Abstract
Glycan-binding receptors known as lectins represent a class of potential therapeutic targets. Yet, the therapeutic potential of targeting lectins remains largely untapped due in part to limitations in tools for building glycan-based drugs. One group of desirable structures is proteins with noncanonical glycans. Cell-free protein synthesis systems have matured as a promising approach for making glycoproteins that may overcome current limitations and enable new glycoprotein medicines. Yet, this approach has not been applied to the construction of proteins with noncanonical glycans. To address this limitation, we develop a cell-free glycoprotein synthesis platform for building noncanonical glycans and, specifically, clickable azido-sialoglycoproteins (called GlycoCAP). The GlycoCAP platform uses an Escherichia coli-based cell-free protein synthesis system for the site-specific installation of noncanonical glycans onto proteins with a high degree of homogeneity and efficiency. As a model, we construct four noncanonical glycans onto a dust mite allergen (Der p 2): α2,3 C5-azido-sialyllactose, α2,3 C9-azido-sialyllactose, α2,6 C5-azido-sialyllactose, and α2,6 C9-azidosialyllactose. Through a series of optimizations, we achieve more than 60% sialylation efficiency with a noncanonical azido-sialic acid. We then show that the azide click handle can be conjugated with a model fluorophore using both strain-promoted and copper-catalyzed click chemistry. We anticipate that GlycoCAP will facilitate the development and discovery of glycan-based drugs by granting access to a wider variety of possible noncanonical glycan structures and also provide an approach for functionalizing glycoproteins by click chemistry conjugation.
Keywords: bacterial glycoengineering, cell-free protein synthesis, click chemistry, lectins, noncanonical glycans
Graphical Abstract
INTRODUCTION
Lectins are glycan-binding receptors that represent a promising class of therapeutic targets. Sialic acid-binding immunoglobulin-like lectins (Siglecs) are one example of a lectin family that has shown particular promise as targets in treating allergic inflammation and preventing anaphylaxis.1–14 Siglecs are transmembrane receptors that bind sialic acid-containing glycans and are differentially expressed on immune cells such that each immune cell has its own Siglec expression profile.15–19 Most, but not all, Siglecs have immune suppressive activity by virtue of possessing cytoplasmic immunoreceptor tyrosine-based inhibitory motifs that recruit phosphatases.19 Therefore, Siglecs not only offer an avenue for immune cell inhibition but also provide an approach for cells-pecific targeting.
Despite their promise, the full potential of lectin-targeting therapeutics, like Siglecs, remains untapped. This is because lectins specifically bind glycans, and we lack adequate tools to rapidly build and manufacture a multiplicity of structures needed for biochemical, mechanistic, and structural studies to create glycan-based, lectin-binding drugs. Current approaches often utilize mammalian cell culture, which yields heterogeneous glycan populations,20–23 or chemoenzymatic synthesis, which produces homogeneous glycan populations but can be expensive and difficult to scale up.5,7,11,12,24–26
Cell-free protein synthesis (CFPS) systems from Escherichia coli-based extracts have emerged as an approach to rapidly make glycoproteins27–29 and could overcome the challenges of current manufacturing methods. Making glycoproteins in CFPS systems involves expressing glycosyltransferases from other organisms and supplementing the reaction with appropriate sugar substrates (e.g., UDP-glucose, lipid-linked oligosaccharides) to install glycans onto proteins. Cell-free glycoprotein synthesis systems have several advantages over conventional cellular manufacturing strategies. First, because E. coli lacks endogenous glycosylation machinery, site-specific glycosylation occurs without competition from other host glycosyltransferases. Second, cell-free systems are not limited by viability requirements, thus avoiding constraints arising from noncanonical substrates.30 Third, the lack of a cell membrane circumvents the need for membrane transport of provided sugar substrates. Finally, cell-free systems are amenable to scale-up for manufacturing, having demonstrated linear scalability over a six order of magnitude range in volume to reach the 100 L milestone.31,32
To date, cell-free glycoprotein synthesis systems have been developed for making human and bacterial glycoproteins,33–42 designing protein glycosylation sites,43,44 and modularly constructing glycosylation pathways.45 However, while cell-free systems offer promise for making glycoproteins, there is one group of glycan structures that has shown therapeutic promise but has, to our knowledge, never been made in an integrated cell-free system: that group is noncanonical glycans.
Noncanonical glycans and, more specifically, clickable azido-sialoglycans have been used to optimize lectin-binding pocket interactions by chemically modifying sialic acid-containing glycans through click chemistry conjugation of small chemical groups.46–48 This approach has shown great promise in targeting Siglecs to treat allergy and autoimmune disease.5,11–14,49–52 Clickable azido-sialoglycans also offer opportunities for creating dual function therapeutics, functionalizing glycoprotein with favorable properties, and facilitating scientific research by providing methods for labeling and localizing proteins of interest.53–55
Here, we set out to leverage the advantages of a cell-free system to harness the promise of noncanonical glycans. Toward this aim, we developed a bacterial-based, cell-free glycosylation platform that enables site-specific installation of clickable azido-sialoglycans onto proteins (called GlycoCAP) (Figure 1A). The key idea was to create cell-free biosynthesis “units” made from crude cell lysates that are selectively enriched with pathway enzymes expressed directly in lysates by CFPS. Then, these units are assembled modularly, in a mix-and-match fashion, to build defined glycosylation pathways harboring noncanonical glycans (Figure 1B). As proof-of-concept, we apply GlycoCAP for the synthesis of four Siglec ligands, which are amenable to strain-promoted and copper-catalyzed click chemistry modification. The GlycoCAP platform contributes a set of tools for constructing noncanonical glycans onto proteins, which could open opportunities for therapeutically targeting Siglecs, among other lectins, and offers strategies for protein labeling, development of dual function therapeutics, and protein localization.
Figure 1.
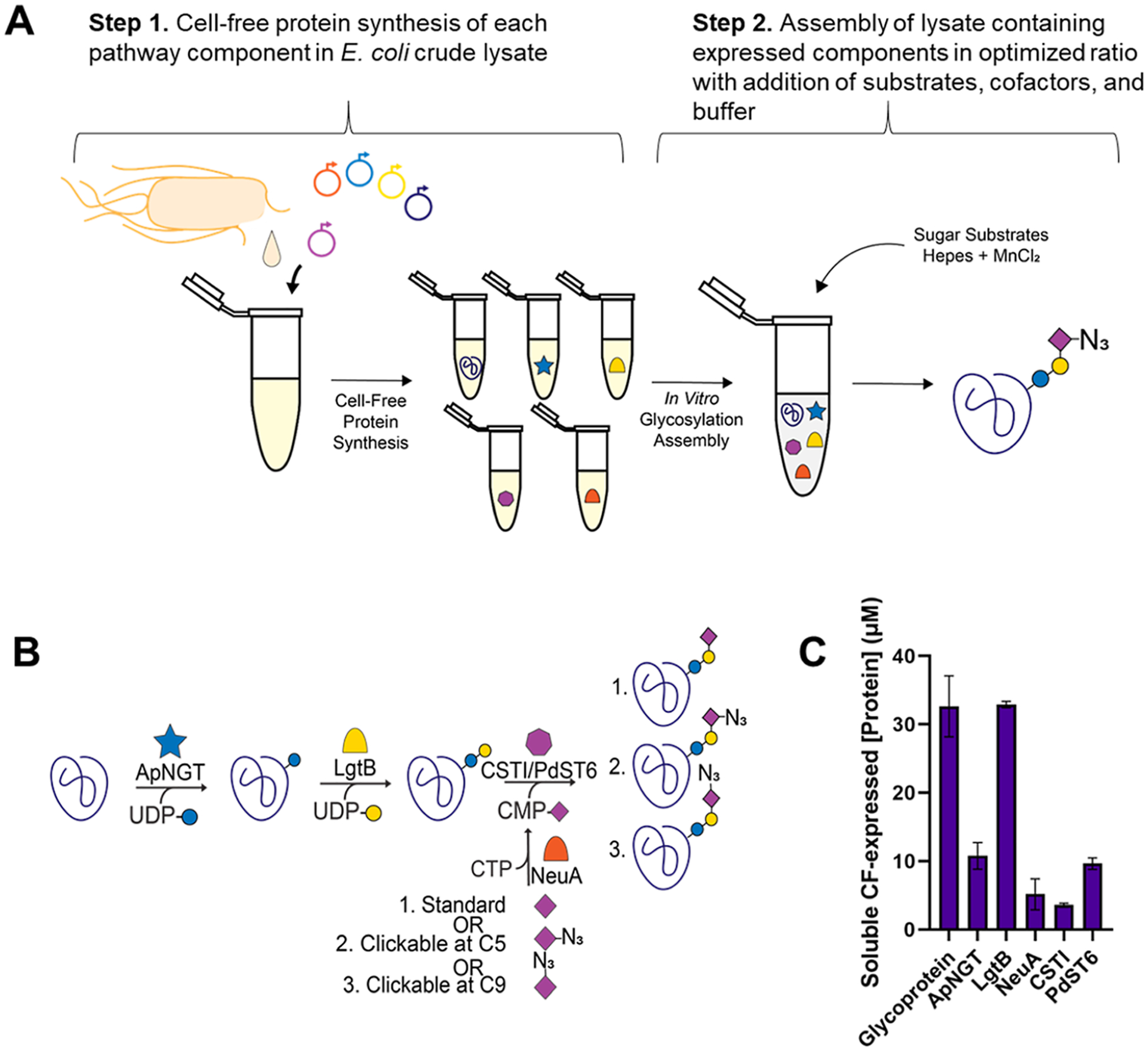
GlycoCAP platform: a cell-free glycosylation platform for building clickable azido-glycoproteins. (A) Plasmids encoding each component of the glycosylation pathway are supplemented in lysate harvested from E. coli cells along with necessary cofactors and building blocks for transcription and translation. After overnight expression, cell-free expressed components are mixed in optimized proportions along with sugar substrates and cofactors in an in vitro glycosylation (IVG) reaction. (B) Components of the pathway include (i) a target glycoprotein designed with an amino acid sequence recognized by (ii) ApNGT for modification with glucose by N-linked glycosylation; (iii) LgtB elaborates galactose onto glucose in a β1,4 linkage, and either (iv) CSTI or PdST6 can be added to elaborate a sialic acid on galactose in an α2,3 or α2,6 linkage, respectively. Finally, (v) NeuA is required to convert noncanonical sugars, such as C5-azido-sialic acid or C9-azido-sialic acid, into activated sugar substrates for CSTI/PdST6 by conjugating the sugars to a phosphonucleotide. (C) Soluble concentration of each component in the CFPS reaction after expression is detected by radioactive incorporation of [14C]-leucine (n = 3; error bars = standard deviation).
