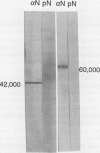
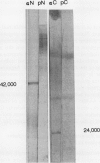
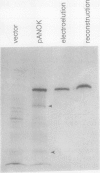
Abstract
We have examined soluble oat (Avena sativa) arginine decarboxylase by probing its structure with polyclonal antibodies that separately recognize amino-terminal and carboxyl-terminal antigens and with a monoclonal antibody that immunoprecipitates enzyme activity. These experiments indicated that oat arginine decarboxylase is clipped from a 66,000-D precursor polypeptide into 42,000- and 24,000-D produce polypeptides. Both of these are found in the enzyme and may be held together by disulfide bonds. A full-length precursor protein could not be detected in plants but could be produced by expression of the cDNA in Escherichia coli. Analysis of the expression of the cDNA in E. coli, with antibodies and using pulse labeling with [35S]methionine, indicated that the bulk of the expressed protein was the full-length 66,000-D form. Small amounts of 42,000- and 24,000-D polypeptides could also be detected. A reconstruction experiment, adding a radioactively labeled full-length protein isolated from E. coli to powdered oat leaves, supported the idea that the protein extraction method used for western blots was not likely to result in artifactual proteolytic degradation.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bell E., Malmberg R. L. Analysis of a cDNA encoding arginine decarboxylase from oat reveals similarity to the Escherichia coli arginine decarboxylase and evidence of protein processing. Mol Gen Genet. 1990 Dec;224(3):431–436. doi: 10.1007/BF00262438. [DOI] [PubMed] [Google Scholar]
- Bitonti A. J., Casara P. J., McCann P. P., Bey P. Catalytic irreversible inhibition of bacterial and plant arginine decarboxylase activities by novel substrate and product analogues. Biochem J. 1987 Feb 15;242(1):69–74. doi: 10.1042/bj2420069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradford M. M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976 May 7;72:248–254. doi: 10.1006/abio.1976.9999. [DOI] [PubMed] [Google Scholar]
- Buch J. K., Boyle S. M. Biosynthetic arginine decarboxylase in Escherichia coli is synthesized as a precursor and located in the cell envelope. J Bacteriol. 1985 Aug;163(2):522–527. doi: 10.1128/jb.163.2.522-527.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiatt A. C., McIndoo J., Malmberg R. L. Regulation of polyamine biosynthesis in tobacco. Effects of inhibitors and exogenous polyamines on arginine decarboxylase, ornithine decarboxylase, and S-adenosylmethionine decarboxylase. J Biol Chem. 1986 Jan 25;261(3):1293–1298. [PubMed] [Google Scholar]
- Hiatt A., Malmberg R. L. Utilization of putrescine in tobacco cell lines resistant to inhibitors of polyamine synthesis. Plant Physiol. 1988 Feb;86(2):441–446. doi: 10.1104/pp.86.2.441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunkapiller M. W., Lujan E., Ostrander F., Hood L. E. Isolation of microgram quantities of proteins from polyacrylamide gels for amino acid sequence analysis. Methods Enzymol. 1983;91:227–236. doi: 10.1016/s0076-6879(83)91019-4. [DOI] [PubMed] [Google Scholar]
- Koerner T. J., Hill J. E., Myers A. M., Tzagoloff A. High-expression vectors with multiple cloning sites for construction of trpE fusion genes: pATH vectors. Methods Enzymol. 1991;194:477–490. doi: 10.1016/0076-6879(91)94036-c. [DOI] [PubMed] [Google Scholar]
- Moore R. C., Boyle S. M. Nucleotide sequence and analysis of the speA gene encoding biosynthetic arginine decarboxylase in Escherichia coli. J Bacteriol. 1990 Aug;172(8):4631–4640. doi: 10.1128/jb.172.8.4631-4640.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polson A., Coetzer T., Kruger J., von Maltzahn E., van der Merwe K. J. Improvements in the isolation of IgY from the yolks of eggs laid by immunized hens. Immunol Invest. 1985 Aug;14(4):323–327. doi: 10.3109/08820138509022667. [DOI] [PubMed] [Google Scholar]
- Ramakrishna S., Adiga P. R. Arginine decarboxylase from Lathyrus sativus seedlings. Purification and properites. Eur J Biochem. 1975 Nov 15;59(2):377–386. doi: 10.1111/j.1432-1033.1975.tb02465.x. [DOI] [PubMed] [Google Scholar]
- Studier F. W., Rosenberg A. H., Dunn J. J., Dubendorff J. W. Use of T7 RNA polymerase to direct expression of cloned genes. Methods Enzymol. 1990;185:60–89. doi: 10.1016/0076-6879(90)85008-c. [DOI] [PubMed] [Google Scholar]
- Wallace R. A., Jared D. W., Nelson B. L. Protein incorporation by isolated amphibian oocytes. I. Preliminary studies. J Exp Zool. 1970 Nov;175(3):259–269. doi: 10.1002/jez.1401750302. [DOI] [PubMed] [Google Scholar]