Abstract
Understanding the operation of biological and artificial networks remains a difficult and important challenge. To identify general principles, researchers are increasingly interested in surveying large collections of networks that are trained on, or biologically adapted to, similar tasks. A standardized set of analysis tools is now needed to identify how network-level covariates—such as architecture, anatomical brain region, and model organism—impact neural representations (hidden layer activations). Here, we provide a rigorous foundation for these analyses by defining a broad family of metric spaces that quantify representational dissimilarity. Using this framework, we modify existing representational similarity measures based on canonical correlation analysis and centered kernel alignment to satisfy the triangle inequality, formulate a novel metric that respects the inductive biases in convolutional layers, and identify approximate Euclidean embeddings that enable network representations to be incorporated into essentially any off-the-shelf machine learning method. We demonstrate these methods on large-scale datasets from biology (Allen Institute Brain Observatory) and deep learning (NAS-Bench-101). In doing so, we identify relationships between neural representations that are interpretable in terms of anatomical features and model performance.
1. Introduction
The extent to which different deep networks or neurobiological systems use equivalent representations in support of similar task demands is a topic of persistent interest in machine learning and neuroscience [1–3]. Several methods including linear regression [4, 5], canonical correlation analysis (CCA; [6, 7]), representational similarity analysis (RSA; [8]), and centered kernel alignment (CKA; [9]) have been used to quantify the similarity of hidden layer activation patterns. These measures are often interpreted on an ordinal scale and are employed to compare a limited number of networks—e.g., they can indicate whether networks and are more or less similar than networks and . While these comparisons have yielded many insights [4–12], the underlying methodologies have not been extended to systematic analyses spanning thousands of networks.
To unify existing approaches and enable more sophisticated analyses, we draw on ideas from statistical shape analysis [13–15] to develop dissimilarity measures that are proper metrics—i.e., measures that are symmetric and respect the triangle inequality. This enables several off-the-shelf methods with theoretical guarantees for classification (e.g. k-nearest neighbors, [16]) and clustering (e.g. hierarchical clustering [17]). Existing similarity measures can violate the triangle inequality, which complicates these downstream analyses [18–20]. However, we show that existing dissimilarity measures can often be modified to satisfy the triangle inequality and viewed as special cases of the framework we outline. We also describe novel metrics within this broader family that are specialized to convolutional layers and have appealing properties for analyzing artificial networks.
Moreover, we show empirically that these metric spaces on neural representations can be embedded with low distortion into Euclidean spaces, enabling an even broader variety of previously unconsidered supervised and unsupervised analyses. For example, we can use neural representations as the inputs to linear or nonlinear regression models. We demonstrate this approach on neural representations in mouse visual cortex (Allen Brain Observatory; [21]) in order to predict each brain region’s anatomical hierarchy from its pattern of visual responses—i.e., predicting a feature of brain structure from function. We demonstrate a similar approach to analyze hidden layer representations in a database of 432K deep artificial networks (NAS-Bench-101; [22]) and find a surprising degree of correlation between early and deep layer representations.
Overall, we provide a theoretical grounding which explains why existing representational similarity measures are useful: they are often close to metric spaces, and can be modified to fulfill metric space axioms precisely. Further, we draw new conceptual connections between analyses of neural representations and established research areas [15, 23], utilize these insights to propose novel metrics, and demonstrate a general-purpose machine learning workflow that scales to datasets with thousands of networks.
2. Methods
This section outlines several workflows (Fig. 1) to analyze representations across large collections of networks. After briefly summarizing prior approaches (sec. 2.1), we cover background material on metric spaces and discuss their theoretical advantages over existing dissimilarity measures (sec. 2.2). We then present a class of metrics that capture these advantages (sec. 2.3) and cover a special case that is suited to convolutional layers (sec. 2.4). We then demonstrate the practical advantages of these methods in Section 3, and demonstrate empirically that Euclidean feature spaces can approximate the metric structure of neural representations, enabling a broad set of novel analyses.
Figure 1:
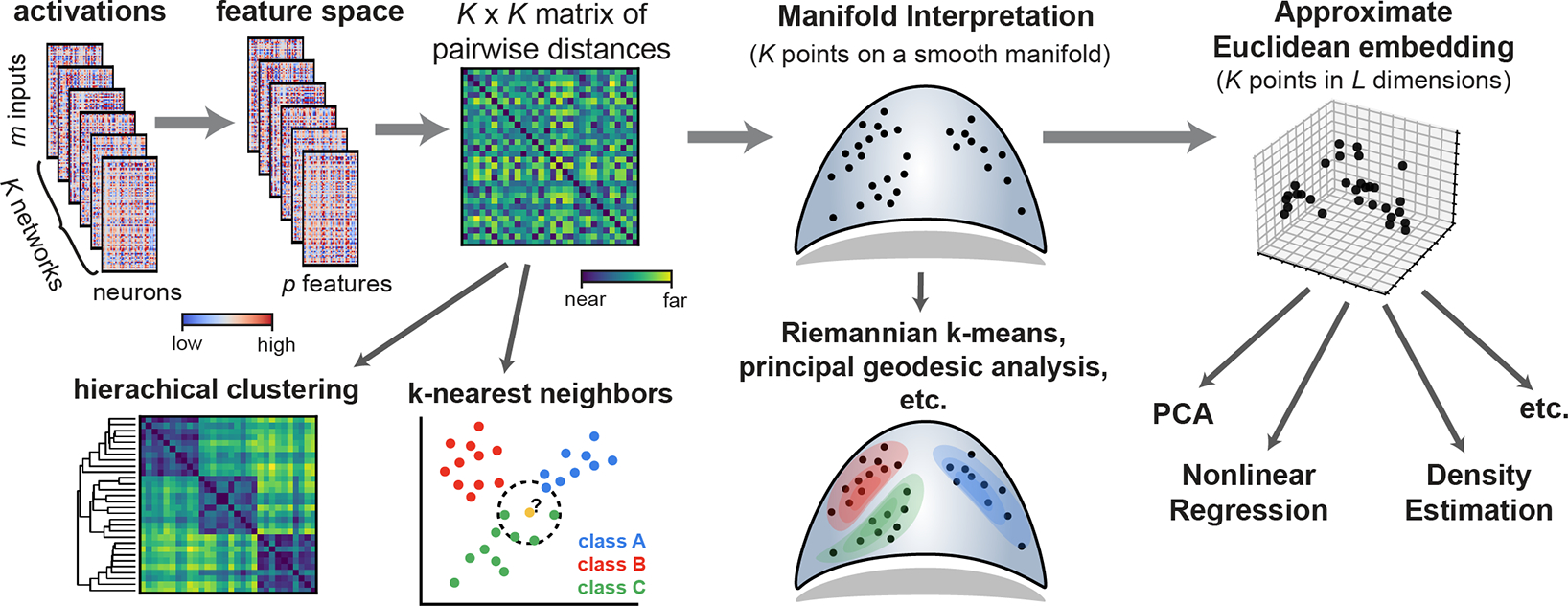
Machine learning workflows enabled by generalized shape metrics.
