Abstract
The nitrate and nitrite reductases of Bacillus subtilis have two different physiological functions. Under conditions of nitrogen limitation, these enzymes catalyze the reduction of nitrate via nitrite to ammonia for the anabolic incorporation of nitrogen into biomolecules. They also function catabolically in anaerobic respiration, which involves the use of nitrate and nitrite as terminal electron acceptors. Two distinct nitrate reductases, encoded by narGHI and nasBC, function in anabolic and catabolic nitrogen metabolism, respectively. However, as reported herein, a single NADH-dependent, soluble nitrite reductase encoded by the nasDE genes is required for both catabolic and anabolic processes. The nasDE genes, together with nasBC (encoding assimilatory nitrate reductase) and nasF (required for nitrite reductase siroheme cofactor formation), constitute the nas operon. Data presented show that transcription of nasDEF is driven not only by the previously characterized nas operon promoter but also from an internal promoter residing between the nasC and nasD genes. Transcription from both promoters is activated by nitrogen limitation during aerobic growth by the nitrogen regulator, TnrA. However, under conditions of oxygen limitation, nasDEF expression and nitrite reductase activity were significantly induced. Anaerobic induction of nasDEF required the ResDE two-component regulatory system and the presence of nitrite, indicating partial coregulation of NasDEF with the respiratory nitrate reductase NarGHI during nitrate respiration.
Changes in oxygen tension and reduced nitrogen supply are common to the habitat of the soil bacterium Bacillus subtilis. When external oxygen is limited, B. subtilis is able to use an alternative electron acceptor, namely, nitrate or nitrite, and grows by anaerobic respiration (for reviews, see references 11 and 15). Nitrite reduction catalyzed by B. subtilis nitrite reductase does not result in a proton gradient and coupled ATP generation. Instead, nitrite enhances anaerobic growth by serving as an electron sink. However, this process can still be designated as respiration in a broad sense according to the simple definition of a process utilizing an inorganic electron acceptor. A respiratory nitrate reductase responsible for nitrate respiration is encoded by the narGHJI operon, the expression of which is highly induced by oxygen limitation (1, 5, 8). The induction of narGHJI depends on an anaerobic regulatory protein, FNR (1, 16). Oxygen-sensitive nitrite reductase activity was also detected in B. subtilis cells grown anaerobically in the presence of nitrate and nitrite (4). The nitrite reductase activity is dependent on the ResD-ResE two-component signal transduction system (16, 20) but not on FNR (1), as shown by examining nitrite-dependent anaerobic growth of resDE and fnr mutant cultures (4).
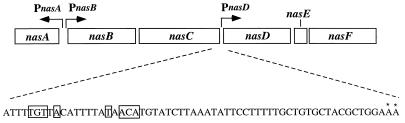
The complete Bacillus genome sequence was recently reported (7), and B. subtilis appears to have only one nitrite reductase that is encoded by nasD and nasE. The NasDE nitrite reductase was previously identified as an assimilatory enzyme (17). nasD and nasE are part of an operon also containing two upstream genes, nasB and nasC, that encode subunits of assimilatory nitrate reductase and a downstream nasF gene required for biosynthesis of siroheme, a cofactor of nitrite reductase. The nasA gene, which is divergently transcribed from the nas operon, is thought to encode a nitrate transporter (Fig. 1) (17). Both nasB and nasC are transcribed from the nasB promoter, and transcription possibly extends through nasD to nasF (17). Both nasA and nasB promoter activities are high during nitrogen-limited growth and repressed in the presence of a good nitrogen source (14). The transcriptional activation of nas as well as that of other genes during nitrogen-limited growth was shown to be mediated by the TnrA regulatory protein, which binds to an upstream cis-acting site of nitrogen-regulated genes (22, 23).
FIG. 1.
Map of the nas region. The open boxes represent open reading frames that encode NasA, -B, -C, -D, -E, and -F. The locations of the promoters are shown by P, and arrows represent the directions of transcription. The nucleotide sequences of the nasD promoter region are shown beneath the operon diagram. Asterisks indicate transcription start sites determined by the primer extension analysis in Fig. 4. Nucleotides in boxes are the putative TnrA binding site.
NasBC nitrate reductase is not detected at all in cells grown anaerobically in rich medium (3), suggesting that transcription from the nasB promoter is repressed under these nitrogen-excess conditions. If nitrite reductase encoded by nasDE is indeed the enzyme required for nitrite respiration, the expression of nasDEF must be directed from another promoter during anaerobiosis. As reported herein, NasDE nitrite reductase functions as both an assimilatory and a dissimilatory enzyme, and an intergenic promoter specific for nasDEF transcription is activated both by oxygen limitation and by nitrogen limitation.
MATERIALS AND METHODS
Bacterial strains and plasmids.
The B. subtilis strains used in this study are derivatives of JH642 (trpC2 pheA1) (Table 1). A nasF-lacZ transcriptional fusion was constructed as follows. A 730-bp fragment containing the 3′ end of nasE and the 5′ end of nasF was isolated by digesting plasmid pMMN217 (17) with restriction endonucleases. The fragment thus obtained was inserted into promoter-probe vector pTKlac (6) to create pMMN224. Plasmid pMMN224 was introduced by transformation into JH642 with selection for chloramphenicol resistance (Cmr). In the transformant (LAB1848) where pMMN224 was integrated into the nasF gene by Campbell-type recombination, the lacZ gene is transcribed from the nasB and nasD promoters as described in Results. pMMN224 was also introduced into nasB (LAB1727) and nasD (LAB1972) mutants to generate strains LAB2012 and LAB2008, respectively. The nasB and the nasD deletion mutants were constructed by replacing internal regions of the nasB and the nasD genes with antibiotic resistance markers as previously described (17). In order to construct a nasC-lacZ fusion, plasmid pMMN391 was first constructed. pMMN391 is a derivative of pTKlac (6) that carries a 970-bp internal fragment of nasC in front of the promoterless lacZ gene. LAB2844 cells carrying nasC-lacZ were generated by transformation of JH642 with pMMN391. A 350-bp fragment containing the nasC and nasD intergenic region along with the nasD promoter was obtained and inserted into pTKlacZ upstream of the promoterless lacZ gene. The resultant plasmid, pMMN392, was introduced by transformation into an SPβ lysogen, where it integrated into the resident SPβ prophage. A phage lysate carrying the nasD-lacZ fusion was introduced into wild-type and various mutant strains as described previously (24). The construction of ΔresDE (16, 20), fnr (1, 16), and ΔnarGH (5, 10) mutants was described elsewhere. A tnrA mutant (SF62) was obtained from Susan Fisher (22).
