Significance
Meiotic crossovers (COs) between homologs result in genetic diversity among offspring. The majority of COs are interference sensitive class (class I) CO in most organisms. Class I COs are regulated by a group of proteins including HEI10 (Human Enhancer of Invasion-10), which is hypothesized to diffuse along synapsed homologous chromosomes and concentrate at CO sites in a process known as coarsening. The mechanisms that facilitate and regulate coarsening are elusive. Here, we show that HEI10 forms condensates and directly facilitates the ubiquitination-dependent degradation of Replication Protein A (RPA1a). Since sexually reproducing species typically depend on COs to accurately segregate their chromosomes during meiosis, this mechanism involved in HEI10 and RPA1a is likely to be broadly relevant among most eukaryotes.
Keywords: meiosis, crossover, phase separation, RPA1a, HEI10
Abstract
Reciprocal exchanges of DNA between homologous chromosomes during meiosis, or crossovers (COs), shuffle genetic information in gametes and progeny. In many eukaryotes, the majority of COs (class I COs) are sensitive to a phenomenon called interference, which influences the occurrence of closely spaced double COs. Class I COs depend on a group of factors called ZMM (Zip, Msh, Mer) proteins including HEI10 (Human Enhancer of Invasion-10). However, how these proteins are recruited to class I CO sites is unclear. Here, we show that HEI10 forms foci on chromatin via a liquid–liquid phase separation (LLPS) mechanism that relies on residue Ser70. A HEI10S70F allele results in LLPS failure and a defect in class I CO formation. We further used immunoprecipitation–mass spectrometry to identify RPA1a (Replication Protein A 1) as a HEI10 interacting protein. Surprisingly, we find that RPA1a also undergoes phase separation and its ubiquitination and degradation are directly regulated by HEI10. We also show that HEI10 is required for the condensation of other class I CO factors. Thus, our results provide mechanistic insight into how meiotic class I CO formation is controlled by HEI10 coupling LLPS and ubiquitination.
During meiotic recombination, DNA is reciprocally exchanged between homologous chromosomes to form crossovers (COs) to facilitate chromosome segregation and generate allelic diversity in gametes and progeny (1, 2). Many organisms commonly have two types of CO: class I and class II. Class I COs are sensitive to a phenomenon called interference which influences how closely two COs can occur on the same chromosome (3). Class I COs are mediated by a group of factors known as ZMM proteins which include ZIP4, MER3, PTD, SHOC1, MSH4, MSH5, HEI10 (Human Enhancer of Invasion-10), and HEIP1 in plants (4, 5). Class II COs are insensitive to interference and depend on MUS81 or FANCD2 (6–8). However, the underlying mechanism that recruits these proteins to CO sites and differentiates between the two CO pathways is elusive.
HEI10 is a conserved eukaryotic really interesting new gene (RING)-domain containing protein that is required for class I CO formation in several species (9–12). During meiosis, hundreds of HEI10 foci form along chromosomes at zygotene, and microscopic images suggest that HEI10 protein concentrates at CO sites at late pachytene (13, 14). HEI10 is also known to colocalize with several class I CO proteins. In Sordaria, HEI10 colocalizes with ZIP4, MER3, and MSH4 (13). In mice and Arabidopsis thaliana, HEI10 colocalizes with MLH1 (15, 16); and rice HEI10 colocalizes with HEIP1 (5). The colocalization implies that they may have a role in promoting CO formation. Recent studies have hypothesized that HEI10 diffuses along the synaptonemal complex (SC) and drives a coarsening process that controls CO numbers and interference in a dosage-dependent manner (14, 15). In addition, HEI10 has a conserved RING domain and has been shown to have ubiquitin E3 ligase activity in humans (17). In mice, HEI10 together with RNF212 act in a small ubiquitin-like modifier (SUMO)-ubiquitin-proteasome pathway to regulate recombination during CO/non-CO differentiation (16, 18). However, the HEI10 ubiquitinated targets remain to be determined.
Liquid–liquid phase separation (LLPS) generates membraneless condensates that often serve as cellular reaction hotspots. LLPS can concentrate various components to promote biochemical reactions such as transcription, chromatin organization, and DNA damage response (19–21). To date, few studies have explored the correlation between meiotic progression and biomolecular condensates. In Saccharomyces cerevisiae, RMM (Rec114–Mei4 and Mer2) proteins recruit the Spo11 (Sporulation 11) core complex to DSB (double stand break) hotspots by forming condensates on DNA (22). In Caenorhabditis elegans, the SC exhibiting liquid crystal-like behaviors are inhibited by 1,6-hexanediol, which can disrupt hydrophobic interaction and prevent phase separation (23). In addition, the formation of condensates by LLPS is biologically regulated by mechanisms including posttranslational modification such as ubiquitination. The yeast E3 ubiquitin ligase Bre1 binds the scaffold protein Lge1 to form condensates that facilitate the ubiquitination of substrate H2B (24).
Here, we demonstrated that Arabidopsis HEI10 undergoes phase separation in vitro and in vivo and that residue Ser70 of HEI10 is required for its condensation and its meiotic recombination activity. We verified that HEI10 acts as a ubiquitin E3 ligase and directly interacts with RPA1a to promote its ubiquitination and degradation. Strikingly, we provided evidence that class I CO proteins form phase-separated behaviors in a HEI10-dependent manner. Thus, we provide mechanistic insight for how phase separation helps to determine meiotic class I CO formation.
Results
HEI10 Can Undergo LLPS.
HEI10 proteins form foci on meiotic chromosomes that are hypothesized to diffuse and join to form larger clusters in a process known as coarsening (13, 14). Since the dynamics of HEI10 accumulation and localization are similar to those seen in protein LLPS (19), we examined the amino acid sequence of HEI10 and found a predicted intrinsically disordered region (IDR) in its C terminus (SI Appendix, Fig. S1A). IDRs are thought to promote phase separation (25). To examine whether HEI10 can undergo LLPS, we expressed HEI10-Flag in tobacco cells to observe its localization with or without exposure to MMS (methyl methanesulfonate), an inducer of DNA double-strand breaks (26). We found that MMS treatment obviously induces HEI10-Flag proteins to form bright puncta in nuclei (Fig. 1A). We used an in vivo FRAP (Fluorescence Recovery After Photobleaching) assay to assess the liquidity of HEI10 puncta by expressing HEI10-YFP in tobacco treated with MMS (Fig. 1B and SI Appendix, Fig. S1B). After laser bleaching, we observed a gradual recovery of brightness in the HEI10-YFP puncta from 37.12 ± 6.47% intensity to 55.33 ± 8.06% (n = 10) over a 90-s time course (Fig. 1 B and D), supporting the idea that the HEI10-YFP puncta are phase-separated droplets. We also performed FRAP with purified recombinant mRFP-HEI10 protein and observed spherical red droplets in a 10% (w/v) PEG8000 solution (Fig. 1C and SI Appendix, Fig. S1C) that gradually recover after photobleaching from 34.54 ± 5.12% to 70.21 ± 12.47% (n = 11) over a 210-s time course (Fig. 1E). In addition, we are able to observe the gradual fusion of the HEI10 droplets over time (Fig. 1F). These results demonstrate that HEI10 proteins have the ability to undergo LLPS.
Fig. 1.
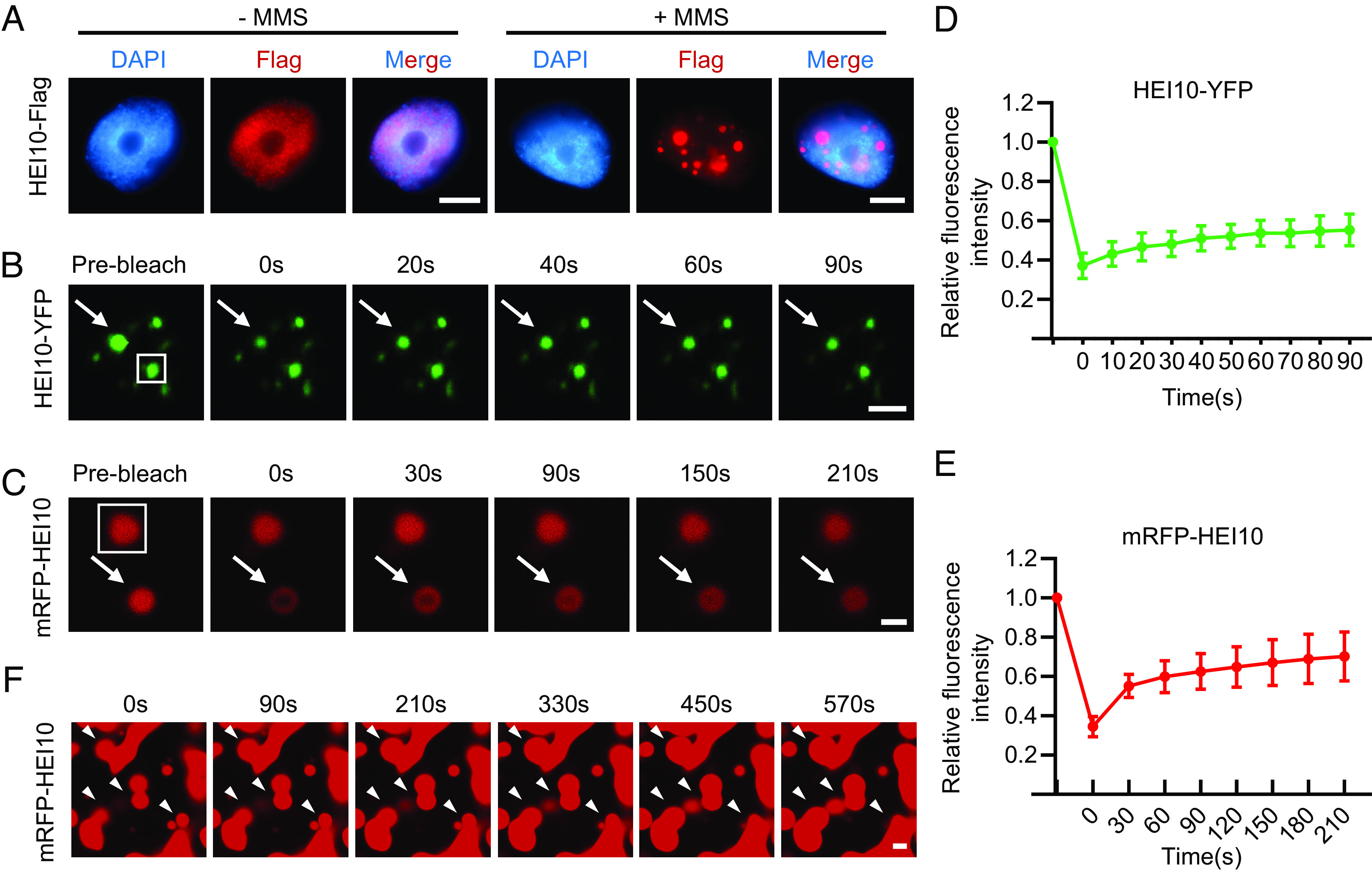
HEI10 can undergo LLPS. (A) Immunostaining of tobacco nuclei expressing HEI10-Flag with or without MMS treatment. HEI10-Flag forms condensates in nuclei when the leaves are treated with MMS for 1.5 h. (Scale bar, 5 μm.) (B) FRAP of HEI10-YFP nuclear bodies in tobacco cells after treated with MMS. 0 s is the time stop photobleaching. (Scale bar, 2 μm.) The white box indicates the unbleached control, and arrows indicate the bleached droplet. (C) FRAP assay for mRFP-HEI10 droplets in the presence of 10% (w/v) PEG8000. 0 s is the time stop photobleaching. (Scale bar, 2 μm.) The white box indicates the unbleached control, and arrows indicate the bleached droplet. (D) Plot showing the time course of the recovery after photobleaching HEI10-YFP nuclear bodies in tobacco. Data are presented as mean ± SD (n = 10). (E) Plot showing the time course of the recovery after photobleaching mRFP-HEI10 droplets. Data are presented as mean ± SD (n = 11). (F) Fusion of mRFP-HEI10 droplets in the presence of 10% (w/v) PEG8000. (Scale bar, 2 μm.) Arrowheads indicate the fusion protein.
