Abstract
Alignment of sigma 54-dependent promoters indicates conservation of two sequence elements. Six nucleotides in the downstream −12 element were mutated individually to each nonconsensus nucleotide. mRNA levels were measured in vivo for each promoter under strongly activating conditions. The results showed that the consensus sequence was not the strongest promoter. Instead, the −12 consensus element consists of two subregions that behave differently when mutated. Single changes in the upstream TTT consensus subregion can lead to increases in transcription, whereas single changes in the downstream GC(A/T) can lead to decreases in transcription. Selected double mutations with changes in both subregions were constructed and studied in vivo. No double mutation increased promoter strength, and some decreased it. Mutant promoters were also assayed under nonactivating conditions in vivo. No mRNA was detected in 23 of the 24 promoters tested. However, one double mutant showed substantial levels of transcript, indicating that the −12 sequence was capable of specifying basal transcription under nonactivating conditions. Overall, the results show that the −12 region has multiple roles in transcription in vivo, including modulating both basal and induced RNA levels.
Sigma 54 is an alternative sigma factor that binds the common bacterial core RNA polymerase and converts it into an enhancer-dependent enzyme (15, 16, 35). It targets promoters with recognition elements centered near positions −24 and −12 (19, 20). When sigma 54 holoenzyme initially binds to such promoters, it fails to melt the DNA and thus remains in an inactive state (2, 21, 22, 26). Activation occurs when a modified activator protein binds the remote enhancer and loops to the bound polymerase, causing it to melt the DNA (3, 8, 22, 25, 29). Transcription then proceeds from this open complex.
DNA binding by sigma 54 polymerase is complex. Sigma 54 alone has a low affinity for most promoters but can bind to the Rhizobium meliloti nifH promoter in the absence of core RNA polymerase (2). Core polymerase association increases the DNA-binding affinity, allowing occupancy on other promoters (5). The main DNA-binding determinants are in the C-terminal portion of sigma 54 (27, 37). However, several other parts of the protein make important contributions to DNA recognition (7). Candidate subregions of sigma involved in DNA binding have been identified (6, 11, 13, 30). These regions of sigma have not been assigned definitively to the recognition of particular elements within the promoter.
Recognition of the promoter −24 element appears to be dominant for DNA binding. That is, there are no examples where sigma 54 mutant complexes form but do not protect the −24 region in a footprinting experiment (14, 37). By contrast numerous mutations exist, both in sigma and in DNA, that allow binding to occur but interfere specifically with protection of the −12 element. These bound complexes that are defective in −12-element recognition are typically of lower affinity (33). Despite the occupancy of the promoter by the holoenzyme, such complexes often are defective in function, implying that −12 interactions have functions in addition to binding to DNA.
Other lines of evidence confirm that the −12 region has multiple effects in sigma 54-dependent transcription. Changes in this region are known to be associated with changes in transcription level (1, 4, 17, 28). The sequence at −12 can determine how sensitive the promoter is in vivo to a particular activator (4, 23). In vitro transcription studies showed that certain −12 sequences tend to promote leaky “bypass” expression in the absence of an activator (34). Consistent with these multiple potential roles of the −12 region is the fact that several separated parts of the sigma 54 polypeptide appear to cooperate to recognize the −12 region (7, 27). Despite the apparent multifunctional role of the −12 region there have not been systematic studies of the effects of changing −12 region sequences.
For these reasons, we have investigated the effects of changing each residue in the −12 region to each other residue. The effects of the changes were assayed by measuring mRNA levels in vivo. The results showed that the consensus −12 sequence was only intermediate in promoter strength. The data also showed that the −12 region consists of two subelements, each of which could have an independent role in transcriptional control.
MATERIALS AND METHODS
Plasmid construction and site-directed mutagenesis.
The plasmid pBR-M12 was constructed by replacing the tetracycline gene of pBR322 with the M12 promoter fragment from pFC50-M12 (9). Plasmid pBR322 was cut with EcoRI and AvaI (New England Biolabs) to separate the 1,008-bp tetracycline gene region from the main body. The large fragment of pBR322 was purified by agarose gel electrophoresis, and the AvaI site was made blunt by using Klenow enzyme (New England Biolabs). Plasmid pFC50-M12 was digested with EcoRI and EheI (New England Biolabs) to release a 1,236-bp fragment containing the functional glnH promoter region, initial coding sequences, and the previously inserted T7 early terminator. The purified fragment was ligated with the pBR322 main-body fragment to create plasmid pBR-M12. The M12 mutant promoter was created previously by point mutation of T to G at −14 in the glnHp2 promoter (9).