TABLE 1.
B. subtilis strains used in this study
| Strain | Relevant characteristics | Source or reference |
|---|---|---|
| JH642 | trpC2 pheA1 | J. A. Hoch |
| LAB1727 | trpC2 pheA1 ΔnasB::erm | 17 |
| LAB1848 | trpC2 pheA1 nasF::pMMN224 | This study |
| LAB1943 | trpC2 pheA1 ΔnasE::phleo Pspac-nasF | 17 |
| LAB1972 | trpC2 pheA1 ΔnasD::phleo | 17 |
| LAB1982 | trpC2 pheA1 ΔnasD::phleo Pspac-nasEF | 17 |
| LAB2008 | trpC2 pheA1 ΔnasD::phleo nasF::pMMN224 | This study |
| LAB2012 | trpC2 pheA1 ΔnasB::erm nasF::pMMN224 | This study |
| LAB2135 | trpC2 pheA1 ΔresDE::tet | 16 |
| LAB2136 | trpC2 pheA1 fnr::spc | 16 |
| LAB2844 | trpC2 pheA1 nasC::pMMN391 | This study |
| LAB2854 | trpC2 pheA1 SPβc2del2::Tn917::pMMN392 | This study |
| LAB2862 | trpC2 pheA1 ΔresDE::tet SPβc2del2::Tn917:: pMMN392 | This study |
| LAB2863 | trpC2 pheA1 fnr::spc SPβc2del2::Tn917:: pMMN392 | This study |
| LAB2908 | trpC2 pheA1 tnrA::Tn917 | This study |
| LAB2911 | trpC2 pheA1 tnrA::Tn917 SPβc2del2::Tn917:: pMMN392 | This study |
| LAB2408 | trpC2 pheA1 ΔnarGH::phleo | 10 |
| LAB2966 | trpC2 pheA1 ΔnarGH::phleo SPβc2del2::Tn917:: pMMN392 | This study |
| MH5081 | trpC2 pheA1 ΔresDE::tet | 20 |
| THB1 | trpC2 pheA1 ΔnarGH::tet | 5 |
| THB2 | trpC2 pheA1 fnr::spc | 4 |
Examination of growth phenotype of nasD and nasE mutants.
B. subtilis cells were grown anaerobically at 37°C in supplemented Luria-Bertani medium containing 10 mM (NH4)2SO4 and NaNO2 as described elsewhere (4, 5). Cells were also grown aerobically at 37°C in minimal medium containing 3.4 mM Na3 citrate, 0.8 mM MgSO4, 20 mM K3PO4 (pH 7.0), 1 mM tryptophan, 0.8 mM phenylalanine, 1 mM glucose, and 10 mM (NH4)2SO4 or NaNO2 as nitrogen source. Cells grown overnight in the minimal medium containing 10 mM (NH4)2SO4 and 10 mM NaNO2 were washed with the same medium without nitrogen source and were diluted 1:100 in fresh medium supplemented with 10 mM (NH4)2SO4 or NaNO2. LAB1982 and LAB1943 cells were grown in the presence of 1 mM IPTG (isopropyl-β-d-thiogalactopyranoside). Growth was monitored by measuring the optical density at 578 nm.
Measurement of nitrite reductase activity.
B. subtilis cells used for measurement of nitrite reductase activity were grown aerobically or anaerobically as described above, except that aerobically grown cells were not washed before inoculating the fresh medium to support residual growth of the tnrA and the nasD/E mutant strains. Cells were harvested at late exponential growth phase, washed with 10 mM Tris-HCl (pH 7.4) and 10 mM MgCl2, and disrupted with a French press. The crude lysate was centrifuged at 5,000 × g for 5 min at 4°C to remove intact cells and cell debris. The cell extract was used for enzyme assays. Subcellular fractionation was performed by centrifugation of the cell extract at 100,000 × g for 45 min at 4°C to separate the membrane (pellet) and cytosolic (supernatant) fractions. The membrane fraction was washed twice with distilled water to avoid cytosolic contamination. Contamination of both fractions was checked by measuring 5-aminolevulinic acid dehydratase as a marker enzyme for the cytosolic fraction and respiratory nitrate reductase as a marker for the membrane fraction, and no significant contamination of either fraction was observed. All procedures were done under strict anaerobic conditions for extracts prepared from both anaerobic and aerobic cultures because nitrite reductase is sensitive to oxygen, as previously described (4). Nitrite reductase activity of the cell extracts was measured under nitrogen by the nitrite-dependent oxidation of NADH in a cuvette containing 50 mM Tris-HCl (pH 7.5), 1 mM NADH, 1 mM KNO2, and 50 to 200 μl of cell extract (containing 20 mg of protein per ml) at room temperature. Reactions were started by the addition of nitrite, and oxidation of NADH was monitored spectrophotometrically at 366 nm. Control reaction mixtures were incubated in the absence of external electron acceptors to determine the endogenous rate of NADH oxidation; this small background activity was subtracted from activities with nitrite to obtain net activity. One unit of enzyme activity corresponds to the reduction of 1 mmol of nitrite per min.
Measurement of β-galactosidase activity.
B. subtilis cells grown overnight on DS (Difco sporulation) agar medium were used to inoculate 2× YT (yeast extract-tryptone) liquid medium (12) supplemented with 1% glucose and 20 mM phosphate buffer (pH 7.0), which contained either 0.2% KNO3 or 10 mM KNO2, to an optical density at 600 nm of 0.02. Alternatively, cells grown aerobically overnight in TSS liquid medium (18) with 0.2% NH4Cl were used to inoculate TSS medium containing 0.2% KNO3 with or without 0.2% NH4Cl. Anaerobic growth of cultures was carried out as described previously (16). To obtain cells grown aerobically with various nitrogen sources, cells grown overnight in TSS liquid medium with 0.2% NH4Cl were used to inoculate TSS liquid medium with 0.2% glutamate, 0.2% NH4Cl, 0.2% KNO3, or 10 mM KNO2. Samples were withdrawn at 1-h intervals, and maximum activities attained at late exponential to early stationary growth are shown in the tables. Assays for β-galactosidase were carried out as previously described (9).