2.1. Prior work and problem setup
Neural network representations are often summarized over a set of reference inputs (e.g. test set images). Let and denote the responses of two networks (with and neurons, respectively) to a collection of these inputs. Quantifying the similarity between and is complicated by the fact that, while the inputs are the same, there is no direct correspondence between the neurons. Even if , the typical Frobenius inner product, , and metric, , fail to capture the desired notion of dissimilarity. For instance, let denote some permutation matrix and let . Intuitively, we should consider and to be identical in this case since the ordering of neurons is arbitrary. Yet, clearly , except in very special cases.
One way to address this problem is to linearly regress over the neurons to predict from . Then, one can use the coefficient of determination as a measure of similarity [4, 5]. However, this similarity score is asymmetric—if one instead treats as the dependent variable that is predicted from , this will result in a different . Canonical correlation analysis (CCA; [6, 7]) and linear centered kernel alignment (linear CKA; [9, 24]) also search for linear correspondences between neurons, but have the advantage of producing symmetric scores. Representational similarity analysis (RSA; [8]) is yet another approach, which first computes an matrix holding the dissimilarities between all pairs of representations for each network. These representational dissimilarity matrices (RDMs), are very similar to the kernel matrices computed and compared by CKA. RSA traditionally quantifies the similarity between two neural networks by computing Spearman’s rank correlation between their RDMs. A very recent paper by Shahbazi et al. [25], which was published while this manuscript was undergoing review, proposes to use the Riemannian metric between positive definite matrices instead of Spearman correlation. Similar to our results, this establishes a metric space that can be used to compare neural representations. Here, we leverage metric structure over shape spaces [13–15] instead of positive definite matrices, leading to complementary insights.
In summary, there are a diversity of methods that one can use to compare neural representations. Without a unifying theoretical framework it is unclear how to choose among them, use their outputs for downstream tasks, or generalize them to new domains.
2.2. Feature space mapping, metrics, and equivalence relations
Our first contribution will be to establish formal notions of distance (metrics) between neural representations. To accommodate the common scenario when the number of neurons varies across networks (i.e. when ), we first map the representations into a common feature space. For each set of representations, , we suppose there is a mapping into a -dimensional feature space, , where . In the special case where all networks have equal size, , we can express the feature mapping as a single function , so that . When networks have dissimilar sizes, we can map the representations into a common dimension using, for example, PCA [6].
Next, we seek to establish metrics within the feature space, which are distance functions that satisfy:
| (1) |
| (2) |
| (3) |
for all , and in the feature space. The symbol ‘~’ denotes an equivalence relation between two elements. That is, the expression means that “ is equivalent to .” Formally, distance functions satisfying Eqs. (1) to (3) define a metric over a quotient space defined by the equivalence relation and a pseudometric over (see Supplement A). Intuitively, by specifying different equivalence relations we can account for symmetries in network representations, such as permutations over arbitrarily labeled neurons (other options are discussed below in sec. 2.3).
Metrics quantify dissimilarity in a way that agrees with our intuitive notion of distance. For example, Eq. (2) ensures that the distance from to is the same as the distance from to . Linear regression is an approach that violates this condition: the similarity measured by depends on which network is treated as the dependent variable.
Further, Eq. (3) ensures that distances are self-consistent in the sense that if two elements ( and ) are both close to a third , then they are necessarily close to each other. Many machine learning models and algorithms rely on this triangle inequality condition. For example, in clustering, it ensures that if and are put into the same cluster as , then and cannot be too far apart, thus implying that they too can be clustered together. Intuitively, this establishes an appealing transitive relation for clustering, which can be violated when the triangle inequality fails to hold. Existing measures based on CCA, RSA, and CKA, are symmetric, but do not satisfy the triangle inequality. By modifying these approaches to satisfy the triangle inequality, we avoid potential pitfalls and can leverage theoretical guarantees on learning in proper metric spaces [16–20].
2.3. Generalized shape metrics and group invariance
In this section, we outline a new framework to quantify representational dissimilarity, which leverages a well-developed mathematical literature on shape spaces [13–15]. The key idea is to treat if and only if there exists a linear transformation within a set of allowable transformations , such that . Although only contains linear functions, nonlinear alignments between the raw representations can be achieved when the feature mappings are chosen to be nonlinear. Much of shape analysis literature focuses on the special case where and is the special orthogonal group , meaning that and are equivalent if there is a -dimensional rotation (without reflection) that relates them. Standard shape analysis further considers each to be a mean-centered and normalized version of the raw landmark locations held in (an assumption that we will relax). That is, the feature map transforms the raw landmarks onto the hypersphere, denoted , of matrices with unit Frobenius norm. In this context, is called a “pre-shape.” By removing rotations from a pre-shape, for pre-shape , we recover its “shape.”
To quantify dissimilarity in neural representations, we generalize this notion of shape to include other feature mappings and alignments. The minimal distance within the feature space, after optimizing over alignments, defines a metric under suitable conditions (Fig. 2A). This results in a broad variety of generalized shape metrics (see also, ch. 18 of [15]), which fall into two categories as formalized by the pair of propositions below. Proofs are provided in Supplement B.
Figure 2:
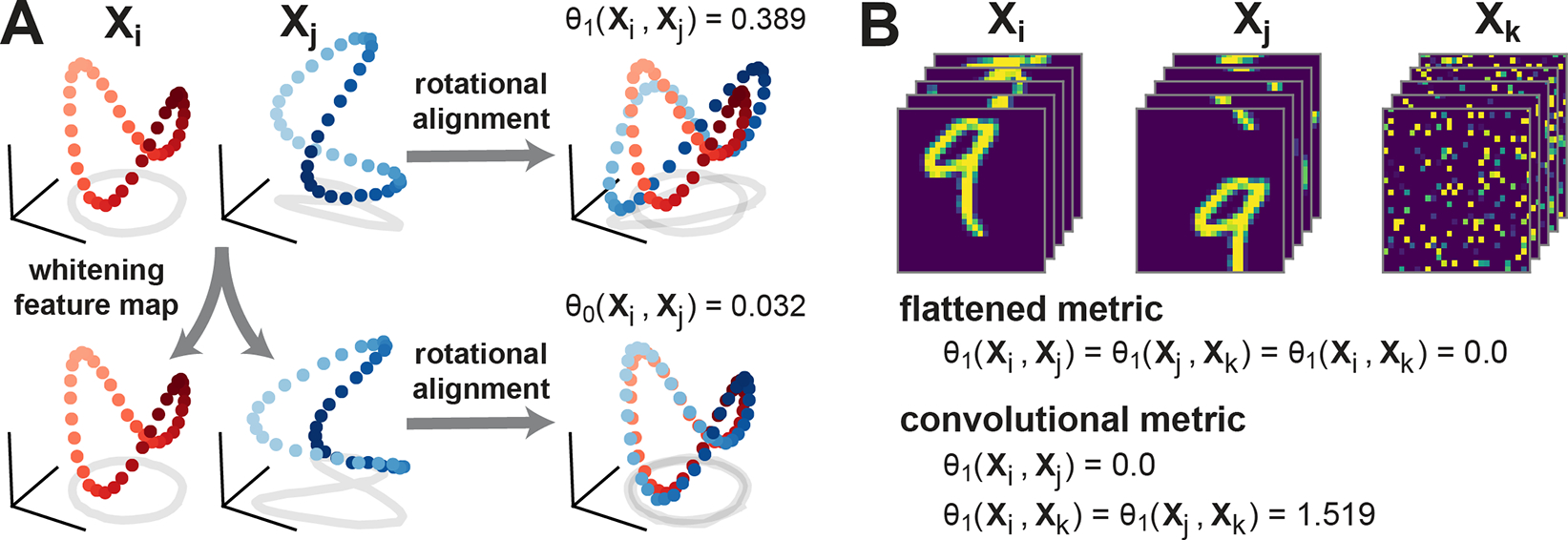
(A) Schematic illustration of metrics with rotational invariance (top), and linear invariance (bottom). Red and blue dots represent a pair of network representations and , which correspond to points in -dimensional space. (B) Demonstration of convolutional metric on toy data. Flattened metrics (e.g. [6, 9]) that ignore convolutional layer structure treat permuted images (, right) as equivalent to images with coherent spatial structure ( and , left and middle). A convolutional metric, Eq. (11), distinguishes between these cases while still treating and as equivalent (obeying translation invariance).
