °C for 5 min and 0.5× SSC at room temperature for 10 min, DAPI (Servicebio, G1012, Wuhan, China) staining solution was added to the sections and incubated in the dark for 8 min. Anti-fluorescence quenching mounting medium was added before the sections were mounted. The tissues were observed using an upright fluorescence microscope (NIKON DS-U3, Japan), and the images were scanner using a panoramic slide scanner (3DHISTECH, PANNORAMIC DESK/MIDI/250/1000, Hungary). The EUB338 probe used in this study was 5′-GCTGCCTCC CGTAGG AGT-3′1. All consumables and containers used in this experiment were steriled to avoid any bacterial contamination from the environment.
All FISH tissue sections were viewed using CaseViewer2.4 software (3DHISTECH, CaseViewer2.4, Hungary) and fluorescent signals were imaged at 200 times magnification, with the background light of each photo adjusted to be consistent. The raw FISH images were analyzed by the Image-Pro Plus 6.0 (Media Cybernetics, USA) analysis software to convert the green/red fluorescent monochrome photos into black and white images. An Olympus confocal microscope FV1000MPE was also used to take photos.
Quantitative reverse transcription PCR (qRT–PCR)
One milliliter of TRIzol Reagent (Thermo Fisher, USA) was added to the collected tissue or fibroblasts for 5 min at room temperature. Next, 0.2 mL of chloroform was added to each tube and shaken vigorously for 15 s. The solution was left to stand at room temperature for 2–3 min and then centrifuged at 12,000 × g for 15 min at 4 °C. Then 0.2 mL of supernatant was placed into a new tube, and 0.5 mL of isopropanol was added to each tube. The solution was mixed by gently pipetting a few times, incubated at room temperature for 10 min, and then centrifuged at 12,000 × g for 10 min at 4 °C. The supernatant was removed, and the pellet was washed with 75% ethanol solution, and then air dried for 5–10 min. An appropriate amount of RNase-free H2O was added to dissolve the RNA precipitate.
Next, RNA was quantified using the Nanodrop 8000 spectrometer (Thermo Fisher Scientific). RNA purity was assessed and was found to have an average absorbance of 1.99 (A260/A280) with a standard deviation of 0.18. The equivalent of 100 ng RNA from each tissue type selected was analyzed by RT-qPCR amplified with 25 μL reaction volume using the SuperScript III Platinum One-Step qRT-PCR kit (Thermo Fisher). The qPCR reaction was carried out on the CFX96 Real-Time PCR Detection System (Bio-Rad), with 95 °C for 30 s, followed by 40 cycles of 95 °C 5 s, 54 °C 10 s, and 72 °C 20 s. A positive result was called if the Ct value <40 and a sigmoidal plot is observed. Each PCR reaction was performed in duplicate. The CAT forward primer was 5′-TGGAGCTGGTAACCCAGTAGG-3′, and the reverse primer was 5′-CCTTTGCCTTGGAGTATTTGGTA-3′. The β-actin forward primer was 5′-ATGCGTTGTTACAGGAAGT-3′, and the reverse primer was 5′-GCACGAAGGCTCATCATT-3′.
Immunofluorescence staining
Each tissue block was cut to 0.5 cm, and the tissue was sectioned and fixed. Samples were washed 3 times with PBS for 5 min each. The sections were incubated with penetrant for 15 min, washed with PBS, and sealed for 30 min. Sections were incubated with primary antibody for 1 h at room temperature and then rinsed with PBS. Sections were incubated with the secondary antibody for 1 h at room temperature, and then the sections were sealed by dropwise addition of sealing slips (Thermo Fisher, USA). The intensity of positive markers was calculated using Image J (version 1.8.0) for quantitative results.
Flow cytometry
The primary fibroblasts were grown in logarithmic phase. Their cell cycle profiles were examined by flow cytometry and the results were analyzed with BD Accuri C6 Plus software to calculate the G1/S ratio.
CCK8 assay
The cell suspension (100 μL/well) was seeded in 96-well plates. The plate was placed in an incubator for preincubation (37 °C, 5% CO2). Ten microliters of CCK8 solution (Beyotime, C0037, Shanghai, China) was added to each well. The plate was incubated for 1–4 h. The absorbance at 450 nm was measured with a microplate reader.
Fabrication of microfluidic chip
The equipment was created and produced utilizing the methods outlined earlier, using poly(dimethyl-siloxane) (PDMS) and the soft lithography rapid-prototyping technique. The channels in the equipment ranged in height about 40 μm, and the inlets and outlets were punctured. Aquapel (PGW Auto Glass, USA) was applied to the microfluidic channels for the purpose of modifying the surfaces to be hydrophobic.
Bacteria encapsulation in droplets
To generate droplets for encapsulating single cells, microbial suspension samples were diluted to approximately 3 × 106 CFU/mL in BHI medium and passed through a droplet maker device, which had a throughput of 1000 drops per second, resulting in droplets of approximately 100 pL in size. These droplets were then collected into 0.2 test tubes and incubated for 3–7 days before evenly applying the bacterial solution to the Petri dish.
Statistical analysis
Statistical analysis was done using SPSS 24.0 software and plotted using GraphPad Prism 8 software. An unpaired T test was used. Box plot of the Shannon index was analyzed by Wilcox Test. P value of <0.05 was considered that the difference was statistically significant.
Role of the funding source
The sponsors did not have any role in the study design, data collection, data analyses, interpretation, or writing of the manuscript.
Result
Bacteria in keloid
Exploring specific bacterial types in keloid
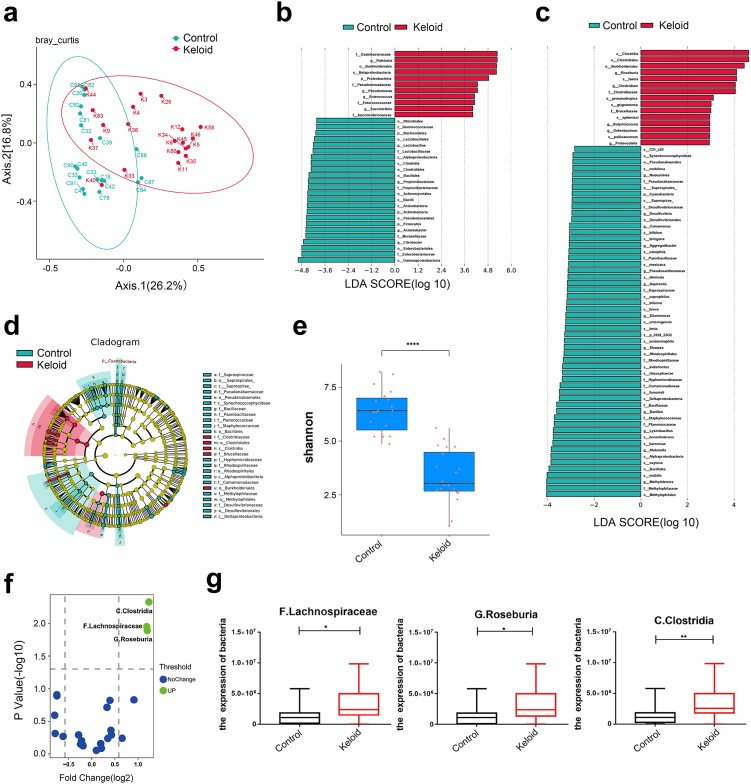
To determine whether there are bacteria in keloid, the microbiome and other multi-omics approaches were used (Fig. 1, Supplementary Fig. 1). To explore the species of bacteria that differed between the two groups, we performed amplicon sequencing on both keloid and normal full-thickness skin tissue to explore the differences in species of bacteria between keloid and normal skin tissue. The sequencing data had good repeatability and could be used for subsequent analysis (Fig. 1a). At the genus level, Clostridium, Oxalobacteraceae, Ralstonia, Burkholderiales, Succinivibrio and Enterococcus was abundant in keloid, while Gammaproteobacteria, Firmicutes, Actinobacteria, Propionibacterium, and Lysinibacillus were abundant in control (Fig. 1b, Table 2). At the species level, Clostridia, Roseburia, Brucellaceae and Burkholderiales were abundant in keloid, while Desulfovibrionaceae, Methylophilaceae, Comamonadaceae, Lysinibacillus and others were abundant in control (Fig. 1c and d). The bacteria enriched in keloid were mostly negative by the catalase test, but the opposite was observed for bacteria in control (Fig. 1b–d, Table 2). There were not only bacterial composition differences between the keloid and control groups but also microbiome diversities. The alpha diversity index is an analysis of species diversity in a sample, including two factors, the richness and evenness of the species composition. Beta diversity compares the composition of microbial communities between different samples.19 The Shannon index is usually used to evaluate the species diversity of a sample (Fig. 1e). Based on the higher Shannon index of control, this group of samples exhibited higher diversity than that of keloid samples (P < 0.05, Wilcox test). Metaproteomics sequencing data showed that C. Clostridia (P = 0.0047, unpaired t test), F. Lachnospiraceae (P = 0.0111, unpaired t test), G. Roseburia (P = 0.0129, unpaired t test) increased in keloid (Fig. 1f and g).