Residue S70 of HEI10 Is Required for Phase Separation and Meiotic Recombination.
To investigate the roles of HEI10 in meiotic recombination, we screened an ethylmethane sulfonate mutagenized population of wild-type Arabidopsis (Col-0) and identified several mutants with meiotic recombination defects (27). We used bulked-segregant analysis to map a mutant harboring a missense mutation in the fourth exon of HEI10, which changes a threonine-cysteine-threonine codon to threonine-threonine-threonine, leading to a substitution of serine (S) with phenylalanine (F) at amino acid position 70 (SI Appendix, Fig. S2A). Sequence alignment showed that HEI10 Ser70 is conserved among vascular plants (SI Appendix, Fig. S2B). hei10S70F plants have reduced fertility, with shorter siliques, and fewer viable pollen grains compared to wild type (SI Appendix, Fig. S2 C–E). In hei10S70F meiocytes, the early stages of meiosis (until diplotene) are indistinguishable from those of Col-0 (SI Appendix, Fig. S2F). At diakinesis, the mutants have some unpaired univalents instead of the five bivalents observed in Col-0. After the second meiotic division, the tetrad-stage microspores contain aberrant numbers of chromosomes (SI Appendix, Fig. S2F). To validate that the phenotypes we observed are caused by the S70F mutation, we crossed hei10S70F with heterozygotes of a T-DNA insertional allele hei10-2 (9). The resulting heteroallelic heterozygotes (hei10S70F−/hei10-2−) have meiotic phenotypes similar to both hei10S70F and hei10-2 (SI Appendix, Fig. S2F).
We examined the meiotic localization of HEI10 and HEI10S70F using a HEI10 antibody with meiocytes from mutant and Col-0 plants. In Col-0, HEI10 forms an average of 9.51 foci (n = 39) at diakinesis, which are thought to correspond to class I CO sites (9, 28). In comparison, HEI10S70F foci remain numerous at pachytene and do not coalesce by diakinesis in hei10S70F meiocytes (SI Appendix, Fig. S2G). We validated the mislocalization of HEI10S70F by analyzing stable HEI10S70F-Flag Arabidopsis transgenic lines. The number of HEI10S70F-Flag foci remains aberrantly high in late pachytene and diplotene, and it does not coalesce at CO sites in diakinesis as is observed with HEI10-Flag (Fig. 2A). Consistent with these observations, when HEI10S70F is expressed in tobacco nuclei, it does not form puncta even when treated with MMS (Fig. 2B). In addition, mRFP-HEI10S70F cannot form droplets in vitro, in contrast to mRFP-HEI10 (Fig. 2C). Together, these results demonstrate that S70 is required for LLPS and normal meiotic localization of HEI10.
Fig. 2.
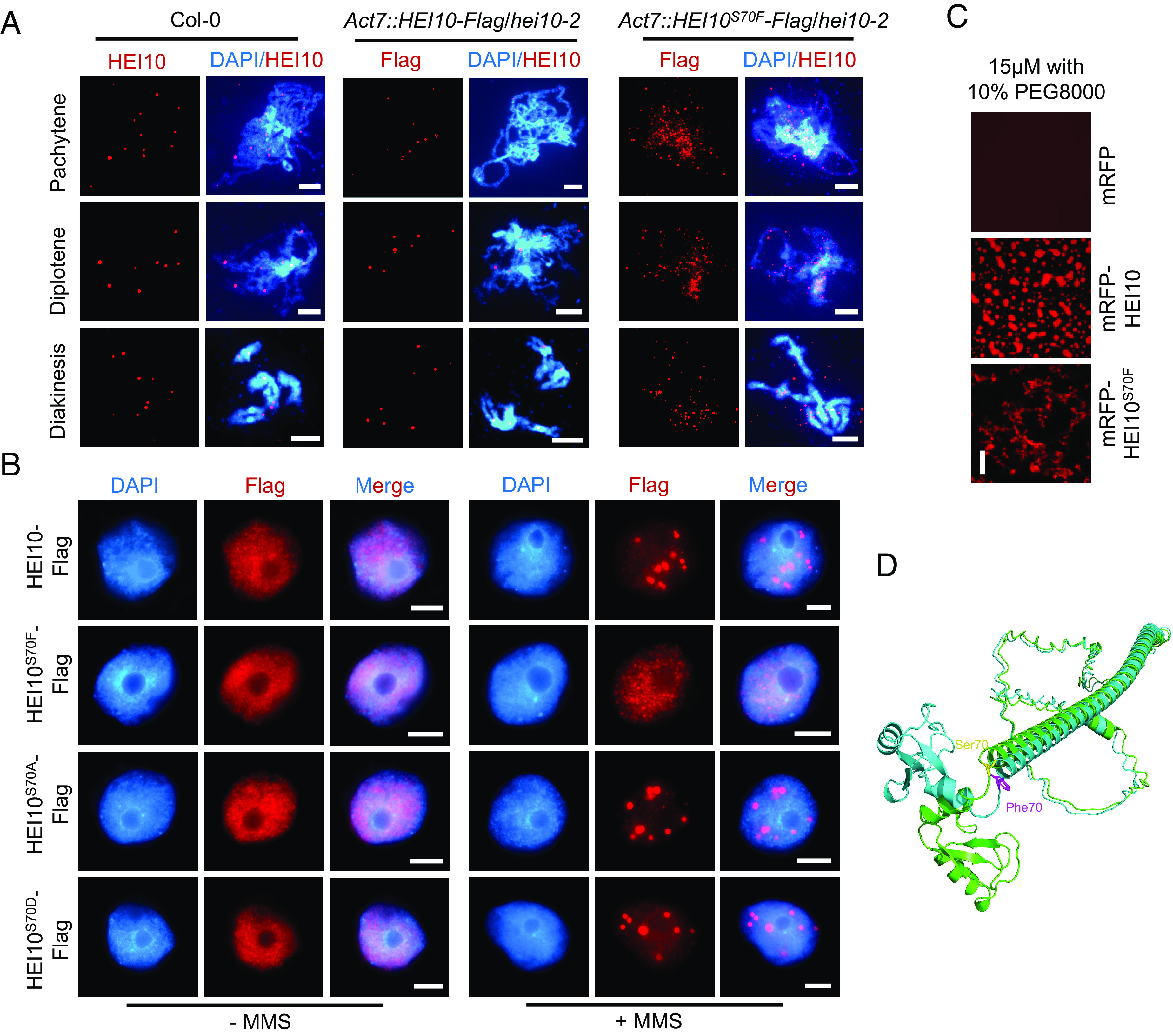
Mutation of S70F affects the phase separation of HEI10. (A) Immunostaining of HEI10 at pachytene, diplotene, and diakinesis in Col-0, HEI10-Flag, and HEI10S70F-Flag transgenic plants. (Scale bar, 5 μm.) (B) Immunostaining of HEI10-Flag and HEI10S70F/A/D-Flag in tobacco nuclei with or without MMS treatment. The leaves are treated with MMS for 1.5 h. HEI10S70A and HEI10S70D simulate the phospho-dead and phospho-mimetic proteins. (Scale bar, 5 μm.) (C) In vitro phase separation assay of mRFP, mRFP-HEI10, and mRFP-HEI10S70F recombinant proteins, with protein concentrations of 15 μM and 10% PEG8000. (Scale bar, 20 μm.) (D) The 3D structures of HEI10 (green) and HEI10S70F (blue), predicted by ESMFold. The Ser70 (yellow) and the Phe70 (purple) residues are shown at the junction between Ring domain and α-helix by stick representation.