The QuikChange site-directed mutagenesis kit (Stratagene) was used to introduce single mutations in the −12 consensus sequence of the M12 promoter in Epicurian Coli XL1-Blue supercompetent cells. The presence of mutations was confirmed by sequencing.
Cell growth and RNA analysis.
pBR-M12 and its derivatives were transformed into a PcnB− strain (with the pcnB gene deleted) to maintain low copy numbers. Cells were grown overnight in Luria broth (LB) medium and then diluted 1:20 into 3 ml of either LB or G-gln (500 ml of G-gln medium contains W salts [5.25 g of K2HPO4, 2.25 g of KH2PO4, 0.215 ml of 1 M MgSO4], 0.4% glucose, 1.0 ml of thiamine [10.0 mg/ml], and 1 g of l-glutamine) containing 100 μg of ampicillin/ml. When the cells reached an optical density at 600 nm of 0.4 to 0.6, they were collected and RNA was extracted with the RNeasy kit (Qiagen) and eluted in a total volume of 30 μl. Sample volumes were adjusted slightly to use the same number of cells in separate experiments.
Samples (12 μl) were taken for mRNA analysis. Primer extension with reverse transcriptase (RT) (Promega) was done with the 32P-labeled oligonucleotide primer GCCAGGGTCAGTGCAGCCA, complementary to the glnH transcript. The 86-nucleotide radioactive cDNA was sized on a 6% urea polyacrylamide gel with sequencing markers and corresponded to expectations based on in vitro transcription (34). Amounts were quantified with a PhosphorImager (Molecular Dynamics).
RESULTS
Derivation of the consensus and construction of mutant promoters.
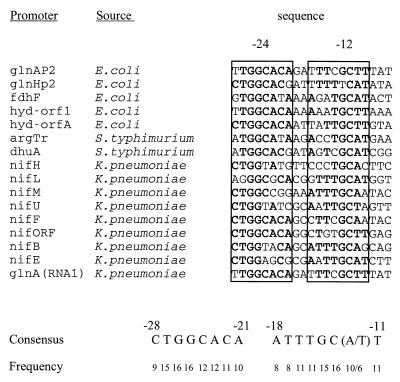
To update the consensus sequence for sigma 54 promoters, we aligned the sequences for 16 confirmed promoters from Escherichia coli, Salmonella typhimurium, and Klebsiella pneumoniae (4, 10, 12) (Fig. 1). The lower portion of the figure shows those nucleotides that are conserved in at least half of the promoters. The analysis confirms that recognition is likely to involve two sequence-conserved blocks. These are CTGGCACA, from −21 to −28 (the −24 region), and ATTTGC(A/T)T, from −11 to −18 (the −12 region). (The numbering system used here is based on glnHp2 and differs from that used for some individual promoters due to minor variations in the location of the transcription start site.) We note that the centers of these two blocks are separated by 10 bp, suggesting that they are centered on the same face of the DNA helix.
FIG. 1.
Alignment of 16 available sigma 54-dependent promoters from three related bacteria (1, 10, 12, 19). The consensus nucleotides and their frequency of appearance are indicated below the alignment. The −24 and −12 regions that contain these consensus elements are boxed, with the consensus nucleotides shown in boldface.
This is consistent with prior alignments (19) but suggests more extended conservation of sequence. In this study of the −12 region we chose to modify the central hexanucleotide sequence. This includes bases identified previously as “consensus,” with the addition of a single upstream T that prior studies have implicated in sigma recognition (2).
The starting plasmid (pFC50-M12) for this work contains a promoter with the hexanucleotide consensus (9). This M12 promoter is a derivative of glnHp2 in which the T at −14 has been changed to the consensus G. Nucleotide −12 in this promoter is an A, which is slightly better conserved than T at this position in the collection shown in Fig. 1. The M12 promoter retains the natural upstream binding sites for activator NtrC (also called NR1) as well as the intervening IHF binding sites. A segment of the initial glnH coding region is also retained.