Fig. 1.
Microbiomic analysis of keloid. (a) Principal co-ordinates analysis. The points in the principal coordinate analysis diagram represent the samples, and the distance between the points indicates the degree of difference in the bacterial flora of the samples. The higher the similarity of the microbial community structure was, the smaller the difference. Keloid: 20 keloid tissues. Control: 19 normal tissues. (b and c) From the genus and species levels, each horizontal column represents a species, the length of the column corresponds to the linear discriminant analysis (LDA) value, and the higher the LDA value is, the greater the difference. The color of the column corresponds to which group of characteristic microorganisms the species belongs, and the characteristic microorganisms indicate that the abundance in the corresponding group is high. (d) From the inside to the outside, the cladogram map corresponds to the different classification levels of the genus and family, and the lines between the levels represent the affiliation. Each circle node represents a species; if the node is yellow, it means that the difference between the groups is not significant, and if the node is not yellow, it means that the species is a characteristic microorganism of the corresponding color group. The colored fan-shaped area marks the subclassification interval of the characteristic microorganism. (e) Box plot of the Shannon index of control and keloid. Wilcox Test was used. (f) Volcano map analysis of differentially expressed bacteria by metaproteomics. P < 0.05, Fold change (FC) threshold >1.5. (g) Detection of the expression levels of C. Clostridia, F. Lachnospiraceae and G. Roseburia by metaproteome. Keloid (n = 18), Control (n = 19). Unpaired t test was used. ∗P < 0.05, ∗∗P < 0.01, ∗∗∗P < 0.001, ∗∗∗∗P < 0.0001.
Table 2.
The features of bacteria identified in keloid.
| Phylum | Class | Order | Family | Genus | Relative Abundance (%) |
log2 Fold Change | P valueb | Bacterial isolation and culture | CAT testa | |
|---|---|---|---|---|---|---|---|---|---|---|
| Control | Keloid | |||||||||
| Proteobacteria | Gammaproteobacteria | Aeromonadales | Succinivibrionaceae | Succinivibrio | 0.64 | 1.33 | 1.06 | 0.53 | / | − |
| Firmicutes | Bacilli | Lactobacillales | Enterococcaceae | Enterococcus | 0.18 | 1.85 | 3.36 | 0.32 | / | − |
| Proteobacteria | Betaproteobacteria | Methylophilales | Methylophilaceae | Methylotenera | 0.29 | 3.19 × 10−3 | −6.51 | 0.004 | / | / |
| Firmicutes | Clostridia | Clostridiales | Clostridiaceae | Clostridium | 1.69 × 10−3 | 0.1 | 5.89 | 0.004 | Clostridium butyricum | − |
| Proteobacteria | Alphaproteobacteria | Rhizobiales | Brucellaceae | Pseudochrobactrum | 0 | 5.5 × 10−5 | / | / | / | − |
| Firmicutes | Bacilli | Bacillales | Planococcaceae | Lysinibacillus | 0.11 | 0.01 | −3.46 | 0.002 | Bacillus subtilis | + |
| Firmicutes | Bacilli | Bacillales | Staphylococcaceae | Staphylococcus | 3.6 | 4.8 | 0.42 | 0.06 | Staphylococcus aureus; Staphylococcus capitis; Staphylococcus epidermidis | + |
| Actinobacteria | Actinobacteria | Actinomycetales | Propionibacteriaceae | Propionibacterium | 4.5 | 1.8 | −1.32 | 0.13 | Propionibacterium acnes;Propionibacterium Avidum | + |
| Firmicutes | Clostridia | Clostridiales | Lachnospiraceae | Roseburia | 0.28 | 0.37 | 0.40 | 0.039 | Rothia mucilaginosa | − |
Catalase test; +: catalase positive; −: catalase negative; /: unknown or unspecific.
Unpaired t test was used to calculate P value.
The representative sequences of Operational Taxonomic Units (OTUs) of interest were selected for phylogenetic analysis (one OTU with the highest abundance was selected for each genus as the representative OTU) to draw an evolutionary relationship tree diagram, and the absolute abundance of OTUs in each group was combined to create the heatmap (Supplementary Fig. 2a). A branching evolutionary tree of Bacteroidetes, Synergistetes, Proteobacteria, and Actinobacteria is displayed. Propionibacterium and Corynebacterium were on the same branch (Supplementary Fig. 2a). Methylobacterium, Rhodobacter, Devosia, and Brevundimonas were close in evolutionary distance and were highly expressed in control and expressed at low levels in keloid (Supplementary Fig. 2a). The interaction relationship between microbes is shown in Supplementary Fig. 2b. Close interaction was observed among Proteobacteria, Actinobacteria and Firmicutes (Supplementary Fig. 2b).
The verification and isolation of specific bacteria in keloids
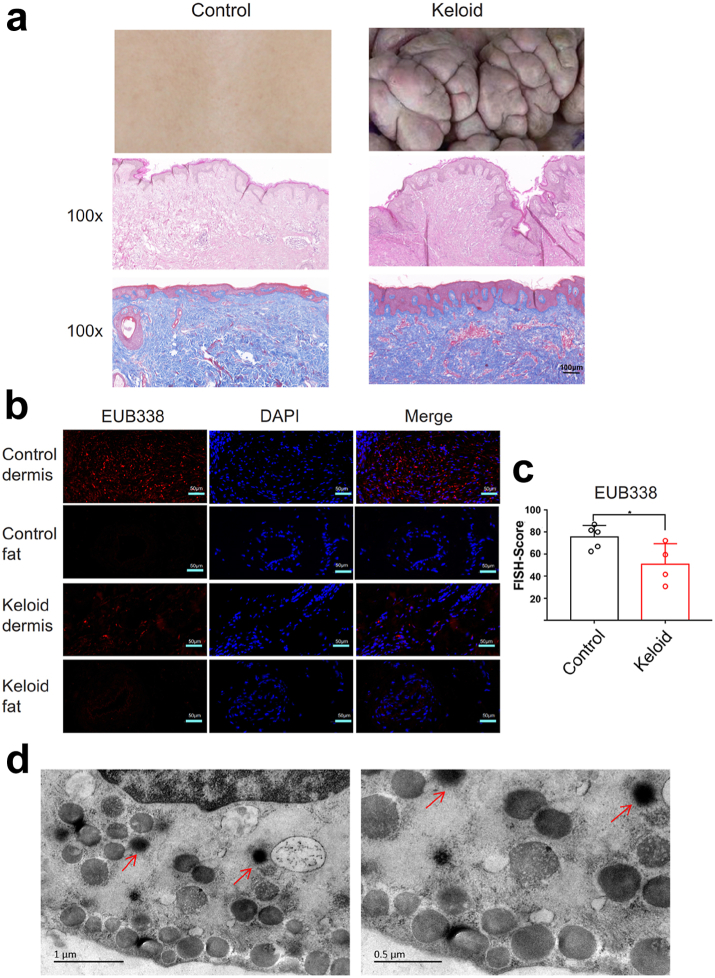
Morphologically, the epidermis of keloids is significantly thicker than that of normal skin (Fig. 2a). The dermal fiber bundles of keloids are abnormally thick and dense, with a large number of blood vessels, while fiber bundles are smaller and sparser in normal skin dermal tissue (Fig. 2a). Fluorescence in situ hybridization (FISH) analysis was performed on the dermis and fat layer of skin samples (Fig. 2b). Positive bacterial staining was observed in dermis layer of keloid and control, and it showed more bacteria existed in control (Fig. 2b and c; P = 0.0384, unpaired t test). However, both the fat layers of the two groups had almost no positive bacterial staining (Fig. 2b). Electron microscopy results verified that there were bacteria in the cytoplasm of keloid fibroblast cells (Fig. 2d).
Fig. 2.
Bacteria were observed in the dermis of keloid. (a) HE and Masson's trichrome representative staining of keloid tissue and normal skin tissue, 100×. Scale bar = 100 μm. (b) Fluorescence in situ hybridization experiments were performed on the dermis and fat layer of skin. Keloid: keloid tissues. Control: normal tissues. 100×. Scale bar = 50 μm. (c) Quantitative statistics of EUB338 expression via FISH experiments. Unpaired t test was used. ∗P < 0.05, ∗∗P < 0.01, ∗∗∗P < 0.001, ∗∗∗∗P < 0.0001. Keloid (n = 4), Control (n = 5). (d) Electron microscopic view of bacteria in the dermis of keloids, red arrows. Scale bar = 0.5 μm.