RESULTS
GlycoCAP Platform.
The goal of this work was to create a bacterial-based, cell-free platform for installing noncanonical glycans onto desired target glycoproteins. Specifically, we focused on site-specific installation of azido-sialyllactose onto proteins as the azide handle on the glycan grants an avenue for conjugation by click chemistry for Siglec therapeutics. To achieve our goal, we (i) established a base pathway for installing sialyllactose onto proteins, (ii) implemented and optimized a pathway required for activating and incorporating azido-glycan substrates, and (iii) applied our pathway to produce azide-containing glycoproteins that could be modified through bioorthogonal chemistry.
Establishing Conditions for Installing Sialyllactose.
Initially, we set out to construct a minimal trisaccharide structure terminating in sialic acid on a model protein target. This involved two steps. First, we individually expressed multiple components of a bacterial glycosylation pathway we previously established45 by CFPS in the lysate of an engineered E. coli strain.56 Pathway components include (i) the target glycoprotein, which has been designed with an amino acid sequence43 recognized for N-linked glycosylation (Table S1), (ii) ApNGT, which is a glycosyltransferase from Actinobacillus pleuropneumoniae and installs glucose onto an asparagine residue,57,58 (iii), LgtB, which is from Neisseria gonorrhoeae and elaborates a galactose onto the glucose in a β1,4 linkage, and (iv) CSTI, from Campylobacter jejuni, which elaborates a sialic acid onto the lactose in an α2,3 linkage, or PdST6, from Photobacterium damselae, which elaborates a sialic acid onto the lactose in an α2,6 linkage. Soluble expression and yield of pathway components were quantified by [14C]-leucine incorporation (Figures 1C and S1).
In step 2, following the expression of pathway components, we assembled in vitro glycosylation (IVG) reactions such that the reactions were composed of an optimized percent volume of each CF-expressed component and optimized molar concentrations of substrates and cofactors based on parameters previously published.45 IVG reactions included CF-expressed (i) target glycoprotein (45% by volume), (ii) ApNGT (2%), (iii) LgtB (7.4%), and (iv) CSTI (13%) or PdST6 (13%) along with (v) activated sugar donors UDP-glucose (2.5 mM), UDP-galactose (2.5 mM), and CMP-sialic acid (2.5 mM) and MnCl2 (10 mM) and HEPES (23 mM) buffer. As a model protein, we used dust mite allergen (Der p 2) for glycan installation.59,60 IVG reactions were incubated overnight, resupplemented with sugar substrate, and incubated for an additional 24 h. Glycosylated allergen was isolated by affinity-tag purification, and liquid chromatography–mass spectrometry (LC–MS) was used to calculate the glycosylation efficiency. We determined a base case standard for glycosylation efficiency, which we determined to be 67 ± 7% (Figure 2A).
Figure 2.
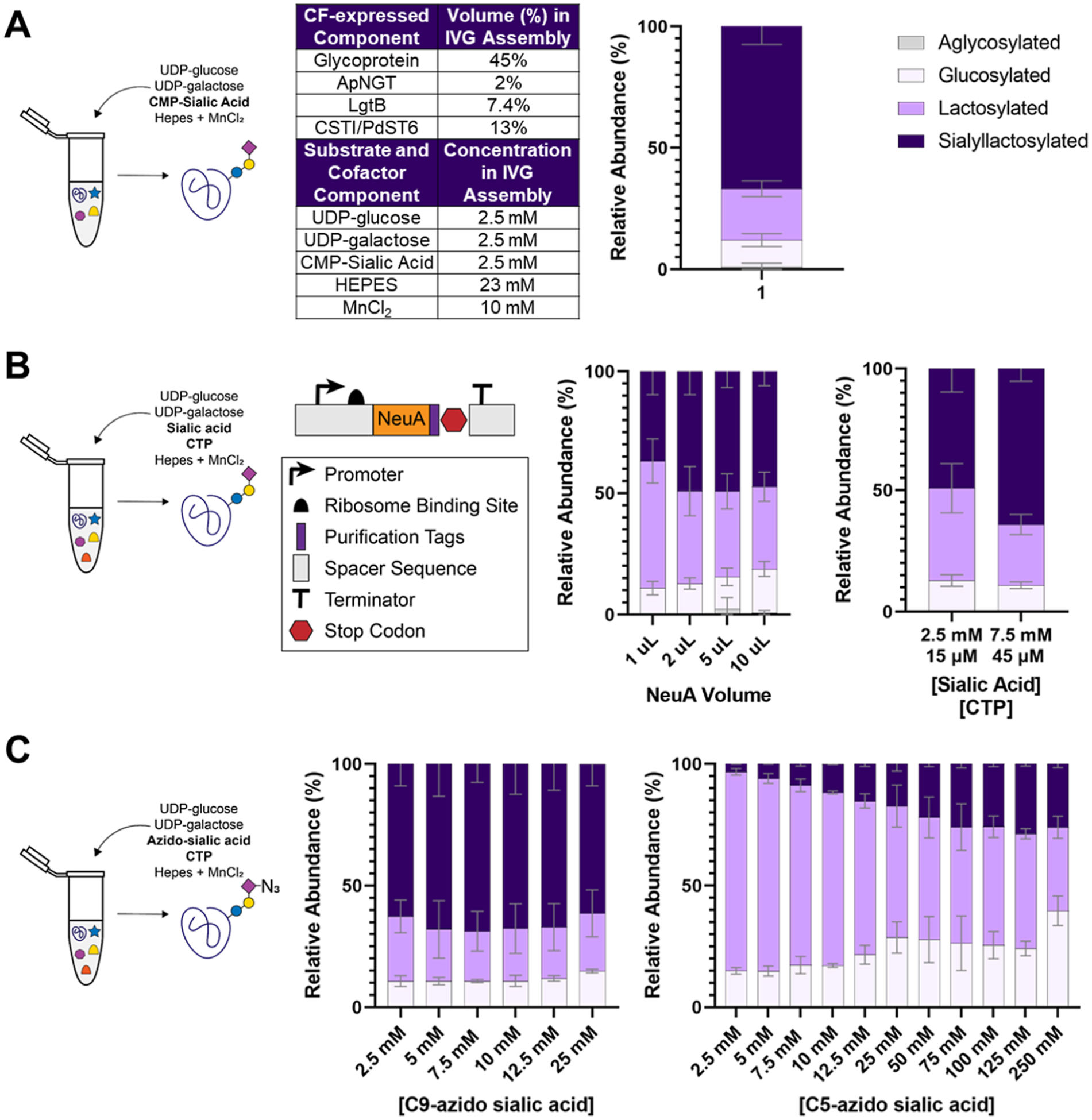
Optimization of GlycoCAP for noncanonical sugar incorporation. (A) IVG reactions were assembled based on optimized ratios (i.e., where 13% of the final reaction volume was made up of either crude lysate expressing PdST6 if an α2,6 linkage was desired or crude lysate expressing CSTI if an α2,3 linkage was desired) to achieve glycosylation when activated CMP-sialic acid is supplemented directly and NeuA is not required to install sialyllactose onto the target glycoprotein. Under these conditions, 67 ± 7% glycosylation efficiency is achieved (n = 2; error bars = average error). The color-coded legend applies to all bar graphs shown. (B) Plasmid encoding NeuA was designed for CFPS. Once expressed, NeuA was assembled into the IVG reaction with its needed substrates: nonactivated sialic acid and CTP. Titration of NeuA was performed to determine 2 μL (which corresponds to 4.5% volume) as the optimal volume to add to the IVG assembly (n = 4; error bars = standard deviation). An additional titration of standard sialic acid shows that increasing [sialic acid] to 7.5 mM and [CTP] to 45 μM enhances the glycosylation efficiency to 64 ± 5% (n = 4; error bars = standard deviation). (C) Titration of noncanonical sialic acids shows that C9-azido-sialic acid is as efficiently incorporated as standard sialic acid, with 7.5 mM sialic acid and 45 μM CTP being the optimal concentration to achieve 69 ± 8% efficiency. A concentration of 125 mM C5-azido-sialic acid is required to achieve the optimal glycosylation efficiency of 29 ± 1% (n = 2; error bars = average error). All glycosylation efficiencies are determined by MS of trypsinized samples and estimated by the area under the curve of each pathway intermediate’s extracted ion chromatogram based on the theoretical mass of the resulting glycopeptide.