HEI10 Ser70 is predicted to be a phosphorylation site by GPS 5.0, a kinase-specific phosphorylation site prediction tool (29). To test whether HEI10S70 phosphorylation is required for its localization, we generated HEI10S70A phospho-dead and HEI10S70D phospho-mimetic alleles. Unexpectedly, the localization and aggregation of HEI10S70A and HEI10S70D are indistinguishable from wild-type HEI10 (Fig. 2B), suggesting that S70 phosphorylation is not required for HEI10 phase separation. We also used ESMFold to predict the structures of HEI10 and HEI10S70F (30). The general predicted shape of the HEI10S70F resembles that of wild-type HEI10; however, the S70F mutation distorts the α-helix within the C-terminal IDR and induces position and orientation changes of the N terminus (rotation of a larger angle approximately 85°) (Fig. 2D). This may result from the weakened interactions between the helix and the loop caused by the benzene ring of Phe70. While the Ser70 residue can interact with Gln72, Ile73, Leu74, and Met75, the Phe70 residue can only interact with Ile73 and Leu74. (SI Appendix, Fig. S1D). These predicted structural changes provide an explanation for why the S70F mutation in HEI10 impacts its phase separation activity.
HEI10 Interacts with RPA1a In Vitro and In Vivo.
To explore the mechanism for HEI10 LLPS during meiosis, we used immunoprecipitation–mass spectrometry (IP-MS) with proteins extracted from inflorescences of Act7::HEI10-Flag/hei10-2 transgenic plants, to identify proteins involved in the HEI10 phase separation condensates (SI Appendix, Fig. S3A). We identified 1,068 proteins after removing background interactions using Act7::Flag transgenic plants (SI Appendix, Fig. S3B and Dataset S1). KEGG pathway enrichment analysis reveals that DNA replication and DNA repair pathways are enriched among the candidate proteins (SI Appendix, Fig. S3C and Table S1), including several known meiotic proteins such as RPA1a (replication protein A) (31). We used yeast two-hybrid (Y2H), pull down and split luciferase complementation assays to confirm that HEI10 interacts with RPA1a in vitro and in vivo (Fig. 3 A–C).
Fig. 3.
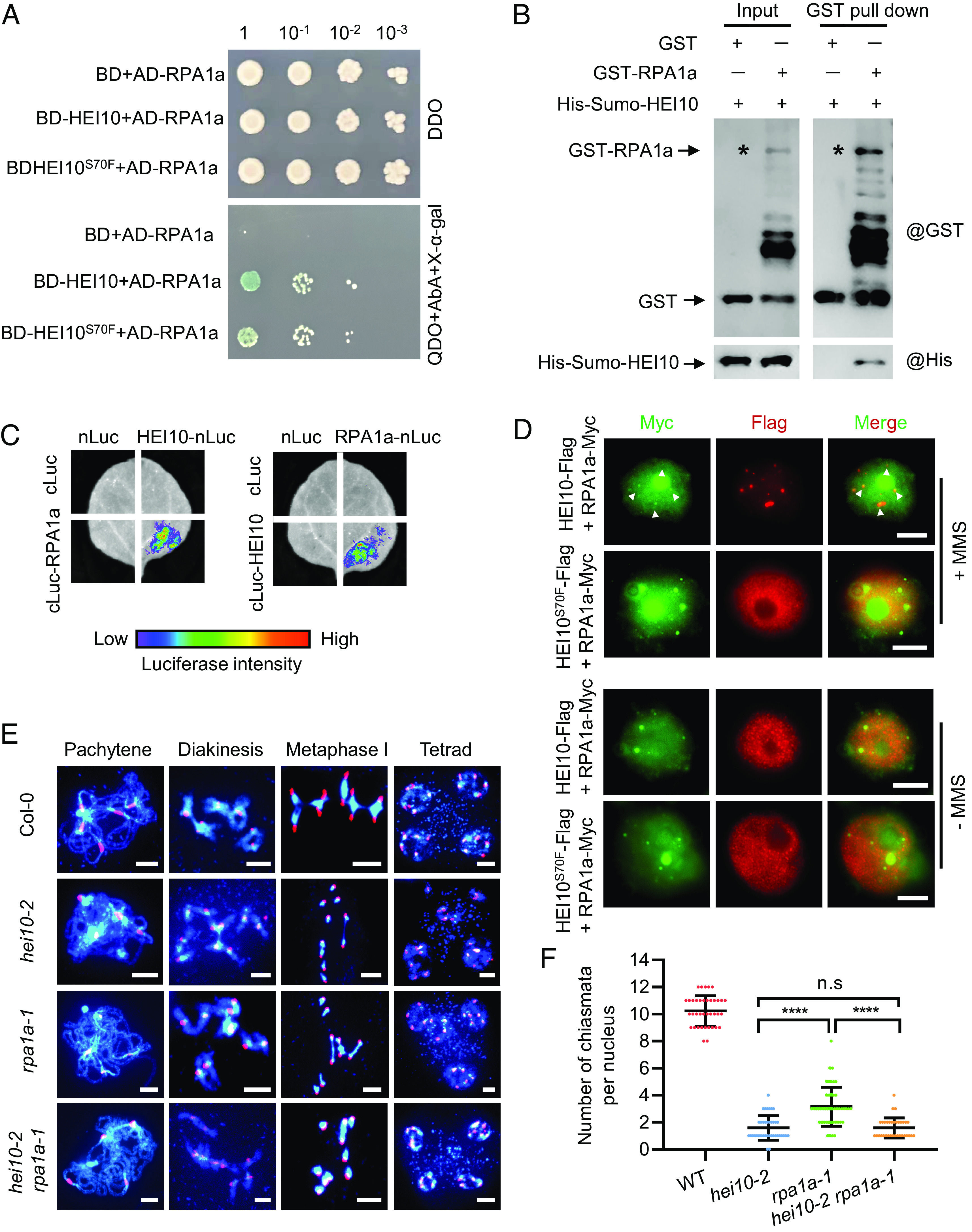
HEI10 interacts with RPA1a in vitro and in vivo. (A) Interactions of HEI10 with RPA1a by yeast two-hybrid using gradient dilution. The initial concentration is OD600 ≈ 1, diluted 10-, 100-, and 1,000-fold, spotted on DDO (double synthetic dropout media lacking leucine and tryptophan) and QDO (quadra synthetic dropout media lacking leucine, tryptophan, histidine, and adenine) for 3 d. (B) Validation of interaction between HEI10 and RPA1a using the affinity purification GST pull-down assay. Asterisks indicate the GST-RPA1a band. (C) Split luciferase complementation imaging assay examines the interaction between HEI10 and RPA1a in tobacco cells. cLUC or nLUC are used as controls. (D) Colocalization of HEI10-Flag with RPA1a-Myc in tobacco nuclei. (Scale bar, 5 μm.) White arrows indicate the RPA1a-Myc signals that merged with HEI10-Flag. (E) Meiotic chromosome phenotypes of Col-0, hei10-2, rpa1a-1, and hei10-2 rpa1a-1 using centromere FISH. (Scale bar, 5 μm.) (F) Statistical analysis of the chiasmata at metaphase I in Col-0 and mutants (****P < 0.0001, two-tailed Student's t test).
We analyzed the colocalization of RPA1a and HEI10 by transiently coexpressing HEI10-Flag or HEI10S70F-Flag with RPA1a-Myc in tobacco cells. HEI10-Flag colocalizes with RPA1a-Myc to form puncta in nuclei after MMS treatment, but HEI10S70F-Flag does not (Fig. 3D). We examined the protein structure of RPA1a and identified a short IDR between its DBD-A and DBD-F domains (SI Appendix, Fig. S4A), which suggests that RPA1a may have the ability to undergo LLPS. We found that RPA1a appears to be able to form condensates in tobacco nuclei independent of HEI10, even in the absence of MMS treatment (SI Appendix, Fig. S4 B and C). This is consistent with a recent study showing that mammalian RPA can undergo phase separation, which is stimulated by single-stranded DNA binding (32). RPA1a is required for class I CO formation in Arabidopsis (31), and rpa1a-1 mutants have multiple univalents at diakinesis and decreased COs (Fig. 3E). However, we found that the localization and condensation of HEI10 on chromosomes at pachytene in rpa1a-1 is indistinguishable compared with wild type (SI Appendix, Fig. S4 D and E). These results are consistent with previous observations in rice rpa1a (33). Additionally, although hei10 and rpa1a both have reduced class I COs (9, 31), our genetic analyses show that hei10-2 rpa1a-1 double mutants have a similar reduction in COs compared to hei10-2 alone and fewer COs than rpa1a-1 alone (Fig. 3 E and F). These results suggest that HEI10 and RPA1a function in the same pathway for class I CO formation.
HEI10 Facilitates the Ubiquitination of RPA1a.