In the experimental scheme for making changes in the consensus sequence, a large set of mutant promoters are studied, each carried in by the vector, which is a multicopy plasmid. In order to minimize potential titration of required transcription factors by the plasmid, two conditions were used. First, a PcnB− host strain was used, which greatly reduces copy numbers (18). Second, the promoter insert of pFC50-M12 was transferred to pBR322 to form pBR-M12, which has a lower copy number than the original pUC-based vector. Two experiments demonstrated that the low copy number was achieved. First, PcnB− cells transformed with pFC50-M12 could not grow when plated on 1/10 the amount of ampicillin required to prevent wild-type growth. Second, agarose gel analysis showed a 10-fold reduction in the amount of DNA (pFC50-M12 or pBR-M12) isolated from the PcnB− host. Based on these and other experiments we estimate that there are two or three copies of the plasmid per cell. As this is substantially less than the number of sigma 54 regulatory regions on the chromosome, we infer that there will be little perturbation of metabolism by factor titration.
Seventeen changes were introduced by site-directed mutagenesis into the central TTTGCA of the −12 region of the M12 promoter contained on this vector (Table 1). These included changes to each nonconsensus nucleotide at each position (position −12 was not changed from the more conserved A to the less conserved T). The goal was to measure the mRNA levels associated with each of these 17 promoters and thereby deduce the importance of each nucleotide.
TABLE 1.
In vivo mRNA levels from promoter M12 and its derivativesa
| Promoter | −12 region | mRNA ratio (± SD) |
|---|---|---|
| M12 | 5′…TTTGCA…3′ | 1 |
| T-17G | 5′…GTTGCA…3′ | 1.8 ± 0.1 |
| T-17C | 5′…CTTGCA…3′ | 0.9 ± 0.2 |
| T-17A | 5′…ATTGCA…3′ | 1.0 ± 0.1 |
| T-16G | 5′…TGTGCA…3′ | 1.6 ± 0.2 |
| T-16C | 5′…TCTGCA…3′ | 1.1 ± 0.3 |
| T-16A | 5′…TATGCA…3′ | 1.0 ± 0.3 |
| T-15G | 5′…TTGGCA…3′ | 1.6 ± 0.9 |
| T-15C | 5′…TTCGCA…3′ | 1.8 ± 0.3 |
| T-15A | 5′…TTAGCA…3′ | 2.6 ± 0.7 |
| G-14C | 5′…TTTCCA…3′ | 0.5 ± 0.4 |
| G-14A | 5′…TTTACA…3′ | 0.9 ± 0.2 |
| G-14T | 5′…TTTTCA…3′ | 0.6 ± 0.4 |
| C-13G | 5′…TTTGGA…3′ | 1.1 ± 0.2 |
| C-13A | 5′…TTTGAA…3′ | 1.1 ± 0.1 |
| C-13T | 5′…TTTGTA…3′ | 0.3 ± 0.3 |
| A-12G | 5′…TTTGCG…3′ | 0.4 ± 0.3 |
| A-12C | 5′…TTTGCC…3′ | 0.9 ± 0.2 |
a Data are normalized to M12 levels. The means and standard deviations shown are from three to seven independent RNA preparations. Changes to nonconsensus nucleotides are underlined.
mRNA production from singly mutated promoters.
We first measured mRNA from cells transformed with the consensus glnH-M12 promoter in different media. The purpose was to identify the media giving the highest and lowest mRNA signals. The transformed cells were diluted from overnight cultures in rich media. The cells were grown to mid-log phase in four media that differ in nitrogen availability: LB medium with excess nitrogen, glucose minimal medium supplemented with either arginine (W-Arg) (31) or glutamine (G-Gln) (24), and glycerol minimal medium supplemented with glutamate (36). The amount of promoter-specific mRNA was estimated by hybridizing a small DNA primer complementary to the initial glnH mRNA sequence and copying it with RT. Because the cells are expected to require glnH operon function for growth on certain media, the endogenous glnH promoter is present on the chromosome; the RT products include those from this single-copy gene. Figure 2 shows the signal obtained on G-Gln medium for nontransformed cells (lane host) and cells transformed with the glnH-M12 fusion (lane M12). The signal from the M12 promoter on the plasmid is taken as the difference between these two signals.
FIG. 2.
Autoradiograph of primer extension products of mRNAs with various promoters in strongly activating G-Gln medium. “Host” refers to nontransformed cells, and the signal is from the endogenous glnH gene.
PhosphorImager analysis was used to determine the amount of promoter M12 transcription in the various media. Transcription was highest on G-Gln medium and lowest (undetectable) on LB, consistent with expectations based on the availability of nitrogen. In the initial studies we use the high-activation potential G-Gln medium.