Extending the Pathway for Incorporation of Nonactivated Sialic Acids.
With a base case glycoprotein synthesis system in place, we turned our efforts toward incorporating noncanonical glycans. Since noncanonical sialic acids are only available in their nonactivated form (not conjugated to a phosphonucleotide), incorporation of a noncanonical sialic acid requires the addition of N-acylneur-aminate cytidylyltransferase (NeuA) (Figure S2 and Table S2), which catalyzes the formation of CMP-sialic acid from canonical or noncanonical sialic acid and cytidine triphosphate (CTP) to produce the appropriate, activated sugar donor for CSTI/PdST6 enzymes. We performed a titration of CFPS-expressed NeuA into a series of IVG reactions where 2.5 mM standard sialic acid and 15 μM CTP were supplemented. NeuA was added at a volume of 1, 2, 5, or 10 μL. Adding 2 μL (corresponding to 4.5% of the reaction volume) was enough to achieve the optimal glycosylation efficiency of 49 ± 10%, meaning the relative abundance of the final desired sialylated product is ~49% and the remaining species are earlier pathway intermediates (Figure 2B).
To determine whether the lower efficiency compared to the base case standard is a result of the IVG assembly being substrate limited, we titrated additional sialic acid and CTP. Increasing [sialic acid] to 7.5 mM and [CTP] to 45 μM achieved a glycosylation efficiency of 64 ± 5%, which is on par with the base case standard using the activated sialic CMP-acid (Figure 2B).
Incorporation of C9- and C5-Azido-Sialic Acids.
We next performed a titration of azido-sialic acids where the azide group is either at the 9th carbon of sialic acid (C9-azido-sialic acid) or where an azidoacetyl group is on the 5th carbon of sialic acid (C5-azido-sialic acid) (Figure S3). The goal was to test whether the enzymes of our pathway are permissive to an additional chemical group on their sialic acid substrate. C9-azido-sialic acid was incorporated at an equal or better efficiency than canonical sialic acid, where 7.5 mM C9-azido-sialic acid and 15 μM CTP led to 69 ± 8% glycosylation efficiency (Figure 2C). C5-azido-sialic acid could also be incorporated. However, higher concentrations of C5-azido-sialic acid were required and still did not match the base case efficiency with the canonical sialic acid (Figure 2A), indicating that the pathway enzymes are less permissive to C5-azido-sialic acid than to C9-azido-sialic acid, possibly due to disrupted binding pocket interactions caused by the azide group at the 5th carbon position. By adding 125 mM C5-azido-sialic acid and 750 μM CTP, we were able to achieve a glycosylation efficiency of 29 ± 1% (Figure 2C).
GlycoCAP Production of α2,3-C5-Azido-sialyllactose, α2,3-C9-Azido-sialyllactose, α2,6-C5-Azido-sialyllactose, and α2,6-C9-Azido-sialyllactose.
With optimal conditions established for noncanonical azido-sialic acid incorporation, we installed four different clickable azido-sialoglycans onto proteins (Figure 3A). Under optimized GlycoCAP conditions, α2,3-C5-azido-sialyllactose was produced at 23 ± 7%, efficiency, α2,3-C9-azido-sialyllactose was produced at 80 ± 8%, efficiency, α2,6-C5-azido-sialyllactose was produced at 23 ± 10%, efficiency, and α2,6-C9-azido-sialyllactose was produced at 64 ± 1% efficiency (Figure 3B). MS confirmed the production of these species and demonstrated the relative proportion of reaction intermediates and the final product present in the final reaction (Figure 3C).
Figure 3.
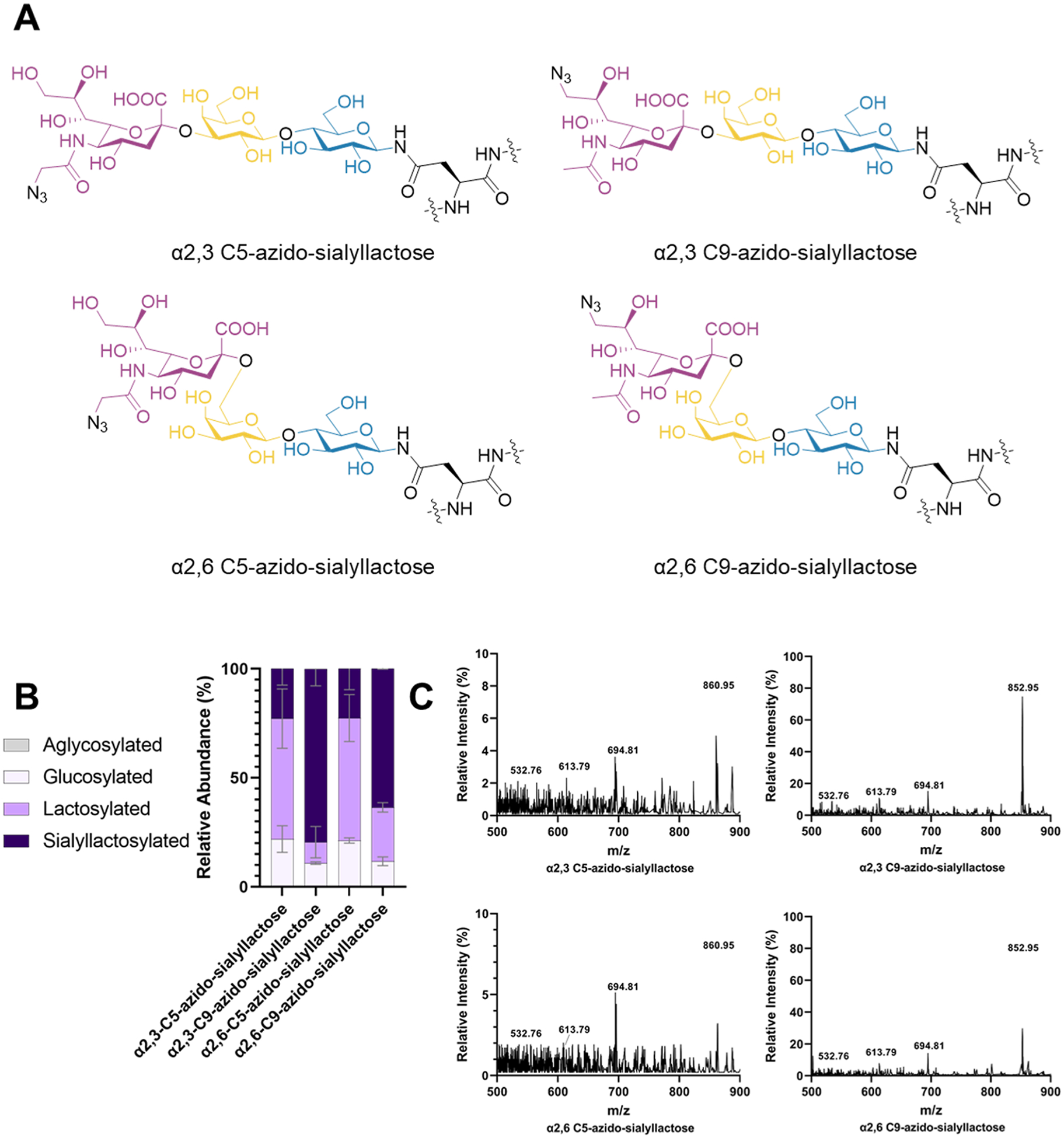
Structures made with optimized conditions of GlycoCAP. (A) Chemical structures installed onto proteins in GlycoCAP. (B) Efficiencies for each species are represented as a bar graph (n = 2; error bars = average error). (C) MS confirmation of each species is shown in relation to the peaks of its pathway intermediates in the +2 charge state. MS data show a representative spectra selected from n = 2 experiments.
Preparative Scale Production of Azido-Sialoglycoproteins.
As a representative synthesis to demonstrate the scalability of the GlycoCAP platform, we performed 1 mL IVGs for each of the four clickable azido-sialoglycoproteins. After purification, we characterized the protein yield by UV absorbance at 280 nm. The abundance of azido-sialylated protein relative to other pathway intermediates was determined by MS and applied as a percentage to the total protein yield to estimate the final product yield. Preparation at this scale yielded approximately 70 μg of C5-azido-α2,3-sialyllactose, 200 μg of C9-azido-α2,3-sialyllactose, 50 μg of C5-azido-α2,6-sialyllactose, and 150 μg of C5-azido-α2,3-sialyllactose (Table 1 and Figure S4).
Table 1.
Data from Preparative Scale Production of Azido-Sialoglycoproteins Where IVGs Were Performed at a 1 mL Scale
| scale up of GlycoCAP azido-sialoglycoprotein production | ||||
|---|---|---|---|---|
| Glycan identity | C5-azido α2,3-sialyllactose | C9-azido α2,3-sialyllactose | C5-azido α2,6-sialyllactose | C9-azido α2,6-sialyllactose |
| Total elution volume (mL) | 1.2 | 1.2 | 1.2 | 1.2 |
| Protein yield (μg/mL) | 347 | 196 | 265 | 194 |
| Total protein (μg) | 416 | 235 | 318 | 233 |
| Azido-sialylation (%) | 18 | 85.0 | 16 | 64 |
| Final product yield (μg) | 73 | 200 | 50 | 148 |
Click Chemistry Conjugation of Clickable Azido-Sialoglycoproteins.