Proposition 1.
Let , and let be a group of linear isometries on . Then,
| (4) |
defines a metric, where if and only if there is a such that .
Proposition 2.
Let , and let be a group of linear isometries on . Then,
| (5) |
defines a metric, where if and only if there is a such that .
Two key conditions appear in these propositions. First, must be a group of functions. This means is a set that contains the identity function, is closed under composition ( for any and ), and whose elements are invertible by other members of the set (if then ). Second, every must be an isometry, meaning that for all and all elements of the feature space. On and , all linear isometries are orthogonal transformations. Further, the set of orthogonal transformations, , defines a well-known group. Thus, the condition that is a group of isometries is equivalent to being a subgroup of —i.e., a subset of satisfying the group axioms.
Intuitively, by requiring to be a group of functions, we ensure that the alignment procedure is symmetric—i.e. it is equivalent to transform to match , or transform the latter to match the former. Further, by requiring each to be an isometry, we ensure that the underlying metric (Euclidean distance for Proposition 1; angular distance for Proposition 2) preserves its key properties.
Together, these propositions define a broad class of metrics as we enumerate below. For simplicity, we assume that in the examples below, with the understanding that a PCA or zero-padding preprocessing step has been performed in the case of dissimilar network sizes. This enables us to express the metrics as functions of the raw activations, i.e. functions .
Permutation invariance
The most stringent notion of representational similarity is to demand that neurons are one-to-one matched across networks. If we set the feature map to be the identity function, i.e., for all , then:
| (6) |
defines a metric by Proposition 1 since the set of permutation matrices, , is a subgroup of . To evaluate this metric we must optimize over the set of neuron permutations to align the two networks. This can be reformulated (see Supplement C) as a fundamental problem in combinatorial optimization known as the linear assignment problem [26]. Exploiting an algorithm due to Jonker and Volgenant [27, 28] we can solve this problem in time. The overall runtime for evaluating Eq. (6) is , since we must evaluate to formulate the assignment problem.
Rotation invariance
Let denote an centering matrix, and consider the feature mapping which mean-centers the columns, . Then,
| (7) |
defines a metric by Proposition 1, and is equivalent to the Procrustes size-and-shape distance with reflections [15]. Further, by Proposition 2,
| (8) |
defines another metric, and is closely related to the Riemannian distance on Kendall’s shape space [15]. To evaluate Eqs. (7) and (8), we must optimize over the set of orthogonal matrices to find the best alignment. This also maps onto a fundamental optimization problem known as the orthogonal Procrustes problem [29, 30], which can be solved in closed form in time. As in the permutation-invariant metric described above, the overall runtime is .
Linear invariance
Consider a partial whitening transformation, parameterized by :
| (9) |
Note that is the empirical covariance matrix of . Thus, when , Eq. (9) corresponds to ZCA whitening [31], which intuitively removes invertible linear transformations from the representations. Thus, when the metric outlined below treats if there exists an affine transformation that relates them: for some and . When , Eq. (9) reduces to the mean-centering feature map used above.
Using orthogonal alignments within this feature space leads to a metric that is related to CCA. First, let denote the singular values of . One can show that
| (10) |
and we can see from Proposition 2 that this defines a metric for any . When , the values are proportional to the canonical correlation coefficients, with being the factor of proportionality. When , these values can be viewed as ridge regularized canonical correlation coefficients [32]. See Supplement C for further details. Past works [6, 7] have used the average canonical correlation as a measure of representational similarity. When , the average canonical correlation is given by . Thus, if we apply to the average canonical correlation, we modify the calculation to produce a proper metric (see Fig. 4A). Since the covariance is often ill-conditioned or singular in practice, setting to regularize the calculation is also typically necessary.
Figure 4:
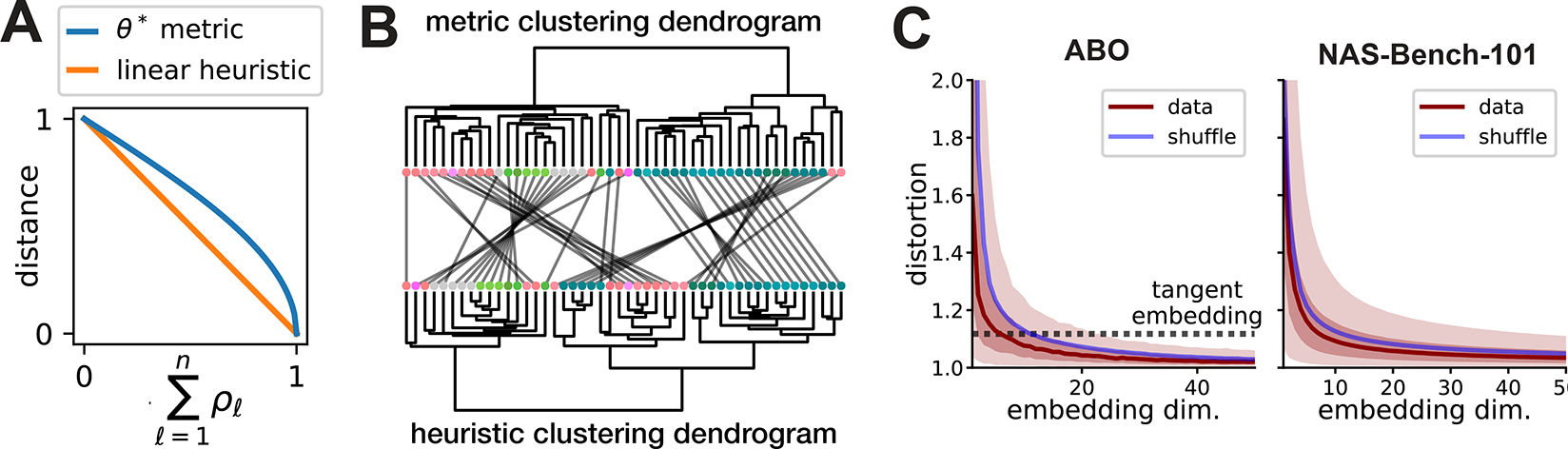
(A) Comparison of metric and linear heuristic. (B) Metric and linear heuristic produce discordant hierarchical clusterings of brain areas in the ABO dataset. Leaves represent brain areas that are clustered by representational similarity (see Fig. 1C), colored by Allen reference atlas, and ordered to maximize dendrogram similarities of adjacent leaves. In the middle, grey lines connect leaves corresponding to the same brain region across the two dendrograms. (C) ABO and NAS-Bench-101 datasets can be accurately embedded into Euclidean spaces. Dark red line shows median distortion. Light red shaded region corresponds to 5th to 95th percentiles of distortion, dark red shaded corresponds to interquartile range. The mean distortion of a null distribution over representations (blue line) was generated by shuffling the inputs independently in each network.