The 17 promoters were individually transformed, and mRNA production was studied as described above. Examples of mRNA assays from six promoters are shown in Fig. 2. As can be seen in these examples, some −12-region changes from the M12 consensus lead to decreases in transcription. Obvious examples are G-14C (i.e., the G at position 14 changed to a C), C-13T, and A-12G. Each of these mRNA signals is less intense than M12 and only slightly above the background signal from the endogenous host glnH gene. Other mutant promoters behave differently. Mutation C-13A shows little change in mRNA level. T-17G shows an increased mRNA level compared to that of the M12 consensus promoter.
The mRNA signals were somewhat variable, and for that reason mRNA was assayed from each of the 17 promoters between three and seven times. Identical numbers of cells were used for each experiment, and each experiment included a parallel sample from the M12 promoter plasmid. The mRNA signal levels for the various promoters were normalized to the M12 levels from the same experiment. The resulting mean (± standard deviation) signals are collected in Table 1.
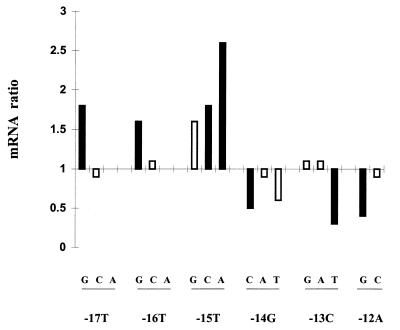
The results show that at least four promoter mutations cause increases in transcription: T-17G, T-16G, T-15C, and T-15A. In addition, at least three promoter mutations cause decreases in transcription: G-14C, C-13T, and A12-G. It is obvious from these data that the consensus sequence is neither the best nor the worst promoter at directing high mRNA levels. The variation caused by the introduction of single-point mutations is eight- to ninefold, increased about 2.6-fold by T-15A and decreased 3.3-fold by C-13T.
The effect of changing each consensus nucleotide is shown in Fig. 3. The data indicate that changing the upstream and downstream parts of the −12 consensus leads to quite different consequences. When the upstream TTT segment is mutated the resulting promoter either produces more mRNA or is unaffected. When the downstream GCA segment is mutated the resulting promoter either produces less mRNA or is unaffected. We infer that the −12 consensus sequence element contains two subelements which have the potential to behave differently in mRNA production.
FIG. 3.
The effect on mRNA levels of single substitutions at each position in the M12 consensus promoter. Changes that increase transcription are shown above the axis, and those that decrease transcription are shown below the axis; the height of each bar represents the ratio of RNA to that of M12. The solid bars represent statistically significant differences from M12, as indicated in Table 1. The nucleotide change associated with each mutant promoter is indicated below each bar.
Effects of double mutations in which both subelements are mutated.
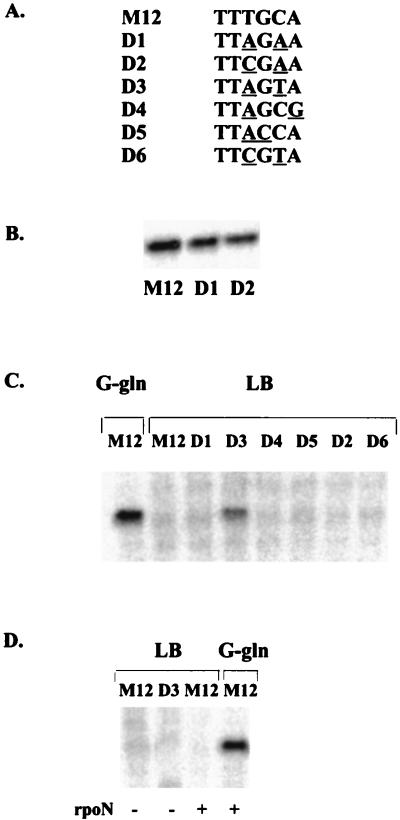
Some of the above-mentioned results are expected based on prior studies, and some are not. The data indicate that the effects of the T stretch may be particularly complex in that changes could lessen sigma binding but enhance transcription (1). The deleterious effect of mutating the highly conserved −13C to T is unsurprising. However, the lack of effect of G and A substitutions at this most highly conserved position is unexpected. One possible explanation is that these −13 substitutions show no effect because they are made in the context of an otherwise consensus promoter. To test this idea, we made double mutants D1 and D2, in which the C-13A mutation was coupled with substitutions for the highly conserved T at position −15 (the sequence is shown in Fig. 4A). For these mutants, and other double mutants discussed below, the mRNA was compared to that of the M12 promoter. This allowed the comparison of levels from double and single mutations.
FIG. 4.