To show that installed azido-sialoglycans are amenable to click chemistry conjugation, we next performed both strain-promoted click chemistry with a fluorophore containing a dibenzocyclooctyne (DBCO) group (Figure 4A, top) and copper-catalyzed click chemistry with a fluorophore containing a standard alkyne (Figure 4A, bottom). In the absence of copper, steric strain around the alkyne is required for successful conjugation with DBCO. Copper-catalyzed reactions yield a smaller intervening triazole ring. Samples were run on a NuPAGE Bis-Tris gel and imaged under fluorescent excitation such that only fluorescent bands are visible. Click conjugation was successful with both methods as indicated by the presence of fluorescent bands at the expected masses for the monomer form of the Der p 2 glycoprotein (~17 kDa) and the dimer form (~34 kDa) (Figure 4B). Additional weight from glycosylation and click modification in addition to incomplete denaturation of Der p 2 due to heat restrictions imposed by the fluorophore’s heat sensitivity and the allergen’s intrinsic stability likely influence its appearance on the gel slightly above its expected mass. The high intensity band at ~34 kDa for the copper-catalyzed click reaction indicates that these conditions favor dimer formation of the target glycoprotein.
Figure 4.
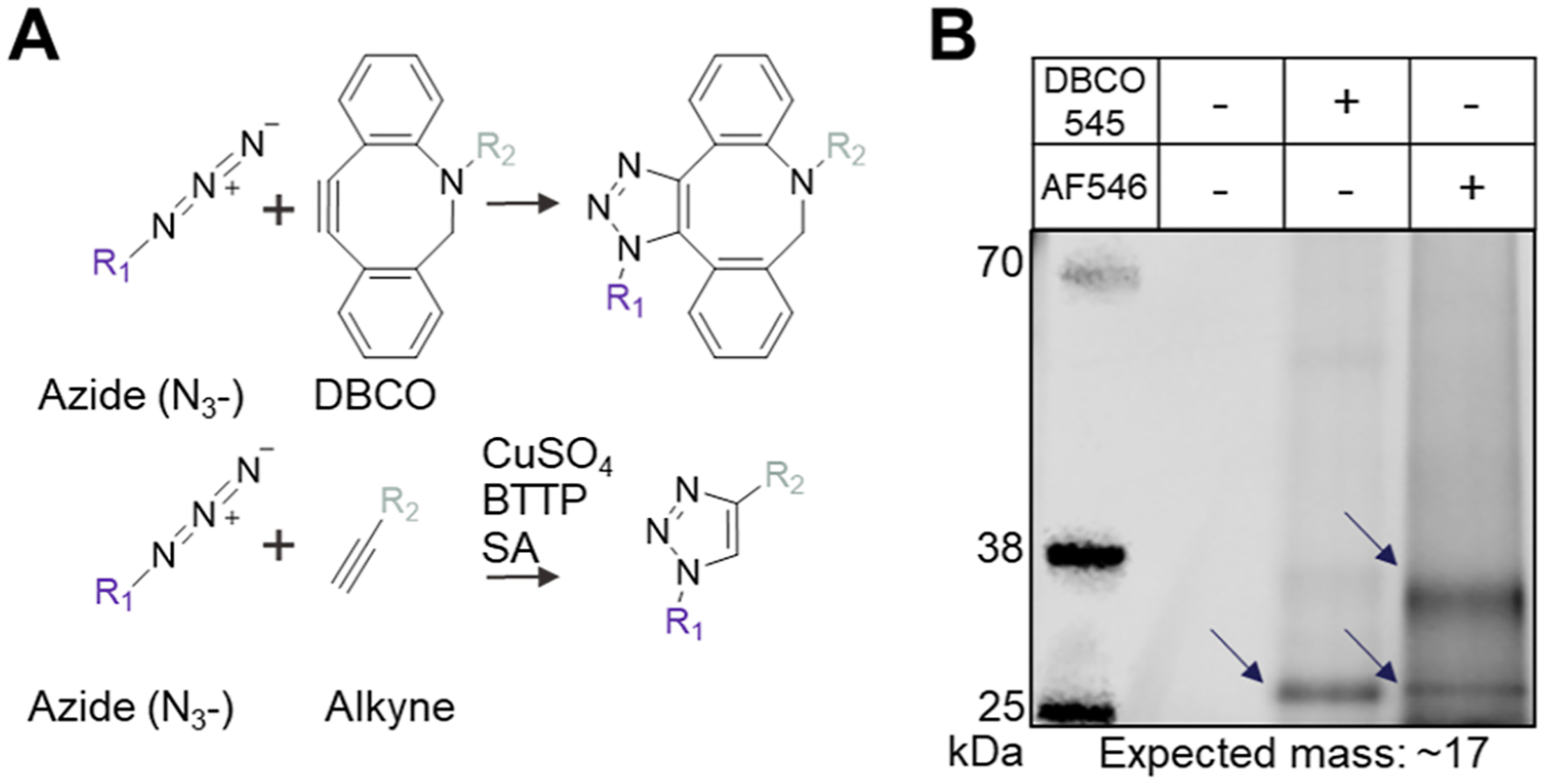
Click chemistry conjugation of azido-sialoglycoproteins. (A) Conjugation by click chemistry with either a strained alkyne (top) or standard alkyne (bottom) is possible with the azido-sialoglycan structures built by GlycoCAP. SA = sodium ascorbate. (B) Fluorescent gel demonstrates successful click conjugation onto a C9-azido-sialoglycoprotein with either a fluorescent strained alkyne (DBCO 545) by strain-promoted click chemistry (lane 3) or a fluorescent standard alkyne (AF546) by copper-catalyzed click chemistry (lane 4). Fluorescent bands are visible at the expected masses for Der p 2 in its monomer form (~17 kDa) and dimer form (~34 kDa) except in the lane where azido-sialoglycoprotein was loaded without any fluorophore conjugation as a negative control (lane 2). Gel representative of n = 3 experiments.
DISCUSSION
Here, we developed the GlycoCAP platform. GlycoCAP enables installation of noncanonical glycans onto proteins in a cell-free glycoprotein synthesis system. We demonstrated the utility of GlycoCAP by producing four noncanonical glycans: α2,3 C5-azido-sialyllactose, α2,3 C9-azido-sialyllactose, α2,6 C5-azido-sialyllactose, and α2,6 C9-azido-sialyllactose, all of which can be functionalized by click chemistry.
GlycoCAP offers several potential advantages for incorporating noncanonical glycans onto proteins. For example, freedom from constraints imposed by living cells may grant access to a wider repertoire of noncanonical glycans than is possible in living cells. As an example, we observed that culturing living E. coli with increasing [C9-azido-sialic acid] led to decreasing optical densities under otherwise equivalent culture conditions (Figure S5). The observation that C9-azido-sialic acid incorporated as efficiently as standard sialic acid in our cell-free system provides proof of concept that our approach can overcome cell toxicity challenges and may be able to build structures otherwise inaccessible.
While GlycoCAP offers some benefits, there remains room for improvement. For example, yields of the desirable products were not 100%. To address this limitation and make the approach suitable for longer, more complex glycans, future efforts can seek to identify glycosyltransferases with higher efficiencies. Reduced permissivity toward C5-azido-sialicacid—possibly due to disrupted binding pocket interactions in NeuA caused by the position of the azide group—could be overcome by future efforts to engineer enzymes to alter promiscuity and efficiency. The GlycoCAP platform could be used to quickly screen a wide range of enzymes toward these efforts.
The GlycoCAP platform offers a range of potential applications. As an example, higher affinity glycan ligands for Siglec binding to generate novel allergy treatments could be pursued. Studies have shown that coengagement of a Siglec receptor with the high affinity IgE receptor (FcεRI) on the surface of mast cells results in robust inhibition of mast cell activation and anaphylaxis.5,12 Siglec-3 (CD33) is expressed on the surface of mast cells and basophils and its endogenous ligand is α2,3 sialyllactose and α2,6 sialyllactose—both of which form the basis of the structures made here. GlycoCAP could be used to install clickable azido-sialoglycans onto allergen, which would engage allergen-specific IgE attached to FcεRI. The azido-sialoglycan could then be chemically modified by clicking on small chemical groups to improve CD33 binding pocket interactions and increase ligand affinity.49,51,52 This would generate a dual-specific therapeutic that coengages IgE/FcεRI and CD33 to inhibit the allergic response. Siglec-8 has been targeted by liposomes decorated with click chemistry modified, sulfated sialoglycans to treat eosinophilic and mast cell diseases,7 and Siglecs have been shown to play a role in neurodegenerative disease61 and cancer.62,63 The tools produced here could aid in the therapeutic development for a range of pathologies.
Looking forward, GlycoCAP provides a potentially powerful approach to discover, study, and optimize glycosylation pathways for noncanonical sugars. This could facilitate basic research and open the door for functionalizing glycoproteins, including conjugating a small-molecule drug to an antibody for localized delivery, modulating drug properties by conjugating property-endowing moieties, or barcoding or labeling proteins of interest with a nucleotide sequence or fluorophore. In addition, GlycoCAP could broaden capabilities for targeting lectins and Siglecs, paving the way for a novel class of medicines that are glycan-based therapeutics.
MATERIALS AND METHODS
Plasmid and DNA Design.