Nonlinear invariances
We discuss feature maps that enable nonlinear notions of equivalence, and which relate to kernel CCA [33] and CKA [9], in Supplement C.
2.4. Metrics for convolutional layers
In deep networks for image processing, each convolutional layer produces a array of activations, whose axes respectively correspond to image height, image width, and channels (number of convolutional filters). If stride-1 circular convolutions are used, then applying a circular shift along either spatial dimension produces the same shift in the layer’s output. It is natural to reflect this property, known as translation equivariance [23], in the equivalence relation on layer representations. Supposing that the feature map preserves the shape of the activation tensor, we have for neural networks indexed by . Letting denote the group of -dimensional circular shifts (a subgroup of the permutation group) and ‘’ denote the Kronecker product, we propose:
| (11) |
for some , as the desired equivalence relation. This relation allows for orthogonal invariance across the channel dimension but only shift invariance across the spatial dimensions. The mixed product property of Kronecker products, , ensures that the overall transformation maintains the group structure and remains an isometry. Figure 2B uses a toy dataset (stacked MNIST digits) to show that this metric is sensitive to differences in spatial activation patterns, but insensitive to coherent spatial translations across channels. In contrast, metrics that ignore the convolutional structure (as in past work [6, 9]) treat very different spatial patterns as identical representations.
Evaluating Eq. (11) requires optimizing over spatial shifts in conjuction with solving a Procrustes alignment. If we fit the shifts by an exhaustive brute-force search, the overall runtime is , which is costly if this calculation is repeated across a large collection of networks. In practice, we observe that the optimal shift parameters are typically close to zero (Fig. 3A). This motivates the more stringent equivalence relation:
| (12) |
which has a more manageable runtime of . To evaluate the metrics implied by Eq. (12), we can simply reshape each from a tensor into a matrix and apply the Procrustes alignment procedure as done above for previous metrics. In contrast, the “flattened metric” in Fig. 2B reshapes the features into a matrix, resulting in a more computationally expensive alignment that runs in time.
Figure 3:
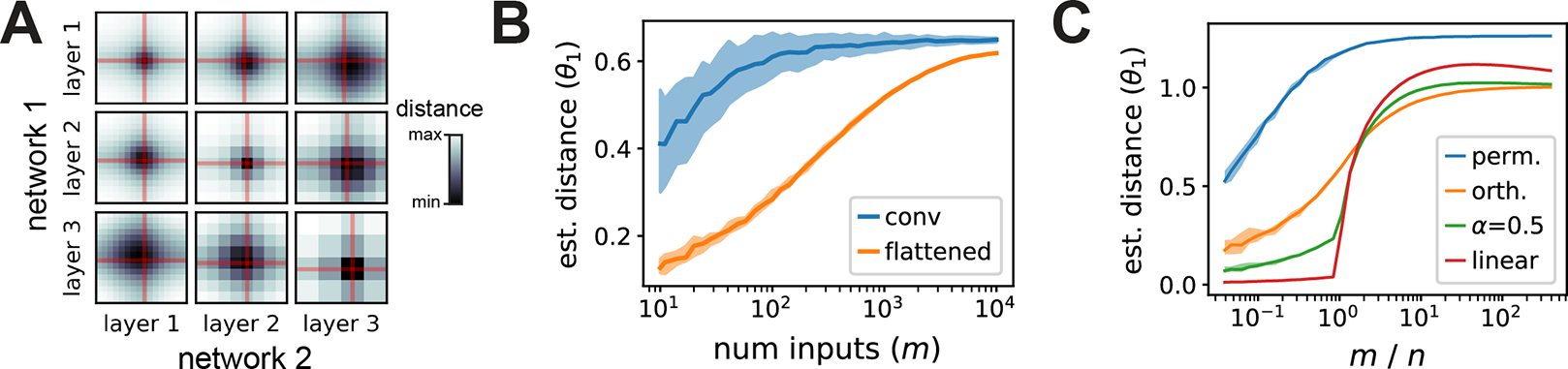
(A) Each heatmap shows a brute-force search over the shift parameters along the width and height dimensions of a pair of convolutional layers compared across two networks. The optimal shifts are typically close to zero (red lines). (B) Impact of sample size, , on flattened and convolutional metrics with orthogonal invariance. The convolutional metric approaches its final value faster than the flattened metric, which is still increasing even at the full size of the CIFAR-10 test set . (C) Impact of sample density, , on metrics invariant to permutation, orthogonal, regularized linear , and linear transformations. Shaded regions mark the 10th and 90th percentiles across shuffled repeats. Further details are provided in Supplement E.
2.5. How large of a sample size is needed?
An important issue, particularly in neurobiological applications, is to determine the number of network inputs, , and neurons, , that one needs to accurately infer the distance between two network representations [12]. Reasoning about these questions rigorously requires a probabilistic perspective of neural representational similarity, which is missing from current literature and which we outline in Supplement D for generalized shape metrics. Intuitively, looser equivalence relations are achieved by having more flexible alignment operations (e.g. nonlinear instead of linear alignments). Thus, looser equivalence relations require more sampled inputs to prevent overfitting. Figure 3B–C show that this intuition holds in practice for data from deep convolutional networks. Metrics with looser equivalence relations—the “flattened” metric in panel B, or e.g. the linear metric in panel C—converge slower to a stable estimate as is increased.
2.6. Modeling approaches and conceptual insights
Generalized shape metrics facilitate several new modeling approaches and conceptual perspectives. For example, a collection of representations from neural networks can, in certain cases, be interpreted and visualized as points on a smooth manifold (see Fig. 1). This holds rigorously due to the quotient manifold theorem [34] so long as is not a finite set (e.g. corresponding to permutation) and all matrices are full rank in the feature space. This geometric intuition can be made even stronger when corresponds to a connected manifold, such as . In this case, it can be shown that the geodesic distance between two neural representations coincides with the metrics we defined in Propositions 1 and 2 (see Supplement C, and [15]). This result extends the well-documented manifold structure of Kendall’s shape space [35].