Transcription from double mutants in nonactivating and activating media. (A) Sequences of the mutants. The changed nucleotides in each of the double mutants are underlined. (B) Analysis of RNA from double mutants D1 and D2 in activating G-Gln medium. (C) Analysis of RNA from double mutants in nonactivating LB medium. The induced transcripts from the consensus M12 promoter in activating G-Gln medium are shown for comparison. (D) Analysis of RNA in a strain lacking (−) or containing (+) sigma 54 (rpoN).
The results show that in the context of a nonconsensus promoter the C-13A change has a strong effect and leads to significant reductions in transcription (Fig. 4B). This is true for both mutants D1 and D2, which have approximately two-thirds the RNA level of the M12 consensus promoter. The reduction due to the −13 change is even stronger than that illustrated here, as it is made in the context of nonconsensus promoters that have mRNA levels higher than that of the consensus M12 promoter. That is, the single −13 position substitution leads to a three- to fourfold lowering of mRNA levels in the context of the nonconsensus promoters T-15C and T-15A (Table 1). Thus, the −13 nucleotide is clearly important, as expected from its conservation. Apparently, its importance can be masked in the context of an otherwise consensus promoter. The complex relationship between the two halves of the sequence from −17 to −12 will be discussed further below.
Previously, we raised the possibility that these −12-region sequences may play an additional role beyond specifying the level of RNA made during activating conditions. In vitro-transcription experiments showed that a nonconsensus promoter could be leakier than a consensus promoter; that is, in the absence of an activator the nonconsensus promoter produced more RNA, albeit at rather low levels (34). To see if this could be true in vivo, we measured RNA levels from all the promoters by using LB medium, where the consensus promoter produced no detectable RNA (Fig. 4, lanes M12). No single-point mutations led to detectable RNA under these conditions, indicating that none of the sequences give leaky transcription (data not shown).
We extended these experiments to learn if double mutations could lead to leaky transcription in vivo. Four additional double mutations were made, each including a substitution at position −15 and an additional change in the downstream half of the promoter element (Fig. 4A). Each of these promoters was used to drive mRNA production on nonactivating LB medium. The results with the six double mutations are shown in Fig. 4.
The results show that one double mutation (Fig. 4C, lane D3) leads to production of mRNA under nonactivating conditions. To confirm that this mRNA is sigma 54 dependent, we transformed D3 and the M12 control into strain ymc109, which lacks sigma 54. Figure 4D shows that the D3 signal is not produced in this strain in LB medium, in contrast to the result obtained in the strain containing sigma 54 and as expected for a sigma 54-dependent transcript.
Quantitation of D3 mRNA in LB medium shows that the level is fairly substantial, as it is about 1/10 of the amount of RNA made by the consensus M12 promoter under fully activated conditions (Fig. 4C, G-Gln). This D3 promoter, as well as the promoters D4, D5, and D6, produces mRNA amounts on fully activated G-Gln medium that do not differ dramatically from that of M12 (data not shown). Thus, promoter D3 is still inducible but the ratio is only about 10-fold, as the basal level is high. Because mRNA from M12 is not detectable under nonactivating conditions (Fig. 4C and D), we cannot quantify the normal induction ratio; our best estimate is that it should be at least 100-fold, but this is very uncertain. In any case, the D3 double mutation creates a promoter with a high basal level and a roughly normal induced level. This demonstrates a new in vivo role for the −12-region promoter element: it can control the level of basal expression independently of the level of induced expression.
DISCUSSION
These results imply that the −12 regions of sigma 54 promoters have multiple and complex roles in transcription. In the initial set of experiments each nucleotide in a central consensus sequence was changed to each possible nonconsensus nucleotide. mRNA production in vivo from each of the singly substituted promoters was then measured. The results showed that the consensus promoter sequence did not direct the highest level of transcription.
This result raises the question of why promoter −12 elements resemble TTTGC(A/T) more than any other sequence. Of the 16 natural promoters surveyed, 3 matched this consensus exactly, 7 had a single mismatch, and 6 had a double mismatch. Each of these promoters needs to bind sigma 54 polymerase, and the −12 sequence plays an important secondary role in this binding (see the introduction). We speculate that the various promoters retain significant resemblance to the consensus so that they will have a sufficient number of recognition determinants to achieve this binding. If so, the consensus promoter would represent the tightest binder of sigma (2). This, however, need not correlate with the highest level of transcription. That is, as long as sigma can become fully bound, other sequences would influence how much RNA would be produced by the bound sigma 54 holoenzyme.