The protein sequence of Der p 2 allergen was retrieved from Uniprot (accession ID: ALL2_DERPT), signaling sequence removed, and then codon optimized for E. coli K12 strains into a DNA sequence using the IDT Codon Optimization Tool. Additional synthetic sequences were added to optimize expression and to enable site-specific glycosylation and affinity purification (Table S1). This insert as well as the NeuA inserts (Table S2) were synthesized into a pJL1 backbone at NdeI and SalI restriction sites by Twist Biosciences.
Plasmid DNA used in CFPS reactions was purified from glycerol stocks provided by Twist Biosciences using the ZymoPURE Midi Kit (Zymo Research D4200). Plasmids have been made available on Addgene (Table S3).
Synthesis of C5- and C9-Azido-Sialic Acids.
To make C5-azido-sialic acid, N-Azidoacetylmannosamine (317 mg, 1.21 mmol) was dissolved in H2O (12.1 mL), followed by addition of sodium pyruvate (665 mg, 5 equiv). Neu5Ac aldolase (8 U mg−1) was added, and the mixture was heated to 37 °C and stirred overnight. The solvents were evaporated, and the crude product was purified by column chromatography (0–20% 0.04 M HCl in MeCN, v/v), yielding the product (344.6 mg, 81.4%) as a white solid. 1H NMR (500 MHz, D2O): δ 4.16–4.06 (m, 4H), 4.01 (t, J = 10.3 Hz, 1H), 3.84 (dd, J = 11.8, 2.7 Hz, 1H), 3.75 (ddd, J = 9.1, 6.3, 2.7 Hz, 1H), 3.62 (dd, J = 11.9, 6.3 Hz, 1H), 3.56 (dd, J = 9.3, 1.2 Hz, 1H), 2.33 (dd, J = 13.1, 5.0 Hz, 1H), 1.89 (dd, J = 13.1, 11.5 Hz, 1H). 13C NMR (126 MHz, D2O): δ 173.5, 171.0, 95.4, 70.2, 70.1, 68.2, 66.5, 63.1, 52.1, 51.9, 38.9 (Figure S3).
C9-azido-sialic acid was synthesized as described previously.64
Harvest and Processing of E. coli Lysate for CFPS.
E. coli lysate was prepared using previously published methods.30 A highly productive engineered strain of MG1655-derived E. coli strain C321.ΔA.75956 was grown in a Sartorius Biostat C+ 10 L fermentor in 10 liters of 2xYTPG media (yeast extract 10 g/L, tryptone 16 g/L, NaCl 5 g/L, K2HPO4 7 g/L, KH2PO4 3 g/L, and glucose 18 g/L, pH = 7.2) at 34 °C with rotation at 250 rpm. T7 RNA polymerase expression was induced with 1 mM isopropyl β-d-1-thiogalactopyranoside (Sigma-Aldrich) at OD = 0.6. At OD = 3.0, cells were put on ice and centrifuged at 8000g for 5 min at 4 °C in a chilled JLA 8.1 rotor in a Beckman Avanti J-25I refrigerated centrifuge. The supernatant was discarded, and the cell pellets were washed three times with S30 buffer (10 mM trisacetate pH 8.2, 14 mM magnesium acetate, 60 mM potassium acetate) and pelleted by centrifugation at 10,000g for 2 min at 4 °C in a Thermo Heraeus Multifuge X3R centrifuge. Cell pellets were flash frozen and stored at −80 °C. After thawing, cells were resuspended in 0.8 mL of S30 buffer per gram of wet cell mass, distributed in 1.4 mL aliquots, and sonicated (Qsonica) at 50% amplitude for a cycle of 45 s on, 59 s off until ~950 Joules was reached. Lysate was kept on ice and spun at 12,000g for 10 min at 4 °C. The supernatant (820 μL) was collected in fresh tubes and incubated at 37° with shaking at 250 rpm for 1 h. Tubes were then spun at 12,000g for 10 min at 4 °C, and the supernatant collected from this final spin (500 μL) comprises the cell-free extract used to assemble CFPS reactions. The extract was flash frozen and stored at −80 °C until ready for use.
Assembly of CFPS Reactions.
CFPS reactions were assembled based on previously published methods with the following building blocks and reagents:30 6 mM magnesium glutamate; 10 mM ammonium glutamate; 130 mM potassium glutamate; 2.646 mM ATP; 1.874 mM each of GTP, UTP, and CTP; 0.075 mg/mL folinic acid, 0.376 mg/mL E. coli tRNA mixture from strain MRE600 (Roche Applied Science); 0.33 mM nicotinamide adenine dinucleotide; 0.27 mM coenzyme-A (CoA); 4 mM oxalic acid; 1 mM putrescine; 1.5 mM spermidine; 57 mM HEPES at pH = 7.2; 2 mM of each of the 20 standard amino acids; 30 mM phosphoenolpyruvate (Roche Applied Science); 13.3 μg/mL designed gene construct in the pJL1 vector; and 27% v/v of E. coli crude lysate (prepared above). Reagents were purchased from Sigma-Aldrich unless otherwise stated. CSTI was expressed under oxidizing conditions, which was assembled as described above with an additional 30 min preincubation of the extract in 14.3 μM iodoacetamide before supplementation into the CFPS reaction and with the additional following reagents: 4 mM oxidized l-glutathione GSSG, 1 mM reduced l-glutathione, and 3 μM purified DsbC from E. coli. CFPS reactions were incubated for 20 h at room temperature, with the exception of LgtB, which was flash frozen after 6 h to prevent protease degradation.
Radioactive Quantification of Proteins Expressed by CFPS.
Radioactive quantification was performed based on previously published methods.30,65 In brief, CFPS reactions were assembled as described above with the addition of 10.67 μM radioactive amino acid, 14C-leucine. A negative control CFPS reaction where no DNA was added was included as a reference control. After 20 h of incubation, the samples were mixed with 0.5 N KOH—to precipitate the protein—in a 1:1 volume ratio (i.e., 5 μL of 0.5 N KOH and 5 μL of CFPS reaction) to quantify the total protein expression. Samples were then spun at 12,000g for 10 min, and the supernatant was mixed with 0.5 N KOH in a 1:1 volume ratio to quantify the soluble protein expression. All CFPS + KOH samples were incubated for 20 min at 37 °C. From each of these mixtures, 4 μL was dispensed onto a filtermat (PerkinElmer, 1450–421) and duplicated on a second filtermat and allowed to dry for 20 min. One filtermat for each set of samples was washed 3× in 5% trichloroacetic acid for 15 min at 4 °C, while the other filtermat was left alone. A sheet of scintillation wax was melted on a hot plate over the filtermat, and the radioactive signal was read from each sample by the Microbeta2 scintillation counter.
Protein yields (μg/mL) are calculated by the following formula:
Protein yields (μg/mL) can be converted to (μM) by the following formula:
Assembly of IVG Reactions.
CFPS reactions were centrifuged at 12,000g for 10 min, and the supernatant was transferred to the final IVG reaction. Lysates were mixed in optimized proportions, where the target glycoprotein comprised 45% of the final reaction volume, ApNGT comprised 2%, LgtB 7.4%, CSTI/PdST6 13%, and NeuA (when included) 4.5%. Activated sugar substrates (UDP-glucose, UDP-galactose, and CMP-sialic acid) were supplemented at a concentration of 2.5 mM. Nonactivated sialic acid and CTP were supplemented at 7.5 mM and 45 μM, respectively. For incorporation of azido-sialic acids, C9-azido-sialic acid should be supplemented at 7.5 mM with CTP at 45 μM and C5-azido-sialic acid should be supplemented at 125 mM with CTP at 750 μM. IVG assemblies were incubated at 30 °C for 24 h, at which time sugar substrates were resupplemented at their optimized concentration and the reactions were incubated for another 24 h at 30 °C.
Glycoprotein Purification.
IVG reactions were diluted in 50 mM NaH2PO4, 300 mM NaCl, pH = 8.0 (Buffer 1) and centrifuged at 12,000g for 10 min. His-tagged, glycosylated Der p 2 allergen was isolated by affinity-tag purification with Ni-NTA magnetic beads (Invitrogen Dynabeads His-tag Isolation and Pulldown, 10104D). Beads were equilibrated with Buffer 1, then resuspended in the IVG reaction supernatant, and allowed to bind for >10 min at 4 °C with end-over-end rotation. Samples were then washed twice with Buffer 1 and eluted with 500 mM imidazole in Buffer 1 with 10 min of end-over-end mixing at room temperature.
The elution was desalted using Zeba spin columns (Thermo Scientific 89883) according to manufacturer’s instructions and collected in nuclease free water (Ambion, AM9937).
LC–MS Analysis of Glycoprotein.
After the samples were trypsinized with 0.0044 μg/μL MS grade trypsin (Thermo Fisher Scientific, PI90057) with an incubation of at least 4 h at 37 °C, the samples were run on the Bruker Impact II ESI QTOF. Samples were first passed through an ACUITY UPLC Peptide BEH C18 column (300 Å, 1.7 μM, 2.1 mm × 100 mM, 186003686 Waters Corp.) equipped with a 10 mm guard column of identical packing (186004629 Waters Corp.). Then, the samples were eluted at a flow rate of 0.5 mL/min and a 40 °C column temperature starting at a 1 min hold of 95% solvent A (100% water and 0.1% formic acid) and 5% solvent B (100% acetonitrile and 0.1% formic acid) with a linear gradient up to 0% solvent A and 100% solvent B for a total run time of 9 min.