Primer extension analysis.
B. subtilis LAB2854 was grown aerobically or anaerobically in 2× YT medium supplemented with 1% glucose, 20 mM phosphate buffer (pH 7.0), and 0.2% KNO3 to obtain cells for RNA purification. LAB2854 was also cultured under aerobic conditions in TSS medium with 0.2% NH4Cl or 0.2% KNO3. Total RNA was extracted from cells harvested at mid- to late exponential growth as previously described (13). The 5′ end of nasD mRNA was localized by primer extension analysis with the Promega system. Forty micrograms of total RNA and 2 pmol of an end-labeled primer (5′ CCCGGCCATTCCATTACCA 3′) were combined in a hybridization mix containing primer extension buffer (50 mM Tris-HCl [pH 8.3], 50 mM KCl, 10 mM MgCl2, 10 mM dithiothreitol, 1 mM deoxynucleoside triphosphates, and 0.5 mM spermidine, supplied by the manufacturer). Primer-RNA hybridization was facilitated by heating for 1 min at 90°C, followed by a 2-min incubation at 60°C and a slow cooling of the hybridization mix to room temperature. Primer extension was performed at 42°C for 30 min with avian myeloblastosis virus reverse transcriptase. After treatment with RNase, the extension products were separated by electrophoresis in 6% polyacrylamide–8 M urea gels. The precise size of the primer extension product was determined by comparison with DNA sequencing reactions by the dideoxy chain-termination method (19) and the same oligonucleotide used for primer extension.
RESULTS
B. subtilis cells possess a nasDE-dependent nitrite reductase activity during aerobic and anaerobic growth.
Previous results strongly suggest that nasD and nasE encode subunits of nitrite reductase (17). We examined whether aerobic nitrite reductase activity is indeed detected in wild-type cells but not present in nasD or nasE mutants. NADH-dependent nitrite reductase activity was detected in a cell extract prepared from the wild-type strain grown with nitrite, but activity with NADPH was much lower; no activity was detected when ammonium was used as sole nitrogen source (Table 2). However, a cell extract prepared from a nasD mutant (LAB1972) grown with nitrite had no detectable nitrite reductase activity. To eliminate the possibility of polar effects caused by the nasD mutation on the expression of the downstream nasE and nasF genes, we used two strains that were constructed previously (17). In these strains, a Pspac promoter was integrated downstream of the mutated genes, allowing induction of the genes residing 3′ to the mutation (LAB1943, ΔnasE Pspac-nasF; and LAB1982, ΔnasD Pspac-nasEF) by IPTG. Previous results showed that expression of nasE and nasF from the Pspac promoter is sufficient to support growth with nitrite as sole nitrogen source (17). However, extracts prepared from LAB1943 and LAB1982 grown with nitrite or ammonium in the presence of IPTG had no significant nitrite reductase activity, indicating that both nasD and nasE genes are required for enzyme activity. Since it was previously shown that NasD and NasE have high homology to subunits of nitrite reductases (17), together these results confirm that the nasD and the nasE genes encode subunits of assimilatory nitrite reductase.
TABLE 2.
NADH-dependent nitrite reductase activities of various B. subtilis strains grown under aerobic and anaerobic conditions
| Strain | Nitrite reductase activity ± SD (10−4 U/mg of protein)a
|
||
|---|---|---|---|
| Anaerobic growth | Aerobic growth
|
||
| Nitrite | Ammonium | ||
| JH642 (wild type) | 334 ± 12b | 146 ± 14b | ND |
| THB1 (ΔnarGH) | 351 ± 21 | 164 ± 18 | ND |
| LAB1943 (ΔnasE Pspac-nasF)c | ND | ND | ND |
| LAB1972 (ΔnasD) | ND | ND | ND |
| LAB1982 (ΔnasD Pspac-nasEF)c | ND | ND | ND |
| MH5081 (ΔresDE) | ND | ND | ND |
| THB2 (fnr) | 527 ± 21 | 348 ± 24 | ND |
NADH-dependent nitrite reductase activities were measured with cell extracts prepared from B. subtilis strains grown aerobically in minimal medium with nitrite and anaerobically in supplemented Luria-Bertani medium with nitrate and nitrite as described in Materials and Methods. Values reported are the averages of at least three experiments performed in triplicate. ND, not detectable (less than 40 × 10−4 U/mg of protein).
Activity with NADPH was less than 10% of that with NADH.
Cells were grown in the presence of 1 mM IPTG.
The possibility that NasDE nitrite reductase also functions in anaerobic respiration was examined by measuring nitrite reductase activity in cell extracts prepared from anaerobic cultures of the wild-type and nas mutant strains (Table 2). NADH-dependent activity was detected in wild-type, but not in LAB1943 and LAB1982, strains. This result clearly indicates that NasDE nitrite reductase is the dissimilatory nitrite reductase involved in anaerobic respiration. No nitrite reductase activity was detected in the membrane fraction, whereas nearly all activity was present in the cytosolic fraction, indicating that the NasDE enzyme is a cytoplasmic protein.