This view is supported by experiments showing that the upstream and downstream halves of a consensus −12-region promoter have the potential to function differently. When the upstream TTT stretch was mutated a number of changes led to increases in transcription. Interestingly, such an effect was observed previously, although it seemed to depend on the nature of the activator-promoter combination (1). This was never observed in the downstream GCA stretch; the sequence behaved more according to expectation in that changes to nonconsensus nucleotides often led to decreases in transcription. It is known that a change to create the consensus TTT sequence can increase the affinity of sigma for DNA (2). One possible explanation for these properties of the TTT sequence is that the sequence can help to attract sigma but the binding could be too tight for optimal function (32) in the context of a fully consensus promoter.
When mutations in the upstream and downstream halves of the −12 region were combined, a variety of effects were observed. None of the doubly mutated promoters showed mRNA levels that exceeded consensus levels, even though the single T-stretch mutations on which they were based had this property. Some of the double mutations led to significantly lower RNA levels. In view of the above considerations, it may be that some minimal match to the consensus is needed to fully bind the sigma 54 holoenzyme and some double mutations fail in this regard. In vitro studies are under way to test these ideas.
Study of the double mutations also revealed a striking effect of the −12 region on the regulation of induction. All of the above studies applied to the functioning of the promoters on G-Gln medium, which is the most strongly activating medium of those surveyed. When the very rich medium LB was used no mRNA was detected, as expected on such nitrogen-rich medium. One double mutant, however, failed to fully restrict RNA synthesis under these conditions of full nitrogen availability. In this case the repressive effects were sufficiently defective to give rise to approximately 10% of the fully induced level of RNA. This occurs despite the fact that the medium is sufficiently repressing to keep NtrC so inactive that it fails to direct any transcription of the consensus promoter. The result demonstrates an additional role for certain −12-region sequences: keeping transcription levels low during conditions of nitrogen availability.
Such leaky transcription has been observed in vitro under specialized conditions and with protocols designed to optimize transcription in the absence of activator activity (34). We proposed in those studies that tight interactions between the −12 region of DNA and the N terminus of sigma 54 might be responsible for keeping basal transcription levels low. The present results support that view, although it would require the construction of many more multiply mutated promoters to begin to define the sequences involved.
Thus, the diversity of sequences associated with the −12 regions of natural promoters can be seen as physiologically appropriate in terms of diverse roles for this element. Promoter −12 elements should exist that support the highest level of activated transcription for genes that require such high levels. The data suggest that such promoters may not match the consensus in the upstream T stretch. Other promoters may diverge further from the consensus in some cases to provide a basal level of transcription for genes that need low-level expression even under conditions of nitrogen sufficiency. The existing −12 sequences may be viewed as providing a balance between the need for RNA upon activation and the need to restrict it under nonactivating conditions. Further studies will be required to learn how the −12-region sequences influence this balance of competing requirements.
ACKNOWLEDGMENTS
This work was supported by NIH grant GM35754.
We thank Robert P. Gunsalus and Paul McNicholas for help with the PcnB− strain and the Gralla research group for their advice.
REFERENCES
- 1.Buck M, Cannon W. Mutations in the RNA polymerase recognition sequence of the Klebsiella pneumoniae nifH promoter permitting transcriptional activation in the absence of NifA binding to upstream activator sequences. Nucleic Acids Res. 1989;17:2597–2612. doi: 10.1093/nar/17.7.2597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Buck M, Cannon W. Activator-independent formation of a closed complex between sigma 54-holoenzyme and nifH and nifU promoters of Klebsiella pneumoniae. Mol Microbiol. 1992;6:1625–1630. doi: 10.1111/j.1365-2958.1992.tb00887.x. [DOI] [PubMed] [Google Scholar]