MS Data Analysis.
MS data were analyzed using the Bruker software. Theoretical masses in the +2 charge state (Table S4), which was determined to be the predominant form, were used to make the extracted ion chromatogram (EIC) for each pathway intermediate and final product. Glycosylation efficiency was determined by taking the area under the curve (AUC) of each intermediate’s EIC and calculating the relative percentage represented by the AUC of a given intermediate relative to summed AUCs of all possible intermediates. Spectra were plotted using m/z values from 500 to 900 at the elution times encompassing EICs and deconvoluted to emphasize species of interest (Table S5).
GlycoCAP at Preparative Scale.
CFPS reactions were assembled using conditions described above at sufficient volumes for addition into IVG reactions. IVG reactions were assembled at a total volume of 1 mL and were comprised of CF-expressed components in optimized relative proportions and the necessary substrates and cofactors at optimum molar concentration as described above (Figure 2). After incubation and supplementation as described for IVG assembly, His-tagged glycoprotein was purified with Ni-NTA magnetic beads (Invitrogen Dynabeads His-tag Isolation and Pulldown, 10104D). Purified samples were desalted as described above and analyzed by MS for glycosylation efficiency. Protein yield was determined by UV absorbance at 280 nM using a Thermo Nanodrop 2000 spectrophotometer (extinction coefficient: 15470, molecular weight: 17376 Da).
Click Chemistry Conjugation.
Click chemistry reactions were assembled using purified glycoprotein at a total volume of 100 μL. For strain-promoted click chemistry, a stock of 3.4 mM DBCO-PEG4-Fluor 545 (Sigma-Aldrich, 760773) suspended in dimethyl sulfoxide was added to the purified and desalted glycoprotein in water (~250 μg/mL) at a concentration of 400 μM and allowed to incubate at 30 °C for 4 h.
For copper-catalyzed click chemistry, ~10 μg of purified, desalted glycoprotein product suspended in 100 μL of 1% RapiGest (Waters, 186008090) was combined with 300 μM CuSO4, 600 μM BTTP (Click Chemistry Tools, 1414–100), and 200 μM Alexa Fluor 546-Alkyne (Jena Bioscience, CLK-1285–1) and allowed to incubate for at least 1 h at room temperature to allow the ligand complex to form. Sodium ascorbate (2.5 mM) was then added the reaction, and the reaction underwent an incubation period of 3.5 h at room temperature.
Fluorescent Gel.
Licor protein sample loading buffer (LICOR Biosciences 928–40004) and 4 mM DTT were added directly to click chemistry conjugation reactions containing the click-modified glycoprotein. Samples were heat denatured at 70 °C for 3 min and then run on a NuPAGE 4–12% Bis-Tris gel (Invitrogen, NP0321BOX) at 150 V with MOPS buffer (Thermo Fisher Scientific, NP0001). Gel was imaged on a LICOR-Odyssey Fc imaging system on the 600 and 800 channels and formatted in ImageStudio.
Supplementary Material
ACKNOWLEDGMENTS
We would like to acknowledge Dr. Ben Owen, Dr. Fernando “Ralph” Tobias, Dr. Saman Shafaie, and Dr. Arsen Gaisin in the Molecular Structure Education and Research Center (IMSERC) at Northwestern University for their service to the project. We would also like to thank Dr. Kevin Bruemmer and Dr. Lindsay Guzmán in Professor Carolyn Bertozzi’s Group for their guidance on click chemistry. We thank Dr. Weston Kightlinger for helpful discussions.
Funding
This work was supported by the National Institutes of Health (1F31AI165279 to A.H.T.), the Defense Threat Reduction Agency (HDTRA12010004 to M.C.J.), the National Science Foundation (MCB 1936789 to M.C.J.; DGE-1842165 to D.A.W.), and an ERC-Stg (Glycoedit, 758913 to T.J.B.).
Footnotes
Supporting Information
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acssynbio.3c00017.
Figure S1: Total and soluble cell-free expression of pathway enzymes, Figure S2: NeuA DNA expression and glycosylation with and without a CAT linker, Figure S3: NMR spectra of synthesized C5-azido-sialic acid, Figure S4: Coomassie-stained protein gel and EIC of scaled up azido-sialoglycoprotein production, Figure S5: C9-azido-sialic acid demonstrating E. coli cell toxicity, Table S1: DNA design of target glycoprotein, Table S2: NeuA DNA design with and without a CAT linker, Table S3: Addgene IDs for plasmid accession, Table S4: Theoretical m/z values, and Table S5: Peak lists used to generate mass spectra (PDF)
Complete contact information is available at: https://pubs.acs.org/10.1021/acssynbio.3c00017
The authors declare the following competing financial interest(s): B.S.B. receives remuneration for serving on the scientific advisory board of Allakos, Inc. and owns stock in Allakos. He receives consulting fees from Third Harmonic Bio, Lupagen, Sanofi, and Acelyrin. He receives publication-related royalty payments from Elsevier and UpToDate. He is a co-inventor on existing Siglec-8-related patents and thus may be entitled to a share of royalties received by Johns Hopkins University during development and potential sales of such products. B.S.B. is also a co-founder of Allakos, Inc. which makes him subject to certain restrictions under university policy. The terms of this arrangement are being managed by Johns Hopkins University and Northwestern University in accordance with their conflict-of-interest policies. M.C.J. has a financial interest in SwiftScale Biologics, Gauntlet Bio, Pearl Bio, Inc., Design Pharmaceutics, and Stemloop Inc. M.C.J.s interests are reviewed and managed by Northwestern University in accordance with their competing interest policies. All other authors declare no competing interests.
Contributor Information
Ariel Helms Thames, Medical Scientist Training Program, Northwestern University Feinberg School of Medicine, Chicago, Illinois 60611, United States; Center for Synthetic Biology, Northwestern University, Evanston, Illinois 60208, United States; Interdisciplinary Biological Sciences Program, Northwestern University, Evanston, Illinois 60208, United States; Division of Allergy and Immunology, Department of Medicine, Northwestern University Feinberg School of Medicine, Chicago, Illinois 60611, United States.
Sam J. Moons, Synvenio B.V., Nijmegen 6525ED, The Netherlands
Derek A. Wong, Center for Synthetic Biology, Northwestern University, Evanston, Illinois 60208, United States; Department of Chemical and Biological Engineering, Northwestern University, Evanston, Illinois 60208, United States
Thomas J. Boltje, Institute for Molecules and Materials, Radboud University Nijmegen, Nijmegen 6525AJ, The Netherlands
Bruce S. Bochner, Medical Scientist Training Program, Northwestern University Feinberg School of Medicine, Chicago, Illinois 60611, United States; Division of Allergy and Immunology, Department of Medicine, Northwestern University Feinberg School of Medicine, Chicago, Illinois 60611, United States
Michael C. Jewett, Medical Scientist Training Program, Northwestern University Feinberg School of Medicine, Chicago, Illinois 60611, United States; Center for Synthetic Biology, Northwestern University, Evanston, Illinois 60208, United States; Interdisciplinary Biological Sciences Program, Northwestern University, Evanston, Illinois 60208, United States; Department of Chemical and Biological Engineering, Northwestern University, Evanston, Illinois 60208, United States; Simpson Querrey Institute, Northwestern University, Chicago, Illinois 60611, United States
REFERENCES
- (1).Ghannadan M; Hauswirth AW; Schernthaner GH; Müller MR; Klepetko W; Schatzl G; Sperr WR; Bühring HJ; Valent P Detection of Novel CD Antigens on the Surface of Human Mast Cells and Basophils. Int. Arch. Allergy Immunol 2002, 127, 299–307. [DOI] [PubMed] [Google Scholar]
- (2).Yokoi H; Choi OH; Hubbard W; Lee HS; Canning BJ; Lee HH; Ryu SD; von Gunten S; Bickel CA; Hudson SA; Macglashan DW Jr.; Bochner BS Inhibition of FcepsilonRI-dependent mediator release and calcium flux from human mast cells by sialic acid-binding immunoglobulin-like lectin 8 engagement. J Allergy Clin Immunol 2008, 121, 499. [DOI] [PubMed] [Google Scholar]
- (3).O’Reilly MK; Paulson JC Siglecs as targets for therapy in immune-cell-mediated disease. Trends Pharmacol. Sci 2009, 30, 240–248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (4).Mizrahi S; Gibbs BF; Karra L; Ben-Zimra M; Levi-Schaffer F Siglec-7 is an inhibitory receptor on human mast cells and basophils. J Allergy Clin Immunol 2014, 134, 230–233.e3. [DOI] [PubMed] [Google Scholar]