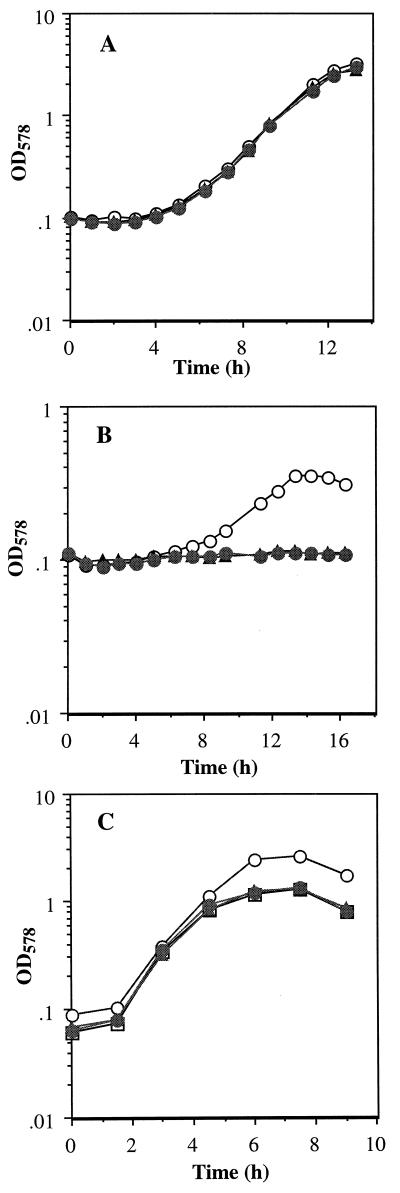
In order to examine effects of nasD and nasE mutations on aerobic nitrite assimilation, LAB1982 (nasD) and LAB1943 (nasE) strains were grown aerobically in minimal medium containing nitrite or ammonium as sole nitrogen source. No significant difference in growth was observed between the wild type and the mutants grown with ammonium as sole nitrogen source (Fig. 2A); however, the mutants were unable to grow when nitrite was used as sole nitrogen source (Fig. 2B). This result is consistent with the previously observed growth defects of the mutants (17). B. subtilis grows anaerobically by fermentation in rich medium in the absence of an electron acceptor (10). When nitrite was present, the anaerobic growth was enhanced as reported previously (4) and as shown in Fig. 2C. The growth behavior of the nasD and the nasE mutants in the presence of nitrite was identical to the fermentative growth of the wild-type cells. These results demonstrate that the NasDE enzyme functions both aerobically and anaerobically.
FIG. 2.
Growth of B. subtilis wild-type, nasD, and nasE mutant strains: JH642 (wild type) (open circles), LAB1982 (ΔnasD Pspac-nasEF) (closed circles), and LAB1943 (ΔnasE Pspac-nasF) (closed triangles). The mutant strains were cultured in the presence of 1 mM IPTG. (A and B) Aerobic growth in minimal medium supplemented with 10 mM (NH4)2SO4 (A) or with 10 mM NaNO2 (B). (C) Anaerobic growth in rich medium containing 10 mM NaNO2 and 10 mM (NH4)2SO4. Open squares indicate growth of JH642 with ammonium only. OD578, optical density at 578 nm.
Effect of mutations in resDE and fnr on aerobic and anaerobic NasDE activity.
ResD/E and FNR are known to be required for the transcription of genes that are expressed in response to oxygen limitation in B. subtilis (for reviews, see references 11 and 15). The two-component signal transduction system comprising the response regulator ResD and its cognate sensor kinase ResE (20) is required for induction of fnr transcription upon oxygen limitation and, thus, for the activation of other anaerobically induced genes requiring FNR for transcription (16). To determine if a resDE or an fnr mutation affects anaerobic nitrite reductase levels, nitrite reductase activity was examined in resDE and fnr mutant cells. A resDE mutant did not possess anaerobic NADH-dependent nitrite reductase activity, which confirmed the previous results with benzyl viologen as the electron donor (4). Furthermore, no nitrite reductase activity was detected in cell extracts of the resDE mutant prepared from cultures grown aerobically in minimal medium supplemented with ammonia or nitrite as sole nitrogen source. This result indicates that ResDE is required for both assimilatory and respiratory nitrite reductase activities.
Increased nitrite reductase activity was observed in cell extracts prepared from anaerobically grown fnr mutant (THB2) cultures compared to that of the wild type (4). Table 2 shows that the fnr mutant also possessed higher nitrite reductase activity during aerobic growth with nitrite than that of the wild-type strain, suggesting a slight negative effect of FNR on nitrite reductase activity during aerobic and anaerobic growth.
Regulation of nitrate and nitrite reductase genes.
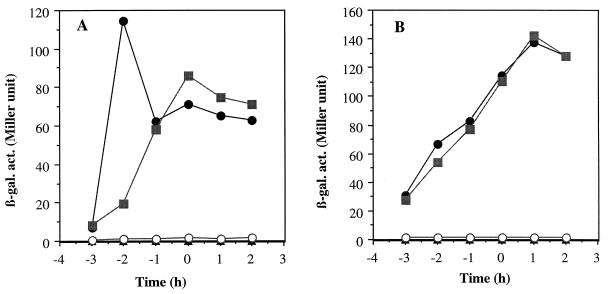
The results described above indicated that nitrite reductase encoded by nasD and nasE is essential for nitrite respiration during anaerobic growth. Our previous results showed that nitrate reductase encoded by nasB and nasC located upstream of the nasD gene functions only in nitrate assimilation during aerobic growth (3, 17). B. subtilis has another nitrate reductase (NarGHJI) that functions as a respiratory enzyme (1, 5, 8). Since NasBC is necessary only during aerobic growth under nitrogen-limited conditions and NasDE nitrite reductase must be produced in response to either nitrogen or oxygen limitation, it is very likely that expression of nasDEF is regulated differently from that of nasBC. In an attempt to analyze the regulation of nitrate reductase (nasBC) and nitrite reductase (nasDEF) genes, a promoterless lacZ gene was inserted into the nasC or nasF gene, and expression of nasC-lacZ and nasF-lacZ was examined under different culture conditions. Expression of nasC and nasF during growth in TSS minimal medium with various nitrogen sources was examined (Table 3). It was previously shown that nasB operon transcription was derepressed during growth in the presence of poor nitrogen sources such as glutamate, proline, and nitrate (nitrogen-limited conditions) and repressed when growth media were supplemented with a good nitrogen source such as ammonium or glutamine (nitrogen-excess conditions). Table 3 shows that expression of nasC from the nasB promoter is regulated by the nitrogen source in the same manner as nasB expression (14). nasF expression is also observed to be derepressed by nitrogen limitation and repressed under conditions of nitrogen excess, which corresponds well to the observed nitrite reductase activities (Table 2). nasC expression is twofold higher in glutamate-containing medium than in nitrate- or nitrite-containing medium. However, nasF expression is equally derepressed in the presence of glutamate, nitrate, or nitrite. The significance of the difference is unknown; however, it may reflect nasF expression from the nasD promoter, which is higher in the presence of nitrate or nitrite than of glutamate as described below (Table 4). This result demonstrates that nasC and nasF are both under nitrogen regulation. Indeed, neither nasC nor nasF was expressed when cells were grown aerobically in a rich medium such as 2× YT (data not shown). When cells were cultured in 2× YT medium under anaerobic conditions, however, nasF expression was highly induced. In contrast, nasC was still severely repressed (Fig. 3). These results indicate that the nitrate reductase (nasBC) genes are activated only by nitrogen limitation but that the nitrite reductase (nasDEF) genes are induced by oxygen limitation as well as by nitrogen limitation. The pattern of nasC and nasF expression corresponds well to the function of their products.