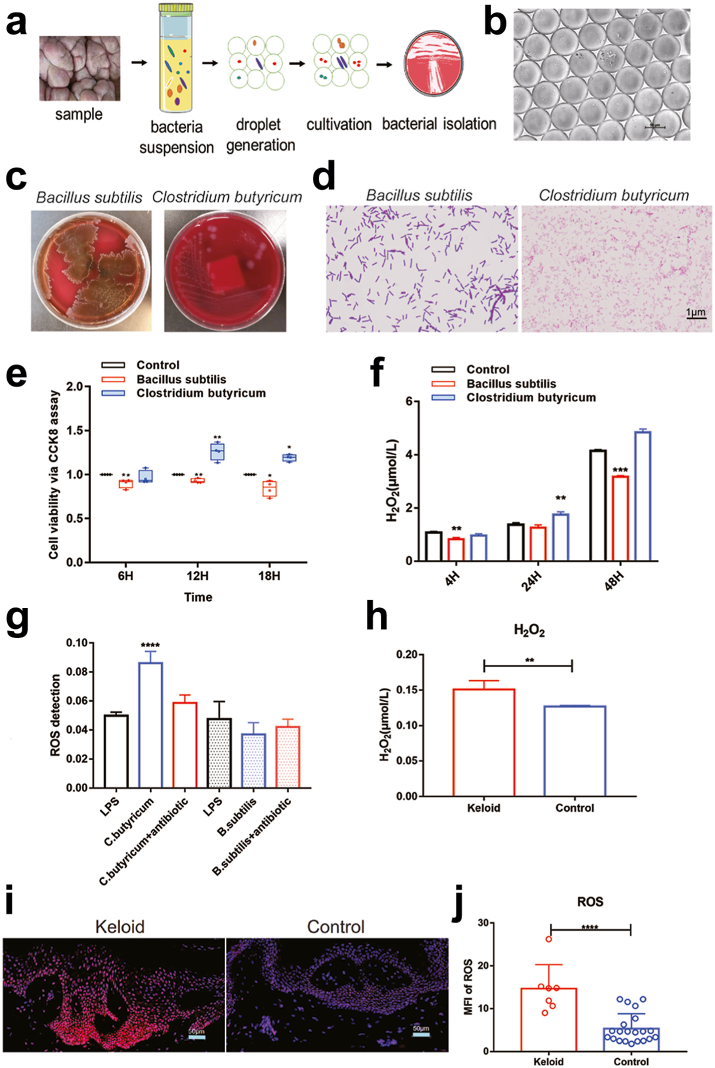
To identify the specific types of bacteria in keloid, bacteria isolation and culture experiments were carried out. With the ordinary bacteria cultivation method, the slow-growing bacteria will be inhibited due to the competitive growth of dominant bacteria. Therefore, we adopted the picodroplet microfluidic cultivation in order to obtain more abundant bacteria (Fig. 3a and b). Overall, seven species of bacteria were isolated, including Bacillus subtilis, Clostridium butyricum (Fig. 3c and d), Staphylococcus epidermidis, Propionibacterium acnes, Propionibacterium Avidum, Staphylococcus capitis, and Staphylococcus aureus (Supplementary Fig. 3a, Table 2).
Fig. 3.
Isolation of bacteria and co-culture with the primary fibroblast cells of keloid. (a) Flow chart of picodroplet microfluidic cultivation. (b) After 120 h, the microfluidic droplets remain stable during cultivation. (c and d) Isolation of Clostridium butyricum and Bacillus subtilis from keloid tissue. Scale bar = 1 μm. (e) Cell count kit 8 was utilized to detect proliferation of fibroblasts at 6, 12 and 18 h (n = 4). (f) The detection of hydrogen peroxide (H2O2) production from the supernatant of fibroblasts at 6, 12 and 18 h (n = 3). (g) The production of ROS in the co-culture cells was detected via flow cytometry (n = 4). (h) Determination of H2O2 production in keloid tissue and normal skin tissue (n = 3). (i and j) Detection of ROS in keloid tissue and normal tissue via immunofluorescence assy. Scale bar = 50 μm. Keloid (n = 7), Control (n = 21). Unpaired t test was used. ∗P < 0.05, ∗∗P < 0.01, ∗∗∗P < 0.001, ∗∗∗∗P < 0.0001.
Since we found Clostridium butyricum and Bacillus subtilis were specific dominant bacteria in keloid and control groups respectively, we selected them to co-cultured with keloid fibroblast cells (Supplementary Fig. 3b), and observed their effects on the of fibroblast cells. The existence of Clostridium butyricum stimulated the proliferation of fibroblast cells (Fig. 3e), H2O2 (Fig. 3f) and ROS (Fig. 3g) production, while Bacillus subtilis produced the opposite phenomena (Fig. 3e–g). This suggests that the existence of catalase-negative bacteria may promote the growth of fibroblast cells through increased H2O2 and ROS. Furthermore, both H2O2 (Fig. 3h) and ROS (Fig. 3i and j) levels were significantly elevated in the keloid tissues of patients, which suggest that H2O2 and ROS may also play important roles on the growth of keloid tissue.
The correlation between microbiome and metabolome
Metabolites are functional outputs of host–microbe interactions that provide timely responses in the physiological state for extremely complex multibiological systems.20 In our previous study,20 we found that differential metabolites in keloid and control included 1-methylnicotinamide, diacetyl, 4-hydroxyproline, beta-alanyl-l-arginine, heptanoic acid, 5-hydroxylysine, galactaric acid, cytidine monophosphate N-acetylneuraminic acid, l-prolinamide, beta-glycerophosphoric acid, etc. The abundance of Oxalobacteraceae, Ralstonia, and Proteobacteria in keloid increased significantly. The abundance of Citrobacter, Moraxellaceae, Firmicutes, and Actinobacteria in control increased significantly (Fig. 1b, Table 2). The correlation analysis between metabolites and microbes is shown in Supplementary Fig. 4. Proteomics and metabolomics data showed that 15-deoxy-Δ12,14-prostaglandin J2 was highly expressed in normal skin tissues (Supplementary Fig. 4b, P = 0.0028, unpaired t test), and COX-2 also showed an increasing trend in normal skin tissues (Supplementary Fig. 4c). A variety of ROS-related metabolites are enriched in keloid tissue, such as 4-hydroxyproline, 5-hydroxylysine, beta-alanyl-l-arginine, and 1-methylnicotinamide.21, 22, 23
Proteomics shows CAT is a distinctive protein in keloid
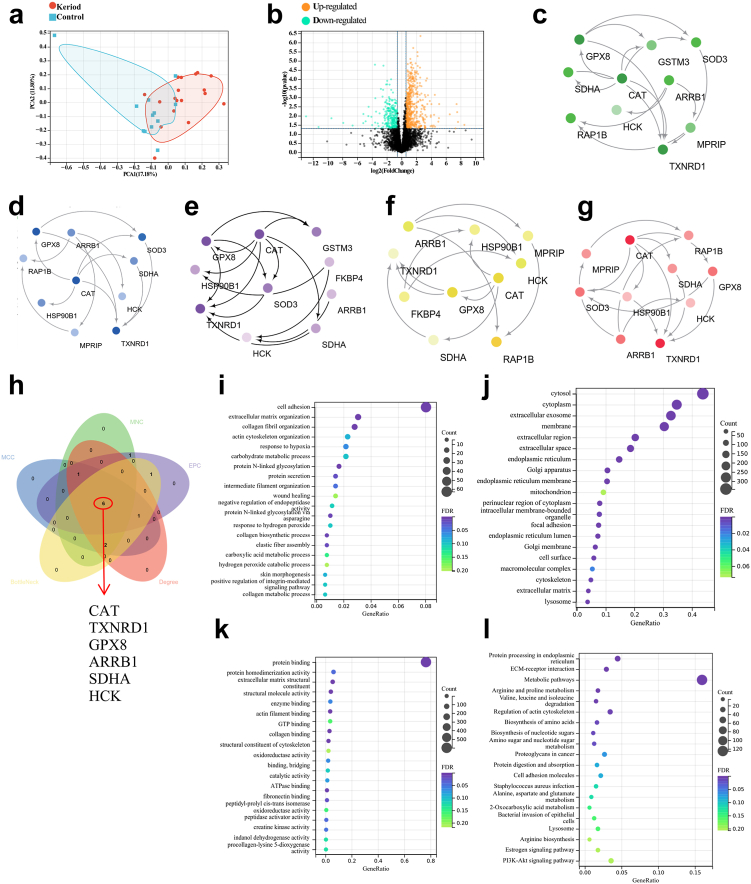
The samples of keloid and normal tissues were distinctly divided into two groups, with good intragroup consistency (Fig. 4a). There was a clear demarcation between the two groups of DEPs, suggesting that keloids are closely associated with these DEPs (Fig. 4b). When the FC was ≥1.5 and the P value was ≤0.05, the upregulated proteins were screened, and when the FC was ≤−1.5 and P value was ≤0.05, the downregulated proteins were screened. There were 775 DEPs between the two groups. A total of 516 DEPs were upregulated, and 259 DEPs were downregulated (Fig. 4b).