- (5).Duan S; Koziol-White CJ; Jester WF Jr.; Nycholat CM; Macauley MS; Panettieri RA Jr.; Paulson JC CD33 recruitment inhibits IgE-mediated anaphylaxis and desensitizes mast cells to allergen. J. Clin. Invest 2019, 129, 1387–1401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (6).Altrichter S; Staubach P; Pasha M; Rasmussen HS; Singh B; Chang AT; Bernstein JA; Siebenhaar F; Maurer M Efficacy and safety data of AK002, an anti-Siglec-8 monoclonal antibody, in patients with multiple forms of uncontrolled chronic urticaria (CU): results from an open-label phase 2a study. Allergy 2019, 70, 120. [Google Scholar]
- (7).Nycholat CM; Duan S; Knuplez E; Worth C; Elich M; Yao A; O’Sullivan J; McBride R; Wei Y; Fernandes SM; Zhu Z; Schnaar RL; Bochner BS; Paulson JC A Sulfonamide Sialoside Analogue for Targeting Siglec-8 and -F on Immune Cells. J. Am. Chem. Soc 2019, 141, 14032–14037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (8).Kerr SC; Gonzalez JR; Schanin J; Peters MC; Lambrecht BN; Brock EC; Charbit A; Ansel KM; Youngblood BA; Fahy JV An anti-siglec-8 antibody depletes sputum eosinophils from asthmatic subjects and inhibits lung mast cells. Clin. Exp. Allergy 2020, 50, 904–914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (9).Schanin J; Gebremeskel S; Korver W; Falahati R; Butuci M; Haw TJ; Nair PM; Liu G; Hansbro NG; Hansbro PM; Evensen E; Brock EC; Xu A; Wong A; Leung J; Bebbington C; Tomasevic N; Youngblood BA A monoclonal antibody to Siglec-8 suppresses non-allergic airway inflammation and inhibits IgE-independent mast cell activation. Mucosal Immunol. 2021, 14, 366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (10).Dellon ES; Peterson KA; Murray JA; Falk GW; Gonsalves N; Chehade M; Genta RM; Leung J; Khoury P; Klion AD; Hazan S; Vaezi M; Bledsoe AC; Durrani SR; Wang C; Shaw C; Chang AT; Singh B; Kamboj AP; Rasmussen HS; Rothenberg ME; Hirano I Anti-Siglec-8 Antibody for Eosinophilic Gastritis and Duodenitis. N. Engl. J. Med 2020, 383, 1624–1634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11).Duan S; Arlian BM; Nycholat CM; Wei Y; Tateno H; Smith SA; Macauley MS; Zhu Z; Bochner BS; Paulson JC Nanoparticles Displaying Allergen and Siglec-8 Ligands Suppress IgE-FcepsilonRI-Mediated Anaphylaxis and Desensitize Mast Cells to Subsequent Antigen Challenge. J. Immunol 2021, 206, 2290–2300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (12).Islam M; Arlian BM; Pfrengle F; Duan S; Smith SA; Paulson JC Suppressing Immune Responses Using Siglec Ligand-Decorated Anti-receptor Antibodies. J. Am. Chem. Soc 2022, 144, 9302–9311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (13).Bochner BS; O’Sullivan JA; Chang AT; Youngblood BA Siglecs in allergy and asthma. Mol. Aspect. Med 2023, 90, 101104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (14).Anesi SD; Tauber J; Nguyen QD; Chang P; Berdy GJ; Lin CC; Chu DS; Levine HT; Fernandez AD; Roy N; Asbell PA; Kantor AM; Chang AT; Singh B; Youngblood BA; Jeng BH; Jhanji V; Rasmussen HS; Foster CS Lirentelimab for severe and chronic forms of allergic conjunctivitis. J Allergy Clin Immunol 2022, 150, 631–639. [DOI] [PubMed] [Google Scholar]
- (15).Paul SP; Taylor LS; Stansbury EK; McVicar DW Myeloid specific human CD33 is an inhibitory receptor with differential ITIM function in recruiting the phosphatases SHP-1 and SHP-2. Blood 2000, 96, 483–490. [PubMed] [Google Scholar]
- (16).Macauley MS; Crocker PR; Paulson JC Siglec-mediated regulation of immune cell function in disease. Nat. Rev. Immunol 2014, 14, 653–666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (17).Hernandez-Caselles T; Miguel RC; Ruiz-Alcaraz AJ; Garcia-Penarrubia P CD33 (Siglec-3) Inhibitory Function: Role in the NKG2D/DAP10 Activating Pathway. J. Immunol. Res 2019, 2019, 6032141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (18).O’Sullivan JA; Chang AT; Youngblood BA; Bochner BS Eosinophil and mast cell Siglecs: From biology to drug target. J. Leukoc. Biol 2020, 108, 73–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (19).Duan S; Paulson JC Siglecs as Immune Cell Checkpoints in Disease. Annu. Rev. Immunol 2020, 38, 365–395. [DOI] [PubMed] [Google Scholar]
- (20).North SJ; Huang HH; Sundaram S; Jang-Lee J; Etienne AT; Trollope A; Chalabi S; Dell A; Stanley P; Haslam SM Glycomics profiling of Chinese hamster ovary cell glycosylation mutants reveals N-glycans of a novel size and complexity. J. Biol. Chem 2010, 285, 5759–5775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (21).Meuris L; Santens F; Elson G; Festjens N; Boone M; Dos Santos A; Devos S; Rousseau F; Plets E; Houthuys E; Malinge P; Magistrelli G; Cons L; Chatel L; Devreese B; Callewaert N GlycoDelete engineering of mammalian cells simplifies N-glycosylation of recombinant proteins. Nat. Biotechnol 2014, 32, 485–489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (22).Yang Z; Wang S; Halim A; Schulz MA; Frodin M; Rahman SH; Vester-Christensen MB; Behrens C; Kristensen C; Vakhrushev SY; Bennett EP; Wandall HH; Clausen H Engineered CHO cells for production of diverse, homogeneous glycoproteins. Nat. Biotechnol 2015, 33, 842–844. [DOI] [PubMed] [Google Scholar]
- (23).Johannssen T; Lepenies B Glycan-Based Cell Targeting To Modulate Immune Responses. Trends Biotechnol. 2017, 35, 334–346. [DOI] [PubMed] [Google Scholar]
- (24).Wang LX; Amin MN Chemical and chemoenzymatic synthesis of glycoproteins for deciphering functions. Chem. Biol 2014, 21, 51–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (25).Rodrigues E; Jung J; Park H; Loo C; Soukhtehzari S; Kitova EN; Mozaneh F; Daskhan G; Schmidt EN; Aghanya V; Sarkar S; Streith L; St Laurent CD; Nguyen L; Julien JP; West LJ; Williams KC; Klassen JS; Macauley MS A versatile soluble siglec scaffold for sensitive and quantitative detection of glycan ligands. Nat. Commun 2020, 11, 5091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (26).Jung J; Enterina JR; Bui DT; Mozaneh F; Lin PH; NitinKuo CW; Kuo CW; Rodrigues E; Bhattacherjee A; Raeisimakiani P; Daskhan GC; St Laurent CD; Khoo KH; Mahal LK; Zandberg WF; Huang X; Klassen JS; et al. Carbohydrate Sulfation As a Mechanism for Fine-Tuning Siglec Ligands. ACS Chem. Biol 2021, 16, 2673–2689. [DOI] [PubMed] [Google Scholar]
- (27).Hershewe J; Kightlinger W; Jewett MC Cell-free systems for accelerating glycoprotein expression and biomanufacturing. J. Ind. Microbiol. Biotechnol 2020, 47, 977–991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (28).Jaroentomeechai T; Taw MN; Li M; Aquino A; Agashe N; Chung S; Jewett MC; DeLisa MP Cell-Free Synthetic Glycobiology: Designing and Engineering Glycomolecules Outside of Living Cells. Front. Chem 2020, 8, 645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (29).Kightlinger W; Warfel KF; DeLisa MP; Jewett MC Synthetic Glycobiology: Parts, Systems, and Applications. ACS Synth. Biol 2020, 9, 1534–1562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (30).Martin RW; Des Soye BJ; Kwon YC; Kay J; Davis RG; Thomas PM; Majewska NI; Chen CX; Marcum RD; Weiss MG; Stoddart AE; Amiram M; Charna AKR; Patel JR; Isaacs FJ; Kelleher NL; Hong SH; Jewett MC Cell-free protein synthesis from genomically recoded bacteria enables multisite incorporation of noncanonical amino acids. Nat. Commun 2018, 9, 1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (31).Zawada JF; Yin G; Steiner AR; Yang J; Naresh A; Roy SM; Gold DS; Heinsohn HG; Murray CJ Microscale to manufacturing scale-up of cell-free cytokine production–a new approach for shortening protein production development timelines. Biotechnol. Bioeng 2011, 108, 1570–1578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (32).Silverman AD; Karim AS; Jewett MC Cell-free gene expression: an expanded repertoire of applications. Nat. Rev. Genet 2020, 21, 151–170. [DOI] [PubMed] [Google Scholar]
- (33).Jaroentomeechai T; Zheng X; Hershewe J; Stark JC; Jewett MC; DeLisa MP A Pipeline for Studying and Engineering Single-Subunit Oligosaccharyltransferases. Methods Enzymol. 2017, 597, 55–81. [DOI] [PubMed] [Google Scholar]
- (34).Jaroentomeechai T; Stark JC; Natarajan A; Glasscock CJ; Yates LE; Hsu KJ; Mrksich M; Jewett MC; DeLisa MP Single-pot glycoprotein biosynthesis using a cell-free transcriptiontranslation system enriched with glycosylation machinery. Nat. Commun 2018, 9, 2686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (35).Schoborg JA; Hershewe JM; Stark JC; Kightlinger W; Kath JE; Jaroentomeechai T; Natarajan A; DeLisa MP; Jewett MC A cell-free platform for rapid synthesis and testing of active oligosaccharyltransferases. Biotechnol. Bioeng 2018, 115, 739–750. [DOI] [PubMed] [Google Scholar]