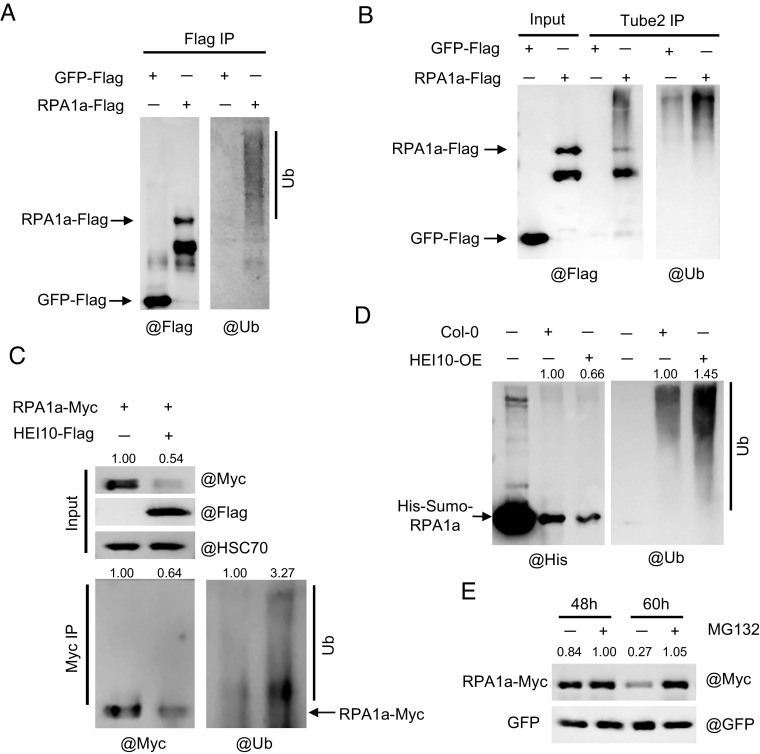
Human HEI10 has E3 ubiquitin ligase activity, and mouse HEI10 is required for turnover of recombination factors (17, 18), suggesting that HEI10 mediated ubiquitination targets proteins for proteolysis. Notably, when HEI10-Flag is coexpressed with RPA1a-Myc in MMS-treated tobacco leaves, the RPA1a-Myc signal is relatively attenuated, compared to its expression alone or when coexpression with HEI10S70F-Flag (Fig. 3D and SI Appendix, Fig. S4B). This suggests that HEI10 may influence the stability of RPA1a. To examine the E3 ubiquitin ligase activity of Arabidopsis HEI10, we used western blots probed with anti-Flag and anti-UBQ11 to detect ubiquitinated forms of HEI10 in protein extracts from tobacco leaves that transiently express HEI10-Flag and Arabidopsis plants that are stably transformed with HEI10-Flag. In both expression systems, we observed a smear of signal above the main HEI10 band with either Flag or UBQ11 antibodies, which may represent ubiquitinated HEI10 (SI Appendix, Fig. S5 A and B). Tube2 agarose beads have specific affinity to ubiquitin and can enrich ubiquitin-modified proteins (34). We used Tube2-IP coupled with the anti-Flag and anti-UBQ11 western blotting and observed similar HEI10 ubiquitination patterns (SI Appendix, Fig. S5C). The ubiquitinated bands of HEI10S70F resemble those of HEI10, indicating that the S70F mutation does not affect the ubiquitination activity of HEI10 (SI Appendix, Fig. S5B). To test whether RPA1a is a ubiquitination target of HEI10, we first demonstrated that a polyubiquitinated form of RPA1a can be detected with anti-UBQ11 antibody using the Flag-IP and Tube2-IP assays from transient expression in tobacco (Fig. 4 A and B). Then, we coexpressed HEI10 and RPA1a in tobacco leaves and found that the protein level of RPA1a decreases, but the ubiquitination level increases, compared to that of RPA1a expressed alone (Fig. 4C). To validate this in an independent assay, we incubated RPA1a recombinant protein extracts from Arabidopsis inflorescences of wild type and HEI10 overexpressing plants (HEI10-OE, Act7::HEI10-Flag/Col-0, SI Appendix, Fig. S3A). After coincubation for 2 h, we found that the ubiquitination level of RPA1a is significantly higher with extracts from HEI10-OE compared with wild type (Fig. 4D). These results support the idea that HEI10 facilitates the ubiquitination of RPA1a.
Fig. 4.
HEI10 promotes RPA1a ubiquitination. (A) Measurement of ubiquitination levels of RPA1a immunoprecipitated from tobacco cell protein extracts by anti-Flag and anti-UBQ11 antibodies. GFP-Flag is used as the negative control. (B) Tube2 IP tests the enrichment of ubiquitin-modified RPA1a-Flag proteins in tobacco cells. GFP-Flag is used as the negative control. (C) Detection of the ubiquitination levels in protein extracts from tobacco cells expressing RPA1a alone or with HEI10. The RPA1a protein level is determined with anti-Myc antibody; the ubiquitination level is determined with anti-UBQ11 antibody. (D) In vitro ubiquitination assay, the recombinant His-Sumo-RPA1a proteins are incubated with the extracts from central inflorescences of Col-0 and HEI10 overexpressing plants (HEI10-OE) at room temperature for 2 h by adding 10 mM ATP plus 50 μM MG132. The samples are analyzed by western blot using anti-His and anti-UBQ11 antibodies. (E) Investigation of the stability of RPA1a proteins treated by 26S proteasome inhibitor MG132 in tobacco cells. The samples are collected at 48 h and 60 h after agrobacterium injection. 50 μM MG132 is injected into the leaves for 12 h before sampling. The protein level of RPA1a-Myc is determined with anti-Myc antibody. GFP is coexpressed with RPA1a as the expression control.
HEI10 Promotes the Degradation of RPA1a in a 26S Proteasome-Dependent Manner.
Polyubiquitination is commonly required for protein degradation by the 26S proteasome (35). To test whether RPA1a is degraded in response to ubiquitination, we expressed RPA1a-Myc in tobacco and found that RPA1a degradation is significantly inhibited by MG132, a 26S proteasome inhibitor (Fig. 4E). We also examined the endogenous RPA1a levels in protein extracts from HEI10-OE, Col-0, hei10S70F, and hei10-2 Arabidopsis inflorescences and found that RPA1a and HEI10 levels are negatively correlated (Fig. 5A). Additionally, the RPA1a level in hei10S70F is higher than Col-0, suggesting that HEI10S70F is compromised in its ability to target RPA1a for degradation. Furthermore, incubation of RPA1a with HEI10 protein expressed in tobacco showed that RPA1a degrades more rapidly when mixed with HEI10, in comparison to the mock control and HEI10S70F (Fig. 5B). In a cell-free degradation assay, the degradation of recombinant RPA1a proteins are also quicker when incubated with proteins extracted from HEI10-OE inflorescences than Col-0 and hei10-2 (Fig. 5C). In all these experiments, HEI10S70F is less effective in degrading RPA1a (Fig. 5 A–C). Thus, we conclude that phase separation may have an important role for facilitating HEI10’s ability to participate in ubiquitination-mediated degradation during meiosis.
Fig. 5.
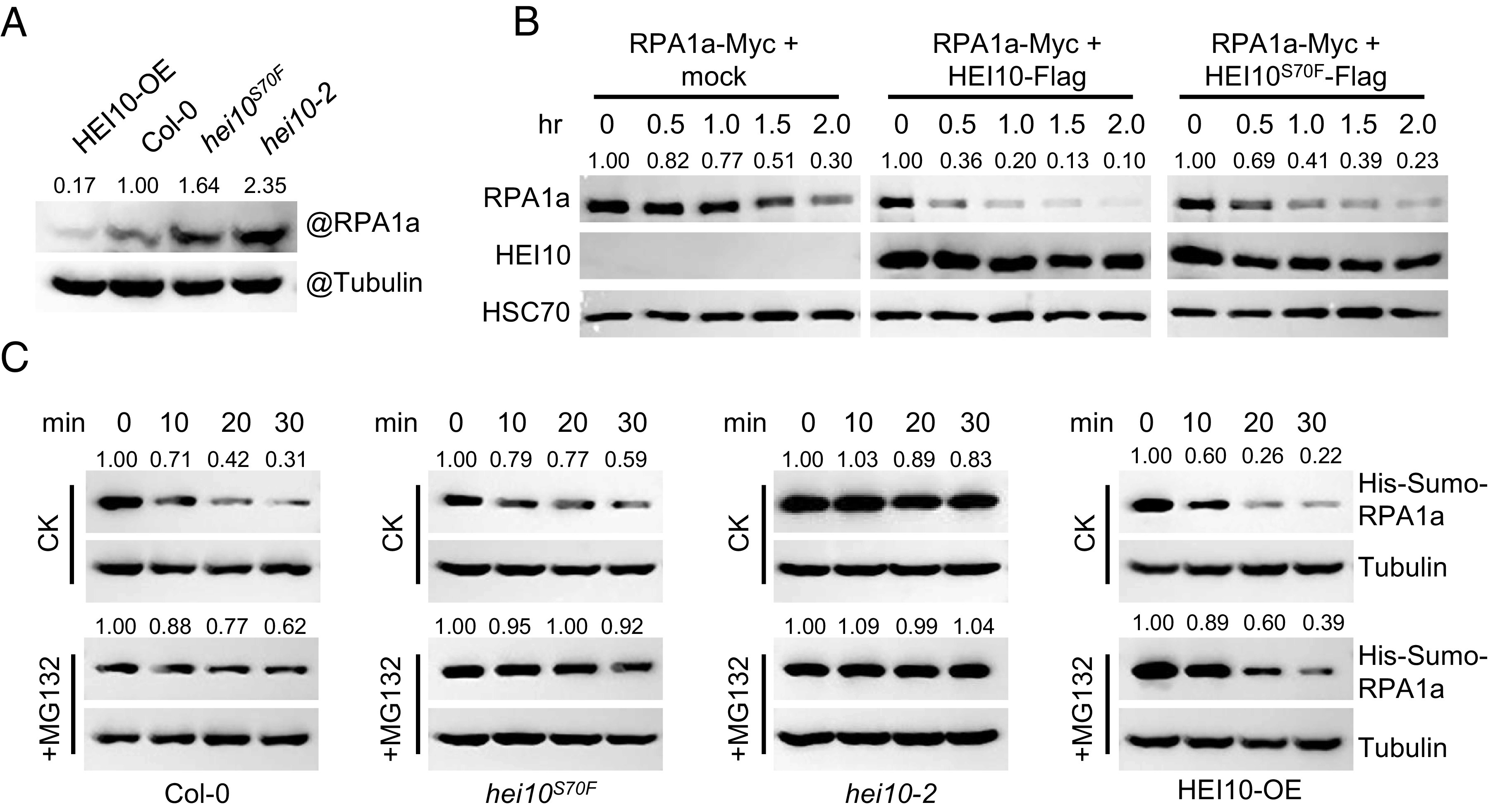
HEI10 phase separation affects the degradation of RPA1a. (A) Measurement of endogenous RPA1a protein level in HEI10-OE (Act7::HEI10-Flag/Col-0), Col-0, hei10S70F, and hei10-2 backgrounds with anti-RPA1a antibody. Tubulin is the loading control. (B) Semi-in vivo protein degradation assay, RPA1a proteins extracted from tobacco cells are mixed with HEI10-Flag, HEI10S70F-Flag, and mock control in a volume ratio of 1:2. The mixtures are incubated at room temperature for different time courses. (C) Cell-free degradation assay, recombinant His-Sumo-RPA1a proteins are incubated with equal amount of central inflorescence extractions of HEI10-OE, Col-0, hei10S70F, and hei10-2, and 100 μM cycloheximide (a protein biosynthesis inhibitor) and 10 mM ATP are added into the buffer and the samples are incubated at room temperature. For MG132 treatment, 50 μM MG132 is added. The protein level of His-Sumo-RPA1a is determined with anti-His antibody. Tubulin is the loading control.