Viewing neural representations as points on a manifold is not a purely theoretical exercise—several models can be adapted to manifold-valued data (e.g. principal geodesic analysis [36] provides a generalization of PCA), and additional adaptions are an area of active research [37]. However, there is generally no simple connection between these curved geometries and the flat geometries of Euclidean or Hilbert spaces [38].1 Unfortunately, the majority of off-the-shelf machine learning tools are incompatible with the former and require the latter. Thus, we can resort to a heuristic approach: the set of representations can be embedded into a Euclidean space that approximately preserves the pairwise shape distances. One possibility, employed widely in shape analysis, is to embed points in the tangent space of the manifold at a reference point [41, 42]. Another approach, which we demonstrate below with favorable results, is to optimize the vector embedding directly via multi-dimensional scaling [43, 44].
3. Applications and Results
We analyzed two large-scale public datasets spanning neuroscience (Allen Brain Observatory, ABO; Neuropixels - visual coding experiment; [21]) and deep learning (NAS-Bench-101; [22]). We constructed the ABO dataset by pooling recorded neurons from anatomically defined brain regions across all sessions; each was a dimensionally reduced matrix holding the neural responses (summarized by principal components) to movie frames (120 second clip, “natural movie three”). The full NAS-Bench-101 dataset contains 423,624 architectures; however, we analyze a subset of networks for simplicity. In this application each is a representation from a specific network layer, with . Here, corresponds to the number of channels and is the product of the number of test set images (105) and the height and width dimensions of the convolutional layer—i.e., we use equivalence relation in Eq. (12) to evaluate dissimilarity.
Triangle inequality violations can occur in practice when using existing methods.
As mentioned above, a dissimilarity measure based on the mean canonical correlation, , has been used in past work [7, 10]. We refer to this as the “linear heuristic.” A slight reformulation of this calculation, , produces a metric that satisfies the triangle inequality (see Eq. (10)). Figure 4A compares these calculations as a function of the average (regularized) canonical correlation: one can see that is approximately linear when the mean correlation is near zero, but highly nonlinear when the mean correlation is near one. Thus, we reasoned that triangle inequality violations are more likely to occur when is large and when many network representations are close to each other. Both ABO and NAS-Bench-101 datasets satisfy these conditions, and in both cases we observed triangle inequality violations by the linear heuristic with full regularization network pairs in the ABO dataset had at least one triangle inequality violation, while 10128/100000 randomly sampled network pairs contained violations in the NAS-Bench-101 Stem layer dataset. We also examined a standard version of RSA that quantifies similarity via Spearman’s rank correlation coefficient [8]. Similar to the results above, we observed violations in 14/1128 pairs of networks in the ABO dataset.
Overall, these results suggest that generalized shape metrics correct for triangle inequality violations that do occur in practice. Depending on the dataset, these violations may be rare (~1% occurrence in ABO) or relatively common (~10% in the Stem layer of NAS-Bench-101). These differences can produce quantitative discrepancies in downstream analyses. For example, the dendrograms produced by hierarchical clustering differ depending on whether one uses the linear heuristic or the shape distance (~85.1% dendrogram similarity as quantified by the method in [45]; see Fig. 4B).
Neural representation metric spaces can be approximated by Euclidean spaces.
Having established that neural representations can be viewed as elements in a metric space, it is natural to ask if this metric space is, loosely speaking, “close to” a Euclidean space. We used standard multidimensional scaling methods (SMACOF, [43]; implementation in [46]) to obtain a set of embedded vectors, , for which for . The embedding dimension is a user-defined hyperparameter. This problem admits multiple formulations and optimization strategies [44], which could be systematically explored in future work. Our simple approach already yields promising results: we find that moderate embedding dimensions is sufficient to produce high-quality embeddings. We quantify the embedding distortions multiplicatively [47]:
| (13) |
for each pair of networks . Plotting the distortions as a function of (Fig. 4C), we see that they rapidly decrease, such that 95% of pairwise distances are distorted by, at most, ~5% (ABO data) or 10% (NAS-Bench-101) for sufficiently large . Past work [10] has used multidimensional scaling heuristically to visualize collections of network representations in dimensions. Our results here suggest that such a small value of , while being amenable to visualization, results in a highly distorted embedding. It is noteworthy that the situation improves dramatically when is even modestly increased. While we cannot easily visualize these higher-dimensional vector embeddings, we can use them as features for downstream modeling tasks. This is well-motivated as an approximation to performing model inference in the true metric space that characterizes neural representations [47].
Anatomical structure and hierarchy is reflected in ABO representations.
We can now collect the -dimensional vector embeddings of network representations into a matrix . The results in Fig. 4C imply that the distance between any two rows, , closely reflects the distance between network representations and in shape space. We applied PCA to to visualize the brain regions and found that anatomically related brain regions indeed were closer together in the embedded space (Fig. 5A): cortical and sub-cortical regions are separated along PC 1, and different layers of the same region (e.g. layers 2/3, 4, 5, and 6a of VISp) are clustered together. As expected from Fig. 4C, performing multidimensional scaling directly to a low-dimensional space (, as done in [10]) results in a qualitatively different outcome with distorted geometry (see Supplement E). Additionally, we used to fit an ensembled kernel regressor to predict an anatomical hierarchy score (defined in [48]) from the embedded vectors (Fig. 5B). Overall, these results demonstrate that the geometry of the learned embedding is scientifically interpretable and can be exploited for novel analyses, such as nonlinear regression. To our knowledge, the fine scale anatomical parcellation used here is novel in the context of representational similarity studies.