TABLE 3.
β-Galactosidase activities of nasC/nasF-lacZ fusions grown aerobically with various nitrogen sources
| Nitrogen sourcea | β-Galactosidase activity (Miller units)b
|
|
|---|---|---|
| LAB2844 (nasC-lacZ) | LAB1848 (nasF-lacZ) | |
| Glutamate | 53 | 22 |
| Nitrate | 19 | 20 |
| Nitrite | 19 | 22 |
| Ammonium | 0.4 | 0.9 |
Cells were grown aerobically in TSS minimal medium with 0.5% glucose containing the indicated nitrogen sources
Maximum activities during growth are shown (averages of three or more experiments). The standard error was less than 25%.
TABLE 4.
β-Galactosidase activities of nasD-lacZ fusions in wild-type and mutant strains grown under aerobic conditions
| Nitrogen sourcea | β-Galactosidase activity (Miller units)b
|
|||
|---|---|---|---|---|
| LAB2854 (wild type) | LAB2862 (ΔresDE) | LAB2863 (fnr) | LAB2911 (tnrA) | |
| Glutamate | 14 | 13 | 20 | 0.4 |
| Nitrate | 40 | 21 | 43 | 0.6 |
| Nitrite | 88 | 29 | 84 | <0.1 |
| Ammonium | 0.7 | 0.6 | 0.6 | 0.4 |
| Nitrate plus ammonium | 1.2 | NEc | NE | NE |
| Nitrite plus ammonium | 2.3 | NE | NE | NE |
Cells were grown aerobically in TSS minimal medium with 0.5% glucose containing the indicated nitrogen sources.
Maximum activities during growth are shown (averages of three or more experiments). The standard error was less than 25%.
NE, not examined.
FIG. 3.
Expression of nasC-lacZ and nasF-lacZ fusions. Cells were grown in 2× YT medium containing 1% glucose and 20 mM phosphate buffer (pH 7.0) with 0.2% KNO3 (A) or 10 mM KNO2 (B) under anaerobic conditions. Time zero indicates the end of exponential growth. Closed circles, LAB1848 (wild type, nasF-lacZ); closed triangles, LAB2844 (wild type, nasC-lacZ); closed squares, LAB2012 (ΔnasB nasF-lacZ); open circles, LAB2008 (ΔnasD nasF-lacZ).
An intergenic nasD promoter is essential for anaerobic induction of nitrite reductase genes.
The results described above suggest that nasDEF is transcribed from an internal promoter under anaerobic conditions. Introduction of a nasB mutation had no significant effect on anaerobic nasF expression; however, a polar mutation in nasD abolished expression (Fig. 3). This suggests that there is likely an intergenic promoter upstream of nasD that is responsible for anaerobic nasDEF transcription. In order to test this possibility, the nasC and the nasD intergenic region was inserted into promoter-probe vector pTKlac (6), and the resulting lacZ fusion (called nasD-lacZ) introduced into the SPβ prophage was observed to be activated both during nitrogen-limited growth (Table 4) and by anaerobiosis (Table 5) as was the case with nasF-lacZ. These findings indicate that nasD, nasE, and nasF are all transcribed from the nasD promoter. Anaerobic nasD expression was slightly but reproducibly higher when cells were grown in the presence of nitrite than when nitrate was present (Table 5). The expression of nasD-lacZ during aerobic growth in TSS medium with nitrite was two- and sixfold higher than that observed in cells grown with nitrate or glutamate, respectively (Table 4). However, previous work showed that nasB expression is slightly higher when glutamate is the sole nitrogen source than when nitrate is present (14). To determine if nitrite reductase gene expression is induced by its substrate nitrite under conditions of nitrogen excess, β-galactosidase activity was examined in LAB2854 cells grown in the presence of ammonium and nitrite (Table 4). β-Galactosidase activity was threefold higher in cells grown in TSS medium containing ammonium and nitrite than in cells grown without nitrite; however, the level was much lower than that observed in cells grown with nitrite alone, and addition of nitrate in the TSS-ammonium medium hardly stimulated nasD expression. These results suggest that aerobic nasDEF expression is not significantly induced by nitrate or nitrite under nitrogen-excess conditions.
TABLE 5.
β-Galactosidase activities of nasD-lacZ fusions in wild-type and mutant strains grown under anaerobic conditions
| Strain | β-Galactosidase activity (Miller units) in mediuma:
|
|||
|---|---|---|---|---|
| 2× YT
|
TSS
|
|||
| Nitrate | Nitrite | Nitrate | Nitrate plus ammonium | |
| LAB2854 (wild type) | 339 | 566 | 394 | 296 |
| LAB2862 (ΔresDE) | 3.5 | 4.2 | 3.3 | 9.6 |
| LAB2863 (fnr) | 32 | 493 | NEb | NE |
| LAB2911 (tnrA) | 225 | 371 | 377 | 317 |
| LAB2966 (ΔnarGH) | 30 | 384 | NE | NE |
Maximum activities during growth are shown (averages of three or more experiments). The standard error was less than 25%. Cells were grown anaerobically in TSS minimal medium with 0.5% glucose or in 2× YT medium containing the indicated nitrogen sources.
NE, not examined.
Effect of mutations in fnr, resDE, and respiratory nitrate reductase genes on nasD expression.