HEI10 and Class I CO Proteins Mutually Promote Each Other’s Condensation.
HEI10 can colocalize with other class I CO proteins such as ZIP4, MSH4, and MLH1 (13, 15, 16). We speculate that the colocalization of these proteins may depend on HEI10 LLPS. To test this hypothesis, we used immunofluorescence assays to examine the localization patterns of ZIP4, MSH5, and MLH3 in tobacco nuclei and found that none forms large condensates even when treated with MMS (Fig. 6A). Using a Y2H assay, we show that HEI10 interacts weakly with MLH3, but not with ZIP4 or MSH5 (SI Appendix, Fig. S6A). However, coexpression of HEI10 with these proteins without MMS treatment results in varying degrees of HEI10 condensation compared to HEI10 expressed alone (Figs. 1A and 6B). In particular, MLH3 and HEI10 clearly form cocondensates when coexpressed (Fig. 6B). Importantly, these class I CO proteins can colocalize with HEI10 to form cocondensates with MMS treatment (Fig. 6B). These results suggest that these class I CO proteins and HEI10 can mutually facilitate each other’s condensation.
Fig. 6.
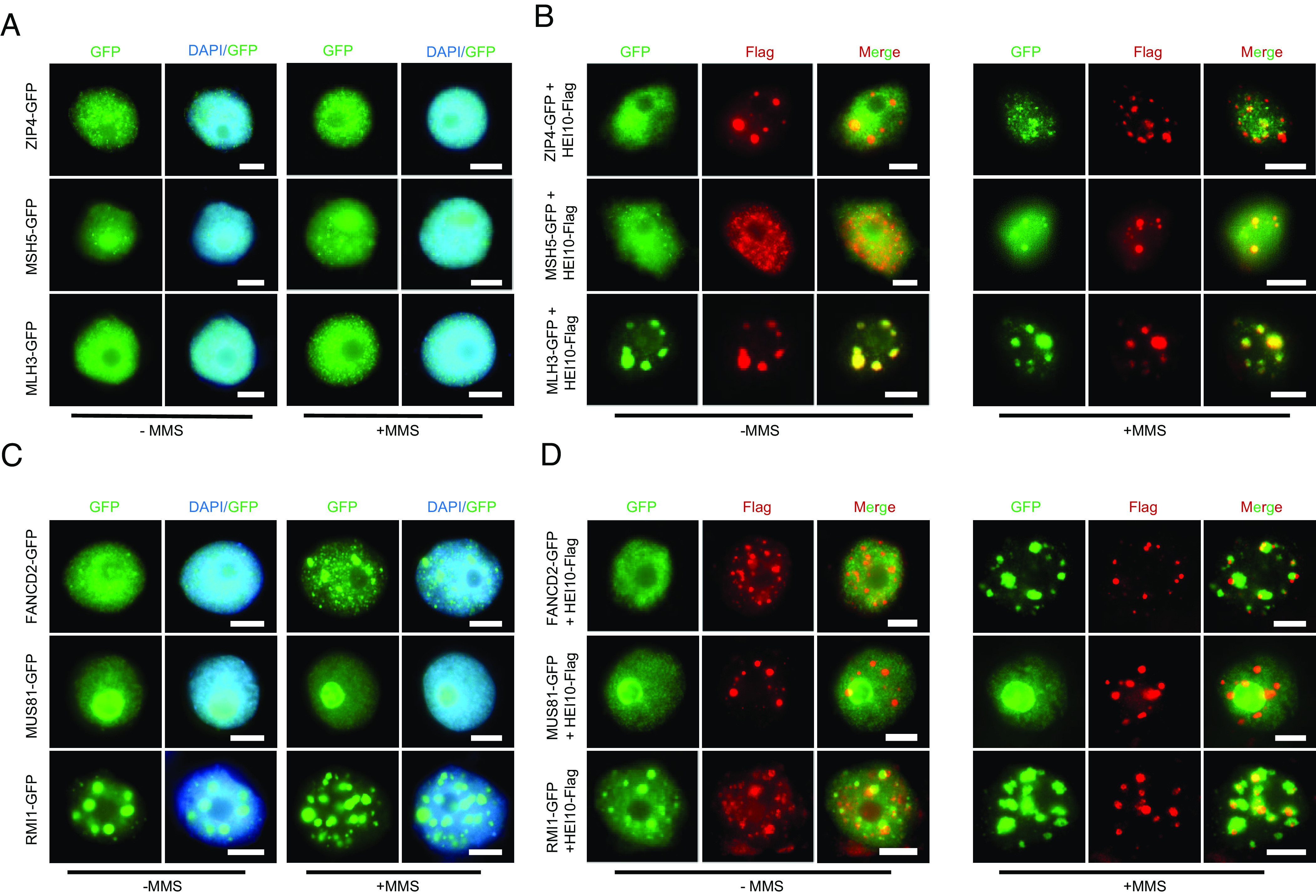
HEI10 and other class I CO proteins mutually promote their condensation. (A) The localization of ZIP4, MSH5, and MLH3 in tobacco nuclei with or without MMS treatment. (B) The colocalization of HEI10 and ZIP4, MSH5, MLH3 in tobacco nuclei with or without MMS treatment. (C) The localization of FANCD2, MUS81, and RMI1 in tobacco nuclei with or without MMS treatment. (D)The colocalization of HEI10 and FANCD2, MUS81, RMI1 in tobacco nuclei with or without MMS treatment. (Scale bar, 5 μm.)
We also investigated the class II CO regulator MUS81 (6), FANCD2 (8), and anti-CO factor RMI1 (36). All three are predicted to have IDRs more or less (SI Appendix, Fig. S6B). When expressed alone in tobacco nuclei, RMI1 can form puncta with or without MMS treatment, FANCD2 can only do so with MMS treatment, while MUS81 seems to lack this ability (Fig. 6C), suggesting that RMI1 and FANCD2 may undergo phase separation, but MUS81 does not. When coexpressed with HEI10, in the absence of MMS, RMI1, MUS81, and FANCD2 can facilitate HEI10 condensation, but not vice versa (Fig. 6D). Strikingly, we found that the localization of these proteins and HEI10 is also mutually unaffected when treated with MMS (Fig. 6D). Although HEI10 condensates seem to partially overlap with FANCD2 and RMI1 signals, their localization patterns are different from the nearly coincident foci of HEI10 and class I proteins following MMS treatment (Fig. 6B). All together, these results suggest that HEI10 LLPS is required for the condensation of class I CO proteins but not for proteins associated with class II COs.
Discussion
In many eukaryotes, the majority of COs are interference-sensitive. Previous studies have identified several proteins that participate in class I CO formation, but how those proteins are concentrated at meiotic CO sites remains unclear. Here, we reveal a LLPS mechanism that coordinates multiple proteins to promote class I CO formation, in which HEI10 is an important regulator of phase separation (Figs. 1 and 2). Interestingly, we found that HEI10 interacts with RPA1a to facilitate its ubiquitination-dependent degradation and that HEI10S70F which is compromised in phase separation is also less efficient at mediating degradation (Figs. 3–5). Moreover, we demonstrated that other class I CO factors contribute to HEI10 condensates, which in turn trigger the condensation of these proteins (Fig. 6). Thus, our results offer insights into how HEI10 plays a role in the formation of class I COs by phase separation by facilitating ubiquitination–degradation processes and by forming cocondensates with class I but not class II CO proteins.
Although phase separation is an exciting emerging mechanism for the formation of protein condensates involved in multiple biological processes, our understanding of its role in meiosis is just beginning. Recent studies found that meiotic DSB formation and chromosome synapsis involve phase separation. In S. cerevisiae, DNA-driven RMM condensates self-assemble on chromosome axes through punctate clusters to create centers of DSB activity (22). In C. elegans, the SCs exhibit liquid crystal-like behaviors indicative of phase separation (23). However, the linkage between phase separation and meiotic recombination is unclear. Previous studies described HEI10’s behavior in meiosis as a diffusion-mediated coarsening model (14), raising a possibility for biomolecular condensates of HEI10. Here, we provide the physical interpretation of HEI10 coarsening phenomenon by LLPS. We found that HEI10 not only undergoes phase separation but also stimulates accumulation of other class I CO proteins in tobacco cells (Fig. 6). Interestingly, other class I CO proteins also promote HEI10 condensates (Fig. 6C). Consistent with these observations, MLH1-MLH3 (MutLγ) is required for the stable accumulation of HEI10 in mouse (16). These findings suggest that HEI10 and class I CO proteins mutually promote each other's accumulation, which may have a dosage effect that inhibits the formation of adjacent condensates, thus providing an explanation for CO interference. Additionally, our results suggest that two other factors involved in the anti-CO and class II CO pathways could also form condensates (SI Appendix, Fig. S6B). These results indicate that phase separation may be a common mechanism in the regulation of meiotic recombination. However, MMS-induced DSBs in somatic cells may not be equivalent to meiotic DSBs. Therefore, further studies that examine these protein dynamics in association with meiosis-specific DSBs are needed.
In the mouse, HEI10 deficiency results in a significant decrease in ubiquitinated proteins along the chromosome (18), suggesting that HEI10 mediates ubiquitin modification during meiosis. Here, we demonstrated that HEI10 directly interacts with RPA1a to promote its ubiquitination for subsequent degradation by 26S proteasome (Figs. 4 and 5). RPA is a conserved protein complex that binds to single-stranded DNA and protects the 3′ end of recombination intermediates from degradation (37). Unlike yeast and mammals which have single copies of RPA1, RPA2, and RPA3, plants have multiple copies of each RPA. In Arabidopsis, there are five paralogs of RPA1 (RPA1a-e), two of RPA2 (RPA2a and b) and two of RPA3 (RPA3a and b), but only RPA1a is known to be required for class I COs formation in meiosis (38) and is proposed to act in second-end capture to enable double Holliday junction (dHJ) formation (31). It has been reported that during DNA damage response, ubiquitination and timely removal of RPA is crucial for the progression of homologous recombination (39, 40). In somatic cells, RPA can act as a cofactor of the BTR complex in dHJ dissolution and the suppression of homologous recombination (41). We found that RPA1a may form condensates in nuclei in a HEI10-independent manner and that HEI10 and RPA1a may form cocondensates by LLPS. Therefore, we propose that during meiosis the precise removal or degradation of RPA1a, probably mediated by HEI10, is important for CO formation.