Fig. 4.
Metaproteomics analysis of keloid samples. (a) The abscissa PC1 and ordinate PC2 in the figure represent the scores of the first and second principal components, respectively, and the scatter color represents the sample grouping. 30 samples (18 keloid tissue vs 12 normal skin tissue) were used for metaproteomics. (b) Differential Protein Volcano Plot. The horizontal axis represents the fold change (log2 value) of the DEPs, the vertical axis represents the P value (-log10 value), black represents the proteins with no significant difference, red represents the upregulated proteins, and green represents the downregulated proteins. P < 0.05, Fold change (FC) threshold >1.5. (c–g) Five algorithms of MNC, MCC, EPC, BottleNeck, and degree were used to screen hub proteins through Cytoscape software. (h) Venn diagrams for MNC, MCC, EPC, BottleNeck, and degree algorithms. (i–k) Gene Ontology (GO) analysis (biological process, cellular component and molecular function). (l) Kyoto Encyclopedia of Genes and Genomes (KEGG) analysis.
Cytoscape (version 3.5.1) software was used to further refine DEPs from the differential protein interaction network. Five algorithms of MNC, MCC, EPC, BottleNeck, and degree were used to screen hub proteins (Fig. 4c–g). A Venn diagram was used to find the intersecting proteins calculated by five algorithms (Fig. 4h). Six proteins were obtained by the intersection of proteins calculated by these algorithms, and their corresponding gene names are CAT, TXNRD1, GPX8, ARRB1, SDHA and HCK (Fig. 4h).
GO analysis included biological process (BP), cellular component (CC) and molecular function (MF) terms. The enrichment for BP included hydrogen peroxide catabolic process, skin morphogenesis and response to hydrogen peroxide (Fig. 4i). The enriched CCs were cytosol and membrane (Fig. 4j). The enriched MFs were oxidoreductase activity and catalytic activity (Fig. 4k). The enriched KEGG pathways were bacterial invasion of epithelial cells, staphylococcus aureus infection and PI3K-Akt signaling pathway (Fig. 4l). Functional enrichment is mainly related to oxidative stress, and the key molecules affecting oxidative stress may play a key role in keloid.
CAT plays an important role in the pathology of keloid fibroblasts
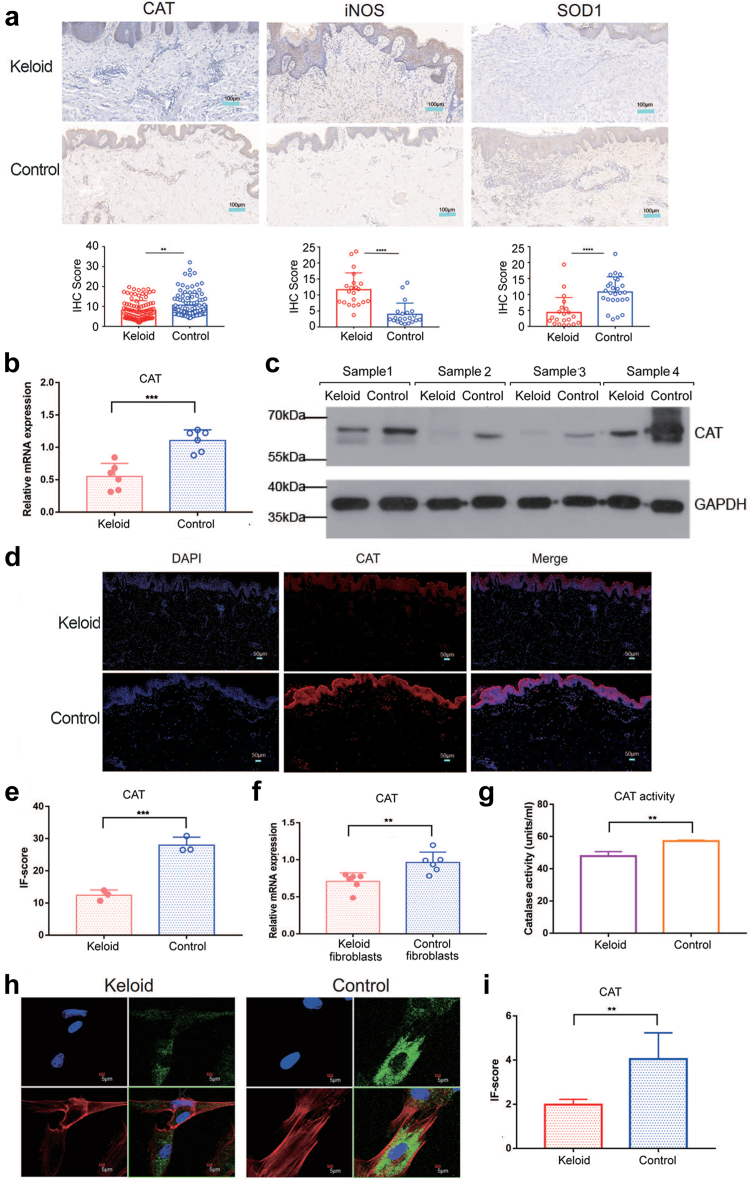
Catalase (CAT), superoxide dismutase 1 (SOD1) and Nitric oxide synthase (iNOS), all significantly contribute to oxidative stress. The level of CAT (Fig. 5a, P = 0.0015) and SOD1 (Fig. 5a, P < 0.0001, unpaired t test) were significantly higher in normal skin tissue as detected by IHC, while iNOS (Fig. 5a, P < 0.0001, unpaired t test) was significantly elevated in keloid tissue. The relatively low expression of CAT was verified in keloid tissue by qRT–PCR (Fig. 5b, P = 0.0004, unpaired t test).
Fig. 5.
The expression of ROS-related genes in keloid. (a) Relative expression of CAT, iNOS and SOD1 in keloid tissue and normal skin tissue according to IHC. Scale bar = 100 μm. (b) The mRNA expression of CAT in keloid and normal tissues (n = 6). (c) The protein expression of CAT in keloid and normal tissues. (d) The expression of CAT between keloid and normal tissues via IF. Scale bar = 50 μm. (e) Relative expression of CAT according to the IF score (n = 3). (f) The mRNA expression of CAT in keloid fibroblasts and normal skin fibroblasts (n = 6). (g) Enzyme activity detection of keloid fibroblasts and normal skin fibroblasts (n = 3). (h and i) Fluorescence confocal microscopic image of keloid fibroblasts and normal skin fibroblasts. Blue indicates the nucleus. Red indicates phalloidin. Green indicates CAT (n = 3). Scale bar = 5 μm. Unpaired t test was used. ∗P < 0.05, ∗∗P < 0.01, ∗∗∗P < 0.001, ∗∗∗∗P < 0.0001.
The western blotting (Fig. 5c) and immunofluorescence assay (Fig. 5d and e; P = 0.0008, unpaired t test) also showed that CAT expression levels decreased in keloid tissue. As we known, H2O2 produced by the human body is cleared by CAT.24 CAT is the main scavenging enzyme of H2O2 in human fibroblasts.24 Next, to determine the role of CAT in keloid fibroblast function, we isolated and cultured primary fibroblasts from keloid tissue for subsequent experiments. The expression of CAT in fibroblasts was verified by qRT–PCR (Fig. 5f, P = 0.0066, unpaired t test). The catalase activity of keloid fibroblasts was lower than that of the control (Fig. 5g, P = 0.0046, unpaired t test). Fluorescence confocal microscopy also confirmed that CAT was expressed at low levels in keloid fibroblasts (Fig. 5h and i; P = 0.0328, unpaired t test).