The results described above demonstrate that ResDE is indispensable for nitrite reductase activity and that FNR has a small negative effect on enzyme activity. To determine if the effect of resDE and fnr on nitrite reductase activity is caused by an effect on expression of nasD, nasD-lacZ expression was examined in resDE and fnr mutant cells. The maximal levels of β-galactosidase activity in resDE and fnr mutants grown anaerobically in 2× YT medium with nitrate were 1 and 10%, respectively, of that detected in wild-type cells (Table 5). When cells were cultured in the presence of nitrite, however, the level of nasD-lacZ expression in the fnr mutant was restored to that of the wild-type strain. In contrast, nitrite had no stimulatory effect on nasD expression in the resDE mutant. FNR is required for transcription of anaerobically induced genes including the respiratory nitrate reductase (narGHJI) genes. To determine if the role of FNR in nasD expression is to transcribe narGHJI and thus to produce nitrite, an inducer for nasD transcription, nasD-lacZ expression was examined in a narGH mutant grown anaerobically in the presence of nitrate or nitrite. Table 5 shows that β-galactosidase activity in the narGH mutant was as low as that of the fnr mutant when cells were grown in the presence of nitrate, and the activity was restored by the addition of nitrite. This result argues that FNR and the respiratory nitrate reductase are required for anaerobic nasD expression solely to produce nitrite while ResD and ResE have a more direct role in nasDEF regulation.
In contrast to the expression observed during anaerobic growth, aerobic nasD expression was not significantly affected by resDE or fnr mutations except that nasD expression in the resDE mutant was two- to threefold lower than that in the wild-type cells when grown in the presence of nitrate or nitrite (Table 4). These results show that ResDE and nitrite are indispensable for anaerobic induction of nasD expression but that ResDE and FNR play no major role in nasD expression under nitrogen-limited aerobic growth.
Effect of the tnrA mutation on nasD expression.
Transcription of many nitrogen-regulated genes in B. subtilis including nasB is activated by TnrA, which responds to nitrogen-limited conditions (22). Table 4 shows that TnrA is also responsible for activation of nasD expression when cells are grown aerobically under poor nitrogen conditions (with glutamate, nitrate, or nitrite as nitrogen source). The tnrA mutant is unable to grow with nitrate or nitrite as sole nitrogen source; however, the growth defect per se is not responsible for the reduced nasD expression. In fact, LAB2844 is also unable to grow with nitrate as sole nitrogen source because the nasC gene is inactivated by the integration of the lacZ fusion; however, high lacZ expression was observed during incubation with nitrate (Table 3). In contrast to the aerobic expression, the tnrA mutation resulted in only slightly lower β-galactosidase levels in cells grown anaerobically in 2× YT medium containing either nitrate or nitrite (Table 5). These results demonstrate that nasDEF transcription from the nasD promoter is activated by TnrA during nitrogen-limited growth and by ResDE and nitrite under anaerobiosis.
ResDE, not TnrA, is required for nasD transcription during nitrogen-limited anaerobic growth.
The results noted above raised a question whether TnrA or ResDE (or both) is involved in activation of nasDEF during nitrogen-limited anaerobic growth. In an attempt to answer this question, β-galactosidase activity from nasD-lacZ was examined in LAB2854 (wild type), LAB2862 (ΔresDE), and LAB2911 (tnrA) strains grown anaerobically in TSS medium containing nitrate or nitrate plus ammonium (Table 5). β-Galactosidase activity in the wild-type cells grown in the presence of nitrate and ammonium was comparable to the activity in the absence of ammonium, confirming that nasD expression is not repressed by conditions of nitrogen excess during anaerobic growth. However, mutation in resDE but not in trnA abolished β-galactosidase activity under both nitrogen-excess and nitrogen-limited growth conditions. This result clearly shows that ResDE is mainly (if not exclusively) responsible for nasDEF expression during anaerobic growth regardless of nitrogen sources.
Identification of nasD transcriptional start sites.
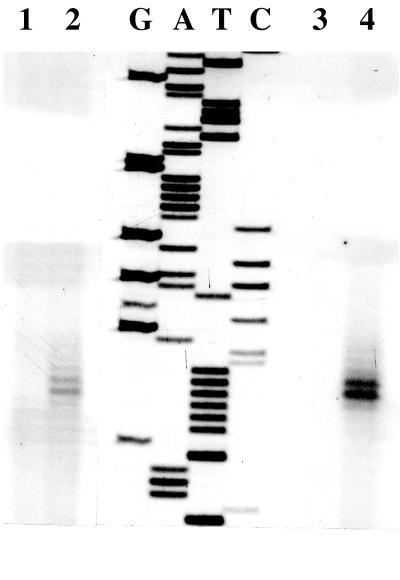
Primer extension analysis was carried out to identify the transcriptional start site for the nasD gene and to determine whether nasD is transcribed from the same start site when activated by nitrogen limitation as when activated by oxygen limitation. No primer extension product was observed with RNA isolated from cells grown aerobically in 2× YT medium or in TSS medium supplemented with a good nitrogen source (ammonium) (Fig. 4). When RNA was isolated from cells cultured anaerobically in 2× YT medium or aerobically in TSS medium with a poor nitrogen source (nitrate), primer extension products which corresponded to RNA with 5′ ends located 30 and 31 bp upstream of the nasD start codon were detected. There is a consensus TnrA box (TGTNAN7TNACA) centered 47 to 48 bases upstream of the transcriptional start site (Fig. 1). This result shows that nasDEF is most likely transcribed from the same transcriptional start sites during aerobic and anaerobic growth.
FIG. 4.
Determination of the transcription start site of nasD by primer extension analysis. Total RNA was isolated from LAB2854 cells grown aerobically in TSS medium with 0.2% NH4Cl (lane 1) or 0.2% KNO3 (lane 2). RNA was also prepared from LAB2854 cells grown in 2× YT with 1% glucose, 20 mM phosphate buffer (pH 7.0), and 20 mM KNO3 under aerobic (lane 3) or anaerobic (lane 4) conditions. The same oligonucleotide primer used for the primer extension analysis was used for sequencing by the dideoxy chain termination method (lanes G, A, T, and C).