In conclusion, we present a working model in which phase separation mediates meiotic interference-sensitive CO formation (Fig. 7). Based on the classical meiotic DSB repair model, following DSB formation and resection, RPA1a binds to the 3′ single-strand DNA end to protect it from degradation. RPA1a is then replaced by the recombinases RAD51 and DMC1, which facilitate single-end invasion to search for homologous DNA template. DNA synthesis, D-loop extension, and second-end capture ultimately form dHJs, a class I CO intermediate. During this process, HEI10 localizes along the SC initially in a randomly distributed manner. As HEI10 diffuses along the SC, it accumulates as described in the coarsening model (12, 14) forming larger foci at CO sites through LLPS. After second end capture, HEI10 has a role in removal of RPA1a through ubiquitination and degradation. HEI10 condensates also recruit other class I CO proteins via mutually reinforcing LLPS-mediated condensate formation. Consequently, MSH4-MSH5 (MutSγ) stabilizes dHJs, and are then replaced by MLH1-MLH3 (MutLγ) for DNA cleavage, thus resulting in the formation of class I COs (42). Our results provide insights into the mechanisms for interference-sensitive CO formation by phase separation.
Fig. 7.
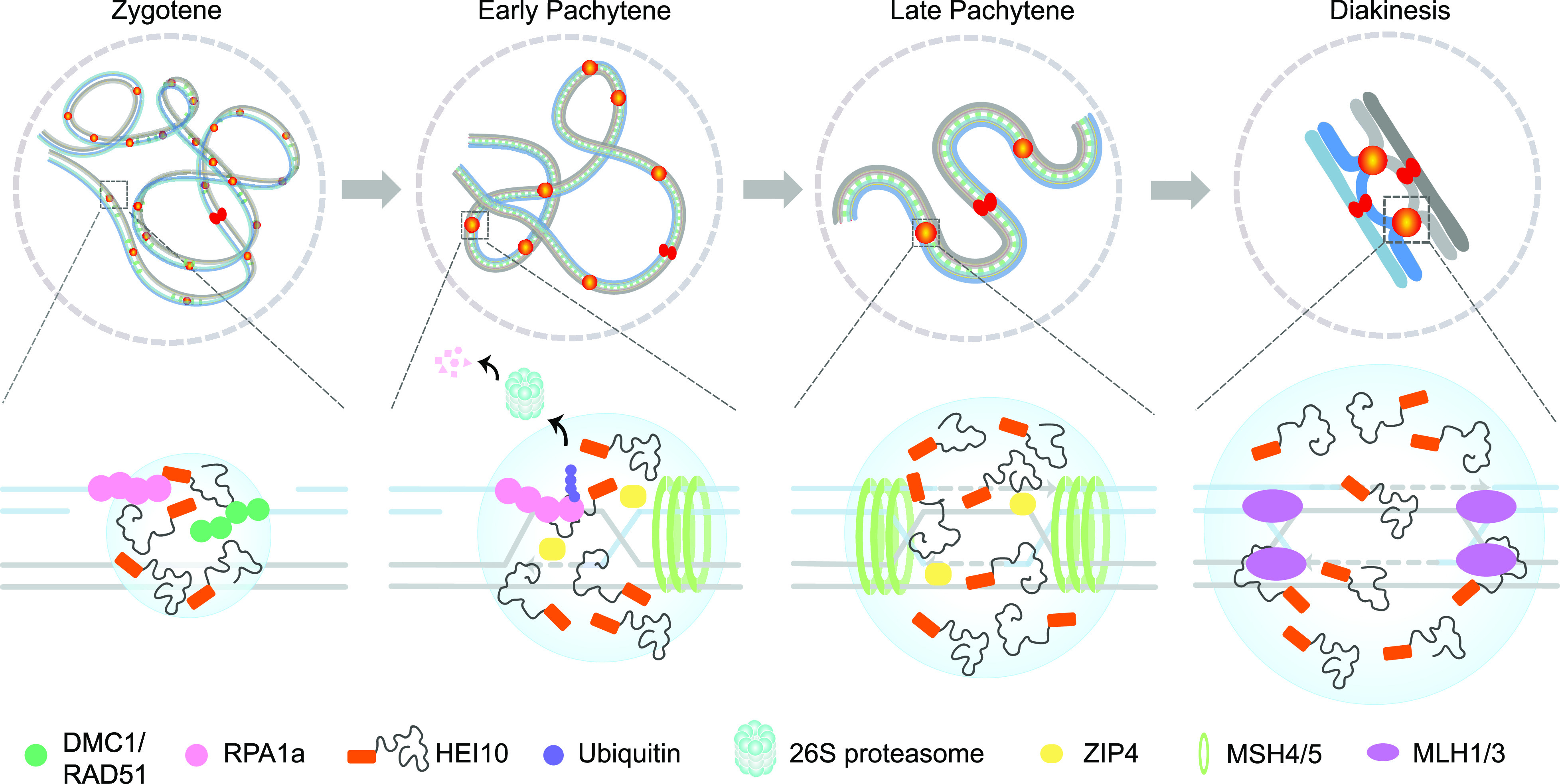
A working model for phase separation determining meiotic Interference-sensitive CO formation. Upper: Homologous chromosome interaction at four stages during meiotic prophase I is shown. Hundreds of HEI10 (orange cycle) foci localize to chromatin following meiotic DSB formation and condense as larger foci at COs sites in pachytene and diakinesis. Bottom: We show a CO-designated DSB site, HEI10 and RPA1a condensed at the DSB and RPA1a binding to single-strand DNA ends. Following second-end capture mediated by RPA1a, within the droplet, HEI10 directly interacts RPA1a to facilitate its ubiquitination-degradation via the 26S proteasome. HEI10 also recruits other class I CO proteins such as ZIP4, MSH4-MSH5 (MutSγ), and MLH1-MLH3 (MutLγ) to these sites through LLPS, leading to the formation and resolution of dHJs to yield class I CO.
Methods
Plant Materials and Transgenic Lines.
All Arabidopsis mutant and transgenic lines are in the Columbia (Col-0) ecotype. Mutant lines hei10-2 (SALK_014624) and rpa1a-1 (SALK_017580) were obtained from the Arabidopsis Biological Resource Center (https://abrc.osu.edu/). All the plants were grown in long-day conditions at 22 °C (16 h light/8 h dark). All transgenic constructs were derived from the pCAMBIA2300 vector. HEI10 CDS fused with 3×Flag was expressed from the Actin7 promoter. T2/T3 transgenic plants were used for experiments. The primers are listed in SI Appendix, Table S2.
The tobacco lines used for transient assays were wild-type Nicotiana benthamiana, grown in the same condition as Arabidopsis.
Microscopy and FRAP Assay.
For in vivo visualization, tobacco leaves were infiltrated with Agrobacterium tumefaciens strain GV3101 containing a construct that expresses 35S::HEI10-YFP. RFAP assay was performed on a Zeiss LSM880 microscope using a 63x oil-immersion objective. HEI10 puncta were bleached using a 100% laser intensity at 514 nm with 10 iterations.
For in vitro observation of HEI10 droplets, HEI10 CDS fused with an N-terminal mRFP tag was cloned into the pET50b+ vector. The observations were carried out with a Zeiss LSM 880 microscope. Droplets were bleached using a 100% laser intensity at 543 nm with 10 iterations. For time-lapse microscopy of mRFP-HEI10 droplet fusion, images were acquired every 30 s. All images were obtained using ZEN software. Image analysis was using FIJI/ImageJ. The primers are listed in SI Appendix, Table S2.
IP-MS.
The Arabidopsis central inflorescences including bud stages 1 to 9 were collected and ground in liquid nitrogen. Total proteins were extracted using extraction buffer (50 mM Tris–HCl pH 7.5, 150 mM NaCl, 0.5% Nonidet P-40, and 1 mM PMSF) at 4 °C for 1 h. Then, the sample was centrifuged at 4 °C, and the supernatant was collected and incubated with anti-Flag (Sigma, M8823) affinity beads 4 h for immunoprecipitation. After washing the beads three times, protein complexes were eluted by boiling 1×SDS loading buffer. NanoLC-MS/MS analysis was conducted utilizing an EASY-nLC 1200 system (Thermo Fisher Scientific, USA) coupled with an Orbitrap Fusion Lumos mass spectrometer (Thermo Fisher Scientific, USA).
Morphological and Cytological Analyses.
Meiotic chromosome spreading, centromere FISH, and immunofluorescence assays were conducted as described previously (43, 44). Chiasmata were counted in male meiocytes at metaphase I as described previously (45, 46). Rod bivalents represented a chiasmata on one chromosome arm, and a ring bivalent represents a chiasmata on both arms (47). The anti-AtHEI10 antibody was produced by raising HEI10 protein peptides PKDEIWPARQNS and immunized in rabbits (Ango Biotechnology, Shanghai, China) (45, 46). The anti-AtHEI10 and anti-Flag (GNI, GNI4110-FG) antibody were diluted 1:200, the secondary antibodies of Alexa Fluor 488 Goat Anti-Mouse IgG (H+L) (Invitrogen, A-11001) was used with 1:500 dilutions, and Alexa Fluor 555 Goat Anti-Rabbit IgG (H+L) (Invitrogen, A-21428) was used with 1:1,000 dilutions. Images were captured with a Zeiss Axio Scope A2 microscope (Zeiss, Heidelberg, Germany).