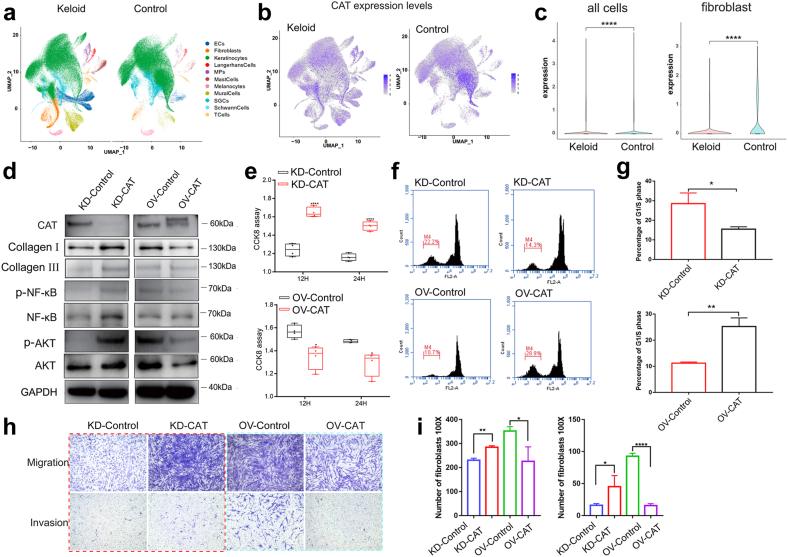
To further explore the characteristics of the local microenvironment of keloids and determine cellular heterogeneity, single-cell RNA sequencing was used with 6 keloid tissues and 6 matched adjacent normal tissues (Supplementary Table 1). The 12 samples were dissociated into 215,795 cells (K: 110,835; N: 104,960) (Fig. 6a). Based on lineage-specific marker genes,25 11 cell lineages were categorized (Fig. 6a, Supplementary Table 3). Fibroblasts number (marked by DCN, COL1A1, and COL1A2) was significantly greater in keloid tissue (6.18%) than normal tissue (0.58%) (Fig. 6a, Supplementary Fig. 5a). Based on the primary pathogenic role of fibroblasts in the growth of keloids, we performed an unsupervised clustering analysis of fibroblasts in all 12 samples. Fibroblasts can be divided into four subpopulations,25 including proinflammatory (PF, marked by APOE, CCL19, CXCL3), mesenchymal (MF, marked by ASPN, POSTN, COMP), secretory-papillary (SPF, marked by APCDD1, COL18A1, COL13A1), and secretory-reticular (SRF, marked by WISP2, ANGPTL1, MFAP5) (Supplementary Fig. 5b). The proportion of MF increased significantly in keloid, while SPF and SRF were significantly increased in the control (Supplementary Fig. 5a). Oxidative stress-related genes were enriched in MFs such as ASPN26 and POSTN27 (Supplementary Fig. 5b). Collagen-related genes were also enriched in MF, such as COL11A1, FN1, COL1A1, and COL12A1 (Supplementary Fig. 5b). In the face of oxidative stress, the fibroblast death score was higher in the keloid than in the normal tissue (Supplementary Fig. 5c). The GO pathway related to oxidative stress was significantly different between keloid and normal tissue (Supplementary Fig. 5c–g, Table 3), including cellular oxidant detoxification, cell death in response to oxidative stress, response to hypoxia, and cellular response to decreased oxygen levels.
Fig. 6.
Single-cell transcriptome analysis and functional study of CAT in keloid fibroblasts. (a) Twelve samples were used for single-cell sequencing. Uniform manifold approximation and projection (UMAP) of unbiased clustering of cells reveals 11 cellular clusters. Clusters are distinguished by different colors. SGCs: sweat gland cells; ECs: vascular endothelial cells; MPs: mononuclear phagocytes. (b) Expression of CAT in keloid tissue and normal tissue via single-cell sequencing. (c) Expression of CAT in overall cells and fibroblasts between keloid tissue and normal tissue. (d) The protein levels of CAT and correlated factors in the dissected tissues masses. (e) The proliferation of cell via CCK8 assay (n = 4). (f and g) Flow cytometry detection of the cell cycle. Unpaired t test was used (n = 3). (h and i) Cell migration and invasion experiments (n = 3). Quantitative calculation by cell migration experiment was performed through 100× field of vision. Unpaired t test was used. ∗P < 0.05, ∗∗P < 0.01, ∗∗∗P < 0.001, ∗∗∗∗P < 0.0001.
Table 3.
Differences in ssGSEA scores of cell death in response to oxidative stress.
| Cell type | Mean ssGSEA score in normal tissue | Mean ssGSEA score in keloid tissue | P Valuea |
|---|---|---|---|
| ECs | −0.108 | −0.151 | 0.024 |
| Fibroblast | −0.112 | −0.223 | <0.0001 |
| Keratinocytes | −0.127 | −0.109 | 0.173 |
| Langerhans cells | −0.068 | −0.077 | 0.453 |
| Mast cells | −0.206 | −0.192 | 0.317 |
| Melanocytes | −0.064 | −0.086 | 0.057 |
| MPs | −0.133 | −0.123 | 0.496 |
| Mural cells | −0.144 | −0.165 | 0.168 |
| T cells | −0.282 | −0.265 | 0.1151 |
GSEA; Gene Set Enrichment Analysis.
P value: unpaired t test.
The single cell RNA sequencing shows CAT is specifically and lowly expressed in the fibroblasts of keloid (Fig. 6b and c). Therefore, we further studied the function of CAT in the primary fibroblast cells (Fig. 6d–i). Lentivirus was used to establish CAT knockdown or overexpression stable cell lines with primary keloid fibroblasts (Fig. 6d). The fibroblasts with CAT knockdown produced more collagen I and collagen III (Fig. 6d), which are both the continuous accumulation of ECM components in keloid.28 The expression levels of the important signaling pathway proteins NF-κB and AKT were increased in CAT knockdown cells but decreased in CAT-overexpressing cells (Fig. 6d). In addition, we also detected the independent effects of antibiotics on the primary fibroblasts. We found antibiotics could inhibit the proliferation of keloid fibroblasts (Supplementary Fig. 6a). Mechanically, antibiotics could decreased the production of H2O2 (Supplementary Fig. 6b) and ROS (Supplementary Fig. 6c and d).
The viability of fibroblasts was increased after CAT knockdown (Fig. 6e, P < 0.0001, unpaired t test), while the proliferative ability of fibroblasts was decreased after CAT overexpression (Fig. 6e, P < 0.05, unpaired t test). The ratio of fibroblasts in G1/S phase was decreased after CAT knockdown (P = 0.0156, unpaired t test), while it was increased after CAT overexpression (Fig. 6f and g; P = 0.0018, unpaired t test). The cell cycle was blocked by CAT overexpression, which affected the proliferation of keloid fibroblasts. Furthermore, we found invasive capacity of fibroblasts with CAT knockdown was also increased (Fig. 6h and i; P < 0.05, unpaired t test), whereas the invasive capacity of fibroblasts with CAT overexpression was decreased (Fig. 6h and i; P < 0.0001, unpaired t test).
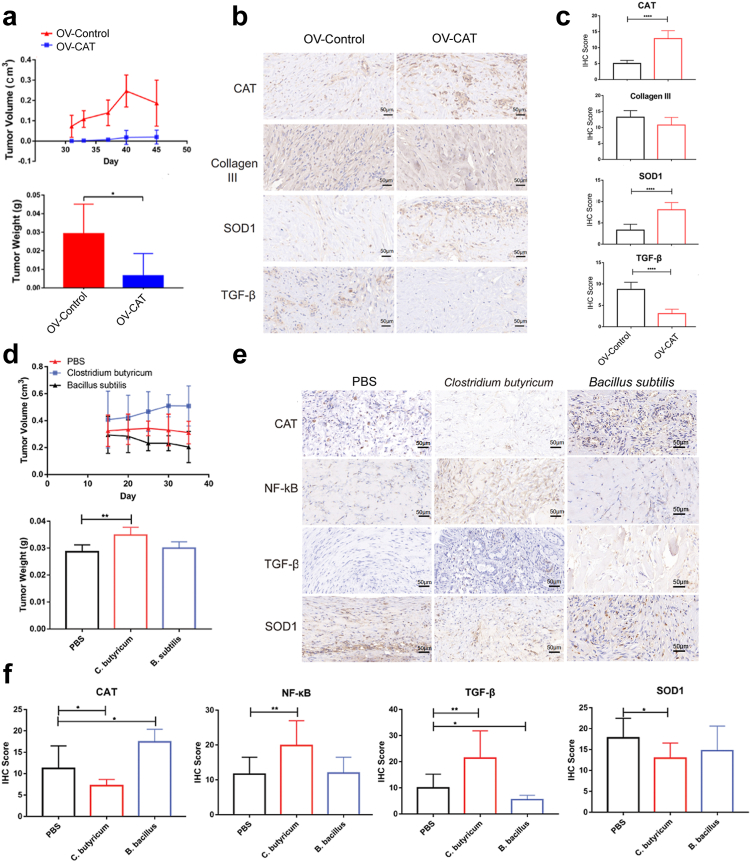
The function study of specific bacteria in keloid growth with cell-line-derived xenograf (CDX) model
Currently, there are no existing animal models for physiological research on keloids.29,30 Here, we used CDX model to imitate the keloid growth in vivo. Well-defined white tissue masses were formed after injection of patient fibroblast cells into the lower abdomen of the nude mice more than one month. We found CAT overexpression inhibited keloid formation and growth in nude mice (Fig. 7a, Supplementary Fig. 6b and c). Futhermore, we dissected tissue masses and found CAT and SOD1 was higher in the tissues injected with OV-CAT fibroblast cells, whereas Collagen III and TGF-β was lower (Fig. 7b and c). ROS were also significantly decreased in OV-CAT tissues compared to control (Supplementary Fig. 6c and d; P < 0.05, unpaired t test). The dissected tissue masses from mice were digested with collagenase and then cultured with antibiotics for 24 h. Interestingly, the production of ROS was lower than that in the group without antibiotics (Supplementary Fig. 6e and f; P = 0.0359, unpaired t test).