DISCUSSION
B. subtilis can utilize nitrate or nitrite as sole nitrogen source, and nitrate and nitrite are also utilized as electron acceptors for anaerobic respiration. While nitrate reduction is catalyzed by an aerobic assimilatory (NasBC) or an anaerobic respiratory (NarGHJI) enzyme, this paper shows that nitrite reduction is performed by a single soluble NADH-dependent reductase (NasDE). We also identified a nasD promoter utilized for nasDEF) transcription that is induced by oxygen limitation via ResDE. Moreover, transcription of nasDEF was shown to be activated by TnrA during nitrogen-limited aerobic growth.
The TnrA-regulated promoters (nrgAB, gabP P2, nasA, and nasB) were shown to have a common dyad symmetry sequence (TGTNAN7TNACA) upstream of the transcriptional start site (2, 14, 21). Recent studies have further demonstrated that the consensus dyad symmetry sequences function as TnrA binding sites and that high-level activation of the nrgAB promoter occurs only when the TnrA site is centered 49 to 51 bp upstream of the transcriptional start site (23). The TnrA site in the nasD promoter is located 47 to 48 bp, instead of 49 to 51 bp, upstream of the transcription start site, but it is not known at present if the TnrA-dependent control of nasD is significantly different from that of nrgAB. For the poor nitrogen sources tested, nasD-lacZ expression was highest when nitrite was present, with nitrate next, followed by glutamate. The stimulatory effect of nitrite (and nitrate probably after reduction to nitrite) is likely to be mediated mainly by ResDE since the resDE mutant exhibited much-reduced nitrite-dependent activation of nasD expression (Table 4). Furthermore, the positive effect of nitrite on the nasD expression is completely dependent on TnrA. These results suggest a possible interaction of TnrA and ResD in the presence of nitrite that is required for full induction of nasD transcription during nitrogen-limited aerobic growth. Since nasC expression is not increased during aerobic growth with nitrite (Table 3), the interaction between TnrA and ResD, if it exists, could be specific for nasD transcription among the TnrA-dependent promoters. As discussed below, under anaerobic conditions, ResDE and the presence of nitrite are sufficient to activate nasD transcription. Although significant levels of aerobic nasD expression were observed in the resDE mutant, aerobic nitrite reductase activity was not detected, suggesting that ResDE may affect nitrite reductase activity during aerobic growth. Alternatively, this difference may be attributed to the sensitivity limits of the nitrite reductase assay. As noted in Table 2, this spectroscopic assay cannot detect activity less than 40 × 10−4 U/mg of protein, indicating that the resDE mutant could have 30% of the wild-type activity, which corresponds to the result that nasD-lacZ activity in the resDE mutant is 30% of that in the wild-type strain during aerobic growth with nitrite (Table 4). In fact, the resDE mutant is able to grow at a reduced rate with nitrite as sole nitrogen source, indicating that the low nitrite reductase activity in the resDE mutant is sufficient to allow cells to assimilate nitrite.
Anaerobically induced transcription of nasDEF during nitrogen-limited growth as well as under conditions of nitrogen excess is dependent on the ResDE two-component signal transduction system. The mutation in resDE nearly abolished nasD expression, indicating that TnrA fails to activate nasDEF expression under anaerobic conditions even when nitrogen is limited. There are some possible explanations for this latter result that can be tested experimentally. For example, expression of tnrA may be repressed during anaerobic growth; conversely, TnrA protein may not respond to nitrogen limitation or the activity of TnrA may be inhibited under anaerobic conditions. A previous study suggested that TnrA receives a signal for nitrogen availability that is generated by glutamine synthetase (22). Possibly, glutamine synthetase activity is affected by anaerobic respiration, resulting in loss of TnrA activity under these conditions. The tnrA mutant is unable to grow aerobically with nitrate or nitrite as sole nitrogen source, which is at least partly due to the inability to activate the nas genes (22). In contrast, the tnrA mutant can grow well anaerobically with nitrate or nitrite as sole nitrogen source (data not shown). This indicates that the respiratory nitrate reductase (NarGHJI) functions together with NasDE nitrite reductase during nitrogen-limited anaerobic growth. In fact, the narGH mutant was unable to grow anaerobically with nitrate as sole nitrogen source, but the nasBC mutant grew well (data not shown).
How is ResDE involved in transcriptional activation of nasDEF by oxygen limitation? ResD (response regulator) and ResE (kinase sensor) are known to be required for aerobic and anaerobic respiration in B. subtilis (20). Three anaerobically induced genes are known to be positively controlled by ResDE: namely, fnr (16), hmp (8), and nasDEF. Whether ResD phosphorylated by the ResE kinase binds directly to these promoters remains to be determined. Analysis of ResDE-controlled promoters is in progress, and the possibility that ResD directly binds to these promoters is under examination in order to elucidate ResDE-dependent anaerobic regulation in B. subtilis.
ACKNOWLEDGMENTS
We thank Peter Zuber and Mitsuo Ogura for valuable discussions and R. K. Thauer for continuous support. We thank Peter Zuber for critical reading of the manuscript. We also thank Lewis Wray and Susan Fisher for providing the tnrA mutant.
The research at LSUMC was supported by NSF grant MCB9722885, and the research at Albert-Ludwigs-Universität Freiburg was supported by grants from the Sonderforschungsbereich 395 of the Deutsche Forschungsgemeinschaft, the Max Planck Society, the University of Freiburg, and the Fonds der Chemischen Industrie.