- (36).Natarajan A; Jaroentomeechai T; Cabrera-Sanchez M; Mohammed JC; Cox EC; Young O; Shajahan A; Vilkhovoy M; Vadhin S; Varner JD; Azadi P; DeLisa MP Engineering orthogonal human O-linked glycoprotein biosynthesis in bacteria. Nat. Chem. Biol 2020, 16, 1062–1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (37).Hershewe JM; Warfel KF; Iyer SM; Peruzzi JA; Sullivan CJ; Roth EW; DeLisa MP; Kamat NP; Jewett MC Improving cell-free glycoprotein synthesis by characterizing and enriching native membrane vesicles. Nat. Commun 2021, 12, 2363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (38).Stark JC; Jaroentomeechai T; Moeller TD; Hershewe JM; Warfel KF; Moricz BS; Martini AM; Dubner RS; Hsu KJ; Stevenson TC; Jones BD; DeLisa MP; Jewett MC Ondemand biomanufacturing of protective conjugate vaccines. Sci. Adv 2021, 7, No. eabe9444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (39).Jaroentomeechai T; Kwon YH; Liu Y; Young O; Bhawal R; Wilson JD; Li M; Chapla DG; Moremen KW; Jewett MC; Mizrachi D; DeLisa MP A universal glycoenzyme biosynthesis pipeline that enables efficient cell-free remodeling of glycans. Nat. Commun 2022, 13, 6325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (40).Warfel KF; Williams A; Wong DA; Sobol SE; Desai P; Li J; Chang YF; DeLisa MP; Karim AS; Jewett MC A Low-Cost, Thermostable, Cell-Free Protein Synthesis Platform for On-Demand Production of Conjugate Vaccines. ACS Synth. Biol 2023, 12, 95–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (41).Williams AJ; Warfel KF; Desai P; Li J; Lee J-J; Wong DA; Nguyen PM; Qin Y; Sobol SE; Jewett MC; Chang Y-F; DeLisa MP A low-cost recombinant glycoconjugate vaccine confers immunogenicity and protection against enterotoxigenic Escherichia coli infections in mice. Front. Mol. Biosci 2023, 10, 2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (42).Warfel KF; Laigre E; Sobol SE; Gillon E; Varrot A; Renaudet O; Dejeu J; Jewett MC; Imberty A Cell-free expression and characterization of multivalent rhamnose-binding lectins using biolayer interferometry. Glycobiology 2023, DOI: 10.1093/glycob/cwad018. [DOI] [PubMed] [Google Scholar]
- (43).Kightlinger W; Lin L; Rosztoczy M; Li W; DeLisa MP; Mrksich M; Jewett MC Design of glycosylation sites by rapid synthesis and analysis of glycosyltransferases. Nat. Chem. Biol 2018, 14, 627–635. [DOI] [PubMed] [Google Scholar]
- (44).Lin L; Kightlinger W; Prabhu SK; Hockenberry AJ; Li C; Wang LX; Jewett MC; Mrksich M Sequential Glycosylation of Proteins with Substrate-Specific N-Glycosyltransferases. ACS Cent. Sci 2020, 6, 144–154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (45).Kightlinger W; Duncker KE; Ramesh A; Thames AH; Natarajan A; Stark JC; Yang A; Lin L; Mrksich M; DeLisa MP; Jewett MC A cell-free biosynthesis platform for modular construction of protein glycosylation pathways. Nat. Commun 2019, 10, 5404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (46).Kolb HC; Finn MG; Sharpless KB Click Chemistry: Diverse Chemical Function from a Few Good Reactions. Angew. Chem., Int. Ed 2001, 40, 2004–2021. [DOI] [PubMed] [Google Scholar]
- (47).Sletten EM; Bertozzi CR Bioorthogonal chemistry: fishing for selectivity in a sea of functionality. Angew. Chem., Int. Ed. Engl 2009, 48, 6974–6998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (48).Baskin JM; Bertozzi CR Bioorthogonal Click Chemistry: Covalent Labeling in Living Systems. QSAR Comb. Sci 2007, 26, 1211–1219. [Google Scholar]
- (49).Rillahan CD; Schwartz E; McBride R; Fokin VV; Paulson JC Click and pick: identification of sialoside analogues for Siglec-based cell targeting. Angew. Chem., Int. Ed. Engl 2012, 124, 11176–11180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (50).Macauley MS; Pfrengle F; Rademacher C; Nycholat CM; Gale AJ; von Drygalski A; Paulson JC Antigenic liposomes displaying CD22 ligands induce antigen-specific B cell apoptosis. J. Clin. Invest 2013, 123, 3074–3083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (51).Rillahan CD; Macauley MS; Schwartz E; He Y; McBride R; Arlian BM; Rangarajan J; Fokin VV; Paulson JC Disubstituted Sialic Acid Ligands Targeting Siglecs CD33 and CD22 Associated with Myeloid Leukaemias and B Cell Lymphomas. Chem. Sci 2014, 5, 2398–2406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (52).Bull C; Heise T; van Hilten N; Pijnenborg JFA; Bloemendal VRLJ; Gerrits L; Kers-Rebel ED; Ritschel T; den Brok MH; Adema GJ; Boltje TJ Steering Siglec-Sialic Acid Interactions on Living Cells using Bioorthogonal Chemistry. Angew. Chem., Int. Ed. Engl 2017, 129, 3357–3361. [DOI] [PubMed] [Google Scholar]
- (53).Baskin JM; Prescher JA; Laughlin ST; Agard NJ; Chang PV; Miller IA; Lo A; Codelli JA; Bertozzi CR Copper-free click chemistry for dynamic in vivo imaging. Proc. Natl. Acad. Sci. U. S. A 2007, 104, 16793–16797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (54).Hudak JE; Barfield RM; de Hart GW; Grob P; Nogales E; Bertozzi CR; Rabuka D Synthesis of heterobifunctional protein fusions using copper-free click chemistry and the aldehyde tag. Angew. Chem., Int. Ed. Engl 2012, 124, 4237–4241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (55).Dehnert KW; Baskin JM; Laughlin ST; Beahm BJ; Naidu NN; Amacher SL; Bertozzi CR Imaging the sialome during zebrafish development with copper-free click chemistry. Chembiochem 2012, 13, 353–357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (56).Des Soye BJ; Gerbasi VR; Thomas PM; Kelleher NL; Jewett MC A Highly Productive, One-Pot Cell-Free Protein Synthesis Platform Based on Genomically Recoded Escherichia coli. Cell Chem. Biol 2019, 26, 1743–1754.e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (57).Naegeli A; Neupert C; Fan YY; Lin CW; Poljak K; Papini AM; Schwarz F; Aebi M Molecular analysis of an alternative N-glycosylation machinery by functional transfer from Actinobacillus pleuropneumoniae to Escherichia coli. J. Biol. Chem 2014, 289, 2170–2179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (58).Schwarz F; Fan YY; Schubert M; Aebi M Cytoplasmic Nglycosyltransferase of Actinobacillus pleuropneumoniae is an inverting enzyme and recognizes the NX(S/T) consensus sequence. J. Biol. Chem 2011, 286, 35267–35274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (59).Chua KY; Doyle CR; Simpson RJ; Turner KJ; Stewart GA; Thomas WR Isolation of cDNA Coding for the Major Mite Allergen Der p II by IgE Plaque Immunoassay. Int. Arch. Allergy Immunol 1990, 91, 118–123. [DOI] [PubMed] [Google Scholar]
- (60).Derewenda U; Li J; Derewenda Z; Dauter Z; Mueller G; Rule G; Benjamin D The Crystal Structure of a Major Dust Mite Allergen Der p 2, and its Biological Implications. J. Mol. Biol 2002, 318, 189–197. [DOI] [PubMed] [Google Scholar]
- (61).Miles LA; Hermans SJ; Crespi GAN; Gooi JH; Doughty L; Nero TL; Markulic J; Ebneth A; Wroblowski B; Oehlrich D; Trabanco AA; Rives ML; Royaux I; Hancock NC; Parker MW Small Molecule Binding to Alzheimer Risk Factor CD33 Promotes Abeta Phagocytosis. iScience 2019, 19, 110–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (62).Landolina N; Zaffran I; Smiljkovic D; Serrano-Candelas E; Schmiedel D; Friedman S; Arock M; Hartmann K; Pikarsky E; Mandelboim O; Martin M; Valent P; Levi-Schaffer F Activation of Siglec-7 results in inhibition of in vitro and in vivo growth of human mast cell leukemia cells. Pharmacol. Res 2020, 158, 104682. [DOI] [PubMed] [Google Scholar]
- (63).Wisnovsky S; Mockl L; Malaker SA; Pedram K; Hess GT; Riley NM; Gray MA; Smith BAH; Bassik MC; Moerner WE; Bertozzi CR Genome-wide CRISPR screens reveal a specific ligand for the glycan-binding immune checkpoint receptor Siglec-7. Proc. Natl. Acad. Sci. U. S. A 2021, 118, DOI: 10.1073/pnas.2015024118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (64).Han S; Collins BE; Bengtson P; Paulson JC Homomultimeric complexes of CD22 in B cells revealed by protein-glycan cross-linking. Nat. Chem. Biol 2005, 1, 93–97. [DOI] [PubMed] [Google Scholar]
- (65).Swartz JR; Jewett MC; Woodrow KA Cell-Free Protein Synthesis With Prokaryotic Combined Transcription-Translation. In Methods in Molecular Biology; Balbás P, Lorence A, Eds.; Humana Press: Totowa, NJ, 2004, pp 169–182. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