Fig. 7.
The CDX model shows CAT plays important roles in keloid formation. (a) Volume and weight of the tissue masses were measured after executing the mice (n = 6). (b) The expression of CAT, collagen III, SOD1 and TGF-β in the CDX model via immunohistochemistry. Scale bar = 50 μm. (c) The expressions of CAT, collagen III, SOD1 and TGF-β in CDX model were qualitatively calculated by immunohistochemistry. (d) The CDX model of Clostridium butyricum (C. butyricum) and Bacillus subtilis (B. subtilis) infection. Volume and weight of the tissue masses were measured after executing the mice (n = 6). (e) The immunohistochemistry experiment of CAT, NF-κB, TGF-β and SOD1 in the dissected tissue masses via CDX model. Scale bar = 50 μm. (f) The expressions of CAT, NF-κB, TGF-β and SOD1 were qualitatively calculated. Unpaired t test was used. ∗P < 0.05, ∗∗P < 0.01, ∗∗∗P < 0.001, ∗∗∗∗P < 0.0001.
Then, we mixed primary keloid fibroblasts with specific bacteria: Clostridium butyricum and Bacillus subtilis, and subcutaneously injected into the low abdomen of nude mice. We found that the Clostridium butyricum could significantly promote the mass growth of keloid fribroblasts (Fig. 7d, Supplementary Fig. 6g), while Bacillus subtilis showed a certain inhibitory effect (Fig. 7d, Supplementary Fig. 6g). FISH experiment verified that a considerable amount of bacteria existed in dissected tissues (Supplementary Fig. 6h). The expression of CAT and SOD1 was lower in Clostridium butyricum group but higher in Bacillus subtilis group, while TGF-β and NF-κB were higher in Clostridium butyricum group but lower in Bacillus subtilis group (Fig. 7e and f). These results together suggest CAT-negative bacteria Clostridium butyricum may be able to promote keloid growth in vivo.
Discussion
Keloid is a common benign tumor of the body surface often caused by skin damage and irritation, trauma, insect bites, burns, surgery, vaccinations, skin perforations, acne, folliculitis, chickenpox, shingles infections, and other conditions.2,31 The occurrence and growth of keloid—begin with abnormal wound healing, which causes a fibroproliferative inflammatory response and, finally, the formation of keloid tissue.1,2 Keloids resemble the histological features of hypertrophic scars but have unique growth characteristics that manifest as persistent scarring beyond the wound margins and generally do not resolve spontaneously.32,33 The lesion site exhibits unique histological features, characterized by a large number of irregularly oriented and thickened hyaline collagens commonly referred to as keloid collagen and increased levels of inflammatory cells and fibroblasts.34,35 In addition, proinflammatory cytokines such as IL-1α, IL-1β, IL-6 and TNF-α are upregulated in keloid tissue, which indicates that the occurrence and growth of keloids are closely related to infection and inflammation.2 Previous studies have found that the occurrence of inflammation in many other skin diseases is closely related to the existence of microorganisms.36,37 However, the certain types and roles of the microorganisms in keloid has not been well illuminated so far.
In this study, we observed bacteria in keloid tissue by FISH and transmission electron microscopy. The results revealed that there are a considerable number of bacteria existed in keloid tissue. Usually, bacteria can cause skin continuous infections and inflammation, and polymorphonuclear leukocytes (PMNs) may be recruited by proinflammatory factors to kill and ingest invading bacteria.38 However, if the infection exceeds the regular inflammatory pathological process, the infection site will expand.38 For example, acne vulgaris (AV) is a chronic skin ailment that involves the gram-positive bacterium anaerobic P. acnes, which resides on skin microflora.39 Superficial bacterial folliculitis (SBF) is usually caused by Staphylococcus pseudintermedius.40 In our study, we also found P. acnes and Staphylococcus epidermidis existed in the samples. This indicates that PMN did not effective clear all the bacteria in the infectious site after the acute inflammatory stage. Some specific bacteria are persistent and may cause continuous chronic inflammation, which beneficial for the keloid growth. Therefore, we used amplicon sequencing to explore different bacteria between keloid and normal skin. We found the bacterial diversity of normal skin tissue was higher than that of keloid. The decrease of bacterial diversity in keloid indicate there might be specific profile of microorganisms in the disease. Clostridiaceae, Brucellaceae and Burkholderiales were abundant in keloid tissue, and these bacterial pathogens may have the ability to evade the host innate immune response to promote disease by producing specific molecules.
The production of superoxide radicals and other reactive oxygen species (ROS) is a crucial step of PMN bactericidal mechanisms.38,41,42 We found the levels of ROS and ROS-related metabolites were higher in keloid compared with the normal tissues. The GO analysis of single-cell RNA sequencing also showed that oxidative stress was enriched in keloid tissue. Oxidative stress is a double-edged sword for cell growth. Excessive oxidative stress may also be the reason for cell apoptosis and death.43 However, moderate oxidative stress promotes cell proliferation,44 which may be suitable for explaining the pathogenesis of keloid disease.
CAT is an antioxidant enzyme in the body that plays an important role in scavenging ROS and maintaining the redox state balance.45 CAT widely exists in various tissues of mammals and is an important antioxidant enzyme that can convert toxic H2O2 into nontoxic water and constitutes the body's first antioxidant defense line.45 A large number of studies have found that CAT expression is downregulated in esophageal cancer, lung cancer, liver cancer and other tumor tissues, suggesting that CAT may be a tumor suppressor.46, 47, 48 Overexpression of CAT inhibits tumor proliferation and metastasis.24,47 The same results were found in keloids in our study. The proliferative, migratory, and invasive abilities of fibroblasts were reduced after CAT overexpression, and the phenotype of CAT knockdown exhibited the opposite. These results indicate that although keloid is a skin benign tumor, like other malignant tumors, its growth is closely related to catalse activity. CAT metabolism is associated with tissue-specific bacteria. Interestingly, we found CAT positive bacteria such as Bacillaceae and Saprospirales have a higher proportion in the control, while CAT negative bacteria such as Clostridia and Roseburia have a considerable higher proportion in keloid. Furthermore, we found that normal tissues were rich in aerobic bacteria, while keloid tissues were relatively rich in anaerobic bacteria. As we known, CAT can be produced in almost all aerobic bacteria49, 50, 51, 52 but lacked in some anaerobes.53 Thus, our results show that the microenvironment of keloid may be more conducive to the survival of anaerobic bacteria, and not conducive to the production of CAT. The specific types of bacteria may also influence the own CAT metabolism of host cell and involve in the bacterium-host cell interaction process. In addition, CAT expression changes may be associated with related cellular signaling pathways in keloid, such as NF-κB and AKT. NF-κB is a redox-sensitive transcription factor that plays a role in regulating immune responses.54 Cong et al.55 found that overexpression of catalase downregulated the activation of the ROS-dependent NF-κB signaling pathway. AKT (also known as protein kinase B) is a proto-oncogene that regulates various cellular functions, including metabolism, growth, proliferation, transcription, and protein synthesis.56 The downregulation of cellular catalase depends on the PI3K/Akt/mTOR signaling pathway.57 In our study, the activities of NF-κB and AKT were increased in CAT knockdown cells but decreased in CAT overexpressing cells, which indicates that bacteria may regulate the signaling pathway of keloid growth through CAT.
In summary, our study systematically explored bacterial diversities and roles on keloid disease. We use the CDX animal model to study the growth of keloid in vivo. Specific types of bacteria affected the expression of CAT in keloid, which in turn resulted in ROS scavenging and affected keloid proliferation. Therefore, our finding could potentially provide as a new strategy for keloid treatment. As for limitations, although we found specific bacteria can affect the overall cellular level of CAT in keloid fibroblasts, the source of CAT was still uncertain. Since bacteria might secrete CAT in the environment and affect the expression of CAT in the host cells simultaneously, it is difficult to exclude either possibility. Thus, it is worth further researched with more advanced technologies to distinguish the source of CAT in the future. In addition, we mainly collected samples in our hospital for this study. There may be heterogeneity between individuals. Region, age, and sex may also have some impacts. More comparative multicenter studies are needed to evaluate the specific roles of bacteria on the keloid disease in the future.