REFERENCES
- 1.Cruz Ramos H, Boursier L, Moszer I, Kunst F, Danchin A, Glaser P. Anaerobic transcription activation in Bacillus subtilis: identification of distinct FNR-dependent and -independent regulatory mechanisms. EMBO J. 1995;14:5984–5994. doi: 10.1002/j.1460-2075.1995.tb00287.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ferson A E, Wray L V, Fisher S H. Expression of the Bacillus subtilis gabP gene is regulated independently in response to nitrogen and amino acid availability. Mol Microbiol. 1996;22:693–701. doi: 10.1046/j.1365-2958.1996.d01-1720.x. [DOI] [PubMed] [Google Scholar]
- 3.Glaser P, Danchin A, Kunst F, Zuber P, Nakano M M. Identification and isolation of a gene required for nitrate assimilation and anaerobic growth of Bacillus subtilis. J Bacteriol. 1995;177:1112–1115. doi: 10.1128/jb.177.4.1112-1115.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hoffmann T, Frankenberg N, Marino M, Jahn D. Ammonification in Bacillus subtilis utilizing dissimilatory nitrite reductase is dependent on resDE. J Bacteriol. 1998;180:186–189. doi: 10.1128/jb.180.1.186-189.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hoffmann T, Troup B, Szabo A, Hungerer C, Jahn D. The anaerobic life of Bacillus subtilis: cloning of the genes encoding the respiratory nitrate reductase system. FEMS Microbiol Lett. 1995;131:219–225. doi: 10.1111/j.1574-6968.1995.tb07780.x. [DOI] [PubMed] [Google Scholar]
- 6.Kenny T J, Moran C P., Jr Genetic evidence for interaction of ςA with two promoters in Bacillus subtilis. J Bacteriol. 1991;173:3282–3290. doi: 10.1128/jb.173.11.3282-3290.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kunst F, Ogasawara N, Moszer I, Albertini A M, Alloni G, Azevedo V, Bertero M G, Bessieres P, Bolotin A, Borchert S, Borriss R, Boursier L, Brans A, Braun M, Brignell S C, Bron S, Brouillet S, Bruschi C V, Caldwell B, Capuano V, Carter N M, Choi S K, Codani J J, Connerton I F, Danchin A E A. The complete genome sequence of the Gram-positive bacterium Bacillus subtilis. Nature. 1997;390:249–256. doi: 10.1038/36786. [DOI] [PubMed] [Google Scholar]
- 8.LaCelle M, Kumano M, Kurita K, Yamane K, Zuber P, Nakano M M. Oxygen-controlled regulation of flavohemoglobin gene in Bacillus subtilis. J Bacteriol. 1996;178:3803–3808. doi: 10.1128/jb.178.13.3803-3808.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Miller J H. Experiments in molecular genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1972. [Google Scholar]
- 10.Nakano M M, Dailly Y P, Zuber P, Clark D P. Characterization of anaerobic fermentative growth of Bacillus subtilis: identification of fermentation end products and genes required for growth. J Bacteriol. 1997;179:6749–6755. doi: 10.1128/jb.179.21.6749-6755.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nakano M M, Hulett F M. Adaptation of Bacillus subtilis to oxygen limitation. FEMS Microbiol Lett. 1997;157:1–7. doi: 10.1111/j.1574-6968.1997.tb12744.x. [DOI] [PubMed] [Google Scholar]
- 12.Nakano M M, Marahiel M A, Zuber P. Identification of a genetic locus required for biosynthesis of the lipopeptide antibiotic surfactin in Bacillus subtilis. J Bacteriol. 1988;170:5662–5668. doi: 10.1128/jb.170.12.5662-5668.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nakano M M, Xia L, Zuber P. Transcription initiation region of the srfA operon, which is controlled by the comP-comA signal transduction system in Bacillus subtilis. J Bacteriol. 1991;173:5487–5493. doi: 10.1128/jb.173.17.5487-5493.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nakano M M, Yang F, Hardin P, Zuber P. Nitrogen regulation of nasA and the nasB operon, which encode genes required for nitrate assimilation in Bacillus subtilis. J Bacteriol. 1995;177:573–579. doi: 10.1128/jb.177.3.573-579.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nakano M M, Zuber P. Anaerobic growth of a “strict aerobe” (Bacillus subtilis) Annu Rev Microbiol. 1998;52:165–190. doi: 10.1146/annurev.micro.52.1.165. [DOI] [PubMed] [Google Scholar]
- 16.Nakano M M, Zuber P, Glaser P, Danchin A, Hulett F M. Two-component regulatory proteins ResD-ResE are required for transcriptional activation of fnr upon oxygen limitation in Bacillus subtilis. J Bacteriol. 1996;178:3796–3802. doi: 10.1128/jb.178.13.3796-3802.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ogawa K, Akagawa E, Yamane K, Sun Z-W, LaCelle M, Zuber P, Nakano M M. The nasB operon and nasA gene are required for nitrate/nitrite assimilation in Bacillus subtilis. J Bacteriol. 1995;177:1409–1413. doi: 10.1128/jb.177.5.1409-1413.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rosenkrantz M S, Dingman D W, Sonenshein A L. Bacillus subtilis citB gene is regulated synergistically by glucose and glutamine. J Bacteriol. 1985;164:155–164. doi: 10.1128/jb.164.1.155-164.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sanger F, Nicklen S, Coulson A R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sun G, Sharkova E, Chesnut R, Birkey S, Duggan M F, Sorokin A, Pujic P, Ehrlich S D, Hulett F M. Regulators of aerobic and anaerobic respiration in Bacillus subtilis. J Bacteriol. 1996;178:1374–1385. doi: 10.1128/jb.178.5.1374-1385.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wray L V, Jr, Atkinson M R, Fisher S H. The nitrogen-regulated Bacillus subtilis nrgAB operon encodes a membrane protein and a protein highly similar to the Escherichia coli glnB-encoded PII protein. J Bacteriol. 1994;176:108–114. doi: 10.1128/jb.176.1.108-114.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wray L V, Jr, Ferson A E, Rohrer K, Fisher S H. TnrA, a transcription factor required for global nitrogen regulation in Bacillus subtilis. Proc Natl Acad Sci USA. 1996;93:8841–8845. doi: 10.1073/pnas.93.17.8841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wray L V, Jr, Zalieckas J M, Ferson A E, Fisher S H. Mutational analysis of the TnrA-binding sites in the Bacillus subtilis nrgAB and gabP promoter regions. J Bacteriol. 1998;180:2943–2949. doi: 10.1128/jb.180.11.2943-2949.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zuber P, Losick R. Role of AbrB in Spo0A- and Spo0B-dependent utilization of a sporulation promoter in Bacillus subtilis. J Bacteriol. 1987;169:2223–2230. doi: 10.1128/jb.169.5.2223-2230.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]