- 3.Buck M, Cannon W, Woodcock J. Transcriptional activation of the Klebsiella pneumoniae nitrogenase promoter may involve DNA loop formation. Mol Microbiol. 1987;1:243–249. doi: 10.1111/j.1365-2958.1987.tb00518.x. [DOI] [PubMed] [Google Scholar]
- 4.Buck M, Khan H, Dixon R. Site-directed mutagenesis of the Klebsiella pneumoniae nifL and nifH promoters and in vivo analysis of promoter activity. Nucleic Acids Res. 1985;13:7621–7638. doi: 10.1093/nar/13.21.7621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cannon W, Claverie-Martin F, Austin S, Buck M. Core RNA polymerase assists binding of the transcription factor sigma 54 to promoter DNA. Mol Microbiol. 1993;8:287–298. doi: 10.1111/j.1365-2958.1993.tb01573.x. [DOI] [PubMed] [Google Scholar]
- 6.Cannon W, Claverie-Martin F, Austin S, Buck M. Identification of a DNA-contacting surface in the transcription factor sigma-54. Mol Microbiol. 1994;11:227–236. doi: 10.1111/j.1365-2958.1994.tb00303.x. [DOI] [PubMed] [Google Scholar]
- 7.Cannon W V, Chaney M K, Wang X, Buck M. Two domains within sigmaN (sigma54) cooperate for DNA binding. Proc Natl Acad Sci USA. 1997;94:5006–5011. doi: 10.1073/pnas.94.10.5006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Carmona M, Claverie-Martin F, Magasanik B. DNA bending and the initiation of transcription at sigma54-dependent bacterial promoters. Proc Natl Acad Sci USA. 1997;94:9568–9572. doi: 10.1073/pnas.94.18.9568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Claverie-Martin F, Magasanik B. Positive and negative effects of DNA bending on activation of transcription from a distant site. J Mol Biol. 1992;227:996–1008. doi: 10.1016/0022-2836(92)90516-m. [DOI] [PubMed] [Google Scholar]
- 10.Collado-Vides J, Magasanik B, Gralla J D. Control site location and transcriptional regulation in Escherichia coli. Microbiol Rev. 1991;55:371–394. doi: 10.1128/mr.55.3.371-394.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Coppard J R, Merrick M J. Cassette mutagenesis implicates a helix-turn-helix motif in promoter recognition by the novel RNA polymerase sigma factor sigma 54. Mol Microbiol. 1991;5:1309–1317. doi: 10.1111/j.1365-2958.1991.tb00777.x. [DOI] [PubMed] [Google Scholar]
- 12.Gralla J D, Collado-Vides J. Organization and function of transcription regulatory elements. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. IV. Regulation of gene expression. Washington, D.C: ASM Press; 1996. pp. 1232–1245. [Google Scholar]
- 13.Guo Y, Gralla J D. DNA-binding determinants of sigma 54 as deduced from libraries of mutations. J Bacteriol. 1997;179:1239–1245. doi: 10.1128/jb.179.4.1239-1245.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hsieh M, Gralla J D. Analysis of the N-terminal leucine heptad and hexad repeats of sigma 54. J Mol Biol. 1994;239:15–24. doi: 10.1006/jmbi.1994.1347. [DOI] [PubMed] [Google Scholar]
- 15.Kustu S, North A K, Weiss D S. Prokaryotic transcriptional enhancers and enhancer-binding proteins. Trends Biochem Sci. 1991;16:397–402. doi: 10.1016/0968-0004(91)90163-p. [DOI] [PubMed] [Google Scholar]
- 16.Magasanik B. The regulation of nitrogen utilization in enteric bacteria. J Cell Biochem. 1993;51:34–40. doi: 10.1002/jcb.240510108. [DOI] [PubMed] [Google Scholar]
- 17.Martin-Verstraete I, Debarbouille M, Klier A, Rapoport G. Mutagenesis of the Bacillus subtilis “−12, −24” promoter of the levanase operon and evidence for the existence of an upstream activating sequence. J Mol Biol. 1992;226:85–99. doi: 10.1016/0022-2836(92)90126-5. [DOI] [PubMed] [Google Scholar]
- 18.Masters M, Colloms M D, Oliver I R, He L, Macnaughton E J, Charters Y. The pcnB gene of Escherichia coli, which is required for ColE1 copy number maintenance, is dispensable. J Bacteriol. 1993;175:4405–4413. doi: 10.1128/jb.175.14.4405-4413.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Merrick M J. In a class of its own—the RNA polymerase sigma factor ς54 (sigma N) Mol Microbiol. 1993;10:903–909. doi: 10.1111/j.1365-2958.1993.tb00961.x. [DOI] [PubMed] [Google Scholar]