Contributors
Mengjie Shan performed major experiments in this study. Yongsheng Huang, Meng Xiao, Jiyu Xu, Wei Sun, Zerui Wang, Wenbin Du, Xiaoyu Liu, Meng Nie, Lin Wang, Xing wang and Zhi Zheng were involved in some experimental technical supports. Zhengyun Liang, Hao Liu, Yan Hao,Yijun Xia, Lin Zhu, Kexin Song, Cheng Feng, Tian Meng, Zhi Wang and Weifang Cao were involved in collecting clinical samples and clinical data information. Youbin Wang and Yongsheng Huang directed the project and wrote the manuscript. Mengjie Shan, Youbin Wang and Yongsheng Huang verified the underlying data. All authors read and approved the final manuscript.
Data sharing statement
All relevant data are within the paper and its supplementary files. The data used to support the findings of this study are available from the corresponding author. The raw data used and/or analysed during the study are deposited in the iProX repository, accession number IPX0004021000.
Declaration of interests
The authors declare no conflicts of interest related to this research.
Acknowledgements
This study was supported by the National High Level Hospital Clinical Research Funding (LY22B0400004653), National Natural Science Foundation of China (81871538), Beijing Municipal Commission of Science and Technology (Z191100006619009) and Beijing Natural Science Foundation (7232105).
Footnotes
Supplementary data related to this article can be found at https://doi.org/10.1016/j.ebiom.2023.104904.
Contributor Information
Youbin Wang, Email: wybenz@sina.com.
Yongsheng Huang, Email: yongsheng@ibms.pumc.edu.cn.
Appendix A. Supplementary data
References
- 1.Andrews J.P., Marttala J., Macarak E., Rosenbloom J., Uitto J. Keloids: the paradigm of skin fibrosis - pathomechanisms and treatment. Matrix Biol. 2016;51:37–46. doi: 10.1016/j.matbio.2016.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ogawa R. Keloid and hypertrophic scars are the result of chronic inflammation in the reticular dermis. Int J Mol Sci. 2017;18(3):606. doi: 10.3390/ijms18030606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Huang C., Ogawa R. Pharmacological treatment for keloids. Expert Opin Pharmacother. 2013;14(15):2087–2100. doi: 10.1517/14656566.2013.826651. [DOI] [PubMed] [Google Scholar]
- 4.Al Aboud D.M., Badri T. StatPearls [Internet] StatPearls Publishing; Treasure Island (FL): 2023. Acne keloidalis nuchae. [PubMed] [Google Scholar]
- 5.Mimouni-Bloch A., Metzker A., Mimouni M. Severe folliculitis with keloid scars induced by wax epilation in adolescents. Cutis. 1997;59(1):41–42. [PubMed] [Google Scholar]
- 6.Ferček I., Lugović-Mihić L., Tambić-Andrašević A., et al. Features of the skin microbiota in common inflammatory skin diseases. Life (Basel) 2021;11:962. doi: 10.3390/life11090962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Berman B., Maderal A., Raphael B. Keloids and hypertrophic scars: pathophysiology, classification, and treatment. Dermatol Surg. 2017;43 Suppl 1:S3–S18. doi: 10.1097/DSS.0000000000000819. [DOI] [PubMed] [Google Scholar]
- 8.Shan M., Liu H., Song K., Liu S., Hao Y., Wang Y. Immune-related gene expression in skin, inflamed and keloid tissue from patients with keloids. Oncol Lett. 2022;23(2):72. doi: 10.3892/ol.2022.13192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang C.H., Shan M.J., Liu H., et al. Hyperbaric oxygen treatment on keloid tumor immune gene expression. Chin Med J (Engl) 2021;134(18):2205–2213. doi: 10.1097/CM9.0000000000001780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lu Y.Y., Wu C.H., Hong C.H., Chang K.L., Lee C.H. GLUT-1 enhances glycolysis, oxidative stress, and fibroblast proliferation in keloid. Life (Basel) 2021;11(6):505. doi: 10.3390/life11060505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Alfonso-Prieto M., Biarnés X., Vidossich P., Rovira C. The molecular mechanism of the catalase reaction. J Am Chem Soc. 2009;131(33):11751–11761. doi: 10.1021/ja9018572. [DOI] [PubMed] [Google Scholar]
- 12.van Dissel J.T., Stikkelbroeck J.J., van Furth R. Differences in the rate of intracellular killing of catalase-negative and catalase-positive Listeria monocytogenes by normal and interferon-gamma-activated macrophages. Scand J Immunol. 1993;37:443–446. doi: 10.1111/j.1365-3083.1993.tb03316.x. [DOI] [PubMed] [Google Scholar]
- 13.Wu Y., Jiang S., Fu Z. Employment of teicoplanin-coated magnetic particles for quantifying gram-positive bacteria via catalase-catalyzed hydrolysis reaction of H(2)O(2) Talanta. 2020;211 doi: 10.1016/j.talanta.2020.120728. [DOI] [PubMed] [Google Scholar]
- 14.Gankande T.U., Wood F.M., Edgar D.W., et al. A modified Vancouver Scar Scale linked with TBSA (mVSS-TBSA): inter-rater reliability of an innovative burn scar assessment method. Burns. 2013;39(6):1142–1149. doi: 10.1016/j.burns.2013.01.014. [DOI] [PubMed] [Google Scholar]
- 15.Nejman D., Livyatan I., Fuks G., et al. The human tumor microbiome is composed of tumor type-specific intracellular bacteria. Science. 2020;368(6494):973–980. doi: 10.1126/science.aay9189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang W., Zhai S., Xia Y., et al. Ochratoxin A induces liver inflammation: involvement of intestinal microbiota. Microbiome. 2019;7(1):151. doi: 10.1186/s40168-019-0761-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Callahan B.J., McMurdie P.J., Rosen M.J., Han A.W., Johnson A.J., Holmes S.P. DADA2: high-resolution sample inference from Illumina amplicon data. Nat Methods. 2016;13(7):581–583. doi: 10.1038/nmeth.3869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Qiao Y., Zhao X., Zhu J., et al. Fluorescence-activated droplet sorting of lipolytic microorganisms using a compact optical system. Lab Chip. 2017;18(1):190–196. doi: 10.1039/c7lc00993c. [DOI] [PubMed] [Google Scholar]
- 19.Kelly B.J., Gross R., Bittinger K., et al. Power and sample-size estimation for microbiome studies using pairwise distances and PERMANOVA. Bioinformatics. 2015;31(15):2461–2468. doi: 10.1093/bioinformatics/btv183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shan M., Liu H., Hao Y., et al. Metabolomic profiling reveals that 5-hydroxylysine and 1-methylnicotinamide are metabolic indicators of keloid severity. Front Genet. 2021;12 doi: 10.3389/fgene.2021.804248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Goncalves R.L., Bunik V.I., Brand M.D. Production of superoxide/hydrogen peroxide by the mitochondrial 2-oxoadipate dehydrogenase complex. Free Radic Biol Med. 2016;91:247–255. doi: 10.1016/j.freeradbiomed.2015.12.020. [DOI] [PubMed] [Google Scholar]
- 22.Kim-Shapiro D.B., Gladwin M.T. Arginine for mitochondrial oxidative enzymopathy. Blood. 2020;136:1376–1378. doi: 10.1182/blood.2020006732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Xie X., Yu H., Wang Y., et al. Nicotinamide N-methyltransferase enhances the capacity of tumorigenesis associated with the promotion of cell cycle progression in human colorectal cancer cells. Arch Biochem Biophys. 2014;564:52–66. doi: 10.1016/j.abb.2014.08.017. [DOI] [PubMed] [Google Scholar]
- 24.Glorieux C., Calderon P.B. Catalase, a remarkable enzyme: targeting the oldest antioxidant enzyme to find a new cancer treatment approach. Biol Chem. 2017;398(10):1095–1108. doi: 10.1515/hsz-2017-0131. [DOI] [PubMed] [Google Scholar]