Figure 5:
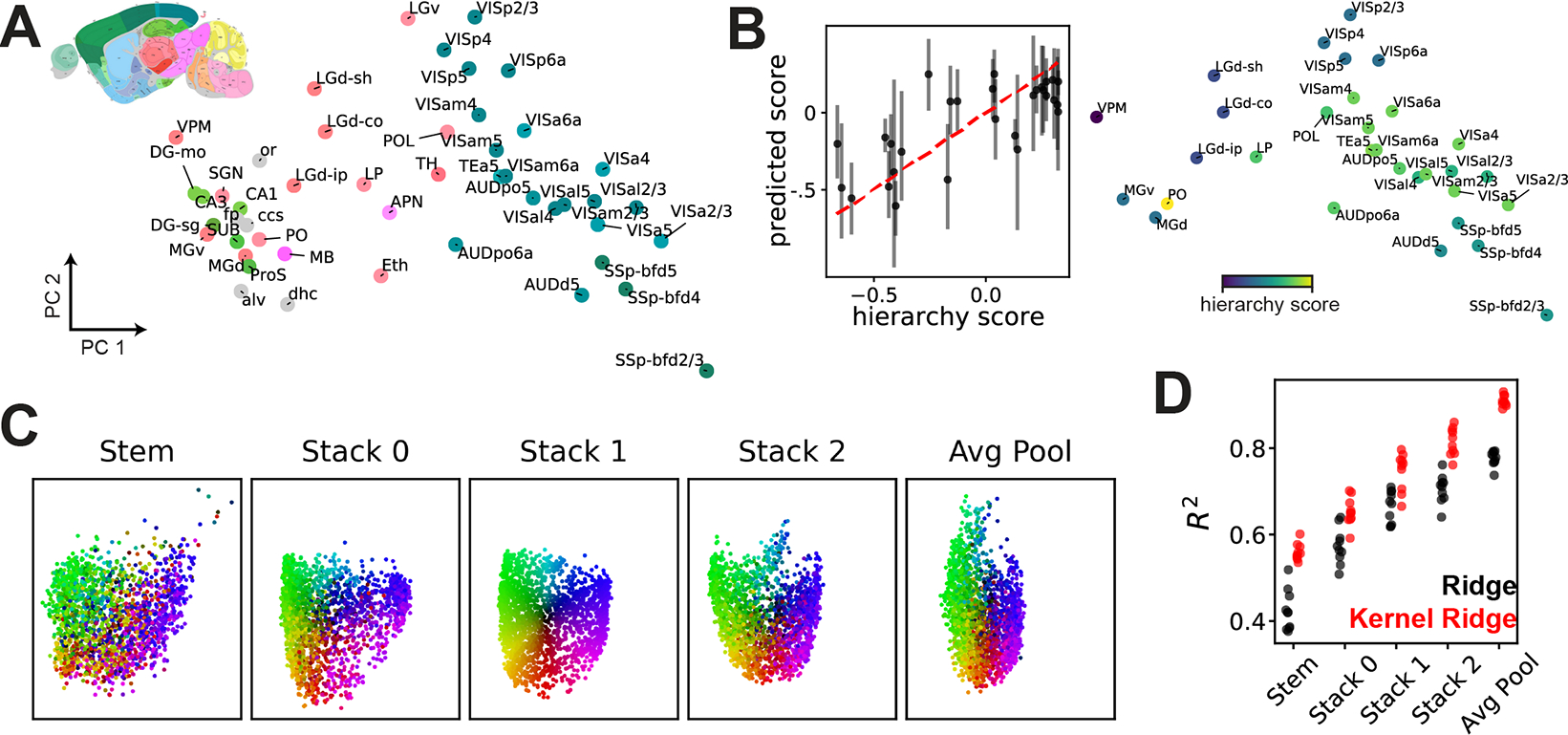
(A) PCA visualization of representations across 48 brain regions in the ABO dataset. Areas are colored by the reference atlas (see inset), illustrating a functional clustering of regions that maps onto anatomy. (B) Left, kernel regression predicts anatomical hierarchy [48] from embedded representations (see Supplement E). Right, PCA visualization of 31 areas labeled with hierarchy scores. (C) PCA visualization of 2000 network representations (a subset of NAS-Bench-101) across five layers, showing global structure is preserved across layers. Each network is colored by its position in the “Stack 1” layer (the middle of the architecture). (D) Embeddings of NAS-Bench-101 representations are predictive of test set accuracy, even in very early layers.
NAS-Bench-101 representations show persistent structure across layers.
Since we collected representations across five layers in each deep network, the embedded representation vectors form a set of five matrices, . We aligned these embeddings by rotations in via Procrustes analysis, and then performed PCA to visualize the network representations from each layer in a common low-dimensional space. We observe that many features of the global structure are remarkably well-preserved—two networks that are close together in the Stack1 layer are assigned similar colors in Fig. 5C, and are likely to be close together in the other four layers. This preservation of representational similarity across layers suggests that even early layers contain signatures of network performance, which we expect to be present in the AvgPool layer. Indeed, when we fit ridge and RBF kernel ridge regressors to predict test set accuracy from representation embeddings, we see that even early layers support moderately good predictions (Fig. 5D). This is particularly surprising for the Stem layer. This is the first layer in each network, and its architecture is identical for all networks. Thus, the differences that are detected in the Stem layer result only from differences in backpropagated gradients. Again, these results demonstrate the ability of generalized shape metrics to incorporate neural representations into analyses with greater scale ( corresponding to thousands of networks) and complexity (nonlinear kernel regression) than has been previously explored.
4. Conclusion and Limitations
We demonstrated how to ground analyses of neural representations in proper metric spaces. By doing so, we capture a number of theoretical advantages [16–20]. Further, we suggest new practical modeling approaches, such as using Euclidean embeddings to approximate the representational metric spaces. An important limitation of our work, as well as the past works we build upon, is the possibility that representational geometry is only loosely tied to higher-level algorithmic principles of network function [10]. On the other hand, analyses of representational geometry may provide insight into lower-level implementational principles [49]. Further, these analyses are highly scalable, as we demonstrated by analyzing thousands of networks—a much larger scale than is typically considered.
We used simple metrics (extensions of regularized CCA) in these analyses, but metrics that account for nonlinear transformations across neural representations are also possible as we document in Supplement C. The utility of these nonlinear extensions remains under-investigated and it is possible that currently popular linear methods are insufficient to capture structures of interest. For example, the topology of neural representations has received substantial interest in recent years [50–53]. Generalized shape metrics do not directly capture these topological features, and future work could consider developing new metrics that do so. A variety of recent developments in topological data analysis may be useful towards this end [54–56].
Finally, several of the metrics we described can be viewed as geodesic distances on Riemannian manifolds [35]. Future work would ideally exploit methods that are rigorously adapted to such manifolds, which are being actively developed [37]. Nonetheless, we found that optimized Euclidean embeddings, while only approximate, provide a practical off-the-shelf solution for large-scale surveys of neural representations.
Supplementary Material
Acknowledgments
We thank Ian Dryden (Florida International University), Søren Hauberg (Technical University of Denmark), and Nina Miolane (UC Santa Barbara) for fruitful discussions. A.H.W. was supported by the National Institutes of Health BRAIN initiative (1F32MH122998-01), and the Wu Tsai Stanford Neurosciences Institute Interdisciplinary Scholar Program. E. K. was supported by the Wu Tsai Stanford Neurosciences Institute Interdisciplinary Graduate Fellows Program. S.W.L. was supported by grants from the Simons Collaboration on the Global Brain (SCGB 697092) and the NIH BRAIN Initiative (U19NS113201 and R01NS113119).
Footnotes
Contributor Information
Alex H. Williams, Statistics Department, Stanford University
Erin Kunz, Electrical Engineering Department, Stanford University.
Simon Kornblith, Google Research, Toronto.