- 20.Morett E, Buck M. In vitro studies on the interactions of DNA polymerase-s54 with the Klebsiella pneumoniae and Rhizobium meliloti nifH promoters. The role of NifA in the formation of an open promoter complex. J Mol Biol. 1989;210:65–77. doi: 10.1016/0022-2836(89)90291-x. [DOI] [PubMed] [Google Scholar]
- 21.Ninfa A J, Reitzer L J, Magasanik B. Initiation of transcription at the bacterial glnAp2 promoter by purified E. coli components is facilitated by enhancers. Cell. 1987;50:1039–1046. doi: 10.1016/0092-8674(87)90170-x. [DOI] [PubMed] [Google Scholar]
- 22.Popham D L, Szeto D, Keener J, Kustu S. Function of a bacterial activator protein that binds to transcriptional enhancers. Science. 1989;243:629–635. doi: 10.1126/science.2563595. [DOI] [PubMed] [Google Scholar]
- 23.Ray L, Claverie-Martin F, Weglenski P, Magasanik B. Role of the promoter in activation of transcription by nitrogen regulator I phosphate in Escherichia coli. J Bacteriol. 1990;172:818–823. doi: 10.1128/jb.172.2.818-823.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Reitzer L J, Magasanik B. Expression of glnA in Escherichia coli is regulated at tandem promoters. Proc Natl Acad Sci USA. 1985;82:1979–1983. doi: 10.1073/pnas.82.7.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Reitzer L J, Magasanik B. Transcription of glnA in E. coli is stimulated by activator bound to sites far from the promoter. Cell. 1986;45:785–792. doi: 10.1016/0092-8674(86)90553-2. [DOI] [PubMed] [Google Scholar]
- 26.Sasse-Dwight S, Gralla J D. Probing the Escherichia coli glnALG upstream activation mechanism in vivo. Proc Natl Acad Sci USA. 1988;85:8934–8938. doi: 10.1073/pnas.85.23.8934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sasse-Dwight S, Gralla J D. Role of eukaryotic-type functional domains found in the prokaryotic enhancer receptor factor sigma 54. Cell. 1990;62:945–954. doi: 10.1016/0092-8674(90)90269-k. [DOI] [PubMed] [Google Scholar]
- 28.Stigter J, Schneider M, de Bruijn F J. Azorhizobium caulinodans nitrogen fixation (nif/fix) gene regulation: mutagenesis of the nifA −24/−12 promoter element, characterization of a ntrA(rpoN) gene, and derivation of a model. Mol Plant-Microbe Interact. 1993;6:238–252. doi: 10.1094/mpmi-6-238. [DOI] [PubMed] [Google Scholar]
- 29.Su W, Porter S, Kustu S, Echols H. DNA-looping and enhancer activity: association between DNA-bound NtrC activator and RNA polymerase at the bacterial glnA promoter. Proc Natl Acad Sci USA. 1990;87:5504–5508. doi: 10.1073/pnas.87.14.5504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Taylor M, Butler R, Chambers S, Casimiro M, Badii F, Merrick M. The RpoN-box motif of the RNA polymerase sigma factor sigma N plays a role in promoter recognition. Mol Microbiol. 1996;22:1045–1054. doi: 10.1046/j.1365-2958.1996.01547.x. [DOI] [PubMed] [Google Scholar]
- 31.Tintut Y, Gralla J D. PCR mutagenesis identifies a polymerase-binding sequence of sigma 54 that includes a sigma 70 homology region. J Bacteriol. 1995;177:5818–5825. doi: 10.1128/jb.177.20.5818-5825.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tintut Y, Wang J T, Gralla J D. A novel bacterial transcription cycle involving sigma(54) Genes Dev. 1995;9:2305–2313. doi: 10.1101/gad.9.18.2305. [DOI] [PubMed] [Google Scholar]
- 33.Wang J T, Gralla J D. The transcription initiation pathway of sigma 54 mutants that bypass the enhancer protein requirement: implications for the mechanism of activation. J Biol Chem. 1996;271:32707–32713. doi: 10.1074/jbc.271.51.32707. [DOI] [PubMed] [Google Scholar]
- 34.Wang J T, Syed A, Gralla J D. Multiple pathways to bypass the enhancer requirement of sigma 54 RNA polymerase: roles for DNA and protein determinants. Proc Natl Acad Sci USA. 1997;94:9538–9543. doi: 10.1073/pnas.94.18.9538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang J T, Syed A, Hsieh M, Gralla J D. Converting Escherichia coli RNA polymerase into an enhancer-responsive enzyme: role of an NH2-terminal leucine patch in sigma 54. Science. 1995;270:992–994. doi: 10.1126/science.270.5238.992. [DOI] [PubMed] [Google Scholar]
- 36.Willis R C, Iwata K K, Furlong C E. Regulation of glutamine transport in Escherichia coli. J Bacteriol. 1975;122:1032–1037. doi: 10.1128/jb.122.3.1032-1037.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wong C, Tintut Y, Gralla J D. The domain structure of sigma 54 as determined by analysis of a set of deletion mutants. J Mol Biol. 1994;236:81–90. doi: 10.1006/jmbi.1994.1120. [DOI] [PubMed] [Google Scholar]