- 25.Feng C., Shan M., Xia Y., et al. Single-cell RNA sequencing reveals distinct immunology profiles in human keloid. Front Immunol. 2022;13 doi: 10.3389/fimmu.2022.940645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sasaki Y., Takagane K., Konno T., et al. Expression of asporin reprograms cancer cells to acquire resistance to oxidative stress. Cancer Sci. 2021;112:1251–1261. doi: 10.1111/cas.14794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sun X.J., Ma W.Q., Zhu Y., Liu N.F. POSTN promotes diabetic vascular calcification by interfering with autophagic flux. Cell Signal. 2021;83 doi: 10.1016/j.cellsig.2021.109983. [DOI] [PubMed] [Google Scholar]
- 28.Wulandari E., Jusman S.W., Moenadjat Y., Jusuf A.A., Sadikin M. Expressions of collagen I and III in hypoxic keloid tissue. Kobe J Med Sci. 2016;62(3):E58–E69. [PMC free article] [PubMed] [Google Scholar]
- 29.Ramos M.L., Gragnani A., Ferreira L.M. Is there an ideal animal model to study hypertrophic scarring. J Burn Care Res. 2008;29:363–368. doi: 10.1097/BCR.0b013e3181667557. [DOI] [PubMed] [Google Scholar]
- 30.van den Broek L.J., Limandjaja G.C., Niessen F.B., Gibbs S. Human hypertrophic and keloid scar models: principles, limitations and future challenges from a tissue engineering perspective. Exp Dermatol. 2014;23:382–386. doi: 10.1111/exd.12419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bagabir R., Byers R.J., Chaudhry I.H., Müller W., Paus R., Bayat A. Site-specific immunophenotyping of keloid disease demonstrates immune upregulation and the presence of lymphoid aggregates. Br J Dermatol. 2012;167(5):1053–1066. doi: 10.1111/j.1365-2133.2012.11190.x. [DOI] [PubMed] [Google Scholar]
- 32.Limandjaja G.C., Niessen F.B., Scheper R.J., Gibbs S. The keloid disorder: heterogeneity, histopathology, mechanisms and models. Front Cell Dev Biol. 2020;8:360. doi: 10.3389/fcell.2020.00360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.McGinty S., Siddiqui W.J. StatPearls [Internet] StatPearls Publishing; Treasure Island (FL): 2023. Keloid. [Google Scholar]
- 34.Ketchum L.D., Cohen I.K., Masters F.W. Hypertrophic scars and keloids. A collective review. Plast Reconstr Surg. 1974;53(2):140–154. doi: 10.1097/00006534-197402000-00004. [DOI] [PubMed] [Google Scholar]
- 35.Shan M., Wang Y. Viewing keloids within the immune microenvironment. Am J Transl Res. 2022;14(2):718–727. [PMC free article] [PubMed] [Google Scholar]
- 36.Egert M., Simmering R., Riedel C.U. The association of the skin microbiota with health, immunity, and disease. Clin Pharmacol Ther. 2017;102:62–69. doi: 10.1002/cpt.698. [DOI] [PubMed] [Google Scholar]
- 37.Hay R.J., Johns N.E., Williams H.C., et al. The global burden of skin disease in 2010: an analysis of the prevalence and impact of skin conditions. J Invest Dermatol. 2014;134:1527–1534. doi: 10.1038/jid.2013.446. [DOI] [PubMed] [Google Scholar]
- 38.Kobayashi S.D., Malachowa N., DeLeo F.R. Pathogenesis of Staphylococcus aureus abscesses. Am J Pathol. 2015;185(6):1518–1527. doi: 10.1016/j.ajpath.2014.11.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kanwar I.L., Haider T., Kumari A., Dubey S., Jain P., Soni V. Models for acne: a comprehensive study. Drug Discov Ther. 2018;12(6):329–340. doi: 10.5582/ddt.2018.01079. [DOI] [PubMed] [Google Scholar]
- 40.Hillier A., Lloyd D.H., Weese J.S., et al. Guidelines for the diagnosis and antimicrobial therapy of canine superficial bacterial folliculitis (Antimicrobial Guidelines Working Group of the International Society for Companion Animal Infectious Diseases) Vet Dermatol. 2014;25(3):163-e43. doi: 10.1111/vde.12118. [DOI] [PubMed] [Google Scholar]
- 41.Kim J., Kim J., Bae J.S. ROS homeostasis and metabolism: a critical liaison for cancer therapy. Exp Mol Med. 2016;48 doi: 10.1038/emm.2016.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Perillo B., Di Zazzo E., Giovannelli P., et al. ROS in cancer therapy: the bright side of the moon. Exp Mol Med. 2020;52:192–203. doi: 10.1038/s12276-020-0384-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gao L., Loveless J., Shay C., Teng Y. Targeting ROS-mediated crosstalk between autophagy and apoptosis in cancer. Adv Exp Med Biol. 2020;1260:1–12. doi: 10.1007/978-3-030-42667-5_1. [DOI] [PubMed] [Google Scholar]
- 44.Reuter S., Gupta S.C., Chaturvedi M.M., Aggarwal B.B. Oxidative stress, inflammation, and cancer: how are they linked. Free Radic Biol Med. 2010;49:1603–1616. doi: 10.1016/j.freeradbiomed.2010.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Goyal M.M., Basak A. Human catalase: looking for complete identity. Protein Cell. 2010;1(10):888–897. doi: 10.1007/s13238-010-0113-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cobanoglu U., Demir H., Duran M., Şehitogullari A., Mergan D., Demir C. Erythrocyte catalase and carbonic anhydrase activities in lung cancer. Asian Pac J Cancer Prev. 2010;11(5):1377–1382. [PubMed] [Google Scholar]
- 47.Galasso M., Gambino S., Romanelli M.G., Donadelli M., Scupoli M.T. Browsing the oldest antioxidant enzyme: catalase and its multiple regulation in cancer. Free Radic Biol Med. 2021;172:264–272. doi: 10.1016/j.freeradbiomed.2021.06.010. [DOI] [PubMed] [Google Scholar]
- 48.Liang Y., Jiang Y., Jin X., et al. Neddylation inhibition activates the protective autophagy through NF-κB-catalase-ATF3 axis in human esophageal cancer cells. Cell Commun Signal. 2020;18(1):72. doi: 10.1186/s12964-020-00576-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lu Z., Sethu R., Imlay J.A. Endogenous superoxide is a key effector of the oxygen sensitivity of a model obligate anaerobe. Proc Natl Acad Sci U S A. 2018;115(14):E3266–E3275. doi: 10.1073/pnas.1800120115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pascual J., Foesel B.U., Geppert A., et al. Roseisolibacter agri gen. nov., sp. nov., a novel slow-growing member of the under-represented phylum Gemmatimonadetes. Int J Syst Evol Microbiol. 2018;68(4):1028–1036. doi: 10.1099/ijsem.0.002619. [DOI] [PubMed] [Google Scholar]
- 51.Satapute P., Paidi M.K., Kurjogi M., Jogaiah S. Physiological adaptation and spectral annotation of Arsenic and Cadmium heavy metal-resistant and susceptible strain Pseudomonas taiwanensis. Environ Pollut. 2019;251:555–563. doi: 10.1016/j.envpol.2019.05.054. [DOI] [PubMed] [Google Scholar]
- 52.Zhang J., Shi Q., Fan S., Zhang Y., Zhang M., Zhang J. Distinction between Cr and other heavy-metal-resistant bacteria involved in C/N cycling in contaminated soils of copper producing sites. J Hazard Mater. 2021;402 doi: 10.1016/j.jhazmat.2020.123454. [DOI] [PubMed] [Google Scholar]
- 53.Harmon S.M., Kautter D.A. Recovery of clostridia on catalase-treated plating media. Appl Environ Microbiol. 1977;33(4):762–770. doi: 10.1128/aem.33.4.762-770.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Han W., Fessel J.P., Sherrill T., Kocurek E.G., Yull F.E., Blackwell T.S. Enhanced expression of catalase in mitochondria modulates NF-κB-dependent lung inflammation through alteration of metabolic activity in macrophages. J Immunol. 2020;205(4):1125–1134. doi: 10.4049/jimmunol.1900820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Cong W., Ruan D., Xuan Y., et al. Cardiac-specific overexpression of catalase prevents diabetes-induced pathological changes by inhibiting NF-κB signaling activation in the heart. J Mol Cell Cardiol. 2015;89(Pt B):314–325. doi: 10.1016/j.yjmcc.2015.10.010. [DOI] [PubMed] [Google Scholar]
- 56.Song M., Bode A.M., Dong Z., Lee M.H. AKT as a therapeutic target for cancer. Cancer Res. 2019;79(6):1019–1031. doi: 10.1158/0008-5472.CAN-18-2738. [DOI] [PubMed] [Google Scholar]
- 57.Glorieux C., Zamocky M., Sandoval J.M., Verrax J., Calderon P.B. Regulation of catalase expression in healthy and cancerous cells. Free Radic Biol Med. 2015;87:84–97. doi: 10.1016/j.freeradbiomed.2015.06.017. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.