Immunostaining of tobacco leaf cells was similar to the method used in Arabidopsis inflorescences with some modifications. The leaves were cut into small pieces with scissors and treated with MMS for 1.5 h. Then the samples were fixed in 2% (w/v) paraformaldehyde with 0.1% (v/v) Triton X-100 and vacuum infiltrated for 20 to 30 min. After fixation, the leaves were digested using an enzyme cocktail (30 g/L cellulase, 50 g/L snailase, and 30 g/L macerozyme R-10, dissolved in 0.01M citrate buffer, pH 4.5) for 1 to 1.5 h at 37 °C. After washing with ddH2O, the leaves were placed on slides and crushed using a tweezer. After covering the sample with a slip, the slides were dipped in liquid nitrogen, and then, the cover slips were removed. The sample can be placed in −80 °C for storage or did the immunostaining immediately. The anti-Flag antibody (Sigma, SAB4301135) and anti-GFP antibody (GNI, GNI4110-GP) were diluted 1:200. The secondary antibodies of Alexa Fluor 555 Goat Anti-Mouse IgG (H+L) (Invitrogen, A-21422) was used with 1:500 dilutions, and Alexa Fluor 488 Goat Anti-Rabbit IgG (H+L) (Invitrogen, A-11008) was used with 1:1,000 dilutions. The Images were captured by a Zeiss Axio Scope A2 microscope (Zeiss, Heidelberg, Germany).
Recombinant Protein Expression and Pull Down Assay.
HEI10 fused with a His-Sumo tag was cloned into the pET28a vector. RPA1a fused with a glutathione S-transferase (GST) tag was cloned into pGEX4T-1 (GE Healthcare). The primers used are listed in SI Appendix, Table S2. The constructs were transformed into Escherichia coli Rosetta (DE3); the proteins were induced by IPTG (isopropyl β-D-1-thiogalactopyranoside) at 18 °C for 16 h. The proteins were purified by His-Tag purification Resin (Roche) and GST-bind Resin (Merck, 70541), respectively. Then, 2 μg proteins were mixed together in pull-down buffer (50 mM Tris–HCl pH 7.5, 150 mM NaCl, 0.1 mM imidazole, 0.1% Nonidet P-40, and 1 mM PMSF) at 4 °C for 5 h. GST agarose beads were added into the mixture and incubated at 4 °C for 2 h. Beads were washed with pull-down buffer for three times and eluted with SDS loading buffer. The eluted proteins were separated by Sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) and detected with anti-GST (Abmart, M20007) and anti-His (Abmart, M20001) antibodies.
Split Luciferase Complementation Imaging Assay.
HEI10 and RPA1a CDS were cloned into JW771 (nLUC) and JW772 (cLUC) plasmids and were introduced into A. tumefaciens GV3101 strain. Subsequently, the strains harboring HEI10 or RPA1a fused with nLUC or cLUC were co-infiltrated into N. benthamiana leaves in pairs, while nLUC and cLUC served as experimental controls. After 36 h, 1 mM luciferin was sprayed onto the leaves to detect the LUC activity. The experiment was done by using LB985 NightShade (Berthhold Tech). The primers used are listed in SI Appendix, Table S2.
Yeast Two-Hybrid Assay.
The GAL4-based Matchmaker Gold Yeast Two-Hybrid System (Clontech) was used for testing the interaction of proteins. Full-length CDS of the genes were cloned into pGBKT7 and pGADT7 vectors. These constructs were transformed into Y2H gold and Y187 yeast strain, respectively. The transformed strains were mated on YPDA medium for 24 h and then transferred to DDO (double synthetic dropout media lacking leucine and tryptophan) and QDO (quadra synthetic dropout media lacking leucine, tryptophan, histidine, and adenine) with X-α-Gal and Aureobasidin A plates to test for positive interactions. The primers are listed in SI Appendix, Table S2
MG132 Treatment and In Vivo Ubiquitination Assay.
To verify whether RPA1a protein degradation is mediated by the 26S proteasome, A. tumefaciens strain GV3101 carrying 35S::RPA1a‐Myc was infiltrated into tobacco leaves, and samples were collected at different time points for immunoblot analysis. The leaves were treated with 50 µM MG132 (Sigma-Aldrich, M7449) for 12 h before being sampled.
The ubiquitinated form of the protein was detected as described previously (48, 49). The 35S::HEI10‐Flag, 35S::RPA1a‐Flag/Myc constructs were infiltrated separately or coinfiltrated into the tobacco leaves using A. tumefaciens GV3101. The primers used are listed in SI Appendix, Table S2. Total proteins were extracted by native extraction buffer (50 mM Tris–MES pH 8.0, 0.5 M sucrose, 1 mM MgCl2, 10 mM EDTA, 5 mM DTT and protease inhibitor cocktail) and incubated with anti‐Flag (Sigma, M8823) or anti‐Myc (Sigma, A7470-1ML) affinity beads 4 h for immunoprecipitation. The beads were then washed three times, and protein complexes were eluted by adding 1×SDS loading buffer for immunoblot analysis. The proteins were detected by anti‐Flag antibody (GNI, GNI4110-FG), anti‐Myc antibody (PROMOTER, P-MYC), and anti‐UBQ11 antibody (Agrisera, AS08 307A).
Tube2 agarose (Life Sensors, UM402) was used to enrich the polyubiquitin-modified proteins. The samples were treated by protein extraction buffer; then, the supernatant was incubated with Tube2 agarose for 2h at 4 °C. The agarose was washed three times before boiling in SDS loading buffer. Then, the ubiquitinated protein were detected with anti-Flag antibody.
To detect the endogenous RPA1a protein level, the anti-RPA1a antibody was generated by raising peptides CETDTEAQKTFSGTGNIPPPN in rabbits (GL Biochem Ltd, Shanghai, China) and used with 1:500 dilutions for western blot analysis.
In Vitro Ubiquitination Assay.
The in vitro ubiquitination assay was performed as described previously with some modifications (50). The His-Sumo-RPA1a protein was incubated with His-Tag purification Resin at 4 °C for 2 h, and the beads were washed three times with binding buffer containing 50 mM Tris–HCl pH 8.0, 300 mM NaCl, and 1 mM PMSF. Total proteins of Arabidopsis central inflorescences were extracted with protein extraction buffer (50 mM Tris–HCl pH 8.0, 150 mM NaCl, 0.5% Nonidet P-40, and 1 mM PMSF) as described previously (48). Equal amounts of recombinant protein were incubated with equal amounts of Col-0 and HEI10-OE extracts at room temperature for 2 h by adding 10 mM ATP (Roche, 10127523001) and 50 μM MG132 (Sigma-Aldrich, M7449). The beads were washed four times with extraction buffer and boiled in SDS loading buffer. Then, the western blots were probed by anti‐His and anti‐UBQ11 antibody. The primers used are listed in SI Appendix, Table S2.
Semi‐In Vivo Protein Degradation.
The semi‐in vivo protein degradation experiment was performed as previously described (48). A. tumefaciens GV3101 strains carrying 35S::RPA1a‐Myc, 35S::HEI10‐Flag construct were infiltrated into tobacco leaves separately. Samples were collected 36 h after infiltration. Total proteins were extracted in native extraction buffer as described (48). Then, 10 mM ATP was added to the sample to preserve the 26S proteasome function. The RPA1a‐Myc extracts were then mixed with HEI10‐Flag or mock extracts in a volume ratio of 1:2. The mixtures were incubated at room temperature for 2 h. Samples were taken at different time points for immunoblot analysis. Anti‐Myc and anti‐Flag antibodies were used for western blot analysis. The primers used are listed in SI Appendix, Table S2.
Cell-Free Decay Assay.
The cell-free decay assay was performed as described previously with minor modifications (51). The Arabidopsis central inflorescences were dissolved with two volumes of lysis buffer (50 mM Tris–HCl pH 7.5, 150 mM NaCl, 0.5% Nonidet P-40, and 1 mM PMSF) at 4 °C for 40 min. Total protein extracts were adjusted to equal concentrations with lysis buffer and add 100 µM Cycloheximide (CHX) and 10 mM ATP to inhibit protein synthesis and maintain the activity of 26S proteasome. Then, 1 µg purified recombinant proteins of His-Sumo-RPA1a were incubated at room temperature with equal amount of the Col-0, hei10-2, hei10S70F and HEI10-OE lysates. For MG132 treatment, 50 µM MG132 was added as indicated. The reaction was stopped by adding 2 ×SDS-PAGE loading buffer, and the extracts were resolved by 10% SDS-PAGE after boiling. Proteins were detected with anti-His antibody.
Supplementary Material
Appendix 01 (PDF)
Dataset S01 (XLSX)
Acknowledgments
We thank Jianhua Gan at Fudan University for the pET50b+ vector. This research was supported by grants from the National Natural Science Foundation of China (Grant Nos. 31925005 and 32000246), Guangdong Laboratory for Lingnan Modern Agriculture (NG2022002), and Double first-class discipline promotion project of South China Agricultural University (2023B10564004).
Author contributions
T.W., C.Y., C.W., and Y.W. designed research; T.W. and H.W. performed research; T.W., Q.L., Q.J., and C.W. analyzed data; and T.W., G.P.C., C.W., and Y.W. wrote the paper.
Competing interests
The authors declare no competing interest.
Footnotes
This article is a PNAS Direct Submission.
Contributor Information
Gregory P. Copenhaver, Email: gcopenhaver@bio.unc.ed.
Cong Wang, Email: wangc@scau.edu.cn.
Yingxiang Wang, Email: yx_wang@fudan.edu.cn.
Data, Materials, and Software Availability
The genes mentioned in this study can be found in the Arabidopsis Information Resource (https://www.arabidopsis.org/) with the following accession numbers, HEI10 (AT1G53490) (9), RPA1a (AT2G06510) (31), ZIP4 (AT5G48390) (52), MSH5 (AT3G20475) (53), MLH3 (AT4G35520) (42), MUS81 (AT4G30870) (6), FANCD2 (AT4G14970) (8), and RMI1 (AT5G63540) (36). All other data are included in the manuscript and/or supporting information.