Scott W. Linderman, Statistics Department, Stanford University
References
- [1].Barrett David GT, Morcos Ari S, and Macke Jakob H. “Analyzing biological and artificial neural networks: challenges with opportunities for synergy?” Current Opinion in Neurobiology 55 (2019). Machine Learning, Big Data, and Neuroscience, pp. 55–64. [DOI] [PubMed] [Google Scholar]
- [2].Kriegeskorte Nikolaus and Wei Xue-Xin. “Neural tuning and representational geometry”. Nature Reviews Neuroscience (2021). [DOI] [PubMed] [Google Scholar]
- [3].Roeder Geoffrey, Metz Luke, and Kingma Durk. “On Linear Identifiability of Learned Representations”. Proceedings of the 38th International Conference on Machine Learning. Ed. by Meila Marina and Zhang Tong. Vol. 139. Proceedings of Machine Learning Research. PMLR, 2021, pp. 9030–9039. [Google Scholar]
- [4].Yamins Daniel L. K., Hong Ha, Cadieu Charles F., Solomon Ethan A., Seibert Darren, and DiCarlo James J. “Performance-optimized hierarchical models predict neural responses in higher visual cortex”. Proceedings of the National Academy of Sciences 111.23 (2014), pp. 8619–8624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Cadena Santiago A., Sinz Fabian H., Muhammad Taliah, Froudarakis Emmanouil, Cobos Erick, Walker Edgar Y., Reimer Jake, Bethge Matthias, Tolias Andreas, and Ecker Alexander S.. “How well do deep neural networks trained on object recognition characterize the mouse visual system?” NeurIPS Workshop Neuro AI (2019). [Google Scholar]
- [6].Raghu Maithra, Gilmer Justin, Yosinski Jason, and Sohl-Dickstein Jascha. “SVCCA: Singular Vector Canonical Correlation Analysis for Deep Learning Dynamics and Interpretability”. Advances in Neural Information Processing Systems 30. Ed. by Guyon I, Luxburg UV, Bengio S, Wallach H, Fergus R, Vishwanathan S, and Garnett R. Curran Associates, Inc., 2017, pp. 6076–6085. [Google Scholar]
- [7].Morcos Ari, Raghu Maithra, and Bengio Samy. “Insights on representational similarity in neural networks with canonical correlation”. Advances in Neural Information Processing Systems 31. Ed. by Bengio S, Wallach H, Larochelle H, Grauman K, Cesa-Bianchi N, and Garnett R. Curran Associates, Inc., 2018, pp. 5727–5736. [Google Scholar]
- [8].Kriegeskorte Nikolaus, Mur Marieke, and Bandettini Peter. “Representational similarity analysis - connecting the branches of systems neuroscience”. Frontiers in Systems Neuroscience 2 (2008), p. 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Kornblith Simon, Norouzi Mohammad, Lee Honglak, and Hinton Geoffrey. “Similarity of Neural Network Representations Revisited”. Ed. by Chaudhuri Kamalika and Salakhutdinov Ruslan. Vol. 97. Proceedings of Machine Learning Research. Long Beach, California, USA: PMLR, 2019, pp. 3519–3529. [Google Scholar]
- [10].Maheswaranathan Niru, Williams Alex, Golub Matthew, Ganguli Surya, and Sussillo David. “Universality and individuality in neural dynamics across large populations of recurrent networks”. Advances in Neural Information Processing Systems 32. Ed. by Wallach H, Larochelle H, Beygelzimer A, d’Alché-Buc F, Fox E, and Garnett R. Curran Associates, Inc., 2019, pp. 15629–15641. [PMC free article] [PubMed] [Google Scholar]
- [11].Nguyen Thao, Raghu Maithra, and Kornblith Simon. Do Wide and Deep Networks Learn the Same Things? Uncovering How Neural Network Representations Vary with Width and Depth. 2020. [Google Scholar]
- [12].Shi Jianghong, Shea-Brown Eric, and Buice Michael. “Comparison Against Task Driven Artificial Neural Networks Reveals Functional Properties in Mouse Visual Cortex”. Advances in Neural Information Processing Systems 32. Ed. by Wallach H, Larochelle H, Beygelzimer A, d’Alché-Buc F, Fox E, and Garnett R. Curran Associates, Inc., 2019, pp. 5764–5774. [Google Scholar]
- [13].Small Christopher G.. The statistical theory of shape. Springer series in statistics. New York: Springer, 1996. [Google Scholar]
- [14].Kendall David George, Barden Dennis, Carne Thomas K, and Le Huiling . Shape and shape theory. New York: Wiley, 1999. [Google Scholar]
- [15].Dryden Ian L. and Mardia Kantilal. Statistical shape analysis with applications in R. Chichester, UK Hoboken, NJ: John Wiley & Sons, 2016. [Google Scholar]
- [16].Yianilos Peter N. “Data structures and algorithms for nearest neighbor search in general metric spaces”. Proceedings of the fourth annual ACM-SIAM Symposium on Discrete algorithms. 1993, pp. 311–321. [Google Scholar]
- [17].Dasgupta Sanjoy and Long Philip M. “Performance guarantees for hierarchical clustering”. Journal of Computer and System Sciences 70.4 (2005), pp. 555–569. [Google Scholar]
- [18].Baraty Saaid, Simovici Dan A., and Zara Catalin. “The Impact of Triangular Inequality Violations on Medoid-Based Clustering”. Foundations of Intelligent Systems. Ed. by Kryszkiewicz Marzena, Rybinski Henryk, Skowron Andrzej, and Raś Zbigniew W.. Berlin, Heidelberg:Springer Berlin Heidelberg, 2011, pp. 280–289. [Google Scholar]
- [19].Wang Fei and Sun Jimeng. “Survey on distance metric learning and dimensionality reduction in data mining”. Data Mining and Knowledge Discovery 29.2 (2015), pp. 534–564. [Google Scholar]
- [20].Chang C, Liao W, Chen Y, and Liou L. “A Mathematical Theory for Clustering in Metric Spaces”. IEEE Transactions on Network Science and Engineering 3.1 (2016), pp. 2–16. [Google Scholar]
- [21].Siegle Joshua H. et al. “Survey of spiking in the mouse visual system reveals functional hierarchy”. Nature 592.7852 (2021), pp. 86–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Ying Chris, Klein Aaron, Christiansen Eric, Real Esteban, Murphy Kevin, and Hutter Frank. “NAS-Bench-101: Towards Reproducible Neural Architecture Search”. Proceedings of the 36th International Conference on Machine Learning. Ed. by Kamalika Chaudhuri and Ruslan Salakhutdinov. Vol. 97. Proceedings of Machine Learning Research. PMLR, 2019, pp. 7105–7114. [Google Scholar]
- [23].Cohen Taco and Welling Max. “Group Equivariant Convolutional Networks”. Proceedings of The 33rd International Conference on Machine Learning. Ed. by Maria Florina Balcan and Kilian Q. Weinberger. Vol. 48. Proceedings of Machine Learning Research. New York, New York, USA: PMLR, 2016, pp. 2990–2999. [Google Scholar]
- [24].Cortes Corinna, Mohri Mehryar, and Rostamizadeh Afshin. “Algorithms for learning kernels based on centered alignment”. The Journal of Machine Learning Research 13.1 (2012), pp. 795–828. [Google Scholar]
- [25].Shahbazi Mahdiyar, Shirali Ali, Aghajan Hamid, and Nili Hamed. “Using distance on the Riemannian manifold to compare representations in brain and in models”. NeuroImage 239 (2021), p. 118271. [DOI] [PubMed] [Google Scholar]