Supporting Information
References
- 1.Wang Y. X., Copenhaver G. P., Meiotic recombination: Mixing it up in plants. Annu. Rev. Plant Biol. 69, 577–609 (2018). [DOI] [PubMed] [Google Scholar]
- 2.Gray S., Cohen P. E., Control of meiotic crossovers: From double-strand break formation to designation. Annu. Rev. Genet. 50, 175–210 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sturtevant A. H., The behavior of the chromosomes as studied through linkage. Z. indukt. Abstamm. Vererbungsl. 13, 234–287 (1915). [Google Scholar]
- 4.Mercier R., Mezard C., Jenczewski E., Macaisne N., Grelon M., The molecular biology of meiosis in plants. Annu. Rev. Plant Biol. 66, 297–327 (2015). [DOI] [PubMed] [Google Scholar]
- 5.Li Y., et al. , HEIP1 regulates crossover formation during meiosis in rice. Proc. Natl. Acad. Sci. U.S.A. 115, 10810–10815 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Berchowitz L. E., Francis K. E., Bey A. L., Copenhaver G. P., The role of AtMUS81 in interference-insensitive crossovers in A-thaliana. PLoS Genet. 3, 1355–1364 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Higgins J. D., Buckling E. F., Franklin F. C. H., Jones G. H., Expression and functional analysis of AtMUS81 in Arabidopsis meiosis reveals a role in the second pathway of crossing-over. Plant J. 54, 152–162 (2008). [DOI] [PubMed] [Google Scholar]
- 8.Kurzbauer M. T., et al. , Arabidopsis thaliana FANCD2 promotes meiotic crossover formation. Plant Cell 30, 415–428 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chelysheva L., et al. , The Arabidopsis HEI10 is a new ZMM protein related to Zip3. PLoS Genet. 8, e1002799 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang K., et al. , The role of rice HEI10 in the formation of meiotic crossovers. PLoS Genet. 8, e1002809 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ward J. O., et al. , Mutation in mouse hei10, an e3 ubiquitin ligase, disrupts meiotic crossing over. PLoS Genet. 3, e139 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Girard C., Zwicker D., Mercier R., The regulation of meiotic crossover distribution: A coarse solution to a century-old mystery? Biochem. Soc. Trans. 51, 1179–1190 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.De Muyt A., et al. , E3 ligase Hei10: A multifaceted structure-based signaling molecule with roles within and beyond meiosis. Genes Dev. 28, 1111–1123 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Morgan C., et al. , Diffusion-mediated HEI10 coarsening can explain meiotic crossover positioning in Arabidopsis. Nat. Commun. 12, 4674 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Durand S., et al. , Joint control of meiotic crossover patterning by the synaptonemal complex and HEI10 dosage. Nat. Commun. 13, 5999 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Qiao H., et al. , Antagonistic roles of ubiquitin ligase HEI10 and SUMO ligase RNF212 regulate meiotic recombination. Nat. Genet. 46, 194–199 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Toby G. G., Gherraby W., Coleman T. R., Golemis E. A., A novel RING finger protein, human enhancer of invasion 10, alters mitotic progression through regulation of cyclin B levels. Mol. Cell Biol. 23, 2109–2122 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rao H. B. D. P., et al. , A SUMO-ubiquitin relay recruits proteasomes to chromosome axes to regulate meiotic recombination. Science 355, 403–407 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Banani S. F., Lee H. O., Hyman A. A., Rosen M. K., Biomolecular condensates: Organizers of cellular biochemistry. Nat. Rev. Mol. Cell Bio. 18, 285–298 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shin Y., Brangwynne C. P., Liquid phase condensation in cell physiology and disease. Science 357, eaaf4382 (2017). [DOI] [PubMed] [Google Scholar]
- 21.Li P., et al. , Phase transitions in the assembly of multivalent signalling proteins. Nature 483, 336–340 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Claeys Bouuaert C., et al. , DNA-driven condensation assembles the meiotic DNA break machinery. Nature 592, 144–149 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rog O., Kohler S., Dernburg A. F., The synaptonemal complex has liquid crystalline properties and spatially regulates meiotic recombination factors. Elife 6, e21455 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gallego L. D., et al. , Phase separation directs ubiquitination of gene-body nucleosomes. Nature 579, 592–597 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Martin E. W., Holehouse A. S., Intrinsically disordered protein regions and phase separation: Sequence determinants of assembly or lack thereof. Emerg. Top. Life Sci. 4, 307–329 (2020). [DOI] [PubMed] [Google Scholar]
- 26.Li J., et al. , Pathways and assays for DNA double-strand break repair by homologous recombination. Acta Biochim. Biophys. Sin. (Shanghai) 51, 879–889 (2019). [DOI] [PubMed] [Google Scholar]
- 27.Wang H., et al. , The cohesin loader SCC2 contains a PHD finger that is required for meiosis in land plants. PLoS Genet. 16, e1008849 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hurel A., et al. , A cytological approach to studying meiotic recombination and chromosome dynamics in Arabidopsis thaliana male meiocytes in three dimensions. Plant J. 95, 385–396 (2018). [DOI] [PubMed] [Google Scholar]
- 29.Wang C., et al. , GPS 5.0: An update on the prediction of kinase-specific phosphorylation sites in proteins. Genomics Proteomics Bioinformatics 18, 72–80 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lin Z., et al. , Evolutionary-scale prediction of atomic-level protein structure with a language model. Science 379, 1123–1130 (2023). [DOI] [PubMed] [Google Scholar]
- 31.Osman K., Sanchez-Moran E., Mann S. C., Jones G. H., Franklin F. C., Replication protein A (AtRPA1a) is required for class I crossover formation but is dispensable for meiotic DNA break repair. EMBO J. 28, 394–404 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Spegg V., et al. , Phase separation properties of RPA combine high-affinity ssDNA binding with dynamic condensate functions at telomeres. Nat. Struct. Mol. Biol. 30, 451–462 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Miao Y., et al. , Replication protein A large subunit (RPA1a) limits chiasma formation during rice meiosis. Plant Physiol. 187, 1605–1618 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lopitz-Otsoa F., et al. , Integrative analysis of the ubiquitin proteome isolated using Tandem Ubiquitin Binding Entities (TUBEs). J. Proteomics 75, 2998–3014 (2012). [DOI] [PubMed] [Google Scholar]
- 35.Komander D., Rape M., The ubiquitin code. Annu. Rev. Biochem. 81, 203–229 (2012). [DOI] [PubMed] [Google Scholar]
- 36.Seguela-Arnaud M., et al. , RMI1 and TOP3 alpha limit meiotic CO formation through their C-terminal domains. Nucleic Acids Res. 45, 1860–1871 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wold M. S., Replication protein A: A heterotrimeric, single-stranded DNA-binding protein required for eukaryotic DNA metabolism. Annu. Rev. Biochem. 66, 61–92 (1997). [DOI] [PubMed] [Google Scholar]
- 38.Aklilu B. B., Soderquist R. S., Culligan K. M., Genetic analysis of the replication protein A large subunit family in Arabidopsis reveals unique and overlapping roles in DNA repair, meiosis and DNA replication. Nucleic Acids Res. 42, 3104–3118 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Elia A. E., et al. , RFWD3-dependent ubiquitination of RPA regulates repair at stalled replication forks. Mol. Cell 60, 280–293 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Inano S., et al. , RFWD3-mediated ubiquitination promotes timely removal of both RPA and RAD51 from DNA damage sites to facilitate homologous recombination. Mol. Cell 66, 622–634.e8 (2017). [DOI] [PubMed] [Google Scholar]
- 41.Xue X. Y., Raynard S., Busygina V., Singh A. K., Sung P., Role of replication protein A in double Holliday junction dissolution mediated by the BLM-topo III alpha-RMI1-RMI2 protein complex. J. Biol. Chem. 288, 14221–14227 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cannavo E., et al. , Regulation of the MLH1-MLH3 endonuclease in meiosis. Nature 586, 618–622 (2020). [DOI] [PubMed] [Google Scholar]
- 43.Wang Y., Cheng Z., Lu P., Timofejeva L., Ma H., Molecular cell biology of male meiotic chromosomes and isolation of male meiocytes in Arabidopsis thaliana. Methods Mol. Biol. 1110, 217–230 (2014). [DOI] [PubMed] [Google Scholar]
- 44.Wang C., et al. , DNA polymerase epsilon interacts with SUVH2/9 to repress the expression of genes associated with meiotic DSB hotspot in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 119, e2208441119 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Li X., et al. , Fanconi anemia ortholog FANCM regulates meiotic crossover distribution in plants. Plant Physiol. 186, 344–360 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Li X., et al. , Regulation of interference-sensitive crossover distribution ensures crossover assurance in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 118, e2107543118 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Moran E. S., Armstrong S. J., Santos J. L., Franklin F. C. H., Jones G. H., Chiasma formation in Arabidopsis thaliana accession Wassileskija and in two meiotic mutants. Chromosome Res. 9, 121–128 (2001). [DOI] [PubMed] [Google Scholar]
- 48.Liu L., et al. , An efficient system to detect protein ubiquitination by agroinfiltration in Nicotiana benthamiana. Plant J. 61, 893–903 (2010). [DOI] [PubMed] [Google Scholar]
- 49.Yan J., et al. , FKF1 F-box protein promotes flowering in part by negatively regulating DELLA protein stability under long-day photoperiod in Arabidopsis. J. Integr. Plant Biol. 62, 1717–1740 (2020). [DOI] [PubMed] [Google Scholar]
- 50.Wang L. L., et al. , The ATR-WEE1 kinase module inhibits the MAC complex to regulate replication stress response. Nucleic Acids Res. 49, 1411–1425 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Garcia-Cano E., Zaltsman A., Citovsky V., Assaying proteasomal degradation in a cell-free system in plants. J. Vis. Exp. 85, e51293 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chelysheva L., et al. , Zip4/Spo22 is required for class I CO formation but not for synapsis completion in Arabidopsis thaliana. PLoS Genet 3, e83 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Higgins J. D., et al. , AtMSH5 partners AtMSH4 in the class I meiotic crossover pathway in Arabidopsis thaliana, but is not required for synapsis. Plant Journal 55, 28-39 (2008). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix 01 (PDF)
Dataset S01 (XLSX)
Data Availability Statement
The genes mentioned in this study can be found in the Arabidopsis Information Resource (https://www.arabidopsis.org/) with the following accession numbers, HEI10 (AT1G53490) (9), RPA1a (AT2G06510) (31), ZIP4 (AT5G48390) (52), MSH5 (AT3G20475) (53), MLH3 (AT4G35520) (42), MUS81 (AT4G30870) (6), FANCD2 (AT4G14970) (8), and RMI1 (AT5G63540) (36). All other data are included in the manuscript and/or supporting information.