- [26].Burkard Rainer, Mauro Dell’Amico, and Silvano Martello. Assignment Problems. Society for Industrial and Applied Mathematics, 2012. [Google Scholar]
- [27].Jonker Rand Volgenant A. “A shortest augmenting path algorithm for dense and sparse linear assignment problems”. Computing 38.4 (1987), pp. 325–340. [Google Scholar]
- [28].Crouse David F.. “On implementing 2D rectangular assignment algorithms”. IEEE Transactions on Aerospace and Electronic Systems 52.4 (2016), pp. 1679–1696. [Google Scholar]
- [29].Schönemann Peter H. . “A generalized solution of the orthogonal procrustes problem”. Psychometrika 31.1 (1966), pp. 1–10. [Google Scholar]
- [30].Gower JC and Garmt B Dijksterhuis. Procrustes problems. Oxford New York: Oxford University Press, 2004. [Google Scholar]
- [31].Kessy Agnan, Lewin Alex, and Strimmer Korbinian. “Optimal whitening and decorrelation”. The American Statistician 72.4 (2018), pp. 309–314. [Google Scholar]
- [32].Vinod Hrishikesh D. “Canonical ridge and econometrics of joint production”. Journal of econometrics 4.2 (1976), pp. 147–166. [Google Scholar]
- [33].Lai PL and Fyfe C. “Kernel and Nonlinear Canonical Correlation Analysis”. International Journal of Neural Systems 10.05 (2000), pp. 365–377. [DOI] [PubMed] [Google Scholar]
- [34].Lee John M.. Introduction to smooth manifolds. 2nd ed. Graduate texts in mathematics 218. New York ; London: Springer, 2013. [Google Scholar]
- [35].Kendall David G.. “Shape Manifolds, Procrustean Metrics, and Complex Projective Spaces”. Bulletin of the London Mathematical Society 16.2 (1984), pp. 81–121. [Google Scholar]
- [36].Thomas Fletcher P and Joshi Sarang. “Riemannian geometry for the statistical analysis of diffusion tensor data”. Signal Processing 87.2 (2007). Tensor Signal Processing, pp. 250–262. [Google Scholar]
- [37].Miolane Nina, Guigui Nicolas, Alice Le Brigant Johan Mathe, Hou Benjamin, Thanwerdas Yann, Heyder Stefan, Peltre Olivier, Koep Niklas, Zaatiti Hadi, Hajri Hatem, Cabanes Yann, Gerald Thomas, Chauchat Paul, Shewmake Christian, Brooks Daniel, Kainz Bernhard, Donnat Claire, Holmes Susan, and Pennec Xavier. “Geomstats: A Python Package for Riemannian Geometry in Machine Learning”. Journal of Machine Learning Research 21.223 (2020), pp. 1–9.34305477 [Google Scholar]
- [38].Feragen Aasa, Lauze Francois, and Hauberg Soren. “Geodesic Exponential Kernels: When Curvature and Linearity Conflict”. Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition (CVPR). 2015. [Google Scholar]
- [39].Feragen Aasa and Hauberg Søren. “Open Problem: Kernel methods on manifolds and metric spaces. What is the probability of a positive definite geodesic exponential kernel?” 29th Annual Conference on Learning Theory. Ed. by Feldman Vitaly, Rakhlin Alexander, and Shamir Ohad. Vol. 49. Proceedings of Machine Learning Research. Columbia University, New York, New York, USA: PMLR, 2016, pp. 1647–1650. [Google Scholar]
- [40].Jayasumana Sadeep, Salzmann Mathieu, Li Hongdong, and Harandi Mehrtash. “A Framework for Shape Analysis via Hilbert Space Embedding”. Proceedings of the IEEE International Conference on Computer Vision (ICCV). 2013. [Google Scholar]
- [41].Dryden Ian L and Mardia Kanti V. “Multivariate shape analysis”. Sankhyā: The Indian Journal of Statistics, Series A (1993), pp. 460–480. [Google Scholar]
- [42].James Rohlf F. “Shape Statistics: Procrustes Superimpositions and Tangent Spaces”. Journal of Classification 16.2 (1999), pp. 197–223. [Google Scholar]
- [43].Borg Ingwer and Groenen Patrick JF. Modern multidimensional scaling: Theory and applications. Springer Science & Business Media, 2005. [Google Scholar]
- [44].Agrawal Akshay, Ali Alnur, and Boyd Stephen. “Minimum-Distortion Embedding”. arXiv (2021). [Google Scholar]
- [45].Gates Alexander J., Wood Ian B., Hetrick William P., and Ahn Yong-Yeol. “Element-centric clustering comparison unifies overlaps and hierarchy”. Scientific Reports 9.1 (2019), p. 8574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Pedregosa F, Varoquaux G, Gramfort A, Michel V, Thirion B, Grisel O, Blondel M, Prettenhofer P, Weiss R, Dubourg V, Vanderplas J, Passos A, Cournapeau D, Brucher M, Perrot M, and Duchesnay E. “Scikit-learn: Machine Learning in Python”. Journal of Machine Learning Research 12 (2011), pp. 2825–2830. [Google Scholar]
- [47].Leena Chennuru Vankadara and von Luxburg Ulrike. “Measures of distortion for machine learning”. Advances in Neural Information Processing Systems. Ed. by Bengio S, Wallach H, Larochelle H, Grauman K, Cesa-Bianchi N, and Garnett R. Vol. 31. Curran Associates, Inc., 2018. [Google Scholar]
- [48].Harris Julie A. et al. “Hierarchical organization of cortical and thalamic connectivity”. Nature 575.7781 (2019), pp. 195–202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Hamrick Jess B. and Mohamed Shakir. “Levels of Analysis for Machine Learning”. Proceedings of the ICLR 2020 Workshop on Bridging AI and Cognitive Science. 2020. [Google Scholar]
- [50].Rybakken Erik, Baas Nils, and Dunn Benjamin. “Decoding of Neural Data Using Cohomological Feature Extraction”. Neural Computation 31.1 (2019), pp. 68–93. [DOI] [PubMed] [Google Scholar]
- [51].Chaudhuri Rishidev, Berk Gerçek Biraj Pandey, Peyrache Adrien, and Fiete Ila. “The intrinsic attractor manifold and population dynamics of a canonical cognitive circuit across waking and sleep”. Nature Neuroscience 22.9 (2019), pp. 1512–1520. [DOI] [PubMed] [Google Scholar]
- [52].Rouse Tevin C., Ni Amy M., Huang Chengcheng, and Cohen Marlene R.. “Topological insights into the neural basis of flexible behavior”. bioRxiv (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Gardner Richard J., Hermansen Erik, Pachitariu Marius, Burak Yoram, Baas Nils A., Dunn Benjamin A., Moser May-Britt, and Moser Edvard I.. “Toroidal topology of population activity in grid cells”. bioRxiv (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Kusano Genki, Fukumizu Kenji, and Hiraoka Yasuaki. “Kernel Method for Persistence Diagrams via Kernel Embedding and Weight Factor”. Journal of Machine Learning Research 18.189 (2018), pp. 1–41. [Google Scholar]
- [55].Moor Michael, Horn Max, Rieck Bastian, and Borgwardt Karsten. “Topological Autoencoders”. Proceedings of the 37th International Conference on Machine Learning. Ed. by Daumé Hal III and Singh Aarti. Vol. 119. Proceedings of Machine Learning Research. PMLR, 2020, pp. 7045–7054. [Google Scholar]
- [56].Jensen Kristopher, Kao Ta-Chu, Tripodi Marco, and Hennequin Guillaume. “Manifold GPLVMs for discovering non-Euclidean latent structure in neural data”. Advances in Neural Information Processing Systems. Ed. by Larochelle H, Ranzato, Hadsell R, Balcan MF, and Lin H. Vol. 33. Curran Associates, Inc., 2020, pp. 22580–22592. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


